Abstract
UvsW protein from bacteriophage controls the transition from origin-dependent to origin-independent initiation of replication through the unwinding of R-loops bound to the T4 origins of replication. UvsW has also been implicated through genetic and biochemical experiments to play a role in DNA repair processes such as replication fork regression and Holliday junction branch migration. UvsW is capable of unwinding a wide variety of substrates, many of which contain only duplex DNA without single-stranded regions. Based on this observation, it has been suggested that UvsW is a dsDNA translocase. In this work we examine the ability of UvsW to translocate on ssDNA. Kinetic analysis indicates that the rate of ATP hydrolysis is strongly dependent on the length of the ssDNA lattice, whereas the KM-DNA remains relatively constant, demonstrating that UvsW translocates on ssDNA in an ATP-dependent fashion. Experiments using streptavidin blocks or poly-dT sequences located at either end of the ssDNA substrate indicate that UvsW translocates in a 3’ to 5’ direction. Mutant competition and heparin trapping experiments reveal that UvsW is extremely processive during ATP-driven translocation with a half-life on the order of several minutes. Finally, functional assays provide evidence that UvsW is monomeric while translocating on ssDNA. The ability of UvsW to unwind DNA duplexes is likely to be mechanistically linked to its ability to processively translocate on ssDNA in a 3’ to 5’ unidirectional fashion.
Keywords: T4 DNA replication, helicase, strand annealing, DNA repair
DNA helicases couple the energy of ATP hydrolysis to the energetically unfavorable process of unwinding the DNA double helix (1). UvsW protein is one of three helicases from bacteriophage T4 and based on sequence identity is a member of the super family 2 (SF2) helicases (2). UvsW has several roles in T4 phage DNA replication and repair. UvsW governs the mode of replication initiation by unwinding the stably bound RNA transcripts (R-loops) from the T4 origins of replication (3). The removal of R-loops eliminates origin-dependent initiation, causing origin independent (recombinogenic) initiation to become the dominant mode of initiation (4). UvsW knockout mutations are sensitive to UV light or hydroxyurea, indicating a role in the processing of damaged DNA and stalled replication forks (5–7). In vitro, UvsW unwinds branched DNA substrates, suggesting that UvsW is responsible for the residual branch resolution activity in the absence of gp49 (8–10). In addition to branch migration activity, UvsW unwinds DNA substrates resembling recombination intermediates and stalled replication forks (11). UvsW unwinds D-loops that are formed from the ssDNA strand invasion into homologous dsDNA and is able to unwind both the leading and lagging strand arms of a model stalled replication fork (11). This led to the proposal that UvsW participates in both synthesis dependent strand annealing (SDSA) and replication fork regression DNA repair pathways (11).
It was recently discovered that UvsW has strand annealing activity (11). Strand annealing activity is a common property among eukaryotic RecQ helicases such as Werner’s and Bloom’s syndrome helicases. It is thought that strand annealing is a physiologically important property, as only RecQ helicases with strand annealing activity are capable of catalyzing replication fork regression (12, 13). The annealing activity of UvsW does not require ATP but is increased in its presence. ATP hydrolysis accompanies the activation of ssDNA annealing since non-hydrolyzable ATPγS is an inhibitor rather than an activator of the reaction. The proposed mechanism for strand annealing in the absence of ATP involves the independent binding of two ssDNA strands at separate locations on UvsW, followed by spontaneous annealing of the properly aligned ssDNA strands (11, 14). To explain the ATP activation, it was proposed that ATP-driven translocation of UvsW along one of the two ssDNA strands is necessary in situations where annealing is prohibited due to non-complementarity or misalignment of the ssDNA. T4 ssDNA binding protein (gp32) attenuates strand annealing, as does UvsW.1. UvsW.1 is a small (9 kD) protein that physically associates with UvsW (11). A recent NMR structure of UvsW.1 revealed a highly electronegative four-helix bundle that resembles the HRDC domain from RecQ helicases (15). The precise role of UvsW.1 is not known, however based on analogy to the HRDC domain, it has been suggested that it may assist UvsW in substrate recognition (15).
The structure of the ligand-free form of UvsW has been determined using X-ray crystallography (15, 16). The structure reveals two RecA-like motor domains, a variable domain at the N-terminal region and a small subdomain located near the C-terminus. The N-terminal variable domain resembles a “double wing” motif and contains a putative substrate binding site (17). However, the structure does not contain a DNA substrate or resemble other helicases outside of the RecA-like domains, making predictions regarding substrate recognition difficult. ATP is predicted to bind in the interface between the two RecA-like domains and a model for the ATP and ssDNA bound form has been generated (15). An inchworm model of DNA unwinding has been proposed for UvsW based on the similarity between the UvsW structure and the structures of other monomeric helicases, mainly the SF1 helicase PcrA (18). The inchworm model involves the repeated movements of the RecA-like motor domains that are driven by the hydrolysis and release of ATP and ADP + Pi, respectively. These movements are conformationally coupled to DNA translocation, which drive the helicase into the DNA duplex where it acts as a “wedge” to separate the DNA strands. The crystal structure of ligand-free form of UvsW is monomeric, however, the oligomeric state has not been investigated in solution.
The mechanism of DNA unwinding is likely to be closely linked to the mechanism of DNA translocation (18). Indeed, most theoretical models of DNA unwinding are based primarily on the ability of the helicase to translocate on single-stranded (ss) or double-stranded (ds) DNA (19–21, 18). Consequently, a complete understanding of ssDNA translocation is necessary to accurately describe the mechanism of dsDNA unwinding at the molecular level. Here, we investigate the properties of UvsW translocation on an ssDNA lattice. We find that UvsW translocates on ssDNA uni-directionally and in an ATP-dependent fashion. UvsW is highly processive during ssDNA translocation and is able to resist inhibition by both an inactive mutant UvsW and a heparin trap. We also provide functional evidence indicating that UvsW is monomeric during ssDNA translocation. Together, these data represent a first step toward the mechanistic understanding of ssDNA translocation by UvsW.
Experimental Procedures
Oligonucleotide substrates were purchased from Integrated DNA Technologies (Coralville, IA) and used as received. Single-stranded M13 DNA (ssM13) from the M13mp18 vector was prepared using standard protocols (22). The pet28a vector was purchased from Novagen. Phosphoenolpyruvate (tricyclohexylammonium salt), ATP (disodium salt), NADH, streptavidin and a buffered aqueous glycerol solution of phosphoenolpyruvate kinase/lactate dehydrogenase were obtained from Sigma.
Mutagenesis, Expression, and Purification of wild-type and K141A UvsW proteins
The K141A mutant was introduced into UvsW-pet28 expression vector (11). The mutation was made using the Quickchange mutagenesis method (Stratagene) and the entire UvsW open reading frame was sequenced using the dideoxy terminator method. The sequence of the mutagenic primer was as follows with the bold underlined letters indicating the mutation site (reverse primer is the reverse complement of the forward): 5’-CTTCCAACATCTGCAGGTGCGTCTTTAATTCAAGCTTTGC-3’.
Expression and purification of wild-type and the K141A mutant was performed essentially as described (11). Briefly, transformed BL21 (DE3) cells were grown at 37°C to an OD600 of 0.8, followed by cooling to 18°C and induction with 0.1 mM of IPTG. The cultures were then incubated for 16 h before being pelleted by centrifugation. The cell pellet was resuspended in 60 mL of 20 mM Tris-HCl, 500 mM NaCl, and 5 mM imidazole (pH 8.0), lysed using sonication, and centrifuged at 17000g for 45 min. The lysate was then loaded onto a nickel nitrilotriacetic acid-agarose column and washed with 50 volumes of lysis buffer containing 40 mM imidazole and 1M NaCl. The protein was eluted with lysis buffer containing 100 mM imidazole. The eluted protein was diluted 3-fold with P11-A buffer (20 mM Tris-HCl, 400 mM NaCl, pH 7.5) and loaded onto a 20 ml P11 phosphocelluose column. The column was washed with 10 column volumes of the same buffer before eluting UvsW with a 0.4 – 1.5 M NaCl gradient. Fractions containing UvsW were pooled and concentrated to 22 µM using an Amicon centrifugation device and frozen in aliquots at −80°C. Protein concentration was calculated based on an extinction coefficient of 73920 M−1 cm−1.
DNA sequences
The sequences of oligonucleotides used are as follows:
d(N)5: 5’-GACAC-3’
d(N)10: 5' - ATGACACAGT - 3'
d(N)15: 5' - AGATGACACAGTAAG - 3'
d(N)20: 5' - ACACAGTAAGAGATGACACA - 3'
d(N)30: 5' - AGATGACACAGTAAGAGATGACACAGTAAG - 3'
d(N)45: 5' - ACAGTAAGAGATGACACAGTAAGAGATGACACAGTAAGAGATGAC -3'
d(N)60: 5' - AGATGACACAGTAAGAGATGACACAGTAAGAGATGACACAGTAAGAGATGACACAGTAAG - 3'
d(N)90: 5' - AGATGA CACAGTAAGAGATGACACAGTAAGAGATGACACAGTAAGAGATGACACAGTAAGAGATGACACAGTAAGAGATGACACAGTAAG - 3'
d(N)150: 5' - AGATGACACAGTAAGAGATGACACAGTAAGAGATGACACAGTAAGAGATGACACAGTAAGAGATGACACAGTAAGAGATGACACAGTAAGAGATGACACAGTAAGAGATGACACAGTAAGAGATGACACAGTAAGAGATGACACAGTAAG - 3'
ATP Hydrolysis Assays
In most cases, UvsW ATPase activity was monitored spectrophotometrically with the coupled NADH oxidation assay (23). All assays were performed at 25 °C in replication buffer [25 mM Tris-OAc, pH 7.8, 125 mM potassium acetate, 10 mM magnesium acetate] containing 1.0 mM ATP, 10 units/ml phosphoenolpyruvate kinase, 5 units/ml lactate dehydrogenase, 2 mM phosphenolpyruvate, 0.2 mM NADH. ATP hydrolysis rates were followed by measuring the conversion of NADH to NAD+ at 340 nm with an extinction coefficient of 6220 M−1 cm−1. The oxidation of 1 mol of NADH corresponds to the hydrolysis of 1 mol of ATP. In general, each time course lasted approximately 5 minutes with at least 2 minutes of linear activity being used to determine the initial rate.
Where indicated, UvsW ATPase activity was monitored using thin layer chromatography (TLC). For the TLC assay, the reactions were carried out in replication buffer containing 1 mM ATP and 10 µCi of [α-32P] ATP in a total volume of 20 µL. At appropriate time points, 2 µL aliquots were removed and quenched with 1 µL of 0.5 M EDTA. Each time point was spotted onto a PEI-cellulose TLC plates and developed with 300 mM KPi, pH 7.0. After drying, the TLC plate was exposed to a phosphorimager cassette for 16 hours and quantitated using the phosphorimager. Initial rate data were fitted to the Michaelis-Menten equation using either IgorPro or Kaleidagraph software.
Results
UvsW ATPase Activity Depends on Length of ssDNA Lattice
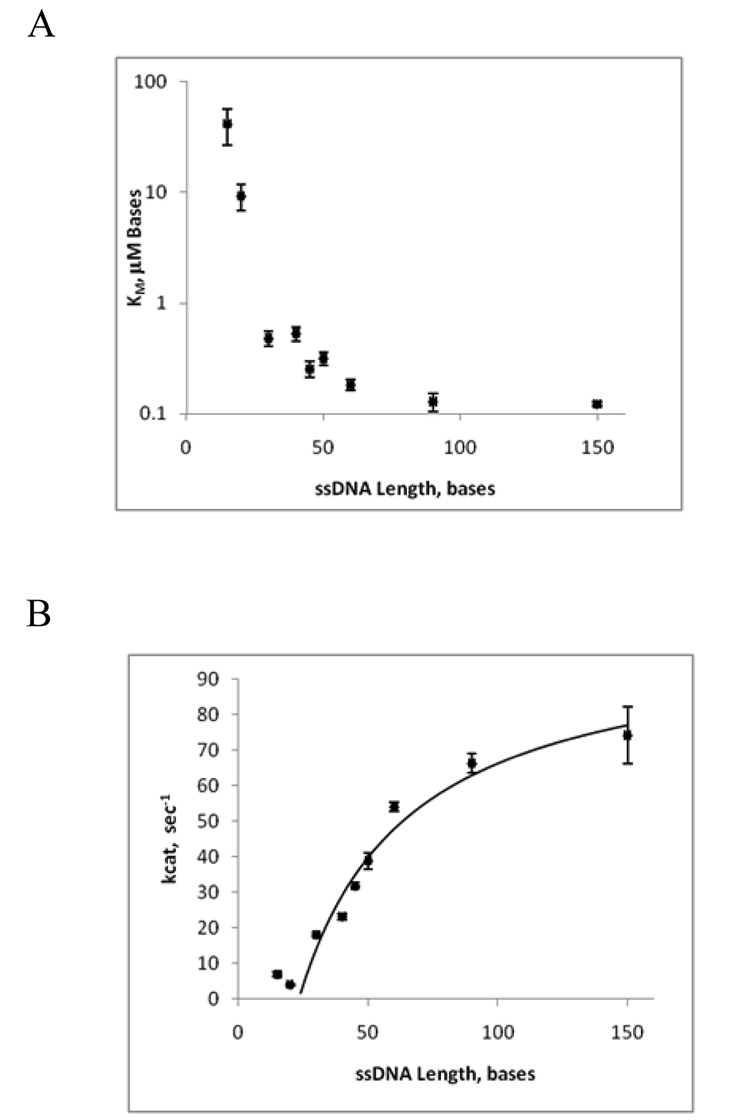
It was previously shown that UvsW is a DNA-dependent ATPase (8, 11). We set out to determine if the ATP hydrolysis activity of UvsW is coupled to translocation. A formal theory has been developed to allow determination of translocation based on the steady-state kinetics of the ATPase activity of a given protein (24, 25). The basis of the theory is that if a protein translocates on an ssDNA lattice, its ATPase activity should depend on the length of that lattice. We determined the KM-DNA and kcat parameters for UvsW on ssDNA oligonucleotides with lengths ranging from 10 to 150 bases (Fig. 1). The sequences of the d(N)10–150 oligonucleotides were rationally designed to eliminate any possible secondary structure (hairpins) or inter-strand duplex formation. The KM-DNA, which can be thought of as a measure of the affinity of UvsW for the ssDNA substrate drops sharply as the length of the oligonucleotide is increased from 10 to 30 bases (>50-fold decrease) whereas at lengths greater than 30 bases, the KM-DNA remains relatively constant (<2fold change) (Fig 1A). This suggests that the optimal binding site size for activating the translocation activity of UvsW is approximately 20–30 bases. The kcat for UvsW increases with increasing oligonucleotide length (Fig. 1B). The kcat remains relatively constant at DNA lengths where KM-DNA is not optimum and then increases in a hyberbolic fashion from 20 to 150 bases, indicating that even on oligonucleotides where the binding affinity has reached a maximum, the velocity of ATP turnover continues to increase with lattice length. This is highly indicative of an ATP-driven ssDNA translocase. This data was tested against a number of different kinetic schemes with the best fitting shown as Scheme I [Scheme IIIb in (24)].
Figure 1. UvsW kinetic parameters as a function of ssDNA length.
A. The KM-ssDNA of UvsW as a function of the ssDNA length. B. The kcat of the ATPase activity of UvsW as a function of ssDNA length. The solid line represents a fit to the data using Equation 1. The data for the ssDNA lengths of 10 and 15 bases were not included in the fit. The error bars in A and B represent the standard deviation of the fit to the Michaelis-Menten equation.
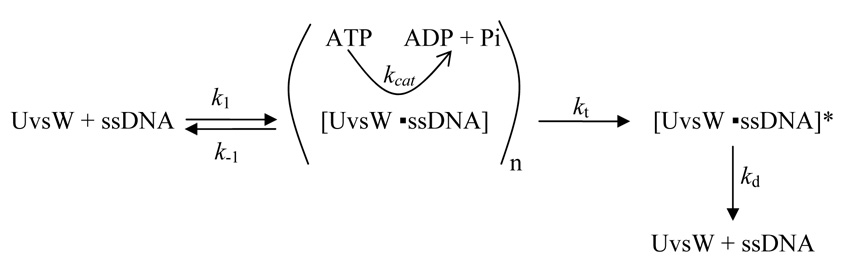
SCHEME 1.
The kcat values from DNA lengths of 20–150 bases were fitted to the following equation:
| Equation 1 |
where:
| Equation 2 |
where V is the kcat on infinitely long ssDNA, λ is the length of the ssDNA, n is the minimal length of ssDNA required for maximal binding (the length where KM-DNA approaches a minimum), and k1, k−1, kt, and kd are the rate constants shown in Scheme I. The rate constant for UvsW translocation to the end of the ssDNA (kt) is related to the intrinsic rate of translocation of a single base (ki) by the following relationship:
| Equation 3 |
The values for DNA lengths of 10 and 15 bases were not included in the fit because these lengths are below the minimum optimal length for UvsW binding (Fig. 1A). The values obtained from fitting the data shown in Fig. 1B to Eq. 1 are V = 102.6 ± 16.0 sec−1, KG = 65.3 ± 9.5 bases, and n = 23 ± 4.0 bases.
Directionality of UvsW ssDNA Translocation
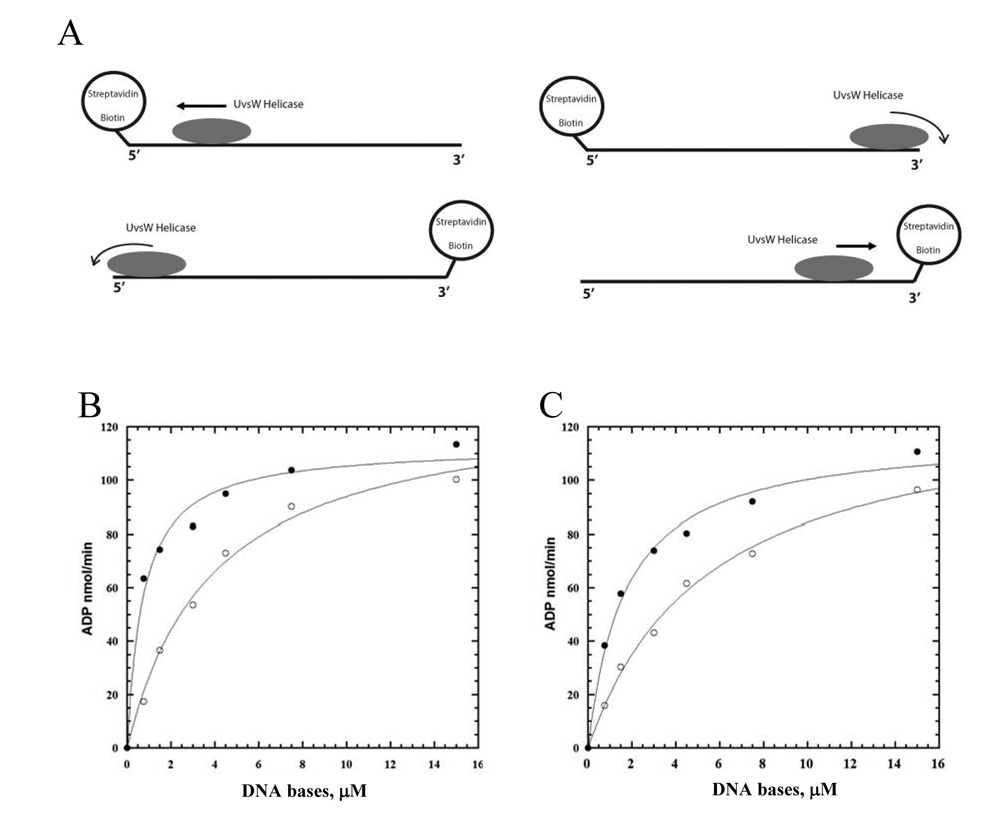
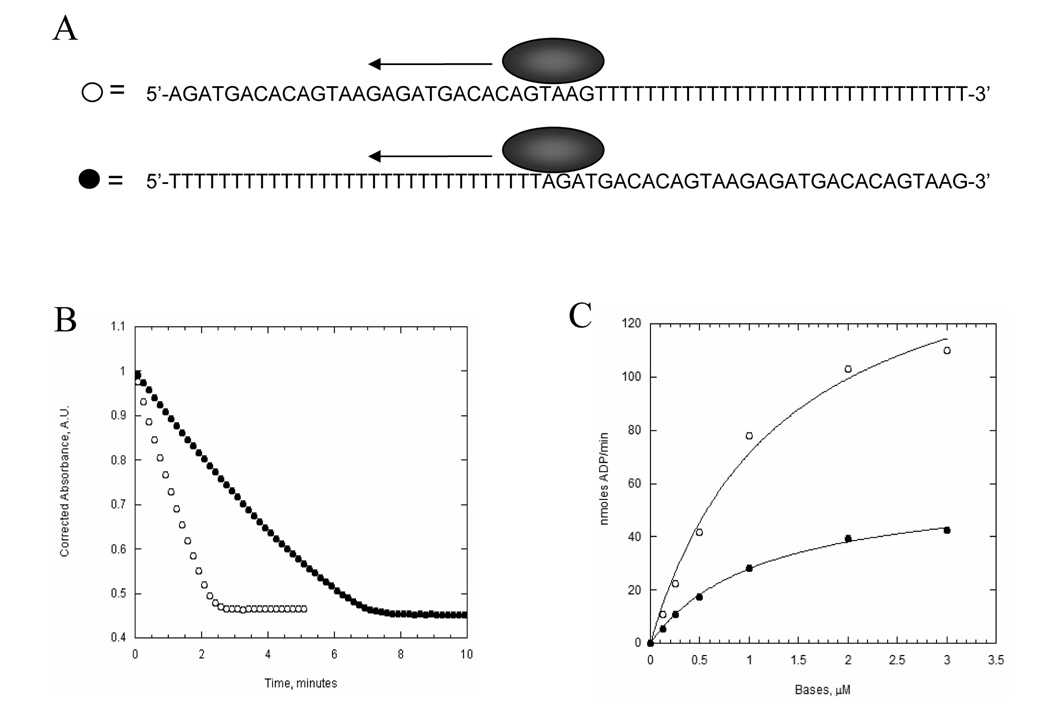
To determine the directionality of UvsW translocation, we placed biotin groups at either the 3’ or 5’ end of a 30 base oligonucleotide (26). Initially, we attempted to use the streptavidin displacement assay (27), however no displacement was observed. Evidently, UvsW does not generate as much force as the other T4 phage helicases (Dda and gp41). Even without displacement, streptavidin can be used to determine directionality (28). In the presence of streptavidin, translocation of UvsW off the blocked end should be greatly reduced (Fig. 2A) and it is assumed this will manifest itself in altered ATPase kinetics (e.g., an increased residence time for UvsW will decrease the KM-DNA). The dependence of UvsW ATPase activity as a function of the concentration of ssDNA with biotin/streptavidin at either the 5’ or 3’ end is shown in Figure 2B. When the block is at the 5’ end of the DNA substrate, the KM-DNA decreases approximately 9-fold and the kcat remains unchanged relative to the unblocked substrate (Table I). A block at the 3’ end results in a more modest decrease in the KM-DNA (3-fold) and a similar kcat value relative to the unblocked substrate (Table I). The greater difference in KM-DNA when the block is located at the 5’-end of the ssDNA suggests that UvsW translocates in the 3’ to 5’ direction. However, because there is a decrease in the in KM-DNA with the block at the 3’-end (albeit less than a 5’ block) we developed an additional assay for directionality.
Figure 2. The streptavidin-block assay for translocation directionality.
A. An illustration depicting the translocation of UvsW helicase on the biotin/streptavidin bearing oligonucleotide. Translocation away from the streptavidin block leads to rapid dissociation from the end of the DNA, whereas dissociation is prevented if translocation occurs in the direction of the streptavidin block. ATPase activity of UvsW (22 nM) as a function of the 5’-biotin-labeled 30mer (B) and 3’-biotin-labeled 30mer (C) in the presence (closed circles) and absence (open circles) of streptavidin (1 µM).
Table 1.
KM and kcat for ssDNA Substrates used for Directionality Assays
| Substrate | KM(µM) | kcat(sec−1) |
|---|---|---|
| 5’-bio-d(N)30 | 4.5 ± 0.4 | 19.8 ± 0.8 |
| 5’-SA-bio-d(N)30 | 0.5 ± 0.1 | 16 ± 0.4 |
| d(N)30-bio-3’ | 5.4 ± 0.6 | 19.7 ± 0.8 |
| d(N)30-bio-SA-3’ | 1.7 ± 0.2 | 17.7 ± 0.7 |
| 5’-d(T)30-d(N)30-3’ | 1.1 ± 0.1 | 16.5 ± 1.3 |
| 5’-d(N)30-d(T)30-3’ | 1.3 ± 0.2 | 45.6 ± 3.9 |
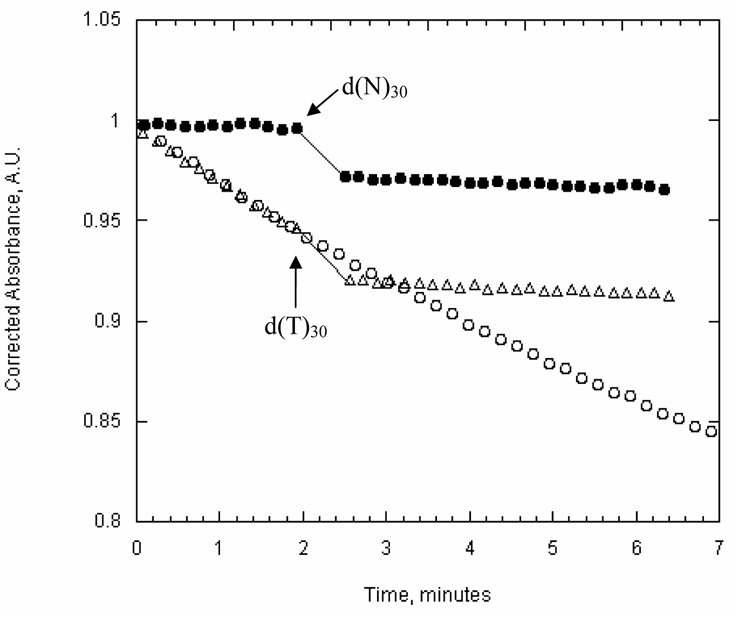
During the course of our characterization of UvsW, we discovered that the ATPase activity of UvsW is reduced when homopolymeric oligonucleotides are used as substrates (Fig. 3; Table II). This reduction is not due to a decrease in binding affinity since poly-(dT)30 effectively competes with the d(N)30 oligonucleotide for the binding of UvsW (Fig. 3). This property enabled us to determine the direction of translocation by placing d(T)30 on either the 5’ or 3’ end of the d(N)30 ssDNA substrate (Fig. 4A). The rate of ATP hydrolysis is approximately 3-fold greater when poly-d(T)30 is placed on the 3’ as compared to the 5’ end (Fig. 4B). The ATPase activity of UvsW as a function of oligonucleotide concentration reveals that the higher activity is due to an increase in the maximal turnover rate (kcat) and not a change in the KM-DNA. These data are consistent with the results from the streptavidin block assay and indicate that UvsW translocates in a 3’ to 5’ direction on ssDNA substrates.
Figure 3. UvsW ATPase activity in the presence of d(N)30 and d(T)30 ssDNA substrates.
ATP hydrolysis of UvsW (10 nM) measured by the coupled spectrophotometric method as described in “Materials and Methods”. The open circles represent the rate of ATP hydrolysis in the presence of 30 µM d(N)30 alone. The closed circles represent the rate of ATP hydrolysis in the presence of 150 µM d(T)30 and the arrow indicates the addition of d(N)30 at concentration of 30 µM to the ongoing reaction. The open triangles represent the rate of ATP hydrolysis in the presence of 30 µM d(N)30 and the arrow indicates the addition of d(T)30 at concentration of 150 µM to the ongoing reaction.
Table 2.
KM and kcat for Various ssDNA Substrates
| Substrate | KM(µM) | kcat(sec−1) |
|---|---|---|
| d(N)30 | 5.8 ± 0.5 | 19.7 ± 0.3 |
| d(T)30 | 3.8 ± 0.2b | 0.47 ± 0.06b |
| d(A)30 | N.D.a | 0.02 ± 0.01bc |
| d(C)30 | N.D.a | 0.05 ± 0.03bc |
| d(AC)15 | 8.7 ± 1.2 | 5.4 ± 0.2 |
| d(TC)15 | 5.6 ± 1.5 | 3.2 ± 0.2 |
| ssM13 | 1.9 ± 0.3 | 75 ± 3 |
| ----d | N.D.a | 0.00 ± 0.02 |
Not determined
Value obtained using TLC plate assay.
The kcat reported is an apparent kcat at saturating ATP and ssDNA concentrations
no addition of ssDNA
Figure 4. The poly-dT assay for translocation directionality.
A. An illustration depicting the 3’ to 5’ translocation of UvsW helicase on ssDNA containing d(T)30 on either the 5’ or 3’ end of d(N)30. UvsW will bind at random positions throughout the ssDNA with an average position in the center of the substrate. Because activation of ATP hydrolysis is minor with d(T)30 as compared to d(N)30, translocation away from the d(T)30 segment leads to higher ATPase activity than translocation into the d(T)30 segment. B. Time courses of ATP hydrolysis by UvsW (20 nM) in the presence of 2 µM 5’-d(T)30-d(N)30-3’ (closed circles) or 5’-d(N)30-d(T)30-3’ (open circles). C. Rate of ATP hydrolysis by UvsW as a function of 5’-d(T)30-d(N)30-3’ (closed circles) or 5’-d(N)30-d(T)30-3’ (open circles) concentration. The solid lines are fits to the data using the Michaelis-Menten equation and these values are found in Table II.
Processivity of UvsW ssDNA Translocation
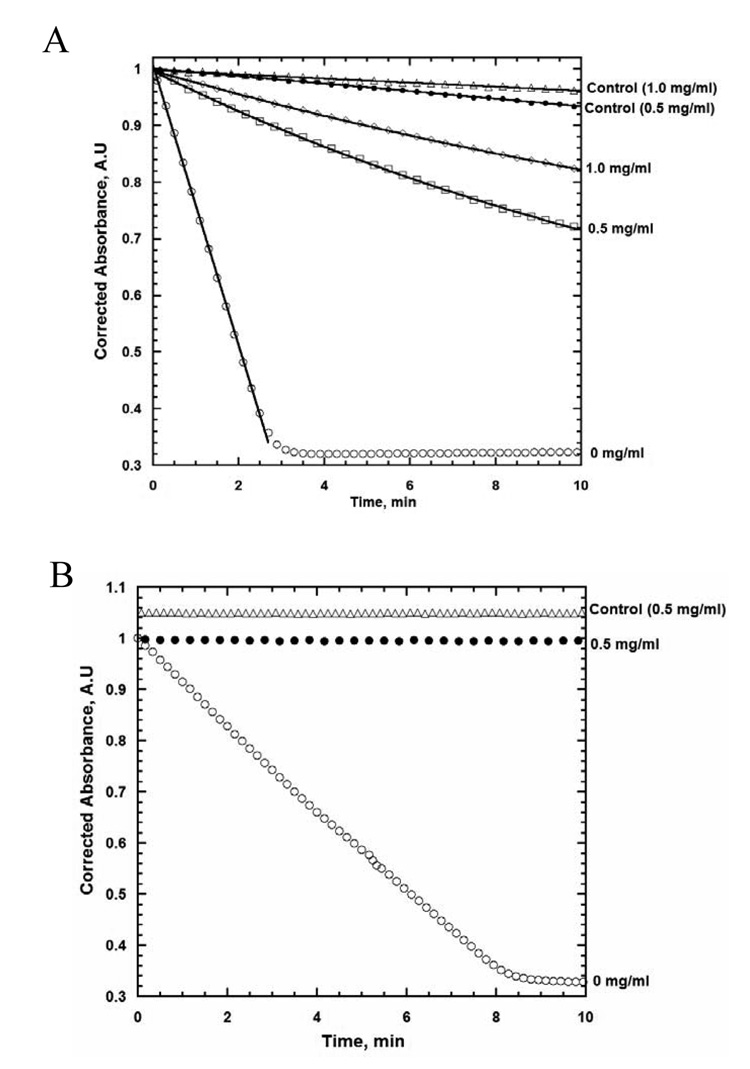
To determine the processivity of UvsW as it translocates on ssDNA, we utilized the ability of heparin bind UvsW in competition with ssDNA. Heparin has been commonly used as a passive trap for protein that bind nucleic acids (29, 30). To determine the dissociation rate of UvsW, the enzyme is preincubated with ssDNA for several minutes followed by the simultaneous addition of ATP and heparin. The dissociation of UvsW from ssDNA is monitored by the time-dependent decrease in ATPase activity. Figure 5 shows the results of this experiment. Two different concentrations of heparin were used. First, as a control, UvsW and ATP are added simultaneously to a reaction mix containing ssDNA and heparin (0.5 or 1 mg/ml). This experiment demonstrates the ability of heparin to successfully outcompete ssDNA for UvsW binding and that the ATP hydrolysis rate of the heparin-bound form of UvsW is negligible. A slight increase in ATPase hydrolysis rate is observed when the heparin concentration is lowered from 1 mg/ml (slope = 0.02 A.U./sec) to 0.5 mg/ml (slope = 0.036 A.U./sec), indicating that at 0.5 mg/ml heparin is not a 100% effective trap. When UvsW is preincubated with ssDNA followed by the addition of ATP and heparin (0.5 mg/ml final), an immediate reduction (slope = 2.16 A.U./sec over the first minute) in the rate of ATPase hydrolysis is seen (as compared to the reaction without heparin which has a slope of 14.8 A.U./sec) followed by a very slow decrease in the rate of ATP hydrolysis over a period of 10 minutes. When the concentration of heparin is increased to 1 mg/ml, a further immediate reduction in ATPase activity is observed (slope = 1.53 A.U./sec over the first minute), again followed by a slow decrease in the rate of ATP hydrolysis. The time-dependent decrease in ATP hydrolysis rate at heparin concentrations of 0.5 and 1.0 mg/ml can be fitted to a single exponential equation with (dissociation) rate constants of 0.062 ± 0.001 and 0.053 ± 0.009 min−1, respectively. The immediate reduction in ATP hydrolysis rate upon addition of heparin is likely due to the trapping of unbound and/or non-productively bound UvsW at the time of heparin addition. The fact that the dissociation rate constants at both concentrations of heparin are similar indicates that heparin is unable to trap UvsW that is actively translocating along the ssDNA. The greater immediate reduction in ATPase activity at the higher heparin concentration suggests that heparin is able to trap non-productively bound UvsW. In this scenario, UvsW exists in two forms on ssDNA, one that is susceptible to heparin and one that is resistant. It is conceivable that these forms are able to interconvert without dissociating from the ssDNA (see below). To ensure that UvsW was indeed translocating on the ssDNA, as opposed to hydrolyzing ATP while remaining stationary, we repeated the same order of addition reaction on a 60 base oligonucleotide. With this short linear ssDNA substrate, UvsW should very rapidly translocate off the 5’-end of the DNA and be trapped by the heparin. As shown, regardless of order of addition, no ATP hydrolysis is observed in the presence of heparin when the d(N)60 oligonucleotide is used.
Figure 5. Effect of Heparin trap on the ATP hydrolysis of UvsW in the presence of circular single-stranded M13 DNA and 60mer oligonucleotide.
ATP hydrolysis of UvsW measured by the coupled spectrophotometric method with different order of additions. A. In control reactions, UvsW (47 nM) and ATP (1 mM) were preincubated together followed by the simultaneous addition of ssM13 DNA (25 µM bases) and heparin at the indicated concentrations. Otherwise, UvsW was preincubated with ssM13 DNA (25 µM bases) followed by the simultaneous addition of ATP (1 mM) and heparin at the indicated concentrations. B. Same order of addition as A. except that d(N)60 was used in place of ssM13 and only a single concentration of heparin was used.
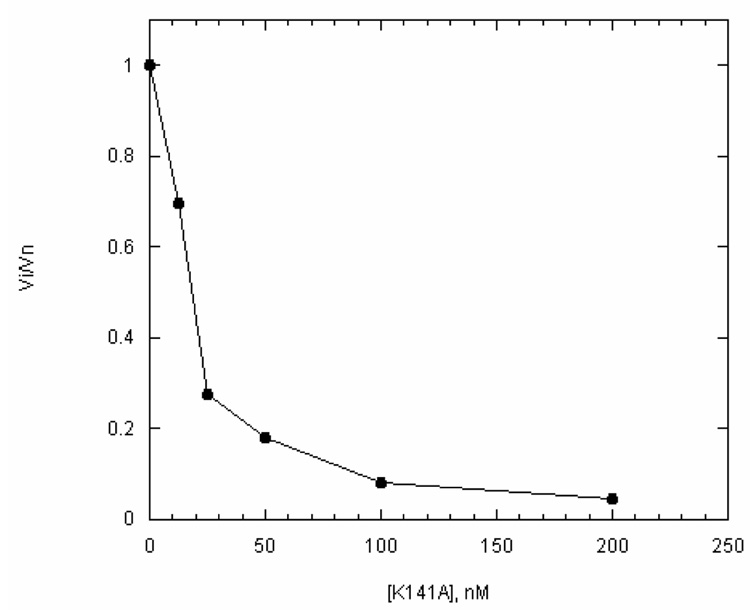
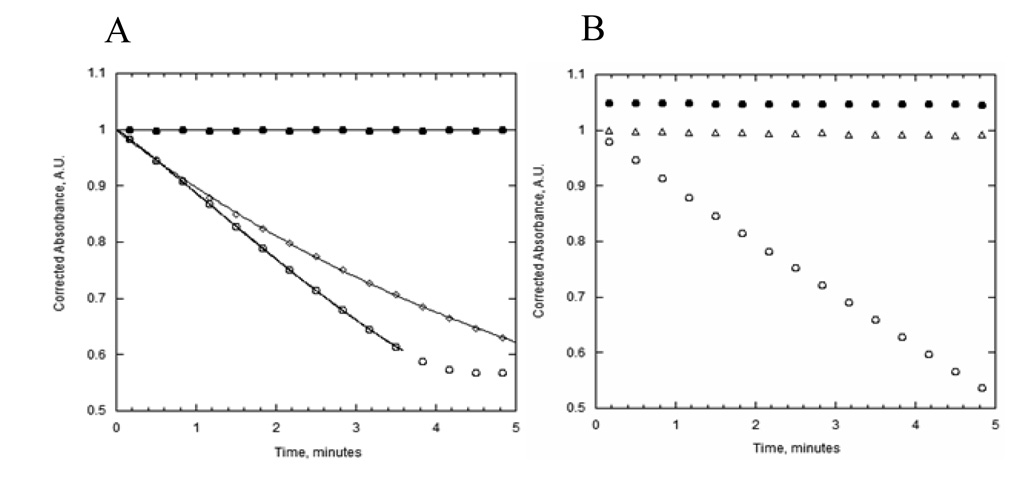
Because heparin appears to bind to a ssDNA-bound form of UvsW, we engineered a protein trap that would compete with the wild-type enzyme for the ssDNA substrate. An essential lysine residue, Lys141, was mutated to Ala. This residue is predicted to interact with the γ-phosphate of UvsW-bound ATP and has been shown to be necessary for ATP hydrolysis (8, 15). To confirm that the trap mutant was able to compete with the wild-type protein for ssDNA, we added increasing amounts of the trap protein to a reaction containing a fixed amount of wt-UvsW (0.1 µM ) and a near-limiting amount of ssDNA (4 µM) (Fig. 6). K141A effectively out-competes the wild-type protein for the available ssDNA binding sites, as seen by the decrease in ATP hydrolysis at increasing concentrations of K141A. This indicates that K141A is able to bind and sequester ssDNA, which prevents the binding of wt-UvsW. It also suggests that once bound, K141A is able to block the translocation of the wild-type enzyme, since 90% inhibition is observed when the ratio of wild-type to K141A is 1:1. K141A was used in a similar manner as the heparin order of addition experiments (Fig. 7). When a 10-fold molar excess of K142A (over wt-UvsW) is simultaneously added with wt-UvsW to a reaction containing ATP and ssDNA, no ATP hydrolysis is observed (slope = 0.006 A.U./sec), consistent with the data shown in Fig. 6. However, when wt-UvsW is pre-incubated with the ssDNA substrate and K141A and ATP are added simultaneously, the rate of ATP hydrolysis over the first ~60 seconds of the reaction is nearly equivalent to a reaction performed without K141A (slope = 6.12 and 6.96 A.U./sec, respectively). The decrease in ATP hydrolysis rate fits well to a single-exponential equation with a dissociation constant of 0.162 ± 0.004 min−1. It is unclear why the dissociation constant is greater when K141A is used as a trap; however, one likely explanation is that because K141A is able to block translocation of the wild-type enzyme the dissociation constant is over-estimated. Regardless, even using the more rapid dissociation constant, the half-life of actively translocating UvsW is > 6 minutes. With this dissociation constant and assuming that ATPase hydrolysis is strictly coupled to translocation the average number of distance translocated in a single binding event is 38 kb. Another interesting difference between the heparin and K141A traps is that only heparin causes the immediate decrease in ATP hydrolysis rate. A likely explanation for this difference is that unlike heparin, K141A is not able to trap nonproductively bound UvsW. If wild-type UvsW that is bound to ssDNA is able to interconvert between productive and nonproductive forms, it is possible that K141A is unable to displace or trap the nonproductive (heparin sensitive) form, allowing time for conversion into the translocating form. It is likely that binding of ATP would facilitate the interconversion. Again, as a control experiment to ensure that ATP hydrolysis is linked to UvsW translocation, we repeated the same order of addition experiment using a d(N)60 oligonucleotide and observed complete inhibition of ATP hydrolysis even when wt-UvsW is preincubated with the DNA substrate. This result is expected if UvsW quickly translocates of the ssDNA, which is then bound by the excess of K141A in solution.
Figure 6. Effect of K141A on the rate of ATP hydrolysis by wt-UvsW.
A plot of normalized initial velocities of ATP hydrolysis versus the concentration of K141A. Vi and Vn represent the rate of ATP hydrolysis in the absence and presence of the indicated concentration of K141A, respectively. All reactions were carried out using the coupled spectrophotometric method with a fixed concentration of UvsW (100 nM) and a limiting amount of ssM13 (4 µM).
Figure 7. Effect of K141A trap on the ATP hydrolysis of UvsW in the presence of circular single-stranded M13 DNA and 60mer oligonucleotide.
A. ATP hydrolysis of UvsW (100 nM) measured by the coupled spectrophotometric method with ssM13 DNA (4 µM bases) with different order of additions. UvsW was preincubated with ssM13 for 30 minutes prior to the initiation of the reaction with ATP (1 mM) and either buffer (open circles) or 1 µM K141A (open triangles). As a control to confirm the effectiveness of the trap protein, all components were added to the reaction mix simultaneously (closed circles). B. Same order of addition as A. except that d(N)60 was used in place of ssM13.
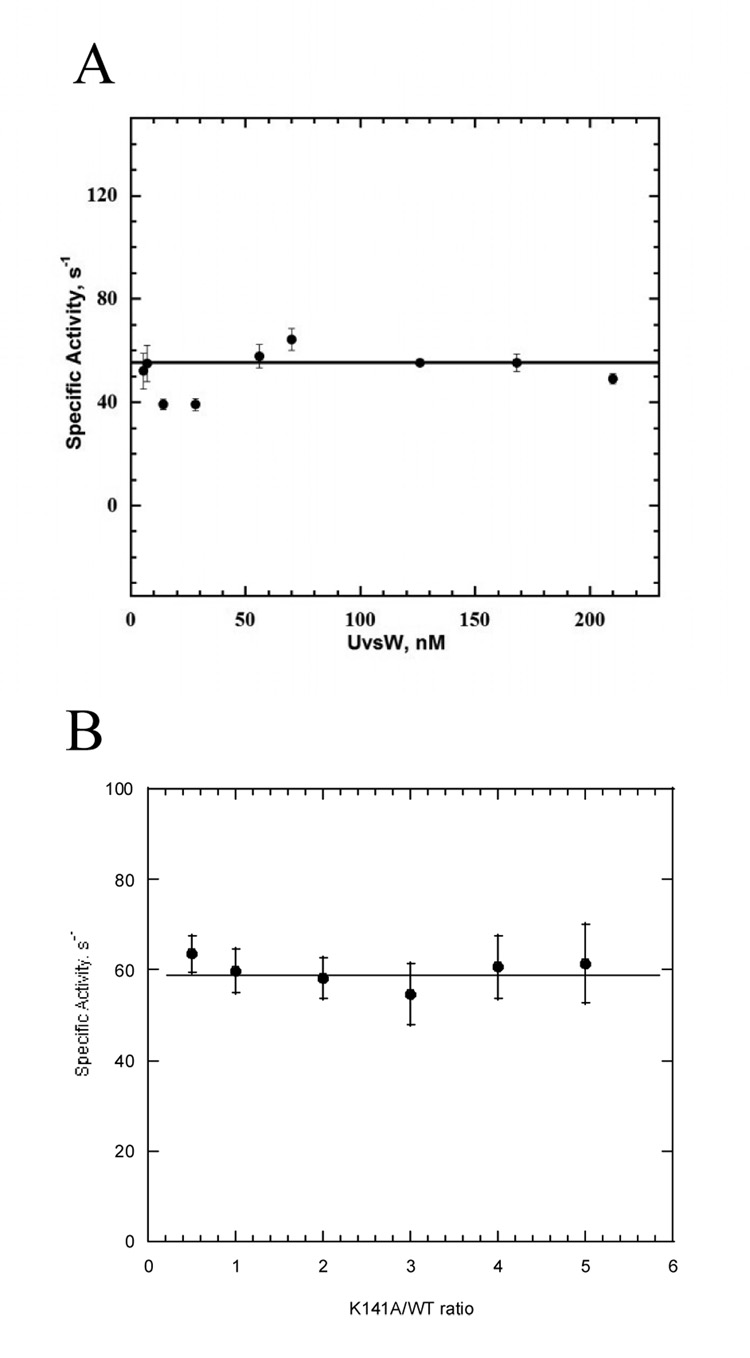
The high processivity of UvsW prompted us to determine the oligomeric state of UvsW as it translocates along the ssDNA substrates. Often, highly processive helicases are oligomeric, although there are several exceptions to this rule. We began by attempting analytical ultracentrifugation in the presence of the d(N)30 oligonucleotide and the non-hydrolyzable ATP analog ATPγS. Unfortunately, we were unable to utilize this method due to rapid precipitation of the protein at concentrations necessary to obtain a suitable absorbance signal (data not shown). For this reason, we turned to functional assays which report on the oligomeric status of the protein during active translocation on ssDNA. We first analyzed the relationship of protein concentration to the kcat of ATP hydrolysis (Fig. 8A). Over a range of 2 to 200 nM UvsW, we observe no dependence of the rate of ATP hydrolysis on UvsW concentration. This indicates that either UvsW is monomeric while translocating on ssDNA or the Kd for UvsW subunit association is less than 2 nM. To rule out the latter possibility, we examined the effect of different K141A to wt -UvsW ratios on the ATP hydrolysis rate under conditions of excess ssDNA (Fig 8B). In these experiments, wt-UvsW and K141A were preincubated in the absence of ATP and ssDNA for 120 minutes prior to the initiation of the reaction. Under these conditions, if UvsW is able to form high-order oligomeric states, then heterooligomers should occur between wt-UvsW and K141A subunits (31). Since K141A cannot hydrolyze ATP, if UvsW translocates in an oligomeric state we would expect a reduction in the ATP hydrolysis rate as the ratio of K141A to wt-UvsW is increased. This analysis assumes that the individual subunits within the oligomer do not function completely independent of each other, which is the case with all characterized oligomeric helicases (32, 33). The activity of wt-UvsW is unaffected by an increase in the K141A to wt-UvsW ratio when ssDNA is in excess of protein. This strongly suggests that UvsW is monomeric during ATP-fueled ssDNA translocation.
Figure 8. Functional assays for the determination of oligomeric state.
A. Concentration dependent DNA stimulated ATPase activity of UvsW. ATPase activity was measured by the coupled spectrophotometric method as described in the experimental section. 60mer oligonucleotide in 25 mM Tris acetate (pH: 7.8), 125 mM KOAc, 10 mM magnesium acetate was mixed with varying concentrations of UvsW (5–210 nM). Specific activity of ATP hydrolysis is plotted as a function of UvsW enzyme concentration. B. DNA stimulated ATPase activity with varying ratios of K141A mutant to wild type UvsW. The reaction was initiated by the addition of varying ratios of the K141A mutant and the wild-type UvsW to a mixture of 60-mer ssDNA and ATP (1 mM) in ATPase buffer. Shown here is a plot of the amount of ADP produced as a function of K141A mutant/wt UvsW.
Discussion
T4 phage encodes for all the proteins necessary for the replication and maintenance of its genome. Remarkably, T4 is able to carry out these vital functions using far fewer proteins than higher systems such as E. coli or S. cerevisiae. For this reason, T4 phage is an ideal model system for understanding the basic mechanisms of DNA replication and repair. The T4 replication apparatus, the replisome, has been extensively examined over the past three decades (34, 35). In contrast, the activities and mechanisms of the T4 repair proteins are relatively unknown. However, it is clear that DNA repair processes are tightly linked with DNA replication (36). For example, it has been estimated that the E. coli replication machinery stalls several times during each cycle of chromosome replication (37). Restart of the stalled replisome is dependent on several DNA repair proteins (38). Clearly, a complete understanding of DNA replication requires an understanding of DNA repair and how these two processes are intertwined. One protein implicated in the restart of the stalled T4 phage replisome is UvsW helicase. UvsW is one of three helicases found in T4 phage and is thought to play a variety of roles during the T4 phage lifecycle including regulating the switch between origin-dependent and origin-independent initiation of replication, the processing of stalled replication forks, and Holliday junction branch migration (4, 8, 9, 11). In this study, we have begun our mechanistic characterization of UvsW by determining its directionality, processivity, and oligomeric state during ssDNA translocation. We have found that UvsW translocates on ssDNA in an ATP-dependent fashion in a 3’ to 5’ direction. UvsW displays remarkably high processivity during translocation and it does so as a monomeric protein.
Until this work, the ability of UvsW to translocate on ssDNA was not clear. UvsW can unwind a variety of DNA substrates, many of which do not contain any regions of ssDNA. Additionally, several SF2 family helicases translocate on dsDNA rather than ssDNA. These two observations led to the proposal that UvsW, like Rad54, translocates exclusively on dsDNA (39, 40). The increase in kcat as a function of ssDNA length clearly demonstrates that UvsW is capable of translocating on ssDNA in an ATP-dependent manner. Using the kinetic theory for translocating ATPases developed by Von Hippel and colleagues (24), we can discern several properties of UvsW ssDNA translocation. First, the data indicate that UvsW translocates in a unidirectional fashion (as opposed to bidirectional). This property is evident by the hyperbolic dependence of kcat on ssDNA length (using ssDNA lengths where the KM -DNA is constant). The data also enable us to quantitatively determine the minimal optimal ssDNA length required for maximal binding and translocation (approximately 24 bases). This value contrasts with the determined optimal binding site size for ssDNA, which is 10–12 bases (Perumal and Benkovic, unpublished data). Clearly, the KM-DNA does not only reflect binding affinity, but also depends on the rates of translocation and dissociation from the DNA end. The constant KG has been used as a measure of processivity (41, 25); however, in the translocation mechanism given in Scheme I, KG is a combination of the intrinsic translocation rate (ki), the rate of dissociation from the end of the DNA template (kd), and the rate of dissociation from an internal site on the DNA template (k−1) (see Eq. 2). The heparin and mutant trapping experiments indicate that k−1 is extremely small, simplifying Eq. 2 and 3 to KG = 2ki/kd. Determination of ki from steady-state kinetic data is not possible; however, if we assume that ATP hydrolysis is completely coupled to the translocation of a single base, then ki = V = 102.6 bases▪sec−1 and since KG = 65.3 bases then kd = 3.1 sec−1, a value for which the kcat is approached using very short ssDNA templates (Fig. 1B).
UvsW translocation is biased in the 3’ to 5’ direction. Two assays were used to determine directionality. The first was a well-established assay using biotin-streptavidin blocks at either the 5’ or 3’ ends of the DNA template (28). Several criteria must be fulfilled for the output of this assay to be useful (i.e., a change in activity upon addition of streptavidin). First, the helicase must dissociate from the streptavidin-blocked DNA end with a rate that is different than an unblocked end. Second, the protein must be adequately processive to reach the streptavidin block. Third, either helicase binding to the DNA substrate or dissociation from the end of the DNA must be the rate-determining step of the overall catalytic cycle. If these criteria are not met, then a change in ATP hydrolysis activity will not be observed. We observe an increase in ATPase activity upon addition of streptavidin, indicating that UvsW satisfies all three criteria. More specifically, this result indicates that when UvsW reaches the streptavidin block it continues to hydrolyze ATP. The precise mechanism for the continued ATP hydrolysis is not clear. It is conceivable that when UvsW reaches the impediment, translocation and ATP hydrolysis become uncoupled so that the UvsW remains immobile but continues to undergo ATP hydrolysis. On the other hand, UvsW could reach the block, undergo one dimensional diffusion (i.e., sliding) in the reverse direction, and then translocate toward the block in an ATP-dependent process. This process could be repeated until UvsW dissociates from an internal site on the DNA substrate. In addition to the 9-fold decrease in KM-DNA when the streptavidin block was placed on the 5’-end of the DNA substrate, we observed a 3-fold decrease when the block was placed at the 3’-end. This result could have two possible origins. The first is that UvsW is able to translocate bidirectionally with a bias in the 3’ to 5’ direction. The second is that when UvsW binds near the 3’ end of the ssDNA, it occasionally slides off the end before it initiates translocation in the 3’ to 5’ direction. In this way, the streptavidin block at the 3’-end of the DNA substrate would serve to increase the affinity of UvsW for the DNA. Bidirectional translocation is extremely rare amongst helicases and is ruled out by the hyperbolic dependence of kcat on the length ssDNA (24), suggesting that UvsW can indeed undergo ATP-independent backward sliding to some degree.
Our second assay for translocation directionality is analogous to the streptavidin block assay, but relies on protein translocation into a region of DNA that is known to inhibit translocation activity (poly-d(T)30). We feel this assay is likely to be as general as the streptavidin-block assay but is more convenient to carry out. Any polymer that supports protein binding but not translocation can be used. For example, most DNA helicases can bind to but not translocate on ssRNA (42). A DNA-RNA chimeric substrate with the RNA placed at either the 3’ or 5’-end of the substrate would be suitable for determining translocation direction. Compared to the streptavidin-block assay, the number of kcat/KM determinations is reduced by half (four in the case of streptavidin-block versus two for this assay). The results from this assay performed with UvsW confirm the findings from the streptavidin-block assay. The Vmax for ATP hydrolysis is approximately 3-fold higher when poly-(dT)30 is placed on the 3’-end of the ssDNA compared to the 5’-end. The simplest explanation for this data is that UvsW translocates in a 3’ to 5’ direction.
It is not clear why UvsW cannot translocate on (dT)30 DNA. We also observe very little ATP hydrolysis on poly-(dA)30 and poly-(dC)30 ssDNA templates. However, simple repeats of poly-(dAC)15 and poly-(dTC)15 support a moderate amount of ATPase activity (Table II). The affinity of UvsW for d(T)30, d(AC)15 and d(TC)15 DNA are very similar to d(N)30 (Table II). From these observations, it is clear that UvsW recognizes the sugar-phosphate backbone of the ssDNA for binding affinity but utilizes the composition of the bases for ssDNA translocation. Consistent with this hypothesis is the fact that etheno modification of ssDNA severely reduces the kcat for ATP hydrolysis but does not affect the KM-DNA (unpublished observations). Significant differences in ATPase activity as a function of nucleotide composition are not uncommon among helicases. The replicative helicase from T4 phage, gp41, displays 4-fold higher activity on ssM13 as compared to poly(dA)n ssDNA (25). The DnaB helicase from E. coli is 2.5-fold more active on poly-(dA)n than on poly-(dT)n(43). These differences, however, are relatively minor when compared to UvsW. A full understanding of the origin of the large difference between, for example, poly-(dA)30 and poly-(dAC)15 will likely require structural analysis of UvsW in the presence of DNA substrates of varying base composition.
It had been assumed that UvsW is a 3’ to 5’ helicase. This assignment was based mainly on the fact that UvsW is an SF2 helicase and most SF2 helicases are 3’ to 5’ helicases (e.g., RecG, RecQ, Rad54). Additionally, UvsW displays weak helicase activity on dsDNA substrates that contain overhanging 3’ ssDNA ends (11). However, the assignment of UvsW as a 3’ to 5’ helicase has been somewhat tenuous because not all characterized SF2 helicases display this directionality (1) and the weak helicase activity on dsDNA with 3’ ssDNA overhangs is only apparent under certain conditions (9). Based on the 3’ to 5’ ssDNA translocation determined here and its probable mechanistic linkage to helicase activity, it is very likely that UvsW is a typical SF2 helicase that unwinds DNA with a 3’ to 5’ directionality.
Of great relevance to the in vivo functioning of UvsW is its processivity. Based on heparin and mutant trapping experiments, UvsW is a highly processive ssDNA translocase. In vivo, UvsW is responsible for unwinding R-loops that are used for the initiation of DNA replication during origin-dependent DNA replication. An R-loop is a stably bound mRNA transcript that is used both as a scaffold for replisome assembly and primer for the initiation of leading strand DNA synthesis. The length of R-loops in vivo is variable, however in the case of uvs(Y) origin, a minimal size has been determined to be close to 50 bp (44). Additionally, UvsW has been proposed to participate in the regression of stalled replication forks and in the branch migration of Holliday junctions (9, 11). These types of DNA structures require translocation of hundreds to thousands of bases. Presumably, it would be advantageous for UvsW to carry out this unwinding in a single binding event and would thus require relatively high processivity.
UvsW also possesses ssDNA annealing activity (11). The proposed model for UvsW-catalyzed ssDNA annealing involves the ATP-driven translocation of UvsW along ssDNA bound to the RecA-like motor domains to achieve proper alignment to a second ssDNA bound to the variable domain. The annealing activity was proposed to play a role in the annealing of the unwound nascent leading and lagging strands during fork regression. It is also possible that UvsW participates in the formation of recombination intermediates by virtue of its ssDNA annealing (45). These ssDNA segments can be produced through the action of a nuclease or incomplete replication of the ends of the T4 genome by the replisome (45). In the latter case, the average amount of ssDNA will be half the length of an Okazaki fragment (0.5 to 1 kb). For the purposes of strand annealing, a highly processive enzyme would be required since the alignment of complementary ssDNA strands would have to restart if UvsW were to dissociate from one of the ssDNA strands.
The high processivity of UvsW during ATP-driven translocation on ssDNA suggests that it is a processive helicase. This conclusion is consistent with a recent report that calculated the rate of branch migration catalyzed by UvsW to be 20 bp/sec.(9). This calculation assumed a 100% processive enzyme. A reduction in processivity would require the rate of branch migration to be much higher than 20 bp/sec due to the fact that following each dissociation event, the enzyme has an equal probability of performing branch migration in either direction. A rate of 20 bp/sec is very typical of other helicases with branch migration activity, ranging from 8 to 40 bp/sec, suggesting that the assumption of 100% processivity is valid (discussed in ref. 10). It is important to note, however, that high processivity during ssDNA translocation does not necessarily indicate high processivity during dsDNA unwinding (46). PcrA helicase was determined to be a processive ssDNA translocase but unable to unwind duplex DNA (47). Thus, it remains to be determined if UvsW is a processive helicase.
Given the high processivity of UvsW during ssDNA translocation, we found it important to determine the oligomeric state during ssDNA translocation since many processive proteins are oligomeric. The crystal structures of UvsW are monomeric, however these do not contain DNA substrate or nucleotide (15, 16). As mentioned above, our attempts to characterize the oligomeric status of UvsW while bound to ssDNA with and without a non-hydrolyzable ATP analogue were unsuccessful due to precipitation of the protein at the concentrations necessary for biophysical techniques. For this reason, we utilized an UvsW mutant protein (K141A) that could bind to DNA normally but was unable to hydrolyze ATP. Under conditions of excess ssDNA, the ratio of wt-UvsW to K141A was widely varied and no change in the ATPase rate was observed. This strongly suggests that UvsW does not associate with the UvsW mutant and is therefore monomeric during ssDNA translocation. Alternatively, the introduction of the active site mutation has prevented association between the wild-type and mutant proteins. We feel this possibility is highly unlikely as the structure of the mutant UvsW is not perturbed, as evidenced by its ability to bind ssDNA normally. A second alternative is that UvsW is oligomeric but the subunits are completely independent of each other. If this were true, then incorporation of an inactive subunit into the oligomer would have no effect. This possibility is also unlikely since every characterized oligomeric motor protein has a rather sophisticated mechanism for coupling the mechanical energy between its individual subunits (48, 49).
In conclusion, we have determined the overall features of UvsW as it translocates on ssDNA templates. We have shown that UvsW is a 3’ to 5’ ssDNA translocase that exhibits very high processivity. We have also provided strong evidence that UvsW is a monomeric helicase during active ssDNA translocation. This represents a first step toward the mechanistic understanding of UvsW ssDNA translocation, which is necessary accurately describe its unwinding mechanism.
Abbreviations used are
- T4
bacteriophage T4
- ssDNA
single-stranded DNA
- dsDNA
double-stranded DNA
- A.U.
Absorbance units.
Footnotes
This work was supported by National Institutes of Health Grant GM013306 (to S.J.B.).
References
- 1.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 2.Tuteja N, Tuteja R. Unraveling DNA helicases. Motif, structure, mechanism and function. Eur. J. Biochem. 2004;271:1849–1863. doi: 10.1111/j.1432-1033.2004.04094.x. [DOI] [PubMed] [Google Scholar]
- 3.Mosig G, et al. Multiple initiation mechanisms adapt phage T4 DNA replication to physiological changes during T4's development. FEMS Microbiol Rev. 1995;17:83–98. doi: 10.1111/j.1574-6976.1995.tb00190.x. [DOI] [PubMed] [Google Scholar]
- 4.Dudas KC, Kreuzer KN. UvsW protein regulates bacteriophage T4 origin-dependent replication by unwinding R-loops. Mol Cell Biol. 2001;21:2706–2715. doi: 10.1128/MCB.21.8.2706-2715.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Derr LK, Drake JW. Isolation and genetic characterization of new uvsW alleles of bacteriophage T4. Mol. Gen. Genet. 1990;222:257–264. doi: 10.1007/BF00633826. [DOI] [PubMed] [Google Scholar]
- 6.Conkling MA, Drake JW. Isolation and characterization of conditional alleles of bacteriophage T4 genes uvsX and uvsY. Genetics. 1984;107:505–523. doi: 10.1093/genetics/107.4.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamlett NV, Berger H. Mutations altering genetic recombination and repair of DNA in bacteriophage T4. Virology. 1975;63:539–567. doi: 10.1016/0042-6822(75)90326-8. [DOI] [PubMed] [Google Scholar]
- 8.Carles-Kinch K, George JW, Kreuzer KN. Bacteriophage T4 UvsW protein is a helicase involved in recombination, repair and the regulation of DNA replication origins. EMBO J. 1997;16:4142–4151. doi: 10.1093/emboj/16.13.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Webb MR, et al. The phage T4 protein UvsW drives Holliday junction branch migration. J Biol Chem. 2007;282:34401–34411. doi: 10.1074/jbc.M705913200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.George JW, Kreuzer KN. Repair of double-strand breaks in bacteriophage T4 by a mechanism that involves extensive DNA replication. Genetics. 1996;143:1507–1520. doi: 10.1093/genetics/143.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nelson SW, Benkovic SJ. The T4 phage UvsW protein contains both DNA unwinding and strand annealing activities. J Biol Chem. 2007;282:407–416. doi: 10.1074/jbc.M608153200. [DOI] [PubMed] [Google Scholar]
- 12.Ralf C, Hickson ID, Wu L. The Bloom's syndrome helicase can promote the regression of a model replication fork. J. Biol. Chem. 2006;281:22839–22846. doi: 10.1074/jbc.M604268200. [DOI] [PubMed] [Google Scholar]
- 13.Machwe A, Xiao L, Groden J, Orren DK. The Werner and Bloom syndrome proteins catalyze regression of a model replication fork. Biochemistry. 2006;45:13939–13946. doi: 10.1021/bi0615487. [DOI] [PubMed] [Google Scholar]
- 14.de Jager M, et al. DNA-binding and strand-annealing activities of human Mre11: implications for its roles in DNA double-strand break repair pathways. Nucleic Acids Res. 2001;29:1317–1325. doi: 10.1093/nar/29.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kerr ID, et al. Crystallographic and NMR analyses of UvsW and UvsW.1 from bacteriophage T4. J Biol Chem. 2007;282:34392–34400. doi: 10.1074/jbc.M705900200. [DOI] [PubMed] [Google Scholar]
- 16.Sickmier EA, Kreuzer KN, White SW. The crystal structure of the UvsW helicase from bacteriophage T4. Structure. 2004;12:583–592. doi: 10.1016/j.str.2004.02.016. [DOI] [PubMed] [Google Scholar]
- 17.Li N, et al. The MotA transcription factor from bacteriophage T4 contains a novel DNA-binding domain: the 'double wing' motif. Mol. Microbiol. 2002;43:1079–1088. doi: 10.1046/j.1365-2958.2002.02809.x. [DOI] [PubMed] [Google Scholar]
- 18.Lohman TM, Tomko EJ, Wu CG. Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nat. Rev. Mol. Cell Biol. 2008;9:391–401. doi: 10.1038/nrm2394. [DOI] [PubMed] [Google Scholar]
- 19.Xie P. Model for helicase translocating along single-stranded DNA and unwinding double-stranded DNA. Biochim. Biophys. Acta. 2006;1764:1719–1729. doi: 10.1016/j.bbapap.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 20.Lohman TM, Bjornson KP. Mechanisms of helicase-catalyzed DNA unwinding. Annu. Rev. Biochem. 1996;65:169–214. doi: 10.1146/annurev.bi.65.070196.001125. [DOI] [PubMed] [Google Scholar]
- 21.Moore KJ, Lohman TM. Helicase-catalyzed DNA unwinding: energy coupling by DNA motor proteins. Biophys. J. 1995;68:180S–184S. discussion 184S–185S. [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook JF, Maniatis T, Sambrook JF. Molecular Cloning: A Laboratory Manual: 1, 1989. Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 23.Gilbert SP, Mackey AT. Kinetics: a tool to study molecular motors. Methods. 2000;22:337–354. doi: 10.1006/meth.2000.1086. [DOI] [PubMed] [Google Scholar]
- 24.Young MC, Kuhl SB, von Hippel PH. Kinetic theory of ATP-driven translocases on one-dimensional polymer lattices. J Mol Biol. 1994;235:1436–1446. doi: 10.1006/jmbi.1994.1099. [DOI] [PubMed] [Google Scholar]
- 25.Young MC, Schultz DE, Ring D, von Hippel PH. Kinetic parameters of the translocation of bacteriophage T4 gene 41 protein helicase on single-stranded DNA. J Mol Biol. 1994;235:1447–1458. doi: 10.1006/jmbi.1994.1100. [DOI] [PubMed] [Google Scholar]
- 26.Morris PD, Tackett AJ, Raney KD. Biotin-streptavidin-labeled oligonucleotides as probes of helicase mechanisms. Methods. 2001;23:149–159. doi: 10.1006/meth.2000.1116. [DOI] [PubMed] [Google Scholar]
- 27.Morris PD, Raney KD. DNA helicases displace streptavidin from biotin-labeled oligonucleotides. Biochemistry. 1999;38:5164–5171. doi: 10.1021/bi9822269. [DOI] [PubMed] [Google Scholar]
- 28.Raney KD, Benkovic SJ. Bacteriophage T4 Dda helicase translocates in a unidirectional fashion on single-stranded DNA. J Biol Chem. 1995;270:22236–22242. doi: 10.1074/jbc.270.38.22236. [DOI] [PubMed] [Google Scholar]
- 29.Slocum SL, Buss JA, Kimura Y, Bianco PR. Characterization of the ATPase activity of the Escherichia coli RecG protein reveals that the preferred cofactor is negatively supercoiled DNA. J. Mol. Biol. 2007;367:647–664. doi: 10.1016/j.jmb.2007.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Korangy F, Julin DA. Kinetics and processivity of ATP hydrolysis and DNA unwinding by the RecBC enzyme from Escherichia coli. Biochemistry. 1993;32:4873–4880. doi: 10.1021/bi00069a024. [DOI] [PubMed] [Google Scholar]
- 31.Morris PD, et al. Evidence for a functional monomeric form of the bacteriophage T4 DdA helicase. Dda does not form stable oligomeric structures. J. Biol. Chem. 2001;276:19691–19698. doi: 10.1074/jbc.M010928200. [DOI] [PubMed] [Google Scholar]
- 32.Xie P. On translocation mechanism of ring-shaped helicase along single-stranded DNA. Biochim. Biophys. Acta. 2007;1774:737–748. doi: 10.1016/j.bbapap.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 33.Patel SS, Picha KM. Structure and function of hexameric helicases. Annu. Rev. Biochem. 2000;69:651–697. doi: 10.1146/annurev.biochem.69.1.651. [DOI] [PubMed] [Google Scholar]
- 34.Benkovic SJ, Valentine AM, Salinas F. Replisome-mediated DNA replication. Annu Rev Biochem. 2001;70:181–208. doi: 10.1146/annurev.biochem.70.1.181. [DOI] [PubMed] [Google Scholar]
- 35.Chastain PD, Makhov AM, Nossal NG, Griffith J. Architecture of the replication complex and DNA loops at the fork generated by the bacteriophage t4 proteins. J Biol Chem. 2003;278:21276–21285. doi: 10.1074/jbc.M301573200. [DOI] [PubMed] [Google Scholar]
- 36.Cox MM. Historical overview: searching for replication help in all of the rec places. Proc. Natl. Acad. Sci. U.S.A. 2001;98:8173–8180. doi: 10.1073/pnas.131004998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cox MM, et al. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- 38.Heller RC, Marians KJ. Replisome assembly and the direct restart of stalled replication forks. Nat. Rev. Mol. Cell Biol. 2006;7:932–943. doi: 10.1038/nrm2058. [DOI] [PubMed] [Google Scholar]
- 39.Amitani I, Baskin RJ, Kowalczykowski SC. Visualization of Rad54, a chromatin remodeling protein, translocating on single DNA molecules. Mol. Cell. 2006;23:143–148. doi: 10.1016/j.molcel.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 40.Mahdi AA, et al. A model for dsDNA translocation revealed by a structural motif common to RecG and Mfd proteins. EMBO J. 2003;22:724–734. doi: 10.1093/emboj/cdg043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bianco PR, Hurley EM. The type I restriction endonuclease EcoR124I, couples ATP hydrolysis to bidirectional DNA translocation. J. Mol. Biol. 2005;352:837–859. doi: 10.1016/j.jmb.2005.07.055. [DOI] [PubMed] [Google Scholar]
- 42.Shin J, Kelman Z. The replicative helicases of bacteria, archaea, and eukarya can unwind RNA-DNA hybrid substrates. J. Biol. Chem. 2006;281:26914–26921. doi: 10.1074/jbc.M605518200. [DOI] [PubMed] [Google Scholar]
- 43.Arai K, Kornberg A. Mechanism of dnaB protein action. II. ATP hydrolysis by dnaB protein dependent on single- or double-stranded DNA. J. Biol. Chem. 1981;256:5253–5259. [PubMed] [Google Scholar]
- 44.Menkens AE, Kreuzer KN. Deletion analysis of bacteriophage T4 tertiary origins. A promoter sequence is required for a rifampicin-resistant replication origin. J Biol Chem. 1988;263:11358–11365. [PubMed] [Google Scholar]
- 45.Mosig G. Recombination and recombination-dependent DNA replication in bacteriophage T4. Annu Rev Genet. 1998;32:379–413. doi: 10.1146/annurev.genet.32.1.379. [DOI] [PubMed] [Google Scholar]
- 46.Tomko EJ, Fischer CJ, Niedziela-Majka A, Lohman TM. A nonuniform stepping mechanism for E. coli UvrD monomer translocation along single-stranded DNA. Mol. Cell. 2007;26:335–347. doi: 10.1016/j.molcel.2007.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Niedziela-Majka A, Chesnik MA, Tomko EJ, Lohman TM. Bacillus stearothermophilus PcrA monomer is a single-stranded DNA translocase but not a processive helicase in vitro. J. Biol. Chem. 2007;282:27076–27085. doi: 10.1074/jbc.M704399200. [DOI] [PubMed] [Google Scholar]
- 48.Liao J, et al. Mechanochemistry of t7 DNA helicase. J. Mol. Biol. 2005;350:452–475. doi: 10.1016/j.jmb.2005.04.051. [DOI] [PubMed] [Google Scholar]
- 49.Nishizaka T, et al. Chemomechanical coupling in F1-ATPase revealed by simultaneous observation of nucleotide kinetics and rotation. Nat. Struct. Mol. Biol. 2004;11:142–148. doi: 10.1038/nsmb721. [DOI] [PubMed] [Google Scholar]