Abstract
Flowering plants often prevent selfing through mechanisms of self-incompatibility (S.I.). The loss of S.I. has occurred many times independently, because it provides short-term advantages in situations where pollinators or mates are rare. The genus Capsella, which is closely related to Arabidopsis, contains a pair of closely related diploid species, the self-incompatible Capsella grandiflora and the self-compatible Capsella rubella. To elucidate the transition to selfing and its relationship to speciation of C. rubella, we have made use of comparative sequence information. Our analyses indicate that C. rubella separated from C. grandiflora recently (≈30,000–50,000 years ago) and that breakdown of S.I. occurred at approximately the same time. Contrasting the nucleotide diversity patterns of the 2 species, we found that C. rubella has only 1 or 2 alleles at most loci, suggesting that it originated through an extreme population bottleneck. Our data are consistent with diploid speciation by a single, selfing individual, most likely living in Greece. The new species subsequently colonized the Mediterranean by Northern and Southern routes, at a time that also saw the spread of agriculture. The presence of phenotypic diversity within modern C. rubella suggests that this species will be an interesting model to understand divergence and adaptation, starting from very limited standing genetic variation.
Many flowering plant species are obligate outcrossers that cannot self-fertilize because of self-incompatibility (S.I.), often determined by a single S-locus (1–3). Differences in the underlying mechanisms indicate that S.I. has evolved independently at least 10 times. However, loss of S.I. is even more common, and is thought to be most prevalent when mating opportunities are limited because of low population densities or absence of pollinators, situations most likely to occur at the edges of a species' range or on islands (3–9). Loss of obligatory outcrossing in flowering plants is often associated with subsequent appearance of differences in a variety of reproductive traits, such as flowering time and floral morphology (10). These, together with chromosomal rearrangements, reduce gene flow between populations with different mating systems, and may eventually lead to reproductive isolation and speciation (11). Therefore, the relationship between the loss of S.I. and speciation is of particular interest.
In the Brassicaceae, sporophytic S.I. is the ancestral condition. The self-incompatibility (S)-locus in this family consists of 2 determinant genes, SRK and SCR, which are normally not separated by recombination. The transmembrane receptor kinase encoded by SRK is expressed at the stigmatic surface of the female, whereas the small soluble SCR ligand is deposited in the pollen wall of the male. When SCR binds to SRK from the same haplotype, the S.I. response is initiated, preventing self-pollination through a series of downstream events (reviewed in ref. 12, 13). S.I. has been lost repeatedly within the Brassicaceae, even within the same genus and/or species (13, 14). Arabidopsis thaliana, the work horse for much of plant molecular genetics, has become self-compatible relatively recently, apparently by the gradual fixation of multiple, independent mutations that weakened or disabled the S.I. system throughout its geographical range (15, 16).
We set out to investigate the breakdown of S.I. in Capsella rubella to test the generality of the pattern described for A. thaliana. The genus Capsella includes the 2 diploid species C. rubella and Capsella grandiflora (2n = 16) and the tetraploid species C. bursa-pastoris (2n = 32) (17, 18). The 2 diploid species show striking morphological differences, particularly for flower size. That they are also genetically diverged can be concluded from the observation that even when experimental crosses do not fail completely, F1 hybrids are often sterile (17). The self-incompatible C. grandiflora has the narrowest distribution and is found in western Greece, some of the Greek islands, Albania and, rarely, in northern Italy. The self-compatible C. rubella occurs throughout the Mediterranean, and has occasionally followed European settlers to the Americas and to Australia. By far the most successful species is the self-compatible C. bursa-pastoris, an invasive weed with an impressive ecological range that is found throughout the world (17, 19–21). Selfers are often better pioneers, because the ability to self-fertilize allows the establishment of new populations by individual plants (6–9, 22, 23). The potential to spread and become a cosmopolitan species therefore often appears higher for selfers than outcrossers. In accordance with this, C. rubella, like its selfing congener C. bursa-pastoris, has a larger distribution range than the outcrossing C. grandiflora.
The S-locus of C. grandiflora is very polymorphic because of strong frequency dependent selection, and comprises at least 38 haplotypes (24, 25), which is very similar to what has been reported for A. lyrata (26). The origin of C. rubella might have been associated with the breakdown of S.I. in a C. grandiflora population. An obvious candidate for sustaining the causative mutation is the S-locus itself. However, although the S-locus has been implicated in the breakdown of S.I. in A. thaliana (15, 16, 27), whether it is any more likely to play a central role in the loss of S.I. than other genes that are required for SRK/SCR activity is unknown. In fact, A. thaliana has been shown to harbor variation for a gene that is closely linked to the S-locus and that can modify expression of SRK (28).
Here, we present data suggesting that breakdown of S.I. was associated with the origin of C. rubella ≈30,000–50,000 years ago. S-locus diversity is compatible with a selective sweep, but diversity at other loci is also very low, indicating that the transition to self-compatibility has been through an extreme bottleneck. Because all loci we have examined have only 1 or 2 haplotypes in C. rubella, we hypothesize that this species has been founded by a single C. grandiflora individual that had become self-compatible. Our data furthermore indicate that breakdown of S.I. occurred near Greece, and spread with agriculture to the rest of Europe.
Results
Nucleotide diversity in C. rubella and C. grandiflora.
To determine nucleotide diversity in C. rubella, we analyzed 23 accessions from throughout its European range, and 1 accession each from Argentina and Australia (Table S1). We compared them to 7 C. grandiflora individuals, each representing different populations in Greece. We sequenced genomic fragments representing 17 nuclear loci: the 2 major S-locus genes, SRK and SCR; 5 and 6 loci flanking the S-locus on each side, exploiting synteny with the A. thaliana reference genome (29, 30); 4 unlinked loci, ALCOHOL DEHYDROGENASE (ADH), FRIGIDA (FRI), FLOWERING LOCUS C (FLC), and PHYTOCHROME C (PHYC). All of these are single copy genes in the A. thaliana reference genome. In addition, we sequenced a chloroplast gene, matK. As gene names for linked genes, we used the identifiers of the A. thaliana orthologs (Table S2).
The sequenced fragments range from 737 bp (At4g21580) to 1,279 bp (At1g77120) for nuclear genes and 2,282 bp for matK, and the total length of aligned sequences across the 2 species for the 16 genes excluding SCR and SRK is 15,388 bp. Nucleotide diversity in C. rubella is generally much lower than in C. grandiflora (Fig. 1 and Table S3). All 25 C. rubella accessions share very closely related SRK sequences and nearly identical SCR sequences, whereas the 7 C. grandiflora individuals contained at least 12 different S alleles. The SRK sequences of C. rubella are very similar to that found in a single S-locus haplotype of C. grandiflora [(average divergence is 0.0043; net divergence 0.0017 (6 differences in 3496 base pairs)], suggesting a common origin from the same functional haplotype in the ancestral species. For SCR, only the first of 2 exons are found in C. rubella, and the sequences of different accessions are nearly identical (2 segregating sites in 805 base pairs, see Table S3). The C. grandiflora SCR sequence overlap with C. rubella sequences by 224 bp only, with only a single bp difference.
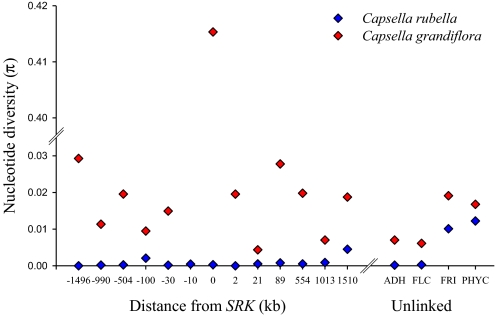
Fig. 1.
Nucleotide diversity in C. rubella and C. grandiflora. Distance from SRK is for the syntenic region from A. thaliana, because exact information is only available for C. rubella from close to SRK (see Table S2). Distance is not to scale. Note the break in the ordinate, to accommodate the nucleotide diversity value for SRK from C. grandiflora.
Although nucleotide diversity (π) in C. grandiflora is generally high (mean ≈2%), there is no apparent peak of diversity around SRK (Fig. 1). This is in contrast to the situation in the self-incompatible relatives Arabidopsis lyrata and A. halleri (31–33). Diversity in C. rubella is very low at 14 out of 18 loci examined in this study. The 4 exceptions are FRI, PHYC, At4g21150 and At4g25100. These genes contain 2 divergent clusters of haplotypes, which have similar counterparts in C. grandiflora (Fig. 2). This suggests that most of the polymorphism at these genes is transspecific, i.e., already existed in the common ancestor of C. rubella and C. grandiflora. This conclusion is supported by the analysis of sequences from 3 previously studied loci (Table S3) (18), 2 of which are monomorphic and one of which features 2 main classes of divergent haplotypes (Fig. 2, Bottom Left). There are very few fixed differences between C. rubella and C. grandiflora in nuclear genes excluding SRK and SCR (Table S3), with only 13 fixed synonymous substitutions in 7,300 silent sites. No fixed differences were found in the chloroplast matK gene. C. rubella had 2 segregating sites in the chloroplast matK gene, both of which were distinct from the 4 segregating sites in C. grandiflora.
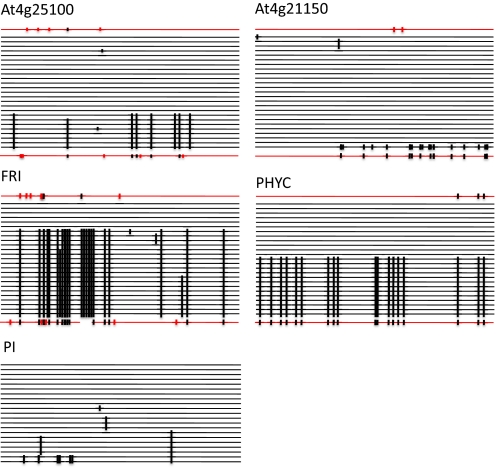
Fig. 2.
C. rubella loci with 2 divergent haplotypes. Each allele is shown as a horizontal line. C. rubella alleles (black) have been sorted by similarity, with the 2 closest C. grandiflora alleles (red) shown above and below the C. rubella alleles. Polymorphisms are shown as vertical lines. Most polymorphisms in C. rubella are also found in C. grandiflora. Although for PI only C. rubella sequences (18) were available, these also seem to fall into 2 dominant haplotypes.
We compared variation at the ADH locus in C. rubella and C. grandiflora with publicly available data for 3 relatives, C. bursa-pastoris, A. thaliana and A. lyrata (18, 34–36) (Table S4). Among the 5 species, nucleotide diversity in C. rubella (π = 0.0002) was by far the lowest, with C. bursa-pastoris and A. lyrata being intermediate (π = 0.0008 and π = 0.0036, respectively), and the other 2 species having similarly high diversity (π = 0.0081 to 0.0085).
To test whether C. rubella sequences evolve neutrally, we calculated Tajima's D (37). Tajima's D is close to zero for most loci, although this is not very informative, because they have few segregating sites (Fig. S1). Among the 5 genes with 2 divergent haplotypes, both PHYC and FRI have large positive values, 3.46 and 1.42, respectively, reflecting that the 2 haplotypes are found in approximately equal frequencies. However, only the PHYC value is statistically significant (P < 0.001). At4g25100 has a significant negative value, reflecting that 1 of the 2 haplotypes is rare (Fig. 2).
Evolution of SRK.
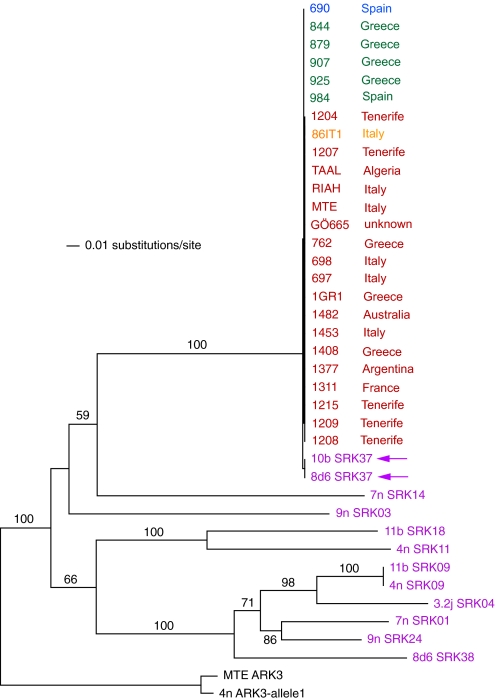
C. grandiflora has several SRK alleles that group in pairs with alleles identified in the genus Arabidopsis, in agreement with transspecific evolution (Fig. S2). Two of the SRK sequences from C. grandiflora (SRK37) group with all 25 SRK sequences of C. rubella, which are very similar to each other (Fig. S3 and Fig. 3). We assume that these represent the same allele in the ancestral species. A related S allele, AlSRK30, is found in A. lyrata, where it is believed to belong to the most dominant class of S alleles (16) (Fig. S2).
Fig. 3.
Phylogenetic tree of SRK alleles from Capsella. C. grandiflora sequences are in purple; arrows indicate 2 C. grandiflora sequences grouping with all C. rubella sequences. Green, C. rubella sequences with intact ORF; blue, frame shift because of a 599-bp fragment insertion into pos. 66 of exon 6; ochre, premature stop codon because of insertion of a T at pos. 58 of exon 2; red, a premature stop codon because of insertion of a T at pos. 912 of exon 1.
There are 3 slightly different types of this SRK allele in C. rubella with truncated ORFs. One, with a 1 base pair insertion, was found in the majority of lines studied, in 18 accessions. Another 1-bp insertion and a 599-bp insertion were found in 1 accession each (Fig. 3). However, there are also 5 accessions that can encode a full-length SRK protein based on the related, likely functional allele of C. grandiflora, suggesting that the 3 SRK types sustained nonsense mutations only after fixation of the single SRK allele in C. rubella.
Dating the origin of Capsella rubella.
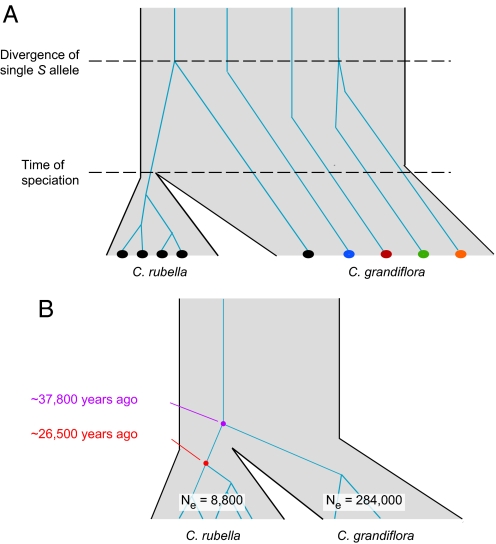
Our main assumption is that all variation within the S-locus in C. rubella arose after speciation, because it seems unlikely that the very same S allele became independently fixed more than once in C. rubella (Fig. 4A). The per-base pair scaled mutation rate θ estimated from C. rubella SRK sequences was 0.000518 (95% confidence interval: 0.000262–0.001046) and for SCR 0.000697 (0.000249–0.002488). By comparing C. grandiflora SRK sequences with closely related SRK sequences from A. lyrata (13 pairs in total), and assuming Capsella and Arabidopsis separated 6–10 million years ago (38), we estimated a mutation rate of 1.46 × 10−8 (1.31 × 10−08 to 1.61 × 10−08) per site per year, with a generation time of 2 years. This estimate is within the generally accepted range of spontaneous mutation rates for multicellular organisms (39). These values were used to estimate the effective population size of C. rubella since fixation of the SRK allele, which was found to be close to 10,000, depending on assumptions about generation time (Table 1).
Fig. 4.
Modeling time of divergence and effective population sizes. (A) Model of the speciation of C. rubella, which assumes that C. rubella originated from C. grandiflora. The genealogy of the S-locus embedded in the species tree illustrates that the time to coalescence after fixation in C. rubella provides a minimum estimate, and the time to coalescence of the shared allele provides a maximum estimate of speciation time. (B) Resulting estimates based on the C. rubella SRK sequences and the 2 closely related SRK sequences from C. grandiflora. See Table 1 for details.
Table 1.
Estimation of time to MRCA [assuming a substitution rate, μ, of 1.46 × 10−08 (1.31–1.61 × 10−08)]
| Ne | MLE of θ per gene | MLE of TMRCA scaled in 2Negenerations | TMRCA in years* | |
|---|---|---|---|---|
| SRK | ||||
| C. rubella | 8,806 (5,246–12,742) | 1.98 (1–4) | 1.50 (1.02–1.92) | 26,418 (13,826–54,983) |
| C. rubella and grandiflora | 7,243† (3,683–11,179) | 5.01 (3–9) | 1.74 (1.08–2.58) | 37,809 (20,530–64,984) |
| SCR | ||||
| C. rubella | 11,630 (5,330–18,796) | 0.56 (0.2–2) | 1.14 (0.78–1.26) | 26,516 (8,331–83,057) |
*Assuming generation time of one year in C. rubella and two years in C. grandiflora. The conversion from generations to years in the analysis using SRK sequences from both C. rubella and C. grandiflora was done assuming an averaged generation time of 1.5 years.
†Calculated as (284,000/50 + 8,806)/2, i.e., mean of estimate for C. grandiflora and C. rubella, assuming 50 SRK alleles in C. grandiflora.
We used a method implemented in the program Genetree (40) to date the origin of the existing variation of the C. rubella S-locus (Fig. 4B). The time to the most recent common ancestor (TMRCA) was estimated to be 1.50 (1.02–1.92) scaled in units of 2Ne generations, corresponding to 26,418 (13,826–54,983) years for SRK, and 1.14 (0.78–1.26) corresponding to 26,516 (8,331–83,057) years for SCR (Table 1). Applying Genetree to the sequences of this SRK allele from both C. rubella and C. grandiflora yielded an estimate of 37,809 years (20,530–64,984) (Table 1).
A rough estimate of the present effective population size of C. grandiflora, based on θ values (calculated with DnaSP) of 9 loci (At1g77120, At4g17760, At4g18975, At4g20130, At4g21150, At4g21580, At4g22720, At4g23840, At4g25100) and the synonymous substitution rate derived above, is 284,000 (252,000–314,000) (Table 1). This estimate is probably downward biased because the substitution rate used is for synonymous changes only. Using these data and assuming that the effective population size of C. grandiflora before speciation was the same as it is today, we can then estimate the average time to coalescence for an S allele in the ancestral species as 2N/number of S alleles (41). Assuming 50 S alleles and constant population size, this leads to an estimate of the average time to coalescence of 11,000 generations or 22,000 years. Because the S allele found in C. rubella belongs to the most dominant class (16), this estimate is likely somewhat upward biased. Nevertheless, given the large standard errors of the underlying components, this estimate is consistent with the origin of C. rubella S-locus diversity being in the same range as the time of speciation.
An alternative maximum estimate of species divergence can be obtained by using the number of fixed differences for all loci except SCR, SRK and matK, which is 13 in 7,300 bp (Table S3). Assuming again a substitution rate of 1.46 × 10−8, this corresponds to ≈120,000 years of evolution, or ≈60,000 years of separation. For SRK, there are 6 fixed differences in 3,824 base pairs, which yielded a point estimate of separation of 53,700 years. These point estimates can be overestimates because they are inflated by the (unknown) polymorphism in the ancestral species.
Phylogeography.
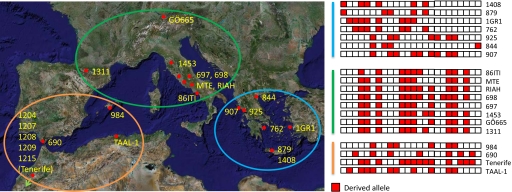
Full-length C. rubella SRK sequences were found mainly in Greece (Fig. 3). Because this is apparently the ancestral allele, this would suggest Greece as the birthplace of the species. This conclusion is supported by the geographic distribution of genetic variation in C. rubella (Fig. 5). We divided the individuals into 3 clusters, East, North, and West Mediterranean. Excluding polymorphic sites shared with C. grandiflora, polymorphism is highest in the Eastern group, followed by Western and then Northern accessions (Table 2). The distribution of genetic diversity is consistent with an origin of the species in Greece and a relatively recent dispersal, perhaps following separate Northern and Southern routes into the rest of its modern range.
Fig. 5.
The geographical pattern of variation in C. rubella. The provenance of GÖ665 is unknown. All segregating sites are shown.
Table 2.
Nucleotide diversity (π) and segregating sites (S) in Eastern (predominantly Greek), Northern and Southern Mediterranean accessions (only sites not polymorphic in C. grandiflora are included)
| Population | π | S |
|---|---|---|
| Eastern | 0.00057 | 25 |
| Northern | 0.00015 | 8 |
| Southern | 0.00045 | 23 |
| Northern and southern | 0.00033 | 27 |
See also Fig. 5.
Discussion
The transition from outcrossing to selfing has occurred repeatedly within the Brassicaceae (13). In the selfer A. thaliana, several S-locus haplotypes have been found. Common to all is that SRK has become a pseudogene, whereas SCR shows a range of states, including having been lost, having become a pseudogene, or possibly still being functional (15, 42, 43). In C. rubella, SCR appears to be a pseudogene in all accessions studied, whereas apparently functional versions of SRK have persisted, although several independent knockout mutations have occurred as well.
Coalescent simulations based on S-locus sequences suggest that C. rubella arose as a new species recently, likely in Greece from C. grandiflora. During or after speciation, most genetic variation was lost from C. rubella, so that today it is much less diverse than the highly polymorphic C. grandiflora. We estimate that a single S allele in C. rubella was fixed at least 27,000 years ago, and that the coalescent of this allele and the similar C. grandiflora S allele occurred 30,000 to 60,000 years ago. These estimates are consistent with breakdown of S.I. having played a causal role in founding of the species, although it is also possible that loss of S.I. merely led to loss of genetic diversity in a population that had already split from the rest of C. grandiflora. Slotte and colleagues (18) recently reported evidence for introgression of C. rubella sequences into C. bursa-pastoris starting 10,000 years ago in Europe, which is consistent with a recent origin of C. rubella.
A great advantage of using the S-locus to assess the history of C. rubella is that very likely the present variation arose after the fixation of a single copy of an S allele. The basis of this argument is that S alleles within a species are in general very different from each other, because they are old and shared not only between species within the same genus, but also across genera (24). The diversity is maintained by frequency-dependent selection, and each of the S alleles occurs only in low frequencies in a self-incompatible species such as C. grandiflora. We found only 7 segregating sites in all 25 C. rubella SRK sequences (3,825 bp), which is in stark contrast with 2 of the most similar alleles in C. grandiflora, which feature 234 segregating sites in 2,034 bp, excluding much of the introns, where they align only poorly. The very low diversity of C. rubella SRK is a strong indicator that these alleles descended from the same functional allele in C. grandiflora. The C. grandiflora S allele that became fixed in C. rubella is no exception to the rule that individual alleles are rare in C. grandiflora; among ≈160 C. grandiflora chromosomes, only 2 had the allele found also in C. rubella. It is therefore most likely that the present variation coalesces in a single copy of a single allele close to the mating system shift. Another great advantage of using the S-locus for inferring the history of C. rubella is that, after the S-locus had lost its function, present variation has very likely been predominantly shaped by neutral forces, which is one of the main assumptions in the Genetree analysis.
The haplotype structure in C. rubella shows a remarkable pattern of either almost no variation (at 14 loci) or a small amount of variation divided into 2 divergent haplotypes (at 4 loci) (see Fig. 2). These haplotypes are to a large extent transspecifically shared with C. grandiflora (Fig. 2) (At4g21150, At4g25100, FRI, and PHYC; PI sequences are not available for C. grandiflora). Thus, the data are compatible with an extremely strong bottleneck during speciation, which removed all variation at the majority of loci and allowed 2 haplotypes to persist at some loci. An alternative hypothesis is that selection has reduced variation across the C. rubella genome. However, linkage disequilibrium between putatively unlinked loci in C. rubella is low (ref. 44; see also SI Text and Fig. S4), indicating that recombination rate is sufficiently high that strong selective sweeps would likely extend over <1 Mb. Therefore, a very large number of strong selective sweeps would be needed to produce the observed reduction in variation at unlinked loci. We believe that a more parsimonious explanation is a single, strong population bottleneck. Because at most 2 divergent haplotypes are found, it is tempting to hypothesize that this bottleneck indeed constituted a single individual that for some reason was able to self and produce fertile offspring. If the progeny mated with each other, they would have preserved some of the variation from the founder, which is presumed to have had 2 quite different haplotypes at most loci, as is observed in present day individuals of C. grandiflora.
In summary, we found that C. rubella separated from C grandiflora recently and that breakdown of S.I. occurred at approximately the same time. C. rubella has only 1 or 2 alleles at most loci, suggesting that speciation was associated with a strong bottleneck. There is already considerable phenotypic differentiation, including in adaptive characters such as flowering time (20). C. rubella should therefore be an interesting model to understand how limited standing genetic variation supports phenotypic diversity, either through new mutations or through new allelic combinations.
The near absence of variation at the S-locus in C. rubella could suggest a selective sweep at this locus, as has been proposed to have partially occurred in A. thaliana (27). The presence of only a single S allele, however, does not necessarily point to a mutation at the S-locus itself having been causal for speciation, because the pattern of only a single allele having been maintained is shared with the majority of loci in C. rubella. A number of other genes are known to be required for S.I. (reviewed in ref. 12), and studies of whole-genome sequence variation will be very informative in the search for loci that have sustained a knockout mutation early on in the history of C. rubella, and that are alternative candidates for having had causal roles in C. rubella population divergence.
Materials and Methods
Plant Material, PCR, and Sequencing.
The origin and accession names of samples analyzed are given in Table S1. C. grandiflora individuals were chosen to represent many different S-locus haplotypes to maximize the probability of observing shared polymorphism with C. rubella. Seeds were germinated in growth chambers and genomic DNA was extracted from fresh leaf material using either the QIAGEN Dneasy Plant Kit (Qiagen) or the CTAB method (45).
PCR primers were designed based on a C. rubella S-locus BAC sequence for the 7 loci closest to SRK, including SCR and SRK. A. thaliana genomic sequences and chloroplast genome were used for the remaining 9 genes. SCR and a fragment flanking full-length SRK were sequenced in C. rubella, whereas in C. grandiflora we sequenced only part of exon 1 of SRK because of difficulty in amplifying the highly variable full-length sequences. Additional sequence data from 3 loci also unlinked to the S-locus (18) was downloaded from GenBank. Note that the accessions used by Slotte and colleagues (18) overlap only partially with the ones used here.
For PCR amplification, Pfu polymerase (Fermentas) was used to amplify genomic DNA. PCR products of C. rubella were sequenced directly. Because of the diversity in C. grandiflora individuals, PCR products were cloned into the pGEM-T Vector (Promega). Three to ten clones were sequenced from each sample to minimize PCR errors. Sequences have been deposited in GenBank under accession nos. FJ649697–FJ650362).
For diversity comparisons with other species, we obtained ADH sequences from GenBank for A. thaliana (19 sequences), A. lyrata (11 sequences), and C. bursa-pastoris (8 sequences) (18, 34–36).
Diversity Studies.
DnaSP version 4.10.9 (46) was used to determine the following population genetic parameters: levels of nucleotide diversity per site (π) (47), θW (48), and Tajima's D was estimated for each locus using all data (37).
Phylogeography.
PAUP* version 4.0b10 (49) was used to reconstruct phylogenetic trees using the Neighbor-joining (NJ) method based on the Kimura 2-parameter model. Topological robustness was assessed by bootstrap analysis with 1,000 replicates, using simple taxon addition (50).
Dating the origin of Capsella rubella.
The program Genetree (40) provided a minimum estimate of the time since origination of C. rubella, based on fixation of the S haplotype. This was done both for SRK and SCR. We also estimated the time to coalescent of C. rubella SRK sequences and the 2 similar C. grandiflora copies either by using Genetree or by estimating the average pairwise synonymous divergence between C. rubella and C. grandiflora sequences, and by using the synonymous substitution rate estimated for the SRK S domain (see below) to calculate the time to coalescent in the ancestral species. This is a maximum estimate of the time since origination of C. rubella.
The scaled mutation rate θ (4Neμ, where Ne is the effective population size and μ is the synonymous substitution rate per site per year) was first estimated using Genetree. To derive Ne, we estimated the synonymous substitution rate based on Ks estimates from 13 similar pairs of SRK alleles from A. lyrata and C. grandiflora, assuming that each pair descended from a single S haplotype in the ancestor, and a separation time between Arabidopsis and Capsella of 8 million years (30). The 95% confidence interval of Ne was obtained by bootstrapping over the empirical distributions of the separation time, substitution rate and θ to generate a distribution of Ne. The empirical distribution of θ was obtained by approximating a normal distribution to the likelihoods obtained by Genetree, of Ks by nonparametric bootstrap replicates over the single Ks estimates, and of separation time by assuming normal distribution and the confidence intervals reported by (38) (6.2–9.8). Depending on the assumed separation time of Arabidopsis and Capsella, the estimated substitution rate, effective population size of SCR and SRK and the TMRCA in years must be changed accordingly. Here, we assume 8 million years; for 10 million years the substitution rate must be multiplied by 0.8 and the effective population size of SCR and SRK and the TMRCA in years must be multiplied by 1.25.
The time to most recent common ancestor (TMRCA) scaled in 2Ne generations was estimated using Genetree. This was converted to generation using the estimate of Ne. Confidence interval was obtained by bootstrapping over the obtained distribution of Ne and a distribution of TMRCA scaled in 2Ne obtained from mean and standard deviation estimates of TMRCA assuming normal distribution. See Fig. 4B for details.
Supplementary Material
Acknowledgments.
We thank Stephen Wright and colleagues for discussion and sharing unpublished information and Thomas Bataillon for discussions regarding data analysis. This work was supported by a European Research Area in Plant Genomics grant ARelatives (to B.N., M.H.S., and D.W.); the Liljewalch and Sernander foundations at Uppsala University (T.S.); the Swedish Research Council for Environmental, Agricultural Sciences and Spatial Planning (M.L.); a Gottfried Wilhelm Leibniz Award (Deutsche Forschungsgemeinschaft) (to D.W.), and the Max Planck Society (D.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. S.C.H.B. is a guest editor invited by the Editorial Board.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. FJ649697–FJ650362).
This article contains supporting information online at www.pnas.org/cgi/content/full/0808012106/DCSupplemental.
References
- 1.Barrett SC. The evolution of plant sexual diversity. Nat Rev Genet. 2002;3:274–284. doi: 10.1038/nrg776. [DOI] [PubMed] [Google Scholar]
- 2.Igic B, Kohn JR. Evolutionary relationships among self-incompatibility RNases. Proc Natl Acad Sci USA. 2001;98:13167–13171. doi: 10.1073/pnas.231386798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Igic B, Lande R, Kohn JR. Loss of self-incompatibility and its evolutionary consequences. Int J Plant Sci. 2008;169:93–104. [Google Scholar]
- 4.Busch JW, Schoen DJ. The evolution of self-incompatibility when mates are limiting. Trends Plants Sci. 2008:128–136. doi: 10.1016/j.tplants.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 5.Wright S. The distribution of self-sterility alleles in populations. Genetics. 1939;24:538–552. doi: 10.1093/genetics/24.4.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker H. Self-compatibility and establishment after “long-distance” dispersal. Evolution. 1955;9:347–349. [Google Scholar]
- 7.Stebbins G. Self fertilization and population variability in the higher plants. Am Nat. 1957;91:337–354. [Google Scholar]
- 8.Jain S. The evolution of inbreeding in plants. Annu Rev Ecol Syst. 1976;7:469–495. [Google Scholar]
- 9.Pannell JR, Barrett SCH. Baker's law revisited: Reproductive assurance in a metapopulation. Evolution. 1998;52:657–668. doi: 10.1111/j.1558-5646.1998.tb03691.x. [DOI] [PubMed] [Google Scholar]
- 10.Charlesworth D, Vekemans X. How and when did Arabidopsis thaliana become highly self-fertilising. Bioessays. 2005;27:472–476. doi: 10.1002/bies.20231. [DOI] [PubMed] [Google Scholar]
- 11.Rieseberg LH, Willis JH. Plant speciation. Science. 2007;317:910–914. doi: 10.1126/science.1137729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rea AC, Nasrallah JB. Self-incompatibility systems: Barriers to self-fertilization in flowering plants. Int J Dev Biol. 2008;52:627–636. doi: 10.1387/ijdb.072537ar. [DOI] [PubMed] [Google Scholar]
- 13.Fobis-Loisy I, Miege C, Gaude T. Molecular evolution of the S locus controlling mating in the Brassicaceae. Plant Biol. 2004;6:109–118. doi: 10.1055/s-2004-817804. [DOI] [PubMed] [Google Scholar]
- 14.Mable BK, et al. Breakdown of self-incompatibility in the perennial Arabidopsis lyrata (Brassicaceae) and its genetic consequences. Evolution. 2005;59:1437–1448. [PubMed] [Google Scholar]
- 15.Tang C, et al. The evolution of selfing in Arabidopsis thaliana. Science. 2007;317:1070–1072. doi: 10.1126/science.1143153. [DOI] [PubMed] [Google Scholar]
- 16.Bechsgaard JS, et al. The transition to self-compatibility in Arabidopsis thaliana and evolution within S-haplotypes over 10 Myr. Mol Biol Evol. 2006;23:1741–1750. doi: 10.1093/molbev/msl042. [DOI] [PubMed] [Google Scholar]
- 17.Hurka H, Neuffer B. Evolutionary processes in the genus Capsella (Brassicaceae) Pl Syst Evol. 1997;206:295–316. [Google Scholar]
- 18.Slotte T, Huang H, Lascoux M, Ceplitis A. Polyploid speciation did not confer instant reproductive isolation in Capsella (Brassicaceae) Mol Biol Evol. 2008;25:1472–1481. doi: 10.1093/molbev/msn092. [DOI] [PubMed] [Google Scholar]
- 19.Neuffer B, Hirschle S, Jäger S. The colonizing history of Capsella in Patagonia (South America)—molecular and adaptive significance. Folia Geobotanica. 2001;34:435–450. [Google Scholar]
- 20.Neuffer B, Hoffrogge R. Ecotypic and allozyme variation of Capsella bursa-pastoris and C. rubella (Brassicaceae) along latitude and altitude gradients on the Iberian peninsula. Anales Jard Bot Madrid. 2000;57:299–315. [Google Scholar]
- 21.Neuffer B, Hurka H. Colonization history and introduction dynamics of Capsella bursa-pastoris (Brassicaceae) in North America: Isozymes and quantitative traits. Mol Ecol. 1999;8:1667–1681. doi: 10.1046/j.1365-294x.1999.00752.x. [DOI] [PubMed] [Google Scholar]
- 22.Hurka H, Bleeker W, Neuffer B. Evolutionary processes associated with biological invasions in the Brassicaceae. Biol Invasions. 2003;5:281–292. [Google Scholar]
- 23.van Kleunen M, Johnson S. Effects of self-compatibility on the distribution range of invasive European plants in North America. Conserv Biol. 2007;21:1537–1544. doi: 10.1111/j.1523-1739.2007.00765.x. [DOI] [PubMed] [Google Scholar]
- 24.Paetsch M, Mayland-Quellhorst S, Neuffer B. Evolution of the self-incompatibility system in the Brassicaceae: Identification of S-locus receptor kinase (SRK) in self-incompatible Capsella grandiflora. Heredity. 2006;97:283–290. doi: 10.1038/sj.hdy.6800854. [DOI] [PubMed] [Google Scholar]
- 25.Nasrallah JB, et al. Epigenetics mechanisms for breakdown of self-incompatibility in inter-specific hybrids. Genetics. 2007;175:1965–1973. doi: 10.1534/genetics.106.069393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Castric V, Vekemans X. Evolution under strong balancing selection: How many codons determine specificity at the female self-incompatibility gene SRK in Brassicaceae? BMC Evol Biol. 2007;7:132. doi: 10.1186/1471-2148-7-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shimizu KK, Shimizu-Inatsugi R, Tsuchimatsu T, Purugganan MD. Independent origins of self-compatibility in Arabidopsis thaliana. Mol Ecol. 2008;17:704–714. doi: 10.1111/j.1365-294X.2007.03605.x. [DOI] [PubMed] [Google Scholar]
- 28.Liu P, Sherman-Broyles S, Nasrallah ME, Nasrallah JB. A cryptic modifier causing transient self-incompatibility in Arabidopsis thaliana. Curr Biol. 2007;17:734–740. doi: 10.1016/j.cub.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boivin K, et al. The Arabidopsis genome sequence as a tool for genome analysis in Brassicaceae. A comparison of the Arabidopsis and Capsella rubella genomes. Plant Physiol. 2004;135:735–744. doi: 10.1104/pp.104.040030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koch MA, Kiefer M. Genome evolution among cruciferous plants: A lecture from the comparison of the genetic maps of three diploid species—Capsella rubella, Arabidopsis lyrata subsp petraea, and A thaliana. Am J Bot. 2005;92:761–767. doi: 10.3732/ajb.92.4.761. [DOI] [PubMed] [Google Scholar]
- 31.Kamau E, Charlesworth B, Charlesworth D. Linkage disequilibrium and recombination rate estimates in the self-incompatibility region of Arabidopsis lyrata. Genetics. 2007;176:2357–2369. doi: 10.1534/genetics.107.072231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kamau E, Charlesworth D. Balancing selection and low recombination affect diversity near the self-incompatibility loci of the plant Arabidopsis lyrata. Curr Biol. 2005;15:1773–1778. doi: 10.1016/j.cub.2005.08.062. [DOI] [PubMed] [Google Scholar]
- 33.Ruggiero MV, Jacquemin B, Castric V, Vekemans X. Hitch-hiking to a locus under balancing selection: High sequence diversity and low population subdivision at the S-locus genomic region in Arabidopsis halleri. Genet Res. 2008;90:37–46. doi: 10.1017/S0016672307008932. [DOI] [PubMed] [Google Scholar]
- 34.Miyashita N, Kawabe A, Innan H, Terauchi R. Intra- and interspecific DNA variation and codon bias of the Alcohol Dehydrogenase (Adh) Locus in Arabis and Arabidopsis Species. Mol Biol Evol. 1998;15:1420–1429. doi: 10.1093/oxfordjournals.molbev.a025870. [DOI] [PubMed] [Google Scholar]
- 35.Savolainen O, Langley CH, Lazzaro BP, Fréville H. Contrasting patterns of nucleotide polymorphism at the alcohol dehydrogenase locus in the outcrossing Arabidopsis lyrata and the selfing Arabidopsis thaliana. Mol Biol Evol. 2000;17:645–655. doi: 10.1093/oxfordjournals.molbev.a026343. [DOI] [PubMed] [Google Scholar]
- 36.Innan H, Tajima F, Terauchi R, Miyashita NT. Intragenic recombination in the Adh locus of the wild plant Arabidopsis thaliana. Genetics. 1996;143:1761–1770. doi: 10.1093/genetics/143.4.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Acarkan A, Rossberg M, Koch M, Schmidt R. Comparative genome analysis reveals extensive conservation of genome organisation for Arabidopsis thaliana and Capsella rubella. Plant J. 2000;23:55–62. doi: 10.1046/j.1365-313x.2000.00790.x. [DOI] [PubMed] [Google Scholar]
- 39.Lynch M. The Origins of Genome Architecture. Sunderland, MA: Sinauer; 2007. [Google Scholar]
- 40.Bahlo M, Griffiths RC. Inference from gene trees in a subdivided population. Theor Pop Biol. 2000;57:79–95. doi: 10.1006/tpbi.1999.1447. [DOI] [PubMed] [Google Scholar]
- 41.Vekemans X, Slatkin M. Gene and allelic genealogies at a gametophytic self-incompatibility locus. Genetics. 1994;137:1157–1165. doi: 10.1093/genetics/137.4.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kusaba M, et al. Self-incompatibility in the genus Arabidopsis: Characterization of the S locus in the outcrossing A. lyrata and its autogamous relative A. thaliana. Plant Cell. 2001;13:627–643. [PMC free article] [PubMed] [Google Scholar]
- 43.Sherman-Broyles S, et al. S locus genes and the evolution of self-fertility in Arabidopsis thaliana. Plant Cell. 2007;19:94–106. doi: 10.1105/tpc.106.048199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Foxe JP, et al. Recent speciation associated with the evolution of selfing in Capsella. Proc Natl Acad Sci USA. 2008 doi: 10.1073/pnas.0807679106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Doyle JJ, Doyle JL. A rapid DNA isolation procedure from small quantities of fresh leaf tissues. Phytochem Bull. 1987;19:11–15. [Google Scholar]
- 46.Rozas J, Sánchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- 47.Nei M. Molecular Evolutionary Genetics. New York, NY: Columbia Univ Press; 1987. [Google Scholar]
- 48.Watterson GA. On the number of segregating sites in genetical models without recombination. Theor Pop Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- 49.Swofford DL. PAUP*. Phylogenetic Analysis Using Parsimony (* and Other Methods): Version 4. Sunderland, Massachusetts: Sinauer; 2003. [Google Scholar]
- 50.Felsenstein J. Confidence-limits on phylogenies—an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.