Abstract
In common with a range of environmental and biological stresses, heat shock results in the accumulation of misfolded proteins and a collection of downstream consequences for cellular homeostasis and growth. Within this complex array of responses, the sensing of and responses to misfolded proteins in specific subcellular compartments involves specific chaperones, transcriptional regulators, and expression profiles. Using biological (ectopic protein expression and virus infection) and chemical triggers for misfolded protein accumulation, we have profiled the transcriptional features of the response to misfolded protein accumulation in the cytosol (i.e., the cytoplasmic protein response [CPR]) and identified the effects as a subcomponent of the wider effects induced by heat shock. The CPR in Arabidopsis thaliana is associated with the heat shock promoter element and the involvement of specific heat shock factors (HSFs), notably HSFA2, which appears to be regulated by alternative splicing and non-sense-mediated decay. Characterization of Arabidopsis HSFA2 knockout and overexpression lines showed that HSFA2 is one of the regulatory components of the CPR.
INTRODUCTION
Protein homeostasis is central to normal cellular function. In sessile plants, this is a major challenge since they are subjected to a plethora of diverse biotic and abiotic challenges, many of which impact upon protein stability. To deal with this, plants have evolved a complex regulatory network of transcription factors that interact with specific promoter elements to transcribe multiple families of heat shock proteins (HSPs) that, in turn, act as chaperones for protein repair or targeted degradation. These are not ubiquitous processes within the cell but are compartmentalized. Hence, the unfolded protein response (UPR), which is induced by the accumulation of misfolded proteins in the endoplasmic reticulum (ER), recruits specific genes and pathways to regulate protein repair in that compartment (Ron and Walter, 2007), and a parallel process, the cytosolic protein response (CPR), operates in the cytosol (Aparicio et al., 2005). Although these processes are essential for maintaining normal cellular function under adverse conditions, how they are integrated into the wider response has been little investigated in higher eukaryotes. For example, heat shock is a pervasive stress with diverse impacts on cellular organization and function. It results in the denaturation of extant proteins, a block to further protein translation, destabilization of cellular membranes, and the activation of oxidative stresses (Morimoto, 1998). It is widely held that the flux of denatured proteins acts as a trigger for many of these effects (Craig and Gross, 1991), but how the various functional aspects relate to different subcellular compartments is not well understood.
The eukaryotic heat shock response involves the induction of HSPs and the action of specific subsets of their transcriptional regulators, heat shock factors (HSFs) (Miller and Mittler, 2006; Swindell et al., 2007). The main role of HSPs in prokaryotes, plants, and animals relates to the protective repair of proteins, although the association of some HSPs with ubiquitin-mediated protein degradation (Qian et al., 2006) also points to a function with respect to protein turnover. Protein homeostasis is maintained through an interaction between HSPs and HSFs, which is disturbed when HSPs are recruited by misfolded proteins. The release of HSFs allows them to trimerize to form the active transcription factor complex required for induced expression of HSPs. Transcription is activated following the binding of the HSF complex to promoter upstream elements, notably the heat shock element (HSE; a palindromic motif of nGAAn). Unlike budding yeast (Saccharomyces cerevisiae) and fruitfly (Drosophila melanogaster), in which a single HSF regulates the expression of heat shock response genes (e.g., HSP70) following heat shock or other stresses, in plants there are multiple HSFs; for example, Arabidopsis thaliana encodes 21 HSF-like genes. These are categorized into three classes by their structures. Class A HSF proteins consist of a DNA binding domain, an oligomerization domain, nuclear localization domains, and transcriptional activation domains (Nover et al., 2001). By contrast, classes B and C lack activation domains. It is thought that this proliferation of HSFs in plants is an adaptation to a sessile lifestyle coping with a dynamic environment (Busch et al., 2005). In addition to the large number of HSFs, further flexibility is provided by the use of both homo- and heterotrimers in the formation of the transcription factor complex (Nover et al., 2001; Bharti et al., 2004; Baniwal et al., 2007).
The UPR is triggered experimentally by treatment with the antibiotic tunicamycin that blocks N-terminal glycosylation, interfering with correct protein folding. Tunicamycin treatment results in the recruitment of BiP (ER-located homologs of HSP70) by misfolded proteins and the induction of larger suites of corrective chaperones. Processes for the proteosome-mediated degradation of terminally misfolded proteins are also activated (Meusser et al., 2005). The CPR is induced by the accumulation of unstable or misfolded proteins in the cytosol and is associated with the induction of a specific subset of HSP70 genes. For example, virus infection in the cytosol in plant or animal cells triggers the induction of HSPs (Whitham et al., 2003), a process that is exacerbated when the virus-encoded proteins are unstable (Jockusch et al., 2001; Jockusch and Wiegand, 2003). The Arabidopsis genome encodes five soluble cytosolic HSP70s, HSP70 (referred to as HSP70A in this article) and HSP70B, and three constitutively expressed HSPs, HSC70-1, HSC70-2, and HSC70-3. All of these genes are induced by heat shock treatment, whereas all but HSP70B are induced in the CPR (Aparicio et al., 2005).
We hypothesized that it should be possible to identify specific responses to misfolded proteins within the overall heat shock response and to use this information to dissect the transcriptional features of the CPR. Using the proline analog l-azetidine-2-carboxylic acid (AZC) to generate misfolded proteins (Trotter et al., 2002) throughout the cell, we compared the transcript profiles following heat shock treatment with profiles associated with AZC (inducing the UPR and CPR) and tunicamycin treatment (UPR alone). From our analyses, we identified the HSE as being essential for the induction of the CPR and that the process was partly regulated by HSFA2. In addition, we identified a novel alternative splicing product of HSFA2 RNA with the potential to modulate HSFA2 activity.
RESULTS
Transcript Profiling the CPR
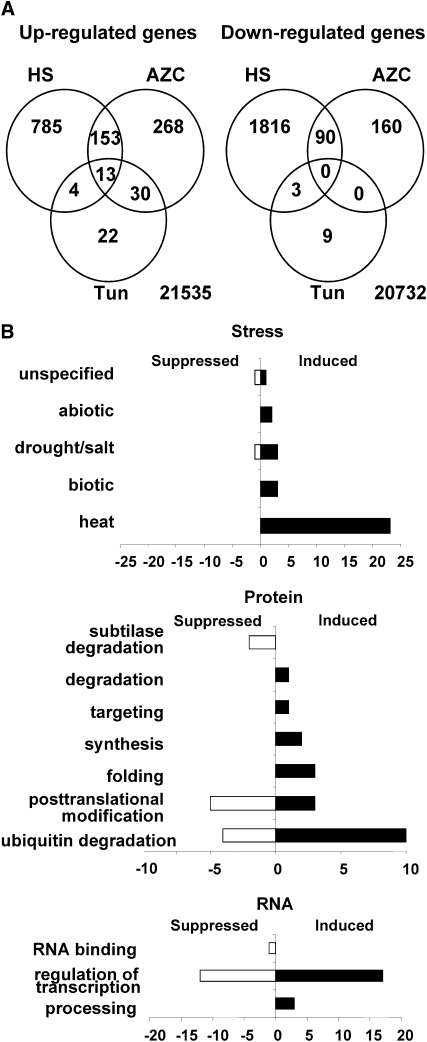
Previously, the CPR has been induced by ectopic protein accumulation following agroinfiltration-mediated transient expression or by virus infection. To avoid the ancillary biological effects associated with these treatments, we created misfolding of nascent polypeptides in vivo by treating Arabidopsis leaves with AZC. In parallel, leaves were treated with tunicamycin or heat shock at 37°C. The optimal timing for these treatments with respect to known responses was assessed using quantitative RT-PCR (qRT-PCR) for HSP70A, luminal binding protein 3, (BiP3), and basic leucine zipper 60 (βZIP60) RNAs (see Supplemental Figure 1 online). HSP70A RNA, typically induced in the CPR and in response to heat shock, showed maximal accumulation after 1 and 3 h for heat shock and AZC treatments, respectively. BiP3 RNA, typically increased in response to misfolded proteins in the ER, showed maximum accumulation 3 h after treatment with tunicamycin and >6 h after treatment with AZC. βZIP60, a key regulator of the UPR (Iwata and Koizumi, 2005), appeared to show an earlier but weaker response to tunicamycin than BiP. Since we were primarily interested in the contribution the CPR and its regulatory pathway made to the overall heat shock response, 1 h (heat shock) and 3 h (AZC and tunicamycin) were selected as treatment periods prior to the transcript analysis using Arabidopsis ATH1 microarrays. Untreated, and proline or dimethyl formamide (tunicamycin solvent) treated tissues were included as controls. Thus, we reasoned that changes from control samples in common between heat and AZC but not present in the tunicamycin treatments would identify a subclass of transcriptional responses that represented the CPR as a subcomponent of the heat shock response. Three biological replicates for each treatment were analyzed. As tunicamycin-treated samples showed a relatively high degree of variation between replicates, we performed hybridizations with two further biological replicates for tunicamycin and control treatments. The microarray data are summarized in Venn diagram form in Figure 1A; detailed lists of transcripts showing significant (P < 0.05 and ≥ twofold change) alteration are included in Supplemental Data Set 1 online; P values were adjusted using the Benjamini-Hochberg (false discovery) method.
Figure 1.
The Differentially Regulated Genes after Three Treatments.
(A) Venn diagram of the differentially regulated genes after three treatments. The numbers of genes showing altered regulation following heat shock (HS), AZC, or tunicamycin treatment in comparison to their respective controls are shown in Venn diagrams. Numbers at the bottom right indicate genes showing no significant (P ≥ 0.05) changes for any of the treatments.
(B) Functional categorization of differentially regulated genes by CPR. CPR upregulated or downregulated genes were grouped according to their known or predicted function using the MapMan functional classification system (Thimm et al., 2004). The three main categories (stress, protein, and RNA) belong to MapMan level 1, while the others belong to level 3. Black bars represent upregulated genes, and white bars represent downregulated genes.
Heat shock, AZC, and tunicamycin treatments showed decreasing impact on transcript levels, assessed as the total number of genes showing upregulation and downregulation in each case (Figure 1A). Specific to the CPR (common area between heat shock and AZC response minus tunicamycin response), there were 153 upregulated genes and 90 downregulated genes. Intuitively, we speculated that positive responses to applied stresses might involve the recruitment of new biological processes seen as transcriptional upregulation, whereas downregulated genes might represent more the consequences of the imposed stress. Focusing on the specific overlap between heat shock and AZC (minus tunicamycin) treatments, we found that the 153 upregulated genes fell into several broad functional classes. Notably, for genes with functional annotation, the classes included (1) a large set of chaperones, including many HSPs (categorized as heat stress genes; with respect to the CPR, these included inducible HSP70 genes and excluded HSP70B); (2) a group of transcription and splicing factors; and (3) a group of genes involved in protein degradation, especially the ubiquitin-mediated degradation system (Figure 1B; see Supplemental Table 1 online). Of the remaining genes, the majority had no known functional annotation.
To validate our strategy and data, we used three approaches. First, we compared our microarray data with a published microarray data set of heat-treated Arabidopsis shoots using a cluster analysis (see Supplemental Figure 2 online). Our data showed that the heat shock and AZC-treated samples clustered with the published responses at 1 and 3 h after treatment, while the tunicamycin-treated samples tended to be closer to the earliest times (0.25 and 0.5 h) of the heat shock response. This may indicate that heat is sensed first within the ER, but more experimentation will be needed to verify this. Second, we asked whether genes known to be induced in the UPR (Urade, 2007) appeared in the overlapping zone between heat shock, AZC, and tunicamycin. Of the 13 upregulated genes, six were previously reported as genes involved in the UPR. This included βZIP60. Interestingly, the upregulated genes for the UPR excluded all of the Arabidopsis HSFs, so identifying the heat shock–induced HSFs as part of the transcriptional activation response restricted to the cytosol. Third, we selected 13 genes from the transcription/splicing factor and protein degradation classes of genes upregulated in response to heat shock and AZC and performed qRT-PCR using three biological replicates (see Supplemental Table 2 online). Our qRT-PCR data confirmed the microarray data with most of the genes being significantly induced by both heat shock and AZC treatment. For most of these genes, the extent of change tended to be greater following heat shock treatment.
A Canonical HSE Promoter Element Is Involved in the CPR
A key feature of the CPR is the selective induction of four of the five HSP70 genes. These four genes have the canonical HSE palindromic nGAAn motif in their upstream promoter regions, whereas this is missing in the noninduced HSP70B, although all these genes are induced by heat. Hence, we asked whether the HSE was highly represented within the promoters of the heat shock/AZC (minus tunicamycin) upregulated class of genes. Bioinformatic analysis of sequences 1000 bp upstream of the transcription start site of genes showed that the HSE was significantly overrepresented in this class (15.03%), as opposed to all genes in the heat-shock-only class (4.46%) or all genes in the Arabidopsis genome (1.66%) (see Supplemental Table 3 online). Gene ontology analysis of the 15.03% of genes showed that 38% of them encoded transcription factors.
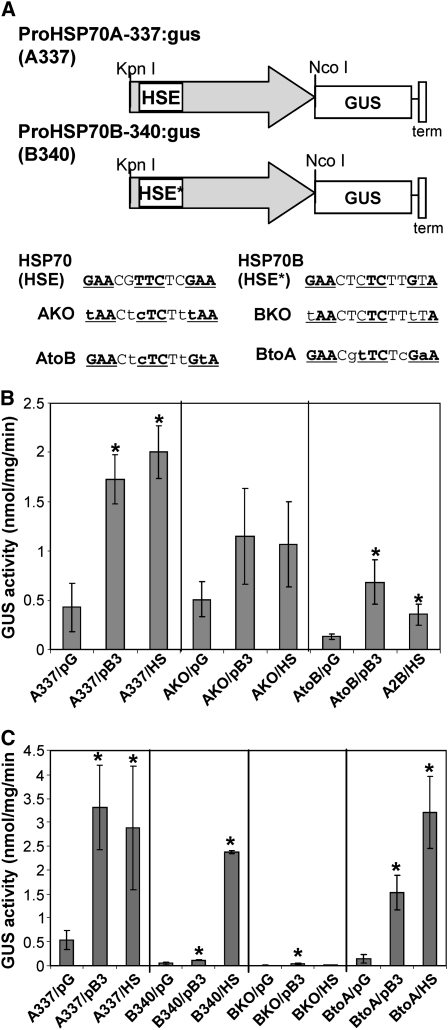
To test the apparent relationship between the CPR and HSEs more formally, we undertook a comparative functional analysis of the promoter regions of HSP70A and HSP70B. The promoters of the two genes were examined using promoter:reporter constructs expressing β-glucuronidase (GUS). Previously, an ∼2 kb upstream promoter region from each gene was fused to GUS within an Agrobacterium tumefaciens binary vector and was assayed following transient expression after infiltration of Agrobacterium carrying this plasmid into Nicotiana benthamiana leaves. The GUS activity faithfully reported the response of these promoters to heat, virus infection, and ectopic protein expression (Aparicio et al., 2005). (It has not been possible in our hands to obtain quantitatively reproducible ectopic protein expression by agroinfiltration into the leaves of Arabidopsis [Aparicio et al., 2005].) We focused our attention on smaller promoter fragments for HSP70A and HSP70B that similarly faithfully reported the heat shock and misfolded protein response in the same transient expression system; an ∼340-bp promoter region upstream of the start codon of HSP70A (proHSP70A337:GUS) and HSP70B (proHSP70B340:GUS) showed the same degree and pattern of responses to heat and protein accumulation, suggesting that the regions contain the essential elements for the HSP70A and HSP70B regulation. This minimal HSP70A promoter (337 bp) includes a canonical HSE (GAACGTTCTCGAA), while the minimal HSP70B promoter (340 bp) includes only an imperfect HSE (HSE*; GAACTcTCTTGtA; Figure 2A). When the canonical HSE in the HSP70A promoter was mutated to tAACGcTCTCtAA (A knockout), the induced response to either heat or protein overexpression was lost (P value > 0.05) (Figures 2A and 2B). However, the mutation did not completely abolish the response of the promoter to heat or protein stress, indicating that there are other sequences that contribute to the activation of the promoter. The minimal HSP70B promoter (proHSP70B340:GUS) shows a basal response to protein accumulation. Changing the imperfect HSE* to tAACTcTCTTtTA (B knockout) showed that HSE* was required for the heat shock response (Figure 2C) and showed no change in the basal response to protein accumulation. Interestingly, replacement of the HSP70B HSE* with HSP70A HSE (BtoA) furnished the HSP70B promoter with the ability to respond to protein accumulation and heat shock (see BtoA; Figure 2C). Replacement of the canonical HSE in the HSP70A promoter with HSE* reduced the response of the HSP70A promoter to both heat and protein accumulation but did not completely abolish the response to either stress (see AtoB; Figure 2B). The results show that a complete HSE is required for the response to protein accumulation. They also indicate that different factors are required for the complete heat or CPR responses and that specific factors required for the latter cannot recognize the HSP70B promoter due to its imperfect HSE*.
Figure 2.
Response of HSP70A and HSP70B Promoters and Their Variants to Heat Shock and Protein Stress.
(A) Promoter:reporter constructs and HSE mutants. Promoter:reporter Agrobacterium binary constructs were made for HSP70A (A337) and HSP70B (B340) using 337 and 340 bp, respectively, of the upstream genomic DNA (gray arrow) fused to GUS and the CaMV 19S terminator (term). Promoter variants were created by site-directed mutagenesis of the HSE in the HSP70A promoter and the HSE-like (HSE*) sequence of the HSP70B promoter. The original sequences are shown at the top. The consensus sequence of HSEs is shown in bold, and the relative positions are underlined. The mutated nucleotides are shown in lowercase.
(B) Promoter responses in N. benthamiana measured as GUS activity following Agrobacterium infiltration of the constructs. Promoter constructs were coinfiltrated with pB3, which overexpresses a cytosolically targeted Arabidopsis protein (At5g08290) to induce the CPR or with an empty vector (pG; nonstressed) as a negative control. Infiltrated tissues were also heat shocked (HS) as a positive control for HSP70A and HSP70B induction. The responses of the HSP70A HSE knockout (AKO) mutation or the HSP70A promoter mutant that substituted HSE with HSE* (AtoB) to the three treatments in comparison with the wild-type minimal promoter for HSP70A (A337) are shown. Assays were performed in triplicate on individual extracts of two infiltrated leaves. Bars indicate ±sd.
(C) Similar experiments to those in (B) but showing the responses to the three treatments of the HSE* knockout mutation in the HSP70B promoter (BKO) or the substitution of the HSE* with HSE in the HSP70B promoter (BtoA) in comparison with the HSP70B (B340) promoter. Responses of the A337 promoter to protein expression (pB3) or heat shock (HS) were included (left panel) as positive controls in the same experiment. Assays were performed in triplicate on individual extracts of two infiltrated leaves. Bars indicate ±sd. Statistical comparisons were made with the coinfiltrated empty plasmid (pG) in each case. P values < 0.05 are indicated with asterisks.
HSFA7, HSFA2, and Alternatively Spliced HSFA2 RNAs Are Induced in the CPR
Our microarray data (see Supplemental Data Set 1 online) showed that of 15 class A HSFs in Arabidopsis, only HSFA2 and HSFA7a were induced by the CPR (common to both AZC and heat shock); class B HSFs (HSFB1, HSFB2a, and HSFB2b) were also induced. In addition, HSFA3, HSFA1e, HSFA1d, and HSFA7b were induced by heat shock only. With the exception of HSFA1e, these data confirm previous reports of heat shock induction of HSFs in aerial tissues (Schramm et al., 2008).
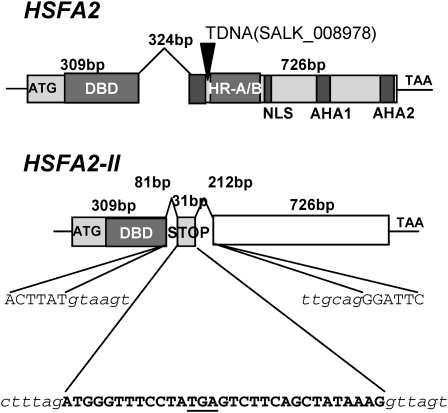
To confirm the nature of the class A HSFs induced in the CPR, we concentrated on the two CPR-induced HSFs and cloned and sequenced the cDNAs for HSFA2 and A7a from heat shock and AZC-treated tissues. HSFA7a cDNAs from both treatments and HSFA2 cDNA from heat shock tissues revealed protein coding sequences conforming to predictions based upon the presence of a single intron within all HSFs (Nover et al., 2001). By contrast, the HSFA2 cDNA from AZC-treated tissues revealed some clones with a novel sequence (see Supplemental Figure 3 online), generated by alternative splicing of the intron (Figure 3). This novel splicing pattern introduced an additional 31 bp mini-exon from within the conserved intron in the DNA binding domain. The mini-exon resulted from new 3′ (tttag:a) and 5′ (g:gttagt) splice sites (Figure 3). The additional mini-exon introduced an in-frame stop codon that would terminate translation after the N-proximal DNA binding domain.
Figure 3.
HSFA2 Is Alternatively Spliced upon AZC Treatment.
The structural organizations of the two forms of the mature HSFA2 transcript are shown. In the HSFA2 mRNA, the single 324-nucleotide intron is removed. By contrast, the HSFA2-II RNA has an additional mini-exon (uppercase bold) included through the recognition of additional splice sites (lowercase) within the intron. The new stop codon is underlined. DBD, DNA binding domain; HR-A/B, oligomerization domain; NLS, nuclear localization signal; AHA, activation domain, notation taken from (Nover et al., 2001). Inverted triangle marks the position of the T-DNA insertion in the Arabidopsis insertional knockout (SALK_008978) line.
To determine whether there was a temporal correlation in the expression patterns for these HSFs and HSP70A, as a marker of the CPR, heat shock– and AZC-treated Arabidopsis plants were analyzed by qRT-PCR (Figure 4). Specific primer sets were used to differentiate the AZC form of HSFA2 (HSFA2-II) from the normally spliced HSFA2 RNA. Despite some quantitative variability in the response, the accumulations of HSFA2, HSFA7a, and HSP70A were consistent across multiple experiments. After heat shock, HSFA7a had increased rapidly at 30 min then decreased to the end of the experiment (3 h). Changes in HSFA2 and HSFA2-II RNAs were delayed relative to HSFA7a, showing maximum accumulation at 1 h after treatment (Figure 4), matching the changes in HSP70A (see Supplemental Figure 1 online). The pattern of induction indicated that the heat stress is sensed most strongly during the first 60 min of treatment and that during the subsequent period (up to 3 h of total treatment) the tissues acclimate to survival at the higher temperature. Features of this early response include a rapid and transient induction of HSFA7a and a subsequent differential accumulation of HSFA2 and HSFA2-II RNA. The latter accumulated to a maximum of 15% of the level seen for HSFA2 RNA. In AZC-treated tissues, HSF RNAs reached a maximum in parallel with the peak of HSP70A RNA accumulation at 3 h after treatment (cf. Figure 4 and Supplemental Figure 1 online). Most notable in the comparisons with heat shock treatment was that the relative proportions of HSFA2 and HSFA2-II RNAs were reversed following AZC treatment, with the level of HSFA2 RNA at only 17% of HSFA2-II RNA. Extremely low levels of HSFA2 expression prevented an accurate determination of the ratio for untreated tissues. Since AZC activates the HSP response primarily through the induction of misfolded proteins and HSP70A was not induced in response to tunicamycin (UPR), it appeared that the presence of HSFA2, HSFA7a, and accumulation of HSFA2-II identified the CPR.
Figure 4.
Time Course of HSF Induction during Heat and AZC Treatment.
(A) Expression level of selected HSFs, relative to EF1a, were examined by qRT-PCR. Following AZC infiltration, leaf samples (triplicate) were harvested after the indicated incubation times. Bars inicate ±sd.
(B) Similar experiment to that in (A), but samples (triplicate) were harvested after heat shock treatment. Bars indicate ±sd.
HSFA2 Is Functionally Important for the CPR
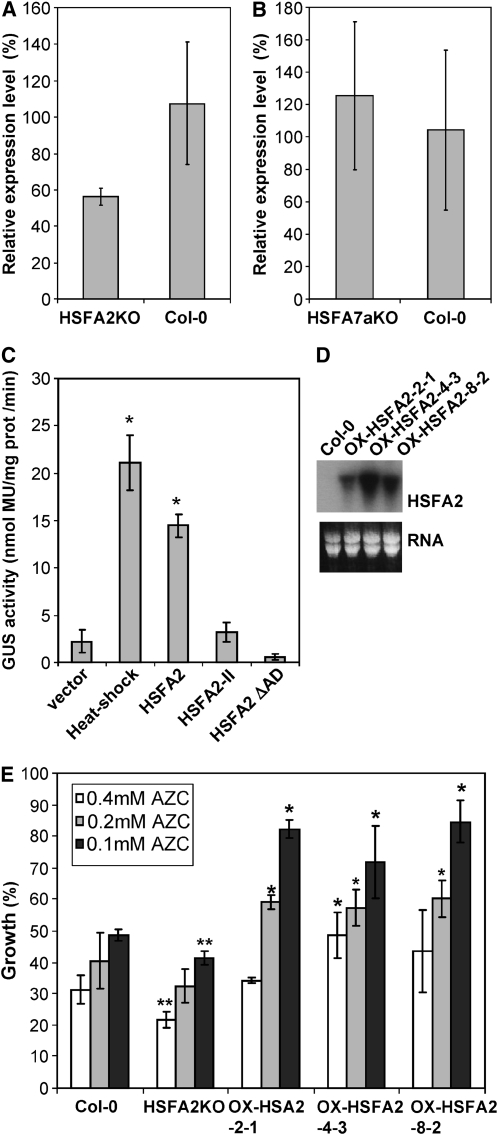
Our transcriptional data identified the profile of HSFA2, HSFA7a, and HSFA2-II RNA accumulation as a signature of the CPR. Since HSFA2 and HSFA7a are Class A HSFs that have transcription activation domains, they might regulate the induction of other CPR-associated genes (e.g., HSP70s and other chaperones) that correct the accumulation of misfolded proteins. To examine the regulatory effect of HSFA2 and HSFA7a during the CPR, we have examined the induction of HSP70A in insertional mutant lines. Using qRT-PCR to assay HSP70A expression, an Arabidopsis insertional mutant knockout line (SALK_008979) for HSFA2 (Alonso et al., 2003; Nishizawa et al., 2006) and a mutant line for HSFA7a (SALK_080138; T-DNA insertion just upstream of the AHA transcriptional activation domain) (Larkindale and Vierling, 2008) were compared with wild-type Arabidopsis Columbia-0 (Col-0) after treatment with AZC. In repeated experiments, the analysis showed a strong trend (P value=0.057) toward weaker induction of HSP70A following AZC treatment of the hsfA2 mutant line when compared with wild-type Arabidopsis Col-0 (Figure 5A); the reduction of HSP70A induction in the hsfA2 mutant was statistically significant (P < 0.05) at 6 h after AZC infiltration (see Supplemental Figure 4 online). By contrast, HSP70A expression in the hsfA7a mutant was unchanged (Figure 5B).
Figure 5.
Functional Roles of HSFA2 and HSFA2-II RNAs.
(A) Expression analysis (qRT-PCR) of HSP70A in the insertional null (KO) mutant for HSFA2 (Nishizawa et al., 2006) and a wild-type (Col-0) control plant 3 h after treatment with AZC. The data, expressed as values relative to the expression of EF1a, are the mean (±sd) of three biological and three technical replicates. HSP70A induction in the HSFA2 mutant was ∼50% of that in the control, significant at P value = 0.057. Significant differences were observed at later times (6 h after infiltration with AZC; see Supplemental Figure 4 online).
(B) Similar experiment to that in (A) but for the HSFA7a insertional mutant. The data, expressed as values relative to the expression of EF1a, are the mean (±sd) of three biological and three technical replicates. In this case, there was no significant difference from the control plant.
(C) The ability of different HSFs to induce the HSP70A 337-bp promoter was tested. N. benthamiana leaves were infiltrated with mixed cultures of Agrobacterium carrying proHSP70A337:GUS and a CaMV pro35S:HSF construct. Induction of each promoter was measured as GUS activity; data are the mean of three biological replicates; bars indicate ±sd; asterisks show significant (P value < 0.05) differences from the empty vector treated control. HS, noninfiltrated heat shock control; HSFA2ΔAD, HSFA2 truncated before the AD. The experiment was repeated twice with similar results.
(D) RNA gel blot detection (top panel) of HSFA2 RNA in the transgenic Arabidopsis lines (2-1, 4-3, and 8-2) overexpressing pro35S:HSFA2. Bottom panel shows rRNA staining used as a loading control.
(E) The growth of Arabidopsis lines (Col-0 wild type, HSFA2 knockout mutant, and three HSFA2 overexpression lines) in media with the indicated concentration of AZC. The percentage of growth was calculated by comparison to the growth of the lines cultured without AZC. The data show the average of three sets of plants; bars indicate ±sd. The lines showing significant change in growth rate with each concentration of AZC compared with Col-0 (Student's t test; P value < 0.05) are indicated by asterisks. Two asterisks show significant growth reduction, and one asterisk shows significant growth increase.
The accumulation of HSFA2-II RNA is also a feature of the CPR. The location of a translational stop codon that eliminated essential functions downstream of the N-proximal DNA binding domain made it unlikely that HSFA2-II RNA could be involved in activating HSP70A expression. To confirm this, a complementary experiment in N. benthamiana was performed where HSFA2, HSFA2-II, or an artificial construct HSFA2ΔAD with a termination codon before the C-terminal activation domain, were expressed from the cauliflower mosaic virus (CaMV) 35S promoter. Activation of the proHSP70A337:GUS reporter, as a measure of HSP70A induction, was tested after cotransient expression with each of the HSF constructs following agroinfiltration into leaf tissues. Again, HSFA2 expression showed significantly (P value < 0.05) increased HSP70A induction to a level close to that seen for heat shock itself, while expression of genes for the truncated HSFA2 proteins showed no change (Figure 5C).
HSFA2 is induced by heat shock and a diverse range of other stresses (Miller and Mittler, 2006), and HSFA2-overexpressing lines can confer improved tolerance to high temperature and light conditions (Nishizawa et al., 2006). Hence, we also tested whether transgenic lines ectopically expressing HSFA2 conferred protection against the damaging effects of AZC treatment by measuring growth in the presence of sublethal concentrations of AZC (Figure 5E). While the HSFA2 knockout line showed less tolerance to AZC, three independent homozygous HSFA2 transgenic lines with varying levels of transgene expression (Figure 5D) showed higher tolerance (Figure 5E). HSFA7a knockout and overexpression lines showed variable and mostly insignificant changes in tolerance to AZC (see Supplemental Figure 5 online).
HSFA2-II RNA Is Not Active and Is Degraded by Non-Sense-Mediated Decay
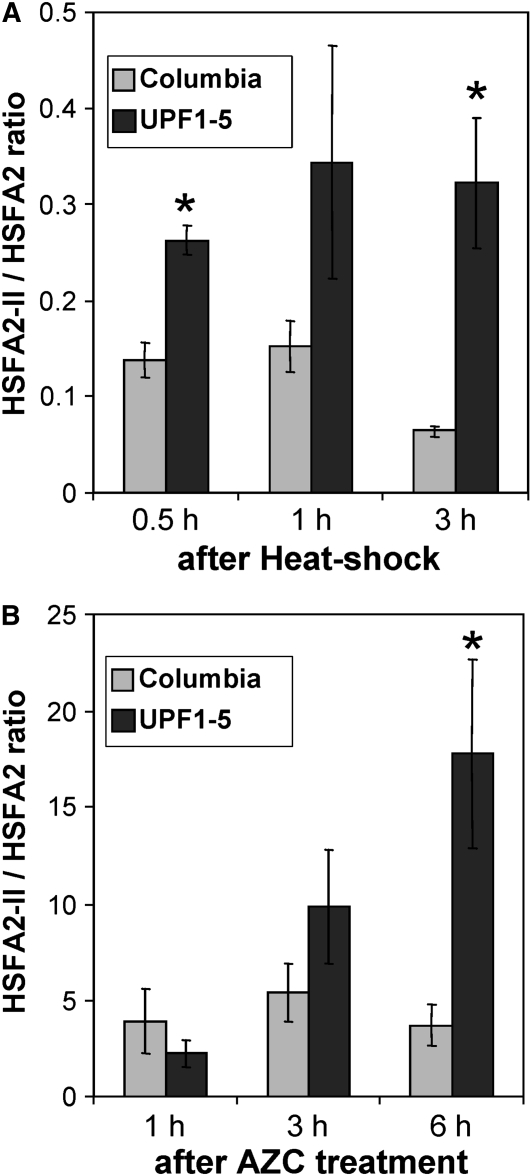
HSFA2-II is specifically induced in but does not itself induce the CPR (Figures 4 and 5C). The introduction of the premature termination codon downstream of the N-proximal activation domain would probably result in a nonfunctional protein. This organization of HSFA2-II RNA has features in common with transcripts processed via the non-sense-mediated decay (NMD) pathway. NMD is a conserved RNA quality control system in eukaryotes that degrades mRNAs with a premature termination codon. In plants, mRNAs that have an extended noncoding distance between the premature termination codon and 3′ end of the mRNA, or which have a downstream splice junction, are recognized by the NMD system and targeted for degradation (Kertesz et al., 2006; Hori and Watanabe, 2007; Kerenyi et al., 2008). To test whether HSFA2-II is subject to NMD, a T-DNA insertion mutant in the NMD machinery (upf1-5; Arciga-Reyes et al., 2006) was compared with wild-type plants for the induction of HSFA2 and HSFA2-II RNAs following heat or AZC treatment. If HSFA2-II was a substrate for NMD, the ratio of HSFA2-II to HSFA2 RNAs would be higher in the upf1-5 mutant. Following heat treatment for 1 h or AZC treatment for 3 h, RNA levels were analyzed by qRT-PCR. In both situations, the ratios of HSFA2-II to HSFA2 RNAs were higher in the NMD mutant line (Figure 6; Supplemental Figure 6 online shows the quantitative transcript values).
Figure 6.
HSFA2-II Is Degraded by NMD.
(A) The time course of relative accumulation of HSFA2-II to HSFA2 RNA after heat shock in Col-0 wild type and upf1-5 mutant. The upf1-5 mutant lacks an essential component of the NMD machinery. The upf1-5 mutant showed higher accumulation of HSFA2-II compared with the wild type. Transcript accumulation was measured by qRT-PCR relative to the expression of EF1a.
(B) Similar experiment to that in (A) except that plants were infiltrated with AZC. Again the upf1-5 mutant showed higher accumulation of HSFA2-II compared with the wild type.
The data are the mean of three replicates. Bars indicate ±sd. Asterisks show significant (P value < 0.05) differences from the wild-type Col-0 control.
The reversed ratio in the accumulation of HSFA2-II and HSFA2 RNAs after heat shock and AZC could point to differential regulation of HSFA2 after the two treatments or a stimulation or suppression of NMD after heat shock or AZC treatment, respectively. To test the latter hypothesis, we examined the effect of heat shock or AZC treatment on the accumulation of two specific RNAs (transcripts from pseudogenes At5g40920 and At1g01060) shown to be regulated by the NMD (Arciga-Reyes et al., 2006). In neither case was the accumulation significantly affected by the treatments when compared with controls (see Supplemental Figure 7 online).
Biological Induction of the CPR Also Induces HSFA2, HSFA2-II, and HSFA7a
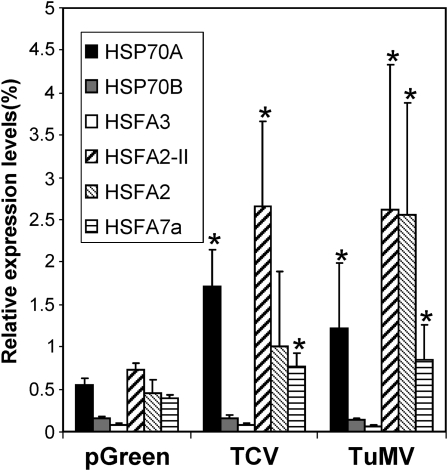
AZC is pervasive in its effects and generates terminally misfolded proteins throughout the cell. Consequences of AZC action are the induction of a large number of genes, among which are genes associated with transcription and transcript splicing (Figure 1). Accordingly, HSFA2, HSFA7a, and HSFA2-II were identified following AZC treatment. Although our analyses relate these RNAs to the CPR, it remains possible that pleiotropic effects associated with AZC have led to the induction of these HSFs and especially to the formation of HSFA2-II RNA. Induction of the CPR, characterized by the differential induction of HSP70 genes, occurs in cells actively involved in virus genome replication and virus gene expression. To test whether the appearance of HSFA2-II RNA correlated with the CPR rather than being a secondary consequence of AZC incorporation, we assayed HSF expression pattern in virus-infected Arabidopsis tissues. Leaves were infected with Turnip mosaic virus (TuMV; genus Potyvirus) or Turnip crinkle virus (TCV; genus Carmovirus), and the newly emerged systemically infected leaves were harvested 7 d after inoculation. The samples were first subjected to RT-PCR to confirm the virus infection and then subjected to qRT-PCR to quantify the expression level of HSFs. The CPR response associated with virus infection is highly localized, being focused on cells actively involved in virus multiplication and virus and virus genome expression (Aranda et al., 1996). As such, the level of induction of HSFs was less than observed in response to AZC or heat shock. Nevertheless, as shown in Figure 7, virus-infected Arabidopsis leaves showed strong accumulation of HSFA2-II, variable induction of HSFA2, and slight induction of HSFA7a but no significant induction of HSP70B or HSFA3; in our microarray experiments, HSFA3 was also not induced in the CPR (using AZC), although it was induced by heat (see Supplemental Data Set 1 online). The same gene induction patterns were observed in both TuMV- and TCV-infected tissues. The relatively high ratio of HSFA2-II to HSFA2 RNAs by virus infection mimics the response to AZC and confirms that alternative splicing of HSFA2 is an integral part of the CPR.
Figure 7.
Virus Infection and AZC Induce the Same HSFs.
Arabidopsis plants systemically infected with TuMV or TCV were analyzed by qRT-PCR for the accumulation of HSP70A, HSP70B, HSFA2, HSFA2-II, HSFA3, and HSFA7a RNAs 7 d after inoculation. The data, which are means of two biological and three technical replicates (±sd), are expressed as values relative to the expression of EF1a. In parallel with the specific induction of HSP70A noted previously (Aparicio et al., 2005), virus-infected tissue showed the clear induction of HSFA2-II and slight induction of HSFA7a. HSFA3, which is an indicator of the heat shock response, and HSF70B were not induced. Compared with the control sample inoculated with Agrobacterium carrying an empty binary vector (see Methods), genes showing significant induction compared with the vector control (Student's t test; P value <0.05) are indicated by asterisks. The experiment was performed twice with similar results.
DISCUSSION
Eukaryotes have evolved complex defense pathways to cope with biotic and abiotic stresses, and in many cases these stress responses overlap. This is particularly true for responses involving genes for HSPs and their HSF transcriptional regulators where specific subsets of gene classes may be involved uniquely or collectively in responding to biotic and abiotic stresses (Miller and Mittler, 2006; Swindell et al., 2007). A good example is the range of complex consequences and responses associated with temperature stress, which impacts plant cells and tissues independent of spatial organization. A primary consequence of heat stress is the accumulation of misfolded proteins, which trigger responses, notably the CPR and UPR, located within discrete subcellular compartments. The aim of this work was to identify the separate contribution of the CPR to the overall heat stress response and to identify key transcriptional features of the CPR.
To aid the dissection of the CPR, we employed the proline analog AZC. Incorporation of AZC leads to terminally misfolded proteins in both the soluble and ER compartments of the cytosol but is distinct from UPR induction by tunicamycin in that the latter fails to induce genes for the cytosolic HSP70s or HSFs. Therefore, unlike AZC, the effect of tunicamycin is spatially restricted. Therefore, comparison of the effects of AZC and tunicamycin treatment allowed us to identify factors and processes that were attributable to the CPR alone.
CPR as a Subcomponent of the Heat Shock Response
By integrating the transcript profiling data from heat shock (CPR + UPR + pleiotropic effects), AZC treatment (CPR + UPR + other pleiotropic effects), and tunicamycin treatment (UPR + pleiotropic effects), we were able to identify groups of genes that were upregulated or downregulated in association with the CPR within the heat shock response (Figure 1; see Supplemental Data Set 1 online). Existing data for heat shock–treated Arabidopsis (AtGenExpress, http://www.Arabidopsis.org/info/expression) (Schramm et al., 2008) are based upon different experimental approaches from ours. Nevertheless, there was a good correspondence between our data set and the public data sets. For the UPR, we identified fewer changes in expression than previously reported (Martinez and Chrispeels, 2003). However, both the experimental approaches and technical array platform differed. Nevertheless, the appearance of βZIP60 and five other known UPR genes, among the 13 genes overlapping between heat shock, AZC, and tunicamycin treatment, gave us confidence that the microarray analysis provided an accurate picture of changes associated with the UPR.
Analysis of the correspondence between the treatments identified 153 upregulated genes associated with the CPR within the heat shock response (see Supplemental Table 1 online). Appropriately, this list included CPR-induced HSP70 genes but not HSP70B. It also included genes for chaperones, HSFs, and components of the splicing machinery, together with a number of unannotated genes (27 expressed or hypothetical proteins). The list provides a key resource for the further dissection of the CPR in relation to other compartmentalized stress responses (e.g., UPR) or responses to more general stress conditions (e.g., heat shock).
CPR Is Partly Regulated through Canonical HSE
A large number of genes that respond to heat shock contain the canonical palindromic motif of nGAAn in their upstream regulatory domains, but the preponderance of the motif in the CPR class of genes identified in the microarray analysis (see Supplemental Table 3 online) implies that this regulatory sequence is more related to the sensing and transduction of responses to misfolded protein than it is to other aspects of the broader heat shock response. Of the 15% of CPR upregulated genes that contain an HSE in the promoter region, ∼40% encode transcription factors. Among these transcription factors were several HSFs, including HSFA2 and HSFA7a. We would predict that some of the other induced factors would in turn activate their own downstream targets, some of which could be in the 85% of induced genes that lack an HSE. This remains to be tested.
The induction of HSP70A, but not HSP70B, in response to ectopic protein accumulation provided the opportunity to experimentally analyze the transcriptional elements required for the CPR. Mutagenesis showed that the HSE within the HSP70A minimal promoter was necessary and that it could confer protein responsiveness upon HSP70B (Figure 2). Since HSP70B is also heat inducible, it raises further the question as to whether the canonical HSE is primarily a heat- or a protein-responsive element.
CPR Induces a Unique Spectrum of HSFs
Arabidopsis deals with the impact of imposed stresses through the recruitment of different combinations of HSFs. Of 15 Class A HSF genes in our microarray analysis, few showed significant induction in response to any of the stresses applied. Only HSFA2, A3, and A7a were significantly induced in response to heat shock, while only HSFA2 and HSFA7a were induced in response to AZC, and none were induced in response to tunicamycin (UPR). Intriguingly, in contrast with the situation following AZC treatment, HSF expression changes in response to heat shock appeared to be phased: HSFA7a showed the earlier response and HSFA2/A2-II a later response (Figure 4). The role of HSFA7a in the heat shock response is reported to be involved in heat acclimation (Larkindale and Vierling, 2008). Unlike HSFA7a, HSFA2 induction was strongly correlated with the induction of HSP70A. However, considering AZC was used as a specific activator of the CPR, it might be seen as surprising that HSFA2 was more strongly induced by heat than by AZC (Figure 4). This may relate to the misfolding of extant proteins by heat shock but, for AZC, only the misfolding of nascent polypeptides during translation. Accordingly, the HSFA2/HSFA2-II ratio, which potentially affects the amount of available active HSFA2, was high in heat shocked samples and low in AZC-treated samples. Crucially, however, the same profile of HSF induction (i.e., induction of HSFA7a, HSFA2, and HSFA2-II and no induction of HSFA3) was observed following biological activation of the CPR by virus infection (Figure 7).
Potential Regulation of the CPR by Alternative Splicing of HSFA2
Overall, HSFA2 is the most widely induced HSF in response to environmental stresses, indicating its central role in cell homeostasis (Miller and Mittler, 2006). hsfA2 knockout plants show reduced tolerance to heat shock treatment (Schramm et al., 2006) and, in our experiments, reduced responsiveness to misfolded protein in the cytosol as measured by the induction of the HSP70A promoter. Although we experienced a large variation between biological replicates, probably due to the pressure infiltration of AZC, we observed the reduction of HSP70A in the hsfA2 knockout plant (Figure 5). In turn, our HSFA2-overexpressing lines showed increased tolerance to the toxic effects of AZC, an effect not seen in the HSFA2-II–overexpressing lines. Many of the genes induced in HSFA2-overexpressing plants (Nishizawa et al., 2006) are also represented in the category of genes overexpressed in response to heat shock and AZC in our microarray experiment. These functions of HSFA2 have a biological impact as shown by the altered growth characteristics of the knockout and overexpressing lines in the presence of AZC (Figure 5). Therefore, we could conclude that HSFA2 partially regulates the CPR. What then is the purpose of the alternatively spliced HSFA2-II RNA, apparently induced uniquely as part of the CPR?
The novel mini-exon within HSFA2-II introduces a stop codon toward the end of the DNA binding domain, hence excluding a complete DNA binding domain, the nuclear localization signal, and the oligomerization and activation domains, and likely removing any functionality from the encoded protein (Figure 2). However, the spliced RNA has features of aberrant mRNAs that make it a substrate for NMD and, accordingly, HSFA2-II RNA accumulated to a higher level in the NMD upf-1 mutant (Figure 6). The specific NMD-mediated degradation of HSFA2-II RNA was confirmed from our experiments that showed that the NMD pathway was not altered generally by heat shock or AZC treatments (see Supplemental Figure 7 online). Therefore, we favor the hypothesis that alternate splicing of HSFA2 RNA coupled with NMD represents a mechanism for posttranscriptional regulation for the production of active HSFA2 protein. Accordingly, our transcript profile analysis showed that the CPR induced a number of genes encoding splicing factors (Figure 1). Since HSFA2 is induced in response to many environmental stresses, this posttranscriptional regulation offers a subtle second level of control. Why then should heat show a lower ratio of HSFA2-II to HSFA2 RNA? Heat denatures proteins throughout the cell, while only newly synthesized proteins are misfolded in AZC-treated cells. Thus, our interpretation is that heat-shocked cells induced and required larger amounts of active HSFA2, while AZC- treated cells had a lesser demand for HSFA2 and therefore showed higher levels of alternative splicing to give HSFA2II RNA.
In parallel, we also examined the importance of HSFA7a function in CPR. We did not detect a reduction in HSP70A expression in the hsfA7a knockout line, and HSFA7a overexpression lines did not show increase of tolerance to AZC (see Supplemental Figure 5 online). However, we cannot completely exclude some involvement of HSFA7a in the regulation of the CPR. The induction of HSFA7a in the CPR and the fact that the hsfA2 knockout line did not eliminate HSP70A reporter expression suggests that HSFA7a may play some role. It is possible that HSFA7a plays a contributory role (with HSFA2) in the CPR.
HSF RNA Alternative Splicing
Alternative splicing of Arabidopsis HSF RNAs has been recorded elsewhere as EST sequences (http://www.plantgdb.org/ASIP/EnterDB.php) although none of these events has been associated with functions. Many ESTs reveal intron retention (e.g., HSFA3, HSFA7b, HSFB2a, and HSFC1) or 3′ untranslated region (e.g., HSFA1b, HSFA1e, and HSFB1) alternative splicing events. HSFA1d RNA has a similar alternative splicing pattern to HSFA2-II. The alternative splicing of HSFA1d (mRNA from hormone-treated callus) has the same donor site in the DNA binding domain region as normal splicing but introduces a new exon and intron and a new acceptor site, resulting in multiple new stop codons. Medicago sativa HSF1 RNA is also alternatively spliced within the conserved intron in the DNA binding domain coding region. This introduces one or two small exons, including premature stop codons. The expression levels of these transcripts were low and were thought to be degraded by NMD (He et al., 2007). The features in common with HSFA2-II suggest that alternative splicing of the HSF intron may be a conserved mechanism in HSF regulation. The combination of alternative splicing and NMD seems to be a broadly conserved regulatory system in eukaryotes (Jaillon et al., 2008). In plants, ∼20% of the expressed genes are alternatively spliced, and 36% (rice) to 43% (Arabidopsis) of alternatively spliced RNAs are predicted to be degraded by NMD (Wang and Brendel, 2006). Alternative splicing (HSFA2-II) is a notable feature of the response to heat shock (Figure 4) and correlates with the upregulation of a number of splicing-related genes as common responses to both heat shock and AZC treatment in our microarray analysis.
CPR for Protein Homeostasis in Eukaryotes
Protein misfolding in the soluble cytosol occurs as a consequence of a range of induced stresses. These can be environmental, chemical, or a consequence of ectopic protein expression associated, for example, with virus infection. The parallels between our data and related observations in yeast (Trotter et al., 2002) point to the CPR as being a fundamental process in wider eukaryotes designed to maintain protein homeostasis in the cytosol. In both cases, the CPR is shown to be distinct from the UPR in the use of HSF- and HSE-mediated induction of HSP70 genes. Although heat shock has some impact upon the ER, the spatial and functional separation of the CPR and UPR provides additional precision in cellular regulation especially when protein misfolding in the ER is commonly associated with posttranslational modification and maturation.
METHODS
Plants, Bacteria, and Viruses
Arabidopsis thaliana Col-0 and Nicotiana benthamiana plants were grown in a growth cabinet at 20°C with a photoperiod of 8 h light/16 h dark.
All the intermediate DNA constructs were maintained in Escherichia coli DH5a cells. Binary plasmids were transformed into Agrobacterium tumefaciens strain C58C1 with (for pGreen-based constructs) and without (for pB7WG2-based constructs) the pSoup helper plasmid (Hellens et al., 2000). Virus infections for TuMV and TCV were as described previously (Aparicio et al., 2005). Briefly, Agrobacterium binary clones pGreen-TuMV, containing a full-length TuMV genome (Dunoyer et al., 2004), and pBIN61-TCV, containing a full-length genome of TCV (Oh et al., 1995), were grown in Luria broth and resuspended in 10 mM MgCl2. The optical density of the suspension was adjusted to 0.5 at 600 nm, and the bacterial suspension was infiltrated into six leaves of 4- to 5-week-old Arabidopsis plants. Six newly emerged and systemically infected leaves per plant were harvested 1 week after the infiltration and subjected to cDNA synthesis.
Promoter Activity Assays
To assay HSP70 promoter activities, Agrobacterium carrying the HSP70A or HSP70B promoter:GUS constructs (Aparicio et al., 2005), or the empty vector, were grown in Luria broth overnight with appropriate antibiotics, and the cells were resuspended in 10 mM of MgCl2 solution at an optical density of 0.5 at 600 nm (OD600) before infiltrating into fully expanded leaves from 5- to 6-week-old N. benthamiana plants using a needleless syringe. For protein expression constructs, the cells were resuspended in 10 mM MgCl2 solution at an OD600 of 0.5 and mixed in a 1:1 ratio by volume with cells containing the reporter construct before infiltration. The plants were maintained under the same growth conditions, and the leaves were harvested 3 d after the infiltration. Two harvested leaves were ground in liquid nitrogen and extracted using 400 μL of extraction buffer (50 mM sodium phosphate, pH 7.0, 10 mM Triton X-100, 10 mM N-lauroylsarcosine, and 1 mM 2-mercaptoethanol). The samples were centrifuged for 5 min at 4°C, and supernatants were recovered for GUS assay. GUS activity was assayed using a buffer containing 4-methylumbelliferyl-β-glucuronide (Sigma-Aldrich) (Aparicio et al., 2005). Fluorescence was measured by a Wallac 1420 VICTOR multilabel counter fluorometer (Wallac Oy) fitted with an excitation filter of 355 nm and an emission filter of 460 nm.
Physical and Chemical Inducers
AZC (10 mM) or l-proline (10 mM), tunicamycin (2 mg/L in 50% DMF), or 50% DMF (control) were infiltrated into expanded Arabidopsis leaves. For heat shock, detached plant leaves were incubated at 37° or 20°C (as a control) for the indicated times.
RNA Analysis
Four to six Arabidopsis leaves were used for RNA extraction using TRI reagent (Sigma-Aldrich) following the manufacturer's instructions. Five micrograms of total RNA was treated with DNase I, and 1 μg of total RNA was subjected to cDNA synthesis using M-MLV reverse transcriptase (Invitrogen). RT-PCR was performed using the primers listed in Supplemental Table 4 online. Real-time PCR was performed using a DNA Engine Opticon 2 (MJ Research) and SYBR Green JumpStart Taq ReadyMix (Sigma-Aldrich). (Primer sequences for all this work are listed in Supplemental Table 4 online.) Each reaction was triplicated, and an average threshold cycle (Ct) was used to determine the fold change of gene expression. Expression level of elongation factor 1-alpha (EF1a; At5g60390) was used as an internal control. For the validation of microarray data, At2g15130 was used as an internal control.
RNA gel blot analysis was performed as described (Jones et al., 1998). Six micrograms of total RNA was separated by 1% agarose gel electrophoresis and transferred to Hybond-NX membranes. The HSFA2 probe was PCR amplified using primers attB1HsfA2FLF and attB1HsfA2FLR, and the HSFA2 full-length cDNA was cloned into pDonor207. The amplified PCR fragment was labeled with 32P-dCTP and used for hybridization.
Promoter Mutant Analysis
To create the constructs pHSP70A-337:GUS and pHSP70B-340:GUS, 337 and 340 bp of genomic DNA upstream from the ATG codon of the Arabidopsis HSP70A and HSP70B coding region, respectively, were amplified by PCR using the primers FA27 and FA6, and FA35 and FA8, respectively. The primers included NcoI and KpnI sites to enable direct replacement of the 2-kb promoter of the pHSP70:GUS vector (Aparicio et al., 2005). To create the promoter mutants, the minimal promoter fragments were cloned into the pGEM-T vector (Promega). Mutations were generated by PCR using the Quickchange site-directed mutagenesis kit (Stratagene) and the primers specified in Supplemental Table 4 online. The promoter regions of the pHSP70A-337:GUS and pHSP70B-340:GUS constructs were replaced by the mutated promoter fragments using the NcoI and KpnI sites.
HSF Constructs
The HSF constructs were made following Gateway technology (Karimi et al., 2002). Specific primers were designed to include a partial sequence of the attB1 or attB2 Gateway recombination sites. HSF full-length cDNA fragments were amplified by PCR using a cDNA library generated from heat- or AZC-treated Arabidopsis leaves. The fragments were further amplified using primers that added the complete attB1 and attB2 sequences at the ends of the fragments. The fragments were polyethylene glycol precipitated and cloned into pDONOR 207 (Invitrogen) using Gateway BP Clonase II (Invitrogen). The fragments in pDONOR 207 were then cloned into the pB7WG2 expression vector (Karimi et al., 2002) using Gateway LR Clonase II (Invitrogen). The HSF clones were sequenced and compared with the published sequences in GenBank. The pB7WG2-HSFA2 construct was used to transform Arabidopsis to create HSFA2 overexpression lines.
Growth Inhibition Assay
Sterile Arabidopsis seedlings were grown on Murashige and Skoog media at 22°C with 16 h of lighting. After 7 d, plantlets were transferred to 2 mL of liquid media with or without AZC. Plantlet weight was determined after a further 7 d of incubation at 22°C, with an 18-h light cycle. The weights of five sets of two plantlets (total 10 plantlets) were taken for each condition tested, but highest and lowest data for each line were discarded.
Transcript Profile Analysis
Total RNA was extracted from treated and control plants and was processed according to the GeneChip standard protocol (Eukaryotic Target Preparation; Affymetrix). Statistical analysis was done using the open source software Bioconductor (Gentleman et al., 2004). Raw data were normalized with the Robust Multichip Average function implemented in the affy package (Irizarry et al., 2003). This function background-corrected the perfect match values using the nonlinear Robust Multichip Average method, normalized them using quantile normalization, and finally summarized them resulting in a set of log2-transformed expression measures. Comparisons between treated plants and their relative controls were performed using the Limma package (Smith, 2005). Differentially expressed genes were identified using the false discovery rate corrected t test (Benjamini and Hochberg, 1995). Raw and normalized data are available from http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=rxinhueeqiuaenkandacc=GSE11758. Cluster analysis of our microarray data with public data of responses following heat shock (Kilian et al., 2007; Gene Expression Omnibus [GEO] accession number GSE5628; Control. GEO accession number GSE5620) was computed by multiscale bootstrap resampling (10,000 iterations; Shimodaira, 2004). The distance matrix was evaluated using the Pearson correlation coefficient, and clusters were created using the complete linkage method.
Promoter Analysis
The 1000-bp regions located upstream of the genes of interest were used for analysis with the Patmatch tool available from The Arabidopsis Information Resource website (http://www.Arabidopsis.org/cgi-bin/patmatch/nph-patmatch.pl).
Gene Ontology Analysis
Gene ontologies were taken from the MapMan functional classification (Thimm et al., 2004).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: HSFA2, NM_128173; HSFA2-II, EG494778; and HSFA7a, NM_115050.
Supplemental Data
The following materials are available in the online version of this article
Supplemental Figure 1. Time Course of CPR and UPR Marker Gene Expression in Response to Heat Shock.
Supplemental Figure 2. Clustal Analysis of Public and Recorded Microarray Data Sets.
Supplemental Figure 3. Nucleotide Sequence of Alternatively Spliced HSFA2-II.
Supplemental Figure 4. Later HSP70A Responses in the hsfa2 Knockout Line Showing CPR.
Supplemental Figure 5. Characterization of HSFA7a Knockout and Overexpression Lines.
Supplemental Figure 6. Time Course of HSFA2 and HSFA2-II Transcript Accumulation in Wild-Type and NMD Defective Plants.
Supplemental Figure 7. Expression of NMD Target RNAs after Heat Shock or AZC Treatment.
Supplemental Table 1. List of the Genes Upregulated by CPR.
Supplemental Table 2. Validation of Selected Microarray Gene Expression Data by qRT-PCR.
Suplemental Table 3. HSE Representation in Genes Upregulated by Both HS and AZC.
Supplemental Table 4. Methods: Primers Used for PCR and Cloning.
Supplemental Data Set 1. List of Genes Showing Differential Responses to HS, AZC, and Tunicamycin Treatment.
Supplementary Material
Acknowledgments
We thank John Brown and Phil Wigge for their critical reviews of the article prior to submission, Shigeoka Shegeru for seeds of the HSFA2 knockout line, and Peter Shaw for seeds of upf1-5 line. A.S. was funded from the European Union Marie Curie IIF Programme (Contract MIF-CT-2006-022085). F.A. was funded by a Spanish Government Fellowship. The John Innes Centre is grant-aided by the Biotechnology and Biological Sciences Research Council.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Andy J. Maule (andy.maule@bbsrc.ac.uk).
Online version contains Web-only data.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Aparicio, F., Thomas, C.L., Lederer, C., Niu, Y., Wang, D., and Maule, A.J. (2005). Virus induction of heat shock protein 70 reflects a general response to protein accumulation in the plant cytosol. Plant Physiol. 138 529–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda, M.A., Escaler, M., Wang, D., and Maule, A.J. (1996). Induction of HSP70 and polyubiquitin expression associated with plant virus replication. Proc. Natl. Acad. Sci. USA 93 15289–15293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arciga-Reyes, L., Wootton, L., Kieffer, M., and Davies, B. (2006). UPF1 is required for nonsense-mediated mRNA decay (NMD) and RNAi in Arabidopsis. Plant J. 47 480–489. [DOI] [PubMed] [Google Scholar]
- Baniwal, S.K., Chan, K.Y., Scharf, K.D., and Nover, L. (2007). Role of heat stress transcription factor HsfA5 as specific repressor of HsfA4. J. Biol. Chem. 282 3605–3613. [DOI] [PubMed] [Google Scholar]
- Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate—a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57 289–300. [Google Scholar]
- Bharti, K., Von Koskull-Doring, P., Bharti, S., Kumar, P., Tintschl-Korbitzer, A., Treuter, E., and Nover, L. (2004). Tomato heat stress transcription factor HsfB1 represents a novel type of general transcription coactivator with a histone-like motif interacting with the plant CREB binding protein ortholog HAC1. Plant Cell 16 1521–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch, W., Wunderlich, M., and Schoffl, F. (2005). Identification of novel heat shock factor-dependent genes and biochemical pathways in Arabidopsis thaliana. Plant J. 41 1–14. [DOI] [PubMed] [Google Scholar]
- Craig, E.A., and Gross, C.A. (1991). Is hsp70 the cellular thermometer? Trends Biochem. Sci. 16 135–140. [DOI] [PubMed] [Google Scholar]
- Dunoyer, P., Thomas, C., Harrison, S., Revers, F., and Maule, A. (2004). A cysteine-rich plant protein potentiates Potyvirus movement through an interaction with the virus genome-linked protein VPg. J. Virol. 78 2301–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman, R.C., et al. (2004). Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5 R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Z.S., Xie, R., Zou, H.S., Wang, Y.Z., Zhu, J.B., and Yu, G.Q. (2007). Structure and alternative splicing of a heat shock transcription factor gene, MsHSF1, in Medicago sativa. Biochem. Biophys. Res. Commun. 364 1056–1061. [DOI] [PubMed] [Google Scholar]
- Hellens, R.P., Edwards, E.A., Leyland, N.R., Bean, S., and Mullineaux, P.M. (2000). pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42 819–832. [DOI] [PubMed] [Google Scholar]
- Hori, K., and Watanabe, Y. (2007). Context analysis of termination codons in mRNA that are recognized by plant NMD. Plant Cell Physiol. 48 1072–1078. [DOI] [PubMed] [Google Scholar]
- Irizarry, R.A., Hobbs, B., Collin, F., Beazer-Barclay, Y.D., Antonellis, K.J., Scherf, U., and Speed, T.P. (2003). Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4 249–264. [DOI] [PubMed] [Google Scholar]
- Iwata, Y., and Koizumi, N. (2005). An Arabidopsis transcription factor, AtbZIP60, regulates the endoplasmic reticulum stress response in a manner unique to plants. Proc. Natl. Acad. Sci. USA 102 5280–5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon, O., et al. (2008). Translational control of intron splicing in eukaryotes. Nature 451 359–362. [DOI] [PubMed] [Google Scholar]
- Jockusch, H., and Wiegand, C. (2003). Misfolded plant virus proteins: Elicitors and targets of ubiquitylation. FEBS Lett. 545 229–232. [DOI] [PubMed] [Google Scholar]
- Jockusch, H., Wiegand, C., Mersch, B., and Rajes, D. (2001). Mutants of tobacco mosaic virus with temperature-sensitive coat proteins induce heat shock response in tobacco leaves. Mol. Plant Microbe Interact. 14 914–917. [DOI] [PubMed] [Google Scholar]
- Jones, A.L., Johansen, I.E., Bean, S.J., Bach, I., and Maule, A.J. (1998). Specificity of resistance to pea seed-borne mosaic potyvirus in transgenic peas expressing the viral replicase (Nlb) gene. J. Gen. Virol. 79 3129–3137. [DOI] [PubMed] [Google Scholar]
- Karimi, M., Inze, D., and Depicker, A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7 193–195. [DOI] [PubMed] [Google Scholar]
- Kerenyi, Z., Merai, Z., Hiripi, L., Benkovics, A., Gyula, P., Lacomme, C., Barta, E., Nagy, F., and Silhavy, D. (2008). Inter-kingdom conservation of mechanism of nonsense-mediated mRNA decay. EMBO J. 27 1585–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz, S., Kerenyi, Z., Merai, Z., Bartos, I., Palfy, T., Barta, E., and Silhavy, D. (2006). Both introns and long 3′-UTRs operate as cis-acting elements to trigger nonsense-mediated decay in plants. Nucleic Acids Res. 34 6147–6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian, J., Whitehead, D., Horak, J., Wanke, D., Weinl, S., Batistic, O., D'Angelo, C., Bornberg-Bauer, E., Kudla, J., and Harter, K. (2007). The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 50 347–363. [DOI] [PubMed] [Google Scholar]
- Larkindale, J., and Vierling, E. (2008). Core genome responses involved in acclimation to high temperature. Plant Physiol. 146 748–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez, I.M., and Chrispeels, M.J. (2003). Genomic analysis of the unfolded protein response in Arabidopsis shows its connection to important cellular processes. Plant Cell 15 561–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meusser, B., Hirsch, C., Jarosch, E., and Sommer, T. (2005). ERAD: The long road to destruction. Nat. Cell Biol. 7 766–772. [DOI] [PubMed] [Google Scholar]
- Miller, G., and Mittler, R. (2006). Could heat shock transcription factors function as hydrogen peroxide sensors in plants? Ann. Bot. (Lond.) 98 279–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto, R.I. (1998). Regulation of the heat shock transcriptional response: Cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 12 3788–3796. [DOI] [PubMed] [Google Scholar]
- Nishizawa, A., Yabuta, Y., Yoshida, E., Maruta, T., Yoshimura, K., and Shigeoka, S. (2006). Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. Plant J. 48 535–547. [DOI] [PubMed] [Google Scholar]
- Nover, L., Bharti, K., Doring, P., Mishra, S.K., Ganguli, A., and Scharf, K.D. (2001). Arabidopsis and the heat stress transcription factor world: How many heat stress transcription factors do we need? Cell Stress Chaperones 6 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, J.W., Kong, Q.Z., Song, C.Z., Carpenter, C.D., and Simon, A.E. (1995). Open reading frames of turnip crinkle virus involved in satellite symptom expression and incompatibility with Arabidopsis thaliana ecotype Dijon. Mol. Plant Microbe Interact. 8 979–987. [DOI] [PubMed] [Google Scholar]
- Qian, S.B., McDonough, H., Boellmann, F., Cyr, D.M., and Patterson, C. (2006). CHIP-mediated stress recovery by sequential ubiquitination of substrates and Hsp70. Nature 440 551–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron, D., and Walter, P. (2007). Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 8 519–529. [DOI] [PubMed] [Google Scholar]
- Schramm, F., Ganguli, A., Kiehlmann, E., Englich, G., Walch, D., and von Koskull-Doring, P. (2006). The heat stress transcription factor HsfA2 serves as a regulatory amplifier of a subset of genes in the heat stress response in Arabidopsis. Plant Mol. Biol. 60 759–772. [DOI] [PubMed] [Google Scholar]
- Schramm, F., Larkindale, J., Kiehlmann, E., Ganguli, A., Englich, G., Vierling, E., and von Koskull-Doring, P. (2008). A cascade of transcription factor DREB2A and heat stress transcription factor HsfA3 regulates the heat stress response of Arabidopsis. Plant J. 53 264–274. [DOI] [PubMed] [Google Scholar]
- Shimodaira, H. (2004). Approximately unbiased tests of regions using multistep-multiscale bootstrap resampling. Ann. Stat. 32 2616–2641. [Google Scholar]
- Smith, G.K. (2005). Limma: Linear Models for Microarray Data. (New York: Springer).
- Swindell, W.R., Huebner, M., and Weber, A.P. (2007). Transcriptional profiling of Arabidopsis heat shock proteins and transcription factors reveals extensive overlap between heat and non-heat stress response pathways. BMC Genomics 8 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm, O., Blasing, O., Gibon, Y., Nagel, A., Meyer, S., Kruger, P., Selbig, J., Muller, L.A., Rhee, S.Y., and Stitt, M. (2004). MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37 914–939. [DOI] [PubMed] [Google Scholar]
- Trotter, E.W., Kao, C.M., Berenfeld, L., Botstein, D., Petsko, G.A., and Gray, J.V. (2002). Misfolded proteins are competent to mediate a subset of the responses to heat shock in Saccharomyces cerevisiae. J. Biol. Chem. 277 44817–44825. [DOI] [PubMed] [Google Scholar]
- Urade, R. (2007). Cellular response to unfolded proteins in the endoplasmic reticulum of plants. FEBS J. 274 1152–1171. [DOI] [PubMed] [Google Scholar]
- Wang, B.B., and Brendel, V. (2006). Genomewide comparative analysis of alternative splicing in plants. Proc. Natl. Acad. Sci. USA 103 7175–7180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitham, S.A., Quan, S., Chang, H.S., Cooper, B., Estes, B., Zhu, T., Wang, X., and Hou, Y.M. (2003). Diverse RNA viruses elicit the expression of common sets of genes in susceptible Arabidopsis thaliana plants. Plant J. 33 271–283. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.