Abstract
We studied molecular and functional properties of Arabidopsis phosphomannose isomerase isoenzymes (PMI1 and PMI2) that catalyze reversible isomerization between d-fructose 6-phosphate and d-mannose 6-phosphate (Man-6P). The apparent Km and Vmax values for Man-6P of purified recombinant PMI1 were 41.3 ± 4.2 μm and 1.89 μmol/min/mg protein, respectively, whereas those of purified recombinant PMI2 were 372 ± 13 μm and 22.5 μmol/min/mg protein, respectively. Both PMI1 and PMI2 were inhibited by incubation with EDTA, Zn2+, Cd2+, and l-ascorbic acid (AsA). Arabidopsis PMI1 protein was constitutively expressed in both vegetative and reproductive organs under normal growth conditions, whereas the PMI2 protein was not expressed in any organs under light. The induction of PMI1 expression and an increase in the AsA level were observed in leaves under continuous light, whereas the induction of PMI2 expression and a decrease in the AsA level were observed under long term darkness. PMI1 showed a diurnal expression pattern in parallel with the total PMI activity and the total AsA content in leaves. Moreover, a reduction of PMI1 expression through RNA interference resulted in a substantial decrease in the total AsA content of leaves of knockdown PMI1 plants, whereas the complete inhibition of PMI2 expression did not affect the total AsA levels in leaves of knock-out PMI2 plants. Consequently, this study improves our understanding of the molecular and functional properties of Arabidopsis PMI isoenzymes and provides genetic evidence of the involvement of PMI1, but not PMI2, in the biosynthesis of AsA in Arabidopsis plants.
Humans and several other mammalian species are unable to synthesize l-ascorbic acid (vitamin C, AsA)2 and thus they are dependent on dietary AsA as an essential nutrient mainly from vegetables and fruits. In photosynthetic organisms, including higher plants, AsA is one of the major soluble carbohydrates and antioxidants, and it plays roles in essential physiological processes such as photosynthesis, trans-membrane electron transport, cell division and growth, and stress resistance (1, 2).
In higher plants, AsA is synthesized from hexose phosphates such as d-glucose-1 or 6-phosphate and d-fructose 6-phosphate (Fru-6P) via GDP-d-mannose (GDP-Man), by the d-mannose/l-galactose (d-Man/l-Gal) pathway (3, 4). GDP-Man is synthesized from Fru-6P in three steps catalyzed by the enzymes, phosphomannose isomerase (PMI), phosphomannomutase (PMM), and GDP-Man pyrophosphorylase (GMP) (5–7). GDP-Man is successively converted to GDP-l-Gal, l-Gal-1-phosphate, l-Gal, l-galactono-1,4-lactone, and finally to AsA. In addition to this pathway, alternative pathways may contribute to AsA biosynthesis. Overexpression of a rat l-gulono-1,4-lactone oxidase resulted in 4–7-fold increase in the AsA content (8). GDP-Man-3′,5′-epimerase (GME), which is in the d-Man/l-Gal pathway, produces not only GDP-l-Gal but also GDP-l-gulose from GDP-Man (9). l-Gulose, as well as l-Gal, can be oxidized to AsA (10), but it is not clear what the relative contributions of these pathways are in vivo. Although myo-inositol was not considered as a major precursor to AsA, the constitutive expression of a myo-inositol oxygenase enhanced the AsA content of the Arabidopsis leaves 2–3-fold (11). Moreover, it has been reported that overexpression of a purple acid phosphatase gene, AtPAP15, which is involved in myo-inositol metabolism, resulted in a 2-fold increase in the AsA content of the Arabidopsis (12), suggesting that this myo-inositol pathway may also be operating in planta.
Screens for ozone-sensitive (13) or AsA-deficient (14) mutants in Arabidopsis thaliana led to the identification of five loci (VTC1, VTC2, VTC3, VTC4, and VTC5) involved in the maintenance of the AsA pool. Characterization of the vtc1 (6) and vtc4 (15) mutants, as well as biochemical studies (16), have allowed the identification of two of the enzymes required for AsA biosynthesis through the d-Man/l-Gal pathway. VTC1 and VTC4 encode GMP and l-Gal-1-phosphate phosphatase, respectively. Recently, GDP-l-Gal phosphorylase/l-Gal guanylyltransferase was identified as the VTC2 and VTC5 genes (17–19). The double mutant of VTC2 and VTC5 showed seedling arrest immediately upon germination without supplementation of AsA or l-Gal (19). This genetic evidence has confirmed that the d-Man/l-Gal pathway is a significant route to AsA biosynthesis in higher plants (3–6, 15, 17, 18).
The mechanism regulating the size of the AsA pool and the expression and regulatory properties of the enzymes involved are the next important subjects of study (4, 20). Leaves exposed to intense light contain more AsA than leaves in the shade (19, 21). Recently, we have reported that the transcript levels of GMP, VTC4, and the VTC2 genes changed in parallel with the changes in leaf AsA pool size during dark and light periods, and the photosynthetic electron transport (PET) was closely linked to the regulation of the expression of these genes and AsA levels in leaves of Arabidopsis plants (22, 23). Pallanca and Smirnoff (24) have reported that the rate of AsA biosynthesis from d-[U-14C]glucose decreased linearly with the increase in the pool size of AsA in embryonic pea seedlings. In fact, l-Gal dehydrogenase and GME, which are in the d-Man/l-Gal pathway, were inhibited by AsA (9, 25). These results suggest that the AsA level in plant cells is regulated at several steps by a feedback control. In addition, it has been reported that other biological factors, such as jasmonic acid, contribute to the accumulation of AsA (26–28).
To date, the enzymes involved in the d-Man/l-Gal pathway have been identified and characterized, except for PMI, which catalyzes the reversible isomerization of Fru-6P and d-Man-6-phosphate (Man-6P) as the first step of the GDP-Man and AsA biosynthetic pathway. Earlier reports have indicated that PMI was lacking in many plants, and even if it was present, the enzyme activity was very low (29). Additionally, an alternative pathway of GDP-Man and AsA synthesis via a GDP-d-glucose-2-epimerase, which does not involve PMI, GMP, and PMM, has been suggested in higher plants (30). However, Sharples and Fry (31) demonstrated by a dual radiolabeling analysis that in Arabidopsis plants, the action of PMI significantly predominates over that of the GDP-d-glucose 2-epimerase. The Arabidopsis vtc1 mutant had reduced GMP activity, and a null mutation of this gene resulted in death by arresting embryonic development at a very early stage (32). Moreover, pmm-12 mutant with markedly decreased activity of PMM had reduced levels of AsA in Arabidopsis (33). Although these biochemical and genetic findings suggest that the pathway from hexose phosphates to GDP-Man involved in PMI, GMP, and PMM is essential for the biosynthesis of AsA in higher plants, very little is known about PMI in higher plants. Two putative genes are identified in A. thaliana (PMI1, At3g02570, and PMI2, At1g67070) based on sequence homology. In this study, we studied the molecular characterization of recombinant PMI1 and PMI2 proteins purified from Escherichia coli and expression patterns of PMI1 and PMI2 in Arabidopsis leaves under light or darkness. Next, using reverse genetic approaches, we examined the physiological function of each enzyme, particularly with respect to AsA biosynthesis, in Arabidopsis plants. To our knowledge, this is the first study to characterize PMI isoenzymes in plants and represents the final piece in the puzzle of how AsA is produced in the d-Man/l-Gal pathway.
EXPERIMENTAL PROCEDURES
Plant Materials and Growth Conditions—Surface-sterilized A. thaliana wild-type (Col-0) and transgenic seeds were sown on Murashige and Skoog (MS) medium plus 3% sucrose. Plates were stratified in darkness for 2 days at 4 °C and then transferred to a growth chamber kept at 25 °C during 16 h of light (100μmol/m2/s) and at 22 °C during 8 h of darkness. After 7 days, seedlings were potted in soil and grown under the same conditions. A T-DNA insertion line for PMI2 in the Col-0 background (SALK_070922) was obtained from the Arabidopsis Biological Resource Center. The continuous light irradiation or dark application was started 4 h after the illumination. Cycloheximide (CHI) at 10 μm and 3-(3,4-dichlorophenyl)-ll-dimethylurea (DCMU) at 10 μm were used for detached leaf experiments. The detached leaves were floated on CHI or DCMU solution in Petri dishes.
Preparation of Total RNA and cDNA Synthesis—Total RNA was semi-automatically purified from various organs of Arabidopsis plants using a QuickGene RNA cultured cell kit and Quick-Gene-Mini80 (FujiFilm Corp., Tokyo, Japan). The first strand cDNA was synthesized using ReverTra Ace (reverse transcriptase; Toyobo) with an oligo(dT) primer. These analyses were performed according to the manufacturer's instructions.
Expression and Purification of Recombinant Arabidopsis PMI Isoenzymes—The open reading frames of PMI1 and PMI2 were amplified from the first strand cDNAs, using the following primer sets: PMI1-SacI-F (5′-GAGCTCATGGAGATCGCTACTGTCGT-3′), PMI1-XbaI-R (5′-TCTAGACTATAGAGGAAACAAAAACCTACTG-3′), PMI2-NdeI-F (5′-CATATGGGAGCAGACGCAATCCA-3′), and PMI2-SalI-F (5′-GTCGACTAGCCTACAATGTTTGG-3′). The amplified DNA fragments were ligated into the vector pZErO-2 (Invitrogen). DNA sequencing was performed using the dideoxy chain terminator method with an automatic DNA sequencer (ABI PRISM™ 310, Applied Biosystems). The resulting constructs were digested with SacI/XbaI for PMI1 or NdeI/SalI for PMI2, and ligated into the expression vector, pCold II (Takara), to produce histidine-tagged proteins. The E. coli strain BL21 (DE3) pLysS transformed with pCold II/PMI1 and -2 was grown in 50 ml of LB medium containing 50 μg/ml ampicillin and 34 μg/ml chloramphenicol. After an overnight culture at 37 °C, the cultures were transferred to 500 ml of LB medium (with the antibiotics) and grown to an A600 between 0.4 and 0.6, and then cells were kept for 30 min at 16 °C without shaking. Isopropyl 1-thio-β-d-galactopyranoside was added to the cultures at a concentration of 0.2 mm, and the cells were further incubated for 24 h at 16 °C with shaking. The harvested cells were resuspended in 20 mm Tris-HCl, pH 8.0, containing 0.5 m NaCl, 5 mm imidazole, and 1 mm β-mercaptoethanol (buffer A), sonicated (10 kHz) using 20-s strokes with 30-s intervals, and centrifuged at 15,000 × g for 15 min. The hexa-histidine-tagged recombinant PMI proteins were purified from the soluble fraction using a HiTrap chelating HP column (Amersham Biosciences) according to the manufacturer's instructions. Briefly, the column was washed with 10 ml of distilled water, charged with 0.5 ml of 0.1 mm NiCl2, and washed again with 5 ml of distilled water. After preparation, the column was equilibrated with 10 volumes of buffer A. The sample was applied using a syringe, and the column was washed with 10 volumes of buffer A containing 20 mm imidazole. The recombinant protein was eluted with 5–10 volumes of buffer A containing 120 mm imidazole, and collected in a 1-ml fraction. Protein concentrations were determined following the method of Bradford (34). SDS-PAGE was carried out on 12.5% (w/v) polyacrylamide slab gels. The gels were stained with 0.06% (w/v) Coomassie Brilliant Blue R-250 in 50% (v/v) methanol for 5 min. Samples were denatured by boiling for 10 min in 0.1% SDS in the presence of 5% (v/v) β-mercaptoethanol.
Enzyme Assay—The activity of PMI was measured at 25 °C using a coupled assay in which the product Fru-6P is converted into d-glucose 6-phosphate. This is then oxidized to 6-phosphogluconate, and the reduction of NADP+ was measured by the change in absorption at 340 nm (35). The coupling enzyme, phosphoglucose isomerase or glucose 6-phosphate dehydrogenase, was normally used at 1 unit in the enzyme assay (total volume, 1 ml), except at the highest or lowest temperature and pH, and at the highest concentration of several inhibitors, where five times this quantity was used (5 units per assay). Careful control experiments were conducted to check this assumption by adding further excess of the coupling enzymes in the presence of the inhibitor. The buffer used was 50 mm Tris-HCl, pH 7.5, with 0.5 mm NADP+. Following the preincubation of various compounds, including inhibitors such as zinc ion, at 25 °C for 30 min in the reaction mixture, the reaction was initiated by the addition of Man-6P. The optimum pH was determined at 25 °C in 50 mm Tris-HCl buffer, pH 6.5–9.0, and 50 mm MOPS buffer, pH 5.5–7.0. The optimum temperature was determined by incubation between 25 and 65 °C with 50 mm Tris-HCl, pH 7.5. The total PMI activity in Arabidopsis was measured according to the method of Dowdle et al. (19) with minor modifications, and Man-6P was used at a final concentration of 1 mm.
The activity of Fru-6P isomerization was determined by high performance anion exchange chromatography coupled with pulsed amperometric detection system using an ICS-3000 ion chromatography system (Dionex, Sunnyvale, CA). The mixture of the enzymatic products (10 μl) was automatically injected onto the column of CarboPac PA1 guard (2 × 50 mm) and CarboPac PA1 (2 × 250 mm) and eluted with NaOH/sodium acetate gradient. The sodium acetate gradient was increased from 0 to 200 mm between 0 and 10 min and to 400 mm between 10 and 24 min with a flow rate set at 0.25 ml/min. The detection was carried out by pulsed amperometry cell using electrochemical detector equipped with a working gold electrode and a combined pH-Ag/AgCl reference electrode.
Quantitative RT-PCR Analysis—The quantitative RT-PCR analysis was performed according to Nishizawa et al. (36). Primer pairs for the quantitative RT-PCR were designed using Primer Express software (Applied Biosystems), and primer sequences were as follows: Actin2-F (5′-GGCAAGTCATCACGATTGG-3′), Actin2-R (5′-CAGCTTCCATTCCCACAAAC-3′); PMI1-rF (5′-GTGGAGCCTCGACGGTGTT-3′), PMI1-rR (5′-TCCTTCACCTTGGAGTACAAGCA-3′); and PMI2-rF (5′-AGCAAGTGGTTTCTCGGATGAA-3′), PMI2-rR (5′-AACCAACTTCTCTTTCTCCGACAGT-3′). Gene-specific primers were chosen such that the resulting PCR product was ∼100 bp long. The quantitative RT-PCR was performed with an Applied Biosystems 7300 Real Time PCR System (Applied Biosystems), using the SYBR Premix Ex Taq (Takara). Actin2 mRNA, set to 100%, was used as an internal standard in all experiments. The quantitative RT-PCR experiments were repeated at least three times for cDNA prepared from three batches of plants.
Western Blot Analysis—Polyclonal mouse antibodies against PMI1 (anti-PMI1) and PMI2 (anti-PMI2) protein were prepared using His-tagged recombinant proteins. Mice were injected with 50 mg of purified recombinant PMI1 or PMI2 proteins emulsified with Freund's complete adjuvant followed by three subcutaneous injections. After bleedings, the antisera were separated from the blood (37). Western blot analysis was carried out as described previously (33). The protein bands were detected by using specific antibodies as the primary antibody, and anti-mouse IgG-horseradish peroxidase conjugate (Bio-Rad) as the secondary antibody.
Measurement of AsA and Dehydroascorbic Acid Levels—AsA and DHA were measured as described by Yabuta et al. (22). The different tissues of Arabidopsis plants (0.5 g wet weight) frozen in liquid N2 were ground using a mortar and pestle with 5 ml of 6% (v/v) HClO4 and centrifuged at 10,000 × g for 10 min at 4 °C. A 100-μl aliquot of the obtained leaf extract was added directly to 900 μl of a 200 mm succinate buffer (pH 12.7, adjusted with NaOH) in the spectrophotometer. The final pH was very near 6.0. The A265 was immediately recorded once and again 5 min after the addition of 5 units of AsA oxidase from Cucurbita sp. (Wako, Osaka, Japan). For the determination of total AsA, the leaf extract was adjusted to pH 6.0 with 1.25 m K2CO3 and centrifuged at 10,000 × g for 5 min. The supernatant was incubated with 10 mm dithiothreitol (DTT) dissolved in 50 mm HEPES-KOH buffer, pH 7.5, for 30 min at 25 °C. A 100-μl aliquot of the solution was directly added to 900 μl of 200 mm succinate buffer, pH 6.0, in the spectrophotometer. The resultant solution was assayed as described above. The difference between the total AsA and AsA contents was taken to be the DHA content.
Generation of Transgenic Plants—The 5′-end of PMI1 cDNA was cloned into the donor vector, pDONR201, and then recloned into the destination vector, pGWB80. The specific primers with attB1 and attB2 sequences were as follows: 5′-PMI1-attB1 (5′-GGCGAAGCCGTCGTCGATCC-3′) and 5′-PMI1-attB2 (5′-GAGAAAAGCTTTGAGTCTTC-3′). PCR and in vitro BP and LR recombinations were carried out according to the manufacturer's instructions (Invitrogen). Agrobacterium tumefaciens, which was transformed with the obtained construct by electroporation, was used to infect Arabidopsis via the vacuum infiltration method (38). T1 seedlings were selected on basic MS medium in Petri dishes containing 3% sucrose and 20 mg/liter hygromycin for 2 weeks and transferred to the soil. T2 seeds were harvested and used for the experiments.
RESULTS
Characterization of Deduced Amino Acid Sequences of Arabidopsis PMI1 and PMI2—To date, three classes of PMI have been defined on the basis of sequence similarities (39). The first class (type I) includes all known eukaryotic PMI with a consensus sequence, YXDXNHKPE, in their primary structure (40). The type I PMI genes have occurred in bacteria, yeasts, and animals such as Aspergillus nidulans (41), E. coli (42), Candida albicans (40, 43), Saccharomyces cerevisiae (39, 44), and humans (39). A search for PMI genes in the National Center for Biotechnology Information Data Base (www.ncbi.nlm.nih) showed that two PMI genes (PMI1 and -2) exist in A. thaliana. The cDNAs of PMI1 and PMI2 of Arabidopsis contained 432 and 441 amino acid residues, respectively (supplemental Fig. S1). The predicted molecular masses for PMI1 and PMI2 were 48.5 and 49.2 kDa, respectively, which were similar to those of S. cerevisiae PMI40 (48.2 kDa), C. albicans PMI (48.8 kDa), and human PMI (46.6 kDa) (39, 43). No potential signal peptides of PMI1 and PMI2 were found. The deduced amino acid sequence of PMI1 showed 64, 37, 39, and 41% identity, respectively, to Arabidopsis PMI2 and other PMIs from S. cerevisiae, C. albicans, and humans. On the other hand, the deduced amino acid sequence of PMI2 showed 36, 38, and 40% identity, respectively, to S. cerevisiae, C. albicans, and human PMIs. Arabidopsis PMI1 and -2 possessed the consensus sequence of type I PMIs, YXDXNHKPE (40).
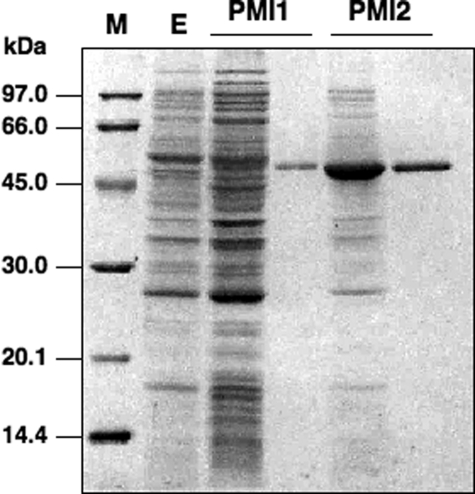
Properties and Kinetic Parameters of PMI1 and PMI2 Isoenzymes—To determine the properties and kinetic parameters of PMI1 and PMI2, histidine-tagged Arabidopsis PMI isoenzymes were expressed in E. coli cells and then purified to homogeneity using nickel chelate affinity chromatography, as judged from a SDS-PAGE analysis (Fig. 1).
FIGURE 1.
Purification of recombinant PMI1 and PMI2 proteins. Recombinant PMI1 and PMI2 proteins were expressed in E. coli, purified with Ni2+ affinity chromatography, and verified using SDS-PAGE with Coomassie Blue staining. The experimental conditions are described under”Experimental Procedures.“The left lane and right lane of each PMI contain 15 μg of crude extract and 2 μg of purified recombinant protein, respectively. M, molecular mass standards (Amersham Biosciences) as indicated on the left. E, empty pCold II vector.
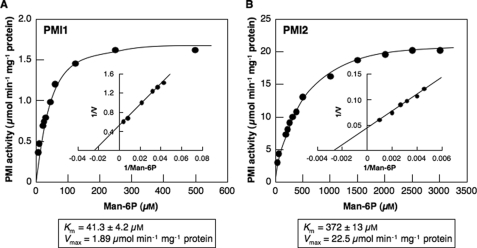
The activities of PMI isoenzymes as the substrate of Man-6P were determined at 25 °C by measuring the amount of NADPH generated using coupling enzymes, phosphoglucose isomerase and d-glucose-6-phosphate dehydrogenase as described under “Experimental Procedures.“The optimum pH and temperature of PMI1 were 7.5 and 52 °C, respectively, whereas those of PMI2 were 7.5 and 48 °C, respectively. PMI1 and PMI2 obeyed Michaelis-Menten type kinetics toward Man-6P (Fig. 2). The apparent Km and Vmax values for Man-6P of PMI1 estimated from Lineweaver-Burk plots were 41.3 ± 4.2 μm and 1.89 μmol/min/mg protein, respectively, whereas those of PMI2 were 372 ± 13 μm and 22.5 μmol/min/mg protein, respectively.
FIGURE 2.
Affinity of recombinant Arabidopsis PMI1 (A) and PMI2 (B) for Man-6P. Initial velocities were assayed as described under”Experimental Procedures“with varied concentrations of Man-6P. The kinetic constants were calculated from the Lineweaver-Burk plots (insets) based on the best fit lines. The data represent mean values from three repetitive experiments.
Type I PMIs have been found to be a zinc-dependent metalloenzyme (45–47). In the C. albicans PMI, the essential zinc has been liganded by four side chains, Gln-111, Glu-138, His-285, and His-113, and a water molecule (43). These amino acid residues were also conserved in the Arabidopsis PMI1 and PMI2 as well as other type I PMIs (supplemental Fig. S1). These findings suggest Arabidopsis PMI isoenzymes require a zinc ion for their activities. On incubation with 2.5 mm EDTA for 30, 60, and 240 min at 25 °C, PMI1 was inhibited 46, 56, and 80%, respectively, whereas PMI2 was inhibited 14, 18, and 25%, respectively (Table 1).
TABLE 1.
Effect of various compounds on Arabidopsis PMI1 and PMI2
Each enzyme was incubated with various compounds for 30 min at 25 °C, and then the activity was measured as described under “Experimental Procedures.” The abbreviations used are as follows: β-ME, β-mercaptoethanol; pCMB, p-chloromercuribenzoate.
| Compound | Concentration | PMI1 activity | PMI2 activity |
|---|---|---|---|
| mm | % | % | |
| None | 100 | 100 | |
| ZnCl2 | 1 | 0 | 0 |
| CdCl2 | 1 | 0 | 0 |
| MgCl2 | 1 | 80 | 100 |
| MnCl2 | 1 | 55 | 94 |
| CaCl2 | 1 | 73 | 93 |
| CoCl2 | 1 | 44 | 64 |
| EDTA | 2.5 | 54 | 86 |
| DTT | 1 | 73 | 84 |
| 5 | 60 | 40 | |
| β-ME | 1 | 73 | 90 |
| 5 | 65 | 56 | |
| pCMB | 0.05 | 33 | 39 |
| H2O2 | 1 | 61 | 79 |
| 5 | 54 | 55 | |
| AsA | 1 | 77 | 77 |
| 5 | 48 | 47 |
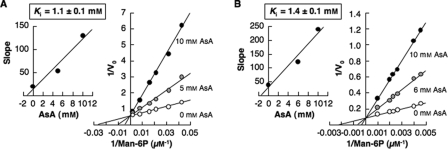
The activities of PMI1 and PMI2 were completely inhibited in a competitive manner by 1 mm Zn2+ or Cd2+ (Table 1). Ki values for Zn2+ of PMI1 and PMI2 were 32 ± 5.1 and 2.1 ± 0.2 μm, respectively, whereas those for Cd2+ were 11 ± 2.3 and 7.8 ± 1.0 μm, respectively (supplemental Fig. S2). However, the activities of PMI1 and PMI2 were not significantly affected by other divalent cations such as Mg2+, Mn2+, Ca2+, Ni2+, and Co2+ (Table 1). The activities of PMI1 were inhibited 40, 35, 67, and 46%, respectively, by the preincubation with 5 mm DTT, 5 mm β-mercaptoethanol, 0.05 mm p-chloromercuribenzoate, and 5 mm H2O2 for 30 min (Table 1). Similar results were also obtained for the activities of PMI2 (Table 1). These findings suggest that sulfhydryl groups in PMI1 and PMI2 participate in the reaction. Interestingly, PMI1 and PMI2 were inhibited by the incubation with AsA in a competitive manner. Ki values for AsA of PMI1 and PMI2 were 1.1 ± 0.1 and 1.4 ± 0.1 mm, respectively (Fig. 3).
FIGURE 3.
Inhibition of recombinant Arabidopsis PMI1 (A) and PMI2 (B) by AsA. Initial velocities were assayed as described under”Experimental Procedures“with varied concentrations of AsA. The Man-6P concentration was held constant at 1 mm. Reciprocals of relative values of the initial velocities are plotted for each AsA concentration. The inset figure is the secondary plot of the slopes against the AsA concentration. The data represent mean values from three repetitive experiments.
To detect the Fru-6P isomerization activity of Arabidopsis PMI isoenzymes, we incubated Fru-6P at a concentration of 1 mm for 10 min (25 °C) with PMI1 or PMI2. Man-6P as a product of Fru-6P isomerization was detected at 20.6 min of retention time (data not shown). On the other hand, Fru-6P was detected as a product of Man-6P isomerization at 22.2 min. In the same way, d-glucose 6-phosphate (20.1 min) was slightly detected as a product of Fru-6P isomerization. However, in the coupled assay, when phosphoglucose isomerase was absent from the reaction mixture, no NADPH was generated (data not shown). These findings indicated that Arabidopsis PMI isoenzymes had slight or negligible phosphoglucose isomerase activity.
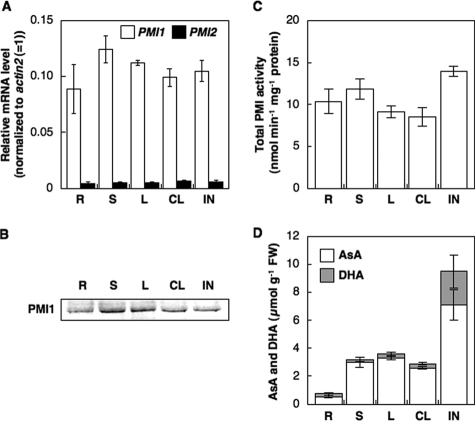
Expression of PMI 1 and PMI 2 in Different Organs—We determined the expression and total activity levels of PMI1 and PMI2 in the leaf, stem, root, cauline leaf, and inflorescence, which were harvested from 5-week-old Arabidopsis plants at 4 h after the illumination. As shown Fig. 4, the transcript of PMI1 was detected in all organs with no difference. The transcript of PMI2 was also detected in five organs with no difference; however, at very low levels compared with that of PMI1. Polyclonal antibodies against the recombinant PMI1 (anti-PMI1) and PMI2 (anti-PMI2) were produced, respectively. Each antibody was specifically cross-reacted with the respective PMI isoenzyme (supplemental Fig. S3). Western blot analysis using anti-PMI1 showed the accumulation of PMI1 protein in all organs with no difference (Fig. 4). In contrast, we could not detect the PMI2 protein in crude extracts from any organs at any growth conditions using anti-PMI2 because of very low expression (data not shown). Total activity of PMI1 and PMI2 was detected in these organs, possibly dependent on the activity of PMI 1, with no difference. The total AsA (AsA + DHA) level was higher in the inflorescence and fruit than in the other organs, and the total AsA level in roots was the lowest.
FIGURE 4.
Expressions and total activity levels of Arabidopsis PMI isoenzymes and AsA concentrations in different organs of A. thaliana. Various organs, including the leaf (L), stem (S), root (R), cauline leaf (CL), and inflorescence (IN) of 5-week-old Arabidopsis plants were harvested at 4 h after the illumination. A, relative levels of PMI1 and PMI2 were determined by real time PCR. Relative levels of the transcripts were normalized to Actin2 mRNA (= 1%). Error bars indicate S.D. (n = 3). B, Western blot of PMI1 protein. C, total PMI activity in crude extracts of each organ was measured by the coupling reaction as described under”Experimental Procedures.“Error bars indicate S.D. (n = 3). D, AsA and DHA concentrations in each organ. Error bars indicate S.D. (n = 3).
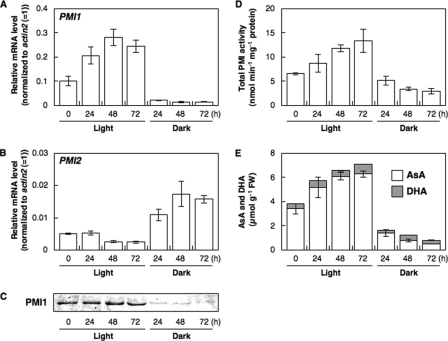
Responses of PMI1 and PMI2 to Light or Dark—To understand the relationship between light irradiation and the expression of PMI isoenzymes in leaves, 3-week-old Arabidopsis plants grown under a 16-h daily photoperiod were moved to continuous light or dark conditions for 24 h, and then the levels of PMI isoenzymes were measured (Fig. 5). The transcript and protein levels of PMI1 as well as total level of AsA strongly increased under light and decreased in the dark. In contrast, the transcript levels of PMI2 were not changed under light but increased in the dark as reported previously (48, 49), being very low compared with those of PMI1. Therefore, we could not detect the PMI2 protein in crude extracts from leaves using anti-PMI2. The total activity levels of PMI isoenzymes increased under light and decreased in the dark. Treatment with CHI, an inhibitor of de novo protein synthesis, inhibited light induction of the total activity levels of PMI isoenzymes (data not shown). Interestingly, there was a highly significant correlation among the total AsA level, total activity levels of PMI isoenzymes, and the level of expression of PMI1.
FIGURE 5.
Expressions and total activity levels of Arabidopsis PMI isoenzymes and AsA concentrations in A. thaliana leaves under continuous light or dark conditions. Three-week-old Arabidopsis plants were moved to continuous light or dark conditions for 72 h, and then leaves were harvested at the indicated time. Relative levels of PMI1 (A) and PMI2 (B) were determined by real time PCR as described in Fig. 4. Error bars indicate S.D. (n = 3). C, Western blot of PMI1 protein. D, total PMI activity in crude extracts of Arabidopsis leaves. Error bars indicate S.D. (n = 3). E, AsA and DHA concentrations in Arabidopsis leaves. Error bars indicate S.D. (n = 3).
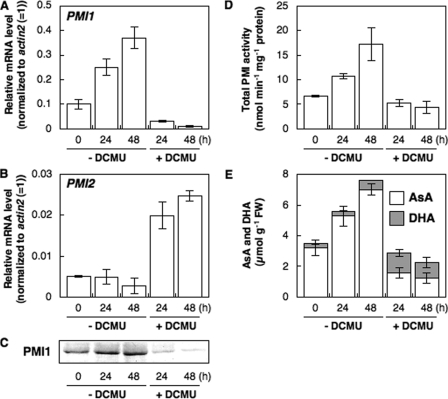
Effect of an Inhibitor of Photosynthetic Electron Transport on Expression of PMI1 and PMI2—To investigate the relationship between PET and the expression of PMI isoenzymes, Arabidopsis leaves were treated with 10 μm DCMU, which blocks the flow of electrons toward plastoquinones in photosystem II, for 48 h under light. This treatment completely diminished the photosystem II quantum yield (data not shown). The transcript and protein levels of PMI1 were decreased by the treatment with DCMU (Fig. 6). Correspondingly, the total activities of PMI isoenzymes and the total AsA levels were decreased. In contrast, the transcription of PMI2 was induced by the treatment. However, it was not correlated with the total activities of PMI isoenzymes.
FIGURE 6.
Expressions and total activity levels of Arabidopsis PMI isoenzymes and AsA concentrations in A. thaliana leaves treated with an inhibitor of the photosynthetic electron transport chain. Three-week-old Arabidopsisplants were moved to continuous light or dark conditions for 24 h, and then leaves were harvested at the indicated time. Relative levels of PMI1 (A) and PMI2 (B) were determined by real time PCR as described in Fig. 4. Error bars indicate S.D. (n = 3). C, Western blot of PMI1 protein. D, total PMI activity in crude extracts of Arabidopsis leaves. Error bars indicate S.D. (n = 3). E, AsA and DHA concentrations in Arabidopsis leaves. Error bars indicate S.D. (n = 3).
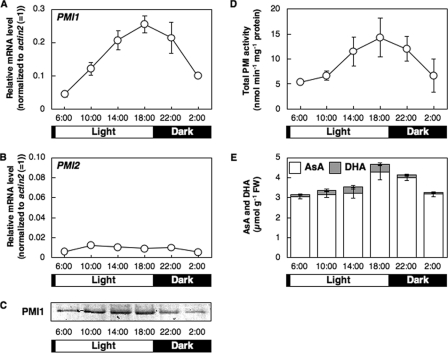
Diurnal Changes of Levels of PMI Isoenzymes and AsA in Arabidopsis Leaves—To further assess the relationship between the expression of PMI isoenzymes and the total AsA level, we analyzed the diurnal changes in these parameters in Arabidopsis leaves. The level of PMI1 transcript was increased during the day and peaked at 18:00, reflecting the protein level (Fig. 7). The diurnal changes in the total activities of PMI isoenzymes and the total AsA levels were similar to those in the expression of PMI1. However, the transcript level of PMI2 did not change in relation to the time of day, and thus the protein was not detected at any time of day.
FIGURE 7.
PMI1 and PMI2 expression, total PMI activity, and AsA concentrations in A. thaliana leaves in relation to the time of day. The leaves of 3-week-old Arabidopsis plants were harvested at the indicated times through a 24-h light/dark cycle. Relative levels of PMI1 (A) and PMI2 (B) were determined by real time PCR as described in Fig. 4. Error bars indicate S.D. (n = 3). C, Western blot of PMI1 protein. D, total PMI activity in crude extracts of Arabidopsis leaves. Error bars indicate S.D. (n = 3). E, AsA and DHA concentrations in Arabidopsis leaves. Error bars indicate S.D. (n = 3).
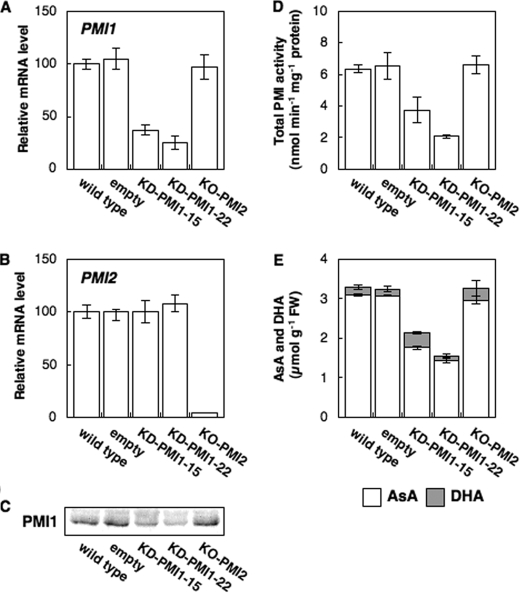
Effects of Suppression of PMI1 or PMI2 Expression in Arabidopsis Plants on AsA Levels—To study in situ how PMI1 and PMI2 contribute to the biosynthesis of AsA, we generated Arabidopsis plants (KD-PMI1) transformed with the RNA interference construct using the 5′-end sequence of PMI1 mRNA, and we obtained an Arabidopsis line (KO-PMI2) containing a T-DNA insert in the fourth exon of the PMI2 gene from the SIGnAL project (Fig. 8). Among a number of lines, two lines of KD-PMI1 plants (KD-PMI1–15, -22) with reduced expression of PMI1 were obtained. The transcript levels of PMI1 in the KD-PMI1–15 and KD-PMI1–22 plants were decreased to ∼37 and 25% of those in the control plants (transformed with empty vector), respectively, which well correlated with the protein levels of PMI1, respectively (Fig. 8). The total PMI activity level in KD-PMI1–15 and KD-PMI1–22 plants was ∼57 and 32% that in control plants, respectively. The suppression of PMI1 expression did not affect the expression of PMI2. The total AsA levels in KD-PMI1–15 and KD-PMI1–22 plants were decreased to 65 and 47% of those in control plants, respectively.
FIGURE 8.
Effect of suppression of PMI1 or PMI2 expression on the total PMI activity and the total AsA level in Arabidopsis leaves. The leaves of 3-week-old wild-type, control, KD-PMI1–15, KD-PMI1–22, and KO-PMI2 plants were harvested at 4 h after illumination. Relative levels of PMI1 (A) and PMI2 (B) were determined by real time PCR as described in Fig. 4. Error bars indicate S.D. (n = 3). C, Western blot of PMI1 protein. D, total PMI activity in crude extracts of Arabidopsis leaves. Error bars indicate S.D. (n = 3). E, AsA and DHA concentrations in Arabidopsis leaves. Error bars indicate S.D. (n = 3).
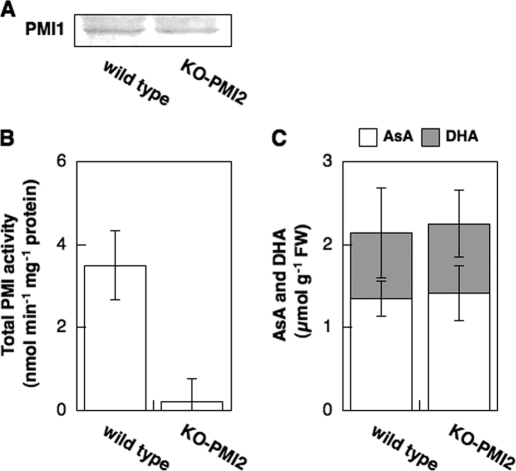
The disruption of the PMI2 gene did not affect PMI1 expression, total PMI activity, or the total AsA level in leaves under normal conditions (4 h after illumination) (Fig. 8). Because PMI2 was expressed only under long term darkness, the KO-PMI2 and wild-type plants were moved into the dark for 48 h. Dark-induced increases in the detectable levels of the PMI2 transcript in the wild-type plants were completely diminished in the KO-PMI2 plants (data not shown). Interestingly, compared with the wild-type plants, a drastic decrease in total PMI activity in leaves of the KO-PMI2 plants was observed after 48 h exposure to darkness (Fig. 9). However, there was no difference in the total AsA level between the KO-PMI2 and wild-type plants.
FIGURE 9.
Expressions and total activity levels of PMI isoenzymes and total AsA levels in wild-type and KO-PMI2 plants under prolonged darkness. Three-week-old Arabidopsis plants were moved to continuous dark conditions for 48 h, and then leaves were harvested. A, Western blot of PMI1 protein. B, total PMI activity in crude extracts of Arabidopsis leaves. Error bars indicate S.D. (n = 3). C, AsA and DHA concentration in Arabidopsis leaves. Error bars indicate S.D. (n = 3).
DISCUSSION
To explore the relationship between the roles of PMI isoenzymes and AsA biosynthesis, first we isolated the genes of Arabidopsis PMI1 and PMI2 and characterized the enzymological properties of purified recombinant proteins. Arabidopsis PMI1 and PMI2 could catalyze the reversible isomerization of Fru-6P and Man-6P (data not shown). Although the Vmax value for Man-6P of the recombinant PMI1 was ∼12-fold lower than that of the recombinant PMI2, the Km value of recombinant PMI1 was ∼10-fold lower than that of the recombinant PMI2 (Fig. 2), indicating their catalytic efficiencies (kcat/ Km) to be nearly equal to each other (Fig. 2).
Type I PMIs in animal and fungal organisms have been isolated as metalloenzymes that require a Zn2+ ion for their activities (essential zinc), and thus the activities of the enzymes were significantly inhibited by the incubation with EDTA (45–47). On the other hand, the activities of PMIs were inhibited by the application of Zn2+ (inhibitory zinc) (35, 40, 43). Arabidopsis PMI1 and PMI2 were both inhibited by the incubation with EDTA or by the application of Zn2+ (Table 1 and supplemental Fig. S2), confirming the view that the sites of action for PMI between essential and inhibitory zinc are different, because the essential zinc participates in the folding of the mature PMI protein as well as its enzymatic reaction (46) and the inhibitory zinc competes with the substrate of PMI protein (supplemental Fig. S2). Moreover, the activities of PMI1 and PMI2 were inhibited by the application of Cd2+ or several reducing compounds, such as DTT (Table 1 and supplemental Fig. S2), which was in agreement with a previous report (50).
It has been demonstrated that the capacity to produce AsA is regulated by several mechanisms, including the negative feedback control by AsA itself (4). The rate of AsA biosynthesis from d-[U-14C]glucose decreased linearly with the increase in the pool size of AsA in embryonic pea seedlings (24). On incubation with 1 mm AsA, the GME of Arabidopsis plants was inhibited 15% (9). Recently, we have reported that l-Gal dehydrogenase activity was inhibited reversibly by AsA in a linear-competitive manner (Ki value of 133 μm) (25). PMI1 and PMI2 were competitively inhibited by the incubation with AsA, having a Ki of 1.1 ± 0.1 and 1.4 ± 0.2 mm, respectively (Fig. 3). Because the cytosolic concentration of AsA is as much as 20 mm (51), it seems likely that the feedback inhibition of PMIs by AsA also occurs in situ. The present results and those reported to date indicated that the AsA-mediated feedback inhibition for these three enzymes is one of the most important regulatory mechanisms in the d-Man/l-Gal pathway.
Changes in the transcript and protein levels of PMI1 in roots, stems, leaves, and flowers of Arabidopsis plants showed that PMI1 is constitutively expressed during both vegetative and reproductive growth (Fig. 4), consistent with the case of PMM (7). Furthermore, the data in the Arabidopsis eFP browser (52) also indicated that PMM and GMP are both highly and constitutively expressed in the major vegetative and reproductive organs of Arabidopsis plants. These data indicated that there may be co-regulation in the expression of PMI1, PMM, and GMP, all of which are required for the biosynthesis of GDP-Man as well as AsA. AsA biosynthesis is regulated by light (19, 21). Recently, we have found that the levels of the VTC2, GMP, and VTC4 transcripts respond to continuous exposure to light in parallel with the changes in the leaf AsA content (22). Interestingly, the responses of these enzymes required PET activity, suggesting the biosynthesis of AsA to be regulated by a retrograde signaling pathway via a PET redox state from chloroplasts (22). It has been reported that the expression of VTC2 was regulated by the circadian clocks (19). In this study, the expression of PMI1, but not PMI2, was induced by continuous exposure to light and blocked by the treatment with DCMU (Fig. 5 and 6). The total level of activity of PMI isoenzymes was increased by the continuous exposure and decreased by the treatment with DCMU. The expression of PMI1 and the total activity level of PMI isoenzymes showed a diurnal change and peaked at the end of day (Fig. 7). Under light, the patterns of PMI1 expression are closely linked to the change in total PMI activity and the total AsA level. Thus it is likely that the expression pattern of PMI1 is similar to those of several AsA biosynthesis-related enzymes, especially VTC2, indicating the involvement of PMI1 expression in the light-based regulation of AsA biosynthesis in leaves of Arabidopsis plants.
To explore the physiological functions of Arabidopsis PMI isoenzymes in vivo, we used the KD-PMI1 and KO-PMI2 plants. The total PMI activity and total AsA levels in KD-PMI1–15 and KD-PMI1–22 plants were drastically decreased compared with those in control plants. There was a high correlation among the PMI1 expression, the total PMI activity, and the total AsA level in leaves. In contrast, the disruption of the PMI2 gene did not affect the total AsA level in Arabidopsis plants even under prolonged darkness (Figs. 8 and 9). These results suggest that under normal growth conditions, the total PMI activity in Arabidopsis leaves is dominated by PMI1 and not by PMI2. Therefore, PMI1 is essential for the biosynthesis of AsA in leaves, and the present findings strongly support the importance of d-Man/l-Gal pathway in Arabidopsis, in the complex AsA network.
It has been reported that GDP-Man contributes to the synthesis of different structural carbohydrates in plant cell walls (53), i.e. GDP-Man is essential for post-translational modifications such as protein glycosylation and glycosylphosphatidyl-inositol anchoring in eukaryotes, because mannose, derived from GDP-Man, is a crucial building block of the core glycan chain attached to the modified proteins (33, 54, 55). It has been reported recently that the Arabidopsis pmm-12 mutants had an impairment of protein glycosylation as well as a reduction in AsA content and, interestingly, showed lethal phenotypes because of the impaired glycosylation rather than the AsA depletion under mild heat stress (33). These findings lead us to ponder whether PMI1 would have consequences for not only AsA biosynthesis, but also glycosylation, cell wall biogenesis, and/or environmental stress response. However, KD-PMI1 plants as well as KO-PMI2 plants showed similar phenotypes to wild-type plants under normal growth conditions (data not shown). Pagnussat et al. (56) performed a large scale screening for mutants with defects in female gametophyte development and function. Among them, MEE31 mutant lacking the PMI1 gene showed endosperm development arrested. Therefore the most plausible explanation is that the residual PMI activity may be sufficient for normal growth and development of transgenic plants. These findings should facilitate the carrying out of further experiments in these new directions using KD-PMI1 plants.
PMI2 has been previously isolated as a dark inducible gene (Din9), which had undetectable transcript levels in photosynthesizing leaves under light, but substantially increased levels in plants removed from the light for more than 24 h (48, 49). In addition, the PMI2 transcript accumulated in sugar-starved suspension cells (48). Likewise, the expression of PMI2 was induced by long term darkness or treatment with DCMU (Figs. 5 and 6). Moreover, at 48 h after the application of prolonged darkness, the total PMI activity completely depended on PMI2 expression (Fig. 9). However, under these conditions the expression of most enzymes involved in the biosynthesis of AsA was drastically suppressed (22). Accordingly, we concluded that PMI2 does not contribute to the biosynthesis. As described above, mannose is one of the major hexose components of plant cell constituents such as cell wall polysaccharides and many types of glycoproteins (33, 53–55). Under sugar-starved conditions, the degradation of cell walls and proteins was increased in plant cells (57–59). Mannose can be readily phosphorylated by hexokinase and further metabolized by PMI to Fru-6P, which can enter into glycolysis. Therefore, it is possible that PMI2 is involved in the utilization of Man-derived carbohydrates as an energy source in leaves under sugar-starved conditions, such as prolonged darkness and/or senescence.
Supplementary Material
Acknowledgments
We are grateful to Dr. Tsuyoshi Nakagawa (Shimane University) for donation of pGWB80 vector.
This work was supported by Scientific Research for Plant Graduate Students from the Nara Institute of Science and Technology (to T. M.), Grant-in-aid for Scientific Research 19208031 from the Ministry of Education, Culture, Sports, Science and Technology, Japan, and the “Academic Frontier” Project for Private Universities matching fund subsidy from Ministry of Education, Culture, Sports, Science and Technology 2004–2008 (to S. S.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
Footnotes
The abbreviations used are: AsA, l-ascorbic acid; PMI, phosphomannose isomerase; Man-6P, d-mannose 6-phosphate; Fru-6P, d-fructose 6-phosphate; PMM, phosphomannomutase; GMP, GDP-Man pyrophosphorylase; GME, GDP-mannose-3′,5′-epimerase; DCMU, 3-(3,4-dichlorophenyl)-ll-dimethylurea; CHI, cycloheximide; DTT, dithiothreitol; PET, photosynthetic electron transport; DHA, dehydroascorbic acid; MOPS, 3-(N-morpholino)propanesulfonic acid; RT, reverse transcription.
References
- 1.Smirnoff, N., and Wheeler, G. L. (2000) Crit. Rev. Biochem. Mol. Biol. 35 291–314 [DOI] [PubMed] [Google Scholar]
- 2.Ishikawa, T., and Shigeoka, S. (2008) Biosci. Biotechnol. Biochem. 72 1143–1154 [DOI] [PubMed] [Google Scholar]
- 3.Wheeler, G. L., Jones, M. A., and Smirnoff, N. (1998) Nature 393 365–369 [DOI] [PubMed] [Google Scholar]
- 4.Ishikawa, T., Dowdle, J., and Smirnoff, N. (2006) Physiol. Plant. 126 343–355 [Google Scholar]
- 5.Conklin, P. L., Pallanca, J. E., Last, R. L., and Smirnoff, N. (1997) Plant Physiol. 115 1277–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conklin, P. L., Norris, S. R., Wheeler, G. L., Williams, E. H., Smirnoff, N., and Last, R. L. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 4198–4203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qian, W., Yu, C., Qin, H., Liu, X., Zhang, A., Johansen, I. E., and Wan, D. (2007) Plant J. 49 399–413 [DOI] [PubMed] [Google Scholar]
- 8.Jain, A. K., and Nessler, C. L. (2000) Mol. Breed. 6 73–78 [Google Scholar]
- 9.Wolucka, B. A., and Van Montagu, M. (2003) J. Biol. Chem. 278 47483–47490 [DOI] [PubMed] [Google Scholar]
- 10.Davey, M. W., Gilot, C., Persiau, G., Østergaard, J., Han, Y., Bauw, G. C., and van Montagu, M. (1999) Plant Physiol. 121 535–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lorence, A., Chevone, B., Mendes, P., and Nessler, C. L. (2004) Plant Physiol. 134 1200–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang, W., Gruszewski, H. A., Chevone, B. I., and Nessler, C. L. (2008) Plant Physiol. 146 431–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conklin, P. L., Williams, E. H., and Last, R. L. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 9970–9974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conklin, P. L., Saracco, S. A., Norris, S. R., and Last, R. L. (2000) Genetics 154 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Conklin, P. L., Gatzek, S., Wheeler, G. L., Dowdle, J., Raymond, M. J., Rolinski, S., Isupov, M., Littlechild, J. A., and Smirnoff, N. (2006) J. Biol. Chem. 281 15662–15670 [DOI] [PubMed] [Google Scholar]
- 16.Laing, W. A., Bulley, S., Wright, M., Cooney, J., Jensen, D., Barraclough, D., and MacRae, E. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 16976–16981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Linster, C. L., Gomez, T. A., Christensen, K. C., Adler, L. N., Young, B. D., Brenner, C., and Clarke, S. G. (2007) J. Biol. Chem. 282 18879–18885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laing, W. A., Wright, M. A., Cooney, J., and Bulley, S. M. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 9534–9539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dowdle, J., Ishikawa, T., Gatzek, S., Rolinski, S., and Smirnoff, N. (2007) Plant. J. 52 673–689 [DOI] [PubMed] [Google Scholar]
- 20.Smirnoff, N., Conklin, P. L., and Loewus, F. A. (2001) Annu. Rev. Plant. Physiol. Plant. Mol. Biol. 52 437–467 [DOI] [PubMed] [Google Scholar]
- 21.Bartoli, C. G., Yu, J., Gómez, F., Fernández, L., McIntosh, L., and Foyer, C. H. (2006) J. Exp. Bot. 57 1621–1631 [DOI] [PubMed] [Google Scholar]
- 22.Yabuta, Y., Mieda, T., Rapolu, M., Nakamura, A., Motoki, T., Maruta, T., Yoshimura, K., Ishikawa, T., and Shigeoka, S. (2007) J. Exp. Bot. 58 2661–2671 [DOI] [PubMed] [Google Scholar]
- 23.Yabuta, Y., Maruta, T., Nakamura, A., Mieda, T., Yoshimura, K., Ishikawa, T., and Shigeoka, S. (2008) Biosci. Biotechnol. Biochem., in press [DOI] [PubMed]
- 24.Pallanca, J. E., and Smirnoff, N. (2000) J. Exp. Bot. 345 669–674 [PubMed] [Google Scholar]
- 25.Mieda, T., Yabuta, Y., Madhusudhan, R., Motoki, T., Takeda, T., Yoshimura, K., Ishikawa, T., and Shigeoka, S. (2004) Plant Cell Physiol. 45 1271–1279 [DOI] [PubMed] [Google Scholar]
- 26.Reinbothe, S., Mollenhauer, B., and Reinbothe, C. (1994) Plant Cell 6 1197–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turner, J. G., Ellis, C., and Devoto, A. (2002) Plant Cell 14 S153–S164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wolucka, B. A., Goossens, A., and Inzé, D. (2005) J. Exp. Bot. 56 2527–2538 [DOI] [PubMed] [Google Scholar]
- 29.Herold, A., and Lewis, D. H. (1977) New Phytol. 79 1–40 [Google Scholar]
- 30.Dalessandro, G., Piro, G., and Northcote, D. H. (1986) Planta 169 564–574 [DOI] [PubMed] [Google Scholar]
- 31.Sharples, S. C., and Fry, S. C. (2007) Plant. J. 52 252–262 [DOI] [PubMed] [Google Scholar]
- 32.Lukowitz, W., Nickle, T. C., Meinke, D. W., Last, R. L., Conklin, P. L., and Somerville, C. R. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 2262–2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoeberichts, F. A., Vaeck, E., Kiddle, G., Coppens, E., van de Cotte, B., Adamantidis, A., Ormenese, S., Foyer, C. H., Zabeau, M., Inzé, D., Périlleux, C., Van Breusegem, F., and Vuylsteke, M. (2008) J. Biol. Chem. 283 5708–5718 [DOI] [PubMed] [Google Scholar]
- 34.Bradford, M. (1976) Anal. Biochem. 72 248–254 [DOI] [PubMed] [Google Scholar]
- 35.Wells, T. N. C., Coulin, F., Payton, M. A., and Proudfoot, A. E. I. (1993) Biochemistry 32 1294–1301 [DOI] [PubMed] [Google Scholar]
- 36.Nishizawa, A., Yabuta, Y., Yoshida, E., Maruta, T., Yoshimura, K., and Shigeoka, S. (2006) Plant J. 48 535–547 [DOI] [PubMed] [Google Scholar]
- 37.Yoshimura, K., Miyao, K., Gaber, A., Takeda, T., Kanaboshi, H., Miyasaka, H., and Shigeoka, S. (2004) Plant J. 37 21–33 [DOI] [PubMed] [Google Scholar]
- 38.Bechtold, N., and Pelletier, G. (1998) Methods Mol. Biol. 82 259–266 [DOI] [PubMed] [Google Scholar]
- 39.Proudfoot, A. E. I., Turcatti, G., Wells, T. N. C., Payton, M. A., and Smith, D. J. (1994) Eur. J. Biochem. 219 415–423 [DOI] [PubMed] [Google Scholar]
- 40.Coulin, F., Magnenat, E., Proudfoot, A. E. I., Payton, M. A., Scully, P., and Wells, T. N. C. (1993) Biochemistry 32 14139–14144 [DOI] [PubMed] [Google Scholar]
- 41.Smith, D. J., and Payton, M. A. (1994) Mol. Cell. Biol. 14 6030–6038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miles, J. S., and Guest, J. R. (1984) Gene (Amst.) 32 41–48 [DOI] [PubMed] [Google Scholar]
- 43.Cleasby, A., Wonacott, A., Skarzynski, T., Hubbard, R. E., Davies, G. J., Proudfoot, A. E. I., Bernard, A. R., Payton, M. A., and Wells, T. N. C. (1996) Nat. Struct. Biol. 3 470–479 [DOI] [PubMed] [Google Scholar]
- 44.Proudfoot, A. E. I., and Payton, M. A. (1994) J. Protein Chem. 13 619–627 [DOI] [PubMed] [Google Scholar]
- 45.Gracy, R. W., and Noltman, E. A. (1968) J. Biol. Chem. 243 4109–4116 [PubMed] [Google Scholar]
- 46.Proudfoot, A. E. I., Goffin, L., Payton, M. A., Wells, T. N. C., and Bernard, A. R. (1996) Biochem. J. 318 437–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roux, C., Gresh, N., Perera, L. E., Piquemal, J. P., and Salmon, L. (2007) J. Comput. Chem. 28 938–957 [DOI] [PubMed] [Google Scholar]
- 48.Fujiki, Y., Ito, M., Nishida, I., and Watanabe, A. (2000) Plant Physiol. 124 1139–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fujiki, Y., Yoshikawa, Y., Sato, T., Inada, N., Ito, M., Nishida, I., and Watanabe, A. (2001) Physiol. Plant. 111 345–352 [DOI] [PubMed] [Google Scholar]
- 50.Gracy, R. W., and Noltman, E. A. (1968) J. Biol. Chem. 243 5410–5419 [PubMed] [Google Scholar]
- 51.Foyer, C. H., and Lelandais, M. (1996) J. Plant. Physiol. 148 391–398 [Google Scholar]
- 52.Schmid, M., Davison, T. S., Henz, S. R., Pape, U. J., Demar, M., Vingron, M., Scholkopf, B., Weigel, D., and Lohmann, J. U. (2005) Nat. Genet. 37 501–506 [DOI] [PubMed] [Google Scholar]
- 53.Seifert, G. J. (2004) Curr. Opin. Plant. Biol. 7 277–284 [DOI] [PubMed] [Google Scholar]
- 54.Lerouge, P., Cabanes-Macheteau, M., Rayon, C., Fischette-Lainé, A. C., Gomord, V., and Faye, L. (1998) Plant. Mol. Biol. 38 31–48 [PubMed] [Google Scholar]
- 55.Spiro, R. G. (2002) Glycobiology 12 R43–R56 [Google Scholar]
- 56.Pagnussat, G. C., Yu, H. J., Ngo, Q. A., Rajani, S., Mayalagu, S., Johnson, C. S., Capron, A., Xie, L. F., Ye, D., and Sundaresan, V. (2005) Development (Camb.) 132 603–614 [DOI] [PubMed] [Google Scholar]
- 57.Brouquisse, R., Rolin, D., Cortés, S., Gaudillére, M., Evrard, A., and Roby, C. (2007) Planta 225 693–709 [DOI] [PubMed] [Google Scholar]
- 58.Contento, A. L., Kim, S. J., and Bassham, D. C. (2004) Plant. Physiol. 135 2330–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee, E. J., Matsumura, Y., Soga, K., Hoson, T., and Koizumi, N. (2007) Plant Cell Physiol. 48 405–413 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.