Abstract
The heterohexameric origin recognition complex (ORC) has been implicated in many cellular activities, including DNA replication, transcriptional control, heterochromatin assembly, centromere and telomere function, and so on. Here, we report a new function for ORC in mediating histone methylation. Using the yeast two-hybrid system, we identify a physical interaction between Orc2p and Spp1p, a member of the Set1 complex, and we demonstrate the interaction between the endogenous ORC and Spp1p by co-immunoprecipitation from yeast extracts. Furthermore, we find that Orc2p physically interacts with trimethylated histone 3 lysine 4 (H3K4) on chromatin by co-immunoprecipitation. Finally, we show that the trimethylation of H3K4 is decreased in orc2-1 cells and abolished in orc2-1, spp1Δ double mutants. Our data reveal a novel facet of ORC in mediating histone methylation in collaboration with Spp1p and demonstrate a connection between ORC and chromatin structure via the Set1 complex.
The origin recognition complex (ORC)2 is a multifunctional protein complex that was identified by its affinity to the autonomously replicating sequence (ARS) consensus sequence (ACS) in DNA replication origins (1, 2). ORC serves as a landing pad for recruitment of downstream replication-initiation proteins like Noc3p, Cdc6p, Cdt1p, and Mcm2–7p, forming a prereplicative complex (3, 4). A genome-wide chromatin immunoprecipitation-on-chip assay identified ∼500 ORC binding sites, with the majority of them being co-localized with MCM binding sites and replication origins (5). It is inarguable that ORC is a central player in DNA replication. Further studies have revealed the functions of ORC in many other cellular processes, such as the establishment of transcriptionally suppressed mating type loci (HMR and HML) (6–8), transcriptional regulation, and chromatin structure (1, 2).
Diverse lines of evidence have emerged to implicate the involvement of ORC in chromatin structure in eukaryotes (1, 2). ORC localizes in telomere repeats, and depletion of ORC leads to dysfunctional telomeres (9). In Saccharomyces cerevisiae, the N-terminal part of Orc1p recruits the chromatin silencing protein Sir1p (10). ORC is responsible for positioning nucleosomes at ARS1 (11), and recent work has elucidated a physical interaction between ORC and histone acetyltransferase (Hat1p and Hat2p) (12). In Drosophila, ORC is enriched at heterochromatin regions and interacts directly with HP1, a key component of heterochromatin (13). Furthermore, mutations in ORC2 result in mislocalization of HP1. The interaction between ORC and HP1 is also identified in Xenopus and mammalian cells. In human cells, small interfering RNA knockdown of ORC2 leads to delocalization of HP1 and chromosome condensation defects (14). An interesting physical interaction has also been revealed between Orc1p and Hbo1p, a histone acetyltransferase (15, 16). However, it has never been explored whether ORC has any function in histone methylation.
In budding yeast, histone 3 lysine residues 4, 36, and 79 are methylated by Set1p (KMT2), Set2p (KMT3), and Dot1p (KMT4), respectively (17). Each modified lysine has three forms: mono-, di-, and trimethylation. The Set1 complex, or COMPASS (complex associated with Set1), is composed of Set1p, Bre2p (Cps60p), Swd1p (Cps50p or Saf 49p), Swd2p (Cps35p or Saf 37p), Swd3p (Cps30p or Saf 35p), Spp1p (Cps40p or Saf41p), Shg1p (Cps15p), and Sdc1p (Cps25p or Saf19p) (18–21). This complex adds 1–3 methyl groups specifically to H3K4 with different components contributing differently. Set1p, Swd1p, and Swd3p are required for any H3K4 methylation, Bre2p and Sdc1p are not essential for, but contribute to, mono-, di-, and trimethylation, and Spp1p is only important for trimethylation (22, 23).
Methylation of H3K4 in budding yeast usually defines an active chromatin state for gene expression (24–28). Genome-wide chromatin immunoprecipitation-on-chip assays have revealed that the trimethylation of H3K4 co-localizes with promoters and the 5′ end of transcribed genes (29). The H3K4 methylation has also been implicated in position-dependent gene silencing at telomeres (30) and the mating type locus (31), heterochromatin formation (31), and ribosomal DNA silencing (32).
From a genome-wide yeast two-hybrid screen to identify human proteins that interact with human DNA replication-initiation proteins, we found that hCGBP (CpG island-binding protein) physically interacts with several DNA replication-initiation proteins including ORC.3 Because budding yeast is a more amendable system for genetic and functional studies, we set out to investigate whether Spp1p, the yeast homolog of hCGBP, interacts with budding yeast replication-initiation proteins. Indeed, we find a physical interaction between Spp1p and Orc2p. Here, we report that ORC mediates histone methylation through cooperation with Spp1p.
EXPERIMENTAL PROCEDURES
Plasmids, Strains, and Antibodies—The primers used to construct pGBKT7-SPP1 were 5′-GGAATTCATGTCATTACCACAATGGTGTCC-3′ (containing an EcoRI site) and 5′-GCGGATCCTTACAAACCTCTTCTTAAAATTTC-3′ (containing a BamHI site). The spp1Δ strain was generated in W303-1a by PCR-mediated one-step disruption using forward primer 5′-ATTTGGAAAAGGCTACTTCGACCTCAATAATTTCTCAGCCTATCTTTCTACGGATCCCCGGGTTAATTAA-3′ and reverse primer 5′-CGAAGTATATATATATGTAGAAACTGATATTTGATTAGGCTCCAACGCCGGAATTCGAGCTCGTTTAAAC-3′. The SPP1-HA-tagged strain was constructed using forward primer 5′-AAAAGCAACTAAATATACAATACTATGAGGAAATTTTAAGAAGAGGTTTGCGGATCCCCGGGTTAATTAA-3′ and the same reverse primer as for SPP1 deletion. The ORC2-TAP strain is provided by Andrew Emili (University of Toronto). The anti-histone 3, anti-dimethylated, and trimethylated H3K4 antibodies were purchased from AbCam (ab1791, ab7766, and ab8580, respectively). The anti-HA (12CA5) is from Roche Applied Science. Anti-ORC antibodies are from Bruce Stillman (Cold Spring Harbor Laboratory).
Yeast Two-hybrid Assay—The yeast two-hybrid system employed was the MATCHMAKER GAL4 two-hybrid system 3 (Clontech), and the assay was conducted according to the manufacturer's instructions.
Co-IP Assay—Co-IP assay was performed as described previously (33) with modifications. In brief, 100 ml of log phase cells (A600 ∼ 2) were harvested for each sample, and total protein extracts were obtained by bead-beating in 400 μl of IP/lysis buffer-100 (100 mm (NH4)2SO4; other components as in Ref. 33). After harvesting the extract, the pellet was extracted again with 400 μl of the IP/lysis buffer-200 (200 mm (NH4)2SO4 containing 200 μg/ml ethidium bromide; other components as in Ref. 33). After the extract was harvested, the pellet was rinsed with 400 μl of IP/lysis buffer-0 (no (NH4)2SO4; other components as in Ref. 33). The three extracts were combined as total protein extract for co-IP. Some 25 μl of protein G beads (Zymed Laboratories Inc.) were added to preclear the protein extract at 4 °C for 1 h. Afterward, the samples were incubated with 5–10 μg of antibody or control mouse IgG (Pierce) for 1–1.5 h and then with 25 μl of protein G beads for 1 h. After extensive wash with IP/lysis buffer-100 (100 mm (NH4)2SO4), the beads were boiled in 25 μl of 2× Laemmli's buffer for 3 min.
Chromatin Binding Assay—A chromatin binding assay to separate total yeast proteins into the crude chromatin pellet and supernatant fractions and the stringent chromatin binding assay involving the use of micrococcal nuclease (MNase) to partially digest the crude chromatin followed by ultracentrifugation to pellet the released chromatin were performed essentially as described previously (34).
Co-IP with MNase-digested Chromatin Fraction—The crude chromatin pellet isolated using the chromatin binding assay was digested with 1000 IU of MNase in 300 μl of MNase buffer (1 mm boric acid; 5 mm NaCl; 5 mm CaCl2; adjusted to pH 8.8) at 37 °C for 10 min. (It was confirmed that the resultant DNA fragments were less than 200 bp.) The supernatant was adjusted to standard IP/lysis buffer, and the co-IP assay was performed as described above.
Cell Synchronization and FACS—Cells were arrested in G1 phase with 5 μg/ml α factor for 2.5–3 h and then released into fresh YPD medium. Cells were harvested at various time points, and FACS analysis was performed as described (33).
RESULTS
Spp1p Interacts with ORC through Orc2p—We performed yeast two-hybrid assays using SPP1 as the bait to test its possible interactions with replication-initiation proteins. We co-transformed the host AH109 cells with pGBKT7-SPP1 (SPP1 constructed in the DNA binding domain vector pGBKT7) and individual plasmids expressing replication-initiation proteins in the activation domain pGADT7 vector. The transformants were streaked on SCM-2 (synthetic complete medium, lacking leucine and tryptophan), SCM-3 (lacking leucine, tryptophan, and histidine), and SCM-4 (lacking leucine, tryptophan, histidine, and adenine) plates. We found that cells containing pGBKT7-SPP1 and pGADT7-ORC2, and those containing the positive control plasmids (pGBKT7-ORC3 with pGADT7-ORC2, and pGBKT7-p53 with pGADT7-T antigen) were able to grow on both SCM-3 and SCM-4 plates (Fig. 1A). The negative control strains containing pGBKT7-SPP1 and the pGADT7 vector, or the pGBKT7 vector and pGADT7-ORC2, could not grow on SCM-3 or SCM-4 plates (Fig. 1A). These data suggest that Spp1p interacts with Orc2p. The cells co-transformed with pGBKT7-SPP1, and individual plasmids expressing other replication-initiation proteins (Orc1p, 3p-6p, Noc3p, Cdc6p, Mcm2p-7p, Cdc45p) or Lamin C in pGADT7 could not grow on SCM-3 or SCM-4 plates (data not shown). These data suggest that there is a specific physical interaction between Spp1p and Orc2p. The interaction between Spp1p and Orc2p in the yeast two-hybrid system was confirmed by a co-immunoprecipitation assay using yeast protein extracts expressing the two-hybrid proteins (data not shown).
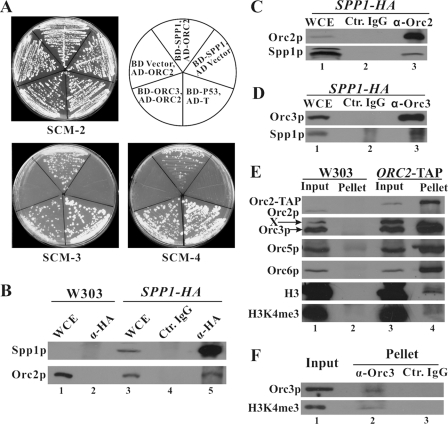
FIGURE 1.
ORC physically interacts with Spp1p and trimethylated H3K4. A, Spp1p interacts with Orc2p by the yeast two-hybrid system. pGBK7-SPP1 expressing Spp1p fused to the DNA binding domain (BD-SPP1) was co-transformed into AH109 cells together with pGADT7-ORC2 expressing Orc2p fused to the activation domain (AD-ORC2). Cells expressing BD and AD-ORC2, and those expressing BD-SPP1 and AD, were used as negative controls. Positive controls included cells expressing BD-ORC3 and AD-ORC2 and those expressing BD-p53 and AD-T antigen. The transformants were streaked on SCM-2 (synthetic complete medium, lacking leucine and tryptophan), SCM-3 (lacking leucine, tryptophan, and histidine), and SCM-4 (lacking leucine, tryptophan, histidine, and adenine) plates. B, co-immunoprecipitation assay confirmed the interaction between ORC and Spp1p with protein extracts from SPP1-HA-tagged strains. Spp1p-HA was immunoprecipitated by anti-HA antibody, and the immunoprecipitates were probed by anti-Orc2 antibody. WCE, whole cell extracts; Ctr. IgG, control IgG. C and D, reverse co-IP confirms the physical interaction. Anti-Orc2 (C) and -Orc3 (D) antibodies were employed to immunoprecipitate yeast extracts, and Spp1p in the precipitates was detected by anti-HA antibody. E, Orc2p interacts with trimethylated H3K4. The proteins from affinity precipitation in ORC2-TAP strain were immunoblotted by various antibodies against Orc2p, -3p, -5p, -6p, H3, and trimethylated H3K4, respectively. X, cross-reaction band by the anti-Orc3 antibody. F, co-IP assay with MNase-released chromatin fraction suggests that ORC interacts with trimethylated H3K4 on chromatin. Log phase W303-1a cells were harvested, and the chromatin fraction was completely digested with MNase. The supernatant containing the proteins from digested chromatin was diluted into the standard IP/lysis buffer, and co-IP assay was performed.
To test the interaction between the endogenous Orc2p and Spp1p, we performed reciprocal co-IP assays using protein extracts from SPP1-HA-tagged yeast cells. Spp1p-HA was immunoprecipitated by anti-HA antibody, and the immunoprecipitates were probed by anti-Orc2 and -Orc3 antibodies. Orc2p and Orc3p could be co-immunoprecipitated with Spp1p (Fig. 1B, lane 5), whereas no signal was detected in the control mouse IgG pellet (lane 4) or untagged strain control (lane 2). In the reverse co-IP, we detected Spp1p-HA in the precipitates pulled down by anti-Orc2 antibody (Fig. 1C) and anti-ORC3 antibody (Fig. 1D). Together with the yeast two-hybrid data, the co-IP results strongly suggest that Spp1p physically interacts with ORC through Orc2p in vivo.
Genetic interactions were also investigated between Spp1p and replication-initiation proteins or replication regulators. The SPP1 deletion has a synthetic sickness phenotype with orc2-1 and orc5-1, respectively, but not with mcm2-1, mcm3-1, mcm5-1, cdc6-1, cdc7-4, or cdc28-4 (data not shown), suggesting that ORC and Spp1p participate in a common pathway.
ORC Interacts with Histone 3 When Histone 3 Lysine 4 Is Trimethylated—Spp1p is a component of the Set1 complex, which is responsible for methylation of H3K4. Having determined that ORC physically interacts with Spp1p, we examined whether ORC interacts with histone 3. The affinity precipitates from the ORC2-TAP strain were blotted by various antibodies (anti-Orc2, -Orc3, -Orc5, -Orc6, -H3, and -trimethylated H3K4). The results show that not only Orc3p, -5p, and -6p, but also H3 and trimethylated H3K4, associated with Orc2p (Fig. 1E, lane 4). None of them was detectable in the untagged control strain (Fig. 1E, lane 2). Correlating the recent work demonstrating a connection between ORC and histone acetyltransferase (12), our data suggest that ORC may dynamically communicate with nucleosomes that contain an intricate combination of modifications.
Because ORC and histone 3 are both chromatin-binding proteins, the co-IP of ORC with histone 3 might be bridged by DNA. To address this concern, we used ethidium bromide to release proteins from DNA in the co-IP assays described above. To further exclude the possibility of DNA/chromatin-mediated co-IP, we employed co-IP assay with proteins released from completely digested chromatin. The anti-Orc3 immunoprecipitates were blotted by anti-Orc3 and anti-trimethylated H3K4 antibodies (Fig. 1F). The trimethylated H3K4 was co-immunoprecipitated with Orc3p, lending support to the conclusion that the association of ORC with trimethylated H3K4 is through protein-protein interactions, which can occur on chromatin and can be maintained after the proteins have been released from chromatin.
ORC and Spp1p Cooperate to Mediate Histone 3 Lysine 4 Trimethylation—To search for the functional significance of the associations among ORC, Spp1p, and histone 3, the H3K4 methylation pattern was analyzed in various strains (W303-1a, spp1Δ, orc2-1, and orc2-1, spp1Δ double mutants). Whole cell extracts from log phase cells were subject to immunoblotting with antibodies against different ORC subunits and di-, and trimethylated H3K4 (Fig. 2A). With SPP1 deletion, the trimethylation of H3K4 was reduced to a third at 25 °C, in line with previous results (22). When SPP1 deletion was combined with orc2-1, the trimethylation of H3K4 was greatly reduced at 25 °C. At 37 °C, H3K4 trimethylation was mostly lost in spp1Δ and orc2-1 single mutants and completely abolished in the orc2-1, spp1Δ double mutant. Together with data showing the interactions among ORC, Spp1p, and histone 3, our results suggest that ORC and Spp1p cooperatively mediate H3K4 trimethylation. In addition, notice that dimethylation of H3K4 was also lowered in spp1Δ and orc2-1 single mutants at 37 but not 25 °C and in the double mutant at both 25 °C and 37 °C.
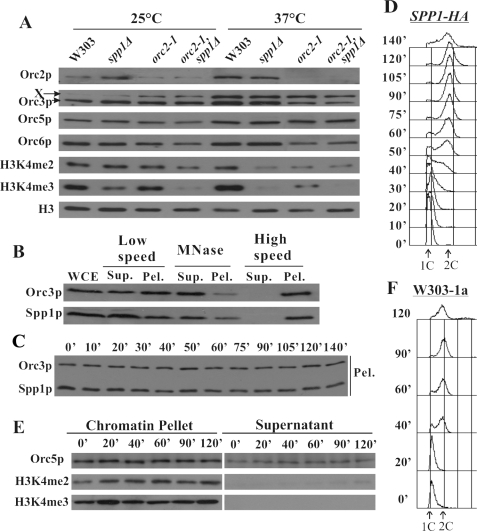
FIGURE 2.
ORC and Spp1p collaboratively mediate histone 3 lysine 4 trimethylation. A, spp1Δ, orc2-1 single and double mutants are defective for H3K4 trimethylation. Cells of various strains (W303-1a, spp1Δ, orc2-1, and orc2-1, spp1Δ double mutants) were incubated at 25 °C to log phase or shifted to 37 °C for 2 h before harvesting. Whole cell extracts were separated by PAGE and immunoblotted with anti-Orc2, -Orc3, -Orc5, -Orc6, -H3, -dimethylated H3K4, and -trimethylated H3K4 antibodies, respectively. X, cross-reaction band by the anti-Orc3 antibody. B, Spp1p is a chromatin-binding protein. The stringent chromatin binding assay was conducted using SPP1-HA-tagged strain. Orc3p was used as a reference. WCE, whole cell extracts; Sup., supernatant; Pel., pellet. C, Spp1p binds to chromatin throughout the cell cycle. SPP1-HA-tagged cells were harvested at different time points released from α factor block, and chromatin binding assay was performed. D, the FACS analysis of the SPP1-HA-tagged cells used in C. E, the overall level of trimethylated H3K4 on chromatin is constant in the cell cycle. The chromatin binding assay was performed using W303-1a cells released from the α factor block. The chromatin pellet and supernatant fractions were immunoblotted by anti-Orc5, -dimethylated H3K4, and -trimethylated H3K4 antibodies. F, FACS analysis of W303-1a cells used in E.
Spp1p Is a Chromatin-binding Protein and Binds to Chromatin throughout the Cell Cycle—As a histone methylase, Spp1p together with other members of the Set1 complex should in principle bind to chromatin. We investigated the chromatin associations of Spp1p using the stringent chromatin binding assay (Fig. 2B) Similar to Orc3p, Spp1p is a chromatin-binding protein as it was detectable in low speed pellet, MNase supernatant, and high speed pellet fractions. To examine the chromatin binding pattern of Spp1p along the cell cycle, we harvested cells at different time points after they were released from α factor block and performed a chromatin binding assay (Fig. 2, C and D). Spp1p was found to bind chromatin constitutively throughout the cell cycle.
Constant Binding of Di- and Trimethylated H3K4 to Chromatin along the Cell Cycle—As Orc2p and Spp1p bind chromatin throughout the cell cycle and cooperatively regulate the trimethylation of H3K4, it is worthwhile to examine the chromatin binding pattern of H3K4 trimethylation in the cell cycle. The W303-1a cells released from α factor were harvested at designated time points. The overall level of histone 3, dimethylated H3K4, and trimethylated H3K4 on chromatin was more-or-less constant in the cell cycle (Fig. 2, E and F).
DISCUSSION
We have identified physical associations of ORC with Spp1p and histone 3 and revealed the functional role of ORC in H3K4 methylation in collaboration with the Set1 complex through Spp1p. The significance of the interactions may have implications in various cellular processes.
Replication Origin Selection—An unsolved puzzle in DNA replication is how replication origins are selected. ORC in budding yeast is commonly thought to be directed to replication origins by the conserved ACS (1). From sequence analysis in the S. cerevisiae genome, the number of matches to ACS is overwhelmingly larger than de facto ORC binding sites and replication origins (5, 35). Apparently, ACS is necessary but not sufficient for ORC binding. Additionally, biochemical studies suggest that the ability of ORC to distinguish specific from nonspecific sequences is limited (3). It has been widely speculated that ORC may recognize a unique chromatin structure, apart from ACS (3). The ORC-interacting Set1 complex may be one of the elusive epigenetic determinants of origin selection. How the interaction between ORC and Spp1p is modulated along the cell cycle and whether Spp1p is connected to replication origins are of instant interests and warrant further exploration.
Functional Overlap of ORC and the Set1 Complex—The functions of ORC and the Set1 complex overlap in diverse cellular processes. The accumulated data on the cellular functions of the Set1 complex and ORC point to a surprising coincidence (2, 23). To summarize, they both participate in transcriptional regulation, heterochromatin formation, telomere and centromere functions (9, 36), and S phase checkpoint (37–39). The connections of the two protein complexes in these cellular activities need to be vigorously tested and will be a fertile area of future research.
Regulation of H3K4 Trimethylation in the Cell Cycle—There is no discernible variation of the global level of ORC and trimethylated H3K4 on chromatin along the cell cycle. However, as the chromatin binding assay detects the overall level of chromatin association of proteins, it is possible that H3K4 trimethylation may be spatially regulated along the chromosomes in the cell cycle, and the association between ORC and Spp1p may occur at certain sites of chromosomes. Bearing in mind the genome-wide distribution of ORC, we postulate a spatial modulation of trimethylation of H3K4 regulated by ORC and Spp1p when they encounter each other on chromosomes, exerting epigenetic controls over a variety of cellular processes.
Acknowledgments
We greatly appreciate the gift of mouse anti-ORC antibodies from Bruce Stillman (Cold Spring Harbor Laboratory) and ORC2-TAP strain from Andrew Emili (University of Toronto).
This work was supported by a grant from the Hong Kong Research Grants Council (Grant HKUST6114/04M). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: ORC, origin recognition complex; ARS, autonomously replicating sequence; ACS, ARS consensus sequence; FACS, fluorescence activated cell sorting; IP, immunoprecipitation; co-IP, co-immunoprecipitation; SCM, synthetic complete medium; HA, hemagglutinin; MNase, micrococcal nuclease; TAP, tandem affinity purification.
L. Zou, J. Kan, and C. Liang, unpublished results.
References
- 1.Bell, S. P. (2002) Genes Dev. 16 659–672 [DOI] [PubMed] [Google Scholar]
- 2.Sasaki, T., and Gilbert, D. M. (2007) Curr. Opin. Cell Biol. 19 337–343 [DOI] [PubMed] [Google Scholar]
- 3.Bell, S. P., and Dutta, A. (2002) Annu. Rev. Biochem. 71 333–374 [DOI] [PubMed] [Google Scholar]
- 4.Mendez, J., and Stillman, B. (2003) BioEssays 25 1158–1167 [DOI] [PubMed] [Google Scholar]
- 5.Wyrick, J. J., Aparicio, J. G., Chen, T., Barnett, J. D., Jennings, E. G., Young, R. A., Bell, S. P., and Aparicio, O. M. (2001) Science 294 2357–2360 [DOI] [PubMed] [Google Scholar]
- 6.Shore, D. (2001) Curr. Biol. 11 R816–R819 [DOI] [PubMed] [Google Scholar]
- 7.Foss, M., McNally, F. J., Laurenson, P., and Rine, J. (1993) Science 262 1838–1844 [DOI] [PubMed] [Google Scholar]
- 8.Micklem, G., Rowley, A., Harwood, J., Nasmyth, K., and Diffley, J. F. X. (1993) Nature 366 87–89 [DOI] [PubMed] [Google Scholar]
- 9.Deng, Z., Dheekollu, J., Broccoli, D., Dutta, A., and Lieberman, P. M. (2007) Curr. Biol. 17 1989–1995 [DOI] [PubMed] [Google Scholar]
- 10.Hou, Z. G., Bernstein, D. A., Fox, C. A., and Keck, J. L. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 8489–8494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lipford, J. R., and Bell, S. P. (2001) Mol. Cell 7 21–30 [DOI] [PubMed] [Google Scholar]
- 12.Suter, B., Pogoutse, O., Guo, X., Krogan, N., Lewis, P., Greenblatt, J. F., Rine, J., and Emili, A. (2007) BMC Biol. 5 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pak, D. T. S., Pflumm, M., Chesnokov, I., Huang, D. W., Kellum, R., Marr, J., Romanowski, P., and Botchan, M. R. (1997) Cell 91 311–323 [DOI] [PubMed] [Google Scholar]
- 14.Prasanth, S. G., Prasanth, K. V., Siddiqui, K., Spector, D. L., and Stillman, B. (2004) EMBO J. 23 2651–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iizuka, M., and Stillman, B. (1999) J. Biol. Chem. 274 23027–23034 [DOI] [PubMed] [Google Scholar]
- 16.Burke, T. W., Cook, J. G., Asano, M., and Nevins, J. R. (2001) J. Biol. Chem. 276 15397–15408 [DOI] [PubMed] [Google Scholar]
- 17.Shilatifard, A. (2008) Curr. Opin. Cell Biol. 20 341–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roguev, A., Schaft, D., Shevchenko, A., Pijnappel, W., Wilm, M., Aasland, R., and Stewart, A. F. (2001) EMBO J. 20 7137–7148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller, T., Krogan, N. J., Dover, J., Erdjument-Bromage, H., Tempst, P., Johnston, M., Greenblatt, J. F., and Shilatifard, A. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 12902–12907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Briggs, S. D., Bryk, M., Strahl, B. D., Cheung, W. L., Davie, J. K., Dent, S. Y. R., Winston, F., and Allis, C. D. (2001) Genes Dev. 15 3286–3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagy, P. L., Griesenbeck, J., Kornberg, R. D., and Cleary, M. L. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 90–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dehe, P. M., Dichtl, B., Schaft, D., Roguev, A., Pamblanco, M., Lebrun, R., Rodriguez-Gil, A., Mkandawire, M., Landsberg, K., Shevchenko, A., Rosaleny, L. E., Tordera, V., Chavez, S., Stewart, A. F., and Geli, V. (2006) J. Biol. Chem. 281 35404–35412 [DOI] [PubMed] [Google Scholar]
- 23.Dehe, P. M., and Geli, V. (2006) Biochem. Cell Biol. 84 536–548 [DOI] [PubMed] [Google Scholar]
- 24.Schneider, J., Wood, A., Lee, J. S., Schuster, R., Dueker, J., Maguire, C., Swanson, S. K., Florens, L., Washburn, M. P., and Shilatifard, A. (2005) Mol. Cell 19 849–856 [DOI] [PubMed] [Google Scholar]
- 25.Ng, H. H., Robert, F., Young, R. A., and Struhl, K. (2003) Mol. Cell 11 709–719 [DOI] [PubMed] [Google Scholar]
- 26.Bernstein, B. E., Humphrey, E. L., Erlich, R. L., Schneider, R., Bouman, P., Liu, J. S., Kouzarides, T., and Schreiber, S. L. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 8695–8700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Santos-Rosa, H., Schneider, R., Bannister, A. J., Sherriff, J., Bernstein, B. E., Emre, N. C. T., Schreiber, S. L., Mellor, J., and Kouzarides, T. (2002) Nature 419 407–411 [DOI] [PubMed] [Google Scholar]
- 28.Morillon, A., Karabetsou, N., Nair, A., and Mellor, J. (2005) Mol. Cell 18 723–734 [DOI] [PubMed] [Google Scholar]
- 29.Pokholok, D. K., Harbison, C. T., Levine, S., Cole, M., Hannett, N. M., Lee, T. I., Bell, G. W., Walker, K., Rolfe, P. A., Herbolsheimer, E., Zeitlinger, J., Lewitter, F., Gifford, D. K., and Young, R. A. (2005) Cell 122 517–527 [DOI] [PubMed] [Google Scholar]
- 30.Krogan, N. J., Dover, J., Khorrami, S., Greenblatt, J. F., Schneider, J., Johnston, M., and Shilatifard, A. (2002) J. Biol. Chem. 277 10753–10755 [DOI] [PubMed] [Google Scholar]
- 31.Santos-Rosa, H., Bannister, A. J., Dehe, P. M., Geli, V., and Kouzarides, T. (2004) J. Biol. Chem. 279 47506–47512 [DOI] [PubMed] [Google Scholar]
- 32.Bryk, M., Briggs, S. D., Strahl, B. D., Curcio, M. J., Allis, C. D., and Winston, F. (2002) Curr. Biol. 12 165–170 [DOI] [PubMed] [Google Scholar]
- 33.Zhang, Y. X., Yu, Z. L., Fu, X. R., and Liang, C. (2002) Cell 109 849–860 [DOI] [PubMed] [Google Scholar]
- 34.Liang, C., and Stillman, B. (1997) Genes Dev. 11 3375–3386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu, W. H., Aparicio, J. G., Aparicio, O. M., and Tavare, S. (2006) BMC Genomics 7 1471–2164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wood, A., and Shilatifard, A. (2005) Nat. Struct. Mol. Biol. 12 839–840 [DOI] [PubMed] [Google Scholar]
- 37.Corda, Y., Schramke, V., Longhese, M. P., Smokvina, T., Paciotti, V., Brevet, V., Gilson, E., and Geli, V. (1999) Nat. Genet. 21 204–208 [DOI] [PubMed] [Google Scholar]
- 38.Schramke, V., Neecke, H., Brevet, V., Corda, Y., Lucchini, G., Longhese, M. P., Gilson, E., and Geli, V. (2001) Genes Dev. 15 1845–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shimada, K., Pasero, P., and Gasser, S. M. (2002) Genes Dev. 16 3236–3252 [DOI] [PMC free article] [PubMed] [Google Scholar]