Summary
Chromosome ends, known as telomeres, have to be distinguished from DNA double-strand breaks (DSBs) that activate the DNA damage checkpoint. In budding yeast, the ATM homolog Tel1 associates preferentially with short telomeres and promotes telomere addition. Here we show that the telomeric proteins Rif1 and Rif2 attenuate Tel1 recruitment to DNA ends through distinct mechanisms. Both Rif1 and Rif2 inhibit the localization of Tel1, but not the Mre11-Rad50-Xrs2 (MRX) complex, to adjacent DNA ends. Rif1 function is weaker at short telomeric repeats compared with Rif2 function, and is partly dependent on Rif2. Rif2 competes with Tel1 for binding to the C-terminus of Xrs2. Once Tel1 is delocalized, MRX does not associate efficiently with Rap1-covered DNA ends. These results reveal a mechanism by which telomeric DNA sequences mask DNA ends from Tel1 recognition for the regulation of telomere length.
Introduction
Telomeres are nucleoprotein complexes at the ends of linear eukaryotic chromosomes, which are distinguished from DNA double-strand breaks (DSBs). To maintain genomic integrity, all organisms respond to DSBs by promptly launching the DNA damage response. This response involves the recruitment of DNA repair factors to sites of DNA damage and the activation of signal transduction pathways, often termed DNA damage checkpoint pathways (Zhou and Elledge, 2000). Telomeres are protected from checkpoint activation (de Lange, 2005; Longhese, 2008). Paradoxically, checkpoint proteins are essential for telomere length maintenance.
Checkpoint signals are initiated through two large protein kinases, ataxia-telangiectasia mutated (ATM) and ATM-Rad3-related (ATR) (Abraham, 2001; Zhou and Elledge, 2000). ATM and ATR are highly conserved among eukaryotes. In budding yeast, ATM and ATR correspond to Tel1 and Mec1, respectively. Several lines of evidence indicate that the Mre11-Rad50-Nbs1 (Xrs2 in budding yeast) complex is the primary sensor that recruits ATR/Mec1 and ATM/Tel1 to DSBs (Falck et al., 2005; Nakada et al., 2004; Nakada et al., 2003a; You et al., 2005). In budding yeast, the Mre11-Rad50-Xrs2 (MRX) complex collaborates with exonucleases in the generation of single-stranded DNA (ssDNA) at DSB ends (Krogh and Symington, 2004). Tel1 interacts with the C-terminus of Xrs2 to localize to DNA ends (Falck et al., 2005; Nakada et al., 2003a; You et al., 2005). Once ssDNA is generated, replication protein A (RPA) recognizes ssDNA (Krogh and Symington, 2004; Wold, 1997), and recruits Mec1 to DNA ends (Nakada et al., 2005; Zou and Elledge, 2003). Mec1 and Tel1 activate the downstream Rad53 kinase (Nakada et al., 2003b; Sanchez et al., 1996; Schwartz et al., 2002; Sun et al., 1996; Sweeney et al., 2005; Usui et al., 2001), thereby leading to transient cell-cycle arrest and transcriptional activation of genes involved in DNA repair.
Telomeres contain a double-stranded DNA region of tandem repeats (e.g. human, T2AG3; budding yeast, TG1-3) and a 3′ protruding ssDNA region of the G-rich strand (Smogorzewska and de Lange, 2004; Vega et al., 2003). Single-stranded tails on telomeres are bound by sequence-specific ssDNA binding proteins, such as Cdc13 in budding yeast (Lin and Zakian, 1996; Nugent et al., 1996). Cdc13 acts as a telomere cap to protect telomeres from degradation (Garvik et al., 1995; Nugent et al., 1996; Pennock et al., 2001), thereby inhibiting RPA recruitment and subsequent Mec1 accumulation (Hirano and Sugimoto, 2007; Rouse and Jackson, 2002). However, Cdc13-mediated capping does not affect accumulation of Tel1 or MRX complex at DNA ends (Hirano and Sugimoto, 2007). Recent evidence indicates that Tel1 associates preferentially with short telomeres and promotes telomere addition (Bianchi and Shore, 2007; Chang et al., 2007; Hector et al., 2007; Sabourin et al., 2007; Viscardi et al., 2007). However, it has not been determined how Tel1 is inhibited from localizing to telomeres of normal length. Double-stranded telomeric DNA repeats are bound by the sequence-specific binding protein Rap1, which recruits Rif1 and Rif2 proteins via its C-terminal domain (Conrad et al., 1990; Hardy et al., 1992; Lustig et al., 1990; Wotton and Shore, 1997). The Rap1-Rif1-Rif2 complex creates a negative feedback loop that regulates telomere length (Levy and Blackburn, 2004; Marcand et al., 1997; Wotton and Shore, 1997), but the molecular details of this feedback mechanism are not fully understood.
In this study, we defined the functions of Rif1 and Rif2 using a system that tethers telomeric proteins adjacent to non-telomeric DNA ends. We show that Rif1 and Rif2 proteins inhibit Tel1 association, but not MRX association, with non-telomeric DNA ends. Rif2 interacts with the Xrs2 C-terminus to inhibit MRX-Tel1 interaction, whereas Rif1 appears to act through a different mechanism. We also show that the MRX complex does not efficiently associate with Rap1-bound DNA ends in the absence of Tel1. These data provide a model in which Rif1 and Rif2 cooperate with Rap1 in regulation of MRX and Tel1 localization at telomeres.
Results
Decreased MRX and Tel1 accumulation at DNA ends containing a longer TG sequence
The 81 bp TG81 sequence acts as a seed for the addition of telomere sequence at DNA ends (Diede and Gottschling, 1999, 2001). Although the TG81 sequence decreases Mec1 localization, it does not affect Tel1 localization to the nearby DNA end (Hirano and Sugimoto, 2007). Indeed, Tel1 associates effectively with short telomeres (Bianchi and Shore, 2007; Chang et al., 2007; Hector et al., 2007; Sabourin et al., 2007; Viscardi et al., 2007). To understand the mechanism by which longer telomeric sequence prevents Tel1 from localizing to DNA ends, we placed two copies of TG81 (the TG162 sequence) in tandem adjacent to the HO cleavage site (Fig. 1A). We first compared telomere addition at TG81 and TG162 ends (Fig. 1B). Cells transformed with the GAL-HO plasmid were grown in sucrose to prevent activation of the GAL promoter and arrested with nocodazole at G2/M. After arrest, cells were incubated with galactose to induce HO expression. DNA samples were collected at various times and examined by Southern blot analysis. DSBs were generated within one hour after HO expression, as shown by the appearance of cleaved fragments (CUT) and disappearance of intact DNA fragments (PRE). As found previously (Hirano and Sugimoto, 2007), the CUT fragments were elongated if the DNA end contained the TG81 sequence. However, no telomere addition was detected at TG162 ends over the course of the experiment. This observation is consistent with the recent finding that 125 bp or shorter telomeres are elongated more extensively in vivo (Chang et al., 2007).
Fig. 1. Effect of the 162 bp TG repeats on HO-induced DNA ends.
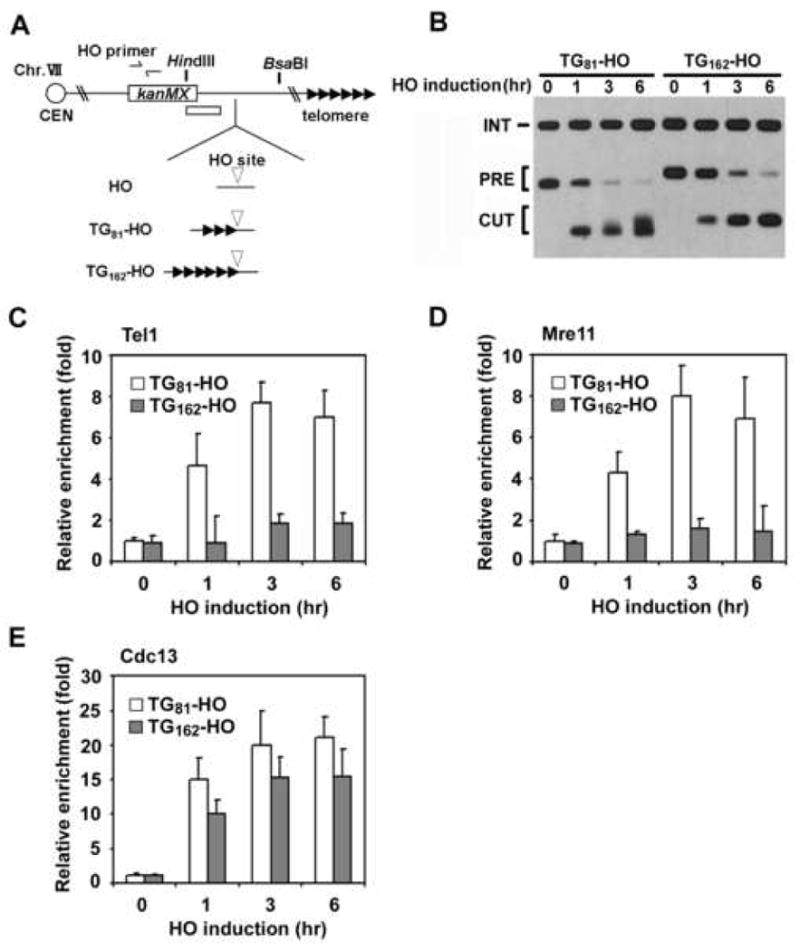
(A) Schematic of the HO cleavage site and the TG repeats at the ADH4 locus on chromosome VII-L. In HO cells, the ADH4 locus was replaced with the cassette containing the KanMX gene and an HO cleavage site (inverted triangle). 81 bp or 162 bp of TG repeat sequence (three or six repetitive arrowheads) was placed centromere-proximal to the HO site in TG81-HO cells or TG162-HO cells, respectively. Centromere is shown as a circle on the left (CEN). The white bar indicates a probe used to monitor telomere synthesis by Southern blot. The HO primer pair was designed to amplify a region 1 kb away from the HO site.
(B) Effect of the 162 bp TG repeats on telomere addition. TG81-HO and TG162-HO cells were transformed with the GAL-HO plasmid. Transformed cells were grown in sucrose and synchronized at G2/M with nocodazole. After arrest, galactose was added to the culture to induce HO expression. Aliquots of cells were collected at the indicated times after HO expression. Genomic DNA was digested with HindIII and BsaBI and then analyzed by Southern blot using the probe shown in A. The band labeled PRE indicates the 1 kb or 1.1 kb HindIII-BsaBI fragment containing the HO site. After cleavage with HO, this band is converted into a new band (0.6 or 0.7 kb, marked CUT). Telomere addition retards migration of the CUT fragment. The probe also detects a 1.8 kb HindIII fragment from the SMC2 locus on chromosome VI. This band is marked INT and serves as a loading control.
(C-E). Association of Tel1, Mre11 and Cdc13 with TG81 or TG162 ends. TG81-HO and TG162-HO cells expressing Tel1-HA (C), Mre11-myc (D) or Cdc13-myc (E) were transformed with the GAL-HO plasmid, and cultured as in B. Aliquots of cells were collected at the indicated times after HO expression and subjected to ChIP assay. Co-precipitated DNA was analyzed by real-time PCR using the HO primer pair shown in A and the SMC2 primer pair for the control SMC2 locus on chromosome VI. Relative enrichment was determined from three independent experiments, and bars represent averages with standard deviations indicated by lines above.
Tel1 associates with short telomeres in an MRX-dependent manner and promotes telomere addition at DNA ends (Bianchi and Shore, 2007; Chang et al., 2007; Hector et al., 2007; Sabourin et al., 2007). We next compared Mre11 and Tel1 binding to TG81 and TG162 ends (Fig. 1C and 1D). As found previously (Hirano and Sugimoto, 2007), Mre11 and Tel1 associated efficiently with TG81 ends. However, Mre11 and Tel1 association was greatly reduced at TG162 ends. MRX is required for efficient ssDNA accumulation at DNA ends (Lee et al., 1998; White and Haber, 1990). Cdc13 proteins bind to telomeric ssDNA for end protection and telomere addition (Lin and Zakian, 1996; Nugent et al., 1996; Pennock et al., 2001). We expected that Cdc13 accumulation would also be decreased at TG162 ends compared with TG81 ends. However, Cdc13 associated similarly with TG81 and TG162 ends (Fig. 1E), suggesting that high accumulation of the MRX complex is not required for creation of ssDNA tracts at TG162 ends. Cdc13 binds to normal length telomeres as efficiently as short telomeres (Bianchi and Shore, 2007; Chang et al., 2007; Sabourin et al., 2007). These results suggest that TG162 ends are converted to DNA ends that behave similarly to a normal length telomere.
Effect of rif1Δ or rif2Δ mutation on MRX and Tel1 accumulation at TG162 ends
Rif1 and Rif2, which interact with the C-terminus of Rap1, are known to act synergistically as negative regulators of telomere elongation (Hardy et al., 1992; Wotton and Shore, 1997). However, increased telomere elongation at TG81 ends was detected in rif2Δ cells, whereas telomere addition at TG81 ends was not observed in rif1Δ cells (data not shown; Diede and Gottschling, 1999; Frank et al., 2006). We therefore tested the effects of the rif1Δ or rif2Δ mutations on telomere synthesis on TG162 ends (Fig. 2A). Telomere synthesis was restored at TG162 ends in rif1Δ or rif2Δ cells, although the rif1Δ mutation exhibited a weak effect. In addition, telomere addition in rif1Δ rif2Δ double mutants was significantly greater than in either of the single mutants.
Fig. 2. Effect of rif1Δ or rif2Δ mutation on association of Tel1 and Mre11 with TG162 ends.
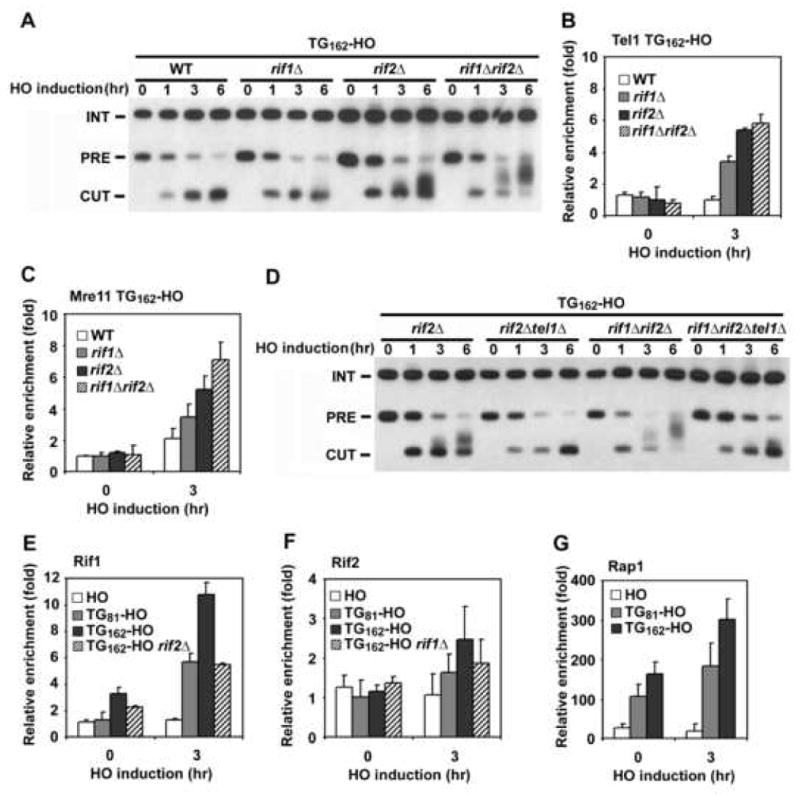
(A) Effect of rif1Δ or rif2Δ mutation on telomere addition at TG162 ends. TG162-HO cells were analyzed by Southern blot as in Fig. 1B.
(B, C) Effect of rif1Δ or rif2Δ mutation on association of Tel1 and Mre11 with TG162 ends. TG162-HO cells expressing Tel1-HA (B) or Mre11-myc (C) were analyzed by ChIP assay as in Fig. 1C.
(D) Effect of tel1Δ mutation on telomere addition at TG162 ends in rif2Δ or rif1Δ rif2Δ mutants. TG162-HO cells were analyzed by Southern blot as in A.
(E) Association of Rif1 with TG81 or TG162 ends. HO, TG81-HO, TG162-HO and TG162-HO rif2Δ cells expressing Rif1-HA were analyzed by ChIP assay as in B.
(F) Association of Rif2 with TG81 or TG162 ends. HO, TG81-HO, TG162-HO and TG162-HO rif1Δ cells expressing Rif2-HA were analyzed by ChIP assay as in B.
(G) Association of Rap1 with TG81 or TG162 ends. HO, TG81-HO, TG162-HO and TG162-HO cells expressing Rap1-HA were analyzed by ChIP assay as in B.
We then examined the effect of rif1Δ or rif2Δ mutation on Mre11 and Tel1 localization to TG162 ends (Fig. 2B and 2C). Consistent with a model in which MRX and Tel1 act in the same pathway and control telomere length (Ritchie and Petes, 2000), rif1Δ or rif2Δ mutations increased association of Mre11 and Tel1 proteins with TG162 ends. Again, the rif2Δ mutation conferred stronger effects than the rif1Δ mutation. The rif1Δ rif2Δ double mutation exhibited greater effects on Mre11 association than either of the singe mutations, whereas it did not apparently enhance Tel1 association compared with the rif2Δ single mutation. Tel1 accumulation might be therefore saturated at TG162 ends in the absence of Rif2. Consistent with inhibitory roles of Rif1 and Rif2 in Tel1 binding, deletion of TEL1 significantly decreased telomere addition in rif2Δ single or rif1Δ rif2Δ double mutants (Fig. 2D). Thus, Rif1 and Rif2 are involved in attenuation of Tel1 and MRX localization to TG162 ends.
Since Rif1 and Rif2 play inhibitory roles at TG82 and TG162 ends, Rif1 and Rif2 should associate with these ends. We therefore addressed whether Rif1 or Rif2 binds to the TG81 and TG162 sequences before or after DSB induction (Fig. 2E and 2F). As a control, we analyzed the localization of Rif1 or Rif2 proteins to HO-induced DNA ends containing no TG sequence (HO ends). Rif1 associated with the TG162 sequence more efficiently than the TG81 sequence. Association of Rif1 with the TG162 repeat was detected before HO expression and increased three-fold after HO expression. Rif1 association is in part dependent on Rif2, because Rif1 association was decreased in the absence of Rif2. No apparent Rif2 binding was detected at the TG162 sequence before HO expression (Supp. Fig. 1S), and unlike Rif1, Rif2 was only modestly enriched at the TG sequence following HO expression. Rif2 association was also TG-length dependent; Rif2 bound more abundantly to TG162 than to TG81. However, these results do not exclude the possibility that Rif2 binds to the TG tracts before HO expression, as previous studies have shown that detection of Rif2 association with telomeres by ChIP assay is lower compared with that of Rif1 association (Sabourin et al., 2007; Smith et al., 2003). Rap1 binds double-stranded TG repeats and recruits Rif1 and Rif2 to telomeric DNA regions (Conrad et al., 1990; Hardy et al., 1992; Lustig et al., 1990; Wotton and Shore, 1997). We found that Rap1 associated with the TG81 and TG162 repeats before HO expression and that Rap1 association at the TG162 increased two-fold after HO expression (Fig. 2G). Not all proteins bind to the binding sites more efficiently after DSB induction nearby. For example, association of LacI proteins with the lacO sequence was not affected after HO expression (Supp. Fig. 2S). No significant association of Rap1, Rif1 or Rif2 proteins was detected at HO ends (Fig. 2E, 2F and 2G). Together, these results support the idea that Rif1 and Rif2 promote the inhibition of MRX and Tel1 binding at telomeric DNA ends.
Inhibition of Tel1 localization to DNA ends by tethering of Rif1 or Rif2 nearby
The above results indicated that Rif1 and Rif2 are required to decrease MRX and Tel1 association at DNA ends containing TG sequences. However, it is not clear whether Rif1 or Rif2 directly inhibits localization of the MRX complex or Tel1. Since Rif1 and Rif2 proteins localize to double-stranded TG sequences in a Rap1-dependent manner, they could stimulate functions of Rap1 proteins and vice versa. To further explore Rif1 and Rif2 function, we set up a system to tether Rif1 and Rfi2 proteins adjacent to DSBs independent of Rap1 (Fig. 3A). We constructed TetR-Rif1 and TetR-Rif2 fusion genes, both of which were found to be fully functional. TetR-Rif1 and TetR-Rif2 restored wild-type telomere length to rif1Δ and rif2Δ mutants, respectively (data not shown). To target these chimeric proteins, we placed different numbers of the TetR-binding sequence (TetO) adjacent to the HO cleavage site. We observed that TetR fusion protein binding is proportional to number of the TetO sequences (Supp. Fig. 3S). To exclude the possibility that Rap1 is recruited by interacting with tethered Rif1 or Rif2, we introduced a C-terminal truncation rap1 (rap1-ΔC) mutation into the strain. We first examined whether expression of TetR-Rif1 or TetR-Rif2 fusion proteins inhibits Mre11 and Tel1 binding to DNA ends containing the 8×TetO repeat (TetO8 ends) (Fig. 3B and Fig. 3C). Both TetR-Rif1 and TetR-Rif2 fusion proteins decreased Tel1 binding to TetO8 ends (Fig. 3B). Curiously, however, neither TetR-Rif1 nor TetR-Rif2 fusion proteins interfered with Mre11 binding (Fig. 3C). No inhibition of Tel1 binding was detected if cells contained no TetO repeat near the HO cleavage site (Supp. Fig. 4S) or expressed the TetR protein by itself (Supp. Fig. 5S). Tel1 localizes to DSBs by interacting with the C-terminus of Xrs2 (Nakada et al., 2003a). It is therefore possible that Rif1 or Rif2 disrupts Tel1 localization by inhibiting MRX complex formation. However, neither TetR-Rif1 nor TetR-Rif2 decreased Xrs2 association with TetO8 ends (Fig. 3D), suggesting against this possibility. Furthermore, this Rif1/Rif2-dependent inhibition appears to be specific to Tel1, as no inhibition of Mec1 was observed (Supp. Fig. 6S). Together, these results indicate that tethering of Rif1 and Rif2 near DSB ends inhibited Tel1 accumulation but not MRX accumulation. We further addressed whether there was a dosage effect of TetR-Rif1 or TetR-Rif2 by examining Tel1 and Mre11 binding at 4×TetO (TetO4) or 2×TetO (TetO2) ends (Fig. 3E and 3F). Both TetR-Rif1 and TetR-Rif2 decreased Tel1 binding at TetO4 ends as efficiently as at TetO8 ends, whereas neither of them inhibited Tel1 binding at TetO2 ends. No attenuation of Mre11 binding by TetR-Rif1 or TetR-Rif2 was detected at TetO2 or TetO4 ends (Supp. Fig. 7S). These results suggest that Rif1 and Rif2, once recruited to DNA ends, can operate independently of Rap1, and support the view that Rif1 and Rif2 act as dosage-dependent inhibitors of telomere lengthening (Levy and Blackburn, 2004). Our results, however, do not exclude the possibility that Rap1-recruited Rif1 and Rif2 proteins can directly decrease MRX binding at DNA ends.
Fig. 3. Effect of Rif1 or Rif2 tethering on association of Tel1 and Mre11 with nearby DSBs.
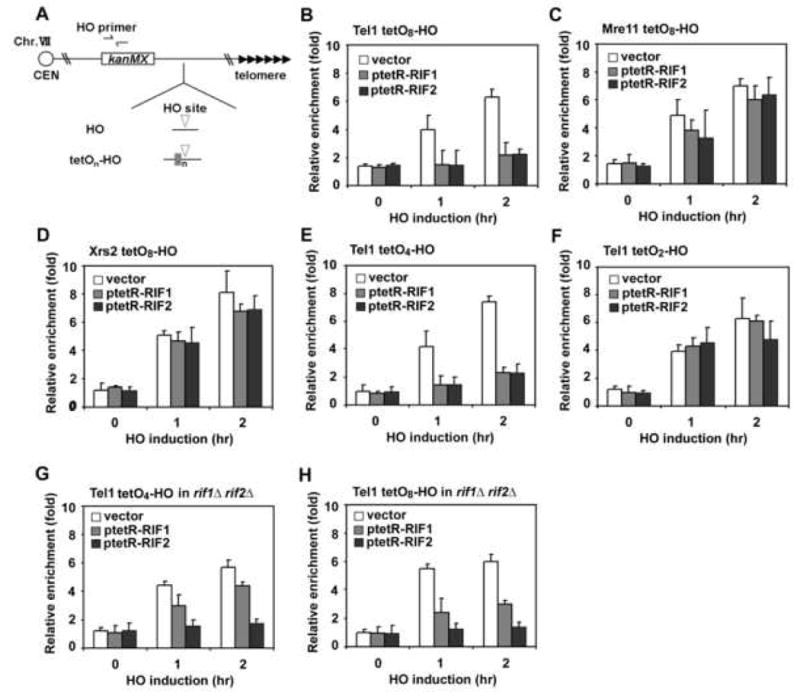
(A) Schematic of the HO cleavage site and the adjacent TetO arrays at the ADH4 locus on chromosome VII-L. The ADH4 locus was replaced with the cassettes containing the KanMX gene, an HO cleavage site (inverted triangle) and different copies (n) of the TetO sequence (a grey box). The RAP1 gene was replaced with a rap1-ΔC mutation. The HO primer pair was designed to amplify a region 1 kb away from the HO site (see Fig. 1A).
(B, C) Effect of TetR-Rif1 or TetR-Rif2 expression on association of Tel1 and Mre11 with TetO8 ends. TetO8-HO cells expressing Tel1-HA (B) or Mre11-myc (C) were transformed with ptetR-RIF1, ptetR-RIF2 or the control vector, together with the GAL-HO plasmid, and analyzed by ChIP assay as in Fig. 1C.
(D) Effect of TetR-Rif1 or TetR-Rif2 expression on association of Xrs2 with TetO8 ends. TetO8-HO cells expressing Xrs2-myc were analyzed as in B.
(E, F) Effect of TetR-Rif1 or TetR-Rif2 expression on association of Tel1 with TetO2 ends or TetO4 ends. TetO4-HO (E) or TetO2-HO cells (F) expressing Tel1-HA were analyzed by ChIP assay as in B.
(G, H) Effect of TetR-Rif1 or TetR-Rif2 expression on association of Tel1 in the absence of Rif1 and Rif2. TetO4-HO rif1Δ rif2Δ (G) or TetO8-HO rif1Δ rif2Δ cells (H) expressing Tel1-HA were analyzed as in B.
As shown above, Rif1 recruitment to TG ends partially depends on Rif2 function (see Fig. 2E). Therefore, inhibition of Tel1 binding by Rif1 might require Rif2 function, and vice versa. To uncover Rif1- or Rif2-specific functions, we investigated the effect of TetR-Rif1 or TetR-Rif2 at TetO4 or TetO8 ends in rif1Δ rif2Δ double mutants (Fig. 3G and 3H). Inhibition of Tel1 binding by TetR-Rif1 was attenuated in rif1Δ rif2Δ mutant cells. Attenuation of the inhibition was more noticeable at TetO4 ends than at TetO8 ends. Introduction of the rif1Δ mutation did not alter TetR-Rif1-dependent inhibition, whereas the rif2Δ mutation exhibited the same effect as the rif1Δ rif2Δ mutation (Supp. Fig. 8S). In contrast, TetR-Rif2-dependent inhibition was not affected by the rif1Δ rif2Δ mutation. Thus, inhibition by TetR-Rif1 becomes dependent on Rif2 at DNA ends containing a short TetO array, although TetR-Rif2 exerts its function independently of Rif1. These results also suggest that Rif1 is a weaker inhibitor of Tel1 binding than Rif2.
Effect of MRX-Tel1 interaction on MRX accumulation at TG ends
Tethered Rif1/Rif2 proteins decreased Tel1 binding but not MRX binding to DSB ends. In contrast, Rif1/Rif2 proteins are required for delocalization of both Tel1 and MRX from TG ends. One explanation could be that localization of MRX to TG ends is dependent on Tel1. We therefore compared Mre11 accumulation at HO ends and TG81 ends in the absence of Tel1 (Fig. 4A and 4B). Mre11 associated with HO-ends in tel1Δ mutants as efficiently as in wild-type cells. Although detectable, Mre11 association with TG81 ends was significantly decreased in tel1Δ mutants. It is possible that Tel1-dependent phosphorylation regulates association of MRX with TG81 ends. However, Mre11 association was not lost in a strain harboring a kinase-deficient tel1 (tel1-KN) mutation (Fig. 4C). As kinase activity is dispensable for Tel1 localization to short telomeres (Hector et al., 2007), the tel1-KN mutation did not affect Tel1 localization to TG81 ends (Fig. 4D). We also addressed whether Tel1 is required for efficient MRX localization to actual telomeric ends. Similarly to Tel1, the MRX complex binds preferentially to shortened telomeres (Viscardi et al., 2007). Telomere shortening occurs equally in cells carrying a tel1Δ or tel1-KN mutation (Supp. Fig. 9S)(Greenwell et al., 1995; Mallory and Petes, 2000). We therefore compared the association of Mre11 with telomeres VI-R and XV-L in tel1Δ and tel1-KN mutants (Fig. 4E and 4F). Mre11 association was increased in both tel1Δ and tel1-KN mutants compared with wild-type cells. However, MRX association was more pronounced in tel1-KN mutants than in tel1Δ mutants. It seems unlikely that the tel1Δ or tel1-KN mutation increases MRX binding at DNA ends containing longer TG tracts, because introduction of the tel1Δ mutation did not increase Mre11 localization to TG162 ends (Supp. Fig. 10S). These results indicate that Tel1 binding promotes MRX accumulation at telomeric DNA ends, and explain why Rif1 and Rif2 are required for inhibition of MRX association with TG ends.
Fig. 4. Effect of Tel1-MRX interaction on Mre11 association with telomeric DNA ends.
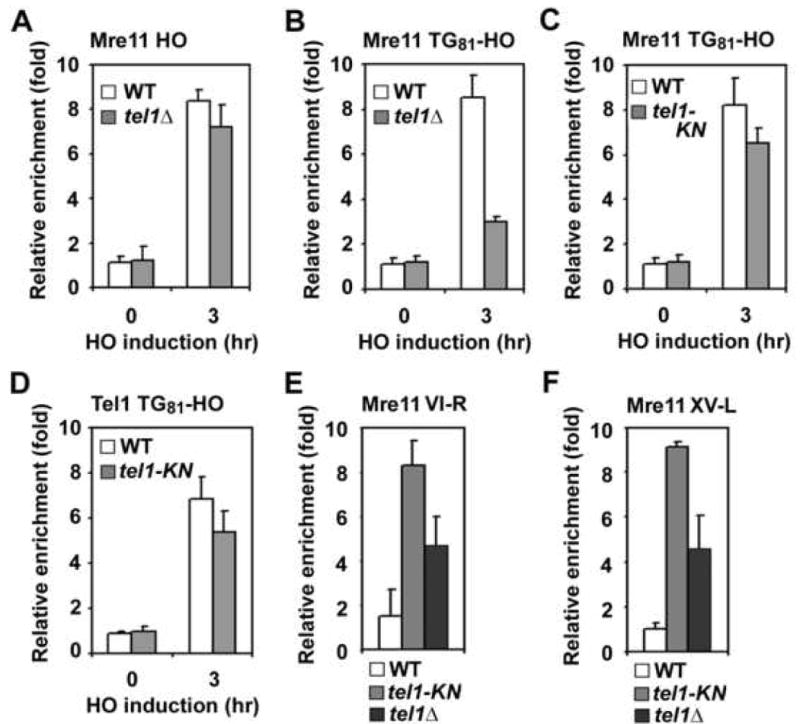
(A, B) Effect of tel1Δ mutation on Mre11 association with HO-induced DSBs or TG81 ends. HO (A) or TG81-HO (B) cells expressing Mre11-myc were analyzed by ChIP assay as in Fig. 1C. Strains used carried the wild-type TEL1 gene or tel1Δ mutation.
(C, D) Effect of tel1-KN mutation on Mre11 association with TG81 ends. TG81-HO cells expressing Mre11-myc and wild-type Tel1-HA or mutant Tel1-KN-HA proteins were analyzed by ChIP assay as in A to monitor Mre11 association (C) or Tel1 association (D).
(E, F) Effect of tel1-KN or tel1Δ mutation on Mre11 association with the telomere VI-R or XV-L. Wild-type, tel1-KN or tel1Δ cells expressing Mre11-myc were synchronized at G2/M with nocodazole and subjected to ChIP assay as in A. Mre11 association with the telomere VI-R (E) or XV-L (F) were monitored.
Rap1-dependent inhibition of MRX accumulation at DNA ends in tel1Δ mutants
Since Rap1 directly binds telomeric DNA sequences, it is possible that Rap1 decreases localization of MRX to DNA ends in the absence of Tel1. We investigated whether the TetR-Rap1 fusion protein decreases Mre11 association with a DSB near the TetO sequence (Fig. 5). Rap1 contributes to the formation of heterochromatin by interacting with Sir2, Sir3 and Sir4 proteins (Grunstein, 1997). We introduced a sir2Δ mutation in TetO8 cells, because expression of TetR-Rap1 partially decreased efficiency of HO cleavage at the TetO8-HO cassette in a SIR2-dependent manner (data not shown). The C-terminus of Rap1 interacts with and recruits Rif1 and Rif2 (Conrad et al., 1990; Hardy et al., 1992; Lustig et al., 1990; Wotton and Shore, 1997). As shown above, tethered Rif1 or Rif2 protein decreases Tel1 localization to adjacent DNA ends. TetR-Rap1 might recruit Rif1 or Rif2 protein and thereby decrease MRX association with the DNA end. As expected, TetR-Rap1 expression partially reduced Mre11 association with TetO8 ends in wild-type cells, whereas no reduction was observed in rif1Δ rif2Δ mutants (Fig. 5A). Likewise, TetR-Rap1 expression diminished Tel1 binding in wild-type cells but not in rif1Δ rif2Δ mutants (Supp. Fig. 11S). We then examined the effect of TetR-Rap1 expression on MRX association in tel1Δ mutants (Fig. 5B). Mre11 association was further decreased in tel1Δ mutants compared with wild-type cells. The observed TetR-Rap1-mediated effect in tel1Δ mutants did not require Rif1 or Rif2 function; Mre11 association was decreased similarly in tel1Δ single and tel1Δ rif1Δ rif2Δ triple mutants (Fig. 5B). Thus, MRX associates less efficiently with Rap1-bound DNA ends in tel1Δ mutants. As discussed above, Rif1 and Rif2 attenuate Tel1 association with adjacent DNA ends. Together, these results support the idea that once Rif1 and Rif2 delocalize Tel1, Rap1 decreases MRX binding at telomeric DNA ends.
Fig. 5. Inhibitory effect of TetR-Rap1 expression on Mre11 accumulation at TetO8 ends.
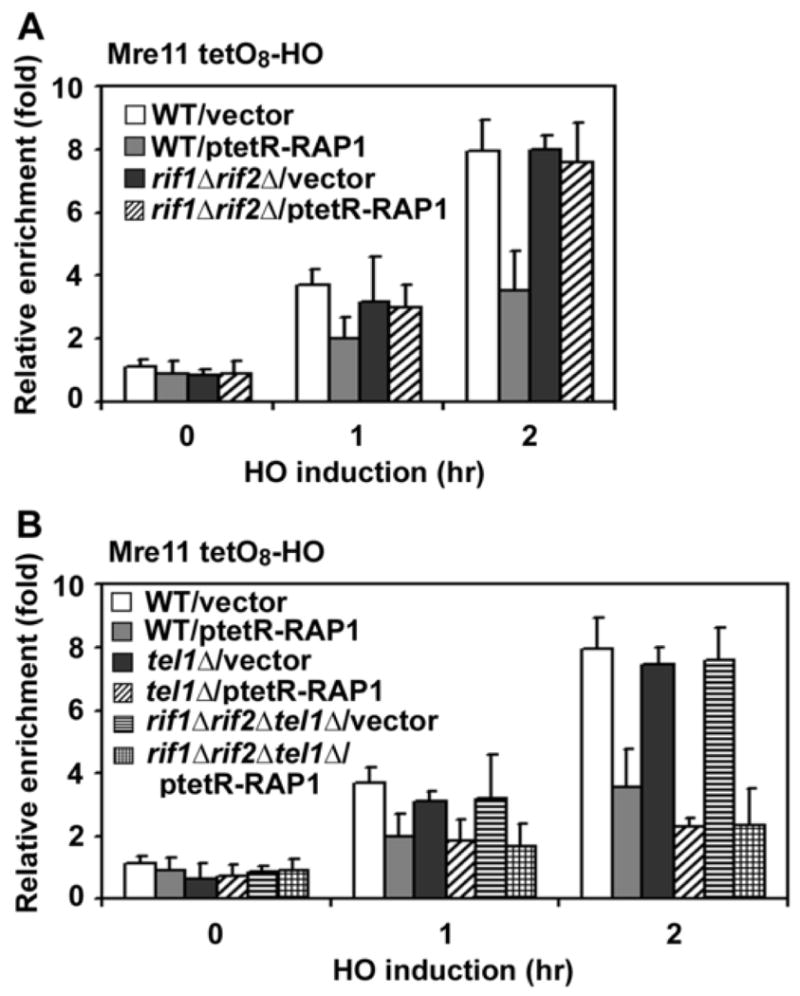
(A) Effect of TetR-Rap1 expression on Mre11 association in rif1Δ rif2Δ cells. TetO8-HO or TetO8-HO rif1Δ rif2Δ cells expressing Mre11-myc were transformed with ptetR-RAP1 or the control vector, together with the GAL-HO plasmid, and analyzed as in Fig. 1C to monitor Mre11 association. The strains used here contained a sir2Δ mutation to suppress inefficient HO cleavage.
(B) Effect of TetR-Rap1 expression on Mre11 association in tel1Δ or tel1Δ rif1Δ rif2Δ cells. TetO8-HO, TetO8-HO tel1Δ or TetO8-HO tel1Δ rif1Δ rif2Δ cells expressing Mre11-myc were examined as in A.
Rif2 binds to the C-terminus of Xrs2 and inhibits the Tel1-Xrs2 interaction
To further understand the mechanism by which Rif1/Rif2 proteins inhibit Tel1 localization to DNA ends, we attempted to determine biochemical activities of Rif1 and Rif2 proteins. Tel1 interacts with the C-terminus of Xrs2 to localize to DNA ends (Nakada et al., 2003a). We therefore examined whether Rif1 or Rif2 interacts with the Xrs2 C-terminus by pull-down assay (Fig. 6A). A GST fusion of the Xrs2 C-terminus (GST-Xrs2C) or GST alone was captured on glutathione beads and incubated with extracts from cells expressing HA-tagged Rif1 or Rif2 proteins. Bound proteins were then analyzed by immunoblotting with anti-HA antibodies. Rif2-HA was precipitated by GST-Xrs2C but not GST alone. In contrast, no specific interaction between Rif1 and the Xrs2 C-terminus was detected. Furthermore, Rif1 was not precipitated with full-length Xrs2 proteins (data not shown). Thus, Rif2 interacts with the C-terminus of Xrs2, whereas Rif1 does not.
Fig. 6. Interaction of Rif2 with Xrs2 C-terminus.
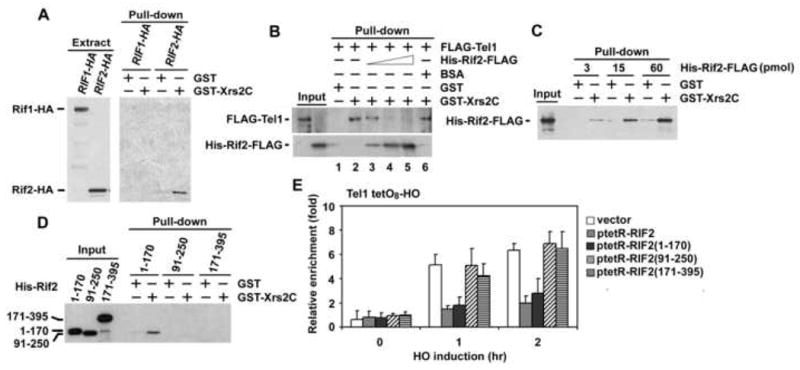
(A) Interaction of Rif1 and Rif2 with Xrs2 C-terminus. GST or GST-Xrs2C was immobilized on CNBr-activated Sepharose, and incubated with yeast extracts (2 mg of protein) containing Rif1-HA or Rif2-HA. One fifth of the eluted proteins were resolved on a SDS-gel for immunoblotting analysis with anti-HA antibodies (Pull-down), whereas cell extracts (7 μg of protein) were analyzed as control.
(B) Effect of Rif2 on Tel1 interaction with the C-terminus of Xrs2. FLAG-Tel1 or His-Rif2-FLAG was incubated with GST or GST-Xrs2C and glutathione beads. Proteins bound to glutathione beads were subjected to immunoblotting analysis with anti-FLAG antibodies. 0.1 pmol of FLAG-Tel1 and 3 pmol of GST or GST-Xrs2C were used for the binding assay. His-Rif2-FLAG was added in the reaction at the dose of 3, 15 or 60 pmol (lanes 3-5). 3 μg of BSA, which is equivalent to 60 pmol of His-Rif2-FLAG, was added in the reaction as a control (lane 6).
(C) Interaction of Rif2 with the C-terminus of Xrs2. The same amounts of His-Rif2-FLAG as those used in B were incubated with 3 pmol of GST or GST-Xrs2C, and pulled down with glutathione beads. Bound proteins were analyzed by immunoblotting with anti-FLAG antibodies.
(D) Interaction of the Rif2 N-terminus with the Xrs2 C-terminus. 3 pmol of His-Rif2 fragments were incubated with 3 pmol of GST or GST-Xrs2C and then precipitated by glutathione beads. All the His-tagged proteins contain a T7 epitope at the N-terminus. Bound proteins were analyzed by immunoblotting with anti-T7 antibodies.
(E) Association of Tel1 with TetO8 ends in cells expressing TetR-Rif2 fusion proteins. TetO8-HO cells expressing Tel1-HA were transformed with ptetR-RIF2, ptetR-RIF2(1-170), ptetR-RIF2(91-250), ptetR-RIF2(171-395) or the control vector, together with the GAL-HO plasmid, and analyzed by ChIP assay as in Fig. 1C.
It is possible that Rif2 interacts directly with the Xrs2 C-terminus and inhibits the Tel1-Xrs2 interaction. To test this hypothesis, we first set up an in vitro assay to detect the Tel1-Xrs2 interaction (Fig. 6B). FLAG-tagged Tel1 protein was incubated with GST-Xrs2C or GST alone and precipitated by glutathione beads. Bound proteins were then analyzed by immunoblotting with anti-FLAG antibodies. Consistent with a model in which Tel1 interacts with the C-terminus of Xrs2, Tel1 protein was specifically pulled-down by the C-terminus of Xrs2. We then asked whether Rif2 directly inhibits the Tel1-Xrs2 interaction in vitro (Fig. 6B). We purified His-FLAG-tagged Rif2 proteins to near homogeneity from E. coli by tandem affinity column chromatography (Supp. Fig. 12S). Inclusion of Rif2 decreased Tel1-Xrs2 interaction, whereas control BSA had no effect. Moreover, higher concentrations of Rif2 increased the Rif2-Xrs2 interaction. We also found that Rif2 interacts with GST-Xrs2C but not GST alone (Fig. 6C). Thus, Rif2 competes with Tel1 for binding to the Xrs2 C-terminus in vitro.
We mapped the region of Rif2 involved in Xrs2 binding (Fig. 6D). Purified N-terminal (amino acids 1-170), central (amino acids 91-250) or C-terminal (amino acids 171-395) fragments of Rif2 protein were incubated with GST-Xrs2C or GST and interaction was analyzed by pull-down assay as above. The N-terminal fragment was found to bind to the Xrs2 C-terminus. However, no interaction with Xrs2 was observed for the central or C-terminal fragment. We also found that tethering of the Rif2 N-terminus inhibited Tel1 localization to DNA ends, whereas the central or C-terminal region did not (Fig. 6E). These results indicate that the N-terminus of Rif2 plays a key role in the interaction with the Xrs2 C-terminus. Taken together, our results support a model in which Rif2 binds to the Xrs2 C-terminus and blocks the recruitment of Tel1 to the MRX complex at DNA ends.
Discussion
As natural DNA ends, telomeres must escape recognition by checkpoint proteins that activate the DNA damage response pathway. In budding yeast, Cdc13-mediated telomere capping inhibits Mec1 localization to the telomeric DNA end, but does not affect Tel1 accumulation. Recent evidence supports a model in which Tel1 preferentially associates with short telomeres and promotes telomere synthesis. However, it has not been determined how Tel1 accumulation at extended telomeres is inhibited. Rap1 binds double-stranded TG repeats and recruits Rif1 and Rif2 to telomeres via its C-terminal domain (Conrad et al., 1990; Hardy et al., 1992; Lustig et al., 1990; Wotton and Shore, 1997). In this study, we show that binding of multiple Rif1 and Rif2 proteins decreases localization of Tel1 to adjacent DNA ends through two different mechanisms. First, Rif1 and Rif2 inhibit recruitment of Tel1 to MRX at DNA ends. Second, in the absence of Tel1, Rap1 attenuates MRX association with DNA ends. Together, these results indicate that Rif1 and Rif2 co-operate with Rap1 and decrease Tel1 accumulation at extended telomeres (Fig. 7).
Fig. 7. Model for localization of MRX and Tel1 to telomeric DNA ends.
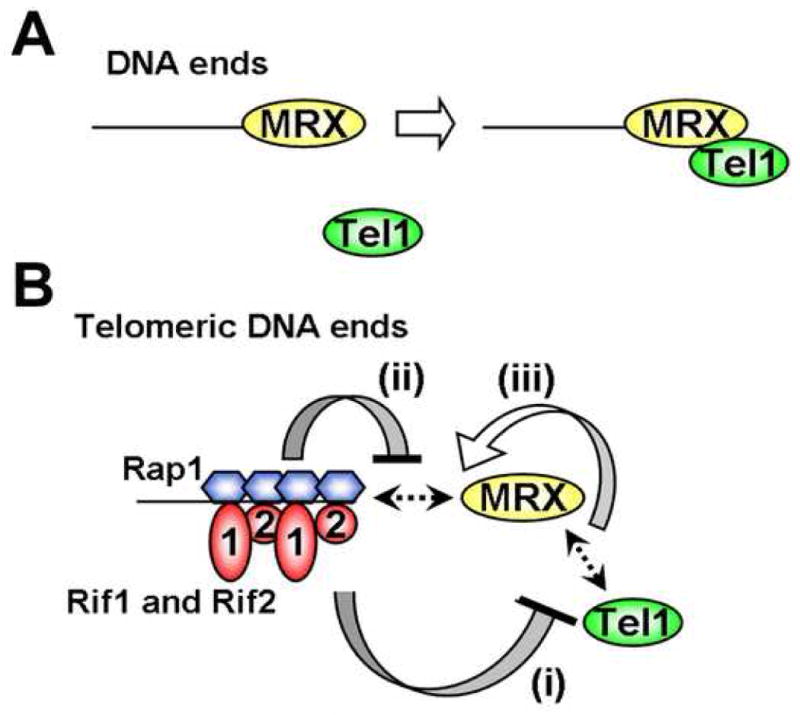
(A) MRX-dependent localization of Tel1 to DNA ends. MRX recognizes DNA ends and then recruits Tel1 to the DNA ends. MRX associates with DNA ends independently of Tel1.
(B) Inhibitory network for MRX and Tel1 localization at telomeric DNA ends.
(i) Rif1 and Rif2 by themselves inhibit MRX-dependent Tel1 localization to DNA ends. Rif2 competes with Tel1 for binding to the C-terminus of Xrs2. It is not known how Rif1 inhibits Tel1 localization to DNA ends.
(ii) MRX does not efficiently localize to Rap1-bound DNA ends in the absence of Tel1.
(iii) MRX-Tel1 interaction could stabilize MRX association with Rap1-bound DNA ends. If Rif1 and Rif2 disrupt Tel1-MRX interaction at DNA ends, MRX could no longer associate efficiently with Rap1-bound DNA ends.
Tel1 localizes to DNA ends by interacting with the C-terminus of Xrs2 (Nakada et al., 2003a; Sabourin et al., 2007). It is possible that Rif1 and Rif2 decrease Tel1 localization to DNA ends by interfering with the Tel1-Xrs2 interaction. Supporting this hypothesis, Rif2 directly binds to the C-terminus of Xrs2 and inhibits the Tel1-Xrs2 interaction in vitro (see Fig. 6). Even though Rif1 and Xrs2 interact with each other, the Rif2-Xrs2 interaction may be weak compared with the Tel1-Xrs2 interaction. For example, MRX recruits Tel1, but not Rif2, to DSBs (see Fig. 2). Moreover, TetR-Rif2 does not recruit MRX to a region adjacent to the TetO8 sequence (see Fig. 3). These observations support a model in which Rif2 inhibits the Tel1-Xrs2 interaction by covering the Tel1 binding site on Xrs2 if multiple Rif2 proteins are placed nearby. It has not been determined whether Rif2 and Tel1 interact with the same sequence on the Xrs2 C-terminus; it therefore remains possible that Rif2 recognizes a different sequence from the Tel1 binding site and stimulates dissociation of Tel1 from MRX at telomeric ends. Interaction of Rif1 with Xrs2 has not been detected; thus, it remains unclear how Rif1 inhibits Tel1 localization at DNA ends.
Rif1 and Rif2 appear to have different functions, consistent with the fact that they are unrelated. First, Rif2 is a more potent inhibitor of Tel1 localization than Rif1, as shown by the observation that TetR-Rif2 fusion decreases Tel1 localization at TetO ends more effectively than the TetR-Rif1 fusion (see Fig. 3G and 3H). Initial telomere addition occurs at TG162 ends more quickly in rif2Δ mutants than rif1Δ mutants (see Fig. 2A). However, like native telomeres, TG162 ends become longer in rif1Δ mutants than in rif2Δ mutants (data not shown; Wotton and Shore, 1997). Thus, Rif1 plays a more important role at longer TG repeats, suggesting that Rif1 acts in a more dosage-dependent manner than Rif2. Secondly, Rif1 function is partially dependent on Rif2; inhibition of Tel1 localization by TetR-Rif1 is decreased in the absence of Rif2 (see Fig. 3G and 3H). These different properties of Rif1 and Rif2 could explain why decreased binding of Rif2 alone leads to Tel1 enrichment at short telomeres (Sabourin et al., 2007). Several lines of evidence indicate that Rif1 association with telomeres fluctuates during cell-cycle progression, peaking at G2/M (Sabourin et al., 2007; Smith et al., 2003). Increased Rif1 binding at the TG162 sequence after DSB induction might result from telomere formation at the TG ends in G2/M-arrested cells (see Fig. 2E).
Rif1 homologs have been identified and characterized in fission yeast and human (Kanoh and Ishikawa, 2001; Silverman et al., 2004; Xu and Blackburn, 2004). Although human Rif1 is not involved in telomere length control, fission yeast Rif1 localizes to telomeres and negatively regulates telomere length, suggesting that Rif1 also controls Tel1 localization at telomeres in fission yeast. There are no recognizable Rif2 homologs in other eukaryotes. Even though the N-terminal half of Rif2 can interact with the Xrs2 C-terminus (see Fig. 6D), it does not share meaningful similarities with other proteins. The C-termini of Nbs1 and Xrs2 are involved in binding to ATM and Tel1, respectively (Falck et al., 2005; Nakada et al., 2003a; You et al., 2005). However, the Nbs1/Xrs2 proteins are not highly conserved compared with Mre11 and Rad50 proteins. It is possible that Rif2 has evolved by adapting to the diversification of Nbs1/Xrs2 proteins.
Rif1 and Rif2 inhibit Tel1 localization, but not MRX localization, to DNA ends independent of Rap1 function (see Fig. 3). In the absence of Tel1, however, Rap1 decreases MRX accumulation at DNA ends independent of Rif1 or Rif2 (see Fig. 5). It is therefore possible that once Rif1 and Rif2 delocalize Tel1 from Rap1-covered telomeric DNA ends, Rap1 could promote the dissociation of MRX from the ends. Since Rif2 interacts with the C-terminus of Xrs2, it might transiently tether MRX to telomeric DNA ends; however, Rap1 could subsequently remove. Supporting this model, Rif2 overexpression has been shown to inhibit MRX association with telomeres (Viscardi et al., 2007). Thus, the Rap1-Rif1-Rif2 complex appears to act as a highly organized machinery to remove MRX and Tel1 from extended telomeres. At present, it is not known why MRX requires Tel1 protein to associate efficiently with telomeric DNA ends. We speculate that MRX might associate more stably with histone-bound DNA than Rap1-covered DNA.
Rap1 can recruit Rif1 and Rif2 not only to telomeric DNA ends but also to intra-chromosomal regions (see Fig. 2E and 2F). On natural chromosomes, however, extensive Rap1 binding is observed mostly at telomeric DNA regions (Smith et al., 2003). Telomeres might possess specific chromatin structures that promote Rap1 binding, since Rap1 binding at TG repeats increases slightly after DSB induction (see Fig. 2G). Telomeres are extended to 4 kb, but not significantly longer, in cells expressing C-terminal truncated Rap1 proteins or in cells carrying rif1Δ rif2Δ double mutation (Kyrion et al., 1992; Wotton and Shore, 1997). It is thus possible that extensive binding of Rap1 on longer telomeres blocks MRX association before recruiting Tel1 to the DNA end. In this situation, Rif1 or Rif2 would not be required for inhibition of MRX localization. Consistent with this view, binding of multiple Rap1 proteins has been shown to decrease MRX accumulation at DNA ends independently of the Rap1 C-terminus (Negrini et al., 2007). In budding yeast, MRX plays an essential role in non-homologous end-joining (NHEJ) as well as homologous recombination repair of DSBs (D'Amours and Jackson, 2002; Krogh and Symington, 2004). In agreement with an inhibitory role of MRX localization, Rap1 prevents telomere fusions by NHEJ (Pardo and Marcand, 2005). Rap1 homologs have been identified as telomeric proteins across species (Smogorzewska and de Lange, 2004; Vega et al., 2003). Human RAP1 is also involved in protecting telomeres from NHEJ (Bae and Baumann, 2007). The conservation of Rap1/RAP1 suggests that Rap1/RAP1 proteins universally protect telomeres from DSB repair machinery.
In summary, we have shown that Rif1 and Rif2 cooperate with Rap1 to shelter telomeric DNA ends from Tel1 recognition. In mammals, the telomeric DNA binding protein TRF2 is involved in inhibition of ATM activation at telomeres (Karlseder et al., 1999). TRF2 interacts with the Mre11-Rad50-Nbs1 complex (Zhu et al., 2000) and recruits other telomere binding proteins to telomeres (Chen et al., 2008). It will be interesting to see whether telomere binding proteins play a similar role in inhibition of ATM localization in mammalian cells.
Experimental Procedures
Strains and plasmids
Cells containing the TG162-HO, tetO2-HO, tetO4-HO or tetO8-HO cassette were generated by the plasmid pTG162-HO, ptetO2-HO, ptetO4-HO or ptetO8-HO, respectively. Cells carrying the TG81-HO cassette were described previously (Hirano and Sugimoto, 2007). The rap1-ΔC strain was constructed by transformation with the PCR fragment amplified from pDL106 as described (Levy and Blackburn, 2004). All the strains used in this study are isogenic to KSC1516 (Nakada et al., 2003a). Details of the strain and plasmid construction are described in Supplemental Information.
Chromatin immunoprecipitation (ChIP) assay
Chromatin immunoprecipitation was performed using anti-HA (16B2) or anti-myc (9E10) antibodies as previously described (Hirano and Sugimoto, 2007). Quantification of immunoprecipitated DNAs was achieved by using a real-time PCR detection system (Bio-Rad). Relative enrichment was determined by normalizing signals from a region near the DSB (HO), telomere VI-R or telomere XV-L to control signals from a region in SMC2. The signals were then normalized to input signals for each primer set. Enrichment is defined as accumulation of proteins at the DSB or the telomere relative to that at the SMC2 locus. Sequence of the PCR primers is described in Supplementary Information.
Pull-down assay
FLAG-Tel1 proteins were purified from budding yeast cells. GST, GST-Xrs2C, His-Rif2-FLAG or His-Rif2 fragments were purified from E. coli. Details for purification were described in Supplementary information. The interaction of Rif1 or Rif2 with the Xrs2 C-terminus was examined using extracts from cells expressing Rif1-HA or Rif2-HA. Purified GST or GST-Xrs2C was coupled to CNBr-activated Sepharose 4B. Extracts were prepared in the lysis buffer by glass-bead beating as described in Supplementary information, and incubated with GST- or GST-Xrs2C-coupled Sepharose beads. Since Rif2-HA is expressed two-fold more than Rif1-HA, extracts from Rif2-HA expressing cells were mixed with extracts from rif2Δ cells. After incubation for 120 min at 4°C, Sepharose beads were washed three times and bound proteins were analyzed by immunoblotting. Interaction of purified Tel1 and Rif2 with Xrs2 was examined as follows. Purified GST or GST-Xrs2C was bound to glutathione sepharose beads and then incubated with purified FLAG-Tel1 or purified Rif2 proteins in the binding buffer (20 mM Tris-HCl at pH 8.0, 100 mM NaCl, 0.01% Triton X-100) at 4°C for 1 hr. Bound proteins were released from the beads by addition of glutathione (reduced form) at a final concentration of 10 mM, and characterized by immunoblotting analysis.
Other methods
Immunoblotting analysis was performed using anti-FLAG, anti-T7 or anti-HA antibodies as described (Hirano and Sugimoto, 2007). Southern blotting was performed as previously described (Hirano and Sugimoto, 2007).
Supplementary Material
Acknowledgments
We thank C. Newlon and H. Araki for critical reading and discussion; Y. Tanaka for construction of pGAL-FLAG-Tel1; D. Nakada for construction of pGST-XRS2C; H. Araki, E. Blackburn, A. Matsuura, D. Shore, A. Straight and V. Zakian for materials. This work was supported by National Institutes of Health grant GM073876.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes & Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- Bae NS, Baumann P. A RAP1/TRF2 complex inhibits nonhomologous end-joining at human telomeric DNA ends. Mol Cell. 2007;26:323–334. doi: 10.1016/j.molcel.2007.03.023. [DOI] [PubMed] [Google Scholar]
- Bianchi A, Shore D. Increased association of telomerase with short telomeres in yeast. Genes Dev. 2007;21:1726–1730. doi: 10.1101/gad.438907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M, Arneric M, Lingner J. Telomerase repeat addition processivity is increased at critically short telomeres in a Tel1-dependent manner in Saccharomyces cerevisiae. Genes Dev. 2007;21:2485–2494. doi: 10.1101/gad.1588807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Yang Y, van Overbeek M, Donigian JR, Baciu P, de Lange T, Lei M. A shared docking motif in TRF1 and TRF2 used for differential recruitment of telomeric proteins. Science. 2008;319:1092–1096. doi: 10.1126/science.1151804. [DOI] [PubMed] [Google Scholar]
- Conrad MN, Wright JH, Wolf AJ, Zakian VA. RAP1 protein interacts with yeast telomeres in vivo: overproduction alters telomere structure and decreases chromosome stability. Cell. 1990;63:739–750. doi: 10.1016/0092-8674(90)90140-a. [DOI] [PubMed] [Google Scholar]
- D'Amours D, Jackson SP. The Mre11 complex: at the crossroads of dna repair and checkpoint signalling. Nature Rev Mol Cell Biol. 2002;3:317–327. doi: 10.1038/nrm805. [DOI] [PubMed] [Google Scholar]
- de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- Diede SJ, Gottschling DE. Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases alpha and delta. Cell. 1999;99:723–733. doi: 10.1016/s0092-8674(00)81670-0. [DOI] [PubMed] [Google Scholar]
- Diede SJ, Gottschling DE. Exonuclease activity is required for sequence addition and Cdc13p loading at a de novo telomere. Curr Biol. 2001;11:1336–1340. doi: 10.1016/s0960-9822(01)00400-6. [DOI] [PubMed] [Google Scholar]
- Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- Frank CJ, Hyde M, Greider CW. Regulation of telomere elongation by the cyclin-dependent kinase CDK1. Mol Cell. 2006;24:423–432. doi: 10.1016/j.molcel.2006.10.020. [DOI] [PubMed] [Google Scholar]
- Garvik B, Carson M, Hartwell L. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol Cell Biol. 1995;15:6128–6138. doi: 10.1128/mcb.15.11.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwell PW, Kronmal SL, Porter SE, Gassenhuber J, Obermaier B, Petes TD. TEL1, a gene involved in controlling telomere length in S. cerevisiae, is homologous to the human ataxia telangiectasia gene. Cell. 1995;82:823–829. doi: 10.1016/0092-8674(95)90479-4. [DOI] [PubMed] [Google Scholar]
- Grunstein M. Molecular model for telomeric heterochromatin in yeast. Curr Opin Cell Biol. 1997;9:383–387. doi: 10.1016/s0955-0674(97)80011-7. [DOI] [PubMed] [Google Scholar]
- Hardy CF, Sussel L, Shore D. A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev. 1992;6:801–814. doi: 10.1101/gad.6.5.801. [DOI] [PubMed] [Google Scholar]
- Hector RE, Shtofman RL, Ray A, Chen BR, Nyun T, Berkner KL, Runge KW. Tel1p preferentially associates with short telomeres to stimulate their elongation. Mol Cell. 2007;27:851–858. doi: 10.1016/j.molcel.2007.08.007. [DOI] [PubMed] [Google Scholar]
- Hirano Y, Sugimoto K. Cdc13 telomere capping decreases Mec1 association but does not affect Tel1 association with DNA ends. Mol Biol Cell. 2007;18:2026–2036. doi: 10.1091/mbc.E06-12-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanoh J, Ishikawa F. spRap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr Biol. 2001;11:1624–1630. doi: 10.1016/s0960-9822(01)00503-6. [DOI] [PubMed] [Google Scholar]
- Karlseder J, Broccoli D, Dai Y, Hardy S, de Lange T. p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science. 1999;283:1321–1325. doi: 10.1126/science.283.5406.1321. [DOI] [PubMed] [Google Scholar]
- Krogh BO, Symington LS. Recombination proteins in yeast. Annu Rev Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- Kyrion G, Boakye KA, Lustig AJ. C-terminal truncation of RAP1 results in the deregulation of telomere size, stability, and function in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:5159–5173. doi: 10.1128/mcb.12.11.5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SE, Moore JK, Holmes A, Umezu K, Kolodner RD, Haber JE. Saccharomyces Ku70, Mre11/Rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- Levy DL, Blackburn EH. Counting of Rif1p and Rif2p on Saccharomyces cerevisiae telomeres regulates telomere length. Mol Cell Biol. 2004;24:10857–10867. doi: 10.1128/MCB.24.24.10857-10867.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JJ, Zakian VA. The Saccharomyces CDC13 protein is a single-strand TG1-3 telomeric DNA-binding protein in vitro that affects telomere behavior in vivo. Proc Natl Acad Sci U S A. 1996;93:13760–13765. doi: 10.1073/pnas.93.24.13760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhese MP. DNA damage response at functional and dysfunctional telomeres. Genes Dev. 2008;22:125–140. doi: 10.1101/gad.1626908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig AJ, Kurtz S, Shore D. Involvement of the silencer and UAS binding protein RAP1 in regulation of telomere length. Science. 1990;250:549–553. doi: 10.1126/science.2237406. [DOI] [PubMed] [Google Scholar]
- Mallory JC, Petes TD. Protein kinase activity of Tel1p and Mec1p, two Saccharomyces cerevisiae proteins related to the human ATM protein kinase. Proc Natl Acad Sci USA. 2000;97:13749–13754. doi: 10.1073/pnas.250475697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcand S, Gilson E, Shore D. A protein-counting mechanism for telomere length regulation in yeast. Science. 1997;275:986–990. doi: 10.1126/science.275.5302.986. [DOI] [PubMed] [Google Scholar]
- Nakada D, Hirano Y, Sugimoto K. Requirement of the Mre11 complex and exonuclease 1 for activation of the Mec1 signaling pathway. Mol Cell Biol. 2004;24:10016–10025. doi: 10.1128/MCB.24.22.10016-10025.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada D, Hirano Y, Tanaka Y, Sugimoto K. Role of the C terminus of mec1 checkpoint kinase in its localization to sites of DNA damage. Mol Biol Cell. 2005;16:5227–5235. doi: 10.1091/mbc.E05-05-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada D, Matsumoto K, Sugimoto K. ATM-related Tel1 associates with double-strand breaks through an Xrs2-dependent mechanism. Genes & Dev. 2003a;17:1957–1962. doi: 10.1101/gad.1099003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada D, Shimomura T, Matsumoto K, Sugimoto K. The ATM-related Tel1 protein of Saccharomyces cerevisiae controls a checkpoint response following phleomycin treatment. Nucleic Acids Res. 2003b;31:1715–1724. doi: 10.1093/nar/gkg252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrini S, Ribaud V, Bianchi A, Shore D. DNA breaks are masked by multiple Rap1 binding in yeast: implications for telomere capping and telomerase regulation. Genes Dev. 2007;21:292–302. doi: 10.1101/gad.400907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent CI, Hughes TR, Lue NF, Lundblad V. Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science. 1996;274:249–252. doi: 10.1126/science.274.5285.249. [DOI] [PubMed] [Google Scholar]
- Pardo B, Marcand S. Rap1 prevents telomere fusions by nonhomologous end joining. EMBO J. 2005;24:3117–3127. doi: 10.1038/sj.emboj.7600778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennock E, Buckley K, Lundblad V. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell. 2001;104:387–396. doi: 10.1016/s0092-8674(01)00226-4. [DOI] [PubMed] [Google Scholar]
- Ritchie KB, Petes TD. The Mre11p/Rad50p/Xrs2p complex and the Tel1p function in a single pathway for telomere maintenance in yeast. Genetics. 2000;155:475–479. doi: 10.1093/genetics/155.1.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse J, Jackson SP. Lcd1p recruits Mec1p to DNA lesions in vitro and in vivo. Mol Cell. 2002;9:857–869. doi: 10.1016/s1097-2765(02)00507-5. [DOI] [PubMed] [Google Scholar]
- Sabourin M, Tuzon CT, Zakian VA. Telomerase and Tel1p preferentially associate with short telomeres in S. cerevisiae. Mol Cell. 2007;27:550–561. doi: 10.1016/j.molcel.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Y, Desany BA, Jones WJ, Liu Q, Wang B, Elledge SJ. Regulation of RAD53 by the ATM-like kinase MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science. 1996;271:357–360. doi: 10.1126/science.271.5247.357. [DOI] [PubMed] [Google Scholar]
- Schwartz MF, Duong JK, Sun Z, Morrow JS, Pradhan D, Stern DF. Rad9 phosphorylation sites couple Rad53 to the Saccharomyces cerevisiae DNA damage checkpoint. Mol Cell. 2002;9:1055–1065. doi: 10.1016/s1097-2765(02)00532-4. [DOI] [PubMed] [Google Scholar]
- Silverman J, Takai H, Buonomo SB, Eisenhaber F, de Lange T. Human Rif1, ortholog of a yeast telomeric protein, is regulated by ATM and 53BP1 and functions in the S-phase checkpoint. Genes Dev. 2004;18:2108–2119. doi: 10.1101/gad.1216004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CD, Smith DL, DeRisi JL, Blackburn EH. Telomeric protein distributions and remodeling through the cell cycle in Saccharomyces cerevisiae. Mol Biol Cell. 2003;14:556–570. doi: 10.1091/mbc.E02-08-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A, de Lange T. Regulation of telomerase by telomeric proteins. Annu Rev Biochem. 2004;73:177–208. doi: 10.1146/annurev.biochem.73.071403.160049. [DOI] [PubMed] [Google Scholar]
- Sun Z, Fay DS, Marini F, Foiani M, Stern DF. Spk1/Rad53 is regulated by Mec1-dependent protein phosphorylation in DNA replication and damage checkpoint pathways. Genes Dev. 1996;10:395–406. doi: 10.1101/gad.10.4.395. [DOI] [PubMed] [Google Scholar]
- Sweeney FD, Yang F, Chi A, Shabanowitz J, Hunt DF, Durocher D. Saccharomyces cerevisiae Rad9 acts as a Mec1 adaptor to allow Rad53 activation. Curr Biol. 2005;15:1364–1375. doi: 10.1016/j.cub.2005.06.063. [DOI] [PubMed] [Google Scholar]
- Usui T, Ogawa H, Petrini JH. A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol Cell. 2001;7:1255–1266. doi: 10.1016/s1097-2765(01)00270-2. [DOI] [PubMed] [Google Scholar]
- Vega LR, Mateyak MK, Zakian VA. Getting to the end: telomerase access in yeast and humans. Nat Rev Mol Cell Biol. 2003;4:948–959. doi: 10.1038/nrm1256. [DOI] [PubMed] [Google Scholar]
- Viscardi V, Bonetti D, Cartagena-Lirola H, Lucchini G, Longhese MP. MRX-dependent DNA damage response to short telomeres. Mol Biol Cell. 2007;18:3047–3058. doi: 10.1091/mbc.E07-03-0285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White CI, Haber JE. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 1990;9:663–673. doi: 10.1002/j.1460-2075.1990.tb08158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold MS. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- Wotton D, Shore D. A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev. 1997;11:748–760. doi: 10.1101/gad.11.6.748. [DOI] [PubMed] [Google Scholar]
- Xu L, Blackburn EH. Human Rif1 protein binds aberrant telomeres and aligns along anaphase midzone microtubules. J Cell Biol. 2004;167:819–830. doi: 10.1083/jcb.200408181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Z, Chahwan C, Bailis J, Hunter T, Russell P. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol Cell Biol. 2005;25:5363–5379. doi: 10.1128/MCB.25.13.5363-5379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou BBS, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- Zhu XD, Kuster B, Mann M, Petrini JH, de Lange T. Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nat Genet. 2000;25:347–352. doi: 10.1038/77139. [DOI] [PubMed] [Google Scholar]
- Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


