Abstract
The evolution from outcrossing to predominant self-fertilization represents one of the most common transitions in flowering plant evolution. This shift in mating system is almost universally associated with the “selfing syndrome,” characterized by marked reduction in flower size and a breakdown of the morphological and genetic mechanisms that prevent self-fertilization. In general, the timescale in which these transitions occur, and the evolutionary dynamics associated with the evolution of the selfing syndrome are poorly known. We investigated the origin and evolution of selfing in the annual plant Capsella rubella from its self-incompatible, outcrossing progenitor Capsella grandiflora by characterizing multilocus patterns of DNA sequence variation at nuclear genes. We estimate that the transition to selfing and subsequent geographic expansion have taken place during the past 20,000 years. This transition was probably associated with a shift from stable equilibrium toward a near-complete population bottleneck causing a major reduction in effective population size. The timing and severe founder event support the hypothesis that selfing was favored during colonization as new habitats emerged after the last glaciation and the expansion of agriculture. These results suggest that natural selection for reproductive assurance can lead to major morphological evolution and speciation on relatively short evolutionary timescales.
Keywords: divergence population genetics, reproductive assurance, bottleneck, selfing syndrome, mating system evolution
Selfing plants benefit from 2 distinct advantages over their outcrossing competitors (1–3). First, because selfers are 100% related to their progeny and can also act as outcross pollen donors for seed produced by other individuals, they have an inherent transmission advantage over outcrossers (4). A second major advantage conferred by selfing, first discussed by Darwin (5), is the ability to reproduce when pollinators or potential mates are limited (reproductive assurance). One important aspect of reproductive assurance is the ability of selfing lineages to colonize new habitats from a very small founding population. Evolutionary theory predicts that selfing will evolve when these advantages outweigh the costs associated with inbreeding depression, reduced fitness caused by increased homozygosity of deleterious recessive alleles (6).
In most cases the timescale over which the selfing syndrome has evolved remains difficult to ascertain. In the model genus Arabidopsis, for example, the selfing Arabidopsis thaliana diverged from its closest outcrossing relatives ≈5 million years ago (7), and patterns of diversity have suggested a possibly complicated picture of the breakdown of outcrossing over a period of more than a million years (7, 8). Thus, it remains unclear how rapidly the selfing syndrome can arise, and what historical conditions have favored mating system evolution. Capturing and characterizing a recent transition is important for inferring this timescale, and the relative importance of various forces favoring its evolution. For example, if selfing is initially favored through the genetic transmission advantage, we would expect that self-compatibility alleles would invade individual populations that were ancestrally outcrossing, and the recently derived selfing species should maintain considerable levels of genetic variation at regions unlinked to the locus causing selfing (9). In contrast, selection for colonization ability in small, founding populations should lead to a severe reduction in genetic variation genome-wide in the derived selfing lineage.
Here, we investigate the evolution of selfing in diploid species in the genus Capsella. Capsella rubella is characterized by a high rate of self-fertilization (10) and shows the typical morphological characteristics of a selfing syndrome (11). From genetic marker studies, the selfing rate in C. rubella has been estimated as 1, with a lower-bound estimate of 0.7 (10). In comparison with its self-incompatible congener Capsella grandiflora, there has been a derived breakdown of the self-incompatibility mechanism, and its floral organ sizes are highly reduced (Fig. 1). The breakdown of self-incompatibility is also associated with an expansion of geographic range; C. grandiflora is restricted to Western Greece and Albania and locally in Northern Italy, whereas C. rubella has expanded into much of southern Europe extending to Middle Europe, Northern Africa, and into Australia and North and South America (11, 12). Interspecific crossing experiments suggest that, in addition to mating system evolution, there is considerable postpollination reproductive isolation between the species, with only a small proportion of crosses producing viable seed (refs. 11 and 13). We characterized multilocus patterns of neutral DNA sequence diversity across the genome to investigate the evolutionary history associated with this transition to selfing. We estimated the parameters of a model of isolation with or without migration, and tested the goodness of fit of the data to models incorporating species-specific recombination rate. Our approach provides a detailed examination of the evolutionary history associated with the origins of selfing Capsella.
Fig. 1.
Floral organs and petals are reduced in C. rubella (Left) compared with C. grandiflora (Right).
Results
Patterns of Polymorphism.
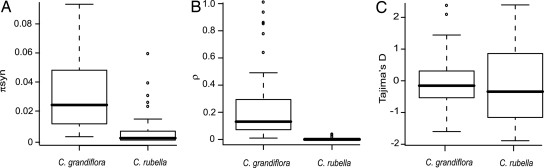
We estimated levels of synonymous and nonsynonymous diversity in a geographically broad sample of both C. rubella (14 individuals) and C. grandiflora (20 diploid individuals) by using direct sequencing of 39 nuclear genes. We identified a total of 587 synonymous single-nucleotide polymorphisms (SNPs) and 343 nonsynonymous SNPs in C. grandiflora. In C. rubella, diversity was highly reduced with 81 synonymous and 41 nonsynonymous SNPs. Fig. 2A illustrates this major reduction in synonymous diversity in C. rubella when compared with C. grandiflora. Over one-third of loci (36%) were devoid of any variation in C. rubella, and almost half (46%) lacked synonymous site variation. In contrast, all loci were highly variable in C. grandiflora.
Fig. 2.
Comparison of polymorphism patterns between C. grandiflora and C. rubella. Bars represent the median, boxes the interquartile range, and whiskers extend out to 1.5 times the interquartile range. (A) π synonymous, where π is the average pairwise differences (16). (B) The population recombination estimator ρ per base pair, using the composite likelihood estimator of Hudson (32). (C) Tajima's D (16) in C. grandiflora and C. rubella.
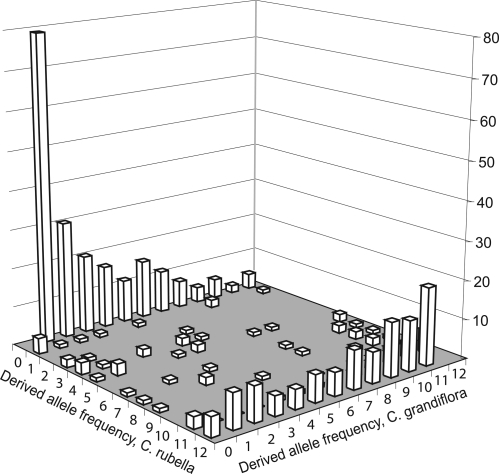
Although a reduction in sequence diversity in selfing species is predicted under models of long-term equilibrium (14), the diversity reduction seen here is extreme (15), and our sequence data imply a very close relationship between C. rubella and C. grandiflora. At the majority of loci (74%), we identify sequence haplotypes shared between the species, and sequence divergence, when present, is very low. To illustrate the patterns of polymorphism, we plot the joint derived SNP frequency spectrum from 12 samples of each species (Fig. 3). More than 80% of the segregating sites found in C. rubella are shared with C. grandiflora, and thus, there is a very small proportion of private polymorphisms present in this species (Fig. 3; private polymorphisms to C. rubella have a derived frequency of 0 in C. grandiflora). We also identify a significant fraction of the segregating sites found in C. grandiflora to be fixed derived SNPs in C. rubella (Fig. 3; derived allele frequency of 12 in C. rubella). Similarly, only 3 fixed differences were identified in our dataset in a total of >20,000 bp surveyed for this study. This suggests that the sequence haplotypes present in C. rubella were mostly subsampled from existing variation in C. grandiflora.
Fig. 3.
Derived SNP frequencies in C. grandiflora and C. rubella calculated by using A. thaliana as an outgroup. For this plot, we randomly subsampled the data to include 12 individuals from each species. Values where the derived allele frequencies are >0 and <12 would represent polymorphisms, a frequency of 12 is a derived fixation, and a frequency of zero implies a complete absence of the derived SNP. For example, values with 0 or 12 for one species and not in the other represent unique polymorphism in the other species, whereas derived frequencies >0 and <12 in both species represent shared polymorphisms.
Analysis of the frequency distribution of polymorphisms and linkage disequilibrium suggest that C. grandiflora represents a large, stable equilibrium population. In particular, the derived frequency spectrum of synonymous SNPs matches closely to the expected distribution under long-term constant population size (Fig. 2C, Fig. S1). Furthermore, linkage disequilibrium decays rapidly with distance (Fig. S2) giving a high estimate of the effective rate of recombination (Fig. 2B), consistent with expectations from neutral equilibrium (10, 16). In contrast C. rubella shows much greater within-locus linkage disequilibrium and very little effective recombination within genes, consistent with a severe bottleneck and recent transition toward high selfing (Fig. 2B; Fig. S2). Nevertheless, a lack of linkage disequilibrium between loci (Fig. S2) suggests that C. rubella does some level of outcrossing, uncoupling coalescent history among distant loci. The site frequency spectrum, measured by Tajima's D (16), shows an elevated variance across loci (Fig. 2) relative to the predictions of stable equilibrium in C. rubella (simulation results, P < 0.05; Fig. 2), providing further evidence for recent changes in population size (17).
Evidence for Single Origin of C. rubella.
The extent of shared haplotypes and polymorphism between these species contrasts with the 5 million year divergence of the selfing species A. thaliana from its closest extant outcrossing relatives (7), and suggests one of three possibilities; very recent evolution of selfing, multiple origins of C. rubella, and/or ongoing hybridization. We first sought to test whether selfing C. rubella was derived from a single speciation event, or whether we can detect evidence for multiple origins or recent hybridization. A Bayesian clustering algorithm with the entire dataset (18) found the most likely number of clusters to be 2, in which all C. rubella and C. grandiflora individuals cluster with their own species, with no evidence for very recent hybridization affecting this sample (Fig. S3). If we analyze the species on their own, we find no evidence for geographic substructure in our sample of C. grandiflora, whereas C. rubella subdivides into three distinct clusters, broadly consistent with the geographic origins of our samples (Fig. S3). Given the wide geographic sampling for this study, the combined results suggest that C. rubella has evolved once, with subsequent geographic expansion and subdivision.
Demographic Model Fitting.
Our results provide evidence for a single recent origin of C. rubella from C. grandiflora, with little or no recent hybridization affecting our samples. To investigate the dynamics of the speciation event in detail, we used a Markov Chain Monte Carlo (MCMC) approach based on coalescent simulations to fit a model of isolation with and without migration to the data (19). The approach makes use of the observed information from each locus on the number of shared and unique polymorphisms, as well as fixed differences. For our analysis, we restrict the inference to synonymous sites, to avoid potential effects of selection on the nonsynonymous variants. The model assumes that a single ancestral population of size Na split into two at time τ, and the two derived populations have distinct effective population sizes (N1 and N2).
Although our clustering results suggest little or no hybridization, it remains possible that historical gene flow has contributed to the shared polymorphism between the species. To estimate demographic parameters, we therefore investigated a series of models that varied in the inclusion of symmetrical, asymmetrical, or no migration, and varied whether or not the ancestral population size was constrained to be the same size as present-day C. grandiflora. We estimated historical demographic parameters from a total of 4 alternative models (Table 1, Tables S1–S4). To evaluate the fit of these models to the data, we used goodness-of-fit tests. In particular, we simulated datasets from both the posterior distributions and the point parameter estimates of each model, and estimated the probability of seeing our observed multilocus summary statistics, including additional data summaries not used in parameter estimation (see SI Text).
Table 1.
Modes of parameter estimates under a range of MIMAR models, with 90% HPD intervals in parentheses
| Model | Ne(Cg)* | Ne(Cr)* | Ne(A)* | MCg-Cr† | MCr-Cg‡ | T§ |
|---|---|---|---|---|---|---|
| 1. Ancestral size constrained, no migration | 488.2¶ (412.2, 536.0) | 3.0 (0.5, 11.5) | 488.2¶ (412.2, 536.0) | – | – | 13.6 (1.5, 51.7) |
| 2. Ancestral size constrained, symmetrical migration | 463.6¶ (352.4, 602.2) | 4.7 (2.0, 17.5) | 463.6¶ (352.4, 602.2) | 1.9¶ (0.007, 4.2) | 1.9¶ (0.007, 4.2) | 15.0 (0.8, 3546.8) |
| 3. Ancestral size constrained, asymmetrical migration | 487.5¶ (400.4, 540.9) | 0.3 (0.06, 6.0) | 487.5¶ (400.4, 540.9) | 1.5 (0.009, 3.1) | 50.9 (7.0, 336.9) | 21.7 (3.5 3579.5) |
| 4. Ancestral size unconstrained, asymmetrical migration | 525.0 (410.4, 652.1) | 0.4 (0.06, 5.5) | 28.3 (16.7, 508.5) | 2.0 (0.01, 4.2) | 73.0 (13.3, 399.8) | 1395.0‖ (13.3, 3156.1) |
*Effective population size (effective number of individuals × 10−3) for C. rubella (Cr), C. grandiflora (Cg), and their ancestor (A).
†Migration rate (4Nem) from C. grandiflora to C. rubella.
‡Migration rate (4Nem) from C. rubella to C. grandiflora.
§Time (ka) of the split of C. rubella and C. grandiflora.
¶Constrained.
‖Bimodal distribution, with second mode <30,000 years. See also goodness-of-fit tests in SI Text.
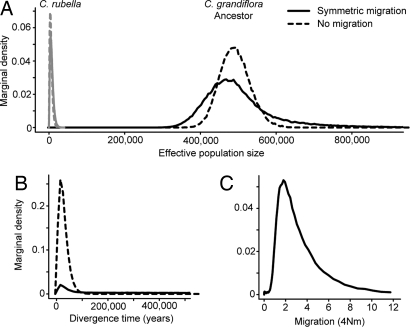
Our parameter estimates from these models are consistent with an extremely recent speciation event associated with a major reduction in effective population size in C. rubella. Fig. 4 shows the parameter estimates from two of the best-fit models, based on the results of goodness-of-fit tests (Table 1 and SI Text). Under the model with no migration, our most likely estimate of divergence time is ≈13,500 years ago with a 90% posterior density from 1,500 to 51,000 years, suggesting species divergence since the last glacial maximum ≈21,000 years ago (20). Similarly, models with migration provide estimates of divergence time of 15,000 or 22,000 years ago. The boundaries on the divergence time estimate across all models examined depend on whether migration is included in the model (Table 1); in particular, allowing for a wide prior probability on migration and time leads to a long tail in the posterior density which is small but nonzero for older divergence (Fig. 4B). However, the best model is clearly a recent divergence in the past 25,000 years. Estimates of population size parameters suggest that C. grandiflora has maintained a large population size since its divergence from the ancestor, whereas we infer an ≈100- to 1,500-fold smaller effective population size in C. rubella (Table 1; Fig. 4B). Taken together, these results clearly indicate recent speciation associated with the breakdown of self-incompatibility from a small number of founding lineages.
Fig. 4.
Smoothed marginal posterior distributions of speciation parameters estimated by MIMAR, for two models with posterior modes showing good fit to data summaries, assuming either symmetric migration (solid lines) or no migration (dashed lines) and equal effective population sizes in the ancestor as in present-day C. grandiflora. (A) Marginal densities for effective population size (individuals) in C. rubella (gray) and C. grandiflora/the ancestral species. (B) Marginal densities for divergence time (years). (C) Marginal density for migration (4Nm), where N is the effective population size of C. grandiflora.
The one exception is a 1.4-million-year divergence time estimate from the fully unconstrained model (Table 1, SI); this model also infers a very high migration rate from C. rubella to C. grandiflora, an extremely low effective population size in C. rubella and a very low ancestral population size. The inferred level of gene flow under this model seems implausibly high given the mating system, our Structure results, and the evidence for postpollination reproductive isolation between the species. Furthermore, goodness-of-fit tests and comparisons of likelihoods suggest that this unconstrained model provides a poorer fit to our data (SI Text). However, because some models with and without migration appear to provide comparable fits to the observed data, divergence may be too recent to have sufficient power to infer whether or not there has been ongoing gene flow.
One potential concern with the approach used is that the model assumes a constant rate of recombination since the species split. Because the transition to selfing is expected to lead to a dramatic shift in the effective recombination rate (12), this assumption is violated. However, the method has been shown to be robust to inaccuracies in estimation of recombination (17), and given the very recent timescale and massive population bottleneck inferred here, effective recombination rates are suppressed even in the absence of this effect. Consistent with this, goodness-of-fit tests using simulations that allow for a lineage-specific change in recombination rate to zero in C. rubella show that the best-fit models provide an equally adequate fit to the data when this shift is incorporated (Table S4).
Discussion
The timescale of speciation suggested by our analysis is consistent with selfing evolving after the last glacial maximum. C. grandiflora is distributed in the Balkans, which was a glacial refugium for many plant species (11), and this is consistent with our inference of long-term equilibrium for the outcrossing species. Furthermore, the massive loss of diversity in C. rubella suggests that selfing alleles did not spread through a previously outcrossing population because of their transmission advantage. Instead selection for reproductive assurance, either due to a lack of pollinators or via selection for colonization ability, seems the most likely explanation for the evolution of selfing. Under either mode of selection for reproductive assurance, the enhanced colonization ability likely contributed to the geographic spread of selfing. As the glaciers receded, agriculture spread and Europe warmed, new habitats emerged, and this would favor colonization by selfing genotypes that can reproduce in the absence of pollinators and/or available mates (21–23). Our inference of a very severe population bottleneck associated with the recent evolution of selfing is consistent with this model for the evolution and spread of selfing Capsella. This result is in line with mounting ecological data suggesting that reproductive assurance is a major factor favoring the evolution and maintenance of selfing (24–26).
If, as our results suggest, C. rubella experienced recent evolution of selfing associated with a severe population bottleneck, we would predict a similar severe loss of variation with little sequence divergence at the self-incompatibility locus. Recent results confirm this prediction (41), providing support for our inference of recent speciation tied to the breakdown of self-incompatibility.
In A. thaliana, the transition to selfing appears to have been gradual, and occurred considerably further back in time (≈1 Myr or more), providing ample time for the evolution of morphological traits associated with the selfing habit (8). In contrast, our inference of recent speciation in Capsella implies that extensive phenotypic evolution has occurred on a very short evolutionary timescale. Given the evidence for rapid morphological evolution, one possible alternative explanation to a severe bottleneck in C. rubella is the occurrence of positive selection reducing diversity genome-wide. Although it is likely that the evolution of selfing and spread through Europe has lead to selective pressures, our observed low between-locus linkage disequilibrium (Fig. S2) implies that this would likely be restricted to a small subset of the genome even in this highly selfing species, unless the extent of positive selection is very high. Furthermore, positive selection at a subset of loci might be expected to increase the variance in diversity across loci beyond the bottleneck expectations, but goodness-of-fit tests suggest that our models are sufficient to explain the observed variance (Tables S3 and S4). Finally, the observed retention of shared ancestral polymorphism in C. rubella is unlikely under a model of genome-wide selective sweeps (Fig. S4).
Given the extreme population bottleneck, this nevertheless implies that rapid evolution has occurred from small amounts of standing variation. Understanding the genomic extent and genetic basis of these changes, and testing the extent to which phenotypic evolution occurred via positive selection or relaxation of stabilizing selection, will be of considerable interest in future investigations.
Methods
Species Sampling and DNA Sequencing.
We collected nucleotide polymorphism data from 39 large exons (see Table S5) in C. grandiflora and C. rubella. A total of 20 diploid C. grandiflora and 14 C. rubella individuals were used for this study. The C. grandiflora samples originated from 8 natural localities in Greece. The plants from which sequences were obtained were 4 individuals from Ioaninna, 3 individuals from the Katara Pass; 2 individuals from Metsovo, 4 individuals from Sokraki on the island of Corfu; 3 individuals from Doukades, Corfu; 2 individuals from Votonosi, Corfu; 1 individual from Paleokastritsas, and 1 individual from Pantokrator, Corfu. The C. rubella samples originated from 4 natural localities. Four individuals were from Buenos Aires, Argentina; four were from Cumbre Dorsal, Tenerife, Spain; three were from Caldera de Taburiente, La Palma, Spain; and three were from Ioaninna, Greece. Outgroup data from Boechera stricta were obtained by using DNA kindly provided by B. H. Song and T. Mitchell-Olds (Duke University, Hillsborough, NC).
Seeds were placed at 4 °C on wet filter paper for 14 days before being allowed to germinate at room temperature. The seedlings were grown at 20 °C under conditions of 18 h of light and 6 h of darkness. After 6 weeks of growth, DNA was extracted from leaf material by using a FastDNA Kit and the FastPrep Instrument (Qbiogene).
PCR primers for the large exons were designed as described by Wright et al. (27) and Ross-Ibarra et al. (28). In brief, primers were designed to amplify 650–700 bp from single large exons based on the A. thaliana genome sequence, chosen with no a priori expectation as to their function or the action of selection on these genes. Each exon was used as a BLAST query against the shotgun genome sequence of Brassica oleracea and homologous regions were used to design primers by using PrimerQuest (Integrated DNA Technologies). PCRs were carried out in 25-μL volumes on an Eppendorf Mastercycler. The cycles were as follows: 2 min at 94 °C, 20 sec at 94 °C, 20 sec at 55 °C, 40 sec at 72 °C, for 35 cycles, with a final extension time of 4 min at 72 °C.
These products were sequenced directly by using ABI sequencing by Cogenics. Chromatograms were checked manually for heterozygous sites, using Sequencher version 4.7 (Gene Codes), with the aid of the “Call secondary peaks” option. Sequences were aligned by using Genedoc (29). Consistent with high levels of selfing, no heterozygous sites were identified in our C. rubella dataset. This complete lack of heterozygosity in C. rubella also allowed us to confirm that we were sequencing single-copy regions only. Nucleotide sequences are deposited in GenBank (accession nos. FJ182244–FJ183352).
Sequence Statistics and Analysis.
Sequence-based summary statistics θ (30) and π (31) synonymous and nonsynonymous, as well as frequency data, were calculated by using a modified version of Perl code (Polymorphurama) written by D. Bachtrog and P. Andolfatto (University of California at San Diego). The frequency spectra of derived polymorphic variants, and the number of shared derived polymorphisms, unique polymorphisms, and fixed differences were calculated by using Perl scripts written by S.I.W.
Population recombination rate estimates were calculated by using the composite likelihood approach of Hudson (32), the maxdip program for diploid unphased data for C. grandiflora, and ldhat for C. rubella (33) with the data-processing code written by J. Ross-Ibarra (University of California, Irvine). Linkage disequilibrium, calculated as the squared correlation coefficient r2 was calculated by using Weir's algorithm (34) for unphased data by using R script kindly provided by Stuart MacDonald (University of Kansas). DNAsp (version 4.0) (35) was used to calculate linkage disequilibrium in C. rubella. To assess significance of the mean and variance of Tajima's D estimates, we used Jody Hey's (Rutgers University) HKA program.
To infer haplotype data in C. grandiflora we used PHASE 2.1, which implements a Bayesian statistical method to reconstruct haplotypes from diploid data (36, 37). Haplotypes with the highest posterior probabilities were used for cluster analysis performed with the program STRUCTURE (18). The program was run under the haploid model assuming values of k (population number) from 1 to 7, each with 1,000,000 repetitions and a burn-in period of 100,000.
Coalescent Simulations.
Coalescent simulations were conducted by using MIMAR, which estimates the parameters of an isolation–migration model based on Hudson's ms (38). Because MIMAR makes use of outgroup information to infer the number of derived SNP fixations, we made every effort to minimize errors associated with ancestral state inference, as described in the SI. Simulations using a modified version of MIMAR that does not rely on outgroup inference provide comparable estimates under all models (Table S6).
Simulations were conducted by using the 25 loci for which we had sequence data from 3 outgroup species for ancestral state inference. Furthermore, sites with >2 segregating bases were excluded from the analysis. To allow for locus-specific mutation rates, the mutation rate scalar was estimated by using θsyn for each of the 26 loci, dividing by the mean θsyn. We ran a series of MIMAR coalescent simulations that differed in the degree of constraint on migration rates and effective population sizes (Table 1). Migration rates were either unconstrained (asymmetric migration), constrained to be symmetrical, or set to zero (no migration), whereas effective population sizes were either unconstrained or assumed to be identical in C. grandiflora and the ancestor of C. rubella and C. grandiflora.
Prior limits for the Bayesian procedure implemented in MIMAR were set based on initial runs using wide priors. Priors for θ were uniform 0.001–0.1 for both C. grandiflora and the ancestral species, and uniform 0–0.0025 for C. rubella. All runs assumed an exponentially distributed prior with rate 1 for ρ/θ, and a mutation rate per bp of 1.5 × 10−8 (39). Migration rate priors were log uniform −5–2.5 for migration from C. grandiflora to C. rubella (forward in time) and log uniform −5–6 for migration from C. rubella to C. grandiflora. The prior for the time of the split between C. rubella and C. grandiflora was uniform 0–4 × 106.
Each simulation was run for a total of 20,160 min (2 weeks) with a burn-in of 100,000 steps, and we performed 3 sets of simulations with different random seeds for each model. Mixing was monitored by assessing parameter autocorrelation over runs and we considered that MIMAR reached convergence when the posterior distributions from independent runs were highly similar (19). The mode of the marginal posterior probability distribution was considered as a point estimate for each parameter, and we calculated 90% highest posterior density (HPD) intervals from the MIMAR output by using the boa package in R 2.6.2 (40).
Supplementary Material
Acknowledgments.
We thank C. Becquet for assistance and advice with running Mimar, and S.C.H. Barrett and A. Cutter for comments on the manuscript. This work was supported by a Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery grant, an Early Researcher Award from the Ontario Ministry of Research and Innovation, and an Alfred P. Sloan Foundation Fellowship (S.I.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0807679106/DCSupplemental.
References
- 1.Charlesworth D. Evolution of plant breeding systems. Curr Biol. 2006;16:R726–R735. doi: 10.1016/j.cub.2006.07.068. [DOI] [PubMed] [Google Scholar]
- 2.Stebbins GL. Variation and Evolution in Plants. New York: Columbia Univ Press; 1950. [Google Scholar]
- 3.Barrett SCH. The evolution of plant sexual diversity. Nat Rev. 2002;3:274–284. doi: 10.1038/nrg776. [DOI] [PubMed] [Google Scholar]
- 4.Fisher RA. Average excess and average effect of a gene substitution. Ann Eugen. 1941;11:53–63. [Google Scholar]
- 5.Darwin CR. The Effects of Cross and Self-fertilization in the Vegetable Kingdom. London: John Murray; 1876. [Google Scholar]
- 6.Charlesworth D, Charlesworth B. Inbreeding depression and its evolutionary consequences. Annu Rev Ecol Systematics. 1987;18:237–268. [Google Scholar]
- 7.Koch MA, Haubold B, Mitchell-Olds T. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae) Mol Biol Evol. 2000;17:1483–1498. doi: 10.1093/oxfordjournals.molbev.a026248. [DOI] [PubMed] [Google Scholar]
- 8.Tang C, et al. The evolution of selfing in Arabidopsis thaliana. Science. 2007;317:1070–1072. doi: 10.1126/science.1143153. [DOI] [PubMed] [Google Scholar]
- 9.Schoen DJ, Morgan MT, Bataillon T. How does self-pollination evolve? Inferences from floral ecology and molecular genetic variation. Philos Trans R Soc Lond B. 1996;351:1281–1290. [Google Scholar]
- 10.Hurka H, Freundner S, Brown AH, Plantholt U. Aspartate aminotransferase isozymes in the genus Capsella (Brassicaceae): Subcellular location, gene duplication, and polymorphism. Biochem Genet. 1989;27:77–90. doi: 10.1007/BF00563019. [DOI] [PubMed] [Google Scholar]
- 11.Hurka H, Neuffer B. Evolutionary processes in the genus Capsella (Brassicaceae)*. Plant Systematics Evol. 1997;206:295–316. [Google Scholar]
- 12.Paetsch M, Maryland-Quellhorst S, Neuffer B. Evolution of the self-incompatibility system in the Brassicaceae: Identification of S-locus receptor kinase (SRK) in self-incompatible Capsella grandiflora. Heredity. 2006;97:283–290. doi: 10.1038/sj.hdy.6800854. [DOI] [PubMed] [Google Scholar]
- 13.Koch M, Kiefer M. Genome evolution among cruciferous plants: A lecture from the of the genetic maps of three diploid species—Capsella rubella, Arabidopsis lyrata subsp. petraea, and A thaliana. Am J Bot. 2005;92:761–767. doi: 10.3732/ajb.92.4.761. [DOI] [PubMed] [Google Scholar]
- 14.Charlesworth D, Wright SI. Breeding systems and genome evolution. Curr Opin Genet Dev. 2001;11:685–690. doi: 10.1016/s0959-437x(00)00254-9. [DOI] [PubMed] [Google Scholar]
- 15.Wright SI, Ness RW, Foxe JP, Barrett SCH. Genomic consequences of outcrossing and selfing in plants. Int J Plant Sci. 2008;169:105–118. [Google Scholar]
- 16.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wright SI, Gaut BS. Molecular population genetics and the search for adaptive evolution in plants. Mol Biol Evol. 2005;22:506–519. doi: 10.1093/molbev/msi035. [DOI] [PubMed] [Google Scholar]
- 18.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Becquet C, Przeworski M. A new approach to estimate parameters of speciation models with application to apes. Genome Res. 2007;17:1505–1519. doi: 10.1101/gr.6409707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kageyama M, et al. The Last Glacial Maximum climate over Europe and western Siberia: A PMIP comparison between models and data. Climate Dyn. 2001;17:23–43. [Google Scholar]
- 21.Ammerman AJ Cavalli-Sforza LL. The Neolithic Transition and the Genetics of Populations in Europe. Princeton, NJ: Princeton Univ Press; 1984. [Google Scholar]
- 22.Baker G. Prehistoric Farming in Europe. Cambridge, UK: Cambridge Univ Press; 1985. [Google Scholar]
- 23.Pinhasi R, Fort J, Ammerman AJ. Tracing the origin and spread of agriculture in Europe. PLoS Biol. 2005;3:e410. doi: 10.1371/journal.pbio.0030410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baker HG. Self-compatibility and establishment after long-distance dispersal. Evolution (Lawrence, Kans) 1955;9:347–349. [Google Scholar]
- 25.Pannell JR, Barrett SCH. Baker's law revisited: Reproductive assurance in a metapopulation. Evolution (Lawrence, Kans) 1998;5:657–668. doi: 10.1111/j.1558-5646.1998.tb03691.x. [DOI] [PubMed] [Google Scholar]
- 26.Eckert CG, Samis KE, Dart S. Reproductive assurance and the evolution of uniparental reproduction in flowering plants. In: Harder LD, Barrett SCH, editors. Ecology and Evolution of Flowers. Oxford, UK: Oxford Univ Press; 2006. pp. 183–203. [Google Scholar]
- 27.Wright SI, et al. Testing for effects of recombination rate on nucleotide diversity in natural populations of Arabidopsis lyrata. Genetics. 2006;174:1421–1430. doi: 10.1534/genetics.106.062588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ross-Ibarra J, et al. Patterns of Polymorphism and Demographic History in Natural Populations of Arabidopsis lyrata. PloS One. 2008;3:e2411. doi: 10.1371/journal.pone.0002411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nicholas KB, Nicholas HB, Jr, Deerfield DW., II GeneDoc: Analysis and visualization of genetic variation. EMBNEWNEWS. 1997;4:14. [Google Scholar]
- 30.Watterson GA. On the number of segregating sites in genetical models without recombination. Theor Popul Biol. 1975;7:188–193. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- 31.Tajima F. Measurement of DNA polymorphism. In: Takahata N, Clark A, editors. Mechanisms of Molecular Evolution. Tokyo: Japan Scientific Societies Press; 1993. pp. 37–60. [Google Scholar]
- 32.Hudson RR. Two-locus sampling distributions and their application. Genetics. 2001;159:1805–1817. doi: 10.1093/genetics/159.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McVean G, Awadalla P, Fearnhead P. A coalescent-based method for detecting and estimating recombination from gene sequences. Genetics. 2002;160:1231–1241. doi: 10.1093/genetics/160.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weir BS. Genetic Data Analysis. Sunderland, MA: Sinauer; 1990. [Google Scholar]
- 35.Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- 36.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hudson RR. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics. 2002;18:337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- 39.Koch M, Haubold B, Mitchell-Olds T. Molecular systematics of the Brassicaceae: Evidence from coding plastidic matK and nuclear Chs sequences. Am J Bot. 2001;88:534–544. [PubMed] [Google Scholar]
- 40.Smith BJ. boa: An R Package for MCMC Output Convergence Assessment and Posterior Inference. J Stat Software. 2007;21:1–37. [Google Scholar]
- 41.Guo Y, et al. Recent specification of Capsella rubella from C. grandiflora, associated with loss of self-incompatibility and an extreme bottleneck. Proc Natl Acad Sci USA. 2009 doi: 10.1073/pnas0808012106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.