Abstract
Sequencing DNA from several organisms has revealed that duplication and drift of existing genes have primarily molded the contents of a given genome. Though the effect of knocking out or over-expressing a particular gene has been studied in many organisms, no study has systematically explored the effect of adding new links in a biological network. To explore network evolvability, we constructed 598 recombinations of promoters (including regulatory regions) with different transcription or σ-factor genes in Escherichia coli, added over a wild-type genetic background. Here we show that ~95% of new networks are tolerated by the bacteria, that very few alter growth, and that expression level correlates with factor position in the wild-type network hierarchy. Most importantly, we find that certain networks consistently survive over the wild-type under various selection pressures. Therefore new links in the network are rarely a barrier for evolution and can even confer a fitness advantage.
The Escherichia coli genome contains ~300 transcription factors (TFs)1,2, organized hierarchically, with few master regulators3-5 (Fig. 1). Only nine regulatory proteins (CRP, FNR, IHF, FIS, ArcA, NarL, H-NS, Fur, and Lrp) control over half of all genes, through direct and indirect interactions6,7. Lower-tier nodes are more sparsely connected and the network structure has a scale-free power-law degree distribution8,9. It has been argued that such networks are particularly robust to random errors since only a few nodes are highly-connected hubs, whose perturbation would affect the network drastically10. This conclusion is based on the effects of deleting or overexpressing individual nodes. However, the addition of new interactions is thought to be an equally important process for evolution, and the network responses to such changes remain to be systematically explored.
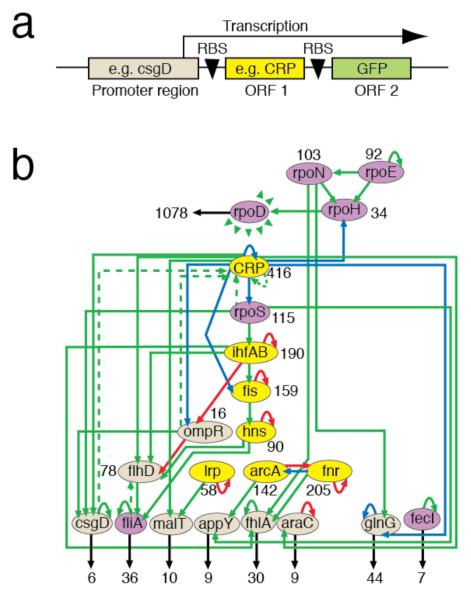
Figure 1. Promoter-ORF network rewiring.
a, Example of construct (csgD-CRP), with two ribosome binding sites (RBS). b, Network diagram of the major transcription factor and σ-factor genes used. Green, red and blue arrows denote direct activating, repressing and dual interactions, respectively, from RegulonDB6,7. σ-factors, master regulators and lower-tier regulators are in purple, yellow and beige. Black numbers denote the total number of direct downstream ORF-gene interactions per node. The housekeeping σ-factor RpoD can activate all other nodes. Dotted arrows illustrate two rewired constructs (fliA-flhD and csgD-CRP; e.g. CRP, RpoS, OmpR and CsgD all regulate csgD, thus connecting 4 nodes to CRP in csgD-CRP).
Genomes are molded by gene duplication, transfer, mutation and loss. Duplication occurs rapidly in all species11,12 and through mutation serves as material for innovation. This drives cellular network evolution13,14, even though relatively few duplications become fixed in populations11,12. We therefore chose to reconstruct events where an open reading frame (ORF) or gene is duplicated and subsequently becomes linked to a new regulatory input. Thus, promoter region-ORF fusions were constructed on high copy number plasmids and a subset were stably integrated in the E. coli chromosome. Although evolution is unlikely to take such a direct approach, except in rare cases such as gene fusions in chromosomal rearrangements, our approach provides a systematic way to sample the viability of new connectivity. By adding new connections to the existing framework across different levels in the network hierarchy, including hub genes, we created a map of the network’s robustness to change.
Rewired constructs and network robustness
598 reconnected gene networks were constructed using the genes for 7 master TFs, 7 σ factors, and 8 downstream TFs5 (Fig. 1). Each construct creates network paths which inherit the inputs to the regulatory region and connect these to the downstream outputs of the ORF. As new connections are added to the wild-type network, they can generate new network motifs5, such as simple feedback loops. For example, if node-A activates node-B then a promoter-B:ORF-A fusion gives a direct positive feedback loop (e.g. fliA-flhD; Fig. 1). Highly complex reconnections are also possible (e.g. csgD-CRP, where 4 csgD promoter inputs, CRP, RpoS, OmpR and CsgD, are connected to CRP output, creating over 4 multi-layer feedback loops).
All 598 rewired high-copy plasmids were cloned, except for ~30 which gave either zero PCR positives in three cloning attempts (Fig. 2; black boxes) or gave positive colonies that died (Fig 2; red boxes). Most clones had similar growth yields (37°C in LB media, 16 h; 6 replicates): 94% had mean OD600 within 2 standard deviations (s. d.) of the mean of 23 control plasmid (Co) colonies. Since ~95% of the rewired networks could be maintained in E. coli, most added connections are well-tolerated.
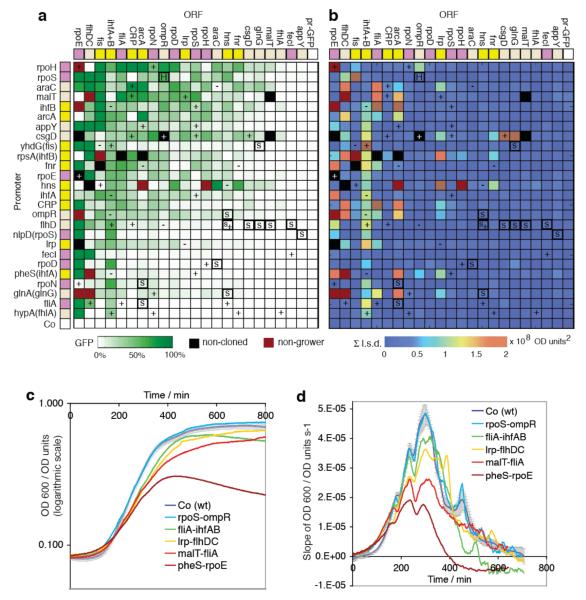
Figure 2. GFP expression and growth signatures of promoter-ORF recombinants.
σ-factors, master regulators and lower-tier regulators are in purple, yellow and beige. a, Green squares show mean GFP expression, ranked by columns, left-to-right, and rows, top-to-bottom (6 repeats, normalised by OD600); non-cloned constructs are black; non-growers (maroon) vary slightly between independent colonies (panels a and b). Controls include promoter-GFP fusions (pr-GFP) and promoter-less GFP (Co). Direct positive and negative feedback loops are marked “+” or “−” and selection results by “S” (serial passaging) or “H” (50°C heat survival). b, OD600 slopes (time-derivatives) give characteristic profiles (growth signatures), displayed by plotting the sum of least squared difference (Σl.s.d.), relative to mean wild-type. c, and d, show selected growth curves (OD600) and signatures. Time = 0 is set at ~7 hours after inoculation, removing lag phase. Error bars show 1 s.d. of 60 Co colonies.
Shuffling connections at the top of the network hierarchy could cause drastic changes, therefore the cells’ tolerance is striking. For example, CRP is the most connected TF in E. coli, directly regulating ~400 genes7, yet changing regulatory inputs is possible (Fig. 2; CRP columns). Similarly, σ factors regulate transcription globally; σ70 and σ54 (RpoD and RpoN) control ~1000 and ~100 genes, respectively7 and also tolerate rewiring. Such hub genes10 could have been less resilient than less-connected genes, but the bacteria can compensate. Therefore, at least when it comes to altering regulatory inputs, the hub genes do not appear to be the Achilles’ heel of the network.
GFP levels and the network structure
The GFP values indirectly measure promoter transcription for all mutants, which can be related back to network properties (Fig. 2a). Spectrophotometer assays showed that 72% expressed GFP over 2 s. d. above mean Co (background). GFP (and OD600) results were also similar in minimal media with glucose, lactose or maltose as the sole carbon source, and in anaerobic conditions (Supplementary Data 1). In control RT-qPCR assays on 84 selected clones, 70% expressed ORF transcripts >12-fold over Co (Mean = 520-fold; Range = 0.4 to 7700-fold; Supplementary Fig. 2). Therefore most constructs are expressed and could potentially establish new network links. As expected, GFP levels vary with promoter region identity (rows, Fig. 2a). Surprisingly, there are also patterns between GFP levels and ORFs (columns, Fig. 2a). Therefore many TFs have associated expression levels that are partially promoter-independent. ANOVA testing confirmed that column GFP means are significantly different (1-way: F-value [21 d. f.] = 8.8; P-value < 2.2 e-16) and that ORFs predict GFP levels better than promoters (2-way; ORFs: F-value [21 d. f.] = 9.8, P-value < 2.2 e-16; Promoters: F-value [25 d. f.] = 3.5, P-value < 5.7 e-08). ORFs could set expression because each could have a particular RNA structure, affecting translation and degradation. Alternatively, the ORF TFs could be widely active, or autoregulating through ORF binding sites. The Ecocyc database15 reports self-regulation for about two-thirds of our 22 TFs, although few ORF binding sites are currently known. Nonetheless, ORFs strongly affect expression in rewired networks.
The lowest ORFs in the wild-type hierarchy often had the lowest GFP expression (Fig. 1, 2). Similarly, higher-tier factors have more interactions and significantly higher GFP (Rank Spearman for GFP versus interactions: r2=0.410; p=0.009); since most network connections are positive, connecting a high-tier ORF to a low-tier promoter may increase the chance of downstream interactions indirectly activating the promoter, creating positive feedback. However, the mean GFP levels for predicted direct positive and negative feedback loops (+ and − in Fig. 2a) were not significantly different (1-sided T-test: p = 0.393). Thus, direct feedback loops can behave unexpectedly in vivo. This itself is informative, suggesting that other levels of network control can counteract direct feedback. Also, plasmid copies increase promoter concentration and thus even weak (non-physiological) TF-promoter interactions might create unpredicted loops. Overall, the results indicate a very complex rewired network response, suggesting that dissection into small network motifs may only lead to useful insights in some cases.
Growth signatures in rewired gene networks
To explore whether acquired network connections affect bacterial growth, OD600 time-courses were measured. The OD time derivative (estimated as linear regression slope for 9 sequential OD readings) gives a characteristic ‘growth signature’, reliably distinguishing between different E. coli strains (C. L., manuscript in preparation). Thus, growth signatures for all 598 constructs were calculated and the sums of least-squared-distances, relative to mean control Co, indicate the scale of perturbations (Fig. 2b). Most constructs have little or no effect on growth: 84% are within the 95% confidence interval of 60 Co colonies (0 - 0.4 e08 OD units2). Therefore only 16% give distinct growth phenotypes (Fig. 2c,d). Interestingly, the corresponding genome-integrated constructs have similar but milder growth signature variations, perhaps because they are expressed 150-fold less on average (Supplementary Information).
Examining the outlier growth signatures, we noticed several patterns. For example, many constructs with ihf A+B ORFs have much-steeper late-growth signatures with reduced late-peaks (Time = ~500 min; Fig. 2d and Supplementary Information). IHF gene products mediate the switch from exponential growth into stationary phase16 and purified IHF binds to regulatory regions in stationary phase genes16. Thus the differently-regulated expression of IHF in the rewired constructs may be affecting stationary phase entry. The ihf A+B clones were studied further using highly-detailed GFP time-courses, as developed by the Alon group; Zaslaver and colleagues recently demonstrated that this could be achieved for 2000 different promoters in E. coli, giving an unprecedented look at E. coli promoter activity17. GFP fluorescence dynamics show distinct expression profiles, with GFP expression peaking during stationary phase transition, and RT-qPCR analysis of different plasmid and integrated clones reveals dose-dependence of the phenotype (Supplementary Information).
To examine ORF overexpression versus rewiring effects, we cloned 21 ORFs into arabinose-inducible pBAD202 Directional TOPO vector (Invitrogen). rpoE did not clone in 3 attempts, which may reflect its apparent toxicity in certain rewired combinations. Different induced expression levels were quantitated using RT-qPCR (Supplementary Information). ihfA+B, rpoD, fliA, appY, and rpoE ORFs show dose-dependence, with higher expression being more deleterious to growth (Supplementary Data 2). Conversely, fis, lrp, rpoS, rpoH, arcA, flhDC, malT, and fhlA ORFs have cases where low or medium expression alter growth more in some promoter-ORF constructs, indicating a dominance of rewiring effects over high expression. ORFs fecI, hns, fnr, araC, glnG, ompR and csgD have very few different growth effects in all conditions. Overall, the growth phenotypes of only 7 of the 22 ORFs tested were explained primarily by overexpression effects. Growth phenotypes are ultimately a mixture of expression levels (dosage), timing and rewiring effects.
Evolvability in rewired gene networks
Since most acquired network connections impact growth minimally, the first step in evolving a new network property is easily accessed. We therefore investigated whether rewired constructs themselves provide any potential for evolution. By pooling all cloned constructs (~570, plus a 23-fold molar excess of wild-type Co) and applying selective pressures, we searched for individuals with specific fitness advantages under three conditions: (i) serial passaging of bacteria in liquid culture; (ii) longevity in extended periods at 37°C; (iii) survival after 50°C heat shock for 1 hour.
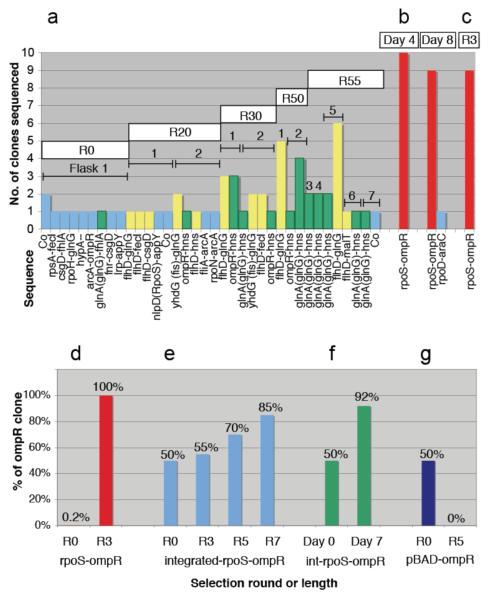
Serial passaging was done in 7 replica flasks, transferring 1 μl of culture mixture into 120 ml fresh medium, every 12-16 hours. After 20 to 55 rounds, 12 network clones were repeatedly selected in independent flasks (Fig. 3a). The clones can be plotted as a ‘selectability map’ (marked “S” in Fig. 2), and are associated with near-wt growth and low (but non-zero) GFP expression. Notably, certain flhD promoter-ORF combinations were enriched. flhD regulates flagellar genes and loss-of-function mutants increase cell division 5-fold18. Flagellar genes are non-essential and cost the cell time, energy and materials. We speculate that unnaturally-connected flhD promoters may repress flagellar biosynthesis, giving a selective advantage. Conversely, expressing FliA flagellar σ factor can disrupt growth (e.g. malT-fliA; Fig. 2d). This correlates with flagellar biosynthesis since 27 of the 30 largest changes in malT-fliA are upregulated flagellar or taxis genes (Supplementary Data 3). Serial passaging can select for mutations and adaptations that optimise the bacteria to their environment19,20, and our results show that reconnected gene networks themselves can provide a substrate for selection.
Figure 3. Selection experiments.
a, Serial passaging of 1 μl of culture mixture into 120 ml fresh medium, every 12-16 h. Replica flasks 1 to 7, and passage rounds R0 to R55, are indicated. Clones related to glnA(glnG)-hns (green) and flhD-glnG (yellow) were selected. b, Clones surviving stationary phase at 37°C (Day 4, Day 8); rpoS-ompR is strongly-selected (red). c, Clones obtained in heat survival (50°C, 1 h) for 3 rounds (R3). d, rpoS-ompR plasmid (650-fold overexpressed transcript), heat selected over the ~570 constructs (3 rounds). e, Integrated rpoS-ompR (2-fold overexpressed transcript), heat selected over integrated Co (7 rounds). f, Integrated rpoS-ompR, selected over wild-type TOP10 cells (7 days, 37°C). g, ompR ORF in pBAD (430-fold overexpressed), is not heat selected over pBAD vector, (5 rounds). b-d, 10 replica flasks. e-g, 5 replica tubes, 5 PCRs per tube.
Two further selection pressures tested the rewired networks. Stationary phase library mixtures were incubated at 37°C, for up to 8 days, in 10 replica flasks. Alternatively, stationary cultures were heat shocked at 42°C (15 min) and then at 50°C (1 hour)21, using three rounds of heat selection, plating and harvesting. In both longevity and heat experiments, virtually all surviving clones were rpoS-ompR (Fig. 3b,c). Since other rpoS-promoter and ompR-ORF clones were never selected, it appears that both are required together. Furthermore, integrated rpoS-ompR is selected over wild-type in heat shock and longevity experiments, despite much lower expression in RT-qPCR: plasmid rpoS-ompR = 650-fold over Co; integrated = 2-fold. Integrated rpoS-ompR heat selection is weaker, requiring more rounds (Fig. 3e), while longevity selection is stronger, reaching 92% after 1 week at 37°C (Fig. 3f). By contrast, 430-fold overexpressed ompR ORF (in pBAD-ompR) is not selected over a pBAD-empty control (Fig. 3g). Therefore selection requires the rewiring combination, functioning even with low expression.
Whilst individual pressures may select for overexpression or new mutations, we have not found evidence of this. Since selections were reproducible in independent tubes, and with different copy numbers, extra mutations are probably not necessary. Therefore, even in a small library space of ~600 networks, acquired connections can themselves provide specific fitness advantages.
DNA chip analysis of rewired gene networks
Affymetrix E. coli Genome 2.0 Arrays were used to get a transcriptome-wide view of rewired networks (3 replicas per sample). The genes were ranked by p-value for different expression between samples. Family-wise error rates (FWER22) and false-discovery rates (FDR23) measured confidence in differential expression (Table 1).
Table 1. DNA chip expression analysis.
a, List of the 23 differentially-expressed genes with lowest p-values, for rpoS-ompR against the control (Co). b, The 8 differentially-expressed genes with lowest p-values for rpoS-ompR (after 3 rounds of heat selection) compared against non heat-treated rpoS-ompR, highlighting downregulation of arginine transport and permeases.
| a | ||||||
|---|---|---|---|---|---|---|
| Rank | P-Value | FDR | FWER | +/− | Gene | Notes |
| 1 | 3E-13 | 1E-7% | 1.1E-9 | + | ompR | osmotic response regulator |
| 2 | 1.4E-9 | 3E-4% | 5.6E-6 | + | nlpD | lipoprotein |
| 3 | 3.1E-5 | 2.3% | 0.13 | + | ycjX | NTH domain |
| 4 | 3.2E-5 | 2.3% | 0.13 | + | ldhA | D-lactate dehydrogenase |
| 5 | 3.2E-5 | 2.3% | 0.13 | + | dnaK | chaperone Hsp70 |
| 6 | 3.4E-5 | 2.3% | 0.14 | − | artM | A3T permease protein |
| 7 | 5.1E-5 | 2.5% | 0.21 | + | groL | GroEL, chaperone Hsp60 |
| 8 | 5.5E-5 | 2.5% | 0.22 | + | maeB | predicted oxidoreductase |
| 9 | 5.5E-5 | 2.5% | 0.23 | + | dnaJ | chaperone with DnaK; Hsp |
| 10 | 7.1E-5 | 2.9% | 0.29 | + | ycjF | conserved inner membrane protein |
| 11 | 1.7E-4 | 6.2% | 0.67 | + | clpB | heat shock protein; Hsp |
| 12 | 1.8E-4 | 6.2% | 0.74 | + | htpG | chaperone Hsp90, Hsp C 62.5 |
| 13 | 2.2E-4 | 6.7% | 0.88 | − | artQ | A3T permease protein |
| 14 | 2.5E-4 | 7.2% | 1 | + | ogrK | prophage P2 ogr protein |
| 15 | 2.7E-4 | 7.3% | 1 | − | cspB | CspB |
| 16 | 3.1E-4 | 7.7% | 1 | + | groS | GroES, Hsp60-binding chaperone |
| 17 | 3.2E-4 | 7.7% | 1 | − | yaiA | ORF for hypothetical protein* |
| 18 | 3.6E-4 | 8.2% | 1 | + | gnty | gluconate transport associated |
| 19 | 4.0E-4 | 8.7% | 1 | + | mfd | mutation frequency decline43 |
| 20 | 4.3E-4 | 8.7% | 1 | − | cspI | cold shock-like protein |
| 21 | 4.9E-4 | 9.1% | 1 | + | hmpA | dihydropteridine reductase |
| 22 | 4.9E-4 | 9.1% | 1 | − | bcsG | gene involved in biofilm formation44 |
| 23 | 5.7E-4 | 9.8% | 1 | + | hslU | Hsp hslVU, chaperone homology |
| b | ||||||
|---|---|---|---|---|---|---|
| Rank | P-Value | FDR | FWER | +/- | Gene | Notes |
| 1 | 1.1E-07 | 0.04% | 0.0004 | − | gadW | Regulator for acid resistance |
| 2 | 2.0E-07 | 0.04% | 0.001 | − | artP | A3T component |
| 3 | 1.1E-06 | 0.1% | 0.004 | − | artQ | A3T permease protein |
| 4 | 2.2E-06 | 0.2% | 0.009 | − | artM | A3T permease protein |
| 5 | 6.2E-06 | 0.4% | 0.025 | − | yfjO | CP4-57 prophage protein* |
| 6 | 6.3E-06 | 0.4% | 0.025 | − | ybcM | DLP12 prophage; AraC type TF* |
| 7 | 7.4E-06 | 0.4% | 0.030 | − | ygiV | DNA gyrase inhibitor paralog |
| 8 | 8.0E-06 | 0.4% | 0.032 | − | yehZ | osmoprotectant (permease)45* |
Abbreviations: FDR = false discovery rate; FWER = family-wise error rate; Hsp = heat shock protein; NTH = nucleoside triphosphate hydrolase; A3T = arginine 3rd transport system
= hypothetical. Shock and permease proteins in yellow and pink, respectively. Up- and down-regulation are in red (+) or green (−).
Comparing rpoS-ompR against Co control, only 13 out of ~4000 genes were differentially expressed with high confidence, including several up-regulated chaperone and shock genes (Table 1a; FWER<1; <1 false positive expected). Extending the list to the 23 most significant differences yields further shock genes (FDR< 10%; 2 expected false positives). After 3 rounds of heat selection, 39 genes changed in rpoS-ompR (FWER<1), 87% being gene down-regulations, including permeases (Table 1b and Supplementary Data 3). RpoS is activated in stationary phase entry, in heat stress and starvation24 and positively regulates genes for acid, heat and salt tolerance25,26. OmpR controls osmoregulation27 and is regulated by several shock pathways to control biofilm formation28,29. Furthermore, endogenous RpoS and OmpR are both positive regulators of csg genes, which are downstream of the cpxA shock signalling pathway30,31. The rpoS-ompR survival mechanism likely includes chaperone and shock gene upregulation, permease downregulation, precise expression timing and refinements after multiple rounds of heat-shock.
The “promoter-only” constructs are interesting, because high-copy promoters could titer out factors that bind to the endogenous promoter, changing overall transcription. However, comparing the rpoS promoter (with GFP ORF) to Co, only 3 high-confidence changes were seen: nlpD(rpoS), cspB and ilvC (FWER <1). In this case, the promoter per se has rather little influence on the transcriptome of the cell.
Unlike the relatively few changes between Co, rpoS, untreated rpoS-ompR and heat-selected rpoS-ompR, 359 genes were differentially expressed in malT-fliA versus Co (FWER<1; Supplementary Information). This clone has a high GFP-level like rpoS-ompR but a contrastingly altered growth signature. Therefore rewiring perturbs ~10% of genes, yet the cell remains viable. Interestingly, integrated malT-fliA has lower transcript expression, relative to Co (11-fold, compared with 67-fold in the plasmid), but it also has perturbed growth, albeit less pronounced (Supplementary Data 2). Since >80% of constructs have near-wt growth characteristics they may be much closer to the rpoS-ompR situation than to malT-fliA, with very few differentially-expressed genes. In that case, the reconnected gene network, even when highly expressed, does not appear to propagate changes across the whole network.
Synthetic biology and gene networks
To understand the forces, hurdles and design principles molding gene network evolution14,32-35 we need to test our understanding by constructing synthetic model systems36-40. The observations described here show that bacteria can both tolerate and exploit radical changes in their circuitry. This raises the exciting possibility that similar experiments could be tried in other organisms, from yeast to mammals, to ascertain whether tolerance towards rewiring is a general feature of evolved biological networks.
It is interesting to compare our rewiring results to those obtained by Sopko and colleagues, who overexpressed 5280 S. cerevisiae genes and found that only 15% cause growth defects41. The effects of rewiring (~16% growth phenotypes) include an element of dosage dependency (overexpression), but also altered timing of expression, and potentially subverting elements in more than one pathway.
For E. coli, it is surprising that rewired clones can have such limited genome-wide transcriptional changes, indicating that bacterial networks have an in-built predisposition to dampen change. E. coli is a complex, tightly coordinated biological system regulated by multiple layers of molecular networks: in tampering with the transcription regulatory network alone, we learn that the static network view provides a map of poor quality to predict the result of genetic perturbations. However, some general trends are ascertainable, such as network hierarchy correlating with expression. Also our results indicate that partition of a network into small modules (negative feedback, feed-forward, etc.) could in some cases be misleading since the behavior of these modules is affected to a large extent by the rest of the network in which they are embedded. The vast majority of added network connections gave no evidence of new phenotypes, even for highly-connected hub genes, yet a few gave selective advantages. This pays tribute to the evolutionary potential provided to the cell by the plasticity of its genome.
METHODS
Cloning
All combinations of the 26 promoter regions with the 22 associated ORFs were cloned into pGLOW-TOPO plasmid (Invitrogen; Fig. 1). Promoter regions were defined as including all upstream TF binding sites annotated in the Ecocyc database15. For ORFs with more than one annotated promoter, both were cloned separately (e.g. rpoS and nlpD promoters for rpoS transcription42; denoted here by nlpD(rpoS)). rpoD has two promoters, and dnaG(rpoD) did not clone successfully (Supplementary Information). Each construct contained the TF or σ-factor coding sequence and a downstream GFP ORF (with separate Shine-Dalgarno sequence; Fig. 1a). A single non-expressing control plasmid (Co) contained a 66 bp non-regulatory DNA sequence upstream of the promoter-less GFP ORF. Full sequences are in Supplementary information.
GFP Measurements
200 μl bacterial cultures (16 h growth) were diluted 20 μl : 180 μl PBS in 96-well plates. 6 independent sample readings (excitation: 485 nm; emission: 520 nm) had Co-background subtracted and were normalised for OD600, with a threshold to remove very low OD readings (background-corrected OD600 <0.03).
Growth signatures
120 μl LB medium (with 100 μg/ml ampicillin and 50 μg/ml streptomycin) were inoculated from 1:200-diluted overnight bacterial cultures using sterile tips. Cultures were grown in 96-well plates. OD600 readings were taken in a Tecan Genios plate reader (XFLUOR4 software; 37°C; 595 nm absorbance; 3 flashes; interval 190 s; Shake duration (orbital low) 130 s; 1000 cycles, ~20 hours; lids on). To avoid edge-effects, only the plates’ central 60 wells were used (outer wells were filled with sterile medium). The assay is sensitive to volume, evaporation and lid condensation; the Tecan machine was optimal (other machines had lid effects). The slope of linear regression of the OD600 readings, over a sliding window of 9 sequential time-points, gave the growth signatures.
RT-qPCR
For Reverse Transcription real time quantitative PCR, RNA was extracted from bacterial cultures with an RNeasy Protect Mini Kit (Qiagen). cDNA was made from 500 ng total RNA, with primer p(dT)15 (Roche) and SuperScript II Reverse Transcriptase (Invitrogen). 0.2 μl cDNA, 0.3 pmols of each primer and 5 μl LightCycler 480 SYBR Green I Master mix (Roche) and a Roche Applied Science LightCycler 480 Instrument (384-wells) were used (10 μl reactions). Samples were normalised for gnd houskeeping gene mRNA and compared to Co expression for fold-difference calculations.
Integrations
~40 representative pGLOW constructs, including Co, were integrated into the E. coli chromosome using manX locus site-directed integration (Gene Bridges Kit K006).
Selection experiments
After serial passaging, 37°C-longevity, or 50°C-heat shock assays, samples were plated onto selective agar media and colonies were picked at random and sequenced or PCR-verified.
Supplementary Material
Acknowledgements
We thank A. Martinez Arias, J. Sharpe, M. Babu, P. Bork and B. Schoenwetter for critical reading of the manuscript; P. Ribeca for RegulonDB analysis; B. Di Ventura & S. Martinez de Pablo for cloning assistance. MI was funded in part by Wellcome Trust Fellowship No. WT066543. CH and ER were funded by European Commission FP6 Netsensor Grant 012948. LS is an ICREA Research Professor (Institució Catalana de Recerca i Estudis Avançats).
ONLINE METHODS
METHODS
Cloning
The 27 promoter regions and 22 ORFs were cloned from Escherichia coli strain Top10 (Invitrogen), using genomic PCR (DNA template extracted with a Genomic-tip 500/G [Qiagen]; 1 μg per PCR). PCR conditions were typically: 97°C, 3 min; (97°C, 30 s; 50°C, 30 s; 72°C, 2 min 30 s) × 10 cycles; (97°C, 30 s; 72°C, 2 min 30 s) × 20 cycles. PCR products were cloned via topoisomerase then FseI-PacI cloning. Full sequences and maps are in Supplementary Information.
The promoter-less control (Co) plasmid was pGLOW-TOPO with a 66 bp DNA fragment in the TOPO cloning site: CGT CGA CGT GGC GCC GCC GGA TAA GGC GTT TAC GTG ACG GCC GGC CCG GGT TCT GGC TTA ATT AAA. This sequence was chosen empirically because it was obtained as an oligonucleotide by-product when cloning promoters into pGLOW-TOPO. The sequence contains no regulatory motifs and has no GFP-inducing activity in bacteria, as measured by fluorimetry and Western blot with anti-GFP antibody.
Site-directed integration into the E. coli chromosome
Approximately 40 of the pGLOW constructs (representing growth phenotypes, selections and Co) were stably integrated into the E. coli chromosome at the manX locus (Gene Bridges Kit K006). The kit uses PCR to provide two 50 bp homology arms, matching the manX locus. Transient expression of the Red/ET recombination proteins provides a highly specific site-directed integration, which is verified by genomic PCR and PCR sequencing. The following generic primers (gel pure) amplify approximately 2.3 kb of plasmid backbone from the original pGLOW constructs (including the ampicillin resistance gene) and add manX homology arms: pGLOW_Amp_F, GTT GAT ACA TGG GGA GGC AGC CCG TTC AAT GCT GCC AGC CGC ATT GTC GTC GCT CAG TGG AAC GAA AAC TC; pGLOW_polyA_R, CGA GCA TTG GAA TGT TAA CGC CTG CAA TGA CTT CAT AAT GCT CTT TGT CGA GCT GGT TCT TTC CGC CTC A.
Library mixtures for selections
The ~570 cloned constructs, plus 23 control (Co) samples, were inoculated from the frozen glycerol stock archive and grown in individual 200 μl wells for 16 hours, as described above. The cultures were then pooled to make a library mixture. Library glycerol stocks were made by adding 4 ml of 50% glycerol to 5 ml of library culture, and storing at −80°C.
Selection by serial passaging
100 μl of library glycerol stock was grown in 120 ml LB medium (with 100 μg/ml ampicillin and 50 μg/ml streptomycin; 37°C and orbital shaking, 300 r.p.m). Culture samples were passaged into fresh medium every 12-16 hours, in 7 replica flasks; for rounds 2 and 3, 100 μl and 10 μl of culture were passaged into fresh medium, respectively; for round 4 onwards, 1 μl samples were passaged (105-106 c.f.u). After 20, 30, 50 and 55 rounds of passaging, samples were plated onto Petri dishes (containing 100 μg/ml ampicillin and 50 μg/ml streptomycin) in order to get single colonies for DNA sequencing (Sequencing primers for pGLOW vector: pGLOW_TOPO_F, TGG CTA GCG TTT AAA CTT AAG C; pGLOW_TOPO_R, GAA TTG GGA CAA CTC CAG TG).
Selection by longevity in stationary phase
1 μl of glycerol stock library mixture inoculated 2 ml LB medium (supplemented with 100 μg/ml ampicillin and 50 μg/ml streptomycin; 10 replica tubes; 37°C, 300 r.p.m. After 24 hrs, 4 days and 8 days, 1 μl samples (diluted in 200 μl LB) were plated onto Petri dishes to get single colonies for DNA sequencing, as above.
Selection by heat shock at 50°C
2 ml library cultures were grown as above. 100 μl samples were then transferred to 0.2 ml PCR tubes on a PCR block, programmed to incubate at 42°C, 15 min, 50°C for 1 hour, and then 4°C, 5 min. 1 μl samples were immediately diluted and plated out onto Petri dishes, as above. Surviving colonies were harvested, grown to stationary phase and the entire procedure was repeated for 3 rounds. One colony was then sequenced per plate, from 10 independent selection tubes.
Affymetrix Chip Analysis
Sample preparation and treatment: Top10 cells containing pGLOW constructs were grown for 16 hours at 37°C (as above), diluted 1000-fold and then grown in 5 ml (pre-warmed) LB medium for 6 hrs (important: cells should not be in stationary phase). 400 μl of culture was used per RNA extraction (Qiagen kit RNeasy). Optical density measurements were taken to quantitate the RNA. Aliquots were checked for RNA degradation by capillary electrophoresis. All subsequent microarray handling was carried out following Affymetrix recommendations. 15 chips (3 per sample) with MG1655 (K12) probesets (Affymetrix E. coli Genome 2.0 Array) were used to test the 5 bacterial populations: Co, rpoS-(no ORF except GFP), rpoS-ompR, rpoS-ompR after heat selection and malT-fliA. To analyse differential expression, a linear model was used through Limma software44 and lists of probabilities of individual genes being differentially expressed were compiled. Holm family-wise error rate45 was used to determine differential expression (FWER<1 as cut-off: fewer than 1 false positive expected in the list of differentially-expressed genes). False discovery rates46 were used as an alternative to calculate the expected number of false positives (fewer than 1 false positive was used as a confidence cut-off to determine significant differential expression).
RT-qPCR
For Reverse Transcription real time quantitative PCR, RNA was extracted from bacterial cultures with an RNeasy Protect Mini Kit (Qiagen). cDNA was made from 500 ng total RNA, with primer p(dT)15 (Roche) and SuperScript II Reverse Transcriptase (Invitrogen). 0.2 μl cDNA, 0.3 pmols of each primer and 5 μl LightCycler 480 SYBR Green I Master mix (Roche) and a Roche Applied Science LightCycler 480 Instrument (384-wells) were used (10 μl reactions). Samples were normalised for gnd houskeeping gene mRNA and compared to Co expression for fold-difference calculations.
- 44.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 45.Holm S. A simple sequentially rejective multiple test procedure. Scand. J. Statist. 1979;6:65–70. [Google Scholar]
- 46.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Statist. Soc. Ser. B. 1995;57:289–300. [Google Scholar]
Footnotes
Supplementary Information accompanies the paper at www.nature.com/nature.
REFERENCES
- 1.Blattner FR, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–74. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 2.Perez-Rueda E, Collado-Vides J. The repertoire of DNA—binding transcriptional regulators in Escherichia coli K—12. Nucleic Acids Res. 2000;28:1838–47. doi: 10.1093/nar/28.8.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salgado H, et al. RegulonDB (version 3.2): transcriptional regulation and operon organization in Escherichia coli K—12. Nucleic Acids Res. 2001;29:72–4. doi: 10.1093/nar/29.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Babu M. Madan, Teichmann SA. Evolution of transcription factors and the gene regulatory network in Escherichia coli. Nucleic Acids Res. 2003;31:1234–44. doi: 10.1093/nar/gkg210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shen-Orr SS, Milo R, Mangan S, Alon U. Network motifs in the transcriptional regulation network of Escherichia coli. Nat. Genet. 2002;31:64–8. doi: 10.1038/ng881. [DOI] [PubMed] [Google Scholar]
- 6.Martinez-Antonio A, Collado-Vides J. Identifying global regulators in transcriptional regulatory networks in bacteria. Curr. Opin. Microbiol. 2003;6:482–9. doi: 10.1016/j.mib.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 7.Salgado H, et al. RegulonDB (version 5.0): Escherichia coli K—12 transcriptional regulatory network, operon organization, and growth conditions. Nucleic Acids Res. 2006;34:D394–7. doi: 10.1093/nar/gkj156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barabasi AL, Albert R. Emergence of scaling in random networks. Science. 1999;286:509–12. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- 9.Guelzim N, Bottani S, Bourgine P, Kepes F. Topological and causal structure of the yeast transcriptional regulatory network. Nat. Genet. 2002;31:60–3. doi: 10.1038/ng873. [DOI] [PubMed] [Google Scholar]
- 10.Albert R, Jeong H, Barabasi AL. Error and attack tolerance of complex networks. Nature. 2000;406:378–82. doi: 10.1038/35019019. [DOI] [PubMed] [Google Scholar]
- 11.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–5. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 12.Hooper SD, Berg OG. On the nature of gene innovation: duplication patterns in microbial genomes. Mol. Biol. Evol. 2003;20:945–54. doi: 10.1093/molbev/msg101. [DOI] [PubMed] [Google Scholar]
- 13.Teichmann SA, Park J, Chothia C. Structural assignments to the Mycoplasma genitalium proteins show extensive gene duplications and domain rearrangements. Proc. Natl. Acad. Sci. U S A. 1998;95:14658–63. doi: 10.1073/pnas.95.25.14658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teichmann SA, Babu MM. Gene regulatory network growth by duplication. Nat. Genet. 2004;36:492–6. doi: 10.1038/ng1340. [DOI] [PubMed] [Google Scholar]
- 15.Keseler IM, et al. EcoCyc: a comprehensive database resource for Escherichia coli. Nucleic Acids Res. 2005;33:D334–7. doi: 10.1093/nar/gki108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mangan MW, et al. The integration host factor (IHF) integrates stationary—phase and virulence gene expression in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 2006;59:1831–47. doi: 10.1111/j.1365-2958.2006.05062.x. [DOI] [PubMed] [Google Scholar]
- 17.Zaslaver A, et al. A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat. Methods. 2006;3:623–8. doi: 10.1038/nmeth895. [DOI] [PubMed] [Google Scholar]
- 18.Pruss BM, Matsumura P. A regulator of the flagellar regulon of Escherichia coli, flhD, also affects cell division. J. Bacteriol. 1996;178:668–74. doi: 10.1128/jb.178.3.668-674.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fong SS, Joyce AR, Palsson BO. Parallel adaptive evolution cultures of Escherichia coli lead to convergent growth phenotypes with different gene expression states. Genome Res. 2005;15:1365–72. doi: 10.1101/gr.3832305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dekel E, Alon U. Optimality and evolutionary tuning of the expression level of a protein. Nature. 2005;436:588–92. doi: 10.1038/nature03842. [DOI] [PubMed] [Google Scholar]
- 21.Delaney JM, Ang D, Georgopoulos C. Isolation and characterization of the Escherichia coli htrD gene, whose product is required for growth at high temperatures. J Bacteriol. 1992;174:1240–7. doi: 10.1128/jb.174.4.1240-1247.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holm S. A simple sequentially rejective multiple test procedure. Scand. J. Statist. 1979;6:65–70. [Google Scholar]
- 23.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Statist. Soc. Ser. B. 1995;57:289–300. [Google Scholar]
- 24.Loewen PC, Hu B, Strutinsky J, Sparling R. Regulation in the rpoS regulon of Escherichia coli. Can. J. Microbiol. 1998;44:707–17. doi: 10.1139/cjm-44-8-707. [DOI] [PubMed] [Google Scholar]
- 25.Cheville AM, Arnold KW, Buchrieser C, Cheng CM, Kaspar CW. rpoS regulation of acid, heat, and salt tolerance in Escherichia coli O157:H7. Appl. Environ. Microbiol. 1996;62:1822–4. doi: 10.1128/aem.62.5.1822-1824.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Foster JW, Moreno M. Inducible acid tolerance mechanisms in enteric bacteria. Novartis Found. Symp. 1999;221:55–69. doi: 10.1002/9780470515631.ch5. discussion 70–4. [DOI] [PubMed] [Google Scholar]
- 27.Pratt LA, Hsing W, Gibson KE, Silhavy TJ. From acids to osmZ: multiple factors influence synthesis of the OmpF and OmpC porins in Escherichia coli. Mol. Microbiol. 1996;20:911–7. doi: 10.1111/j.1365-2958.1996.tb02532.x. [DOI] [PubMed] [Google Scholar]
- 28.Vidal O, et al. Isolation of an Escherichia coli K—12 mutant strain able to form biofilms on inert surfaces: involvement of a new ompR allele that increases curli expression. J. Bacteriol. 1998;180:2442–9. doi: 10.1128/jb.180.9.2442-2449.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prigent-Combaret C, et al. Complex regulatory network controls initial adhesion and biofilm formation in Escherichia coli via regulation of the csgD gene. J. Bacteriol. 2001;183:7213–23. doi: 10.1128/JB.183.24.7213-7223.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cosma CL, Danese PN, Carlson JH, Silhavy TJ, Snyder WB. Mutational activation of the Cpx signal transduction pathway of Escherichia coli suppresses the toxicity conferred by certain envelope—associated stresses. Mol. Microbiol. 1995;18:491–505. doi: 10.1111/j.1365-2958.1995.mmi_18030491.x. [DOI] [PubMed] [Google Scholar]
- 31.Romling U, Bian Z, Hammar M, Sierralta WD, Normark S. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J. Bacteriol. 1998;180:722–31. doi: 10.1128/jb.180.3.722-731.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Babu M. Madan,, Teichmann SA, Aravind L. Evolutionary dynamics of prokaryotic transcriptional regulatory networks. J. Mol. Biol. 2006;358:614–33. doi: 10.1016/j.jmb.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 33.Lozada-Chavez I, Janga SC, Collado-Vides J. Bacterial regulatory networks are extremely flexible in evolution. Nucleic Acids Res. 2006;34:3434–45. doi: 10.1093/nar/gkl423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Babu MM, Luscombe NM, Aravind L, Gerstein M, Teichmann SA. Structure and evolution of transcriptional regulatory networks. Curr. Opin. Struct. Biol. 2004;14:283–91. doi: 10.1016/j.sbi.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 35.Poelwijk FJ, Kiviet DJ, Tans SJ. Evolutionary potential of a duplicated repressor—operator pair: simulating pathways using mutation data. PLoS Comput. Biol. 2006;2:e58. doi: 10.1371/journal.pcbi.0020058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hasty J, McMillen D, Collins JJ. Engineered gene circuits. Nature. 2002;420:224–30. doi: 10.1038/nature01257. [DOI] [PubMed] [Google Scholar]
- 37.Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–8. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 38.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–42. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 39.Isalan M, Lemerle C, Serrano L. Engineering gene networks to emulate Drosophila embryonic pattern formation. PLoS Biol. 2005;3:e64. doi: 10.1371/journal.pbio.0030064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Isalan M, Santori MI, Gonzalez C, Serrano L. Localized transfection on arrays of magnetic beads coated with PCR products. Nat. Methods. 2005;2:113–8. doi: 10.1038/nmeth732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sopko R, et al. Mapping pathways and phenotypes by systematic gene overexpression. Mol. Cell. 2006;21:319–30. doi: 10.1016/j.molcel.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 42.Lange R, Hengge-Aronis R. The nlpD gene is located in an operon with rpoS on the Escherichia coli chromosome and encodes a novel lipoprotein with a potential function in cell wall formation. Mol. Microbiol. 1994;13:733–43. doi: 10.1111/j.1365-2958.1994.tb00466.x. [DOI] [PubMed] [Google Scholar]
- 43.Solano C, et al. Genetic analysis of Salmonella enteritidis biofilm formation: critical role of cellulose. Mol. Microbiol. 2002;43:793–808. doi: 10.1046/j.1365-2958.2002.02802.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.