Abstract
Germline CDH1 point or small frameshift mutations can be identified in 30–50% of hereditary diffuse gastric cancer (HDGC) families. We hypothesized that CDH1 genomic rearrangements would be found in HDGC and identified 160 families with either two gastric cancers in first-degree relatives and with at least one diffuse gastric cancer (DGC) diagnosed before age 50, or three or more DGC in close relatives diagnosed at any age. Sixty-seven carried germline CDH1 point or small frameshift mutations. We screened germline DNA from the 93 mutation negative probands for large genomic rearrangements by Multiplex Ligation-Dependent Probe Amplification. Potential deletions were validated by RT–PCR and breakpoints cloned using a combination of oligo-CGH-arrays and long-range-PCR. In-silico analysis of the CDH1 locus was used to determine a potential mechanism for these rearrangements. Six of 93 (6.5%) previously described mutation negative HDGC probands, from low GC incidence populations (UK and North America), carried genomic deletions (UK and North America). Two families carried an identical deletion spanning 193 593 bp, encompassing the full CDH3 sequence and CDH1 exons 1 and 2. Other deletions affecting exons 1, 2, 15 and/or 16 were identified. The statistically significant over-representation of Alus around breakpoints indicates it as a likely mechanism for these deletions. When all mutations and deletions are considered, the overall frequency of CDH1 alterations in HDGC is ∼46% (73/160). CDH1 large deletions occur in 4% of HDGC families by mechanisms involving mainly non-allelic homologous recombination in Alu repeat sequences. As the finding of pathogenic CDH1 mutations is useful for management of HDGC families, screening for deletions should be offered to at-risk families.
INTRODUCTION
Hereditary diffuse gastric cancer (HDGC) is an autosomal dominant disorder that accounts for <1% of all cases of gastric cancer (GC). Although uncommon, this disease constitutes an important health issue due to its severity, its high penetrance, early age at presentation and the unavailability of effective screening tools. Diffuse gastric cancer (DGC) is the most important cause of cancer lethality in mutation positive HDGC families with lobular breast cancer being a secondary medical concern (1).
Heterozygous germline point or small frameshift mutations in E-cadherin gene (CDH1) [OMIM +192090], a calcium-dependent cell-to-cell adhesion molecule and a tumor suppressor protein, are the only germline molecular defect associated with HDGC (2–4). Overall, carriers of CDH1 germline mutations have a cumulative GC risk, before age 75, of 40–67% for men and 63–83% for women and a risk for lobular breast cancer of 39–52% (5,6). Clinical expression of the disease often occurs before the third decade of life (1), but pre-clinical multifocal tumors, which may have the potential for metastatic spread, are already present in the stomachs of young asymptomatic CDH1 mutation carriers (7,8).
Clinical criteria have been established, in 1999, by the International Gastric Cancer Linkage Consortium (IGCLC) to select families for mutation screening in the CDH1 gene, based mainly on early age of disease presentation and diffuse histology of the tumors (9). These have been later adapted in an attempt to enlarge the population to which this screening could apply, in countries with low incidence of GC (10). Overall, 30–40% of HDGC families harbor CDH1 germline mutations, but this frequency is highly variable between countries with different incidences of GC, with low incidence countries like UK, North America and Canada, displaying nearly 50% of HDGC mutation positive families, while high incidence countries like Portugal and Italy, displaying around 10% of HDGC mutation positive families (4,6,11).
Approximately two-thirds of families, screened worldwide, remain genetically unexplained and the concern related with the management of CDH1 mutation negative gastric cancer families persists. Since the required samples for full genome linkage, in any GC family, are difficult to obtain, a candidate gene approach to identify novel susceptibility genes has been more frequently used. Putative tumor suppressor genes, which are commonly inactivated in sporadic GC and/or associated with GC development, could also represent good candidate susceptibility genes to familial GC. Previous studies including ours, however, have lead us to rule out RUNX3, Caspase-10, SMAD4, HPP1, and Desmoglein 2 as major GC predisposition genes in families with aggregation of gastric carcinoma (12–14). Virtually all diffuse gastric carcinomas, both hereditary and sporadic, independently of whether they carry a CDH1 mutation, display identical morphological features as well as aberrant patterns of expression of E-cadherin (unpublished data). As no other gene has been so far shown to play a major genetic role in the disease, we hypothesize that other genetic events at the CDH1 locus, potentially missed by standard screening techniques, such as large genomic rearrangements, deletions or insertions could be found in families with multiple cases of DGC. Examples of this type of molecular alterations have been recently reported for other cancer associated syndromes. Large genomic rearrangements have been described in hMLH1 and hMSH2 in 10–20% of HNPCC mutation negative families, in APC in 15% of FAP mutation negative families and in lower frequencies in BRCA1 and BRCA2 in breast cancer families initially identified as negative, as well as in CDKN2A in melanoma negative families (15–21).
Based on these observations, we aimed at characterizing CDH1 genomic rearrangements in a large population of HDGC patients, negative for CDH1 point mutations. Moreover, by using a larger series of families screened for CDH1 point mutations and rearrangements, in five reference centers world-wide and arising in countries with different GC incidence rates, we aimed at clarifying the frequency of CDH1 germline alterations and its relationship with GC incidence as well as with clinical criteria for selection of patients at risk.
RESULTS
Multiplex ligation-dependent probe amplification analysis for the CDH1 gene
We collected, from five reference centers, 93 DNA samples from families with aggregation of DGC and negative for CDH1 germline point mutations. These families were selected based on their risk to carry a germline alteration in CDH1 and the clinical criteria described in Materials and Methods section.
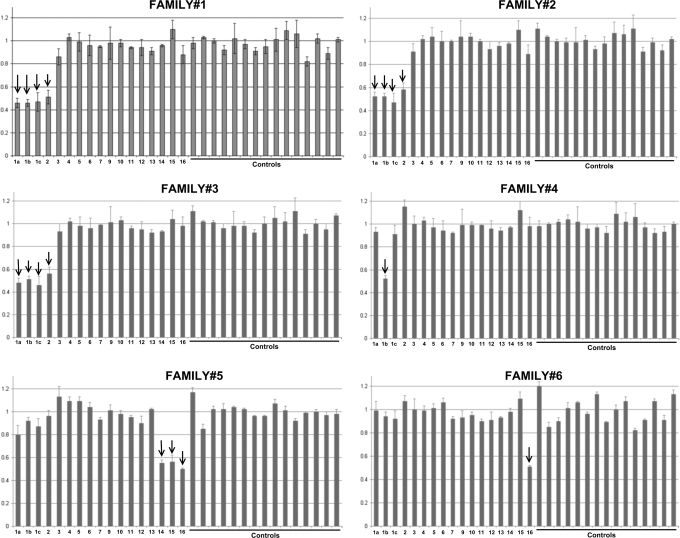
We found that six out of the 93 (6.5%) probands showed abnormal multiplex ligation-dependent probe amplification (MLPA) features when compared with the controls, displaying an ∼70% signal reduction in one or more exons of the CDH1 gene and, suggesting the presence of large deletions affecting the CDH1 locus (Fig. 1, Table 1). No abnormalities were observed in the remaining 87 probands.
Figure 1.
MLPA output from the six HDGC families carrying germline deletions of the CDH1 locus. The first 17 bars represent signal obtained with MLPA probes within the CDH1 gene. Please note that for exon 1, three probes were used (1a, 1b and 1c). CDH1 deletions are represented, in the graph, by smaller bars marked with arrows. Bars on the right hand-side represent results obtained with control probes for MLPA (n = 15).
Table 1.
Table summarizing HDGC families analyzed for large CDH1 rearrangements (gray columns) and compilation of frequency of CDH1 alterations indicating geographic origin grouped by GC incidence rates
| Incidence | Origin (n) | Rearrangement analysis | Deletion (%) | CDH1 positive (%) | CDH1 negative (%) |
|---|---|---|---|---|---|
| Low | North America (92) | 49 | 4 (8.2) | 47 (51.1) | 45 (48.9) |
| UK (33) | 18 | 2 (11.1) | 17 (51.5) | 16 (48.5) | |
| Holland (1) | 0 | 0 | 1 (100.0) | 0 | |
| Total (126) | 67 | 6 (9.0) | 65 (51.6) | 61 (48.4) | |
| Moderate | Germany (16) | 12 | 0 | 4 (25.0) | 12 (75.0) |
| Total (16) | 12 | 0 | (25.0) | 2 (75.0) | |
| High | Portugal (15) | 12 | 0 | 3 (20.0) | 12 (80.0) |
| Italy (3) | 2 | 0 | 1 (33.3) | 2 (66.6) | |
| Total (18) | 14 | (22.2) | 14 (77.8) | ||
| Total (160) | 93 | 6 (6.5)a | 73 (45.6) | 87 (54.4) |
aThis frequency represents 6/93 CDH1 deletions found in negative families. The overall frequency of large CDH1 deletions is 3.8% (6/160).
After reproducing these results in an independent MLPA reaction, we verified that four probands (families 1 to 4) presented deletions at the 5′-end of the gene: in three probands deletions encompassed at least exon 1, intron 1 and exon 2 and, in one proband a smaller deletion that encompassed one of the three probes designed for CDH1 exon 1 was found. The remaining two probands (families 5 and 6) displayed deletions at the further 3’end of the gene: in one proband a deletion of the genomic sequence encompassing exons 14 and 16 was observed, an in the other a deletion of exon 16 only was observed (Fig. 1).
Fine mapping of the deletions breakpoints
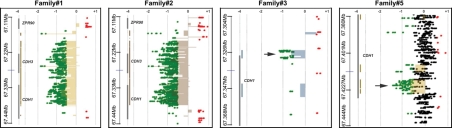
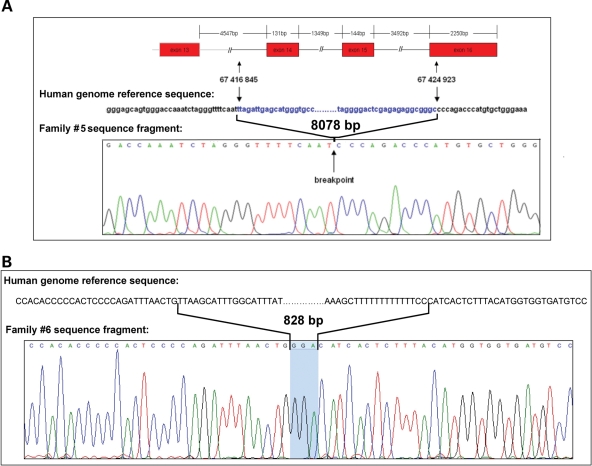
A combination of array CGH and PCR was utilized to map the putative breakpoints in five of the six probands (Fig. 2). The array CGH indicated the regions of chromosome 16 at which the breakpoints were located. PCR primers were then designed to span the breakpoints and the resulting PCR products sequenced (Table 2). Family 1 and family 2 showed identical MLPA patterns as well as virtually identical array CGH results. Sequencing of the breakpoint revealed that these two families harbored an identical 193 593 bp deletion encompassing the full sequence of CDH3 and extending to position IVS2+57 595 in CDH1. Family 3 was found to carry a 5671 bp deletion with breakpoints at position −3809 upstream of the CDH1 transcription start site (TSS) and at position IVS2+742 in CDH1. Family 4 harbored a 150 bp deletion encompassing the TSS of CDH1, with breakpoints located 125 bp upstream and 25 bp downstream of the TSS. Array CGH was not done for this proband since the PCR encompassing CDH1 exon 1 produced both a normal sized and a shorter band that was visible by dHPLC and further used to clone the breakpoints for the deletion by direct sequencing. Family 5 carried an 8078 bp deletion ranging from IVS13-2738 to 20 bp downstream of the stop codon of CDH1, as confirmed by long-range PCR using specific primers flanking the breakpoints (Fig. 3A). Family 6 was found to carry an 828 bp deletion ranging from IVS15+3097 to 223 bp downstream of the stop codon of CDH1 (Fig. 3B).
Figure 2.
Array CGH profiles obtained for HDGC families carrying large CDH1 deletions. Examples are shown for four of the six families. See Tables 2 and 3 for details on the deletion found in each family. Deletions are represented by dense areas of green dots on the left side of each panel. Vertical thick gray bars on the left represent genes in the genomic area surrounding the CDH1 locus and corresponding gene symbols are also shown. Smaller deletions in families#3 and #5 are pointed with black arrows. Please note the scale in Mb on the right, for chromosome position.
Table 2.
Summary of HDGC families displaying CDH1 germline deletions
| ID | Ancestry | GC incidence | Other cancers (age) | Genomic rearrangement |
|---|---|---|---|---|
| Family 1 | Northern European | Low | LBC (61) | Del exon 1–2 (193 593 bp) |
| Family 2 | Canadian | Low | Del exon 1–2 (193 593 bp) | |
| Family 3 | Eastern European | Low | PCa (43) | Del exon 1–2 (5671 bp) |
| Family 4 | Southern European | Low | LCa (nd), PCa (42) | Del 5′-UTR-exon 1 (150 bp) |
| Family 5 | Central European | Low | Unknown | Del exon 14–16 (8078 bp) |
| Family 6 | Central European | Low | Unknown | Del exon 16 (828 bp) |
LBC, lobular breast cancer; PCa, pancreatic cancer; LCa, liver cancer.
Figure 3.
Sequencing chromatogram of the mapped breakpoints in family 5 (A) and family 6 (B). Comparison with the human genome reference sequence. (A) Deletion of 8078 bp without insertion of P or N nucleotides. (B) Deletion of 828 bp and insertion of a triplet at the breakpoint.
Bioinformatics analysis of breakpoints and deletion containing sequences
Assuming that deletions are more likely to occur due to recombination events, we analyzed the genomic sequences flanking the boundaries of these five deletions, which are predicted to constitute target regions for breaks and recombinations to occur. We performed a search for Alu elements which have been reported to mediate non-allelic recombination events in several other malignancies. Evidence for non-homologous end joining (NHEJ) and non-allelic homologous recombination (NAHR) has been described in atypical NF1 microdeletions. These deletions were traced back to short sequences with similarities to Alu elements near the deletion junction (22). Moreover, deletions in the PMP22 gene have been described to be a product of homologous recombination between AluS elements present in intronic regions of the gene (23). We also performed a search for the presence of cryptic sequences recognizable by the RAG1/2 recombinase near the breakpoints and the insertion of P and/or N nucleotides in the rejoining sites. Such sequence features, reminiscent of V(D)J recombination sites, are expected to indicate whether cleavage by the RAG1/2 recombinase may occur near these joining sites (24,25).
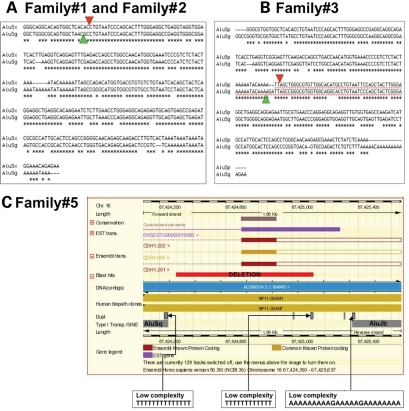
In three families carrying two different deletions we found that deletions overlap Alu repeats (Table 3). In families 1 and 2, bearing the same deletion at chr16:67193822–67387415, both deletion boundaries fall within annotated Alu repeats, AluSp near the start and AluSg near the end. The sequences of the two repeats are highly similar (percentage identity >75%) (Table 3, Fig. 4A). In family 3, carrying a deletion at chr16:67324886–67330557, the start of the deletion maps inside an AluSx repeat while the end map into a repeat AluSg and, the sequences of these two repeats display an identity of ∼80% (Table 3, Fig. 4B).
Table 3.
Deletion coordinates and distance from deletion ends to the closest Alu containing sequences
| Family ID | Deletion coordinates | Closest start Alu | Closest start Alu distance | Closest end Alu | Closest end Alu distance |
|---|---|---|---|---|---|
| Family 1 | chr16:67193822–67387415 | AluSp | overlap | AluSg | overlap |
| Family 2 | chr16:67193822–67387415 | AluSp | overlap | AluSg | overlap |
| Family 3 | chr16:67324886–67330557 | AluSx | overlap | AluSg | overlap |
| Family 4 | chr16:67328695–67328844 | AluJo | −304 | AluJo | 1497 |
| Family 5 | chr16:67416845–67424923 | FLAM_C | −344 | AluJb | 442 |
| Family 6 | chr16:67424298–67425126 | AluSq | −50 | AluJb | 239 |
Figure 4.
Analysis of sequences flanking the deletion breakpoints. (A and B) Homology analysis of AluS type elements flanking deletions in families 1, 2 and 3. (C) Ensembl based scheme displaying the repetitive elements flanking the deletion in family 5.
For the deletion at chr16:67328695–67328844, present in family 4, an AluJo repeat is present 304 bp upstream from the start of the deletion, and another AluJo is present 1497 bp downstream the end of the deletion. The closest repeat external to the deletion is a MIR named MIRb, 1235 bp downstream of the end-point but no sequence identity can be seen in the −150/+50 and −50/+150 bp fragments centered, respectively, on the start and the end of the deletion. An interesting feature of this 150 bp deletion is the presence of microsinteny/microhomology around the breakpoints due to the presence of five nucleotides sequence (CTCTC) between the upstream and downstream breakpoints.
Family 5 displays a deletion at chr16:67416845–67424923 that does not overlap Alu repeats, although it occurs in the proximity of this type of repetitive regions (Table 3). The closest repeats both upstream and downstream are AT rich sequences, ∼300 bp distant from the deletion (Fig. 4C). Moreover, 344 bp upstream of the deletion start a FLAM_C repeat exists and 442 bp downstream of the endpoint there is an AluJb sequence (Table 3). The alignment of the −150/+150 bp surrounding the deletion ends did not show any identity.
The deletion in family 6, at chr16:67424298–67425126, is unlikely to have been produced by homology-dependent mechanisms, since a small (3 bp) sequence insertion at the junction between the deletion breakpoints was identified (Fig. 3B). For this deletion, the closest Alu repeats found on both sides of the deletion are an AluSq 50 bp upstream and an AluJb 239 bp downstream of the deletion’s start and end, which share a sequence identity >77% (Table 3).
In a range of +/−5 Mb flanking the five deletions reported in this study, we did not find evidence for the presence of RAG1/2 consensus sequences accompanied or not by the insertion of P and/or N nucleotides in the rejoining sites. Nevertheless, the presence of Alus overlapping 6 out of the 12 points analyzed (the ends of the six deletion sequences) is statistically significant (z-score 3.3, P-value 0.0005) as tested by performing a randomization analysis with 1000 random sampling of 12 random locations (see Materials and Methods).
Clinical features of deletion-carrying HDGC families
The proband in family 1 was a 61-year-old female of European Irish/German/Scottish/Danish ancestry presenting with lobular breast cancer. Her daughter and son, as well as a grand-daughter developed DGC at the age of 40, 37 and 28, respectively, and are already diseased. The deletion was only looked for in the proband that is currently alive.
In family 2, the proband, of unknown origin but currently living in Canada, was a female who developed DGC at 38 and later at 43 years old and the only patient tested in this family. Her two sisters also developed DGC at the ages of 30 and 35, respectively, but were not tested for the presence of the same deletion.
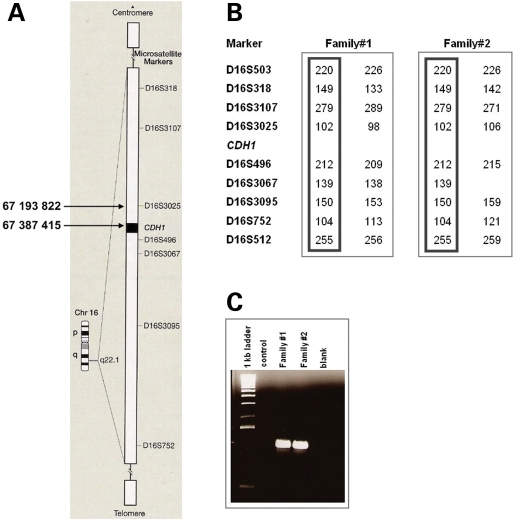
The presence of the same deletion in two different families could be explained two ways. Either of these families had a common ancestor or the deletions arose independently. To clarify this issue, we performed haplotype analysis with seven microsatellite markers surrounding the CDH1 gene and verified that an allele for each of the microsatellite markers was common between the two families, demonstrating the existence of a common ancestor carrying this deletion (Fig. 5).
Figure 5.
Haplotype analysis of family 1 and family 2 probands. (A) Scheme of chromosome 16 with reference to microsatellite markers used in the haplotype analysis and to the breakpoints of the deletion. (B) Common haplotypes displayed by probands of family 1 and family 2 and proving a common ancestry. (C) Agarose gel of the PCR that allowed the mapping of the deletion, showing similar band sizes for probands in both families.
The proband in family 3, was a 34-year-old female of European Lithuanian origin, with DGC. Her mother was also affected by the same disease at uncertain age, and her grand-father developed pancreatic cancer at the age of 43 and none of them was tested for the deletion. The proband in family 4, was a 40-year-old male of Hispanic origin who developed DGC at the age of 40. His 24-year-old brother died from exactly the same disease and his father died at 42 with a pancreatic cancer, neither has been tested for the deletion. Carrier tests have been performed in ten other family members and one of which was positive.
In family 5, the proband was a patient of European ancestry, who developed DGC at 33 years. His two sisters developed DGC at 33 and 35 years. At least one second degree relative developed GC, but no more information was available. So far, only the proband was tested, but five other family members are currently being tested for the presence of the deletion. As for family 6, the proband was a 51-year-old patient of European ancestry who developed DGC. His mother died of GC of unconfirmed histology at age of 52. Two sisters were recently diagnosed with DGC by surveillance endoscopy, although only one of them was tested for the deletion and proved to be a carrier. This sister’s offspring, constituted of two asymptomatic individuals, also tested positive for the deletion. Four other second degree relatives died of cancer, one with DGC, two of GC of unknown histology and one of lobular breast cancer.
Clinical criteria, frequency of CDH1 alterations and GC incidence
In the present study, we analyzed 93 families based on two clinical criteria: (i) three or more DGC in first degree relatives diagnosed at any age or; (ii) two or more GC in first degree relatives with at least one DGC diagnosed before age 50; and found that 6.5% of these families carry large CDH1 germline deletions.
With the purpose of clarifying the relative contribution of germline CDH1 point mutations and deletions in a large series of families fulfilling the aforementioned criteria, we collected data not only on the 93 negative families, herein analyzed for genomic rearrangements, but also on 67 HDGC families carrying CDH1 point mutations, from five different reference centers, and originated from countries with low, moderate and high GC incidence rates.
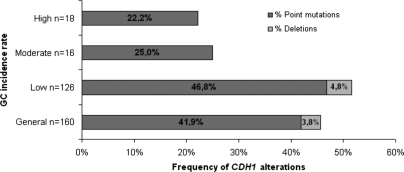
From the series of 160 probands, 73 (45.6%) were proved to carry germline CDH1 alterations: 41.9% point mutations and 3.8% large deletions (Fig. 6).
Figure 6.
Frequency of CDH1 alterations in a series of 160 HDGC families, subdivided by GC incidence rates as well as by type of gene alteration (point or small frameshift mutations and large deletions).
In low incidence countries namely in the North America, Canada, UK and Holland, the frequency of germline CDH1 alterations was 51.6%, while in moderate (Germany) and high (Portugal and Italy) incidence countries the frequency of alterations was 25 and 22.2%, respectively (Fig. 6). Large deletions were only identified in countries with low incidence of GC and account for 9% of all CDH1 alterations in HDGC families from such countries (Table 1).
DISCUSSION
In this report, we demonstrate that germline large deletions affecting the CDH1 locus are one of the mechanisms underlying HDGC. Our data supports the addition of MLPA based screening for such deletions to clinical screening for CDH1 mutations.
The CDH1 locus, like the hMLH1, hMSH2, BRCA1, APC and CDKN2A loci, is a region susceptible to genomic rearrangements leading to increased cancer susceptibility (18,19,21,26).
The analysis of deletions found in our HDGC patients, revealed that two out of five deletions showed extensive sequence identity between the two breakpoints, strongly suggesting that NAHR was the mechanism of their formation (Families 1, 2 and 3). The deletion breakpoints of these two deletions fall within Alu repeats similar to what has been previously reported for hMLH1, hMSH2, BRCA1 and infrequently for APC (18,19,26). Alu repetitive sequences have long been believed to give rise to genomic deletions promoting recombinational instability (27). PMP22 deletions have been described to be a product of homologous recombination between intronic AluS elements in hereditary neuropathies (23), but the type of Alu sequences involved in deleterious deletions underlying cancer-associated syndromes have not been systematically explored in the literature. Nevertheless, at least one of the Alu sequences located at large deletion breakpoints in hMLH1 gene are described to be of the AluSx type (28). In our study, we analyzed the type of Alu sequences associated with CDH1 large deletions in HDGC, and found that six breakpoints overlap AluS elements: three overlap AluSg, two overlap AluSp and one overlaps an AluSx (Table 3).
One of the deletions herein identified occurs in a context of microhomology and could have arisen by microhomology-mediated recombination, a mechanism of NHEJ. This mechanism is associated with very short stretches of sequence identity (a few bp) between the two ends of the breakpoint junctions (29,30), rather than with long stretches of sequence identity at these locations (31,32). Moreover, it involves the double strand breakage of DNA followed by end joining in the absence of extensive sequence homology. A similar microhomology-mediated intra-strand repair mechanism has been previously described to occur either in pathological conditions such as trisomy in a family displaying an inverted duplication/deletion of 2p25.1–25.3 (33) or in recombination events underlying genomic variation during evolution (34). The same putative mechanism underlying the type of deletion and end re-joining, found in this family, has been described in a recent report where the treatment of mice, with specific chemicals or radiation during late spermatogenic stages, generates very short deletions, duplications, or inversions with microhomology around breakpoints. The ligation by NHEJ mediated by microhomology, is raised in the aforementioned report, as the major path of repair (35), and provides insight on how this specific CDH1 deletion may have initially arisen.
Another of our deletions is unlikely to have been produced by homology-dependent mechanisms, since a small sequence insertion (3 bp) at the junction between the deletion breakpoints was identified. Since no extensive homology exists in the vicinity of breakpoints, it is likely that this deletion have been mediated by NHEJ. Identical deletions involving small insertions (3, 4, 6 bp), of unknown origin at the site of the breakpoints, have been previously identified within copy number variants (CNVs) breakpoint-region sequences, in normal population (32,34).
In contrast to all other CDH1 deletions described so far in this report, one does not seem to be caused by NAHR, microhomology-mediated recombination or small sequence insertions at the junction site. This deletion is flanked by upstream and downstream low complexity AT rich sequences, as well as FLAM_C repeat and an AluJb sequence. Similar features surrounding deletions have been previously described to occur in CNVs described in apparently normal individuals (34), but the mechanisms involved in their generation is not known. The AT rich sequences could mediate a microhomology-mediated deletion mechanism based on their base composition bias, though further evidence would be needed to prove this.
Deletions found in HDGC families mainly fall into three separate classes: products of NAHR, in which recombination between Alu containing sequences leads to deletion of the sequence between them; products of microhomology-mediated recombination, a mechanism of NHEJ; and results of small insertions of unknown origin at the site of the breakpoint junction. These deleterious deletions in CDH1 appear as the pathogenic counterpart of apparently normal evolution generated CNVs (32,34).
The probands studied (n = 160) fulfilled clinically used criteria for HDGC (see Materials and Methods). We found that 45.6% (73/160) of them displayed CDH1 alterations (Fig. 6). In comparison, only 7/123 (5.7%) of the probands of families who did not meet the above testing criteria, had point mutations or small frameshift mutations, and none had deletions detectable through MLPA (data not shown). Based on this analysis, we have demonstrated that the clinical criteria herein used (three or more DGC in first degree relatives diagnosed at any age or two or more GC in first degree relatives with at least one DGC diagnosed before age 50) will identify most families with HDGC associated germline CDH1 alterations, with a mutation pick up rate of ∼46%. Based on these criteria, ∼4% of HDGC patients display large CDH1 germline deletions (Fig. 6).
Two of the families shared the same large deletion that included the whole of the CDH3 gene and the first two exons of CDH1. Individuals with homozygous germline truncating mutations in the CDH3 gene present with hypotrichosis with juvenile macular dystrophy (HJMD) syndrome (36,37). Heterozygous carriers of such mutations are phenotypically normal and have not been reported to have increased GC risk. Haplotype analysis supports a shared ancestor as the source of the (chr16:67193822–67387415) in these two families. We have previously used this approach to demonstrate that recurrent HDGC associated CDH1 germline point mutations can arise from a common ancestor (6).
In conclusion, 6.5% of probands from strictly defined HDGC families who previously tested negative for CDH1 mutations carried heterozygous germline deletions. As these can be readily identified using MLPA, an assay frequently used in clinical cancer genetics laboratories, we suggest that MLPA be added to the testing strategy for all families who meet clinical criteria for HDGC.
MATERIALS AND METHODS
Patients and families
The collection of clinical data and peripheral blood samples was approved by the appropriate Ethics Committee from each of the centers participating in this work: University of British Columbia, Vancouver, Canada; Institute of Molecular Pathology and Immunology, University of Porto, Porto, Portugal; Department of Human Pathology and Oncology, Section of Surgical Oncology, Translational Research Laboratory, University of Siena, Italy; Institute of Pathology, Technische Universität München, Munich, Germany; Department of Oncology, University of Cambridge, Cambridge, UK. A series of 160 probands from families with aggregation of DGC was analyzed in this study, based on the following clinical criteria: (i) three or more DGC in first degree relatives diagnosed at any age or; (ii) two or more GC in first degree relatives with at least one DGC diagnosed before age 50 (see Table 1). From these 160 probands, 67 showed point mutations in the CDH1 gene and were used only for analysis of general frequency of CDH1 mutations within HDGC families. The majority of these families have been published elsewhere (3,6,10–13,38–46) except for 15 families from University of British Columbia, Vancouver, Canada (unpublished data DGH), six families from Department of Oncology, University of Cambridge, Cambridge (unpublished data CC) and two families from Technische Universität München, Munich, Germany (unpublished data GK). The remaining 93 probands belonged to families that tested negative for CDH1 germline point mutations. These were analyzed for CDH1 genomic rearrangements.
Peripheral blood leukocytes from the 93 negative probands were referred for analysis at the different centers. High molecular weight genomic DNA was isolated from all patients using standard methodologies.
Multiplex ligation-dependent probe amplification
A series of 93 probands, negative for CDH1 germline point mutations, were tested for the presence of large genomic alterations at the CDH1 locus using MLPA. MLPA is a PCR-based technique for identifying gene dosage alterations, which has been previously described in detail (47). All reagents were provided by the manufacturer in a kit (SALSA P085 CDH1 MLPA kit; MRC-Holland, Amsterdam, Holland), and testing was performed according to the manufacturer’s recommendations. The assay contains probes for all but one of the CDH1 exons. For technical reasons exon 8 has been excluded and exon 1 has three different probes. The assay was done according to manufacturer’s instructions. Briefly, the probes were hybridized overnight at 60°C with 100 ng of germline DNA and subsequently ligated. Ten microliters of the ligation reaction was used in the PCR which contained FAM labeled primers. The PCR products were analyzed on an ABI 3130×L Genetic Analyzer. The output for each patient displayed a peak that corresponded to the amount of amplified ligated probe present for each exon of the CDH1 gene, along with several peaks representative of extragenic regions used for ligation and PCR monitoring controls. Peak heights for fragments corresponding to specific exons and control regions were binned, appended to a table, and then saved as a text file, all within the Applied Biosystems Genotyper software. The resulting data were analyzed using the Coffalyser software downloaded from MRC-Holland. Any exon with a decrease or increase in peak height of 70% was scored as a deletion or duplication, respectively. MLPA assays which showed a variation between peak ratios were repeated.
Comparative genomic hybridization microarray
A CGH array strategy was done to confirm the deletions identified by MLPA and to identify the location of deletion breakpoints. Custom CGH arrays were designed using Agilent’s eArray program. An 8×15K array format was generated with Agilent HD CGH Database Catalog Probes for coverage of chromosome 16: 066880834–067879823. Genomic DNA (1.5 µg) was labeled using the Agilent Genomic DNA Labeling Kit PLUS (Agilent Technologies, Santa Clara, CA). Samples labeled with Cy3 were co-hybridized with Cy5 labeled gender-matched normal reference gDNA (Promega, Madison WI) to custom 8×15K Agilent human CGH microarrays. Microarrays were washed with Agilent CGH wash buffers and scanned on an Agilent DNA Microarray Scanner. Images were analyzed using Agilent Feature Extraction (version 8.5) and data were analyzed using Agilent CGH Analytics (version 3.4.27).
Breakpoint identification
Based on the information obtained from the CGH array, primers were designed spanning the putative breakpoints for each case and used in long-range PCR. A table with primer sequences is supplied as supplementary material (Supplementary Material, Table S1).
Haplotype analysis
Analysis was performed using the following microsatellite markers surrounding the CDH1 gene: D16S318, D16S3107, D16S3025, D16S496, D16S3067, D16S3095 and D16S752. Each marker was amplified and then analyzed on an ABI 3130×L Genetic Analyzer followed by analysis using Genemapper V7.0 software. Primer sequences for the microsatellite markers D16S3025, D16S496, D16S3095 and D16S752 have been previously reported (11). Sequences for the remaining markers, D16S318, D16S3107 and D16S3067 are listed in the genome database (http://www.gdb.org).
Analysis of breakpoints sequence context
Breakpoints were defined as a set of coordinates on the genome spanning the genomic sequence of the deletions. The coordinates have been loaded on the UCSC genome browser as a BED file accessible on http://biodev.cbm.fvg.it/remo/DEL.txt. Both automatic and manual bioinformatics analysis have been carried out in order to analyze the genomic context of these regions.
For each deletion we analyzed the closest repeats in a range of 1500 bp flanking the start/end points. Repeats were obtained from the Ensembl Homo sapiens core database version 50 and UCSC genome browser Human Mar. 2006 Assembly. Sequences in a range of −150/+50 bp flanking the start-point and −50/+150 bp flanking the end-point of the deletions have been collected in order to test the similarity around the breakpoints by using both clustalw and blast2 with default parameters. An interval of 5 Mb around the deletion start/end points has been analyzed in order to look for the presence of the RAG1 binding site on both strands by using a custom perl script and known consensus sequences (CACTGTG.{11,13}GGTTTTTGT,GGTTTTTGT.{11,13}CACTGTG, ACAAAAACC.{11,13}CACAGTG,CACAGTG.{11,13}ACAAAAACC,CACTGTG.{22,24}GGTTTTTGT,GGTTTTTGT.{22,24}CACTGTG,ACAAAAACC.{22,24}CACAGTG,CACAGTG.{22,24}ACAAAAACC). In order to perform randomization analysis we collected 150 000 random points on the human genome and calculated for each of them the distance from the closest Alu. The distribution of the distances has been trimmed in order to avoid considering points in poorly annotated regions and/or in gene deserts and/or in genomic gaps, which resulted nonetheless to be very far from Alus. We trimmed out all the points retaining a distance bigger than 4519 bp (the 1.5*IQR of the distribution of all the distances) from the closest Alu.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the following grants: The National Cancer Institute of Canada (Canadian Cancer Society); The Michael Smith Foundation for Health Research to D.H.; The Portuguese Foundation for Science and Technology—FCT (grant numbers POCTI/SAU-OBS/58111/2004, PTDC/SAU-GMG/72168/2006, Program Ciência 2007 to C.O.); Sixth Framework Programme from European Union FP6 (grant number LSHC-CT-2005-018754); The Deutsche krebshilfe (grant number 10-1345-HÖ2) and Istituto Toscano Tumori (ITT) (Grant 2007 Ref: Gene expression profile and therapeutic implication in GC. From the clinical overview to the translational research). Funding to pay the Open Access charge was provided by National Cancer Institute of Canada Grant #018381 “The effect of E-cadherin mutations on gastric and breast cancer risk: from families to populations.”
Supplementary Material
ACKNOWLEDGEMENTS
We gratefully acknowledge patients, families and their caregivers for their willing participation in this research project and who provided consent regarding use of the information obtained from the study.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Lynch H.T., Kaurah P., Wirtzfeld D., Rubinstein W.S., Weissman S., Lynch J.F., Grady W., Wiyrick S., Senz J., Huntsman D.G. Hereditary diffuse gastric cancer: diagnosis, genetic counseling, and prophylactic total gastrectomy. Cancer. 2008;112:2655–2663. doi: 10.1002/cncr.23501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guilford P., Hopkins J., Harraway J., McLeod M., McLeod N., Harawira P., Taite H., Scoular R., Miller A., Reeve A.E. E-cadherin germline mutations in familial gastric cancer. Nature. 1998;392:402–405. doi: 10.1038/32918. [DOI] [PubMed] [Google Scholar]
- 3.Gayther S.A., Gorringe K.L., Ramus S.J., Huntsman D., Roviello F., Grehan N., Machado J.C., Pinto E., Seruca R., Halling K., et al. Identification of germ-line E-cadherin mutations in gastric cancer families of European origin. Cancer Res. 1998;58:4086–4089. [PubMed] [Google Scholar]
- 4.Oliveira C., Seruca R., Carneiro F. Genetics, pathology, and clinics of familial gastric cancer. Int. J. Surg. Pathol. 2006;14:21–33. doi: 10.1177/106689690601400105. [DOI] [PubMed] [Google Scholar]
- 5.Pharoah P.D., Guilford P., Caldas C. International Gastric Cancer Linkage Consortium. Incidence of gastric cancer and breast cancer in CDH1 (E-cadherin) mutation carriers from hereditary diffuse gastric cancer families. Gastroenterology. 2001;121:1348–1353. doi: 10.1053/gast.2001.29611. [DOI] [PubMed] [Google Scholar]
- 6.Kaurah P., MacMillan A., Boyd N., Senz J., De Luca A., Chun N., Suriano G., Zaor S., Van Manen L., Gilpin C., et al. Founder and recurrent CDH1 mutations in families with hereditary diffuse gastric cancer. JAMA. 2007;297:2360–2372. doi: 10.1001/jama.297.21.2360. [DOI] [PubMed] [Google Scholar]
- 7.Huntsman D.G., Carneiro F., Lewis F.R., MacLeod P.M., Hayashi A., Monaghan K.G., Maung R., Seruca R., Jackson C.E., Caldas C. Early gastric cancer in young, asymptomatic carriers of germ-line E-cadherin mutations. N. Engl. J. Med. 2001;344:1904–1909. doi: 10.1056/NEJM200106213442504. [DOI] [PubMed] [Google Scholar]
- 8.Chun Y.S., Lindor N.M., Smyrk T.C., Petersen B.T., Burgart L.J., Guilford P.J., Donohue J.H. Germline E-cadherin gene mutations: is prophylactic total gastrectomy indicated? Cancer. 2001;92:181–187. doi: 10.1002/1097-0142(20010701)92:1<181::aid-cncr1307>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 9.Caldas C., Carneiro F., Lynch H.T., Yokota J., Wiesner G.L., Powell S.M., Lewis F.R., Huntsman D.G., Pharoah P.D., Jankowski J.A., et al. Familial gastric cancer: overview and guidelines for management. J. Med. Genet. 1999;36:873–880. [PMC free article] [PubMed] [Google Scholar]
- 10.Brooks-Wilson A.R., Kaurah P., Suriano G., Leach S., Senz J., Grehan N., Butterfield Y.S., Jeyes J., Schinas J., Bacani J., et al. Germline E-cadherin mutations in hereditary diffuse gastric cancer: assessment of 42 new families and review of genetic screening criteria. J. Med. Genet. 2004;41:508–517. doi: 10.1136/jmg.2004.018275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suriano G., Yew S., Ferreira P., Senz J., Kaurah P., Ford J.M., Longacre T.A., Norton J.A., Chun N., Young S., et al. Characterization of a recurrent germ line mutation of the E-cadherin gene: implications for genetic testing and clinical management. Clin. Cancer Res. 2005;11:5401–5419. doi: 10.1158/1078-0432.CCR-05-0247. [DOI] [PubMed] [Google Scholar]
- 12.Keller G., Vogelsang H., Becker I., Plaschke S., Ott K., Suriano G., Mateus A.R., Seruca R., Biedermann K., Huntsman D., et al. Germline mutations of the E-cadherin(CDH1) and TP53 genes, rather than of RUNX3 and HPP1, contribute to genetic predisposition in German gastric cancer patients. J. Med. Genet. 2004;41:e89. doi: 10.1136/jmg.2003.015594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oliveira C., Ferreira P., Nabais S., Campos L., Ferreira A., Cirnes L., Alves C.C., Veiga I., Fragoso M., Regateiro F., et al. E-Cadherin (CDH1) and TP53 rather than SMAD4 and Caspase-10 germline mutations contribute to genetic predisposition in Portuguese gastric cancer patients. Eur. J. Cancer. 2004;40:1897–1903. doi: 10.1016/j.ejca.2004.04.027. [DOI] [PubMed] [Google Scholar]
- 14.Biedermann K., Vogelsang H., Becker I., Plaschke S., Siewert J.R., Höfler H., Keller G. Desmoglein 2 is expressed abnormally rather than mutated in familial and sporadic gastric cancer. J. Pathol. 2005;207:199–206. doi: 10.1002/path.1821. [DOI] [PubMed] [Google Scholar]
- 15.Nakagawa H., Hampel H., de la Chapelle A. Identification and characterization of genomic rearrangements of MSH2 and MLH1 in Lynch syndrome (HNPCC) by novel techniques. Hum. Mutat. 2003;22:258. doi: 10.1002/humu.9171. [DOI] [PubMed] [Google Scholar]
- 16.Wijnen J., van der Klift H., Vasen H., Khan P.M., Menko F., Tops C., Meijers Heijboer H., Lindhout D., Møller P., Fodde R. MSH2 genomic deletions are a frequent cause of HNPCC. Nat. Genet. 1998;20:326–328. doi: 10.1038/3795. [DOI] [PubMed] [Google Scholar]
- 17.Charbonnier F., Olschwang S., Wang Q., Boisson C., Martin C., Buisine M.P., Puisieux A., Frebourg T. MSH2 in contrast to MLH1 and MSH6 is frequently inactivated by exonic and promoter rearrangements in hereditary nonpolyposis colorectal cancer. Cancer Res. 2002;62:848–853. [PubMed] [Google Scholar]
- 18.Baudhuin L.M., Ferber M.J., Winters J.L., Steenblock K.J., Swanson R.L., French A.J., Butz M.L., Thibodeau S.N. Characterization of hMLH1 and hMSH2 gene dosage alterations in Lynch syndrome patients. Gastroenterology. 2005;129:846–854. doi: 10.1053/j.gastro.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 19.Michils G., Tejpar S., Thoelen R., van Cutsem E., Vermeesch J.R., Fryns J.P., Legius E., Matthijs G. Large deletions of the APC Gene in 15% of mutation-negative patients with classical polyposis (FAP): a Belgian study. Hum. Mutat. 2005;25:125–134. doi: 10.1002/humu.20122. [DOI] [PubMed] [Google Scholar]
- 20.Thomassen M., Gerdes A.M., Cruger D., Jensen P.K., Kruse T.A. Low frequency of large genomic rearrangements of BRCA1 and BRCA2 in western Denmark. Cancer Genet. Cytogenet. 2006;168:168–171. doi: 10.1016/j.cancergencyto.2005.12.016. [DOI] [PubMed] [Google Scholar]
- 21.Knappskog S., Geisler J., Arnesen T., Lillehaug J.R., Lønning P.E. A Novel Type of Deletion in the CDKN2A Gene Identified in a Melanoma-Prone Family. Genes Chrom. Cancer. 2006;45:1155–1163. doi: 10.1002/gcc.20379. [DOI] [PubMed] [Google Scholar]
- 22.Venturin M., Gervasini C., Orzan F., Bentivegna A., Corrado L., Colapietro P., Friso A., Tenconi R., Upadhyaya M., Larizza L., et al. Evidence for non-homologous end joining and non-allelic homologous recombination in atypical NF1 microdeletions. Hum. Genet. 2004;115:69–80. doi: 10.1007/s00439-004-1101-2. [DOI] [PubMed] [Google Scholar]
- 23.Matejas V., Huehne K., Thiel C., Sommer C., Jakubiczka S., Rautenstrauss B. Identification of Alu elements mediating a partial PMP22deletion. Neurogenetics. 2006;7:119–126. doi: 10.1007/s10048-006-0030-8. [DOI] [PubMed] [Google Scholar]
- 24.Ramsden D.A., McBlane J.F., van Gent D.C., Gellert M. Distinct DNA sequence and structure requirements for the two steps of V(D)J recombination signal cleavage. EMBO J. 1996;15:3197–3206. [PMC free article] [PubMed] [Google Scholar]
- 25.Lewis S.M., Agard E., Suh S., Czyzyk L. Cryptic signals and the fidelity of V(D)J joining. Mol. Cell. Biol. 1997;17:3125–3136. doi: 10.1128/mcb.17.6.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Casilli F., Tournier I., Sinilnikova O.M., Coulet F., Soubrier F., Houdayer C., Hardouin A., Berthet P., Sobol H., Bourdon V., et al. The contribution of germline rearrangements to the spectrum of BRCA2 mutations. J. Med. Genet. 2006;43:e49. doi: 10.1136/jmg.2005.040212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krawczak M., Cooper D.N. Gene deletions causing human genetic disease: mechanisms of mutagenesis and the role of the local DNA sequence environment. Hum. Genet. 1991;86:425–441. doi: 10.1007/BF00194629. [DOI] [PubMed] [Google Scholar]
- 28.Pistorius S., Görgens H., Plaschke J., Hoehl R., Krüger S., Engel C., Saeger H.D., Schackert H.K. Genomic rearrangements in MSH2, MLH1 or MSH6 are rare in HNPCC patients carrying point mutations. Cancer Lett. 2007;248:89–95. doi: 10.1016/j.canlet.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 29.Chan C.Y., Kiechle M., Manivasakam P., Schiestl R.H. Ionizing radiation and restriction enzymes induce microhomology-mediated illegitimate recombination in Saccharomyces cerevisiae. Nucleic Acids Res. 2007;35:5051–5059. doi: 10.1093/nar/gkm442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee K., Lee S.E. Saccharomyces cerevisiae Sae2- and Tel1-dependent single-strand DNA formation at DNA break promotes microhomology-mediated end joining. Genetics. 2007;176:2003–2014. doi: 10.1534/genetics.107.076539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Korbel J.O., Urban A.E., Affourtit J.P., Godwin B., Grubert F., Simons J.F., Kim P.M., Palejev D., Carriero N.J., Du L., et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perry G.H., Ben-Dor A., Tsalenko A., Sampas N., Rodriguez-Revenga L., Tran C.W., Scheffer A., Steinfeld I., Tsang P., Yamada N.A., et al. The fine-scale and complex architecture of human copy-number variation. Am. J. Hum. Genet. 2008;82:685–695. doi: 10.1016/j.ajhg.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bonaglia M.C., Giorda R., Massagli A., Galluzzi R., Ciccone R., Zuffardi O. A familial inverted duplication/deletion of 2p25.1-25.3 provides new clues on the genesis of inverted duplications. Eur. J. Hum. Genet. 2008 doi: 10.1038/ejhg.2008.160. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Smith A.J., Walters R.G., Coin L.J., Steinfeld I., Yakhini Z., Sladek R., Froguel P., Blakemore A.I. Small deletion variants have stable breakpoints commonly associated with alu elements. PLoS ONE. 2008;3:e3104. doi: 10.1371/journal.pone.0003104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elso C., Lu X., Morrison S., Tarver A., Thompson H., Thurkow H., Yamada N.A., Stubbs L. Germline translocations in mice: unique tools for analyzing gene function and long-distance regulatory mechanisms. J. Natl Cancer Inst. Monogr. 2008;39:91–95. doi: 10.1093/jncimonographs/lgn008. [DOI] [PubMed] [Google Scholar]
- 36.Sprecher E., Bergman R., Richard G., Lurie R., Shalev S., Petronius D., Shalata A., Anbinder Y., Leibu R., Perlman I., et al. Hypotrichosis with juvenile macular dystrophy is caused by a mutation in CDH3, encoding P-cadherin. Nat. Genet. 2001;29:134–136. doi: 10.1038/ng716. [DOI] [PubMed] [Google Scholar]
- 37.Kjaer K.W., Hansen L., Schwabe G.C., Marques-de-Faria A.P., Eiberg H., Mundlos S., Tommerup N., Rosenberg T. Distinct CDH3 mutations cause ectodermal dysplasia, ectrodactyly, macular dystrophy (EEM syndrome) J. Med. Genet. 2005;42:292–298. doi: 10.1136/jmg.2004.027821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oliveira C., Bordin M.C., Grehan N., Huntsman D., Suriano G., Machado J.C., Kiviluoto T., Aaltonen L., Jackson C.E., Seruca R., et al. Screening of E-Cadherin in gastric cancer families reveals germ-line mutations only in hereditary diffuse gastric cancer kindred. Hum. Mutat. 2002;19:510–517. doi: 10.1002/humu.10068. [DOI] [PubMed] [Google Scholar]
- 39.Oliveira C., de Bruin J., Nabais S., Ligtenberg M., Moutinho C., Nagengast F.M., Seruca R., van Krieken H., Carneiro F. Intragenic deletion of CDH-1 as the inactivating mechanism of the wild-type allele in a HDGC tumor. Oncogene. 2004;23:9192–9196. doi: 10.1038/sj.onc.1207335. [DOI] [PubMed] [Google Scholar]
- 40.Simões-Correia J., Figueiredo J., Oliveira C., van Hengel J., Seruca R., van Roy F., Suriano G. Endoplasmic reticulum quality control: a new mechanism of E-cadherin regulation and its implication in cancer. Hum. Mol. Genet. 2008;17:3566–3576. doi: 10.1093/hmg/ddn249. [DOI] [PubMed] [Google Scholar]
- 41.Frebourg T., Oliveira C., Hochain P., Karam R., Manouvrier S., Graziadio C., Vekemans M., Hartmann A., Baert-Desurmont S., Alexandre C., et al. Cleft lip/palate and CDH1/E-cadherin mutations in families with hereditary diffuse gastric cancer. J. Med. Genet. 2006;43:138–142. doi: 10.1136/jmg.2005.031385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roviello F., Corso G., Pedrazzani C., Marrelli D., De Falco G., Berardi A., Garosi L., Suriano G., Vindigni C., De Stefano A., et al. Hereditary diffuse gastric cancer and E-cadherin: description of the first germline mutation in an Italian family. Eur. J. Surg. Oncol. 2007;33:448–451. doi: 10.1016/j.ejso.2006.10.028. [DOI] [PubMed] [Google Scholar]
- 43.Keller G., Vogelsang H., Becker I., Hutter J., Ott K., Candidus S., Grundei T., Becker K.F., Mueller J., Siewert J.R., et al. Diffuse type gastric and lobular breast carcinoma in a familial gastric cancer patient with an E-cadherin germline mutation. Am. J. Pathol. 1999;155:337–342. doi: 10.1016/S0002-9440(10)65129-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barber M., Murrell A., Ito Y., Maia A.T., Hyland S., Oliveira C., Save V., Carneiro F., Paterson A.L., Grehan N. Mechanisms and sequelae of E-cadherin silencing in hereditary diffuse gastric cancer. J. Pathol. 2008;216:295–306. doi: 10.1002/path.2426. [DOI] [PubMed] [Google Scholar]
- 45.Avizienyte E., Launonen V., Salovaara R., Kiviluoto T., Aaltonen L. E-cadherin is not frequently mutated in hereditary gastric cancer. J. Med. Genet. 2001;38:49–52. doi: 10.1136/jmg.38.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Richards F.M., McKee S.A., Rajpar M.H., Cole T.R., Evans D.G., Jankowski J.A., McKeown C., Sanders D.S., Maher E.R. Germline E-cadherin gene (CDH1) mutations predispose to familial gastric cancer and colorectal cancer. Hum. Mol. Genet. 1999;8:607–610. doi: 10.1093/hmg/8.4.607. [DOI] [PubMed] [Google Scholar]
- 47.Schouten J.P., McElgunn C.J., Waaijer R., Zwijnenburg D., Diepvens F., Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.