Abstract
We used DNA microarrays to characterize the global gene expression patterns in surface epithelial cancers of the ovary. We identified groups of genes that distinguished the clear cell subtype from other ovarian carcinomas, grade I and II from grade III serous papillary carcinomas, and ovarian from breast carcinomas. Six clear cell carcinomas were distinguished from 36 other ovarian carcinomas (predominantly serous papillary) based on their gene expression patterns. The differences may yield insights into the worse prognosis and therapeutic resistance associated with clear cell carcinomas. A comparison of the gene expression patterns in the ovarian cancers to published data of gene expression in breast cancers revealed a large number of differentially expressed genes. We identified a group of 62 genes that correctly classified all 125 breast and ovarian cancer specimens. Among the best discriminators more highly expressed in the ovarian carcinomas were PAX8 (paired box gene 8), mesothelin, and ephrin-B1 (EFNB1). Although estrogen receptor was expressed in both the ovarian and breast cancers, genes that are coregulated with the estrogen receptor in breast cancers, including GATA-3, LIV-1, and X-box binding protein 1, did not show a similar pattern of coexpression in the ovarian cancers.
INTRODUCTION
Ovarian cancer is the fifth leading cause of cancer deaths in women in the United States, with an incidence of ∼23,000 new cases and 14,000 deaths annually (Greenlee et al., 2001). Carcinomas of the surface epithelium of the ovary comprise the large majority (80–90%) of ovarian cancers (Nap et al., 1996; Auersperg et al., 1999; Yin and Lloyd, 2001). Among these epithelial cancers, the most common morphological subtype is serous papillary, with less common subtypes including clear cell, mucinous, endometrioid, transitional, and undifferentiated. Currently, there are no specific markers that enable the early detection of ovarian carcinomas. CA-125 is the most common marker used in monitoring therapy of this disease, but it is not sufficiently specific and sensitive to be useful as a screening test, with serum values in the normal range in half of the patients with stage I disease (Nagele et al., 1995; Eltabbakh et al., 1999).
The application of DNA microarray technology has enabled the study of gene expression profiles of large numbers of tumor samples and has provided an opportunity to classify different neoplasms based on characteristic expression patterns (Alizadeh et al., 2000; Perou et al., 2000). For example, Alizadeh et al. (2000) profiled diffuse large B-cell lymphoma (DLBCL), a subtype of non-Hodgkin's lymphoma and reported an expression profile that distinguishes DLBCL patients with differential expression of a set of genes that distinguish normal B cells at different stages of development. These two subgroups of DLCBL were found to have statistically significant differences in survival. Perou et al. (2000) similarly defined subclasses of breast cancer, which they termed luminal and basal epithelial subtypes based on differences in global gene expression patterns that parallel differences between the basal and luminal epithelial cells in normal breast. Recently, Sørlie et al. (2001) were able to correlate differences in expression patterns of breast cancers with clinical outcome and identified subclasses having poor prognosis. These and similar studies are beginning to identify novel approaches to classifying cancer based on the patterns of expressed genes (Golub et al., 1999; Garber et al., 2001; van't Veer et al., 2002).
Both ovarian and breast cancers arise from hormonally responsive tissues, comprise several different histopathological subtypes, and display considerable variability in clinical manifestations and prognosis. A number of groups have applied the microarray technology to the study of ovarian cancer (Schummer et al., 1999; Ono et al., 2000; Lassus et al., 2001; Shridhar et al., 2001; Tapper et al., 2001; Wong et al., 2001; Zarrinkar et al., 2001; Haviv and Campbell, 2002). The major objectives of this study were to identify genes associated with histopathologic subtypes and grades of cancer of the ovary as well as genes that differentiate ovarian from breast carcinomas. We describe gene expression signatures that may prove useful in diagnosing both serous and clear cell carcinomas of the ovary. In addition, we report specific gene expression patterns that distinguish breast from ovarian carcinomas.
MATERIALS AND METHODS
Arrays
DNA microarrays are based on IMAGE clones (Lennon et al., 1996) prepared by the Research Genetics Corporation (Huntsville, AL). Three sets of microarrays (9K, 23K, and 42K), representing successive generations of microarray printing, were used for this study. These microarrays comprised, respectively, 9216 elements (9K), representing 7781 unique Unigene clusters, 23,079 elements (23K), representing 18,142 unique Unigene clusters, and 42,749 elements (42K), representing 32,275 unique Unigene clusters (Build no. 158, released on 01-18-2003). Known gene content, as judged by the number of unique Unigene symbols, is 5714 (9K), 8618 (23K), and 11,946 (42K), respectively. All arrays were printed at the Stanford University School of Medicine according to Brown laboratory protocols in either the Brown and Botstein laboratories or the Stanford Functional Genomics Facility. For analysis of the 44 ovarian carcinoma samples and 12 ovarian cell lines, only those specimens analyzed on either the 23K or 42K arrays were used, and the analysis was restricted to the genes on the 23K arrays, which were also all represented on the 42K arrays. For the comparison of the ovarian samples with breast carcinomas, the 9K gene list was used, because the published breast cancer data were obtained using 9K arrays.
Tumor Specimens
The majority of ovarian cancer specimens used in this study were archived at Stanford University, and IRB approval was obtained to analyze them by gene expression profiling. In addition, samples were obtained from the Cooperative Human Tissue Network of the National Cancer Institute and the Norwegian Radium Hospital. A total of 162 archived ovarian cancer samples were used for initial RNA isolation, and 59 of these yielded a sufficient amount or quality of mRNA (Web Supplement, Table 5). The histological subtypes of the ovarian carcinomas included 39 serous papillary carcinomas, 7 clear cell, 2 endometrioid, 4 undifferentiated, and 3 adenocarcinoma from ascites specimens with unspecified subtype. Among the 55 primary ovarian specimens, 10 represented recurrent disease, staging information was unavailable for 8, and the stage distribution for the other 37 cases included one each of stages I and II, 3 stage IV, and 32 stage III patients. Four specimens were serous papillary carcinomas, thought to arise from the extraovarian peritoneal epithelium. These primary peritoneal carcinomas share many biological and clinical features with primary ovarian carcinomas (Dalrymple et al., 1989; Wick et al., 1989; Altaras et al., 1991; Chew et al., 1995; Ben-Baruch et al., 1996; Halperin et al., 2001a, 2001b). For the analysis of clear cell vs. serous papillary subtypes of ovarian carcinomas, 44 specimens from 42 patients were hybridized to either 23K or 42K arrays, with two specimens each from two patients. OV98 was a solid specimen and OV98B the ascites from the same patient at the initial surgery. OV25 and OV25C were both ascites samples from the same patient collected three months apart.
The comparison of the expression profiles of ovarian carcinomas with previously published breast cancers was restricted to the 8102 gene elements that overlapped between all three sets of arrays, because the breast cancer data had used exclusively 9K arrays. In this analysis 57 ovarian carcinomas were used, including the samples from 42 different patients, and an additional 15 specimens that had been hybridized to 9K arrays only or for which adequate data were available for this analysis from 23K arrays.
The analysis of differences in gene expression between low grade (histological grade I or II) and high grade (histological grade III) tumors was restricted to solid specimens of serous papillary carcinomas that were predominantly of one histological grade. There were 19 such specimens, 9 grade I or II and 10 grade III (Web Supplement, Table 5).
Cell Lines
Twelve ovarian carcinoma cell lines were obtained from the following sources: OVCAR-3 and SKOV-3 were purchased from the American Type Culture Collection (ATCC, Rockville, MD). OVCA 429, OVCA 432, HEY, and OVCA 420 were a gift from Dr. Robert Knapp at the Dana-Farber Research Institute (Boston, MA). OVCAR-5, OVCAR-8, OVCAR-4, and IGROV-4 were provided by the National Cancer Institute's tumor repository. The ES-2 and MES-OV lines were developed in our laboratory at Stanford (Lau et al., 1991) and (B.I.S., our unpublished results). All cell lines were grown in complete McCoy's medium supplemented with 10% newborn calf serum, 0.3% mg of glutamine/L, 100 U of penicillin/ml, and 100 mg of streptomycin/L (all from Invitrogen Life Technologies, Carlsbad, CA).
Common Reference, Isolation of RNA, Labeling, and Hybridization
Details of these methods are available on the web supplement. The common reference control consisting of equal amounts of mRNA from 11 human cancer cell lines (Perou et al., 1999, 2000). Each sample was compared with this common reference labeled with Cy3-dUTP as described previously (Alizadeh et al., 1998; Perou et al., 1999; Ross et al., 2000; Whitfield et al., 2002). Complete experimental details may be found at: http://brownlab.stanford.edu/protocols.html.
Data Analysis and Clustering
Data Selection Data were analyzed by using either the GenePix 3.0 (Axon Instruments, Foster City, CA) or ScanAlyze (Eisen, http://rana.lbl.gov) software. Spots with aberrant measurements due to array artifacts or poor quality were manually flagged and removed from further analysis. A filter was applied to omit measurements where fluorescent signal from the DNA spot was <20% above the measured background fluorescence surrounding the printed DNA spot in both the Cy3 and Cy5 channels. Genes that did not meet these criteria for at least 80% of the measurements across the cases were excluded from further analysis. Data were retrieved as log2(Cy5/Cy3). The (Cy5/Cy3) ratio is defined in SMD as the normalized ratio of the background-corrected intensities (Sherlock et al., 2001).
We identified an artifact using Singular Value Decomposition (Alter et al., 2000) that was correlated with 23K and the 42K print batches. To adjust for this systematic bias in the datasets, we mean-centered the measurements across genes in experiments carried out on 23K and those carried out on the 42K arrays separately, using Cluster (Eisen, http://rana.lbl.gov) and then carried out further analysis on the combined datasets. Genes were filtered further to select only the subset whose expression varied significantly across the dataset by the criterion that the expression levels measured in at least three samples differed by at least threefold from the mean expression level for all samples. These criteria resulted in selection of a list of 1558 genes that was used for further analysis of the ovarian cancer specimens and cell lines. Hierarchical clustering was applied to the genes and arrays, using the Pearson r coefficient as the measure of similarity and average linkage clustering, as described previously (Eisen et al., 1998; Alizadeh et al., 2000; Perou et al., 2000; Ross et al., 2000), and the results were visualized using Treeview (Eisen, http://rana.lbl.gov). The complete cluster and the entire dataset may be found at our website: http://genome-www.stanford.edu/ovarian_cancer/.
Data Selection for the Breast and Ovarian Combined Cluster Previously published data for breast cancer (Sørlie et al., 2001) were compared with the ovarian dataset. A total of 125 specimens were used, including 68 breast cancer cases and 57 ovarian cancer cases. Genes were selected for further analysis if they displayed at least a twofold variation from their mean expression value for all samples, in at least two of those samples. Arrays and genes were clustered by Pearson correlation using a noncentered metric. Only spots with fluorescent signal at least twofold greater than the local background were included in the analysis. In addition, genes that did not meet these criteria for at least 80% of the measurements across the cases were excluded from further analysis, resulting in 3363 genes for data analysis. These data are also available at http://genome-www.stanford.edu/ovarian_cancer/.
Data Selection for the Grade Analysis For the grade analysis of 19 serous papillary cases, genes were filtered to include measurements where the fluorescent signal from the DNA spot was at least 2.5 times greater than the measured background fluorescence surrounding the printed DNA spot in both the Cy3 and Cy5 channels. In addition, genes that did not meet these criteria for at least 80% of the measurements across the cases were excluded from further analysis. This resulted in 3053 genes that were used for statistical analyses. Complete details of statistical methods used including Significance Analysis of Microarrays (SAM), nonparametric t test, Rank sum test, and Predictive Analysis of Microarrays (PAM) may be found in the web supplement under their respective subheadings (Tibshirani et al., 2002; Tusher et al., 2001).
Immunohistochemistry
Tissue microarray sections were constructed from the archives of Surgical Pathology at Stanford University. The tissue microarray block contains two 0.6-mm representative cores from each of 162 serous ovarian carcinomas and 34 clear cell carcinomas. All of the cases on the tissue array were reviewed and diagnoses confirmed by a single pathologist (T.L.). The tissue arrays were immunohistochemically stained as previously described (van de Rijn et al., 2002), with monoclonal antibodies specific for WT1 (C-19, DAKO, Carpinteria, CA), Ep-CAM (VU-1D9, LabVision, Fremont, CA), and annexin IV (N-19, Santa Cruz Biotechnology, Inc., Santa Cruz, CA) at dilutions of 1:800, 1:1000, and 1:50, respectively. Staining for HE-4 was performed with a polyclonal antibody (see web supplement for details of polyclonal antibody production) at a dilution of 1:25. Antigen retrieval was achieved by microwaving the slides in citrate buffer at pH 6.0. Staining was performed according to the manufacturer's instructions for DAKO's EnVision™+ System, HRP (DAB) kit except that PBS, rather than Tris, was used as the wash buffer. Immunoreactivity with each of the antibodies was scored by two independent observers (T.L., D.R.) as follows: 0, no staining; 2, weak to moderate staining, and 3, strong staining.
RESULTS
Gene Expression Patterns Among Ovarian Carcinoma Specimens
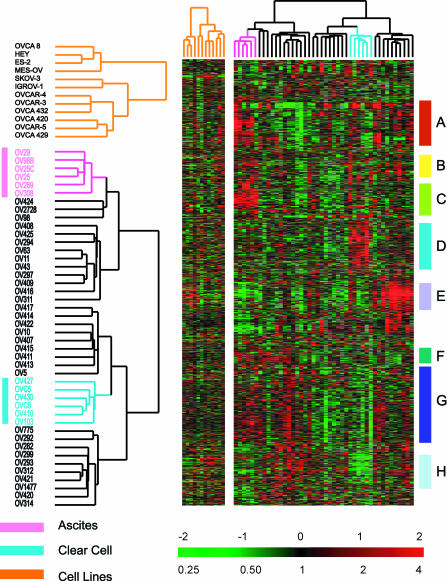
Gene expression profiles of 44 ovarian tumor samples from 42 patients were measured by hybridization to either 23K or 42K element spotted cDNA arrays. Hierarchical cluster analysis revealed multidimensional variation in gene expression by these tumors, including features that appear to be related to specific aspects of ovarian tumor biology (Figure 1). Two distinct subsets of tumors emerged in the nonsupervised clustering, consisting of the six clear cell specimens and the six ascites specimens. Other than these two groups, the other histological subtypes and grades of ovarian cancer specimens were molecularly heterogeneous and intermingled in the hierarchical cluster (Figure 1).
Figure 1.
Unsupervised hierarchical clustering of ovarian cell lines and ovarian cancers. Cell lines were not coclustered with the tumor specimens, because these cell lines have a very prominent proliferation cluster (Perou et al., 1999; Ross et al., 2000) that significantly influences the clustering of the tumor samples if the two sample sets are not analyzed separately. Ovarian cancer specimens and cell lines were clustered based on variation of expression of 1558 genes, as detailed in MATERIALS AND METHODS. Genes were clustered based on similarity in their expression patterns among these cancers. Eight gene clusters are highlighted in this display. (A) Lymphocyte cluster, (B) epithelial/keratin expression, (C) ascites signature, (D) clear cell overexpressed genes, (E) extracellular matrix/stromal cluster, (F) proliferation cluster, (G) heterogeneity across ovarian cases, and (H) clear cell under-expressed genes. The color contrast of the scale bar indicates the fold of gene expression change in log2 space (numbers above the bar).
Lymphocyte Cluster
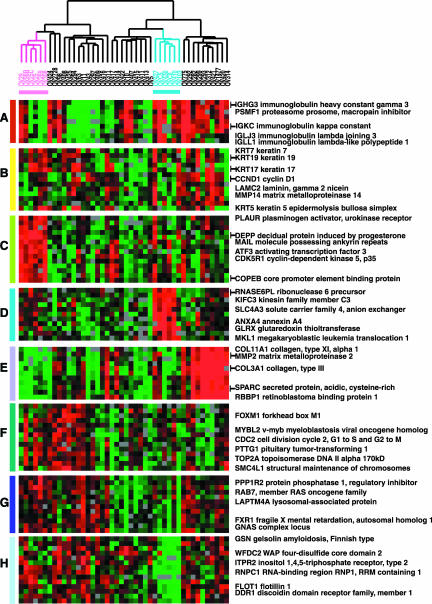
A group of genes characteristic of B-cells and T-cells displayed distinct expression patterns similar to those observed in other tumor classification studies (Figure 2, gene cluster A). This gene cluster included transcripts encoding immunoglobulin genes (IGH3, IGKC, and IGLJ3) members of the class II major histocompatibility complex, several cytokines, and genes regulated by interferon. A small insert from this cluster is shown in Figure 2, gene cluster A, and the entire figure may be viewed in Web Supplement Figure 6.
Figure 2.
Zoomed images of selected regions of Figure 1, which clustered the ovarian specimens based on variation of expression of 1558 genes. (A) Immune cell cluster, (B) epithelial/keratin expression (C) ascites signature, (D) clear cell overexpressed genes, (E) invasion/stromal cluster, (F) proliferation cluster, (G) heterogeneity across ovarian cases, and (H) clear cell underexpressed genes.
Epithelial Cluster
The “epithelial” gene cluster displayed distinct expression patterns across the cases, with very high expression of keratins 5, 7, 17, and 19 in a subset of cases. The expression of two transcripts encoding extracellular matrix proteins, matrix metalloproteinase 14 (MMP14), and laminin C2, shared a similar pattern of expression with these keratins (Figure 2, gene cluster B, Web Supplement Figure 7).
Expression Patterns in Peritoneal Effusions (Ascites)
A cluster of genes including many genes that are characteristically expressed in epithelial cells displayed a unique signature, including relatively abundant transcripts of the “leukocyte” cluster as well as a set of genes more specific to the ascites samples that may reflect the prevalence of activated macrophages in these peritoneal metastases (Figure 2, gene cluster C, Web Supplement Figure 8). In addition, notable overexpression of the transcription factor ATF3 was consistently relatively higher in the ascites samples. ATF3 represses matrix metalloproteinase 2 (MMP2), which is expressed at a relatively low level in the ascites cases.
Extracellular Matrix/Stromal Cluster
A set of genes characteristic of extracellular matrix formation including collagen type III, alpha 1, collagen type VI, alpha 3, collagen type XI, alpha 1, matrix metalloproteinase 2, cadherin 11, type 2 and SPARC, were strongly expressed in a subset of the ovarian cancers (Figure 2, gene cluster E, Web Supplement Figure 9). The 10 tumor specimens in this set were not distinguishable by subtype or grade and included one of the endometrioid subtype and 4 grade I and II serous carcinomas.
Proliferation Cluster
A group of genes whose expression is consistently associated with cell proliferation has previously been reported in studies of global gene expression in tumors, cell lines, and normal tissues (Perou et al., 1999, 2000; Sørlie et al., 2001). This cluster is characterized by a preponderance of cell cycle–regulated genes including CDC2, forkhead box, M1(FOXM1), CDC2, and topoisomerase 2A (Figure 2, gene cluster F, Web Supplement Figure 10; Alizadeh et al., 2000; Perou et al., 2000; Ross et al., 2000; Whitfield et al., 2002).
Ovarian Carcinoma Signature: Heterogeneity among Ovarian Cancers
A very large cluster of genes displayed heterogeneous expression among the ovarian cancers (Figure 2, gene cluster G). The complete list of genes and expanded version of this list is available on the Web Supplement (Figure 11). The biological interpretation of this feature of the expression profiles is not yet clear. However, it is likely to have clinical significance. For example, EPCAM, the target of a mAb that has shown promise in treatment of carcinomas, is among the variably expressed genes in this cluster.
A Gene Expression Signature for the Clear Cell Carcinoma Subtype of Ovarian Cancers
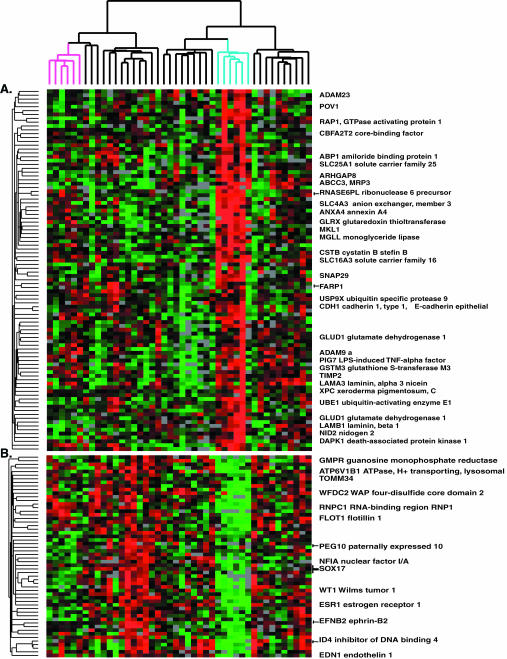
The clear cell subtype of ovarian carcinomas displays a distinct signature of genes that are differentially expressed in clear cell cancers in comparison with other types of ovarian cancers. This signature was readily evident by simple hierarchical clustering of the tumors based on their global expression patterns, which segregated the six clear cell cases into a distinct cluster (Figures 1 and 2, gene clusters D and H, and Figure 3). To more definitively identify transcripts that distinguished clear cell cases from other morphologic subtypes, we used three different statistical approaches for identifying differentially expressed genes. The SAM method identified 84 positive significant genes and 84 negative significant genes (Web Supplement, Table 6 and Figure 12), when the median number of false significant genes was 2.6, and delta 0.676. Differences in gene expression between clear cell and other ovarian cancers were further analyzed using a nonparametric t test with 50,000 permutations (Web Supplement Table 7), and the rank sum test (Web Supplement Tables 8A and 8B). Twenty-five genes were identified as differentially expressed by all three statistical tests that were performed, and the overlapping gene lists are listed in Tables 1 and 2, which include SAM and p-values less than or equal to 0.005 by both the t test and rank sum test (Troyanskaya et al., 2001). Complete gene lists and p-values may be found on the web supplement.
Figure 3.
Clear cell signature determined by hierarchical clustering. Genes were selected as detailed in Figure 1 and in MATERIALS AND METHODS. An expanded view of the gene expression patterns (A) over- or (B) underexpressed in clear cell cancers identified using simple hierarchical clustering from Figure 1 is shown.
Table 1.
Genes that are more highly expressed in clear cell carcinomas than in other ovarian epithelial cancers determined by supervised (SAM, PAM) and unsupervised (hierarchical clustering) analyses
| GLRX, glutaredoxin (thioltransferase) |
| SLC16A3, solute carrier family 16 member 3 (monocarboxylate transporter) |
| MKL1, megakaryoblastic leukemia (translocation) 1 |
| GNE, UDP-N-acetylglucosamine-2-epimerase/N-acetylmannosamine kinase |
| KIFC3, kinesin family member C3 |
| NAP1, pronapsin A |
| ABCC3, ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
| NDRG1, N-myc downstream regulated gene 1 |
| TST, thiosulfate sulfurtransferase (rhodanese) |
| EML2, echinoderm microtubule associated protein like 2 |
| NP, nucleoside phosphorylase |
| RAP1GA1, RAP1, GTPase activating protein 1 |
| AKR1C1, aldo-keto reductase family 1, member C1 |
| IGFBP3, insulin-like growth factor binding protein 3 |
| ARHB, ras homolog gene family, member B |
| IMPA2, inositol(myo)-1(or 4)-monophosphatase 2 |
| COL4A2, collagen, type IV, alpha 2 |
| ANXA4, annexin A4 |
| SLC4A3, solute carrier family 4, anion exchanger, member 3 |
| FGFR4, fibroblast growth factor receptor 4 |
| TFAP2A, transcription factor AP-2 alpha |
| PTPRM, protein tyrosine phosphatase, receptor type, M |
| SMTN, smoothelin |
| ARHGAP8, Rho GTPase activating protein 8 |
| C1QTNF6, C1q and tumor necrosis factor related protein 6 |
Table 2.
Genes less highly expressed in clear cell carcinomas than in other ovarian epithelial cancers determined by supervised (SAM, PAM) and unsupervised (hierarchical clustering) analyses
| ITPR2, inositol 1,4,5-triphosphate receptor, type 2 |
| ESR1, estrogen receptor 1 |
| WFDC2, WAP 4-disulfide core domain |
| FGFRL1, fibroblast growth factor receptor-like 1 |
| NFIA, nuclear factor I/A |
| SELENBP1, selenium binding protein 1 |
| CDH2, **cadherin 2, type 1, N-cadherin (neuronal) |
| PKIB, protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| SCNN1A, sodium channel, nonvoltage-gated 1 alpha |
| IGFBP2, insulin-like growth factor binding protein 2 (36kD) |
| CMAS, CMP-N-acetylneuraminic acid synthase |
| ID4, inhibitor of DNA binding 4, dominant negative helix-loop-helix protein |
| FLOT1, flotillin 1 |
| CYP4B1, cytochrome P450, subfamily IVB, polypeptide 1 |
| UBE2E3, ubiquitin-conjugating enzyme E2E 3 (UBC4/5 homolog, yeast) |
| GAS1, growth arrest-specific 1 |
| WT1, Wilms tumor 1 |
| EFNB2, ephrin-B2 |
| MAP1B, microtubule-associated protein 1B |
| DDR1, discoidin domain receptor family, member 1 |
| APOA1 B1, ATPase, H+ transporting, lysosomal 56/58kD, V1 subunit B, isoform 1 (Ren tubular acidosis with deafness) |
| TSC22, transforming growth factor beta-stimulated protein TSC-22 |
| TRIP7, thyroid hormone receptor interactor 7 |
| EDN1, endothelin 1 |
Several genes with potential roles in drug resistance and metabolism were among those more highly expressed in the clear cell subtype. The gene encoding the redox regulating protein, glutaredoxin, was highly expressed in the clear cell cancers. Glutaredoxin may play a role in resistance to platinum drugs (Nakamura et al., 2000). Several genes encoding transporters that may play a role in drug resistance were more highly expressed in clear cell cancers, including SLC16A3 (monocarboxylic acid solute carrier family 16, member 3), ABCC3 (ATP-binding cassette, subfamily C, member 3), SLC4A3 (solute carrier family 4, anion exchanger, member 3), and ATP11A (ATPase, class VI, type 11A).
Two genes involved in cell-cell adhesion were differentially expressed in the clear cell cancers, with E-cadherin relatively highly expressed and a member of the discoidin domain receptor family (DDR1) expressed at a lower level in clear cell cancers (Figures 2, gene cluster D, and 3). Osteonidogen (nidogen 2), a component of basement membranes, was highly expressed in the clear cell cancers. Both estrogen receptor 1 and cytochrome P450 4B1 were expressed at relatively low levels in clear cell cancers, compared with other ovarian cancers. HE4 (epididymis-specific; WFDC2 WAP four-disulfide core domain 2), which has recently been described as a marker of ovarian cancer (Schummer et al., 1999) was poorly expressed in the clear cell subtype suggesting that cases of this subtype should be considered separately in studies aimed at exploring the utility of HE4 as a marker of ovarian cancers. WT1 (Wilm's tumor1) displayed high expression in the serous papillary cases, but low expression at both the protein and mRNA levels in the clear cell cancers (Figures 2, gene cluster H, and 3).
Gene Expression Patterns Among Ovarian Carcinoma Cell Lines
Twelve ovarian cell lines were similarly analyzed. Variation among the cell lines identified two distinct phenotypes. One set of 4 cell lines (OVCA8, HEY, MES-OV, and ES-2) manifests some mesenchymal features, with high expression of collagens (type VI, alpha 3, type I, alpha 2, type III, and alpha 1), lumican, matrix metalloproteinase 2, and SPARC (Web Supplement, Figure 13). These cell lines lack many of the distinct molecular characteristics of epithelial cells, typically seen in ovarian neoplasms, although expression of MUC1 and mesothelin was notable in these lines. The other set of 8 ovarian cell lines expressed genes typical of epithelial cells including cytokeratins 17 and 19, and claudins 4 and 7. As expected, genes associated with cell proliferation were consistently expressed at higher levels in the cultured cells (our unpublished results; Alizadeh et al., 2000; Perou et al., 2000; Ross et al., 2000).
Analysis of Grade I/II vs. Grade III Tumors
The serous tumors were further analyzed to identify a set of genes that distinguished the low vs. high grade tumors. SAM analysis identified a set of 23 genes differentially expressed in grade I and II vs. grade III tumors (Table 3 and Web Supplement Table 9). Glutathione S-transferases M1, M2, and M4 were among the 20 genes more highly expressed in the grade III tumors. The three undifferentiated carcinomas clustered with the grade III serous cancers when they were clustered based on this set of 22 genes. However, this limited set of genes did not accurately classify the grade of the primary peritoneal specimens (all grade III), nor did it accurately classify the grade of the other histological subtypes. PAM analysis identified a few additional genes that were differentially expressed in low vs. high grade serous papillary carcinomas (Web Supplement Figure 14).
Table 3.
Genes that differ between Grade I and II versus grade III serous papillary carcinomas
| Over-expressed in Grade III | |
| PAI-RBP1 | PAI-RBP1 PAI-1 mRNA-binding protein |
| GSTM1 | Glutathione S-transferase M1 |
| GSTM2 | Glutathione S-transferase M2 |
| GSTM4 | Glutathione S-transferase M4 |
| SLC16A1 | Solute carrier family 16 (monocarboxylic acid transporters), member 1 |
| NUCKS | Similar to rat nuclear ubiquitous casein kinase 2 |
| PNN | Pinin, desmosome associated protein |
| PABN1 | Poly(A) binding protein, nuclear 1 |
| DKC1 | Dyskeratosis congenita 1, dyskerin |
| ATP5F1 | ATP synthase, H+ transporting, subunit b, isoform 1 |
| BAT8 | HLA-B associated transcript 8 |
| MRPS26 | Mitochondrial ribosomal protein S26 |
| MOV10 | Moloney leukemia virus 10, homolog |
| SDHC | Succinate dehydrogenase complex, subunit C, integral membrane protein |
| ZNF265 | Zinc finger protein 265 |
| EIF2S2 | Eukaryotic translation initiation factor 2, subunit 2 beta, 38kDa |
| NUDT3 | Nudix (nucleoside diphosphate linked moiety X)-type motif 3 |
| MEP50 | MEP50 protein |
| FUS | Fusion, derived from t(12;16) malignant liposarcoma |
| T ARBP1 | TAR (HIV) RNA binding protein 1 |
| Homo sapiens full length insert cDNA YU36C09 | |
| Homo sapiens cDNA FLJ34888 fis, clone NT2NE2017332 | |
| Homo sapiens cDNA FLJ38479 fis, clone FEBRA2022787 | |
| Under-expressed in Grade III | |
| COL3A | Collagen, type III, alpha 1 |
| IGHG3 | Immunoglobulin heavy constant gamma 3 |
| AEBP1 | AE binding protein 1 |
Comparison of Expression Patterns in Breast and Ovarian Samples
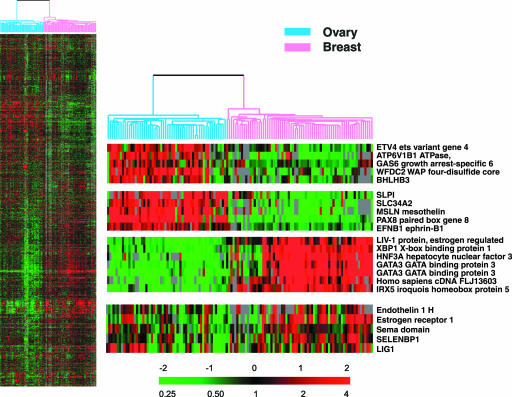
Previously, Sørlie et al. (2001) published a study of the relationship between variation in gene expression patterns and clinical course in breast cancer. We compared the gene expression patterns in breast and ovarian cancers to search for signatures that might differentiate the two groups. Hierarchical clustering based on expression of 3363 genes resulted in almost complete separation of the two cancer types, except for one ovarian cancer (Figure 4). SAM analysis identified 551 genes that were significantly differentially expressed between these two tumor types, with 62 genes more highly expressed in ovarian cancers and 489 genes more highly expressed in breast cancers. PAM analysis identified a minimal set of 61 genes that correctly classified the 68 breast and 57 ovarian cases, with 10 of these genes more highly expressed in ovarian, and 51 in breast cancers (Table 4 and Web Supplement Table 10). Genes more highly expressed among the ovarian carcinomas included PAX8 (paired box gene 8), mesothelin, and ephrin-B1 (EFNB1). Although estrogen receptor was expressed in the ovarian cancers, other genes coordinately expressed with ER-1 in breast cancers, including GATA-3, LIV-1 and X-box binding protein 1, were not similarly expressed in concert with ER-1 in the ovarian cancers.
Figure 4.
Clustering of breast and ovarian carcinoma cases. 68 breast and 57 ovarian cases were co-clustered to discern both similarities and disparities between the two sample sets. An ovarian-specific set of highly expressed transcripts was identified in comparison to breast across the 3363 transcripts. The color contrast of the scale bar indicates the fold of gene expression change in log2 space (numbers above the bar).
Table 4.
Genes that are more highly expressed in ovarian than breast carcinomas, among the 61 genes identified by the PAM method to result in optimal identification of 68 breast and 57 ovarian carcinomas
| PAX8, paired box gene 8 |
| MSLN, mesothelin |
| SLC34A2, solute carrier family 34 (sodium phosphate), member 2 |
| SLPI, secretory leukocyte protease inhibitor (antileukoproteinase) |
| EFNB1, ephrin-B1 |
| EPAC, Rap1 guanine-nucleotide-exchange factor directly activated by cAMP |
| CDH6, cadherin 6, type 2, K-cadherin (fetal kidney) |
| LGALS4, lectin, galactoside-binding, soluble, 4 (galectin 4) |
| FOS, v-fos FBJ murine osteosarcoma viral oncogene homolog |
| ATP6V1B1, ATPase, H+ transporting, lysosomal 56/58kD, V1 subunit B, isoform 1 |
| The complete gene list from is available as Web Supplement Table 10. |
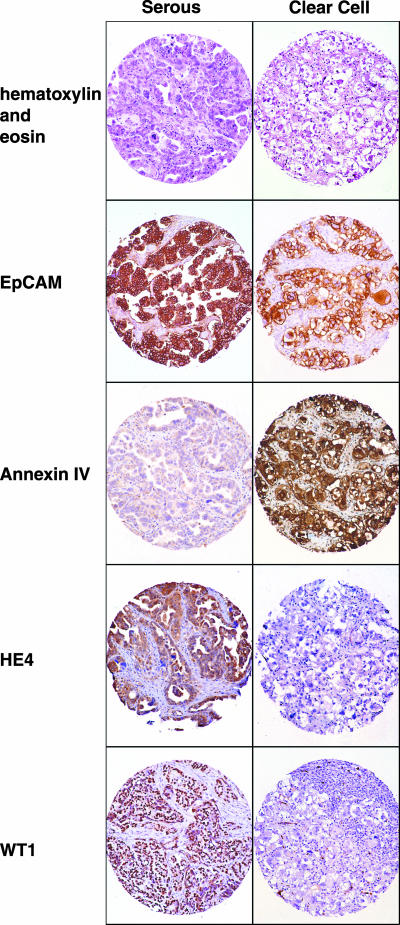
Immunohistochemistry
Immunohistochemistry was carried out with antibodies against four proteins, Annexin IV, HE4, WT1, and EPCAM (TACSTD1), on tissue arrays containing both clear cell and serous ovarian cancers (Figure 5), in order to compare our findings from RNA expression analysis with protein expression data. Staining for HE4 and WT1 paralleled mRNA expression, with high expression of these proteins among the serous cancers and low expression among clear cell cancers (Figure 5). The relatively high Annexin IV mRNA expression in clear cell carcinomas was also consistently reflected in the immunohistochemical staining of the tissue array. Detectable expression was also observed among the serous cancers. However, this staining was much lower relative to that of the clear cell cancers. EPCAM staining was also consistent with RNA expression data, showing heterogeneous protein expression among clear cell and serous cancers (Figure 5).
Figure 5.
Immunohistochemistry: ovarian cancer tissue arrays comprised of both serous (left panel) and clear cell (right panel) ovarian cancers. Hematoxylin and eosin staining is shown in the top panel. Staining of a representative case of serous and clear cell, respectively, were stained with antibodies against EPCAM, annexin IV, HE4, and WT1.
DISCUSSION
In the past few years, a number of published studies have reported cDNA microarray analysis of gene expression from ovarian neoplasms (Schummer et al., 1999; Ono et al., 2000; Hough et al., 2001; Welsh et al., 2001; Wong et al., 2001). Schummer et al. (1999) examined 10 ovarian tumors and 6 normal tissues and identified over one hundred transcripts that were more abundant in tumors of the ovary than ovarian surface epithelium (OSE). HE4, human epididymis gene 4, was identified as a potential marker for ovarian cancer on this basis. Welsh et al. (2001) reported the application of oligonucleotide arrays representing ∼6000 human genes to profile ovarian cancer tissues and compare their expression patterns to the patterns in normal ovarian epithelia. Differences between normal and neoplastic ovarian tissue included expression of a number of cytokeratins, MUC1, and HE4 in the ovarian cancers. Ono et al. (2000) used DNA microarrays to study differences between mucinous and serous ovarian neoplasms. Shridhar et al. (2001) compared early vs. late stage ovarian cancers using cDNA arrays and comparative genomic hybridization (CGH).
One of the major findings of this study was the identification of a distinctive profile of gene expression for clear cell carcinomas of the ovary. Clear cell carcinomas are a distinct histopathological and clinical subtype of ovarian epithelial cancers, characterized by resistance to chemotherapy and a worse clinical prognosis compared with other subtypes (Hameed et al., 1969; Crozier et al., 1989; Jenison et al., 1989; Behbakht et al., 1998; Tammela et al., 1998; Jennings et al., 1999). We identified specific genes that were differentially expressed in six clear cell cancers compared with the other 38 other, predominantly serous papillary, carcinomas (Tables 1 and 2; Web Supplement Tables 7, 8A and 8B; and Web Supplement Figure 12). These genes may provide clues to prognosis and treatment for patients with clear cell ovarian cancer. Of particular interest are several genes involved in drug detoxification that were highly expressed in the clear cell cancers, including annexin IV, glutaredoxin, and ABCC3 (MRP3; Tammela et al., 1998). Annexin IV has been implicated in drug resistance after exposure of cells to paclitaxel (Han et al., 2000). Glutaredoxin (thioltransferase), a redox-regulating protein, has been implicated in resistance to cisplatin (Nakamura et al., 2000; Arner et al., 2001). MRP3 is a transporter that may confer resistance to several chemotherapeutic agents, including topoisomerase inhibitors, platinums, and antimetabolites (Kool et al., 1999; Zeng et al., 1999). The increased AKR1C1 expression in clear cell cancers of the ovary is of interest, because this aldo-keto reductase is also highly expressed in normal kidneys and in renal carcinomas that are of clear cell morphology (O'Connor et al., 1999). Several growth signaling proteins were relatively overexpressed in clear cell carcinomas, including the ras homologue ARHB, the insulin-like growth factor–binding protein IGFBP3, fibroblast growth factor receptor 4, and inositol monophosphatase-2.
Recently, Schwartz et al. (2002) reported a comparison of gene expression patterns between clear cell carcinoma of the ovary and other subtypes of ovarian carcinoma based on results obtained using commercial oligonucleotide arrays containing 7129 probe sets. The authors applied Principal Component Analysis to determine differences between the different subtypes. There is some overlap among the genes identified by our approach and theirs, despite the use of markedly different microarrays and statistical approaches. Notably, annexin A4 and glutaredoxin displayed very high expression in clear cell cases in both datasets. Our arrays allowed us to analyze a larger number of genes (∼23,000 vs. 7000) and thus identify a more comprehensive gene expression signature of clear cell carcinomas (Figure 3, Tables 1 and 2, Web Supplement Tables 6–8).
WFDC2 (HE4) was relatively poorly expressed in the clear cell cancers but was previously suggested to be a good marker of ovarian carcinoma (Schummer et al., 1999). Our findings indicate that this marker is subtype-specific and that clear cell cases should be considered separately in studies of WFDC2 expression as a diagnostic test for ovarian carcinoma. Estrogen receptor 1 displayed lower expression in the clear cell cancers when compared with other ovarian cancers in our study, as shown previously via immunohistochemical staining (Doria et al., 1987).
Because histological grade is an important prognostic factor in ovarian serous papillary carcinomas (Makar et al., 1995; Shimizu et al., 1998; Brun et al., 2000), we analyzed gene expression in grade I/II vs. grade III serous cancers. The analysis was restricted to solid tumor specimens of serous cancers, in order to avoid a bias from the characteristic signature of ascites specimens. Three isoforms of the mu class of glutathione transferases (GST-mu) were more highly expressed in grade III tumors. This class of GST genes has been implicated in resistance to chlorambucil and other bifunctional alkylating agents, which are sometimes used for the therapy of ovarian carcinomas (Horton et al., 1999). In a prior study, patients with ovarian serous carcinomas and low GST-mu expression have been found to survive longer than those with high GST-mu (Matsumoto et al., 1997). Grade III tumors manifested high levels of several genes involved in regulation of gene expression, including NUCKS, a transcription factor involved in regulation of the cell cycle (Ostvold et al., 2001), and pinin, a modulator of RNA splicing (Wang et al., 2002). Collagen type 3A1 is reported to be increased in ovarian carcinomas compared with benign ovarian adenomas (Tapper et al., 2001). In our set of serous papillary carcinomas, the expression of this collagen was decreased in the high-grade compared with the low-grade cancers.
Interestingly, proliferation genes were not a good discriminator of histological grade in our set of 19 serous papillary, solid tumor specimens. Perhaps this is a reflection of the fact that all of our low-grade tumors were also advanced stages III or IV. The relatively small number of graded serous specimens used in the analysis may also have reduced the possibility of finding differences in proliferation related genes among histological grades. We reexamined this question by reanalyzing the graded samples together with the cell lines, isolating the proliferation cluster of genes, and performing an analysis of their expression according to grade I/II vs. grade III, and again found no significant differences in proliferation gene expression (our unpublished results).
The primary peritoneal tumors did not cluster with other grade III serous papillary cancers when we used the list of 23 genes identified by SAM as discriminating the 19 graded, serous ovarian solid tumors. However, they did cluster with the ovarian serous papillary cancers in both the nonsupervised and supervised analyses of breast vs. ovarian cancers. It is possible that a larger gene list generated from a more extensive set of graded specimens would identify the primary peritoneal carcinomas as grade III. Conversely, it is possible that there are biological differences among primary ovarian cancers and primary peritoneal serous cancers to the extent that the latter will not cocluster with corresponding grades of primary ovarian serous cancers.
Ovarian and breast cancers are important diagnostic considerations for women who have metastatic carcinomas of unknown primary site (Greco and Hainsworth, 1994; Greco et al., 2000, 2001). Standard histopathological and immunohistochemical examinations often cannot distinguish between these two tumor types. We have shown that gene expression profiles can be used to accurately discriminate ovarian from breast carcinomas, illustrating the power of this approach both in classifying cancers and identifying genes of biological interest. PAX-8 and EPAC were among the genes more highly expressed in ovarian than breast cancers (Table 4). All four primary peritoneal serous papillary carcinomas coclustered with the known ovarian primary specimens in the nonsupervised hierarchical clustering, which included breast and ovarian cancers. This supports the concept that primary peritoneal cancers share histogenetic and other biological features with carcinomas arising from the ovarian surface epithelium (Dalrymple et al., 1989; Wick et al., 1989; Altaras et al., 1991; Chew et al., 1995; Ben-Baruch et al., 1996; Halperin et al., 2001a, 2001b).
Some of the genes whose expression discriminated between breast and ovarian carcinomas may be useful in immunohistochemical assays to distinguish these entities, including GATA-3. We are currently exploring the diagnostic value of panels of antibodies to some of these candidate genes. Estrogen receptor was expressed in both breast and ovarian specimens. However, the differential expression in breast cancers of genes comprising an estrogen receptor related cluster, including GATA-3, LIV-1, and X-box binding protein 1, suggests that aspects of estrogen receptor biology are fundamentally different between ovarian and breast tissues.
In summary, we have applied DNA microarray technology to examine variations in gene expression profile related to ovarian cancer histological subtypes and grades of differentiation and to differentiate ovarian from breast carcinomas. A set of genes distinguishing low- from high-grade tumors was identified using statistical methods. We found gene expression signatures that differentiate the clear cell subtype of ovarian cancers from the more common serous papillary subtype. Comparison of the breast and ovarian cancers has revealed distinct signatures, including consistent differential expression of estrogen-regulated genes between the two tumor types. The comparison between breast and ovarian cancers may facilitate the differential diagnosis of these diseases and may also reveal insights regarding their underlying biology.
Acknowledgments
This work was supported by National Institutes of Health Grants R33 CA 89830 (B.I.S.), U01 CA 85129 (P.O.B. and D.B.), and T32 CA09302 (Cancer Biology Training Grant, M.E.S.), California Cancer Research Program Grant 99–00561V-10091 (B.I.S.), the Howard Hughes Medical Institute, and the Margaret Fagin and Beatrice Quackenbush Research Funds for Ovarian Cancer.
References
- Alizadeh, A., Eisen, M., Botstein, D., Brown, P.O., and Staudt, L.M. (1998). Probing lymphocyte biology by genomic-scale gene expression analysis. J. Clin. Immunol. 18, 373-379. [DOI] [PubMed] [Google Scholar]
- Alizadeh, A.A. et al. (2000). Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403, 503-511. [DOI] [PubMed] [Google Scholar]
- Altaras, M.M., Aviram, R., Cohen, I., Cordoba, M., Weiss, E., and Beyth, Y. (1991). Primary peritoneal papillary serous adenocarcinoma: clinical and management aspects. Gynecol. Oncol. 40, 230-236. [DOI] [PubMed] [Google Scholar]
- Alter, O., Brown, P.O., and Botstein, D. (2000). Singular value decomposition for genome-wide expression data processing and modeling. Proc. Natl. Acad. Sci. USA 97, 10101-10106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arner, E.S., Nakamura, H., Sasada, T., Yodoi, J., Holmgren, A., and Spyrou, G. (2001). Analysis of the inhibition of mammalian thioredoxin, thioredoxin reductase, and glutaredoxin by cis-diamminedichloroplatinum (II) and its major metabolite, the glutathione-platinum complex. Free Radic. Biol. Med. 31, 1170-1178. [DOI] [PubMed] [Google Scholar]
- Auersperg, N., Pan, J., Grove, B.D., Peterson, T., Fisher, J., Maines-Bandiera, S., Somasiri, A., and Roskelley, C.D. (1999). E-cadherin induces mesenchymal-to-epithelial transition in human ovarian surface epithelium. Proc. Natl. Acad. Sci. USA 96, 6249-6254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behbakht, K., Randall, T.C., Benjamin, I., Morgan, M.A., King, S., and Rubin, S.C. (1998). Clinical characteristics of clear cell carcinoma of the ovary. Gynecol. Oncol. 70, 255-258. [DOI] [PubMed] [Google Scholar]
- Ben-Baruch, G., Sivan, E., Moran, O., Rizel, S., Menczer, J., and Seidman, D.S. (1996). Primary peritoneal serous papillary carcinoma: a study of 25 cases and comparison with stage III-IV ovarian papillary serous carcinoma. Gynecol. Oncol. 60, 393-396. [DOI] [PubMed] [Google Scholar]
- Brun, J.L., Feyler, A., Chene, G., Saurel, J., Brun, G., and Hocke, C. (2000). Long-term results and prognostic factors in patients with epithelial ovarian cancer. Gynecol. Oncol. 78, 21-27. [DOI] [PubMed] [Google Scholar]
- Chew, S., Tham, K.F., Lim, F.K., and Ratnam, S.S. (1995). Papillary serous carcinoma of the peritoneum. J. Obstet. Gynaecol. 21, 341-347. [DOI] [PubMed] [Google Scholar]
- Crozier, M.A., Copeland, L.J., Silva, E.G., Gershenson, D.M., and Stringer, C.A. (1989). Clear cell carcinoma of the ovary: a study of 59 cases. Gynecol Oncol 35, 199-203. [DOI] [PubMed] [Google Scholar]
- Dalrymple, J.C. et al. (1989). Extraovarian peritoneal serous papillary carcinoma. A clinicopathologic study of 31 cases. Cancer 64, 110-115. [DOI] [PubMed] [Google Scholar]
- Doria, M.I., Jr., Adamec, T., and Talerman, A. (1987). Alpha-lactalbumin in “common” epithelial tumors of the ovary. An immunohistochemical study. Am. J. Clin. Pathol. 87, 752-756. [DOI] [PubMed] [Google Scholar]
- Eisen, M.B., Spellman, P.T., Brown, P.O., and Botstein, D. (1998). Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95, 14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eltabbakh, G.H., Yadav, P.R., Morgan, A., and Yadev, P.R. (1999). Clinical picture of women with early stage ovarian cancer. Gynecol. Oncol. 75, 476-479. [DOI] [PubMed] [Google Scholar]
- Golub, T.R. et al. (1999). Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 286, 531-537. [DOI] [PubMed] [Google Scholar]
- Greco, F.A., Burris, H.A., 3rd, Erland, J.B., Gray, J.R., Kalman, L.A., Schreeder, M.T., and Hainsworth, J.D. (2000). Carcinoma of unknown primary site. Cancer 89, 2655-2660. [PubMed] [Google Scholar]
- Greco, F.A., Gray, J., Burris, H.A., 3rd, Erland, J.B., Morrissey, L.H., and Hainsworth, J.D. (2001). Taxane-based chemotherapy for patients with carcinoma of unknown primary site. Cancer J 7, 203-212. [PubMed] [Google Scholar]
- Greco, F.A., and Hainsworth, J.D. (1994). Poorly differentiated carcinoma or adenocarcinoma of unknown primary site: long-term results with cisplatin-based chemotherapy. Semin. Oncol. 21, 77-82. [PubMed] [Google Scholar]
- Greenlee, R.T., Hill-Harmon, M.B., Murray, T., and Thun, M. (2001). Cancer statistics, 2001. CA Cancer J. Clin. 51, 15-36. [DOI] [PubMed] [Google Scholar]
- Halperin, R., Zehavi, S., Hadas, E., Habler, L., Bukovsky, I., and Schneider, D. (2001a). Immunohistochemical comparison of primary peritoneal and primary ovarian serous papillary carcinoma. Int. J. Gynecol. Pathol. 20, 341-345. [DOI] [PubMed] [Google Scholar]
- Halperin, R., Zehavi, S., Langer, R., Hadas, E., Bukovsky, I., and Schneider, D. (2001b). Primary peritoneal serous papillary carcinoma: a new epidemiologic trend? A matched-case comparison with ovarian serous papillary cancer. Int. J. Gynecol. Cancer. 11, 403-408. [DOI] [PubMed] [Google Scholar]
- Hameed, K., Burslem, M.R., and Tupper, W.R. (1969). Clear cell carcinoma of the ovary. Cancer 24, 452-459. [DOI] [PubMed] [Google Scholar]
- Han, E.K., Tahir, S.K., Cherian, S.P., Collins, N., and Ng, S.C. (2000). Modulation of paclitaxel resistance by annexin IV in human cancer cell lines. Br. J. Cancer 83, 83-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haviv, I., and Campbell, I.G. (2002). DNA microarrays for assessing ovarian cancer gene expression. Mol. Cell. Endocrinol. 191, 121-126. [DOI] [PubMed] [Google Scholar]
- Horton, J.K., Roy, G., Piper, J.T., Van Houten, B., Awasthi, Y.C., Mitra, S., Alaoui-Jamali, M.A., Boldogh, I., and Singhal, S.S. (1999). Characterization of a chlorambucil-resistant human ovarian carcinoma cell line overexpressing glutathione S-transferase mu. Biochem. Pharmacol. 58, 693-702. [DOI] [PubMed] [Google Scholar]
- Hough, C.D., Cho, K.R., Zonderman, A.B., Schwartz, D.R., and Morin, P.J. (2001). Coordinately up-regulated genes in ovarian cancer. Cancer Res. 61, 3869-3876. [PubMed] [Google Scholar]
- Jenison, E.L., Montag, A.G., Griffiths, C.T., Welch, W.R., Lavin, P.T., Greer, J., and Knapp, R.C. (1989). Clear cell adenocarcinoma of the ovary: a clinical analysis and comparison with serous carcinoma. Gynecol. Oncol. 32, 65-71. [DOI] [PubMed] [Google Scholar]
- Jennings, T.S., Haddad, M.E., and Smith, T.M. (1999). Clear cell ovarian carcinoma following long-term tamoxifen use. Breast J. 5, 132-135. [DOI] [PubMed] [Google Scholar]
- Kool, M. et al. (1999). MRP3, an organic anion transporter able to transport anti-cancer drugs. Proc. Natl. Acad. Sci. USA 96, 6914-6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassus, H., Laitinen, M.P., Anttonen, M., Heikinheimo, M., Aaltonen, L.A., Ritvos, O., and Butzow, R. (2001). Comparison of serous and mucinous ovarian carcinomas: distinct pattern of allelic loss at distal 8p and expression of transcription factor GATA-4. Lab. Invest. 81, 517-526. [DOI] [PubMed] [Google Scholar]
- Lau, D.H., Lewis, A.D., Ehsan, M.N., and Sikic, B.I. (1991). Multifactorial mechanisms associated with broad cross-resistance of ovarian carcinoma cells selected by cyanomorpholino doxorubicin. Cancer Res. 51, 5181-5187. [PubMed] [Google Scholar]
- Lennon, G., Auffray, C., Polymeropoulos, M., and Soares, M.B. (1996). The IMAGE Consortium: an integrated molecular analysis of genomes and their expression. Genomics 33, 151-152. [DOI] [PubMed] [Google Scholar]
- Makar, A.P., Baekelandt, M., Trope, C.G., and Kristensen, G.B. (1995). The prognostic significance of residual disease, FIGO substage, tumor histology, and grade in patients with FIGO stage III ovarian cancer. Gynecol. Oncol. 56, 175-180. [DOI] [PubMed] [Google Scholar]
- Matsumoto, T., Hayase, R., Kodama, J., Kamimura, S., Yoshinouchi, M., and Kudo, T. (1997). Immunohistochemical analysis of glutathione S-transferase mu expression in ovarian tumors. Eur. J. Obstet. Gynecol. Reprod. Biol. 73, 171-176. [DOI] [PubMed] [Google Scholar]
- Nagele, F., Petru, E., Medl, M., Kainz, C., Graf, A.H., and Sevelda, P. (1995). Preoperative CA 125, an independent prognostic factor in patients with stage I epithelial ovarian cancer. Obstet. Gynecol. 86, 259-264. [DOI] [PubMed] [Google Scholar]
- Nakamura, H. et al. (2000). Expression of thioredoxin and glutaredoxin, redox-regulating proteins, in pancreatic cancer. Cancer Detect. Prev. 24, 53-60. [PubMed] [Google Scholar]
- Nap, M. et al. (1996). Immunohistochemical characterization of 22 monoclonal antibodies against the CA125 antigen: 2nd report from the ISOBM TD-1 Workshop. Tumour Biol. 17, 325-331. [PubMed] [Google Scholar]
- O'Connor, T., Ireland, L.S., Harrison, D.J., and Hayes, J.D. (1999). Major differences exist in the function and tissue-specific expression of human aflatoxin B1 aldehyde reductase and the principal human aldo-keto reductase AKR1 family members. Biochem. J. 343(Pt 2), 487-504. [PMC free article] [PubMed] [Google Scholar]
- Ono, K., Tanaka, T., Tsunoda, T., Kitahara, O., Kihara, C., Okamoto, A., Ochiai, K., Takagi, T., and Nakamura, Y. (2000). Identification by cDNA microarray of genes involved in ovarian carcinogenesis. Cancer Res. 60, 5007-5011. [PubMed] [Google Scholar]
- Ostvold, A.C., Norum, J.H., Mathiesen, S., Wanvik, B., Sefland, I., and Grundt, K. (2001). Molecular cloning of a mammalian nuclear phosphoprotein NUCKS, which serves as a substrate for Cdk1 in vivo. Eur. J. Biochem. 268, 2430-2440. [DOI] [PubMed] [Google Scholar]
- Perou, C.M. et al. (1999). Distinctive gene expression patterns in human mammary epithelial cells and breast cancers. Proc. Natl. Acad. Sci. USA 96, 9212-9217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou, C.M. et al. (2000). Molecular portraits of human breast tumours. Nature 406, 747-752. [DOI] [PubMed] [Google Scholar]
- Ross, D.T. et al. (2000). Systematic variation in gene expression patterns in human cancer cell lines. Nat. Genet. 24, 227-235. [DOI] [PubMed] [Google Scholar]
- Schummer, M. et al. (1999). Comparative hybridization of an array of 21, 500 ovarian cDNAs for the discovery of genes overexpressed in ovarian carcinomas. Gene 238, 375-385. [DOI] [PubMed] [Google Scholar]
- Schwartz, D.R. et al. (2002). Gene expression in ovarian cancer reflects both morphology and biological behavior, distinguishing clear cell from other poor-prognosis ovarian carcinomas. Cancer Res 62, 4722-4729. [PubMed] [Google Scholar]
- Sherlock, G. et al. (2001). The Stanford Microarray Database. Nucleic Acids Res. 29, 152-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu, Y., Kamoi, S., Amada, S., Akiyama, F., and Silverberg, S.G. (1998). Toward the development of a universal grading system for ovarian epithelial carcinoma: testing of a proposed system in a series of 461 patients with uniform treatment and follow-up. Cancer 82, 893-901. [DOI] [PubMed] [Google Scholar]
- Shridhar, V. et al. (2001). Genetic analysis of early- versus late-stage ovarian tumors. Cancer Res. 61, 5895-5904. [PubMed] [Google Scholar]
- Sørlie, T. et al. (2001). Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 98, 10869-10874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tammela, J., Geisler, J.P., Eskew, P.N., Jr., and Geisler, H.E. (1998). Clear cell carcinoma of the ovary: poor prognosis compared to serous carcinoma. Eur. J. Gynaecol. Oncol. 19, 438-440. [PubMed] [Google Scholar]
- Tapper, J., Kettunen, E., El-Rifai, W., Seppala, M., Andersson, L.C., and Knuutila, S. (2001). Changes in gene expression during progression of ovarian carcinoma. Cancer Genet. Cytogenet. 128, 1-6. [DOI] [PubMed] [Google Scholar]
- Tibshirani, R., Hastie, T., Narasimhan, B., and Chu, G. (2002). Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc. Natl. Acad. Sci. USA 99, 6567-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyanskaya, O., Cantor, M., Sherlock, G., Brown, P., Hastie, T., Tibshirani, R., Botstein, D., and Altman, R.B. (2001). Missing value estimation methods for DNA microarrays. Bioinformatics 17, 520-525. [DOI] [PubMed] [Google Scholar]
- Tusher, V.G., Tibshirani, R., and Chu, G. (2001). Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98, 5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van 't Veer, L.J. et al. (2002). Gene expression profiling predicts clinical outcome of breast cancer. Nature 415, 530-536. [DOI] [PubMed] [Google Scholar]
- van de Rijn, M. et al. (2002). Expression of cytokeratins 17 and 5 identifies a group of breast carcinomas with poor clinical outcome. Am. J. Pathol. 161, 1991-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, P., Lou, P.J., Leu, S., and Ouyang, P. (2002). Modulation of alternative pre-mRNA splicing in vivo by pinin. Biochem. Biophys. Res. Commun. 294, 448-455. [DOI] [PubMed] [Google Scholar]
- Welsh, J.B., Zarrinkar, P.P., Sapinoso, L.M., Kern, S.G., Behling, C.A., Monk, B.J., Lockhart, D.J., Burger, R.A., and Hampton, G.M. (2001). Analysis of gene expression profiles in normal and neoplastic ovarian tissue samples identifies candidate molecular markers of epithelial ovarian cancer. Proc. Natl. Acad. Sci. USA 98, 1176-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield, M.L. et al. (2002). Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol. Biol. Cell 13, 1977-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick, M.R., Mills, S.E., Dehner, L.P., Bollinger, D.J., and Fechner, R.E. (1989). Serous papillary carcinomas arising from the peritoneum and ovaries. A clinicopathologic and immunohistochemical comparison. Int. J. Gynecol. Pathol. 8, 179-188. [DOI] [PubMed] [Google Scholar]
- Wong, K.K., Cheng, R.S., and Mok, S.C. (2001). Identification of differentially expressed genes from ovarian cancer cells by MICROMAX cDNA microarray system. Biotechniques 30, 670-675. [DOI] [PubMed] [Google Scholar]
- Yin, B.W., and Lloyd, K.O. (2001). Molecular cloning of the CA125 ovarian cancer antigen: Identification as a new mucin (MUC14). J. Biol. Chem. 21, 21. [DOI] [PubMed] [Google Scholar]
- Zarrinkar, P.P., Mainquist, J.K., Zamora, M., Stern, D., Welsh, J.B., Sapinoso, L.M., Hampton, G.M., and Lockhart, D.J. (2001). Arrays of arrays for high-throughput gene expression profiling. Genome Res. 11, 1256-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, H., Bain, L.J., Belinsky, M.G., and Kruh, G.D. (1999). Expression of multidrug resistance protein-3 (multispecific organic anion transporter-D) in human embryonic kidney 293 cells confers resistance to anticancer agents. Cancer Res 59, 5964-5967. [PubMed] [Google Scholar]