Abstract
The effect of an increment of exposure on disease risk may vary with time since exposure. If the pattern of temporal variation is simple (e.g., a peak then decline in excess risk of disease) then this may be modeled efficiently via a parametric latency function. Estimation of the parameters for such a model can be difficult because the parameters are not a function of a simple summary of the exposure history. Typically such parameters are estimated via an iterative search that requires calculating a different time-weighted exposure for each combination of the latency function parameters. This paper describes a simple approach to fitting logistic regression models that include a parametric latency function. This approach is illustrated using data from a study of the association between radon exposure and lung cancer mortality among underground uranium miners. This approach should facilitate fitting models to assess variation with time since exposure in the effect of a protracted environmental or occupational exposure.
INTRODUCTION
In occupational and environmental research a common approach to summarizing information about a protracted exposure history is to calculate a cumulative metric of exposure. Implicit in the use of a cumulative exposure metric is the assumption that the effects of exposures are additive and the impact of a unit of exposure on disease risk is the same regardless of when it occurred. Recognizing that there is often an induction period (a period of time between exposure and resultant induction of disease) and latent period (a period of time between the induction of disease and its detection by the investigator), data analysts will often lag a cumulative metric of exposure by a fixed interval.1–4 For simplicity in this paper I will use the term latency to refer to the interval between an increment of exposure and a subsequent change in disease risk.
The variation over time in the excess risk of disease following an increment of exposure can be described by a latency function. The approach of lagging a cumulative metric of exposure implies a stepwise latency function; it does not allow a data analyst to evaluate whether the effect of exposure on disease risk persists, increases over time, or eventually diminishes. An assessment of whether the effect of exposure increases or diminishes over time is relatively straightforward in a setting of a population exposed to a single, acute exposure. A data analyst can quantify risks or relative risks at various intervals following the acute exposure. In contrast, in a setting of repeated or protracted exposure different analytical methods are needed to investigate variation over time in the impact of an increment of exposure on disease risk. Exposure time-window analyses offer one method for describing variation in the relative risk of disease as a function of time-since-exposure.1,4 An exposure history is partitioned into intervals; the data analyst then examines separate estimates of the association between disease risk and exposures received in a series of time intervals prior to the current age. The exposure time-window approach provides a piecewise constant model of variation in an exposure’s effect with time since exposure. However, in occupational and environmental studies of low level protracted exposures, reliable estimation of exposure effects may be difficult when exposure is partitioned into multiple time-windows; and, evidence of a temporal pattern may be sensitive to decisions about the number of time-windows and the boundary points between them.
If the exposure effect follows a simple temporal pattern (e.g., a peak then decline in excess risk of disease) then a piecewise constant latency model may be less plausible (and less parsimonious) than a simple parametric model of latency effects. While methods exist for fitting parametric latency models to protracted exposure data, these are seldom employed by occupational and environmental epidemiologists.5,6 One possible reason for the limited use of these methods is that there have been difficulties in fitting such models. Typically, the approach to estimate the parameters for such exposure-time-response models has involved an iterative search.1,7
In this paper I describe an approach to incorporate estimation of model parameters to describe variation in exposure effects with time-since-exposure into a logistic regression model fitting. I illustrate some simple parametric models for exposure-time-response functions and apply these methods to analyses of mortality in a study of radon-exposed uranium miners. The methods are described in the context of a nested case-control study in which density sampling has been used and a large number of controls have been sampled for each case.
METHODS
Consider a nested case-control study with density sampling of controls. Let’s say that yj denotes the case status of individual j in a risk set that is matched on attained age A. The exposure history for each person is recorded as an exposure estimate for each increment (e.g., year) of age, a. Let’s say that xj(a) represents the exposure history for individual j as a function of age, a.
Suppose that we postulate that after an instantaneous exposure increment at age a the observed disease risk at attained age A is proportional to the baseline disease risk at age A (i.e., the risk in the absence of exposure) multiplied by a relative risk that is dependent upon the intensity of the exposure increment and a time-dependent exposure weighting function, w(t), that describe the variation in the exposure’s effect with time-since-exposure, t, where t = (A − a).
Now suppose that the exposure is protracted or repeated over time. The integral, , represents person j’s cumulative exposure accrued through attained age A. The integral of the time-weighted exposures, , represents the cumulative effective exposure accrued by individual j at attained age A. Under the standard approach of lagging exposure assignment by a fixed interval, l, an exposure lag might be expressed as a latency function of the form w(t) = I[l ≤ t], where I[“logical expression”] equals 1 if “logical expression” is true and 0 if it is false.
For studies in which it is appropriate to posit a latency function in which the impact of exposure increases then diminishes with time since exposure, one simple latency function that conforms to this pattern is a bilinear model, consisting of two attached lines that form a triangular function.5 For the first α1 years after exposure, the relative effect of exposure increases linearly to its maximum value; then, the effect diminishes linearly with additional time since exposure, obtaining a weight of zero (no effect) α2 years in the past. This bilinear model may be expressed as a weighting function of the form
A lognormal probability density function (pdf) for the latent period also can describe a rise and subsequent fall in the effect of exposure with time-since-exposure.6,8 This implies a weighting function, w(t) = pdf (Lognormal, t,θ,λ), where θ and λ are the location and scale parameters for this distribution and the modal value of the latency function is given by exp(θ − λ2).
Fitting Latency Models
Consider an analysis of the association between cumulative effective exposure and odds of disease, odds, conducted via fitting of a logistic regression model of the form , where z1j..znj represent covariates (and could include indicator variables for risk sets), and the parameter δ provides an estimate of the change in the log odds of disease per unit weighted cumulative exposure. Alternatively, a linear odds ratio model could be fitted, of the form , where δ represents the excess odds ratio (EOR) per unit weighted cumulative exposure.9
In the above regression models, a person’s weighted exposure is the product of the observed exposure xj(a) and the latency function, w(t), which itself may involve unknown parameter(s). For example, a linear odds ratio model where latency effects are described via a bilinear latency function, corresponding to a regression model of the form:
would imply that the model parameters to be estimated include β0 − βn, δ, and α1 and α2. Maximum likelihood estimates for each of these parameters, along with their associated standard errors, can be obtained via SAS PROC NLMIXED.10 In this way, in a single regression model fitting, the data analyst obtains estimates of the parameters specifying the latency function. The appendix provides a simple SAS macro that may be used to fit a variety of such models.
Pointwise 95% credible intervals may be derived via Monte Carlo methods in order to illustrate the statistical uncertainty in the estimated relative risk per unit dose as a function of time since exposure. This can be done with a slight modification of the SAS code shown in the appendix, invoking the SAS MCMC procedure.10 Parameter estimates are generated via each of a large number (e.g., 10,000) of Monte Carlo iterations, specifying non-informative priors. For each set of parameter estimates, the risk ratio is calculated for each time point over the interval for which one wishes to derive confidence intervals, and the 2.5 and 97.5 percentiles of the distribution of these values for each time point serve as the lower and upper bounds of the credible interval.
Examples
To illustrate this approach to estimating the parameters of a latency function, I use data from a study of underground uranium miners.5,11 The Colorado Plateau cohort includes male workers employed in underground uranium mining operations between January 1, 1950 and December 31, 1960. Vital status was ascertained through December 31, 1990. The outcome of interest, lung cancer mortality, was defined based upon underlying cause of death, coded according to the revision of the International Classification of Diseases (ICD). The primary exposure of interest was defined as cumulative radon exposure, expressed in working level months (WLM), and was computed for each worker as the product of the length of employment in each job in a year by the estimated radon exposure rate for that job. For each lung cancer death, a risk set was formed that included all workers who were alive and eligible to be in the study at the attained age of the index case; controls were also matched to cases on calendar year at risk (defined in five-year categories from <1960 to 1990+). I used the nested case-control data described by Langholz et al.; this data set included 263 lung cancer deaths.5 Up to forty controls were selected for each lung cancer death by random sampling without replacement from all members of the risk set (excluding the index case itself).
In order to account for the matched design of the study, the regression model included 262 binary indicator variables for the 263 strata defined by the risk sets. A linear odds ratio model was fitted, as in previous analyses of these data.5,9 I first estimated the association between cumulative radon dose and lung cancer mortality assuming a time-constant model (i.e., w(t)=1). Next, I fitted logistic regression models that incorporated a bilinear latency function. The results are compared with those previously obtained by Langholz et al. In addition, the results of fitting a model in which cumulative exposure was partitioned into six time-intervals are reported in order to provide a simple description of the shape of the latency function. Lastly, I fitted a logistic regression model that incorporated a lognormal latency function. Plots were generated in order to compare the results of obtained in fitting the bilinear and lognormal latency functions.
RESULTS
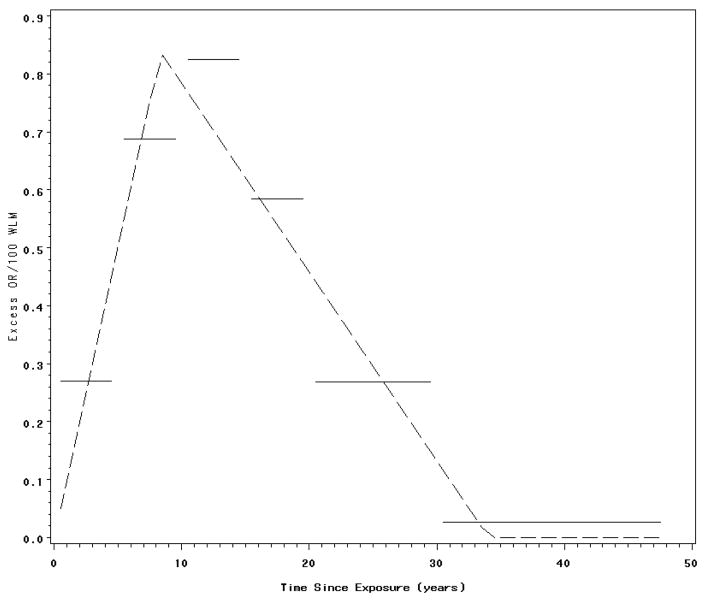
Fitting a linear odds ratio model for lifetime cumulative exposure under a time-constant model (i.e., w(t)=1), led an estimated EOR/100 WLM=0.33 (se=0.10). This time-constant model was compared to a regression model that incorporated a bilinear latency function. The fitted bilinear function obtains a maximal value 8.58 (se=3.72) years after exposure and then declines linearly to a null value 33.55 (se=2.66) years after exposure. Figure 1 illustrates the variation of the EOR/100 WLM with time since exposure as described by the fitted bilinear latency function; also shown for comparison is the excess relative risk as a function of time since exposure as estimated from a fitted piecewise constant model.
Figure 1.
Fitted bilinear and piecewise constant latency functions. Association between lung cancer mortality and radon exposure among Colorado plateau uranium miners.
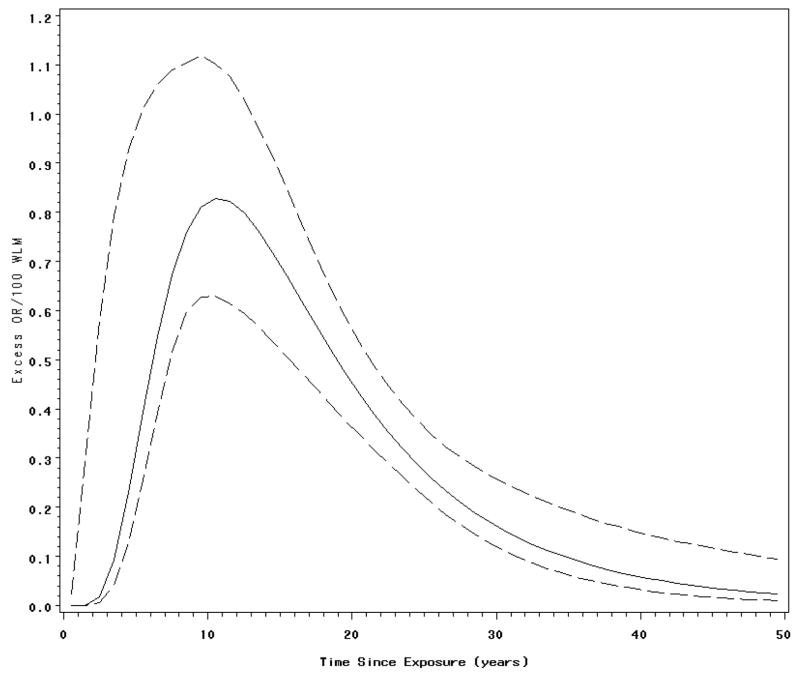
A lognormal latency function also was fitted to the Colorado Plateau data. Figure 2 illustrates the variation of the EOR/100 WLM with time since exposure as described by the fitted lognormal latency function. The fitted lognormal latency model obtains a modal value about 10 years after exposure.
Figure 2.
Fitted lognormal latency functions and associated 95% pointwise credible interval. Association between lung cancer mortality and radon exposure among Colorado plateau uranium miners.
The bilinear latency function provided the best fit to these data (Table 1). While not nested models, the improvement in fit when applying the bilinear latency model is substantial relative to the time-constant model.
Table 1.
Estimated logistic regression coefficients† obtained via SAS PROC NLMIXED. Fittings of sigmoid, bi-linear, and lognormal latency functions to lung cancer mortality among workers in the Colorado Plateau
| Latency function | Parameter | Estimate | Std. Error | Deviance |
|---|---|---|---|---|
| Time-Constant | 2328.1 | |||
| δ | 0.3306 | 0.1018 | ||
| Bilinear | 2316.7 | |||
| α1 | 8.58 | 3.72 | ||
| α2 | 33.55 | 2.66 | ||
| δ | 0.8593 | 0.316 | ||
| Lognormal | 2318.2 | |||
| θ | 2.67 | 0.16 | ||
| λ | 0.58 | 0.13 | ||
| δ | 15.1279 | 5.2410 |
a linear odds rate model of the form: , where z1- zn represent indicators for strata, and δ represents estimated cumulative weighted exposure (in WLM) and w(t) is the latency function..
DISCUSSION
This paper describes an approach for estimation of latency model parameters. While I have focused on two simple latency models, the bilinear and the lognormal latency models, both of which allow a data analyst to describe exposure effects that rise and subsequently diminish with time-since-exposure, other latency models could be developed along similar lines. There are a number of reasons to favor relatively simple latency models such as the bilinear and lognormal models. These include ease of interpretation and communication. The bilinear and lognormal latency models are weighting functions that are bounded by 0 and 1, which facilitates interpretation of the model coefficients. The bilinear model is attractive as the estimated model parameters address questions of public health concern regarding the timing of the maximal effect of exposure and its persistence.5 The lognormal latency model implies that the sum of the weights integrate to unity; and, one potential appeal of the lognormal latency function is that the effects of exposure gradually diminish over time but remain non-zero with protracted time-since-exposure.6 However, the choice of model form for a latency function should not be dictated by these considerations alone, but should follow from examination of the data. As in any modeling exercise, the aim is smoothing, pattern recognition, and summarization. Exposure time-window analyses, such as those reported previously for the analyses of these Colorado Plateau miners, can provide a useful, flexible, piecewise constant description of the exposure-time-response association, guiding the choice of other plausible parametric latency functions that may facilitate summarization of the temporal effects. Spline functions offer another flexible approach to modeling exposure-time-response associations.12,13 Hauptmann et al. proposed the use of cubic B-splines to describe latency functions; this approach is attractive although the modeled association may be sensitive to the choice of the number and location of knots, and there is a tendency for instability in the tails of the fitted function.
Estimation of the parameters for a parametric latency models can be difficult because the parameters are not a function of a simple summary of the exposure history. Typically such parameters have been estimated via an iterative search, which involves computing different time-weighted exposure summaries for each combination of the parameters defining the latency function. For example, for a latency function that involved 2 parameters, this requires a grid-search over the parameter space defined by both parameters.5,14 The use of PROC NLMIXED for estimation of latency model parameters avoids the iterative model searching typically employed in epidemiological analyses of latency effects and provides the data analyst with estimates of the standard error for each model parameter.
I have illustrated this approach using data from a study of mortality among Colorado Plateau miners, a population that has been studied extensively. Consequently, the results obtained from estimation of latency functions via PROC NLMIXED can be contrasted to prior findings.5,9,13 The parameters for the bilinear latency function obtained via PROC NLMIXED are similar to those obtained by Langholz et al. via a grid search over a range of values; Langholz et al. reported a peak in the exposure effect at 8.5 years (se=3.57) and no effect after 34 years (se=2.09). It should be noted, however, that the estimates reported by Langholz et al. were obtained via the EPICURE software package, and required substantial programming to implement the grid search for these model parameters. In the current paper, I obtained nearly identical values for the parameters of the bilinear latency function. One notable difference is that Langholz et al. used a conditional logistic regression model, while I have fitted an unconditional logistic regression model with indicator variables for each stratum defined by the risk sets. These methods should result in similar parameter estimates in scenarios in which the data are not sparse within strata (permitting an unconditional logistic model fitting). Given the need to explicitly model parameters for matching factors in the nested case-control study, this suggests that the approach described in this paper is best suited to studies in which large numbers of controls may be sampled for each case. One such setting is when data are available for the entire study cohort; density sampling in such a setting provides a means of computational reduction when fitting models that approximate a Cox proportional hazards regression. For matched case-control studies in which the number of controls per case is small, an approach employing conditional logistic regression would be necessary.
A similar approach to estimating the parameters of a latency function could be developed using Poisson regression methods. While Poisson regression is often conducted on a grouped data structure, such an approach does not lend itself to an evaluation of latency functions, since person-time and events must be classified into categories of cumulative dose. However, Poisson regression analyses may also be conducted using a counting process formulation of the data (i.e., an ungrouped data structure). In this approach each person is represented by a series of records corresponding to time periods at risk with an indicator of failure status at each interval. Each record has a vector of exposure variables (e.g., annual exposure estimates). Using the latter analytical data structure, PROC NLMIXED could be used to estimate latency parameters via Poisson regression methods in an approach that is directly analogous to that presented in this paper (although much more computationally-intensive).
Extensions of these models may be developed that allow for evaluation of whether the joint effects of exposures conform to a multiplicative model, or to allow for an assessment of whether latency functions differ across categories of a potential effect modifier. Examples of such investigations have been reported previously, although the estimates obtained from such model fittings may be unstable unless there are substantial numbers of cases within each stratum of the potential modifying factor.8,13
NLMIXED allows for a variety of optimization algorithms to be used. As indicated in the appendix, I have used conjugate gradient methods for the optimization approach in these model fittings, an approach well-suited to this sort of large problem. In fact, many large applications of PROC NLMIXED can only be solved by this optimization approach.
This approach overcomes much of the difficulty previously encountered in latency analyses, where models were often fitted in an iterative fashion and, for latency functions that involved more than a single parameter this required large numbers of model fittings and a grid search over the likelihood function. Latency analyses address an important question. This approach should facilitate wider consideration of parametric latency functions.
Acknowledgments
This project was supported by grant R01-CA117841 from the National Cancer Institute, National Institutes of Health.
APPENDIX
Assume that we have data set, DS, which contains information derived from a case-control study. Each record in the data set includes the following main variables: y, which denotes case-control status (1=case, 0 otherwise); a vector of indicator variables zi (i=1…n) which take a value 1 if the subject is in risk set i and 0 otherwise; age, which indicates attained age of the risk set; and r01, r02, …, r80 which represent annual exposures (e.g., r52 is exposure during the age interval 51–52 years.
The SAS macro below can be used to fit a wide variety of models to these data. The model is invoked by the command %latency (data=DS, case=y, exphx=r01-r80, age=age, parms=, lag=, wt=, and regmodel=). The term ‘parms’ of the command invoking the macro allows the user to specify initial values for the regression model parameters; while not required, model convergence may be facilitated by providing plausible starting values for model parameters. By default, PROC NLMIXED assigns all model parameters an initial value of 1. The term ‘lag’ of the command invoking the macro allows the user to specify a lag assumption. This was included in order to replicate analyses by Langholz et al. which lagged exposure assignment by 2 years when computing exposure history summaries. The term ‘wt’ of the command invoking the macro allows the user to specify a parametric latency function which can be defined in terms of time-since-exposure, t. The weight assigned to each annual exposure is calculated as of the midpoint of that year. Lastly, the term ‘regmodel’ of the command invoking the macro allows the user to specify the regression model form, which may include covariates and the variable edose, representing the cumulative effective dose under the specific latency function.

Lifetime cumulative dose analyzed under an exponential odds model
For analyses of lifetime cumulative dose, the latency function is fixed at unity for all values t (i.e., w(t)=1). A log-linear odds model of the form , with 2 covariates for simplicity, z1 and z2, would be fitted as follows:
![]()
Lifetime cumulative dose analyzed under a linear odds ratio model
A linear odds ratio model of the form , where w(t)=1 would be fitted as follows:
![]()
A bilinear latency function analyzed under a linear odds ratio model
A logistic regression model of the form , where , would be fitted by invoking macro %latency as follows:

A lognormal latency function analyzed under a linear odds ratio model
A lognormal latency function can be included. A linear odds ratio model would be fitted by invoking macro %latency as follows:
![]()
References
- 1.Rothman KJ. Induction and latent periods. American Journal of Epidemiology. 1981;114:253–259. doi: 10.1093/oxfordjournals.aje.a113189. [DOI] [PubMed] [Google Scholar]
- 2.Armenian H. Incubation periods of cancer: old and new. J Chron Dis. 1987;40(Suppl 2):9S–15S. doi: 10.1016/s0021-9681(87)80004-8. [DOI] [PubMed] [Google Scholar]
- 3.Salvan A, Stayner L, Steenland K, Smith R. Selecting an exposure lag period. Epidemiology. 1995;6:387–390. doi: 10.1097/00001648-199507000-00010. [DOI] [PubMed] [Google Scholar]
- 4.Checkoway H, Pearce N, Hickey JL, Dement JM. Latency analysis in occupational epidemiology. Archives of Environmental Health. 1990;45:95–100. doi: 10.1080/00039896.1990.9935932. [DOI] [PubMed] [Google Scholar]
- 5.Langholz B, Thomas D, Xiang A, Stram D. Latency analysis in epidemiologic studies of occupational exposures: application to the Colorado Plateau uranium miners cohort. Am J Ind Med. 1999;35:246–256. doi: 10.1002/(sici)1097-0274(199903)35:3<246::aid-ajim4>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 6.Breslow NE, Day NE. Statistical Methods in Cancer Research: The Design and Analysis of Cohort Studies. II. Lyon: International Agency for Research on Cancer; 1987. [PubMed] [Google Scholar]
- 7.Thomas DC. Models for exposure-time-response relationships with applications to cancer epidemiology. Annual Review of Public Health. 1988;9:451–482. doi: 10.1146/annurev.pu.09.050188.002315. [DOI] [PubMed] [Google Scholar]
- 8.Thomas DC, Whittemore AS. Methods for testing interactions, with applications to occupational exposures, smoking, and lung cancer. American Journal of Industrial Medicine. 1988;13:131–147. doi: 10.1002/ajim.4700130109. [DOI] [PubMed] [Google Scholar]
- 9.National Research Council, Committee on the Biological Effects of Ionizing Radiation (BEIR IV) Health Risks of Radon and other Internally Deposited Alpha-emitters. Washington, DC: National Academy Press; 1988. [PubMed] [Google Scholar]
- 10.SAS Institute Inc. SAS OnlineDoc® 9.2. Cary, NC: SAS Institute Inc; 2007. [Google Scholar]
- 11.Hornung RW, Meinhardt TJ. Quantitative risk assessment of lung cancer in U.S. uranium miners. Health Physics. 1987;52:417–430. doi: 10.1097/00004032-198704000-00002. [DOI] [PubMed] [Google Scholar]
- 12.Hauptmann M, Wellmann J, Lubin JH, Rosenberg PS, Kreinenbrock L. Analysis of exposure-time-response relationships using a spline weight function. Biometrics. 2000;56:1105–1108. doi: 10.1111/j.0006-341x.2000.01105.x. [DOI] [PubMed] [Google Scholar]
- 13.Hauptmann M, Berhane K, Langholz B, Lubin J. Using splines to analyse latency in the Colorado Plateau uranium miners cohort. Journal of Epidemiology & Biostatistics. 2001;6:417–424. doi: 10.1080/135952201317225444. [DOI] [PubMed] [Google Scholar]
- 14.Richardson D, Ashmore JP. Investigating Time Patterns of Variation in Radiation-Cancer Associations. Occupational & Environmental Medicine. 2005;62:1–5. doi: 10.1136/oem.2004.017368. [DOI] [PMC free article] [PubMed] [Google Scholar]