Abstract
Integrin activation, the rapid conversion of integrin adhesion receptors from low to high affinity, occurs in response to intracellular signals that act on the short cytoplasmic tails of integrin β subunits. Talin binding to integrin β tails provides one key activation signal, but additional factors are likely to cooperate with talin to regulate integrin activation. The integrin β tail-binding proteins kindlin-2 and kindlin-3 were recently identified as integrin co-activators. Here we report an analysis of kindlin-1 and kindlin-2 interactions with β1 and β3 integrin tails and describe the effect of kindlin expression on integrin activation. We demonstrate a direct interaction of kindlin-1 and -2 with recombinant integrin β tails in pulldown binding assays. Our mutational analysis shows that the second conserved NXXY motif (Tyr795), a preceding threonine-containing region (Thr788 and Thr789) of the integrin β1A tail, and a conserved tryptophan in the F3 subdomain of the kindlin FERM domain (kindlin-1 Trp612 and kindlin-2 Trp615) are required for direct kindlin-integrin interactions. Similar interactions were observed for integrin β3 tails. Using fluorescence-activated cell sorting we further show that transient expression of kindlin-1 or -2 in Chinese hamster ovary cells inhibits the activation of endogenous α5β1 or stably expressed αIIbβ3 integrins. This inhibition is not dependent on direct kindlin-integrin interactions because mutant kindlins exhibiting impaired integrin binding activity effectively inhibit integrin activation. Consistent with previous reports, we find that when co-expressed with the talin head, kindlin-1 or -2 can activate αIIbβ3. This effect is dependent on an intact integrin-binding site in kindlin. Notably however, even when co-expressed with activating levels of talin head, neither kindlin-1 or -2 can cooperate with talin to activate β1 integrins; instead they strongly inhibit talin-mediated activation. We suggest that kindlins are adaptor proteins that regulate integrin activation, that kindlin expression levels determine their effects, and that kindlins may exert integrin-specific effects.
Integrins are a family of αβ heterodimeric transmembrane receptors that mediate cell adhesion to extracellular matrix, cell surface, or soluble protein ligands and modulate a variety of intracellular signaling cascades. A key feature of integrins is their ability to dynamically regulate their affinity for extracellular ligands. In a tightly regulated process generally termed integrin activation, intracellular signals that impinge upon the β subunit cytoplasmic tail induce conformational rearrangements in the integrin extracellular domains, increasing the binding affinity for extracellular ligands (1-3). Ligand-bound integrins then recruit additional signaling, adaptor, and cytoskeletal proteins to the integrin cytoplasmic domains, providing mechanical connections to the actin cytoskeleton and a link to a variety of signal transduction pathways (2-8).
Recent years have seen significant advances in our understanding of integrin activation. Notable among these is the identification of the actin- and integrin-binding protein talin as a key integrin activator (1, 9). The 50-kDa talin head contains the principal integrin-binding site, and expression of the talin head is sufficient to activate β1 and β3 integrins (10, 11). The talin head contains a FERM (four point one ezrin radixin moesin) domain. FERM domains consist of trefoil arrangement of three subdomains (F1, F2, and F3). The phosphotyrosine-binding domain-like F3 subdomain of the talin FERM directly binds a conserved NP(I/L)Y motif in integrin β tails, and this interaction is necessary for integrin activation in vitro and in vivo (10, 12-19). However, although abundant evidence supports the importance of talin binding to integrin β tails during integrin activation, differences in sensitivity of integrins to talin activation and submaximal activation by overexpressed talin suggested that other activating factors may cooperate with talin (10, 20). In an attempt to identify and characterize potential co-activators, we investigated the kindlin family of FERM domain-containing proteins.
Kindlin family proteins (21) were first characterized in nematodes where the sole Caenorhabditis elegans kindlin, UNC-112, was identified in an embryonic screen for defective motility and shown to be essential for the assembly of proper cell-matrix adhesion structures, where it normally co-localized with β integrin (22-24). UNC-112 is conserved across many species, because the nematode, fly, and human homologs are ∼60% similar (∼41% identical) over their entire length (24). Humans express three known homologs of UNC-112: kindlin-1 (Kindlerin, URP1, and FERMT1), kindlin-2 (Mig2 and mig-2), and kindlin-3 (Mig2B and URP2) (25-27). Kindlin-1 and -2 are most closely related, sharing 60% identity and 74% similarity, whereas kindlin-3 shares 53% identity and 69% similarity to kindlin-1 and 49% identity and 67% similarity to kindlin-2 (28). The kindlin proteins all contain a predicted Pleckstrin homology domain and a FERM domain that is most closely related to the talin FERM domain, particularly within the integrin-binding F3 subdomain (29). Based on this sequence similarity we proposed that kindlin FERM domains may directly bind integrin β tails, and we previously showed that kindlin-1 could be pulled down from cell lysates using recombinant integrin β1 and β3 tails and that kindlin-1 co-localized with integrins in focal adhesions (29). A similar localization was reported for kindlin-2 (26, 30), and recent reports provided clear evidence implicating kindlin-2 and kindlin-3 in regulation of integrin activation (31-33). Here, we have used integrin pulldown assays to demonstrate direct binding of full-length kindlin-1 to the cytoplasmic tails of β1A and β3 integrins and to identify key binding residues within the integrin tails and the kindlin F3 subdomain. We confirm that these interactions are important for recruiting kindlin-1 to focal adhesions and show that, contrary to expectations, overexpressed kindlin-1 or -2 inhibit β1 and β3 integrin activation. Overexpressed kindlin-1 or -2 can, however, cooperate with expressed talin head to activate β3 but not β1 integrins. We therefore provide the first data suggesting that kindlin-1 and -2 effects on integrin activation may show β subunit specificity.
EXPERIMENTAL PROCEDURES
Antibodies—Ligand-mimetic anti-αIIbβ3 PAC1 (BD Biosciences), anti-hamster α5β1 PB1 (Developmental Studies Hybridoma Bank), anti-talin 8d4 (Sigma), goat polyclonal anti-human kindlin-2 (Santa Cruz Biotechnology), rabbit polyclonal anti-GST3 (Chemicon), mouse anti-FLAG (Sigma-Aldrich), and goat anti-GFP (Rockland) were purchased. Generation of anti-human kindlin-1 was previously described (29). The anti-αIIbβ3 monoclonal antibody D57 has been described previously (34). The α5β1-specific inhibitor 3F compound was kindly provided by Dr. Kessler (35). GST-fibronectin type III repeats 9-11 (FN9-11) have been described previously (36).
Protein Production—Recombinant His-tagged human integrin cytoplasmic tail model proteins were produced and purified as previously described (37). Human kindlin-1 and -2 full length (amino acids 1-677 and 1-680, respectively), ΔFERM (amino acids 1-181 and 1-183, respectively), and FERM (amino acids 193-677 and 195-680, respectively) constructs were generated by polymerase chain reaction from human cDNAs generously provided by Mary Beckerle (University of Utah) and subcloned into pGEX4T-3 (Amersham Biosciences) and pEGFP-C1 (BD Biosciences), pFLAG-CMV2 (Sigma-Aldrich), or pDsRed monomer (Clontech) mammalian expression vectors. Point mutations were introduced by QuikChange™ site-directed mutagenesis (Stratagene). All of the inserts were verified by DNA sequencing. GST fusion proteins were produced in Escherichia coli BL21 cells or Rosetta cells (Novagen) and purified on glutathione-Sepharose 4B (Amersham Biosciences) according to the manufacturer's instructions. GFP-fused mouse talin1 head (amino acids 1-433) was generated as previously described (10).
Binding Assays and Analysis—Binding assays using recombinant integrin cytoplasmic tails bound to His-bind resin (Novagen) were performed as previously described (37). Briefly, we expressed integrin β tails in a pET15b vector containing an N-terminal His tag sequence followed by a thrombin cleavage site, a cysteine-residue linker, a coiled-coil-forming helical sequence, a four-residue glycine spacer, and the integrin cytoplasmic domain (human β1A residues 751-798 or β3 residues 715-762). Integrin constructs were expressed in BL21 cells (Novagen), purified, and coated to Ni2+-charged nitrilotriacetic acid resin (Novagen) as previously described (37).
For FLAG fusion proteins, Chinese hamster ovary (CHO) cells were transiently transfected with 3 μg of DNA using Lipofectamine™ (Invitrogen), and the cells were harvested 24 h later and lysed. Cell lysates or bacterially purified proteins were incubated with integrin tails bound to beads (24 h for lysates and 2 h for purified proteins), washed, resuspended in SDS sample buffer, heated for 5 min at 95 °C, and analyzed for binding on 4-20% Tris-glycine SDS-polyacrylamide gradient gel (Invitrogen). The loading percentage lanes in the Western blot images represent input lysates or purified protein as a percentage of total mixed with integrin tails and serve as a reference for relative protein amounts loaded into the assay.
Immunofluorescence—NIH 3T3 cells were transiently transfected with 3 μg of indicated cDNAs using Lipofectamine™ (Invitrogen). 24 h after transfection, the cells were detached and allowed to readhere and spread on fibronectin-coated (5 μg/ml) coverslips. After 4 h of plating, the cells were fixed, permeabilized, and stained with anti-vinculin as described previously (38, 39).
Analysis of Integrin Activation—The activation state of endogenous α5β1 or stably overexpressed αIIbβ3 integrins in CHO cells transiently expressing DsRed-tagged kindlins and/or GFP-talin head was assessed in three-color FACS assays using a modification of previously described methods (10, 12, 36). FACS data analysis was carried out using FlowJo FACS analysis software and statistical analysis using Graphpad Prism.
Briefly, to assess α5β1 activation CHO cells were co-transfected with the indicated GFP and DsRed expression constructs using Lipofectamine™ 2000 (Invitrogen), and 24 h later the cells were suspended and incubated with biotinylated recombinant GST-FN9-11 in the presence or absence of integrin inhibitor (1 mm RGD peptide (Sigma) or 0.1 μm 3F α5β1 inhibitor (35)). For each preparation of biotinylated GST-FN9-11, the effective concentration was determined by titration. The cells were washed, and bound FN9-11 was detected with allophycocyanin-conjugated streptavidin. FN9-11 binding to doubly transfected (GFP-positive and DsRed-positive) cells was assessed on a FACSCalibur instrument (BD Biosciences). The α5β1 integrin expression was assessed in parallel by staining with antibody PB1 (40). The α5β1 activation index was defined as AI = (F - Fo)/(Fintegrin), where F is the geometric mean fluorescence intensity (MFI) of FN9-11 binding, Fo is the MFI of FN9-11 binding in presence of inhibitor, and Fintegrin is the normalized MFI of PB1 binding to transfected cells.
The activation state of αIIbβ3 integrins was assessed as described above but using the ligand-mimetic anti-αIIbβ3 monoclonal antibody PAC1 (11, 12, 36, 41) in place of biotinylated FN9-11. αIIbβ3 integrin expression was assessed in parallel by staining with D57 antibody (34). Bound PAC1 was detected using Alexa 647 fluorophore-conjugated goat anti-mouse IgM (Invitrogen). Activation of αIIbβ3 in doubly transfected (GFP-positive and Red-positive) cells was quantified in three-color flow cytometric assays, and an activation index was calculated as defined above where F is the MFI of PAC1 binding, Fo is the MFI of PAC1 binding in presence of RGD peptide, and Fintegrin is the MFI of D57 binding to transfected cells. Integrin expression ratio was defined for double expressing cells as follows: Integrin expression ratio = (Ftrans)/(Funtrans), where Ftrans is the geometric MFI of PB1 or D57 binding to double expressing cells, and Funtrans is the MFI of PB1 or D57 binding to untransfected cells.
RESULTS
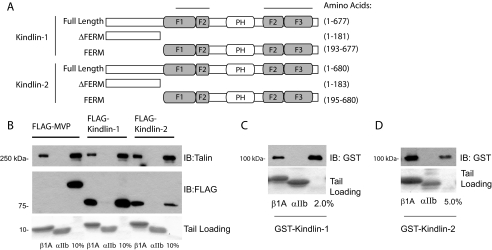
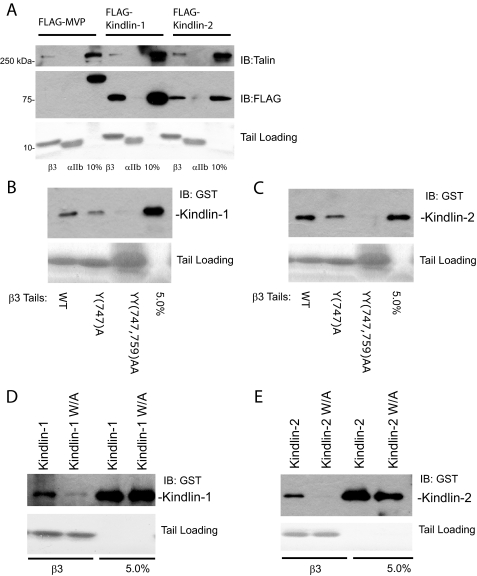
Kindlin-1 and -2 Bind Directly to β1A Integrin Tails—Our initial investigation of kindlin-integrin interactions demonstrated binding of FLAG-tagged full-length kindlin-1 from CHO cell lysates to recombinant models of β1A integrin cytoplasmic tails (29). Here we extended this analysis to kindlin-2 and showed that both full-length kindlin-1 and -2 bound β1A tails specifically because no binding of an irrelevant control FLAG-tagged protein (major vault protein (42)) was observed, and the kindlins did not bind to αIIb tails (Fig. 1B). Binding of endogenous talin from the cell lysate to β1A but not αIIb tails was examined as an additional specificity control (Fig. 1B, upper panel). To determine whether the interaction between kindlins and integrin β tails is direct, we expressed and purified full-length kindlin-1 and -2 as GST fusion proteins from E. coli. Despite using a range of optimization approaches, including testing various bacterial strains, different induction conditions, and different growth temperatures, the yields of soluble GST-fused kindlin proteins were low (∼0.05 mg/liter of bacterial culture). These proteins were nonetheless sufficient for use in protein-protein interaction assays and revealed direct binding of kindlin-1 and -2 to β1A but not αIIb tails (Fig. 1, C and D). We were unable to quantify the affinity of the kindlin-integrin interaction because dose-response curves did not reach saturation at the available protein concentrations (supplemental Fig. S1). Nonetheless these assays suggest that kindlin-integrin interactions are direct but of relatively low affinity.
FIGURE 1.
Kindlin-1 and -2 bind directly to β1A integrin tails. A, schematic diagram of the human kindlin-1 and -2 proteins and fragments generated. The shaded regions correspond to the predicted FERM domain. The two upper lines signify a portion of the FERM domain with high similarity to the FERM domain of talin. The white PH region corresponds to a region with high homology to Pleckstrin homology domains. B, pulldown assays using recombinant β1A tails mixed with CHO cell lysates containing FLAG-tagged kindlin-1 or -2. Binding of kindlins, endogenous talin (positive control), and the major vault protein (negative control), was assessed by Western blotting. αIIb tails serve as controls for binding specificity to the β tails. Loading of each tail protein was judged by protein staining. The 10% lanes represent 10% of the starting material in the binding assay. C and D, direct binding of GST-tagged kindlin-1 and -2 to β1A but not αIIb tails. IB, immunoblot.
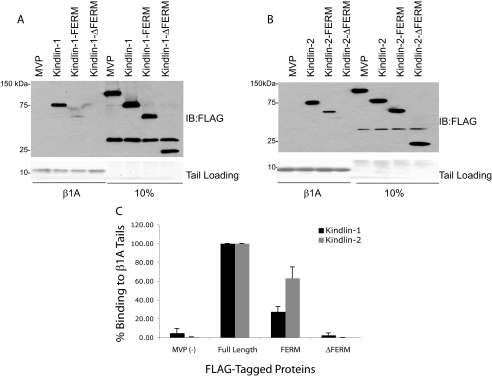
The FERM Domain in Kindlin-1 and -2 Is Required for Binding to Integrin β1A Cytoplasmic Tails—To determine whether the kindlin FERM domain is sufficient for β1A binding, we expressed the isolated FLAG-tagged kindlin-1 and -2 FERM domains in CHO cells. Cell lysate pulldown assays using recombinant β1A tails revealed binding of kindlin-1-FERM and kindlin-2-FERM to β1A tails (Fig. 2, A and B). This binding was specific because no interaction with the αIIb tails was observed (data not shown) but was much less than that seen for the binding of full-length kindlins. The kindlin FERM domain is necessary for integrin binding because deletion of the FERM domain completely abrogated kindlin-1 and -2 binding to β1A tails (Fig. 2C). Thus the FERM domain of kindlin-1 and -2 is required for interaction with β1A integrin tails. This is consistent with the results of Shi et al. (30), who previously reported that the kindlin-2-FERM was required for β tail binding. However, although the kindlin FERM domain may be sufficient for integrin binding, the presence of more N-terminal portions of the protein appears to contribute to the interaction.
FIGURE 2.
The FERM domain in kindlin-1 and -2 is required for binding toβ1A integrin tails. A and B, pulldown assays examining the ability of FLAG-tagged fragments with and without the FERM domain of kindlin-1 and -2 to bind β1A tails. C, kindlin-1 and -2 binding was quantified by densitometry, normalized to starting material loading, and then calculated relative to binding of wild-type full-length constructs (means ± S.E.; n ≥ 4). IB, immunoblot.
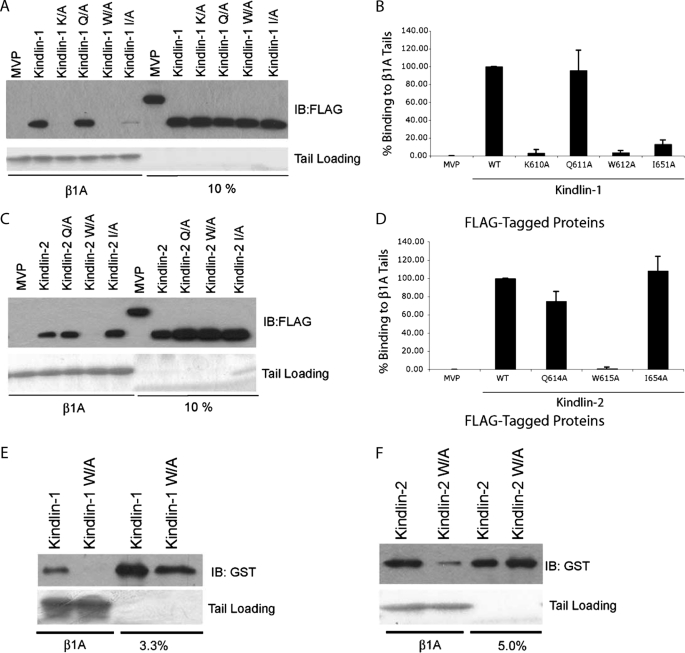
A Partially Conserved Site in the F3 Subdomain of the Kindlin-1 and -2 FERM Domain Is Required for Binding to β1A Tails—Similarity between the talin FERM domain and the kindlin-1 FERM domain, particularly within the F3 subdomain (29), prompted us to test whether kindlin-1 binds integrin β tails via its F3 subdomain, as talin does. We have shown that Arg358, Trp359, and Ile396 in the talin1-F3 subdomain are important for binding to integrin β tails, whereas Lys357 plays a lesser role (10, 12, 16). We therefore mutated the corresponding residues in the F3 subdomain of FLAG-kindlin-1 and tested binding to β1A tails. As shown in Fig. 3 (A and B), K610A, W612A, and I651A mutations strongly inhibited binding to β1A tails, whereas a Q611A substitution (predicted to correspond to talin Arg358) had no effect. Furthermore, when the W612A was introduced into GST-kindlin-1, the mutation was sufficient to strongly inhibit direct kindlin-1 binding to β1A tails (Fig. 3E). We also introduced corresponding mutations into kindlin-2 and tested their effect on β1A binding (Fig. 3, C, D, and F). As was seen for kindlin-1, a Q614A mutation in kindlin-2 did not substantially inhibit β1A binding, whereas W615A mutations strongly inhibited binding. However, in the case of kindlin-2, I654A mutation did not inhibit β1A binding. Together this reveals that, as was seen for talin-β tail interactions, the F3 subdomain is important for kindlin binding to β1A tails and that residues on a similar portion of the molecule are involved. However, the differences in specific residues whose mutation inhibits the interaction suggest subtle differences between the kindlin-1, kindlin-2, and talin interactions with β tails.
FIGURE 3.
A partially conserved site in the F3 subdomain of the kindlin-1 and -2 FERM domains is required for binding to β1A tails. A, pulldown assays mapping the F3 subdomain of kindlin-1 for mutants that inhibit association with β1A tails. Lysates from CHO cells expressing FLAG-tagged kindlin-1 wild-type and mutant constructs were mixed with recombinant β1A tails and assessed for binding. B, the binding of kindlin-1 and mutants was quantified by densitometry, normalized to starting material loading, and then calculated relative to binding of wild-type kindlin-1 (means ± S.E.; n = 10). C, pulldown assays mapping the F3 subdomain of kindlin-2 for mutants that inhibit association with β1A tails. D, the binding of kindlin-2 and mutants was quantified by densitometry, normalized to starting material loading, and then calculated relative to binding of wild-type kindlin-2 (means ± S.E.; n = 3). E and F, purified GST-tagged kindlin-1 and -2 constructs with mutations of a critical tryptophan residue, W612A and W615A for kindlin-1 and -2, respectively, were tested for binding to β1A tails. IB, immunoblot; WT, wild type.
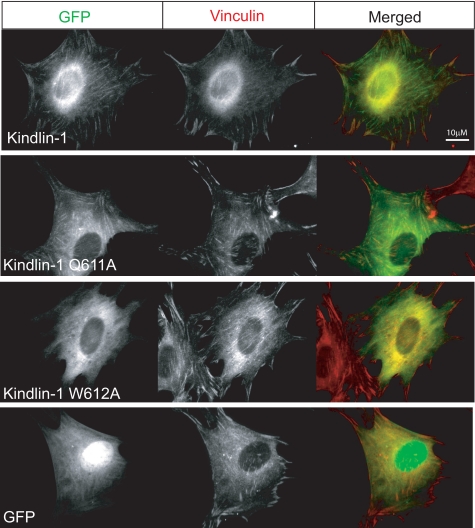
Integrin Binding Activity Correlates with Kindlin-1 Targeting to Focal Adhesions—Focal adhesions are integrin-rich sites where adherent cells make strong contacts with the surrounding matrix (43). Previous studies have shown that kindlin-1 and -2 localize to focal adhesions (26, 29, 30). To assess the importance of kindlin-integrin interactions in focal adhesion targeting, we generated GFP-tagged kindlin-1 constructs with mutations in the F3 subdomain intended to disrupt binding to integrin β tails. In agreement with our biochemical findings (Fig. 3), cells expressing GFP-kindlin-1 with K610A, W612A, and I651A mutations showed diffuse GFP localization, whereas the GFP-kindlin-1 Q611A mutant localized to vinculin-rich focal adhesion sites similar to wild-type kindlin-1 (Fig. 4 and data not shown). Thus cell-based assays support the conclusions from our biochemical findings, emphasizing the importance of the kindlin-1-F3 subdomain interactions with integrin β tails for recruiting to integrin-rich sites.
FIGURE 4.
Integrin binding activity correlates with kindlin-1 targeting to focal adhesions. Images of NIH 3T3 cells transiently transfected with GFP-tagged kindlin-1 wild-type and mutant expression constructs. Kindlin-1 co-localizes with vinculin at focal adhesions. Recruiting kindlin-1 to vinculin-rich focal adhesions is blocked by W612A but not Q612A mutations.
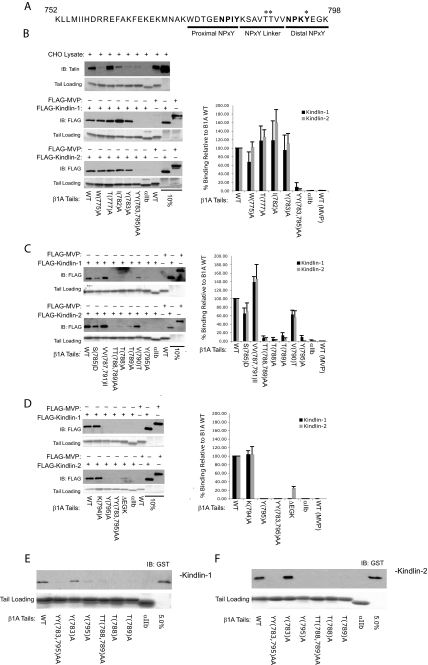
Kindlin-1 and -2 Bind to the Membrane-distal NPKY Motif in β1A Tails—Integrin β tails contain two conserved NPXY motifs, and the more membrane-proximal site is required for interaction with the F3 subdomain of talin (16, 17). We therefore tested kindlin binding to a variety of β1A tails containing mutations within and around the first NPXY motif (Fig. 5B). Consistent with previous reports (10), W775A, I782A, and Y783A all inhibited talin binding; however, they had little or no effect on kindlin-1 or -2 binding. As previously reported (29), kindlin-1 and -2 binding was blocked by a double mutation targeting the tyrosine in each NPXY motif (β1A(Y783A,Y795A)). This suggests that the second rather than the first NPXY motif may be key to kindlin-1 and -2 binding to β1A integrins. We therefore designed a series of mutations within and following the second NPXY motif of β1A. In this case we found that a single Y795A mutation was sufficient to block kindlin-1 and -2 binding, whereas a K794A mutation at the adjacent residue did not impact binding (Fig. 5D). Deletion of the C-terminal three residues also reduced kindlin-1 and -2 binding, although the effect was less pronounced for kindlin-2, again suggesting subtle differences between kindlin-1 and -2 interactions with β1 integrins.
FIGURE 5.
Kindlin-1 and -2 bind to the membrane-distal NPKY motif in β1A tails. A, the protein sequence for the cytoplasmic tail of human integrin β1A. Asterisks denote residues required for kindlin-1 and -2 binding. B-D, pulldown assays mapping the binding site on β1A tails for kindlin-1 and -2 binding. Lysates from CHO cells either untransfected or transiently expressing FLAG-tagged kindlin-1 or -2 were mixed with wild-type and mutant β1A tails and assessed for binding. The binding of kindlin-1 and -2 was quantified by densitometry, normalized to starting material loading, and then calculated relative to binding of wild-type β1A (means ± S.E.; n ≥ 3). E and F, bacterially purified GST-tagged kindlin-1 and -2 were examined for their ability to bind to β1A tail mutants. IB, immunoblot; WT, wild type.
The preceding experiments reveal that kindlin-1 and -2 bind the membrane-distal NPKY motif in β1A tails and that the kindlin-F3 subdomain is required for this interaction. The talin F3-β1 tail NPIY interaction requires additional binding sites 8 residues upstream of the tyrosine, specifically Trp775 (10, 16). We therefore examined the binding of kindlin-1 and -2 to β1A tails containing mutations between the two NPXY motifs. This revealed that two threonines (Thr788 and Thr789) also played key roles in the kindlin-integrin interaction (Fig. 5C). Analysis of direct kindlin-1 and -2 binding to β1A tail mutants (Fig. 5, E and F) further supports the conclusion that the distal threonines and tyrosine are important for β1 tail binding.
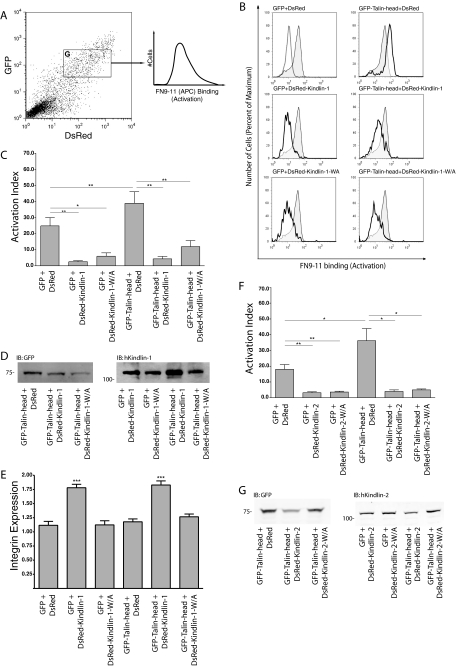
Kindlin Overexpression Suppresses β1 Integrin Activation—Talin binding, via its F3 FERM subdomain, to the membrane-proximal NPXY motif in integrin β tails is a key step in integrin activation. Our biochemical and cell-based experiments indicate that the F3 subdomain of kindlin-1 and -2 mediates direct contacts with the membrane-distal NPXY region of integrin β1A cytoplasmic tails. To test whether this interaction is involved in integrin activation, we adapted a well established FACS-based assay to assess the effect of kindlin-1 overexpression on the activation state of endogenous α5β1 integrins in CHO cells. CHO cells do not express detectible levels of endogenous kindlin-1 (44).4 The assay relies on identifying a population of cells expressing our protein(s) of interest and measuring the binding of the soluble recombinant fibronectin fragment FN9-11 to these transfected cells (Fig. 6A) (36, 45). Integrin expression on an equivalent population of transfected cells is measured in parallel using antibodies that bind in an activation-independent manner. Overexpression of GFP- or DsRed-tagged kindlin-1 but not GFP or DsRed alone significantly inhibited α5β1 activation (Fig. 6, B and C; supplemental Fig. S2; and data not shown). This effect was independent of the integrin binding activity of kindlin-1 because the binding-defective mutant W612A also inhibited integrin activation. Integrin inhibition was cell autonomous, because only transfected cells exhibited inhibition and concentration-dependent as more highly transfected cells exhibited greater reductions in FN9-11 binding (supplemental Fig. S2). Effects are unlikely to be due to the attached fluorescent tag because (i) neither GFP nor DsRed had an effect when expressed alone, (ii) GFP- and DsRed-tagged kindlin-1 still bound integrin tails in pulldown assays (data not shown), and (iii) tagged proteins co-localized with integrins in focal adhesions (Fig. 4 and data not shown).
FIGURE 6.
Kindlin-1 and -2 inhibit talin mediated-activation of α5β1 integrin. A, wild-type CHO cells were co-transfected with GFP-tagged mouse-talin-head (1-433) and DsRed-tagged kindlin-1 cDNAs. Cells expressing both GFP and DsRed were selected (gate G) and assessed for activation of endogenous α5β1 integrin (FN9-11 binding) or α5β1 integrin expression (PB1 binding). B, representative histogram panels of FN9-11 binding to active α5β1 integrin in gated co-expressing CHO cells. Upper left panel, FN9-11 binding to GFP and DsRed gated expressing cells (shaded, native; dashed, in the presence RGD inhibitor). In the remaining panels, the shaded regions represent native FN9-11 binding to gated cells expressing GFP and DsRed, whereas the bold histograms represent FN9-11 binding in the presence of indicated tagged proteins. Careful gating ensured comparable levels of GFP and DsRed fluorescence in the analyzed population. C, activation indices of α5β1 integrin from gated CHO cells co-expressing GFP-talin head and DsRed-kindlin-1 were calculated and normalized for integrin expression (see “Experimental Procedures”). The results represent the means ± S.E. (n ≥ 3). **, p < 0.01; *, p < 0.05. D, total lysates from double transfected CHO cells were separated by SDS-PAGE and analyzed by immunoblot for tagged proteins. E, integrin expression ratios from gated CHO cells co-expressing GFP-talin head and DsRed-kindlin-1 were calculated relative to untransfected cells (see “Experimental Procedures”). The results represent means ± S.E. (n ≥ 3). ***, p < 0.0001; **, p < 0.01; *, p < 0.05. F, activation indices of α5β1 integrin from gated CHO cells co-expressing GFP or GFP-talin head and DsRed or DsRed-kindlin-2 constructs as indicated were calculated and normalized for integrin expression (see “Experimental Procedures”). The results represent the means ± S.E. (n ≥ 3). **, p < 0.01; *, p < 0.05. G, total lysates from double transfected CHO cells were separated by SDS-PAGE and analyzed by immunoblot for tagged proteins.
Inhibition of FN9-11 binding was not due to inhibition of the surface expression of α5β1 integrins because activation indices were normalized for integrin expression (Fig. 6C), and measurement of α5β1 surface expression showed a kindlin-1-dependent increase in α5β1 expression not a decrease (Fig. 6E). Unlike effects on integrin activation, the effect of kindlin-1 on integrin expression was dependent on an intact F3 domain, suggesting that kindlin-1-integrin interactions may impact integrin surface expression.
To test whether kindlin-1 could inhibit talin head-mediated activation of β1 integrins, we co-expressed the integrin-activating talin head with wild-type or mutant kindlin-1 in CHO cells. Integrin activation was assessed by three-color FACS, allowing cells expressing two different fluorophore-tagged proteins (GFP and DsRed) to be identified and assessed for integrin activation in a third channel. Importantly, this system allows accurate monitoring of the expression of each protein and permits gating on cells with defined expression levels of each protein, making comparisons between the effects of mutants more readily interpretable (Fig. 6A). In this way we showed that although talin head (amino acids 1-433) activated α5β1, as previously reported (10), co-expressed kindlin-1 or kindlin-1 W612A strongly inhibited integrin activation (Fig. 6, B and C). This inhibition was not due to effects on GFP-talin head expression because cells were gated to have equivalent levels of this protein. Furthermore, as seen for expression of kindlin-1 alone, the effect is not due to a loss of cell surface integrin expression in kindlin-1-expressing cells because activation indices were normalized to account for changes in integrin levels, and kindlin-1 expression consistently led to an increase rather than a decrease in surface α5β1 levels (Fig. 6E). Western blot analysis was used to confirm expression of full-length fusion proteins (Fig. 6D).
Because a recent report of the kindlin-2 knock-out mouse implicates kindlin-2 in controlling β1 integrin activation (33), we also tested the effect of DsRed-kindlin-2 expression on FN9-11 binding to CHO cells. Kindlin-2 is the only kindlin protein expressed in CHO cells (32, 44). In our hands, cells expressing high levels of DsRed-kindlin-2 strongly suppressed α5β1 integrin activation (Fig. 6F), as seen with kindlin-1-overexpressing cells (Fig. 6C).
Kindlin-1 and -2 Cooperate with Talin to Activate αIIbβ3—As described above, overexpressed kindlin-1 and -2 inhibit β1 integrin activation in a manner that does not require their interaction with the integrin β tail. The activation state of β3 integrins is also tightly regulated, and contrary to our results with β1 integrins, recent publications indicate that kindlin-2 cooperates with talin to activate αIIbβ3 integrins (32, 33). We therefore assessed the binding of kindlin-1 and -2 to β3 integrin tails. As shown in Fig. 7A, β3 tails can selectively pulldown FLAG-tagged kindlin-1 and -2 from CHO cell lysates. Kindlin-1 and -2 also directly bind to β3 integrin tails, and binding is inhibited by point mutations in the second NXXY motif of the β tail (Fig. 7, B and C) or Trp → Ala mutations within the kindlin F3 subdomain (Fig. 7, D and E). Thus kindlins appear to bind β1 and β3 integrins in a comparable manner.
FIGURE 7.
Kindlin-1 and -2 bind directly to β3 integrin tails. A, pulldown assays using recombinant β3 tails mixed with CHO cell lysates containing FLAG-tagged kindlin-1 or -2. Binding of kindlins, endogenous talin (positive control), and the major vault protein (negative control) was assessed by Western blotting. αIIb tails serve as a control for binding specificity to the β tails. Loading of each tail protein was judged by protein staining. The 10% lanes represent 10% of the starting material in the binding assay. B and C, direct binding of GST-tagged kindlin-1 and -2 to wild-type and mutant β3 tails. D and E, purified GST-tagged kindlin-1 and -2 constructs with mutations of a critical tryptophan residue, W612A and W615A for kindlin-1 and -2, respectively, were tested for binding to β3 tails. IB, immunoblot.
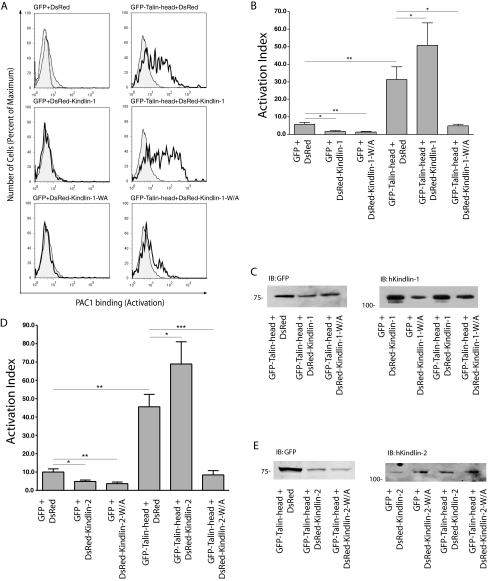
To investigate the effect of kindlin-1 on αIIbβ3 activation, we first overexpressed kindlin-1 in CHO cells stably expressing αIIbβ3 and assessed activation using the ligand-mimetic anti-αIIbβ3 monoclonal antibody PAC1 (34). As observed for β1 integrins, expression of either DsRed-kindlin-1 or DsRed-kindlin-1-W612A inhibited αIIbβ3 activation (Fig. 8, A and B). As expected, based on previous reports (10, 17), overexpression of GFP-talin-head, co-expressed with DsRed as a experimental control, dramatically increased β3 integrin activation. Unlike the case for β1 integrins, when kindlin-1 and talin head were co-expressed, αIIbβ3 activation was further enhanced beyond that seen for talin head alone (Fig. 8, A and B). Thus talin and kindlin-1 cooperate to activate β3 integrins. This effect is likely to be dependent upon integrin binding because the kindlin-1 W612A mutant did not enhance talin head-mediated activation, and instead it strongly inhibited β3 integrin activation to resting levels. Again effects were not due to altered levels of expressed proteins because cells were gated to have comparable levels, and assessment of αIIβ3 integrin expression using antibody D57 (34) allowed correction for effects of kindlin-1 on αIIbβ3 surface expression (Fig. 8B and data not shown). Western blot analysis was used to confirm expression of full-length fusion proteins (Fig. 8C). Similar results were obtained with kindlin-2 (Fig. 8D). In summary, overexpressing kindlin-1 or -2 inhibits activation of β1 and β3 integrins but, in the case of αIIbβ3 integrins, co-expression of talin head with kindlins, but not with integrin-binding deficient kindlin mutants, results in enhanced β3 activation.
FIGURE 8.
Kindlin-1 and -2 cooperate with talin to activate αIIbβ3. CHO cells stably expressing αIIbβ3 integrin were co-transfected with GFP or GFP-tagged mouse-talin-head (1-433) and DsRed or DsRed-tagged kindlin-1 cDNAs as indicated. Co-expressing cells with similar fluorescence of GFP and DsRed tags were assessed for αIIbβ3 integrin activation (PAC1 binding) or αIIbβ3 integrin expression (D57 Binding). A, representative histogram panels of PAC1 binding to active αIIbβ3 integrin in gated co-expressing CHO cells. Upper left panel, PAC1 binding to GFP and DsRed gated expressing cells (shaded, native; dashed, in the presence RGD inhibitor). In the remaining panels the shaded regions represent native PAC1 binding to gated cells expressing GFP and DsRed, and the bold histograms represent PAC1 binding in the presence of indicated tagged proteins. B, activation indices of αIIbβ3 integrin from gated CHO cells co-expressing GFP-talin head and DsRed-kindlin-1 were calculated and normalized for integrin expression (see “Experimental Procedures”). The results represent the means ± S.E. (n ≥ 3). **, p < 0.01; *, p < 0.05. C, total lysates from double transfected CHO cells were separated by SDS-PAGE and analyzed by immunoblot for tagged proteins. D, activation indices of αIIbβ3 integrin from gated CHO cells co-expressing GFP-talin head and DsRed-kindlin-2 were calculated and normalized for integrin expression (see “Experimental Procedures”). The results represent the means ± S.E.(n≥3). **, p < 0.01; *, p < 0.05. E, total lysates from double transfected CHO cells were separated by SDS-PAGE and analyzed by immunoblot for tagged proteins. IB, immunoblot.
DISCUSSION
Here we have investigated the interaction of kindlin-1 and kindlin-2 with β1A and β3 integrin cytoplasmic tails and assessed the effect of transient kindlin-1 or -2 expression on activation of β1 and β3 integrins. We found that (i) kindlin-1 and -2 bind directly to the second conserved NXXY motif and a preceding threonine-containing region in integrin β tails; (ii) the F3 subdomain within the kindlin FERM domain is required for β tail binding, but the isolated FERM domain binds β tails less well than intact kindlin, suggesting that residues in the N-terminal region somehow contribute to the interaction; (iii) transient expression of kindlin-1 or -2 inhibits β1 and β3 integrin activation, but this inhibition does not require an intact integrin-binding site in the kindlin F3 domain; (iv) when co-expressed with the integrin-activating talin head, kindlin-1 and -2 can cooperate with talin to enhance activation of αIIbβ3 but not α5β1 integrins; and (v) the activating effect of kindlins requires an intact integrin-binding site because nonbinding mutants do not synergize with talin, instead they strongly inhibit integrin activation. Taken together, and in the context of recent reports revealing a role for kindlin-2 and -3 in integrin activation (21, 31-33), our data show that kindlin-1, like other kindlins, can modulate integrin activation. However, our data also indicate that kindlin activities may be more complex than initially appreciated, revealing subtle differences between kindlin-1 and -2 interactions with integrins and differential effects on β1 and β3 integrin activation by kindlins.
The kindlin FERM domain is most similar to that of talin, particularly within the F3 subdomain (29). The talin F3 subdomain contains the major integrin-binding site (17), and here we show that in kindlin-1 and -2, the F3 domain is important for binding to β1 and β3 integrins. This is consistent with previous reports on kindlin-2 and -3 interactions with integrins (31, 33). We have been unable to determine the affinity of the kindlin-integrin interaction because the low yields of purified GST-kindlin we obtain are insufficient to saturate binding in our assays. Furthermore, in our hands, purified kindlin-1 FERM domain or F3 fragments did not exhibit specific integrin binding. Unlike talin, where possibly because of release of autoinhibition (46, 47), the isolated F3 or FERM domain binds integrins more tightly than the intact protein, the smaller fragments of kindlin-1 and -2 bind β1A tails less well than the intact kindlin protein does. This suggests that the N-terminal region of kindlins, which did not detectably interact with integrin β tails in isolation, aids integrin binding either by providing a second weaker interaction site or possibly through conformational effects on the FERM domain. Ma et al. (32) recently reported similar results for kindlin-2-β3 integrin interactions. Kindlins also differ from talin in their binding site on β tails. Talin binds to the first of two conserved NPXY motifs in integrin β tails, whereas kindlins bind to the second. We cannot explain why our results differ from our previously reported data (29) pointing to a membrane-proximal NPXY interaction, but note that in that report, as in a recent kindlin-3 report (31), Tyr-Tyr → Ala-Ala mutations were more potent inhibitors of integrin β tail binding than single membrane-proximal Tyr → Ala mutations. The talin F3 subdomain adopts a phosphotyrosine binding-like fold and binds integrin via a variant of the canonical phosphotyrosine binding domain-NPXY motif interaction (16). This interaction also requires a Trp residue upstream of the NPXY motif (12, 16). Despite binding to a different site, the mode of interaction is predicted to be similar for kindlins (21, 29, 30) and requires Thr residues upstream of the NPXY motif. Analysis of kindlin-1 and -2 mutants shows the importance of conserved F3 residues in the interaction; however, there are subtle differences in the importance of specific residues between kindlin-1 and -2 and talin. A detailed explanation of the reasons for these differences will require structural analyses of the interactions.
Integrin activation is central to control of cell adhesion, migration, and signaling and is regulated by the association of intracellular proteins with the cytoplasmic tails of integrin β subunits (1). The binding of the cytoskeletal protein talin to integrin β tails is a key step in integrin activation (12, 15). However, other integrin-binding proteins are likely to cooperate or compete with talin to modulate integrin activation and possibly to provide integrin specificity to activation signals (10, 20, 38). The results described here, along with recent published reports (30-33), indicate that kindlins are important regulators of integrin activation, but the mechanisms by which they exert their effects will require further study.
Kindlin-2 or -3 knock-out results in early embryonic and early post-natal lethality, respectively, and kindlin-2 or -3-deficient cells exhibit a variety of defects including impaired activation of β1 and β3 integrins (31, 33, 48). This indicates that kindlin-2 or -3 are required for integrin activation. The effect of kindlin-2 or -3 overexpression is more variable and apparently somewhat cell type-specific. Kindlin-3 expression enhances β1 activation in RAW264.7 cells but not CHO cells (31), whereas expression of kindlin-2 in αIIbβ3-expressing CHO cells has been reported to both weakly activate (30, 32) and slightly inhibit (33) αIIbβ3 activation. Recent kindlin-1 knock-out experiments in mice have reported skin defects and lethal neonatal intestinal epithelial dysfunction as a consequence of impaired integrin activation (44). In our experiments using two independent reporters of activation for αIIbβ3 and endogenous CHO α5β1, we see strong inhibition of β1 and β3 integrins following expression of either kindlin-1 or kindlin-2. Inhibition is not due to a loss of cell surface integrin because activation indices were normalized to account for changes in integrin levels. Furthermore kindlin-1 expression increased α5β1 expression levels while reducing binding of the α5β1 ligand FN9-11. Effects on β3 integrin levels were not statistically significant but tended in the same direction as those seen for β1 integrins. The in vivo relevance of kindlin effects on integrin levels remains unclear. The inhibitory effect of overexpressed kindlins is also unlikely to be due to competition with talin for binding to integrin β tails because nonbinding kindlin mutants retain inhibitory activity, and the separation between talin- and kindlin-binding sites on the β tails suggests that both proteins may be able to bind simultaneously (32). A clear molecular explanation for the inhibitory effect of overexpressed kindlin-1 and -2 is still lacking but, when considered in light of the kindlin-1, -2, and -3 knock-out results, the data suggest that either removal or overexpression can inhibit activation. Similarly, both knockdown and overexpression of kindlin-2 can inhibit cell migration (30, 32). Such a pattern is consistent with an adaptor or scaffolding role (49) where kindlins link integrin β tails to another kindlin-binding integrin-activating protein and overexpressed kindlin would uncouple that adapter function. The inhibitory effect of overexpressed integrin binding-defective kindlin may also be explained by this model because these mutants would be expected to produce dominant-negative effects. Likewise differences in levels of kindlin-binding proteins may explain the cell type-specific effects of kindlins.
Further work will be required to identify putative kindlin-binding integrin-activating proteins, but consistent with recent reports for kindlin-2 (32, 33), we find that co-expression of kindlin-1 or -2 with the talin head enhances talin-mediated αIIbβ3 integrin activation. This effect is dependent on an intact integrin-binding site in the kindlin F3 domain, and nonintegrin-binding mutants strongly inhibit the talin-mediated integrin activation. A talin-kindlin interaction could therefore play a role in β3 integrin activation; however, preliminary experiments have not yet revealed such an interaction.
We previously found differences in talin-mediated activation of β1 and β3 integrins (10) and now show that even in the presence of activating levels of the talin head, kindlin-1 and -2 continue to strongly suppress α5β1 activation. This supports differences in β1 and β3 activation mechanisms, and it will be important to resolve the basis for these differences.
In summary, we show that kindlin-1, in addition to kindlin-2 and -3, is a regulator of integrin activation. However, effects on activation depend on kindlin expression level and levels of active talin and show integrin β subunit specificity. The details of how kindlins exert their effects on activation remain to be resolved, but we favor a model where kindlins act as adaptor proteins and increasing or reducing expression levels can hence uncouple that activity, resulting in inhibition of activation unless other relevant partners are co-expressed. Whether kindlin-containing complexes stabilize talin binding to the integrin tail, stabilize the integrin in an active conformation, or prevent talin competitors, such as filamin, from binding remains to be determined, and the role of known kindlin-binding proteins, such as migfilin (26) or integrin-linked kinase (33), now also needs to be assessed.
Supplementary Material
Acknowledgments
We thank Dr. Mary Beckerle (Huntsman Cancer Institute, University of Utah, Salt Lake City, UT) for generously providing kindlin cDNAs and anti-kindlin-1 antibody and Drs. Horst Kessler and Dominik Heckmann (Department of Chemistry, Technical University München, Garching, Germany) for providing the α5β1-specific inhibitor 3F. Lastly, we thank Tony Koleske, Yatish Lad, and members of the Calderwood lab for helpful discussions and comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1 GM068600 and R21 HL089433. This work was also supported by a National Science Foundation Graduate Research Fellowship Award (to D. S. H.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
Footnotes
The abbreviations used are: GST, glutathione S-transferase; CHO, Chinese hamster ovary; GFP, green fluorescent protein; FACS, fluorescence-activated cell sorter; MFI, mean fluorescence intensity.
D. S. Harburger, M. Bouaouina, and D. A. Calderwood, unpublished data.
References
- 1.Calderwood, D. A. (2004) J. Cell Sci. 117 657-666 [DOI] [PubMed] [Google Scholar]
- 2.Hynes, R. O. (2002) Cell 110 673-687 [DOI] [PubMed] [Google Scholar]
- 3.Luo, B., Carman, C., and Springer, T. (2007) Annu. Rev. Immunol. 25 619-647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harburger, D., and Calderwood, D. (2009) J. Cell Sci. 122 159-163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zaidel-Bar, R., Itzkovitz, S., Ma'ayan, A., Iyengar, R., and Geiger, B. (2007) Nat. Cell Biol. 9 858-867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu, S., Calderwood, D. A., and Ginsberg, M. H. (2000) J. Cell Sci. 113 3563-3571 [DOI] [PubMed] [Google Scholar]
- 7.Evans, E., and Calderwood, D. (2007) Science 316 1148-1153 [DOI] [PubMed] [Google Scholar]
- 8.Ginsberg, M., Partridge, A., and Shattil, S. (2005) Curr. Opin. Cell Biol. 17 509-516 [DOI] [PubMed] [Google Scholar]
- 9.Critchley, D., and Gingras, A. (2008) J. Cell Sci. 121 1345-1347 [DOI] [PubMed] [Google Scholar]
- 10.Bouaouina, M., Lad, Y., and Calderwood, D. A. (2008) J. Biol. Chem. 283 6118-6125 [DOI] [PubMed] [Google Scholar]
- 11.Calderwood, D. A., Zent, R., Grant, R., Rees, D. J., Hynes, R. O., and Ginsberg, M. H. (1999) J. Biol. Chem. 274 28071-28074 [DOI] [PubMed] [Google Scholar]
- 12.Tadokoro, S., Shattil, S. J., Eto, K., Tai, V., Liddington, R. C., de Pereda, J. M., Ginsberg, M. H., and Calderwood, D. A. (2003) Science 302 103-106 [DOI] [PubMed] [Google Scholar]
- 13.Nieswandt, B., Moser, M., Pleines, I., Varga-Szabo, D., Monkley, S., Critchley, D., and Fässler, R. (2007) J. Exp. Med. 204 3113-3118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petrich, B., Fogelstrand, P., Partridge, A., Yousefi, N., Ablooglu, A., Shattil, S., and Ginsberg, M. (2007) J. Clin. Investig. 117 2250-2259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wegener, K. L., Partridge, A. W., Han, J., Pickford, A. R., Liddington, R. C., Ginsberg, M. H., and Campbell, I. D. (2007) Cell 128 171-182 [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Alvarez, B., de Pereda, J. M., Calderwood, D. A., Ulmer, T. S., Critchley, D., Campbell, I. D., Ginsberg, M. H., and Liddington, R. C. (2003) Mol. Cell 11 49-58 [DOI] [PubMed] [Google Scholar]
- 17.Calderwood, D. A., Yan, B., de Pereda, J. M., Alvarez, B. G., Fujioka, Y., Liddington, R. C., and Ginsberg, M. H. (2002) J. Biol. Chem. 277 21749-21758 [DOI] [PubMed] [Google Scholar]
- 18.Petrich, B. G., Marchese, P., Ruggeri, Z. M., Spiess, S., Weichert, R. A., Ye, F., Tiedt, R., Skoda, R. C., Monkley, S. J., Critchley, D. R., and Ginsberg, M. H. (2007) J. Exp. Med. 204 3103-3111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Czuchra, A., Meyer, H., Legate, K. R., Brakebusch, C., and Fassler, R. (2006) J. Cell Biol. 174 889-899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma, Y. Q., Qin, J., and Plow, E. F. (2007) J. Thromb. Haemostasis 5 1345-1352 [DOI] [PubMed] [Google Scholar]
- 21.Larjava, H., Plow, E. F., and Wu, C. (2008) EMBO Rep. 9 1203-1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams, B. D., and Waterston, R. H. (1994) J. Cell Biol. 124 475-490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mackinnon, A. C., Qadota, H., Norman, K. R., Moerman, D. G., and Williams, B. D. (2002) Curr. Biol. 12 787-797 [DOI] [PubMed] [Google Scholar]
- 24.Rogalski, T. M., Mullen, G. P., Gilbert, M. M., Williams, B. D., and Moerman, D. G. (2000) J. Cell Biol. 150 253-264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weinstein, E. J., Bourner, M., Head, R., Zakeri, H., Bauer, C., and Mazzarella, R. (2003) Biochim. Biophys. Acta 1637 207-216 [DOI] [PubMed] [Google Scholar]
- 26.Tu, Y., Wu, S., Shi, X., Chen, K., and Wu, C. (2003) Cell 113 37-47 [DOI] [PubMed] [Google Scholar]
- 27.Boyd, R. S., Adam, P. J., Patel, S., Loader, J. A., Berry, J., Redpath, N. T., Poyser, H. R., Fletcher, G. C., Burgess, N. A., Stamps, A. C., Hudson, L., Smith, P., Griffiths, M., Willis, T. G., Karran, E. L., Oscier, D. G., Catovsky, D., Terrett, J. A., and Dyer, M. J. (2003) Leukemia 17 1605-1612 [DOI] [PubMed] [Google Scholar]
- 28.Ussar, S., Wang, H. V., Linder, S., Fassler, R., and Moser, M. (2006) Exp. Cell Res. 312 3142-3151 [DOI] [PubMed] [Google Scholar]
- 29.Kloeker, S., Major, M. B., Calderwood, D. A., Ginsberg, M. H., Jones, D. A., and Beckerle, M. C. (2004) J. Biol. Chem. 279 6824-6833 [DOI] [PubMed] [Google Scholar]
- 30.Shi, X., Ma, Y. Q., Tu, Y., Chen, K., Wu, S., Fukuda, K., Qin, J., Plow, E. F., and Wu, C. (2007) J. Biol. Chem. 282 20455-20466 [DOI] [PubMed] [Google Scholar]
- 31.Moser, M., Nieswandt, B., Ussar, S., Pozgajova, M., and Fassler, R. (2008) Nat. Med. 14 325-330 [DOI] [PubMed] [Google Scholar]
- 32.Ma, Y. Q., Qin, J., Wu, C., and Plow, E. F. (2008) J. Cell Biol. 181 439-446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Montanez, E., Ussar, S., Schifferer, M., Bosl, M., Zent, R., Moser, M., and Fassler, R. (2008) Genes Dev. 22 1325-1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Toole, T. E., Katagiri, Y., Faull, R. J., Peter, K., Tamura, R., Quaranta, V., Loftus, J. C., Shattil, S. J., and Ginsberg, M. H. (1994) J. Cell Biol. 124 1047-1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heckmann, D., Meyer, A., Marinelli, L., Zahn, G., Stragies, R., and Kessler, H. (2007) Angew Chem. Int. Ed Engl. 46 3571-3574 [DOI] [PubMed] [Google Scholar]
- 36.Calderwood, D. A., Tai, V., Di Paolo, G., De Camilli, P., and Ginsberg, M. H. (2004) J. Biol. Chem. 279 28889-28895 [DOI] [PubMed] [Google Scholar]
- 37.Lad, Y., Harburger, D. S., and Calderwood, D. A. (2007) Methods Enzymol. 426 69-84 [DOI] [PubMed] [Google Scholar]
- 38.Kiema, T., Lad, Y., Jiang, P., Oxley, C. L., Baldassarre, M., Wegener, K. L., Campbell, I. D., Ylanne, J., and Calderwood, D. A. (2006) Mol. Cell 21 337-347 [DOI] [PubMed] [Google Scholar]
- 39.Lad, Y., Jiang, P., Ruskamo, S., Harburger, D. S., Ylanne, J., Campbell, I. D., and Calderwood, D. A. (2008) J. Biol. Chem. [DOI] [PMC free article] [PubMed]
- 40.Brown, P. J., and Juliano, R. L. (1985) Science 228 1448-1451 [DOI] [PubMed] [Google Scholar]
- 41.O'Toole, T. E., Ylanne, J., and Culley, B. M. (1995) J. Biol. Chem. 270 8553-8558 [DOI] [PubMed] [Google Scholar]
- 42.Kolli, S., Zito, C. I., Mossink, M. H., Wiemer, E. A., and Bennett, A. M. (2004) J. Biol. Chem. 279 29374-29385 [DOI] [PubMed] [Google Scholar]
- 43.Brakebusch, C., and Fassler, R. (2005) Cancer Metastasis Rev. 24 403-411 [DOI] [PubMed] [Google Scholar]
- 44.Ussar, S., Moser, M., Widmaier, M., Rognoni, E., Harrer, C., Genzel-Boroviczeny, O., and Fässler, R. (2008) PLoS Genet. 4 e1000289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hughes, P. E., Oertli, B., Hansen, M., Chou, F. L., Willumsen, B. M., and Ginsberg, M. H. (2002) Mol. Biol. Cell 13 2256-2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan, B., Calderwood, D. A., Yaspan, B., and Ginsberg, M. H. (2001) J. Biol. Chem. 276 28164-28170 [DOI] [PubMed] [Google Scholar]
- 47.Goksoy, E., Ma, Y. Q., Wang, X., Kong, X., Perera, D., Plow, E. F., and Qin, J. (2008) Mol. Cell 31 124-133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dowling, J. J., Gibbs, E., Russell, M., Goldman, D., Minarcik, J., Golden, J. A., and Feldman, E. L. (2008) Circ. Res. 102 423-431 [DOI] [PubMed] [Google Scholar]
- 49.Levchenko, A., Bruck, J., and Sternberg, P. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 5818-5823 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.