Abstract
Plants can metabolize sulfate by two pathways, which branch at the level of adenosine 5′-phosphosulfate (APS). APS can be reduced to sulfide and incorporated into Cys in the primary sulfate assimilation pathway or phosphorylated by APS kinase to 3′-phosphoadenosine 5′-phosphosulfate, which is the activated sulfate form for sulfation reactions. To assess to what extent APS kinase regulates accumulation of sulfated compounds, we analyzed the corresponding gene family in Arabidopsis thaliana. Analysis of T-DNA insertion knockout lines for each of the four isoforms did not reveal any phenotypical alterations. However, when all six combinations of double mutants were compared, the apk1 apk2 plants were significantly smaller than wild-type plants. The levels of glucosinolates, a major class of sulfated secondary metabolites, and the sulfated 12-hydroxyjasmonate were reduced approximately fivefold in apk1 apk2 plants. Although auxin levels were increased in the apk1 apk2 mutants, as is the case for most plants with compromised glucosinolate synthesis, typical high auxin phenotypes were not observed. The reduction in glucosinolates resulted in increased transcript levels for genes involved in glucosinolate biosynthesis and accumulation of desulfated precursors. It also led to great alterations in sulfur metabolism: the levels of sulfate and thiols increased in the apk1 apk2 plants. The data indicate that the APK1 and APK2 isoforms of APS kinase play a major role in the synthesis of secondary sulfated metabolites and are required for normal growth rates.
INTRODUCTION
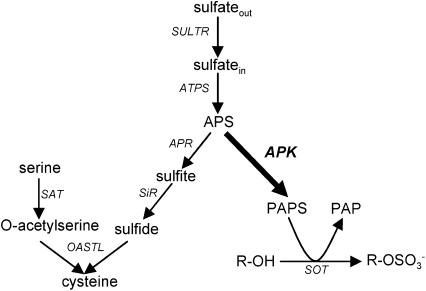
Sulfur is an essential nutrient for plant growth. Plants meet their demand for sulfur by assimilation of inorganic sulfate (Figure 1). Sulfate is taken up via sulfate transporters and incorporated into adenosine 5′-phosphosulfate (APS) by ATP sulfurylase (ATPS). APS can then be used in two ways: it can be sequentially reduced by APS reductase and sulfite reductase to sulfite and sulfide, and incorporated into O-acetylserine to form Cys (for review, see Kopriva, 2006). APS can also be phosphorylated to 3′-phosphoadenosine 5′-phosphosulfate (PAPS) by the action of APS kinase (APK). PAPS is the sulfate donor for the creation of sulfated secondary metabolites, which are formed by sulfotransferase (SOT) enzymes. The SOTs transfer the sulfate group from PAPS onto free hydroxyl groups of appropriate acceptor molecules. Sulfated plant metabolites include glucosinolates, phytosulfokines, and some hormones and flavonoids.
Figure 1.
Sulfate Assimilation Pathways in Plants.
Sulfate can be assimilated in a reduction pathway to Cys (left) or via PAPS to sulfated compounds (right). The enzymes are shown in italics; APK is shown in bold. SULTR, sulfate transporter; SiR, sulfite reductase; SAT, Ser acetyltransferase; OASTL, O-acetylserine (thiol) lyase; SOT, sulfotransferase; PAP, adenosine-3′,5′-bisphosfate; R, acceptor molecule.
The glucosinolates (GLs) are a large group of sulfur-rich amino acid–derived metabolites found mainly in the order Capparales (Fahey et al., 2001; Halkier and Gershenzon, 2006). Within the Capparales are the Brassicaceae, which contain many agriculturally important food crops, such as cabbage (Brassica oleracea var. capitata), broccoli (Brassica oleracea var. botrytis), cauliflower (Brassica oleracea var. botrytis), oilseed rape (Brassica napus), and the model plant Arabidopsis thaliana. GLs are an important form of defense against herbivore and insect attack since their breakdown products are both toxic and deterrent to the attacker (Halkier and Gershenzon, 2006). The GLs are also important nutritionally, as their breakdown products have been shown to have anticarcinogenic activity in humans (Mithen et al., 2003). They can, however, have a negative impact when present in high amounts in animal feed derived from rapeseed (Schöne et al., 1997). GL biosynthesis and its regulation have been well-studied in Arabidopsis (for reviews, see Grubb and Abel, 2006; Halkier and Gershenzon, 2006; Yan and Chen, 2007). The sulfation of the desulfo-glucosinolate (DS-GL) precursors is the final step of GLs synthesis (Underhill et al., 1973). In Arabidopsis, the group VII SOTs, SOT16, 17, and 18, are responsible for the sulfation of DS-GLs (Piotrowski et al., 2004; Klein et al., 2006). The sulfate group is necessary for formation of the biologically active products after GL degradation; indeed, this functional group is targeted by sulfatases produced by crucifer specialist insects to disarm the plant defense (Ratzka et al., 2002).
Other plant metabolites that depend on PAPS for sulfation include hormones such as 12-hydroxyjasmonate and 24-epibrassinolide. The sulfated derivatives of these have been detected in plant tissues, and in Arabidopsis, the corresponding sulfotransferases have been identified (Rouleau et al., 1999; Gidda et al., 2003). Sulfation of these hormones is believed to modulate their biological activity, but the precise roles of these sulfated derivatives have yet to be elucidated. Plants also produce at least two classes of small sulfated peptides (Matsubayashi and Sakagami, 1996; Amano et al., 2007). Phytosulfokines (PSKs) are sulfated pentapeptides that stimulate cell proliferation in culture after binding to a membrane-localized receptor (Yang et al., 2001; Matsubayashi et al., 2006). Disruption of this receptor leads to premature senescence of rosette leaves and gradual loss of the ability to form calluses (Matsubayashi et al., 2006). Another sulfated oligopeptide, named PLANT PEPTIDE CONTAINING SULFATED TYROSINE1 (PSY1), was recently isolated from Arabidopsis, and its receptor was identified (Amano et al., 2007). Plants that lost the ability to bind PSY1 due to disruption of three paralogous receptor kinases displayed a semidwarf phenotype again with a premature senescence. Both peptides require Tyr sulfation for biological activity (Matsubayashi et al., 1996; Amano et al., 2007).
Despite the wide range of sulfated compounds in plants, very little is known about the enzyme providing the activated sulfate donor PAPS, the APK. Previous studies have identified two functional APK isoforms in Arabidopsis (Lee and Leustek, 1998; Lillig et al., 2001). The completion of the Arabidopsis genome sequence has revealed two further predicted isoforms based on sequence homology to the original members (Arabidopsis Genome Initiative, 2000). Here, we investigate the localization and function of these four APK isoforms in Arabidopsis. We show that Arabidopsis has four functional isoforms of APK and that the APK1 and APK2 isoforms not only play a major role in the synthesis of sulfated metabolites but are important for normal vegetative development.
RESULTS
The Arabidopsis Genome Encodes Four Functional APKs
In Arabidopsis, two APK enzymes were previously shown to form PAPS from APS and ATP (Lee and Leustek, 1998; Lillig et al., 2001). In addition to these two isoforms, APK1 (At2g14750) and APK2 (At4g39940), the completed Arabidopsis genome sequence contains two further predicted isoforms, annotated APK3 (At3g03900) and APK4 (At5g67520) (Table 1). The APK isoforms are 62 to 72% identical on the amino acid level, with ∼46% identity to APK from Escherichia coli (see Supplemental Table 1 online). Web-based subcellular prediction programs (ChloroP and TargetP) (Nielsen et al., 1997; Emanuelsson et al., 2000) predict that APK3 is cytosolic and that APK1, -2, and -4 have putative N-terminal plastidic target peptides (Table 1). To determine whether these additional genes encode functional APK isoforms, the cDNAs for each isoform (minus their putative target peptides) were expressed in E. coli using the pET expression vector and the recombinant proteins purified using the His-Tag system. The previously established APK activity assay of Renosto et al. (1984) was used to test the activity of the four recombinant enzymes. All four of the proteins exhibited activity, converting APS to PAPS in the in vitro assay with similar kinetic properties (Table 1). Thus, Arabidopsis contains four functional APK isoforms.
Table 1.
Features of Arabidopsis APK Isoforms
| Isoform | AGI Code | TargetP Prediction/Score | ChloroP Predicted TP Length | MW with Target Peptide | MW without Target Peptide | Km[APS] | Vmax |
|---|---|---|---|---|---|---|---|
| APK1 | At2g14750 | Chloroplast 0.735 | 38 aa | 29.8 kD | 25.9 kD | 5.3 μM | 20.1 units mg protein−1 |
| APK2 | At4g39940 | Chloroplast 0.548 | 59 aa | 32.0 kD | 25.8 kD | 1.0 μM | 10.6 units mg protein−1 |
| APK3 | At3g03900 | Other 0.752 | – | n/a | 23.1 kD | 7.4 μM | 12.4 units mg protein−1 |
| APK4 | At5g67520 | Chloroplast 0.653 | 75 aa | 32.8 kD | 26.3 kD | 2.1 μM | 12.7 units mg protein−1 |
The subcellular localization of APK and the prediction of transite peptide cleavage sites were determined by the web-based program TargetP. The Km and Vmax values were calculated from enzyme activity data obtained with varied (0.5 to 10 μM) concentrations of APS using the standard Michaelis-Menten model. AGI, Arabidopsis Genome Initiative; TP, transite peptide; MW molecular weight; aa, amino acids.
APK Isoforms Localize to Different Subcellular Compartments
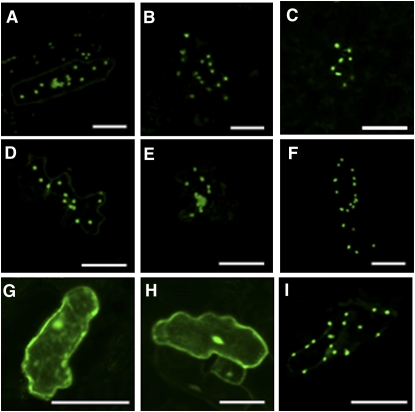
To confirm the predicted subcellular locations of APK1, -2, and -4 (plastid) and APK3 (cytosol), green fluorescent protein (GFP) was fused to the full-length coding sequence of each APK and to the N-terminal portions of APK1, -2, and -4. Transient expression of these fusions following particle bombardment of Arabidopsis leaves showed that APK3 is localized in the cytosol, while the remaining isoforms are localized in the chloroplast. Control constructs included GFP alone (cytosol) and GFP fused to the small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase (plastid) (Figure 2).
Figure 2.
Subcellular Localization of APK Isoforms.
The N terminus putative target peptide region or full-length coding region of APK genes were fused to GFP and transiently expressed in Arabidopsis epidermal cells under the control of the cauliflower mosaic virus 35S promoter. APK1 N terminus (A), APK2 N terminus (B), APK4 N terminus (C), APK1 full length (D), APK2 full length (E), APK4 full length (F), and APK3 full length (G). Control constructs for cytosolic and chloroplastic localization were GFP alone (H) and ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit (I). Bars = 50 μm.
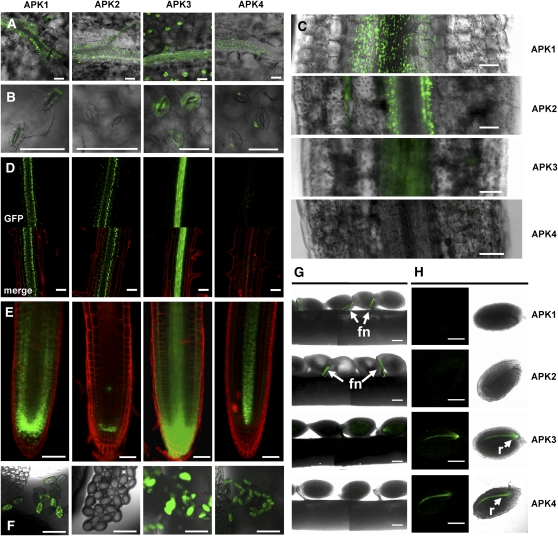
APK Isoforms Show Different Tissue-Specific Expression
To examine the tissue-specific expression of the four isoforms, APKpro:APK:GFP fusion constructs were used to stably transform Arabidopsis. GFP fluorescence was localized to the veins of the leaves of transgenic plants for all four isoforms (Figure 3A). With the exception of APK2, all isoforms showed expression in epidermal cells and in guard cells (Figure 3B). The plastid-specific localization of APK1, -2, and -4 can be seen in these images, confirming the results of the transient assays. In the hypocotyl, APK1, -2, and -3 all are expressed around the vasculature; however, APK4 does not appear to be expressed at this developmental stage (Figure 3C). In the hypocotyl, APK2 appears to be expressed tightly around the perimeter of the vasculature, whereas APK1 seems to be more diffuse but still concentrated around the vein. APK3 shows a similar pattern to APK2. In roots, again, APK1, -2, and -3 are expressed in and around the vasculature, and APK4 expression can be seen but is weak (Figure 3D). In the root tip, APK2 expression appears to be restricted to the quiescent center and APK4 to the vasculature. APK1 and -3 show more general expression in most cell types of the root tip (Figure 3E). APK2 is absent from pollen, but APK1, -3, and -4 are strongly expressed in pollen grains (Figure 3F). In developing seeds, APK1 and -2 appear to be strongly and specifically expressed in the funiculus (Figure 3G), whereas APK3 and -4 are expressed in the radicle of the immature seeds (Figure 3H). Clearly, the individual isoforms show very different expression patterns, indicating only partial redundancy in function.
Figure 3.
Tissue-Specific Expression of APK Isoforms.
DNA fragments encoding from 3000 bp upstream of ATG to the penultimate codon of the coding region of APK isoforms were fused to GFP, cloned into binary vector, and used to stably transform Arabidopsis. Images are as follows: rosette leaves (A), guard cells (B), hypocotyls (C), root (D), root tip (E), pollen grains (F), opened developing silique (G), immature seed (H), right panel, bright field; left panel, all signal apart of GFP filtered. Bars = 50 μm, except (G) and (H), where bars = 200 μm. In (D), GFP alone and merged images are shown. In (D) and (E), the red color is result of staining with propidium iodide to mark the cell walls. Expression in the funiculus (fn) and radicle (r) is indicated by arrows.
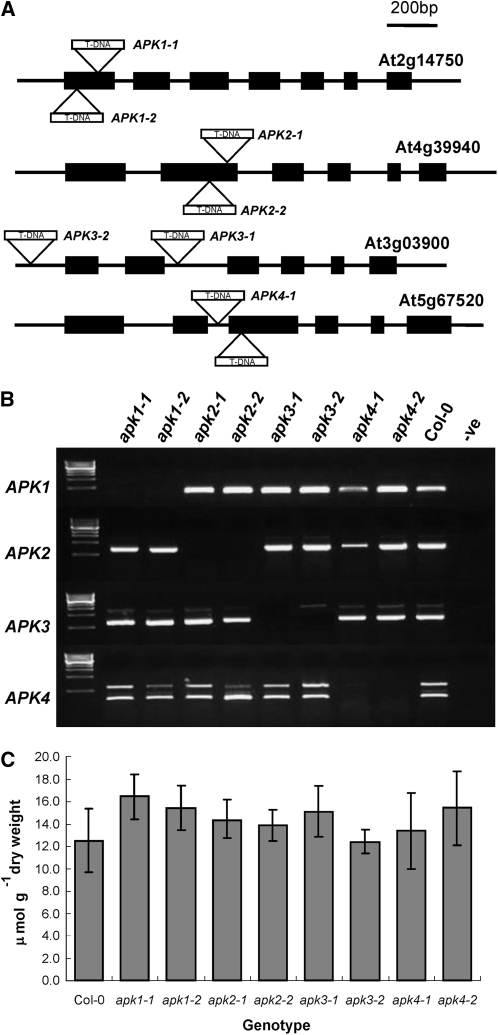
Single Knockouts of APK Do Not Show Visible Phenotypic Changes
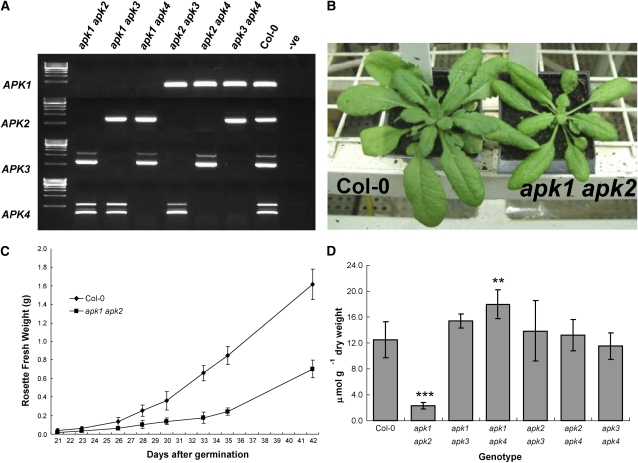
To investigate whether the individual Arabidopsis APK isoforms have specific functions, we used available reverse genetics resources. Two independent SALK T-DNA insertion alleles for each isoform in the Columbia-0 (Col-0) genetic background were obtained from the Nottingham Arabidopsis Stock Centre (NASC) (Figure 4A). Homozygous lines were isolated by PCR, and the positions of the insertions were confirmed (see Supplemental Figure 1 online). The loss of transcripts in the T-DNA lines was established by RT-PCR (Figure 4B). These null T-DNA insertion lines did not show any obvious phenotypic differences compared with Col-0. To determine whether disruption of the genes for individual APK isoforms has any effect on sulfated metabolites, we determined the total GLs levels in rosette leaves of the 5-week-old mutants grown in short days (Figure 4C). There were no significant effects on the levels of total sulfated GLs in any of the single knockout lines. The results thus revealed that none of the four APK isoforms on its own is essential for provision of sufficient PAPS for GL synthesis.
Figure 4.
SALK T-DNA insertions in APK Genes.
(A) The positions of two independent SALK T-DNA insertion alleles for each APK isoform. Insertion sites are indicated by triangles. Exons are indicated by black boxes and introns as thin lines. For exact insertion sites, see Supplemental Figure 1 online.
(B) Confirmation of knockout status of plants homozygous for the T-DNA insertion by RT-PCR. cDNA from knockouts was amplified with primers specific to coding regions of APK isoforms; first lane contains DNA marker, Col-0 is included as positive control, and water is used as negative control (-ve). Note that the presence of contaminating genomic DNA is seen in apk3-2; this is due to the insertion being in the promoter, thus allowing the amplification of a genomic fragment.
(C) Total GL contents in 5-week-old leaves of the single knockout plants. Data represents mean ± sd for five replicates.
Double Knockout of APK1 and APK2 Has a Semidwarf Phenotype and Contains Reduced Levels of GLs
As there is at least some functional redundancy in the APK family, it was necessary to obtain multiple knockout lines. Crosses were performed between single knockout plants to create all six double mutant combinations. The lines apk1-1, apk2-1, apk3-1, and apk4-1 were used as parents. Segregating F2 individuals were PCR genotyped at the parental loci; plants homozygous for the T-DNA insertion at both loci were analyzed by RT-PCR to confirm lack of the corresponding transcripts (Figure 5A). All six possible combinations were isolated. Remarkably, the apk1 apk2 mutant displayed a visible semidwarf phenotype (Figure 5B). The mutant plants were smaller than wild-type plants (∼70% reduction in fresh weight) over the whole growth period in short (10 h) days (Figure 5C); in long days, they initiated flowering ∼1 week later than the wild type. However, flowers appeared normal and seed set and germination were not obviously affected. Analysis of GLs in the rosette leaves of 5-week-old plants of the double insertion lines grown in short days revealed that the total GLs level in the apk1 apk2 mutant was reduced to 20% of wild-type levels (Figure 5D). GL levels in other double mutant combinations were not affected, with the exception of the apk1 apk4 mutant. Surprisingly, this double mutant contained 30% more total GLs (Figure 5D). The lower GL content in apk1 apk2 plants also was observed in 2-week-old seedlings grown in liquid culture (see Supplemental Figure 2 online). Thus, the decrease of the APK1/APK2-catalyzed synthesis of activated sulfate strongly affects the rate of vegetative development.
Figure 5.
Creation and Confirmation of Double Mutants of APK.
Pairs of single knockouts were used to create all six possible double knockout combinations.
(A) PCR amplification of cDNA from homozygous double knockout lines confirmed the lack of appropriate transcript in the mutants. First lane contains DNA marker, Col-0 is included as positive control, and water is used as negative control (-ve).
(B) Growth phenotype of the apk1 apk2 mutant (right) compared with the wild type at 5 weeks in short days in controlled environment room.
(C) Comparison of growth (based on rosette fresh weight) of apk1 apk2 double mutant plants with Col-0 during 6 weeks in short days. The data show the mean ± sd of five rosettes.
(D) Total GL content in 5-week-old rosette leaves of the double knockouts. The data are presented as the mean of five replicates ± sd. Student's t test significantly different to the wild type at ** = P < 0.01 and *** = P < 0.001.
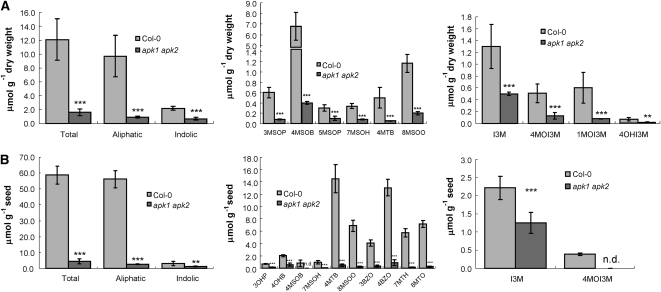
Nine different GLs were identified in mature leaves of the wild-type plants: six aliphatic and three indolic. Individual aliphatic GLs were reduced by between 4- and 15-fold in apk1 apk2, with the most abundant 4-methylsulfinylbutyl-GL being reduced most. Total aliphatic GLs were reduced to <11% of the level of the wild-type leaves (Figure 6A). The indolic GLs were less affected in the rosette leaves of the apk1 apk2 plants; the total indolic GLs content was reduced to 30% of that of control plants. Levels of individual modified indolic GLs were reduced 3- to 7-fold in the leaves of apk1 apk2 compared with wild-type leaves. However, it has to be noted that in other experiments a great variation of the content of indolic GLs was observed. In one out of four experiments, the indolic GLs in leaves of apk1 apk2 were reduced only by 10% because the indol-3-ylmethyl-GL (I3M) content actually increased by 40% (see Supplemental Figure 3 online).
Figure 6.
GL Measurements in the apk1 apk2 Double Knockout.
Comparison of levels of sulfated GLs in Col-0 and apk1 apk2 in rosette leaves (A) and seeds (B). Levels of total, aliphatic, and indolic GLs (left graph) are presented along with the levels of individual aliphatic (middle graph) and indolic GLs (right graph). 3MSOP, 3-methylsulfinylpropyl-GL; 4MSOB, 4-methylsulfinylbutyl-GL; 5MSOP, 5-methylsulfinylpentyl-GL; 7MSOH, 7-methylsulfinylheptyl-GL; 4MTB, 4-methylthiobutyl-GL; 8MSOO, 8-methylsulfinyloctyl-GL; 3OHP, 3-hydroxypropyl-GL; 4OHB, 4-hydroxybutyl-GL; 3BZO, 3-benzoyloxypropyl-GL; 4BZO, 4-benzoyloxybutyl-GL; 7MTH, 7-methylthioheptyl-GL; 8MTO, 8-methylthiooctyl-GL; 4OHI3M, 4-hydroxy-indolyl-3-methyl-GL; 1MOI3M, 1-methoxyindol-3-ylmethyl-GL. Data are presented as mean ± sd for five replicates (A) and eight replicates (B). Student's t test significantly different to the wild type at * P < 0.05, ** P < 0.01, and *** P < 0.001. n.d., not detected.
We also measured GLs in the mature seeds of wild-type and apk1 apk2 plants (Figure 6B). Twelve GLs were identified in the seeds. Total seed GLs in apk1 apk2 were present at 7% of the wild-type level. Individually, the seed GLs were reduced between 1.7-fold for I3M and 35-fold for several aliphatic ones. The 4-methylsulfinylbutyl-GL and 4-methoxyindol-3-methyl-GL (4MOI3M) were undetectable in seeds of apk1 apk2.
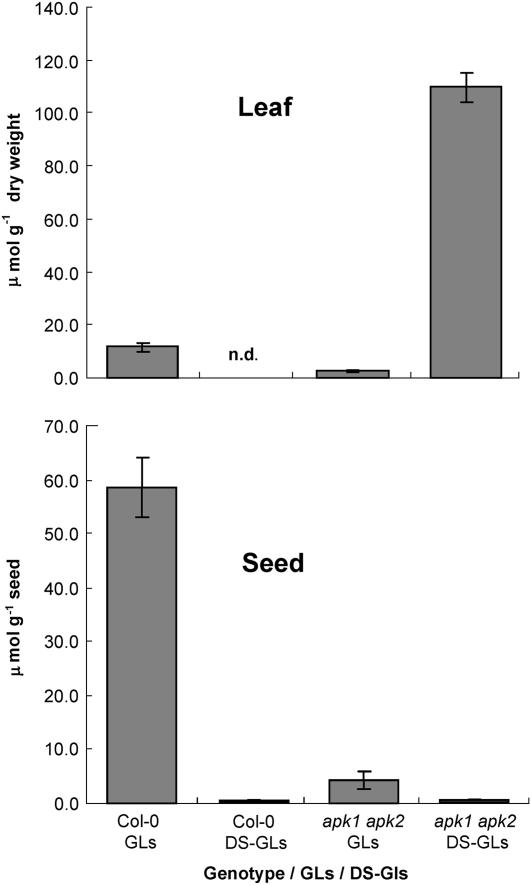
Desulfo-Precursors Accumulate in apk1 apk2 Leaves but Not in Seeds
Levels of DS-GLs, the immediate biosynthetic precursors of GLs, were determined in rosette leaves of 5-week-old apk1 apk2 and wild-type plants grown in short days and in mature seeds. DS-GLs were present at very low levels in leaves and were absent or below detection limit in seeds of wild-type plants (Figure 7). However, in the leaves of the apk1 apk2 mutant, DS-GLs accumulated to 11-fold higher levels than the sulfated GLs in the wild type (Figure 7). This was true for all the DS-GLs measured. In the seeds of the mutants, on the other hand, only very low levels of DS-GLs were detected (Figure 7).
Figure 7.
Levels of DS-GLs in Col-0 and apk1 apk2 Leaves and Seeds.
DS-GLs were quantified in rosette leaves of 5-week-old apk1 apk2 and wild-type plants grown in short days and seeds. Each data point represents mean ± sd for five replicates (leaves) and eight replicates (seeds).
The GL Biosynthetic Pathway Is Constitutively Upregulated in apk1 apk2
The increase in DS-GLs levels in the leaves of apk1 apk2 plants suggested that GL biosynthesis may be upregulated in these plants. We performed a microarray analysis to examine the transcript levels of the GL biosynthetic pathway and overall alterations of transcriptome in the leaves of apk1 apk2 mutant. The microarray analysis revealed that transcript levels of genes involved in GLs synthesis, such as the UGT74B1 (Grubb et al., 2004), CYP83B1 (Bak et al., 2001), SUR1 (Mikkelsen et al., 2004), SOT16, SOT17, and SOT18 (Piotrowski et al., 2004), were significantly increased in the apk1 apk2 plants compared with the wild type (Table 2). Most of these results were verified by quantitative real-time RT-PCR (qRT-PCR), confirming 2- to 18-fold higher mRNA levels in the apk1 apk2 mutants (Table 2; see Supplemental Figure 4 online). The qRT-PCR revealed that the genes encoding MYB factors involved in regulation of aliphatic and indolic GL synthesis—HAG1/MYB28, HAG3/MYB29, and HAG2/MYB76; and HIG1/MYB51, HIG2/MYB122, and HIG3/ATR1/MYB34 (Celenza et al., 2005; Gigolashvili et al., 2007a, 2007b; Hirai et al., 2007)—were highly induced in the mutant plants (Table 2; see Supplemental Figure 4 online). Thus, the reduction in PAPS supply and consequently the reduction in GL levels results in a strong upregulation of the GL biosynthetic pathway.
Table 2.
Fold-Changes of Transcript Levels of Genes Involved in GL Biosynthetis and Its Transcriptional Regulation in apk1 apk2 Plants
| Microarrays
|
qRT-PCR
|
||||||
|---|---|---|---|---|---|---|---|
| Gene | AGI | Description | Fold-Change | P | q | Fold-Change | P |
| APK1 | At2g14750 | APS kinase 1 | 0.0111 | 0.0000002 | 0.00006 | 0.0017 | 0.0004 |
| APK2 | At4g39970 | APS kinase 2 | 0.0294 | 0.00493 | 0.0805 | 0.0013 | 0.0002 |
| APK3 | At3g03900 | APS kinase 3 | 1.070 | 0.568 | 0.532 | 1.147 | 0.0751 |
| APK4 | At5g67520 | APS kinase 4 | 0.548 | 0.233 | 0.418 | 0.911 | 0.665 |
| MAM1 | At5g23010 | Methylthioalkylmalate synthase 1 | 3.13 | 0.000045 | 0.0058 | ||
| MAM3 | At5g23020 | Methylthioalkylmalate synthase 3 | 8.318 | 0.0000073 | 0.0018 | 18.089 | 0.0083 |
| ASA1 | At5g05730 | Anthranilate synthase α-subunit | 1.98 | 0.0170 | 0.142 | ||
| ASB1 | At1g25220 | Anthranilate synthase β-subunit | n/r | – | – | ||
| TSA1 | At4g02610 | Tryptophan synthase α-subunit 1 | 1.24 | 0.0157 | 0.137 | ||
| TSB1 | At5g54810 | Tryptophan synthase β-subunit 1 | n/r | – | – | 4.673 | 0.0073 |
| CYP79A2 | At5g05260 | CYTOCHROME P450 79A2 | n/e | – | – | ||
| CYP79F1/SUS1 | At1g16410 | CYTOCHROME P450 79F1, SUPERSHOOT1 | n/r | – | – | ||
| CYP79F2 | At1g16400 | CYTOCHROME P450 79F2 | n/r | – | – | ||
| CYP79B2 | At4g39950 | CYTOCHROME P450 79B2 | 8.524 | 0.000125 | 0.0121 | ||
| CYP79B3 | At2g22330 | CYTOCHROME P450 79B3 | 4.967 | 0.000268 | 0.0201 | ||
| CYP83A1 | At4g13770 | CYTOCHROME P450 83A1 | 2.313 | 0.000389 | 0.0236 | ||
| CYP83B1/SUR2 | At4g31500 | CYTOCHROME P450 83B1, SUPERROOT2 | 1.918 | 0.000362 | 0.0229 | 4.062 | 0.0083 |
| SUR1 | At2g20610 | C-S lyase, SUPERROOT1 | 2.076 | 0.000212 | 0.0169 | 4.303 | 0.0150 |
| UGT74B1 | At1g24100 | UDP-Glucosyltransferase 74B1 | 2.389 | 0.00246 | 0.0598 | ||
| UGT74C1 | At2g31790 | UDP-Glucosyltransferase 74C1 | 2.225 | 0.0000785 | 0.0132 | ||
| SOT16 | At1g74100 | Sulfotransferase, indolic GLs | 2.097 | 0.00489 | 0.0805 | 4.624 | 0.0082 |
| SOT17 | At1g18590 | Sulfotransferase, aliphatic GLs | 4.186 | 0.000146 | 0.0130 | 7.636 | 0.0075 |
| SOT18 | At1g74090 | Sulfotransferase, aliphatic GLs | 2.291 | 0.000278 | 0.0239 | 5.648 | 0.0120 |
| At1g65880 | Benzoate-CoA ligase | n/e | – | – | |||
| AOP1 | At4g03070 | 2-Oxoglutarate-dependant dioxygenase | 1.09 | 0.476 | 0.494 | ||
| AOP2 | At4g03060 | 2-Oxoglutarate-dependant dioxygenase | 3.15 | 0.000150 | 0.0132 | ||
| AOP3 | At4g03050 | 2-Oxoglutarate-dependant dioxygenase | n/e | – | – | ||
| HAG1/MYB28 | At5g61420 | High Aliphatic GL 1 R2R3-MYB transcription factor | 1.326 | 0.0185 | 0.146 | 2.094 | 0.0405 |
| HAG2/MYB76 | At5g07700 | High Aliphatic GL 2 R2R3-MYB transcription factor | 7.694 | 0.0000208 | 0.0365 | 9.324 | 0.009 |
| HAG3/MYB29 | At5g07690 | High Aliphatic GL 3 R2R3-MYB transcription factor | 2.370 | 0.0277 | 0.178 | 3.591 | 0.0096 |
| HIG1/MYB51 | At1g18570 | High Indolic GL 1 R2R3-MYB transcription factor | 1.325 | 0.060 | 0.246 | 2.000 | 0.0466 |
| HIG2/MYB122 | At1g74080 | High Indolic GL 2 R2R3-MYB transcription factor | n/e | – | – | 3.06 | 0.0352 |
| HIG3/MYB34/ ATR1 | At5g60890 | High Indolic GL 3 R2R3-MYB transcription factor | 15.796 | 0.116 | 0.312 | 3.205 | 0.0357 |
| OBP2 | At1g07640 | DOF transcription factor | 0.870 | 0.222 | 0.410 | ||
| IQD1 | At3g09710 | Calmodulin binding protein | 0.610 | 0.374 | 0.450 | ||
| FRY1/SAL1 | At5g63980 | Fiery1/Sal1, 3′(2′),5′-bisphosphate nucleotidase activity; detox of PAP | 2.044 | 0.000453 | 0.0256 | ||
n/e, not expressed; n/r, not represented on the ATH1 microarray. Transcripts highlighted in bold are significantly altered in the apk1 apk2 mutant compared with Col-0 samples at q < 0.05 or by qRT-PCR at P < 0.01.
The apk1 apk2 Double Mutant Contains Reduced Levels of Sulfated Jasmonate
To find out whether the limitation of PAPS synthesis in the apk1 apk2 double mutant specifically affects levels of GLs or whether it also has broader effects on other sulfated metabolites, we analyzed levels of another sulfated compound, the sulfated 12-hydroxyjasmonate (Gidda et al., 2003). The SOT catalyzing its synthesis (SOT15) exhibits strict substrate specificity in vitro for the hydroxylated jasmonates 11-OH-JA and 12-OH-JA. Since so far among the sulfated jasmonates only sulfated 12-OH-JA was found at remarkably abundant levels in different plant species (Miersch et al., 2008), we were specifically interested whether reduced levels of PAPS lead to an alteration in the level of sulfated 12-OH-JA (12-HSO4-JA). Therefore, levels of 12-HSO4-JA and those of related compounds, such as oxo-phytodienoic acid, jasmonic acid (JA), 11-OH-JA, 12-OH-JA, and 12-O-glucosyl-JA (12-O-Glc-JA), were determined in 5-week-old rosette leaves of the six double mutants and of apk1-1 and apk2-1 single insertion lines that had been crossed to generate the apk1 apk2 double mutant. Similarly to the GLs, levels of 12-HSO4-JA were reduced by 78% in the apk1 apk2 mutant compared with the wild type (Table 3), and there was a corresponding increase in the levels of the 11-OH-JA, 12-OH-JA, and 12-O-Glc-JA. Levels of JA itself were not affected; however, its precursor oxo-phytodienoic acid was decreased by 40%. In contrast with GLs, which were decreased only in the apk1 apk2 mutant, the content of 12-HSO4-JA was also reduced in other mutants, apk1-1, apk1 apk3, and apk1 apk4, however, only by 20 to 30% (Table 3). Surprisingly, in apk2 apk4 and apk3 apk4, the level of 12-HSO4-JA was higher than in the wild-type plants. The precursor of 12-HSO4-JA, 12-OH-JA, accumulated to slightly but significantly higher levels in apk1-1 and most of the double mutants except for apk1 apk3 and apk1 apk4.
Table 3.
Levels of JA and Its Derivatives in apk Mutants
| Genotype | OPDAa | JA | 11-OH-JA | 12-OH-JA | 12-HSO4-JA | 12-O-Glc-JA |
|---|---|---|---|---|---|---|
| Col-0 | 4451.3 ± 1072.8 | 86.8 ± 65.5 | 145.8 ± 62.9 | 180.0 ± 18.8 | 192.3 ± 31.5 | 89.8 ± 104.9 |
| apk1-1 | 5225.3 ± 680.9 | 51.8 ± 35.3 | 146.8 ± 11.0 | 246.8 ± 59.3 | 149.8 ± 24.9 | 106.5 ± 88.1 |
| apk2-1 | 6286.8 ± 658.2 * | 105.0 ± 32.4 | 161.8 ± 11.8 | 163.8 ± 24.8 | 172.5 ± 30.6 | 87.8 ± 65.9 |
| apk1 apk2 | 2639.3 ± 583.4 * | 84.8 ± 32.1 | 254.8 ± 64.3 * | 235.0 ± 13.4 ** | 42.0 ± 12.1 *** | 198.5 ± 27.0 * |
| apk1 apk3 | 5508.0 ± 359.2 | 76.8 ± 48.6 | 127.3 ± 47.3 | 199.3 ± 28.1 | 133.8 ± 31.4 * | 77.0 ± 98.2 |
| apk1 apk4 | 7684.5 ± 1199.4 ** | 94.5 ± 54.2 | 103.3 ± 20.3 | 219.8 ± 74.5 | 146.5 ± 41.5 * | 64.5 ± 74.6 |
| apk2 apk3 | 4929.8 ± 1196.8 | 163.8 ± 123.2 | 171.3 ± 24.6 | 298.3 ± 67.7 ** | 179.8 ± 81.1 | 56.8 ± 113.5 |
| apk2 apk4 | 5482.8 ± 783.4 | 117.8 ± 52.2 | 196.3 ± 38.0 | 312.8 ± 79.5 ** | 295.3 ± 34.6 ** | 110.0 ± 74.6 |
| apk3 apk4 | 5449.0 ± 361.2 | 123.0 ± 52.3 | 148.5 ± 32.2 | 239.0 ± 31.3 ** | 689.5 ± 400.0 * | 42.5 ± 60.1 |
Measurements are expressed as pmol g−1 fresh tissue and are the mean ± sd of four replicates. Asterisks mark values significantly different to the wild type at * P < 0.05, ** P < 0.01, and *** P < 0.001. OPDA, 12-oxo-phytodienoic acid; 11/12-OH-JA, 11/12-hydroxy-jasmonate; 12-HSO4-JA, 12-sulfo-jasmonate; 12-O-Glc-JA, 12-O-glucosyl-jasmonate.
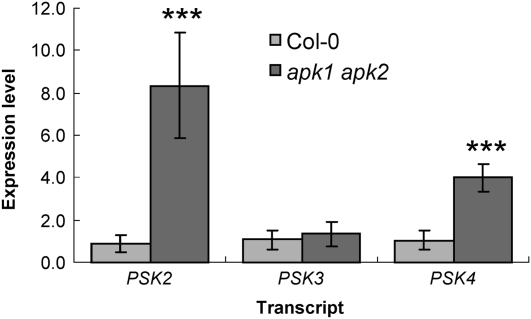
Expression Level of PSK Precursors Is Elevated in the apk1 apk2 Double Mutant
PSK genes encode precursors for the sulfated PSK pentapeptides, which are sulfated at both of their Tyr residues using PAPS as the sulfate donor. We hypothesized that if the levels of the sulfated peptides diminish, the transcript levels of their precursors would be upregulated. Therefore, mRNA level of three PSK precursor genes was examined in the apk1 apk2 double mutant by qRT-PCR (Figure 8) and microarray analysis (see Supplemental Table 2 online). Levels of PSK2 and PSK4 mRNA were approximately eightfold and fivefold increased, respectively, in the apk1 apk2 double mutants, while the other PSK genes were not affected.
Figure 8.
Expression of PSK Precursor Genes in the apk1 apk2 Double Mutant.
qRT-PCR was performed on cDNA synthesized from total RNA extracted from 5-week-old rosette leaves for the PSK precursor genes PSK2, PSK3, and PSK4. Data are presented as mean ± sd for four replicates. Asterisks mark values significantly different to the wild type at P < 0.001.
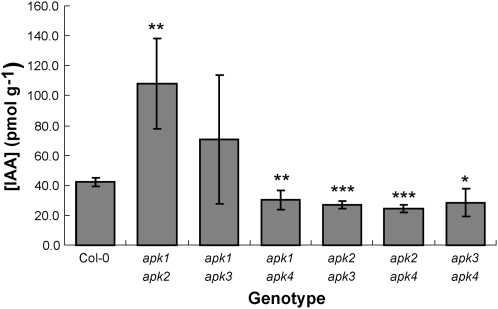
The apk1 apk2 Double Mutant Accumulates Auxin
Mutations in GL biosynthesis often result in accumulation of auxin, since the biosynthetic pathways of indolic GLs and indole-3-acetic acid (IAA) have a common intermediate, the indole acetaldoxime (Bak et al., 2001; Grubb and Abel, 2006). To test whether the decrease in GLs in the apk1 apk2 mutant affects auxin levels, we determined total IAA levels in rosette leaves of 5-week-old plants of all the double knockout lines. Again, only the apk1 apk2 mutant showed a substantial difference in IAA content, which was approximately threefold higher than in the wild type or other mutants (Figure 9). IAA levels in apk1 apk4, apk2 apk3, apk2 apk4, and apk3 apk4 were slightly but significantly lower than in wild-type plants. We compared the root morphology of apk1 apk2 and wild-type plants grown on vertical agar plates as well as their flower morphology, but despite the high IAA levels, the apk1 apk2 plants did not display any typical high-auxin phenotypic alterations.
Figure 9.
Levels of IAA in apk Double Mutants.
IAA content was determined from rosette leaves of 5-week-old Col-0 and all six double apk mutants. The data are presented as means ± sd from four biological replicates. Asterisks mark values significantly different to the wild type at * P < 0.05, ** P < 0.01, and *** P < 0.001.
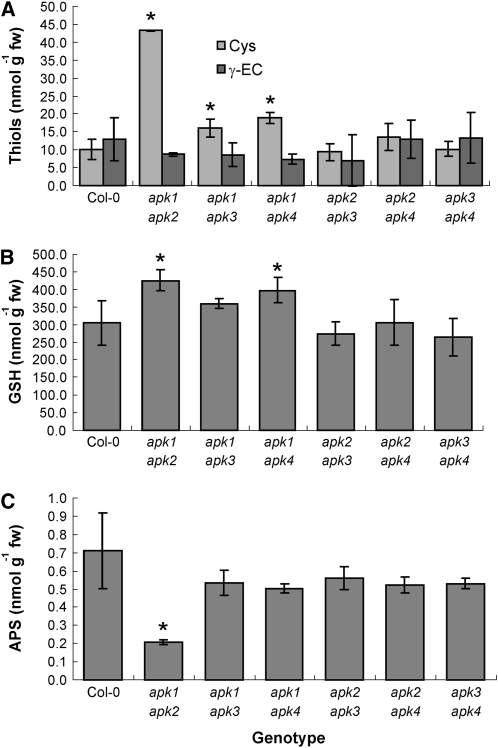
Disruption of APK Strongly Affects Primary Sulfate Assimilation
As APK clearly influences the accumulation of sulfated compounds, we tested whether the selective inactivation of APK1 and APK2 also affects primary sulfate assimilation. Therefore, the levels of thiols were determined in the rosette leaves of 5-week-old plants of all double knockout lines. All genotypes missing APK1 contained significantly higher Cys levels, with the highest one, in apk1 apk2 plants, fourfold increased compared with the wild type (Figure 10). The level of glutathione, which is an order of magnitude more abundant than Cys and γ-glutamylcysteine, was elevated in apk1 apk2 and apk1 apk4 plants. We were also interested whether the reduction in capacity to phosphorylate APS would affect its steady state foliar concentration. Again, the levels of APS were significantly reduced only in the apk1 apk2 plants (Figure 10C). As the thiols and APS in apk1 apk2 were most affected among the apk double mutants, we tested in these plants whether the disruption of APK1 and APK2 has consequences for other components of sulfur metabolism. Indeed, the apk1 apk2 plants accumulated sulfate and O-acetylserine in the leaves to approximately threefold and fourfold higher levels, respectively, than Col-0 (see Supplemental Figure 5 online).
Figure 10.
Levels of Thiols and APS in apk Double Mutants.
Cys and γ-glutamylcysteine (A) glutathione (B) and APS (C) content was determined in rosette leaves of 5-week-old Col-0 and all six double apk mutants. The data are presented as means ± sd from three to four biological replicates. Asterisks denote values significantly (P < 0.05) different from wild-type values.
The microarray analysis confirmed that the primary sulfate assimilation to Cys is disturbed by the disruption of APK1 and APK2. The transcript level of the low affinity sulfate transporter SULTR2;1, which is responsible for root-to-shoot sulfate transport, was increased in apk1 apk2 (see Supplemental Table 2 online). Similarly, the mRNA levels of ATPS1 and ATPS3 isoforms of ATP sulfurylase were significantly higher (see Supplemental Figure 4 and Supplemental Table 2 online). Indeed, the ATP sulfurylase enzyme activity was approximately twofold increased in leaves of apk1 apk2 plants compared with Col-0, while no changes were seen in APS reductase activity (see Supplemental Figure 5 online). Genes encoding components of Cys synthase complex, Ser acetyltransferase, and O-acetylserine (thiol)lyase or enzymes of glutathione biosynthesis were not dramatically affected (see Supplemental Table 2 online).
To identify which other metabolic pathways are affected by the reduction in PAPS synthesis, we determined levels of various metabolites not containing sulfur and explored the microarrays. Although Cys levels were highly increased in apk1 apk2 plants, the levels of other amino acids were not substantially affected (see Supplemental Figure 6 online). Phosphate but not nitrate significantly accumulated in the leaves of apk1 apk2 plants compared with the wild type (see Supplemental Figure 6 online). Other metabolites significantly different in apk1 apk2 plants compared with wild-type plants were ATP and ADP (see Supplemental Figure 6 online). The microarray analysis revealed that despite the semidwarf phenotype of the apk1 apk2 plants, only 176 genes were found to be differentially expressed (fold-change threshold twofold, q value threshold of 0.05). Out of these, 129 genes were more highly expressed in the leaves of apk1 apk2, while 47 were more highly expressed in the wild-type leaves. Of the upregulated genes, 13 genes are part of the GL network (Table 2) and six are involved in sulfur metabolism (see Supplemental Table 2 online), leaving 157 affected genes from other processes. Iterative group analysis using the Aracyc classification of metabolic pathways confirmed that genes of the GL biosynthesis pathway are overrepresented in the upregulated group of genes and revealed that the same is true for genes involved in IAA biosynthesis, Leu biosynthesis, NAD biosynthesis, cytokinin biosynthesis, glucoside biosynthesis, and JA biosynthesis (see Supplemental Table 3 online). The same analysis also revealed several metabolic pathways that are downregulated in the mutant (see Supplemental Table 4 online), namely, pathways for cellulose biosynthesis, cytokinine glucoside biosynthesis, and Gly biosynthesis and degradation.
DISCUSSION
Arabidopsis Has Four Functional APK Isoforms
APK is positioned at a key branch of APS conversion that is directed to sulfur assimilation into reduced compounds or to organic sulfates. To determine the contribution of APK to the regulation of accumulation of sulfated compounds, we investigated the function and subcellular and tissue-specific localization of the four Arabidopsis APK isoforms and the effect of disrupting the genes encoding them. We confirmed that all four genes encode functional enzymes that do not dramatically differ in their catalytical properties (Table 1), thus extending previous findings on APK1 and -2 (Lee and Leustek, 1998; Lillig et al., 2001). The localization of APK1, -2, and -4 in the plastid and APK3 in the cytosol (Figures 2 and 3) is in agreement with two sites of APS formation by ATPS, which is also present in plastids and in the cytosol (Rotte and Leustek, 2000). It also reveals that there are two pools of PAPS in the cell, one in the plastid and one in the cytosol. The SOT enzymes that use PAPS as the sulfate donor for the sulfation of acceptor molecules to generate sulfated secondary metabolites are, however, most probably cytosolic (Klein and Papenbrock, 2004), which is certainly the case for the DS-GLs sulfating SOT16, -17, and -18 (Piotrowski et al., 2004; Klein et al., 2006). Therefore, the function of the plastidic PAPS is not obvious, unless it is exported to the cytosol. This evidently must be the case since the apk3 mutants, which lack the only cytosolic APK isoform, do not show any reduction in sulfated metabolites. PAPS transporters have been identified in Drosophila melanogaster (Kamiyama et al., 2003; Lüders et al., 2003; Goda et al., 2006) and in mammals (Mandon et al., 1994). However, no PAPS transporter has been identified in plants so far.
Since the presence of four APK genes in Arabidopsis may be associated with the relatively high content of sulfated metabolites (i.e., the GLs in this species), we explored the gene family in other plant species. Rice (Oryza sativa) and poplar (Populus trichocarpa) have three APK isoforms, as does the lycophyte Selaginella moellendorffii, while the moss Physcomitrella patens has four and the green algae Chlamydomonas reinhardtii and Ostreococcus tauri contain one APK gene each (Kopriva et al., 2007). APK thus is encoded by small multigene families in most plants analyzed so far. There are, however, differences in the putative targeting of the derived proteins. While rice seems to encode only APK isoforms with a chloroplast targeting peptide, in P. patens, all APKs appear to be cytosolic (see Supplemental Figure 7 online). Interestingly, while rice and poplar genomes encode multiple SOT genes, no homologs to this class of sulfotransferases are present in P. patens or C. reinhardtii.
APK Expression Colocalizes with Sites of GL Synthesis
The localization of the APKs to the vasculature (Figure 3) corresponds to that of several enzymes of the GL biosynthesis pathway that have been shown to be closely associated with vascular tissues in a number of studies (Reintanz et al., 2001; Schuster et al., 2006). A recent report has shown that the distribution of GLs in Arabidopsis leaves is nonuniform (Shroff et al., 2008). Although GLs are present throughout the leaf, the three major GLs are more abundant in the tissues surrounding the main vein and the outer lamina (Shroff et al., 2008). This is believed to be due to a higher level being present at the parts of the leaf most vulnerable to damage by insects: the main vein, which needs to be protected from damage to prevent the disruption of the movement of nutrients and water through the leaf, and the edges of the leaf, which are most accessible to chewing insects. Myrosinase has been shown to be localized specifically in myrosin cells of the phloem parenchyma in Arabidopsis (Andréasson et al., 2001) and in guard cells (Andréasson et al., 2001; Thangstad et al., 2004). Koroleva et al. (2000) also demonstrated a high amount of GLs in specific S-cells in flower stalks associated with vasculature. Localization of PAPS synthesis at these sites would thus enable provision of the activated sulfate in situ without the need for long-distance transport. Interestingly, such specific generation and location of other compounds active in plant stress responses in vascular bundles (e.g., jasmonates and their metabolites) were found also in other plant species (Stenzel et al., 2003; Schilmiller and Howe, 2005).
APK1 and APK2 Are the Major APK Isoforms in Arabidopsis
To determine specific functions of the APK isoforms, we analyzed T-DNA insertion lines for all four genes. The lack of an obvious phenotype in the single mutants indicated that none of the APK isoforms are essential for normal growth and seed set. This is presumably due to at least partial functional redundancy within the family. The double mutants, however, revealed that this redundancy is limited. The apk1 apk2 double mutant showed a clear growth phenotype. The plants were smaller than wild-type plants and showed delayed but normal flower and seed development. As GLs are the major group of sulfated compounds in Arabidopsis, we determined their content in all the mutant plants. Indeed, levels of total sulfated GLs in the apk1 apk2 mutant were reduced 5-fold in mature leaves and 14-fold in the seeds. This indicates that APK1 and APK2 are the major APK isoforms that provide PAPS for synthesis of GLs and other sulfated metabolites. APK1 and -2 on their own are able to provide sufficient PAPS for GLs synthesis, as single knockout alleles of both isoforms contained normal or even slightly elevated levels of sulfated GLs.
These differences in double knockout plants may be caused by specific functions of the individual isoforms or by an overall reduction in the APK activity. Unfortunately, the enzyme activity assay is not sensitive enough to measure the activity in plant extracts. We raised antibodies against recombinant APK1 to assess APK protein accumulation in the mutant plants; however, the antiserum was specific to APK1 and did not cross-react with other recombinant APK isoforms. Also, the transcripts for the individual isoforms are too diverse (see Supplemental Table 1 online) to allow a total APK mRNA quantification. Thus, the only indication that the severity of phenotype in apk1 apk2 is due to a general decrease of PAPS synthesis capacity is the higher abundance of mRNA for APK1 and APK2 compared with APK3 and APK4 as judged from the available microarray experiments and qRT-PCR data.
Manipulation of APK Affects GL Biosynthesis
Many Arabidopsis mutants have been characterized with low levels of total or specific GLs (Bak et al., 2001; Reintanz et al., 2001; Grubb et al., 2004; Mikkelsen et al., 2004; Gigolashvili et al., 2007a, 2007b; Beekwilder et al., 2008). The underlying genes encoded enzymes of GL backbone biosynthesis or transcriptional factors and contributed significantly to the elucidation of the pathway and its regulation. We show here that manipulation of the last step in GL biosynthesis, the sulfation of DS-GLs, also affects GL levels. Disruption of APK1 and -2 leads to a reduced capacity to synthesize PAPS; thus, the activated sulfate is less available. This is similar to the situation during sulfur starvation, which also leads to reduction of GLs content (Falk et al., 2007). This is achieved through coordinated downregulation of several GL biosynthetic genes dependent on the SULFUR LIMITATION1 transcription factor (Maruyama-Nakashita et al., 2006). As the limiting step for GL synthesis in the apk1 apk2 mutant seems to be the provision of PAPS, we expected that the DS-GL precursors may accumulate. Indeed, in leaves of the apk1 apk2 mutant DS-GLs accumulated to very high levels. The content of DS-GLs in apk1 apk2 mutant was 11-fold higher than that of intact GLs in the wild type. The DS-GLs accumulation is much higher than would be expected from a simple reduction of concentration of the second substrate. This indicates that plants recognized the low level of GL products and increased the production of the desulfated precursors. This conclusion is supported by the coordinated upregulation of genes involved in GL biosynthesis. In addition, mRNA levels of several genes encoding transcription factors regulating GL biosynthesis were elevated in apk1 apk2 compared with the wild type. The increased levels of the biosynthetic genes thus may be caused by the induction of the MYB factors, which were expressed up to eightfold higher than in the wild type (Table 2; see Supplemental Figure 4 online). Overexpression of these transcription factors was shown to result in increased levels of mRNA for GL biosynthetic genes and accumulation of GLs (Gigolashvili et al., 2007a, 2007b, 2008). We can thus postulate a feedback regulation of GL biosynthesis by the end products. The mechanism of this regulation and the exact nature of the signal will be the subject of further study.
Surprisingly, in seeds of apk1 apk2 plants, the DS-GLs did not accumulate, suggesting that intact GLs are loaded into the developing seeds and not synthesized there, and the sulfate group is required for loading. This is in agreement with previous reports that concluded that GLs and not DS-GLs are the form that is transported in the phloem (Chen et al., 2001) and that fully formed GLs are loaded into the seeds via phloem from maternal tissue (Magrath and Mithen, 1993). In Sinapis alba, it has been shown that at least one GL destined for the seed tissue is synthesized in the silique wall and transported into the seed (Du and Halkier, 1998). The specific expression of APK1 and APK2 in the funiculus in Arabidopsis may indicate that DS-GLs are synthesized in siliques and sulfated in the funiculus during transport into the developing seeds.
The reduction in total GLs in the apk1 apk2 mutant is, however, not uniformly reflected in the levels of individual GLs. While all aliphatic GLs were reduced at least fivefold, the basic indolic GL I3M was reduced only by 62% in the experiment presented in Figure 6. Analysis of different harvests of these genotypes showed a great variation specifically in the content of the I3M, with one experiment out of four showing even an increase in its level in apk1 apk2 plants. In the remaining two measurements, I3M was reduced by 85 and 75%; however, in both cases, the reduction was less prominent than the reduction in aliphatic GLs. The great variation in indolic GLs content in apk1 apk2 is consistent with their rapid induction by herbivory and other stresses and low stability (Barth and Jander, 2006; Kim and Jander, 2007). Furthermore, recently it could be shown that indolic GLs are also required for the biosynthesis of some novel I3M- and 4MOI3M- derived compounds during plant defense against microbial pathogens and fungi (Bednarek et al., 2009; Clay et al., 2009). Thus, variations between experiments are probably due to small differences in growth conditions and sample handling and/or otherwise unobserved microbial or fungal leaf damage.
The analysis of individual GLs and DS-GLs revealed that the desulfo-precursors of I3M accumulated in leaves, but the DS-GLs corresponding to the modified indolic GLs were undetectable. This corroborates the hypothesis that the modifications of the indolic GLs occur after sulfation to form I3M (Kim and Jander, 2007) and contradicts the findings of Pedras and Montaut (2004), who hypothesized that the aldoxime moiety is modified early in biosynthesis (Pedras and Montaut, 2004).
The fact that I3M was sulfated in apk1 apk2 to higher degree than the aliphatic GLs may be explained by different PAPS pools provided by different APK isoforms for indolic and aliphatic DS-GLs sulfation. It has been shown previously that upon attack by the insect Myzus persicae, I3M is converted rapidly to 4MOI3M, which is less stable than I3M and more likely to break down spontaneously in the insect gut, as this pest inserts its stylet intracellularly, thus avoiding the myrosinase defense system (Kim and Jander, 2007). It is thus plausible to hypothesize that a hierarchy of individual GLs exists with the I3M being synthesized preferentially. When PAPS supply is not limiting, all GLs are made; if, however, it is limiting, such as in the apk1 apk2 mutant, the PAPS is preferentially used to synthesize I3M at the expense of other GLs. This would provide the plant even at a lower total GLs level with the most effective protection against a broad variety of bacterial diseases (Clay et al., 2009), fungal infections (Bednarek et al., 2009), and herbivores.
Disruption of APK1 and APK2 Affects Other Sulfated Compounds
As all four APK isoforms are functional and disruption of APK1 and -2 results in reduction of GLs, we were interested to find out whether levels of other sulfated compounds are also altered. Because not much is known about sulfated compounds in Arabidopsis apart from GLs, we turned to the only sulfated metabolite that can be reliably determined, the 12-HSO4-JA (Miersch et al., 2008). In the apk1 apk2 mutant, the level of 12-HSO4-JA was reduced to the same extent as the levels of GLs, indicating that the SOTs synthesizing this compound use the same PAPS pool (Table 3). The levels of 11-OH-JA and 12-OH-JA, which is the precursor of 12-HSO4-JA, were elevated in these plants but not to such high levels as DS-GLs. This is another indication that the synthesis of DS-GLs is indeed actively upregulated in the apk1 apk2 plants in contrast with simple accumulation of unused substrates. On the other hand, in apk1 apk2 mutant plants 12-O-Glc-JA was increased, showing that when the 12-OH-JA cannot be sulfated, alternative routes of its metabolism are enhanced. This corresponds to data on tomato (Solanum lycopersicum) and tobacco (Nicotiana tabacum), where 12-O-Glc-JA accumulates preferentially in wounded leaves but is shifted into 12-HSO4-JA by constitutive overexpression of the SOT (J. Neumerkel and C. Wasternack, personal communication).
However, in contrast with GLs, we observed significant alterations in 12-HSO4-JA levels also by other mutants in APK. In all genotypes lacking APK1, this compound was significantly reduced, which was mostly accompanied by increase of 12-OH-JA (Table 3). These results indicate either that APK1 is specifically associated with the SOT involved in jasmonate sulfation or that in single apk1 mutants the PAPS synthesis is compromised enough so that it is not sufficient for all sulfated compounds. GLs as major metabolites in defense against biotic stress might thus be synthesized as a priority and the other compounds, such as 12-HSO4-JA, only when enough PAPS is available. The PAPS levels thus might be slightly decreased in the apk1-1, apk1 apk3, and apk1 apk4 plants, where the 12-HSO4-JA is reduced. The explanation for the increase of 12-HSO4-JA in apk2 apk4 and apk3 apk 4 is much more difficult to find. Although we have not seen any major compensation of the disrupted isoforms by other APK genes on mRNA levels (the APK3 mRNA was increased only by ∼40% in the leaves of apk1 apk2 plants), it is not excluded that other levels of regulation exist. This would also explain the slightly increased GLs content in apk1 apk4 mutants.
Interconnection of GL Synthesis and Hormone Homeostasis
Many mutants in GL biosynthesis are also affected in auxin homeostasis (Delarue et al., 1998; Barlier et al., 2000; Bak et al., 2001; Reintanz et al., 2001; Grubb et al., 2004; Mikkelsen et al., 2004). Reduced GLs levels are often accompanied by high IAA concentration because indole-3-acetaldoxime, the precursor of indolic GLs, is also an intermediate in the biosynthesis of IAA (Mikkelsen et al., 2004). The apk1 apk2 plants also possessed significantly lowered GLs levels; however, they did not display any of the typical high auxin phenotypes: elongated hypocotyl, epinastic cotyledons, crinkled leaves, increased number of lateral roots, multiple adventitious roots, enhanced number of lateral shoots, altered flower morphology, or (semi)sterility. Therefore, we did not expect any alterations in auxin levels in these plants. Nevertheless, in 5-week-old rosette leaves of apk1 apk2, but no other double mutant, the levels of IAA were threefold higher than in wild-type leaves. In addition, the microarray analysis revealed that expressions of several genes involved in IAA synthesis were increased in apk1 apk2 leaves compared with the wild type. This shows that even though the GL level was modified by a completely different route, the effect on auxin still remained. The lack of high auxin phenotype, which was observed also in plants accumulating IAA due to overexpression of ATR1/MYB34 transcription regulator (Celenza et al., 2005), can be explained by different extent of IAA accumulation, developmental stage when IAA accumulates, or variation in levels of IAA conjugates found in the sur2 plants deficient in C-S lyase involved in GL synthesis (Barlier et al., 2000; Mikkelsen et al., 2004).
The apk1 apk2 plants, however, displayed a clear morphological phenotype, although different from the typical high auxin one. The plants were smaller throughout the whole vegetative growth phase and set flower later than wild-type plants. Reduction in aliphatic GLs and both indolic and aliphatic GLs in plants with reduced expression of HAG1/MYB28 and HIG1/MYB51 transcription factors had no effect on plant growth (Gigolashvili et al., 2007a, 2007b); therefore, GLs are unlikely to be responsible for this growth phenotype. The key thus might be another sulfated metabolite. Because the 12-HSO4-JA content also was reduced in apk1 apk2 plants, we might reasonably expect that the level of other sulfated molecules would be decreased concurrently and that some might be responsible for the slower growth. The decrease in 12-HSO4-JA itself is unlikely to cause the growth retardation as this metabolite also was reduced in other double mutants that grew normally. Possible candidates are thus the sulfated peptides PSK and PSY1, which have been associated with regulation of growth (Matsubayashi and Sakagami, 1996; Amano et al., 2007). Disruption of receptors for PSK and PSY1 leads to premature senescence of rosette leaves, gradual loss of the ability to form calluses, and a semidwarf phenotype (Matsubayashi et al., 2006; Amano et al., 2007). Although we have not observed premature senescence in apk1 apk2 plants, their smaller size resembles the effect of the lost ability to perceive PSY1 signals. We have not measured PSK or PSY1; however, this hypothesis is strongly supported by the observed increase in mRNA levels for PSK2 and PSK4 precursors in apk1 apk2 plants. As the decrease in GLs is accompanied by an increase in DS-GLs and lower levels of 12-HSO4-JA are accompanied by higher 12-OH-JA levels, it is highly probable that the increase in PSK precursors is linked to the decrease of the mature PSKs. The molecular basis of the growth phenotype of apk1 apk2 plants, as well as general effects of the reduction in APK on plant metabolism, however, have to be investigated in more detail.
Effects of APK Disruption on Primary Metabolism
The disruption of APK1 and APK2 resulted in a decrease in synthesis of sulfated compounds with further consequences for sulfur metabolism. The obvious expectation was that diminishing sulfur flux into PAPS would enable greater flux through the primary sulfate reduction. Indeed, steady state levels of Cys and glutathione were higher in leaves of apk1 apk2 plants than in wild-type leaves. However, the alterations in sulfur metabolism were far more complicated than this simple effect and included potential accumulation of upstream compounds as well as feedback signals from downstream metabolites. Despite the reduction in rate of its metabolization, APS content diminished in the apk1 apk2 plants. This was probably caused by the increase in APS reduction rate as demonstrated by the increase in thiol content. However, this was not seen on the level of APR activity, which was not affected in any of the APK double mutants. The increased thiol content was surprisingly accompanied by an increase in sulfate accumulation. Several genes encoding sulfate transporters and two isoforms of ATPS were induced in the apk1 apk2 plants, presumably in an attempt to increase the provision of PAPS. This agrees with a recent report showing an upregulation of sulfate assimilation due to overexpression of the MYB transcription factors HAG3/MYB29, HAG2/MYB76, HIG1/MYB51, and HIG3/ATR1/MYB34 (Malitsky et al., 2008). The increased expression of the sulfate transporter genes might be responsible for the increased sulfate content despite the higher synthesis of thiols. The increase in O-acetylserine in apk1 apk2 is intriguing, as this compound is considered to be involved in signaling of sulfur status of the plant (Koprivova et al., 2000, Hirai et al., 2003); however, the alterations of the pathway in apk1 apk2 plants are not consistent with the described effects of O-acetylserine on various components of primary sulfate assimilation. The analysis of the apk1 apk2 plants thus revealed an unexpectedly strong and complex interconnection between GL accumulation and primary sulfate metabolism.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana (ecotype Col) plants were used as the wild type in this study. Unless otherwise stated, all plants were grown in a controlled environment chamber under a short-day 10-h-light/14-h-dark cycle at constant temperature of 22°C, 60% relative humidity, and light intensity of 160 μmol m−2 s−1. A minimum of four whole rosettes were harvested at 5 weeks old and immediately frozen in liquid nitrogen. Tissue was then either freeze-dried (for GL and DS-GL analysis) or ground under liquid nitrogen in a pestle and mortar (for RNA extraction, JA, and IAA measurements). Plants for seeds were grown in a standard glasshouse. To compare root phenotypes of the apk1 apk2 mutants and wild-type plants, the seeds were plated on 0.8% agarose plates containing Murashige and Skoog medium supplemented with 3% sucrose and, after 4 d at 4°C in the dark, grown vertically in the controlled environment chamber.
Screening of T-DNA Insertion Mutants
Putative T-DNA insertion mutants of Arabidopsis were obtained from the NASC. All T-DNA lines were in the Col background. Two independent alleles were obtained for each isoform, namely, SALK_053427 (apk1-1), SALK_034586 (apk1-2), SALK_093072 (apk2-1), SALK_077590 (apk2-2), SALK_115182 (apk3-1), SALK_145507 (apk3-1), SALK_035815 (apk4-1), and SALK_068062 (apk4-2). Homozygous mutants were identified using primer LBb1 and an appropriate gene-specific primer. Primer sequences for the identification of homozygous T-DNA insertions are listed in Supplemental Table 5 online.
DNA Extraction and Amplification for Screening T-DNA Insertion Lines
Crude DNA preparations were prepared by homogenizing ∼50 mg young rosette leaves in a 1.5-mL eppendorf tube in 400 μL extraction buffer (200 mM Tris-HCl, pH 7.5, 250 mM NaCl, 25 mM EDTA, and 0.5% SDS). Samples were vortexed and spun at 13,000 rpm for 5 min. Supernatant (300 μL) was transferred to a fresh tube containing 300 μL isopropanol. Samples were mixed and spun at 13,000 rpm for 5 min. Pellets were washed in 70% ethanol, dried, and resuspended in 100 μL SDW. Crude extract (2.5 μL) was used for amplification in a 25-μL reaction by GoTaq Flexi DNA Polymerase (Promega).
RNA Extraction and Expression Analysis
To verify absence of APK transcripts in the T-DNA lines, total RNA was isolated from leaves by phenol:chloroform:isoamylalcohol (25:24:1) extraction and LiCl precipitation (Sambrook et al., 1989). Aliquots of 500 ng cDNA were reverse transcribed using SuperScriptII Reverse Transcriptase (Invitrogen) according to the manufacturer's instructions. PCR was done with GoTaqFlexi DNA Polymerase in 25 μL reaction volume with primers specific to individual isoforms and 35 cycles (see Supplemental Table 6 online). The qRT-PCR to verify the microarray results using the SYBR-Green master kit and a GeneAmp 5700 sequence detection system was performed exactly as described by Gigolashvili et al. (2007a). Relative quantification of expression levels was performed using the comparative Ct method, and the calculated relative expression values were normalized to the wild-type expression level (wild type = 1). Primer sequences for the qRT-PCR are listed in Supplemental Table 7 online.
Microarray Analysis
The RNA was extracted from 5-week old rosette leaves by phenol:chloroform:isoamylalcohol (25:24:1) extraction and LiCl precipitation, treated with DNaseI (Sigma-Aldrich), and repurified using the Qiagen RNeasy plant mini kit according to the manufacturer's instructions. The labeling, hybridization, and detection using three biological replicates of both wild-type and apk1 apk2 plants and ATH1 chip was performed by the NASC's International Affymetrix Service. Only genes flagged by the scanner software as present in all six samples were included in further analysis. The expression data were normalized according to the AtGenExpress recommendations using a global mean normalization excluding the top and bottom 2% of the data. Fold-changes in expression levels were calculated from means of the three biological replicates, and their statistical significance was tested by a t test on the log-transformed signal intensities. In addition, a false discovery rate control was performed according to Storey and Tibshirani (2003), setting the threshold q value at 0.05. Iterative group analysis was employed to identify categories of genes that are overrepresented among the genes differing significantly in expression levels between the two genotypes (Breitling et al., 2004). The Aracyc metabolic pathways were downloaded from the Plant Metabolic Network (http://www.plantcyc.org/). The microarray data are accessible through the NASCArrays service (http://affymetrix.Arabidopsis.info/narrays/experimentbrowse.pl) under accession number NASCARRAYS-457.
Heterologous Expression of Recombinant APK in Escherichia coli
Coding regions of APK isoforms were amplified from cDNA without the putative target peptides and cloned into pET 14b expression vector (Novagen). Primer sequences can be found in Supplemental Table 8 online. Constructs were transformed into BL21(DE3) cells (Novagen), and 50-mL cultures were grown overnight at 28°C in Luria-Bertani containing 50 μg mL−1 ampicillin. Cells were disrupted by sonication and proteins purified using the His-Bind purification kit (Novagen) according to the manufacturer's instructions. Purified protein was quantified with the Bio-Rad protein assay solution (Bio-Rad) and used in activity assays.
Enzyme Assays
APK activity with recombinant proteins was measured using the coupled spectrophotometric assay of Renosto et al. (1984), using ∼5 μg of purified APK per assay. For kinetic characterization of the individual APK isoforms, the assays were performed with varied (0.5 to 10 μM) concentration of APS, and the Km and Vmax values were calculated using the standard Michaelis-Menten model. However, the assay was not sensitive enough to analyze plant extracts. APR activity was determined in leaf extracts as the production of [35S]sulfite, assayed as acid volatile radioactivity formed in the presence of [35S]APS and dithioerythritol as reductant (Koprivova et al., 2000). ATPS was measured in the same extracts as the APS and pyrophosphate-dependent formation of ATP (Cumming et al., 2007). The protein concentrations were determined using the Bio-Rad protein assay solution with BSA as a standard.
Expression of 35Spro:APK:GFP in Arabidopsis
Molecular biological experiments were performed according to the standard protocols (Sambrook et al., 1989). The chimeric gene constructs of 35Spro:APK:GFP were created as follows. Primer sequences can be found in Supplemental Table 9 online. The partial or full-length coding sequences of APK1, APK2, APK3, and APK4 were amplified by PCR from first-strand cDNA. Fragments were cloned into the pTH2 vector (Chiu et al., 1996) and were fully sequenced to confirm their identity. The resulting plasmids were coated onto 1-μm gold particles (Bio-Rad) that were then used to bombard rosette leaves of Arabidopsis ecotype Col plants using a Helios gene gun (Bio-Rad).
Creation of Transgenic Plants Expressing APKpro:APK:GFP
The chimeric gene constructs were created as follows. Oligonucleotide primers (see Supplemental Table 10 online) were designed to amplify genomic DNA fragments from 5′-promoter region 3000 bp upstream of the translational initiation site and terminated just before translational stop site of APK1, APK2, APK3, and APK4. PCR was performed on genomic DNA prepared from Arabidopsis ecotype Col-0 using KOD plus DNA polymerase (Toyobo). The resultant PCR-amplified fragments of APK1, APK2, APK3, and APK4 were cloned into pCR-BluntII-TOPO (Invitrogen) and fully sequenced. Oligonucleotide primers were designed to amplify the sGFP(S65T) coding sequence. PCR was performed on pTH2 plasmid using KOD plus DNA polymerase (Toyobo) and fully sequenced.
For the construction of the fusion gene construct of APK1pro:APK1:GFP, the SpeI-EcoRI fragment of APK1 and EcoRI-SacI fragment of sGFP(S65T) described above were ligated. The resultant SpeI-SacI fragment of the APK1pro:APK1:GFP fusion cassette was inserted between XbaI and SacI sites of the binary plasmid, pBI101 (Clontech), replacing the β-glucuronidase gene. For the construction of fusion gene construct of APK2pro:APK2:GFP, we first constructed a promoterless GFP binary vector, pBI-GFP. BamHI-EcoRI fragment of pTH2 (Chiu et al., 1996) containing the fusion gene cassette of sGFP(S65T) gene and the nopaline synthase terminator was placed into the position of β-glucuronidase gene and the nopaline synthase terminator in the binary plasmid, pBI101 (Clontech). The XhoI-XbaI fragment of APK2 was inserted between SalI and XbaI sites of the resulting pBI-GFP binary plasmid. For the construction of fusion gene construct of APK3pro:APK3:GFP, the SpeI-BamHI fragment of APK3 was inserted between XbaI and BamHI sites of the pBI-GFP binary plasmid. For the construction of fusion gene construct of APK4pro:APK4:GFP, the SalI-ended fragment of APK4 was cloned into the SalI site of the pBI-GFP binary plasmid.
The resulting binary plasmids were transformed to Agrobacterium tumefaciens GV3101 (pMP90) (Koncz and Schell, 1986) by the freeze-thaw method (Höfgen and Willmitzer, 1988). Arabidopsis ecotype Col plants were transformed according to the floral dip method (Clough and Bent, 1998). Transgenic plants were selected on GM agar media (Valvekens et al., 1988) containing 50 mg L−1 kanamycin sulfate. Kanamycin-resistant T2 progenies of eight independent lines were analyzed.
Microscopy and Imaging of GFP
Fluorescence of APK-GFP was observed under an ECLIPSE E600 microscope (Nikon) equipped with a Radiance 2000 AGR-3 confocal laser scanning system (Bio-Rad) and 530- to 560-nm band-pass filter. To stain cell walls in roots of transgenic plants, root tissues were submerged in 10 μg mL−1 solution of propidium iodide (Sigma-Aldrich) for 1 min.
Crossing of apk Mutants
Single mutants apk1-1 and apk2-1, apk1-1 and apk3-1, apk1-1 and apk4-1, apk2-1 and apk3-1, apk2-1 and apk4-1, and apk3-1 and apk4-1 were crossed to generate double mutants. Segregating F2 seeds were screened by PCR at one locus, and plants homozygous for the insertion at this locus were then screened at the second locus. Doubly homozygous individuals were obtained for all six possible mutant combinations.
GL Analysis of apk Mutants
GLs were extracted from 50 mg of crushed freeze-dried leaf material or from 20 mg of seeds. Quantification of intact GLs followed the protocol described by Burow et al. (2006). Briefly, samples were extracted in 80% methanol (v:v) containing the internal standard 4-hydroxybenzyl-GL. After centrifugation, the supernatants were loaded onto columns of DEAE-Sephadex. The columns were washed with 80% methanol (v:v) and water. Bound intact GLs were desulfated with sulfatase overnight. DS-GL extracts after elution of the columns were separated by reverse-phase HPLC and quantified by UV absorption at 229 nm relative to the internal standard using response factors.
Native DS-GLs in the plant tissues were extracted as described for intact GLs above. However, DS-GLs do not bind to DEAE-Sephadex. The flow-through from loading the extract onto DEAE-Sephadex columns and the following washing step with 80% methanol (v:v) were collected, combined, and analyzed by HPLC-UV as described above. DS-GLs were quantified by response curves generated from isolated pure DS-GL standards (Brown et al., 2003).
Identity of intact GLs and of DS-GLs in the plant extracts was confirmed by liquid chromatography–mass spectrometry on a Bruker Esquire 6000 ion trap mass spectrometer (Bruker Daltonics).
JA Analysis of apk Mutants
Jasmonates were quantified from 500 mg of fresh crushed leaf tissue. Extraction, HPLC purification, and quantification by gas chromatography–mass spectrometry and liquid chromatography-electrospray-selected reaction monitoring were performed as described by Miersch et al. (2008).
Isolation, Purification, and Quantification of IAA
Fresh plant material (∼500 mg) was homogenized in 10 mL methanol with appropriate amounts of (13C6)IAA (100 ng; Cambridge Isotope Laboratories) as internal standards. The homogenate was filtered and placed on a cartridge filled with DEAE-Sephadex A-25 (3 mL gel, acetate form, methanol). The column was washed with 3 mL methanol, with 3 mL of 0.1 M HOAc in methanol, and with 3 mL of 1 M acetic acid in methanol. Fractions eluted with 3 mL of 1.5 M and 9 mL of 3 M acetic acid in methanol were combined, evaporated, and separated by RP-18 HPLC.
The HPLC analyses were performed using an LC 1100 series Agilent system equipped with a Lichrospher 100 RP-18 column (250 × 4 mm, 5 μm; Merck). The chromatographic conditions were as follows: flow rate at 1 mL min−1; detection at 210 nm; solvent A, 0.2% (v/v) acetic acid in water; and solvent B, methanol. The gradient was from 40% B to 100% B in 25 min. The fraction corresponding to authentic IAA (10 to 11.5 min) was collected, concentrated in vacuo, methylated with ethereal diazomethane, and stored at −20°C prior to the gas chromatography–mass spectrometry analysis.
The quantitative IAA analyses were performed on a Polariz Q instrument (Thermo-Finnigan) using the following parameters: 100 eV, positive chemical ionization, 1.5 mL methane as ionization gas, ion source temperature 200°C, column: Rtx-MS (Restek) 15 m × 0.25 mm, 0.25 μm film thickness, cross-bond 5% diphenyl to 95% dimethyl polysiloxane, injection temperature: 220°C, interface temperature: 250°C; helium:1 mL min−1; splitless injection; column temperature program: 1 min 60°C, 20°C min−1 to 300°C, 5 min 300°C. The retention times of methyl esters were obtained at 7.92 min for the internal standard (13C6)IAA and for IAA. Fragments at m/z 136 (13C6)IAA-Me and at m/z 130 (IAA-Me) were used for quantification.
Determination of Other Metabolites
The metabolites were extracted from leaves of Arabidopsis plants according to Wirtz and Hell (2003). Thiols and amino acids, including O-acetylserine, were quantified after derivatization with monobromobimane (Calbiochem, EMD Chemicals) and AccQ-Tag reagent (Waters), respectively, and HPLC separation (Wirtz and Hell, 2003). Anions were separated by HPLC and quantified after 10-fold dilution in water according to Wirtz and Hell (2007). APS was derivatized with chloroacetaldehyde and separated by HPLC as described by Bürstenbinder et al. (2007) for adenosine compounds using the same HPLC system. The separation method for APS was developed according to Haink and Deussen (2003).
Sequence Analysis
Sequence analysis was performed with the Biology WorkBench web-based platform (http://workbench.sdsc.edu/) (Subramaniam, 1998). Sequence comparisons to calculate sequence identity were performed with the program ALIGN using a gap opening penalty of −16 and a gap extension penalty of −4.
The subcellular localization of APK was analyzed by the web-based program TargetP (Emanuelsson et al., 2000) for plant proteins and the incorporated function of ChloroP (Nielsen et al., 1997) to predict the cleavage sites.
Phylogenetic Analysis
Protein sequences were obtained from the GenBank or Joint Genome Initiative (JGI) genome assemblies using TBLASTN and APK1 as a query and aligned by ClustalW (Gonnet weight matrix, gap opening penalty −10, gap extension penalty −0.1). The alignment was inspected for needs of manual correction. The phylogenetic tree was constructed by the software MEGA2 version 2.1 (Kumar et al., 2001) using the neighbor-joining method (Poisson correction). APK from E. coli was used as an outgroup. Statistical support for the tree was obtained by bootstrap analysis with 1000 replicates.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL/JGI databases under the following accession numbers: APK1, At2g14750, AY132010; APK2, At4g39940, AY097421; APK3, At3g03900, NM_111261; APK4, At5g67520, NM_126152; Os APK1, Os11g0634800, NM_001074858; Os APK2, Os07g0573100, NM_001066599; Os APK3, AK241470; Pt APK1, XM_002313142; Pt APK2, XM_002298740; Pt APK3, XM_002319658; Sm APK1, e_gw1.8.1396.1; Sm APK2, e_gw1.142.99.1; Sm APK3, e_gw1.10.1027.1; Pp APK1, XM_001755153; Pp APK2, XM_001761511; Pp APK3, XM_001778721; Pp APK4, XM_001782577; Cr APK, XM_001695907; Ot APK, estExt_genewise1.C_Chr_09.00010262; Ol APK, XM_001419771; and Ec APK, CP001164.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. T-DNA Insertion Sites of APK Mutant Alleles.
Supplemental Figure 2. Accumulation of Glucosinolates in 2-Week-Old Plants Grown in Hydroculture.
Supplemental Figure 3. Variation in Levels of Indolic Glucosinolates.
Supplemental Figure 4. Quantitative Real-Time RT-PCR Analysis of Glucosinolate Biosynthetic Genes and ATPS Gene Family Members in Col-0 and the apk1 apk2 Knockout Mutant.
Supplemental Figure 5. Analysis of Sulfate Assimilation in apk1 apk2 Plants.
Supplemental Figure 6. Metabolite Analysis of apk1 apk2 Plants.
Supplemental Figure 7. Phylogenetic Analysis of APS Kinases from Plants and Green Algae.
Supplemental Table 1. Sequence Identity in Percent Identical Residues of Arabidopsis APS Kinase Isoforms and the APK from E. coli.
Supplemental Table 2. Fold-Changes of Transcription Levels of Genes of Primary Sulfur Metabolism and Metabolism of Sulfated Compounds in apk1 apk2 Mutant.
Supplemental Table 3. Metabolic Pathways Overrepresented among the Altered Transcripts: Pathways More Highly Expressed in the apk1 apk2 Mutant.
Supplemental Table 4. Metabolic Pathways Overrepresented among the Altered Transcripts: Pathways More Highly Expressed in the Wild Type.
Supplemental Table 5. Primer Sequences for Identification of T-DNA Insertions in apk Mutants.
Supplemental Table 6. Primer Sequences for RT-PCR Analysis of APK Transcripts in T-DNA Lines.
Supplemental Table 7. Primer Sequences for Expression Analysis by qPCR.
Supplemental Table 8. Primer Sequences for the pET14b Constructs for Heterologous Expression of Recombinant APKs.
Supplemental Table 9. Primer Aequences for the Chimeric Gene Constructs of CaMV35S Promoter-APK cDNA-GFP.
Supplemental Table 10. Primer Sequences for Creation of Transgenic Plants Expressing APK Promoter-Coding Sequence-GFP.
Supplemental Data Set 1. Alignment Used for the Phylogenetic Analysis in Supplemental Figure 6.
Supplementary Material
Acknowledgments
We thank Mrs. C. Kuhnt (Leibniz Institute of Plant Biochemistry, Halle, Germany) for analyses of 12-HSO4-JA and 12-O-Glc-JA. This study was supported in part by a grant from the British Biotechnology and Biological Sciences Research Council (BB/D009596/1 to S.K.), by a Special Postdoctoral Fellowship from RIKEN (to N.Y.), and by Grants-in-Aid for Scientific Research in Priority Areas from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to H.T. and K.S.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Stanislav Kopriva (stanislav.kopriva@bbsrc.ac.uk).
Online version contains Web-only data.
References
- Amano, Y., Tsubouchi, H., Shinohara, H., Ogawa, M., and Matsubayashi, Y. (2007). Tyrosine-sulfated glycopeptide involved in cellular proliferation and expansion in Arabidopsis. Proc. Natl. Acad. Sci. USA 104 18333–18338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andréasson, E., Bolt Jørgensen, L., Höglund, A.S., Rask, L., and Meijer, J. (2001). Different myrosinase and idioblast distribution in Arabidopsis and Brassica napus. Plant Physiol. 127 1750–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815. [DOI] [PubMed] [Google Scholar]
- Bak, S., Tax, F.E., Feldmann, K.A., Galbraith, D.W., and Feyereisen, R. (2001). CYP83B1, a cytochrome P450 at the metabolic branch point in auxin and indole glucosinolate biosynthesis in Arabidopsis. Plant Cell 13 101–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlier, I., Kowalczyk, M., Marchant, A., Ljung, K., Bhalerao, R., Bennett, M., Sandberg, G., and Bellini, C. (2000). The SUR2 gene of Arabidopsis thaliana encodes the cytochrome P450 CYP83B1, a modulator of auxin homeostasis. Proc. Natl. Acad. Sci. USA 97 14819–14824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth, C., and Jander, G. (2006). Arabidopsis myrosinases TGG1 and TGG2 have redundant function in glucosinolate breakdown and insect defense. Plant J. 46 549–562. [DOI] [PubMed] [Google Scholar]
- Bednarek, P., Pilewska-Bednarek, M., Svato, A., Schneider, B., Doubsk, J., Mansurova, M., Humphry, M., Consonni, C., Panstruga, R., Sanchez-Vallet, A., Molina, A., and Schulze-Lefert, P. (2009). A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323 101–106. [DOI] [PubMed] [Google Scholar]
- Beekwilder, J., (2008). The impact of the absence of aliphatic glucosinolates on insect herbivory in Arabidopsis. PLoS One 3 e2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitling, R., Amtmann, A., and Herzyk, P. (2004). Iterative Group Analysis (iGA): A simple tool to enhance sensitivity and facilitate interpretation of microarray experiments. BMC Bioinformatics 5 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, P.D., Tokuhisa, J.G., Reichelt, M., and Gershenzon, J. (2003). Variation of glucosinolate accumulation among different organs and developmental stages of Arabidopsis thaliana. Phytochemistry 62 471–481. [DOI] [PubMed] [Google Scholar]
- Burow, M., Müller, R., Gershenzon, J., and Wittstock, U. (2006). Altered glucosinolate hydrolysis in genetically engineered Arabidopsis thaliana and its influence on the larval development of Spodoptera littoralis. J. Chem. Ecol. 32 2333–2349. [DOI] [PubMed] [Google Scholar]
- Bürstenbinder, K., Rzewuski, G., Wirtz, M., Hell, R., and Sauter, M. (2007). The role of methionine recycling for ethylene synthesis in Arabidopsis. Plant J. 49 238–249. [DOI] [PubMed] [Google Scholar]
- Celenza, J.L., Quiel, J.A., Smolen, G.A., Merrikh, H., Silvestro, A.R., Normanly, J., and Bender, J. (2005). The Arabidopsis ATR1 Myb transcription factor controls indolic glucosinolate homeostasis. Plant Physiol. 137 253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S., Petersen, B.L., Olsen, C.E., Schulz, A., and Halkier, B.A. (2001). Long-distance phloem transport of glucosinolates in Arabidopsis. Plant Physiol. 127 194–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu, W.L., Niwa, Y., Zeng, W., Hirano, T., Kobayashi, H., and Sheen, J. (1996). Engineered GFP as a vital reporter in plants. Curr. Biol. 6 325–330. [DOI] [PubMed] [Google Scholar]
- Clay, N.K., Adio, A.M., Denoux, C., Jander, G., and Ausubel, F.M. (2009). Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323 95–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Cumming, M., Leung, S., McCallum, J., and McManus, M.T. (2007). Complex formation between recombinant ATP sulfurylase and APS reductase of Allium cepa (L.). FEBS Lett. 581 4139–4147. [DOI] [PubMed] [Google Scholar]
- Delarue, M., Prinsen, E., Onckelen, H.V., Caboche, M., and Bellini, C. (1998). Sur2 mutations of Arabidopsis thaliana define a new locus involved in the control of auxin homeostasis. Plant J. 14 603–611. [DOI] [PubMed] [Google Scholar]
- Du, L., and Halkier, B.A. (1998). Biosynthesis of glucosinolates in the developing silique walls. Phytochemistry 48 1145–1150. [Google Scholar]
- Emanuelsson, O., Nielsen, H., Brunak, S., and von Heijne, G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300 1005–1016. [DOI] [PubMed] [Google Scholar]
- Fahey, J.W., Zalcmann, A.T., and Talalay, P. (2001). The chemical diversity and distribution of glucosinolates and isothiocyanates among plants. Phytochemistry 56 5–51. [DOI] [PubMed] [Google Scholar]
- Falk, K.L., Tokuhisa, J.G., and Gershenzon, J. (2007). The effect of sulfur nutrition on plant glucosinolate content: Physiology and molecular mechanisms. Plant Biol. 5 573–581. [DOI] [PubMed] [Google Scholar]
- Gidda, S.K., Miersch, O., Levitin, A., Schmidt, J., Wasternack, C., and Varin, L. (2003). Biochemical and molecular characterization of a hydroxyjasmonate sulfotransferase from Arabidopsis thaliana. J. Biol. Chem. 278 17895–17900. [DOI] [PubMed] [Google Scholar]
- Gigolashvili, T., Berger, B., Mock, H.P., Müller, C., Weisshaar, B., and Flügge, U.I. (2007. a). The transcription factor HIG1/MYB51 regulates indolic glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 50 886–901. [DOI] [PubMed] [Google Scholar]
- Gigolashvili, T., Engqvist, M., Yatusevich, R., Müller, C., and Flügge, U.I. (2008). HAG2/MYB76 and HAG3/MYB29 exert a specific and coordinated control on the regulation of aliphatic glucosinolate biosynthesis in Arabidopsis thaliana. New Phytol. 177 627–642. [DOI] [PubMed] [Google Scholar]
- Gigolashvili, T., Yatusevich, R., Berger, B., Müller, C., and Flügge, U.I. (2007. b). The R2R3-MYB transcription factor HAG1/MYB28 is a regulator of methionine-derived glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 51 247–261. [DOI] [PubMed] [Google Scholar]
- Goda, E., Kamiyama, S., Uno, T., Yoshida, H., Ueyama, M., Kinoshita-Toyoda, A., Toyoda, H., Ueda, R., and Nishihara, S. (2006). Identification and characterization of a novel Drosophila 3′-phosphoadenosine 5′-phosphosulfate transporter. J. Biol. Chem. 281 28508–28517. [DOI] [PubMed] [Google Scholar]
- Grubb, C.D., and Abel, S. (2006). Glucosinolate metabolism and its control. Trends Plant Sci. 11 89–100. [DOI] [PubMed] [Google Scholar]
- Grubb, C.D., Zipp, B.J., Ludwig-Müller, J., Masuno, M.N., Molinski, T.F., and Abel, S. (2004). Arabidopsis glucosyltransferase UGT74B1 functions in glucosinolate biosynthesis and auxin homeostasis. Plant J. 40 893–908. [DOI] [PubMed] [Google Scholar]
- Haink, G., and Deussen, A. (2003). Liquid chromatography method for the analysis of adenosine compounds. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 784 189–193. [DOI] [PubMed] [Google Scholar]
- Halkier, B.A., and Gershenzon, J. (2006). Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol. 57 303–333. [DOI] [PubMed] [Google Scholar]
- Hirai, M.Y., Fujiwara, T., Awazuhara, M., Kimura, T., Noji, M., and Saito, K. (2003). Global expression profiling of sulfur-starved Arabidopsis by DNA macroarray reveals the role of O-acetyl-l-serine as a general regulator of gene expression in response to sulfur nutrition. Plant J. 33 651–663. [DOI] [PubMed] [Google Scholar]
- Hirai, M.Y., et al. (2007). Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc. Natl. Acad. Sci. USA 104 6478–6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höfgen, R., and Willmitzer, L. (1988). Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res. 16 9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiyama, S., et al. (2003). Molecular cloning and identification of 3′-phosphoadenosine 5′-phosphosulfate transporter. J. Biol. Chem. 278 25958–25963. [DOI] [PubMed] [Google Scholar]
- Kim, J.H., and Jander, G. (2007). Myzus persicae (green peach aphid) feeding on Arabidopsis induces the formation of a deterrent indole glucosinolate. Plant J. 49 1008–1019. [DOI] [PubMed] [Google Scholar]
- Klein, M., and Papenbrock, J. (2004). The multi-protein family of Arabidopsis sulfotransferases and their relatives in other plant species. J. Exp. Bot. 55 1809–1820. [DOI] [PubMed] [Google Scholar]
- Klein, M., Reichelt, M., Gershenzon, J., and Papenbrock, J. (2006). The three desulfoglucosinolate sulfotransferase proteins in Arabidopsis have different substrate specificities and are differentially expressed. FEBS J. 273 122–136. [DOI] [PubMed] [Google Scholar]
- Koncz, C., and Schell, J. (1986). The promoter of T-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel types of Agrobacterium binary vector. Mol. Gen. Genet. 204 383–396. [Google Scholar]
- Kopriva, S. (2006). Regulation of sulfate assimilation in Arabidopsis and beyond. Ann. Bot. (Lond.) 97 479–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopriva, S., Wiedemann, G., and Reski, R. (2007). Sulfate assimilation in basal land plants - What does genomic sequencing tell us? Plant Biol. 9 556–564. [DOI] [PubMed] [Google Scholar]
- Koprivova, A., Suter, M., Op den Camp, R., Brunold, C., and Kopriva, S. (2000). Regulation of sulfate assimilation by nitrogen in Arabidopsis. Plant Physiol. 122 737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koroleva, O.A., Davies, A., Deeken, R., Thorpe, M.R., Tomos, A.D., and Hedrich, R. (2000). Identification of a new glucosinolate-rich cell type in Arabidopsis flower stalk. Plant Physiol. 124 599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S., Tamura, K., Jakobsen, I.B., and Nei, M. (2001). MEGA2: Molecular evolutionary genetics analysis software. Bioinformatics 17 1244–1245. [DOI] [PubMed] [Google Scholar]
- Lee, S., and Leustek, T. (1998). APS kinase from Arabidopsis thaliana: Genomic organization, expression, and kinetic analysis of the recombinant enzyme. Biochem. Biophys. Res. Commun. 247 171–175. [DOI] [PubMed] [Google Scholar]
- Lillig, C.H., Schiffmann, S., Berndt, C., Berken, A., Tischka, R., and Schwenn, J.D. (2001). Molecular and catalytic properties of Arabidopsis thaliana adenylyl sulfate (APS)-kinase. Arch. Biochem. Biophys. 392 303–310. [DOI] [PubMed] [Google Scholar]
- Lüders, F., Segawa, H., Stein, D., Selva, E.M., Perrimon, N., Turco, S.J., and Häcker, U. (2003). Slalom encodes an adenosine 3′-phosphate 5′-phosphosulfate transporter essential for development in Drosophila. EMBO J. 22 3635–3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magrath, R., and Mithen, R. (1993). Maternal effects on the expression of individual aliphatic glucosinolates in seeds and seedlings of Brassica napus. Plant Breed. 111 249–252. [Google Scholar]
- Malitsky, S., Blum, E., Less, H., Venger, I., Elbaz, M., Morin, S., Eshed, Y., and Aharoni, A. (2008). The transcript and metabolite networks affected by the two clades of Arabidopsis glucosinolate biosynthesis regulators. Plant Physiol. 148 2021–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandon, E.C., Milla, M.E., Kempner, E., and Hirschberg, C.B. (1994). Purification of the Golgi adenosine 3′-phosphate 5′-phosphosulfate transporter, a homodimer within the membrane. Proc. Natl. Acad. Sci. USA 91 10707–10711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama-Nakashita, A., Nakamura, Y., Tohge, T., Saito, K., and Takahashi, H. (2006). Arabidopsis SLIM1 is a central transcriptional regulator of plant sulfur response and metabolism. Plant Cell 18 3235–3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi, Y., Hanai, H., Hara, O., and Sakagami, Y. (1996). Active fragments and analogs of the plant growth factor, phytosulfokine: Structure-activity relationships. Biochem. Biophys. Res. Commun. 225 209–214. [DOI] [PubMed] [Google Scholar]
- Matsubayashi, Y., Ogawa, M., Kihara, H., Niwa, M., and Sakagami, Y. (2006). Disruption and overexpression of Arabidopsis phytosulfokine receptor gene affects cellular longevity and potential for growth. Plant Physiol. 142 45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi, Y., and Sakagami, Y. (1996). Phytosulfokine, sulfated peptides that induce the proliferation of single mesophyll cells of Asparagus officinalis L. Proc. Natl. Acad. Sci. USA 93 7623–7627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miersch, O., Neumerkel, J., Dippe, M., Stenzel, I., and Wasternack, C. (2008). Hydroxylated jasmonates are commonly occurring metabolites of jasmonic acid and contribute to a partial switch-off in jasmonate signaling. New Phytol. 177 114–127. [DOI] [PubMed] [Google Scholar]
- Mikkelsen, M.D., Naur, P., and Halkier, B.A. (2004). Arabidopsis mutants in the C-S lyase of glucosinolate biosynthesis establish a critical role for indole-3-acetaldoxime in auxin homeostasis. Plant J. 37 770–777. [DOI] [PubMed] [Google Scholar]
- Mithen, R., Faulkner, K., Magrath, R., Rose, P., Williamson, G., and Marquez, J. (2003). Development of isothiocyanate-enriched broccoli, and its enhanced ability to induce phase 2 detoxification enzymes in mammalian cells. Theor. Appl. Genet. 106 727–734. [DOI] [PubMed] [Google Scholar]
- Nielsen, H., Engelbrecht, J., Brunak, S., and von Heijne, G. (1997). Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10 1–6. [DOI] [PubMed] [Google Scholar]
- Pedras, M.S., and Montaut, S. (2004). The biosynthesis of crucifer phytoalexins: Unprecedented incorporation of a 1-methoxyindolyl precursor. Chem. Commun. (Camb.) 21 452–453. [DOI] [PubMed] [Google Scholar]
- Piotrowski, M., Schemenewitz, A., Lopukhina, A., Müller, A., Janowitz, T., Weiler, E.W., and Oecking, C. (2004). Desulfoglucosinolate sulfotransferases from Arabidopsis thaliana catalyze the final step in the biosynthesis of the glucosinolate core structure. J. Biol. Chem. 279 50717–50725. [DOI] [PubMed] [Google Scholar]
- Ratzka, A., Vogel, H., Kliebenstein, D.J., Mitchell-Olds, T., and Kroymann, J. (2002). Disarming the mustard oil bomb. Proc. Natl. Acad. Sci. USA 99 11223–11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reintanz, B., Lehnen, M., Reichelt, M., Gershenzon, J., Kowalczyk, M., Sandberg, G., Godde, M., Uhl, R., and Palme, K. (2001). Bus, a bushy Arabidopsis CYP79F1 knockout mutant with abolished synthesis of short-chain aliphatic glucosinolates. Plant Cell 13 351–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renosto, F., Seubert, P.A., and Segel, I.H. (1984). Adenosine 5′-phosphosulfate kinase from Penicillium chrysogenum. Purification and kinetic characterization. J. Biol. Chem. 259 2113–2123. [PubMed] [Google Scholar]
- Rotte, C., and Leustek, T. (2000). Differential subcellular localization and expression of ATP sulfurylase and 5′-adenylylsulfate reductase during ontogenesis of Arabidopsis leaves indicates that cytosolic and plastid forms of ATP sulfurylase may have specialized functions. Plant Physiol. 124 715–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouleau, M., Marsolais, F., Richard, M., Nicolle, L., Voigt, B., Adam, G., and Varin, L. (1999). Inactivation of brassinosteroid biological activity by a salicylate-inducible steroid sulfotransferase from Brassica napus. J. Biol. Chem. 274 20925–20930. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schilmiller, A.L., and Howe, G.A. (2005). Systemic signaling in the wound response. Curr. Opin. Plant Biol. 8 369–377. [DOI] [PubMed] [Google Scholar]
- Schöne, F., Rudolph, B., Kirchheim, U., and Knapp, G. (1997). Counteracting the negative effects of rapeseed and rapeseed press cake in pig diets. Br. J. Nutr. 78 947–962. [DOI] [PubMed] [Google Scholar]
- Shroff, R., Vergara, F., Muck, A., Svatos, A., and Gershenzon, J. (2008). Nonuniform distribution of glucosinolates in Arabidopsis thaliana leaves has important consequences for plant defense. Proc. Natl. Acad. Sci. USA 105 6196–6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster, J., Knill, T., Reichelt, M., Gershenzon, J., and Binder, S. (2006). Branched-chain aminotransferase4 is part of the chain elongation pathway in the biosynthesis of methionine-derived glucosinolates in Arabidopsis. Plant Cell 18 2664–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenzel, I., Hause, B., Maucher, H., Pitzschke, A., Miersch, O., Ziegler, J., Ryan, C., and Wasternack, C. (2003). Allene oxide cyclase dependence of the wound response and vascular bundle specific generation of jasmonates in tomato - Amplification in wound-signalling. Plant J. 33 577–589. [DOI] [PubMed] [Google Scholar]
- Storey, J.D., and Tibshirani, R. (2003). Statististical significance for genome-wide experiments. Proc. Natl. Acad. Sci. USA 100 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam, S. (1998). The Biology Workbench–A seamless database and analysis environment for the biologist. Proteins 32 1–2. [PubMed] [Google Scholar]
- Thangstad, O.P., Gilde, B., Chadchawan, S., Seem, M., Husebye, H., Bradley, D., and Bones, A.M. (2004). Cell specific, cross-species expression of myrosinases in Brassica napus, Arabidopsis thaliana and Nicotiana tabacum. Plant Mol. Biol. 54 597–611. [DOI] [PubMed] [Google Scholar]
- Underhill, E.W., Wetter, L.R., and Chisholm, M.D. (1973). Biosynthesis of glucosinolates. Biochem. Soc. Symp. 38 303–326. [PubMed] [Google Scholar]
- Valvekens, D., Van Montagu, M., and Van Lijsebettens, M. (1988). Agrobacterium-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc. Natl. Acad. Sci. USA 85 5536–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz, M., and Hell, R. (2003). Production of cysteine for bacterial and plant biotechnology: Application of cysteine feedback-insensitive isoforms of serine acetyltransferase. Amino Acids 24 195–203. [DOI] [PubMed] [Google Scholar]
- Wirtz, M., and Hell, R. (2007). Dominant-negative modification reveals the regulatory function of the multimeric cysteine synthase protein complex in transgenic tobacco. Plant Cell 19 625–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, X., and Chen, S. (2007). Regulation of plant glucosinolate metabolism. Planta 226 1343–1352. [DOI] [PubMed] [Google Scholar]
- Yang, H., Matsubayashi, Y., Nakamura, K., and Sakagami, Y. (2001). Diversity of Arabidopsis genes encoding precursors for phytosulfokine, a peptide growth factor. Plant Physiol. 127 842–851. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.