Abstract
Genome sequencing dramatically increased our ability to understand cellular response to perturbation. Integrating system-wide measurements such as gene expression with networks of protein–protein interactions and transcription factor binding revealed critical insights into cellular behavior. However, the potential of systems biology approaches is limited by difficulties in integrating metabolic measurements across the functional levels of the cell despite their being most closely linked to cellular phenotype. To address this limitation, we developed a model-based approach to correlate mRNA and metabolic flux data that combines information from both interaction network models and flux determination models. We started by quantifying 5,764 mRNAs, 54 metabolites, and 83 experimental 13C-based reaction fluxes in continuous cultures of yeast under stress in the absence or presence of global regulator Gcn4p. Although mRNA expression alone did not directly predict metabolic response, this correlation improved through incorporating a network-based model of amino acid biosynthesis (from r = 0.07 to 0.80 for mRNA-flux agreement). The model provides evidence of general biological principles: rewiring of metabolic flux (i.e., use of different reaction pathways) by transcriptional regulation and metabolite interaction density (i.e., level of pairwise metabolite-protein interactions) as a key biosynthetic control determinant. Furthermore, this model predicted flux rewiring in studies of follow-on transcriptional regulators that were experimentally validated with additional 13C-based flux measurements. As a first step in linking metabolic control and genetic regulatory networks, this model underscores the importance of integrating diverse data types in large-scale cellular models. We anticipate that an integrated approach focusing on metabolic measurements will facilitate construction of more realistic models of cellular regulation for understanding diseases and constructing strains for industrial applications.
Keywords: amino acid stress response, fluxomics, gcn4, systems biology
Metabolic fluxes are informative indicators of cellular physiology and in vivo homeostasis. Fluxes are the model-independent rates of metabolite interconversion that emerge through the interplay of genes, proteins, and metabolites at multiple regulatory levels. Protein–protein interactions (1) and transcription factor binding (2, 3) can be used to predict a cellular behavior at the gene and protein level (4, 5). However, complex metabolite-enzyme interactions (6), translational regulation (7), and posttranscriptional mechanisms prevent direct linkage of transcriptional state and metabolic phenotype. Previous in silico attempts to address this gap included metabolic network and regulatory on/off switch models for growth and viability prediction (8, 9). In addition, experimental studies of posttranscriptional control have measured mRNA expression in conjunction with either 13C-based flux (10) or metabolite level measurements (11). Such studies have proven invaluable for emphasizing the importance of metabolic phenotypes; however, new approaches are needed which, by integrating these diverse data types in biological models, can identify specific mechanisms by which genetic regulatory circuits mediate metabolic flux phenotype.
Toward this goal, we created a pathway model of amino acid biosynthesis that includes genetic regulatory circuits and metabolite-enzyme interactions to simultaneously integrate flux, metabolite, and mRNA data into a functional biological model. Specifically, our investigation focused on native transcriptional control of amino acid biosynthetic enzymes via the Gcn4p-mediated stress response. This collection of pathways has been studied extensively by both classical genetics and genomic approaches (3, 12), and human counterparts of Gcn4p-regulated pathways have been associated with 209 genetic disorders including Phenylketonuria (PKU). Our study aimed to use this model system to demonstrate that integrating metabolic flux phenotypes would yield new and specific insights to a well-studied set of pathways. Here, we present the results of our measurements, a mechanistic model of amino acid biosynthesis, a predictor of metabolic flux changes, and a discussion of what we learned about the metabolic control structure for this model system.
Results
To study the response enabled by Gcn4p, we compared wild-type yeast to a gcn4Δ-knockout strain (i.e., stress response present versus absent) with both strains cultivated in a chemostat under constant pH, controlled growth rate, and starvation-induced stress (Fig. 1A). Chemostat cultivations, which enable growth under a tightly regulated steady-state environment, eliminate variability that is inherent in dynamically changing batch cultures. In shake-flask studies, for example, it is difficult, and in some cases impossible, to control several key cultivation parameters (e.g., growth rate, media composition, dissolved oxygen concentration, pH, etc.), which often obscures the “real” parameter under investigation. This is particularly true when exploring the transcriptional response, which is known to change with specific growth rate (13).
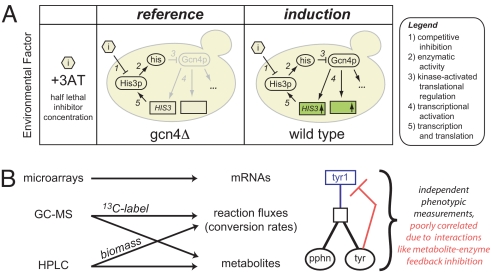
Fig. 1.
Experimental design and measurement strategy. (A) Analyzing the Gcn4p-dependent stress response in a controlled-growth chemostat environment. Wild-type S288C and an isogenic gcn4Δ derivative were cultured in a glucose-limited chemostat diluted at 0.10 h−1 (416 min per doubling) in unsupplemented YNB minimal media regulated to 5 +/− 0.1 pH. Titrated histidine biosynthesis inhibitor levels (10 and 0.1 mM, respectively, of 3-amino-1,2,4-triazole or 3AT) created histidine near starvation (Fig. S1), which causes Gcn4p translational activation and transcriptional activation of hundreds of targets in the wild type (+Gcn4p). Through this modulation of the inhibitor (3AT) concentration, both wild-type and gcn4Δ cultures were grown at the same specific growth rate (0.10 h−1) and achieved similar cell densities and production rates of ethanol and CO2 (Table S3). (B) Multitiered measurement strategy for analyzing large-scale network perturbations. We used microarrays, GC-MS, and HPLC to measure mRNAs, fluxes, and metabolites (see text and SI Text for further detail). These independent measurements give a broad view of the Gcn4p stress response and help to characterize network effects, such as metabolite interaction densities.
Despite similar growth rate conditions and overall macroscopic growth characteristics, the responses of the 2 strains differed markedly at the molecular level. Our gene expression measurements (Dataset S1) confirmed that active Gcn4p in the wild-type population induces transcription of hundreds of target genes within amino acid biosynthesis and other pathways (3, 14), as shown in Fig. 2A. The median enzyme mRNA expression level increased at least 2-fold for the arginine, asparagine, tryptophan, phenylalanine, tyrosine, histidine, isoleucine, leucine, and valine pathways, and at least 1 transcript reached this threshold for the cysteine, methionine, and lysine pathways.
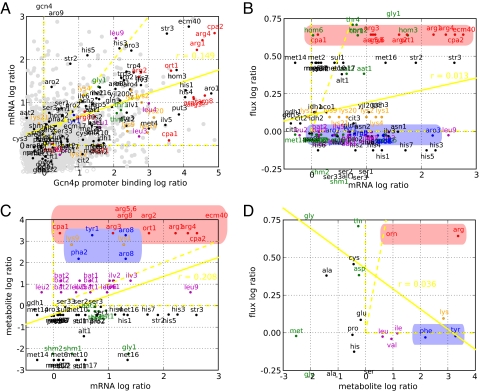
Fig. 2.
Pairwise comparisons of changes often reveal poor correlation in Gcn4p binding, mRNA, flux, and end product metabolite. Induction ratios for the measurements indicated on the axes correspond to Fig. 1. (A) Ratios of mRNAs (measurements described in text) and Gcn4p binding at the upstream promoter under starvation measured with ChIP arrays as described by Harbison, et al. (3) are scattered. As purely Gcn4p-regulated, CPA2 (Upper Right) illustrates a general observation: Stronger binding often leads to increased transcriptional activation. For other genes, the binding relationship gives insight to its transcriptional control and identifies genes (i) bound yet not activated (>2 log2 bound, <0.5 log2 expressed) and (ii) activated yet not bound (<1 log2 bound, >1 log2 expressed). The first class includes genes encoding transcription factors Met4p, Leu3p, and Lys14p, which likely require coactivators, and CPA1, which likely undergoes a uORF-mediated activation of mRNA decay (a metabolite-RNA interaction) (32, 33) by 10-fold increase in free arginine. The second class of genes includes ARO9 that likely have indirect mechanisms of transcriptional activation, for instance, by Aro80p in response to the 2- and 3-fold increases in free phenylalanine (34). (B–D) Log ratios for the mRNA, reaction flux, and metabolite measurements described in the text are associated by pathway and labeled. Arginine and aromatic data, which differ in metabolite interaction densities, are highlighted in red and blue, respectively. Yellow dotted lines highlight the positive x axis, y axis, and diagonal, and the yellow solid line represents the least squares fit of the data. Data points outside the axes were placed at the periphery. (B) Despite similar transcriptional activations, flux measurements diverge for arginine (in red, increased) and aromatic (in blue, invariant) pathways. Generally, transcriptional activation is insufficient in predicting flux activation. (C and D) Free phenylalanine and tyrosine rise 2- and 3- fold upon transcriptional activation, yet aromatic fluxes remains invariant likely because increased product levels modulate pathway activity through a high density of feedback metabolite-enzyme interactions. Overall, a variety of posttranscriptional control (e.g., metabolite interaction density) mechanisms simultaneously modulating pathways confound an effort to correlate mRNA, flux, and metabolite changes.
Despite these drastic changes at the molecular level, the wild type and gcn4Δ chemostat cultures maintain a macroscopic physiological similarity. To explain this, we hypothesized that the cell compensated at the level of metabolism to maintain similar macroscopic growth under the divergent transcriptional programs observed in the wild-type and gcn4Δ cultures. Metabolic compensation can be assessed by monitoring 2 features: metabolite levels—defined as the relative abundances of non-genetically-encoded substrates, intermediates, or products of metabolic pathways (15), and reaction fluxes—defined as the rates of conversion by each reaction in the metabolic network (16) (Fig. 1B). To measure metabolite levels, we used gas chromatography coupled to mass spectrometry (GC-MS) to separate, characterize, and quantify metabolites from whole-cell extracts. Additionally, we used high performance liquid chromatography (HPLC) to obtain more precise measurements of 17 of the 20 free amino acids. In total, we assigned statistical confidences to abundance ratios of 54 observed metabolites in the gcn4Δ versus wild-type cells (Dataset S2).
Reaction fluxes provide a direct measure of metabolite interconversion rates. In the absence of experimental data, values of reaction fluxes are often estimated in silico under the constraints of maximized biomass production, an assumed cellular stoichiometry, a steady state metabolic network and fixed protein composition (17). However, these simulations result in an underdetermined system (more fluxes than measurements), and it is still an open question whether such methods adequately capture regulatory-induced large network perturbations. For this reason, we cultivated cells on 1-13C-glucose and experimentally measured condition-specific biomass amino acid content with HPLC and isotopic labeling patterns with GC-MS to overdetermine the reaction system (more measurements than fluxes) and thus more robustly estimate fluxes (18) that can be found in Dataset S3. Stable isotopic labeling is an established technique for flux determination whereby metabolic conversion of 13C-enriched substrates generates specifically labeled metabolites, i.e., isotope isomers, whose labeling patterns are direct functions of the flux configuration (16). As such, flux can be estimated from measurements (by GC-MS) of isotopic enrichment of metabolites and solving the inverse problem. Overall, our measurements yielded 17 amino acid fluxes into biomass and 250 13C-labeling abundances, thus providing a very overdetermined system (see below) for accurate and robust flux estimation.
To convert these 13C-labeling abundances to intracellular flux values and statistically verify the results (18), we developed a new method based on the yeast metabolic reaction network refined and expanded from Gombert et al. (19). Compared with the 2 existing methods for flux determination (19, 20), we took greater advantage of redundant flux information (i.e., more measurements than fluxes) by (i) using the complete labeling distributions instead of lumping abundances into a summed fractional labeling [relative to the GC-MS method (19)], and (ii) simultaneously fitting the entire measurement set [relative to the NMR method (20)]. We used a network of 83 reactions (with corresponding carbon transitions) distributed among the 3 assigned compartments (cytosolic, mitochondrial, extracellular), using 75 metabolites (1 substrate, 51 balanced intracellular, and 23 products). Combining our measurement set with this network resulted in 105 redundant measurements for our now over determined system (SI Appendix, C13-Based Reaction Flux Determination). By adding experimentally obtained amino acid fluxes into biomass measurements (previously assumed unchanging), we expanded our resolution beyond that of previous methods that only use networks of central carbon metabolism. We also note the exceptionally high quality of fit in Fig. S2 that gives us high confidence in our experimentally measured fluxes. Because metabolite levels and fluxes are independent measurements (21), together they give a more complete view of the metabolic regulatory system.
Consistent with the previous observation that mRNA expression insufficiently predicts protein level (22), biosynthetic mRNA changes demonstrated poor correlation with flux changes (r = 0.02 for log ratios or differences in Fig. 2B, r = 0.07 for deltas or linear differences in Fig. 4B). Indeed, we found little correlation for any pairwise combination of mRNA, flux, and end-product metabolite changes (Fig. 2 B–D). Furthermore, Gcn4p DNA-binding strength did not always translate to transcriptional activation (Fig. 2A).
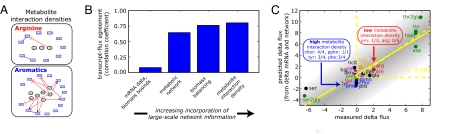
Fig. 4.
Incorporating large-scale biomolecular networks improve flux phenotype understanding. (A) Metabolite interaction densities are illustrated for arginine and aromatics biosythesis. (B) Changes in mRNA measurements were used to predict flux changes as described in the text and SI Text. By adding the statistically verified reaction network, biomass efflux bounds, and metabolite interaction density, the predicted fluxes from mRNA better matched measured fluxes with a correlation coefficient value of 0.80 relative to 0.07 corresponding to the initial assumption set (see SI Appendix, section 3). (C) Measured delta flux for individual reactions is labeled by end product metabolite and plotted versus the flux predicted from mRNA measurements and the 2 parameter model. Notably, adding metabolite interaction densities discriminates arginine and aromatic pathways to predict arginine's up-regulated flux. Without the model incorporating metabolite interaction densities, aromatic pathways would have been wrongly predicted to increase in flux.
We next proposed that a mechanistic model of amino acid biosynthesis capturing regulatory knowledge would better correlate the mRNA, intracellular flux, and metabolite level datasets. Accordingly, we created a biosynthetic network consisting of gene/protein, reaction, and metabolite nodes connected by condition-specific transcription factor binding interactions (3), protein–protein binding interactions (23), and enzyme-reaction and reaction-metabolite interactions. This network contained at its core the metabolic reaction network used previously for (and statistically verified by) flux determination. Additionally, we curated all known metabolite-enzyme interactions (24) as an explicit avenue of enzyme-level regulation. The key feature of our updated model is that we no longer rely on stoichiometry alone. Now, regulatory information (given by interactions within the network) is incorporated that might impact the correlation between mRNA levels and metabolic flux. This expanded network model linking all measurement types was visualized in Cytoscape (25) as shown in Fig. 3 (with a log2 color bar for measurement ratios of the wild type strain relative to gcn4Δ).
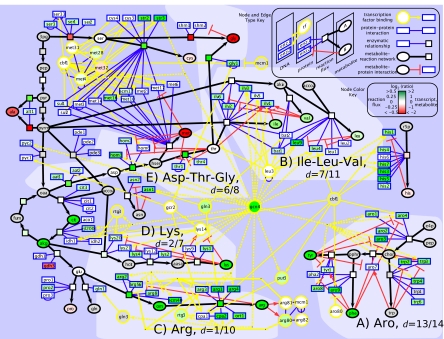
Fig. 3.
Integrated perspective of a large-scale network perturbation on amino acid biosynthesis. The central GCN4 node anchors a biomolecular gene/protein network expanded to include reactions and metabolites. The network contains condition-specific transcription factor binding, protein–protein binding, enzyme-reaction, reaction-metabolite, and metabolite-protein feedback edges (see node type and edge key). High metabolite interaction density is evident in A, B, and E. The dotted yellow edges emanating from the GCN4 node indicate an interaction found only in the wild type condition (i.e., not in gcn4Δ) Measurement ratios of wild type relative to gcn4Δ were visualized with a log2 colorbar (see node color key), and gray coloring indicates the lack of a measurement for that node. Findings for individual pathways are discussed in the text.
To improve the poor mRNA-flux agreement seen in Fig. 2B, we used this expanded network model to construct a predictor of metabolic flux changes as presented in SI Appendix, section 3. A novelty of the model is the introduction of a parameter termed “metabolite interaction density” for each reaction that captures the degree to which the reaction's enzymes are negatively or positively regulated by metabolites as represented in the above pathway model. Specifically, for a pathway this quantity is defined as the ratio of the number of metabolite-enzyme interactions to that of the total reaction enzymes in the pathway, as displayed in Fig. 4A. Thus, pathways with high degree of feedback inhibition (Fig. 3 A, B, and E) and other enzyme-level regulation will have high metabolite interaction density for their constituent reactions.
Using this concept, network flux changes were predicted from changes in mRNA levels and metabolite interaction densities, using the following equation: Δflux = exp(−p1 · dinteraction) . The rationale for this equation is to allow metabolite interaction density (dinteraction) to modulate the extent to which changes in mRNA levels are allowed to propagate to changes in flux, with p1 and p2 determined to minimize the difference between actual and predicted fluxes (SI Appendix, mRNA-Flux Model). According to this equation, the effect of mRNA changes (ΔmRNA) is attenuated by metabolite interaction density in causing changes in the pathway flux (Δflux). The last feature of our model is the additional constraint that fluxes predicted by the above equation must also be consistent with the overall metabolic network stoichiometry and satisfy the steady state reaction network, i.e., the equation S · Δv = 0.
This model represents a new, hybrid approach to correlating mRNA and flux data that combines information from both interaction network models and flux determination models. A core element is that the model preserves metabolic network consistency both in flux determination and in flux prediction from mRNA data. In addition, the model introduces the new concept of metabolite interaction density that embodies a hypothesis that a simpler, less-connected regulatory network often has more correlated mRNA and flux changes.
The model illustrates that flux control (i.e., invariance to perturbation) increases as metabolite—enzyme interaction density increases. In other words, greater interaction density lessens transcriptional control and gives metabolites an increasing role in regulating the flux phenotype. This model supports the body of literature on feedback inhibition in that increased metabolite regulation of enzyme activity results in greater metabolic control. The high interaction density aromatics and isoleucine-leucine-valine pathways (dinteraction = 13/14 and dinteraction = 7/11 in Fig. 3 A and B, respectively) provide an illustrative foil to the low interaction density arginine and lysine pathways (dinteraction = 1/10 and dinteraction = 2/7 in Fig. 3 C and D, respectively) as depicted in Fig. 4A and discussed in SI Discussion. For the high metabolite interaction densities, we hypothesize that because end products are not being increasingly used in protein synthesis, the metabolite levels build up and cause feedback inhibition at the enzyme level, resulting in tightly regulated flux. In contrast, we hypothesize that low metabolite interaction density pathways use alternative strategies. These pathways rely on preferential vacuolar localization of products and activation of transcription factor regulators separate from Gcn4p to compensate for unnecessary metabolite build-up.
Despite high metabolite interaction densities of the aspartate and threonine pathways, dinteraction = 6/8, fluxes are transcriptionally regulated and increase significantly because of rewiring of glycine flux, as revealed by the 13C-labeling patterns (Fig. 3E). Glycine may be produced from 3 independently 13C-enriched precursors: glyoxylate (Agx1p), serine (Shm1p, Shm2p), and threonine (Gly1p). After discarding glyoxylate flux as 0 (within 3% noise) in all conditions, ≈99% of glycine flux comes via serine in the reference condition; however, the presence of activated Gcn4p shifts 44% of that flux to a threonine precursor. We hypothesize that although free aspartate and threonine do not increase appreciably, the significant (FDR <2%) activation of aat2, hom3, and hom2 and near significant (FDR <3% or log2 ratio >1.0) activation of thr1, thr4, and gly1 likely provide an increased biosynthetic capacity. Thus, some glycine biosynthetic flux is rewired away from serine to a route depleting the now more available free aspartate and threonine and increasing fluxes (>30% change), i.e., the up-regulated green reaction nodes in Fig. 3E. Overall, these data suggest that global rewiring of flux through the network may be a mechanism to override high metabolite interaction density.
In total, the predictive network model highlights 5 general observations and 10 new system hypotheses not evident without high resolution metabolic flux data (Tables S1 and S2). Because transcriptional flux rewiring has not been studied extensively and, more specifically, glycine flux rewiring has not previously been reported as a potential conserved physiological for stress response, we chose to examine these observations and hypotheses in greater detail.
To validate the predictive network model for transcriptional changes that drive a rewiring of global network fluxes, we created additional perturbations of yeast transcriptional regulation in the vicinity of glycine biosynthesis (Fig. 3E) with met28Δ, cbf1Δ, met31Δ, and met32Δ knockout strains. We measured mRNA levels for each strain in shake flask cultures and found that met28Δ and cbf1Δ displayed significant changes in amino acid biosynthetic pathways (Figs. S3–S6). Using the measured mRNA levels and the predictive network model from above without any further modification, we determined that both met28Δ and cbf1Δ should exhibit a glycine flux rewiring that favors a threonine precursor.
To experimentally validate the conclusions of the predictive network model, we used the 13C labeling analysis described above to measure glycine biosynthetic fluxes for the wild type, met28Δ and cbf1Δ strains in YNB minimal media supplemented with methionine to correct for the corresponding knockout auxotrophies. As before, we achieved an exceptionally high quality of fit in Fig. S7.
Notably, we observed that the predictive network model correctly identified this experimentally-determined glycine flux rewiring, using the mRNA data. We observed a shifting of glycine flux to a threonine precursor where serine-originating glycine flux decreased from ≈50% in the wild type to ≈25% in met28Δ to 0% in cbf1Δ as shown in Fig. 5. We hypothesize that the increasing shutdown of mRNA levels in the methionine pathways increased the availability of homoserine and thus threonine to yield greater conversion rates of threonine to glycine.
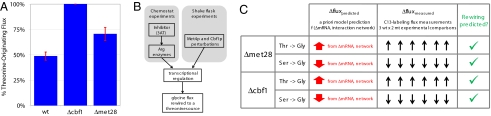
Fig. 5.
Observations in glycine flux rewiring demonstrate a biological principle and validate the predictive network model. (A) Additional transcriptional regulatory perturbations drive a rewiring in glycine flux. Knockout strains for transcriptional regulators in the vicinity of glycine biosynthesis (met28Δ, cbf1Δ, met31Δ, and met32Δ) were constructed, mRNA levels were measured, and biosynthetic fluxes were determined for the 2 strains (met28Δ and cbf1Δ) that displayed significant transcriptional changes. The results for the percentage of threonine-originating glycine flux are displayed in the first panel. Similar to the previous Gcn4p-induced stress response data, we observe a rewiring of glycine flux away from serine precursor in both the met28Δ and cbf1Δ cultures. (B) Convergent rewiring of glycine flux. In both chemostat and shake flask experiments, transcriptional regulatory networks drives a rewiring of glycine flux to a threonine source. (C) The predictive model from Gcn4 experiments uses mRNA changes and the interaction network correctly identified flux rewiring in knockout experiments. mRNA changes from the met28Δ and cbf1Δ knockout strains were input to the existing predictive flux model to assess glycine flux rewiring. For both knockout mRNA profiles, the glycine flux was predicted to be rewired from a serine precursor to threonine. The experimentally measured flux changes of the serine and glycine (displayed in A) verify the prediction of this observed rewiring.
We were especially encouraged by 3 aspects of the glycine flux rewiring observations in our follow-on analysis. First, the 13C high resolution flux determination enabled the experimental observation of a previously unreported mechanism for which an interaction network allows mRNA to mediate a flux rewiring for a cellular response. Second, this glycine rewiring mechanism was experimentally observed for perturbations originating in different network locations indicating a convergent rewiring pathway activated by transcriptional regulation (Fig. 5B). Finally, we demonstrated that mRNA and network data may be used to correctly predict flux changes.
Discussion
Looking forward toward a full fusion of transcriptional and metabolic data, the suspected role of metabolite interaction density and metabolite-triggered activation of ARO9 transcription (among others) demonstrate opportunities to substantially augment the currently available interactome. Despite modulating nearly all pathways observed, cell-wide data on interactions between metabolites and proteins are not yet available unlike protein–protein interactions. Chemical genomics investigations screen small molecule libraries for desired function (e.g., drug efficacy), but the inverse process to enumerate all interactions of a given metabolite is not as common. We anticipate that this metabolite-protein interactome will result from new protein array techniques and iterative interaction network reconstructions of systematically measured fluxes and metabolite levels akin to previous metabolic and genetic network reconstructions (26, 27). Just as Hall et al. (28) demonstrated that metabolic enzyme Arg-5,6p was a transcription factor by globally screening proteins for DNA binding activity, many metabolite-enzyme interactions remain to be discovered in broad screens looking outside a targeted system.
In this investigation, we offer evidence that should place bounds on the interpretation of transcriptional data and provide 5 general observations and 10 new system hypotheses not evident without high resolution metabolic data (Tables S1 and S2). We observe that flux control by metabolites at dense regions of metabolite-enzyme interactions is a key posttranscriptional regulatory strategy that allows cells to balance biosynthetic needs for growth. Additionally, we observe that predictive flux models may be used to correctly identify flux changes, using mRNA levels and interaction networks. We conclude that the satisfactory fusion of metabolomic and fluxomic data for microbial and mammalian systems will not only benefit from improving analytical and computational techniques for metabolite and flux measurement but will also necessarily rely on large-scale interaction networks expanded to include globally-screened data on metabolite interactions.
Materials and Methods
Chemostat Growth Conditions.
Strains were cultivated in aerobic carbon-limited chemostat cultures in 2-L fermentors (Applikon) in standard YNB media obtained from Qbiogene with indicated levels of 3-amino-triazole (3AT). The working volume was 1.0 L and the dilution rate was 0.10 h−1. Cultivations were carried out at 30 °C with an agitation speed of 600 rpm, pH of 5.0, airflow rate of 1.0 standard L·min−1, and dissolved oxygen >80% air saturation. The bioreactors were fitted with cooled condensers (4 °C), and the off-gas was directed to a gas analyzer (INNOVA) for measurement of CO2 and O2. Steady-state samples were taken after 5 volume changes. Biomass concentration, extracellular metabolite concentrations, and carbon dioxide evolution rate were constant over at least 3 measurements taken before sampling.
Biomass Amino Acid C13 Enrichment Analysis.
C13 enrichment biomass was achieved by chemostat experiments performed in in-house-built reactors with a working volume of 200 mL. Cultivations were carried out at 30 °C with an agitation speed of 600 rpm, a dilution rate of 0.1 h−1, a pH of 5.0, and airflow rate of 1.0 standard L·min−1. Steady-state samples were taken after 5 volume changes. 100% of the glucose used was labeled in position 1 (1-13C glucose was from Omicron Biochemicals). The 13C-labeled biomass was harvested by centrifugation at 4,000 × g and 0 °C for 5 min. After centrifugation, the supernatant was poured off, and the cell pellet was frozen instantaneously in liquid nitrogen and stored at −80 °C. Hydrolysis, derivatization, and analysis were carried out as described by Antoniewicz et al. (29).
Metabolic Flux Determination.
Metabolic fluxes were determined using the Metran software, using the elementary metabolite units (EMU) algorithm (30, 31). Metran accepts as input a user-defined metabolic network model consisting of biochemical reactions and atom transitions, and a set of 13C-measurements and external fluxes; it produces as output metabolic fluxes for the entire network, confidence intervals for all fluxes, and statistical analysis of the goodness-of-fit (18).
Additional materials and methods on strains, shake flask growth conditions, whole genome expression analysis, biomass amino acid composition analysis, endometabolome analysis, and exometabolome analysis are in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank Ryan Kelley, Tong Ihn Lee, Scott McCuine, Chris Workman, Jose Aleman, Richard Young, and Gerald Fink for advice on experimental design and analysis. This work was supported by National Institutes of Health Grant 1R01 DK075850-01, the National Science Foundation International Research Fellowship Program, the Singapore–Massachusetts Institute of Technology Alliance, and National Center for Research Resources award 018627.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0811091106/DCSupplemental.
References
- 1.Uetz P, et al. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 2.Lee TI, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- 3.Harbison CT, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ideker T, et al. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science. 2001;292:929–934. doi: 10.1126/science.292.5518.929. [DOI] [PubMed] [Google Scholar]
- 5.Ideker T, Ozier O, Schwikowski B, Siegel AF. Discovering regulatory and signalling circuits in molecular interaction networks. Bioinformatics. 2002;18(Suppl 1):S233–240. doi: 10.1093/bioinformatics/18.suppl_1.s233. [DOI] [PubMed] [Google Scholar]
- 6.Helmstaedt K, Strittmatter A, Lipscomb WN, Braus GH. Evolution of 3-deoxy-d-arabino-heptulosonate-7-phosphate synthase-encoding genes in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2005;102:9784–9789. doi: 10.1073/pnas.0504238102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hinnebusch AG. Translational regulation of yeast GCN4. A window on factors that control initiator-trna binding to the ribosome. J Biol Chem. 1997;272:21661–21664. doi: 10.1074/jbc.272.35.21661. [DOI] [PubMed] [Google Scholar]
- 8.Stelling J, Klamt S, Bettenbrock K, Schuster S, Gilles ED. Metabolic network structure determines key aspects of functionality and regulation. Nature. 2002;420:190–193. doi: 10.1038/nature01166. [DOI] [PubMed] [Google Scholar]
- 9.Covert MW, Knight EM, Reed JL, Herrgard MJ, Palsson BO. Integrating high-throughput and computational data elucidates bacterial networks. Nature. 2004;429:92–96. doi: 10.1038/nature02456. [DOI] [PubMed] [Google Scholar]
- 10.Fong SS, Nanchen A, Palsson BO, Sauer U. Latent pathway activation and increased pathway capacity enable Escherichia coli adaptation to loss of key metabolic enzymes. J Biol Chem. 2006;281:8024–8033. doi: 10.1074/jbc.M510016200. [DOI] [PubMed] [Google Scholar]
- 11.Kresnowati MTAP, et al. When transcriptome meets metabolome: Fast cellular responses of yeast to sudden relief of glucose limitation. Mol Syst Biol. 2006;2:49. doi: 10.1038/msb4100083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hinnebusch AG. Mechanisms of gene regulation in the general control of amino acid biosynthesis in Saccharomyces cerevisiae. Microbiol Rev. 1988;52:248–273. doi: 10.1128/mr.52.2.248-273.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fazio A, et al. Transcription factor control of growth rate dependent genes in Saccharomyces cerevisiae: A three factor design. BMC Genomics. 2008;9:341. doi: 10.1186/1471-2164-9-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Natarajan K, et al. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol Cell Biol. 2001;21:4347–4368. doi: 10.1128/MCB.21.13.4347-4368.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jewett MC, Hofmann G, Nielsen J. Fungal metabolite analysis in genomics and phenomics. Curr Opin Biotechnol. 2006:191–197. doi: 10.1016/j.copbio.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 16.Sauer U. High-throughput phenomics: Experimental methods for mapping fluxomes. Curr Opin Biotechnol. 2004;15:58–63. doi: 10.1016/j.copbio.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 17.Schilling CH, Palsson BO. The underlying pathway structure of biochemical reaction networks. Proc Natl Acad Sci USA. 1998;95:4193–4198. doi: 10.1073/pnas.95.8.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Antoniewicz MR, Kelleher JK, Stephanopoulos G. Determination of confidence intervals of metabolic fluxes estimated from stable isotope measurements. Metab Eng. 2006:324–337. doi: 10.1016/j.ymben.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 19.Gombert AK, Moreira dos Santos M, Christensen B, Nielsen J. Network identification and flux quantification in the central metabolism of Saccharomyces cerevisiae under different conditions of glucose repression. J Bacteriol. 2001;183:1441–1451. doi: 10.1128/JB.183.4.1441-1451.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maaheimo H, et al. Central carbon metabolism of Saccharomyces cerevisiae explored by biosynthetic fractional (13)C labeling of common amino acids. Eur J Biochem. 2001;268:2464–2479. doi: 10.1046/j.1432-1327.2001.02126.x. [DOI] [PubMed] [Google Scholar]
- 21.Rossell S, et al. Unraveling the complexity of flux regulation: A new method demonstrated for nutrient starvation in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2006;103:2166–2171. doi: 10.1073/pnas.0509831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gygi SP, Rochon Y, Franza BR, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol Cell Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alfarano C, et al. The biomolecular interaction network database and related tools 2005 update. Nucleic Acids Res. 2005;33:D418–D424. doi: 10.1093/nar/gki051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones EW, Fink GR. In: The Molecular Biology of the Yeast Saccharomyces. Strathern JN, Jones EW, Broach JR, editors. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1982. pp. 181–289. [Google Scholar]
- 25.Shannon P, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vance W, Arkin A, Ross J. Determination of causal connectivities of species in reaction networks. Proc Natl Acad Sci USA. 2002;99:5816–5821. doi: 10.1073/pnas.022049699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gardner TS, di Bernardo D, Lorenz D, Collins JJ. Inferring genetic networks and identifying compound mode of action via expression profiling. Science. 2003;301:102–105. doi: 10.1126/science.1081900. [DOI] [PubMed] [Google Scholar]
- 28.Hall DA, et al. Regulation of gene expression by a metabolic enzyme. Science. 2004;306:482–484. doi: 10.1126/science.1096773. [DOI] [PubMed] [Google Scholar]
- 29.Antoniewicz MR, Kelleher JK, Stephanopoulos G. Accurate assessment of amino acid mass isotopomer distributions for metabolic flux analysis. Anal Chem. 2007;79:7554–7559. doi: 10.1021/ac0708893. [DOI] [PubMed] [Google Scholar]
- 30.Yoo H, Antoniewicz MR, Stephanopoulos G, Kelleher JK. Quantifying reductive carboxylation flux of glutamine to lipid in a brown adipocyte cell line. J Biol Chem. 2008;283:20621–20627. doi: 10.1074/jbc.M706494200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Antoniewicz MR, Kelleher JK, Stephanopoulos G. Elementary metabolite units (EMU): A novel framework for modeling isotopic distributions. Metab Eng. 2007;9:68–86. doi: 10.1016/j.ymben.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Delbecq P, Calvo O, Filipkowski RK, Pierard A, Messenguy F. Functional analysis of the leader peptide of the yeast gene CPA1 and heterologous regulation by other fungal peptides. Curr Genet. 2000;38:105–112. doi: 10.1007/s002940000140. [DOI] [PubMed] [Google Scholar]
- 33.Gaba A, Jacobson A, Sachs MS. Ribosome occupancy of the yeast CPA1 upstream open reading frame termination codon modulates nonsense-mediated mRNA decay. Mol Cell. 2005;20:449–460. doi: 10.1016/j.molcel.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 34.Iraqui I, Vissers S, Andre B, Urrestarazu A. Transcriptional induction by aromatic amino acids in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:3360–3371. doi: 10.1128/mcb.19.5.3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.