Abstract
The renewed interest in strategies to combat infectious agents with epidemic potential has led to a re-examination of vaccination protocols against smallpox. To help define which antigens elucidate a human antibody response, we have targeted proteins known or predicted to be presented on the surface of the intracellular mature virion (IMV) or the extracellular enveloped virion (EEV). The predicted ectodomains were expressed in a mammalian in vitro coupled transcription/translation reaction using tRNAlys precharged with lysine-ε-biotin followed by solid phase immobilization on 384 well neutravidin-coated plates. The generated array is highly specific and sensitive in a microELISA format. By comparison of binding of vaccinia-immune sera to the reticulocyte lysate-produced proteins and to secreted post-translationally-modified proteins, we demonstrate that for several proteins including the EEV proteins B5 and A33, proper recognition is dependent upon appropriate folding, with little dependence upon glycosylation per se. We further demonstrate that the humoral immune response to vaccinia among different individuals is not uniform in specificity or strength, as different IMV and EEV targets predominate within the group of immunogenic proteins. This heterogeneity likely results from the diversity of HLA Class II alleles and CD4 T helper cell epitopes stimulating B cell antibody production. Our findings have important implications both for design of new recombinant subunit vaccines as well as for methods of assaying the human antibody response utilizing recombinant proteins produced in vitro.
Keywords: vaccinia, protein array, humoral response
Introduction
The capacity for viruses and bacteria to rapidly evolve in response to environmental challenges, either natural or in response to therapeutics, leads to the inescapable conclusion that emerging infectious diseases remain a constant threat [1, 2]. This state has been exacerbated further by the likelihood of using infectious agents maliciously [3]. Raised awareness of a possible re-emergent smallpox epidemic is predicated upon ease of virus transmissibility, high associated mortality and the substantial increase in the susceptible population following cessation of routine vaccination in 1972 in the U.S.A. and by 1980 elsewhere [4]. To address the latter issue, the U.S.A. has an established stockpile of vaccinia virus available for vaccination and a series of guidelines for mass vaccination in the event of an outbreak [5, 6]. Despite the efficacy to induce effective immunity against the far more pathogenic smallpox variola virus by both the current Dryvax formulation derived from the New York City Board of Health vaccinia strain [7] and by the Lister strain used for the global eradication program [8], adverse reactions following vaccinia immunization have been noted. These may be life-threatening and include inadvertent inoculation, ocular or generalized vaccinia, eczema vaccinatum, progressive vaccinia and post-vaccinial encephalitis [9]. It has further been suggested that immunization with vaccinia may be contraindicated for up to 30% of the general population who are at increased risk; in particular, for those with immunocompromising conditions including atopic dermatitis or eczema, AIDS/HIV infection, leukemia, lymphoma or immunosuppressant therapy, as well as in pregnant women, children under 1 year and anyone with an allergic response to any component of the vaccine [10]. More recently, other adverse events observed include myo/pericarditis and dilated cardiomyopathy [11]. These events in general are associated with the use of a live replication-competent virus for vaccination thus development of attenuated/non-replicating virus preparations as vaccines have attracted great interest.
Amongst the prime candidates for vaccinia variants which do not replicate to produce further infectious virus in human cells is modified vaccinia Ankara (MVA) [12]. This form of the virus selected by propagation over multiple passages in chick embryo fibroblasts has adapted to enable replication in these latter cells. Through loss of approximately 15% of the genome, primarily from the 5′ and 3′ termini representing mostly host-range determining genes, the viral inoculate can still infect human cells, produce DNA, RNA and protein and can assemble into an immature particle which is non-infectious [13]. The response to vaccinia is mediated by both the humoral and cell-mediated arms of the immune system where the humoral response to MVA, at least in the early reports, seems to indicate that the breadth of reactivity to specific antigens is not substantially different to that observed in vaccinia-immunized individuals or that of archived sera from smallpox-infected individuals [14]. Likewise, a recent report suggests that the T cell responses generated by MVA are similar to those induced by Dryvax where both result in activation of polyfunctional CD8+ T cells with an unusual memory phenotype [15]. In all such investigations, due to the pathogenicity of variola, the response to vaccinia must be used as a proxy for the response to smallpox virus. Accordingly, amenable targets chosen for study must be substantially shared between studied vaccinia strains and variola.
An alternative to using replication-restricted MVA is to bypass the need for intact virus and administer a vaccine based upon recombinant protein or domain subunits expressing multiple identified epitopes able to induce a neutralizing antibody response [16]. Such an approach requires an informed understanding of the precise determinants to which an individual is responding, and assessment of whether the candidate antigen is a reasonable target for an effective neutralizing humoral immune response. Many studies to date have identified that effective antibody-based neutralization of vaccinia in animal models requires targeting of both intracellular mature virion (IMV) and extracellular enveloped virion (EEV) antigens [17–20]. As a consequence, we have evaluated the antigens encoded by the vaccinia genome in the light of published reports and extensive bioinformatics analysis to build a consensus picture of antigens likely to be presented on the surface of the two major infectious forms of the virus, the IMV involved predominantly in host-to-host-transfer, and the EEV which is more fragile but plays a major role in long range transfer within the host [21]. We have further analyzed predicted topology and organization of the selected proteins to produce the viral ectodomains and express them in a mammalian cytoplasmic-based format believed to likely preserve structure and antigenicity in a moderately high-throughput format for analysis of specific antibody responses. This ELISA-based format allows high sensitivity with minimal background interference and very high reliability and reproducibility. It is apparent from the results that this targeted approach reveals the common antigenicity of several vaccinia antigens while simultaneously demonstrating that human anti-vaccinia humoral immunity consists of significant variations in the level of response against individual antigens within the common antigenic group. We further observe that post-translational assembly, particularly of EEV antigens, is of paramount importance in assessing the antibody response, reinforcing the proposal that appropriate expression systems need to be chosen in producing antigen for utilization in screening arrays.
Materials/Methods
Vaccinia protein fusion constructs
Predicted ectodomains regions upstream or downstream of transmembrane domains and downstream of predicted signal peptides (Table S1) were amplified from a circular plasmid encoding the complete vaccinia Western Reserve (WR) strain genome (VAC-BAC WR10; kindly provided by Dr. B. Moss, NIH; [22]) using the Advantage 2 PCR system (Clontech, Mountain View CA). Following digestion with NotI and HindIII or KpnI, the cleaned amplified fragment was ligated into appropriately-digested pcDNA3.1-myc-His-A (Invitrogen, Carlsbad CA) and sequenced to ensure correct insertion.
Protein expression
Proteins were expressed from vaccinia ORF-pcDNA3.1-myc-His in a rabbit reticulocyte lysate-based coupled transcription/translation mix (TnT; Promega, Madison WI) supplemented with methionine (20μM final) and tRNAlys charged with lysine-ε-biotin (Transcend, Promega). Due to low [Mg2+] in the TnT protocol which did not favor subsequent translation of L1R, A4L and A39R trancripts, these 3 proteins were first transcribed under low Mg2+ conditions and generated mRNA was then used to prime a separate translation reaction with higher [Mg2+] (PROTEINscript II; Applied Biosystems, Foster City CA). Expression was confirmed by protein separation on a 10–20% SDS-PAGE gradient gel and transfer to PVDF followed by blocking in tris-buffered saline (TBS; pH8.0) containing 0.5% Tween 20. Biotinylated expressed protein was detected using streptavidin peroxidase (1:5000 in TBS containing 0.5% Tween 20; MP Biochemicals, Solon OH) followed by washing in TBS containing 0.05% Tween 20 (TBS-T; Sigma, St Louis MO) and chemiluminescent development (ECL system; GE Healthcare, Buckinghamshire UK).
Sera and recombinant proteins
Blood samples were obtained from 24 healthy volunteers immunized with vaccinia (Dryvax) utilizing an Institutional Review Board-approved protocol for that purpose, and after informed consent was obtained from each subject. The isolated sera were grouped as naïve (n=4), as naïve prior to immunization and assayed 1–2 months post-vaccination (n=9), as immunized more than 20 years previously, then boosted and assayed 1–2 months post-vaccination (n=8), as immunized more than 20 years previously and boosted >7 years previously (n=1), and as immunized >20 years previously and boosted ~1 year previously (n=2). Human hyperimmune anti-vaccinia immunoglobulin (VIg) was obtained from the Center for Disease Control (Atlanta GA). Two separate batches (both approximately 50mg/ml) were used which differed slightly in the quantitative reactivity (particularly to D13) but not in qualitative reactivity. Sera against varicella zoster (VZV), Hepatitis B secreted antigen (HBs), rabies, cytomegalovirus and naïve sera were obtained from healthy naïve individuals hyperimmunized with the appropriate vaccine/antigen (NABI Biopharmaceuticals, Rockville MD). All secondary HRP-labeled anti-human immunoglobulin reagents were obtained as F(ab′)2 preparations from Jackson Immunoresearch (West Grove PA). Recombinant A27 (amino acids 1–110), A33 (amino acids 58–185), B5 (amino acids 20–275) and L1 (amino acids 1–185) were obtained as glycosylated secreted proteins produced from a baculovirus construct in insect cells (Biodefense and Emerging Infections Research Resources Laboratory, Manassas VA; [23]). The glycosylated secreted proteins expressed from baculovirus constructs were deglycosylated, removing N-linked oligomannose, hybrid, biantennary and triantennary oligosaccharides while retaining native protein structure using endoglycosidases F1, F2 and F3 according to the manufacturer’s protocol (Sigma). Proteins and vaccinia virus lysates were denatured by addition of 0.5% trifluoroacetic acid.
Immunoprecipitations and serum pre-clearing
Biotinylated recombinant protein in the reticulocyte lysate was incubated with VIg (125μg/ml) followed by incubation at 20°C for 1h in a rotary shaker (900rpm). Following incubation, 20μl of Protein G-sepharose (50% in PBS) was added to the VIg-reticulocyte lysate mixture and incubated again at 20°C for 1h in a rotary shaker. Following 4 washes in TBS-T and one wash in TBS, x2 SDS-PAGE Laemmli buffer was added and separated on a SDS-PAGE 10–20% Tris-glycine gradient gel and processed for Western blotting as described above.
Serum pre-processing
Serum (50μl) was precleared of albumin using Cibacron blue F3GA-agarose pellets (SwellGel Blue; Pierce, Rockville IL) according to the manufacturer’s directions. To purify IgG from the samples, 25μl of serum was mixed with 500μl of high pH, high salt Protein A-binding buffer (Pierce) together with 100μl of Protein A-agarose slurry (50%) in the insert of a Spin-X tube (Costar-Corning, Corning NY) followed by rotation for 1h at 4°C. After centrifugation at 4000rpm for 1min the flowthrough (which could be used as IgG-depleted serum taking into account the 1:25 dilution) was removed and the packed slurry resuspended in 0.7ml binding buffer followed by 5min incubation with rotation at room temperature. After 3 further washes and centrifugations, the insert containing packed slurry was transferred to a clean microcentrifuge tube containing Tris-HCl (50μl; 1M, pH7.6). The beads were resuspended in glycine-HCl elution buffer (pH3), left for 5 min, then centrifuged through the insert into the Tris buffer followed by repetition with a further 100μl of elution buffer. Depending on the individual, 150–400μg of IgG could be recovered from each 25μl serum sample.
Array processing and assay conditions
Lysate containing biotin-labeled protein prepared as described above was diluted 1:15 in TBS-T and 40μl incubated in 384-well neutravidin-coated microELISA plates preblocked with Superblock (Pierce, Rockford IL; approximately 8–10ng labeled protein/well) for 24h after which unbound protein was washed away and the plates were extensively washed with Tris-buffered saline containing 0.05% Tween 20 (TBS-T) followed by incubation with human purified IgG (20μg/μl in TBS-T + 1% BSA) for 1.5h at room temperature. After 3 washes in TBS-T, wells were incubated with goat anti-human IgG alkaline phosphatase (1:10,000 in TBS-T + 1%BSA; Jackson Immunoresearch, West Grove PA) for 1h. After 3 further washes in TBS-T, bound alkaline phosphatase was detected by hydrolysis of pNPP (1 mg/ml in TBS; Sigma) and assayed at 405nm (VICTOR3 1420 multi-label counter; Perkin-Elmer, Shelton CT). Results were plotted as a heatmap generated by the JColorGrid program [24].
Neutralization Assays
Neutralizing antibody responses against vaccinia were measured using a luciferase-based assay in HeLa. This assay measures the reduction in luciferase reporter gene expression in target cells following a single-round of virus infection. The recombinant strain of vaccinia virus strain WR containing a luciferase reporter gene (VV:Luc) was a generous gift from Dr. David Bartlett (University of Pittsburgh). All stocks of vaccinia virus were grown on HeLa cells and purified by sucrose centrifugation as previously described [25]. Vaccinia virus titers were determined by plaque assay on CV-1 cells. For assessment of neutralizing antibody activity, 3-fold serial dilutions of plasma samples were performed in triplicate (96-well flat bottom plate) in 10% D-MEM growth media (100 μl/well). VV:Luc (1×105 pfu) was added to each well in a volume of 50μl and the plates were incubated for 1 hour at 37°C. HeLa cells were then added (1×105/well in 50μl volume) in 10% D-MEM growth medium to achieve a multiplicity of infection (MOI) of 1:1. Cytosine arabinofuranoside (Sigma, St. Louis, MO) was added at a final concentration of 20 μg/ml to prevent secondary rounds of infection. Assay controls included replicate wells of target cells alone (cell control) and target cells with virus (virus control). Following an overnight incubation at 37°C, 100μl of assay medium was removed from each well and 100μl of Bright-Glo luciferase reagent (Promega, Madison, WI) was added. The cells were allowed to lyse for 2 minutes, then 150μl of the cell lysate was transferred to a 96-well black solid plate and luminescence was measured using a Victor 3 luminometer (Perkin Elmer). The 50% inhibitory dose (ID50) titer was calculated as the serum dilution that caused a 50% reduction in relative luminescence units [26] compared to the virus control wells after subtraction of cell control RLUs. VIg was utilized as a positive control reagent for all neutralization assays performed.
To determine inhibition of EEV cell-to-cell infectivity by anti-vaccinia antisera, the comet tail inhibition assay was used [27]. Briefly, BSC-40 cell monolayers were established in 1ml cultures in 12 well plates, and infected with vaccinia virus strain IHDJ at ~100 PFU/well for 2h at 37°C. The inoculum was removed and cells washed with complete medium followed by culture in 1ml DMEM containing 2% FCS with or without anti-vaccinia sera (1:50). After 30h at 37°C, cells were fixed and stained by direct addition to the culture of 0.25ml 0.15% crystal violet/11.1% formaldehyde/4.75% ethanol for 1h at room temperature. The supernatant was removed and the wells washed in distilled water and dried prior to image analysis.
Data processing
For every experiment, background binding to control surfaces was subtracted. These surfaces consisted of untreated wells, wells treated with diluted reticulocyte lysate identical to test samples but no plasmid DNA added, and wells treated with diluted reticulocyte lysate in which biotinylated luciferase was produced. All experiments incorporated up to 4 replicates and if variation around the mean exceeded 10%, and suspect well data could be definitively identified, this result was removed. All results were plotted as heat maps using the JColorgrid javascript module [24]. The significance of the linear regression correlation co-efficient was derived from the Pearson co-efficient using the t statistic.
Results
Selection and production of vaccinia target antigens
To ascertain the antigen specificities of the humoral response to orthopox viruses, several parameters are paramount. First, the specificities represented and assayed in vaccinia should also be represented in variola virus for which vaccinia is a surrogate. Second, the antigens assayed in vitro should mimic as closely as possible the topology of presentation on the orthopox virus. Third, focus should be on determining which antigens are likely to be the targets of a neutralizing antibody response. With these parameters in mind, we selected a subset of 25 of the 218 open reading frames encoded in the vaccinia strain WR genome based on their likelihood of being exposed on the outer surface of the IMV or EEV, or on an already known antigenicity or target for a neutralizing response, or on bioinformatic prediction of likely expression on the virus surface (Figure 1). It is not yet established whether all the potential predicted vaccinia open reading frames (ORF, defined as greater than 50 amino acids) encode subsequently translated protein. Consequently, we focused upon those proteins for which there was well-established evidence for protein translation either by mass spectroscopic analysis of peptides from purified virus preparations, by functional abnormalities in mutant virus, or by antibody-mediated detection (Table S1). Although included initially, F13, A32 and A34 were never seen to be targets of antibody recognition in any of our subsequent assays and thus were omitted in most subsequent testing.
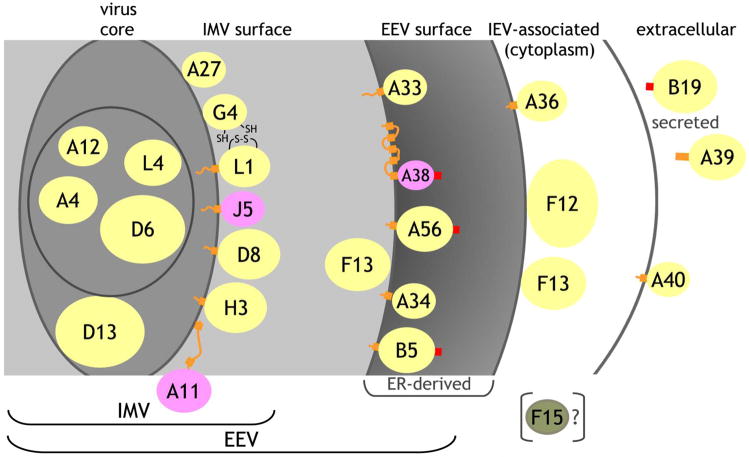
Figure 1. Virus topology of vaccinia proteins used for array.
Based on prior publications and bioinformatics analysis (Table S1), proteins were assigned to the virus core, to the inner membrane of the IMV, to the IMV surface, to the EEV surface, to the outer membrane of the IEV, or as secreted from the host cell. For each protein, the yellow (experimentally demonstrated topology), pink (predicted) or green (F15) ellipse represents the domain synthesized and used in the array where the colored areas are proportional to the spherical size of each protein relative to the others. Not included in the constructed proteins are cytoplasmic domains or short inter-transmembrane regions (orange lines), transmembrane domains (orange blocks), or signal peptides (red blocks). A11 partitions predominantly in the cell but some may associate with the IMV, where the two indicated predicted transmembrane regions do not insert in the IMV membrane. The topology of A38 places it as a Type I integral membrane protein facing into the ER but it has not been unequivocally demonstrated to be on the EEV surface. A39 is secreted in the Copenhagen strain, but is membrane-bound in the vaccinia WR strain. B19 is secreted from the host cell. F15 is not part of the IMV but its topology is uncertain.
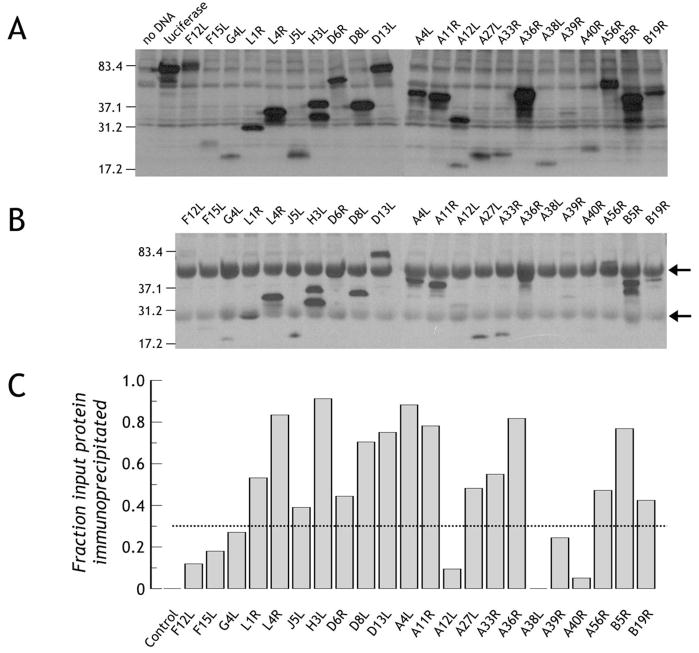
In order to mimic the mammalian cytoplasmic environment used by vaccinia virus for all stages of development [28], we used a rabbit reticulocyte lysate system for protein production, allowing synthesis of soluble protein from DNA within 90 minutes. Many of the vaccinia proteins contain significant hydrophobic domains, either as single or multiple transmembrane domains, or simply as long hydrophobic stretches ([29, 30], Figure 1, Table S1). To maximize solubility and minimize aggregation of the synthesized proteins during synthesis in vitro, predicted hydrophobic signal peptides and transmembrane domains were eliminated from the protein-encoding transcripts (Table S1). The required sequences were amplified from vaccinia strain WR using a forward primer encoding a start codon embedded in an optimum Kozak sequence and products were ligated upstream of a myc-His(6) tag in a mammalian expression vector. Coupled transcription-translation (TnT) in vitro of the products is depicted in Figure 2A. Detection of protein was achieved through use of an HRP-labeled-streptavidin binding to the biotin incorporated during translation as a lysine ε-amino-adduct. Consequently, the strength of signal is predominantly dependent upon both the size of the protein and the relative representation of lysines, where % incorporation of the labeled to unlabeled lysine averages 20% [31]. Indicating labeling efficiency, the A33R construct sequence incorporates only 8 lysines, yet a good signal is detected at the expected Mr of ~20kDa while F12L carries the greatest number of lysines (84) and generates a strong signal at the expected of Mr of ~80kDa (Figure 2A). Incorporation of biotins, tending to be on exposed hydrophilic surfaces, has minimal effect upon protein function and structure [32] allowing use of the biotin not only as a label but also as an affinity tag facilitating downstream array immobilization (described below).
Figure 2. Production of vaccinia proteins and their immunoprecipitation with VIg.
A. Production of biotin-labeled vaccinia protein domains. The desired vaccinia domains were produced incorporating tRNAlys precharged with ε-amino-biotinylated lysine. Synthesized proteins were detected with streptavidin-horse radish peroxidase. B. Immunoprecipitation of synthetically labeled proteins by human anti-vaccinia antibody. Identical amounts of protein to those loaded and detected in panel A were incubated with human anti-vaccinia hyperimmune IgG (VIg) and bound biotinylated protein was immunoprecipitated using Protein G-agarose. Following gel electrophoresis and transfer, bound protein was detected as in panel A. Arrows represent human Ig H and L chains, respectively. C. Broad reactivity of VIg to the synthetic antigen array. For each protein in panel A and panel B, the band relative intensity was normalized to luciferase run on the same gel. Since identical amounts of luciferase were run on each gel, the ratio of immunoprecipitated protein (panel B) to total protein loaded (panel A) gives a broad measure of VIg reactivity to specific antigens. Using a conservative cut-off threshold of 0.35 (indicated by the dotted line, strong reactivity is seen to L1, L4, J5, H3, D6, D8, D13, A4, A11, A27, A33, A36, A56, B5 and B19. Control is luciferase precipitated by VIg.
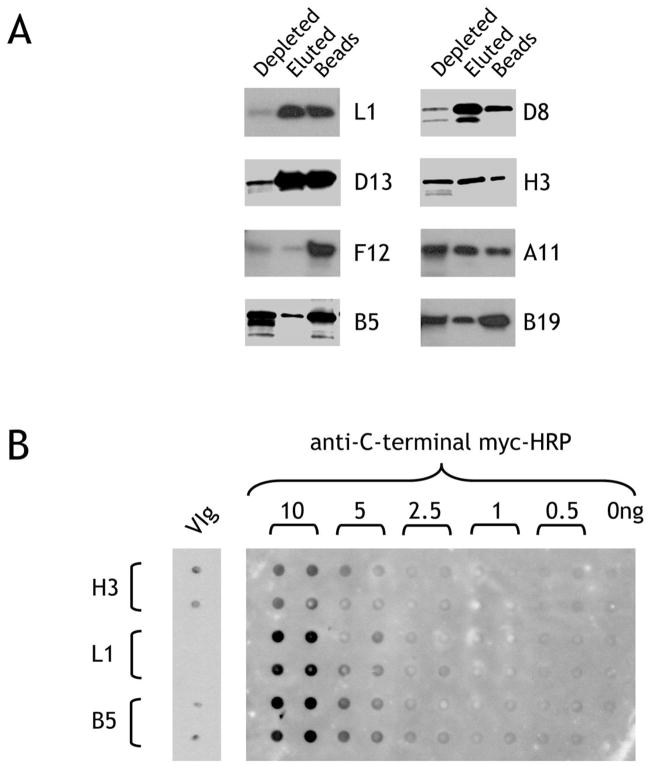
Although all the produced proteins contain a His tag, conventional nickel-based affinity media do not chelate His-tagged proteins efficiently from a reticulocyte lysate due to simultaneous pulldown of hemoglobin to the exclusion of the protein of interest. We therefore used zinc-charged magnetic beads to effectively chelate His-tagged protein with undetectable hemoglobin binding. As depicted in Figure 3A, this worked well for some proteins (L1, D8 and D13, for example) where significant amounts of protein could be eluted from the chelating magnetic beads. For other proteins, however, the lysate could be efficiently depleted but non-specific protein-bead interactions occurred which prevented elution except by harsh denaturing methods (e.g. F12) while for other proteins only a small amount could be efficiently isolated and eluted (e.g. A11, B5, B19 and H3). For efficiently recovered protein, yields were approximately 100–200ng protein/50μl reticulocyte lysate (2–4 μg/ml). To confirm that full-length protein with a C-terminal myc tag was produced, purified protein was immobilized on nitrocellulose and could be detected with anti-myc antibody (Figure 3B).
Figure 3. Purification of His(6)-tagged proteins and reactivity on nitrocellulose.
A. Purification of vaccinia proteins synthesized in vitro. Reticulocyte lysate containing the newly-synthesized proteins were incubated with MagZ magnetic beads to bind the His(6)-tagged proteins. After washing and elution, the depleted lysate, eluate, or beads were run on a reducing SDS-PAGE gel and biotinylated protein detected by binding of streptavidin-peroxidase. B. Immobilization on nitrocellulose may affect the presentation of antigenic epitopes. Recombinant biotin-labeled H3, L1 and B5 vaccinia proteins purified by His-tag affinity were spotted in quadruplicate on nitrocellulose and probed with anti-myc-HRP directed against the C-terminal myc tag followed by chemiluminescent assay (right-hand panel). The same proteins were blotted in duplicate (10ng/spot) on a separate blot, incubated with VIg and detected with anti-human IgG-HRP.
Antigenicity of in vitro-produced proteins
Despite production of full-length protein, binding of a hyperimunized human anti-vaccinia IgG (VIg) preparation to the nitrocellulose-immobilized proteins clearly revealed significant differences: H3 was well detected relative to the total amount immobilized, a relatively small amount of B5 was detected and anti-L1 activity was undetectable (Figure 3B, left-hand panel). Although this could reflect proportions of antibody within the VIg preparation, solid-phase immobilization may effect the 3-dimensional structure of the antigen, mask the epitopes or, alternatively, post-translational modifications may be required to properly duplicate the antigenic determinants displayed on the virus in vivo. To ascertain whether our proteins are originally in a conformation recognizable by antibody but altered on nitrocellulose immobilization, we then tested the ability of VIg to immunoprecipitate the proteins in solution. In this assay, the VIg interacts directly with the biotinylated protein in the reticulocyte lysate followed by immunoprecipitation and Western detection. Under these conditions, and by comparing the relative amount of biotin-labeled immunoprecipitated protein to the amount initially included in the assay, it was demonstrated that the VIg at high concentration (~125μg/ml) was able to efficiently immunoprecipitate 15 of 22 vaccinia proteins (Figure 2B and 2C), including the L1 which could not be detected when immobilized on nitrocellulose. To overcome the possibility that solid-phase immobilization may alter structure or mask epitopes, we examined whether we could duplicate the antigenicity retained in solution using a solid-phase format by immobilizing the biotinylated vaccinia protein with plastic-bound neutravidin, potentially holding the vaccinia protein away from the hydrophobic plastic surface and orienting it towards the aqueous phase. In this assay, the diluted reticulocyte lysate could be added directly to neutravidin-coated pre-blocked wells, capturing the biotinylated protein whereas all non-specific proteins (after dilution, approximately 15mg/ml consisting predominantly of hemoglobin) could be efficiently washed away.
Biotinylated vaccinia proteins retain antigenicity when immobilized though interaction with neutravidin
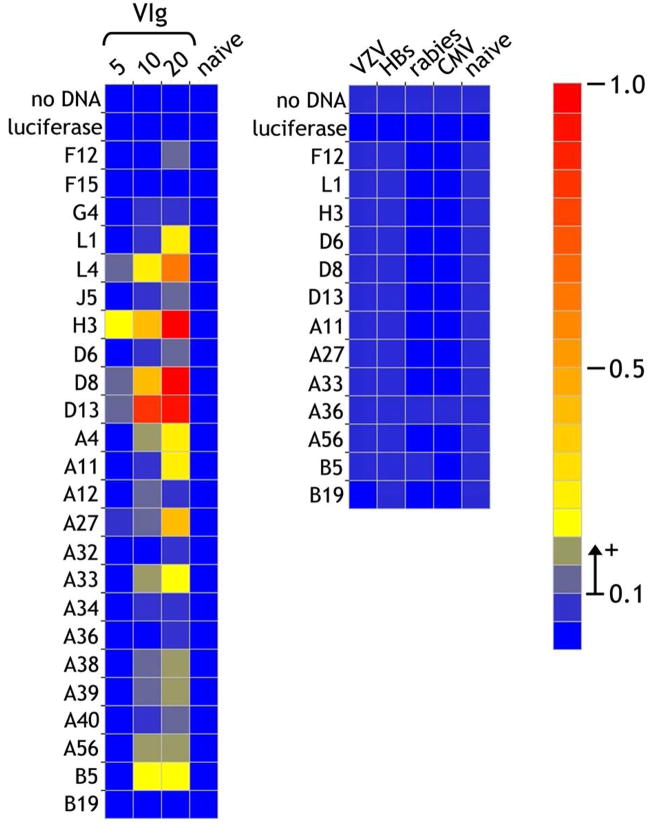
The vaccinia protein-coated surfaces were then incubated with VIg (5–20μg/ml). Following detection of bound antibody, sera from a naïve individual was negative against all antigens arrayed while increasing VIg concentration resulted in a clear dose-response with significant responses against L1, L4, H3, D8, D13, A4, A11, A27, A33, A38, A39, A56 and B5 and weak responses against F12, J5, D6 and A40 (Figure 4). This compares very favorably with the proteins detected by immunoprecipitation where, using VIg at the higher concentration of 125μg/ml, only 2 other antigens (A36 and B19) were detected. Conversely, the only antigen weakly detected by the ELISA technique and not observed by immunoprecipitation was F12. Importantly, known neutralizing antigens such as L1 which could not be detected when bound as native protein to nitrocellulose could be detected in the ELISA format, lending support to the proposal that hydrophobic interactions on the nitrocellulose might alter or mask the antigenic surface. Based on these results, now replicated more than 19 times with a mean standard deviation of 1.787% of the mean for all antigens, we felt that the assay could reliably duplicate the general pattern of reactivity observed using the more labor-intensive and variability-prone immunoprecipitation technique.
Figure 4. Detection of VIg antigenic reactivities using the ELISA protein array.
Vaccinia proteins produced by in transcription/translation in vitro were immobilized on neutravidin-coated plates through the biosynthetically-incorporated biotin tags. In the left-hand panel, VIg was incubated at 5, 10 and 20μg/ml and bound antibody detected by binding of anti-human IgG alkaline phospahatase activity. The highest concentration of 20μg/ml corresponds to a dilution of 1/2,500 from the stock. Immunoglobulin (20μg/ml) from a naïve individual was included as a control. As a further control for specificity, IgG from sera of individuals hyperimmunized against other microorganisms [varicella zoster (VZV), Hepatitis B secreted antigen (HBs), rabies and cytomegalovirus (CMV)] were also tested at 20μg/ml. The scale for the heatmap indicates optical density at 405nm detected in the ELISA assay. A value of >0.1 O.D. units above background was considered as significantly positive, while responses of 0.05–0.1 were considered borderline.
Although reactivity was negative in naïve individuals, it was important to demonstrate selective specificity and that the increased reactivity in hyperimmune individuals was not a manifestation of a general humoral immune response activation. Testing IgG from sera of initially naïve individuals hyper-immunized to varicella zoster, rabies, Hepatitis B secreted antigen and cytomegalovirus established that these individuals mounting a validated micro-organism-specific response did not generate a response to any of the vaccinia antigens used in our ELISA format (Fig. 4).
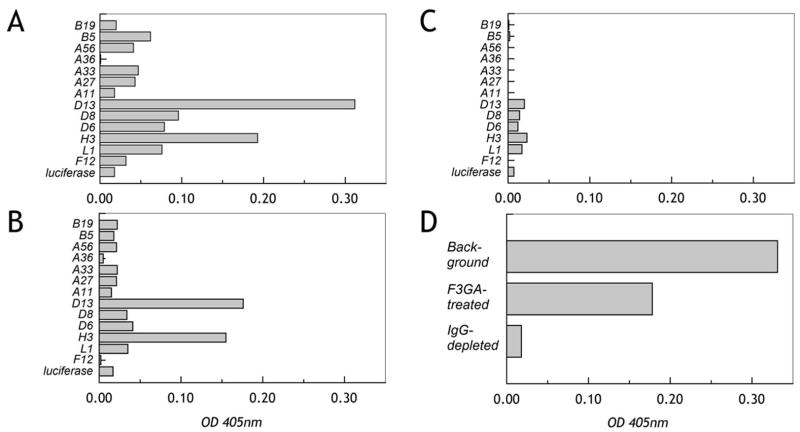
Purified immunoglobulin rather than diluted sera was used in this latter experiment since it was noted using even significantly-diluted sera that there was a consistent individual-specific background binding to the control surface (following incubation with reticulocyte lysate but no vaccinia protein) and an equivalent binding to the luciferase-coated surface produced as a control protein. This background was very apparent at serum dilutions of 1/150 or less. A recent study analyzing vaccinated sera responses to arrayed recombinant vaccinia proteins produced in E. coli found that a suitable working dilution range for sera was 1/50 – 1/100 [33]. Nevertheless, given that significant end-point titers for apparent major vaccinia antigens are in the range 1/200 to 1/10,000, while naïve background levels are already in the 1/50 range [34], it was felt that serum pretreatment/dilution could be tolerated. We found that pre-treatment of small serum samples with Cibacron blue (F3GA)-agarose to remove albumin and other common non-immunoglobulin serum proteins had no effect upon reducing the background and even reduced the specific Ig response to some degree (Figure 5A, B). Conversely, removal of IgG using Protein A-agarose not only completely removed the anti-vaccinia signal but also effectively removed all background (Figure 5C, D). Thus the variable individual background seemed to be a property of the IgG itself and may represent individual variations in natural polyreactive antibodies with hydrophobic interaction properties [35]. In analyzing multiple sera, we found that the average variation around the mean for replicates was <4%; using Protein A-purified IgG, the average variation fell to less than 2% and the smaller variation allowed greater sensistivity and reliability for lower range interaction values after subtracting background. Furthermore, by comparison of sera and isolated IgG, we could confirm that in our assay almost the entire reactivity resided in the IgG rather than IgM compartment.
Figure 5. Sera background binding is an intrinsic property of IgG.
A. Specific response, following background subtraction, to a subset of recombinant vaccinia antigens on the micro-ELISA array using sera from a vaccinated individual (1/300). B. Following Cibacron Blue (F3GA)-agarose treatment of the sera to remove albumin, specific responses with background subtracted are also reduced indicating partial removal of the IgG component. C. Following pretreatment of the sera with Protein A-agarose removing the IgG component, all specific anti-vaccinia protein reactivity is removed. D. For this individual, the background independent of any antigen presence was high; removal of albumin reduced the background by almost 0.5, but reduced the IgG signal by a similar amount (see panel B). Removal of the IgG component alone eliminated the background entirely.
The use of an array of more than 20 antigens is not readily amenable to extensive serial dilution analysis but with selection of an appropriate uniform dilution, can give a panoramic profile of reactivity for a particular individual. Accordingly, we chose a standard rapid protocol for isolation of IgG from 25μl of serum, and used this IgG in all assays at a standard 20μg/ml corresponding to a serum dilution of 1/300 – 1/750 depending upon the donor. The results in our assay thus represent a plane through the spectrum of reactivity where very low borderline reactivities close to background titer may be missed. In one study [34], end-point titers in primary immunized individuals at d21 post-vaccination ranged from 1/126 to 1/1213 while VIg end-point titer hovered between 1:50–1:1000 depending on antigen, whereas in our assay, VIg diluted 1:2,500 or more gave robust results (Figure 4), thus it was felt that our selected dilution range should retain sensitivity while minimizing background.
Detection of antigen reactivities in vaccinia-immunized individuals
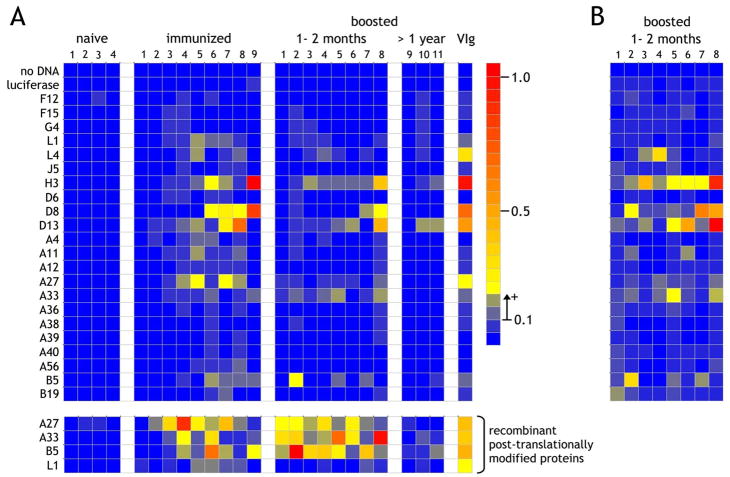
The assay platform was then extended to test responses of a number of individuals classified as naïve (“naïve” in Figure 6A), previously naïve, immunized and assayed 1–2 months post-primary vaccination (“immunized”), and subjects immunized more than 20 years previously, then boosted and assayed 1–2 months post-vaccination (“boosted, 1–2 months”) or assayed >1 year post-vaccination (“boosted, >1 year”). As expected, the naïve individuals were unreactive and only 2 antigen reactivities (H3 and D13) were detectable in the ELISA array for individuals boosted more than a year prior to analysis. In the primary immunized individuals, reactivity was observed against L1, L4, H3, D8, D13, A4, A11, A33, A56, B5 and B19 with maximum intensities not significantly different to those observed using VIg at 10μg/ml. Nevertheless, responses were not observed in all individuals and the array of antigen specificities varied widely from individual to individual.
Figure 6. Detection of anti-vaccinia reactivity from naïve, primary immunized, and immunized and boosted individuals.
A. Purified IgG at 20μg/ml from 4 naïve individuals, 9 immunized individuals and 11 individuals vaccinated in childhood and subsequently boosted were assessed for vaccinia protein reactivity on the micro-ELISA array. Of the boosted individuals, one was tested more than 7 years after the boost and two were tested approximately 1 year after boost. All other vaccinated and boosted individuals were tested one to two months after the last exposure. VIg at 10μg/ml was used as a standard positive control. The lowest 4 rows separated from the main array indicate results for the same IgG preparations against recombinant A27, A33, B5 and L1 produced as secreted post-translationally-modified proteins by insect cells. All 4 antigens were immobilized at 200ng/well on uncoated plates followed by blocking with TBS-BSA (1%), after which the procedure was identical to that used for the reticulocyte lysate-produced proteins. B. The individuals tested at 1–2 months following boost were retested using IgG at 100μg/ml.
Surprisingly, in boosted individuals the responses against the arrayed antigens were low using the standardized concentration of 20 μg/ml where strong recall responses might be expected. To address whether the IgG range we were testing was simply too low, we reperformed the analysis using an IgG concentration of 100μg/ml for the immunized and recently-boosted individuals (Figure 6B). Clearly the initial analysis was both correct and specific since the responses are predominantly amplifications of the previous responses observed using the lower IgG concentration but with signal intensities now within the range of those observed for the primary immunized group. Additionally, with the possible exception of A11 and B19, no new antigenic specificities were detected, further demonstrating the specificity even at higher concentrations. Nevertheless, these results were obtained on considerably amplified backgrounds mandating against general use of high IgG/sera concentrations in this format.
Comparitive antigenicity of lysate-produced and post-translationally modified proteins
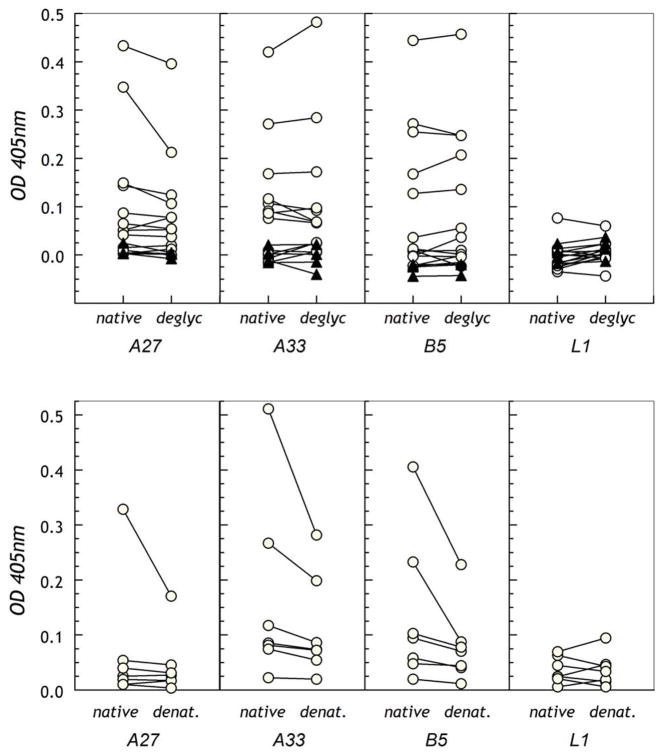
A consistent concern for all studies expressing vaccinia antigens in vitro relates to structural integrity of the proteins expressed during viral morphogenesis [33, 34, 36]. Several studies have used E. coli-produced proteins where degrees of reactivity appear to be maintained. Further, in at least one instance, E. coli-expressed L1R appeared to gain reactivity from an essentially non-reactive state if it was correctly disulfide-bonded [37]. Using any cell-free lysate for protein synthesis, no significant post-translational modifications except for disulfide bonding enabled by the virally-encoded cytoplasmic redox system are expected for proteins expressed on the IMV. Nevertheless, for EEV proteins where the likely virally-expressed ectodomains are processed within the lumen of the ER, extensive disulfide bonding and glycosylation events are unlikely to occur in a cell-free system. To address this issue, we compared the reactivity of the same sera groups on the array against recombinant secreted and post-translationally-modified A27, A33, B5 and L1 (Figure 6A, lower panels). It is readily apparent that the responses against L1 from either source are low and not significantly different (post-translationally modified responses 1.26-fold greater on average). Although an increase against A27, A33 and B5 is detected using protein from either source, the responses against A27, A33 and B5 are stronger using the mature proteins (x4.76, x5.09 and x6.63, respectively) (Figure S1). This suggests that at least for these antigens, the reticulocyte lysate-expressed proteins are detecting only a subset of reactivity, and that a significant component of the immune IgG is directed against post-translationally-added structures or conformational epitopes induced by structured-folding. Further, although responses to the arrayed antigens appear weaker in the boosted individuals, this is not the case for their response to the recombinant post-translationally modified proteins, suggesting that the recall response is focusing down upon those epitopes presented as they might be on the intact virus, particularly the EEV form.
We examined the reactivity of deglycosylated proteins to determine whether loss of glycosylation without denaturing the proteins led to reduction in immune sera reactivity (Figure 7A and Figure S2). In a natural infection, both A33 and B5 are likely to be glycosylated and treatment with endoglycosidase F1 leads to a significant reduction in molecular weights indicating removal of N-linked sugar. Treatment with endoglycosidase F2/F3 to remove complex biantennary and triantennary structures had minimal effect upon any of the proteins, as expected for glycoproteins secreted by insect cells [38]. Although there is some glycosylation of L1 and A27 consequent to being produced as recombinant secreted protein, this is not their natural state. With the exception of a single A27 reactivity, removal of N-linked sugar did not effect antigenicity, indicating that the anti-vaccinia antibody response is predominantly directed against peptidic regions of the antigens rather than against glycosylated regions or segments altered by glycosylation. Nevertheless, denaturation of the proteins with trifluoroacetic acid (TFA) in situ during protein immobilization resulted in a strong reduction in reactivity of those positive sera (Figure 7B). This suggests that structurally-constrained epitopes rather than linear sequence epitopes are a significant component of the humoral immune response against vaccinia proteins.
Figure 7. Optimal antigenic reactivity of sera requires post-translationally-modified or properly-folded target proteins.
A. Untreated or endoglycosidase F1-treated proteins were immobilized on uncoated 384-well micro-ELISA plates and reactivity of purified IgG was assessed. B. Untreated or TFA-treated proteins were immobilized on ELISA plates for testing of reactivity of IgG from selected vaccinated individuals. In both panels, the dotted line indicates the cut-off for positivity where 0.05–0.1 is considered borderline and above 0.1 significantly positive.
To assess relative reactivities against native and denatured virally-incorporated proteins, we probed nitrocellulose-immobilized non-denatured viral lysates and the identical lysates denatured by exposure to TFA (0.5%) with sera from immune and recently-boosted individuals. In all instances, the sera reacted better against native viral lysate than against denatured lysate by densitometric analysis of the signal, with the increase ranging from 29.2% to 73.3% (Figure S3).
Antigen selection in the human humoral response to vaccinia
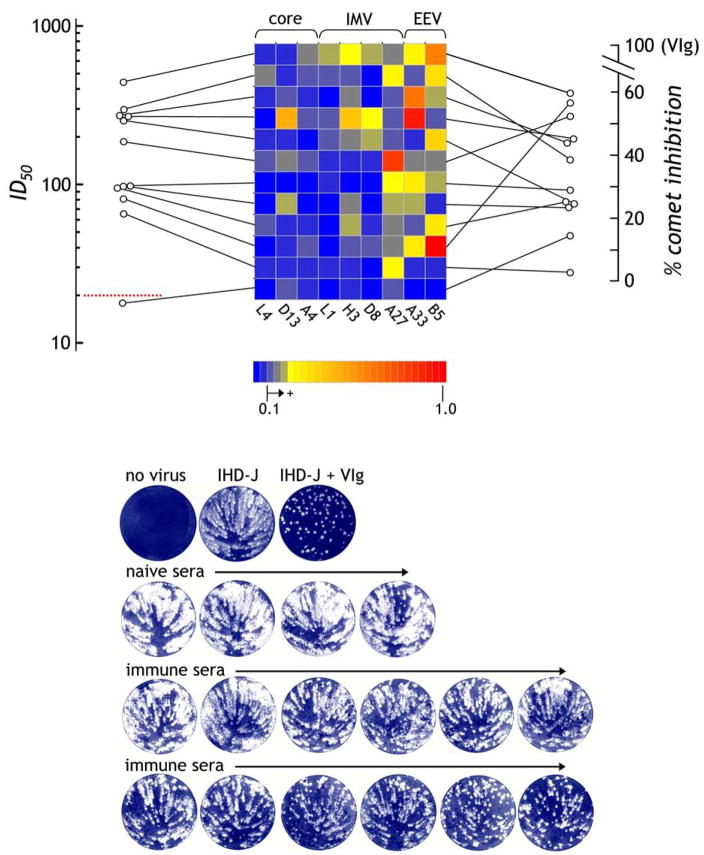
Both using the cell-free-produced protein array and the insect-produced protein, we observed that the individual response to the selected antigens is highly variable among subjects (Figure 6). These data suggest that the human immune system can target multiple determinants where the variable responses, as long as they represent a selection of both IMV and EEV antigens, may lead to effective viral neutralization. Nevertheless, particular antigens stand out as consistent targets for both immune sera and VIg preparations. To examine the individual responses to these targets in further detail, responses to proteins localized to the viral core (L4, D13, A4), to the IMV surface (L1, H3, D8, A27) and to the EEV surface (A33, B5) were correlated to IMV or EEV neutralizing activities (Figure 8). Utilizing a luciferase-encoding vaccinia virus to measure antibody neutralization of IMV, the individual with the broadest reactivity against the 4 IMV antigens has the highest functional serum ID50 titer (Figure 8). In fact, despite the low titers for anti-L1 reactivity, all 4 of the serum samples with detectable anti-L1R reactivity are ranked in the top 5 for neutralizing activity. Nevertheless, at least one individual with good titer has minimal response against L1 but weak to moderate responses against A27 and H3, both of which are known to contribute towards IMV neutralization [39, 40], while another has only responses to A27. Some individuals ranked low for IMV neutralization have good reactivity to EEV antigens. To determine whether this translates into efficient EEV neutralizing activity, inhibition of EEV comet formation was undertaken. As depicted in Figure 8 (right-hand scale), the two individuals with the highest anti-B5 activity rank highest for EEV comet inhibition while the two individuals with no detectable B5 or A33 activity rank lowest. In between the top- and bottom-ranked, however, there is a mix of reactivity profiles with no obvious pattern although the general correlation between IMV and EEV neutralization in vitro, respectively, for these subjects is strong (r = 0.79, P < 0.005).
Figure 8. The human antibody response to vaccinia target antigens is individually diverse but still leads to effective neutralization titers.
Based on data in Figure 6, the responses of vaccinees to the dominantly recognized antigens are depicted, with each representing one individual. For each individual, the responses are connected by a line to the left indicating the corresponding ID50 for vaccinia infection (VV:Luc) of HeLa cells representing IMV neutralization. The red dotted line (ID50 = 20) represents background, where 20–30 ID50 is considered borderline and >30 significantly positive. To the right of each row, a line connects to the % inhibition of comet formation representing EEV activity. Using image densitometry, the VIg responses were set to 100% inhibition and the responses to naïve sera were used to set the zero baseline. The presumed virus topology of each antigen is indicated at the top of array. In the lower panel, the representative comet inhibition responses are depicted where the 12 immune sera are ordered in increasing comet inhibition activity following incubation of BSC-40 cells with IHD-J EEV-forming virus (100pfu).
Discussion
The development of new vaccine strategies for protection against smallpox has accelerated in part due to the realization that large segments of the population have never been immunized, that stocks of the vaccinia vaccine are limited and that the replication-competent formulation is contraindicated for a significant fraction of the population [5]. In addition to investigating attenuated viruses, recent advances in high throughput protein production and protein array screening [41], together with the complete sequencing of the variola, vaccinia and related orthopox genomes, have opened up the possibility of expressing all open reading frames to screen immune sera. Identified targets of specific reactivities can then be experimentally investigated as potential vaccine immunogens. In this report, we take a selective approach to producing vaccinia target antigens known or bioinformatically-predicted to be expressed on the surface of vaccinia virus in either of its infectious forms. By screening for reactivities only against proteins shared between vaccinia and variola, the inference is that a neutralizing target for vaccinia is likely to be a neutralizing target for variola. This latter inference is qualified, however, by the observation that small sequence differences, as observed between vaccinia B5R and variola B6R, may lead to a significant anti-B5 response which neutralizes vaccinia EEV but does not recognize variola B6 [42]. We then progress to show that proteins expressed in an in vitro cell lysate system are produced as full-length proteins which retain antigenicity but may not retain the full antigenicity of protein expressed in the context of virus. Nevertheless, the technique is specific and sensitive and, in the context of appropriate folded and post-translationally modified proteins where relevant, provides a window into the profiles of antigenic reactivity which develop in response to vaccinia. It is clear that the target specificities detected are very much in accordance with those already identified as possible targets in animal models, but unlike these latter studies usually carried out on genetically-related subjects, the individual human response does not readily conform to a pattern. Essentially, each individual selects reactivity from an apparently restricted pool of antigens but where intensities against each antigen are not uniform.
In this study, we find that within the 22 antigens already selected from the more than 200 potentially expressed by the vaccinia virus, hyperimmune human anti-vaccinia IgG reacts strongly with 10 antigens and less so but significantly with a further seven. These antigens can be grouped by topological presentation. Of the viral core antigens studied, we find reactivity against L4, D13, A4 and D6. These antigens are not viable candidates for neutralizing targets being inaccessible to humoral attack against the intact virion. We find reactivity against A11 and A40, where the former is expressed within the cell while the latter is expressed on the cell membrane and neither appears to be expressed on the IMV or EEV [43]. The activity against B19 is of interest since it is not incorporated into the virion but is secreted and likely functions as a decoy secreted IFN-α/β receptor, where inhibition of its function may augment host anti-orthopox immune responses [44]. Nevertheless, reactivity against B19 was infrequent in the vaccinated cohort tested here. Of the IMV antigens assessed, VIg reactivity is observed against L1, H3, D8, A27 and J5. In several models, A27 and L1 have already been established as antibody targets. In contrast, H3 and D8, both likely functioning as glycosaminoglycan receptors involved in IMV cell entry, have only recently become appreciated as possible targets [17, 45, 46]. Interestingly, J5 has not yet been considered as a target; it’s function is unknown but it may be involved in host range restriction. Efforts to express J5 as a recombinant protein secreted from cells have been completely unsuccessful since it appears to be cytotoxic [47]. Regarding EEV antigens, we find VIg reactivity against B5, A33, A38, A39 and A56 where strongest and most prevalent reactivity is directed against B5 and A33, in accordance with previously-published results [48, 49]. The reactivity of sera from immunized or boosted individuals is less strong than that of the pooled hyperimmune sera when compared at equivalent immunoglobulin concentrations. Nevertheless, the pattern of reactivity is very similar and only responses to core antigen D6, IMV antigen J5, possible EEV antigen A38 and cell-surface antigen A40 are missing.
Based on non-human model studies, efficient antibody neutralization of vaccinia requires immunoglobulin specificity directed against both IMV and EEV antigens [17]. Several studies have reported that various combinations of IMV and EEV antigens may be used, usually consisting of a mix of two or more of A27, A33, B5, L1 and, more recently, D8 to induce a protective immunity to orthopox virus infection [17, 19, 50–53]. These studies tend to use genetically-similar animal models when comparing control and treated groups. Given the genetic diversity of the human population, however, where both HLA Class II alleles at the DR, DP and DQ loci as well as immunoglobulin receptor repertoire may have a critical influence upon epitope selection, a minimalist approach looking for the least number of antigens required to induce neutralizing antibody in animal studies may not be applicable to the more diverse human population. This appears to be the case here, where a strong broad response to the antigens assessed provides a good indicator of both IMV and EEV neutralizing activity while a uniformly weak response correlates with poor neutralizing activity (Figure 8). Nevertheless, between these limits, antigen selectivity is not uniform and no consistent pattern is observed. This is very much in accord with a recently-published report suggesting that there is a redundancy and plasticity to the human humoral immune response to vaccinia [36]. Decision tree analysis of our results supports even a weak L1 response as being a primary determinant of IMV neutralization in vitro (Brusic V; personal communication) reinforcing the concept that the qualitative rather than quantitative antibody reactivity is most critical, with D8 reactivity playing a significant secondary role. As found here, it is generally accepted that B5 is probably the strongest target for neutralization of the EEV. In fact, in a recent study where reactivity to 3 EEV antigens was determined (A33, A56 and B5), it was postulated that anti-B5 reactivity was the only important EEV determinant as far as neutralization in vitro is concerned [34]. Against this notion, however, B5R-deleted mutant vaccinia was able to elicit protection equivalent to that of the parental B5R-expressing virus in both monkey and mouse models [54, 55]. Furthermore, as shown in our vaccinated cohort, we find that in at least one individual, an insignificant B5 response in the context of a high A33 response is associated with a strong EEV neutralizing titer. Examination of Figure 1 suggests that likely IMV and EEV neutralizing targets are limited in number and our results are concordant with this prediction.
The observation that boosted individuals appear to have weaker responses relative to primary immunized individuals against the arrayed antigens, including the very immunogenic H3 and D8 IMV proteins, suggests two non-mutually exclusive possibilities. First, that boosting focuses the immune response against EEV rather than IMV proteins, as is seen for the A33 and B5 reactivities using recombinant cell-secreted proteins as targets. Second, that the recall reponse focuses upon the proper topological arrangement of correctly-folded proteins on the viral surface. In this regard, chemical denaturation of target proteins leads to a significant decrease in antibody reactivity. That there remains some reactivity after unfolding could either indicate a mixed antibody response directed at segmented and conformational epitopes or else that some degree of protein refolding occurred after removal of the denaturant prior to antibody incubation.
The issue of antigen integrity on high throughput protein arrays is critical for assessment of humoral reactivity and for subsequent identification of target epitopes. We find evidence here that solid phase immobilization itself may mask epitopes, and consequently chose an immobilized neutravidin format which holds the biotinylated vaccinia protein away from the hydrophobic surface. This assay provided results similar to that observed by immunoprecipitation in solution. Although we used a mammalian cell-free reticulocyte-based protein synthesis procedure to mimic the viral replicative environment, no ER-based post-translational modifications including disulfide bond formation or glycosylation will occur. Several studies have used E. coli-produced proteins where degrees of reactivity appear to be maintained [14, 33, 37]. In this format, E. coli-expressed L1R appeared to gain reactivity from an essentially non-reactive state if it was correctly disulfide-bonded using a modified reaction [37]. In contrast, we find that our L1R produced in vitro is comparable in reactivity to that against the correctly-folded protein (Figure S1). This is in accord with another study demonstrating no difference in immune sera reactivity to E. coli-produced L1 and to correctly-folded L1 used for crystallography, where responses were as low as those observed here [34].
Several platforms have been introduced recently to methodically address the specificity of the human humoral immune response to vaccinia. First, using a regular ELISA assay and appropriately produced ectodomains of three IMV proteins (A27, H3, L1) and three EEV proteins (A33, A56, B5) with proper post-translational folding and glycosylation, responses of a large number of individuals were analyzed with results presented as pooled data masking individual profiles [34]. In this format, it was readily apparent that antibody end-point titers fell rapidly within 6 months of immunization or boosting, in some instances approaching pre-immune levels by 1 year. We also find a rapid decline post-immunization for samples presented here. It is possible that borderline titers may be close enough to that background that our cut-off plane misses them. A further observation from the 6 antigen study mentioned above is that neutralizing titers against whole vaccinia remain much higher for significantly longer than against the individual proteins [34]. This raises possibilities of preferential antigenicity of proteins presented in the context of virus and/or antigenic specificities remaining to be discovered. Against this latter contention, using our expanded platform focused upon viral surface-presented proteins, we do not identify any new candidate antigens outside of those already-discovered, with the possible exception of J5 detected by VIg preparations. Even in this latter instance, although predicted to orient on the IMV membrane, we have no prima facie evidence that J5 is not in fact oriented internally. Despite the rapid fall of anti-vaccinia virus neutralizing or inhibitory antibody titer within a year post-vaccination, titers then seem to plateau for decades [26]. From our result, titers against individual antigens appear to fall rapidly after immunization and are almost undetectable several years later, leaving open to further speculation what might be the causes for differences between the sum of the individual antigen responses on comparison with the antibody response against intact virus.
A second platform recently described has been designed as a genuine high-throughput analysis system with reactivities measured against individual proteins spotted on microarray slides [14, 33, 37]. In this format, unlike the present study, no account is taken of structure, topology or misfolding due to retention of signal peptides and transmembrane domains. The complete sequence of every open reading frame is expressed in its entirety as an E. coli protein. This global analysis increases the likelihood of picking up responses outside of the group of usual suspects but at the expense of missing reactivity against correctly-folded protein. Further, since all humans tend to have an anti-E. coli IgG response, the test sera have to be incubated with E. coli lysate prior to incubation on the protein arrays which may inadvertently remove specificities directed against vaccinia. The microarray format based on the full E. coli-expressed vaccinia proteome requires incubation with antibody at dilutions significantly close to the background titer (1/50–1/100), a level for which we find the background signal may be overwhelming. In this report, we have tried here to find a middle ground between the various assay platforms by making an informed choice of target antigen, thus increasing the pool of likely targets to be tested without the need for introducing E. coli protein. This approach works well for IMV proteins where significant responses can be detected with good sensitivity, but it is clear that for EEV proteins, isolation of appropriately-processed recombinant secreted protein is preferred for development of high throughput assays. Using this system, we clearly demonstrate that the vaccinia antigen reactivity selection by the human immune system is broad with no uniform pattern, and may well be influenced by genetic background, particularly Class II MHC alleles. This is of critical importance for development of recombinant subunit vaccines with the likelihood that the most effective vaccines will be those based on several IMV and EEV proteins.
Supplementary Material
Figure S1. Comparitive antigenicity of recombinant non-post-translationally modified vaccinia proteins and mature secreted recombinant proteins. To compare antigenicity of the reticulocyte lysate-produced proteins to the recombinant baculovirus-expressed secreted post-translationally-modified proteins, for each IgG sample from a vaccinated individual, the response against each of the 4 relevant antigens (A27, A33, B5 and L1) was plotted. Value below 0.05 were considered insignificant, from 0.05 to 0.1 were considered borderline, and above 0.1 were considered significantly positive. Note that the reactivity against L1 was similar (regression line is plotted with a correlation coefficient (r) of 0.8) while for A27, A33 and B5, binding was significantly stronger to the mature proteins.
Figure S2. Deglycosylation of recombinant A27, A33, B5 and L1. As described in the Methods section, secreted recombinant proteins (10μg) were incubated with endoglycosidase F1, or endoclycosidases F2/F3, for 1h at 37°C, then separated by SDS-PAGE (10–20% gradient gel) and stained with Coomassie Blue. As expected, A33 and B5 are extensively N-glycosylated, and these structures could be removed with endoglycosidase F1.
Figure S3. A significant fraction of the human antibody response is directed to vaccinia antigens in their native conformation. VIg binding to denatured vaccinia strain WR (sucrose gradient-purified) 1% NP-40 detergent lysate detected by Western blotting (left panel). Native and TFA-denatured protein lysates of vaccinia strain WR (~10μg) were immobilized on nitrocellulose, blocked and probed with sera (1/1000) from a naïve individual, recently-immunized individuals (2 samples), recently-boosted (2 samples) and VIg (1μg/ml) (right panel). Consistency of viral lysate protein binding was confirmed by Coomassie Blue-staining of the unblocked dot blots.
Comparison of native to denatured reactivities by densitometry:
Donor b (immune): +42.3%
Donor c (immune): +29.2%
Donor d (boosted): +73.3%
Donor e (boosted): +40.4%
Sample f (VIg): +39.3%
Acknowledgments
The following reagents were obtained through the NIH Biodefense and Emerging Infections Research Resources Repository, NIAID, NIH: Vaccinia Virus (WR) A27L, A33R, B5R and L1R proteins with C-terminal histidine tag produced as recombinant baculovirus proteins. This work was supported by NIH grant 5U19-AI057330 (ELR).
Abbreviations
- CEV
cell-associated enveloped virion
- EEV
extracellular enveloped virion
- ER
endoplasmic reticulum
- HBs
Hepatitis B secreted antigen
- HRP
horseradish peroxidase
- IEV
intracellular enveloped virion
- IMV
intracellular mature virion
- ORF
open reading frame
- TBS-T
Tris-buffered saline containing 0.1% Tween 20
- TFA
trifluoroacetic acid
- TnT
coupled transcription-translation
- VIg
Human hyperimmune anti-vaccinia IgG
- VV
Luc, vaccinia virus strain WR expressing a luciferase reporter gene
- VZV
varicella zoster
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- 1.Fauci AS. Emerging infectious diseases: a clear and present danger to humanity. JAMA. 2004 Oct 20;292(15):1887–8. doi: 10.1001/jama.292.15.1887. [DOI] [PubMed] [Google Scholar]
- 2.Morens DM, Folkers GK, Fauci AS. The challenge of emerging and re-emerging infectious diseases. Nature. 2004 Jul 8;430(6996):242–9. doi: 10.1038/nature02759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fauci AS, Touchette NA, Folkers GK. Emerging infectious diseases: a 10-year perspective from the National Institute of Allergy and Infectious Diseases. Emerg Infect Dis. 2005 Apr;11(4):519–25. doi: 10.3201/eid1104.041167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lane JM, Goldstein J. Evaluation of 21st-century risks of smallpox vaccination and policy options. Ann Intern Med. 2003 Mar 18;138(6):488–93. doi: 10.7326/0003-4819-138-6-200303180-00014. [DOI] [PubMed] [Google Scholar]
- 5.Rotz LD, Dotson DA, Damon IK, Becher JA. Vaccinia (smallpox) vaccine: recommendations of the Advisory Committee on Immunization Practices (ACIP), 2001. MMWR Recomm Rep. 2001 Jun 22;50(RR10):1–25. quiz CE1–7. [PubMed] [Google Scholar]
- 6.Henderson DA, Inglesby TV, Bartlett JG, Ascher MS, Eitzen E, Jahrling PB, et al. Smallpox as a biological weapon: medical and public health management. Working Group on Civilian Biodefense. JAMA. 1999 Jun 9;281(22):2127–37. doi: 10.1001/jama.281.22.2127. [DOI] [PubMed] [Google Scholar]
- 7.Frey SE, Newman FK, Cruz J, Shelton WB, Tennant JM, Polach T, et al. Dose-related effects of smallpox vaccine. N Engl J Med. 2002 Apr 25;346(17):1275–80. doi: 10.1056/NEJMoa013431. [DOI] [PubMed] [Google Scholar]
- 8.Strassburg MA. The global eradication of smallpox. Am J Infect Control. 1982 May;10(2):53–9. doi: 10.1016/0196-6553(82)90003-7. [DOI] [PubMed] [Google Scholar]
- 9.Aragon TJ, Ulrich S, Fernyak S, Rutherford GW. Risks of serious complications and death from smallpox vaccination: a systematic review of the United States experience, 1963–1968. BMC Public Health. 2003 Aug 11;3:26. doi: 10.1186/1471-2458-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cono J, Casey CG, Bell DM. Smallpox vaccination and adverse reactions. Guidance for clinicians. MMWR Recomm Rep. 2003 Feb 21;52(RR4):1–28. [PubMed] [Google Scholar]
- 11.Morgan J, Roper MH, Sperling L, Schieber RA, Heffelfinger JD, Casey CG, et al. Myocarditis, pericarditis, and dilated cardiomyopathy after smallpox vaccination among civilians in the United States, January-October 2003. Clin Infect Dis. 2008 Mar 15;46(Suppl 3):S242–50. doi: 10.1086/524747. [DOI] [PubMed] [Google Scholar]
- 12.McCurdy LH, Larkin BD, Martin JE, Graham BS. Modified vaccinia Ankara: potential as an alternative smallpox vaccine. Clin Infect Dis. 2004 Jun 15;38(12):1749–53. doi: 10.1086/421266. [DOI] [PubMed] [Google Scholar]
- 13.Sutter G, Staib C. Vaccinia vectors as candidate vaccines: the development of modified vaccinia virus Ankara for antigen delivery. Curr Drug Targets Infect Disord. 2003 Sep;3(3):263–71. doi: 10.2174/1568005033481123. [DOI] [PubMed] [Google Scholar]
- 14.Davies DH, Wyatt LS, Newman FK, Earl PL, Chun S, Hernandez JE, et al. Antibody profiling by proteome microarray reveals the immunogenicity of the attenuated smallpox vaccine modified vaccinia virus ankara is comparable to that of Dryvax. J Virol. 2008 Jan;82(2):652–63. doi: 10.1128/JVI.01706-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Precopio ML, Betts MR, Parrino J, Price DA, Gostick E, Ambrozak DR, et al. Immunization with vaccinia virus induces polyfunctional and phenotypically distinctive CD8(+) T cell responses. J Exp Med. 2007 Jun 11;204(6):1405–16. doi: 10.1084/jem.20062363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakhatskyy P, Wang S, Zhang C, Chou TH, Kishko M, Lu S. Immunogenicity and protection efficacy of subunit-based smallpox vaccines using variola major antigens. Virology. 2008 Feb 5;371(1):98–107. doi: 10.1016/j.virol.2007.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fogg C, Lustig S, Whitbeck JC, Eisenberg RJ, Cohen GH, Moss B. Protective immunity to vaccinia virus induced by vaccination with multiple recombinant outer membrane proteins of intracellular and extracellular virions. J Virol. 2004 Oct;78(19):10230–7. doi: 10.1128/JVI.78.19.10230-10237.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fogg CN, Americo JL, Lustig S, Huggins JW, Smith SK, Damon I, et al. Adjuvant-enhanced antibody responses to recombinant proteins correlates with protection of mice and monkeys to orthopoxvirus challenges. Vaccine. 2007 Apr 12;25(15):2787–99. doi: 10.1016/j.vaccine.2006.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hooper JW, Custer DM, Thompson E. Four-gene-combination DNA vaccine protects mice against a lethal vaccinia virus challenge and elicits appropriate antibody responses in nonhuman primates. Virology. 2003 Feb 1;306(1):181–95. doi: 10.1016/S0042-6822(02)00038-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sakhatskyy P, Wang S, Chou TH, Lu S. Immunogenicity and protection efficacy of monovalent and polyvalent poxvirus vaccines that include the D8 antigen. Virology. 2006 Nov 25;355(2):164–74. doi: 10.1016/j.virol.2006.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boulter EA, Appleyard G. Differences between extracellular and intracellular forms of poxvirus and their implications. Prog Med Virol. 1973;16:86–108. [PubMed] [Google Scholar]
- 22.Domi A, Moss B. Cloning the vaccinia virus genome as a bacterial artificial chromosome in Escherichia coli and recovery of infectious virus in mammalian cells. Proc Natl Acad Sci U S A. 2002 Sep 17;99(19):12415–20. doi: 10.1073/pnas.192420599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao Y, Aldaz-Carroll L, Ortiz AM, Whitbeck JC, Alexander E, Lou H, et al. A protein-based smallpox vaccine protects mice from vaccinia and ectromelia virus challenges when given as a prime and single boost. Vaccine. 2007 Jan 26;25(7):1214–24. doi: 10.1016/j.vaccine.2006.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joachimiak MP, Weisman JL, May B. JColorGrid: software for the visualization of biological measurements. BMC Bioinformatics. 2006;7:225. doi: 10.1186/1471-2105-7-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kotwal GJ, Abrahams M-R. Growing poxviruses and determining virus titer. In: Isaacs SN, editor. Vaccinia virus and poxvirology: methods and protocols. Totowa NJ: Humana Press Inc; 2004. pp. 101–12. [DOI] [PubMed] [Google Scholar]
- 26.Hammarlund E, Lewis MW, Hansen SG, Strelow LI, Nelson JA, Sexton GJ, et al. Duration of antiviral immunity after smallpox vaccination. Nat Med. 2003 Sep;9(9):1131–7. doi: 10.1038/nm917. [DOI] [PubMed] [Google Scholar]
- 27.Aldaz-Carroll L, Whitbeck JC, Ponce de Leon M, Lou H, Hirao L, Isaacs SN, et al. Epitope-mapping studies define two major neutralization sites on the vaccinia virus extracellular enveloped virus glycoprotein B5R. J Virol. 2005 May;79(10):6260–71. doi: 10.1128/JVI.79.10.6260-6271.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schramm B, Locker JK. Cytoplasmic organization of POXvirus DNA replication. Traffic. 2005 Oct;6(10):839–46. doi: 10.1111/j.1600-0854.2005.00324.x. [DOI] [PubMed] [Google Scholar]
- 29.Katsafanas GC, Moss B. Colocalization of transcription and translation within cytoplasmic poxvirus factories coordinates viral expression and subjugates host functions. Cell Host Microbe. 2007 Oct 11;2(4):221–8. doi: 10.1016/j.chom.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith GL, Vanderplasschen A, Law M. The formation and function of extracellular enveloped vaccinia virus. J Gen Virol. 2002 Dec;83(Pt 12):2915–31. doi: 10.1099/0022-1317-83-12-2915. [DOI] [PubMed] [Google Scholar]
- 31.Kurzchalia TV, Wiedmann M, Breter H, Zimmermann W, Bauschke E, Rapoport TA. tRNA-mediated labelling of proteins with biotin. A nonradioactive method for the detection of cell-free translation products. Eur J Biochem. 1988 Mar 15;172(3):663–8. doi: 10.1111/j.1432-1033.1988.tb13940.x. [DOI] [PubMed] [Google Scholar]
- 32.Promega. Transcend non-radioactive translation detection systems. Technical Bulletin TB182: Promega Corporation, 2006.
- 33.Davies DH, Liang X, Hernandez JE, Randall A, Hirst S, Mu Y, et al. Profiling the humoral immune response to infection by using proteome microarrays: high-throughput vaccine and diagnostic antigen discovery. Proc Natl Acad Sci U S A. 2005 Jan 18;102(3):547–52. doi: 10.1073/pnas.0408782102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Putz MM, Midgley CM, Law M, Smith GL. Quantification of antibody responses against multiple antigens of the two infectious forms of Vaccinia virus provides a benchmark for smallpox vaccination. Nat Med. 2006 Nov;12(11):1310–5. doi: 10.1038/nm1457. [DOI] [PubMed] [Google Scholar]
- 35.Plagemann PG, Rowland RR, Cafruny WA. Polyclonal hypergammaglobulinemia and formation of hydrophobic immune complexes in porcine reproductive and respiratory syndrome virus-infected and uninfected pigs. Viral Immunol. 2005;18(1):138–47. doi: 10.1089/vim.2005.18.138. [DOI] [PubMed] [Google Scholar]
- 36.Benhnia MR, McCausland MM, Su HP, Singh K, Hoffmann J, Davies DH, et al. Redundancy and plasticity of neutralizing antibody responses are cornerstone attributes of the human immune response to the smallpox vaccine. J Virol. 2008 Apr;82(7):3751–68. doi: 10.1128/JVI.02244-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davies DH, Molina DM, Wrammert J, Miller J, Hirst S, Mu Y, et al. Proteome-wide analysis of the serological response to vaccinia and smallpox. Proteomics. 2007 May;7(10):1678–86. doi: 10.1002/pmic.200600926. [DOI] [PubMed] [Google Scholar]
- 38.Hollister J, Grabenhorst E, Nimtz M, Conradt H, Jarvis DL. Engineering the protein N-glycosylation pathway in insect cells for production of biantennary, complex N-glycans. Biochemistry. 2002 Dec 17;41(50):15093–104. doi: 10.1021/bi026455d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davies DH, McCausland MM, Valdez C, Huynh D, Hernandez JE, Mu Y, et al. Vaccinia virus H3L envelope protein is a major target of neutralizing antibodies in humans and elicits protection against lethal challenge in mice. J Virol. 2005 Sep;79(18):11724–33. doi: 10.1128/JVI.79.18.11724-11733.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He Y, Manischewitz J, Meseda CA, Merchlinsky M, Vassell RA, Sirota L, et al. Antibodies to the A27 protein of vaccinia virus neutralize and protect against infection but represent a minor component of Dryvax vaccine--induced immunity. J Infect Dis. 2007 Oct 1;196(7):1026–32. doi: 10.1086/520936. [DOI] [PubMed] [Google Scholar]
- 41.Reid JD, Parker CE, Borchers CH. Protein arrays for biomarker discovery. Curr Opin Mol Ther. 2007 Jun;9(3):216–21. [PubMed] [Google Scholar]
- 42.Aldaz-Carroll L, Xiao Y, Whitbeck JC, de Leon MP, Lou H, Kim M, et al. Major neutralizing sites on vaccinia virus glycoprotein B5 are exposed differently on variola virus ortholog B6. J Virol. 2007 Aug;81(15):8131–9. doi: 10.1128/JVI.00374-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilcock D, Duncan SA, Traktman P, Zhang WH, Smith GL. The vaccinia virus A4OR gene product is a nonstructural, type II membrane glycoprotein that is expressed at the cell surface. J Gen Virol. 1999 Aug;80( Pt 8):2137–48. doi: 10.1099/0022-1317-80-8-2137. [DOI] [PubMed] [Google Scholar]
- 44.Alcami A, Symons JA, Smith GL. The vaccinia virus soluble alpha/beta interferon (IFN) receptor binds to the cell surface and protects cells from the antiviral effects of IFN. J Virol. 2000 Dec;74(23):11230–9. doi: 10.1128/jvi.74.23.11230-11239.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hsiao JC, Chung CS, Chang W. Vaccinia virus envelope D8L protein binds to cell surface chondroitin sulfate and mediates the adsorption of intracellular mature virions to cells. J Virol. 1999 Oct;73(10):8750–61. doi: 10.1128/jvi.73.10.8750-8761.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin CL, Chung CS, Heine HG, Chang W. Vaccinia virus envelope H3L protein binds to cell surface heparan sulfate and is important for intracellular mature virion morphogenesis and virus infection in vitro and in vivo. J Virol. 2000 Apr;74(7):3353–65. doi: 10.1128/jvi.74.7.3353-3365.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zajac P, Spehner D, Drillien R. The vaccinia virus J5L open reading frame encodes a polypeptide expressed late during infection and required for viral multiplication. Virus Res. 1995 Jul;37(2):163–73. doi: 10.1016/0168-1702(95)00025-l. [DOI] [PubMed] [Google Scholar]
- 48.Bell E, Shamim M, Whitbeck JC, Sfyroera G, Lambris JD, Isaacs SN. Antibodies against the extracellular enveloped virus B5R protein are mainly responsible for the EEV neutralizing capacity of vaccinia immune globulin. Virology. 2004 Aug 1;325(2):425–31. doi: 10.1016/j.virol.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 49.Law M, Smith GL. Antibody neutralization of the extracellular enveloped form of vaccinia virus. Virology. 2001 Feb 1;280(1):132–42. doi: 10.1006/viro.2000.0750. [DOI] [PubMed] [Google Scholar]
- 50.Berhanu A, Wilson RL, Kirkwood-Watts DL, King DS, Warren TK, Lund SA, et al. Vaccination of BALB/c mice with Escherichia coli-expressed vaccinia virus proteins A27L, B5R, and D8L protects mice from lethal vaccinia virus challenge. J Virol. 2008 Apr;82(7):3517–29. doi: 10.1128/JVI.01854-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heraud JM, Edghill-Smith Y, Ayala V, Kalisz I, Parrino J, Kalyanaraman VS, et al. Subunit recombinant vaccine protects against monkeypox. J Immunol. 2006 Aug 15;177(4):2552–64. doi: 10.4049/jimmunol.177.4.2552. [DOI] [PubMed] [Google Scholar]
- 52.Hooper JW, Custer DM, Schmaljohn CS, Schmaljohn AL. DNA vaccination with vaccinia virus L1R and A33R genes protects mice against a lethal poxvirus challenge. Virology. 2000 Jan 20;266(2):329–39. doi: 10.1006/viro.1999.0096. [DOI] [PubMed] [Google Scholar]
- 53.Hooper JW, Thompson E, Wilhelmsen C, Zimmerman M, Ichou MA, Steffen SE, et al. Smallpox DNA vaccine protects nonhuman primates against lethal monkeypox. J Virol. 2004 May;78(9):4433–43. doi: 10.1128/JVI.78.9.4433-4443.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morikawa S, Sakiyama T, Hasegawa H, Saijo M, Maeda A, Kurane I, et al. An attenuated LC16m8 smallpox vaccine: analysis of full-genome sequence and induction of immune protection. J Virol. 2005 Sep;79(18):11873–91. doi: 10.1128/JVI.79.18.11873-11891.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saijo M, Ami Y, Suzaki Y, Nagata N, Iwata N, Hasegawa H, et al. LC16m8, a highly attenuated vaccinia virus vaccine lacking expression of the membrane protein B5R, protects monkeys from monkeypox. J Virol. 2006 Jun;80(11):5179–88. doi: 10.1128/JVI.02642-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Comparitive antigenicity of recombinant non-post-translationally modified vaccinia proteins and mature secreted recombinant proteins. To compare antigenicity of the reticulocyte lysate-produced proteins to the recombinant baculovirus-expressed secreted post-translationally-modified proteins, for each IgG sample from a vaccinated individual, the response against each of the 4 relevant antigens (A27, A33, B5 and L1) was plotted. Value below 0.05 were considered insignificant, from 0.05 to 0.1 were considered borderline, and above 0.1 were considered significantly positive. Note that the reactivity against L1 was similar (regression line is plotted with a correlation coefficient (r) of 0.8) while for A27, A33 and B5, binding was significantly stronger to the mature proteins.
Figure S2. Deglycosylation of recombinant A27, A33, B5 and L1. As described in the Methods section, secreted recombinant proteins (10μg) were incubated with endoglycosidase F1, or endoclycosidases F2/F3, for 1h at 37°C, then separated by SDS-PAGE (10–20% gradient gel) and stained with Coomassie Blue. As expected, A33 and B5 are extensively N-glycosylated, and these structures could be removed with endoglycosidase F1.
Figure S3. A significant fraction of the human antibody response is directed to vaccinia antigens in their native conformation. VIg binding to denatured vaccinia strain WR (sucrose gradient-purified) 1% NP-40 detergent lysate detected by Western blotting (left panel). Native and TFA-denatured protein lysates of vaccinia strain WR (~10μg) were immobilized on nitrocellulose, blocked and probed with sera (1/1000) from a naïve individual, recently-immunized individuals (2 samples), recently-boosted (2 samples) and VIg (1μg/ml) (right panel). Consistency of viral lysate protein binding was confirmed by Coomassie Blue-staining of the unblocked dot blots.
Comparison of native to denatured reactivities by densitometry:
Donor b (immune): +42.3%
Donor c (immune): +29.2%
Donor d (boosted): +73.3%
Donor e (boosted): +40.4%
Sample f (VIg): +39.3%