Abstract
Theory predicts that partially asexual organisms may make the “best of both worlds”: for the most part, they avoid the costs of sexual reproduction, while still benefiting from an enhanced efficiency of selection compared to obligately asexual organisms. There is, however, little empirical data on partially asexual organisms to test this prediction. Here we examine patterns of nucleotide diversity at eight nuclear loci in continentwide samples of two species of cyclically parthenogenetic Daphnia to assess the effect of partial asexual reproduction on effective population size and amount of recombination. Both species have high nucleotide diversities and show abundant evidence for recombination, yielding large estimates of effective population sizes (300,000–600,000). This suggests that selection will act efficiently even on mutations with small selection coefficients. Divergence between the two species is less than one-tenth of previous estimates, which were derived using a mitochondrial molecular clock. As the two species investigated are among the most distantly related species of the genus, this suggests that the genus Daphnia may be considerably younger than previously thought. Daphnia has recently received increased attention because it is being developed as a model organism for ecological and evolutionary genomics. Our results confirm the attractiveness of Daphnia as a model organism, because the high nucleotide diversity and low linkage disequilibrium suggest that fine-scale mapping of genes affecting phenotypes through association studies should be feasible.
THE efficacy of natural selection may be severely reduced in asexual compared to sexual organisms due to the absence of recombination and segregation (Fisher 1930; Barton and Charlesworth 1998; Otto and Lenormand 2002; Agrawal 2006). Consequently, asexual populations may adapt more slowly to changing environments (Peck 1994; Orr 2000; Roze and Barton 2006) and suffer from an increased genetic load (Muller 1964; Crow and Kimura 1970; Pamilo et al. 1987; Kondrashov 1988; Charlesworth 1994). Both of these factors may contribute to the rarity of obligate asexuality in eukaryotes (Bell 1982), despite its immediate advantages over sexual reproduction (Maynard Smith 1978). The main reason for the decreased efficiency of selection in asexual organisms is that due to the complete linkage of their genomes, selection cannot operate on different mutations independently (the Hill–Robertson effect, Hill and Robertson 1966). Thus, deleterious mutations anywhere in the genome reduce the effective population size (Ne) experienced by other loci (Hill and Robertson 1966; Felsenstein 1974; Charlesworth 1994; Keightley and Otto 2006), resulting in a predicted reduction in neutral genetic variation and accumulation of slightly deleterious alleles, due to inefficacy of selection on mutations with small selection coefficients.
However, even a small amount of sexual reproduction is predicted to greatly alleviate the disadvantages of pure asexual reproduction while conserving most of its advantages (Lynch and Gabriel 1983; Pamilo et al. 1987; Charlesworth et al. 1993; Green and Noakes 1995; Hurst and Peck 1996). Thus, partially asexual organisms should have Ne and neutral genetic diversity similar to that of obligately sexual organisms of similar body size, abundance, and geographic range. The predominance of obligate sexual life cycles is thus surprising and remains unexplained (Hadany and Beker 2007). Partially asexual life cycles, with only occasional rounds of sexual reproduction, occur in many invertebrates and fungi and may provide insights into the fitness and genomic consequences of occasional sex (Bell 1982). However, few studies of neutral genetic diversity exist for partially asexual organisms, and thus their Ne and levels of recombination are largely unknown (e.g., Hughes and Verra 2001; Delmotte et al. 2002; D'souza and Michiels 2006).
The genus Daphnia belongs to a clade of brachiopod crustaceans, which, on the basis of phylogenetic and paleontological evidence, has a long evolutionary history (>100 MY) of partially asexual reproduction (Taylor et al. 1999). Daphnia generally reproduce by cyclical parthenogenesis, with typically one sexual generation and ∼5–20 asexual generations per year. The two species investigated here, Daphnia magna and D. pulex, inhabit small to medium-sized freshwater ponds. Both species are widely distributed and locally abundant throughout the northern hemisphere, which would suggest a very large population size, and, if they were sexual, a large Ne, comparable to other widely distributed invertebrates such as common Drosophila or outcrossing Caenorhabditis species. Here, we assess the consequences of partial asexual reproduction on nucleotide diversity and recombination, and estimate Ne for Daphnia. Ideally, estimates of diversity, recombination, and Ne would be compared between partially asexual and strictly sexual Daphnia species, but this is impossible, because no strictly sexual Daphnia are known.
We analyzed nucleotide diversity and linkage disequilibrium in eight housekeeping genes in D. magna and D. pulex. Allozyme studies have already shown that Daphnia can have considerable genetic diversity, both within and between populations (e.g., Hebert 1978; Lynch and Spitze 1994; De Meester et al. 2006). However, data on nucleotide diversity are needed to estimate silent and synonymous diversity, which are likely to be neutral or weakly selected, so as to estimate Ne. Although diversity at silent sites can be influenced by selection at linked sites, this will generally reduce diversity (Begun and Aquadro 1992), leading to an underestimation of neutral diversity and thus to an underestimation of Ne, unless there is pervasive long-term balancing selection, which is implausible. Hence, a finding of high diversity is likely to be conservative with respect to Ne. In addition, DNA sequence data allow estimates of recombination and linkage disequilibrium (LD) over the small physical distances at which LD is likely to exist. As Daphnia is being developed as a model organism for ecological and evolutionary genomics and is the first crustacean to have its genome sequenced (draft genome accessible on http://genome.jgi-psf.org/Dappu1/Dappu1.home.html), data on nucleotide diversities and LD are useful because several types of analyses would be impeded by low diversity and long-range linkage disequilibria.
MATERIALS AND METHODS
Origin of samples:
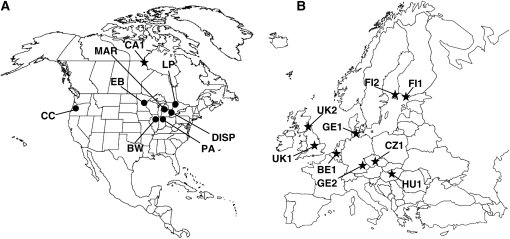
We investigated 10 strains of D. magna, 8 strains of D. pulex, and 1 strain of D. parvula. Each strain originated from a different population (Table 1, Figure1). The strains were isolated as single females and multiplied by clonal reproduction in the laboratory before DNA/RNA extraction. All the strains originated from cyclically parthenogenetic populations.
TABLE 1.
List of the 19 collection sites of the samples used in this study from Europe and North America
| Species | Straina | Locality | Latitude | Longitude |
|---|---|---|---|---|
| D. magna | BE1 | Leuven, Belgium | 50°52′N | 04°41′E |
| D. magna | CA1 | Churchill, MB, Canada | 58°46′N | 94°11′W |
| D. magna | CZ1 | Sedlec, Czech Republic | 48°46.52′N | 16°43.41′E |
| D. magna | FI1 | Tvärminne, Finland | 59°49.43′N | 23°15.15′E |
| D. magna | FI2 | Aland, Finland | 60°01.30′N | 19°54.15′E |
| D. magna | GE1 | Kniphagen, Germany | 54°10.45′N | 10°47.3′E |
| D. magna | GE2 | Ismaning, Germany | 48°12.2′N | 11°41′E |
| D. magna | HU1 | Jaraspuszta, Hungary | 46°48′N | 19°08′E |
| D. magna | UK1 | Cumnor, UK | 51°43.9′N | 01°20.4′W |
| D. magna | UK2 | Leitholm, UK | 55°42.15′N | 02°20.43′W |
| D. pulex | BW102 | Busey Woods, Urbana, IL | 40°07′N | 88°12′W |
| D. pulex | CC1 | Creswell Court, OR | 43°55.5′N | 123°01′W |
| D. pulex | DISP14 | Disputed Road, LaSalle, ON, Canada | 42°13′N | 83°02′W |
| D. pulex | EB1 | Eloise Butler, Minneapolis | 44°58.5′N | 93°19.5′W |
| D. pulex | FAT | Fatties, St-Alexis-des-Monts, QC, Canada | 46°25.40′N | 73°13.44′W |
| D. pulex | PA32 | Portland Arch, Fountain, IN | 40°13′N | 87°20′W |
| D. pulex | LP8 | Long Point, ON, Canada | 42°34′N | 80°15′W |
| D. pulex | MAR | Marion Road, Saline, MI | 42°08.5′N | 83°48.5′W |
| D. parvula | A60 | Acton Lake, OH | 39°57.23′N | 84°74.78′W |
Strains result from a single individual isolated from the wild population, which is subsequently clonally propagated in the lab.
Figure 1.—
Geographical locations of the Daphnia populations surveyed from (A) North America and (B) Europe. Population locations of D. magna are indicated by stars and those of D. pulex by circles. For more details, see Table 1. Maps of North America and Europe are not to scale.
Molecular methods:
We extracted DNA from D. magna strains using the DNeasy Blood and Tissue kit (QIAGEN). Because the D. pulex and D. parvula strains were used in a different project that required cDNA, we extracted RNA from these species with the RNeasy kit (QIAGEN). Reverse transcription was carried out using the Reverse Transcription System (Promega).
We studied eight genes (amino acid identity with Drosophila melanogaster: 44–85%). They include central metabolic genes, a translation initiation factor, and a nuclear receptor protein (Table 2). Daphnia have no sex chromosomes, so all the genes are autosomal. Primers were designed with Primer3 (Rozen and Skaletsky 2000). Primers for D. magna were based on sequences available in GenBank or in the D. magna EST library (Colbourne et al. 2005), and for D. pulex they were based on the draft genome sequence, which we annotated using blast hits of sequences from D. magna (when available) or D. melanogaster. The initial sequence of Mpi in D. magna was obtained using degenerate primers designed with Codehop (Rose et al. 2003). Primers and sizes of amplicons are given in supporting information, Table S1. In a few cases, additional, internal primers (sequences available upon request) were used to verify specific regions or to check for allele-specific amplification in cases of all-homozygous sequences. PCR was carried out using GoTaq polymerase (Promega) and 57° as annealing temperature (in a few instances modified by ±2–4°). Sequences (GenBank accession nos. FJ668030–FJ668168) were obtained by direct sequencing from purified PCR product.
TABLE 2.
List of the eight nuclear loci analyzed in this study
| Locus | Abbreviation | Characterization |
|---|---|---|
| Eukaryotic translation initiation factor 2γ | Eif2γ | Translation initiation factor |
| Enolase | Eno | Metabolic enzyme |
| Glyceraldehyde-3-phosphate dehydrogenase | Gapdh | Metabolic enzyme |
| Glutamine-oxaloacetic transaminase | Got | Metabolic enzyme |
| l-lactate dehydrogenase | Ldh | Metabolic enzyme |
| Mannose-phosphate isomerase | Mpi | Metabolic enzyme |
| Phosphoglucose isomerase | Pgi | Metabolic enzyme |
| Ultraspiracle | Usp | Nuclear receptor protein |
Analysis:
Sequences were aligned using Sequencher version 4.8 (Gene Codes) and BioEdit (Hall 1999). All heterozygous sites were confirmed by resequencing from independent PCR reactions, and primer sequences were removed before analysis. In D. magna, two short regions within Pgi and one within Got could not be sequenced in one individual each, due to two or more length variants within the same amplicon. In D. pulex, Usp had multiple heterozygous length variants and consequently we were unable to obtain high-quality sequences for most individuals. Hence this gene was not included in the D. pulex analysis.
From the aligned heterozygous sequences, we obtained pseudohaplotypes using the program PHASE version 2.1 (Stephens et al. 2001). Since the D. pulex sequences were obtained from cDNA, introns were not sequenced and thus genomic distances were not preserved. We therefore determined the location and length of introns from the draft genome sequence, and inserted an appropriate number of “N”s into the sequences. The pseudohaplotypes obtained (N = 20 for D. magna, N = 14 for D. pulex) were then used in the analyses.
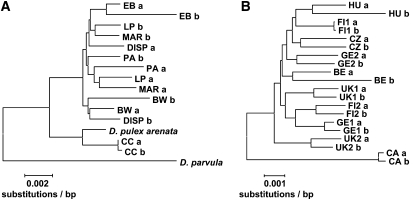
Neighbor-joining trees were constructed using MEGA 4 (Tamura et al. 2007). In addition to our own data, we included a single haplotype obtained from the D. pulex draft genome. The trees indicated that one individual from each species (the D. magna from Canada and the D. pulex from Oregon) might belong to different subspecies (see results). Including different subspecies would inflate measures of diversity; hence, these individuals were excluded from the remaining analyses.
We used DnaSP version 4.50.3 (Rozas et al. 2003) to estimate diversity from pairwise differences (π) and from the number of segregating sites (θ) and for recombination and linkage disequilibrium (LD) analyses. Further analyses of recombination and LD were performed with RecMin (Myers and Griffiths 2003), maxdip (http://genapps.uchicago.edu/maxdip/index.html; Hudson 2001), and LI1AN (Haubold and Hudson 2000). In the analysis of the D. magna data, we distinguished between synonymous sites (indicated by the subscript s), nonsynonymous sites (a), noncoding sites (nc), and silent sites (si, i.e., synonymous and noncoding sites combined). For D. pulex, noncoding sites were not sequenced.
In the analysis of between-species divergence, we report the net divergence, which subtracts π (or average π if polymorphism data from both species is available) from the observed divergence (Nei 1987). To estimate divergence times, T (in units of 2Ne generations), we use the framework of the HKA test (Hudson et al. 1987). To correct for multiple hits at fourfold degenerate sites, we employed the Tamura–Nei correction (Tamura and Nei 1993) using MEGA 4 (Tamura et al. 2007).
RESULTS
Stratification of samples:
Neighbor-joining trees revealed that the D. magna individual from Canada (CA) and the D. pulex individual from Oregon (CC) were highly divergent from all other individuals in their respective species clades (Figure 2), supporting an earlier conclusion from mitochondrial haplotypes that European and American D. magna are highly divergent (De Gelas and De Meester 2005). The D. pulex from Oregon grouped with the sequence from the draft genome (Figure 2), which belongs to the subspecies D. p. arenata (https://dgc.cgb.indiana.edu), which is restricted to a small area in Oregon (e.g., Colbourne et al. 1998; Lynch et al. 1999), whereas D. pulex from other populations included in this study have previously been identified as D. pulex s. str. (Crease et al. 1997). We concluded that these two specimens (CA and CC) potentially belong to different subspecies and thus excluded them from further analysis.
Figure 2.—
Neighbor-joining trees of (A) D. pulex and (B) D. magna. Trees were constructed from concatenated sequence data from seven (D. pulex) and eight (D. magna) genes. Pseudohaplotypes are indicated by the suffix “a” or “b” after the strain name. The D. pulex arenata sequences were obtained from the draft genome available at http://wfleabase.org/. Strains CC (D. pulex) and CA (D. magna) were considered putative subspecies and removed from further analyses.
Nucleotide polymorphism:
Across loci, we obtained 8719 bp of sequence for D. magna and 6576 bp for D. pulex, and found high average silent or synonymous site diversity in both species, although synonymous site diversity in D. pulex is almost twice as high as in D. magna (Table 3). All loci had low diversity at nonsynonymous sites, as expected for highly conserved sequences (Table 3).
TABLE 3.
Number of segregating sites, genetic diversity, and number of length variants in eight nuclear loci in D. magna and seven nuclear loci in D. pulex
| Sequence length (nt)
|
Diversity (π)
|
N length variantse | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Species | Locus | NHapa | Coding | Noncoding | Sb | πs | πnc | πsi | πa | πall | |
| D. magna | Eif2γ | 18 | 429 | 186 | 14 | 0.0210 | 0.0091 | 0.0130 | 0.0013 | 0.0067 | 0 |
| D. magna | Eno | 18 | 438 | 193 | 12 | 0.0092 | 0.0100 | 0.0098 | 0.0016 | 0.0053 | 0 |
| D. magna | Gapdh | 18 | 369 | 70 | 9 | 0.0164 | 0.0220 | 0.0190 | 0.0000 | 0.0068 | 0 |
| D. magna | Got | 18 | 1188 | 348 | 19 | 0.0008 | 0.0130 | 0.0072 | 0.0008 | 0.0034 | 5 |
| D. magna | Ldh | 18 | 999 | 493 | 9 | 0.0004 | 0.0022 | 0.0016 | 0.0004 | 0.0010 | 8 |
| D. magna | Mpi | 18 | 645 | 319 | 14 | 0.0008 | 0.0059 | 0.0043 | 0.0011 | 0.0026 | 3 |
| D. magna | Pgi | 18 | 1611 | 957 | 112c | 0.0280 | 0.0160 | 0.0190 | 0.0024 | 0.0110 | 13 |
| D. magna | Usp | 18 | 333 | 141 | 14 | 0.0440 | 0.0120 | 0.0220 | 0.0000 | 0.0100 | 2 |
| D. pulex | Eif2γ | 12 | 1233 | 0 | 15 | 0.0130 | NA | NA | 0 | 0.0031 | 0 |
| D. pulex | Eno | 14 | 933 | 0 | 14 | 0.0180 | NA | NA | 0.0002 | 0.0043 | 0 |
| D. pulex | Gapdh | 14 | 762 | 0 | 8 | 0.0120 | NA | NA | 0 | 0.0030 | 0 |
| D. pulex | Got | 12 | 852 | 0 | 14 | 0.0170 | NA | NA | 0 | 0.0039 | 0 |
| D. pulex | Ldh | 12 | 672 | 0 | 18 | 0.0260 | NA | NA | 0.0032 | 0.0087 | 0 |
| D. pulex | Mpi | 12 | 759 | 0 | 6 | 0.0062 | NA | NA | 0.0010 | 0.0022 | 0 |
| D. pulex | Pgi | 12 | 1365 | 0 | 78d | 0.0620 | NA | NA | 0.0024 | 0.0164 | 0 |
| D magna | Average | 24.3 | 0.0150 | 0.0110 | 0.0120 | 0.0009 | 0.0059 | ||||
| D. pulex | Average | 21.9 | 0.0220 | NA | NA | 0.0009 | 0.0059 | ||||
s, synonymous; nc, noncoding; si, silent (synonymous and noncoding combined); a, nonsynonymous, NA, not assessed.
Number of haplotypes.
Number of segregating sites.
Includes four triallelic sites.
Includes one triallelic site.
All length variants are in introns.
Tajima's D (Tajima 1989) was significantly negative in D. pulex (one sample t-test for D on the basis of synonymous sites, mean across loci = −0.62, t = −4.76, d.f. = 6, P = 0.0031). In D. magna, Tajima's D also tended to be negative, but was not significant (one sample t-test for D based on synonymous sites, mean = −0.48, t = −1.51, d.f. = 7, P = 0.17, one sample t-test based on all silent sites: mean = −0.35, t = −1.38, d.f. = 7, P = 0.21).
Recombination and linkage disequilibrium:
At least one recombination event was detected in most genes, even with the conservative four-gamete test (Table 4) (Hudson and Kaplan 1985). The per-site recombination parameter ρ estimated from unphased genotypic data (using the program maxdip) varied greatly among loci (Table 4). Average ρ is 0.064 in D. magna and 0.087 in D. pulex (Table 4), but median values are lower (0.015 and 0.039, respectively). When estimated from phased pseudohaplotypes (using DnaSP), the values were somewhat lower in D. magna (mean = 0.019, median = 0.007), and similar in D. pulex (mean = 0.091, median = 0.041).
TABLE 4.
Summary statistics of recombination and linkage disequilibrium estimates in D. magna and D. pulex
| Species | Locus | Rma | Rminb | ρ | IASc | Znsd | Zae | ZZf | P(ZZ) |
|---|---|---|---|---|---|---|---|---|---|
| D. magna | Eif2γ | 1 | 2 | 0.016 | 0.27**** | 0.26 | 0.44 | 0.18 | 0.01 |
| D. magna | Eno | 3 | 2 | 0.025 | 0.27**** | 0.15 | 0.21 | 0.06 | 0.19 |
| D. magna | Gapdh | 0 | 0 | 0 | 0.32**** | 0.30 | 0.26 | −0.03 | 0.65 |
| D. magna | Got | 0 | 0 | 0.002 | 0.21**** | 0.31 | 0.37 | 0.06 | 0.18 |
| D. magna | Ldh | 0 | 0 | 0.001 | 0.39**** | 0.36 | 0.50 | 0.15 | 0.055 |
| D. magna | Mpi | 2 | 2 | 0.440 | 0.11**** | 0.16 | 0.03 | −0.13 | 0.98 |
| D. magna | Pgi | 20 | 31 | 0.017 | 0.02**** | 0.10 | 0.17 | 0.07 | 0.006 |
| D. magna | Usp | 3 | 4 | 0.015 | 0.17**** | 0.25 | 0.33 | 0.08 | 0.10 |
| D. pulex | Eif2γ | 3 | 4 | 0.040 | 0.07**** | 0.15 | 0.20 | 0.05 | 0.26 |
| D. pulex | Eol | 5 | 5 | 0.289 | 0.03**** | 0.13 | 0.24 | 0.11 | 0.10 |
| D. pulex | Gapdh | 1 | 1 | 0.011 | 0.12**** | 0.10 | 0.05 | −0.05 | 0.73 |
| D. pulex | Got | 0 | 0 | 0.008 | 0.10**** | 0.20 | 0.33 | 0.13 | 0.10 |
| D. pulex | Ldh | 3 | 4 | 0.013 | 0.08**** | 0.28 | 0.30 | 0.02 | 0.37 |
| D. pulex | Mpi | 1 | 1 | 0.039 | −0.03 | 0.08 | 0.17 | 0.09 | 0.17 |
| D. pulex | Pgi | 19 | 25 | 0.207 | 0.03**** | 0.14 | 0.18 | 0.05 | 0.13 |
| D. magna | Average | 3.6 | 5.1 | 0.064 | 0.22 | 0.23 | 0.29 | 0.05 | |
| D. pulex | Average | 4.6 | 5.7 | 0.087 | 0.06 | 0.15 | 0.21 | 0.06 |
****P < 0.0001.
Rm, minimum number of recombination events (Hudson and Kaplan, using pseudohaplotypes).
Rmin, minimum number of recombination events (Recmin, using pseudohaplotypes).
IAS, LD summary statistics (LIAN).
Zns, average r2.
Za, average r2 between adjacent polymorphic sites.
ZZ = Za − Zns, tested with coalescence simulations in DnaSP.
LD (assessed with the program LIAN, Haubold and Hudson 2000) was significant but weak at all loci except Mpi in D. pulex (Table 4). LD decreased significantly with distance for some loci (Table 4, evaluated with a ZZ test, Rozas et al. 2001). LD was assessed using phased pseudohaplotypes, rather than from genotypic data, because the latter methods assume Hardy–Weinberg equilibrium, whereas PHASE infers haplotypes very accurately even when this assumption is not met, as is likely in our samples (Smith and Fearnhead 2005).
Effective population size:
Assuming that a population is at equilibrium, Ne can be estimated from either nucleotide polymorphism θ = 4Neμ or the recombination parameter ρ = 4Ner, where μ is the mutation rate, and r is the recombination rate per nucleotide; we refer to these estimates as Ne(θ) and Ne(ρ), respectively. The mutation rate in Daphnia is not yet known, but, at least for microsatellites, it appears to be similar to that of Caenorhabditis elegans and D. melanogaster (Seyfert et al. 2008). We thus used the average genomic mutation rate of D. melanogaster and C. elegans, μ ≈ 10−8 per generation (Denver et al. 2004; Haag-Liautard et al. 2007). For the average recombination rate, we use an estimate based on the D. pulex genetic map (Cristescu et al. 2006) of r ≈ 7.5 × 10−6 cM/bp per sexual generation, or r ≈ 7.5 × 10−7 per generation (sexual and asexual generations combined, assuming one sexual generation per 10 asexual ones). Using these estimates, and the average θsi and ρ values from our data, we obtain estimates of Ne for D. magna Ne(θ) = 311,000 and Ne(ρ) = 2,148,000, and for D. pulex Ne(θ) = 642,000 and Ne(ρ) = 2,890,000. Using the more conservative median values for ρ, Ne(ρ) is 503,000 for D. magna, and 1,286,000 for D. pulex. Although these estimates are highly uncertain, because local mutation and recombination rates may deviate substantially from genomewide averages, the Ne of European D. magna is clearly of the order of 300,000–500,000 and that of North American D. pulex at least twice as high. Using diversity at fourfold degenerate sites, the D. pulex Ne estimate is 3.3 times that of D. magna.
Between-species divergence:
As expected from sequences of mitochondrial genes (e.g., Lehman et al. 1995; Colbourne and Hebert 1996), the net synonymous site divergence between D. magna and D. pulex is high, averaging Ks = 0.494 across loci (Table 5). Multiple mutations at the same site are thus likely, and indeed the Tamura–Nei corrected divergence estimates for fourfold degenerate sites (Table 5) indicate a substantial but (with exception of the Gapdh locus) not extreme underestimation of divergence. Using the geometric mean rather than the arithmetic mean, to reduce any undue influence of Gapdh, we obtained an average divergence of 1.98 substitutions per site (Table 5).
TABLE 5.
Divergence between species and presumed subspecies
|
D. magna to D. pulex
|
European D. magna to Canadian D. magna: | D. pulex to D. parvula: | D. pulex s. str. to D. p. arenata: | ||
|---|---|---|---|---|---|
| Locus | Ks(net)c | Ks(corr)d | Ksi(net)c | Ks(net)c | Ks(net)c |
| Eif2γ | 0.437 | 0.631 | 0.003 | 0.022 | 0.017 |
| Eno | 0.482 | 1.141 | 0.022 | 0.059 | 0.016 |
| Gapdh | 0.426 | 62.123 | 0.002 | 0.048 | 0.028 |
| Got | 0.542 | 3.744 | 0.026 | 0.115 | 0.038 |
| Ldh | 0.620 | 0.968 | 0.022 | 0.093 | 0.019 |
| Mpi | 0.338 | 0.413 | 0 | 0.055 | 0.008 |
| Pgi | 0.541 | 1.792 | 0.003 | 0.035 | 0.007 |
| Uspa | 0.563 | 1.138 | 0.010 | ||
| Average | 0.494 | 1.982b | 0.011 | 0.061 | 0.019 |
s, synonymous; si, silent (synonymous and noncoding combined).
Estimate for Usp corrects for polymorphism in D. magna only, compared to the genome sequence of D. pulex.
Geometric mean.
Net divergence, corrected for within-species polymorphism.
Divergence at fourfold degenerate sites corrected for multiple hits (Tamura–Nei correction).
The other species or putative subspecies pairs show much less sequence divergence. Between D. pulex and D. parvula, net divergence at synonymous sites averaged Ks = 0.061. Between D. pulex s. str. and D. pulex arenata, Ks = 0.019, and between Canadian and European D. magna, silent site divergence Ksi = 0.011 (Table 5). The divergence at nonsynonymous sites was low in all comparisons (data not shown).
Using the polymorphism and divergence data for synonymous sites, and assuming neutrality of synonymous differences, we estimated divergence times T for the different species/putative subspecies pairs in units of 2Ne generations (Nmagna and Npulex denote the Ne of D. magna and D. pulex, respectively). The estimated divergence times for the closely related species/subspecies pairs were 1.8 for D. pulex vs. D. parvula (in units of 2Npulex), and, in the same units, 0.48 for the divergence between D. pulex s. str. and D. p. arenata, and 0.74 for the divergence between European and Canadian D. magna (in units of 2Nmagna, based on silent sites). Note that there are no polymorphism data from the second species/subspecies, so that these estimates are based on assuming equal Ne. Using polymorphism and divergence at fourfold degenerate sites, the divergence time between D. magna and D. pulex is much greater, estimated as 76.1 in units of 2Npulex generations.
DISCUSSION
Abundant diversity and recombination:
Both cyclically parthenogenetic, Daphnia species studied here have considerable silent site diversity (1–2%), frequent intragenic recombination, and low levels of linkage disequilibrium. Compared to other abundant and widely distributed invertebrate species, diversity levels are intermediate between outcrossing and self-fertilizing species of Caenorhabditis (Cutter 2006; Cutter et al. 2006a,b), and similar to or somewhat lower than in Drosophila species (e.g., Hamblin and Aquadro 1999; Andolfatto 2001; Dyer and Jaenike 2004; Maside and Charlesworth 2007). Daphnia species are currently being developed as new model organisms for evolutionary and ecological genomics (Colbourne et al. 2005). Several approaches proposed for such studies, including fine-mapping by association analyses, demand high diversity and low levels of linkage disequilibrium. The results of our study suggest that these approaches might indeed be feasible in Daphnia.
Genetic differentiation among Daphnia populations can be strong (e.g., Hebert 1978; Lynch and Spitze 1994; De Meester et al. 2006). Population subdivision can contribute to high continentwide diversity (Whitlock and Barton 1997; Pannell and Charlesworth 2000; Wakeley 2000), but should also decrease recombination (Nordborg 2000; Wakeley and Aliacar 2001). Under the island model of population subdivision, diversity is increased and recombination decreased by a factor (1 − FST) compared to a panmictic population (Conway et al. 1999; Ingvarsson 2004). Our sample included just one individual per population (a “scattered sample”), and hence our diversity estimate are πT, the continentwide values; this is ideal for estimating LD, as it avoids excess LD that is expected within populations due to recent common ancestry (Wakeley and Aliacar 2001; Song et al. 2009). πT allows us to obtain an estimate of Ne, the “metapopulation effective size,” which determines, for instance, fixation times of neutral alleles (Roze and Rousset 2003; Whitlock 2003). However, a scattered sample does not allow us to estimate within-deme diversity πS. An estimate of πS can, however, be obtained from πS = πT (1 − FST) (Pannell and Charlesworth 2000). Because FST measures the proportion of the total diversity that is found between populations, it is not very sensitive to the type of genetic information used, and thus we can tentatively use earlier allozyme-based estimates of FST ≈ 0.3 in D. magna and D. pulex based on allozymes (Vanoverbeke and De Meester 1997; Lynch et al. 1999). This suggests that removing the effect of population subdivision would reduce our estimate of Ne(θ) by a factor of ∼0.7 and increase the estimate of Ne(ρ) by the inverse of this factor. Despite these potentially substantial effects of population subdivision, within-population diversity (∼1%) and Ne(θ) (>100,000) are still high and recombination abundant.
While it may be questioned whether the above FST-estimates indeed apply to our data, the evidence for abundant recombination (see also below) speaks against highly subdivided populations. Thus Ne is certainly high, even taking into account the effect of population subdivision, and hence selection is expected to be effective in the Daphnia species studied here, even for mutations with small selection coefficients (mutations with selection coefficients s > 1/2Ne are predominantly influenced by selection as opposed to genetic drift). This situation is contrary to that expected in strictly asexual or highly inbreeding organisms, where selection is expected to be inefficient for mutations with small selection coefficients (Hill and Robertson 1966; Felsenstein 1974; Charlesworth 1994; Cutter and Charlesworth 2006; Keightley and Otto 2006).
Has a partially asexual life cycle affected Ne of Daphnia?
Although estimates of Ne(θ) in the Daphnia species we studied are large, they are lower (up to one order of magnitude) than for outcrossing C. remanei (Cutter et al. 2006a) and most Drosophila species (Wall et al. 2002; Yi et al. 2003), especially if corrected for population subdivision. It is, however, unclear whether this is a consequence of the partial asexual life cycle. The census population size Nc (i.e., the actual number of individuals) may also be lower. For example, the obligately sexual invertebrate D. miranda has low diversity, presumably due to low Nc (Bachtrog 2003; Yi et al. 2003).
Most Daphnia, including all the strains we analyzed, reproduce by cyclical parthenogenesis (partial asexuality), while some strains of D. pulex (and a few other Daphnia species) have become obligate asexuals (Hebert and Crease 1980). Observations of low numbers of clones in obligate parthenogenetic Daphnia populations (Weider et al. 1987; Hebert et al. 1989), and a higher rate of accumulation of slightly deleterious mutations than in cyclically parthenogenetic strains (Paland and Lynch 2006), suggest small Ne values in the obligate asexuals, and a comparison of two mitochondrial genes between D. pulex populations with differing breeding systems found slightly lower genetic diversity in obligate asexual populations (Paland et al. 2005). However, the sample may have included several independently arisen asexual lines, and thus no direct conclusions could be made about the relative Ne of populations of obligate and partial asexuals. We did not include obligate asexual strains in the present study because (in addition to the possible inclusion of independently arisen asexual lines) estimates of Ne in obligate asexuals from nuclear gene diversity may be affected by ancestral polymorphism, and possibly, if they have been asexual for long evolutionary times, by divergence between the alleles within lineages, and thus within individuals (Butlin 2002; Balloux et al. 2003).
Recombination:
Per-site recombination parameters in both species of Daphnia are similar to those in outcrossing C. remanei and most Drosophila species (Andolfatto and Przeworski 2000; Yi et al. 2003; Haddrill et al. 2005; Cutter et al. 2006a). In addition, there is clearly much lower linkage disequilibrium in Daphnia than in self-fertilizing species such as Arabidopsis thaliana or C. elegans (Nordborg et al. 2002; Cutter 2006), consistent sexual reproduction in Daphnia being more common than outcrossing in the latter species.
The Ne estimated from the recombination parameters are somewhat larger than values from diversity, but, given the uncertainties about the underlying assumptions, the two estimates of effective population sizes are quite consistent, especially with Ne(ρ) from the median ρ estimates. The consistency between the estimates of effective population size from recombination and from diversity is, however, altered if population structure is taken into account. The ρ/θ ratio is an estimate of r/μ, the relative frequency of recombination compared to mutation. Given the effects of population subdivision, we would expect ρ/θ = (r/μ)(1 − FST)2 (Conway et al. 1999; Nordborg 2000; Ingvarsson 2004). On the basis of our assumed mutation rate μ, recombination rate r, and frequency of sexual reproduction, we have r/μ = 0.75, and thus (using FST = 0.3, as above), we expect ρ/θ = 0.37. However, the observed values are 1.25 in D. magna and 1.75 in D. pulex, even using the more conservative median estimates of ρ. Thus, recombination in Daphnia is surprisingly high, particularly considering the fact that demographic history can also affect diversity and recombination (Hudson 1987; Andolfatto and Przeworski 2000; Wall et al. 2002; Haddrill et al. 2005): Demographic events that produce genomewide negative Tajima's D (as observed in D. pulex, see also Paland et al. 2005), for example, a strong population bottleneck followed by population growth, should increase LD (e.g., Tishkoff et al. 1996; Haddrill et al. 2005) so that the observed low levels of LD are even more surprising.
The unexpectedly high levels of recombination may be explained if our assumptions about the mutation rate, recombination rate, the frequency of sexual reproduction or the FST value are incorrect. However, at least two additional effects might increase the effective recombination rate in Daphnia. First, two recent studies provided evidence that, contrary to our assumptions above, some recombination also takes place during the clonal phase in Daphnia (Omilian et al. 2006; Mctaggart et al. 2007), which would increase the estimate of r per sexual generation. Second, the offspring of heterozygotes may be overrepresented in Daphnia populations because of reduced fitness of homozygous individuals (Ebert et al. 2002; Haag et al. 2002). This could increase the effective recombination rate because crossing-over results in recombination only in double-heterozygous individuals.
Between-species divergence:
D. magna and D. pulex are among the most distantly related species within the genus Daphnia; previous estimates on the basis of a mitochondrial molecular clock suggested a divergence time of ∼200 MY (Colbourne and Hebert 1996). We estimated divergence time as 76 × 2Npulex generations. Assuming 2Npulex = 106, the divergence time estimate becomes 76 × 106 generations, which, assuming 10 generations per year (sexual and asexual generations combined, as also assumed by Paland et al. 2005), is only 7.6 MY. Many factors could contribute to the large discrepancy between these estimates. If the mutation and recombination rates for the studied genes are lower than we have assumed, our estimates of Ne, and consequently divergence times, would be larger. However, a very low mutation rate of μ ≈ 5 × 10−10 would be needed to yield an estimate of 200 MY. This seems unlikely, given that microsatellite mutation rates in Daphnia are similar to those in C. elegans and D. melanogaster (Seyfert et al. 2008). Second, there may be fewer than 10 generations per year, but values that would reconcile the date estimates (less than one generation per year) are inconsistent with the biology of the species (time to first reproduction is 10–15 days at 20°, Ebert 2005). A third possibility is population subdivision, which increases intraspecific polymorphism and decreases interspecific divergence estimates by a factor of 1 − FST (Ingvarsson 2004). Assuming FST = 0.3 increases the divergence time estimate to 15.6 MY. A very high degree of population differentiation would, however, have to be assumed to yield an estimate of 200 MY, and this is incompatible with the high recombination estimates.
Finally, despite correcting for multiple hits, we may have underestimated the divergence between the two species. However, under our assumptions of mutation rates and the number of Daphnia generations per year, the previous estimate of 200 MY would require ∼20 substitutions per site since the two species became reproductively isolated. Our estimates of divergence times for the other two species pairs (with much lower raw divergence estimates than for D. magna and D. pulex, so that multiple hits are much less likely) are also much lower than previous estimates on the basis of mitochondrial sequences (Colbourne and Hebert 1996).
Alternatively, the earlier studies might have overestimated the age of the genus Daphnia. As for our estimates, multiple assumptions were necessary, most notably a constant mitochondrial rate (per year) of nucleotide substitution among animals. Yet, even among mammals this rate can vary by two orders of magnitude (Nabholz et al. 2008) and it is also highly variable among insects (Oliveira et al. 2008). If substitution rates were higher than assumed, this may have led to an overestimation of divergence time. Additionally, polymorphism in the ancestral species could contribute to a potential overestimation of divergence times if the two daughter species received highly divergent copies (Lynch and Jarrell 1993). Finally, because of the linkage of the whole mitochondrial genome, background selection and hitchhiking potentially contribute further difficulties in the interpretation of mitochondrial divergence times (Bazin et al. 2006; Oliveira et al. 2008). Until the relevant data are available for Daphnia, the discrepant divergence time estimates cannot currently be fully explained. However, our data suggest that the genus Daphnia may be considerably younger than previously thought. This might also change earlier conclusions that pairs of morphologically similar Daphnia species have diverged for tens of millions of years without noticeable morphological change (Colbourne and Hebert 1996).
Acknowledgments
We are grateful to D. Ebert, J. Hottinger, M. Lynch, J. Stoeckel, and T. Zumbrunn for contributing samples. We thank M. Lehto for assistance and D. Begun and two anonymous reviewers for helpful comments. This work was supported by a Marie Curie Fellowship, the United Kingdom Natural Environment Research Council, the United Kingdom Welcome Trust, and the Swiss National Science Foundation. This is part of project no. 08574001 at Tvärminne Zoological Station.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.101147/DC1.
References
- Agrawal, A. F., 2006. Evolution of sex: why do organisms shuffle their genotypes. Curr. Biol. 16 R696–R704. [DOI] [PubMed] [Google Scholar]
- Andolfatto, P., 2001. Contrasting patterns of X-linked and autosomal nucleotide variation in Drosophila melanogaster and Drosophila simulans. Mol. Biol. Evol. 18 279–290. [DOI] [PubMed] [Google Scholar]
- Andolfatto, P., and M. Przeworski, 2000. A genomewide departure from the standard neutral model in natural populations of Drosophila. Genetics 156 257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog, D., 2003. Protein evolution and codon usage bias on the neo-sex chromosomes of Drosophila miranda. Genetics 165 1221–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balloux, F., L. Lehmann and T. De Meeûs, 2003. The population genetics of clonal and partially clonal diploids. Genetics 164 1635–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton, N. H., and B. Charlesworth, 1998. Why sex and recombination. Science 281 1986–1990. [PubMed] [Google Scholar]
- Bazin, E., S. Glemin and N. Galtier, 2006. Population size does not influence mitochondrial genetic diversity in animals. Science 312 570–572. [DOI] [PubMed] [Google Scholar]
- Begun, D. J., and C. F. Aquadro, 1992. Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature 356 519–520. [DOI] [PubMed] [Google Scholar]
- Bell, G., 1982. The Masterpiece of Nature: The Evolution and Genetics of Sexuality. University of California Press, Berkeley, CA.
- Butlin, R., 2002. The costs and benefits of sex: new insights from old asexual lineages. Nat. Rev. Genet. 3 311–317. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B., 1994. The effect of background selection against deleterious mutations on weakly selected, linked variants. Genet. Res. 63 213–227. [DOI] [PubMed] [Google Scholar]
- Charlesworth, D., M. T. Morgan and B. Charlesworth, 1993. Mutation accumulation in finite populations. J. Hered. 84 321–325. [Google Scholar]
- Colbourne, J., V. Singan and D. Gilbert, 2005. wFleaBase: the Daphnia genome database. BMC Bioinformatics 6 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbourne, J. K., T. J. Crease, L. J. Weider, P. D. N. Hebert, F. Dufresne et al., 1998. Phylogenetics and evolution of a circumarctic species complex (Cladocera: Daphnia pulex). Biol. J. Linn. Soc. Lond. 65 347–365. [Google Scholar]
- Colbourne, J. K., and P. D. N. Hebert, 1996. The systematics of North American Daphnia (Crustacea: Anomopoda): a molecular phylogenetic approach. Phil. Trans. R. Soc. Lond. B 351 349–360. [DOI] [PubMed] [Google Scholar]
- Conway, D. J., C. Roper, A. M. J. Oduola, D. E. Arnot, P. G. Kremsner et al., 1999. High recombination rate in natural populations of Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 96 4506–4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crease, T. J., S.-K. Lee, S.-L. Yu, K. Spitze, N. Lehman et al., 1997. Allozyme and mtDNA variation in populations of the Daphnia pulex complex from both sides of the Rocky Mountains. Heredity 79 242–251. [Google Scholar]
- Cristescu, M. E. A., J. K. Colbourne, J. Radivojac and M. Lynch, 2006. A microsatellite-based genetic linkage map of the waterflea, Daphnia pulex: on the prospect of crustacean genomics. Genomics 88 415–430. [DOI] [PubMed] [Google Scholar]
- Crow, J. F., and M. Kimura, 1970. An Introduction to Population Genetics Theory. Harper & Row, New York.
- Cutter, A. D., 2006. Nucleotide polymorphism and linkage disequilibrium in wild populations of the partial selfer Caenorhabditis elegans. Genetics 172 171–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter, A. D., and B. Charlesworth, 2006. Selection intensity on preferred codons correlates with overall codon usage bias in Caenorhabditis remanei. Curr. Biol. 16 2053–2057. [DOI] [PubMed] [Google Scholar]
- Cutter, A. D., S. E. Baird and D. Charlesworth, 2006. a High nucleotide polymorphism and rapid decay of linkage disequilibrium in wild populations of Caenorhabditis remanei. Genetics 174 901–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter, A. D., M.-A. Felix, A. Barriere and D. Charlesworth, 2006. b Patterns of nucleotide polymorphism distinguish temperate and tropical wild isolates of Caenorhabditis briggsae. Genetics 173 2021–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Souza, T. G., and N. K. Michiels, 2006. Genetic signatures of occasional sex in parthenogenetic subpopulations of the freshwater planarian Schmidtea polychroa. Freshw. Biol. 51 1890–1900. [Google Scholar]
- De Gelas, K., and L. De Meester, 2005. Phylogeography of Daphnia magna in Europe. Mol. Ecol. 14 753–764. [DOI] [PubMed] [Google Scholar]
- De Meester, L., J. Vanoverbeke, K. De Gelas, R. Ortells and P. Spaak, 2006. Genetic structure of cyclic parthenogenetic zooplankton populations: a conceptual framework. Arch. Hydrobiol. 167 217–244. [Google Scholar]
- Delmotte, F., N. Leterme, J.-P. Gauthier, C. Rispe and J.-C. Simon, 2002. Genetic architecture of sexual and asexual populations of the aphid Rhopalosiphum padi based on allozyme and microsatellite markers. Mol. Ecol. 11 711–723. [DOI] [PubMed] [Google Scholar]
- Denver, D. R., K. Morris, M. Lynch and W. K. Thomas, 2004. High mutation rate and predominance of insertions in the Caenorhabditis elegans nuclear genome. Nature 430 679–682. [DOI] [PubMed] [Google Scholar]
- Dyer, K. A., and J. Jaenike, 2004. Evolutionarily stable infection by a male-killing endosymbiont in Drosophila innubila: molecular evidence from the host and parasite genomes. Genetics 168 1443–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert, D., 2005. Ecology, epidemiology, and evolution of parasitism in Daphnia, pp. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Books.
- Ebert, D., C. Haag, M. Kirkpatrick, M. Riek, J. W. Hottinger et al., 2002. A selective advantage to immigrant genes in a Daphnia metapopulation. Science 295 485–488. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J., 1974. The evolutionary advantage of recombination. Genetics 78 737–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher, R. A., 1930. The Genetical Theory of Natural Selection. Clarendon Press, Oxford.
- Green, R. F., and D. L. G. Noakes, 1995. Is a little bit of sex as good as a lot. J. Theor. Biol. 174 87–96. [Google Scholar]
- Haag-Liautard, C., M. Dorris, X. Maside, S. Macaskill, D. L. Halligan et al., 2007. Direct estimation of per nucleotide and genomic deleterious mutation rates in Drosophila. Nature 445 82–85. [DOI] [PubMed] [Google Scholar]
- Haag, C. R., J. W. Hottinger, M. Riek and D. Ebert, 2002. Strong inbreeding depression in a Daphnia metapopulation. Evolution 56 518–526. [PubMed] [Google Scholar]
- Hadany, L., and T. Beker, 2007. Sexual selection and the evolution of obligatory sex. BMC Evol. Biol. 7 245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddrill, P. R., K. R. Thornton, B. Charlesworth and P. Andolfatto, 2005. Multilocus patterns of nucleotide variability and the demographic and selection history of Drosophila melanogaster populations. Genome Res. 15 790–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, T. A., 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41 95–98. [Google Scholar]
- Hamblin, M. T., and C. F. Aquadro, 1999. DNA sequence variation and the recombinational landscape in Drosophila pseudoobscura: a study of the second chromosome. Genetics 153 859–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haubold, B., and R. R. Hudson, 2000. LIAN 3.0: detecting linkage disequilibrium in multilocus data. Bioinformatics 16 847–849. [DOI] [PubMed] [Google Scholar]
- Hebert, P. D. N., 1978. The population biology of Daphnia (Crustacea, Daphnidae). Biol. Rev. 53 387–426. [Google Scholar]
- Hebert, P. D. N., and T. Crease, 1980. Clonal coexistence in Daphnia pulex (Leydig): another planktonic paradox. Science 207 1363–1365. [Google Scholar]
- Hebert, P. D. N., M. J. M.J. Beaton, S. S. Schwartz and D. J. Stanton, 1989. Polyphyletic origins of asexuality in Daphnia pulex I. Breeding system variation and levels of clonal diversity. Evolution 43 1004–1015. [DOI] [PubMed] [Google Scholar]
- Hill, W. G., and A. Robertson, 1966. The effect of linkage on limits to artificial selection. Genet. Res. 8 269–294. [PubMed] [Google Scholar]
- Hudson, R. R., 1987. Estimating the recombination parameter of a finite population model without selection. Genet. Res. 50 245–250. [DOI] [PubMed] [Google Scholar]
- Hudson, R. R., 2001. Two-locus sampling distributions and their application. Genetics 159 1805–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. R., and N. L. Kaplan, 1985. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111 147–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. R., M. Kreitman and M. Aguade, 1987. A test of neutral molecular evolution based on nucleotide data. Genetics 116 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, A. L., and F. Verra, 2001. Very large long-term effective population size in the virulent human malaria parasite Plasmodium falciparum. Proc. R. Soc. Lond. B 268 1855–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst, L. D., and J. R. Peck, 1996. Recent advances in understanding of the evolution and maintenance of sex. Trends Ecol. Evol. 11 A46–A52. [DOI] [PubMed] [Google Scholar]
- Ingvarsson, P. K., 2004. Population subdivision and the Hudson-Kreitman-Aguade test: testing for deviations from the neutral model in organelle genomes. Genet. Res. 83 31–39. [DOI] [PubMed] [Google Scholar]
- Keightley, P. D., and S. P. Otto, 2006. Interference among deleterious mutations favours sex and recombination in finite populations. Nature 443 89–92. [DOI] [PubMed] [Google Scholar]
- Kondrashov, A. S., 1988. Deleterious mutations and the evolution of sexual reproduction. Nature 336 435–440. [DOI] [PubMed] [Google Scholar]
- Lehman, N., M. E. Pfrender, P. A. Morin, T. J. Crease and M. Lynch, 1995. A hierarchical molecular phylogeny within the genus Daphnia. Mol. Phyl. Evol. 4 395–407. [DOI] [PubMed] [Google Scholar]
- Lynch, M., and W. Gabriel, 1983. Phenotypic evolution and parthenogenesis. Am. Nat. 122 745–764. [Google Scholar]
- Lynch, M., and P. E. Jarrell, 1993. A method for calibrating molecular clocks and its application to animal mitochondrial DNA. Genetics 135 1197–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, M., M. Pfrender, K. Spitze, N. Lehman, J. Hicks et al., 1999. The quantitative and molecular genetic architecture of a subdivided species. Evolution 53 100–110. [DOI] [PubMed] [Google Scholar]
- Lynch, M., and K. Spitze, 1994. Evolutionary genetics of Daphnia, pp. 109–128 in Ecological Genetics, edited by L. A. Real. Princeton University Press, Princeton, NJ.
- Maside, X., and B. Charlesworth, 2007. Patterns of molecular variation and evolution in Drosophila americana and its relatives. Genetics 176 2293–2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith, J., 1978. The Evolution of Sex. Cambridge University Press, Cambridge, UK.
- McTaggart, S. J., J. L. Dudycha, A. Omilian and T. J. Crease, 2007. Rates of recombination in the ribosomal DNA of apomictically propagated Daphnia obtusa lines. Genetics 175 311–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, H. J., 1964. The relation of recombination to mutational advance. Mutat. Res. 1 2–9. [DOI] [PubMed] [Google Scholar]
- Myers, S. R., and R. C. Griffiths, 2003. Bounds on the minimum number of recombination events in a sample history. Genetics 163 375–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabholz, B., S. Glemin and N. Galtier, 2008. Strong variations of mitochondrial mutation rate across mammals–the longevity hypothesis. Mol. Biol. Evol. 25 120–130. [DOI] [PubMed] [Google Scholar]
- Nei, M., 1987. Molecular Evolutionary Genetics. Columbia University Press, New York.
- Nordborg, M., 2000. Linkage disequilibrium, gene trees and selfing: an ancestral recombination graph with partial self-fertilization. Genetics 154 923–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg, M., J. O. Borevitz, J. Bergelson, C. C. Berry, J. Chory et al., 2002. The extent of linkage disequilibrium in Arabidopsis thaliana. Nat. Genet. 30 190–193. [DOI] [PubMed] [Google Scholar]
- Oliveira, D. C. S. G., R. Raychoudhury, D. V. Lavrov and J. H. Werren, 2008. Rapidly evolving mitochondrial genome and directional selection in mitochondrial genes in the parasitic wasp Nasonia (Hymenoptera: Pteromalidae). Mol. Biol. Evol. 25 2167–2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omilian, A. R., M. E. A. Cristescu, J. L. Dudycha and M. Lynch, 2006. Ameiotic recombination in asexual lineages of Daphnia. Proc. Natl. Acad. Sci. USA 103 18638–18643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr, H. A., 2000. The rate of adaptation in asexuals. Genetics 155 961–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto, S. P., and T. Lenormand, 2002. Resolving the paradox of sex and recombination. Nat. Rev. Genet. 3 252–261. [DOI] [PubMed] [Google Scholar]
- Paland, S., J. K. Colbourne and M. Lynch, 2005. Evolutionary history of contagious asexuality in Daphnia pulex. Evolution 59 800–813. [PubMed] [Google Scholar]
- Paland, S., and M. Lynch, 2006. Transitions to asexuality result in excess amino acid substitutions. Science 311 990–992. [DOI] [PubMed] [Google Scholar]
- Pamilo, P., M. Nei and W. Li, 1987. Accumulation of mutations in sexual and asexual populations. Genet. Res. 49 135–146. [DOI] [PubMed] [Google Scholar]
- Pannell, J. R., and B. Charlesworth, 2000. Effects of metapopulation processes on measures of genetic diversity. Phil. Trans. R. Soc. Lond. B 355 1851–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck, J. R., 1994. A ruby in the rubbish: beneficial mutations, deleterious mutations and the evolution of sex. Genetics 137 597–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, T., J. Henikoff and S. Henikoff, 2003. CODEHOP (COnsensus-DEgenerate Hybrid Oligonucleotide Primer) PCR primer design. Nucleic Acids Res. 31 3763–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas, J., M. Gullaud, G. Blandin and M. Aguade, 2001. DNA variation at the rp49 gene region of Drosophila simulans: evolutionary inferences from an unusual haplotype structure. Genetics 158 1147–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas, J., J. C. Sanchez-DelBarrio, X. Messeguer and R. Rozas, 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19 2496–2497. [DOI] [PubMed] [Google Scholar]
- Roze, D., and N. Barton, 2006. The Hill–Robertson Effect and the evolution of recombination. Genetics 173 1793–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roze, D., and F. Rousset, 2003. Selection and drift in subdivided populations: a straightforward method for deriving diffusion approximations and applications involving dominance, selfing and local extinctions. Genetics 165 2153–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen, S., and H. J. Skaletsky, 2000. Primer3 on the WWW for general users and for biologist programmers, pp. 365–386 in Bioinformatics Methods and Protocols: Methods in Molecular Biology, edited by S. Krawetz and S. Misener. Humana Press, Totowa, NJ. [DOI] [PubMed]
- Seyfert, A. L., M. E. A. Cristescu, L. Frisse, S. Schaack, W. K. Thomas et al., 2008. The rate and spectrum of microsatellite mutation in Caenorhabditis elegans and Daphnia pulex. Genetics 178 2113–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, N. G. C., and P. Fearnhead, 2005. A comparison of three estimators of the population-scaled recombination rate: accuracy and robustness. Genetics 171 2051–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, B.-H., A. J. Windsor, K. J. Schmid, S. Ramos-Onsins, M. E. Schranz et al., 2009. Multilocus patterns of nucleotide diversity, population structure and linkage disequilibrium in Boechera stricta, a wild relative of Arabidopsis. Genetics 181 1021–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens, M., N. J. Smith and P. Donnelly, 2001. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 68 978–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima, F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K., J. Dudley, M. Nei and S. Kumar, 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24 1596–1599. [DOI] [PubMed] [Google Scholar]
- Tamura, K., and M. Nei, 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10 512–526. [DOI] [PubMed] [Google Scholar]
- Taylor, D. J., T. J. Crease and W. M. Brown, 1999. Phylogenetic evidence for a single long-lived clade of crustacean cyclic parthenogens and its implications for the evolution of sex. Proc. R. Soc. Lond. B 266 791–797. [Google Scholar]
- Tishkoff, S. A., E. Dietzsch, W. Speed, A. J. Pakstis, J. R. Kidd et al., 1996. Global patterns of linkage disequilibrium at the CD4 locus and modern human origins. Science 271 1380–1387. [DOI] [PubMed] [Google Scholar]
- Vanoverbeke, J., and L. De Meester, 1997. Among-populational genetic differentiation in the cyclical parthenogen Daphnia magna (Crustacea, Anomopoda) and its relation to geographic distance and clonal diversity. Hydrobiologia 360 135–142. [Google Scholar]
- Wakeley, J., 2000. The effects of subdivision on the genetic divergence of populations and species. Evolution 54 1092–1101. [DOI] [PubMed] [Google Scholar]
- Wakeley, J., and N. Aliacar, 2001. Gene genealogies in a metapopulation. Genetics 159 893–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall, J. D., P. Andolfatto and M. Przeworski, 2002. Testing models of selection and demography in Drosophila simulans. Genetics 162 203–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weider, L. J., M. J. Beaton and P. D. N. Hebert, 1987. Clonal diversity in high-artic populations of Daphnia pulex, a polyploid apomictic complex. Evolution 41 1335–1346. [DOI] [PubMed] [Google Scholar]
- Whitlock, M. C., 2003. Fixation probability and time in subdivided populations. Genetics 164 767–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock, M. C., and N. H. Barton, 1997. The effective size of a subdivided population. Genetics 146 427–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, S., D. Bachtrog and B. Charlesworth, 2003. A survey of chromosomal and nucleotide sequence variation in Drosophila miranda. Genetics 164 1369–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]