Abstract
The quorum-sensing pathway in Vibrio cholerae controls the expression of the master regulator HapR, which in turn regulates several important processes such as virulence factor production and biofilm formation. While HapR is known to control several important phenotypes, there are only a few target genes known to be transcriptionally regulated by HapR. In this work, we combine bioinformatic analysis with experimental validation to discover a set of novel direct targets of HapR. Our results provide evidence for two distinct binding motifs for HapR-regulated genes in V. cholerae. The first binding motif is similar to the motifs recently discovered for orthologs of HapR in V. harveyi and V. vulnificus. However, our results demonstrate that this binding motif can be of variable length in V. cholerae. The second binding motif shares common elements with the first motif, but is of fixed length and lacks dyad symmetry at the ends. The contributions of different bases to HapR binding for this second motif were demonstrated using systematic mutagenesis experiments. The current analysis presents an approach for systematically expanding our knowledge of the quorum-sensing regulon in V. cholerae and other related bacteria.
INTRODUCTION
The Gram-negative bacterium Vibrio cholerae is the causative agent of cholera, an acute dehydrating diarrheal disease that is still endemic in many developing countries. Within its human host, V. cholerae must survive harsh conditions in the stomach before reaching its colonization site in the small intestine. Outside of its human host, V. cholerae can be found in marine environments in association with either biotic substrates, such as copepods, or abiotic substrates, such as sediment (1) To survive and disseminate effectively, V. cholerae must be able to sense and respond to transitions between these starkly contrasting environmental conditions. A major regulatory network controlling the response to changing conditions is the pathway involved in ‘quorum sensing’, generally defined as the regulation of gene expression in response to cell density (2,3). Quorum sensing has been shown to regulate a number of physiological functions that are important for V. cholerae pathogenesis and environmental survival (4–10). Microarray experiments have demonstrated that the quorum-sensing pathway brings about global changes in gene expression (7,11). However, the connections between the regulatory elements of the pathway and the regulated targets have been established for only a limited number of targets.
The master regulator of the quorum-sensing pathway is the TetR-family transcriptional regulator HapR (12). Members of the TetR family share sequentially similar N-terminal binding domains containing helix–turn–helix motifs as well as structurally similar but sequentially diverse C-terminal domains usually involved in dimerization (13). The quorum-sensing pathway controls expression of HapR post-transcriptionally using multiple regulatory small RNAs (14,15). At low cell density, the small RNAs are transcribed at high levels and act to destabilize hapR mRNA, effectively repressing its expression. In contrast, small RNA production is significantly reduced at high-cell density, thereby resulting in increased HapR levels. In addition to post-transcriptional regulation via the small regulatory RNAs, the transcription of hapR is regulated by other regulatory components (16,17). Correspondingly, several genes are activated or repressed by HapR, and these regulated targets, in turn, are responsible for the control of important phenotypes. Thus, an important step in connecting the quorum-sensing pathway to the regulated phenotypes is identification of novel target genes directly regulated by HapR. Bioinformatic approaches can potentially identify genes that are regulatory targets under different natural settings and thus, they can complement studies such as microarray analyses and genetic screens, which are performed under specific conditions.
The identification of target genes regulated by HapR can be facilitated by the identification of specific binding sites for HapR, which can be used to characterize the HapR-binding motif. One of the challenges in doing so is the limited number of HapR targets for which specific binding regions have been identified. A known direct target is the transcription factor AphA, which regulates virulence gene expression via tcpP (18). HapR has been shown to repress AphA by binding to a specific site in the aphA promoter (19). Follow-up experiments have also shown that HapR is autoregulatory and binds to its own promoter. However, it was concluded that the binding region upstream of hapR shows only weak conservation when compared to the corresponding binding region in the aphA promoter (20). Additional targets of HapR have been identified in recent studies (21), but their HapR-binding regions have not been characterized in as much detail as those of aphA and hapR. Thus, the challenge is to develop a predictive approach for identifying novel targets of HapR given the limited experimental information currently available.
In this work, we use an integrated approach combining bioinformatic analysis and experiments to discover additional genes regulated by HapR in V. cholerae. The starting point of our analysis is the observation that the binding sites identified upstream of aphA and hapR are actually quite similar when the complementary strand is taken into consideration. Furthermore, similar binding sequences can be found upstream of aphA and hapR orthologs in other Vibrios. Our analysis suggests that the predicted binding sites can be organized into two distinct categories: Motif 1 binding sites are variable in length whereas Motif 2 binding sites can be represented using the standard position specific weight matrix (PSWM) approach (22). We then carried out experiments to test predicted targets in both categories to validate this categorization based on sequence analysis. In summary, our combined analysis provides evidence that HapR binds to two distinct binding motifs and has led to the identification of several novel HapR-regulated targets in V. cholerae.
MATERIALS AND METHODS
Sequence analysis
Genomic upstream regions from –300 bp relative to the translation start site were obtained, and the corresponding analysis was carried out using the sequence analysis tools at: http://rsat.ulb.ac.be/rsat/ (23,24). Multiple sequence alignments were carried out using the TCoffee alignment tool (25). Genome-wide searches for binding sites were carried out using the programs CONSENSUS and PATSER (22), as implemented at: http://rsat.ulb.ac.be/rsat/. Sequence logos were generated using the Weblogo program (26).
Bacterial strains, plasmids and standard culture conditions
All V. cholerae strains used in this study were derived from E1 Tor C6706 (27) and were propagated in Luria broth (LB) media containing appropriate antibiotics at 37°C. A plasmid used to overexpress the maltose binding protein (MBP)–HapR fusion was constructed by PCR-amplifying the HapR coding sequence and cloning into pMal-c2x (New England Biolabs). Plasmids used to measure HapR-regulated transcription were constructed by PCR-amplifying the promoter regions of each putative target gene and cloning into pBBR-lux (28) to make transcriptional fusions with the luxCDABE genes. The resulting plasmids were introduced into V. cholerae wild type and hapR mutants (8) by conjugal transfer.
Gel retardation assays
MBP–HapR fusions were purified through amylose columns according to the manufacturer's instructions (New England Biolabs). Upstream regions of the putative HapR-regulated targets were PCR-amplified while systematic point mutations in the Motif 2-binding site of vca0880 were generated by hybridizing complementary oligonucleotides containing the desired mutations. The DNA fragments were digested with EcoRI and end-labeled using [α-32P]dATP and the Klenow fragment of DNA polymerase I. Binding reactions contained 0.1 ng DNA and various amounts of MBP–HapR protein in a buffer of 10 mM Tris–HCl (pH 7.9), 1 mM EDTA, 1 mM DTT, 60 mM KCl, 30 mg/ml poly(dI–dC) DNA, 20 mg/ml BSA and 10% glycerol. After a 20-min incubation at room temperature, samples were size-fractionated using 5% polyacrylamide gels in 1 × TAE buffer (20 mM Tris–acetate, 1 mM EDTA, pH 8.5) at 4°C. Radioactivity of free DNA and HapR–DNA complexes was visualized using a StormB840 PhosphorImager (Molecular Dynamics).
Luminescence assay of putative HapR-regulated genes
The V. cholerae wild type and hapR mutant strains containing transcriptional lux fusion plasmids were grown in LB with appropriate antibiotics at 37°C overnight, diluted by a ratio of 1:1000 in fresh LB and incubated at 37°C. Luminescence was read at 2-h intervals for 8 h using a Bio-Tek Synergy HT spectrophotometer. Relative Light Units are defined as light unit/OD600.
RESULTS
Sequence analysis of known HapR targets and orthologs
The V. cholerae quorum-sensing regulator HapR is a TetR-family protein that functions both as an activator and as a repressor. Quorum sensing in V. cholerae has been demonstrated to regulate a variety of cellular functions, but HapR-binding sites have only been determined for two targets thus far: aphA and hapR itself. It has been shown that HapR binds to a site from 61 to 82 bp upstream of the transcriptional start site in the aphA promoter (19) and to a site 12 to 33 bp downstream of the transcriptional start site for hapR (20). It is likely that autoregulation of HapR along with virulence regulation via AphA are conserved across the Vibrios, in which case the corresponding binding sites should show strong conservation across species. Thus, we carried out a multiple sequence alignment of orthologous regions across closely related Vibrio species for aphA and its orthologs as well as for hapR and its orthologs. As noted, a previous study had shown that the binding regions upstream of hapR and aphA show only weak conservation when the direct strands are compared (20) To check for similarity in the binding sites when the complementary strands are also considered, we performed a sequence alignment of the direct strand in the region upstream of aphA with the complementary strand to the region upstream of hapR. The alignment results showed that the binding regions for both genes are quite similar when the complementary strand is also considered (see Supplementary Data, Figure S1), indicating that bioinformatic procedures can be used to represent the corresponding binding motif as discussed further below.
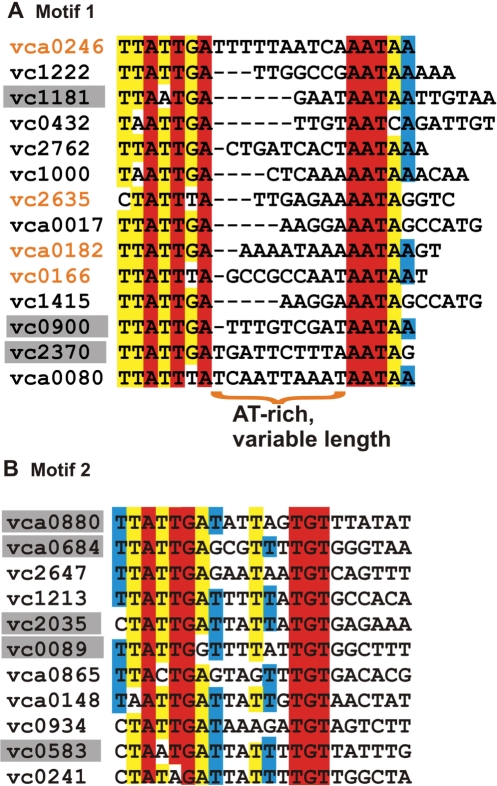
The upstream regions of aphA orthologs and hapR orthologs were then aligned, taking the complementary strands into account, in order to identify a consensus binding sequence. A notable feature of the conserved binding regions (Table 1) is that for a subset of the genes (specifically hapR orthologs opaR, luxR, smcR and litR), it is possible to identify a conserved region with dyad symmetry similar to the binding sites that have recently been identified for HapR orthologs in Vibrio vulnificus and Vibrio harveyi (29,30). These sites are highlighted in Table 1 under the heading Motif 1. A more striking observation is that the binding sites for aphA and its orthologs and for hapR do not follow the same pattern. While the initial sequence of the binding sites of these genes are similar to those in Motif 1, the concluding sequence does not show the corresponding dyad symmetry and has the absolutely conserved sequence ‘TGT’ instead of the Motif 1 consensus ‘AATAR’ (where R represents A/G). Binding sites following this second pattern are referred to as Motif 2 sites (Table 1). Since HapR is able to activate the V. harveyi luminescence operon, which is a direct target of the HapR ortholog, LuxR, we reasoned that these binding sites in closely related species can be used to construct a preliminary representation of binding motifs for HapR itself (6). Thus, the preliminary analysis identifies potential HapR-binding sites and indicates that there are two distinct HapR binding motifs.
Table 1.
Predicted HapR-binding sites upstream of orthologs of hapR and aphA in the Vibrios and upstream of four additional genes in V. cholerae (VCA0865, VC0900, VC2370 and VCA0080) known to be regulated by HapR
 |
Binding sites are categorized according to two distinct motifs.
aBinding sites on complementary strands are highlighted.
The above observations are further strengthened when we consider additional V. cholerae genes that are known to be regulated by HapR, but for which specific binding sites have not yet been identified. These targets include hapA (31) which encodes the protease HapA and 3 genes which encode GGDEF and/or GGDEF/EAL domain proteins: vc0900, vc2370 and vca0080 (21). We identified potential HapR binding sites for these genes by searching their upstream regions for patterns similar to Motif 1 or Motif 2. Specifically, Motif 1-binding sites were identified for vc0900, vc2370 and vca0080 and a Motif 2-binding site was identified upstream of hapA. These sites were included in the initial list of predicted HapR-binding sites (Table 1). It is interesting to note that all the known targets listed in Table 1 with Motif 1-binding sites contain the motif AATAR. In some cases (e.g. litR), there are multiple contiguous occurrences of this motif, in which case the proposed binding site is taken to end at the last occurrence of the AATAR motif. It is noteworthy that for Motif 1 targets listed in Table 1 with a single AATAR concluding motif, the distance between the initial conserved sequence and the concluding AATAR sequence is variable and thus, Motif 1-binding sites appear to be of variable length. As we discuss in the following sections, characterization of the HapR-binding motif according to the two categories outlined in Table 1 leads to testable predictions for novel target genes regulated by HapR. A schematic flowchart outlining the procedure followed is shown in the Supplementary Data (Figure S2).
Motif 1 target predictions
Analysis of the Motif 1-binding sites listed in Table 1 reveals that the motif can be characterized by: (i) an initial 7 bp conserved region represented by the motif YWATTKA (where Y stands for C/T, W stands for A/T, and K stands for G/T); (ii) followed by a region of variable length which is typically AT-rich; and (iii) concluding with the sequence AATAR. This motif is similar to the binding motif recently identified for HapR orthologs in V. vulnificus and V. harveyi, but an important distinction is that our analysis considers the binding length to be variable. Note that while the initial conserved sequence can be summarized by YWATTKA, this would include sequences (e.g. CAATTTA) which are not observed in any of the known targets listed in Table 1. A more restrictive constraint for the initial 7-bp sequence is that it corresponds to one of the three patterns represented by YTATTGA, TAATTGA or YTATTTA. We carried out a genome-wide search for genes whose upstream regions include a potential binding site satisfying the constraints noted above. Since the binding region can be variable in length, we considered binding sites ranging from 16 to 22 bp. In some cases, there are multiple occurrences of the motif AATAR, so the length of the spacer region between the conserved elements at the ends is not well defined (Figure 1A). Nevertheless, even if we consider only those targets with a single occurrence of the concluding motif AATAR, the length of the binding site is seen to vary over the entire range considered (i.e. from 16 to 22 bp). The genome-wide search for potential binding sites yielded 33 intergenic regions. For each binding site length, we picked one to two candidate genes for experimental validation. These genes, along with their predicted binding sites, are shown in Figure 1A. (Note that for the 16-bp targets, we considered targets with 1-bp mutations compared to the patterns noted above, since searching for the exact patterns yielded only one hypothetical gene as a predicted target.) As discussed below, all of the targets listed in Figure 1A were subsequently validated by gel retardation assays. The functions of the regulated target genes along with predictions for the corresponding operons are provided in Table S1. These results demonstrate that the proposed sequence pattern is an effective way of representing a binding motif for HapR and further indicate that there are many more genes that are potentially regulated by HapR.
Figure 1.
Alignment of predicted Motif 1 (A) and Motif 2 (B) binding sites in intergenic regions of V. cholerae. Genes for which predicted binding sites are on the complementary strand are indicated with gray shading. Loci labeled in orange indicate the first of two divergently transcribed genes that contain a HapR binding site in the intergenic region. The 22-bp-binding regions are shown for each binding site. While the predicted binding site length is seen to be variable for Motif 1 genes, Motif 2-binding sites show very weak conservation beyond 16 bp. The alignment demonstrates the distinctions between the two proposed motifs in terms of concluding sequences and binding site lengths.
Motif 2 target predictions
Unlike binding sites corresponding to Motif 1, Motif 2-binding sites contain a strongly conserved 16-bp region that ends with the sequence TGT. This suggests that Motif 2-binding sites can be represented using a PSWM corresponding to a 16-bp binding site. Accordingly, the predicted Motif 2-binding sites (listed in Table 1) were used to generate a preliminary PSWM. This PSWM was used to score potential binding sites in the upstream regions of all genes in the V. cholerae genome. We also specifically considered genes which were shown to be differentially regulated in a hapR mutant as compared to WT in previous microarray studies (7). The five highest-scoring novel binding sites in the whole-genome scan and the highest-scoring target also identified in microarray data (vca0880) were chosen for experimental validation. The predicted targets and the corresponding HapR-binding sites are listed in Figure 1B. Target protein function and operon predictions are provided in Table S1. As discussed further in the experimental results, all the predicted targets except one (vc0241) were validated by gel retardation assays. The binding sites for the experimentally validated targets were used to generate a revised PSWM for HapR based solely on targets in V. cholerae. Using the revised PSWM to scan the V. cholerae genome, additional predictions for high-scoring target genes were made. Out of these, three additional genes were chosen (based on PSWM scores and microarray data) for further experimental validation. All three target genes were confirmed experimentally, and the corresponding binding sites are listed in Figure 1B. It is noteworthy that while the 7-bp initial region of Motif 2-binding sites (with consensus sequence YTATTGA) is similar to the corresponding region in Motif 1-binding sites, the concluding sequence for Motif 2-binding sites is strongly conserved (TGT) and distinct from Motif 1-binding sites (AATAR). Furthermore, alignment of the predicted binding regions (Figure 1B) shows minimal conservation beyond the 16-bp region ending with TGT, indicating the Motif 2-binding sites have a fixed binding length of 16 bp and do not exhibit the dyad symmetry typical of Motif 1-binding sites. Our results thus provide evidence for two distinct binding motifs for HapR and have led to the identification of novel targets of HapR corresponding to each motif as described further below.
HapR binds to the promoter regions of newly identified targets
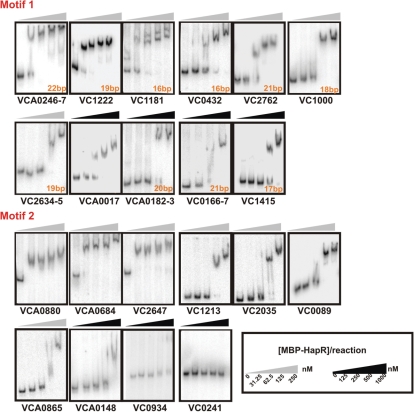
In order to test whether HapR protein can bind to the putative target promoters containing either Motif 1 or Motif 2-binding sites, an MBP-HapR fusion was constructed. This tagged HapR protein was able to complement a hapR deletion mutant when using the HapR-activated heterologous luxCDABE operon from V. harveyi as a bioluminescent reporter (data not shown). For each promoter region that was tested, a 300–400-bp region containing the binding site was PCR amplified and radiolabeled. Gel retardation assays were performed to detect binding of the MBP-HapR protein to each of the promoter regions. MBP-HapR bound to all 11 of the tested promoters containing a Motif 1-binding site and all but one (vc0241) of the eight promoters containing a Motif 2-binding site (Figure 2) Thus, the Motif 1-binding sequence reliably predicts promoter regions capable of being bound by HapR. The original Motif 2 PSWM also predicts HapR-binding sites fairly accurately, and it was further refined based on experimentally validated targets.
Figure 2.
HapR binds to promoters containing either Motif 1- or Motif 2-binding sites. HapR's ability to bind to various promoters containing either of the two consensus binding sites was determined by EMSA in the presence of poly(dI–dC). Due to differences in HapR-binding affinity, two different sets of HapR protein concentrations were used, as indicated by the color of the wedges above the gel shifts. Solid wedges represent four times more protein than unfilled wedges. Since Motif 1 does not have a set length, promoters containing Motif 1-binding sites of varying lengths were tested, and the length of the putative binding site for each promoter is indicated. Consensus binding sequences that fall between two divergently transcribed genes are labeled with both flanking gene locus numbers. The double bands of vc1181 resulted from the nonspecific labeled PCR products of the vc1181 promoter region.
HapR activates or represses expression from newly identified target promoters
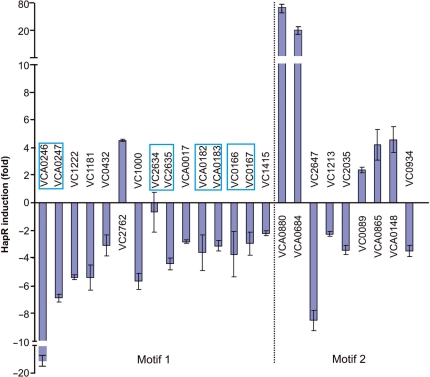
The above gel retardation analysis suggests that the target genes validated experimentally are directly regulated by HapR. In order to test whether regulation by HapR results in activation or repression of transcription for these genes, transcriptional lux fusions were constructed. PCR products used for the electrophoretic mobility shift assays (EMSAs) were cloned upstream of luxCDABE in the pBBR-lux reporter plasmid. Many of these promoters are not functional in Escherichia coli (data not shown). Therefore, the resulting constructs were transformed into wild-type and hapR-deleted strains of V. cholerae, which is their native host. Some binding sites were located between divergently transcribed genes, so these promoters were cloned into the lux reporter in both orientations. All the promoters showed either activation or repression by HapR ranging from ∼2-fold to almost 80-fold (Figure 3). For three of the binding sites located between divergently transcribed genes (vca0246/7, vc0166/7 and vca0182/3), HapR was able to repress expression from the promoters when they were oriented in either direction. On the other hand, for the binding site located between vc2634 and vc2635, HapR only repressed expression from the promoter when it was oriented in the same direction as vc2635, while it had no effect on expression in the other direction. A control plasmid containing Plac-luxCDABE displayed similar Lux activity in wild-type and hapR mutants (data not shown). Therefore, HapR is not only able to bind to promoters containing the Motif 1 or Motif 2 consensus binding sequences, but it is also able to affect transcription levels of genes expressed from these promoters.
Figure 3.
HapR regulates expression from promoters containing either Motif 1 or Motif 2. Promoter-lux fusions were constructed and transformed into WT and hapR-deleted strains. Fold induction by HapR was determined as the ratio between bioluminescence readings from the reporter fusions in WT/ΔhapR. Negative fold induction indicates repression. Each bar represents three data points, and error bars represent standard deviations. Divergently transcribed genes with HapR binding sites located in between are indicated with boxes.
Validation of the PSWM for Motif 2
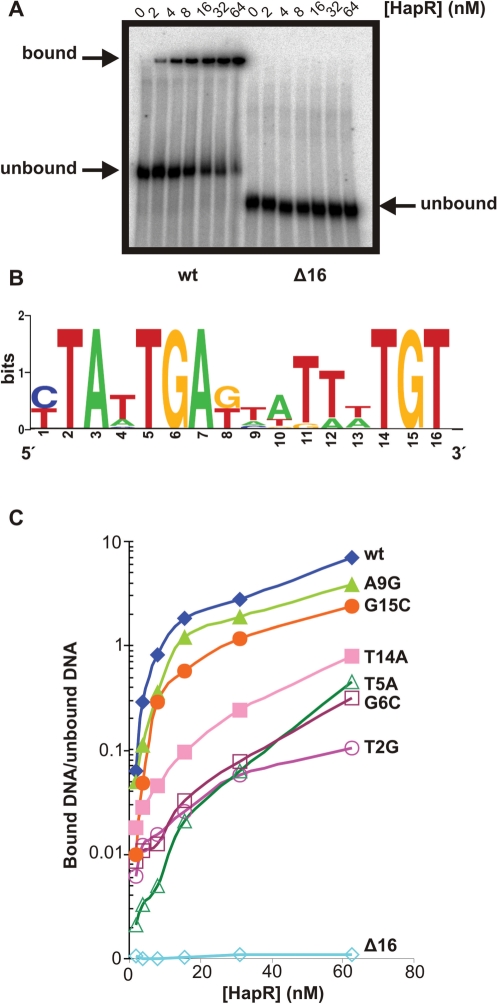
The consensus binding sequence of the HapR ortholog in V. harveyi, LuxR, was studied in detail in a recent paper (30). Since this binding sequence is similar to Motif 1, we focused on a Motif 2-binding site for further analysis in this study. Single-point mutations were made within the Motif 2-binding sequence located in the promoter region of vca0880 to validate the PSWM for Motif 2. The promoter of vca0880 was chosen because it is bound and activated very strongly by HapR (Figures 2 and 3), and would thus allow for better resolution of the binding abilities of various point mutants. EMSAs were performed to determine the extent of binding of MBP-HapR to a 60-bp fragment of the promoter region of vca0880 containing the Motif 2 consensus binding sequence as well as to six other versions of the fragment, each with a different single-point mutation. DNA fragments containing the desired point mutations were generated by hybridizing complementary oligonucleotides. A fragment lacking the entire consensus binding site (Δ16) was used as a negative control. As can be seen in Figure 4A, MBP-HapR bound to the wild-type fragment with a fairly high affinity while it was unable to bind to a fragment that was lacking the consensus binding site. It should be noted, however, that the difference in length due to binding site deletion could impact DNA stiffness, which in turn could potentially be an important factor for HapR binding. Each point mutation affected HapR-binding affinity to varying degrees, and our results agree with the PSWM in that a mutation at position 9, which is not highly conserved across binding sites known to be bound by HapR, affected HapR binding affinity the least out of all the tested mutations, and in fact, bound to HapR almost as well as the wild-type sequence did. The other point mutations altered highly conserved nucleotides, and thus affected HapR binding to a greater degree (Figure 4B and C). The T5A mutation resulted in significant loss of binding, which is consistent with the negative EMSA for the vc0241 promoter, which also contains the same T5A mutation. The G6C mutation also led to a significant loss of binding, which is consistent with previous studies of HapR binding to the aphA promoter (19). While the G15C mutation had a less significant effect on HapR binding than might have been expected based on the PSWM, the change was still more deleterious than the A9G mutation. The T14A mutation had a strong effect on HapR binding, and a previous study (20) showed that the single change, T16C, prevents HapR binding. This suggests that the ‘TGT’ motif is important for HapR binding, although the G15C mutation by itself does not have a strong effect on HapR binding. Taken together, these results suggest that the PSWM that was constructed based on confirmed binding sites is reasonably accurate in terms of the importance of each base's contribution to HapR binding.
Figure 4.
Point mutations in the Motif 2-consensus-binding site alter HapR binding affinity as predicted by the PSWM. (A) EMSAs were performed on a 60-bp region of the vca0880 promoter that contains the Motif 2-binding site as well as the same region with the binding site deleted. (B) Sequence logo for Motif 2-binding sites reflecting the V. cholerae-specific PSWM derived after one iteration of the procedure outlined in Figure S1. (C) HapR-binding affinity was determined by quantifying the amount of bound and unbound DNA and calculating the ratio of bound/unbound. The effect of various point mutations in the binding site on HapR-binding affinity was determined by EMSAs over a range of HapR concentrations.
DISCUSSION
In this paper, we present a sequence-based strategy for uncovering novel genes regulated by the quorum-sensing master regulator HapR in V. cholerae. Previously identified HapR-binding regions in combination with sequence alignments of orthologous regions across closely related species, taking both the direct and reverse complementary strand into account, were used to identify potential binding sites. Potential binding sites that were experimentally verified were then used to further refine the binding motif. The predicted binding sites were classified into two categories, and this scheme of classification led to several novel predictions for regulated targets, some of which have been validated in this study. The results presented in this study are thus an important step towards understanding global regulation by quorum sensing in V. cholerae.
Comparison of Motif 1- versus Motif 2-binding Motifs
To compare the two proposed binding motifs, consider the sequence logos generated using 22-bp sequences from the predicted start positions of the binding sites for all the experimentally validated targets (Figure S3). Note that the initial 7-bp region is quite similar for the two motifs. However, all of the binding sequences used to generate the sequence logo for Motif 1 end with the sequence AATAR (which is the complement of the initial binding site sequence YTATT), while all of the sequences for Motif 2 end with TGT. Additionally, the intervening region between the beginning YTATT and the ending AATAR for Motif 1 is not strongly conserved, and binding sites can range from 16 to 22 bp. Note that, even with the 7-bp variability in the spacer region, the probability for a random sequence to contain the motif AATAR is ∼1/73. In contrast, Motif 2-binding sites show strong conservation of the sequence TGT in positions 14–16 and very little conservation beyond that, indicating that the binding sites are of fixed length and are distinct from those of Motif 1. Another interesting distinction is that all but one of the Motif 1 targets are repressed by HapR, whereas Motif 2 targets can be either repressed or activated. It will be of interest to explore whether the presence of two distinct motifs is related to the action of a small-molecule ligand that was suggested to bind HapR by recent work analyzing the structure of HapR (32). It will also be of interest to explore the mechanism of activation or repression by HapR at target gene promoters. Our results indicate that there is no correlation between binding site orientation (direct or reverse complement) and activation/repression of a regulated target (i.e. HapR can function as an activator or a repressor regardless of whether the target promoter binding site is located on the direct strand or on the complementary strand). It will be of interest to carry out more in-depth studies to identify characteristics of binding sites that determine whether a target gene is activated or repressed by HapR.
A comparison with recent studies analyzing the regulons for the HapR orthologs LuxR (V. harveyi) (30) and SmcR (V. vulnificus) (29) indicates that 22-bp-long Motif 1-binding sites are similar to the binding motif identified for these HapR orthologs. Since the present work has shown that Motif 1-binding sites can vary in length from 16 to 22 bp, it would be of interest to investigate if this feature also holds true for binding sites of LuxR and SmcR. Along the same lines, it will be of interest to investigate whether or not the presence of two distinct binding motifs is specific to HapR in V. cholerae or whether it also applies to HapR orthologs in other Vibrios.
Novel direct targets reveal new roles for HapR-mediated quorum sensing
Not only has our work revealed that HapR binds to two distinct sequence motifs, but it has also identified new direct targets of HapR. Some of these newly discovered HapR-regulated genes reveal novel connections between known components of the HapR regulatory network while others hint at previously unknown roles for HapR in V. cholerae biology and pathogenesis. Examples of targets that affect known network components include vc1213, which encodes VarA, vc0934, which encodes VpsL and vc1222, which encodes IHFα. VarA, the response regulator for the VarS/VarA two-component regulatory system (33), has already been shown to modulate virulence factor expression (34) as well as the expression of HapR itself (15). Here, we uncover a feedback loop in this system by showing that HapR represses expression from the varA promoter, adding yet another layer of complexity to quorum-sensing regulation in V. cholerae. Additionally, HapR has already been shown in numerous works to regulate biofilm formation (5). Previous studies have shown that this regulation is mediated through two transcriptional activators, VpsR and VpsT (11,21). VpsR and VpsT, in turn, are required for expression from the two Vibrio polysaccharide biosynthesis operons vpsA-K and vpsL-Q (35,36). Direct binding of HapR to the vpsT promoter has been shown (21). In that same study, weak binding of HapR to the vpsL promoter was also demonstrated, but it was difficult to conclude whether or not this binding was physiologically relevant. In this study, we confirm the positive EMSA results and go further to identify the HapR binding sequence in the vpsL promoter. This provides further credibility to the proposition that HapR regulates biofilm production not only through the transcriptional regulators, VpsR and VpsT, but also directly by binding to a vps biosynthesis gene promoter. Finally, HapR regulation of the virulence factors, cholera toxin (CT) and the toxin-coregulated pilus (TCP), via AphA has been studied extensively (6,8,37,38). Here, we show that HapR regulates IHFα, which has previously been shown to positively regulate expression from both the tcpA and ctx promoters (39). This indicates that HapR is able to regulate known virulence factors via distinct pathways, which may allow for integration of cell-density information with other sensory inputs to precisely regulate virulence gene expression.
Several newly identified targets indicate completely novel roles for HapR (Table S1). The most strongly induced target that we identified is vca0880, which encodes a hypothetical protein. vca0880 is found in an operon with vca0881, vca0882 and vca0883, which are all annotated as hypothetical proteins. Interestingly, a protein BLAST search showed homology between VCA0883 and the L1 component of hemolysin BL as well as the nonhemolytic enterotoxin NheB, both of which are found in Bacillus cereus (40). This is potentially a new mechanism by which HapR controls the virulence of V. cholerae and warrants further investigation. Additionally, one of the newly identified HapR-regulated genes is vca0183, which is annotated as a ferrisiderophore reductase. In fact, sequence analysis reveals that it belongs to a class of microbial globins known as flavohemoproteins. The prototypic flavohemoprotein, Hmp, found in E. coli and Salmonella enterica serovar typhimurium, has been implicated in responses to nitric oxide (NO) and nitrosative stress (41,42). Transcription from the E. coli hmp promoter has been shown to be activated by the addition of NO, and its gene product acts to detoxify NO (43–46). Interestingly, upstream of and transcribed divergently from vca0183 is vca0182, which encodes a homolog of the NO-responsive transcriptional regulator, NorR. Our results show that both vca0182 and vca0183 are repressed by HapR, thereby suggesting a potentially novel role for HapR in the response of V. cholerae to NO stress.
Generalizing HapR-binding motifs to expand known HapR regulon
All but one of the predicted targets were validated experimentally in this study, which not only indicates that the proposed motifs can successfully predict HapR-binding sites, but also suggests that the actual binding motifs could be more general. A previous study (11) identified a different consensus motif for predicting HapR targets, and this motif shares some common elements with motifs identified in this work but emphasizes the contributions of different bases to HapR binding. It is likely that future work will combine these different approaches to provide evidence for a broader characterization of HapR-binding motifs. Indeed, this is already indicated by sequence analysis of other known targets of HapR in V. cholerae for which the binding regions have not yet been identified but binding of HapR to upstream regions has been demonstrated. These include the transcriptional regulator VpsT (VCA0952) (21), a transcription factor required for competence in V. cholerae (VC1917) (4), the regulator H-NS (VC1130) (47), a GGDEF-domain protein (VCA0074) and an EAL-domain protein (VC1851) (21). For the above regulated targets, we have identified potential binding sites which are very similar to Motif 1-binding sites but with a significant difference being that the predicted binding region is in some cases >22 bp. These predictions, if validated in future studies, would yield important insight into the regulation of critical processes such as biofilm formation and competence by HapR, and they would also provide evidence for a broader range of binding lengths and patterns corresponding to Motif 1 for HapR-binding sites.
Our method of combining computational and experimental approaches for identifying HapR targets allows for the identification of targets that would be difficult to identify by experimental means alone. The traditional approach to discovering the regulon of a particular transcription factor is to compare the gene expression profile of a wild-type strain with that of a strain deleted for the transcription factor, which is often done using microarray experiments. The success of this approach is dependent on the samples being collected under conditions that allow for optimal activation or repression of target genes, which is impossible to ensure when the target genes are not known. Even when the genes are known, it is still difficult to determine what in vitro experimental conditions to use. Thus, the incorporation of a computational approach that focuses on promoter sequences and is unaltered by experimental conditions increases the number of target genes that can be found and opens up new avenues for research on the HapR regulon.
Summary
In conclusion, we have demonstrated that the iterative use of bioinformatic predictions and experimental validation can be used to progressively refine the initial motif representation of the HapR-binding site to successfully predict several novel members of the HapR regulon. It is hoped that future work combining bioinformatic approaches and experiments will uncover even more targets regulated by quorum sensing via HapR and will lead to the discovery of novel regulon members for related global regulatory proteins.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Jeffress Foundation and a seed grant from the Institute for Critical Technology and Applied Science at Virginia Tech (to R.V.K); the National Institutes of Health (R01AI072479 to J.Z); a National Institutes of Health T32 Bacteriology training grant (to A.T); and an award from the China Scholarship Council (to T.C). Funding for open access charge: National Institutes of Health (R01AI072479).
Conflict of interest statement: None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Dr Bonnie Bassler for providing the plasmid pBBR-lux.
REFERENCES
- 1.Vezzulli L, Guzman CA, Colwell RR, Pruzzo C. Dual role colonization factors connecting Vibrio cholerae's lifestyles in human and aquatic environments open new perspectives for combating infectious diseases. Curr. Opin. Biotechnol. 2008;19:254–259. doi: 10.1016/j.copbio.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 2.Fuqua WC, Winans SC, Greenberg EP. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 1994;176:269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 4.Meibom KL, Blokesch M, Dolganov NA, Wu CY, Schoolnik GK. Chitin induces natural competence in Vibrio cholerae. Science. 2005;310:1824–1827. doi: 10.1126/science.1120096. [DOI] [PubMed] [Google Scholar]
- 5.Hammer BK, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol. Microbiol. 2003;50:101–104. doi: 10.1046/j.1365-2958.2003.03688.x. [DOI] [PubMed] [Google Scholar]
- 6.Miller MB, Skorupski K, Lenz DH, Taylor RK, Bassler BL. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell. 2002;110:303–314. doi: 10.1016/s0092-8674(02)00829-2. [DOI] [PubMed] [Google Scholar]
- 7.Zhu J, Mekalanos JJ. Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev. Cell. 2003;5:647–656. doi: 10.1016/s1534-5807(03)00295-8. [DOI] [PubMed] [Google Scholar]
- 8.Zhu J, Miller MB, Vance RE, Dziejman M, Bassler BL, Mekalanos JJ. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl Acad. Sci. USA. 2002;99:3129–3134. doi: 10.1073/pnas.052694299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nielsen AT, Dolganov NA, Otto G, Miller MC, Wu CY, Schoolnik GK. RpoS controls the Vibrio cholerae mucosal escape response. PLoS Pathog. 2006;2:e109. doi: 10.1371/journal.ppat.0020109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vaitkevicius K, Lindmark B, Ou G, Song T, Toma C, Iwanaga M, Zhu J, Andersson A, Hammarstrom ML, Tuck S, et al. A Vibrio cholerae protease needed for killing of Caenorhabditis elegans has a role in protection from natural predator grazing. Proc Natl Acad. Sci. USA. 2006;103:9280–9285. doi: 10.1073/pnas.0601754103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yildiz FH, Liu XS, Heydorn A, Schoolnik GK. Molecular analysis of rugosity in a Vibrio cholerae O1 El Tor phase variant. Mol. Microbiol. 2004;53:497–515. doi: 10.1111/j.1365-2958.2004.04154.x. [DOI] [PubMed] [Google Scholar]
- 12.Jobling MG, Holmes RK. Characterization of hapR, a positive regulator of the Vibrio cholerae HA/protease gene hap, and its identification as a functional homologue of the Vibrio harveyi luxR gene. Mol. Microbiol. 1997;26:1023–1034. doi: 10.1046/j.1365-2958.1997.6402011.x. [DOI] [PubMed] [Google Scholar]
- 13.Ramos JL, Martinez-Bueno M, Molina-Henares AJ, Teran W, Watanabe K, Zhang X, Gallegos MT, Brennan R, Tobes R. The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 2005;69:326–356. doi: 10.1128/MMBR.69.2.326-356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 15.Lenz DH, Miller MB, Zhu J, Kulkarni RV, Bassler BL. CsrA and three redundant small RNAs regulate quorum sensing in Vibrio cholerae. Mol. Microbiol. 2005;58:1186–1202. doi: 10.1111/j.1365-2958.2005.04902.x. [DOI] [PubMed] [Google Scholar]
- 16.Liu Z, Hsiao A, Joelsson A, Zhu J. The transcriptional regulator VqmA increases expression of the quorum-sensing activator HapR in Vibrio cholerae. J. Bacteriol. 2006;188:2446–2453. doi: 10.1128/JB.188.7.2446-2453.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Z, Miyashiro T, Tsou A, Hsiao A, Goulian M, Zhu J. Mucosal penetration primes Vibrio cholerae for host colonization by repressing quorum sensing. Proc. Natl Acad. Sci. USA. 2008;105:9769–9774. doi: 10.1073/pnas.0802241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skorupski K, Taylor RK. A new level in the Vibrio cholerae ToxR virulence cascade: AphA is required for transcriptional activation of the tcpPH operon. Mol. Microbiol. 1999;31:763–771. doi: 10.1046/j.1365-2958.1999.01215.x. [DOI] [PubMed] [Google Scholar]
- 19.Kovacikova G, Skorupski K. Regulation of virulence gene expression in Vibrio cholerae by quorum sensing: HapR functions at the aphA promoter. Mol. Microbiol. 2002;46:1135–1147. doi: 10.1046/j.1365-2958.2002.03229.x. [DOI] [PubMed] [Google Scholar]
- 20.Lin W, Kovacikova G, Skorupski K. Requirements for Vibrio cholerae HapR binding and transcriptional repression at the hapR promoter are distinct from those at the aphA promoter. J. Bacteriol. 2005;187:3013–3019. doi: 10.1128/JB.187.9.3013-3019.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waters CM, Lu W, Rabinowitz JD, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic di-GMP levels and repression of vpsT. J. Bacteriol. 2008;190:2527–2536. doi: 10.1128/JB.01756-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hertz GZ, Stormo GD. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics. 1999;15:563–577. doi: 10.1093/bioinformatics/15.7.563. [DOI] [PubMed] [Google Scholar]
- 23.Thomas-Chollier M, Sand O, Turatsinze JV, Janky R, Defrance M, Vervisch E, Brohee S, van Helden J. RSAT: regulatory sequence analysis tools. Nucleic Acids Res. 2008;36:W119–W127. doi: 10.1093/nar/gkn304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Helden J. Regulatory sequence analysis tools. Nucleic Acids Res. 2003;31:3593–3596. doi: 10.1093/nar/gkg567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poirot O, O'Toole E, Notredame C. Tcoffee@igs: a web server for computing, evaluating and combining multiple sequence alignments. Nucleic Acids Res. 2003;31:3503–3506. doi: 10.1093/nar/gkg522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thelin KH, Taylor RK. Toxin-coregulated pilus, but not mannose-sensitive hemagglutinin, is required for colonization by Vibrio cholerae O1 El Tor biotype and O139 strains. Infect. Immun. 1996;64:2853–2856. doi: 10.1128/iai.64.7.2853-2856.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hammer BK, Bassler BL. Regulatory small RNAs circumvent the conventional quorum sensing pathway in pandemic Vibrio cholerae. Proc. Natl Acad. Sci. USA. 2007;104:11145–11149. doi: 10.1073/pnas.0703860104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee DH, Jeong HS, Jeong HG, Kim KM, Kim H, Choi SH. A consensus sequence for binding of SmcR, a Vibrio vulnificus LuxR homologue, and genome-wide identification of the SmcR regulon. J. Biol. Chem. 2008;283:23610–23618. doi: 10.1074/jbc.M801480200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pompeani AJ, Irgon JJ, Berger MF, Bulyk ML, Wingreen NS, Bassler BL. The Vibrio harveyi master quorum-sensing regulator, LuxR, a TetR-type protein is both an activator and a repressor: DNA recognition and binding specificity at target promoters. Mol. Microbiol. 2008;70:76–88. doi: 10.1111/j.1365-2958.2008.06389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Silva AJ, Pham K, Benitez JA. Haemagglutinin/protease expression and mucin gel penetration in El Tor biotype Vibrio cholerae. Microbiology. 2003;149:1883–1891. doi: 10.1099/mic.0.26086-0. [DOI] [PubMed] [Google Scholar]
- 32.De Silva RS, Kovacikova G, Lin W, Taylor RK, Skorupski K, Kull FJ. Crystal structure of the Vibrio cholerae quorum-sensing regulatory protein HapR. J. Bacteriol. 2007;189:5683–5691. doi: 10.1128/JB.01807-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lapouge K, Schubert M, Allain FH, Haas D. Gac/Rsm signal transduction pathway of gamma-proteobacteria: from RNA recognition to regulation of social behaviour. Mol. Microbiol. 2008;67:241–253. doi: 10.1111/j.1365-2958.2007.06042.x. [DOI] [PubMed] [Google Scholar]
- 34.Wong SM, Carroll PA, Rahme LG, Ausubel FM, Calderwood SB. Modulation of expression of the ToxR regulon in Vibrio cholerae by a member of the two-component family of response regulators. Infect. Immun. 1998;66:5854–5861. doi: 10.1128/iai.66.12.5854-5861.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Casper-Lindley C, Yildiz FH. VpsT is a transcriptional regulator required for expression of vps biosynthesis genes and the development of rugose colonial morphology in Vibrio cholerae O1 El Tor. J. Bacteriol. 2004;186:1574–1578. doi: 10.1128/JB.186.5.1574-1578.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yildiz FH, Dolganov NA, Schoolnik GK. VpsR, a member of the response regulators of the two-component regulatory systems, is required for expression of vps biosynthesis genes and EPS(ETr)-associated phenotypes in Vibrio cholerae O1 El Tor. J. Bacteriol. 2001;183:1716–1726. doi: 10.1128/JB.183.5.1716-1726.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kovacikova G, Lin W, Skorupski K. The virulence activator AphA links quorum sensing to pathogenesis and physiology in Vibrio cholerae by repressing the expression of a penicillin amidase gene on the small chromosome. J. Bacteriol. 2003;185:4825–4836. doi: 10.1128/JB.185.16.4825-4836.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin W, Kovacikova G, Skorupski K. The quorum sensing regulator HapR downregulates the expression of the virulence gene transcription factor AphA in Vibrio cholerae by antagonizing Lrp- and VpsR-mediated activation. Mol. Microbiol. 2007;64:953–967. doi: 10.1111/j.1365-2958.2007.05693.x. [DOI] [PubMed] [Google Scholar]
- 39.Stonehouse E, Kovacikova G, Taylor RK, Skorupski K. Integration host factor positively regulates virulence gene expression in Vibrio cholerae. J. Bacteriol. 2008;190:4736–4748. doi: 10.1128/JB.00089-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Granum PE. Bacillus cereus and its toxins. Soc Appl Bacteriol Symp Ser. 1994;23:61S–66S. [PubMed] [Google Scholar]
- 41.Membrillo-Hernandez J, Coopamah MD, Anjum MF, Stevanin TM, Kelly A, Hughes MN, Poole RK. The flavohemoglobin of Escherichia coli confers resistance to a nitrosating agent, a “nitric oxide releaser,” and paraquat and is essential for transcriptional responses to oxidative stress. J. Biol. Chem. 1999;274:748–754. doi: 10.1074/jbc.274.2.748. [DOI] [PubMed] [Google Scholar]
- 42.Stevanin TM, Poole RK, Demoncheaux EAG, Read RC. Flavohemoglobin Hmp Protects Salmonella enterica Serovar Typhimurium from Nitric Oxide-Related Killing by Human Macrophages. Infect. Immun. 2002;70:4399–4405. doi: 10.1128/IAI.70.8.4399-4405.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poole RK, Anjum MF, Membrillo-Hernandez J, Kim SO, Hughes MN, Stewart V. Nitric oxide, nitrite, and Fnr regulation of hmp (flavohemoglobin) gene expression in Escherichia coli K-12. J. Bacteriol. 1996;178:5487–5492. doi: 10.1128/jb.178.18.5487-5492.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Corker H, Poole RK. Nitric oxide formation by Escherichia coli. Dependence on nitrite reductase, the NO-sensing regulator Fnr, and flavohemoglobin Hmp. J. Biol. Chem. 2003;278:31584–31592. doi: 10.1074/jbc.M303282200. [DOI] [PubMed] [Google Scholar]
- 45.Cruz-Ramos H, Crack J, Wu G, Hughes MN, Scott C, Thomson AJ, Green J, Poole RK. NO sensing by FNR: regulation of the Escherichia coli NO-detoxifying flavohaemoglobin, Hmp. EMBO J. 2002;21:3235–3244. doi: 10.1093/emboj/cdf339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gardner PR, Gardner AM, Martin LA, Salzman AL. Nitric oxide dioxygenase: an enzymic function for flavohemoglobin. Proc. Natl Acad. Sci. USA. 1998;95:10378–10383. doi: 10.1073/pnas.95.18.10378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Svenningsen SL, Waters CM, Bassler BL. A negative feedback loop involving small RNAs accelerates Vibrio cholerae's transition out of quorum-sensing mode. Genes Dev. 2008;22:226–238. doi: 10.1101/gad.1629908. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.