Abstract
Listeria monocytogenes is a human intracellular pathogen able to colonize host tissues after ingestion of contaminated food, causing severe invasive infections. In order to gain a better understanding of the nature of host–pathogen interactions, we studied the L. monocytogenes genome expression during mouse infection. In the spleen of infected mice, ≈20% of the Listeria genome is differentially expressed, essentially through gene activation, as compared to exponential growth in rich broth medium. Data presented here show that, during infection, Listeria is in an active multiplication phase, as revealed by the high expression of genes involved in replication, cell division and multiplication. In vivo bacterial growth requires increased expression of genes involved in adaptation of the bacterial metabolism and stress responses, in particular to oxidative stress. Listeria interaction with its host induces cell wall metabolism and surface expression of virulence factors. During infection, L. monocytogenes also activates subversion mechanisms of host defenses, including resistance to cationic peptides, peptidoglycan modifications and release of muramyl peptides. We show that the in vivo differential expression of the Listeria genome is coordinated by a complex regulatory network, with a central role for the PrfA-SigB interplay. In particular, L. monocytogenes up regulates in vivo the two major virulence regulators, PrfA and VirR, and their downstream effectors. Mutagenesis of in vivo induced genes allowed the identification of novel L. monocytogenes virulence factors, including an LPXTG surface protein, suggesting a role for S-layer glycoproteins and for cadmium efflux system in Listeria virulence.
Author Summary
The facultative intracellular bacterial pathogen Listeria monocytogenes is the etiological agent of a severe foodborne disease. In humans it causes a variety of manifestations ranging from asymptomatic intestinal carriage and gastroenteritis to invasive and disseminated severe diseases. Septicemia, meningoencephalitis, and infection of the foetus in pregnant women are the most serious clinical features of listeriosis. Virulence is a trait that only manifests in a susceptible host, involving a highly coordinated interaction between bacterial factors and host components. This article reports the use of in vivo genome expression profiling as a powerful approach to gain a detailed understanding of the Listeria responses and the molecular cross-talk taking place in infected mice. We showed that, during infection, L. monocytogenes shifts the expression of its entire genome to promote virulence, subverting host defenses and adapting to host conditions. This first analysis of L. monocytogenes gene expression in vivo significantly enhances our understanding of the means by which intracellular pathogens promote infection.
Introduction
Listeria monocytogenes is an intracellular food-borne pathogen that causes listeriosis, an infection characterized by gastroenteritis, meningitis, encephalitis, and maternofetal infections in humans. It has one of the highest fatality rate among food-borne infections (20%–30%) [1]. Our knowledge of the infectious process in vivo mostly derives from infections in various animal models, in particular the mouse model. It is considered that bacteria after crossing the intestinal barrier reach, via the lymph and the blood, the liver and the spleen where they replicate actively. Then, bacteria via hematogenous dissemination, can reach the brain and the placenta. The disease is thus due to the original property of L. monocytogenes to be able to cross three host barriers: the intestinal barrier, the blood brain barrier and the materno-fetal barrier. It is also due to the capacity of Listeria to resist intracellular killing when phagocytosed by macrophages and to invade many non-phagocytic cell types. In the murine model, within minutes after intravenous inoculation, most bacteria can be found in the spleen and the liver [2].
L. monocytogenes ranks among the best-known intracellular pathogens and, until now, 50 genes have been shown to be involved in virulence in the mouse model (Table S1). However, whereas the different steps of the cell infectious process and the virulence factors specifically involved are well described [3], our knowledge of the in vivo infectious process is still fragmentary. Virulence is by definition expressed in a susceptible host, and involves a dynamic cross talk between the host and the pathogen. A detailed understanding of this interaction thus requires global approaches in the context of an in vivo infection. Analysis of the pathogen whole genome expression within the host should allow the identification of new bacterial genes critical for the infectious process, and lead to a better understanding of the molecular events responsible for Listeria infection.
The technology of DNA arrays allows to both study the gene content of different strains and measure gene expression levels on a genome-wide scale under different conditions. The genetic basis of L. monocytogenes pathogenicity was addressed by comparative genomics [4] and transcriptomics [5] using Listeria DNA arrays and various L. monocytogenes strains. Listeria arrays were also used for the analysis of the in vitro global gene expression of Listeria mutants for PrfA, the central regulator of virulence genes [6], and for other transcriptional regulators important for stress response and virulence (σB, σ54, HrcA, CtsR, VirR) [7]–[13]. Recently, this approach was applied to the determination of the intracellular gene expression profile of L. monocytogenes in epithelial and macrophage cell lines [14],[15]. In vivo genome profiles of other pathogens (Streptococcus pneumoniae, S. pyogenes, Mycobacterium tuberculosis, Borrelia burgdoferi, Yersinia pestis) infecting different mouse organs (dermis, soft tissue, lung, blood) were previously performed [16]. However, to our knowledge, the genome expression of a pathogen was never studied in infected mouse spleen.
Here, we present the first “in vivo” transcriptome of L. monocytogenes. We compared expression profiles of L. monocytogenes grown in standard culture medium in exponential phase vs. bacteria recovered from mouse spleens 24, 48 and 72 hours after intravenous infection. We determined the detailed expression kinetics of the complete L. monocytogenes genome in the course of the infection, and identified new Listeria virulence factors whose expression was highly up regulated in vivo.
Results
The in vivo transcriptome approach
We used the DNA macroarray technology to profile the transcriptome of Listeria during mouse infection. We used the previously described L. monocytogenes whole-genome arrays containing 500-bp-long PCR products specific for each gene [6]. Ninety-nine per cent of the 2853 predicted ORFs of the L. monocytogenes EGDe genome are represented on the arrays. They were used to analyze Listeria transcription profiles under in vitro growth in BHI in exponential phase at 37°C under aerobic conditions with shaking (pH 7) (Figure S1), and under in vivo growth conditions (mouse spleen) at 1, 2 and 3 days post intravenous infection (p.i.). Listeria present in spleen were analyzed because this organ is with the liver one of the major sites of L. monocytogenes infection. For unknown reasons, we never succeeded to prepare good quality bacterial RNAs from infected mouse livers. The time points chosen reflect key steps in the Listeria infectious process.
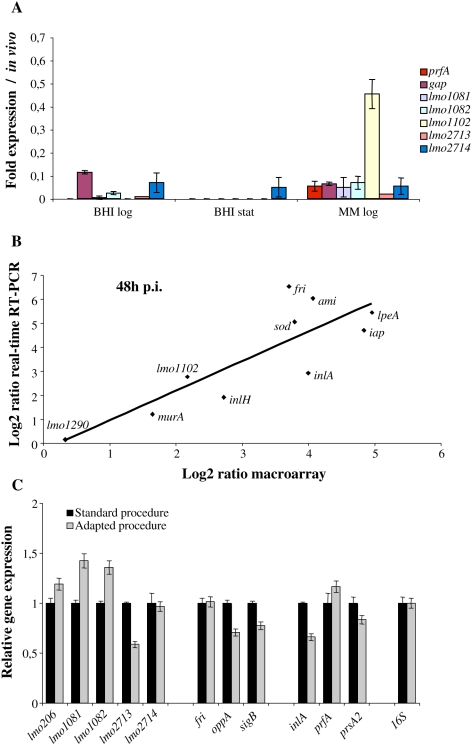
Culture in BHI in exponential phase at 37°C with shaking was chosen as reference conditions because BHI is the Listeria reference growth medium where bacteria divide in exponential growth phase at rates that are comparable to those observed for intracellular growth [17]. In addition, these are the in vitro reference conditions used in all previous studies analyzing the genome expression of L. monocytogenes in vitro or intracellularly [6]–[15]. However, in order to analyze the potential impact of the in vitro culture conditions used as reference on the relative gene expression in vivo, we first analyzed the results obtained comparing transcriptome from in vivo grown bacteria to transcriptomes from bacteria grown in vitro in exponential or stationary phase (Table S2). In addition, expression of known and potential virulence genes was analyzed by quantitative real time-PCR (qPCR) on RNAs extracted from bacteria cultured in BHI at 37°C in exponential or stationary growth phase, or in defined minimal medium [18], and compared to in vivo expression (Figure 1A). Results indicated that culture in exponential growth phase are closer conditions to those met by Listeria in vivo (Table S2). In addition, even if the expression of tested genes behaved differently in function of the in vitro conditions, expression of all the genes was always lower in vitro as compared to in vivo, independently of the in vitro growth conditions (Figure 1A). These experiments supported the choice of exponential growth phase in BHI as reference conditions and minimized the impact of the in vitro growth conditions on the identification of genes differentially expressed in vivo.
Figure 1. Macroarray validations.
(A) Analysis of the impact of the in vitro culture conditions used as reference. The expression of known and potential virulence factors was analyzed in BHI at 37°C in exponential (BHI log) or stationary (BHI stat) growth phase, or in minimal medium in exponential growth phase (MM log) by real-time RT-PCR, and normalized to expression in mouse spleen. (B) Validation of macroarray data by real-time RT-PCR. Fold changes in in vivo gene expression 48 h p.i. compared to that in BHI were measured by macroarray and real-time RT-PCR, log transformed and compared for correlation analysis. (C) Analysis of the effect of the RNA extraction method on L. monocytogenes gene expression. RNAs from bacteria grown in BHI were prepared using the standard and adapted procedures for RNA extraction. The relative expression of potential virulence genes, cold shock genes and known virulence genes was determined by real-time RT-PCR.
The reliability of the macroarray expression data was further assessed by qPCR. We selected a subset of 10 genes and performed qPCR on cDNA from bacteria grown in either standard medium or extracted from mouse spleens 48 h p.i.. qPCR results and array data exhibited a high correlation coefficient (0.7) (Figure 1B). This strong correlation was also observed for other infection time points (Figure S2). However, the differences in gene expression, as measured by qPCR, were generally higher, indicating that in vivo transcriptome data rather underestimate changes in gene expression.
The procedure used for bacterial RNA extraction from infected mouse spleens is an adaptation of the standard procedure originally used for transcriptional analysis of RNA extracted from pure culture. In order to test the effect of the RNA extraction method on gene expression, RNAs from bacteria grown in pure culture were extracted using the two methods. The relative expression of known virulence genes, cold shock genes and potential virulence genes was analyzed by qPCR in the two RNA pools. The results showed that the relative expression of the genes tested is not significantly affected by the RNA extraction procedure (Figure 1C).
For bacteria cultured in BHI at 37°C in exponential phase or extracted from infected mouse spleen at the different times p.i., two different RNA preparations from independent cultures (or infections) were used for cDNA synthesis and subsequent hybridization to two sets of arrays. To identify statistically significant differences in gene expression, we used the Statistical Analysis for Microarrays (SAM) program [19]. Subsequently, all the genes showing statistically significant changes in the expression level and an at least two-fold change in their level of expression were considered in our analysis.
Important global changes in L. monocytogenes gene expression occur during in vivo growth
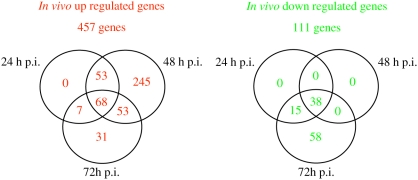
Overall, a total of 568 genes representing ≈20% of the total genome exhibited a differential expression during infection as compared to growth in BHI at 37°C in exponential phase. Among these 568 genes, 457 were up regulated (≈80%) and 111 (≈20%) were down regulated during mouse infection as compared to exponential growth in BHI medium (Table S3).
In order to identify genes potentially implicated in virulence, all the genes differentially regulated in vivo were analyzed for the presence of an ortholog in the nonpathogenic close relative Listeria innocua strain CLIP11262 [20]. This analysis revealed that only 30 of the in vivo regulated genes (25 up and 5 down regulated) were absent from L. innocua (Table 1). Of these 30 genes, 20 were L. monocytogenes “specific” (i.e. also present in L. monocytogenes 1/2a F6854, L. monocytogenes 4b F2365 and H7858 [21], and absent from L. innocua). Interestingly, of these 20 genes, 16 were up regulated in vivo. Among these 16 genes, 11 have been previously implicated in Listeria virulence. The remaining 10 in vivo regulated genes, among which 9 up- and 1 down-regulated in vivo, appeared lineage specific, i.e. present only in the sequenced serovar 1/2a strains (Table 1).
Table 1. L. monocytogenes EGDe genes absent from L. innocua and differentially regulated in the host.
| Gene designation | Gene | Annotation | Homolog in 1/2a F6854 | Homolog in 4b F2365 | Homolog in 4b H7858 | Fold change 24 h | Fold change 48 h | Fold Change 72 h |
| prfA | lmo0200 | listeriolysin positive regulatory protein | LMOf6854_0209 | LMOf2365_0211 | LMOh7858_0220 | 8,09 | 4,00 | |
| plcA | lmo0201 | phosphatidylinositol-specific phospholipase c | LMOf6854_0210 | LMOf2365_0212 | LMOh7858_0221 | 7,20 | 48,31 | 6,70 |
| hly | lmo0202 | listeriolysin O precursor | LMOf6854_0211 | LMOf2365_0213 | LMOh7858_0222 | 35,56 | 118,39 | 15,14 |
| mpl | lmo0203 | zinc metalloproteinase precursor | LMOf6854_0212 | LMOf2365_0214 | LMOh7858_0223 | 3,36 | 18,41 | 4,68 |
| actA | lmo0204 | actin-assembly inducing protein precursor | LMOf6854_0213 | LMOf2365_0215 | LMOh7858_0224 | 6,02 | 15,58 | 4,49 |
| plcB | lmo0205 | phospholipase C | LMOf6854_0214 | LMOf2365_0216 | LMOh7858_0225 | 12,37 | 106,64 | 31,78 |
| lmo0206 | lmo0206 | unknown protein | LMOf6854_0214.1 | LMOf2365_0217 | LMOh7858_0225.1 | 3,11 | ||
| lmo0257 | lmo0257 | unknown protein | LMOf6854_0261.2 | LMOf2365_0265 | LMOh7858_0288 | 2,12 | ||
| inlH | lmo0263 | internalin H | LMOf6854_0275 | LMOf2365_0281 | LMOh7858_0295 | 3,08 | 6,60 | 2,41 |
| inlA | lmo0433 | internalin A | LMOf6854_0469 | LMOf2365_0471 | LMOh7858_0499 | 15,92 | 3,86 | |
| inlB | lmo0434 | internalin B | LMOf6854_0470 | LMOf2365_0472 | LMOh7858_0501.2 | 3,82 | 2,55 | |
| uhpT | lmo0838 | hexose phosphate transport protein | LMOf6854_0883 | LMOf2365_0855 | LMOh7858_0894 | 5,13 | 2,59 | |
| lmo0915 | lmo0915 | similar to phosphotransferase system enzyme IIC | LMOf6854_0962 | LMOf2365_0937 | LMOh7858_0989 | 3,15 | 2,37 | |
| lmo1290 | lmo1290 | similar to internalin proteins, putative peptidoglycan bound protein (LPXTG) | LMOf6854_1332 | LMOf2365_1307 | LMOh7858_1374.2 | 6,24 | ||
| inlC | lmo1786 | internalin C | LMOf6854_1844.2 | LMOf2365_1812 | LMOh7858_1916.1 | 8,56 | 3,57 | |
| lmo2157 | lmo2157 | unknown protein | LMOf6854_2221 | LMOf2365_2189 | LMOh7858_2290.1 | 3,29 | 5,85 | 2,01 |
| lmo2257 | lmo2257 | hypothetical CDS | LMOf6854_2321.1 | LMOf2365_2290 | LMOh7858_2400 | −4,62 | −3,99 | |
| lmo2672 | lmo2672 | weakly similar to transcription regulator | LMOf6854_2788 | LMOf2365_2652 | LMOh7858_2935 | −2,31 | ||
| lmo2733 | lmo2733 | similar to PTS system, fructose-specific IIABC component | LMOf6854_2852 | LMOf2365_2720 | LMOh7858_2997 | −2,35 | ||
| lmo2736 | lmo2736 | unknown protein | LMOf6854_2855 | LMOf2365_2723 | LMOh7858_3000 | −2,48 | ||
| lmo1081 | lmo1081 | similar to glucose-1-phosphate thymidyl transferase | LMOf6854_1134 | 8,25 | 5,99 | 5,17 | ||
| lmo1082 | lmo1082 | similar to dTDP-sugar epimerase | LMOf6854_1135 | 47,21 | 24,06 | 21,01 | ||
| lmo1083 | lmo1083 | similar to dTDP-D-glucose 4,6-dehydratase | LMOf6854_1136 | 3,49 | ||||
| lmo1084 | lmo1084 | similar to DTDP-L-rhamnose synthetase | LMOf6854_1137 | 3,89 | ||||
| lmo2276 | lmo2276 | similar to an unknown bacteriophage protein | LMOf6854_2338 | −2,72 | ||||
| lmo0150 | lmo0150 | unknown protein | 16,53 | |||||
| lmo0471 | lmo0471 | unknown protein | 3,18 | |||||
| lmo1099 | lmo1099 | similar to a protein encoded by Tn916 | 65,42 | 30,42 | 24,04 | |||
| lmo1102 | lmo1102 | similar to cadmium efflux system accessory proteins | 9,71 | 4,51 | 4,72 | |||
| lmo2277 | lmo2277 | unknown protein | 2,61 |
To identify genes regulated during different stages of listeriosis, gene expression levels of spleen-recovered bacteria at different time points p.i. were compared. This analysis revealed a core regulon of 106 genes (68 up and 38 down regulated) whose expression was significantly differentially regulated at all the time points of the infection as compared to bacteria grown in pure culture (Figure 2). No gene appeared specifically differentially regulated at 24 h p.i. At two days p.i., a large proportion (245/457) of genes was up regulated. The largest number of down regulated genes was observed 72 h p.i..
Figure 2. Venn diagrams showing the distribution of the up and down regulated genes at the three in vivo infection time points.
Major virulence regulators and their downstream target genes are highly up regulated in vivo
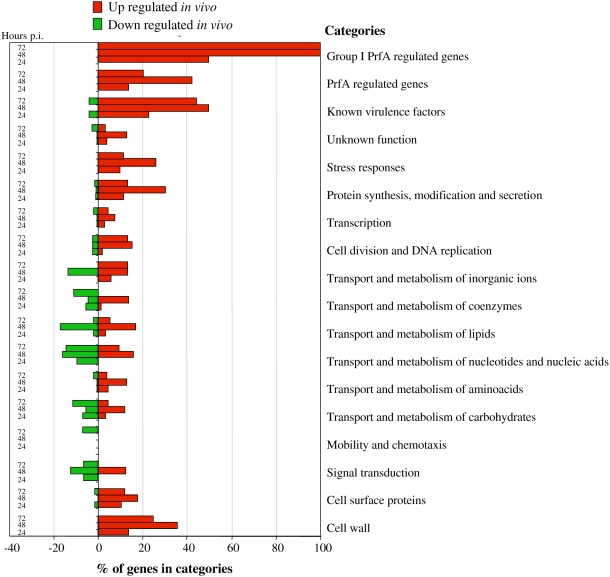
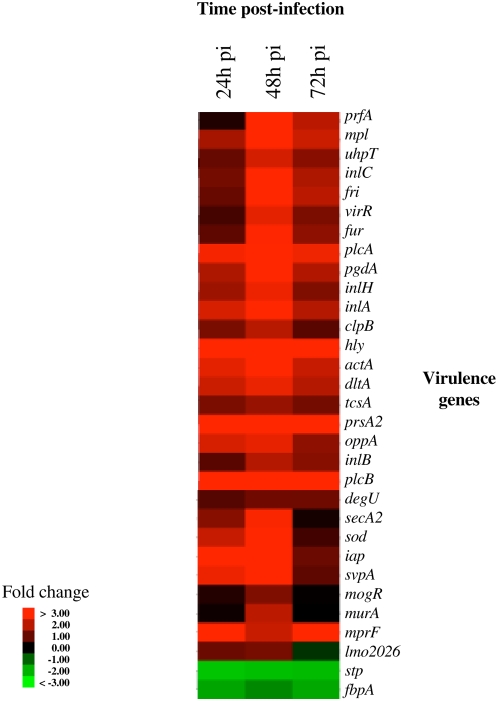
As compared to Listeria grown in BHI at exponential phase, bacteria extracted from mouse spleens showed a differential expression of genes belonging to various functional categories (Figure 3). In particular, analysis of the expression profile of the 50 genes previously implicated in Listeria virulence in the mouse model revealed that 29 were up regulated during infection, and two (stp and fbpA) down regulated in vivo (Figure 4 and Table S1).
Figure 3. Differentially regulated genes of L. monocytogenes EGDe obtained from temporal transcriptome profiling experiments, classified in functional categories.
Figure 4. In vivo expression of virulence genes.
Expression during mouse spleen infection of the 31 known virulence genes differentially regulated in vivo. A peak of expression was observed for the majority of these virulence genes 48 h p.i. All measurements are relative to culture in exponential phase in BHI. Genes were selected for this analysis when their expression deviated from BHI by at least a factor of 2.0 in at least one time point. The image was produced as described in Materials and Methods. Each gene is represented by a single row of colored boxes; each time point is represented by a single column.
We observed that the entire virulence gene cluster of L. monocytogenes comprising the genes prfA, plcA, hly, mpl, actA and plcB was highly activated during the 3 first days of infection (Table 2). In addition to the virulence gene cluster, genes encoding the two major L. monocytogenes factors implicated in entry into eukaryotic cells (inlA and inlB) [22], and uhpT, a gene encoding a sugar phosphate transporter that mediates rapid intracellular proliferation [23] were also activated during infection. PrfA is the principal regulator of the expression of not only these key virulence genes, but also of most other L. monocytogenes genes involved in intracellular survival and virulence [6]. The 12 genes previously reported to be preceded by a PrfA box and positively regulated by PrfA in a transcriptional analysis of the PrfA regulon [6], were all highly up regulated in mouse spleens (Table 2). From the 53 other genes already shown as positively regulated by PrfA [6], 20 were more expressed in vivo. As previously shown [8], 19 of these 20 genes are also controlled by SigB, including the LPXTG internalin-like protein inlH known to be involved in Listeria virulence [24]. Two genes, lmo0206 and lmo0207, recently shown as regulated by PrfA and implicated in L. monocytogenes intracellular survival [14] were also activated in infected mice. Importantly, no gene previously shown under the PrfA positive regulation appeared down regulated during mouse infection.
Table 2. L. monocytogenes EGDe genes positively controlled by PrfA and up regulated in the host.
| Gene designation | Gene | Annotation | Homolog in L. innocua | Fold change 24 h | Fold change 48 h | Fold change 72 h |
| Group I PrfA regulated genes | ||||||
| prfA | lmo0200 | listeriolysin positive regulatory protein | 8,09 | 4,00 | ||
| plcA | lmo0201 | phosphatidylinositol-specific phospholipase C | 7,20 | 48,31 | 6,70 | |
| hly | lmo0202 | listeriolysin O precursor | 35,56 | 118,39 | 15,14 | |
| mpl | lmo0203 | zinc metalloproteinase precursor | 3,36 | 18,41 | 4,68 | |
| actA | lmo0204 | actin-assembly inducing protein precursor | 6,02 | 15,58 | 4,49 | |
| plcB | lmo0205 | phospholipase C | 12,37 | 106,64 | 31,78 | |
| inlA | lmo0433 | internalin A | 15,92 | 3,86 | ||
| inlB | lmo0434 | internalin B | 3,82 | 2,55 | ||
| lmo0788 | lmo0788 | unknown protein | lin0781 | 60,06 | 31,36 | 20,50 |
| uhpT | lmo0838 | hexose phoshate transport protein | 5,13 | 2,59 | ||
| inlC | lmo1786 | internalin C | 8,56 | 3,57 | ||
| prsA2 | lmo2219 | similar to post-translocation molecular chaperone | lin2322 | 12,02 | 24,00 | 8,95 |
| Group III PrfA regulated genes co-controlled by SigB | ||||||
| lmo0169 | lmo0169 | similar to a glucose uptake protein | lin0212 | 3,11 | ||
| inlH | lmo0263 | internalin H | 3,08 | 6,60 | 2,41 | |
| lmo0439 | lmo0439 | weakly similar to a module of peptide synthetase | lin0460 | 2,76 | ||
| lmo0539 | lmo0539 | similar to tagatose-1,6-diphosphate aldolase | lin0543 | 18,71 | 24,03 | |
| lmo0596 | lmo0596 | unknown protein | lin0605 | 23,31 | 68,15 | |
| lmo0781 | lmo0781 | similar to mannose-specific phosphotransferase system (PTS) component IID | lin0774 | 2,57 | ||
| lmo0782 | lmo0782 | similar to mannose-specific phosphotransferase system (PTS) component IIC | lin0775 | 4,69 | ||
| lmo0783 | lmo0783 | similar to mannose-specific phosphotransferase system (PTS) component IIB | lin0776 | 3,82 | ||
| lmo0784 | lmo0784 | similar to mannose-specific phosphotransferase system (PTS) component IIA | lin0777 | 2,89 | ||
| lmo0794 | lmo0794 | similar to B. subtilis YwnB protein | lin0787 | 4,47 | ||
| lmo0796 | lmo0796 | unknown protein | lin0789 | 4,21 | ||
| lmo0994 | lmo0994 | unknown protein | lin0993 | 2,53 | ||
| opuCD | lmo1425 | osmoprotectant transport system permease protein | lin1464 | 2,69 | ||
| lmo1601 | lmo1601 | similar to general stress protein | lin1642 | 2,18 | ||
| lmo1602 | lmo1602 | unknown protein | lin1643 | 4,54 | 2,13 | |
| lmo2157 | lmo2157 | unknown protein | 3,29 | 5,85 | 2,01 | |
| lmo2391 | lmo2391 | conserved hypothetical protein similar to B. subtilis YhfK protein | lin2490 | 4,56 | ||
| lmo2696 | lmo2696 | similar to hypothetical dihydroxyacetone kinase | lin2844 | 2,49 | ||
| lmo2697 | lmo2697 | unknown protein | lin2845 | 3,14 | ||
| Group III PrfA regulated gene not controlled by SigB | ||||||
| lmo0937 | lmo0937 | unknown protein | lin0936 | 3,92 | ||
| Other PrfA regulated genes | ||||||
| lmo0206 | lmo0206 | unknown protein | 3,11 | |||
| lmo0207 | lmo0207 | hypothetical lipoprotein | lin0239 | 5,31 | 3,27 | |
VirR, another key Listeria virulence regulator that mainly controls genes involved in the modification of bacterial surface components, is the response regulator of a two-component system (TCS) implicated in cell invasion and virulence [13]. Using a transcriptomic approach, 17 genes were previously identified as regulated by VirR in vitro [13]. In our study, 13 of the 17 VirR regulated genes, including the dlt operon and mprF, were up regulated in vivo (Table 3). The dlt operon is necessary for D-alanylation of lipoteichoic acid (LTA) and was reported to be important for L. monocytogenes virulence [13]. The VirR regulated mprF encodes a protein shown to be required for lysinylation of phospholipids in the Listeria cytoplasmic membrane and to confer Listeria resistance to cationic antimicrobial peptides (CAMPs) [25]. The virR and virS genes were themselves up regulated, constituting the only TCS whose expression of both components was induced in mouse spleens.
Table 3. L. monocytogenes EGDe genes regulated by VirR and up regulated in the host.
| Gene designation | Gene | Annotation | Homolog in L. innocua | Fold change 24 h | Fold change 48 h | Fold change 72 h |
| lmo0604 | lmo0604 | similar to B. subtilis YvlA protein | lin0613 | 51,45 | 27,43 | |
| dltD | lmo0971 | DltD protein for D-alanine esterification of lipoteichoic acid and wall teichoic acid | lin0970, dltD | 11,98 | 21,95 | 3,73 |
| dltC | lmo0972 | D-alanine–poly(phosphoribitol) ligase subunit 2 DltC | lin0971, dltC | 4,13 | 6,22 | 2,35 |
| dltB | lmo0973 | DltB protein for D-alanine esterification of lipoteichoic acid and wall teichoic acid | lin0972, dltB | 6,48 | 7,58 | 2,73 |
| dltA | lmo0974 | D-alanine–D-alanyl carrier protein ligase DltA | lin0973, dltA | 6,37 | 3,85 | |
| mprF | lmo1695 | similar to MprF protein of S aureus | lin1803 | 9,68 | 4,59 | 24,61 |
| virS | lmo1741 | two-component sensor histidine kinase | lin1852 | 2,76 | ||
| adeC | lmo1742 | highly similar to adenine deaminases | lin1853 | 2,66 | ||
| virR | lmo1745 | two-component response regulator | lin1856 | 5,96 | 2,29 | |
| lmo2114 | lmo2114 | similar to ABC transporter (ATP-binding protein) | lin2219 | 3,68 | 2,93 | |
| lmo2115 | lmo2115 | similar to ABC transporter (permease) | lin2220 | 4,86 | 5,92 | 3,06 |
| lmo2177 | lmo2177 | unknown protein | lin2280 | 3,36 | 6,14 | 2,23 |
| lmo2439 | lmo2439 | unknown protein | lin2533 | 4,44 |
In addition to VirRS, the L. monocytogenes genome contains 15 additional predicted TCS systems [26]. Genes encoding one component of three of these TCS (degU, resD and phoR) were also up regulated in vivo. DegU is an orphan response regulator (absence of the sensor kinase DegS in the L. monocytogenes genome) and a pleiotropic regulatory system previously involved in Listeria virulence [27],[28]. In particular, DegU has been implicated in the regulation of some Listeria secreted proteins (gap, tsf, sod, lmo0644) [26]. Interestingly, the expression of these four genes was also increased in mouse spleens.
Finally, OhrR a transcriptional regulator controlling OhrA, a hydroxyperoxidase implicated in intracellular survival of Listeria [14], as well as several predicted transcriptional regulators were up regulated in infected mouse spleens.
Strong activation of genes encoding cell wall metabolism proteins during infection
In addition to genes already mentioned and involved in LTA modification (dltABCD), we observed that several genes implicated in peptidoglycan (PG) biosynthesis (lmo0516, lmo0540, lmo1438, lmo1521, lmo1855, lmo2522, lmo2526 and pbpB), cell shape determination (mreBC, lmo1713), cell wall peptide synthesis (murC) were up regulated in bacteria growing in mouse spleens (Table 4).
Table 4. L. monocytogenes EGDe genes implicated in cell wall metabolism and differentially regulated in the host.
| Gene designation | Gene | Annotation | Homolog in L. innocua | Fold change 24 h | Fold change 48 h | Fold change 72 h |
| pgdA | lmo0415 | peptidoglycan N-acetylglucosamine deacetylase A | lin0436 | 14,32 | 3,71 | |
| lmo0516 | lmo0516 | similar to Bacillus anthracis encapsulation protein CapA | lin0516 | 14,22 | ||
| lmo0540 | lmo0540 | similar to penicillin-binding protein | lin0544 | 3,78 | 3,97 | 2,28 |
| iap | lmo0582 | P60 extracellular protein, invasion associated protein Iap | lin0591 | 34,54 | 28,64 | 2,04 |
| dltD | lmo0971 | DltD protein for D-alanine esterification of lipoteichoic acid and wall teichoic acid | lin0970, dltD | 11,98 | 21,95 | 3,73 |
| dltC | lmo0972 | D-alanine–poly(phosphoribitol) ligase subunit 2 DltC | lin0971, dltC | 4,13 | 6,22 | 2,35 |
| dltB | lmo0973 | DltB protein for D-alanine esterification of lipoteichoic acid and wall teichoic acid | lin0972, dltB | 6,48 | 7,58 | 2,73 |
| dltA | lmo0974 | D-alanine–D-alanyl carrier protein ligase DltA | lin0973, dltA | 6,37 | 3,85 | |
| lmo1075 | lmo1075 | similar to teichoic acid translocation ATP-binding protein TagH (ABC transporter) | lin1063 | 9,25 | 5,46 | 4,20 |
| lmo1081 | lmo1081 | similar to glucose-1-phosphate thymidyl transferase | 8,22 | 5,98 | 5,17 | |
| lmo1082 | lmo1082 | similar to dTDP-sugar epimerase | 47,18 | 24,08 | 20,97 | |
| lmo1083 | lmo1083 | similar to dTDP-D-glucose 4,6-dehydratase | 3,51 | |||
| lmo1084 | lmo1084 | similar to DTDP-L-rhamnose synthetase | 3,89 | |||
| lmo1291 | lmo1291 | similar to acyltransferase (to B. subtilis YrhL protein) | lin1329 | 4,72 | ||
| uppS | lmo1315 | similar to undecaprenyl diphosphate synthase | lin1352 | 2,16 | ||
| lmo1438 | lmo1438 | similar to penicillin-binding protein | lin1477 | 3,56 | ||
| lmo1521 | lmo1521 | similar to N-acetylmuramoyl-L-alanine amidase | lin1556 | 2,73 | ||
| mreC | lmo1547 | similar to cell-shape determining protein MreC | lin1581 | 2,45 | ||
| mreD | lmo1548 | similar to cell-shape determining protein MreB | lin1582 | 4,08 | ||
| murC | lmo1605 | similar to UDP-N-acetyl muramate-alanine ligase | lin1646 | 2,39 | ||
| mprF | lmo1695 | multiple peptide resistance factor | lin1803 | 9,71 | 4,59 | 24,59 |
| lmo1713 | lmo1713 | similar to cell-shape determining protein MreB | lin1825 | 5,82 | ||
| lspA | lmo1844 | signal peptidase II | lin1958 | −2,01 | ||
| lmo1851 | lmo1851 | similar to carboxy-terminal processing proteinase | lin1965 | −2,12 | ||
| lmo1855 | lmo1855 | similar to similar to D-alanyl-D-alanine carboxypeptidases | lin1969 | 3,32 | ||
| pbpB | lmo2039 | penicilin binding protein 2B | lin2145 | 5,00 | ||
| srtB | lmo2181 | sortase B | lin2285 | 2,44 | ||
| svpA | lmo2185 | unknown protein | lin2289 | 6,61 | 11,11 | |
| lmo2186 | lmo2186 | unknown protein | lin2290 | 2,72 | ||
| lmo2203 | lmo2203 | similar to N-acetylmuramoyl-L-alanine amidase and to internalin B | lin2306 | 4,50 | ||
| prsA2 | lmo2219 | similar to post-translocation molecular chaperone | lin2322 | 12,02 | 24,00 | 8,95 |
| spl | lmo2505 | peptidoglycan lytic protein P45 | lin2648, spl | 12,21 | 2,68 | |
| lmo2522 | lmo2522 | similar to hypothetical cell wall binding protein from B. subtilis | lin2666 | 5,74 | 3,07 | |
| lmo2526 | lmo2526 | UDP-N-acetylglucosamine 1-carboxyvinyltransferase | lin2670 | 2,40 | ||
| murA | lmo2691 | similar to autolysin, N-acetylmuramidase | lin2838 | 4,06 |
The expression of 3 genes encoding virulence factors involved in bacterial cell wall modifications (murA, iap, and pgdA) [29]–[31] was also increased in vivo. MurA and P60, the iap gene product, are two SecA2-secreted autolysins required for Listeria full virulence [29],[30]. pgdA encodes for the PG N-deacetylase of L. monocytogenes that was demonstrated as playing an important role in virulence and evasion from host defenses [31]. In addition, spl [32] and lmo2203 are two other autolysins encoding genes up regulated in vivo, but until now never implicated in virulence.
Moreover, prsA2, a gene encoding a surface protein involved in protein folding and previously shown as implicated in Listeria intracellular survival and virulence [14],[33] was up regulated in vivo. Interestingly, the gene encoding the sortase SrtB that covalently links proteins to the Listeria peptidoglycan, and two genes encoding SrtB substrates (svpA and lmo2186) [34], were also over expressed in vivo (Table 4).
Differential expression of genes encoding specific surface and secreted proteins during infection
Whereas a total of 44 genes encoding potential surface proteins were up regulated in vivo, only three were observed as down regulated during infection (lspA, lmo1851 and lmo2642) (Table 5). In addition, among the 55 proteins previously identified in the cell wall subproteome of L. monocytogenes [35], we found that 23 were up regulated in vivo (Table S4). The L. monocytogenes genome encodes 41 LPXTG surface proteins [20],[36],[37]. This class includes proteins containing leucine rich repeats (LRRs) and belonging to the internalin family. Four LPXTG-protein encoding genes were up regulated in vivo. In addition to InlA and InlH, lmo1290 and lmo2714 are the two other LPXTG encoding genes activated during infection (Table 5). Four genes encoding proteins associated to the cell wall via GW modules were also more expressed in vivo: inlB, the known invasion protein [38], and lmo1521, lmo2203 and lmo2713. actA [39] was the only gene encoding a protein with a carboxyl-terminal hydrophobic tail up regulated in vivo. Genes encoding lipoproteins previously implicated in Listeria virulence (TcsA and OppA) [33],[40] or in cell invasion (LpeA) [41], were over expressed in mouse spleens. In addition, 10 genes predicted to encode other lipoproteins were activated in vivo (Table 5).
Table 5. L. monocytogenes EGDe cell surface encoding genes differentially regulated in the host.
| Gene designation | Gene | Annotation | Homolog in L. innocua | Fold change 24 h | Fold change 48 h | Fold change 72 h |
| Sortase substrates: LPXTG and NXXTX proteins | ||||||
| inlH | lmo0263 | internalin H | 3,07 | 6,59 | 2,41 | |
| inlA | lmo0433 | internalin A | 15,89 | 3,86 | ||
| lmo1290 | lmo1290 | similar to internalin proteins, putative peptidoglycan bound protein (LPXTG motif) | 6,23 | |||
| svpA | lmo2185 | unknown protein | lin2289 | 6,61 | 11,11 | |
| lmo2186 | lmo2186 | unknown protein | lin2290 | 2,72 | ||
| lmo2714 | lmo2714 | peptidoglycan anchored protein (LPXTG motif) | lin2862 | 25,46 | 70,03 | 3,10 |
| Proteins with noncovalent association to cell wall | ||||||
| inlB | lmo0434 | internalin B | 3,84 | 2,55 | ||
| iap | lmo0582 | P60 extracellular protein, invasion associated protein Iap | lin0591, iap | 34,64 | 28,54 | 2,05 |
| lmo1521 | lmo1521 | similar to N-acetylmuramoyl-L-alanine amidase | lin1556 | 2,73 | ||
| murC | lmo1605 | UDP-N-acetylmuramate–L-alanine ligase | lin1646 | 2,40 | ||
| lmo1851 | lmo1851 | similar to carboxy-terminal processing proteinase | lin1965 | −2,12 | ||
| lmo2203 | lmo2203 | similar to N-acetylmuramoyl-L-alanine amidase | lin1556 | 4,50 | ||
| lmo2522 | lmo2522 | similar to hypothetical cell wall binding protein from B. subtilis | lin2666 | 5,74 | 3,07 | |
| lmo2713 | lmo2713 | secreted protein with 1 GW repeat | lin2861 | 17,03 | 2,79 | |
| Proteins with an hydrophobic tail | ||||||
| actA | lmo0204 | actin-assembly inducing protein precursor | 6,02 | 15,56 | 4,50 | |
| Lipoproteins | ||||||
| qoxA | lmo0013 | AA3-600 quinol oxidase subunit II | lin0013 | 2,95 | ||
| lmo0153 | lmo0153 | similar to a probable high-affinity zinc ABC transporter (Zn(II)-binding lipoprotein) | lin0191 | 5,31 | ||
| lmo0207 | lmo0207 | hypothetical lipoprotein | lin0239 | 5,31 | 3,27 | |
| lmo0303 | lmo0303 | putaive secreted, lysin rich protein | lin0331 | 3,18 | ||
| lmo0355 | lmo0355 | similar to Flavocytochrome C Fumarate Reductase chain A | lin0374 | 2,97 | 6,68 | 2,51 |
| lmo0366 | lmo0366 | putative lipoprotein | lin0385 | 2,69 | 2,17 | |
| prs | lmo0509 | similar to phosphoribosyl pyrophosphate synthetase | 2,87 | |||
| lmo0541 | lmo0541 | similar to ABC transporter (binding protein) | lin0545 | 6,19 | 10,27 | 2,07 |
| tcsA | lmo1388 | CD4+ T cell-stimulating antigen, lipoprotein | lin1425 | 2,19 | ||
| lmo1649 | lmo1649 | unknown protein | lin1689 | 3,39 | ||
| lpeA | lmo1847 | similar to adhesion binding proteins and lipoproteins | lin1961 | 6,54 | 31,12 | 10,63 |
| lmo2184 | lmo2184 | similar to ferrichrome ABC transporter (binding protein) | lin2288 | 5,10 | 2,57 | |
| oppA | lmo2196 | similar to pheromone ABC transporter (binding protein) | lin2300 | 5,21 | 6,15 | 2,73 |
| lmo2642 | lmo2642 | unknown protein | lin2791 | −2,13 | ||
| Surface proteins involved in cell wall metabolism | ||||||
| pgdA | lmo0415 | peptidoglycan N-acetylglucosamine deacetylase A | lin0436 | 14,32 | 3,71 | |
| lmo0540 | lmo0540 | similar to penicillin-binding protein | lin0544 | 3,78 | 3,97 | 2,28 |
| iap | lmo0582 | P60 extracellular protein, invasion associated protein Iap | lin0591 | 34,54 | 28,64 | 2,04 |
| lmo1438 | lmo1438 | similar to penicillin-binding protein | lin1477 | 3,56 | ||
| lmo1521 | lmo1521 | similar to N-acetylmuramoyl-L-alanine amidase | lin1556 | 2,73 | ||
| lmo1855 | lmo1855 | similar to similar to D-alanyl-D-alanine carboxypeptidases | lin1969 | 3,32 | ||
| pbpB | lmo2039 | penicilin binding protein 2B | lin2145 | 5,00 | ||
| lmo2203 | lmo2203 | similar to N-acetylmuramoyl-L-alanine amidase and to internalin B | lin2306 | 4,50 | ||
| spl | lmo2505 | peptidoglycan lytic protein P45 | lin2648, spl | 12,21 | 2,68 | |
| lmo2522 | lmo2522 | similar to hypothetical cell wall binding protein from B. subtilis | lin2666 | 5,74 | 3,07 | |
| murA | lmo2691 | similar to autolysin, N-acetylmuramidase | lin2838 | 4,06 | ||
| Surface proteins involved in protein processing, folding and cell surface anchoring | ||||||
| lspA | lmo1844 | signal peptidase II | lin1958, lspA | −2,01 | ||
| lmo1851 | lmo1851 | similar to carboxy-terminal processing proteinase | lin1965 | −2,12 | ||
| srtB | lmo2181 | sortase B | lin2285 | 2,44 | ||
| prsA2 | lmo2219 | similar to post-translocation molecular chaperone | lin2322 | 12,02 | 24,00 | 8,95 |
Protein secretion is of key importance in both the colonization process and virulence of Listeria [42]. Besides L. monocytogenes virulence factors with a signal peptide (ActA, LLO, InlA, InlB, InlC, InlH, Mpl, MurA, PlcA, PlcB, P60 and SvpA), three other virulence proteins (Fri, TcsA and Sod) were also found secreted in the Listeria culture supernatant [43]. All the genes encoding these secreted virulence factors appeared activated in our in vivo approach (Table S5). The analysis of the products present in the Listeria culture supernatant after growth in vitro allowed the identification of 89 additional proteins [43]. 29 of the genes encoding these secreted proteins were up regulated in vivo (Table S5). Most of the Listeria secreted proteins are presumed to be secreted through the Sec translocation system. A gene encoding one component of the predicted Sec system, secE, was observed up regulated in vivo. SecA2 is an auxiliary secretory protein required for persistent colonization of host tissues, and responsible for the secretion of several Listeria virulence factors (MurA, P60, Sod, OppA and TcsA) [29],[30],[44]. We observed an in vivo up regulation of the majority of the genes encoding SecA2-secreted proteins, including all the SecA2-secreted virulence factors (Table S5).
In vivo high expression of genes involved in DNA metabolism, RNA and protein synthesis, cell division and multiplication
We observed an in vivo up regulation of several genes involved in DNA synthesis (dnaX and lmo0162), DNA restriction/modifications and repair (mutL, uvrB, lmo1639 and lmo1782), DNA recombination (recFRX, codV and lmo2267), and DNA packaging and segregation (gyrA, hup, lmo1606 and lmo2794) (Table S6). In addition, the expression of 25 genes encoding ribosomal proteins, as well as genes involved in protein synthesis initiation (infAC), elongation (fus, tsf, lmo1067) and termination (frr) was up regulated during infection. Genes encoding proteins implicated in chromosomal replication and segregation (dnaABC, ssb and divIVA), and cell elongation and division (mreBC, ftsHX and lmo0196) were also up regulated in mouse spleens (Table S6).
Induction of genes implicated in stress responses during infection
In our study, genes belonging to the three principal classes of stress genes were up regulated in the host. Class I genes encode classical chaperones and are controlled by the HrcA repressor. Nine of the 25 genes previously shown as HrcA repressed [11] were activated in vivo, including genes encoding the molecular chaperones DnaK and GroEL respectively also shown as induced in macrophages and required for survival following phagocytosis [45],[46] (Table S7). Inversely, 17 of the 36 genes shown to be indirectly positively regulated by HrcA [11], were up regulated in mouse spleens. This list includes genes encoding ribosomal proteins, as well as a number of DNA replication, transcription or translation related genes.
The class II stress response is mediated by sigma B (SigB). A total of 30 genes that have been recently classified as SigB activated [8] appeared here up regulated in vivo (Table S7). In particular we detected the up regulation of inlH [24], ltrC implicated in response to cold shock [47], and lmo1601 similar to general stress proteins. Interestingly, 40 genes previously classified as down regulated by SigB during the stationary growth phase [8] were detected as activated in vivo (Table S7). These include kat, a catalase involved in the oxidative stress response [48], a large proportion of genes encoding ribosomal proteins or implicated in translation, cell division and cell wall biogenesis. Furthermore, iap, the P60 gene [29], is part of this group. Finally, rsbU and rsbX, two components of the complex regulation system of SigB [49] were also up regulated in mouse spleens (Table S7).
CtsR is a transcriptional repressor involved in the control of class III stress proteins and previously shown to be responsible for the repression of 42 genes [12], 15 of which appeared up regulated in the host (Table S7). In particular, CtsR regulates the expression of Clp proteases required for the degradation of abnormal proteins and implicated in bacterial escape from macrophage vacuoles and virulence in mice [50]. Expression of clpBCE was activated during infection, as well as mcsA and mcsB the modulators of the CtsR regulon.
In some host cells, bacteria are confronted with severe oxidative stress due to the release of reactive oxygen intermediates. We observed the in vivo activation of an important number of oxidative stress resistance mechanisms. The qoxABCD operon that encodes a quinol oxidase important for oxidative stress response, and two major proteins implicated in protection against superoxides and reactive oxygen species (ROS), Kat and Sod, were highly up regulated in vivo (Table S7). Sod was previously shown as required for Listeria full virulence and is a target of Stp, a serine-threonine phosphatase also involved in L. monocytogenes virulence [44],[51], and detected down regulated in the host. A decrease in the level of Stp was previously associated to an increase in phosphorylated Sod, accompanied by the secretion of active non-phosphorylated Sod by the SecA2 system [44],[51]. Furthermore, genes encoding a thioredoxin and two thioredoxin reductases involved in the response to oxidative stress (lmo2152, trxB and lmo2390) were up regulated in our study (Table S7). The ferritin protein Fri, that also provides protection against reactive oxygen species, is essential for virulence and is required for efficient bacterial growth at early stages of the infection process [52],[53]. Fri transcription is directly regulated by Fur, the ferric uptake regulator. The expression of fri and fur was activated during infection. In addition, ohrA and gap were up regulated in vivo and encode two proteins respectively involved in hydroperoxide resistance [54] and in resistance against reactive oxygen species produced by host phagocytic cells in Leishmania [55] (Table S7).
L. monocytogenes metabolism adaptation to in vivo conditions
Remarkably, 30% of the in vivo regulated genes are involved in L. monocytogenes metabolism (99 metabolism-related genes were up and 72 were down regulated) (Table S8). As described above, uhpT is an in vivo highly up regulated virulence gene, regulated by PrfA and that promotes the uptake of phosphorylated hexoses (glucose-1-phosphate and glucose-6-phosphate) [23],[56]. Phosphorylated glucose is the product of glycogen hydrolysis in eukaryotic cells and there is experimental evidence that the PrfA-dependent utilization of this compound has a role in L. monocytogenes virulence [23],[56].
We observed an in vivo up regulation of several genes encoding enzymes involved in the glycolysis, like gap, pgi, fbaA, and pgm. Inversely, we found a down regulation of the expression of four genes involved in the non-oxidative phase of the pentose phosphate pathway (lmo2660, lmo2661, lmo2662 and lmo2674). The final step of glycolysis leads to pyruvate, which is then converted to acetyl-CoA by the pyruvate dehydrogenase complex. We found this complex partly up regulated in vivo, as well as one of its activator, the lipoate ligase protein LplA2 [57],[58]. The citric acid cycle is continuously supplied with acetyl-CoA during aerobic respiration. We observed an up regulation of three citric acid cycle genes (citBCG) (Table S8). The citric acid cycle is followed by oxidative phosphorylation. In this study, we found the up regulation of several genes implicated in biosynthesis and assembly of components of the respiratory chain (menD, lmo1677, qoxABD, ctaA, cydA, cydD, atpD). In addition, genes encoding resD, a regulator of aerobic and anaerobic respiration [59] and rex, a redox-sensing transcriptional repressor [60], were also up regulated in vivo. Genes encoding the pyruvate-formate lyase (pfl) and pyruvate-formate lyase activating enzymes (pflCB) are required for the anaerobic metabolism of pyruvate and were activated in the host (Table S8).
Genes implicated in amino acid biosynthesis were also induced in vivo, in particular aroA and pheA, two genes responsible for aromatic amino acid biosynthesis. Mutations in aroA and pheA were previously shown to induce an attenuation of virulence in the mouse model [61],[62]. Furthermore, the expression of genes implicated in the biosynthetic pathways of branched amino acids (alsS, ilvN and lmo0978), and amino acids of the aspartate and glutamate families (ansB, lmo0594, lmo1006, lmo1011, lmo2413 and glnA, lmo2770, respectively), was also increased in vivo (Table S8).
Significantly, mannose (lmo0781–lmo0784), maltose (lmo0278) and cellobiose (lmo0301 and lmo0915) -specific PTS encoding genes [63] were up regulated in vivo. Inversely, fructose (lmo2733), galactitol (lmo2665) and mannitol (lmo2649) -specific PTS encoding genes appeared down regulated.
Among the genes involved in bacterial ion uptake systems, a potassium-transporting ATPase encoding gene (kdpB) was down regulated in vivo. Cobalt (lmo1207), manganese (lmo1424) and calcium (lmo0841) transporter systems were, inversely, up regulated. As indicated above, the ferritin and ferric uptake protein encoding genes, fri and fur, shown to be activated under low iron concentration [64],[65], appeared highly up regulated in vivo (Table S8).
Detection of potential virulence genes by in vivo transcriptomics
A major goal of this work was the identification of genes that encode proteins that may play a role in the infectious process. To identify such virulence genes and in order to establish a short list, we arbitrarily used several criteria. The gene should be preferentially 1) highly activated during infection; 2) absent in the non pathogenic strain L. innocua and present in other L. monocytogenes strains from diverse serotypes; 3) a member of a specific protein family encoding gene (surface, secreted, stress) possibly involved in virulence; 4) controlled by virulence regulators (PrfA, VirR, CtsR, HrcA, SigB). Several candidates emerged, matching, at least, some of the above criteria (Table 6).
Table 6. L. monocytogenes EGDe genes differentially regulated in the host and potential virulence factors.
| Gene designation | Gene | Annotation | Homolog in L. innocua | Homolog in 1/2a F6854 | Homolog in 4b F2365 | Homolog in 4b H7858 | Fold change 24 h | Fold change 48 h | Fold change 72 h | Regulation | Secreted/Surface protein |
| lmo0206 | lmo0206 | unknown protein | LMOf6854_0214.1 | LMOf2365_0217 | LMOh7858_0225.1 | 3,11 | PrfA | Secreted | |||
| lmo0257 | lmo0257 | unknown protein | LMOf6854_0261.2 | LMOf2365_0265 | LMOh7858_0288 | 2,12 | |||||
| lmo0540 | lmo0540 | similar to penicillin-binding protein | lin0544 | LMOf6854_0581 | LMOf2365_0569 | LMOh7858_0598 | 3,77 | 3,96 | 2,29 | Secreted/Surface | |
| lmo0604 | lmo0604 | similar to B subtilis YvlA protein | lin0613 | LMOf6854_0642.2 | LMOf2365_0633 | LMOh7858_0663.2 | 51,45 | 27,43 | VirR | ||
| lmo0788 | lmo0788 | unknown protein | lin0781 | LMOf6854_0832 | LMOf2365_0804 | LMOh7858_0842 | 60,06 | 31,36 | 20,50 | PrfA | |
| lmo0915 | lmo0915 | similar to phosphotransferase system enzyme IIC | LMOf6854_0962 | LMOf2365_0937 | LMOh7858_0989 | 3,15 | 2,37 | ||||
| lmo1081 | lmo1081 | similar to glucose-1-phosphate thymidyl transferase | LMOf6854_1134 | 8,25 | 5,99 | 5,17 | |||||
| lmo1082 | lmo1082 | similar to dTDP-sugar epimerase | LMOf6854_1135 | 47,21 | 24,06 | 21,01 | |||||
| lmo1099 | lmo1099 | similar to a protein encoded by Tn916 | 65,42 | 30,42 | 24,04 | ||||||
| lmo1102 | lmo1102 | similar to cadmium efflux system accessory proteins | 9,71 | 4,51 | 4,72 | ||||||
| lmo1290 | lmo1290 | similar to internalin proteins (LPXTG motif) | LMOf6854_1332 | LMOf2365_1307 | LMOh7858_1374.2 | 6,24 | Surface | ||||
| lmo1438 | lmo1438 | similar to penicillin-binding protein | lin1477 | LMOf6854_1481 | LMOf2365_1457 | LMOh7858_1533 | 3,55 | Secreted/Surface | |||
| lmo1521 | lmo1521 | similar to N-acetylmuramoyl-L-alanine amidase | lin1556 | LMOf6854_1568 | LMOf2365_1540 | 2,73 | Sig54 | Secreted/Surface | |||
| lmo1601 | lmo1601 | similar to general stress protein | lin1642 | LMOf6854_1653.1 | LMOf2365_1622 | LMOh7858_1707. | 2,18 | PrfA-SigB-SigL | |||
| lmo1602 | lmo1602 | unknown protein | lin1643 | LMOf6854_1653.2 | LMOf2365_1623 | LMOh7858_1707.4 | 4,54 | 2,13 | PrfA-SigB-SigL | Secreted | |
| adeC | lmo1742 | highly similar to adenine deaminases | lin1853 | LMOf6854_1800 | LMOf2365_1767 | LMOh7858_1867 | 2,66 | VirR | |||
| lmo1855 | lmo1855 | similar to similar to D-alanyl-D-alanine carboxypeptidases | lin1969 | LMOf6854_1915 | LMOf2365_1883 | LMOh7858_1980 | 3,31 | Surface | |||
| lmo2048 | lmo2048 | unknown protein | lin2154 | LMOf6854_2109.1 | LMOf2365_2079 | LMOh7858_2176.1 | 4,93 | 2,51 | SigB-HcrA | ||
| lmo2114 | lmo2114 | similar to ABC transporter (ATP-binding protein) | lin2219 | LMOf6854_2178 | LMOf2365_2147 | LMOh7858_2245 | 3,68 | 2,93 | ViR-CtsR-SigL | ||
| lmo2115 | lmo2115 | similar to ABC transporter (permease) | lin2220 | LMOf6854_2179 | LMOf2365_2148 | LMOh7858_2246 | 4,86 | 5,92 | 3,06 | ViR-SigL | |
| lmo2157 | lmo2157 | unknown protein | LMOf6854_2221 | LMOf2365_2189 | LMOh7858_2290.1 | 3,29 | 5,85 | 2,01 | PrfA-SigB | ||
| lmo2177 | lmo2177 | unknown protein | lin2280 | LMOf6854_2241 | LMOf2365_2209 | LMOh7858_2310 | 3,36 | 6,14 | 2,23 | VirR | |
| fabF | lmo2201 | similar to 3-oxoacyl-acyl-carrier protein synthase | lin2304 | LMOf6854_2265 | LMOf2365_2234 | LMOh7858_2335 | 3,90 | 2,03 | SigB | ||
| lmo2203 | lmo2203 | similar to N-acetylmuramoyl-L-alanine amidase | lin2306 | LMOf6854_2266.1 | LMOf2365_2236 | LMOh7858_2337 | 4,50 | Secreted/Surface | |||
| lmo2439 | lmo2439 | unknown protein | lin2533 | LMOf6854_2499.3 | LMOf2365_2411 | LMOh7858_2584.4 | 4,44 | VirR | Secreted | ||
| gap | lmo2459 | glyceraldehyde-3-phosphate dehydrogenase | lin2553 | LMOf6854_2520 | LMOf2365_2432 | LMOh7858_2608 | 4,49 | 2,37 | HcrA | Surface | |
| lmo2522 | lmo2522 | similar to hypothetical cell wall binding protein | lin2666 | LMOf6854_2584 | LMOf2365_2495 | LMOh7858_2674 | 5,72 | 3,07 | Secreted/Surface | ||
| lmo2713 | lmo2713 | secreted protein with 1 GW repeat | lin2861 | LMOf6854_2831.1 | LMOf2365_2693 | LMOh7858_2976.1 | 8,86 | 17,08 | 2,80 | Secreted/Surface | |
| lmo2714 | lmo2714 | peptidoglycan anchored protein (LPXTG motif) | lin2862 | LMOf6854_2833 | LMOf2365_2694 | LMOh7858_2978 | 25,45 | 70,27 | 3,09 | Surface |
lmo0206, lmo0257, lmo0915, lmo1290 and lmo2157 are genes that, as eleven already known virulence factors, are L. monocytogenes species-specific and induced in vivo. lmo0206 and lmo2157 are the only two genes activated in vivo, controlled by PrfA, absent from L. innocua and whose role in virulence was never investigated. lmo0206, orfX [66], is in addition located at the end of the Listeria virulence cluster and was recently implicated in intracellular survival [14]. The expression of lmo2157 was shown to be controlled by PrfA and SigB [6],[8].
lmo1081, lmo1082, lmo1099 and lmo1102 are L. monocytogenes EGDe species-specific genes highly up regulated in vivo over the three time points of the infection (Table 6). Interestingly, these genes encode proteins potentially involved in cell wall metabolism and heavy metal detoxification.
Only two uncharacterized genes encoding LPXTG surface proteins (lmo1290 and lmo2714) and three encoding GW surface proteins (lmo1521, lmo2203 and lmo2713) were up regulated within the host (Table 6). lmo1521 and lmo2203 are in addition predicted autolysins. lmo2713 and lmo2714 seem to be part of a genomic region over expressed at all time points of the infection and Lmo2714 was found in the Listeria culture supernatant [43]. Four genes (lm0540, lmo1438, lmo1855 and lmo2522) predicted to be involved in cell wall metabolism were up regulated in vivo, and similar to pgdA, iap, and murA [29]–[31], could participate in Listeria infection.
Twenty-five uncharacterized genes activated in vivo encode secreted proteins that may interact with the host cells, including Lmo2201, a Tat-secreted protein [42], and GAPDH. GAPDH was previously shown to be part of the Listeria cell wall subproteome [35], and to impair Listeria phagosome maturation [67]. GAPDH seems, in addition, to be implicated in the virulence of several other pathogens [68]–[70].
lmo0788 is highly activated in mouse spleens during infection and is the only gene of the group I PrfA-regulated genes (i.e. preceded by a PrfA-box and positively regulated by PrfA) [6] whose role during infection has never been addressed (Tables 2 and 6). lmo0788 encodes a protein similar to subunits (BadFG) of the benzoyl-CoA reductase used by facultative aerobes in absence of oxygen for reductive aromatic metabolism [71].
VirR appears as the second main regulator of virulence genes and controls lmo0604, lmo1742, lmo2114, lmo2115, lmo2177 and lmo2439, whose expression was activated in the host (Tables 3 and 6). lmo2114 and lmo2115 are in addition part of a transcriptional unit co-regulated by CtsR and SigL [7].
Several stress protein encoding genes that are under the control of different stress regulators were up regulated in vivo. In particular, lmo2048 is a stress protein-encoding gene that is co-controlled by CtsR and HrcA (Table 6). The 19 genes up regulated in vivo and co-controlled by PrfA and SigB (Table 2) could also be important for the infectious process. Among these, lmo1601 and lmo1602 are furthermore regulated by SigL [7].
The use of such arbitrary criteria obviously not guaranteed that a selected gene was a virulence factor, and conversely probably excluded many virulence genes. In particular, it is worth mentioning that 91 genes encoding proteins similar to unknown proteins, and 31 encoding putative proteins with no similarity in public databases were differentially expressed in vivo (Table S3), representing a large reservoir of potential new virulence factors. Of these genes, those highly regulated all over the infectious process could be of special relevance for virulence.
Identification of new L. monocytogenes virulence factors
In order to validate our transcriptomics approach and identify new L. monocytogenes virulence factors, 6 genes (lmo1081, lmo1082, lmo1102, lmo2713, lmo2714 and gap) were selected for mutagenesis using the criteria presented above. As we were unable to produce a gap deletion mutant (probably because GAPDH is an essential protein), we constructed a GAPDH secretion mutant.
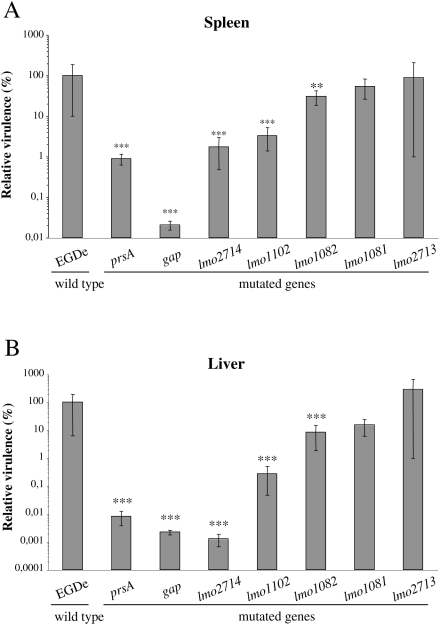
To analyze the potential role of the selected genes in virulence, we performed intravenous inoculations of BALB/c mice with wild type (wt) and mutant strains, and the number of bacteria in the mouse liver and spleen was determined 72 h after infection (Figure 5). Mutants can be classified with respect to their virulence potential. Bacterial counts for lmo1081 and lmo2713 mutants were not significantly changed as compared to the wt strain, suggesting the non-implication of these genes in Listeria virulence in mice. For the lmo1082 mutant, bacterial counts were significantly affected (≈1 log) in mouse livers and at a lesser extent in the spleens. Interestingly, for lmo1102, lmo2714 and gap mutants we observed a remarkable decrease of bacterial counts in both mouse organs as compared to the wt. In particular, the number of bacteria was dramatically impaired in the liver 72 h after inoculation (≈2,5 to 4,5 log). The gap mutant appeared as the most attenuated mutant of our analysis with a considerable virulence decrease in both organs reaching 3,5 log in the spleen and 4,5 log in the liver as compared to the wild type (Figure 5).
Figure 5. In vivo characterization of new Listeria mutants.
BALB/c mice were intravenously inoculated with 104 CFUs. The number of bacteria in the spleen (A) and liver (B) of mice was determined at 72 h post-infection. A prsA mutant was constructed and used as control. Five mice for each bacterial strain. Statistically significant differences are indicated as compared to wild type strain: ** = P<0.01, *** = P<0.001.
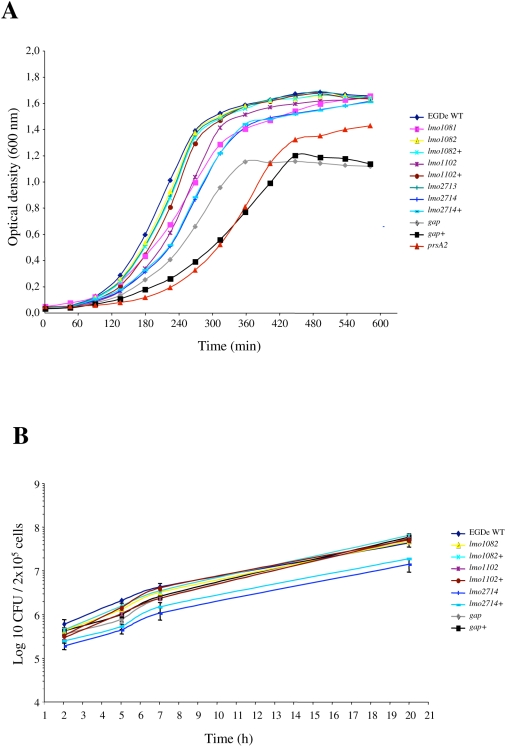
In order to better characterize virulence attenuated strains, mutants for lmo1082, lmo1102, lmo2714 and gap were complemented. The corresponding wild-type gene was inserted as a single copy under the control of its own promoter on the chromosome of the mutant strain, at the PSA bacteriophage attachment site using the pPL2 integration vector [72]. Wild type, mutant and complemented strains were tested for growth in BHI at 37°C and for intracellular behavior after internalization in the murine macrophage cell line J774 (Figure 6).
Figure 6. In vitro behavior of L. monocytogenes mutants.
(A) Growth curves of L. monocytogenes EGDe strains in BHI at 37°C with shaking. (B) Intracellular behavior of L. monocytogenes EGDe strains in J774 cultured cells.
The growth rate observed in BHI at 37°C for the majority of the strains tested was comparable to that of the wild type (Figure 6A). However, the gap secretion mutant exhibited an important in vitro reduced growth rate and reduced density at the stationary phase. The growth defect observed for the gap mutant was even accentuated in the complemented strain (Figure 6A). This is most probably the result of an over expression of intracellular GAPDH, expressed at the same time from the bacterial genome and from the plasmid harbored by this strain. Surprisingly, the prsA2 mutant presented also a notable growth delay. This growth defect was not mentioned in previous studies implicating PrsA2 in intracellular behavior and virulence [14],[33].
Wild type, mutant and complemented strains were also tested for intracellular behavior. As shown in Figure 6B, all the strains grew with similar multiplication rates after internalization in J774 cells, indicating that the slight growth delay observed in BHI at 37°C for some strains has no consequences on intracellular multiplication.
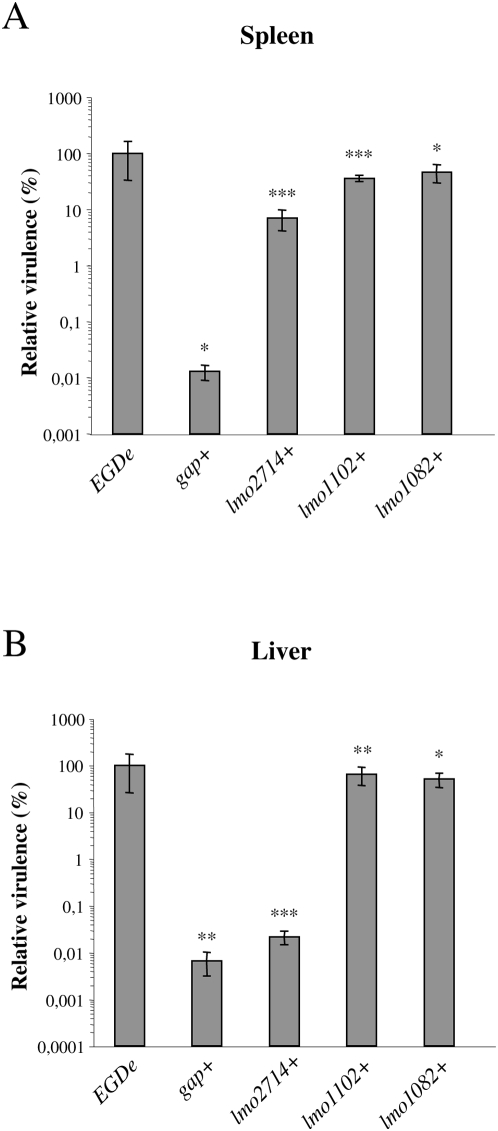
In addition, complemented strains were analyzed after intravenous inoculations of BALB/c mice as compared to wt and mutant strains, and the number of bacteria in the mouse liver and spleen was determined 72 h after infection (Figure 7). The virulence phenotype was restored, albeit partially in the case of lmo2714, in complemented strains, except for the gap mutant. The virulence defect of the gap complemented strain was even more severe in the spleen as compare to the corresponding mutant (Figure 7A). This was in correlation with the increased growth defect observed in BHI at 37°C for the gap complemented strain.
Figure 7. In vivo complementation of new Listeria mutants.
BALB/c mice were intravenously inoculated with 104 CFUs. The number of bacteria in the spleen (A) and liver (B) of mice was determined at 72 h post-infection. Five mice for each bacterial strain. Statistically significant differences are indicated as compared to the corresponding mutant for complemented strains: * = P<0.05, ** = P<0.01, *** = P<0.001.
These results revealed a role for lmo1082 and lmo1102, and at less extent for lmo2714 and gap in Listeria virulence, validating our in vivo transcriptomics approach.
Discussion
Identification of bacterial gene expression patterns during host-pathogen interactions has long been a goal for understanding infectious processes of intracellular pathogens [73]. Here, we undertook the first time course study of the L. monocytogenes in vivo transcriptome by comparing the genomic transcriptional patterns of bacteria grown under laboratory conditions (BHI, 37°C, exponential growth phase, pH 7) with that of in vivo-grown bacteria over three days of infection (mouse spleen). This constitutes also the first genome expression analysis of a pathogen in infected mouse spleens. Our results indicate that a significant part of the Listeria genome is differentially expressed for adaptation to the host environment, essentially through gene activation. We showed an in vivo over expression of an impressive number of genes involved in virulence and subversion of the host immune systems, together with genes involved in adaptation of the bacterial metabolism to host conditions and stress responses. We revealed that all these expression modifications are controlled by a complex regulatory network.
In vivo activation of Listeria virulence mechanisms
Whereas metabolic genes represent only 22% of the in vivo activated genes, they constitute the major part of the down regulated genes (65%). These observations reveal that the modification of the Listeria genome expression during infection is dominated by the activation of virulence specific genes.
A major finding of our study was the demonstration that the majority of genes previously reported as implicated in virulence were highly up regulated in the host. We observed a peak of activation for these genes 48 h p.i., preceding the peak of bacterial loads that occurs at 72 h p.i. when mice are intravenously inoculated with a sub-lethal dose. Our results support the idea that Listeria uses in vivo a complex and coordinated regulatory network that includes virulence and stress regulators in order to tightly control genes that contribute to its survival and progression of the disease. Our study definitively establishes PrfA as the major Listeria virulence regulator and VirR as the second one. These two regulators, as well as a large proportion of the genes they regulate, including known virulence factors and potentially new virulence genes, were strongly activated in vivo. In addition, the co-control by PrfA and SigB of several genes activated in vivo strongly highlights the importance of the interplay between these two regulons during the infectious process. The hypothesis on an in vivo intersecting regulation of SigB and PrfA is in agreement with a very recent study demonstrating the contribution of SigB and PrfA to a regulatory network critical for appropriate regulation of virulence gene expression [74]. The large number of additional regulons and predicted transcriptional regulators differentially regulated inside the host underlines the high degree of regulation required for adaptation of Listeria to the host environment.
Another major aspect observed when Listeria interacts with its host is the active remodeling of the bacterial envelope through activation of the cell wall metabolism and enhanced exposure of virulence proteins at the bacterial surface.
Pathogens have evolved various systems for the secretion of bacterial factors that contribute to the progression of the disease. Listeria uses different secretion systems and a significant number of their products were activated in vivo. It is particularly the case of the SecA2 system, itself activated in vivo, and responsible for the secretion of several proteins lacking a signal peptide and also up regulated in the host, including known virulence factors.
Although competence genes have been found in L. monocytogenes genome [20], Listeriae have never been shown to be naturally competent. Interestingly, we observed several competence genes (comEA, comEB, comGF, comGE, clpC, mecA and degU) up regulated in vivo. This is the first report of a simultaneous activation of a great number of competence genes in Listeria, suggesting that this bacterium could be competent during infection and use this system to incorporate DNA from the host environment in order to acquire new potentialities.
Subversion of host defenses
In addition to the up regulation of a number of virulence factors, L. monocytogenes activates mechanisms of subversion of the host defenses. Lysinylation of phospholipids in Listeria membranes by MprF, and D-alanylation of cell wall TAs and LTAs by the dlt complex lead to a reduced negative charge of the bacterial surface. One of the consequences of this process is the repulsion of cationic antimicrobial peptides [13],[25]. In vivo activation of both dlt and mprF by VirR strongly suggests a regulation of L. monocytogenes resistance to cationic peptides in the host, as previously proposed [25]. Furthermore, surface components e.g. LTAs and PG play a role in the innate immune response through receptors like Nods and Toll-like receptors [75]. LTA modification by VirR regulated factors could thus be used by Listeria to escape the host innate immune response.
SecA2-dependent secretion has been proposed to coordinate PG digestion by the activity of secreted autolysins [30]. The muramyl glycopeptide predicted to be generated by the combined activities of p60 and MurA is known to modify host inflammatory responses [76],[77]. Thus, the strong in vivo induction of SecA2 and SecA2-secreted proteins may activate release of specific peptides that interfere with host pattern recognition.
N-deacetylation by PgdA was shown to be a major modification of Listeria PG, conferring the ability to survive in the gastrointestinal tract, in professional phagocytes, evade the action of host lysozyme, and modulate the inflammatory response [31]. The over expression of pgdA during infection appears as an additional strategy used by Listeria to subvert host pattern recognition and control the host inflammatory responses to promote its own survival.
In the same way, in accordance to the potent proinflammatory activity of flagellin [78], Listeria down regulates flagella related genes (lmo0681 and lmo0697) during the infection by the activation of mogR, a repressor of motility and chemotaxis gene expression [79].
Adaptation of the metabolism to the host environment
Once inside the host, bacteria need to adapt to nutritional changes, including carbon and nitrogen sources. This is illustrated by the high number of metabolic genes differentially regulated in vivo.
UhpT and several enzymes involved in glycolysis were up regulated whereas enzymes implicated in the non-oxidative phase of the pentose phosphate pathway were repressed. These data suggest that phosphorylated glucose transported by UhpT is being metabolized through glycolysis. Glucose or phosphorylated glucose seems thus one of the major carbon sources in vivo, the pentose phosphate cycle appearing not essential for the generation of necessary intermediates and for gluconeogenesis. In addition to genes involved in glycolysis, we observed the in vivo up regulation of numerous genes implicated in the citric acid cycle and in oxidative phosphorylation, indicating that L. monocytogenes is using oxidative phosphorylation to generate energy. However, the activation of genes involved in fermentation, suggests that, even though L. monocytogenes is using oxidative phosphorylation to generate energy, it might be experiencing some level of oxygen starvation in spleen cells. The activation of the glycerol kinase and glycerol-3-phosphate dehydrogenase, indicates that glycerol, probably deriving from the activity of the phospholipases A and B on cellular lipids, is an additional carbon source for intracellular growth.
Metal ions are essential cofactors for functional expression of many proteins in bacterial systems. Thus, alterations in Listeria ion transport genes in vivo reflect the accessibility/inaccessibility of those ions in the intracellular environment, i.e presence of potassium and lack of cobalt, manganese, calcium and iron.
Due to defense mechanisms developed by the host to limit bacterial multiplication, it could be expected a growth rate decrease for invading pathogens in their host [16]. A very interesting discovery resulting from our in vivo transcriptomics analysis was the observation of an active growth status of Listeria in infected mouse spleens. This was demonstrated by the increased expression of numerous genes encoding proteins involved in bacterial growth and multiplication, including genes implicated in DNA replication and cell division. Listeria thus seems to have 24 h p.i. overcome organism defenses and being engaged in an active multiplication phase.
Stress responses to the host environment
Bacterial responses to environmental changes are often characterized by the induction of specific stress responses. The in vivo induction of Listeria stress genes indicated that bacteria are faced with stress within the host. The alternative sigma factor B is the master regulator of stress. Even in the absence of a significant regulation of sigB itself in vivo, an impressive number (70) of genes previously shown to be under SigB-regulation were up regulated in vivo, including numerous virulence factors. In addition, genes up regulated in vivo and under the control of HrcA and CtsR seem also to be particularly relevant for virulence. During the process of host colonization, L. monocytogenes induces a host inflammatory response [80]. This defense is accompanied by the generation of ROS presented to the persistent pathogen. In addition to the traditional ROS combating enzymes like catalase and superoxide dismutase, this transcriptomic analysis revealed the in vivo activation of panoply of genes implicated in the response to oxidative stress, suggesting a special relevance of this response for Listeria pathogenesis/persistence.
Strong difference between in vivo and “in cultured cells” transcriptome data
Our study highlights that the analysis of a host-pathogen interaction in its real context (i.e. the living host) is highly informative. Indeed, it is not currently feasible to reconstruct in vitro the exact environment faced by Listeria in the host. This is illustrated by the strong difference observed between our in vivo transcriptome and previous transcriptomes of Listeria growing inside epithelial cells or macrophages [14],[15], with only 15% and 29% overlap for the up regulated genes, and no more than 3% for the down regulated genes. Furthermore, 25% and 44% of the genes down regulated in epithelial cells and macrophages respectively, were up regulated during mouse infection. One of the main differences between in vitro intracellular growth and in vivo infection was the much higher number of previously known virulence genes activated in vivo (29 against 17), or down regulated intracellularly (8 against 2). Genes identified in this study as up regulated in vivo and implicated in virulence are not regulated in epithelial cells, and not regulated (lmo1102) or even repressed (lmo1082, gap) in macrophages. lmo2714 is the only of these genes that appeared activated both in vivo and in macrophages. Other significant differences between “in cultured cells” and in vivo approaches were observed at the level of genes involved in cell division and cell wall metabolism, repressed in macrophages but activated in the host, suggesting a more active multiplication status of Listeria in mouse organs. Listeria metabolism in the two environments appeared also significantly different, in particular concerning glycolysis and the pentose phosphate pathway that, in contrast to what was observed in cultured cells [14],[15], were respectively activated and repressed in bacteria growing in mouse spleens. Finally, we observed a strong in vivo down regulation of flagella related genes, in accordance to the potent proinflammatory activity of flagellin [78], and inversely to what observed during intramacrophagic growth [14],[15].
Identification of new L. monocytogenes virulence factors by in vivo genome profiling and mutagenesis
In addition to the global analysis of the expression of the entire Listeria genome during infection, a major goal of this study was the identification of new Listeria virulence factors. The in vivo differential expression of a remarkable number of genes previously implicated in Listeria intracellular survival and virulence underscores the relevance of our approach. Our analysis allowed the detection of several potential novel virulence genes. Mutagenesis of 6 of these genes demonstrated the implication of a majority of them in virulence, thus definitively establishing the value of our strategy.
lmo2714 is a gene up regulated during infection and that encode a LPXTG surface protein [35],[36]. The probable implication of this LPXTG protein in L. monocytogenes virulence in mice confirms the importance of this protein family for the Listeria-host interaction and underlines the complexity of the mechanisms developed by this pathogen to reach a maximal infectious capacity. As other known surface virulence determinants [81],[82], Lmo2714 could interact with a specific cellular receptor or ligand that remains to be identified. Lmo2714 was also shown as present in the Listeria supernatant [43], and could thus also act as secreted factor.
gap encodes GAPDH, a glycolytic enzyme involved in bacterial energy generation that is essential for growth in the absence of neoglucogenic substrates. In Listeria, GADPH was previously described as a surface protein present in the cell wall, as well as a secreted protein [35],[43]. As a secreted product, GAPDH was shown to impair Rab5a mediated phagosome–endosome fusion [67]. Interestingly, GAPDH was also recently shown to be a key virulence-associated protein of Streptococcus suis type 2 up regulated in vivo [70]. The impossibility of constructing a gap deletion mutant confirmed the essential role of this protein in the bacterial metabolism as previously shown [70]. Using a gap secretion mutant, we showed that, whereas required for full growth in BHI, secreted GAPDH was not essential for intracellular multiplication. In addition, mouse infection indicated a role for secreted GAPDH in Listeria virulence, probably in part through its ability to retain and inactivate phagosomal Rab5a as previously described [67]. However, the gap secretion mutant exhibited an important in vitro growth defect. In addition, the complemented strain showed an accentuated growth delay in vitro and a more pronounced virulence decrease in vivo. These effects, even most probably due to an over expression of intracellular GAPDH, should be corrected in order to definitively prove the critical role of secreted GAPDH in virulence.
lmo1082 is homolog to rmlC that encodes a dTDP-dehydrorhamnose epimerase potentially implicated in the surface layer (S-layer) glycoprotein synthesis [83]. lmo1082 is furthermore part of a L. monocytogenes EGDe specific chromosomal region that contains two other genes that are homologous to S-layer biosynthesis genes in other organisms, several genes involved in TA biosynthesis, and the autolysin Auto previously implicated in Listeria virulence [84]. S-layers are two-dimensional crystalline arrays that completely cover bacterial cells. In addition to the impaired survival of the lmo1082 mutant in mouse organs, the high in vivo activation of lmo1082 could suggest a role for S-layer glycoproteins in Listeria virulence. S-layers have been shown to be virulence factors of several pathogens. In particular, the S-layer glycoproteins were implicated in mechanisms evolved by pathogenic bacteria to evade host immune systems. However, the examination of several strains using different techniques has so far never demonstrated the presence of S-layers in Listeria [85]. Preliminary experiments by electron microscopy did not allow to confirm the presence of a Listeria S-layer in vivo (data not shown). This aspect thus requires further investigation.
Lmo1102 is similar to CadC, a protein required for cadmium resistance. Cadmium is a heavy metal and its cation is toxic for microbes in the environment. Cadmium resistance in Listeria is an energy-dependent cadmium efflux system, involving two proteins, CadA and CadC. Listeria cadA and cadC genes for cadmium resistance were previously located, in the strains analyzed, on a transposable element (Tn5422) closely related to Tn917 and capable of intramolecular transposition [86],[87]. In the L. monocytogenes EGDe strain, the cadA and cadC genes are part of an EGDe specific chromosomal region located downstream an integrase encoding gene and containing 13 genes similar to Tn916 genes. This seems to indicate that L. monocytogenes EGDe has also acquired resistance to cadmium by transposon insertion. The strong in vivo activation of cadC and the significant impaired virulence of the cadC mutant suggest that this heavy metal resistance system constitutes an advantage for in vivo Listeria survival. The real function of CadC in vivo reserves further investigation.
Concluding remarks
In vivo, bacteria are challenged with unique cues that are difficult to reproduce under in vitro conditions. The molecular analysis of in vivo infectious processes by means of this approach provides the first comprehensive view of how L. monocytogenes adapts to the host environment in the course of the infection. We showed that the remarkable shift of the Listeria genome expression during infection is characterized by the activation of a number of genes involved in virulence and subversion of the host immune systems, and is associated with the adaptation of the bacterial metabolism to host conditions. All these mechanisms are under the control of a complex regulatory network. As confirmed here by the identification of several new virulence genes, this analysis provides a powerful tool for the detection of novel virulence determinants and a better understanding of the complex strategies used by pathogens to promote infections.
It would be now particularly interesting to perform the same in vivo genome profile analysis on different infected organs (intestine, liver, brain) and using different animal models in order to identify organ- or host-specific virulence factors.
Materials and Methods
Bacterial strains and growth conditions
L. monocytogenes EGDe was grown in Brain Heart Infusion (BHI) medium (BD-Difco) or in a defined minimal medium (modified Welshimer's broth [18]) at 37°C, under aerobic conditions with shaking. Erythromycin was included at 5 µg/ml when the bacteria carried pMAD and pAUL-A derivatives. Chloramphenicol was included at 7 µg/ml when the bacteria carried pPL2 derivatives. E. coli strains were grown in LB medium at 37°C, with shaking. Ampicilin and erythromycin were added at 100 µg/ml and 300 µg/ml, respectively, when required.
Isolation of L. monocytogenes EGDe total RNA
From pure culture
Cultures for preparing RNA samples were grown overnight at 37°C under aerobic conditions in liquid medium with shaking. Overnight pre-cultures were diluted in liquid medium and incubated at 37°C under aerobic conditions with shaking. Exponentially growing cells (OD600 = 0.6) or cells in stationary growth phase (OD600 = 1.5) were harvested by centrifugation for 15 min at 4700 rpm at 4°C. Total RNA was extracted as previously described [6]. Quality of RNA was assessed by determining the OD260/280 ratio and by visualization following agarose gel electrophoresis and ethidium bromide stain.
From infected mice organs
Specific pathogen-free female CD1 mice (Charles River) were intravenously infected with L. monocytogenes EGDe. At 24, 48 and 72 h post-infection, the mice were sacrificed and dissected. The livers and spleens were harvested and immediately frozen in liquid nitrogen. The organs were stored at −80°C. Prior to RNA isolation, the organs were thawed on ice and homogenized in 20 ml of an ice-cold solution composed of 0.2 M sucrose/0.01% SDS. The homogenate was gently centrifuged for 20 min at 300 rpm and filtered to remove large tissue debris. The tissue suspension was centrifuged for 20 min at 4000 rpm to pellet the bacteria. Centrifugations were performed at 4°C. Bacterial RNA extraction was performed as previously described [6]. Quality of RNA was assessed by determining the OD260/280 ratio and by visualization following agarose gel electrophoresis and ethidium bromide stain.
Construction of whole-genome macroarrays
The macroarrays used here are described in [6]. Briefly, specific primer pairs were designed for each of the 2853 ORFs of the L. monocytogenes EGDe genome, in order to amplify a fragment of ∼500 bp specific for each ORF. For macroarray preparation, nylon membranes were soaked in 10 mM TE, pH 7.6. Spot blots of ORF-specific PCR products and controls were printed using a Qpix robot. Immediately after spot deposition, membranes were neutralized for 15 min in 0.5 M NaOH, 1.5 M NaCl, washed three times with distilled water and stored wet at −20°C until use.
cDNA synthesis, labeling and hybridization to macroarrays
cDNA synthesis, labeling and hybridization were performed as previously described [6]. Briefly, cDNA was reversely transcribed in the presence of [α-33P]-dCTP. Labeled cDNA was purified using a QIAquick column (Qiagen). Hybridization and washing steps were carried out using SSPE buffer. Macroarrays were pre-wet in 2× SSC and pre-hybridized in hybridization solution (5× SSPE, 2% SDS, 1× Denhardt's reagent, 100 µg of sheared salmon sperm DNA/ml) at 65°C. Hybridization was carried out for 20 h at 65°C. After hybridization, membranes were washed twice at room temperature and twice at 65°C in 0.5× SSC, 0.2% SDS. For each condition, two independent RNA preparations were tested, and two cDNAs from each of the RNA preparations were hybridized to two sets of arrays and analyzed.
Array results are available at the GEO database under the accession number GPL7248 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=jvqvnqwicgkoajy&acc=GSE13057).
Data analysis
Membranes were scanned using a 445SI PhosphoImager. The ARRAYVISION software was used for quantification of the hybridization intensities. The intensity of each spot was normalized according to the median value of the total intensities of all spots on each array. The global background was calculated from the median intensity of 610 “no-DNA” spots homogeneously distributed throughout the membrane. For spots whose intensity value was lower than the median background intensity, the intensity value was replaced by the median background intensity, for analysis purposes.
The significance analysis of microarrays (SAM) program was used for identification of genes with statistically significant changes in expression [19]. SAM was conducted with the following log2 ratios of gene expression values: 1) 24 h post-infection versus pure culture, 2) 48 h post-infection versus pure culture, and 3) 72 h post-infection versus pure culture. One-class responses were chosen to test if the mean level of gene expression differed from a hypothesized mean. A delta value corresponding to a false discovery rate <5% was chosen. Genes with at least a twofold expression change that were significant according to this analysis in at least one time point were taken into account.
For clustering analysis, data was log transformed, median centered and an average-linkage clustering was carried out using CLUSTER software and the results were visualized by TREEVIEW [88].
Quantitative real time RT-PCR analysis
Up to 1 µg of total RNA was reverse-transcribed by using the iScript kit (Bio-Rad). Forward and reverse primers (Table S6) were designed using Primer3 software (http://frodo.wi.mit.edu/) to produce an amplicon length of 70–200 bp. A standard curve was generated for each primer pair by using four ten-fold dilutions of cDNA from L. monocytogenes EGDe, to ensure that PCR efficiency was 100%. Quantitative PCR was performed for 45 cycles with 2 µl of cDNA, 10 µl of 2× SYBR green PCR master mix (Bio-Rad) and 0,25 pM (each) forward and reverse primers in a final volume of 20 µl. For each primer pair, a negative control (water), was included during cDNA quantification. After PCR amplification, a melting curve was generated for every PCR product to check the specificity of the PCR reaction. Data were analyzed by the ΔΔCt method which provides the target gene expression value as unitless fold changes in the unknown sample compared with a calibrator sample [89]. Both unknown and calibrator sample target gene expression data were normalized by the relative expression of 16S rRNA.
Mutagenesis
Construction of deletion mutants (lmo1082, lmo1102)
Two ∼1000 bp fragments flanking the target genes were amplified by PCR from L. monocytogenes EGDe chromosomal DNA. Primers used to generate the flanking regions are shown in Table S9 in the supplemental material (restriction sites are underlined). The purified PCR fragments were digested as stated in Table S9 and coligated in the thermosensitive plasmid pMAD [90]. Plasmid DNA of pMAD bearing the fragments was used to electroporate L. monocytogenes EGDe to generate the chromosomal deletion mutants, as described previously [90]. The deletions were verified by PCR analysis of chromosomal DNA using pairs of primers inside each gene (Table S9).
Construction of insertion mutants (prsA2, lmo1081, lmo2713, lmo2714)
A 500–1000 bp fragment internal to the target genes was amplified by PCR from L. monocytogenes EGDe chromosomal DNA. Primers used to generate the internal regions are shown in Table S9 (restriction sites are underlined). The purified PCR fragments were digested as stated in Table S9 and coligated in the thermosensitive plasmid pAUL-A [91]. Plasmid DNA of pAUL-A bearing the fragments was used to electroporate L. monocytogenes EGDe to generate the chromosomal intertion mutants, as described previously [91]. The insertions were verified by PCR analysis of chromosomal DNA using pairs of primers described in Table S9.
Construction of gap secretion mutant
The prediction of hydrophobicity of a putative hydrophobic tail was determined based on the dense alignment surface (DAS) score as described at the website http://www.sbc.su.se/~miklos/DAS/. The amino acid sequence of a hydrophobic tail to be inserted at the C-terminal end of GAPDH was based on the translated amino acid sequence of the actA gene and was edited using the DAS method to get an optimum DAS score (≥3.0), one indicative of a typical transmembrane location [92]. The gap gene and a ∼1000 bp DNA fragment corresponding to the downstream region of the gap gene was amplified by PCR using primer pairs described in Table S9 (restriction sites are underlined, hydrophobic tail is in boldface). The purified PCR products were cloned in the multiple cloning sites located upstream and downstream of the kanamycin resistance gene in the plasmid pOD23 [93]. pOD23 bearing the fragments was used to electroporate L. monocytogenes EGDe to generate the chromosomal secretion mutant, as described previously [93].
Complementation
For complementation, the entire gene and flanking regions were amplified using primers described in Table S9. PCR products were digested as described in Table S9 and ligated to the site-specific phage integration vector pPL2 [72]. Plasmid DNA of pPL2 bearing the fragments was transformed into E. coli S17-1 and the resulting strain was mated into each mutant strain. Chloramphenicol-resistant transconjugants were tested by PCR for pPL2 integration at the appropriate chromosomal site using primers PL102 (5′-TATCAGACCAACCCAAACCTTCC-3′) and PL95 (5′-ACATAATCAGTCCAAAGTAGATGC-3′). Primers described in Table S9 were used to confirm the presence of each gene in the respective complemented strain.
Virulence studies
Animal experiments were performed as previously described in [94]. Bacterial growth in mice was studied by injecting 6-week-old specific pathogen-free female BALB/c mice (Charles River) intravenously with a sublethal bacterial inoculum, 104 CFUs, of wild type or mutant strains. At 72 h after infection the liver and spleen were sterilely dissected and the number of CFUs was determined by plating serial dilutions of organ (liver and spleen) homogenates on BHI agar medium (five animals for each strain).
Ethics statement
All animals were handled in strict accordance with good animal practice as defined by the relevant national and local animal welfare bodies, and all animal work was approved by the Direcção Geral de Veterinária (FCT-POCI/SAU-MMO/60443/2004, FCT-PTDC/SAU-MII/65406/2006).
Intracellular multiplication assay
Listeria strains were grown to OD600 = 0.6, washed and diluted in DMEM such that the MOI was about 10 bacteria per cell. Bacterial suspensions were added to J774A.1 cells for 45 min. Cells were then washed and non-phagocytosed bacteria were killed by adding 20 µg/ml gentamicin for 1 h15 min. After washing, cells were lysed in 0.2% Triton X-100, at 2 h, 5 h, 7 h and 20 h post-infection and the number of viable bacteria released from the cells was assessed after serial dilutions of the lysates on BHI agar plates. Experiments were repeated two times in triplicate.
Supporting Information
Growth and pH curves of L. monocytogenes EGDe in BHI at 37°C with shaking
(0.07 MB PDF)
Validation of macroarray data by real-time RT-PCR. Fold changes in in vivo gene expression 24 h p.i. (A) or 72 h p.I. (B) compared to that in BHI were measured by macroarray and real-time RT-PCR, log transformed and compared for correlation analysis.
(0.04 MB PDF)
Known L. monocytogenes virulence factors
(0.03 MB PDF)
L. monocytogenes genes regulated in the host as compared to exponential or stationary growth phase in BHI
(0.05 MB PDF)
L. monocytogenes genes differentially regulated in the host as compared to exponential growth in BHI at 37°C
(0.07 MB PDF)
L. monocytogenes EGDe genes encoding proteins of the cell wall subproteome and differentially regulated in the host
(0.03 MB PDF)
L. monocytogenes genes encoding secreted proteins and differentially regulated in the host
(0.03 MB PDF)
L. monocytogenes genes involved in DNA metabolism, RNA and protein synthesis, cell division and multiplication, and up regulated in the host
(0.03 MB PDF)
L. monocytogenes genes involved in stress responses and differentially regulated in the host
(0.04 MB PDF)
L. monocytogenes genes involved in metabolism and differentially regulated in the host
(0.05 MB PDF)
Primers
(0.04 MB PDF)
Footnotes
The authors have declared that no competing interests exist.
This work was supported by grants from the Portuguese Fundação para a Ciência e a Tecnologia, FCT-POCI/SAU-MMO/60443/2004 and FCT-PTDC/SAU-MII/65406/2006, and by an ERANET Pathogenomics grant, FCT-ERA/PTG/0003-SPATELIS (http://alfa.fct.mctes.pt). AC and OR are recipients of FCT doctoral fellowships SFRH/BD/29314/2006 and SFRH/BD/28185/2006. SS is a recipient of an FCT postdoctoral fellowship, SFRH/BPD/21549/2005. PC is a Howard Hughes Medical Institute international research scholar. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Swaminathan B, Gerner-Smidt P. The epidemiology of human listeriosis. Microbes Infect. 2007;9:1236–1243. doi: 10.1016/j.micinf.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 2.Berche P. Bacteremia is required for invasion of the murine central nervous system by Listeria monocytogenes. Microb Pathog. 1995;18:323–336. doi: 10.1006/mpat.1995.0029. [DOI] [PubMed] [Google Scholar]
- 3.Cossart P, Toledo-Arana A. Listeria monocytogenes, a unique model in infection biology: An overview. Microbes Infect. 2008;10:1041–1050. doi: 10.1016/j.micinf.2008.07.043. [DOI] [PubMed] [Google Scholar]
- 4.Doumith M, Cazalet C, Simoes N, Frangeul L, Jacquet C, et al. New aspects regarding evolution and virulence of Listeria monocytogenes revealed by comparative genomics and DNA arrays. Infect Immun. 2004;72:1072–1083. doi: 10.1128/IAI.72.2.1072-1083.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Severino P, Dussurget O, Vencio RZ, Dumas E, Garrido P, et al. Comparative transcriptome analysis of Listeria monocytogenes strains of the two major lineages reveals differences in virulence, cell wall, and stress response. Appl Environ Microbiol. 2007;73:6078–6088. doi: 10.1128/AEM.02730-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Milohanic E, Glaser P, Coppee JY, Frangeul L, Vega Y, et al. Transcriptome analysis of Listeria monocytogenes identifies three groups of genes differently regulated by PrfA. Mol Microbiol. 2003;47:1613–1625. doi: 10.1046/j.1365-2958.2003.03413.x. [DOI] [PubMed] [Google Scholar]
- 7.Arous S, Buchrieser C, Folio P, Glaser P, Namane A, et al. Global analysis of gene expression in an rpoN mutant of Listeria monocytogenes. Microbiology. 2004;150:1581–1590. doi: 10.1099/mic.0.26860-0. [DOI] [PubMed] [Google Scholar]
- 8.Hain T, Hossain H, Chatterjee SS, Machata S, Volk U, et al. Temporal transcriptomic analysis of the Listeria monocytogenes EGD-e sigmaB regulon. BMC Microbiol. 2008;8:20. doi: 10.1186/1471-2180-8-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kazmierczak MJ, Mithoe SC, Boor KJ, Wiedmann M. Listeria monocytogenes sigma B regulates stress response and virulence functions. J Bacteriol. 2003;185:5722–5734. doi: 10.1128/JB.185.19.5722-5734.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kazmierczak MJ, Wiedmann M, Boor KJ. Contributions of Listeria monocytogenes sigmaB and PrfA to expression of virulence and stress response genes during extra- and intracellular growth. Microbiology. 2006;152:1827–1838. doi: 10.1099/mic.0.28758-0. [DOI] [PubMed] [Google Scholar]
- 11.Hu Y, Oliver HF, Raengpradub S, Palmer ME, Orsi RH, et al. Transcriptomic and phenotypic analyses suggest a network between the transcriptional regulators HrcA and sigmaB in Listeria monocytogenes. Appl Environ Microbiol. 2007;73:7981–7991. doi: 10.1128/AEM.01281-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu Y, Raengpradub S, Schwab U, Loss C, Orsi RH, et al. Phenotypic and transcriptomic analyses demonstrate interactions between the transcriptional regulators CtsR and Sigma B in Listeria monocytogenes. Appl Environ Microbiol. 2007;73:7967–7980. doi: 10.1128/AEM.01085-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mandin P, Fsihi H, Dussurget O, Vergassola M, Milohanic E, et al. VirR, a response regulator critical for Listeria monocytogenes virulence. Mol Microbiol. 2005;57:1367–1380. doi: 10.1111/j.1365-2958.2005.04776.x. [DOI] [PubMed] [Google Scholar]
- 14.Chatterjee SS, Hossain H, Otten S, Kuenne C, Kuchmina K, et al. Intracellular gene expression profile of Listeria monocytogenes. Infect Immun. 2006;74:1323–1338. doi: 10.1128/IAI.74.2.1323-1338.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joseph B, Przybilla K, Stuhler C, Schauer K, Slaghuis J, et al. Identification of Listeria monocytogenes genes contributing to intracellular replication by expression profiling and mutant screening. J Bacteriol. 2006;188:556–568. doi: 10.1128/JB.188.2.556-568.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.La MV, Raoult D, Renesto P. Regulation of whole bacterial pathogen transcription within infected hosts. FEMS Microbiol Rev. 2008;32:440–460. doi: 10.1111/j.1574-6976.2008.00103.x. [DOI] [PubMed] [Google Scholar]
- 17.Slaghuis J, Goetz M, Engelbrecht F, Goebel W. Inefficient replication of Listeria innocua in the cytosol of mammalian cells. J Infect Dis. 2004;189:393–401. doi: 10.1086/381206. [DOI] [PubMed] [Google Scholar]
- 18.Premaratne RJ, Lin WJ, Johnson EA. Development of an improved chemically defined minimal medium for Listeria monocytogenes. App Environ Microbiol. 1991;57:3046–3048. doi: 10.1128/aem.57.10.3046-3048.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glaser P, Frangeul L, Buchrieser C, Rusniok C, Amend A, et al. Comparative genomics of Listeria species. Science. 2001;294:849–852. doi: 10.1126/science.1063447. [DOI] [PubMed] [Google Scholar]
- 21.Nelson KE, Fouts DE, Mongodin EF, Ravel J, DeBoy RT, et al. Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res. 2004;32:2386–2395. doi: 10.1093/nar/gkh562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dussurget O, Pizarro-Cerda J, Cossart P. Molecular Determinants of Listeria monocytogenes Virulence. Annu Rev Microbiol. 2004;58:587–610. doi: 10.1146/annurev.micro.57.030502.090934. [DOI] [PubMed] [Google Scholar]
- 23.Chico-Calero I, Suarez M, Gonzalez-Zorn B, Scortti M, Slaghuis J, et al. Hpt, a bacterial homolog of the microsomal glucose- 6-phosphate translocase, mediates rapid intracellular proliferation in Listeria. Proc Natl Acad Sci U S A. 2002;99:431–436. doi: 10.1073/pnas.012363899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schubert WD, Urbanke C, Ziehm T, Beier V, Machner MP, et al. Structure of internalin, a major invasion protein of Listeria monocytogenes, in complex with its human receptor E-cadherin. Cell. 2002;111:825–836. doi: 10.1016/s0092-8674(02)01136-4. [DOI] [PubMed] [Google Scholar]
- 25.Thedieck K, Hain T, Mohamed W, Tindall BJ, Nimtz M, et al. The MprF protein is required for lysinylation of phospholipids in Listerial membranes and confers resistance to cationic antimicrobial peptides (CAMPs) on Listeria monocytogenes. Mol Microbiol. 2006;62:1325–1339. doi: 10.1111/j.1365-2958.2006.05452.x. [DOI] [PubMed] [Google Scholar]
- 26.Williams T, Bauer S, Beier D, Kuhn M. Construction and characterization of Listeria monocytogenes mutants with in-frame deletions in the response regulator genes identified in the genome sequence. Infect Immun. 2005;73:3152–3159. doi: 10.1128/IAI.73.5.3152-3159.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knudsen GM, Olsen JE, Dons L. Characterization of DegU, a response regulator in Listeria monocytogenes, involved in regulation of motility and contributes to virulence. FEMS Microbiol Lett. 2004;240:171–179. doi: 10.1016/j.femsle.2004.09.039. [DOI] [PubMed] [Google Scholar]
- 28.Gueriri I, Cyncynatus C, Dubrac S, Arana AT, Dussurget O, et al. The DegU orphan response regulator of Listeria monocytogenes autorepresses its own synthesis and is required for bacterial motility, virulence and biofilm formation. Microbiology. 2008;154:2251–2264. doi: 10.1099/mic.0.2008/017590-0. [DOI] [PubMed] [Google Scholar]
- 29.Machata S, Hain T, Rohde M, Chakraborty T. Simultaneous deficiency of both MurA and p60 proteins generates a rough phenotype in Listeria monocytogenes. J Bacteriol. 2005;187:8385–8394. doi: 10.1128/JB.187.24.8385-8394.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lenz LL, Mohammadi S, Geissler A, Portnoy DA. SecA2-dependent secretion of autolytic enzymes promotes Listeria monocytogenes pathogenesis. Proc Natl Acad Sci U S A. 2003;100:12432–12437. doi: 10.1073/pnas.2133653100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boneca IG, Dussurget O, Cabanes D, Nahori MA, Sousa S, et al. A critical role for peptidoglycan N-deacetylation in Listeria evasion from the host innate immune system. Proc Natl Acad Sci U S A. 2007;104:997–1002. doi: 10.1073/pnas.0609672104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schubert K, Bichlmaier AM, Mager E, Wolff K, Ruhland G, et al. P45, an extracellular 45 kDa protein of Listeria monocytogenes with similarity to protein p60 and exhibiting peptidoglycan lytic activity. Arch Microbiol. 2000;173:21–28. doi: 10.1007/s002030050003. [DOI] [PubMed] [Google Scholar]
- 33.Port GC, Freitag NE. Identification of novel Listeria monocytogenes secreted virulence factors following mutational activation of the central virulence regulator, PrfA. Infect Immun. 2007;75:5886–5897. doi: 10.1128/IAI.00845-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bierne H, Garandeau C, Pucciarelli MG, Sabet C, Newton S, et al. Sortase B, a new class of sortase in Listeria monocytogenes. J Bacteriol. 2004;186:1972–1982. doi: 10.1128/JB.186.7.1972-1982.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schaumburg J, Diekmann O, Hagendorff P, Bergmann S, Rohde M, et al. The cell wall subproteome of Listeria monocytogenes. Proteomics. 2004;4:2991–3006. doi: 10.1002/pmic.200400928. [DOI] [PubMed] [Google Scholar]
- 36.Cabanes D, Dehoux P, Dussurget O, Frangeul L, Cossart P. Surface proteins and the pathogenic potential of Listeria monocytogenes. Trends Microbiol. 2002;10:238–245. doi: 10.1016/s0966-842x(02)02342-9. [DOI] [PubMed] [Google Scholar]
- 37.Bierne H, Cossart P. Listeria monocytogenes surface proteins: from genome predictions to function. Microbiol Mol Biol Rev. 2007;71:377–397. doi: 10.1128/MMBR.00039-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dramsi S, Bourdichon F, Cabanes D, Lecuit M, Fsihi H, et al. FbpA, a novel multifunctional Listeria monocytogenes virulence factor. Mol Microbiol. 2004;53:639–649. doi: 10.1111/j.1365-2958.2004.04138.x. [DOI] [PubMed] [Google Scholar]
- 39.Lambrechts A, Gevaert K, Cossart P, Vandekerckhove J, Van Troys M. Listeria comet tails: the actin-based motility machinery at work. Trends Cell Biol. 2008;18:220–227. doi: 10.1016/j.tcb.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 40.Borezee E, Pellegrini E, Berche P. OppA of Listeria monocytogenes, an oligopeptide-binding protein required for bacterial growth at low temperature and involved in intracellular survival. Infect Immun. 2000;68:7069–7077. doi: 10.1128/iai.68.12.7069-7077.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reglier-Poupet H, Pellegrini E, Charbit A, Berche P. Identification of LpeA, a PsaA-like membrane protein that promotes cell entry by Listeria monocytogenes. Infect Immun. 2003;71:474–482. doi: 10.1128/IAI.71.1.474-482.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Desvaux M, Hebraud M. The protein secretion systems in Listeria: inside out bacterial virulence. FEMS Microbiol Rev. 2006;30:774–805. doi: 10.1111/j.1574-6976.2006.00035.x. [DOI] [PubMed] [Google Scholar]
- 43.Trost M, Wehmhoner D, Karst U, Dieterich G, Wehland J, et al. Comparative proteome analysis of secretory proteins from pathogenic and nonpathogenic Listeria species. Proteomics. 2005;5:1544–1557. doi: 10.1002/pmic.200401024. [DOI] [PubMed] [Google Scholar]
- 44.Archambaud C, Nahori MA, Pizarro-Cerda J, Cossart P, Dussurget O. Control of Listeria superoxide dismutase by phosphorylation. J Biol Chem. 2006;281:31812–31822. doi: 10.1074/jbc.M606249200. [DOI] [PubMed] [Google Scholar]
- 45.Hanawa T, Fukuda M, Kawakami H, Hirano H, Kamiya S, et al. The Listeria monocytogenes DnaK chaperone is required for stress tolerance and efficient phagocytosis with macrophages. Cell Stress Chaperones. 1999;4:118–128. [PMC free article] [PubMed] [Google Scholar]
- 46.Gahan CG, O'Mahony J, Hill C. Characterization of the groESL operon in Listeria monocytogenes: utilization of two reporter systems (gfp and hly) for evaluating in vivo expression. Infect Immun. 2001;69:3924–3932. doi: 10.1128/IAI.69.6.3924-3932.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chan YC, Boor KJ, Wiedmann M. SigmaB-dependent and sigmaB-independent mechanisms contribute to transcription of Listeria monocytogenes cold stress genes during cold shock and cold growth. Appl Environ Microbiol. 2007;73:6019–6029. doi: 10.1128/AEM.00714-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rea R, Hill C, Gahan CG. Listeria monocytogenes PerR mutants display a small-colony phenotype, increased sensitivity to hydrogen peroxide, and significantly reduced murine virulence. Appl Environ Microbiol. 2005;71:8314–8322. doi: 10.1128/AEM.71.12.8314-8322.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wiedmann M, Arvik TJ, Hurley RJ, Boor KJ. General stress transcription factor sigmaB and its role in acid tolerance and virulence of Listeria monocytogenes. J Bacteriol. 1998;180:3650–3656. doi: 10.1128/jb.180.14.3650-3656.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rouquette C, de Chastellier C, Nair S, Berche P. The ClpC ATPase of Listeria monocytogenes is a general stress protein required for virulence and promoting early bacterial escape from the phagosome of macrophages. Mol Microbiol. 1998;27:1235–1245. doi: 10.1046/j.1365-2958.1998.00775.x. [DOI] [PubMed] [Google Scholar]
- 51.Archambaud C, Gouin E, Pizarro-Cerda J, Cossart P, Dussurget O. Translation elongation factor EF-Tu is a target for Stp, a serine-threonine phosphatase involved in virulence of Listeria monocytogenes. Mol Microbiol. 2005;56:383–396. doi: 10.1111/j.1365-2958.2005.04551.x. [DOI] [PubMed] [Google Scholar]
- 52.Mohamed W, Darji A, Domann E, Chiancone E, Chakraborty T. The ferritin-like protein Frm is a target for the humoral immune response to Listeria monocytogenes and is required for efficient bacterial survival. Mol Genet Genomics. 2006;275:344–353. doi: 10.1007/s00438-005-0090-8. [DOI] [PubMed] [Google Scholar]
- 53.Dussurget O, Dumas E, Archambaud C, Chafsey I, Chambon C, et al. Listeria monocytogenes ferritin protects against multiple stresses and is required for virulence. FEMS Microbiol Lett. 2005;250:253–261. doi: 10.1016/j.femsle.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 54.Fuangthong M, Atichartpongkul S, Mongkolsuk S, Helmann JD. OhrR is a repressor of ohrA, a key organic hydroperoxide resistance determinant in Bacillus subtilis. J Bacteriol. 2001;183:4134–4141. doi: 10.1128/JB.183.14.4134-4141.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Holzmuller P, Hide M, Sereno D, Lemesre JL. Leishmania infantum amastigotes resistant to nitric oxide cytotoxicity: Impact on in vitro parasite developmental cycle and metabolic enzyme activities. Infect Genet Evol. 2006;6:187–197. doi: 10.1016/j.meegid.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 56.Taylor CM, Beresford M, Epton HA, Sigee DC, Shama G, et al. Listeria monocytogenes relA and hpt mutants are impaired in surface-attached growth and virulence. J Bacteriol. 2002;184:621–628. doi: 10.1128/JB.184.3.621-628.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brookfield DE, Green J, Ali ST, Machado RS, Guest JR. Evidence for two protein-lipoylation activities in Escherichia coli. FEBS Lett. 1991;295:13–16. doi: 10.1016/0014-5793(91)81373-g. [DOI] [PubMed] [Google Scholar]
- 58.Keeney KM, Stuckey JA, O'Riordan MX. LplA1-dependent utilization of host lipoyl peptides enables Listeria cytosolic growth and virulence. Mol Microbiol. 2007;66:758–770. doi: 10.1111/j.1365-2958.2007.05956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Larsen MH, Kallipolitis BH, Christiansen JK, Olsen JE, Ingmer H. The response regulator ResD modulates virulence gene expression in response to carbohydrates in Listeria monocytogenes. Mol Microbiol. 2006;61:1622–1635. doi: 10.1111/j.1365-2958.2006.05328.x. [DOI] [PubMed] [Google Scholar]
- 60.Brekasis D, Paget MS. A novel sensor of NADH/NAD+ redox poise in Streptomyces coelicolor A3(2). Embo J. 2003;22:4856–4865. doi: 10.1093/emboj/cdg453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alexander JE, Andrew PW, Jones D, Roberts IS. Characterization of an aromatic amino acid-dependent Listeria monocytogenes mutant: attenuation, persistence, and ability to induce protective immunity in mice. Infect Immun. 1993;61:2245–2248. doi: 10.1128/iai.61.5.2245-2248.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stritzker J, Janda J, Schoen C, Taupp M, Pilgrim S, et al. Growth, virulence, and immunogenicity of Listeria monocytogenes aro mutants. Infect Immun. 2004;72:5622–5629. doi: 10.1128/IAI.72.10.5622-5629.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barabote RD, Saier MH., Jr Comparative genomic analyses of the bacterial phosphotransferase system. Microbiol Mol Biol Rev. 2005;69:608–634. doi: 10.1128/MMBR.69.4.608-634.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Polidoro M, De Biase D, Montagnini B, Guarrera L, Cavallo S, et al. The expression of the dodecameric ferritin in Listeria spp. is induced by iron limitation and stationary growth phase. Gene. 2002;296:121–128. doi: 10.1016/s0378-1119(02)00839-9. [DOI] [PubMed] [Google Scholar]
- 65.Fisher CW, Martin SE. Effects of iron and selenium on the production of catalase, superoxide dismutase, and listeriolysin O in Listeria monocytogenes. J Food Protection. 1999;62:1206–1209. doi: 10.4315/0362-028x-62.10.1206. [DOI] [PubMed] [Google Scholar]
- 66.Vazquez-Boland JA, Kocks C, Dramsi S, Ohayon H, Geoffroy C, et al. Nucleotide sequence of the lecithinase operon of Listeria monocytogenes and possible role of lecithinase in cell-to-cell spread. Infect Immun. 1992;60:219–230. doi: 10.1128/iai.60.1.219-230.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alvarez-Dominguez C, Madrazo-Toca F, Fernandez-Prieto L, Vandekerckhove J, Pareja E, et al. Characterization of a Listeria monocytogenes protein interfering with Rab5a. Traffic. 2008;9:325–337. doi: 10.1111/j.1600-0854.2007.00683.x. [DOI] [PubMed] [Google Scholar]
- 68.Madureira P, Baptista M, Vieira M, Magalhaes V, Camelo A, et al. Streptococcus agalactiae GAPDH is a virulence-associated immunomodulatory protein. J Immunol. 2007;178:1379–1387. doi: 10.4049/jimmunol.178.3.1379. [DOI] [PubMed] [Google Scholar]
- 69.Terao Y, Yamaguchi M, Hamada S, Kawabata S. Multifunctional glyceraldehyde-3-phosphate dehydrogenase of Streptococcus pyogenes is essential for evasion from neutrophils. J Biol Chem. 2006;281:14215–14223. doi: 10.1074/jbc.M513408200. [DOI] [PubMed] [Google Scholar]
- 70.Tan C, Liu M, Jin M, Liu J, Chen Y, et al. The key virulence-associated genes of Streptococcus suis type 2 are upregulated and differentially expressed in vivo. FEMS Microbiol Lett. 2008;278:108–114. doi: 10.1111/j.1574-6968.2007.00980.x. [DOI] [PubMed] [Google Scholar]
- 71.Fuchs G. Anaerobic metabolism of aromatic compounds. Ann N Y Acad Sci. 2008;1125:82–99. doi: 10.1196/annals.1419.010. [DOI] [PubMed] [Google Scholar]
- 72.Lauer P, Chow MY, Loessner MJ, Portnoy DA, Calendar R. Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J Bacteriol. 2002;184:4177–4186. doi: 10.1128/JB.184.15.4177-4186.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bryant PA, Venter D, Robins-Browne R, Curtis N. Chips with everything: DNA microarrays in infectious diseases. Lancet Infect Dis. 2004;4:100–111. doi: 10.1016/S1473-3099(04)00930-2. [DOI] [PubMed] [Google Scholar]
- 74.Ollinger J, Bowen B, Wiedmann M, Boor KJ, Bergholz TM. Listeria monocytogenes sigmaB modulates PrfA-mediated virulence factor expression. Infect Immun. 2009 doi: 10.1128/IAI.01205-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Takeda K, Akira S. Toll-like receptors in innate immunity. Int Immunol. 2005;17:1–14. doi: 10.1093/intimm/dxh186. [DOI] [PubMed] [Google Scholar]
- 76.Girardin SE, Boneca IG, Carneiro LA, Antignac A, Jehanno M, et al. Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science. 2003;300:1584–1587. doi: 10.1126/science.1084677. [DOI] [PubMed] [Google Scholar]
- 77.Chamaillard M, Hashimoto M, Horie Y, Masumoto J, Qiu S, et al. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat Immunol. 2003;4:702–707. doi: 10.1038/ni945. [DOI] [PubMed] [Google Scholar]
- 78.Way SS, Thompson LJ, Lopes JE, Hajjar AM, Kollmann TR, et al. Characterization of flagellin expression and its role in Listeria monocytogenes infection and immunity. Cell Microbiol. 2004;6:235–242. doi: 10.1046/j.1462-5822.2004.00360.x. [DOI] [PubMed] [Google Scholar]
- 79.Shen A, Higgins DE. The MogR transcriptional repressor regulates nonhierarchal expression of flagellar motility genes and virulence in Listeria monocytogenes. PLoS Pathog. 2006;2:e30. doi: 10.1371/journal.ppat.0020030. doi:10.1371/journal.ppat.0020030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zenewicz LA, Shen H. Innate and adaptive immune responses to Listeria monocytogenes: a short overview. Microbes Infect. 2007;9:1208–1215. doi: 10.10110/2/076/j.micinf.2007.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Seveau S, Pizarro-Cerda J, Cossart P. Molecular mechanisms exploited by Listeria monocytogenes during host cell invasion. Microbes Infect. 2007;9:1167–1175. doi: 10.1016/j.micinf.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 82.Cabanes D, Sousa S, Cebria A, Lecuit M, Garcia-del Portillo F, et al. Gp96 is a receptor for a novel Listeria monocytogenes virulence factor, Vip, a surface protein. Embo J. 2005;24:2827–2838. doi: 10.1038/sj.emboj.7600750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Messner P, Steiner K, Zarschler K, Schaffer C. S-layer nanoglycobiology of bacteria. Carbohydr Res. 2008;343:1934–1951. doi: 10.1016/j.carres.2007.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cabanes D, Dussurget O, Dehoux P, Cossart P. Auto, a surface associated autolysin of Listeria monocytogenes required for entry into eukaryotic cells and virulence. Mol Microbiol. 2004;51:1601–1614. doi: 10.1111/j.1365-2958.2003.03945.x. [DOI] [PubMed] [Google Scholar]
- 85.Davies EA, Falahee MB, Adams MR. Involvement of the cell envelope of Listeria monocytogenes in the acquisition of nisin resistance. J Appl Bacteriol. 1996;81:139–146. doi: 10.1111/j.1365-2672.1996.tb04491.x. [DOI] [PubMed] [Google Scholar]
- 86.Lebrun M, Audurier A, Cossart P. Plasmid-borne cadmium resistance genes in Listeria monocytogenes are present on Tn5422, a novel transposon closely related to Tn917. J Bacteriol. 1994;176:3049–3061. doi: 10.1128/jb.176.10.3049-3061.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lebrun M, Audurier A, Cossart P. Plasmid-borne cadmium resistance genes in Listeria monocytogenes are similar to cadA and cadC of Staphylococcus aureus and are induced by cadmium. J Bacteriol. 1994;176:3040–3048. doi: 10.1128/jb.176.10.3040-3048.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 90.Arnaud M, Chastanet A, Debarbouille M. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl Environ Microbiol. 2004;70:6887–6891. doi: 10.1128/AEM.70.11.6887-6891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chakraborty T, Leimeister-Wachter M, Domann E, Hartl M, Goebel W, et al. Coordinate regulation of virulence genes in Listeria monocytogenes requires the product of the prfA gene. J Bacteriol. 1992;174:568–574. doi: 10.1128/jb.174.2.568-574.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cserzo M, Wallin E, Simon I, von Heijne G, Elofsson A. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 1997;10:673–676. doi: 10.1093/protein/10.6.673. [DOI] [PubMed] [Google Scholar]
- 93.Dussurget O, Cabanes D, Dehoux P, Lecuit M, Buchrieser C, et al. Listeria monocytogenes bile salt hydrolase is a PrfA-regulated virulence factor involved in the intestinal and hepatic phases of listeriosis. Mol Microbiol. 2002;45:1095–1106. doi: 10.1046/j.1365-2958.2002.03080.x. [DOI] [PubMed] [Google Scholar]
- 94.Cabanes D, Lecuit M, Cossart P. Animal models of Listeria infection. Curr Protoc Microbiol Chapter. 2008;9:Unit9B 1. doi: 10.1002/9780471729259.mc09b01s10. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Growth and pH curves of L. monocytogenes EGDe in BHI at 37°C with shaking
(0.07 MB PDF)
Validation of macroarray data by real-time RT-PCR. Fold changes in in vivo gene expression 24 h p.i. (A) or 72 h p.I. (B) compared to that in BHI were measured by macroarray and real-time RT-PCR, log transformed and compared for correlation analysis.
(0.04 MB PDF)
Known L. monocytogenes virulence factors
(0.03 MB PDF)
L. monocytogenes genes regulated in the host as compared to exponential or stationary growth phase in BHI
(0.05 MB PDF)
L. monocytogenes genes differentially regulated in the host as compared to exponential growth in BHI at 37°C
(0.07 MB PDF)
L. monocytogenes EGDe genes encoding proteins of the cell wall subproteome and differentially regulated in the host
(0.03 MB PDF)
L. monocytogenes genes encoding secreted proteins and differentially regulated in the host
(0.03 MB PDF)
L. monocytogenes genes involved in DNA metabolism, RNA and protein synthesis, cell division and multiplication, and up regulated in the host
(0.03 MB PDF)
L. monocytogenes genes involved in stress responses and differentially regulated in the host
(0.04 MB PDF)
L. monocytogenes genes involved in metabolism and differentially regulated in the host
(0.05 MB PDF)
Primers
(0.04 MB PDF)