Abstract
Iron is required for survival of mammalian cells. Recently, understanding of iron metabolism and trafficking has increased dramatically, revealing a complex, interacting network largely unknown just a few years ago. This provides an excellent model for systems biology development and analysis. The first step in such an analysis is the construction of a structural network of iron metabolism, which we present here. This network was created using CellDesigner version 3.5.2 and includes reactions occurring in mammalian cells of numerous tissue types. The iron metabolic network contains 151 chemical species and 107 reactions and transport steps. Starting from this general model, we construct iron networks for specific tissues and cells that are fundamental to maintaining body iron homeostasis. We include subnetworks for cells of the intestine and liver, tissues important in iron uptake and storage, respectively; as well as the reticulocyte and macrophage, key cells in iron utilization and recycling. The addition of kinetic information to our structural network will permit the simulation of iron metabolism in different tissues as well as in health and disease.
Keywords: iron, liver, macrophage, reactive oxygen species, red blood cells
Introduction
Understanding of iron metabolism and trafficking in eucaryotes has increased exponentially in recent years. New genes important in cellular and systemic iron homeostasis have been identified, and their roles have been intensively investigated (see1–3, for review). These studies have unveiled a complex, interacting network largely unknown just a few years ago. As such, the cellular iron network provides an excellent model for systems biology development and analysis. The first step in such an analysis is the construction of a network model, which we present here.
Iron metabolism is highly regulated at both the organismal and cellular levels. Iron is an essential element, required for the function of many enzymes and transport proteins. In these proteins, iron functions as a requisite cofactor, frequently in the form of iron-sulfur clusters or heme. Well-known proteins that require iron for their function include hemoglobin and myoglobin, which function in oxygen transport; cytochromes, necessary for both respiration and detoxification; aconitase and other proteins required for oxidative metabolism; ribonucleotide reductase, which catalyzes the rate-limiting step in DNA synthesis, and many others.
Iron is also a redox active element with substantial potential toxicity. It can facilitate the formation of oxygen free radicals through the Fenton reaction, in which reduced iron (FeII) reacts with hydrogen peroxide to produce the hydroxyl radical, a highly reactive radical capable of damaging DNA, lipids and proteins. Iron is also extremely insoluble (10−18 M) in aqueous solution4. Thus the proper distribution and sequestering of iron is a challenge at both the cellular and organismic level.
The importance of appropriate iron homeostasis becomes clear when conditions of iron imbalance are considered. For example, hemochromatosis is a disease of iron overload characterized by excess uptake of dietary iron5. In patients with hemochromatosis, iron can accumulate not only in the liver, its normal storage site, but in the pancreas and heart, where it can lead to diabetes or heart disease. More prevalent than these conditions of iron overload are conditions of iron insufficiency, which can lead to iron-deficiency anemia. Indeed, according to the World Health Organization, iron deficiency is the most common and widespread nutritional disorder in the world: 2 billion people – over 30% of the world’s population – are anemic, many due to iron deficiency6.
At the level of the whole organism, iron homeostasis is regulated at sites of absorption, utilization (and re-utilization) and storage. There is no excretory pathway for iron, although small amounts are lost through the skin, intestinal mucosa, and menstruation in women. Major sites of iron regulation are the duodenum, where dietary iron is absorbed; the bone marrow, where red blood cells are produced; the liver, where excess iron is stored; and the spleen and other cells of the reticuloendothelial system (RES), where red blood cells are catabolized and iron is extracted for reuse. Cell types that exert these functions are the enterocyte in the duodenum; reticulocyte in the bone marrow; and macrophages in the spleen, liver and RES.
At the cellular and molecular level, the regulation of iron is achieved through the concerted action of iron importers, iron storage proteins, and iron efflux pumps. Collectively, these act to deliver iron to its sites of utilization within proteins, while minimizing the ability of iron to participate in oxygen free radical-generating reactions. Within the cell, iron is also shuttled among different compartments. For example, following its uptake through transporters located on the plasma membrane, iron is found in early/recycling/late endosomes. It is subsequently delivered to mitochondria for heme biosynthesis and formation of iron-sulfur clusters. Iron storage proteins, which minimize the formation of ROS (reactive oxygen species), as well as iron regulatory proteins, which provide an important feedback mechanism when iron is low, are located in the cytoplasm. Iron-containing proteins are also present in the lysosome, where they are degraded; the iron from these proteins is then recycled back into the cytoplasm. In our models, we have illustrated these major cellular compartments of iron, as well as iron localized to the extracellular space. This extracellular compartment represents iron delivered through the blood as a complex with the transport protein transferrin, as well as extracellular iron present as other species. To simplify drawing the diagram, we do not include the nucleus and instead depict transcription in the cytoplasm, but the reader should bear in mind that this is of course implicit.
The complexities of iron regulatory pathways suggest that mathematical approaches might be useful in understanding the interactions among iron-dependent species, identifying key regulatory nodes, simulating their response to various stimuli, and understanding how these differ in various cell types. As a first step towards that goal, we present a structural model of iron metabolism in a mammalian cell using CellDesigner v.3.5.2.
CellDesigner7 is a diagram editor for creating biochemical networks and has been used to construct maps of other important pathways8, 9. CellDesigner is not merely a pictorial tool, but a platform for the creation of biochemical network models in the SBML language. The SBML files created by CellDesigner can be used to generate kinetic simulations of the network using a system of ordinary differential equations or through stochastic simulation algorithms with software packages such as COPASI10, 11 and others (see 12 for an example of CellDesigner network construction followed by kinetic simulation through the SBML platform).
Results
General network
Using CellDesigner, we first created a general iron network (Fig. 1). In this network we do not depict the reactions that occur in a particular cell type, but rather a culmination of the reactions occurring in cells of numerous tissue types as described in the published literature. Sometimes, multiple studies report conflicting results, and often the precise reaction details are not yet understood. For those cases, we have made a judgment call for the purpose of our network and indicate the controversy in the detailed Description of Reactions, below.
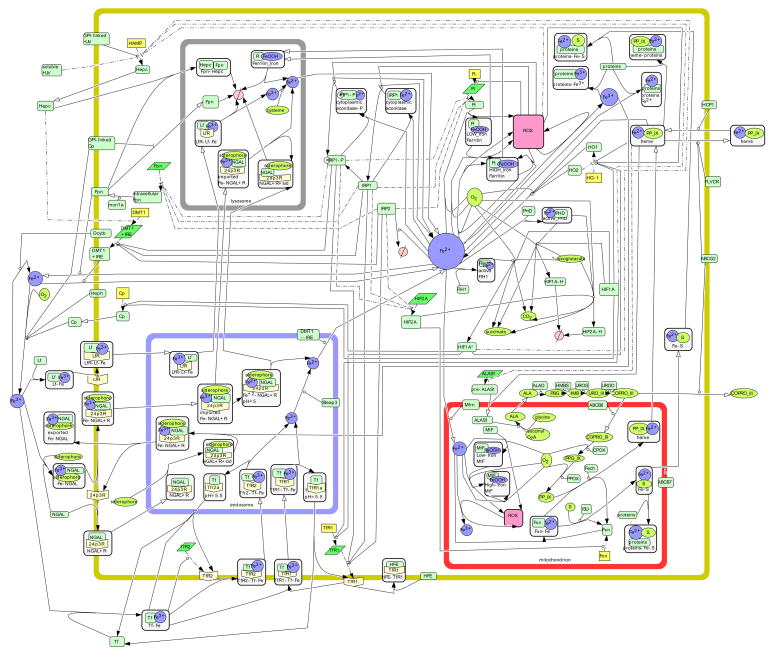
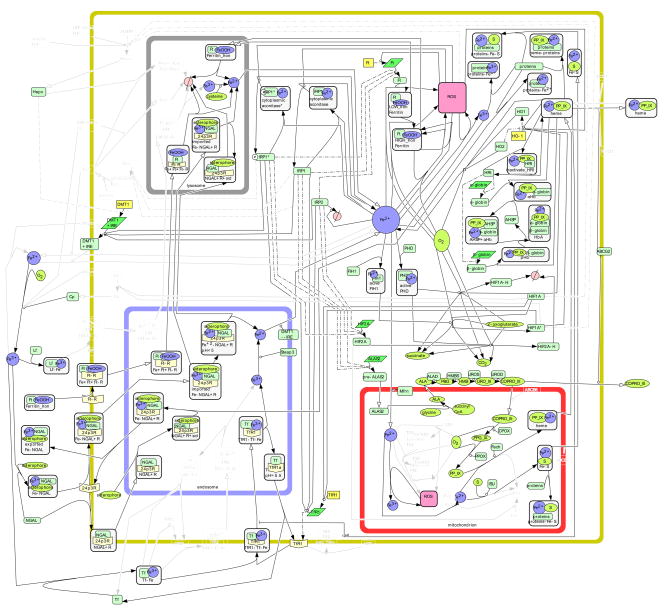
Figure 1. The Iron Metabolic Network.
This structural network contains a culmination of reactions occurring in epithelial cells of numerous tissue types. Symbols used in the network include  protein,
protein,  receptor,
receptor,  gene,
gene,  RNA,
RNA,  simple molecule,
simple molecule,  ion,
ion,  complex,
complex,  stste transition,
stste transition,  transcription,
transcription,  translation,
translation,  transport,
transport,  catalysis,
catalysis,  inhibition,
inhibition,  transcriptional inhibition,
transcriptional inhibition,  translational inhibition, and
translational inhibition, and  degradation. A complete list of the symbols used in CellDesigner can be found on the CellDesigner website (http://www.celldesigner.org).
degradation. A complete list of the symbols used in CellDesigner can be found on the CellDesigner website (http://www.celldesigner.org).
Our general network of iron metabolism contains 151 chemical species and 107 reactions and transport steps. When a chemical species is present in more than one compartment, multiple individual species are present in the map (thus they actually represent “pools”). There are 57 proteins, 7 mRNAs, and 7 genes represented explicitly. Our main interest lies in the protein level, and hence we only represent the corresponding genes or mRNAs if there is an iron-related modification of transcription or translation (i.e. all other transcription/translation events are left implicit). Additionally, we have 46 complexes which include protein-protein and protein-iron complexes. The set of reactive oxygen species (ROS) is also represented by the symbol for complexes but in this case any ROS is meant (i.e. they are not in complexes in the cell). 10 iron ions exist as well as 22 metabolites (“simple molecules” in the notation of CellDesigner), such as the intermediates in porphyrin formation. Degradation occurs in two compartments, which accounts for 2 species.
Tissue- specific networks
We then focused more closely to create subnetworks specific to cells important in iron metabolism: the hepatocyte, reticulocyte, enterocyte, and macrophage. We first assessed the expression of each of the species depicted in the General Network (Fig. 1) in these specific cell types (Table 1). We included those genes and proteins for which there was evidence of expression in relevant mammalian tissues, although in some cases only cell line data was available. The general criteria for the entry of each mRNA or protein in the tissue-specific network are documented in Table 1; specific judgment calls related to individual species are discussed in detail below. Tissue-specific networks created in this way were the hepatocyte (Fig. 2), reticulocyte (Fig. 3), enterocyte (Fig. 4) and macrophage (Fig. 5).
Table 1.
| Liver | Intestine | Macrophage | Reticulocyte | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Protein | RNA | Unigene | Protein | RNA | Unigene | Protein | RNA | Protein | RNA | |
| 24p3R | -- | + | +280 | +280 | ||||||
| ABCB6 | +281, 282 | --281 | --281 | |||||||
| ABCB7 | +149 | +283 | +283 | |||||||
| ABCG2 | +279 | +279 | +284 | +285 | ||||||
| ALAD | + | + | ||||||||
| ALAS1 | + | + | ||||||||
| ALAS2 | +285 | |||||||||
| Cp | +286 | -- | ||||||||
| Cp (GPI) | + | +227 | ||||||||
| Dcytb | +287 | +70 | ||||||||
| DMT no IRE | +288 | +289 | +288, 289 | +290 | +263 | |||||
| DMT+IRE | +288 | +289 | +288, 289 | +290 | +264 | |||||
| Fech | +291 | + | *+292 | +285, 291 | ||||||
| FIH1 | +293 | + | +293 | +285 | ||||||
| FLVCR | +109 | +109 | +109 | +108, 109 | ||||||
| Fpn | +93 | +93 | +93 | +93 | +98 | |||||
| Ft | +294 | +294 | +294 | +264, 285 | ||||||
| Fxn | +295 | *+296 | *+297 | |||||||
| HCP1 | +70 | +70 | +298 | +298 | ||||||
| Hepc | +220 | --220 | +290 | |||||||
| Heph | --105 | +289 | +105, 289 | |||||||
| HFE | +299 | +299 | +299 | +299 | +299 | +299 | ||||
| HIF1A, HIF2A | +300 | +300 | *+301 | |||||||
| HJV | +236 | --236 | ||||||||
| HMBS | + | + | +285 | |||||||
| HO1,2 | +302 | +302 | +302 | +303 | ||||||
| IRP1,2 | +304 | +305 | +305 | +305 | +305 | +264 | ||||
| ISU | + | + | *+306 | *+306 | +285 | |||||
| LfR | +307 | +57 | +57 | +308 | ||||||
| Megalin | --309 | -- | +280 | +280 | ||||||
| Mfrn | +310 | --310 | +135 | 285 | ||||||
| Mon1a | -- | + | +98 | |||||||
| MtF | --311 | --311 | --312 | |||||||
| PHD | +293 | +293 | +293 | +313 | + | |||||
| PPOX | + | + | +285 | |||||||
| Steap3 | +36 | --36 | +36 | |||||||
| TfR1 | +314 | +315 | +314 | +253 | +285 | |||||
| TfR2 | +82 | +82 | --82 | --82 | *--82 | |||||
| UROD | + | + | +285 | |||||||
| UROS | + | + | ||||||||
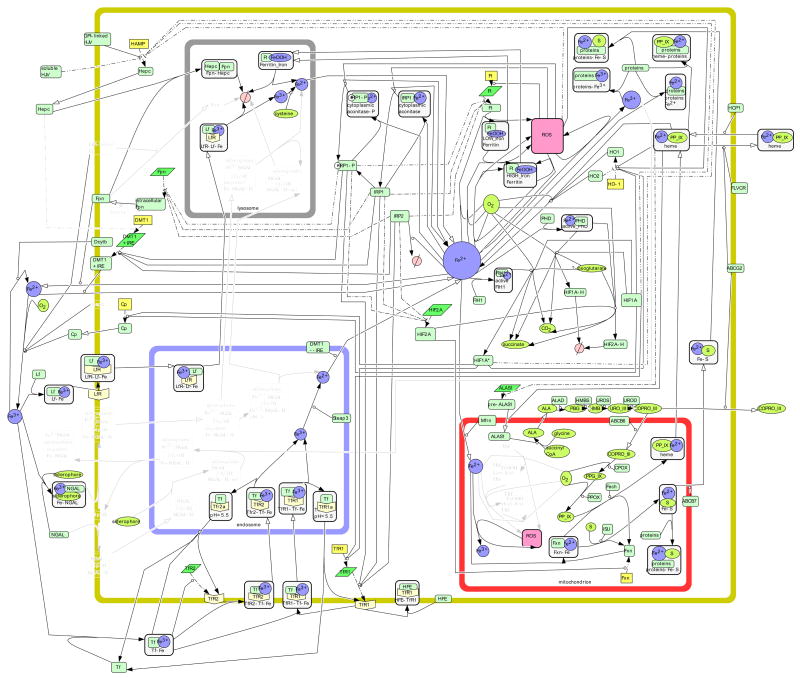
Figure 2. The Liver Cell.
Using gene/protein expression information listed in Table 1, iron metabolism in the liver epithelial cell is illustrated. Species are lightened when there is no published positive evidence of said species in the liver cell, as described in Materials and Methods.
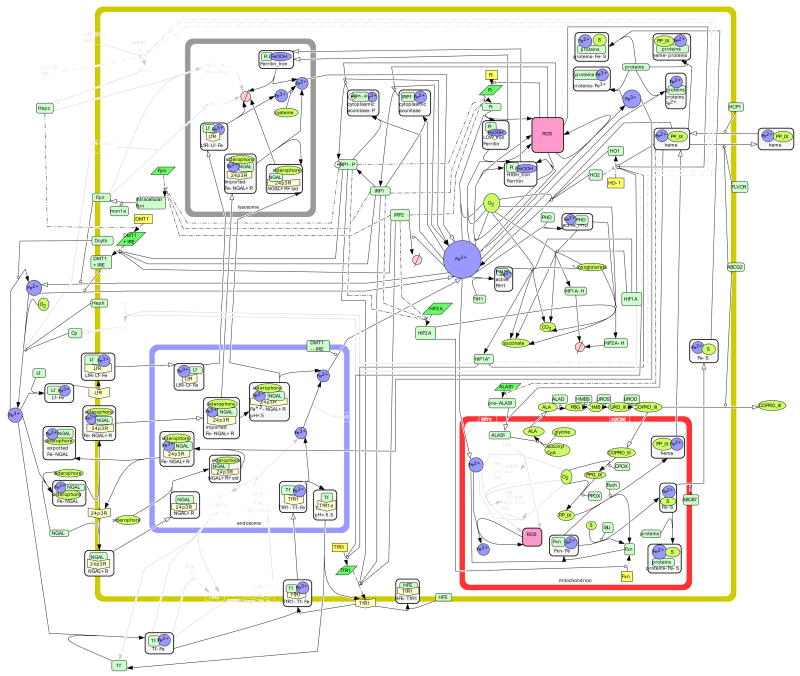
Figure 3. The Intestinal Cell.
Using gene/protein expression information listed in Table 1, iron metabolism in the intestinal epithelial cell is illustrated. Species are lightened when there is no published positive evidence of said species in the intestinal cell.
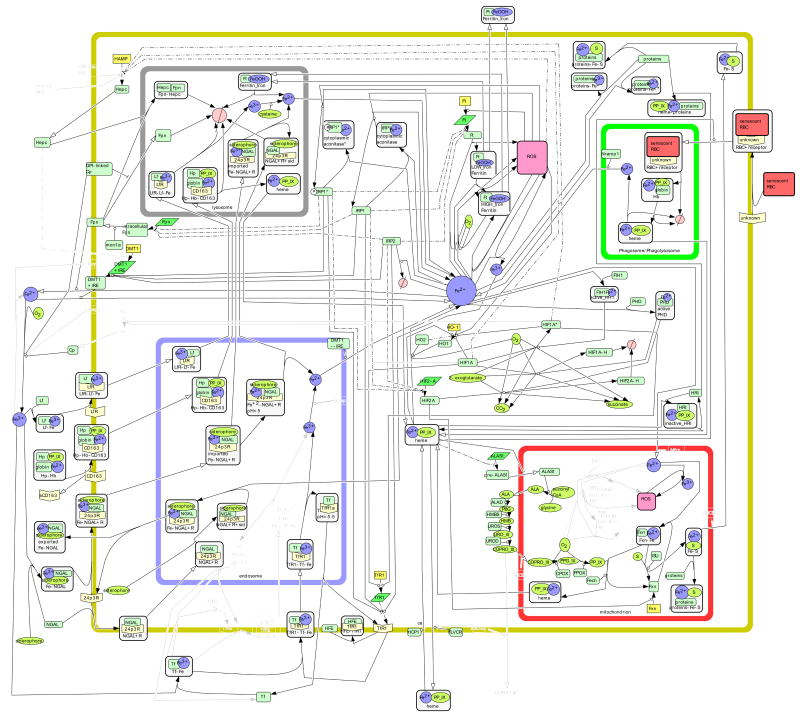
Figure 4. The Macrophage.
Using gene/protein expression information listed in Table 1, iron metabolism in the macrophage is illustrated. Species are lightened when there is no published positive evidence of said species in the macrophage. Additional species and reactions are present that do not occur in epithelial cells and are specific to macrophages, as discussed in the text.
Figure 5. The Reticulocyte.
Using gene/protein expression information listed in Table 1, iron metabolism in the reticulocyte is illustrated. Species are lightened when there is no published positive evidence of said species in the reticulocyte. Additional species and reactions are present that do not occur in epithelial cells and are specific to reticulocytes, as discussed in the text.
Description of reactions in the general network
Reactions included in the general network are those involved in iron/heme import (e.g. TfR1, DMT1, lactoferrin receptor, 24p3R, HCP1); iron/heme export (ferroportin, FLVCR, ABCG2); and iron storage (ferritin). Synthetic reactions involving iron, including those involved in synthesis of iron sulfur clusters (ISU) and heme (ALAS, ferrochelatase), are described under the heading of mitochondria, although some of the reactions in these pathways occur in the cytoplasm. Feedback regulatory reactions that operate at the level of the cell (IRPs) or whole organism (hepcidin) are also included.
A. Iron/heme importers
1. TfR1
Most iron circulates in the blood bound to transferrin, a protein with two binding sites for ferric iron13, 14. In healthy adults, transferrin is approximately 30% saturated with iron15, while increased transferrin saturation levels (>45%) are associated with the iron overload disorder hereditary hemochromatosis16. The binding and release of iron to transferrin is pH dependant13, 17, 18, which allows transferrin to bring iron into cells through a receptor-mediated process. Transferrin receptor 1 (TfR1) is a ubiquitous protein expressed on the cell surface19, 20. TfR1 is a homodimer that binds holo-transferrin in a 2:2 stoichiometry21, and this complex is internalized into an early endosome. As the endosome matures, a proton pump facilitates a drop in acidity. In the acidic environment, transferrin undergoes a conformational change22 and TfR1 facilitates the release of iron from transferrin18, 23. The endosome recycles TfR1 back to the cell surface, and transferrin re-enters the blood. The kinetics of TfR1 receptor-mediated endocytosis of iron bound to transferrin have been the subject of multiple studies17, 22, 24–26.
Holo-transferrin competes with another protein, HFE, for the binding of transferrin receptor. The gene associated with HFE is mutated in type 1 hemochromatosis27, and its protein is expressed in a wide variety of tissue types28. At this point, we understand that HFE can bind with TfR1 to lower its affinity for holo-transferrin21, 29–32. The stoichiometry for the HFE/TfR1 complex can be 2:2 or 1:221, and a complex with holo-transferrin, HFE, and TfR1 might exist30, although this is not shown in our network. Some authors suggest that the HFE/TfR1 complex enters the endosome29, 33 although this is not fully established, and another main study suggesting this was retracted34. Our network does not show HFR/TfR1 inside the endosome, and in general HFE’s role in iron metabolism has not been settled upon35.
2. Divalent metal transporter 1
Once free in the endosome, ferric iron is reduced by STEAP3 36 and is exported into the cytoplasm by DMT1. Divalent metal transporter 1 (DMT1, also called Nramp2, SLC11A2, and DCT1) is a ubiquitous protein which plays an important role in the transport of iron across membranes37, 38. DMT1 is highly expressed in the duodenum37, 39 where it transports ferrous iron into the cell40, 41 assisted by the ferrireductase duodenal cytochrome B (Dcytb)42–45. Dcytb reduces dietary ferric iron to ferrous iron. In many types of cells, DMT1 is located both on the plasma membrane and in the endosome 40. In both locations, DMT1 transports ferrous iron into the labile iron pool46.
3. Lactoferrin Receptor
Lactoferrin is a member of the transferrin family that was originally found in human milk47 and other epithelial secretions. Lactoferrin is also secreted by neutrophils48. Because it tightly sequesters iron, lactoferrin is suggested to have many biological roles including defense against microbial infection and inflammation49. Unlike transferrin, lactoferrin does not lose its iron in mild acidity but instead requires pH<3.5 for iron release47. Lactoferrin also has 2.5–3 times higher affinity for ferric iron than does transferrin50. This fits with the function of lactoferrin being to hold on to iron and transferrin’s function being iron transport. The structure and kinetics of binding and release of ferric iron to lactoferrin have been the focus of many studies50–53. While neither TfR1 nor TfR2 bind to lactoferrin54, a lactoferrin receptor is present on the plasma membrane of intestinal cells55 and many other tissue types56, 57. Once lactoferrin and ferric iron are complexed with lactoferrin receptor, the complex is not affected by the acidic pH of the early and recycling endosome environment. In our network, we show iron being released from lactoferrin only in the lysosome, an iron-rich environment that degrades many iron-containing proteins and whose pH is significantly lower than the early/recycling endosome58.
4. 24p3R
Neutrophil gelatinase-associated lipocalin (NGAL, also called lipocalin 2) was first found in human neutrophils59 and is secreted by multiple types of cells during inflammation60, 61. NGAL is a member of the lipocalin family and binds siderophores, which are high affinity iron chelators62, 63. NGAL serves two functions in the delivery of iron to/from a cell, and both are mediated by a cell surface receptor 24p3R (also called SLC22A17). 24p3R derives its name from the mouse homologue of NGAL, which is called 24p364. First, the holo-NGAL complex consisting of ferric iron, siderophore, and NGAL can bind to 24p3R and enter receptor-mediated endocytosis. Iron does not begin to be released from the NGAL—24p3R complex until the pH decreases to 5, and this occurs slowly until the pH falls lower than 565. In this acidified compartment, ferric iron is reduced to ferrous iron, which does not bind to the acidified siderophore. Hence, ferrous iron dissociates from the NGAL—24p3R complex. This iron then enters the labile iron pool66. Like lactoferrin, holo-NGAL is targeted to the late endosome rather than the early/recycling endosome for transferrin65. Second, 24p3R can uptake apo-NGAL. In this case, apo-NGAL picks up a siderophore and then complexes with ferric iron from within the cell. This iron is exported out of the cell and 24p3R is recycled to the plasma membrane64. It is possible that 24p3R also recycles after mediating the uptake of holo-NGAL65, but this is yet to be shown. Megalin (also called gp330) can facilitate the uptake of NGAL67 and its function is understood to be the same as 24p3R.
5. Heme carrier protein 1
Following the discovery of the iron transporter DMT1 in the duodenum, many researchers speculated that a heme transporter must exist in the duodenum68, 69. Heme carrier protein 1 (HCP1, also called SCL46A1, hPCFT) was identified as a heme importer on the apical membrane of the duodenum70. Later, researchers found that HCP1 transports folate with greater affinity and suggest that folate transport could be this protein’s primary function71–73. Since its discovery, HCP1 has been found in many other human organs including brain, breast, kidney, prostate, and liver74, 75. We show HCP1 transporting heme as it may play a role in iron homeostasis for many tissue types. Once heme is imported into a cell, heme oxygenase 176–79 and heme oxygenase 280 break down heme and the iron can enter the labile iron pool. While our network only shows heme and oxygen as reactants with ferrous iron as a product, biliverdin is also produced. In a second reaction, biliverdin is converted to bilirubin, a powerful antioxidant. We have chosen to omit both biliverdin and bilirubin as our main interest lies in the metabolism of iron.
6. TfR2
Holo-transferrin can also bind to transferrin receptor 2 (TfR2)81, 82, a homologue of TfR1 that has only recently come into the picture82. The role of TfR2 is not fully determined, but we do know that TfR2 is essential for iron homeostasis: a defective TfR2 gene leads to a non-HFE form of hemochromatosis83–87. Expression of TfR2 is largely restricted to the liver82, 85, 88. Like TfR1, TfR2 does mediate the endocytosis of holo-transferrin89, 90, but there are known differences between the two receptors. TfR2 has an affinity for holo-transferrin that is 25-fold lower than of TfR1 and does not bind to HFE81. It has been suggested that TfR2 acts primarily as an ‘iron sensor’ rather than an iron importer: its protein expression is increased by holo-transferrin both in vitro16, 86, 91, 92 and in vivo 91. The exact mechanism of this regulation is not known, but it is likely to be post-transcriptional92. A recent study suggests that holo-transferrin might facilitate the recycling of TfR289.
B. Iron/heme exporters
1. Ferroportin
Ferroportin (also called MTP1, IREG1, SLC40A1) is a mammalian iron exporter located on the plasma membrane and is expressed in a wide variety of human tissue types93–97. Once translated, intracellular ferroportin is transported to the plasma membrane by mon1a98 where it can function as an iron exporter. Ferroportin exports ferrous iron that is then oxidized to ferric iron by a ferroxidase. This oxidation is catalyzed by either ceruloplasmin, a circulating protein produced by the liver99–102, or hephaestin40, 44, 103–105 in the duodenum.
2. FLVCR and ABCG2
While ferroportin is the only known iron exporter, we currently know of two heme exporters located on the cell surface: feline leukemia virus C receptor (FLVCR) and ATP binding cassette protein G2 (ABCG2, also BCRP)69. FLVCR was first cloned as a receptor for feline leukemia virus subgroup C106, 107 and was later shown to export heme from the cell108. High levels of both the mRNA and protein for FLVCR are present in small intestine, liver, and red blood cells, and FLVCR is also expressed in many other tissue types108. A recent study showed FLVCR is necessary for iron homeostasis and exports heme in vivo109. ABCG2 was cloned from MCF-7 cells as a multi-drug resistance transporter110. Like other ABC transporters, ABCG2 can transport substrates against a concentration gradient111. ABCG2 interacts with both heme and porphyrins and prevents heme/porphyrin build up within the cell111, 112. The expression of ABCG2 is highest in the placenta with high expression levels also in the liver and small intestine110, 113. ABCG2 is expressed at different levels in other tissue types but is not a ubiquitous protein113. While the story is not completely understood for either FLVCR or ABCG2, our network presents both proteins acting as heme exporters.
C. Storage of iron
Ferritin
Excess free ferrous iron can be toxic to the cell. In the presence of hydrogen peroxide, ferrous iron can be oxidized to ferric iron via the Fenton reaction which forms dangerous hydroxyl radicals114, 115. The ferritin protein can mineralize and store excess ferrous iron to prevent hydroxyl radical production. Each molecule of ferritin can hold 4500 iron atoms inside a chamber with a diameter of 8nm116. Ferritin is comprised of 24 subunits of two distinct types: H117, 118 and L119, 120; the number of ferritin H versus L subunits depends on tissue-type121. Ferritin H and L are the products of distinct genes. The subunits have different functions in the storage of iron: H-chain ferritin has ferroxidase activity while L-chain does not122. Many studies have explored the kinetics and molecular mechanisms of the storage of iron in ferritin116, 123–128. Recently, a research group found that poly (rC)-binding protein 1 (PCBP1) might deliver cystolic iron to ferritin129; however, since this work was performed primarily in yeast, we do not depict PCBP1 in our map. Iron is stored as mineral core (FeOOH) in ferritin by at least three reactions, as listed below128.
| A |
| B |
| C |
In reaction A, ferrous iron is oxidized at the ferroxidase site of H-chain ferritin. This reaction occurs almost exclusively when iron levels in ferritin are low and continues at lower levels throughout the iron loading process126, 128. Reaction C governs iron loading in the middle stages of iron loading while reaction B dominates when iron in ferritin is high128. Since the kinetics of reaction A changes from low (< 48Fe/Protein) to intermediate/high (>48 Fe/Protein) iron in ferritin126 and reaction B is the sum of reactions A and C, we represent two distinct species of the ferritin-iron complex in our network: Low iron ferritin (<48 Fe/Protein) and High iron ferritin (>48 Fe/Protein) and depict reactions A and C. Once iron is stored in ferritin, it is tightly sequestered, and some studies suggest that ferritin must be degraded in the lysosome for iron to be released130, 131. Another group of researchers propose that iron chelators and reducers can reach the iron mineral core through the eight pores in the ferritin protein132–134. The specific details of iron moving out of the pores are not understood, but this is thought to occur in iron deficiency and hemoglobin production133. Additionally, ferritin pores are mostly closed both in vivo and in vitro134. Because we lack definitive information regarding the ferritin pores, we exclude this mechanism of releasing iron from ferritin in our map.
D. Mitochondria and iron
Mitochondria are essential in cellular iron homeostasis as most metabolic processes involving iron depend on the mitochondrion. Ferrous iron enters a mitochondrion from the labile iron pool. Mitoferrin (also called SLC25a37) was recently proposed to be an iron importer located on the membrane of the mitochondrion135.
1. Mitochondrial ferritin
Like cystolic ferritin, mitochondrial ferritin stores iron as mineral core to control the amount of free ferrous iron present. While mitochondrial ferritin is also comprised of 24 subunits, this protein is a homopolymer whose subunits behave somewhat like ferritin H subunits125, 136, 137. Both mitochondrial ferritin and H-ferritin contain a ferroxidase center that oxidizes ferrous iron in the process of mineral core formation136–138. In mitochondrial ferritin, 12 of the 24 ferroxidase sites are inoperative125 and mitochondrial ferritin has a slower rate of mineral core formation125, 137. Moreover, the switch in kinetics occurs at 24 Fe/Protein rather than 48 Fe/Protein125. Thus, in our network “Low Iron Mtf” is <24Fe/Protein and “High Iron Mtf” is >24Fe/Protein. Reaction C listed for cystolic ferritin does occur for mitochondrial ferritin in high iron loading, but the overall mineral core formation is governed by two reactions instead of three. The stoichiometry is slightly different for the reaction corresponding to H-ferrtin’s reaction A, and there is no reaction consuming hydrogen peroxide as in reaction B125.
2. Frataxin, ferrochelatase, ISU
Frataxin was discovered through its connection with the neurodegenerative disease Fredreich ataxia139, 140. Researchers are not in full agreement on the precise cellular functions of frataxin in mammalian cells, and many studies focus on the yeast homolog Yfh1p. We do understand that frataxin is a mitochondrial protein that facilitates the formation of iron-sulfur clusters and heme140–147. Specifically, frataxin binds ferrous iron and delivers it to ISU142, 143, 146 for the formation of iron-sulfur clusters. The iron-sulfur clusters are then either used in mitochondrial iron-sulfur cluster proteins or exported to the cytoplasm for use throughout the cell. The ABC transporter ABCB7, a functional orthologue of yeast Atm1p148–150, is thought to facilitate this transport148, 151. While some studies suggest that iron-sulfur cluster formation can also occur in the cytoplasm152, other authors stress the importance of the mitochondrion in iron-sulfur cluster formation151. Moreover, as far as frataxin is concerned, the mature protein is located solely in mitochondria143, 153, and a recent study showed that deletion of frataxin in mice affected three extra-mitochondrial iron-sulfur proteins143. We have elected to show iron-sulfur formation occurring only in the mitochondrion, and our network represents a simplified picture of the complex iron-sulfur biogenesis process151.
3. Heme synthesis
Frataxin also appears to deliver iron to ferrochelatase for the production of heme141, 147. Ferrochelatase catalyzes the final step in heme production154–157, which involves the insertion of iron into protoporphyrn IX (PPIX). We have included all of the steps of heme biosynthesis in our network. Before iron is added, a series of reactions occurs, beginning and ending in the mitochondrion and with middle steps occurring in the cytoplasm. The heme biosynthesis pathway has been well understood for some time now and can be found in textbooks158, pathway databases159, 160, and other sources161. One detail worth mentioning is the transport of coproporphyrinogen III (COPRO III) from the cytoplasm into the mitochondrion, which is catalyzed by the transporter ABCB6162. After the final step in heme production, heme is exported out of the mitochondrion into the cytoplasm for use in heme proteins. The protein responsible for this transport has not been determined although speculations exist163. Once in the cytoplasm, heme can also be degraded by heme oxygenase 1 and 2, as previously discussed.
Intracellular heme provides important regulation of its own production and degradation. Heme’s regulation of its production occurs via delta aminolevulinate synthase (ALAS), which catalyzes the first step in the formation of the porphyrin ring164. ALAS comes from two different genes: ALAS2 in erythroid and ALAS1 in non-erythroid cells165. We only depict ALAS1 in the general epithelial network. In mammals, heme inhibits ALAS1 translationally166 as well as at the protein level. The regulation at the protein level involves an inhibition of the transport of ALAS1 protein from the cytoplasm into the mitochondrion167–170. Heme also regulates its own degradation by inducing heme oxygenase 171. Heme oxygenase 1 is also induced by reactive oxygen species78, 172, 173 and by decreased levels of heme oxygenase 2174.
E. Feedback regulation
1. Iron regulatory proteins
In mammalian cells, a fundamental mechanism for controlling iron metabolism occurs post-transcriptionally through iron regulatory proteins (IRPs) and iron responsive elements (IREs)175. The IRE is a nucleotide hairpin located in the untranslated region (UTR) of mRNAs encoding certain proteins of iron metabolism176–179. Two different iron regulatory proteins, IRP1 and IRP2, bind to the IRE with high affinity when intracellular iron levels are low180. The IRE can be located in the 5′ UTR in which case the binding of IRPs blocks translation175, 181. When the IRE is located in the 3′ UTR, the binding of IRPs to IRE stabilizes the mRNA and hence more protein is translated175. IRP1 and IRP2 have distinct IRE binding sites182, 183 and are regulated by iron levels differently184. IRP1 is active when iron levels are low and functions as cystolic aconitase when iron levels are high185–189. The cystolic aconitase form of IRP1 contains a 4Fe-4S iron-sulfur cluster, and one iron molecule detaches from this cluster to activate IRP1185, 190. This activation occurs in response to low intracellular iron levels and oxidative stress187, 190, 191. IRP1 can be phosphorylated by protein kinase C, which increases its affinity for IRE binding192–194. IRP2, which shares 57% homology to IRP1195, also binds to IRE elements. Increased levels of iron cause the ubiquitination and degradation of IRP2196–199. Two studies found that IRP2 can be phosphorylated193, 200, although they give conflicting statements as to the effect of phosphorylation on IRE binding. Due to the lack of agreed upon evidence, we do not depict the phosphorylation of IRP2.
For the genes in our network, the mRNA for ferritin, ferroportin, and HIF2-alpha contain IREs in the 5′ UTR while TfR1 and DMT1 mRNA contain IREs in the 3′ UTR. It is well-accepted that the IREs in ferritin mRNA and TfR1 mRNA are functional201–203. Despite their similarity to ferritin and TfR1, the mRNA of mitochondrial ferritin and TfR2 lack IREs138. At the transcriptional level, ferritin is induced by reactive oxygen species204 and inflammatory mediators205, 206. The IRE in the mRNA for HIF2-alpha has only recently been discovered207. The ability of iron regulatory proteins to affect translation of DMT1 and ferroportin is disputed. Iron regulatory proteins (IRPs) can bind to the IRE is ferroportin mRNA96, and ferroportin’s IRE is known to be functional in certain tissue-types208, 209. Moreover IRP activity is necessary to obtain the required ferroportin levels in the duodenum210, and ferroportin expression is increased in an iron-rich environment211. We do depict IRPs modifying ferroportin translation in our iron metabolic network.
The human DMT1 gene includes alternative 3′ exons: one contains an IRE in the 3′ UTR and one does not212. Alternative 5′ exons were also discovered213 yielding four different isoforms of DMT1214. The isoform expressed varies depending on tissue-type and subcellular location213–216, however all four isoforms operate as iron transporters with the same efficiency214. Thus, in our model we only distinguish between +IRE and −IRE forms of DMT1. The +IRE isoform of DMT1 is typically located on the plasma membrane of epithelial cells while the −IRE isoform is found on the membrane of the endosome215. The functionality of the IRE in the +IRE DMT1 mRNA could depend on tissue-type210, 217. In the duodenum, DMT1 is greatly increased in iron-starved animals37, 39, 218, and intracellular iron levels could affect the transcription of DMT1219, although we do not depict this in our network.
2. Hepcidin
Hepcidin is an antimicrobial peptide that is produced and secreted by the liver in response to systemic iron levels220, 221. Overexpression of hepcidin leads to low plasma iron levels and anemia while underexpression of hepcidin leads to high plasma iron levels and hemochromatosis222. Hepcidin circulating in the blood can regulate plasma iron levels in two ways. The first is through its interaction with ferroportin. Hepcidin binds to ferroportin and causes its internalization and degradation222–225. When ferroportin levels are reduced, less iron is exported into the blood. Until very recently, this was the only known regulatory mechanism of hepcidin. However, two studies suggest that in the duodenum, hepcidin does not affect ferroportin but rather inhibits DMT1 production transcriptionally223, 226. In cells expressing GPI-linked ceruloplasmin, this protein prevents internalization and degradation of ferroportin in a hepcidin-independent pathway227.
The production of hepcidin is regulated at the level of transcription and involves an intricate BMP signaling cascade222, 228–231. Normally, hemojuvelin is GPI-linked and this form activates this BMP signaling cascade228, 229, 231, 232. In abnormal states, soluble hemojuvelin inhibits the BMP signal229, 233, 234. In our network, we omit the BMP signaling cascade and represent both forms of hemojuvelin. Hemojuvelin and its gene HFE2 are fundamental to iron metabolism since defects in HFE2 cause juvenile hemochromatosis235–237. HFE and TfR2 are suspected of participating in the regulation of hepcidin, possibly through a signaling process, but we have not found direct evidence of this85, 222, 238, 239.
3. HIF Pathway
HIF (hypoxia inducible factor) is a transcription factor that plays a central role in the cellular adaptation to low oxygen levels (see240 for review). Consequences of HIF activity are profound: HIF is a transcription factor, and its stabilization induces the transcriptional activation of over 100 genes. Key targets of HIF include EPO (erythropoietin), which controls red blood cell production, VEGF (vascular endothelial growth factor), which regulates angiogenesis, and many others. HIF activity is regulated by iron, and moreover HIF activity affects levels of iron.
First, iron controls the stability of HIF. HIF is a heterodimer composed of an unstable alpha subunit (e.g. HIF1alpha, HIF2alpha) and a stable beta subunit (e.g. HIF1beta). Under normoxic conditions, the alpha subunit of HIF is degraded. Degradation occurs due to the action of members of the prolyl hydroxylase domain (PHD) family. These proteins hydroxylate HIF at conserved proline residues, creating a recognition site for ubiquitin ligases that target HIF for degradation. The PHD proteins are Fe(II) and 2-oxoglutarate-dependent dioxygenases whose activity is dependent on oxygen as well as iron. Thus, under conditions of low oxygen or low iron, prolyl hydroxylase activity is decreased, and HIF becomes stabilized.
Iron also controls the activity of HIF. In addition to prolyl hydroxylation, HIF1alpha activity is regulated by asparaginyl hydroxylation, a modification that decreases the activity of HIF by reducing its affinity for coactivators. Asparaginyl hydroxylation is also catalyzed by a PHD family member termed FIH1241 (also called HIF1AN). HIF2alpha (also called EPAS1) is more resistant to this mechanism of inactivation242. As previously mentioned, the mRNA for HIF2alpha contains an IRE, and thus iron controls the translation of HIF. This mechanism has been postulated to act as a feedback loop to restrict HIF2alpha expression when iron is scarce.
HIF, in turn, affects the transcription of genes in the iron metabolic network. There is evidence for HIF1alpha modifying transcription for genes heme oxygenase 1243, hepcidin244, ceruloplasmin245, and transferrin receptor246. Moreover, HIF2alpha induces the frataxin gene247.
Description of reactions in specialized cells
Most of the body’s iron is found in hemoglobin, where it carries out oxygen transport. Two types of specialized cells play a key role in this fundamental aspect of body iron metabolism: the reticulocyte and the macrophage. Many of the species and reactions from our general mammalian cell model are present in these cells, as shown in Table 1. However, the reticulocyte and macrophage also express additional genes that are involved in reactions that do not occur in other mammalian cells.
A. Reticulocyte
The reticulocyte is an immature erythroid cell that still has mitochondria and ribosomes. Once mature, a circulating erythroid cell transports both oxygen and carbon dioxide and contains mostly hemoglobin. In our network, we show the synthesis of hemoglobin A (HbA), the type primarily synthesized in adults. HbA is composed of α and β peptide globin chains and is regulated by heme-regulated eIF2α kinase (HRI) and α hemoglobin-stabilizing protein (AHSP). Activated HRI inhibits globin synthesis through phosphorylation of eIF2α, and HRI is regulated by heme248. When intracellular heme levels are high, heme binds to and deactivates HRI. In heme deficiency, heme detaches from HRI which activates the protein248, 249. Thus, HRI ensures that the amount of globin produced matches the amount of heme present. The mechanism by which AHSP regulates hemoglobin production is different. AHSP binds to α hemoglobin (αHb), stabilizing αHb for hemoglobin synthesis250. In certain conditions, for instance in β thalassemia, excess αHb accumulates in the reticulocyte and AHSP can bind to this αHb to protect the cell250.
The main way that reticulocytes obtain iron for hemoglobin production is through the Tf-TfR pathway251–253. A cell-surface receptor for ferritin, however, also exists in this cell type254. FtR likely mediates endocytosis of soluble ferritin255. Once internalized, the soluble ferritin is degraded by the lysosome256 and the iron enters the labile iron pool, affecting IRP activity255, 257. There are some major differences in the regulation of proteins involved in iron metabolism of the reticulocyte. First, ALAS2, the erythroid-specific form of ALAS, contains an IRE in the 5′ UTR258, 259 and neither its synthesis nor its activity is inhibited by heme260. In reticulocytes, heme inhibits the internalization of transferrin by TfR1261, 262. The non-IRE form of DMT1 dominates in reticulocytes and is found on the membranes of endosomes containing TfR1263. DMT1 is not thought to import iron directly into the reticulocyte, but evidence of the +IRE isoform does exist at the mRNA level264. We show DMT1+IRE on the plasma membrane just like in the general model. However, we do not show it transporting iron, as we have no evidence of this. DMT1+IRE could possibly be on the membrane of the endosome as well.
B. Macrophage
In terms of iron homeostasis, the main function of the macrophage is to recycle iron from hemoglobin back into ferrous iron for circulation and use throughout the body. Macrophages are present in most tissues throughout the body, and we do not distinguish between different types of macrophages in our model. The two ways that the macrophage ingests hemoglobin are CD163-mediated endocytosis of haptoglobin:hemoglobin (Hp-Hb) complexes265, 266 and phagocytosis of senescent erythroid cells267. CD163 is a member of the scavenger receptor cystein-rich (SRCR) family of receptors expressed in macrophages. CD163 is a cell-surface receptor that mediates the endocytosis of Hb-Hp complexes265, 268. The Hb-Hp complex enters the lysosome where heme is released and subsequently degraded by heme oxygenase 1 and 2269. CD163 can detach from the plasma membrane of macrophages to contribute to the soluble CD163 (sCD163) in the plasma, but the function of sCD163 is not known266, 270.
An important function of the macrophage in iron metabolism is phagocytosis of senescent erythroid cells. This process begins when cell-surface receptor binds to the senescent red blood cell267, which activates the receptor. The receptor signals the macrophage to initiate phagocytosis, and the red blood cell is absorbed into a vesicle called a phagosome. This phagosome fuses with a lysosome, forming a phagolysosome267. In our network, we depict the phagosome and phagolysosome as a single compartment. At this point, hydrolytic enzymes degrade the red blood cell and then hemoglobin is degraded to release heme271. We show heme being exported into the cytoplasm for degradation by the heme oxygenase system. Additionally, heme could be degraded in the phagolysosome. In macrophages, Nramp1 is expressed on the membrane of the phagosome and, like DMT1 (Nramp2), transports ferrous iron. Nramp1 transports recycled iron out of the phagosome272. It is possible that Nramp1 transports iron bidirectionally and moreover affects other proteins involved in iron metabolism273, but this is still under debate and is not depicted in our network. HRI is present in macrophages at a much lower level than in erythroid cells and this protein is functional133. The role of HRI in the iron metabolism of macrophages is to stimulate phagocytosis133. Macrophages recycle iron primarily through ferroportin, but some studies suggest that macrophages secrete ferritin255. As previously discussed, this soluble ferritin may deliver iron to reticulocytes for hemoglobin production.
Discussion
We believe we are the first to use CellDesigner to map one metabolic network in multiple types of cells. In the construction of the tissue/cell specific networks, we see both redundancy and uniqueness. The redundancies are essential elements in the metabolism of iron for different types of cells. The unique species and reactions give a cell the ability to perform its special functions. A number of insights can be gained from inspection of these networks.
In graph theoretic terms, we may use the degree (number of edges) of a node as a measure of a species’ significance in the overall network274, 275. In all networks, the nodes for LIP, cystolic heme, and cystolic ROS have the highest degrees (19, 8, and 11, respectively, for the general network). Interestingly, the ratio of reactions involving LIP to those involving heme shifts according to cell type: the LIP is involved in over 2.3 times as many reactions as cystolic heme in epithelial cell types, while involved in only 1.6 and 1.7 times as many reactions in the macrophage and reticulocyte, respectively (the ratio of reactions involving LIP to those involving heme is 19/8 for epithelial cells, 20/12 for macrophages, and 17/10 for reticulocytes). This may illustrate the fact that the macrophage and reticulocyte have a greater need for heme, as compared to the epithelial cell.
A main observation gathered from the iron metabolic network is the large number of feedback loops involving the labile iron pool (LIP) and intracellular species (see Fig. 6). IRP activity participates in multiple negative feedback loops beginning and ending at the LIP, whereas HO1 participates in positive feedback loops for the LIP. Perhaps surprisingly, given the extensive differences between hepatocytes, enterocytes, and macrophages, the feedback loops in all these cell types are virtually identical (Fig. 6A). However, the feedback loops for the erythroid cell are different (Fig. 6B).
The four cell types exhibit a different distribution of species in their compartments. For instance, the macrophage has the smallest percentage of species in the mitochondrion and the largest percentage of species in the lysosome, as compared to the other cell types. The macrophage also has the largest number of reactions (112 in total) of any cell type and the highest percentage of transport reactions involving iron (20.5%). The additional transport is due, in part, to the sixth compartment—the phagosome/phagolysosome—that exists only in the macrophage. The reticulocyte has significantly fewer membrane-bound proteins—both in number (8) and in percentage of proteins in network (19%)—participating in iron metabolism, but this could be due to the lack of expression data for certain genes/proteins in the reticulocyte. The general network has an equal number of transcriptional and translational modifications. However, for any given tissue type, there are more translational modifications in the iron metabolic network. For the general network as well as all four tissue types, there are more modifers of transcription than modifiers of translation. This is due in part to the fact that most translational modification coming from IRE/IRP system. The reaction with the greatest number of modifiers is the transcription of the HO1 gene, which is affected by five different species in all tissue types. In the liver and general network, transcription of the HAMP gene also has five modifiers.
However, a limitation of this interpretation is that the number of times a reaction or species is studied in a given cell type may influence its apparent importance in that cell’s network. A natural next step in this modeling activity will therefore be to include kinetic information about each of the reactions in the iron metabolism map. In an advanced stage, this will then lead to different models of iron metabolism specific for the different cell types. These models would allow us to quantitatively determine the importance of the various species and reactions for the different cell types. Perhaps this analysis would support our frequency observations above, but surely one would learn new information regarding the metabolism of iron. A perceived problem with this path is the lack of kinetic data for many of the reactions depicted in these maps, even though for many reactions such information is available, as indicated in the text above. An alternative to using detailed enzyme kinetics data is to use a generic rate law274 to substitute kinetics that are not known in detail, as has been used previously in metabolic models275. Once kinetic information has been added to the model, it can then be simulated using a software package like COPASI11. The modeling effort is facilitated by the fact that CellDesigner stores the model using the Systems Biology Markup Language (SBML276), which is also understood by simulation software like COPASI. We hope to further understand, through modeling efforts, how the common and distinguishing mechanisms interlock with one another to produce cells that are functionally very different in terms of iron metabolism. Additionally, one could use a modeling framework to shed light on how iron is metabolized differently in various states (e.g. health versus inflammation or malignancy).
Materials and Methods
Construction of the general iron network
The general iron network was created in CellDesigner, a diagram editor for drawing biological networks7. CellDesigner involves Kitano’s graphical notation276 and stores files in Systems Biology Markup Language (SBML277). Networks created with CellDesigner can be exported as a picture file for network visualization, and the models can also be simulated or analyzed by software packages like Copasi11 and others. CellDesigner features a graphical, point-and-click user interface with different symbols for the various types of molecular species and reactions. The specific symbols we used in the iron network are listed in the legend of Fig. 1. A complete list of symbols used in CellDesigner is available on the CellDesigner website (http://www.celldesigner.org). For clarity, we note that the transport arrow is used to signify movement of a species from one compartment to another, or movement between the interior and transmembrane locations of a single compartment.
The reactions occurring in the network are based on the published literature. A PubMed search was performed using broad search terms such as iron metabolism, heme production, iron transport, Fenton reaction, iron and gene expression, and iron regulation. In addition, PubMed searches were performed for each specific species identified in Fig. 1. Searches were also conducted for gene expression in specific cell types (macrophages, hepatocytes, enterocytes, reticulocytes). Reference lists from recent articles were used as a secondary method to assure the completeness of the literature reviewed. Due to space limitations, not all key references used in the construction of this network are cited in the text.
Construction of Table 1 and tissue-specific networks
To create tissue-specific networks, we considered each species in the general network, and evaluated evidence for its expression in specific tissues. Our first choice for data in determining positive or negative gene expression was data from tissues including human, mouse, and rat. When this was unavailable, we looked for expression in cell lines. For the liver and intestine, we also relied upon NCBI’s database Unigene278 when no other data was available. Unigene is based on expressed sequence tags (ESTs) and reports the number of transcripts per million for many tissue types including the liver and intestine. For the macrophage, we included evidence from all types of macrophages, for instance Kupffer cells, histiocytes, or cultured macrophage cell lines. For the reticulocyte, we included evidence from erythroid precursors, erythroids, reticulocyte lysates, erythroid colonies (CFUE), and erythroblasts. We did not, however, use expression data from cultured erythroleukemia cell lines, like K562 or Friend cells, because these are disease states and may not represent true gene expression in healthy reticulocytes.
Note that “--” in Table 1 does not necessarily indicate absent expression. Some studies report quantitative measurements of RNA for both the liver and intestine 36, 220, 279, and moreover Unigene also gives a quantitative report for the liver and intestine. Since very low levels of gene expression often give rise to proteins that are not functionally present, we report “--” if the level of one tissue type is less than 8% of the level measured in the other. The 8% cut off is somewhat arbitrary, although it was chosen so that, according to Unigene, ferrochelatase would be present in both the liver and intestine, and hephaestin would not be present in the liver (matching the tissue data 105). Sometimes we have no evidence (positive or negative) of a gene’s expression in a certain tissue/cell type. This is shown in the table when all the entries are blank for a given gene and tissue/cell type. In our tissue-specific networks, we exclude the species for which we have no evidence of expression. The exception to this rule involves species in the heme biosythesis pathway, since it is believed that it occurs in almost all types of mammalian cells. For the macrophage, we found evidence for presence of ferrochelatase, frataxin, and ISU, three mitochondrial proteins involved in heme production. We thus infer the presence of the other proteins in the heme biosythesis pathway. Similarly, we did not find any published evidence for each protein involved in heme biosynthesis in the reticulocyte, but these are included in the reticulocyte network. Another exception involves HIF in reticulocytes. We have postive evidence of PHD and FIH1 in reticulocytes, and we infer the presence of HIF1alpha and HIF2alpha. Currently, little is known about gene expression in the reticulocyte, and perhaps in the future more of the species will prove to be present in this cell type.
In depicting the tissue-specific networks, we used a lighter color to shade the species for which we found no positive evidence of expression. In most cases, we also lightened all subsequent reactions involving said species as we assume those reactions do not occur. Sometimes, however, we kept the reactions in our network, for instance ABCB6 was not detected in the intestine. While the ABCB6 protein is lightened, we still show the transport of coproporphyrinogen III (COPRO III) from the cytoplasm into the mitochondrion during heme biosynthesis as we assume another protein must catalyze this reaction in the intestine. As mentioned in the discussion, hepcidin has two possible interactions: the internalization and degradation of ferroportin and the inhibition of DMT1 translation. The first is the standard, accepted interaction which does not occur in the intestine and the second has been found only in the intestine. We therefore lightened the pathway depicting hepcidin’s internalization and degradation of ferroportin in the intestinal cell network, and lightened the inhibition of DMT1 in all other tissue-specific networks.
Supplementary Material
Acknowledgments
This work was supported in part by grants T32 CA079448 (V.H.), R37 DK042412 (F.M.T.), R01CA120170 (V.S), and R01 GM080219 (P. M.). P. Mendes also acknowledges financial support from the U.K. EPSRC. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
References
- 1.Muckenthaler MU, Galy B, Hentze MW. Systemic iron homeostasis and the iron-responsive element/iron-regulatory protein (IRE/IRP) regulatory network. Annu Rev Nutr. 2008 doi: 10.1146/annurev.nutr.28.061807.155521. [DOI] [PubMed] [Google Scholar]
- 2.Wrighting DM, Andrews NC. Iron homeostasis and erythropoiesis. Curr Top Dev Biol. 2008;82:141–67. doi: 10.1016/S0070-2153(07)00006-3. [DOI] [PubMed] [Google Scholar]
- 3.Andrews NC, Schmidt PJ. Iron homeostasis. Annu Rev Physiol. 2007;69:69–85. doi: 10.1146/annurev.physiol.69.031905.164337. [DOI] [PubMed] [Google Scholar]
- 4.Theil EC, Matzapetakis M, Liu X. Ferritins: iron/oxygen biominerals in protein nanocages. J Biol Inorg Chem. 2006;11(7):803–10. doi: 10.1007/s00775-006-0125-6. [DOI] [PubMed] [Google Scholar]
- 5.Beutler E. Hemochromatosis: genetics and pathophysiology. Annu Rev Med. 2006;57:331–47. doi: 10.1146/annurev.med.57.121304.131310. [DOI] [PubMed] [Google Scholar]
- 6.World Health Organization Micronutrient deficiencies: Iron deficiency anaemia. [July 2, 2008]; http://www.who.int/nutrition/topics/ida/en/
- 7.Funahashi A, Morohashi M, Kitano H, Tanimura N. CellDesigner: a process diagram editor for gene-regulatory and biochemical networks. Biosilico. 2003;1(5):159–162. [Google Scholar]
- 8.Calzone L, Gelay A, Zinovyev A, Radvanyi F, Barillot E. A comprehensive modular map of molecular interactions in RB/E2F pathway. Mol Syst Biol. 2008;4:174. doi: 10.1038/msb.2008.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oda K, Matsuoka Y, Funahashi A, Kitano H. A comprehensive pathway map of epidermal growth factor receptor signaling. Mol Syst Biol. 2005;1:2005.0010. doi: 10.1038/msb4100014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klipp E, Liebermeister W, Helbig A, Kowald A, Schaber J. Systems biology standards--the community speaks. Nat Biotechnol. 2007;25(4):390–1. doi: 10.1038/nbt0407-390. [DOI] [PubMed] [Google Scholar]
- 11.Hoops S, Sahle S, Gauges R, Lee C, Pahle J, Simus N, Singhal M, Xu L, Mendes P, Kummer U. COPASI--a complex pathway simulator. Bioinformatics. 2006;22(24):3067–3074. doi: 10.1093/bioinformatics/btl485. [DOI] [PubMed] [Google Scholar]
- 12.Dampier W, Tozeren A. Signaling perturbations induced by invading H. pylori proteins in the host epithelial cells: A mathematical modeling approach. J Theor Biol. 2007;248(1):130–144. doi: 10.1016/j.jtbi.2007.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wally J, Halbrooks PJ, Vonrhein C, Rould MA, Everse SJ, Mason AB, Buchanan SK. The crystal structure of iron-free human serum transferrin provides insight into inter-lobe communication and receptor binding. J Biol Chem. 2006;281(34):24934–24944. doi: 10.1074/jbc.M604592200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang F, Lum JB, McGill JR, Moore CM, Naylor SL, Bragt PHV, Baldwin WD, Bowman BH. Human transferrin: cDNA characterization and chromosomal localization. Proc Nat Acad Soc. 1984;81(9):2752–2756. doi: 10.1073/pnas.81.9.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huebers HA, Finch CA. The physiology of transferrin and transferrin receptors. Physiol Rev. 1987;67(2):520–582. doi: 10.1152/physrev.1987.67.2.520. [DOI] [PubMed] [Google Scholar]
- 16.Jacobs EM, Verbeek AL, Kreeftenberg HG, van Deursen CT, Marx JJ, Stalenhoef AF, Swinkels DW, de Vries RA. Changing aspects of HFE-related hereditary haemochromatosis and endeavors to early diagnosis. Neth J Med. 2007;65(11):419–424. [PubMed] [Google Scholar]
- 17.Hemadi M, Ha-Duong N-T, El Hage Chahine J-M. The mechanism of iron release from the transferrin-receptor 1 adduct. J Mol Biol. 2006;358(4):1125–1136. doi: 10.1016/j.jmb.2006.02.055. [DOI] [PubMed] [Google Scholar]
- 18.Bali PK, Zak O, Aisen P. A new role for the transferrin receptor in the release of iron from transferrin. Biochemistry. 1991;30(2):324–328. doi: 10.1021/bi00216a003. [DOI] [PubMed] [Google Scholar]
- 19.McClelland A, Kuhn LC, Ruddle FH. The human transferrin receptor gene: genomic organization, and the complete primary structure of the receptor deduced from a cDNA sequence. Cell. 1984;39(2 Part 1):267–274. doi: 10.1016/0092-8674(84)90004-7. [DOI] [PubMed] [Google Scholar]
- 20.Schneider C, Owen MJ, Banville D, Williams JG. Primary structure of human transferrin receptor deduced from the mRNA sequence. Nature. 1984;311(5987):675–678. doi: 10.1038/311675b0. [DOI] [PubMed] [Google Scholar]
- 21.West AP, Giannetti AM, Herr AB, Bennett MJ, Nangiana JS, Pierce JR, Weiner LP, Snow PM, Bjorkman PJ. Mutational analysis of the transferrin receptor reveals overlapping HFE and transferrin binding sites. J Mol Biol. 2001;313(2):385–397. doi: 10.1006/jmbi.2001.5048. [DOI] [PubMed] [Google Scholar]
- 22.Chahine J-MEH, Pakdaman R. Transferrin, a mechanism for iron release. Eur J Biochem. 1995;230(3):1102–1110. doi: 10.1111/j.1432-1033.1995.tb20661.x. [DOI] [PubMed] [Google Scholar]
- 23.Sipe DM, Murphy RF. Binding to cellular receptors results in increased iron release from transferrin at mildly acidic pH. J Biol Chem. 1991;266(13):8002–8007. [PubMed] [Google Scholar]
- 24.Schuler J, Frank J, Trier U, Schafer-Korting M, Saenger W. Interaction kinetics of tetramethylrhodamine transferrin with human transferrin receptor studied by fluorescence correlation spectroscopy. Biochemistry. 1999;38(26):8402–8408. doi: 10.1021/bi9819576. [DOI] [PubMed] [Google Scholar]
- 25.Hemadi M, Kahn PH, Miquel G, ElHageChahine JM. Transferrin’s mechanism of interaction with receptor 1. Biochemistry. 2004;43(6):1736–1745. doi: 10.1021/bi030142g. [DOI] [PubMed] [Google Scholar]
- 26.Giannetti AM, Halbrooks PJ, Mason AB, Vogt TM, Enns CA, Bjorkman PJ. The molecular mechanism for receptor-stimulated iron release from the plasma iron transport protein transferrin. Structure. 2005;13(11):1613–1623. doi: 10.1016/j.str.2005.07.016. [DOI] [PubMed] [Google Scholar]
- 27.Feder JN, Gnirke A, Thomas W, Tsuchihashi Z, Ruddy DA, Basava A, Dormishian F, Domingo R, Ellis MC, Fullan A, Hinton LM, Jones NL, Kimmel BE, Kronmal GS, Lauer P, Lee VK, Loeb DB, Mapa FA, McClelland E, Meyer NC, Mintier GA, Moeller N, Moore T, Morikang E, Prass CE, Quintana L, Starnes SM, Schatzman RC, Brunke KJ, Drayna DT, Risch NJ, Bacon BR, Wolff RK. A novel MHC class I-like gene is mutated in patients with hereditary haemochromatosis. Nat Genet. 1996;13(4):399–408. doi: 10.1038/ng0896-399. [DOI] [PubMed] [Google Scholar]
- 28.Parkkila S, Parkkila AK, Waheed A, Britton RS, Zhou XY, Fleming RE, Tomatsu S, Bacon BR, Sly WS. Cell surface expression of HFE protein in epithelial cells, macrophages, and monocytes. Haematologica. 2000;85(4):340–345. [PubMed] [Google Scholar]
- 29.Roy CN, Penny DM, Feder JN, Enns CA. The hereditary hemochromatosis protein, HFE, specifically regulates transferrin-mediated iron uptake in HeLa cells. J Biol Chem. 1999;274(13):9022–9028. doi: 10.1074/jbc.274.13.9022. [DOI] [PubMed] [Google Scholar]
- 30.Lebron JA, West AP, Bjorkman PJ. The hemochromatosis protein HFE competes with transferrin for binding to the transferrin receptor. J Mol Biol. 1999;294(1):239–245. doi: 10.1006/jmbi.1999.3252. [DOI] [PubMed] [Google Scholar]
- 31.Gross CN, Irrinki A, Feder JN, Enns CA. Co-trafficking of HFE, a nonclassical major histocompatibility complex class I protein, with the transferrin receptor implies a role in intracellular iron regulation. J Biol Chem. 1998;273(34):22068–22074. doi: 10.1074/jbc.273.34.22068. [DOI] [PubMed] [Google Scholar]
- 32.Feder JN, Penny DM, Irrinki A, Lee VK, Lebron JA, Watson N, Tsuchihashi Z, Sigal E, Bjorkman PJ, Schatzman RC. The hemochromatosis gene product complexes with the transferrin receptor and lowers its affinity for ligand binding. Proc Nat Acad Sci. 1998;95(4):1472–1477. doi: 10.1073/pnas.95.4.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davies PS, Zhang A-S, Anderson EL, Roy CN, Lampson MA, McGraw TE, Enns CA. Evidence for the interaction of the hereditary haemochromatosis protein, HFE, with the transferrin receptor in endocytic compartments. Biochem J. 2003;373(1):145–153. doi: 10.1042/BJ20030202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramalingam TS, West AP, Lebron JA, Nangiana JS, Hogan TH, Enns CA, Bjorkman PJ. Retraction. Binding to he transferrin receptor is required for the endocytosis of HFE and regulation of iron homeostasis. Nat Cell Biol. 2003;5(7):680. doi: 10.1038/35046611. [DOI] [PubMed] [Google Scholar]
- 35.Chorney MJ, Yoshida Y, Meyer PN, Yoshida M, Gerhard GS. The enigmatic role of the hemochromatosis protein (HFE) in iron absorption. Trends Mol Med. 2003;9(3):118–125. doi: 10.1016/s1471-4914(03)00023-6. [DOI] [PubMed] [Google Scholar]
- 36.Ohgami RS, Campagna DR, Greer EL, Antiochos B, McDonald A, Chen J, Sharp JJ, Fujiwara Y, Barker JE, Fleming MD. Identification of a ferrireductase required for efficient transferrin-dependent iron uptake in erythroid cells. Nat Genet. 2005;37(11):1264–1269. doi: 10.1038/ng1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gunshin H, Mackenzie B, Berger UV, Gunshin Y, Romero MF, Boron WF, Nussberger S, Gollan JL, Hediger MA. Cloning and characterization of a mammalian proton-coupled metal-ion transporter. Nature. 1997;388(6641):482–488. doi: 10.1038/41343. [DOI] [PubMed] [Google Scholar]
- 38.Vidal S, Belouchi AM, Cellier M, Beatty B, Gros P. Cloning and characterization of a second human NRAMP gene on chromosome 12q13. Mamm Genome. 1995;6(4):224–230. doi: 10.1007/BF00352405. [DOI] [PubMed] [Google Scholar]
- 39.Canonne-Hergaux F, Gruenheid S, Ponka P, Gros P. Cellular and subcellular localization of the Nramp2 iron transporter in the intestinal brush border and regulation by dietary iron. Blood. 1999;93(12):4406–4417. [PubMed] [Google Scholar]
- 40.Ma Y, Yeh M, Yeh K-y, Glass J. Iron imports. V. transport of iron through the intestinal epithelium. Am J Physiol Gastrointest Liver Physiol. 2006;290(3):G417–422. doi: 10.1152/ajpgi.00489.2005. [DOI] [PubMed] [Google Scholar]
- 41.Worthington MT, Browne L, Battle EH, Luo RQ. Functional properties of transfected human DMT1 iron transporter. Am J Physiol Gastrointest Liver Physiol. 2000;279(6):G1265–1273. doi: 10.1152/ajpgi.2000.279.6.G1265. [DOI] [PubMed] [Google Scholar]
- 42.Oakhill JS, Marritt SJ, Gareta EG, Cammack R, McKie AT. Functional characterization of human duodenal cytochrome b (Cybrd1): Redox properties in relation to iron and ascorbate metabolism. Biochim Biophys Acra. 2008;1777(3):260–268. doi: 10.1016/j.bbabio.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 43.Turi JL, Wang X, McKie AT, Nozik-Grayck E, Mamo LB, Crissman K, Piantadosi CA, Ghio AJ. Duodenal cytochrome b: a novel ferrireductase in airway epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2006;291(2):L272–280. doi: 10.1152/ajplung.00342.2005. [DOI] [PubMed] [Google Scholar]
- 44.Zoller H, Theurl I, Koch RO, McKie AT, Vogel W, Weiss G. Duodenal cytochrome B and hephaestin expression in patients with iron deficiency and hemochromatosis. Gastroenterology. 2003;125(3):746–754. doi: 10.1016/s0016-5085(03)01063-1. [DOI] [PubMed] [Google Scholar]
- 45.McKie AT, Barrow D, Latunde-Dada GO, Rolfs A, Sager G, Mudaly E, Mudaly M, Richardson C, Barlow D, Bomford A, Peters TJ, Raja KB, Shirali S, Hediger MA, Farzaneh F, Simpson RJ. An iron-regulated ferric reductase associated with the absorption of dietary iron. Science. 2001;291(5509):1755–1759. doi: 10.1126/science.1057206. [DOI] [PubMed] [Google Scholar]
- 46.Picard V, Govoni G, Jabado N, Gros P. Nramp 2 (DCT1/DMT1) expressed at the plasma membrane transports iron and other divalent cations into a calcein-accessible cytoplasmic pool. J Biol Chem. 2000;275(46):35738–35745. doi: 10.1074/jbc.M005387200. [DOI] [PubMed] [Google Scholar]
- 47.Montreuil J, Tonnelat J, Mullet S. Preparation et proprietes de la lactosiderophiline (lactotransferrine) du lait de femme. Biochim Biophys Acta. 1960;45:413–421. doi: 10.1016/0006-3002(60)91478-5. [DOI] [PubMed] [Google Scholar]
- 48.Masson PL, Heremans JF, Schonne E. Lactoferrin, an iron-binding protein in neutrophilic leukocytes. J Exp Med. 1969;130(3):643–658. doi: 10.1084/jem.130.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ward PP, Paz E, Conneely OM. Multifuntional roles of lactoferrin: a critical overview. Cel Mol Life Sci. 2005;62(22):2540–2548. doi: 10.1007/s00018-005-5369-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pakdaman R, Petitjean M, El Hage Chahine J-M. Transferrins. A mechanism for iron uptake by lactoferrin. Eur J Biochem. 1998;254(1):144–153. doi: 10.1046/j.1432-1327.1998.2540144.x. [DOI] [PubMed] [Google Scholar]
- 51.Peterson NA, Anderson BF, Jameson GB, Tweedie JW, Baker EN. Crystal Structure and Iron-Binding Properties of the R210K Mutant of the N-Lobe of Human Lactoferrin: Implications for Iron Release from Transferrins. Biochemistry. 2000;39(22):6625–6633. doi: 10.1021/bi0001224. [DOI] [PubMed] [Google Scholar]
- 52.Baker HM, Baker EN. Lactoferrin and iron: structural and dynamic aspects of binding and release. BioMetals. 2004;17:209–216. doi: 10.1023/b:biom.0000027694.40260.70. [DOI] [PubMed] [Google Scholar]
- 53.Abdallah FB, El Hage Chahine J-M. Transferrins: iron release from lactoferrin. J Mol Biol. 2000;303(2):255–266. doi: 10.1006/jmbi.2000.4101. [DOI] [PubMed] [Google Scholar]
- 54.Kawabata H, Tong X, Kawanami T, Wano Y, Hirose Y, Sugai S, Phillip Koeffler H. Analyses for binding of the transferrin family of proteins to the transferrin receptor 2. Br J Haematol. 2004;127(4):464–473. doi: 10.1111/j.1365-2141.2004.05224.x. [DOI] [PubMed] [Google Scholar]
- 55.Ashida K, Sasaki H, Suzuki YA, Lönnerdal B. Cellular internalization of lactoferrin in intestinal epithelial cells. BioMetals. 2004;17(3):311–315. doi: 10.1023/b:biom.0000027710.13543.3f. [DOI] [PubMed] [Google Scholar]
- 56.Suzuki Y, Lopez V, Lönnerdal B. Lactoferrin. Cell Mol Life Sci. 2005;62(22):2560–2575. doi: 10.1007/s00018-005-5371-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suzuki Y, Shin K, Lonnerdal B. Molecular cloning and functional expression of a human intestinal lactoferrin receptor. Biochemistry. 2001;40(51):15771–15779. doi: 10.1021/bi0155899. [DOI] [PubMed] [Google Scholar]
- 58.Kurz T, Terman A, Gustafsson B, Brunk UT. Lysosomes in iron metabolism, ageing and apoptosis. Histochem Cell Biol. 2008;129(4):389–406. doi: 10.1007/s00418-008-0394-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kjeldsen L, Johnsen AH, Sengelov H, Borregaard N. Isolation and primary structure of NGAL, a novel protein associated with human neutrophil gelatinase. J Biol Chem. 1993;268(14):10425–10432. [PubMed] [Google Scholar]
- 60.Cowland JB, Sorensen OE, Sehested M, Borregaard N. Neutrophil gelatinase-associated lipocalin is up-regulated in human epithelial cells by IL-1{beta}, but not by TNF-{alpha} J Immunol. 2003;171(12):6630–6639. doi: 10.4049/jimmunol.171.12.6630. [DOI] [PubMed] [Google Scholar]
- 61.Nielsen BS, Borregaard N, Bundgaard JR, Timshel S, Sehested M, Kjeldsen L. Induction of NGAL synthesis in epithelial cells of human colorectal neoplasia and inflammatory bowel diseases. Gut. 1996;38(3):414–420. doi: 10.1136/gut.38.3.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goetz DH, Holmes MA, Borregaard N, Bluhm ME, Raymond KN, Strong RK. The neutrophil lipocalin NGAL Is a bacteriostatic agent that Interferes with siderophore-mediated iron acquisition. Molecular Cell. 2002;10(5):1033–1043. doi: 10.1016/s1097-2765(02)00708-6. [DOI] [PubMed] [Google Scholar]
- 63.Borregaard N, Cowland JB. Neutrophil gelatinase-associated lipocain, a siderophore-binding eukaryotic protein. BioMetals. 2006;19:211–215. doi: 10.1007/s10534-005-3251-7. [DOI] [PubMed] [Google Scholar]
- 64.Devireddy LR, Gazin C, Zhu X, Green MR. A cell-surface receptor for lipocalin 24p3 selectively mediates apoptosis and iron uptake. Cell. 2005;123(7):1293–1305. doi: 10.1016/j.cell.2005.10.027. [DOI] [PubMed] [Google Scholar]
- 65.Yang J, Goetz D, Li JY, Wang W, Mori K, Setlik D, Du T, Erdjument-Bromage H, Tempst P, Strong R, Barasch J. An iron delivery pathway mediated by lipocain. Mol Cell. 2002;10:1045–1056. doi: 10.1016/s1097-2765(02)00710-4. [DOI] [PubMed] [Google Scholar]
- 66.Li J-Y, Ram G, Gast K, Chen X, Barasch K, Mori K, Schmidt-Ott K, Wang J, Kuo H-C, Savage-Dunn C, Garrick MD, Barasch J. Detection of intracellular iron by its regulatory effect. Am J Physiol Cell Physiol. 2004;287(6):C1547–1559. doi: 10.1152/ajpcell.00260.2004. [DOI] [PubMed] [Google Scholar]
- 67.Hvidberg V, Jacobsen C, Strong RK, Cowland JB, Moestrup SK, Borregaard N. The endocytic receptor megalin binds the iron transporting neutrophil-gelatinase-associated lipocalin with high affinity and mediates its cellular uptake. FEBS Letters. 2005;579(3):773–777. doi: 10.1016/j.febslet.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 68.Rouault TA. The intestinal heme transporter revealed. Cell. 2005;122(5):649–651. doi: 10.1016/j.cell.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 69.Krishnamurthy P, Xie T, Schuetz JD. The role of transporters in cellular heme and porphyrin homeostasis. Pharmacol Therapeut. 2007;114(3):345–358. doi: 10.1016/j.pharmthera.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 70.Shayeghi M, Latunde-Dada GO, Oakhill JS, Laftah AH, Takeuchi K, Halliday N, Khan Y, Warley A, McCann FE, Hider RC, Frazer DM, Anderson GJ, Vulpe CD, Simpson RJ, McKie AT. Identification of an intestinal heme transporter. Cell. 2005;122(5):789–801. doi: 10.1016/j.cell.2005.06.025. [DOI] [PubMed] [Google Scholar]
- 71.Andrews NC. When is a heme transporter not a heme transporter? When it’s a folate transporter. Cell Metab. 2007;5(1):5–6. doi: 10.1016/j.cmet.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 72.Nakai Y, Inoue K, Abe N, Hatakeyama M, Ohta K-y, Otagiri M, Hayashi Y, Yuasa H. Functional characterization of human proton-coupled folate transporter/heme carrier protein 1 heterologously expressed in mammalian cells as a folate transporter. J Pharmacol Exp Ther. 2007;322(2):469–476. doi: 10.1124/jpet.107.122606. [DOI] [PubMed] [Google Scholar]
- 73.Qiu A, Jansen M, Sakaris A, Min SH, Chattopadhyay S, Tsai E, Sandoval C, Zhao R, Akabas MH, Goldman ID. Identification of an intestinal folate transporter and the molecular basis for hereditary folate malabsorption. Cell. 2006;127(5):917–928. doi: 10.1016/j.cell.2006.09.041. [DOI] [PubMed] [Google Scholar]
- 74.Latunde-Dada GO, Takeuchi K, Simpson RJ, McKie AT. Haem carrier protein 1 (HCP1): Expression and functional studies in cultured cells. FEBS Letters. 2006;580(30):6865–6870. doi: 10.1016/j.febslet.2006.11.048. [DOI] [PubMed] [Google Scholar]
- 75.Sharma S, Dimasi D, Broer S, Kumar R, Della NG. Heme carrier protein 1 (HCP1) expression and functional analysis in the retina and retinal pigment epithelium. Exp Cell Res. 2007;313(6):1251–1259. doi: 10.1016/j.yexcr.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 76.Poss KD, Tonegawa S. Heme oxygenase 1 is required for mammalian iron reutilization. Proc Natl Acad Sci. 1997;94(20):10919–10924. doi: 10.1073/pnas.94.20.10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schuller DJ, Wilks A, Ortiz de Montellano PR, Poulos TL. Crystal structure of human heme oxygenase-1. Nat Struct Mol Biol. 1999;6(9):860–867. doi: 10.1038/12319. [DOI] [PubMed] [Google Scholar]
- 78.Shibahara S, Sato M, Muller RM, Yoshida T. Structural organization of the human heme oxygenase gene and the function of its promoter. Eur J Biochem. 1989;179(3):557–563. doi: 10.1111/j.1432-1033.1989.tb14583.x. [DOI] [PubMed] [Google Scholar]
- 79.Tenhunen R, Marver HS, Schmid R. Microsomal heme oxygenase. characterization of the enzyme. J Biol Chem. 1969;244(23):6388–6394. [PubMed] [Google Scholar]
- 80.McCoubrey WK, Ewing JF, Maines MD. Human heme oxygenase-2: Characterization and expression of a full-length cDNA and evidence suggesting that the two HO-2 transcripts may differ by choice of polyadenylation signal. Arch Biochem Biophys. 1992;295(1):13–20. doi: 10.1016/0003-9861(92)90481-b. [DOI] [PubMed] [Google Scholar]
- 81.West AP, Jr, Bennett MJ, Sellers VM, Andrews NC, Enns CA, Bjorkman PJ. Comparison of the interactions of transferrin receptor and transferrin receptor 2 with transferrin and the hereditary hemochromatosis protein HFE. J Biol Chem. 2000;275(49):38135–38138. doi: 10.1074/jbc.C000664200. [DOI] [PubMed] [Google Scholar]
- 82.Kawabata H, Yang R, Hirama T, Vuong PT, Kawano S, Gombart AF, Koeffler HP. Molecular cloning of transferrin receptor 2. a new member of the transferrin receptor-like family. J Biol Chem. 1999;274(30):20826–20832. doi: 10.1074/jbc.274.30.20826. [DOI] [PubMed] [Google Scholar]
- 83.Roetto A, Totaro A, Piperno A, Piga A, Longo F, Garozzo G, Cali A, De Gobbi M, Gasparini P, Camaschella C. New mutations inactivating transferrin receptor 2 in hemochromatosis type 3. Blood. 2001;97(9):2555–2560. doi: 10.1182/blood.v97.9.2555. [DOI] [PubMed] [Google Scholar]
- 84.Wallace DF, Summerville L, Crampton EM, Subramaniam VN. Defective trafficking and localization of mutated transferrin receptor 2: implications for type 3 hereditary hemochromatosis. Am J Physiol Cell Physiol. 2008;294(2):C383–390. doi: 10.1152/ajpcell.00492.2007. [DOI] [PubMed] [Google Scholar]
- 85.Wallace DF, Summerville L, Subramaniam VN. Targeted disruption of the hepatic transferrin receptor 2 gene in mice leads to iron overload. Gastroenterology. 2007;132(1):301–310. doi: 10.1053/j.gastro.2006.11.028. [DOI] [PubMed] [Google Scholar]
- 86.Deaglio S, Capobianco A, Cali A, Bellora F, Alberti F, Righi L, Sapino A, Camaschella C, Malavasi F. Structural, functional, and tissue distribution analysis of human transferrin receptor-2 by murine monoclonal antibodies and a polyclonal antiserum. Blood. 2002;100(10):3782–3789. doi: 10.1182/blood-2002-01-0076. [DOI] [PubMed] [Google Scholar]
- 87.Camaschella C, Roetto A, Cali A, De Gobbi M, Garozzo G, Carella M, Majorano N, Totaro A, Gasparini P. The gene TFR2 is mutated in a new type of haemochromatosis mapping to 7q22. Nat Genet. 2000;25(1):14–15. doi: 10.1038/75534. [DOI] [PubMed] [Google Scholar]
- 88.Kawabata H, Germain RS, Ikezoe T, Tong X, Green EM, Gombart AF, Koeffler HP. Regulation of expression of murine transferrin receptor 2. Blood. 2001;98(6):1949–1954. doi: 10.1182/blood.v98.6.1949. [DOI] [PubMed] [Google Scholar]
- 89.Johnson MB, Chen J, Murchison N, Green FA, Enns CA. Transferrin receptor 2: evidence for ligand-induced stabilization and redirection to a recycling pathway. Mol Biol Cell. 2007;18(3):743–754. doi: 10.1091/mbc.E06-09-0798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Graham RM, Reutens GM, Herbison CE, Delima RD, Chua ACG, Olynyk JK, Trinder D. Transferrin receptor 2 mediates uptake of transferrin-bound and non-transferrin-bound iron. J Hepatol. 2008;48(2):327–334. doi: 10.1016/j.jhep.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 91.Robb A, Wessling-Resnick M. Regulation of transferrin receptor 2 protein levels by transferrin. Blood. 2004;104(13):4294–4299. doi: 10.1182/blood-2004-06-2481. [DOI] [PubMed] [Google Scholar]
- 92.Johnson MB, Enns CA. Diferric transferrin regulates transferrin receptor 2 protein stability. Blood. 2004;104(13):4287–4293. doi: 10.1182/blood-2004-06-2477. [DOI] [PubMed] [Google Scholar]
- 93.Donovan A, Brownlie A, Zhou Y, Shepard J, Pratt SJ, Moynihan J, Paw BH, Drejer A, Barut B, Zapata A, Law TC, Brugnara C, Lux SE, Pinkus GS, Pinkus JL, Kingsley PD, Palis J, Fleming MD, Andrews NC, Zon LI. Positional cloning of zebrafish ferroportin1 identifies a conserved vertebrate iron exporter. Nature. 2000;403(6771):776–781. doi: 10.1038/35001596. [DOI] [PubMed] [Google Scholar]
- 94.Abboud S, Haile DJ. A novel mammalian iron-regulated protein involved in intracellular iron metabolism. J Biol Chem. 2000;275(26):19906–19912. doi: 10.1074/jbc.M000713200. [DOI] [PubMed] [Google Scholar]
- 95.Fleming MD, Trenor CC, Su MA, Foernzler D, Beier DR, Dietrich WF, Andrews NC. Microcytic anaemia mice have a mutation in Nramp2, a candidate iron transporter gene. Nat Genet. 1997;16(4):383–386. doi: 10.1038/ng0897-383. [DOI] [PubMed] [Google Scholar]
- 96.McKie AT, Marciani P, Rolfs A, Brennan K, Wehr K, Barrow D, Miret S, Bomford A, Peters TJ, Farzaneh F, Hediger MA, Hentze MW, Simpson RJ. A novel duodenal iron-regulated transporter, IREG1, implicated in the basolateral transfer of iron to the circulation. Mol Cell. 2000;5(2):299–309. doi: 10.1016/s1097-2765(00)80425-6. [DOI] [PubMed] [Google Scholar]
- 97.Donovan A, Lima CA, Pinkus JL, Pinkus GS, Zon LI, Robine S, Andrews NC. The iron exporter ferroportin/Slc40a1 is essential for iron homeostasis. Cell Metab. 2005;1(3):191–200. doi: 10.1016/j.cmet.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 98.Wang F, Paradkar PN, Custodio AO, McVey Ward D, Fleming MD, Campagna D, Roberts KA, Boyartchuk V, Dietrich WF, Kaplan J, Andrews NC. Genetic variation in Mon1a affects protein trafficking and modifies macrophage iron loading in mice. Nat Genet. 2007;39(8):1025–1032. doi: 10.1038/ng2059. [DOI] [PubMed] [Google Scholar]
- 99.Osaki S. Kinetic studies of ferrous ion oxidation with crystalline human ferroxidase (ceruloplasmin) J Biol Chem. 1966;241(21):5053–5059. [PubMed] [Google Scholar]
- 100.Osaki S, Walaas O. Kinetic studies of ferrous ion oxidation with crystalline human ferroxidase II. Rate constants at various steps and formation of a possible enzyme-substrate complex. J Biol Chem. 1967;242(11):2653–2657. [PubMed] [Google Scholar]
- 101.Yang F, Naylor SL, Lum JB, Cutshaw S, McCombs JL, Naberhaus KH, McGill JR, Adrian GS, Moore CM, Barnett DR, Bowman BH. Characterization, mapping, and expression of the human ceruloplasmin gene. Proc Nat Acad Soc. 1986;83(10):3257–3261. doi: 10.1073/pnas.83.10.3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yang FM, Friedrichs WE, Cupples RL, Bonifacio MJ, Sanford JA, Horton WA, Bowman BH. Human ceruloplasmin. Tissue-specific expression of transcripts produced by alternative splicing. J Biol Chem. 1990;265(18):10780–10785. [PubMed] [Google Scholar]
- 103.Okhee Han E-YK. Colocalization of ferroportin-1 with hephaestin on the basolateral membrane of human intestinal absorptive cells. J Cell Biochem. 2007;101(4):1000–1010. doi: 10.1002/jcb.21392. [DOI] [PubMed] [Google Scholar]
- 104.Syed BA, Beaumont NJ, Patel A, Naylor CE, Bayele HK, Joannou CL, Rowe PSN, Evans RW, Srai SKS. Analysis of the human hephaestin gene and protein: comparative modelling of the N-terminus ecto-domain based upon ceruloplasmin. Protein Eng. 2002;15(3):205–214. doi: 10.1093/protein/15.3.205. [DOI] [PubMed] [Google Scholar]
- 105.Vulpe CD, Kuo Y-M, Murphy TL, Cowley L, Askwith C, Libina N, Gitschier J, Anderson GJ. Hephaestin, a ceruloplasmin homologue implicated in intestinal iron transport, is defective in the sla mouse. Nat Genet. 1999;21(2):195–199. doi: 10.1038/5979. [DOI] [PubMed] [Google Scholar]
- 106.Quigley JG, Burns CC, Anderson MM, Lynch ED, Sabo KM, Overbaugh J, Abkowitz JL. Cloning of the cellular receptor for feline leukemia virus subgroup C (FeLV-C), a retrovirus that induces red cell aplasia. Blood. 2000;95(3):1093–1099. [PubMed] [Google Scholar]
- 107.Tailor CS, Willett BJ, Kabat D. A putative cell surface receptor for anemia-inducing feline leukemia virus subgroup C is a member of a transporter superfamily. J Virol. 1999;73(8):6500–6505. doi: 10.1128/jvi.73.8.6500-6505.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Quigley JG, Yang Z, Worthington MT, Phillips JD, Sabo KM, Sabath DE, Berg CL, Sassa S, Wood BL, Abkowitz JL. Identification of a human heme exporter that Is essential for erythropoiesis. Cell. 2004;118(6):757–766. doi: 10.1016/j.cell.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 109.Keel SB, Doty RT, Yang Z, Quigley JG, Chen J, Knoblaugh S, Kingsley PD, De Domenico I, Vaughn MB, Kaplan J, Palis J, Abkowitz JL. A Heme Export Protein Is Required for Red Blood Cell Differentiation and Iron Homeostasis. Science. 2008;319(5864):825–828. doi: 10.1126/science.1151133. [DOI] [PubMed] [Google Scholar]
- 110.Doyle LA, Yang W, Abruzzo LV, Krogmann T, Gao Y, Rishi AK, Ross DD. A multidrug resistance transporter from human MCF-7 breast cancer cells. Proc Nat Acad Sci. 1998;95(26):15665–15670. doi: 10.1073/pnas.95.26.15665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Krishnamurthy P, Schuetz JD. Role of ABCG2/BCRP in biology and medicine. Ann Rev Pharmacol Toxicol. 2006;46(1):381–410. doi: 10.1146/annurev.pharmtox.46.120604.141238. [DOI] [PubMed] [Google Scholar]
- 112.Krishnamurthy P, Ross DD, Nakanishi T, Bailey-Dell K, Zhou S, Mercer KE, Sarkadi B, Sorrentino BP, Schuetz JD. The Stem Cell Marker Bcrp/ABCG2 Enhances Hypoxic Cell Survival through Interactions with Heme. J Biol Chem. 2004;279(23):24218–24225. doi: 10.1074/jbc.M313599200. [DOI] [PubMed] [Google Scholar]
- 113.Austin Doyle L, Ross DD. Multidrug resistance mediated by the breast cancer resistance protein BCRP (ABCG2) Oncogene. 22(47):7340–7358. doi: 10.1038/sj.onc.1206938. [DOI] [PubMed] [Google Scholar]
- 114.Rush JD, Bielski BHJ. Pulse radiolytic studies of the reaction of perhydroxyl/superoxide O2- with iron(II)/iron(III) ions. The reactivity of HO2/O2- with ferric ions and its implication on the occurrence of the Haber-Weiss reaction. J Phys Chem. 1985;89(23):5062–5066. [Google Scholar]
- 115.Qian SY, Buettner GR. Iron and dioxygen chemistry is an important route to initiation of biological free radical oxidations: an electron paramagnetic resonance spin trapping study. Free Radic Biol Med. 1999;26(11–12):1447–1456. doi: 10.1016/s0891-5849(99)00002-7. [DOI] [PubMed] [Google Scholar]
- 116.Chasteen ND, Harrison PM. Mineralization in ferritin: an efficient means of iron storage. J Struct Biol. 1999;126(3):182–194. doi: 10.1006/jsbi.1999.4118. [DOI] [PubMed] [Google Scholar]
- 117.Costanzo F, Santoro C, Colantuoni V, Bensi G, Raugei G, Romano V, Cortese R. Cloning ans sequencing of a full length cDNA coding for a human apoferritin H chain: evidence for a multigene family. EBMO Journal. 1984;3(1):23–27. doi: 10.1002/j.1460-2075.1984.tb01756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hentze MW, Keim S, Papadopoulos P, O’Brien S, Modi W, Drysdale J, Leonard WJ, Harford JB, Klausner RD. Cloning, characterization, expression, and chromosomal localization of a human ferritin heavy-chain gene. Proc Nat Acad Soc. 1986;83(19):7226–7230. doi: 10.1073/pnas.83.19.7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dorner MH, Salfeld J, Will H, Leibold EA, Vass JK, Munro HN. Structure of human ferritin light subunit messenger RNA: comparison with heavy subunit message and functional implications. Proc Nat Acad Soc. 1985;82(10):3139–3143. doi: 10.1073/pnas.82.10.3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Santoro C, Marone M, Ferrone M, Costanzo F, Colombo M, Minganti C, Cortese R, Silengo L. Cloning of the gene coding for human L apoferritin. Nucl Acids Res. 1986;14(7):2863–2876. doi: 10.1093/nar/14.7.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Boyd D, Vecoli C, Belcher DM, Jain SK, Drysdale JW. Structural and functional relationships of human ferritin H and L chains deduced from cDNA clones. J Biol Chem. 1985;260(21):11755–11761. [PubMed] [Google Scholar]
- 122.Levi S, Yewdall SJ, Harrison PM, Santambrogio P, Cozzi A, Rovida E, Albertini A, Arosio P. Evidence of H- and L-chains have co-operative roles in iron-uptake mechanism of human ferrtin. Biochem J. 1992;288:591–596. doi: 10.1042/bj2880591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bou-Abdallah F, Arosio P, Santambrogio P, Yang X, Janus-Chandler C, Chasteen ND. Ferrous ion binding to recombinant human H-chain ferritin. An isothermal titration calorimetry study. Biochemistry. 2002;41(37):11184–11191. doi: 10.1021/bi020215g. [DOI] [PubMed] [Google Scholar]
- 124.Bou-Abdallah F, Carney E, Chasteen ND, Arosio P, Viescas AJ, Papaefthymiou GC. A comparative Mossbauer study of the mineral cores of human H-chain ferritin employing dioxygen and hydrogen peroxide as iron oxidants. Biophys Chem. 2007;130(3):114–121. doi: 10.1016/j.bpc.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bou-Abdallah F, Santambrogio P, Levi S, Arosio P, Chasteen ND. Unique iron binding and oxidation properties of human mitochondrial ferritin: a comparative analysis with human H-chain ferritin. J Mol Biol. 2005;347(3):543–554. doi: 10.1016/j.jmb.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 126.Bou-Abdallah F, Zhao G, Mayne HR, Arosio P, Chasteen ND. Origin of the unusual kinetics of iron deposition in human H-chain ferritin. J Am Chem Soc. 2005;127(11):3885–3893. doi: 10.1021/ja044355k. [DOI] [PubMed] [Google Scholar]
- 127.Sun S, Arosio P, Levi S, Chasteen ND. Ferroxidase kinetics of human liver apoferritin, recombinant H-chain apoferritin, and site-directed mutants. Biochemistry. 1993;32(36):9362–9369. doi: 10.1021/bi00087a015. [DOI] [PubMed] [Google Scholar]
- 128.Zhao G, Bou-Abdallah F, Arosio P, Levi S, Janus-Chandler C, Chasteen ND. Multiple pathways for mineral core formation in mammalian apoferritin. The role of hydrogen peroxide. Biochemistry. 2003;42(10):3142–3150. doi: 10.1021/bi027357v. [DOI] [PubMed] [Google Scholar]
- 129.Shi H, Bencze KZ, Stemmler TL, Philpott CC. A cytosolic iron chaperone that delivers iron to ferritin. Science. 2008;320(5880):1207–1210. doi: 10.1126/science.1157643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Radisky DC, Kaplan J. Iron in cytosolic ferritin can be recycled through lysosomal degradation in human fibroblasts. Biochem J. 1998;336(1):201–205. doi: 10.1042/bj3360201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kidane TZ, Sauble E, Linder MC. Release of iron from ferritin requires lysosomal activity. Am J Physiol Cell Physiol. 2006;291(3):C445–455. doi: 10.1152/ajpcell.00505.2005. [DOI] [PubMed] [Google Scholar]
- 132.Jin W, Takagi H, Pancorbo B, Theil EC. “Opening” the Ferritin Pore for Iron Release by Mutation of Conserved Amino Acids at Interhelix and Loop Sites. Biochemistry. 2001;40(25):7525–7532. doi: 10.1021/bi002509c. [DOI] [PubMed] [Google Scholar]
- 133.Liu S, Suragani RN, Wang F, Han A, Zhao W, Andrews NC, Chen JJ. The function of heme-regulated eIF2alpha kinase in murine iron homeostasis and macrophage maturation. J Clin Invest. 2007;117(11):3296–3305. doi: 10.1172/JCI32084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Liu X, Jin W, Theil EC. Opening protein pores with chaotropes enhances Fe reduction and chelation of Fe from the ferritin biomineral. Proc Nat Acad Soc. 2003;100(7):3653–3658. doi: 10.1073/pnas.0636928100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Shaw GC, Cope JJ, Li L, Corson K, Hersey C, Ackermann GE, Gwynn B, Lambert AJ, Wingert RA, Traver D, Trede NS, Barut BA, Zhou Y, Minet E, Donovan A, Brownlie A, Balzan R, Weiss MJ, Peters LL, Kaplan J, Zon LI, Paw BH. Mitoferrin is essential for erythroid iron assimilation. Nature. 2006;440(7080):96–100. doi: 10.1038/nature04512. [DOI] [PubMed] [Google Scholar]
- 136.Corsi B, Cozzi A, Arosio P, Drysdale J, Santambrogio P, Campanella A, Biasiotto G, Albertini A, Levi S. Human mitochondrial ferritin expressed in HeLa cells incorporates iron and affects cellular iron metabolism. J Biol Chem. 2002;277(25):22430–22437. doi: 10.1074/jbc.M105372200. [DOI] [PubMed] [Google Scholar]
- 137.Langlois d’Estaintot B, Santambrogio P, Granier T, Gallois B, Chevalier JM, Precigoux G, Levi S, Arosio P. Crystal structure and biochemical properties of the human mitochondrial ferritin and its mutant Ser144Ala. J Mol Biol. 2004;340(2):277–293. doi: 10.1016/j.jmb.2004.04.036. [DOI] [PubMed] [Google Scholar]
- 138.Levi S, Corsi B, Bosisio M, Invernizzi R, Volz A, Sanford D, Arosio P, Drysdale J. A human mitochondrial ferritin encoded by an intronless gene. J Biol Chem. 2001;276(27):24437–24440. doi: 10.1074/jbc.C100141200. [DOI] [PubMed] [Google Scholar]
- 139.Campuzano V, Montermini L, Molto MD, Pianese L, Cossee M, Cavalcanti F, Monros E, Rodius F, Duclos F, Monticelli A, Zara F, Canizares J, Koutnikova H, Bidichandani SI, Gellera C, Brice A, Trouillas P, De Michele G, Filla A, De Frutos R, Palau F, Patel PI, Di Donato S, Mandel J-L, Cocozza S, Koenig M, Pandolfo M. Friedreich’s Ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271(5254):1423–1427. doi: 10.1126/science.271.5254.1423. [DOI] [PubMed] [Google Scholar]
- 140.Rotig A, de Lonlay P, Chretien D, Foury F, Koenig M, Sidi D, Munnich A, Rustin P. Aconitase and mitochondrial iron-sulphur protein deficiency in Friedreich ataxia. Nat Genet. 1997;17(2):215–217. doi: 10.1038/ng1097-215. [DOI] [PubMed] [Google Scholar]
- 141.Bencze KZ, Yoon T, Millan-Pacheco C, Bradley PB, Pastor N, Cowan JA, Stemmler TL. Human frataxin: iron and ferrochelatase binding surface. Chem Commun. 2007;(18):1798–1800. doi: 10.1039/b703195e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bulteau A-L, O’Neill HA, Kennedy MC, Ikeda-Saito M, Isaya G, Szweda LI. Frataxin acts as an iron chaperone protein to modulate mitochondrial aconitase activity. Science. 2004;305(5681):242–245. doi: 10.1126/science.1098991. [DOI] [PubMed] [Google Scholar]
- 143.Martelli A, Wattenhofer-Donze M, Schmucker S, Bouvet S, Reutenauer L, Puccio H. Frataxin is essential for extramitochondrial Fe S cluster proteins in mammalian tissues. Hum Mol Genet. 2007;16(22):2651–2658. doi: 10.1093/hmg/ddm163. [DOI] [PubMed] [Google Scholar]
- 144.Schoenfeld RA, Napoli E, Wong A, Zhan S, Reutenauer L, Morin D, Buckpitt AR, Taroni F, Lonnerdal B, Ristow M, Puccio H, Cortopassi GA. Frataxin deficiency alters heme pathway transcripts and decreases mitochondrial heme metabolites in mammalian cells. Hum Mol Genet. 2005;14(24):3787–3799. doi: 10.1093/hmg/ddi393. [DOI] [PubMed] [Google Scholar]
- 145.Stehling O, Elsasser H-P, Bruckel B, Muhlenhoff U, Lill R. Iron-sulfur protein maturation in human cells: evidence for a function of frataxin. Hum Mol Genet. 2004;13(23):3007–3015. doi: 10.1093/hmg/ddh324. [DOI] [PubMed] [Google Scholar]
- 146.Yoon T, Cowan JA. Iron-sulfur cluster biosynthesis. Characterization of frataxin as an iron donor for assembly of [2Fe-2S] clusters in ISU-type proteins. J Am Chem Soc. 2003;125(20):6078–6084. doi: 10.1021/ja027967i. [DOI] [PubMed] [Google Scholar]
- 147.Yoon T, Cowan JA. Frataxin-mediated iron delivery to ferrochelatase in the final step of heme biosynthesis. J Biol Chem. 2004;279(25):25943–25946. doi: 10.1074/jbc.C400107200. [DOI] [PubMed] [Google Scholar]
- 148.Bekri S, Kispal G, Lange H, Fitzsimons E, Tolmie J, Lill R, Bishop DF. Human ABC7 transporter: gene structure and mutation causing X-linked sideroblastic anemia with ataxia with disruption of cytosolic iron-sulfur protein maturation. Blood. 2000;96(9):3256–3264. [PubMed] [Google Scholar]
- 149.Csere P, Lill R, Kispal G. Identification of a human mitochondrial ABC transporter, the functional orthologue of yeast Atm1p. FEBS Letters. 1998;441(2):266–270. doi: 10.1016/s0014-5793(98)01560-9. [DOI] [PubMed] [Google Scholar]
- 150.Shimada Y, Okuno S, Kawai A, Shinomiya H, Saito A, Suzuki M, Omori Y, Nishino N, Kanemoto N, Fujiwara T, Horie M, Takahashi E. Cloning and chromosomal mapping of a novel ABC transporter gene (hABC7), a candidate for X-linked sideroblastic anemia with spinocerebellar ataxia. J Human Genet. 1998;43(2):115. doi: 10.1007/s100380050051. [DOI] [PubMed] [Google Scholar]
- 151.Lill R, Muhlenhoff U. Iron-sulfur protein biogenesis in eukaryotes: components and mechanisms. Annu Rev Cell Dev Biol. 2006;22(1):457–486. doi: 10.1146/annurev.cellbio.22.010305.104538. [DOI] [PubMed] [Google Scholar]
- 152.Tong W-H, Rouault TA. Functions of mitochondrial ISCU and cytosolic ISCU in mammalian iron-sulfur cluster biogenesis and iron homeostasis. Cell Metab. 2006;3(3):199–210. doi: 10.1016/j.cmet.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 153.Condo I, Ventura N, Malisan F, Rufini A, Tomassini B, Testi R. In vivo maturation of human frataxin. Hum Mol Genet. 2007;16(13):1534–1540. doi: 10.1093/hmg/ddm102. [DOI] [PubMed] [Google Scholar]
- 154.Nakahashi Y, Taketani S, Okuda M, Inoue K, Tokunaga R. Molecular cloning and sequence analysis of cDNA encoding human ferrochelatase. Biochemical and Biophysical Research Communications. 1990;173(2):748–755. doi: 10.1016/s0006-291x(05)80099-3. [DOI] [PubMed] [Google Scholar]
- 155.Sellers VM, Wu CK, Dailey TA, Dailey HA. Human ferrochelatase: characterization of substrate-iron binding and proton-abstracting residues. Biochemistry. 2001;40(33):9821–9827. doi: 10.1021/bi010012c. [DOI] [PubMed] [Google Scholar]
- 156.Wu C-K, Dailey HA, Rose JP, Burden A, Sellers VM, Wang B-C. The 2.0 A structure of human ferrochelatase, the terminal enzyme of heme biosynthesis. Nat Struct Mol Biol. 2001;8(2):156–160. doi: 10.1038/84152. [DOI] [PubMed] [Google Scholar]
- 157.Ferreira GC, Franco R, Lloyd SG, Moura I, Moura JJG, Huynh BH. Structure and function of ferrochelatase. J Bioenerg Biomembr. 1995;27(2):221–229. doi: 10.1007/BF02110037. [DOI] [PubMed] [Google Scholar]
- 158.Stryer L. Biochemistry. 4. W. H. Freeman and Company; New York: 1995. [Google Scholar]
- 159.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Caspi R, Foerster H, Fulcher CA, Kaipa P, Krummenacker M, Latendresse M, Paley S, Rhee SY, Shearer AG, Tissier C, Walk TC, Zhang P, Karp PD. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucl Acids Res. 2008;36(suppl1):D623–631. doi: 10.1093/nar/gkm900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ferreira GC. Heme biosythesis: biochemistry, molecular biology, and relationship to disease. J Bioenerg Biomembr. 1995;27(2):147–150. doi: 10.1007/BF02110029. [DOI] [PubMed] [Google Scholar]
- 162.Krishnamurthy PC, Du G, Fukuda Y, Sun D, Sampath J, Mercer KE, Wang J, Sosa-Pineda B, Murti KG, Schuetz JD. Identification of a mammalian mitochondrial porphyrin transporter. Nature. 2006;443(7111):586–589. doi: 10.1038/nature05125. [DOI] [PubMed] [Google Scholar]
- 163.Dunn LL, Rahmanto YS, Richardson DR. Iron uptake and metabolism in the new millennium. Trends Cell Biol. 2007;17(2):93–100. doi: 10.1016/j.tcb.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 164.Ferreira GC, Gong J. 5-Aminolevulinate synthase and the first step of heme biosythesis. J Bioenerg Biomembr. 1995;27(2):151–159. doi: 10.1007/BF02110030. [DOI] [PubMed] [Google Scholar]
- 165.Bishop DF. Two different genes encode {delta}-aminolevulinate synthase in humans: nucleotide sequences of cDNAs for the housekeeping and erythroid genes. Nucl Acids Res. 1990;18(23):7187–7188. doi: 10.1093/nar/18.23.7187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Yamamoto M, Hayashi N, Kikuchi G. Translational inhibition by heme of the synthesis of hepatic [delta]-aminolevulinate synthase in a cell-free system. Biochem Biophys Res Commun. 1983;115(1):225–231. doi: 10.1016/0006-291x(83)90993-2. [DOI] [PubMed] [Google Scholar]
- 167.Yamamoto M, Hayashi N, Kikuchi G. Evidence for the transcriptional inhibition by heme of the synthesis of [delta]-aminolevulinate synthase in rat liver. Biochem Biophys Res Commun. 1982;105(3):985–990. doi: 10.1016/0006-291x(82)91067-1. [DOI] [PubMed] [Google Scholar]
- 168.Yamauchi K, Hayashi N, Kikuchi G. Translocation of delta-aminolevulinate synthase from the cytosol to the mitochondria and its regulation by hemin in the rat liver. J Biol Chem. 1980;255(4):1746–1751. [PubMed] [Google Scholar]
- 169.Munakata H, Sun J-Y, Yoshida K, Nakatani T, Honda E, Hayakawa S, Furuyama K, Hayashi N. Role of the heme regulatory motif in the heme-mediated inhibition of mitochondrial import of 5-aminolevulinate synthase. J Biochem. 2004;136(2):233–238. doi: 10.1093/jb/mvh112. [DOI] [PubMed] [Google Scholar]
- 170.Dailey TA, Woodruff JH, Dailey HA. Examination of mitochondrial protein targeting of haem synthetic enzymes: in vivo identification of three functional haem-responsive motifs in 5-aminolaevulinate synthase. Biochem J. 2005;386(2):381–386. doi: 10.1042/BJ20040570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Yoshida T, Biro P, Cohen T, Muller RM, Shibahara S. Human heme oxygenase cDNA and induction of its mRNA by hemin. Eur J Biochem. 1988;171(3):457–461. doi: 10.1111/j.1432-1033.1988.tb13811.x. [DOI] [PubMed] [Google Scholar]
- 172.Keyse SM, Applegate LA, Tromvoukis Y, Tyrrell RM. Oxidant stress leads to transcriptional activation of the human heme oxygenase gene in cultured skin fibroblasts. Mol Cell Biol. 1990;10(9):4967–4969. doi: 10.1128/mcb.10.9.4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Applegate LA, Luscher P, Tyrrell RM. Induction of heme oxygenase: a general response to oxidant stress in cultured mammalian cells. Cancer Res. 1991;51(3):974–978. [PubMed] [Google Scholar]
- 174.Ding Y, Zhang YZ, Furuyama K, Ogawa K, Igarashi K, Shibahara S. Down-regulation of heme oxygenase-2 is associated with the increased expression of heme oxygenase-1 in human cell lines. FEBS Journal. 2006;273(23):5333–5346. doi: 10.1111/j.1742-4658.2006.05526.x. [DOI] [PubMed] [Google Scholar]
- 175.Haile DJM. Regulation of genes of iron metabolism by the iron-response proteins. Am J Med Sci. 1999;318(4):230. doi: 10.1097/00000441-199910000-00003. [DOI] [PubMed] [Google Scholar]
- 176.Henderson BR, Menotti E, Bonnard C, Kuhn LC. Optimal sequence and structure of iron-responsive elements. Selection of RNA stem-loops with high affinity for iron regulatory factor. J Biol Chem. 1994;269(26):17481–17489. [PubMed] [Google Scholar]
- 177.Hirling H, Emery-Goodman A, Thompson N, Neupert B, Seiser C, Kuhn LC. Expression of active iron regulatory factor from a full length human cDNA by in vitro transcription/translation. Nucl Acids Res. 1992;20(1):33–39. doi: 10.1093/nar/20.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Rouault TA, Tang CK, Kaptain S, Burgess WH, Haile DJ, Samaniego F, McBride OW, Harford JB, Klausner RD. Cloning of the cDNA encoding an RNA regulatory protein-the human iron-responsive element-binding protein. Proc Nat Acad Sci. 1990;87(20):7958–7962. doi: 10.1073/pnas.87.20.7958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Addess KJ, Basilion JP, Klausner RD, Rouault TA, Pardi A. Structure and dynamics of the iron responsive element RNA: implications for binding of the RNA by iron regulatory binding proteins. J Mol Biol. 1997;274(1):72–83. doi: 10.1006/jmbi.1997.1377. [DOI] [PubMed] [Google Scholar]
- 180.Wallander ML, Leibold EA, Eisenstein RS. Molecular control of vertebrate iron homeostasis by iron regulatory proteins. Biochim Biophys Acta. 2006;1763(7):668–689. doi: 10.1016/j.bbamcr.2006.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Menotti E, Henderson BR, Kuhn LC. Translational regulation of mRNAs with distinct IRE sequences by iron regulatory proteins 1 and 2. J Biol Chem. 1998;273(3):1821–1824. doi: 10.1074/jbc.273.3.1821. [DOI] [PubMed] [Google Scholar]
- 182.Henderson BR, Menotti E, K’hn LC. Iron regulatory proteins 1 and 2 bind distinct sets of RNA target sequences. J Biol Chem. 1996;271(9):4900–4908. doi: 10.1074/jbc.271.9.4900. [DOI] [PubMed] [Google Scholar]
- 183.Butt J, Kim H-Y, Basilion JP, Cohen S, Iwai K, Philpott CC, Altschul S, Klausner RD, Rouault TA. Differences in the RNA binding sites of iron regulatory proteins and potential target diversity. Proc Nat Acad Soc. 1996;93(9):4345–4349. doi: 10.1073/pnas.93.9.4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Meyron-Holtz EG, Ghosh MC, Iwai K, LaVaute T, Brazzolotto X, Berger UV, Land W, Ollivierre-Wilson H, Grinberg A, Love P, Rouault TA. Genetic ablations of iron regulatory proteins 1 and 2 reveal why iron regulatory protein 2 dominates iron homeostasis. EBMO Journal. 2004;23:386–395. doi: 10.1038/sj.emboj.7600041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Haile DJ, Rouault TA, Tang CK, Chin J, Harford JB, Klausner RD. Reciprocal control of RNA-binding and aconitase activity in the regulation of the iron-responsive element binding protein: role of the iron-sulfur cluster. Proc Nat Acad Sci. 1992;89(16):7536–7540. doi: 10.1073/pnas.89.16.7536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Kaptain S, Downey WE, Tang C, Philpott C, Haile D, Orloff DG, Harford JB, Rouault TA, Klausner RD. A regulated RNA binding protein also possesses aconitase activity. Proc Nat Acad Soc. 1991;88(22):10109–10113. doi: 10.1073/pnas.88.22.10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Volz K. The functional duality of iron regulatory protein 1. Curr Opin Struct Biol. 2008;18(1):106–111. doi: 10.1016/j.sbi.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Tang CK, Chin J, Harford JB, Klausner RD, Rouault TA. Iron regulates the activity of the iron-responsive element binding protein without changing its rate of synthesis or degradation. J Biol Chem. 1992;267(34):24466–24470. [PubMed] [Google Scholar]
- 189.Gray NK, Quick S, Goossen B, Constable A, Hirling H, Kuhn LC, Hentze MW. Recombinant iron-regulatory factor functions as an iron-responsive-element-binding protein, a translational repressor and an aconitase. A functional assay for translational repression and direct demonstration of the iron switch. Eur J Biochem. 1993;218(2):657–667. doi: 10.1111/j.1432-1033.1993.tb18420.x. [DOI] [PubMed] [Google Scholar]
- 190.Haile DJ, Rouault TA, Harford JB, Kennedy MC, Blondin GA, Beinert H, Klausner RD. Cellular regulation of the iron-responsive element binding protein: disassembly of the cubane iron-sulfur cluster results in high-affinity RNA binding. Proc Nat Acad Sci. 1992;89(24):11735–11739. doi: 10.1073/pnas.89.24.11735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Pantopoulos K, Mueller S, Atzberger A, Ansorge W, Stremmel W, Hentze MW. Differences in the Regulation of Iron Regulatory Protein-1 (IRP-1) by Extra- and Intracellular Oxidative Stress. J Biol Chem. 1997;272(15):9802–9808. doi: 10.1074/jbc.272.15.9802. [DOI] [PubMed] [Google Scholar]
- 192.Eisenstein RS, Tuazon PT, Schalinske KL, Anderson SA, Traugh JA. Iron-responsive element-binding protein. Phosphorylation by protein kinase C. J Biol Chem. 1993;268(36):27363–27370. [PubMed] [Google Scholar]
- 193.Schalinske KL, Eisenstein RS. Phosphorylation and activation of both iron regulatory proteins 1 and 2 in HL-60 cells. J Biol Chem. 1996;271(12):7168–7176. doi: 10.1074/jbc.271.12.7168. [DOI] [PubMed] [Google Scholar]
- 194.Eisenstein RS. Iron regulatory proteins and the molecular control of iron metabolism. Annu Rev Nutr. 2000;20(1):627–662. doi: 10.1146/annurev.nutr.20.1.627. [DOI] [PubMed] [Google Scholar]
- 195.Samaniego F, Chin J, Iwai K, Rouault TA, Klausner RD. Molecular characterization of a second iron-responsive element binding protein, iron regulatory protein 2. Structure, function, and post-translational regulation. J Biol Chem. 1994;269(49):30904–30910. [PubMed] [Google Scholar]
- 196.Wang J, Chen G, Muckenthaler M, Galy B, Hentze MW, Pantopoulos K. Iron-mediated degradation of IRP2, an unexpected pathway involving a 2-oxoglutarate-dependent oxygenase activity. Mol Cell Biol. 2004;24(3):954–965. doi: 10.1128/MCB.24.3.954-965.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Iwai K, Drake SK, Wehr NB, Weissman AM, LaVaute T, Minato N, Klausner RD, Levine RL, Rouault TA. Iron-dependent oxidation, ubiquitination, and degradation of iron regulatory protein 2: Implications for degradation of oxidized proteins. Proc Nat Acad Soc. 1998;95(9):4924–4928. doi: 10.1073/pnas.95.9.4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Hanson ES, Rawlins ML, Leibold EA. Oxygen and Iron Regulation of Iron Regulatory Protein 2. J Biol Chem. 2003;278(41):40337–40342. doi: 10.1074/jbc.M302798200. [DOI] [PubMed] [Google Scholar]
- 199.Guo B, Phillips JD, Yu Y, Leibold EA. Iron regulates the intracellular degradation of iron regulatory protein 2 by the proteasome. J Biol Chem. 1995;270(37):21645–21651. doi: 10.1074/jbc.270.37.21645. [DOI] [PubMed] [Google Scholar]
- 200.Wallander ML, Zumbrennen KB, Rodansky ES, Romney SJ, Leibold EA. Iron-independent phosphorylation of iron regulatory protein 2 regulates ferritin during the cell cycle. J Biol Chem. 2008 doi: 10.1074/jbc.M803005200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Koeller DM, Casey JL, Hentze MW, Gerhardt EM, Chan L-NL, Klausner RD, Harford JB. A cytosolic Protein Binds to Structural Elements within the Iron Regulatory Region of the Transferrin Receptor mRNA. Proc Nat Acad Sci. 1989;86(10):3574–3578. doi: 10.1073/pnas.86.10.3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Caughman SW, Hentze MW, Rouault TA, Harford JB, Klausner RD. The ironresponsive element is the single element responsible for iron-dependent translational regulation of ferritin biosynthesis. Evidence for function as the binding site for a translational repressor. J Biol Chem. 1988;263(35):19048–19052. [PubMed] [Google Scholar]
- 203.Casey JL, Hentze MW, Koeller DM, Caughman SW, Rouault TA, Klausner RD, Harford JB. Iron-responsive elements: regulatory RNA sequences that control mRNA levels and translation. Science. 1988;240(4854):924–928. doi: 10.1126/science.2452485. [DOI] [PubMed] [Google Scholar]
- 204.Tsuji Y, Ayaki H, Whitman SP, Morrow CS, Torti SV, Torti FM. Coordinate transcriptional and translational regulation of ferritin in response to oxidative stress. Mol Cell Biol. 2000;20(16):5818–5827. doi: 10.1128/mcb.20.16.5818-5827.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Kwak EL, Larochelle DA, Beaumont C, Torti SV, Torti FM. Role for NF-kappa B in the regulation of ferritin H by tumor necrosis factor-alpha. J Biol Chem. 1995;270(25):15285–93. doi: 10.1074/jbc.270.25.15285. [DOI] [PubMed] [Google Scholar]
- 206.Torti SV, Kwak EL, Miller SC, Miller LL, Ringold GM, Myambo KB, Young AP, Torti FM. The molecular cloning and characterization of murine ferritin heavy chain, a tumor necrosis factor-inducible gene. J Biol Chem. 1988;263(25):12638–44. [PubMed] [Google Scholar]
- 207.Sanchez M, Galy B, Muckenthaler MU, Hentze MW. Iron-regulatory proteins limit hypoxia-inducible factor-2[alpha] expression in iron deficiency. Nat Struct Mol Biol. 2007;14(5):420–426. doi: 10.1038/nsmb1222. [DOI] [PubMed] [Google Scholar]
- 208.Liu X-b, Hill P, Haile DJ. Role of the ferroportin iron-responsive element in iron and nitric oxide dependent gene regulation. Blood Cell Mol Dis. 2002;29(3):315–326. doi: 10.1006/bcmd.2002.0572. [DOI] [PubMed] [Google Scholar]
- 209.Lymboussaki A, Pignatti E, Montosi G, Garuti C, Haile DJ, Pietrangelo A. The role of the iron responsive element in the control of ferroportin1/IREG1/MTP1 gene expression. J Hepatol. 2003;39(5):710–715. doi: 10.1016/s0168-8278(03)00408-2. [DOI] [PubMed] [Google Scholar]
- 210.Galy B, Ferring-Appel D, Kaden S, Grone H-J, Hentze MW. Iron Regulatory Proteins Are Essential for Intestinal Function and Control Key Iron Absorption Molecules in the Duodenum. Cell Metabolism. 2008;7(1):79–85. doi: 10.1016/j.cmet.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 211.Yang F, Wang X, Haile DJ, Piantadosi CA, Ghio AJ. Iron increases expression of iron-export protein MTP1 in lung cells. Am J Physiol Lung Cell Mol Physiol. 2002;283(5):L932–939. doi: 10.1152/ajplung.00114.2002. [DOI] [PubMed] [Google Scholar]
- 212.Lee PL, Gelbart T, West C, Halloran C, Beutler E. The human Nramp2 gene: characterization of the gene structure, alternative splicing, promoter region and polymorphisms. Blood Cells Mol Dis. 1998;24(2):199–215. doi: 10.1006/bcmd.1998.0186. [DOI] [PubMed] [Google Scholar]
- 213.Hubert N, Hentze MW. Previously uncharacterized isoforms of divalent metal transporter (DMT)-1: Implications for regulation and cellular function. Proc Nat Acad Soc. 2002;99(19):12345–12350. doi: 10.1073/pnas.192423399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Mackenzie B, Takanaga H, Hubert N, Rolfs A, Hediger MA. Functional properties of multiple isoforms of human divalent metal-ion transporter 1 (DMT1) Biochem J. 2007;403(1):59–69. doi: 10.1042/BJ20061290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Lam-Yuk-Tseung S, Gros P. Distinct targeting and recycling properties of two isoforms of the iron transporter DMT1 (NRAMP2, Slc11A2) Biochemistry. 2006;45(7):2294–2301. doi: 10.1021/bi052307m. [DOI] [PubMed] [Google Scholar]
- 216.Tabuchi M, Tanaka N, Nishida-Kitayama J, Ohno H, Kishi F. Alternative splicing regulates the subcellular localization of divalent metal transporter 1 isoforms. Mol Biol Cell. 2002;13(12):4371–4387. doi: 10.1091/mbc.E02-03-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Gunshin H, Allerson CR, Polycarpou-Schwarz M, Rofts A, Rogers JT, Kishi F, Hentze MW, Rouault TA, Andrews NC, Hediger MA. Iron-dependent regulation of the divalent metal ion transporter. FEBS Letters. 2001;509(2):309–316. doi: 10.1016/s0014-5793(01)03189-1. [DOI] [PubMed] [Google Scholar]
- 218.Fleming RE, Migas MC, Zhou X, Jiang J, Britton RS, Brunt EM, Tomatsu S, Waheed A, Bacon BR, Sly WS. Mechanism of increased iron absorption in murine model of hereditary hemochromatosis: Increased duodenal expression of the iron transporter DMT1. Proc Nat Acad Sci. 1999;96(6):3143–3148. doi: 10.1073/pnas.96.6.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Zoller H, Theurl I, Koch R, Kaser A, Weiss G. Mechanisms of iron mediated regulation of the duodenal iron transporters divalent metal transporter 1 and ferroportin 1. Blood Cell Mol Dis. 2002;29(3):488–497. doi: 10.1006/bcmd.2002.0587. [DOI] [PubMed] [Google Scholar]
- 220.Krause A, Neitz S, Margert H-J, Schulz A, Forssman W-G, Schulz-Knappe P, Adermann K. LEAP-1, a novel highly disulfide-bonded human peptide, exhibits antimicrobial activity. FEBS letters. 2000;480(2):147–150. doi: 10.1016/s0014-5793(00)01920-7. [DOI] [PubMed] [Google Scholar]
- 221.Park CH, Valore EV, Waring AJ, Ganz T. Hepcidin, a urinary antimicrobial peptide synthesized in the liver. J Biol Chem. 2001;276(11):7806–7810. doi: 10.1074/jbc.M008922200. [DOI] [PubMed] [Google Scholar]
- 222.De Domenico I, Ward DM, Langelier C, Vaughn MB, Nemeth E, Sundquist WI, Ganz T, Musci G, Kaplan J. The molecular mechanism of hepcidin-mediated ferroportin down-regulation. Mol Biol Cell. 2007;18(7):2569–2578. doi: 10.1091/mbc.E07-01-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Chaston T, Chung B, Mascarenhas M, Marks J, Patel B, Srai SK, Sharp P. Evidence for differential effects of hepcidin In macrophages and intestinal epithelial cells. Gut. 2007:gut.2007.131722. doi: 10.1136/gut.2007.131722. [DOI] [PubMed] [Google Scholar]
- 224.Delaby C, Pilard N, Goncalves AS, Beaumont C, Canonne-Hergaux F. Presence of the iron exporter ferroportin at the plasma membrane of macrophages is enhanced by iron loading and down-regulated by hepcidin. Blood. 2005;106(12):3979–3984. doi: 10.1182/blood-2005-06-2398. [DOI] [PubMed] [Google Scholar]
- 225.Nemeth E, Tuttle MS, Powelson J, Vaughn MB, Donovan A, Ward DM, Ganz T, Kaplan J. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science. 2004;306(5704):2090–2093. doi: 10.1126/science.1104742. [DOI] [PubMed] [Google Scholar]
- 226.Mena NP, Esparza A, Tapia V, Valdes P, Nunez MT. HEPCIDIN INHIBITS APICAL IRON UPTAKE IN INTESTINAL CELLS. Am J Physiol Gastrointest Liver Physiol. 2007:00122.2007. doi: 10.1152/ajpgi.00122.2007. [DOI] [PubMed] [Google Scholar]
- 227.De Domenico I, Ward DM, di Patti MC, Jeong SY, David S, Musci G, Kaplan J. Ferroxidase activity is required for the stability of cell surface ferroportin in cells expressing GPI-ceruloplasmin. EBMO Journal. 2007;26:2823–2831. doi: 10.1038/sj.emboj.7601735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Babitt JL, Huang FW, Wrighting DM, Xia Y, Sidis Y, Samad TA, Campagna JA, Chung RT, Schneyer AL, Woolf CJ, Andrews NC, Lin HY. Bone morphogenetic protein signaling by hemojuvelin regulates hepcidin expression. Nat Genet. 2006;38(5):531–539. doi: 10.1038/ng1777. [DOI] [PubMed] [Google Scholar]
- 229.Babitt JL, Huang FW, Xia Y, Sidis Y, Andrews NC, Lin HY. Modulation of bone morphogenetic protein signaling in vivo regulates systemic iron balance. J Clin Invest. 2007;117(7):1933–1939. doi: 10.1172/JCI31342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Babitt JL, Zhang Y, Samad TA, Xia Y, Tang J, Campagna JA, Schneyer AL, Woolf CJ, Lin HY. Repulsive Guidance Molecule (RGMa), a DRAGON homologue, is a bone morphogenetic protein co-receptor. J Biol Chem. 2005;280(33):29820–29827. doi: 10.1074/jbc.M503511200. [DOI] [PubMed] [Google Scholar]
- 231.Truksa J, Peng H, Lee P, Beutler E. Bone morphogenetic proteins 2, 4, and 9 stimulate murine hepcidin 1 expression independently of Hfe, transferrin receptor 2 (Tfr2), and IL-6. Proc Nat Acad Sci. 2006;103(27):10289–10293. doi: 10.1073/pnas.0603124103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Xia Y, Babitt JL, Sidis Y, Chung RT, Lin HY. Hemojuvelin regulates hepcidin expression via a selective subset of BMP ligands and receptors independently of neogenin. Blood. 2008:blood-2007-09–111567. doi: 10.1182/blood-2007-09-111567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Lin L, Goldberg YP, Ganz T. Competitive regulation of hepcidin mRNA by soluble and cell-associated hemojuvelin. Blood. 2005;106(8):2884–2889. doi: 10.1182/blood-2005-05-1845. [DOI] [PubMed] [Google Scholar]
- 234.Silvestri L, Pagani A, Fazi C, Gerardi G, Levi S, Arosio P, Camaschella C. Defective targeting of hemojuvelin to plasma membrane is a common pathogenetic mechanism in juvenile hemochromatosis. Blood. 2007;109(10):4503–4510. doi: 10.1182/blood-2006-08-041004. [DOI] [PubMed] [Google Scholar]
- 235.Niederkofler V, Salie R, Arber S. Hemojuvelin is essential for dietary iron sensing, and its mutation leads to severe iron overload. J Clin Invest. 2005;115(8):2180. doi: 10.1172/JCI25683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Papanikolaou G, Samuels ME, Ludwig EH, MacDonald MLE, Franchini PL, Dube M-P, Andres L, MacFarlane J, Sakellaropoulos N, Politou M, Nemeth E, Thompson J, Risler JK, Zaborowska C, Babakaiff R, Radomski CC, Pape TD, Davidas O, Christakis J, Brissot P, Lockitch G, Ganz T, Hayden MR, Goldberg YP. Mutations in HFE2 cause iron overload in chromosome 1q-linked juvenile hemochromatosis. Nat Genet. 2004;36(1):77–82. doi: 10.1038/ng1274. [DOI] [PubMed] [Google Scholar]
- 237.Huang FW, Pinkus JL, Pinkus GS, Fleming MD, Andrews NC. A mouse model of juvenile hemochromatosis. J Clin Invest. 2005;115(8):2187. doi: 10.1172/JCI25049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Vujic Spasic M, Kiss J, Herrmann T, Galy B, Martinache S, Stolte J, Grone H-J, Stremmel W, Hentze MW, Muckenthaler MU. Hfe acts in hepatocytes to prevent hemochromatosis. Cell Metab. 2008;7(2):173–178. doi: 10.1016/j.cmet.2007.11.014. [DOI] [PubMed] [Google Scholar]
- 239.Bridle KR, Frazer DM, Wilkins SJ, Dixon JL, Purdie DM, Crawford DHG, Subramaniam VN, Powell LW, Anderson GJ, Ramm GA. Disrupted hepcidin regulation in HFE-associated haemochromatosis and the liver as a regulator of body iron homoeostasis. The Lancet. 2003;361(9358):669–673. doi: 10.1016/S0140-6736(03)12602-5. [DOI] [PubMed] [Google Scholar]
- 240.Kaelin WG, Jr, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30(4):393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 241.Kaelin WG. Proline hydroxylation and gene expression. Annu Rev Biochem. 2005;74(1):115–128. doi: 10.1146/annurev.biochem.74.082803.133142. [DOI] [PubMed] [Google Scholar]
- 242.Yan Q, Bartz S, Mao M, Li L, Kaelin WG., Jr The hypoxia-inducible factor 2{alpha} N-terminal and C-terminal transactivation domains cooperate to promote renal tumorigenesis in vivo. Mol Cell Biol. 2007;27(6):2092–2102. doi: 10.1128/MCB.01514-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Lee PJ, Jiang B-H, Chin BY, Iyer NV, Alam J, Semenza GL, Choi AMK. Hypoxia-inducible factor-1 mediates transcriptional activation of the heme oxygenase-1 gene in response to hypoxia. J Biol Chem. 1997;272(9):5375–5381. [PubMed] [Google Scholar]
- 244.Peyssonnaux C, Zinkernagel AS, Schuepbach RA, Rankin E, Vaulont S, Haase VH, Nizet V, Johnson RS. Regulation of iron homeostasis by the hypoxia-inducible transcription factors (HIFs) J Clin Invest. 2007;117(7):1926–1932. doi: 10.1172/JCI31370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Mukhopadhyay CK, Mazumder B, Fox PL. Role of hypoxia-inducible factor-1 in transcriptional activation of ceruloplasmin by iron deficiency. J Biol Chem. 2000;275(28):21048–21054. doi: 10.1074/jbc.M000636200. [DOI] [PubMed] [Google Scholar]
- 246.Bianchi L, Tacchini L, Cairo G. HIF-1-mediated activation of transferrin receptor gene transcription by iron chelation. Nucl Acids Res. 1999;27(21):4223–4227. doi: 10.1093/nar/27.21.4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Oktay Y, Dioum E, Matsuzaki S, Ding K, Yan L-J, Haller RG, Szweda LI, Garcia JA. Hypoxia-inducible factor 2{alpha} regulates expression of the mitochondrial aconitase chaperone protein frataxin. J Biol Chem. 2007;282(16):11750–11756. doi: 10.1074/jbc.M611133200. [DOI] [PubMed] [Google Scholar]
- 248.Han AP, Yu C, Lu L, Fujiwara Y, Browne C, Chin G, Fleming M, Leboulch P, Orkin SH, Chen JJ. Heme-regulated eIF2alpha kinase (HRI) is required for translational regulation and survival of erythroid precursors in iron deficiency. EMBO J. 2001;20(23):6909–6918. doi: 10.1093/emboj/20.23.6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Crosby JS, Lee K, London IM, Chen JJ. Erythroid expression of the heme-regulated eIF-2 alpha kinase. Mol Cell Biol. 1994;14(6):3906–3914. doi: 10.1128/mcb.14.6.3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Yu X, Kong Y, Dore LC, Abdulmalik O, Katein AM, Suiping Z, Choi JK, Gell D, Mackay JP, Gow AJ, Weiss MJ. An erythroid chaperone that facilitates folding of alpha-globin subunits for hemoglobin synthesis. J Clin Invest. 2007;117(7):1856–1865. doi: 10.1172/JCI31664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Lesley J, Hyman R, Schulte R, Trotter J. Expression of transferrin receptor on murine hematopoietic progenitors. Cell Immunol. 1984;83(1):14–25. doi: 10.1016/0008-8749(84)90220-x. [DOI] [PubMed] [Google Scholar]
- 252.Iacopetta BJ, Morgan EH, Yeoh GCT. Transferrin receptors and iron uptake during erythroid cell development. Biochim Biophys Acta. 1982;687(2):204–210. doi: 10.1016/0005-2736(82)90547-8. [DOI] [PubMed] [Google Scholar]
- 253.Harding C, Heuser J, Stahl P. Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J Cell Biol. 1983;97(2):329–339. doi: 10.1083/jcb.97.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Meyron-Holtz EG, Fibach E, Gelvan D, Konijn AM. Binding and uptake of exogenous isoferritins by cultured human eythroid precursor cells. Br J Haematol. 1994;86(3):635–641. doi: 10.1111/j.1365-2141.1994.tb04797.x. [DOI] [PubMed] [Google Scholar]
- 255.Leimberg MJ, Prus E, Konijn AM, Fibach E. Macrophages function as a ferritin iron source for cultured human erythroid precursors. J Cell Biochem. 2008;103(4):1211–1218. doi: 10.1002/jcb.21499. [DOI] [PubMed] [Google Scholar]
- 256.Vaisman B, Fibach E, Konijn AM. Utilization of intracellular ferritin iron for hemoglobin sythesis in developing human erythroid precursors. Blood. 1997;90(2):831–838. [PubMed] [Google Scholar]
- 257.Meyron-Holtz EG, Vaisman B, Cabantchik ZI, Fibach E, Rouault TA, Hershko C, Konijn AM. Regulation of intracellular iron metabolism in human erythroid precursors by internalized extracellular ferritin. Blood Cells Mol Dis. 1999;94(9):3205–3211. [PubMed] [Google Scholar]
- 258.Bhasker CR, Burgiel G, Neupert B, Emery-Goodman A, Kuhn LC, May BK. The putative iron-responsive element in the human erythroid 5- aminolevulinate synthase mRNA mediates translational control. J Biol Chem. 1993;268(17):12699–12705. [PubMed] [Google Scholar]
- 259.Cox TC, Bawden MJ, Martin A, May BK. Human erythroid 5-aminolevulinate sythase: promoter analysis and identification of an iron-responsive element in the mRNA. EMBO J. 1991;10(7):1891–1902. doi: 10.1002/j.1460-2075.1991.tb07715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Ponka P. Cell biology of heme. Am J Med Sci. 1999;318(4):241–256. doi: 10.1097/00000441-199910000-00004. [DOI] [PubMed] [Google Scholar]
- 261.Cox TM, O’Donnell MW, Aisen P, London IM. Hemin inhibits internalization of transferrin by reticulocytes and promotes phosphorylation of the membrane transferrin receptor. Proc Nat Acad Sci. 1985;82(15):5170–5174. doi: 10.1073/pnas.82.15.5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Ponka P, Schulman HM. Regulation of heme synthesis in erythroid cells: hemin inhibits transferrin iron utilization but not protoporphyrin synthesis. Blood. 1985;65(4):850–857. [PubMed] [Google Scholar]
- 263.Canonne-Hergaux F, Zhang A-S, Ponka P, Gros P. Characterization of the iron transporter DMT1 (NRAMP2/DCT1) in red blood cells of normal and anemic mk/mk mice. Blood. 2001;98(13):3823–3830. doi: 10.1182/blood.v98.13.3823. [DOI] [PubMed] [Google Scholar]
- 264.Kato J, Kobune M, Ohkubo S, Fujikawa K, Tanaka M, Takimoto R, Takada K, Takahari D, Kawano Y, Kohgo Y, Niitsu Y. Iron/IRP-1-dependent regulation of mRNA expression for transferrin receptor, DMT1 and ferritin during human erythroid differentiation. Exp Hematol. 2007;35(6):879–887. doi: 10.1016/j.exphem.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 265.Kristiansen M, Graversen JH, Jacobsen C, Sonne O, Hoffman HJ, Law SK, Moestrup SK. Identification of the haemoglobin scavenger receptor. Nature. 2001;409(6817):198–201. doi: 10.1038/35051594. [DOI] [PubMed] [Google Scholar]
- 266.Fabriek BO, Dijkstra CD, van den Berg TK. The macrophage scavenger receptor CD163. Immunobiology. 2005;210(2–4):153–160. doi: 10.1016/j.imbio.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 267.Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson JD. Molecular Biology of the Cell. 3. Garland Publishing Inc; New York: 1994. p. 1294. [Google Scholar]
- 268.Graversen JH, Madsen M, Moestrup SK. CD163: a signal receptor scavenging haptoglobin-hemoglobin complexes from plasma. Int J Biochem Cell Biol. 2002;34(4):309–314. doi: 10.1016/s1357-2725(01)00144-3. [DOI] [PubMed] [Google Scholar]
- 269.Madsen M, Graversen JH, Moestrup SK. Haptoglobin and CD163: captor and receptor gaiting hemoglobin to macrophage lysosomes. Redox Rep. 2001;6(6):386–388. doi: 10.1179/135100001101536490. [DOI] [PubMed] [Google Scholar]
- 270.Droste A, Sorg C, Högger P. Shedding of CD163, a novel regulatory mechanism for a member of the scavenger receptor cysteine-rich family. Biochem Biophys Res Commun. 1999;256(1):110–113. doi: 10.1006/bbrc.1999.0294. [DOI] [PubMed] [Google Scholar]
- 271.Knutson M, Weslling-Resnick M. Iron metabolism in the reticuloendothelial system. Crit Rev Biochem Mol Biol. 2003;38(1):61–88. doi: 10.1080/713609210. [DOI] [PubMed] [Google Scholar]
- 272.Soe-Lin S, Sheftel AD, Wasyluk B, Ponka P. Nramp1 equips macrophages for efficient iron recycling. Exp Hematol. 2008 doi: 10.1016/j.exphem.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 273.Fritsche G, Nairz M, Theurl I, Mair S, Bellmann-Weiler R, Barton HC, Weiss Gn. Modulation of macrophage iron transport by Nramp1 (Slc11a1) Immunobiology. 2008;212(9–10):751–757. doi: 10.1016/j.imbio.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 274.Fraser HB, Hirsh AE, Steinmetz LM, Scharfe C, Feldman MW. Evolutionary rate in the protein interaction network. Science. 2002;296(5568):750–2. doi: 10.1126/science.1068696. [DOI] [PubMed] [Google Scholar]
- 275.Han JD, Bertin N, Hao T, Goldberg DS, Berriz GF, Zhang LV, Dupuy D, Walhout AJ, Cusick ME, Roth FP, Vidal M. Evidence for dynamically organized modularity in the yeast protein-protein interaction network. Nature. 2004;430(6995):88–93. doi: 10.1038/nature02555. [DOI] [PubMed] [Google Scholar]
- 276.Kitano H, Funahashi A, Matsuoka Y, Oda K. Using process diagrams for the graphical representation of biological networks. Nat Biotech. 2005;23(8):961–966. doi: 10.1038/nbt1111. [DOI] [PubMed] [Google Scholar]
- 277.Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, Arkin AP, Bornstein BJ, Bray D, Cornish-Bowden A, Cuellar AA, Dronov S, Gilles ED, Ginkel M, Gor V, Goryanin II, Hedley WJ, Hodgman TC, Hofmeyr JH, Hunter PJ, Juty NS, Kasberger JL, Kremling A, Kummer U, Le Novere N, Loew LM, Lucio D, Mendes P, Minch E, Mjolsness ED, Nakayama Y, Nelson MR, Nielsen PF, Sakurada T, Schaff JC, Shapiro BE, Shimizu TS, Spence HD, Stelling J, Takahashi K, Tomita M, Wagner J, Wang J the rest of the SF. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19(4):524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- 278.Pontius JU, Wagner L, Schuler GD. The NCBI Handbook. National Center for Biotechnology Information; Bethesda, MD: 2003. UniGene: a unified view of the transcriptome. [Google Scholar]
- 279.Doyle AL, Ross DD. Multidrug resistance mediated by the breast cancer resistance protein BCRP (ABCG2) Oncogene. 2003;22(47):7340–7358. doi: 10.1038/sj.onc.1206938. [DOI] [PubMed] [Google Scholar]
- 280.Miharada K, Hiroyama T, Sudo K, Danjo I, Nagasawa T, Nakamura Y. Lipocalin 2-mediated growth suppression is evident in human erythroid and monocyte/macrophage lineage cells. J Cell Physiol. 2008;215(2):526–537. doi: 10.1002/jcp.21334. [DOI] [PubMed] [Google Scholar]
- 281.Melaine N, Satie A-P, Lassurguere J, Desmots S, Jegou B, Samson M. Molecular cloning of several rat ABC transporters including a new ABC transporter, Abcb8, and their expression in rat testis. Int J Androl. 2006;29(3):392–399. doi: 10.1111/j.1365-2605.2005.00616.x. [DOI] [PubMed] [Google Scholar]
- 282.Emadi-Konjin HP, Zhang H, Anandan V, Sun D, Schuetz J, Furuya KN. Isolation of a genomic clone containing the promoter region of the human ATP binding cassette (ABC) transporter, ABCB6. Biochim Biophys Acta. 2002;1574(2):117–130. doi: 10.1016/s0167-4781(01)00340-2. [DOI] [PubMed] [Google Scholar]
- 283.Pondarre C, Antiochos BB, Campagna DR, Clarke SL, Greer EL, Deck KM, McDonald A, Han A-P, Medlock A, Kutok JL, Anderson SA, Eisenstein RS, Fleming MD. The mitochondrial ATP-binding cassette transporter Abcb7 is essential in mice and participates in cytosolic iron-sulfur cluster biogenesis. Human Mol Genet. 2006;15(6):953–964. doi: 10.1093/hmg/ddl012. [DOI] [PubMed] [Google Scholar]
- 284.Zhou S, Schuetz JD, Bunting KD, Colapietro A-M, Sampath J, Morris JJ, Lagutina I, Grosveld GC, Osawa M, Nakauchi H, Sorrentino BP. The ABC transporter Bcrp1/ABCG2 is expressed in a wide variety of stem cells and is a molecular determinant of the side-population phenotype. Nat Med. 2001;7(9):1028–1034. doi: 10.1038/nm0901-1028. [DOI] [PubMed] [Google Scholar]
- 285.Goh S-H, Josleyn M, Lee YT, Danner RL, Gherman RB, Cam MC, Miller JL. The human reticulocyte transcriptome. Physiol Genomics. 2007;30(2):172–178. doi: 10.1152/physiolgenomics.00247.2006. [DOI] [PubMed] [Google Scholar]
- 286.Grimes A, McArdle HJ, Mercer JFB. A total dot plot hybridization procedure for mRNA quantitation in small samples of tissues and cultured cells. Anal Biochem. 1988;172:436–443. doi: 10.1016/0003-2697(88)90466-6. [DOI] [PubMed] [Google Scholar]
- 287.Latunde-Dada GO, Van der Westhuizen J, Vulpe CD, Anderson GJ, Simpson RJ, McKie AT. Molecular and functional roles of duodenal cytochrome B (Dcytb) in iron metabolism. Blood Cells Mol Dis. 2002;29(3):356–360. doi: 10.1006/bcmd.2002.0574. [DOI] [PubMed] [Google Scholar]
- 288.Tchernitchko D, Bourgeois M, Martin M-E, Beaumont C. Expression of the two mRNA isoforms of the iron transporter Nramp2/DMTI in mice and function of the iron responsive element. Biochem J. 2002;363(3):449–455. doi: 10.1042/0264-6021:3630449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Kong WN, Chang YZ, Wang SM, Zhai XL, Shang JX, Li LX, Duan XL. Effect of erythropoietin on hepcidin, DMT1 with IRE, and hephaestin gene expression in duodenum of rats. J Gastroenterol. 2008;43(2):136–143. doi: 10.1007/s00535-007-2138-5. [DOI] [PubMed] [Google Scholar]
- 290.Kong WN, Zhao S-E, Duan X-L, Yang Z, Qian Z-M, Chang Y-Z. Decreased DMT1 and increased ferroportin 1 expression is the mechanisms of reduced iron retention in macrophages by erythropoietin in rats. J Gastroenterol. 2008;104(2):629–641. doi: 10.1002/jcb.21654. [DOI] [PubMed] [Google Scholar]
- 291.Chan RY, Schulman HM, Ponka P. Expression of ferrochelatase mRNA in erythroid and non-erythroid cells. Biochem J. 1993;292(2):343–349. doi: 10.1042/bj2920343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Furukawa T, Kohno H, Tokunaga R, Taketani S. Nitric oxide-mediated inactivation of mammalian ferrochelatase in vivo and in vitro: possible involvement of the iron sulfur cluster enzyme. Biochem J. 1995;310(2):533–538. doi: 10.1042/bj3100533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Soilleux EJ, Tian HTYM, Pugh CW, Gatter KC, Harris AL. Use of novel monoclonal antibodies to determine the expression and distribution of the hypoxia regulatory factors PHD-1, PHD-2, PHD-3 and FIH in normal and neoplastic human tissues. Histopathology. 2005;47(6):602–610. doi: 10.1111/j.1365-2559.2005.02280.x. [DOI] [PubMed] [Google Scholar]
- 294.Mason DY, Taylor CR. Distribution of transferrin, ferritin, and lactoferrin in human tissues. J Clin Pathol. 1978;31(4):316–327. doi: 10.1136/jcp.31.4.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Koutnikova H, Campuzano V, Foury F, Dolle P, Cazzalini O, Koenig M. Studies of human, mouse and yeast homologues indicate a mitochondrial function for frataxin. Nat Genet. 1997;16(4):345–351. doi: 10.1038/ng0897-345. [DOI] [PubMed] [Google Scholar]
- 296.Acquaviva F, De Biase I, Nezi L, Ruggiero G, Tatangelo F, Pisano C, Monticelli A, Garbi C, Acquaviva AM, Cocozza S. Extra-mitochondrial localisation of frataxin and its association with IscU1 during enterocyte-like differentiation of the human colon adenocarcinoma cell line Caco-2. J Cell Sci. 2005;118(17):3917–3924. doi: 10.1242/jcs.02516. [DOI] [PubMed] [Google Scholar]
- 297.Becker EM, Greer JM, Ponka P, Richardson DR. Erythroid differentiation and protoporphyrin IX down-regulate frataxin expression in Friend cells: characterization of frataxin expression compared to molecules involved in iron metabolism and hemoglobinization. Blood. 2002;99(10):3813–3822. doi: 10.1182/blood.v99.10.3813. [DOI] [PubMed] [Google Scholar]
- 298.Schaer CA, Vallelian F, Imhof A, Schoedon G, Schaer DJ. Heme carrier protein (HCP-1) spatially interacts with the CD163 hemoglobin uptake pathway and is a target of inflammatory macrophage activation. J Leukoc Biol. 2008;83(2):325–333. doi: 10.1189/jlb.0407226. [DOI] [PubMed] [Google Scholar]
- 299.Bastin JM, Jones M, O’Callaghan CA, Schimanski L, Mason DY, Townsend ARM. Kupffer cell staining by an HFE-specific monoclonal antibody: implications for hereditary haemochromatosis. Br J Haematol. 1998;103(4):931–941. doi: 10.1046/j.1365-2141.1998.01102.x. [DOI] [PubMed] [Google Scholar]
- 300.Tian H, McKnight SL, Russell DW. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev. 1997;11(1):72–82. doi: 10.1101/gad.11.1.72. [DOI] [PubMed] [Google Scholar]
- 301.Mi Z, Rapisarda A, Taylor L, Brooks A, Creighton-Gutteridge M, Melillo G, Varesio L. Synergystic induction of HIF1-alpha transcriptional activity by hypoxia and lipopolysaccharide in macrophages. Cell Cycle. 2008;7(2):232–241. doi: 10.4161/cc.7.2.5193. [DOI] [PubMed] [Google Scholar]
- 302.Fernandez M, Bonkovsky HL. Increased heme oxygenase-1 gene expression in liver cells and splanchnic organs from portal hypertensive rats. Hepatology. 1999;29(6):1672–1679. doi: 10.1002/hep.510290621. [DOI] [PubMed] [Google Scholar]
- 303.Ibrahim NG, Lutton JD, Levere RD. The role of haem biosynthetic and degradative enzymes in erythroid colony development: the effect of haemin. Br J Haematol. 1982;50(1):17–28. doi: 10.1111/j.1365-2141.1982.tb01886.x. [DOI] [PubMed] [Google Scholar]
- 304.Guo B, Brown FM, Phillips JD, Yu Y, Leibold EA. Characterization and expression of iron regulatory protein 2 (IRP2) J Biol Chem. 1995;270(28):16529–16535. doi: 10.1074/jbc.270.28.16529. [DOI] [PubMed] [Google Scholar]
- 305.Recalcati S, Alberghini A, Campanella A, Gianelli U, De Camilli E, Conte D, Cairo G. Iron regulatory proteins 1 and 2 in human monocytes, macrophages and duodenum: expression and regulation in hereditary hemochromatosis and iron deficiency. Haematologica. 2006;91(3):303–310. [PubMed] [Google Scholar]
- 306.Canal F, Fosset C, Chauveau M-J, Drapier J-C, Bouton C. Regulation of the cysteine desulfurase Nfs1 and the scaffold protein IscU in macrophages stimulated with interferon-[gamma] and lipopolysaccharide. Arch Biochem Biophys. 2007;465(1):282–292. doi: 10.1016/j.abb.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 307.Ziere GJ, van Dijk MC, Bijsterbosch MK, van Berkel TJ. Lactoferrin uptake by the rat liver. Characterization of the recognition site and effect of selective modification of arginine residues. J Biol Chem. 1992;267(16):11229–11235. [PubMed] [Google Scholar]
- 308.van Snick JL, Markowetz B, Masson PL. The ingestion and digestion of human lactoferrin by mouse peritoneal macrophages and the transfer of its iron into ferritin. J Exp Med. 1977;146(3):817–827. doi: 10.1084/jem.146.3.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Lundgren S, Carling T, Hjalm G, Juhlin C, Rastad J, Pihlgren U, Rask L, Akerstrom G, Hellman P. Tissue distribution of human gp330/Megalin, a putative Ca2+-sensing protein. J Histochem Cytochem. 1997;45(3):383–392. doi: 10.1177/002215549704500306. [DOI] [PubMed] [Google Scholar]
- 310.Haitina T, Lindblom J, Renström T, Fredriksson R. Fourteen novel human members of mitochondrial solute carrier family 25 (SLC25) widely expressed in the central nervous system. Genomics. 2006;88(6):779–790. doi: 10.1016/j.ygeno.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 311.Drysdale J, Arosio P, Invernizzi R, Cazzola M, Volz A, Corsi B, Biasiotto G, Levi S. Mitochondrial ferritin: a new player in iron metabolism. Blood Cells Mol Dis. 2002;29(3):376–383. doi: 10.1006/bcmd.2002.0577. [DOI] [PubMed] [Google Scholar]
- 312.Cazzola M, Invernizzi R, Bergamaschi G, Levi S, Corsi B, Travaglino E, Rolandi V, Biasiotto G, Drysdale J, Arosio P. Mitochondrial ferritin expression in erythroid cells from patients with sideroblastic anemia. Blood. 2003;101(5):1996–2000. doi: 10.1182/blood-2002-07-2006. [DOI] [PubMed] [Google Scholar]
- 313.Masson N, Appelhoff RJ, Tuckerman JR, Tian Y-m, Demol H, Puype M, Vandekerckhove J, Ratcliffe PH, Pugh CW. The HIF prolyl hydroxylase PHD3 is a potential substrate of the TRiC chaperonin. FEBS Letters. 2004;570:166–170. doi: 10.1016/j.febslet.2004.06.040. [DOI] [PubMed] [Google Scholar]
- 314.Gatter KC, Brown G, Trowbridge IS, Woolston RE, Mason DY. Transferrin receptors in human tissues: their distribution and possible clinical relevance. J Clin Pathol. 1983;36(5):539–545. doi: 10.1136/jcp.36.5.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Panaccio M, Zalcberg JR, Thompson CH, Leyden MJ, Sullivan JR, Lichtenstein M, McKenzie IF. Heterogeneity of the human transferrin receptor and use of anti-transferrin receptor antibodies to detect tumors in vivo. Immunol Cell Biol. 1987;65(6):461–472. doi: 10.1038/icb.1987.55. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.