Abstract
The T cell receptor (TCR) determines the cellular response to antigens, which are presented on the surface of target cells in the form of a peptide bound to a product of the major histocompatibility complex (pepMHC). The response of the T cell depends on the affinity of the TCR for the pepMHC, yet many TCRs have been shown to be of low affinity, and some naturally occurring T cell responses are poor due to low affinities. Accordingly, engineering the TCR for increased affinity for pepMHC, particularly tumor-associated antigens, has become an increasingly desirable goal, especially with the advent of adoptive T cell therapies. For largely technical reasons, to date there have been only a handful of TCRs engineered in vitro for higher affinity using well established methods of protein engineering. Here we report the use of a T cell display system, using a retroviral vector, for generating a high affinity TCR from the mouse T cell clone 2C. The method relies on the display of the TCR, in its normal, signaling competent state, as a CD3 complex on the T cell surface. A library in the CDR3α of the 2C TCR was generated in the MSCV retroviral vector and transduced into a TCR-negative hybridoma. Selection of a high affinity, CD8-independent TCR was accomplished after only two rounds of flow cytometric sorting using the pepMHC SIYRYYGL/Kb (SIY/Kb). The selected TCR contained a sequence motif in the CDR3α with characteristics of several other TCRs previously selected by yeast display. In addition, it was possible to directly use the selected T cell hybridoma in functional assays without the need for sub-cloning, revealing that the selected TCR was capable of mediating CD8-independent activity. The method may be useful in the direct isolation and characterization of TCRs that could be used in therapies with adoptive transferred T cells.
Keywords: T cell receptors, affinity, peptide-MHC, protein engineering, retroviral vectors
1. Introduction
The T cell receptor (TCR) is composed of two chains that pair on the surface of the T cell to form the αβ heterodimeric receptor. Each chain is composed of two domains. Constant domains (C) that anchor the protein in the cell membrane and that associate with invariant subunits of the CD3 signaling apparatus and variable domains (V) that confer antigen recognition through six loops, called complementarity determining regions (CDR), three in each of the V domains. These CDRs interact with a complex between an antigenic peptide bound to a protein encoded by the major histocompatibility complex (pepMHC) (Garcia et al., 1999). Class I MHC bind to peptides of 8 to 9 amino acids in length and can be expressed on any nucleated cell in the body. The interaction of a TCR and a pepMHC can drive the T cell into various states of activation, depending on the affinity (or dissociation rate) of binding (Matsui et al., 1994; Sykulev et al., 1994; Davis et al., 1998). The TCR recognition process also allows a T cell to discriminate between a normal, healthy cell and one that has become transformed via a virus or malignancy.
T cells that develop in the thymus are selected on self-pepMHC ligands, and accordingly their affinity must be sufficiently low in order to prevent autoreactivity upon export of the mature T cell. Indeed, those T cells that express receptors that bind too strongly to self-pepMHC are deleted (Kappler et al., 1987; Hogquist et al., 2005; Gallegos and Bevan, 2006). The process of selection leaves the normal repertoire of TCRs with low affinity for foreign peptides bound to MHC (KD = 1 – 100 µM) (Alam et al., 1996; Davis et al., 1998; van der Merwe and Davis, 2003; Daniels et al., 2006). This affinity range poses a problem when T cells are confronted with a malignancy: many self-antigens are expressed on tumor tissue and the most effective T cells against these antigens would have been deleted (Dudley and Rosenberg, 2003; Ho et al., 2003). This leaves the individual with T cells that express TCRs of sub-optimal affinity for recognition and destruction of the tumor cells. Furthermore, these T cells may be less efficient at clearing tumors because they can not recognize tumor cell variants that might express lower levels of the antigenic peptide/MHC complexes (Spiotto et al., 2002; Zhang et al., 2008). This is because there is an inverse relationship between the number of pepMHC required for T cell activation, and the affinity of the TCR (i.e. the lower the affinity, the greater the number of ligands required) (Schodin et al., 1996). One method to overcome this issue has been to engineer higher affinity TCRs in vitro (Boulter and Jakobsen, 2005; Richman and Kranz, 2007), with the hope of introducing the novel α and β chain genes into T cells for adoptive transfer into patients, in order to fight the malignancy (i.e. in vitro TCR gene therapy followed by adoptive T cell therapy) (Schumacher, 2002; Blattman and Greenberg, 2004; Engels and Uckert, 2007; Rosenberg et al., 2008). A recent study using a wild type anti-tumor antigen TCR has shown that such an approach is feasible in patients with melanoma, but to date there are no reports of clinical applications that used in vitro engineered TCRs (Morgan et al., 2006).
One method of in vitro protein engineering, yeast display, allows for the protein of interest to be expressed on the surface of yeast as an Aga2-fusion (Boder and Wittrup, 1997). While this system has been successful in the engineering of higher affinity single-chain antibodies, fibronectin, and other proteins (Boder and Wittrup, 1997; Gai and Wittrup, 2007; Lipovsek et al., 2007; Tereshko et al., 2008), TCRs have posed a more formidable challenge. In the yeast display system, the TCR has been displayed as a stabilized single-chain protein, in the Vβ-linker-Vα form (Kieke et al., 1999; Shusta et al., 2000). Two mouse TCRs have been engineered for higher affinity using this system: 3.L2 (MHC class-II restricted) and 2C (MHC class-I restricted) (Holler et al., 2000; Weber et al., 2005). The major drawback to using yeast display for engineering high-affinity TCRs has involved the need to mutate the TCR V domains in order to allow the scTCR to be expressed on the surface of yeast before affinity maturation (Richman and Kranz, 2007). To date, the only TCRs that have been amenable to this system are those with clonotypic antibodies that allowed for the stabilization and surface display of the single-chain format.
A second system, phage display, attaches the protein of interest on the surface of the phage through fusion to the N-terminus of a viral coat protein. Two TCRs, A6 and 1G4 (both MHC class-I restricted), have been engineered for higher affinity using this method (Li et al., 2005). Phage display of these TCRs was enabled by introduction of a non-native disulfide bond between the two C domains in order to promote pairing of the α and β chains (Boulter and Jakobsen, 2005). An advantage of this format is that it allowed for the engineering of the full-length (VαCα/VβCβ) protein without the need for prior selection of V domain stabilizing mutations. However, in studies using the full-length format in yeast display, the expression levels are low and variable among different TCRs (manuscript submitted and unpublished data).
A third system that has been reported for the engineering of TCRs is mammalian cell display (Kessels et al., 2000). This system used a retroviral vector to introduce the TCR α and β-chains into a TCR-negative T cell hybridoma. The introduced TCR was expressed on the surface in its native conformation, complexed with CD3 subunits, allowing for a fully functional T cell (signaling competent). Retroviral mediated gene transfer and display does not require mutation of the TCR for proper surface expression as it takes advantage of the normal assembly of the CD3 accessory molecules involved in surface expression (Call and Wucherpfennig, 2005). Although TCRs have not been engineered for higher affinity using this method, there has been one reported study in which the specificity of a TCR was altered (Kessels et al., 2000).
In order to determine the potential for using retroviral display of TCRs for engineering improvements in TCR affinity, here we have examined the parameters that govern generation and selection of TCR mutant libraries. In addition, we report the generation and isolation of a high affinity receptor, called m100, from the mouse T cell clone 2C. 2C recognizes the foreign pepMHC SIYRYYGL/Kb (SIY/Kb) in a CD8-dependent manner, and the 2C TCR has a relatively low affinity (KD = 30 µM) for SIY/Kb (Udaka et al., 1996; Garcia et al., 1997; Daniels and Jameson, 2000; Degano et al., 2000; Cho et al., 2001). This allowed for the identification of mutants that would bind to SIY/Kb with higher affinity and thereby confer T cell activity in the absence of the co-receptor CD8 (by analogy to the goal of identifying TCRs with high affinity for a tumor antigen, such that they would mediate activity in helper T cells, which are CD8 negative). In fact, the SIY peptide has now been used as a model tumor antigen system by a number of labs (Blank et al., 2004; Zhang et al., 2007; Thomas et al., 2008). The αβ−, CD8−, CD4− T cell hybridoma 58−/− (Letourneur and Malissen, 1989) was transduced with DNA from a 2C TCR mutant library that was degenerate at five codons that encode the third complementarity determining region of the α-chain (CDR3α). Transduced T cells were sorted for TCR expression and binding to SIY/Kb. After only two sorts, almost all of the cells were positive for SIY/Kb binding at concentrations that the wild type 2C TCR was negative. Limiting dilution of the positive population, followed by RNA isolation, RT-PCR and sequencing analysis identified a novel TCR with mutations in the CDR3α. Soluble single-chain TCR of the mutant revealed that it had a 15-fold higher affinity for SIY/Kb. The selected T cell hybridoma was stimulated to release IL-2 when SIY was added to Kb-positive antigen presenting cells in a CD8-independent manner, allowing the direct testing of TCRs without the need for subcloning (e.g. as is necessary in yeast or phage display approaches). This method should prove useful in identifying TCRs that could be used in adoptive T cell therapies directed against tumor antigens, or for use as soluble TCRs against various MHC-restricted targets (Molloy et al., 2005; Richman and Kranz, 2007).
2. Materials and methods
2.1. Peptides, antibodies, and pepMHC tetramer production
SIYRYYGL (SIY) and SIINFEKL (OVA) peptides were synthesized by the Macromolecular Core Facility of the Section of Research Sources, Penn State College of Medicine. Peptides were purified by reverse phase chromatography using a C-18 column and mass was confirmed by MALDI. The 2C clonotypic antibody 1B2 was purified from hybridoma supernatant by ammonium sulfate precipitation followed by Protein G chromatography (Kranz et al., 1984). Biotinylation of purified antibody was performed using the EZ-Link Suflo-NHS-LC-Biotinylation Kit (Pierce). The antibody used for staining surface levels of TCR was biotinylated H57–597 which recognizes the TCR Cβ chain (BD-Pharmingen).
For SIY/Kb tetramer production, Kb-heavy chain, with a C-terminal biotinylation signal sequence and β2m were expressed as inclusion bodies in E. coli. Kb was biotinylated in vivo by co-expression of biotin ligase (Zhang et al., 2007). Monomeric complexes were obtained by refolding solubilized Kb and β2m in the presence of SIY peptide. Complexes were purified by anion exchange and size exclusion chromatography. Fluorescent tetramers of SIY/Kb were produced by adding streptavidin:phycoerythrin (BD-Pharmingen) to the biotinylated monomeric complexes in a 1:4 molar ratio.
2.2. Cell lines
The retroviral packaging cell line, EcoPack 2–293 (Clonetech) was cultured in DMEM supplemented with 10% fetal calf serum, L-glutamine, penicillin and streptomycin. T2-Kb, a TAP-deficient lymphoblastoid cell line, and the αβ negative T cell hybridoma 58−/− (Letourneur and Malissen, 1989) were maintained in RPMI-1640 supplemented with 10% fetal calf serum, L-glutamine, penicillin and streptomycin.
2.3. Construction of 2C CDR3α DNA library
2Cα and 2Cβ cDNAs, were cloned into the murine stem cell virus (MSCV) retroviral vector in a bicistronic configuration of 2Cα-IRES-2Cβ (kindly provided by Phil Holler and Jianzhu Chen, MIT). The 5 codon amino acid degeneracy library in CDR3α of the 2C TCR (GFASA, positions 99–103) was created by overlap extension PCR (SOE-PCR - overlap regions between two preSOE products are underlined. AgeI upstream primer, sense strand: 5’ GGAATTAGATCCACCGGTCGCCGCCATGCTCCTGGACTCCTCCCAGTGCTGGGG 3’; CDR3α 5 codon degeneracy primer, anti-sense strand: 5’ caatgacttttgtgccagatccaaatgtCAGSNNSNNSNNSNNSNNgctCACAGCACAGAAGTACACAGCCGAGTCGCTCC 3’ and CDR3α SOE Overlap Primer, sense strand: 5’ GGATCTGGCACAAAAGT CATTGTTCTACCATACATCCAGAACCCAGAACC 3’; Calpha NotI reverse primer, anti-sense strand: 5’ CGGAATTCTAGAGTCGCGGCCGCTTTACTTGTACATTATC AACTGGACCACAGCCTCAG 3’ (Integrated DNA Technologies). The 869 base-pair SOE product VαCα cassette with 5 codon degeneracy spanning CDR3α was ligated into the MSCV vector as a AgeI-NotI fragment. Ligation was performed with T4 DNA ligase (New England Biolabs) with a 2:1 insert to vector ratio. After incubation for 1 hr at room temperature, the reaction was diluted 5-fold with Tris-HCl (pH=7.4) and 1 mM EDTA, and one-sixth of this mixture was added to a 100 uL aliquot of DH5α MAX Efficiency E. coli Competent Cells (Invitrogen). Cells were incubated for 30 min on ice and subsequently placed at 42°C for 45 seconds. Following a 2 minute recovery on ice, cells were quenched with 0.9 mL of SOC medium (Invitrogen) and allowed to recover for 1 hr. at 37°C. Each transformation yielded 1.6 × 104 transformants. Thirty transformations were pooled to give ~ 5 × 105 transformants relative to vector only transformation. Sequencing of individual E. coli colonies showed that ~70% contained in-frame sequences with CDR3α diversity and ~30% contained non-productive TCR ligation products. The transformed cells were pooled and expanded for 8 hours in 250 mL of Luria Broth. Vector DNA was isolated by Maxiprep kit (Qiagen).
2.4. Retroviral transduction of T cells, flow cytometry analysis and cell sorting
MSCV retroviral DNA was added to the packaging line EcoPack 2–293 for transient transfection. Twenty-four hours prior to transfection, 2–293 cells were plated at 3 × 105 cells/well in a 6-well plate (Corning). Four µg of DNA was added to the CalPhos Mammalian Transfection Kit (Clonetech) and incubated at room temperature for 20 minutes. The transfection mixture was then added to the packaging cells for 6 hours after which the cells were washed twice with fresh RPMI-1640 to remove any residual traces of the mixture. Cells were replenished with fresh RPMI-1640 and incubated for 48 hrs at 37°C, 5% CO2.
Supernatant containing the packaged, replication-incompetent retrovirus was harvested, filtered and added to 58−/− cells (105) with 8 µg/mL of polybrene (Sigma). Cells were centrifuged at 1200 × g for 45 minutes at room temperature then placed at 37°C, 5% CO2 for 3 days. Transduction efficiencies were determined by assaying for TCRβ expression using 5 µg/mL of biotinylated H57–597 anti-TCR Cβ chain (BD Pharmingen) followed by 1:200 dilution of Streptavidin:PE (BD Pharmingen), and analysis on a BD FACScanto cytometer.
For library selections, transduced T cells were dual stained with biotinylated anti-mouse TCR Cβ (H57–597) SAv:APC and SIY/Kb:PE tetramer. Sorting was performed using a Cytomation MoFlo (Dako Cytomation, Ft. Colllins, CO) at room temperature. For tetramer binding analysis, T cells were incubated with varying concentrations of phycoerythrin-labeled SIY/Kb tetramer for 1 hour at 4°C. Cells were washed 3 times with ice-cold 1% PBS-BSA and analyzed on a BD FACScanto cytometer.
2.5. T cell limiting dilution and reverse-transcriptase polymerase chain reaction
To isolate clonal populations of T cells positive for SIY/Kb binding, limiting dilution cloning was performed using a feeder layer consisting of C57/Bl6 splenocytes (5 × 104 cells per well) pre-treated with 10 µg/mL Mitomycin C (Fisher). After expansion of wells from single cell cultures, 2 × 106 cells were pelleted and total RNA extracted using the TRIzol RNA Isolation kit (Invitrogen). Genomic DNA was removed and reverse transcription of RNA was carried out using the Quantitect Reverse Transcription kit (Qiagen). Amplification of the α-chain cDNA was performed by nested PCR and products were sequenced.
2.6. Soluble single-chain TCR production and binding analysis
The variable domains of m100 were cloned into the pET28a E. coli expression vector as a Vβ-linker-Vα construct and transformed into BL21 (DE3) (Stratagene) for production as insoluble inclusion bodies, as described previously (Jones et al., 2006). Briefly, the washed inclusion bodies were added to 400 mL of denaturing solution (3 M Urea, 50 mM Tris, pH 8.0) for 2 hours. Dilution buffer (200 mM NaCl, 50 mM Tris, pH 8.0) was added dropwise over a 36 hour period. Refolded single-chain TCR was collected by adding Ni-NTA Agarose beads (Qiagen) using a scintered funnel and eluted with 500 mM imidizole. Eluted protein was purified by size-exclusion chromatography.
To measure the equilibrium binding constants of the scTCR, a competitive binding experiment was performed. T2-Kb were loaded with excess SIY (>10 µM) for 2 hours at 37°C, 5% CO2. The SIY-loaded T2-Kb cells (5 × 104) were incubated with a biotinylated high-affinity scTCR m67 (KD = 16 nM) at a concentration that yielded sub-maximal binding and varying concentrations of unlabeled scTCRs for 1 hour at 4°C. Cells were washed 3 times with 1% PBS-BSA and streptavidin:PE was added to detect bound m67. Cells were analyzed for fluorescence on a Coulter Epics-XL. KD values of the unlabeled scTCR inhibitors were calculated using the Cheng/Prusoff (Cheng and Prusoff, 1973) equation: KD = IC50/(1 + ([m67]/KDm67)), where IC50 equals the concentration of inhibitor that yielded 50% inhibition, the concentration of m67 was 12.5 nM, and the KD of m67 was 16 nM.
2.7. Measurement of IL-2 secretion
For T cell activation assays, 105 transduced T cell hybridoma cells were incubated with 105 T2-Kb with varying concentrations of SIY peptide for 24 hours at 37°C and 5% CO2. IL-2 in supernatants was assayed using a quantitative capture ELISA as follows: 96-well plates (Immulon 2HB) were coated with 2.5 mg/mL of rat anti-mouse IL-2 capture antibody (BD Pharmingen) in coating buffer (0.1M Na2HPO4, pH 9) for 2 hrs. at room temperature. Plates were washed 3 times with PBS containing 0.05% Tween-20 (PBS-T) and blocked with 1% PBS-BSA for 12 hrs. at 4°C. Plates were washed once with PBS-T before the addition of 50 µL of supernatant for 2 hrs at room temperature. Plates were washed 3 times with PBS-T then incubated with 6.7 mg/mL of biotinylated anti-mouse IL-2 (BD Pharmingen) in PBS for 1 hr. After washing 3 times with PBS-T, streptavidin:HRP at a 1:10,000 dilution in PBS was added for 30 minutes. Plates were once again washed 3 times with PBS-T, followed by development with 50 µL of TMB substrate (KPL). The reaction was stopped once a color change was seen in the control wells by adding 50 µL of 1N H2SO4 and the absorbance of each well measured at 450nm with a microplate reader.
3. Results
3.1. Analysis of the number of retrovirus integrants per T cell
In order for T cell display using a retroviral vector to be a viable alternative for engineering TCRs, the system must minimize the number of target T cells that contain multiple virions integrated into their genome. This is because T cells with multiple TCR integrants would express lower levels of a potential higher affinity TCR, making detection and selection difficult. Using two different TCRs expressed in retroviruses we evaluated the ratio of single versus multiple infections on a per cell basis. The TCRs included the wild type 2C TCR for which we have a clonotypic monoclonal antibody (mAb) called 1B2 (Kranz et al., 1984), and a mutant of this TCR called m33 that has a 1000-fold higher affinity for the antigen SIY/Kb (Holler et al., 2003). CD8-negative T cells transduced with the 2C TCR do not stain with SIY/Kb tetramers at the concentrations used, as it has been shown that CD8 is required for detection of bound SIY/Kb tetramer on 2C T cells (Daniels and Jameson, 2000; Cho et al., 2001). T cells transduced with the m33 TCR stain very efficiently with SIY/Kb tetramers in the absence of CD8. In contrast, the mutations in m33 have eliminated detectable binding to mAb 1B2. Thus, the properly associated α and β chains of 2C and m33 can be uniquely identified using mAb 1B2 and SIY/Kb tetramer, respectively.
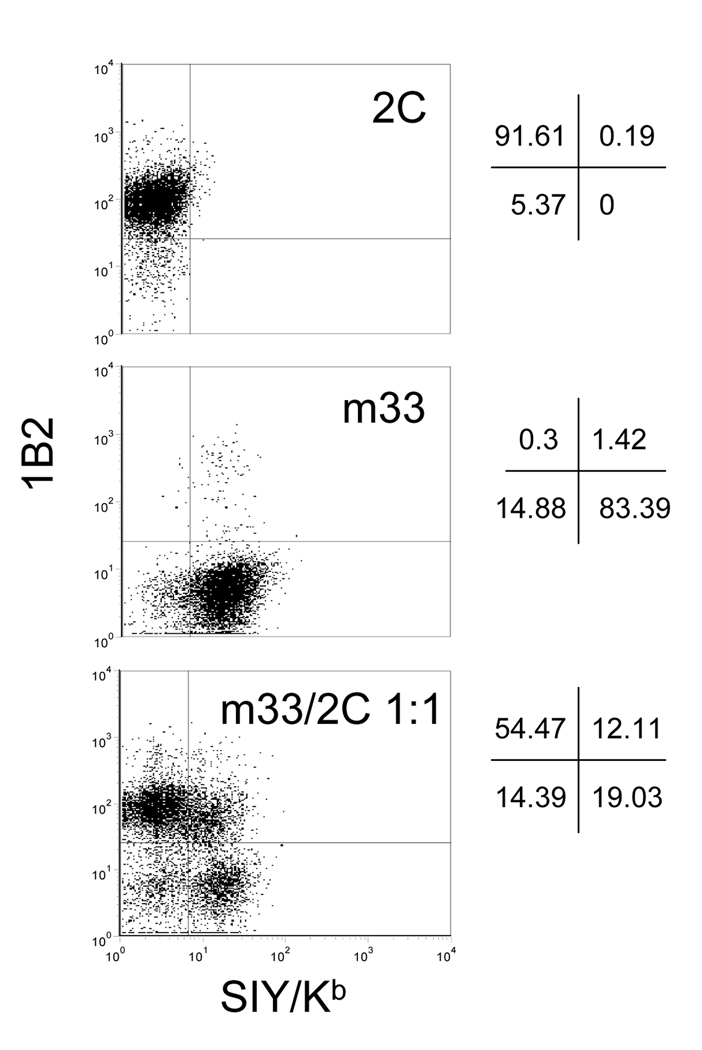
To assess the level of multiple infections on a per cell basis, m33 and 2C TCRs, cloned into the MSCV retroviral vector, were separately packaged into infectious virions using EcoPack 2–293 cells by transient transfection. The 2C and m33 supernatants were harvested 48 hours post-transfection and half of the 2C supernatant was added to 105 58−/− cells and half of the m33 supernatant was added to a separate population of 105 58−/− cells. The remaining supernatants were combined at a 1:1 ratio and added to of 105 58−/− cells. The transduction efficiencies for the three TCR preparations, determined using the anti-Cβ mAb, were 19% (2C alone), 21% (m33 alone), and 30% (m33/2C at 1:1). After five days, the three cell samples were enriched for cells with TCR expression using the anti- Cβ mAb and the resulting TCR-positive populations were analyzed by dual-staining with the TCR-specific reagents 1B2 and SIY/Kb tetramers (Figure 1). The population of T cells transduced with only 2C retrovirus was 92% positive for 1B2 and 0.2% positive for SIY/Kb. In contrast, T cells transduced with only m33 retrovirus were 84% positive for SIY/Kb and 1.4% positive for 1B2. The population of T cells that received the 1:1 mixture of 2C and m33 retroviruses showed three distinct populations corresponding to: 2C (1B2 positive, SIY/Kb negative, 54%), m33 (1B2 negative, SIY/Kb positive, 19%) and the dual expression of 2C and m33 (1B2 positive, SIY/Kb positive, 12%). It can be concluded for these retroviral transductions that the frequency of a single virion infecting an individual T cell was approximately 70%, whereas the frequency of two virions infecting an individual T cell was approximately 12%. This level of multiple virion integration is sufficiently low to allow the isolation of T cells that express mutant TCRs. It is also worth noting that, consistent with our expectation that T cells expressing multiple virions would have a lower expression level of each TCR, the population which was double positive showed approximately a two-fold reduction in signal (MFU) for both the 1B2 and SIY/Kb staining. This is consistent with approximately equal expression of the two TCRs and the action of other factors (e.g. CD3 subunit levels) that control maximum expression (Kearse et al., 1995; Kuhns et al., 2006).
Figure 1. Flow cytometry analysis of dual T cell receptor expression.
The 58−/− TCR-negative T cell hybridoma was transduced with retrovirus containing either wild-type 2C TCR, mutant m33, or both 2C and m33 at a 1:1 ratio. TCR positive cells were sorted after transduction with biotinylated anti-mouse TCR Cβ (H57–597) followed by streptavidin:PE. The enriched TCR positive populations were then dual stained with biotinylated 1B2 and streptavidin:APC to detect the 2C TCR and SIY/Kb-PE labeled tetramer to detect the m33 TCR.
3.2. Isolation of T cells that express a high-affinity TCR from a large population of T cells that express a wild type TCR
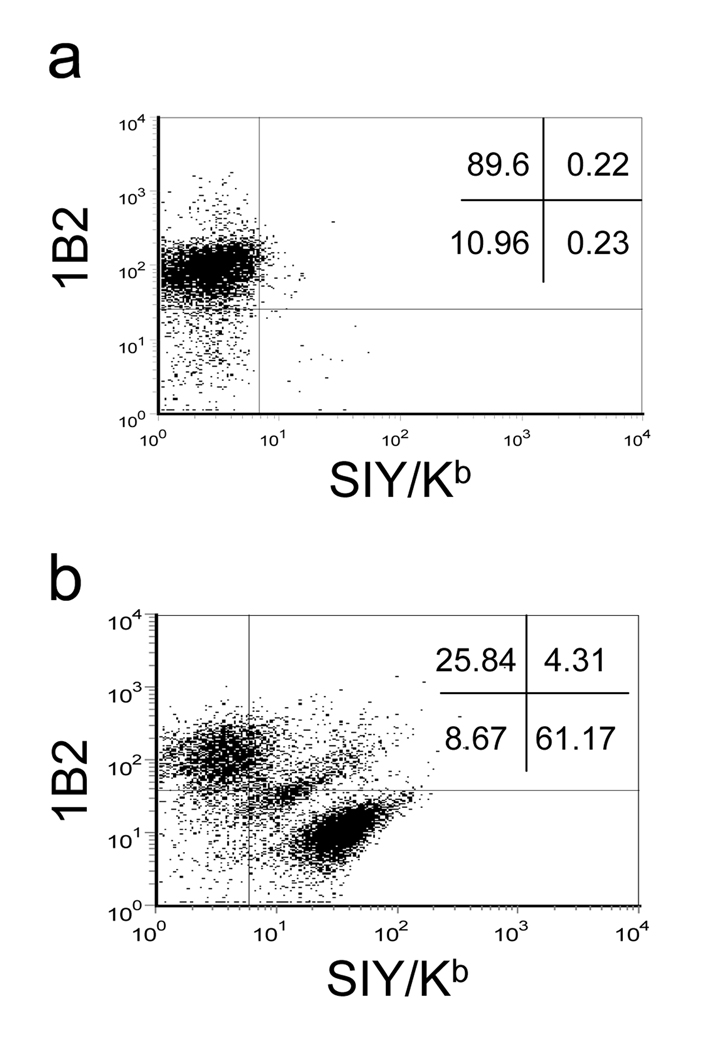
The majority of DNA from a degenerative sequence library will encode TCRs that are not high affinity. To determine whether flow sorting using pepMHC tetramers could be used to select a minor population of T cells with high affinity among a great excess of other T cells (i.e. the “needle in the haystack”), we mimicked the experiment that would be performed with degenerate TCR libraries, again using the 2C and m33 TCR systems. Packaging cells were transfected with a mixture containing 4 µg of 2C DNA and 0.04 µg of m33 DNA and the retroviral supernatant was harvested 48 hours post-transfection and added to 105 58−/− T cells, yielding an efficiency of TCR transduction of 12%. Cells were sorted for positive TCR Cβ expression five days after transduction in order to enrich for transduced cells. When the TCR positive population was dual-stained for both 2C and m33, the large majority of T cells expressed only 2C as indicated by 89% positive 1B2 staining and 0.23% SIY/Kb tetramer staining. However, 0.23% was higher than that observed for SIY/Kb tetramer staining of a population of 2C cells alone (Figure 1).
To determine whether the small fraction of SIY/Kb positive cells could be isolated by cell sorting, the cells were stained with the same concentration of SIY/Kb tetramer (60 nM) and the positive population (0.23%) was collected. After one round of sorting, the SIY/Kb positive population increased to more than 60%, corresponding to over 200-fold enrichment (Figure 2b). The 2C population decreased from 89% to 25% and a third distinct population of double positive cells also emerged after a single sort. This population most likely corresponds to cells that expressed multiple virions including both 2C and m33, as the intensity of 1B2 and SIY/Kb staining was again approximately one-half of the respective single TCR expressing populations.
Figure 2. Isolation of higher affinity receptor within larger wild-type population.
The 58−/− TCR-negative T cell hybridoma was transduced with retrovirus containing 100-fold more wild-type TCR 2C than m33. Cells positive for TCR expression were enriched by sorting using a biotinylated anti-mouse Cβ (H57–597) with streptavidin:PE. (a) Cells were dual stained with the biotinylated 1B2 and streptavidin:APC to detect T cells displaying the 2C TCR and SIY/Kb-PE labeled tetramer to detect T cells displaying the m33 TCR. (b) Cells from (a) were sorted on the m33 positive (SIY/Kb) population and re-analyzed with both 1B2 and SIY/Kb.
3.3. Generation of a CDR3α library in the retroviral vector
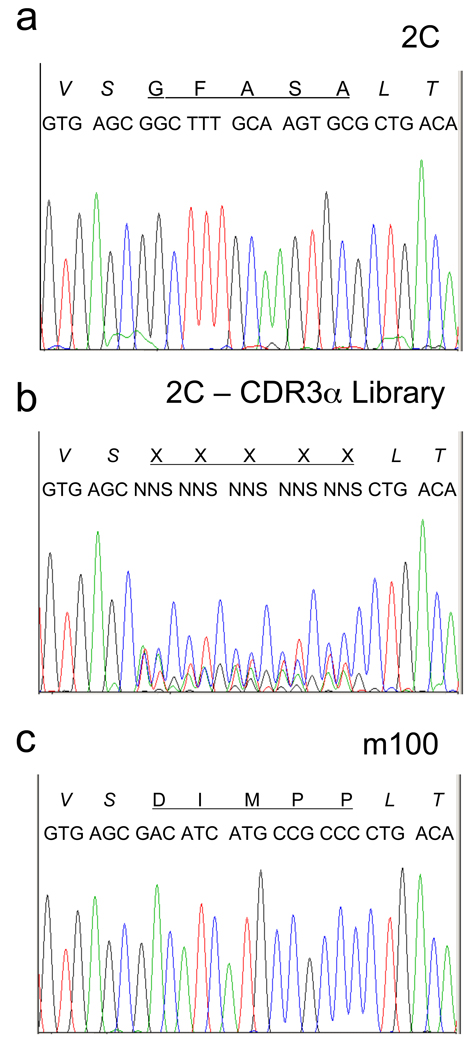
The high affinity mutant m33 was generated by yeast display using a mutated single-chain form of the 2C TCR and a degenerative library targeted to the CDR3α loop (Holler et al., 2003). To be able to compare our retroviral strategy with this previous study, we generated an independent degenerative library of the same five codons in the CDR3α in the retroviral vector MSCV (see Materials and Methods 2.1). Transformation of the library into E. coli yielded approximately 5 × 105 transformants. DNA from the transformed population was sequenced and the region that encodes the CDR3 was compared to the region from the original clone 2C (Figure 3b and a, respectively). Analogous sequencing of TCR genes, called spectratyping, has been used to assess the oligoclonality of distinct T cell populations (Gorski et al., 1994). In the retroviral display library of 2C, there was clear degeneracy at the five codons comprising the CDR3α loop, although the third position of each codon showed a strong cytosine signal (from the expected C/G bases incorporated at these positions), possibly due to sequencing bias. In fact, DNA sequences from ten random colonies showed that C and G were both represented at these positions and that of the ten clones, seven contained in-frame TCRs with diverse CDR3α. This extrapolates to a functional library size from E. coli of about 3 × 105.
Figure 3. CDR3α DNA sequencing analysis.
DNA sequencing profiles of the CDR3α region for (a) wild-type 2C, (b) 2C-CDR3α degenerate library and (c) m100 TCR.
3.4. Selection of high affinity T cells from the CDR3α library
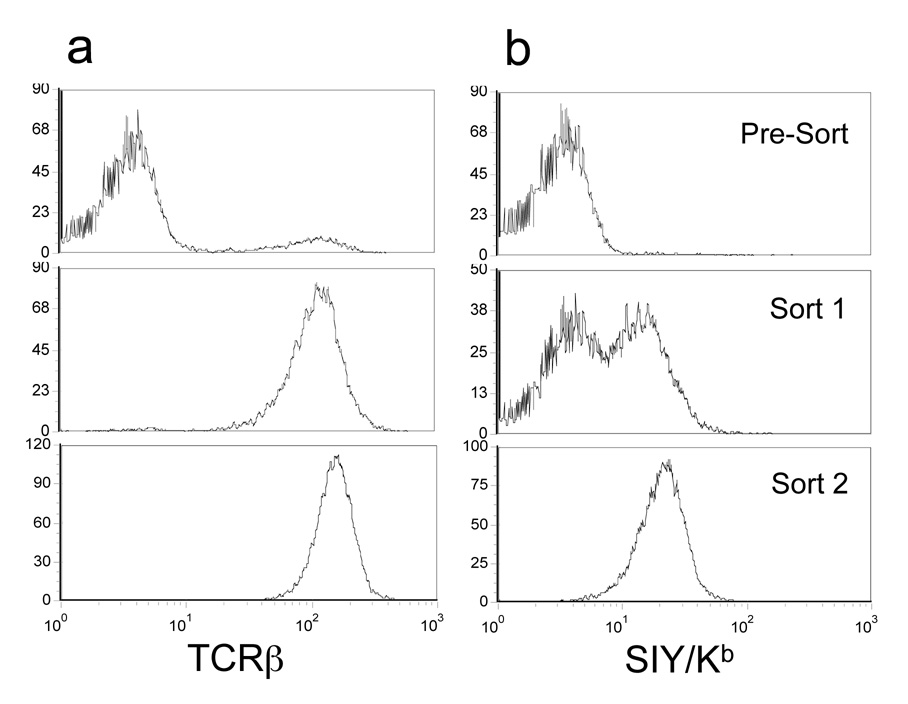
Vector DNA from the CDR3α library was used to transfect the packaging line EcoPack 2–293 for production of infectious, replication-incompetent retrovirus and after 48 hours, supernatant was harvested and added to 105 58−/− T cells. TCRβ expression, analyzed after 3 days, showed that approximately 10% of the cells were positive (Figure 4a, top panel). Thus, we estimate that the maximum size of the retrovirus CDR3α library was about 104 (10% of the 105 58−/− cell). As expected, staining with the SIY/Kb tetramer did not yield a distinct population (Figure 4b, top panel).
Figure 4. Analysis of T cells transduced with the 2C-CDR3α library.
The 58−/− TCR-negative T cell hybridoma was transduced with retrovirus from the 2C-CDR3α library and various populations were examined for: (a) TCR expression using the anti-Cβ mAb and (b) binding to SIY/Kb-PE tetramer. These analyses were performed on cells immediately after the initial transduction (pre-sort) and after both rounds of sorting (sort 1 and sort 2), as described in the Materials and Methods.
To sort the library, cells were dual-stained with the anti-TCRβ antibody and 60 nM of SIY/Kb-PE labeled tetramer. The top 0.7% double positive cells were sorted and cultured for 5 days. Analysis of the sorted population (Figure 4a, b, middle panel) showed that most of the cells expressed a TCR and that approximately 50% of the population was positive for SIY/Kb tetramer staining. A second round of sorting was performed, collecting the top 20% of the SIY/Kb positive cells. After culturing, this population now showed almost 100% positive cells for both TCR and SIY/Kb tetramer staining (Figure 4a, b, bottom panel). Thus, only two rounds of sorting were required to isolate a population of cells that bound to SIY/Kb even in the absence of CD8.
3.5. Isolation and characterization of a high-affinity TCR clone
Limiting dilution cloning was performed to isolate single cell clones. Staining of the clones showed that they all bound to SIY/Kb tetramer at the concentration used for sorting (data not shown). RT-PCR was used with α-chain primers to sequence the clones, and the same CDR3α sequence, represented by mutant m100, was obtained for all clones (Figure 3c). This DNA sequence encoded a CDR3α amino acid sequence of DIMPP, distinctly different from the wild type sequence GFASA (Figure 3a). The sequence also differed from a collection of TCR mutants against SIY/Kb, including m33, that were previously isolated by yeast display. However, m100 contained the invariant proline found in all of the yeast display isolates and in m33 (LHRPA) (Holler et al., 2003), further indicating that this residue is critical for achieving high-affinity for SIY/Kb.
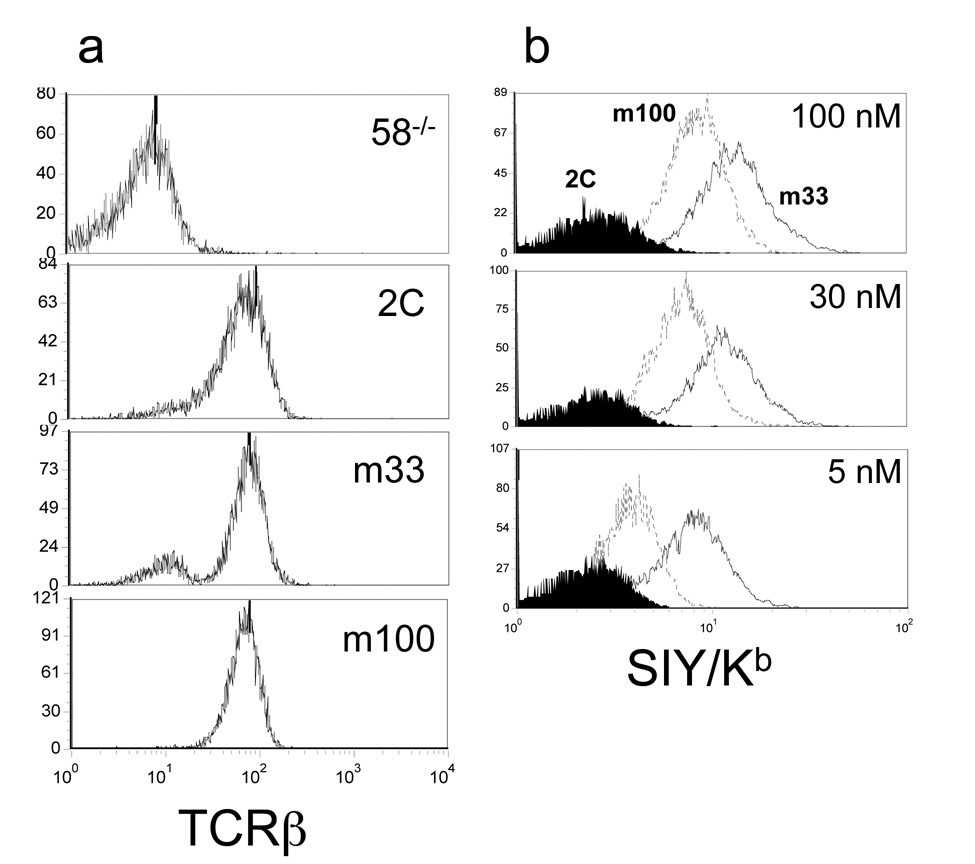
To characterize the tetramer binding properties of m100, SIY/Kb tetramer binding of m100 transduced cells were compared to 2C and m33. All three transductants expressed similar TCR expression levels (Figure 5a). Titrations with 5 nM, 30 nM, and 100 nM SIY/Kb tetramer showed distinct binding profiles for all three TCRs (Figure 5b). As expected, wild-type 2C had no discernable binding at each of these concentrations. T cells that expressed m33 (KD = 30 nM SIY/Kb) had the greatest level of staining with SIY/Kb tetramer at all three concentrations. T cells that expressed m100 were positive for SIY/Kb tetramer binding at all concentrations but at lower intensity levels than for m33. Thus, as all three populations had equal levels of total TCR, we estimated the order of binding affinities to be m33 > m100 > 2C.
Figure 5. Binding of SIY/Kb tetramers to various T cell hybridomas.
T cells expressing the wild-type 2C, m33 and m100 TCRs were assayed for (a) TCR expression using the anti-Cβ mAb and (b) binding to SIY/Kb-PE labeled tetramer at 100, 30, and 5 nM. Histograms for tetramer binding in (b) were for 2C (shaded), m100 (dotted line) and m33 (solid line).
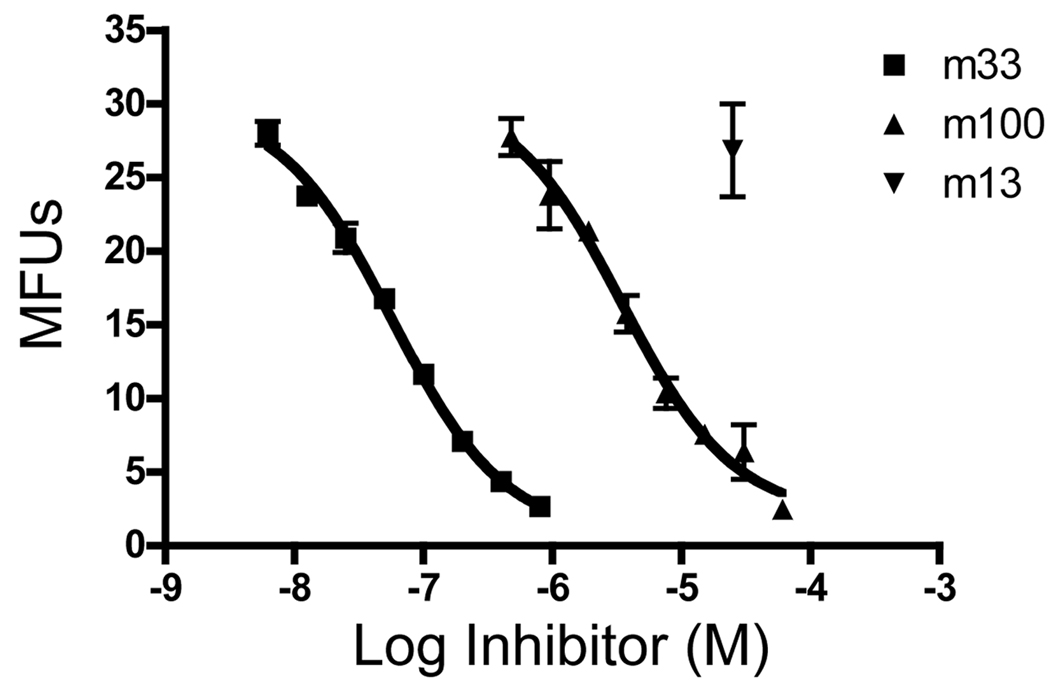
To determine the equilibrium binding constant of the m100:SIY/Kb interaction, we cloned the CDR3α mutations into the 2C single-chain TCR for expression in E. coli. Purified m100 and m33 proteins were used in a competition format that allows calculation of KD values (Cheng and Prusoff, 1973). In this assay, a biotin-labeled high-affinity scTCR, m67, was incubated together with various concentrations of unlabeled m33 and m100 scTCRs purified from E. coli (Figure 6). T2-Kb cells were loaded with excess SIY peptide and incubated with the various scTCR mixtures. The concentration of scTCR that yielded 50% maximum inhibition can be used to calculate the KD value of the competitor. Using this assay format, the m33 scTCR was shown to have an affinity of 32 nM, very similar to the value determined from surface plasmon resonance and other approaches (Holler and Kranz, 2003). TCR m100 was shown to have an affinity of 1,900 nM. This measurement, consistent with the qualitative order of binding seen using SIY/Kb tetramers, is 15-fold higher than the affinity reported for the wild type 2C TCR measured by surface plasmon resonance and 4-fold higher than the affinity measured for the 2C scTCR using the competition assay (Jones et al., 2008).
Figure 6. Binding affinity analysis of soluble single-chain TCR m100 using a competition assay.
T2-Kb antigen presenting cells were loaded with excess SIY peptide and incubated with a fixed concentration of soluble biotinylated m67 and varying concentrations of soluble m33 scTCR, m100 scTCR or an irrelevant scTCR m13 (known not to bind to SIY/Kb). Remaining bound biotinylated m67 was detected by streptavidin:PE. IC50 values were calculated using GraphPad Prism 4 and KD values were determined as described in Materials and Methods.
3.6. Functional activity mediated by the m100 mutant TCR
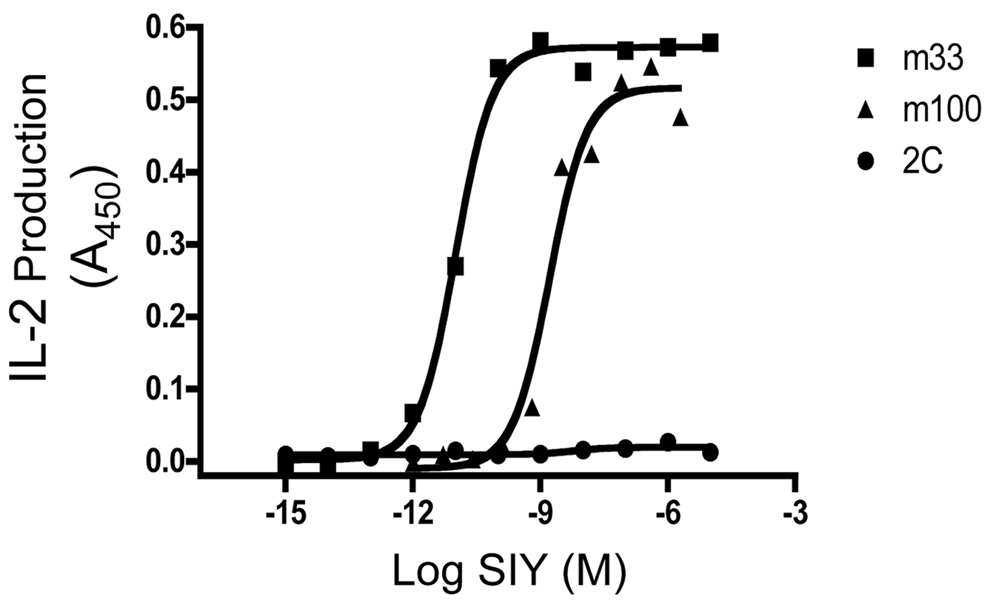
A potential advantage of the T cell display method as described here, is that the selected clones can be used directly in assays to assess the functional stimulatory ability of the pepMHC used for selections. As the 58−/− line lacks CD8, both the sorting strategies with class I tetramers and the functional assay avoid the complication associated with the well-known ability of CD8 to enhance signaling and binding, even from very low affinity interactions (e.g. 2C CD8+ cells are potently stimulated by SIY/Kb despite the low affinity, 30 µM, of the TCR:SIY/Kb interaction (Daniels and Jameson, 2000; Cho et al., 2001). To examine the ability of m100 to respond to antigen in a CD8-independent manner, the three CD8-negative T cell lines, 2C, m33, and m100 were incubated in the presence of the antigen presenting cell line (APC) T2-Kb with varying concentrations of SIY peptide (Figure 7). As expected, wild-type 2C was not activated under these conditions (as it requires the co-receptor CD8), while m33 is strongly stimulated in the absence of CD8 (Holler et al., 2003). Like the m33 TCR, the m100 TCR also mediated IL-2 release in a CD8-independent manner, with the half-maximal release occurring at a higher concentration of SIY peptide than for m33. This result is consistent with the lower affinity of m100 compared to m33, but the results show that it is possible to isolate TCRs using the T cell/retroviral method that are specific for class I products and that could function in T helper cells (i.e. CD4 positive, CD8 negative cells).
Figure 7. Antigenic peptide stimulation of T cells expressing the m100 TCR.
T cell hybridomas that express the m100, m33 and 2C TCRs were incubated with T2-Kb loaded with various concentrations of SIY peptide for 24 hours and assayed for IL-2 production.
4. Discussion
In vitro engineering of TCRs for increased affinity has been accomplished using two different methods (yeast and phage display), but so far with limited success (Boulter and Jakobsen, 2005; Richman and Kranz, 2007). The major obstacle to success has been the difficulties associated with the expression of the αβ heterodimer in heterologous display systems, a feature that has for many decades plagued efforts to express soluble TCRs (Rudolph et al., 2006). Here we have explored a system that takes advantage of the normal ability of T cells to assemble the αβ heterodimer with its requisite subunits as a CD3 complex on the surface of a transformed T cell hybridoma. Under these conditions, most TCRs can be expressed at high surface levels (e.g. 50,000 molecules/cell) without the need for mutations in either the α or β chains (Wegener et al., 1992; Holler et al., 2001). We show here that it is possible to isolate TCR affinity mutants in the T cell display system using pepMHC tetramers and to directly assess the functional ability of the TCRs.
The use of retrovirus vectors for direct protein display on the viral particle has been documented previously (Russell et al., 1993; Urban et al., 2005). The protein of interest was fused to the retroviral Env protein for incorporation and display in the viral membrane. Winter and colleagues were able to display a functional single-chain antibody fragment (scFv) on the surface of retroviral particles (Russell et al., 1993) while a group led by Buchholz created a scFv library using the same method (Urban et al., 2005). While these groups used the retroviral particle itself as the display platform, we used the particle as a vector to introduce the genes that encode a TCR library for display on the surface of T cells. Schumacher and colleagues were able to use this method to alter the specificity of a TCR for its cognate pepMHC (Kessels et al., 2000). However, there has not been a high affinity TCR engineered using this method of retroviral mammalian display, raising the question of what technical obstacles have prevented its wider spread use for this purpose.
Several technical challenges have most likely made it difficult to use T cell display with retroviral vectors for successful engineering of high-affinity TCRs. First, the issue of multiple virions per T cell would reduce the surface levels of a specific TCR, thereby making current selection strategies with flow sorting more difficult. Here, we show that using transduction conditions that yield 10 to 20% efficiency, libraries can be generated where the majority of transduced cells express a single virion. In this regard, it is interesting that optimization of transduction efficiencies may actually increase the problems associated with multiple virions, and thus it may be best to keep transduction efficiencies at a minimal level (i.e. that needed for generation of adequate size libraries). Our findings raise the question of whether activated human or mouse T cells could be used directly as a system for TCR engineering. Human T cells are more readily transduced than mouse T cells, but the challenges to use as a T cell display system may be the difficulty in post-transduction selection and expansion (i.e. growth properties of hybridomas compared to non-transformed cells), and the possible complications due to CD4 and CD8 enhancement of pepMHC binding.
A second, and most important, issue has to do with the size of the libraries. Here we estimated from transduction efficiencies and the number of T cells transduced that the maximum size of the T cell library was 104 but the actual size may have been lower than this based on other estimates. For example, we approximate that 0.3% of the library appears to have been represented by the DIMPP clone (i.e. ~0.7% of the sorted library yielded almost 50% positive cells after a single sort, and all of the isolated clones appeared to have the same sequence, DIMPP). This might argue that the library was closer to 103 in diversity. An independent approach to estimating the size examined what sequence motifs might be required to generate the high-affinity TCRs in this system. In this approach, we assumed that the minimal “high-affinity” solution for the 2C CDR3α is a proline at position 102, with a few other flanking residues that have some preferences ((Holler et al., 2003) and the present report). Proline would be at this position in 2 out of the 32 possible codons encoded by the library (6.2%). If several of the other residues in this sequence contributed to the higher affinity, but were not required, one could argue that the DIMPP solution represented a clone from among 102 to 103 possible sequences. Accordingly, the DIMPP TCR (m100) maintained higher affinity binding to SIY/Kb, but other proline-containing solutions with equal or higher affinity (such as m33) were not present in the original library. This would explain why m100 was the only clone selected even though it had lower affinity than m33 or other proline-containing TCRs obtained from the yeast display library, which had a size of 5 × 105.
This analysis suggests that if a TCR has a high degree of sequence restriction (especially across multiple residues) in order to achieve higher affinity, it will be difficult to generate libraries of the appropriate sizes. However, we believe that it is possible to generate library sizes that are at least one to two orders of magnitude beyond what was achieved here, and to do so without adding the complications of multiple virions per cell. For example, using up to 107 host T cells, instead of 105 as used here, would already provide a library of 100-fold greater diversity. In addition, recent results have suggested that the solutions to generating TCRs with affinities that are CD8-independent may be relatively prevalent since simple single amino acid substitutions of a TCR generated such leads (Robbins et al., 2008). This result is no doubt related to the observation that the affinities necessary to generate CD8 independence are in the high nanomolar range (Holler and Kranz, 2003) (as with m100) rather than the low nanomolar range (as with m33) and the frequency of such mutants in a library can be expected to be higher than the very high-affinity TCRs. Finally, even though the size of the T cell libraries may be relatively small compared to other systems, this approach can thus be used to assess several orders of magnitude more sequence space than could be achieved by single-site mutagenesis (Robbins et al., 2008).
In summary, we believe that it should be possible to apply the T cell display approach used here to engineer a wider range of TCRs than has currently been possible by other methods. The isolates can be directly assessed for function in the context of the selecting cell. The method has added promise since it appears that the affinity increases needed to attain CD8-independent activity are modest, to approximately 500 to 2,000 nM KD values, and we show that it is possible to use the tetramer sorting strategy to identify TCRs that are in this range. The most significant advantage of using TCRs with higher affinities may be the ability to recruit CD4-positive T cells against tumor or viral antigen/class I targets. In addition, affinity increases in the TCR could increase the sensitivity of T cells and allow the T cells to recognize mutated target antigens.
Acknowledgement
We thank Phil Holler for advice and discussions, Natlie Bowerman for providing the biotinylated m67 TCR, and the staff of the University of Illinois Biotechnology Center Flow Cytometry Facility for assistance with flow cytometry. This work was supported by NIH grant GM55767 (to D.M.K.). D.H.A was supported as a trainee by NIH grant T32 GM07283.
Abbreviations
- β2m
β2 microglobulin
- BSA
bovine serum albumin
- C
constant
- CDR
complementarity determining region
- mAb
monoclonal antibody
- MHC
major histocompatibility complex
- OVA
SIINFEKL peptide
- PBS
phosphate buffered saline
- pep
peptide
- SIY
SIYRYYGL peptide
- TCR
T cell receptor
- V
variable
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alam SM, Travers PJ, Wung JL, Nasholds W, Redpath S, Jameson SC, Gascoigne NR. T-cell-receptor affinity and thymocyte positive selection. Nature. 1996;381:616–620. doi: 10.1038/381616a0. [DOI] [PubMed] [Google Scholar]
- Blank C, Brown I, Peterson AC, Spiotto M, Iwai Y, Honjo T, Gajewski TF. PD-L1/B7H-1 inhibits the effector phase of tumor rejection by T cell receptor (TCR) transgenic CD8+ T cells. Cancer Res. 2004;64:1140–1145. doi: 10.1158/0008-5472.can-03-3259. [DOI] [PubMed] [Google Scholar]
- Blattman JN, Greenberg PD. Cancer immunotherapy: a treatment for the masses. Science. 2004;305:200–205. doi: 10.1126/science.1100369. [DOI] [PubMed] [Google Scholar]
- Boder ET, Wittrup KD. Yeast surface display for screening combinatorial polypeptide libraries. Nat Biotechnol. 1997;15:553–557. doi: 10.1038/nbt0697-553. [DOI] [PubMed] [Google Scholar]
- Boulter JM, Jakobsen BK. Stable, soluble, high-affinity, engineered T cell receptors: novel antibody-like proteins for specific targeting of peptide antigens. Clin Exp Immunol. 2005;142:454–460. doi: 10.1111/j.1365-2249.2005.02929.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Call ME, Wucherpfennig KW. The T cell receptor: critical role of the membrane environment in receptor assembly and function. Annu Rev Immunol. 2005;23:101–125. doi: 10.1146/annurev.immunol.23.021704.115625. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Prusoff WH. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- Cho BK, Lian KC, Lee P, Brunmark A, McKinley C, Chen J, Kranz DM, Eisen HN. Differences in antigen recognition and cytolytic activity of CD8(+) and CD8(−) T cells that express the same antigen-specific receptor. Proc Natl Acad Sci U S A. 2001;98:1723–1727. doi: 10.1073/pnas.98.4.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels MA, Jameson SC. Critical role for CD8 in T cell receptor binding and activation by peptide/major histocompatibility complex multimers. J Exp Med. 2000;191:335–346. doi: 10.1084/jem.191.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels MA, Teixeiro E, Gill J, Hausmann B, Roubaty D, Holmberg K, Werlen G, Hollander GA, Gascoigne NR, Palmer E. Thymic selection threshold defined by compartmentalization of Ras/MAPK signalling. Nature. 2006;444:724–729. doi: 10.1038/nature05269. [DOI] [PubMed] [Google Scholar]
- Davis MM, Boniface JJ, Reich Z, Lyons D, Hampl J, Arden B, Chien Y. Ligand recognition by alpha beta T cell receptors. Annu Rev Immunol. 1998;16:523–544. doi: 10.1146/annurev.immunol.16.1.523. [DOI] [PubMed] [Google Scholar]
- Degano M, Garcia KC, Apostolopoulos V, Rudolph MG, Teyton L, Wilson IA. A functional hot spot for antigen recognition in a superagonist TCR/MHC complex. Immunity. 2000;12:251–261. doi: 10.1016/s1074-7613(00)80178-8. [DOI] [PubMed] [Google Scholar]
- Dudley ME, Rosenberg SA. Adoptive-cell-transfer therapy for the treatment of patients with cancer. Nat Rev Cancer. 2003;3:666–675. doi: 10.1038/nrc1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels B, Uckert W. Redirecting T lymphocyte specificity by T cell receptor gene transfer--a new era for immunotherapy. Mol Aspects Med. 2007;28:115–142. doi: 10.1016/j.mam.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Gai SA, Wittrup KD. Yeast surface display for protein engineering and characterization. Curr Opin Struct Biol. 2007;17:467–473. doi: 10.1016/j.sbi.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallegos AM, Bevan MJ. Central tolerance: good but imperfect. Immunol Rev. 2006;209:290–296. doi: 10.1111/j.0105-2896.2006.00348.x. [DOI] [PubMed] [Google Scholar]
- Garcia KC, Tallquist MD, Pease LR, Brunmark A, Scott CA, Degano M, Stura EA, Peterson PA, Wilson IA, Teyton L. Alphabeta T cell receptor interactions with syngeneic and allogeneic ligands: affinity measurements and crystallization. Proc Natl Acad Sci U S A. 1997;94:13838–13843. doi: 10.1073/pnas.94.25.13838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia KC, Teyton L, Wilson IA. Structural basis of T cell recognition. Annu Rev Immunol. 1999;17:369–397. doi: 10.1146/annurev.immunol.17.1.369. [DOI] [PubMed] [Google Scholar]
- Gorski J, Yassai M, Zhu X, Kissela B, Kissella B, Keever C, Flomenberg N. Circulating T cell repertoire complexity in normal individuals and bone marrow recipients analyzed by CDR3 size spectratyping. Correlation with immune status. J Immunol. 1994;152:5109–5119. [PubMed] [Google Scholar]
- Ho WY, Blattman JN, Dossett ML, Yee C, Greenberg PD. Adoptive immunotherapy: engineering T cell responses as biologic weapons for tumor mass destruction. Cancer Cell. 2003;3:431–437. doi: 10.1016/s1535-6108(03)00113-2. [DOI] [PubMed] [Google Scholar]
- Hogquist KA, Baldwin TA, Jameson SC. Central tolerance: learning self-control in the thymus. Nat Rev Immunol. 2005;5:772–782. doi: 10.1038/nri1707. [DOI] [PubMed] [Google Scholar]
- Holler PD, Chlewicki LK, Kranz DM. TCRs with high affinity for foreign pMHC show self-reactivity. Nat Immunol. 2003;4:55–62. doi: 10.1038/ni863. [DOI] [PubMed] [Google Scholar]
- Holler PD, Holman PO, Shusta EV, O'Herrin S, Wittrup KD, Kranz DM. In vitro evolution of a T cell receptor with high affinity for peptide/MHC. Proc Natl Acad Sci U S A. 2000;97:5387–5392. doi: 10.1073/pnas.080078297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holler PD, Kranz DM. Quantitative analysis of the contribution of TCR/pepMHC affinity and CD8 to T cell activation. Immunity. 2003;18:255–264. doi: 10.1016/s1074-7613(03)00019-0. [DOI] [PubMed] [Google Scholar]
- Holler PD, Lim AR, Cho BK, Rund LA, Kranz DM. CD8(−) T cell transfectants that express a high affinity T cell receptor exhibit enhanced peptide-dependent activation. J Exp Med. 2001;194:1043–1052. doi: 10.1084/jem.194.8.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones LL, Brophy SE, Bankovich AJ, Colf LA, Hanick NA, Garcia KC, Kranz DM. Engineering and characterization of a stabilized alpha1/alpha2 module of the class I major histocompatibility complex product Ld. J Biol Chem. 2006;281:25734–25744. doi: 10.1074/jbc.M604343200. [DOI] [PubMed] [Google Scholar]
- Jones LL, Colf LA, Stone JD, Garcia KC, Kranz DM. Distinct CDR3 conformations in T cell receptors determine the level of cross-reactivity for diverse antigens, but not the docking orientation. J Immunol. 2008 doi: 10.4049/jimmunol.181.9.6255. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappler JW, Roehm N, Marrack P. T cell tolerance by clonal elimination in the thymus. Cell. 1987;49:273–280. doi: 10.1016/0092-8674(87)90568-x. [DOI] [PubMed] [Google Scholar]
- Kearse KP, Roberts JL, Singer A. TCR alpha-CD3 delta epsilon association is the initial step in alpha beta dimer formation in murine T cells and is limiting in immature CD4+ CD8+ thymocytes. Immunity. 1995;2:391–399. doi: 10.1016/1074-7613(95)90147-7. [DOI] [PubMed] [Google Scholar]
- Kessels HW, van Den Boom MD, Spits H, Hooijberg E, Schumacher TN. Changing T cell specificity by retroviral T cell receptor display. Proc Natl Acad Sci U S A. 2000;97:14578–14583. doi: 10.1073/pnas.97.26.14578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieke MC, Shusta EV, Boder ET, Teyton L, Wittrup KD, Kranz DM. Selection of functional T cell receptor mutants from a yeast surface-display library. Proc Natl Acad Sci U S A. 1999;96:5651–5656. doi: 10.1073/pnas.96.10.5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz DM, Tonegawa S, Eisen HN. Attachment of an anti-receptor antibody to non-target cells renders them susceptible to lysis by a clone of cytotoxic T lymphocytes. Proc Natl Acad Sci U S A. 1984;81:7922–7926. doi: 10.1073/pnas.81.24.7922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhns MS, Davis MM, Garcia KC. Deconstructing the form and function of the TCR/CD3 complex. Immunity. 2006;24:133–139. doi: 10.1016/j.immuni.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Letourneur F, Malissen B. Derivation of a T cell hybridoma variant deprived of functional T cell receptor alpha and beta chain transcripts reveals a nonfunctional alpha-mRNA of BW5147 origin. Eur J Immunol. 1989;19:2269–2274. doi: 10.1002/eji.1830191214. [DOI] [PubMed] [Google Scholar]
- Li Y, Moysey R, Molloy PE, Vuidepot AL, Mahon T, Baston E, Dunn S, Liddy N, Jacob J, Jakobsen BK, Boulter JM. Directed evolution of human T-cell receptors with picomolar affinities by phage display. Nat Biotechnol. 2005;23:349–354. doi: 10.1038/nbt1070. [DOI] [PubMed] [Google Scholar]
- Lipovsek D, Lippow SM, Hackel BJ, Gregson MW, Cheng P, Kapila A, Wittrup KD. Evolution of an interloop disulfide bond in high-affinity antibody mimics based on fibronectin type III domain and selected by yeast surface display: molecular convergence with single-domain camelid and shark antibodies. J Mol Biol. 2007;368:1024–1041. doi: 10.1016/j.jmb.2007.02.029. [DOI] [PubMed] [Google Scholar]
- Matsui K, Boniface JJ, Steffner P, Reay PA, Davis MM. Kinetics of T-cell receptor binding to peptide/I-Ek complexes: correlation of the dissociation rate with T-cell responsiveness. Proc Natl Acad Sci U S A. 1994;91:12862–12866. doi: 10.1073/pnas.91.26.12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molloy PE, Sewell AK, Jakobsen BK. Soluble T cell receptors: novel immunotherapies. Curr Opin Pharmacol. 2005;5:438–443. doi: 10.1016/j.coph.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Morgan RA, Dudley ME, Wunderlich JR, Hughes MS, Yang JC, Sherry RM, Royal RE, Topalian SL, Kammula US, Restifo NP, Zheng Z, Nahvi A, de Vries CR, Rogers-Freezer LJ, Mavroukakis SA, Rosenberg SA. Cancer regression in patients after transfer of genetically engineered lymphocytes. Science. 2006;314:126–129. doi: 10.1126/science.1129003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman SA, Kranz DM. Display, engineering, and applications of antigen-specific T cell receptors. Biomol Eng. 2007;24:361–373. doi: 10.1016/j.bioeng.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Robbins PF, Li YF, El-Gamil M, Zhao Y, Wargo JA, Zheng Z, Xu H, Morgan RA, Feldman SA, Johnson LA, Bennett AD, Dunn SM, Mahon TM, Jakobsen BK, Rosenberg SA. Single and dual amino acid substitutions in TCR CDRs can enhance antigen-specific T cell functions. J Immunol. 2008;180:6116–6131. doi: 10.4049/jimmunol.180.9.6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg SA, Restifo NP, Yang JC, Morgan RA, Dudley ME. Adoptive cell transfer: a clinical path to effective cancer immunotherapy. Nat Rev Cancer. 2008;8:299–308. doi: 10.1038/nrc2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph MG, Stanfield RL, Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annu Rev Immunol. 2006;24:419–466. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- Russell SJ, Hawkins RE, Winter G. Retroviral vectors displaying functional antibody fragments. Nucleic Acids Res. 1993;21:1081–1085. doi: 10.1093/nar/21.5.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schodin BA, Tsomides TJ, Kranz DM. Correlation between the number of T cell receptors required for T cell activation and TCR-ligand affinity. Immunity. 1996;5:137–146. doi: 10.1016/s1074-7613(00)80490-2. [DOI] [PubMed] [Google Scholar]
- Schumacher TN. T-cell-receptor gene therapy. Nat Rev Immunol. 2002;2:512–519. doi: 10.1038/nri841. [DOI] [PubMed] [Google Scholar]
- Shusta EV, Holler PD, Kieke MC, Kranz DM, Wittrup KD. Directed evolution of a stable scaffold for T-cell receptor engineering. Nat Biotechnol. 2000;18:754–759. doi: 10.1038/77325. [DOI] [PubMed] [Google Scholar]
- Spiotto MT, Yu P, Rowley DA, Nishimura MI, Meredith SC, Gajewski TF, Fu YX, Schreiber H. Increasing tumor antigen expression overcomes "ignorance" to solid tumors via crosspresentation by bone marrow-derived stromal cells. Immunity. 2002;17:737–747. doi: 10.1016/s1074-7613(02)00480-6. [DOI] [PubMed] [Google Scholar]
- Sykulev Y, Brunmark A, Jackson M, Cohen RJ, Peterson PA, Eisen HN. Kinetics and affinity of reactions between an antigen-specific T cell receptor and peptide-MHC complexes. Immunity. 1994;1:15–22. doi: 10.1016/1074-7613(94)90005-1. [DOI] [PubMed] [Google Scholar]
- Tereshko V, Uysal S, Koide A, Margalef K, Koide S, Kossiakoff AA. Toward chaperone-assisted crystallography: protein engineering enhancement of crystal packing and X-ray phasing capabilities of a camelid single-domain antibody (VHH) scaffold. Protein Sci. 2008;17:1175–1187. doi: 10.1110/ps.034892.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DL, Kranz DM, Roy EJ. Experimental manipulations of afferent immune responses influence efferent immune responses to brain tumors. Cancer Immunol Immunother. 2008;57:1323–1333. doi: 10.1007/s00262-008-0467-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udaka K, Wiesmuller KH, Kienle S, Jung G, Walden P. Self-MHC-restricted peptides recognized by an alloreactive T lymphocyte clone. J Immunol. 1996;157:670–678. [PubMed] [Google Scholar]
- Urban JH, Schneider RM, Compte M, Finger C, Cichutek K, Alvarez-Vallina L, Buchholz CJ. Selection of functional human antibodies from retroviral display libraries. Nucleic Acids Res. 2005;33:e35. doi: 10.1093/nar/gni033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Merwe PA, Davis SJ. Molecular interactions mediating T cell antigen recognition. Annu Rev Immunol. 2003;21:659–684. doi: 10.1146/annurev.immunol.21.120601.141036. [DOI] [PubMed] [Google Scholar]
- Weber KS, Donermeyer DL, Allen PM, Kranz DM. Class II-restricted T cell receptor engineered in vitro for higher affinity retains peptide specificity and function. Proc Natl Acad Sci U S A. 2005;102:19033–19038. doi: 10.1073/pnas.0507554102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegener AM, Letourneur F, Hoeveler A, Brocker T, Luton F, Malissen B. The T cell receptor/CD3 complex is composed of at least two autonomous transduction modules. Cell. 1992;68:83–95. doi: 10.1016/0092-8674(92)90208-t. [DOI] [PubMed] [Google Scholar]
- Zhang B, Bowerman NA, Salama JK, Schmidt H, Spiotto MT, Schietinger A, Yu P, Fu YX, Weichselbaum RR, Rowley DA, Kranz DM, Schreiber H. Induced sensitization of tumor stroma leads to eradication of established cancer by T cells. J Exp Med. 2007;204:49–55. doi: 10.1084/jem.20062056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Karrison T, Rowley DA, Schreiber H. IFN-gamma- and TNF-dependent bystander eradication of antigen-loss variants in established mouse cancers. J Clin Invest. 2008;118:1398–1404. doi: 10.1172/JCI33522. [DOI] [PMC free article] [PubMed] [Google Scholar]