Abstract
The genome of epithelial tumors is characterized by numerous chromosomal aberrations, DNA base sequence changes, and epigenetic abnormalities. The epigenome of cancer cells has been most commonly studied at the level of DNA CpG methylation. In squamous cell carcinomas of the lung, CpG methylation patterns undergo substantial changes relative to normal lung epithelium. Using a genome-scale mapping technique for CpG methylation (MIRA-chip), we characterized CpG island methylation and methylation patterns of entire chromosome arms at a level of resolution of ∼100 bp. In individual stage I lung carcinomas, several hundred and probably up to a thousand CpG islands become methylated. Interestingly, a large fraction (almost 80%) of the tumor-specifically methylated sequences are targets of the Polycomb complex in embryonic stem cells. Homeobox genes are particularly overrepresented and all four HOX gene loci on chromosomes 2, 7, 12, and 17 are hotspots for tumor-associated methylation because of the presence of multiple methylated CpG islands within these loci. DNA hypomethylation at CpGs in squamous cell tumors preferentially affects repetitive sequence classes including SINEs, LINEs, subtelomeric repeats, and segmental duplications. Since these epigenetic changes are found in early stage tumors, their contribution to tumor etiology should be considered as well as their potential usefulness as diagnostic or prognostic biomarkers of the disease.
1. Mammalian DNA methylation
The only known enzymatic modification of DNA bases in mammalian cells is the post-replicative addition of a methyl group to position 5 of cytosines. The methylated cytosines are almost exclusively formed at the CpG (5′) dinucleotide sequence. CpG methylation is catalyzed by DNA methyltransferase proteins (DNMTs). DNA methyltransferase 1 (DNMT1) is responsible for faithful copying of the preexisting cellular DNA methylation patterns following DNA replication. DNMT3A and DNMT3B are primarily thought of as de novo DNA methyltransferases responsible for methylation of previously unmethylated CpG sites [1] although all DNMTs are generally important for the maintenance of methylation patterns [2]. The DNMT2 protein was initially characterized as a DNA methyltransferase [3] but more recently has been shown to mediate tRNA methylation [4]. Removal of methyl groups from DNA cytosines can be accomplished by a passive ‘dilution’ process involving DNA replication in the absence of DNMT proteins. Alternatively, the methyl group or the entire methylated base may be removed in an active enzymatic pathway. However, the exact nature of the putative mammalian DNA demethylase has remained obscure and controversial [5]. The distribution of CpG sequences along mammalian chromosomes is not uniform. Sequences near transcription start sites, and often including the first exon and intron of a gene, have a much higher frequency of CpG dinucleotides than the rest of the genome. These sequences are called CpG islands [6]. Only about half or less of all CpG islands, however, are associated with protein-coding genes leading to the assumption that CpG islands may have other regulatory roles. CpG islands are thought to remain completely unmethylated in the germ line thus avoiding mutational erosion and CpG loss due to methylation-associated mutagenic mechanisms [7]. Methylation of CpG islands near promoters leads to gene inactivation by several known mechanisms. The binding of certain transcription factors is directly prevented by DNA CpG methylation [8]. Methylated DNA sequences are bound by specialized proteins that have a high affinity for methylated DNA. Examples are MeCP2, MBD1, MBD2. These methyl-CpG binding proteins have the ability to recruit histone deacetylase complexes upon binding to mCpG DNA [9]. The CpG-methylated DNA is often associated with inactive chromatin marks, including deacetylated histones H3 and H4, histone H3 lysine 9 (H3K9) methylation and histone H3 lysine 27 (H3K27) methylation, chromatin configurations, which reinforce the inactive gene expression state.
2. DNA methylation changes in cancer
Changes in DNA methylation patterns are one of the most frequent events that occur in human tumors, and altered CpG methylation patterns discriminate tumor tissue from its nonmalignant counterpart tissue or normal adjacent tissue [10]. Two types of methylation changes are most commonly observed: hypermethylation of CpG islands and a more global hypomethylation of DNA in tumors. The literature now contains thousands of reports that have documented methylation of CpG islands associated with hundreds of different genes, including almost every type of human solid tumor or hematological malignancy. It is unlikely that all of these methylation changes play a causative role in tumorigenesis, and it is a challenge today to pinpoint those crucial genes that are susceptible to methylation-associated gene silencing and are functionally important in preventing tumorigenesis. Tumor-specific methylation may provide a means for detection and early diagnosis of cancer. Identification of methylated CpG islands in easily accessible biological materials such as serum, sputum or urine has the potential to be useful for the early diagnosis of lung cancer and other malignancies [11-13]. If methylation of CpG islands were a critical parameter in tumor maintenance or progression, it would be desirable to reverse DNA hypermethylation. This can be accomplished, at least transiently and in vitro, by treatment of cells with inhibitors of DNA methylation. The prototype of such inhibitors is 5-azacytidine [14]. The potential clinical use of 5-azacytidine and other more recently developed DNA methylation inhibitors as anti-cancer drugs is now being explored by many investigators.
Repetitive DNA elements, such as short and long interspersed nuclear elements (SINEs and LINEs) and other repeat sequences are often hypomethylated in tumors [15-23]. While it seems plausible that methylation-induced silencing of a critical tumor suppressor gene can be an important event in tumorigenesis, the biological significance of tumor-associated DNA hypomethylation is less clear [17,22]. Mouse models of DNA hypomethylation have suggested that loss of methylation can lead to tumor formation. Mice carrying a hypomorphic allele of DNMT1 are susceptible to the development of aggressive T cell lymphomas [24]. One hypothesis that links hypomethylation mechanistically to tumorigenesis is the induction of genomic instability by hypomethylation, either by reactivation of transposable elements or by chromosome rearrangement events directly associated with hypomethylation [25-28].
The mechanisms of CpG island hypermethylation in cancer are mostly unknown. Specific DNA sequences within CpG islands may be associated with the methylation process [29,30]. Whether these sequences are associated with DNA binding proteins in vivo that somehow attract methylation is not known. Others have proposed that gene inactivity imposed by changes in chromatin structure or histone modification predisposes to DNA methylation [31-34]. Specific chromatin configurations may either protect from methylation or may promote DNA methylation at CpG islands. Trimethylation of histone H3 lysine 4 (H3K4me3) is associated with active or potentially active genes and unmethylated CpG islands [35]. This modification interferes with binding of the de novo DNA methyltransferase DNMT3L/DNMT3A complex [36,37]. Trimethylation of histone H3 lysine 27 (H3K27me3), the histone modification mark established by Polycomb repressive complexes [38], is often associated with repression of developmentally regulated genes. Presence of this mark in stem cells has been associated with DNA methylation of the same sequences in human cancers [39-42] (see also below). Although this connection is mostly based on indirect comparisons between different cell types, the striking coincidence of H3K27me3 and DNA CpG methylation suggests a mechanistic connection that could explain methylation patterns in tumor cells. Our lack of understanding of these mechanisms is at least in part related to a lack of a comprehensive picture of genome-wide chromatin structure and DNA hypermethylation events in tumors, somatic stem cells and tumor progenitor cells.
3. DNA methylation and lung cancer
Lung cancer is the leading cause of cancer death in the United States and most other countries [43]. Its causation by cigarette smoking is unquestionable [44]. Lung cancer accounts for about 30% of all deaths from cancer and at least 1.5 million annual deaths from lung cancer are projected worldwide by 2010. The high (>80%) mortality rate associated with lung cancer is at least in part related to suboptimal therapeutic strategies and the lack of an efficient screening approach for early detection. In comparison, breast, colon or prostate cancers, for which early detection approaches exist, have much higher survival rates. Lung cancers are divided into small cell (SCLC) and non-small cell lung carcinomas (NSCLC) depending on histology and cellular origin. Non-small cell lung cancers are further classified on the basis of histological parameters into three subtypes: squamous cell carcinoma (SCC), adenocarcinoma (ADC) and large cell carcinoma (LCC). Squamous cell carcinomas often affect the central airways while adenocarcinomas arise in the peripheral areas of the lung.
During tumorigenesis, both alleles of a tumor suppressor gene need to be inactivated, for example by chromosomal deletions or loss-of-function mutations in the coding region of a gene. As an alternative mechanism, hypermethylation of CpG islands spanning the promoter regions of tumor suppressor genes (for example, RB, p16, VHL, APC, MLH1, RASSF1A and BRCA1) is a common and important mechanism in carcinogenesis [45-50]. Since hypermethylation generally leads to permanent inactivation of gene expression, this epigenetic alteration is considered to be a key pathway for long-term silencing of tumor suppressor genes.
The importance of CpG island methylation in functional inactivation of lung cancer suppressor genes is becoming increasingly recognized. From initial analysis of a subset of genes, it has been estimated that between 0.5% and 3% of all genes carrying CpG-rich promoter sequences may be silenced by DNA methylation in advanced stage lung cancer [46,51]. We recently reported that several hundred (∼200-800) CpG islands are methylated in individual stage I squamous cell carcinoma of the lung [23]. Some of the hypermethylated genes may be bona fide tumor suppressor genes, but in other cases the methylation event may be a consequence of preexisting tissue-specific gene silencing or may somehow be associated with tumor formation rather than being a cause of tumorigenesis. Several specific CpG-island-associated genes are methylated in lung cancer including, for example, p16, RASSF1A, RARbeta, MGMT, GSTP1, CDH13, APC, DAPK, TIMP3, and many others [52-58]. The methylation frequency (i.e., the percentage of tumors analyzed that carry methylated alleles) ranges from only a few percent to more than 80% for these genes. These methylation frequency numbers often differ substantially depending on the study population, tumor histology, and/or methodology used to assess CpG island methylation. In our laboratory, the tumor suppressor gene RASSF1A has been identified and characterized [47]. RASSF1A, which is localized at 3p21.3 in an area of common deletion or heterozygous loss in lung cancer, is inactivated by promoter methylation in about 30-40% of non-small cell lung cancers (37% of squamous cell carcinomas) and in close to 80% of small cell lung cancers [47,59-61].
4. DNA methylation detection methods
The field of DNA methylation analysis is moving fast towards genome-wide characterization rather than studying methylation of individual genes in tumors. Diverse technical approaches for large-scale methylation analysis have been developed [62]. The first group of techniques is based on methylation-sensitive restriction endonuclease cleavage of the target sequences (e.g., HpaII, NotI) [63,46,64]. These techniques are useful but are limited by the occurrence of the respective recognition sequences within a CpG island. One other variation of current methylation microarray approaches includes the use of the methylation-dependent restriction enzyme McrBC, which cleaves methylated DNA but not unmethylated DNA [65,66]. Another group of techniques employ bisulfite-induced cytosine to uracil conversion [67,68]. After bisulfite treatment of genomic DNA, the unmethylated cytosines are converted to uracils by deamination, while methylated cytosine residues can hardly react with this agent and remain intact. Bisulfite-based sequencing approaches are not yet cost-effectively applicable to genome-wide DNA methylation screening in mammalian systems but specific chromosomal areas have been sequenced [69]. Genome complexity is effectively reduced to three DNA bases (G, A, U or T) in addition to the rare base 5-methylcytosine after bisulfite conversion of cytosines, which makes it difficult to design specific probes for microarrays or to map unique sequence tags to the genome.
An indirect way to find methylated genes is to use gene expression arrays to identify genes reactivated by treatment with the DNA methylation inhibitor 5-aza-deoxycytidine [70-73]. However, this approach can only be used with cell lines and some CpG-methylated genes may be refractory to 5-aza-dC-induced reactivation.
An antibody against 5-methylcytosine has been used in immunoprecipitation experiments combined with microarrays [74,30] and this method, termed methylated DNA immunoprecipitation or MeDIP, is now becoming used quite commonly. However, this antibody is a resource that is difficult to standardize. The antibody requires single-stranded DNA for recognition, which is often not easily achieved in GC-rich DNA.
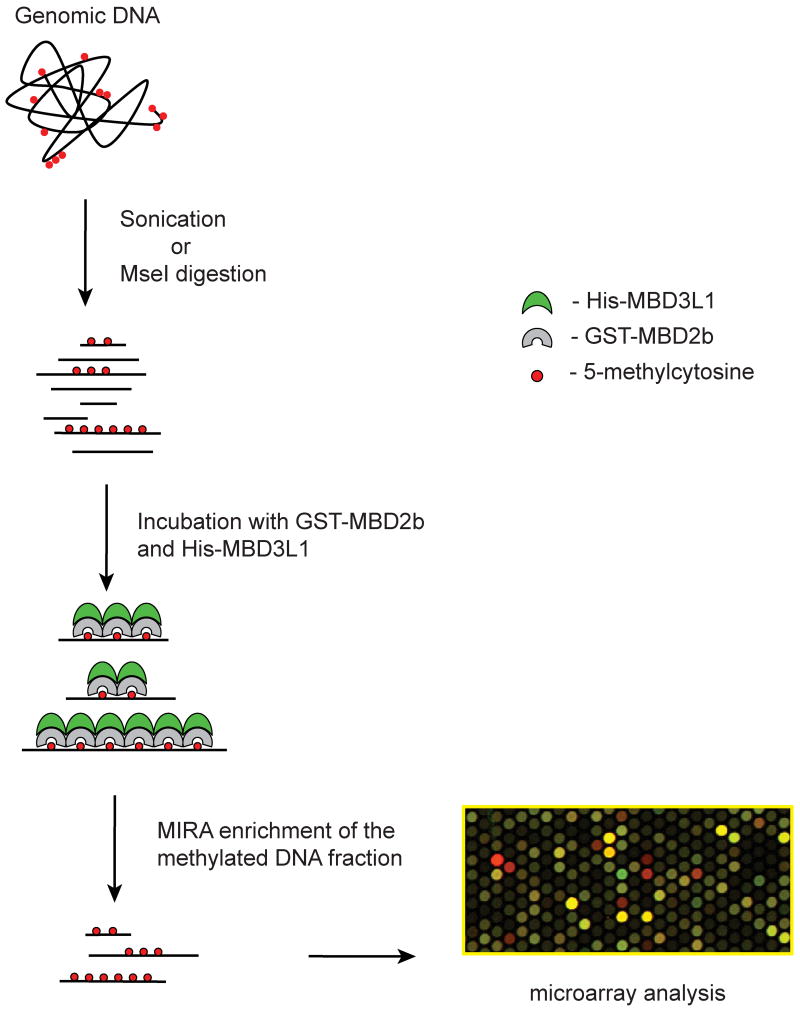
We have developed a sensitive methylation detection method, the methylated-CpG island recovery assay (MIRA), which is based on the high affinity of the MBD2/MBD3L1 complex for double-stranded CpG-methylated DNA (Figure 1). This method can be used to analyze the DNA methylation status of large numbers of genes simultaneously using microarray-based approaches or high-throughput sequencing. Methyl-CpG binding domain (MBD) proteins, such as MBD2, have the capacity to bind specifically to methylated DNA sequences but have little affinity to unmethylated DNA. MBD2b, the shorter isoform translated from the MBD2 mRNA, forms a heterodimer with a related protein, MBD3L1, which was identified and characterized in our laboratory [75,76]. MBD3L1 strongly increases the affinity of recombinant MBD2 for methylated DNA [77,78]. In the MIRA procedure, sonicated or restriction-cut genomic DNA isolated from normal or malignant tissue is incubated with GST-tagged MBD2b and His-MBD3L1. The bound methylated DNA is eluted from a glutathione affinity matrix, and used for hybridization to microarrays (MIRA-chip). The MIRA procedure has a high specificity for enriching methylated DNA and unmethylated DNA molecules stay in the supernatant (Rauch et al., 2005). MIRA requires two mCpG sites for efficient pulldown [78] and the signal depends on the number of methylated CpGs. In collaboration with Dr. Huidong Shi (Univ. of Missouri) and working with a 454 sequencer, we derived ∼500,000 sequencing reads of MIRA pulldown fragments derived from a lung cancer cell line. The MIRA-enriched fragments underwent bisulfite conversion before sequencing. As expected, >98% of the sequenced molecules were derived from CpG-rich methylated DNA including CpG islands and repetitive DNA (unpublished data). Thus, the efficiency of the MIRA approach depends on mCpG density and the approach seems to be ideally suited for examining CpG methylation patterns on a genome-wide scale. The MIRA method is several times more sensitive than MeDIP allowing analysis with less DNA (T.A. Rauch and G.P. Pfeifer, unpublished results). MIRA is compatible with several types of microarrays including those manufactured by Agilent, Affymetrix and NimbleGen. The MIRA-enriched DNA can be analyzed by high-throughput massive parallel sequencing as determined with a Roche 454 sequencer (H. Shi, T.A. Rauch, and G.P. Pfeifer, unpublished data) and a Solexa/Illumina G1 sequencer (H. Gao, T.A. Rauch, and G.P. Pfeifer, unpublished data). This variation of the MIRA technique, termed MIRA-seq, has potential to provide complete genome coverage and may supplement or replace the array-based MIRA-chip technique.
Figure 1. Outline of the MIRA-chip procedure.
Genomic DNA is fragmented to an average size of a few hundred base pairs, either by sonication of by cleavage with the restriction enzyme MseI. Methylated DNA molecules are enriched by incubation with the MBD2b-MBD3L1 protein complex. Input DNA and MIRA-enriched (methylated) fractions are labeled with different dyes, mixed, and hybridized to microarray slides. Alternatively, MIRA-enriched DNA from normal and tumor cells can be compared directly.
5. Analysis of lung carcinoma genomes by MIRA-chip analysis
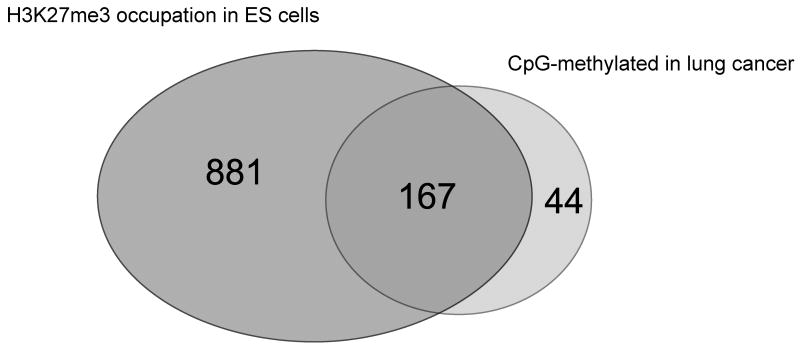
Initially, we used spotted CpG island arrays to analyze methylation patterns of the lung cancer cell line A549. Using the data obtained from such arrays, a list of genes was compiled that show hypermethylation in A549 lung cancer cells relative to normal human bronchial epithelial (NHBE) cells [78]. Cancer cell line-specific methylation and lack of methylation in NHBE cells was confirmed by bilsulfite-based analysis for several of the targets identified by the microarrays. Among the 25 targets verified at random in the list of the top 50 of the methylation targets, we have not identified any false positives. This indicates that the false positive discovery rate of MIRA microarrays is low (<4%). For analysis of primary lung cancers, we have been using CpG island microarrays from Agilent Technologies. These arrays contain 237,000 probes covering 27,800 CpG islands. We used Agilent CpG island arrays to characterize the methylated CpG islands in lung tumors in a comprehensive manner [40,23]. In primary stage I lung squamous cell carcinomas, we found that the number of methylated CpG islands ranged from ∼200 to more than 800 per each individual tumor [23]. This is likely an underestimation since some sequences may not amplify efficiently due to a long distance between MseI sites and other CpG-rich sequences may not be represented on the arrays. A large number of the genes methylated in lung tumors, in both adenocarcinomas and squamous cell carcinomas, were homeobox genes [40]. We found that all four HOX gene clusters on chromosomes 2, 7, 12, and 17 are preferential targets for DNA methylation in cancer cell lines and also in early stage lung tumors. CpG islands associated with many other homeobox genes, such as SIX, LHX, PAX, DLX and ENGRAILED, were highly methylated as well. Together, more than half (104 of 192) of all CpG island-associated homeobox genes in the lung cancer cell line A549 were methylated. The finding of widespread methylation of homeobox genes in lung cancers lends support to the hypothesis that a substantial fraction of genes methylated in cancer are targets of the Polycomb complex [78,39-42,34]. We analyzed the relationship between Polycomb marking in human ES cells and DNA CpG methylation in lung tumors for five lung squamous cell carcinomas. We considered a gene as a methylation target when 2 of the 5 tumors were methylated. This resulted in a list of 364 methylated CpG islands. For 211 of these genes, data on H3K27me occupancy in human ES cells was available [79]. Of these 211 methylated CpG islands, 167 (= 79%) were Polycomb targets as determined from published data on Polycomb marks in human embryonic stem cells (Figure 2). This is an astonishingly high percentage, and the correlation supports the Polycomb-DNA methylation connection. For the remaining 197 methylated genes, 153 contained the CpG island outside of promoters, and for those genes, Polycomb data are not available since a promoter array was used in the previous study [79]. Thus, it is quite possible that non-promoter CpG islands are also Polycomb targets and are therefore susceptible to methylation.
Figure 2. A large fraction of the methylated CpG islands in lung cancer is associated with the Polycomb complex in embryonic stem cells.
A CpG island was scored as methylation positive when it was methylated by MIRA microarray analysis in at least two of five squamous cell carcinomas. This list of genes was compared with the genes occupied by H3K27me3 in human embryonic stem cells according published data [79]. A Venn diagram shows the degree of overlap. Seventy-nine percent of the methylated promoter-associated CpG islands are Polycomb targets in embryonic stem cells.
In a recent study, we have used the MIRA method in combination with CpG island and genomic tiling arrays to characterize at high resolution the DNA methylation changes that occur in the genome of early stage lung squamous cell carcinomas. A number of new tumor-specific CpG island DNA methylation markers were identified as well as a specific defect in methylation of repetitive DNA elements [23]. Interestingly, normal tissues from different individuals showed overall very similar DNA methylation patterns at the level of resolution of these tiling arrays (∼100 bp). Each tumor contained several hundred CpG islands that were hypermethylated relative to the corresponding normal lung tissue. We identified and confirmed 12 CpG islands that were methylated in 70 to 100% of the 20 SCC tumors tested and these hold promise as effective biomarkers for early detection of lung cancer [23]. These markers included the CpG islands of the OTX1, BARHL2, MEIS1, OC2, PAX6, IRX2, TFAP2A, and EVX2 genes. These markers were highly specific for tumor-associated methylation, i.e. these CpG islands showed very little CpG methylation in blood DNA and in lung tissue of lung cancer-free individuals.
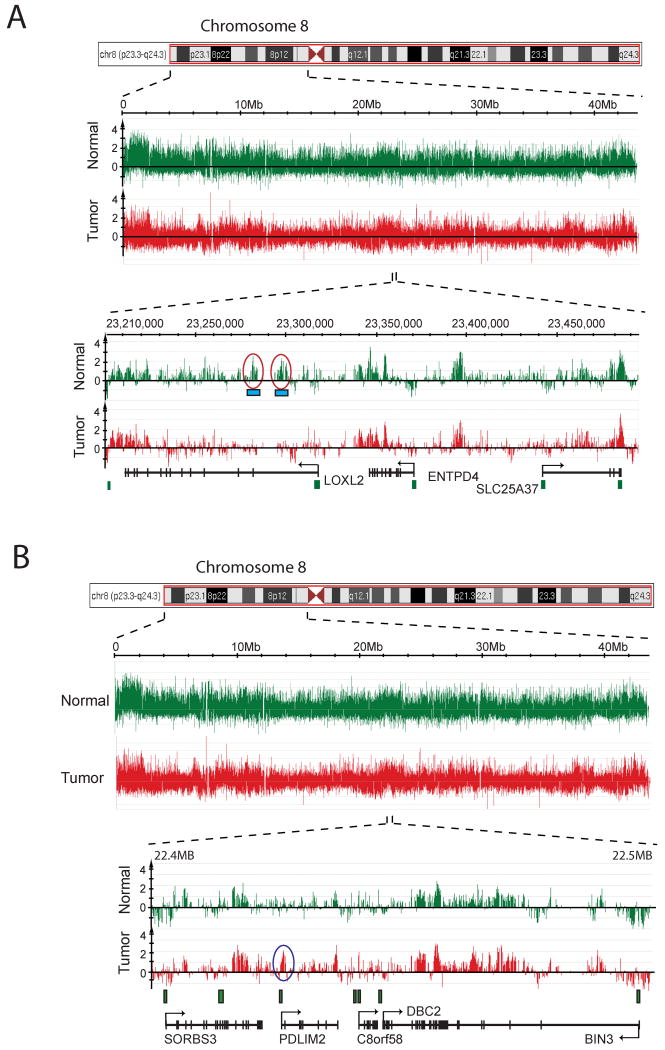
To analyze tumor-associated DNA methylation changes outside of CpG islands, we used chromosome tiling arrays from NimbleGen (Figure 3). On these tiling arrays, it was apparent that sequences near the centromeres and telomeres of individual chromosomes are more highly methylated than other parts of the chromosome. When zooming in towards much higher resolution, we could identify several hypermethylated CpG islands on each chromosome arm. These methylated islands were entirely consistent with those obtained from the Agilent CpG island arrays. When searching for cancer-associated DNA hypomethylation on chromosomes 7 and 8 and parts of chromosome 6, we found that extensive DNA hypomethylation in lung tumors occurred specifically at repetitive sequences, including SINE, LINE, and LTR elements, segmental duplications, and subtelomeric regions, but single copy sequences rarely became demethylated. The results are consistent with a specific defect in methylation of repetitive DNA sequences in human cancer [23].
Figure 3. DNA methylation patterns along the short arm of chromosome 8.
This Figure shows a schematic display of a chromosome 8 region and the methylation signals (log2 of MIRA-enriched DNA signal versus input DNA signal) in a normal lung tissue (green) and a matching stage I squamous cell lung tumor (red). A. The areas circled in red shows an example of hypomethylated DNA sequences that are associated with SINE and LINE elements (blue boxes). B. The area circled in blue shows an example of a hypermethylated CpG island at the PDLIM2 gene. PDLIM2 encodes a protein that may be involved in cytoskeleton formation and might contribute to tumor cell adhesion and migration [84]. In addition, PDLIM2 deficiency results in larger amounts of nuclear p65, a subunit of NF-kappaB, defective p65 ubiquitination and augmented production of proinflammatory cytokines [85].
The mechanism of tumor-associated DNA hypomethylation of repetitive DNA remains unknown. One possibility is that repetitive DNA is actively demethylated in cancer cells, perhaps through reactivation of a DNA demethylase gene that normally is expressed only at the zygote stage or in embryonic germ cells. However, the nature of the mammalian DNA demethylase has remained obscure [5]. Alternatively, CpG methylation of repetitive DNA may be lost during rapid DNA replication in cancer cells. Although the maintenance DNA methyltransferase DNMT1 is primarily responsible for copying CpG methylation patterns there is no evidence for a diminished DNMT1 function in cancer tissue. DNMT3A and DNMT3B are de novo DNA methyltransferases, and a related protein, DNMT3L, which is devoid of methyltransferase activity by itself, is capable of stimulating the activity of DNMT3A and DNMT3B [80]. DNMT3L regulates DNA methylation at imprinted sequences and at repeat sequences in mouse germ cells [81] but the role of these proteins in maintaining methylation of repeat sequences in somatic cells is not clear. Instead of invoking a defect in DNA methyltransferases, another possibility is that the accessibility of DNMTs to repetitive DNA is impeded during tumorigenesis. Further, small-RNA-based mechanisms that guide methylation to repetitive DNA through heterochromatin formation is also a formal possibility that has not been investigated and a defect in this pathway may be associated with cancer.
6. The significance of epigenetic changes in cancer
From recent large scale sequencing of human tumor DNA, it has become clear that recurrent changes in the DNA sequence, such as point mutations, insertions, or deletions within specific genes are quite uncommon in human tumors [82,83]. Most mutations seem to be rather stochastic and are rarely selected in specific genes (exception TP53, RAS genes, a few others). Chromosomal aberrations involving loss of genetic material, e.g. loss of heterozygosity, chromosomal deletions, copy number changes, or translocations are common in tumors, but recurrent clonal chromosomal aberrations are generally rare with the exception of leukemias. However, epigenetic changes are frequent events in all human tumors including lung cancers. These changes affect several hundred genes in individual tumors, and some of them quite frequently and repetitively in a patient population. Future studies will be aimed at determining the causative significance of these epigenetic alterations in tumorigenesis.
7. The challenges that lie ahead
The analysis of lung cancer epigenomes should be expanded to include the determination of genome-wide changes of histone modification patterns. How are these changes in chromatin correlated to changes in the DNA methylation pattern?
It is important to establish a temporal sequence of epigenomic alterations during the initiation and progression of lung squamous cell cancers. Early lesions including hyperplasia, dysplasia and carcinoma in situ should be analyzed to see which types of epigenetic events are the earliest ones that can be identified. This might provide important clues for the etiology of lung SCCs and the contribution of epigenetic changes to disease initiation and/or progression. For example, it could be hypothesized that mutational changes in critical genes are initiating events and epigenetic changes are events necessary for tumor promotion. The relative contribution of genetic and epigenetic changes as well as gross chromosomal aberrations (i.e. aneuploidy) to the different stages of lung tumorigenesis needs to be investigated.
The mechanisms of tumor-specific methylation changes remains to be determined. This applies to both CpG island hypermethylation and hypomethylation of repetitive DNA. It is unknown if perhaps a specific defect in the epigenetic machinery is responsible for both defects. An important question is if and how exposure of the lung tissue to cigarette smoke components can modulate DNA methylation patterns and other epigenetic chromatin marks. Squamous cell carcinomas almost always occur in current and former smokers. However, adenocarcinomas of the lung can be found in smokers as well as in nonsmokers and may provide a better system to investigate the effect of tobacco smoking on the epigenome.
The exact contribution of epigenetic aberrations, specifically of DNA methylation changes, to lung tumor etiology is not understood. More mechanistically based studies including mouse models of epigenomic abnormalities are required. The identification of epigenetically altered genes that truly contribute to lung carcinogenesis will be an important area of research.
The diagnosis of lung cancer at its earliest stages is important for the application of curative treatment regimens including surgery. Methylation biomarkers have great potential to become sensitive diagnostic tools for detecting early disease in blood serum or sputum. Although several promising methylation markers have now been identified, the development of sensitive and reliable techniques for detecting rare methylated DNA molecules in sputum, serum, or circulating tumor cells should now be a top priority.
Acknowledgments
The work of the authors has been supported by grants from the National Cancer Institute to G.P.P. (CA084469 and CA128495).
Footnotes
Conflict of interest statement: The authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chen T, Li E. Structure and function of eukaryotic DNA methyltransferases. Curr Top Dev Biol. 2004;60:55–89. doi: 10.1016/S0070-2153(04)60003-2. [DOI] [PubMed] [Google Scholar]
- 2.Chen ZX, Riggs AD. Maintenance and regulation of DNA methylation patterns in mammals. Biochem Cell Biol. 2005;83:438–448. doi: 10.1139/o05-138. [DOI] [PubMed] [Google Scholar]
- 3.Kunert N, Marhold J, Stanke J, Stach D, Lyko F. A Dnmt2-like protein mediates DNA methylation in Drosophila. Development. 2003;130:5083–5090. doi: 10.1242/dev.00716. [DOI] [PubMed] [Google Scholar]
- 4.Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X, Golic KG, Jacobsen SE, Bestor TH. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006;311:395–398. doi: 10.1126/science.1120976. [DOI] [PubMed] [Google Scholar]
- 5.Ooi SK, Bestor TH. The colorful history of active DNA demethylation. Cell. 2008;133:1145–1148. doi: 10.1016/j.cell.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 6.Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 7.Pfeifer GP. Mutagenesis at methylated CpG sequences. Curr Top Microbiol Immunol. 2006;301:259–281. doi: 10.1007/3-540-31390-7_10. [DOI] [PubMed] [Google Scholar]
- 8.Tate PH, Bird AP. Effects of DNA methylation on DNA-binding proteins and gene expression. Curr Opin Genet Dev. 1993;3:226–231. doi: 10.1016/0959-437x(93)90027-m. [DOI] [PubMed] [Google Scholar]
- 9.Bird AP, Wolffe AP. Methylation-induced repression--belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 10.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer. 2003;3:253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- 12.Belinsky SA. Gene-promoter hypermethylation as a biomarker in lung cancer. Nat Rev Cancer. 2004;4:707–717. doi: 10.1038/nrc1432. [DOI] [PubMed] [Google Scholar]
- 13.Ushijima T. Detection and interpretation of altered methylation patterns in cancer cells. Nat Rev Cancer. 2005;5:223–231. doi: 10.1038/nrc1571. [DOI] [PubMed] [Google Scholar]
- 14.Jones PA, Taylor SM. Cellular differentiation, cytidine analogs and DNA methylation. Cell. 1980;20:85–93. doi: 10.1016/0092-8674(80)90237-8. [DOI] [PubMed] [Google Scholar]
- 15.Burtseva NN, Azizov YM, Itkin BZ, Vanyushin BF. Changes in methylation of cattle lymphocyte DNA during chronic lymphoid leukosis. Biokhimiya. 1977:1690–1696. [Google Scholar]
- 16.Lapeyre JN, Becker FF. 5-Methylcytosine content of nuclear DNA during chemical hepatocarcinogenesis and in carcinomas which result. Biochem Biophys Res Commun. 1979;87:698–705. doi: 10.1016/0006-291x(79)92015-1. [DOI] [PubMed] [Google Scholar]
- 17.Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21:5400–5413. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
- 18.Weisenberger DJ, Campan M, Long TI, Kim M, Woods C, Fiala E, Ehrlich M, Laird PW. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res. 2005;33:6823–6836. doi: 10.1093/nar/gki987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cadieux B, Ching TT, Vandenberg SR, Costello JF. Genome-wide Hypomethylation in Human Glioblastomas Associated with Specific Copy Number Alteration, Methylenetetrahydrofolate Reductase Allele Status, and Increased Proliferation. Cancer Res. 2006;66:8469–8476. doi: 10.1158/0008-5472.CAN-06-1547. [DOI] [PubMed] [Google Scholar]
- 20.Rodriguez J, Frigola J, Vendrell E, Risques RA, Fraga MF, Morales C, Moreno V, Esteller M, Capella G, Ribas M, Peinado MA. Chromosomal Instability Correlates with Genome-wide DNA Demethylation in Human Primary Colorectal Cancers. Cancer Res. 2006;66:8462–9468. doi: 10.1158/0008-5472.CAN-06-0293. [DOI] [PubMed] [Google Scholar]
- 21.Estecio MR, Gharibyan V, Shen L, Ibrahim AE, Doshi K, He R, Jelinek J, Yang AS, Yan PS, Huang TH, Tajara EH, Issa JP. LINE-1 Hypomethylation in Cancer Is Highly Variable and Inversely Correlated with Microsatellite Instability. PLoS ONE. 2007;2:e399. doi: 10.1371/journal.pone.0000399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson AS, Power BE, Molloy PL. DNA hypomethylation and human diseases. Biochim Biophys Acta. 2007;1775:138–162. doi: 10.1016/j.bbcan.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 23.Rauch TA, Zhong X, Wu X, Wang M, Kernstine KH, Wang Z, Riggs AD, Pfeifer GP. High-resolution mapping of DNA hypermethylation and hypomethylation in lung cancer. Proc Natl Acad Sci U S A. 2008;105:252–257. doi: 10.1073/pnas.0710735105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaudet F, Hodgson JG, Eden A, Jackson-Grusby L, Dausman J, Gray JW, Leonhardt H, Jaenisch R. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300:489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- 25.Almeida A, Kokalj-Vokac N, Lefrancois D, Viegas-Pequignot E, Jeanpierre M, Dutrillaux B, Malfoy B. Hypomethylation of classical satellite DNA and chromosome instability in lymphoblastoid cell lines. Hum Genet. 1993;91:538–546. doi: 10.1007/BF00205077. [DOI] [PubMed] [Google Scholar]
- 26.Roman-Gomez J, Jimenez-Velasco A, Agirre X, Cervantes F, Sanchez J, Garate L, Barrios M, Castillejo JA, Navarro G, Colomer D, Prosper F, Heiniger A, Torres A. Promoter hypomethylation of the LINE-1 retrotransposable elements activates sense/antisense transcription and marks the progression of chronic myeloid leukemia. Oncogene. 2005;24:7213–7223. doi: 10.1038/sj.onc.1208866. [DOI] [PubMed] [Google Scholar]
- 27.Yamada Y, Jackson-Grusby L, Linhart H, Meissner A, Eden A, Lin H, Jaenisch R. Opposing effects of DNA hypomethylation on intestinal and liver carcinogenesis. Proc Natl Acad Sci U S A. 2005;102:13580–13585. doi: 10.1073/pnas.0506612102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Howard G, Eiges R, Gaudet F, Jaenisch R, Eden A. Activation and transposition of endogenous retroviral elements in hypomethylation induced tumors in mice. Oncogene. 2008;27:404–408. doi: 10.1038/sj.onc.1210631. [DOI] [PubMed] [Google Scholar]
- 29.Feltus FA, Lee EK, Costello JF, Plass C, Vertino PM. Predicting aberrant CpG island methylation. Proc Natl Acad Sci U S A. 2003;100:12253–12258. doi: 10.1073/pnas.2037852100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keshet I, Schlesinger Y, Farkash S, Rand E, Hecht M, Segal E, Pikarski E, Young RA, Niveleau A, Cedar H, Simon I. Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat Genet. 2006;38:149–153. doi: 10.1038/ng1719. [DOI] [PubMed] [Google Scholar]
- 31.Song JZ, Stirzaker C, Harrison J, Melki JR, Clark SJ. Hypermethylation trigger of the glutathione-S-transferase gene (GSTP1) in prostate cancer cells. Oncogene. 2002;21:1048–1061. doi: 10.1038/sj.onc.1205153. [DOI] [PubMed] [Google Scholar]
- 32.Bachman KE, Park BH, Rhee I, Rajagopalan H, Herman JG, Baylin SB, Kinzler KW, Vogelstein B. Histone modifications and silencing prior to DNA methylation of a tumor suppressor gene. Cancer Cell. 2003;3:89–95. doi: 10.1016/s1535-6108(02)00234-9. [DOI] [PubMed] [Google Scholar]
- 33.Strunnikova M, Schagdarsurengin U, Kehlen A, Garbe JC, Stampfer MR, Dammann R. Chromatin inactivation precedes de novo DNA methylation during the progressive epigenetic silencing of the RASSF1A promoter. Mol Cell Biol. 2005;25:3923–3933. doi: 10.1128/MCB.25.10.3923-3933.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hahn MA, Hahn T, Lee DH, Esworthy RS, Kim BW, Riggs AD, Chu FF, Pfeifer GP. Methylation of Polycomb target genes in intestinal cancer is mediated by inflammation. Cancer Res. 2008;68 doi: 10.1158/0008-5472.CAN-08-1957. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barrera LO, Li Z, Smith AD, Arden KC, Cavenee WK, Zhang MQ, Green RD, Ren B. Genome-wide mapping and analysis of active promoters in mouse embryonic stem cells and adult organs. Genome Res. 2008;18:46–59. doi: 10.1101/gr.6654808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature. 2007;449:248–251. doi: 10.1038/nature06146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, Cheng X, Bestor TH. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 39.Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, Mohammad HP, Chen W, Daniel VC, Yu W, Berman DM, Jenuwein T, Pruitt K, Sharkis SJ, Watkins DN, Herman JG, Baylin SB. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rauch T, Wang Z, Zhang X, Zhong X, Wu X, Lau SK, Kernstine KH, Riggs AD, Pfeifer GP. Homeobox gene methylation in lung cancer studied by genome-wide analysis with a microarray-based methylated CpG island recovery assay. Proc Natl Acad Sci U S A. 2007;104:5527–5532. doi: 10.1073/pnas.0701059104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, Eden E, Yakhini Z, Ben-Shushan E, Reubinoff BE, Bergman Y, Simon I, Cedar H. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 42.Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I, Laird PW. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 43.Edwards BK, Brown ML, Wingo PA, Howe HL, Ward E, Ries LA, Schrag D, Jamison PM, Jemal A, Wu XC, Friedman C, Harlan L, Warren J, Anderson RN, Pickle LW. Annual report to the nation on the status of cancer, 1975-2002, featuring population-based trends in cancer treatment. J Natl Cancer Inst. 2005;97:1407–1427. doi: 10.1093/jnci/dji289. [DOI] [PubMed] [Google Scholar]
- 44.Pfeifer GP, Denissenko MF, Olivier M, Tretyakova N, Hecht SS, Hainaut P. Tobacco smoke carcinogens, DNA damage and p53 mutations in smoking-associated cancers. Oncogene. 2002;21:7435–7451. doi: 10.1038/sj.onc.1205803. [DOI] [PubMed] [Google Scholar]
- 45.Issa JP. Aging, DNA methylation and cancer. Crit Rev Oncol Hematol. 1999;32:31–43. doi: 10.1016/s1040-8428(99)00019-0. [DOI] [PubMed] [Google Scholar]
- 46.Costello JF, Fruhwald MC, Smiraglia DJ, Rush LJ, Robertson GP, Gao X, Wright FA, Feramisco JD, Peltomaki P, Lang JC, Schuller DE, Yu L, Bloomfield CD, Caligiuri MA, Yates A, Nishikawa R, Su Huang H, Petrelli NJ, Zhang X, O'Dorisio MS, Held WA, Cavenee WK, Plass C. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat Genet. 2000;24:132–138. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- 47.Dammann R, Li C, Yoon JH, Chin PL, Bates S, Pfeifer GP. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nature Genet. 2000;25:315–319. doi: 10.1038/77083. [DOI] [PubMed] [Google Scholar]
- 48.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 49.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 50.Nephew KP, Huang TH. Epigenetic gene silencing in cancer initiation and progression. Cancer Lett. 2003;190:125–133. doi: 10.1016/s0304-3835(02)00511-6. [DOI] [PubMed] [Google Scholar]
- 51.Shiraishi M, Sekiguchi A, Terry MJ, Oates AJ, Miyamoto Y, Chuu YH, Munakata M, Sekiya T. A comprehensive catalog of CpG islands methylated in human lung adenocarcinomas for the identification of tumor suppressor genes. Oncogene. 2002;21:3804–3813. doi: 10.1038/sj.onc.1205454. [DOI] [PubMed] [Google Scholar]
- 52.Zochbauer-Muller S, Fong KM, Virmani AK, Geradts J, Gazdar AF, Minna JD. Aberrant promoter methylation of multiple genes in non-small cell lung cancers. Cancer Res. 2001;61:249–255. [PubMed] [Google Scholar]
- 53.Toyooka S, Maruyama R, Toyooka KO, McLerran D, Feng Z, Fukuyama Y, Virmani AK, Zochbauer-Muller S, Tsukuda K, Sugio K, Shimizu N, Shimizu K, Lee H, Chen CY, Fong KM, Gilcrease M, Roth JA, Minna JD, Gazdar AF. Smoke exposure, histologic type and geography-related differences in the methylation profiles of non-small cell lung cancer. Int J Cancer. 2003;103:153–160. doi: 10.1002/ijc.10787. [DOI] [PubMed] [Google Scholar]
- 54.Yanagawa N, Tamura G, Oizumi H, Takahashi N, Shimazaki Y, Motoyama T. Promoter hypermethylation of tumor suppressor and tumor-related genes in non-small cell lung cancers. Cancer Sci. 2003;94:589–592. doi: 10.1111/j.1349-7006.2003.tb01487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Franklin WA. Premalignant evolution of lung cancer: Gilles F Filley lecture. Chest. 2004;125:90S–94S. doi: 10.1378/chest.125.5_suppl.90s. [DOI] [PubMed] [Google Scholar]
- 56.Topaloglu O, Hoque MO, Tokumaru Y, Lee J, Ratovitski E, Sidransky D, Moon CS. Detection of promoter hypermethylation of multiple genes in the tumor and bronchoalveolar lavage of patients with lung cancer. Clin Cancer Res. 2004;10:2284–2288. doi: 10.1158/1078-0432.ccr-1111-3. [DOI] [PubMed] [Google Scholar]
- 57.Dammann R, Strunnikova M, Schagdarsurengin U, Rastetter M, Papritz M, Hattenhorst UE, Hofmann HS, Silber RE, Burdach S, Hansen G. CpG island methylation and expression of tumour-associated genes in lung carcinoma. Eur J Cancer. 2005;41:1223–1236. doi: 10.1016/j.ejca.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 58.Kim YT, Park SJ, Lee SH, Kang HJ, Hahn S, Kang CH, Sung SW, Kim JH. Prognostic implication of aberrant promoter hypermethylation of CpG islands in adenocarcinoma of the lung. J Thorac Cardiovasc Surg. 2005;130:1378. doi: 10.1016/j.jtcvs.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 59.Burbee DG, Forgacs E, Zochbauer-Muller S, Shivakumar L, Fong K, Gao B, Randle D, Kondo M, Virmani A, Bader S, Sekido Y, Latif F, Milchgrub S, Toyooka S, Gazdar AF, Lerman MI, Zabarovsky E, White M, Minna JD. Epigenetic inactivation of RASSF1A in lung and breast cancers and malignant phenotype suppression. J Natl Cancer Inst. 2001;93:691–699. doi: 10.1093/jnci/93.9.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dammann R, Takahashi T, Pfeifer GP. The CpG island of the novel tumor suppressor gene RASSF1A is intensely methylated in primary small cell lung carcinomas. Oncogene. 2001;20:3563–3567. doi: 10.1038/sj.onc.1204469. [DOI] [PubMed] [Google Scholar]
- 61.Dammann R, Schagdarsurengin U, Seidel C, Strunnikova M, Rastetter M, Baier K, Pfeifer GP. The tumor suppressor RASSF1A in human carcinogenesis: an update. Histol Histopathol. 2005;20:645–663. doi: 10.14670/HH-20.645. [DOI] [PubMed] [Google Scholar]
- 62.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 63.Ushijima T, Morimura K, Hosoya Y, Okonogi H, Tatematsu M, Sugimura T, Nagao M. Establishment of methylation-sensitive-representational difference analysis and isolation of hypo- and hypermethylated genomic fragments in mouse liver tumors. Proc Natl Acad Sci U S A. 1997;94:2284–2289. doi: 10.1073/pnas.94.6.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yan PS, Perry MR, Laux DE, Asare AL, Caldwell CW, Huang TH. CpG island arrays: an application toward deciphering epigenetic signatures of breast cancer. Clin Cancer Res. 2000;6:1432–1438. [PubMed] [Google Scholar]
- 65.Nouzova M, Holtan N, Oshiro MM, Isett RB, Munoz-Rodriguez JL, List AF, Narro ML, Miller SJ, Merchant NC, Futscher BW. Epigenomic changes during leukemia cell differentiation: analysis of histone acetylation and cytosine methylation using CpG island microarrays. J Pharmacol Exp Ther. 2004;311:968–981. doi: 10.1124/jpet.104.072488. [DOI] [PubMed] [Google Scholar]
- 66.Lippman Z, Gendrel AV, Colot V, Martienssen R. Profiling DNA methylation patterns using genomic tiling microarrays. Nat Methods. 2005;2:219–224. doi: 10.1038/nmeth0305-219. [DOI] [PubMed] [Google Scholar]
- 67.Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci U S A. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Clark SJ, Harrison J, Paul CL, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Eckhardt F, Lewin J, Cortese R, Rakyan VK, Attwood J, Burger M, Burton J, Cox TV, Davies R, Down TA, Haefliger C, Horton R, Howe K, Jackson DK, Kunde J, Koenig C, Liddle J, Niblett D, Otto T, Pettett R, Seemann S, Thompson C, West T, Rogers J, Olek A, Berlin K, Beck S. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Suzuki H, Gabrielson E, Chen W, Anbazhagan R, van Engeland M, Weijenberg MP, Herman JG, Baylin SB. A genomic screen for genes upregulated by demethylation and histone deacetylase inhibition in human colorectal cancer. Nat Genet. 2002;31:141–149. doi: 10.1038/ng892. [DOI] [PubMed] [Google Scholar]
- 71.Yamashita K, Upadhyay S, Osada M, Hoque MO, Xiao Y, Mori M, Sato F, Meltzer SJ, Sidransky D. Pharmacologic unmasking of epigenetically silenced tumor suppressor genes in esophageal squamous cell carcinoma. Cancer Cell. 2002;2:485–495. doi: 10.1016/s1535-6108(02)00215-5. [DOI] [PubMed] [Google Scholar]
- 72.Sato N, Fukushima N, Maitra A, Matsubayashi H, Yeo CJ, Cameron JL, Hruban RH, Goggins M. Discovery of novel targets for aberrant methylation in pancreatic carcinoma using high-throughput microarrays. Cancer Res. 2003;63:3735–3742. [PubMed] [Google Scholar]
- 73.Shi H, Wei SH, Leu YW, Rahmatpanah F, Liu JC, Yan PS, Nephew KP, Huang TH. Triple analysis of the cancer epigenome: an integrated microarray system for assessing gene expression, DNA methylation, and histone acetylation. Cancer Res. 2003;63:2164–2171. [PubMed] [Google Scholar]
- 74.Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 75.Jiang CL, Jin SG, Lee DH, Lan ZJ, Xu X, O'Connor TR, Szabo PE, Mann JR, Cooney AJ, Pfeifer GP. MBD3L1 and MBD3L2, two new proteins homologous to the methyl-CpG-binding proteins MBD2 and MBD3: characterization of MBD3L1 as a testis-specific transcriptional repressor. Genomics. 2002;80:621–629. doi: 10.1006/geno.2002.7001. [DOI] [PubMed] [Google Scholar]
- 76.Jiang CL, Jin SG, Pfeifer GP. MBD3L1 is a transcriptional repressor that interacts with methyl-CpG-binding protein 2 (MBD2) and components of the NuRD complex. J Biol Chem. 2004;279:52456–52464. doi: 10.1074/jbc.M409149200. [DOI] [PubMed] [Google Scholar]
- 77.Rauch T, Pfeifer GP. Methylated-CpG island recovery assay: a new technique for the rapid detection of methylated-CpG islands in cancer. Lab Invest. 2005;85:1172–1180. doi: 10.1038/labinvest.3700311. [DOI] [PubMed] [Google Scholar]
- 78.Rauch T, Li H, Wu X, Pfeifer GP. MIRA-assisted microarray analysis, a new technology for the determination of DNA methylation patterns, identifies frequent methylation of homeodomain-containing genes in lung cancer cells. Cancer Res. 2006;66:7939–7947. doi: 10.1158/0008-5472.CAN-06-1888. [DOI] [PubMed] [Google Scholar]
- 79.Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, Koseki H, Fuchikami T, Abe K, Murray HL, Zucker JP, Yuan B, Bell GW, Herbolsheimer E, Hannett NM, Sun K, Odom DT, Otte AP, Volkert TL, Bartel DP, Melton DA, Gifford DK, Jaenisch R, Young RA. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006b;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen ZX, Mann JR, Hsieh CL, Riggs AD, Chedin F. Physical and functional interactions between the human DNMT3L protein and members of the de novo methyltransferase family. J Cell Biochem. 2005;95:902–917. doi: 10.1002/jcb.20447. [DOI] [PubMed] [Google Scholar]
- 81.Bourc'his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- 82.Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, Edkins S, O'Meara S, Vastrik I, Schmidt EE, Avis T, Barthorpe S, Bhamra G, Buck G, Choudhury B, Clements J, Cole J, Dicks E, Forbes S, Gray K, Halliday K, Harrison R, Hills K, Hinton J, Jenkinson A, Jones D, Menzies A, Mironenko T, Perry J, Raine K, Richardson D, Shepherd R, Small A, Tofts C, Varian J, Webb T, West S, Widaa S, Yates A, Cahill DP, Louis DN, Goldstraw P, Nicholson AG, Brasseur F, Looijenga L, Weber BL, Chiew YE, DeFazio A, Greaves MF, Green AR, Campbell P, Birney E, Easton DF, Chenevix-Trench G, Tan MH, Khoo SK, Teh BT, Yuen ST, Leung SY, Wooster R, Futreal PA, Stratton MR. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, Silliman N, Szabo S, Dezso Z, Ustyanksky V, Nikolskaya T, Nikolsky Y, Karchin R, Wilson PA, Kaminker JS, Zhang Z, Croshaw R, Willis J, Dawson D, Shipitsin M, Willson JK, Sukumar S, Polyak K, Park BH, Pethiyagoda CL, Pant PV, Ballinger DG, Sparks AB, Hartigan J, Smith DR, Suh E, Papadopoulos N, Buckhaults P, Markowitz SD, Parmigiani G, Kinzler KW, Velculescu VE, Vogelstein B. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 84.Loughran G, Healy NC, Kiely PA, Huigsloot M, Kedersha NL, O'Connor R. Mystique is a new insulin-like growth factor-I-regulated PDZ-LIM domain protein that promotes cell attachment and migration and suppresses Anchorage-independent growth. Mol Biol Cell. 2005;16:1811–1822. doi: 10.1091/mbc.E04-12-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tanaka T, Grusby MJ, Kaisho T. PDLIM2-mediated termination of transcription factor NF-kappaB activation by intranuclear sequestration and degradation of the p65 subunit. Nat Immunol. 2007;8:584–591. doi: 10.1038/ni1464. [DOI] [PubMed] [Google Scholar]