Abstract
Secretion of cholera toxin and other virulence factors from Vibrio cholerae is mediated by the type II secretion (T2S) apparatus, a multiprotein complex composed of both inner and outer membrane proteins. To better understand the mechanism by which the T2S complex coordinates translocation of its substrates, we are examining the protein-protein interactions of its components, encoded by the extracellular protein secretion (eps) genes. In this study, we took a cell biological approach, observing the dynamics of fluorescently tagged EpsC and EpsM proteins in vivo. We report that the level and context of fluorescent protein fusion expression can have a bold effect on subcellular location and that chromosomal, intraoperon expression conditions are optimal for determining the intracellular locations of fusion proteins. Fluorescently tagged, chromosomally expressed EpsC and EpsM form discrete foci along the lengths of the cells, different from the polar localization for green fluorescent protein (GFP)-EpsM previously described, as the fusions are balanced with all their interacting partner proteins within the T2S complex. Additionally, we observed that fluorescent foci in both chromosomal GFP-EpsC- and GFP-EpsM-expressing strains disperse upon deletion of epsD, suggesting that EpsD is critical to the localization of EpsC and EpsM and perhaps their assembly into the T2S complex.
The type II secretion (T2S) pathway is widely used by pathogenic gram-negative bacteria for delivery of virulence factors into the extracellular milieu (11, 17, 46). Proteins destined for release through this pathway are first translocated across the cytoplasmic membrane via the Sec (24, 42) or Tat (59) machinery. Following folding and assembly in the periplasm, the proteins are transported across the outer membrane via the T2S machinery, a complex composed of 12 to 16 different gene products, depending on the species. In Vibrio cholerae, the elements of the T2S apparatus are encoded by the extracellular protein secretion (eps) genes, epsC through epsN and pilD (vcpD) (18, 31, 39, 49, 50). Together these proteins coordinate the outer membrane translocation of the major virulence factor, cholera toxin, as well as chitinase, lipase, hemagglutinin/protease, and other proteases (12, 27, 49). Our studies are focused on better understanding how the T2S complex assembles in the cell envelope of V. cholerae to begin to elucidate the mechanism by which extracellular secretion is accomplished.
The T2S apparatus is modeled as an envelope-spanning complex with subcomplexes in the inner and outer membranes (see Fig. S1 in the supplemental material). The precise stoichiometry and juxtaposition of the Eps proteins are not known, but accumulating biochemical, genetic, and molecular studies continue to refine our understanding of complex assembly and function (for a review, see reference 25). A trimolecular complex consisting of cytoplasmic protein EpsE and inner membrane proteins EpsL and EpsM has been identified. EpsL and EpsM have been shown to coimmunoprecipitate and participate in mutual stabilization interactions in vivo by protecting each other from proteolysis (34, 41, 43, 48). Homologs of inner membrane protein EpsC have been implicated in interactions with the aforementioned inner membrane subcomplex (20, 29, 57), as well as homologs of outer membrane protein EpsD, which form oligomeric rings through which the secreted substrates, it is hypothesized, exit the cell (1, 10, 36, 38). More specifically, EpsC homologs in Pseudomonas aeruginosa and Klebsiella oxytoca are sensitive to proteolysis or unable to oligomerize in the absence of EpsD homologs (2, 40); however, direct interactions between these two proteins in their full-length forms have not been shown by coimmunoprecipitation or copurification. Although yeast two-hybrid analysis of the periplasmic domains of the Erwinia chrysanthemi EpsC and EpsD homologs also did not reveal interaction (15), recently it was shown that periplasmic subdomains of EpsC and EpsD homologs of Vibrio vulnificus copurified (28). It seems likely that EpsC, having interactions with both inner and outer membrane subcomplexes, plays a crucial role in complex function by connecting the inner membrane components to the outer membrane EpsD pore. Furthermore, it has been speculated that EpsC homologs impart specificity to the various T2S systems by directly interacting with proteins to be secreted (3).
We have taken a cell biology approach to characterizing Eps protein interactions, observing the dynamics of green fluorescent protein (GFP)-tagged components of the Eps complex in live cells by fluorescence microscopy. This method permits study of Eps protein assembly in the context of the complete apparatus, situated in both membranes, without the disruptive procedures required for many in vitro molecular and biochemical analyses of protein-protein interactions. Here we present data illustrating the importance of expressing GFP fusions for localization studies with all other interacting components, preserving wild-type stoichiometry and expression levels. In particular, we note that GFP-EpsM does not appear to be focused at the polar membrane as previously described (53), when expressed in balance with its interacting proteins. Chromosomal replacement of epsM and epsC with gfp-tagged versions instead reveals a more distributed pattern, with punctate fluorescent foci along the full length of the cell. We have exploited these chromosomal gfp-eps strains to further dissect the interactions and requirements for localization of EpsC and EpsM by systematically deleting other eps genes in the operon.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacterial strains and plasmids used in this study are summarized in Table 1. The primers used are listed in Table 2.
TABLE 1.
Strains and plasmids
| Strain, genotype, or plasmid | Features | Reference or source |
|---|---|---|
| V. cholerae strains or genotypes | ||
| TRH7000 | El Tor strain, wild type for T2S | 23 |
| PBAD::eps | TRH7000 PBAD::eps | 55 |
| ΔepsC | In-frame replacement of epsC with aph-3 (Kmr) | This study |
| ΔepsD | In-frame replacement of epsD with aph-3 (Kmr) | This study |
| ΔepsG | In-frame replacement of epsG with cat (Cmr) | This study |
| ΔepsL | In frame replacement of epsL with aph-3 (Kmr) | 55 |
| PU3 (epsM mutant) | Tn5 insertion in epsM at position 247 (Kmr) | 39 |
| gfp-epsC | TRH7000 with gfp-epsC replacement at epsC locus | This study |
| gfp-epsC ΔepsD | gfp-epsC with replacement of epsD with aph-3 (Kmr) | This study |
| gfp-epsC ΔepsL | gfp-epsC with replacement of epsL with aph-3 (Kmr) | This study |
| gfp-epsC epsM mutant | PU3 with replacement of epsC with gfp-epsC | This study |
| gfp-epsM | TRH7000 with gfp-epsM replacement at epsM locus | This study |
| gfp-epsM ΔepsC | gfp-epsM with replacement of epsC with aph-3 (Kmr) | This study |
| gfp-epsM ΔepsD | gfp-epsM with replacement of epsD with aph-3 (Kmr) | This study |
| gfp-epsM ΔepsL | gfp-epsM with replacement of epsL with aph-3 (Kmr) | This study |
| PBAD::eps gfp-epsC | TRH7000 PBAD::eps with gfp-epsC replacement at epsC locus | This study |
| PBAD::eps gfp-epsM | TRH7000 PBAD::eps with gfp-epsM replacement at epsM locus | This study |
| PBAD::eps gfp-epsC ΔepsD | PBAD::eps gfp-epsC with replacement of epsD with aph-3 (Kmr) | This study |
| PBAD::eps mcherry-epsC gfp-epsM | TRH7000 PBAD::eps with mcherry-epsC replacement at epsC locus and gfp-epsM replacement at epsM locus | This study |
| E. coli strains | ||
| MC1061 | F−lac mutant; K-12 laboratory strain | 9 |
| MM294/pRK2013 | Helper strain for conjugations | 33 |
| SY327 λpir | λpir lysogen; permits replication of pCVD442 | 35 |
| Plasmids | ||
| pMMB66 | Low-copy, IPTG-inducible vector (Apr) | 19 |
| pMMB67 | Low-copy, IPTG-inducible vector (Apr) | 19 |
| pCVD442 | Suicide vector containing sacB (Apr) | 14 |
| pUC18K | aphA-3 gene (Kmr, Apr) | 32 |
| pMMB68 | etxB (heat-labile enterotoxin B subunit gene) (Apr) | 47 |
| pBAD33 | cat gene (Cmr) | 22 |
| pK18mobsacB | Suicide vector containing sacB (Kmr) | 51 |
| pEpsC | pMMB67EH-epsC | This study |
| pEpsCD | pMMB67-epsC-epsD | This study |
| pEpsD | pMMB67EH-epsD | This study |
| pEpsG | pMMB67EH-epsG | This study |
| pMS44 (pEpsL) | pMMB67HE-epsL | 47 |
| pMMB524 (pEpsM) | pMMB67EH-epsM | 39 |
| pGFP-EpsM | pMMB-gfp-epsM | 53 |
| pmCherry-EpsCD | pMMB67EH-mcherry-epsCD | This study |
| pVC1200 | pMMB66-vc1200 | This study |
TABLE 2.
Primers used for plasmid constructions
| Primer | Sequence (5′-3′) | Feature(s) | Construct(s) generated |
|---|---|---|---|
| epsC01 | CGGATCCGAATTTAAACAACTTCCTC | BamHI | pGFP-EpsC, pGFP-EpsCD |
| epsC02 | TCTGCAGGTCGTTACTCGCCTTAG | PstI | pGFP-EpsC |
| epsC03 | CTCTAGATGGCAATATGAAGCGTA | XbaI | pBAD33-EpsC |
| epsD01 | ATCTAGATCGGTCATTGCTTGGCTTC | XbaI | pGFP-EpsCD |
| epsD03 | GGATCCGAGTAACGACGCCTTA | BamHI | pEpsD |
| epsD04 | GCATGCGCTGGAGAGATCACCA | SphI | pEpsD |
| epsG-OEup | GGCGAATTCGACATAGTGTGGA | EcoRI | pEpsG |
| epsG-OEdn | TTCCGTCGACTACCGCTAATTAG | SalI | pEpsG |
| epsC-ko1 | CCGAGCTCACCTTGATAGCGC | SacI | ΔepsC vector |
| epsC-ko2 | CGGAGGTACCTGTTTAAATTCCATA | KpnI | ΔepsC vector |
| epsC-ko3 | GATGGATCCCAACATGATGTAT | BamHI | ΔepsC vector |
| epsC-ko4 | CTCTAGATTACGTTTTGAAGTG | XbaI | ΔepsC vector |
| epsD-ko1 | CTGAGCTCGTGTTATATTGCGATG | SacI | ΔepsD vector |
| epsD-ko2 | CTAAGGTACCGTTACTCGCCTTA | KpnI | ΔepsD vector |
| epsD-ko3 | CTTTATGGATCCGATGGAAGCCAAG | BamHI | ΔepsD vector |
| epsD-ko4 | CCGTCTAGAAATCACGAATTTCC | XbaI | ΔepsD vector |
| epsD-ko1gfpC | CTTTATTGTCTAGATGGAAGCCAAG | XbaI | gfp-epsC ΔepsD vector |
| epsD-ko2gfpC | GCCGCATGCAAATCACGAATTTC | SphI | gfp-epsC ΔepsD vector |
| gfp03 | CCTGAGCTCTTACCAGACAACCA | SacI | gfp-epsC ΔepsD vector (in gfp) |
| CmR-1 | GTTGTCGACATTTTCAGGAGCTAA | SalI | ΔepsG vector |
| CmR-2 | AGCTGCAGGCGTTTAAGGGCA | PstI | ΔepsG vector |
| epsG-ko1 | CAGTCTAGAAAACCGCGGATTCG | XbaI | ΔepsG vector |
| epsG-ko2 | GGGTCGACCCCGTTTGTTTAC | SalI | ΔepsG vector |
| epsG-ko3 | GGTAACTGCAGTATCCAAGATTTTC | PstI | ΔepsG vector |
| epsG-ko4 | TACGCATGCTCACGACTGGG | SphI | ΔepsG vector |
| epsL-ko1 | GAGCTCTTAATTGTGATTCTGCTCCT | SacI | ΔepsL vectors |
| epsL-ko2 | GGTACCAAACCAGCCAAGGGATATC | KpnI | ΔepsL vectors |
| epsL-ko3 | TCTAGAGTTTGTGGTGAAGCCCAAG | XbaI | ΔepsL, ΔepsL-gfpM vectors |
| epsL-ko4 | GTCGACAAGCTAAGCTGCCTTCG | SalI | ΔepsL vector |
| epsL-ko5gfpM | GTCGACTTGAACTGACGCGTCGA | SalI | ΔepsL-gfpM vector |
| gfpCchr1 | AGTTCTTCTCCTTTACTCATAAATTTCCACGTTATTCCTT | epsC rbs and gfp junction | Vector for creation of gfp-epsC strain |
| gfpCchr2 | AAGGAATAACGTGGAAATTTATGAGTAAAGGAGAAGAACT | epsC rbs and gfp junction | Vector for creation of gfp-epsC strain |
| gfpCchr3 | GGCCCGGGACTCGTTTGCCAT | SmaI | Vector for creation of gfp-epsC strain |
| epsL21 | GTCAGCATGCAAATATGCTGCC | SphI | Vector for creation of gfp-epsM strain |
| epsM28 | ATGCAGGTCTGGATCCAACC | BamHI (natural) | Vector for creation of gfp-epsM strain |
| epsN03 | CCAAGCTGCAGCGCACCAAT | PstI (natural) | Vector for creation of gfp-epsM strain |
| gfpMchr1 | AGTTCTTCTCCTTTACTCATTTCTCCTTACTTGGGCTTCA | epsM rbs and gfp junction | Vector for creation of gfp-epsM strain |
| gfpMchr2 | TGAAGCCCAAGTAAGGAGAAATGAGTAAAGGAGAAGAACT | epsM rbs and gfp junction | Vector for creation of gfp-epsM strain |
| epsN04 | CCAAGCCCGGGCGCACCAAT | SmaI | Vector for creation of gfp-epsM strain |
| mcherry-up | GGTACCATGGTAAGCAAGGGCGA | KpnI | pmCherry |
| mcherry-dn | GGATCCCTTGTACAGCTCGTCCAT | BamHI | pmCherry |
| PBAD-mcherry overlap | AAGGAATAACGTGGAAATTTATGGTAAGCAAGGGCGAGGA | PBAD promoter region and mCherry junction | Vector for creation of PBAD::eps mcherry-epsC gfp-epsM |
| PBAD-mg-up | TCTAGAACTGCTGGCGGAAAAGATGTGAC | XbaI | Vector for creation of PBAD::eps gfp-epsC and PBAD::eps gfp-epsM |
| EpsC-mg-down | GCATGCAAACTGCCCGCCAACAGCCAT | SphI | Vector for creation of PBAD::eps gfp-epsC |
| PBAD::gfpepsC overlap | GAATAACGTGGAAATTTATGAGTAAAGGAGAAG | PBAD promoter region and gfp junction | Vector for creation of PBAD::eps gfp-epsC |
| VC1200-f | GAATTCAGGATTGAGTTGTGCATCAG | EcoRI | pVC1200 |
| VC1200-r | CTGCAGATCTCGGATAGGTAATCAAG | PstI | pVC1200 |
Plasmid construction.
All PCRs were performed using Phusion high-fidelity DNA polymerase (New England BioLabs), and further amplification of PCR products was performed using the PCR-Script Amp cloning kit (Stratagene). Ampicillin and carbenicillin were used interchangeably for maintenance of plasmids containing the bla gene for selection.
The pMMB-based EpsC plasmid, pEpsC, was created by cloning the epsC-containing BamHI-PstI fragment from a pBAD33-EpsC clone, which was generated from PCR amplification from Vibrio cholerae strain TRH7000 with the primer pair epsC02/epsC03. The plasmid mCherry-EpsCD, which encodes an in-frame fusion of the red fluorescent protein mCherry with EpsC, was first constructed by PCR amplification of the gene encoding mCherry from pKS mCherry using primers mcherry-up and mcherry-dn. The mCherry PCR product was cloned into pCRScript SK+ and then pMMB67, resulting in pmCherry. pmCherry-EpsCD was then formed by ligating a fragment containing epsCD from pEpsCD into pmCherry using the restriction enzymes BamHI and XbaI.
pVC1200 was created by PCR amplification of the native VC1200 gene directly from N16961 chromosomal DNA and cloning into low-copy-vector pMMB66 using the primer pairs and restriction sites indicated in Table 1. All other plasmids for complementation of the various deletion strains were constructed by PCR amplification of the native genes directly from TRH7000 chromosomal DNA and cloning into low-copy-vector pMMB66 or pMMB67 using the primer pairs and restriction sites indicated in Table 1. pGFP-EpsC and pGFP-EpsCD were constructed by ligating epsC and epsC-epsD, amplified by primers epsC01/epsC02 and epsC01/epsD01, respectively, in frame into pGFP (53).
Creation of gfp-epsC and gfp-epsM strains.
For replacement of epsC and epsM on the chromosome with gfp-tagged versions of the genes, suicide vectors containing approximately 1 kb of homologous sequence upstream and downstream of the gfp insertion were constructed. Overlapping primers at the junctions between the preceding sequences and gfp were designed to retain the native ribosome binding sites of each eps gene for expression of the fusion. For the chromosomal gfp-epsC strain, the upstream homologous region was amplified from TRH7000 chromosomal DNA, using primers gfpC-ko1 and gfpCchr1. The gfp-epsC gene fusion plus the first 83 nucleotides of epsD was amplified from the pGFP-EpsCD plasmid using gfpCchr2 and gfpCchr3 primers. The products from both PCRs were gel purified, combined, and together used as a template for a third PCR using primers gfpC-ko1 and gfpCchr3, taking advantage of the overlapping sequences at the eps promoter-gfp junction engineered with primers gfpCchr1 and gfpCchr2. This final 2.8-kb PCR product, containing gfp-epsC with 1 kb of homologous sequence up and downstream of the gfp gene, was gel purified, cloned into PCRScript SK+ for further amplification, and then ligated into pCVD442.
A similar approach was taken for construction of the gfp-epsM suicide vector. First, primers epsM28 and epsN03 were used to amplify a fragment from TRH7000 chromosomal DNA containing partial sequences of epsM and epsN. This fragment was cloned into pGFP-EpsM with naturally occurring BamHI and PstI to extend the downstream sequence a total of 1 kb past the gfp gene. This new pGFP-EpsMN′ plasmid was then used as a template for amplification of gfp-epsM, with a total of ∼1 kb of homologous sequence downstream of the end of gfp, using primers gfpMchr2 and epsN04. The 1 kb upstream of epsM was amplified from the TRH7000 chromosome using primers epsL21 and epsMchr1. These two PCR products were used as a template for another PCR using primers epsL21 and epsN04. This product containing gfp-epsM, with 1 kb of homologous sequence up- and downstream of the gfp gene, was gel purified, cloned into PCRScript SK+ for further amplification, and then ligated into pCVD442.
To construct PBAD::eps gfp-epsC, a suicide vector containing approximately 1 kb of homologous sequence upstream and downstream of the gfp insertion was constructed from the V. cholerae strain PBAD::eps, which contains the entire eps operon under the control of the arabinose-inducible pBAD promoter (55). The upstream homologous region was amplified from PBAD::eps using primers PBAD::mg-up and PBAD::gfpepsC overlap reverse. The gfp-epsC gene fusion plus a portion of epsD was amplified from the pGFP-EpsCD plasmid using PBAD::gfpepsC overlap and epsC-mg-down primers. The products from both PCRs were used as a template for a third PCR using primers PBAD::mg-up and epsC-mg-down. This final PCR product was gel purified, cloned into pCRScript SK+ for further amplification, and then ligated into pCVD442.
The suicide vectors were propagated in Escherichia coli strain SY327 λpir and conjugated into TRH7000 or PBAD::eps with the assistance of MM294/pRK2013. Carbenicillin-resistant transconjugants resulting from integration of the suicide vectors onto the chromosome were isolated as described previously (55). Colonies were screened by PCR for the replacement of native epsC or epsM with gfp-tagged versions.
To construct PBAD::eps mcherry-epsC gfp-epsM, a suicide vector containing approximately 1 kb of homologous sequence upstream and downstream of the site of the mcherry insertion was constructed from PBAD::eps. The upstream homologous region was amplified from PBAD::eps using primers PBAD::mg-up and PBAD-mcherry overlap reverse. The mcherry-epsC gene fusion plus a portion of epsD was amplified from the pmCherry-EpsCD plasmid using PBAD-mcherry overlap and epsC-mg-down primers. The products from both PCRs were used as a template for a third PCR using primers PBAD::mg-up and epsC-mg-down. This final PCR product was cloned into PCRScript SK+ for further amplification and then ligated into pCVD442. It was then introduced into the PBAD::eps gfp-epsM strain by conjugation. Colonies were screened by PCR for the replacement of native epsC with the mcherry-tagged version.
Construction of deletion strains.
To attain the gfp-epsC epsM mutant, the suicide vector used to construct the gfp-epsC strain was introduced into previously described transposon mutant PU3, which contains a Tn5 disruption of epsM (39), and replacement of native epsC was performed as described in the previous section. The ΔepsC, ΔepsD, and ΔepsL strains were constructed as described previously (55) using the primers indicated in Table 1.
The ΔepsG strain was generated similarly, replacing epsG with the cat gene, conferring chloramphenicol resistance. The disruption construct was generated by first amplifying 1 kb upstream and downstream of epsG in TRH7000 and the cat gene from pBAD33 using the primers indicated in Table 2. All three fragments were cloned stepwise into pK18mobsacB (51) using the restriction sites listed in Table 2 and conjugated into TRH7000. Isolates that were kanamycin sensitive, chloramphenicol resistant, and negative for secretion were further analyzed by PCR and protease secretion assays.
Detection of secreted protease activity.
Activity of secreted proteases in culture supernatants from overnight cultures grown in LB was detected as described previously using the substrate N-tert-butoxy-carbonyl-Gln-Ala-Arg-7-amido-4-methyl-coumarin (Sigma) (26, 55). Upon proteolytic cleavage of the substrate, fluorescence was measured using the excitation and emission wavelengths 385 nm and 440 nm, respectively. For determination of activity of VC1200 protease following overexpression in mid-log phase, strains containing plasmid pVC1200 were grown in M9 medium containing 0.4% Casamino Acids, 0.2% glycerol, and 100 μg/ml thymine and induced with 100 μΜ IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 h prior to analysis. Emission rates were normalized to a culture optical density at 600 nm (OD600) of 1.0 for comparison and presented as the change in fluorescence units (ΔFU)/min/OD600.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting procedures.
Frozen cell pellets were resuspended in loading buffer containing 50 mM dithiothreitol, boiled for 5 min, and the equivalent of 10 μl at an OD600 of 2.0 was loaded onto NuPAGE 4 to 12% Bis-Tris gradient gels and immunoblotted as described previously (26, 55).
Microscopy.
Cultures of V. cholerae were grown overnight at 37°C in M9 medium containing 0.4% Casamino Acids, either 0.4% glucose or 0.2% glycerol, and 100 μg/ml thymine; diluted 1:50 into fresh medium; and grown to mid-log phase before observation, unless otherwise noted. Plasmids were maintained with 50 and 200 μg/ml carbenicillin in log-phase and overnight cultures, respectively. Plasmid expression was induced with IPTG as described above. For fluorescence microscopy of live cells, cultures were mounted on 1.5% low-melt-agarose pads prepared with M9 glucose medium supplemented with 50 μg/ml carbenicillin and IPTG where appropriate. All microscopy was performed with a Nikon Eclipse 90i fluorescence microscope equipped with a Nikon Plan Apo VC 100× (1.4 numerical aperture) oil immersion objective and a CoolSNAPHQ2 digital camera. A GFP HC HiSN zero shift filter cube, with a 450- to 490-nm excitation filter and a 500- to 550-nm barrier filter, was used for visualizing GFP fluorescence and Alexa Fluor 488 F(ab′)2 goat anti-rabbit immunoglobulin G (IgG) staining for immunofluorescence. For visualization of mCherry fluorescence, a 530- to 560-nm excitation filter and a 590- to 650-nm barrier filter were used. Captured images were analyzed with NIS-Elements imaging software (Nikon). For quantitation of fluorescent foci, an average of 200 cells from three separate experiments was reported. For PBAD::eps mcherry-epsC gfp-epsM colocalization, an average of 20 cells from three separate experiments (totaling 600 foci) was counted. For presentation, image input levels were adjusted with Adobe PhotoShop CS2 to compensate for variations in expression levels in which the fusions were overexpressed via IPTG induction or upregulated due to increased expression from the eps promoter (see Fig. 1, 6, and 8).
FIG. 1.
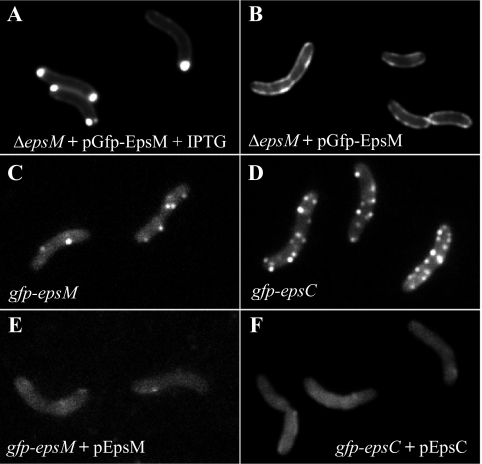
Distribution of GFP chimeras varies with expression level and context. Plasmid-borne GFP-EpsM in live cells of V. cholerae epsM mutant PU3 was polarly localized when overexpressed with 10 μM IPTG (A) but circumferentially distributed when not induced (B). Both patterns differed from that of chromosomally expressed GFP-EpsM, balanced with the other Eps proteins, which formed fluorescent foci along the cell membranes (C). (D) GFP-EpsC, expressed from the chromosome, similarly displayed fluorescent foci along the full lengths of the cells. (E and F) Both GFP-EpsM and GFP-EpsC fluorescent foci dissipated upon coexpression of IPTG-induced, plasmid-encoded native EpsM and EpsC, respectively.
FIG. 6.
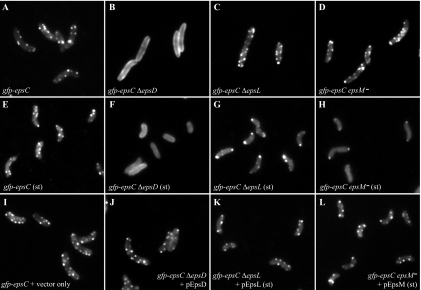
Differential localization of GFP-EpsC in the absence of EpsD, EpsL and EpsM. Localization of chromosomally expressed GFP-EpsC was examined in ΔepsD (B and F), ΔepsL (C and G), and epsM mutant (D and H) backgrounds in log- and stationary-phase (st) cultures and compared with its localization in an otherwise wild-type background (A and E) by fluorescence microscopy. GFP-EpsC displayed a continuous membrane localization in the gfp-epsC ΔepsD strain (B) compared to the otherwise wild-type background (A). Punctate fluorescence was restored when the gfp-epsC ΔepsD strain was complemented with the pEpsD plasmid in the presence of 10 μM IPTG (J). Both the gfp-epsC ΔepsL strain (C) and gfp-epsC epsM mutant (D) retained punctate fluorescence, with subtle accumulation at the polar membrane. In stationary-phase cultures, this phenotype appeared to be magnified, as there is a distinct accumulation at the poles in both the gfp-epsC ΔepsL strain (G) and gfp-epsC epsM mutant (H). Introduction of the pEpsL and pEpsM plasmids to the epsC ΔepsL strain (K) and gfp-epsC epsM mutant (L), respectively, restored the patterns to that of the wild-type strain containing a vector control in the stationary-phase cultures (I).
FIG. 8.
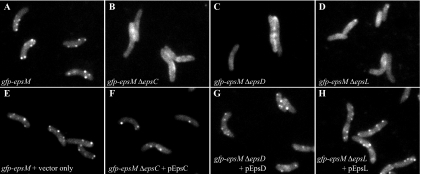
Fluorescence microscopy studies of gfp-epsM deletion strains. Chromosomally expressed GFP-EpsM localization was examined by fluorescence microscopy in live cells of wild-type (wt) (A), ΔepsC (B), ΔepsD (C), and ΔepsL (D) backgrounds. Fluorescent GFP-EpsM foci, not apparent in the mutant backgrounds, were restored by complementation with plasmids expressing the missing proteins EpsC (F), EpsD (G) and EpsL (H) and compared with the foci present in the gfp-epsM strain containing a vector control only (E). Complementation with pEpsD in the gfp-epsM ΔepsD strain required the addition of 10 μM IPTG.
Immunofluorescence.
The equivalent of 1 ml culture at an OD600 of 1.0 for each sample was pelleted at 3,300 × g and resuspended in 0.5 ml fixative composed of 1% paraformaldehyde and 0.1% glutaraldehyde in phosphate-buffered saline (PBS) (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4). Samples were fixed for 30 min at room temperature, pelleted, and washed three times with 1 ml PBS, pH 7.2. Fixed samples were then resuspended in 0.5 ml GTE (50 mM glucose, 20 mM Tris [pH 7.5], 1 mM EDTA [pH 8.0]) with 0.2 mg/ml lysozyme and incubated at room temperature for 20 min. Samples were washed three more times with 1 ml PBS, resuspended in 0.5 ml 2% bovine serum albumin (Sigma) in PBS, and incubated for 30 min. The cells were then pelleted and resuspended in 1 ml anti-EpsG antisera diluted 1:10,000 with 2% bovine serum albumin in PBS and rocked for 1 h. Samples were washed three times with 1 ml PBS and then resuspended in 1 ml of a 1:5,000 dilution of Alexa Fluor 488 F(ab′)2 goat anti-rabbit IgG (Molecular Probes) in 2% goat serum (Gibco/Invitrogen) in PBS. After 1 h, the samples were washed three final times with 1 ml PBS, mounted on slides with 1.5% low-melt agarose made with PBS, and observed by fluorescence microscopy.
Effect of MreB inhibitor A22 on GFP-EpsC fluorescent foci and toxin secretion.
Cultures of the gfp-epsC strain containing pMMB68, which expresses EtxB, the B subunit of the E. coli heat-labile enterotoxin, from an IPTG-inducible promoter, were grown overnight in M9 growth medium containing 200 μg/ml carbenicillin. Overnight cultures were diluted 1:100 and grown with 50 μg/ml carbenicillin and 100 μΜ IPTG until the OD600 was 0.4, and then A22 (Calbiochem) was added to the culture at a final concentration of 10 μg/ml. Samples for Western blot analysis and microscopy were collected 30, 60, and 120 min after addition of A22. Microscopy samples were mounted on slides containing 1.5% low-melt agarose prepared with M9 glucose medium containing 10 μg/ml A22, where appropriate. Supernatants containing secreted EtxB were separated from cells by centrifugation at 16,000 × g. SDS loading buffer without dithiothreitol was added, and supernatant and pellet samples were boiled for 5 min and analyzed by SDS-PAGE and Western blotting with monoclonal anti-EtxB antibody 118-8 as described previously (55).
RESULTS
Plasmid-borne GFP-EpsM distribution varies based on expression level.
Previously, we examined the localization pattern of plasmid-borne GFP-EpsM via fluorescence microscopy and observed that the fusion, expressed in either V. cholerae or E. coli, localized predominantly to the polar membrane (53). In these experiments, GFP-EpsM expression was induced with 10 μM IPTG to enhance visualization. Recently, a more sensitive fluorescence microscope and digital camera enabled us to examine plasmid-borne GFP-EpsM uninduced. With this setup, we first repeated the localization experiments under previous growth conditions, inducing GFP-EpsM expression for 90 min with 10 μM IPTG. GFP-EpsM in V. cholerae epsM mutant PU3 appeared as it did previously upon induction, with the bulk of the fluorescent signal at the polar membrane (Fig. 1A). Uninduced, however, GFP-EpsM fluorescence was distributed around the periphery of the cell membrane, with occasional patches of more intense fluorescence (Fig. 1B). The nonpolar distribution of GFP-EpsM under these conditions is concordant with the recently published localization pattern for the Klebsiella oxytoca EpsM homolog PulM, with which GFP-PulM was observed along the entire cell circumference when it was expressed from the λ attachment site of the E. coli chromosome and induced with 10 μM IPTG (4). These data suggest that the polarity of the GFP-EpsM fusion previously observed likely does not reflect the distribution of the native EpsM protein under wild-type expression conditions and may instead be due to overexpression.
Creation of V. cholerae strains with chromosomally expressed GFP-EpsM and GFP-EpsC fusions.
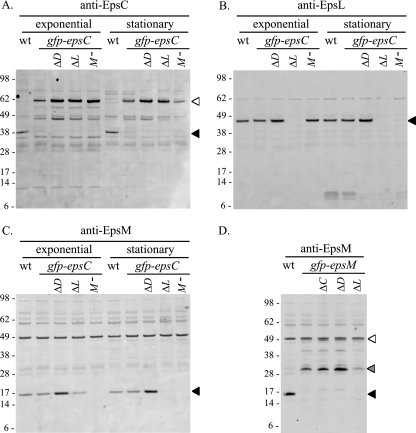
It is unclear from the localization studies whether the variable localization of plasmid-borne GFP-EpsM is due to the increased expression level or more specifically, the overproduction of the GFP fusion without concomitant overproduction of its interaction partners in the Eps complex. To begin to address this, we replaced chromosomal copies of epsM and epsC in V. cholerae with gfp-tagged copies of each gene, such that they would be expressed under the control of the eps promoter, translated from their respective ribosome binding sites, and produced in conjunction with the full complement of eps gene products. GFP-EpsC was detected at a level comparable to that of the wild-type protein, and while a significant amount of full-length GFP-EpsM was detected, a degradation product was also observed (Fig. 2A and D).
FIG. 2.
Western blot analyses of gfp-epsC and gfp-epsM deletion strains. Cell extracts of log-phase and stationary-phase cultures from the wild-type (wt), gfp-epsC, and ΔepsD (ΔD), ΔepsL (ΔL), and epsM::Tn5 (M−) strains were separated by SDS-PAGE and analyzed by Western blotting with detection by anti-EpsC (A), anti-EpsL (B), and anti-EpsM (C) antisera. (D) Log-phase culture samples of the wt, gfp-epsM, and ΔepsC (ΔC), ΔepsD (ΔD), and ΔepsL (ΔL) strains were immunoblotted with anti-EpsM antisera. Molecular weight markers (in thousands) for all blots are indicated to the left, and the positions of the native proteins and GFP fusions are indicated with black and white triangles, respectively. Full-length GFP-EpsM (white triangle) (D) is partially obscured by a cross-reactive band also present in the wild-type strain. A degradation product of the GFP-EpsM fusion is indicated with a gray triangle (D).
The gfp-epsM and gfp-epsC strains were also tested for protease secretion to determine whether the GFP fusions were functional components of the T2S complex and to verify that the intraoperon insertion of the gfp tag did not interfere with the expression of the other eps genes. Protease activities in the supernatants of the gfp-epsM and gfp-epsC strains were comparable to those of wild-type V. cholerae, indicating that these fusions support T2S (Table 3).
TABLE 3.
Protease secretion assays
| Strain or genotype | ΔFU/min per OD600a | Complementation | ΔFU/min per OD600d |
|---|---|---|---|
| TRH7000 | 77.7 ± 2.3 | ||
| ΔepsC | 0 ± 0.3 | ΔepsC + pEpsC | 78.0 ± 4.4 |
| ΔepsD | 1.3 ± 0.7 | ΔepsD + pEpsD | 53.9 ± 5.0 |
| ΔepsD + pEpsD + IPTGb | 71.7 ± 3.8 | ||
| ΔepsG | 2.6 ± 0.6 | ΔepsG + pEpsG | 90.8 ± 10.0 |
| ΔepsL | 0 ± 0.7 | ΔepsL + pEpsL | 65.1 ± 3.6 |
| PU3 (epsM mutant) | 6.0 ± 1.1 | PU3 (epsM mutant) + pEpsM | 71.8 ± 1.5 |
| gfp-epsC | 76.4 ± 3.5 | ||
| gfp-epsC ΔepsD | 0.7 ± 1.1 | gfp-epsC ΔepsD + pEpsD | 46.6 ± 2.5 |
| gfp-epsC ΔepsD + pEpsD + IPTGb | 76.5 ± 4.8 | ||
| gfp-epsC ΔepsL | 0 ± 0.5 | gfp-epsC ΔepsL + pEpsL | 71.9 ± 2.4 |
| gfp-epsC epsM mutant | 5.4 ± 2.1 | gfp-epsC epsM mutant + pEpsM | 73.8 ± 1.7 |
| gfp-epsM | 76.3 ± 5.7 | ||
| gfp-epsM ΔepsC | 1.3 ± 1.2 | gfp-epsM ΔepsC + pEpsC | 80.0 ± 2.0 |
| gfp-epsM ΔepsD | 0 ± 0.9 | gfp-epsM ΔepsD + pEpsD | 42.5 ± 2.3 |
| gfp-epsM ΔepsD + pEpsD + IPTGb | 65.9 ± 4.5 | ||
| gfp-epsM ΔepsL | 0.8 ± 1.2 | gfp-epsM ΔepsL + pEpsL | 93.6 ± 2.8 |
| PBAD::epsc | 70.3 ± 0.7 | ||
| PBAD::eps gfp-epsCc | 65.9 ± 2.3 | ||
| PBAD:: eps gfp-epsC ΔepsDc | 0 ± 3.9 | PBAD::eps gfp-epsC ΔepsD + pEpsDc | 26.8 ± 0.5 |
| PBAD::eps gfp-epsC ΔepsD + pEpsD + IPTGb,c | 63 ± 2.0 | ||
| PBAD:: eps gfp-epsMc | 62.9 ± 2.6 | ||
| PBAD:: eps mcherry-epsC gfp-epsMc | 63.4 ± 4.5 |
ΔFU indicates the protease activity detected in the supernatants of overnight cultures for TRH7000 and deletion mutant strains. Protease activity in overnight culture supernatants was assayed by measuring methylcoumarin fluorescence generated from Boc-Gln-Ala-Arg-7-amido-4-methylcoumarin hydrolysis, and rates were normalized to an OD600. The average of at least three experiments is presented ± the standard error of the mean.
10 μM IPTG.
0.01% arabinose.
ΔFU indicates the protease activity for the TRH7000 and gfp fusion mutant strains upon introduction of plasmids.
Chromosomally expressed GFP-EpsM and GFP-EpsC form discrete points of fluorescence along the cell periphery.
Having demonstrated that the native epsM and epsC genes were correctly replaced with fully functional gfp-tagged versions, we examined the spatial distributions of these fusion proteins by fluorescence microscopy. For both the gfp-epsM and gfp-epsC strains, we observed discrete spots of fluorescence all along the cell membranes, a dramatically different pattern than that observed with plasmid-borne GFP-EpsM (Fig. 1C and D). Overall, there were more fluorescent foci visible in the GFP-EpsC-expressing strain (5.3 foci/cell) than in the GFP-EpsM-expressing strain (3.2 foci/cell), and the fluorescence of the GFP-EpsC foci appeared to be slightly brighter than that of GFP-EpsM. This may reflect the relative instability of GFP-EpsM, as it has a higher ratio of degradation product to full-length protein via Western blotting (Fig. 2A and D).
Both gfp fusion strains displayed a pattern distinctly different than that of plasmid-borne GFP-EpsM, confirming that the context of expression of these fusion proteins has a profound effect on intracellular localization. Stoichiometric expression of the GFP fusions with their interacting proteins appears to be critical for determining their spatial distribution by fluorescence microscopy. To assess if the fluorescent foci observed were a result of incorporation of the GFP fusions into T2S complexes, plasmids expressing native EpsM and EpsC were introduced into the gfp-epsM and gfp-epsC strains, respectively, and induced with 50 μM IPTG. The patterns of punctate fluorescence dissipated in the gfp-epsM and gfp-epsC strains upon coexpression of the corresponding native proteins (Fig. 1E and F). The fluorescent signal of GFP-EpsM and GFP-EpsC was dispersed in the membrane and cytoplasm, and a fraction of the fusion proteins were likely subjected to proteolysis, as their stabilizing protein partners within the T2S complex became limited under these conditions (not shown). Taken together, the results from these coexpression studies imply that the GFP fusion proteins were outcompeted and replaced by the native proteins and suggest that the fluorescent foci may represent assembled T2S complexes. Alternatively, the fluorescent foci may represent GFP fusion aggregates into which native Eps proteins insert when overexpressed, thereby diluting the fluorescence signal. Although a possibility, this latter scenario is less likely, as the fusion proteins are functional and support secretion.
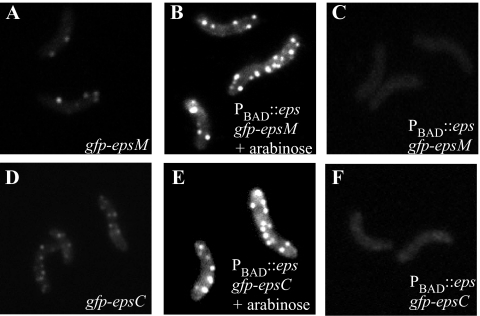
To further verify that the stoichiometric ratio of GFP chimeras and their interaction partners is critical for observing valid localization patterns and that this may be more important than the absolute level of GFP fusion production, the chimeric genes gfp-epsC and gfp-epsM were introduced into the PBAD::eps strain (55). In these strains the entire eps operon, including the gfp chimeras, is under the control of the arabinose-inducible pBAD promoter. Upon addition of arabinose, not only is the GFP fusion protein induced and expressed at higher levels than those from the native eps promoter, but so are all other Eps proteins, thereby maintaining the balance of Eps components in the cell. As seen in Fig. 3, although induction with 0.01% arabinose resulted in an increase in the number and brightness of fluorescent foci per cell, the patterns of fluorescence observed in both PBAD::eps gfp-epsC and PBAD::eps gfp-epsM were very similar to those seen when the fusion proteins were expressed from the native eps promoter (compare Fig. 3A and D to B and E). Without addition of arabinose, no fluorescent foci were observed (Fig. 3C and F), and protease secretion was not detected.
FIG. 3.
Simultaneous overexpression of the entire eps operon maintains the punctate distribution of GFP chimeras. GFP-EpsM (A) and GFP-EpsC (D) expressed from the native V. cholerae promoter form fluorescent foci. The intensity and number of GFP-labeled foci were increased in the PBAD::eps gfp-epsM (B) and PBAD::eps gfp-epsC (E) strains when induced with 0.01% arabinose. Without the additon of arabinose, no fluorescent foci were observed with either PBAD::eps gfp-epsM (C) or PBAD::eps gfp-epsC (F). All images are shown at the same exposure level to facilitate comparison of expression levels.
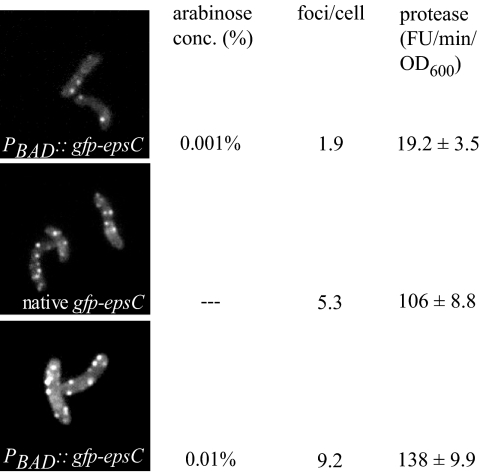
Quantification studies of the PBAD::eps gfp-epsC strain showed that the number of foci present per cell increased with the level of arabinose induction. At low levels of induction (0.001% arabinose), the average cell contained approximately two visible foci (Fig. 4, top). With the addition of higher levels of arabinose (0.01%), an average of nine foci were observed per cell (Fig. 4, bottom), a greater frequency than that observed with native expression of gfp-epsC (Fig. 4, center).
FIG. 4.
Number of fluorescent foci correlates with extracellular protease activity. V. cholerae gfp-epsC and PBAD::eps gfp-epsC cells producing the type II-dependent protease VC1200 were supplemented with IPTG at a final concentration of 100 μM and grown to mid-log stage. In the case of PBAD::eps gfp-epsC (pVC1200), 0.001% or 0.01% arabinose was added to the cultures. The GFP-EpsC-expressing cells were analyzed by fluorescence microscopy, and for each expression condition, the number of fluorescent foci in 200 cells was scored. Protease activity in mid-log culture supernatants was assayed by measuring methylcoumarin fluorescence generated from Boc-Gln-Ala-Arg-7-amido-4-methylcoumarin hydrolysis, and rates were normalized to OD600. The average of at least three experiments is presented ± the standard error of the mean.
To examine whether growth in the presence of increasing arabinose concentrations also resulted in increased protease secretion, the extracellular protease activity was measured with PBAD::eps gfp-epsC cultures grown with 0%, 0.001%, and 0.01% arabinose and compared with the level of secreted protease activity from the gfp-epsC strain. To each of the cultures, IPTG was added to overexpress plasmid-encoded VC1200 protease, and following growth into mid-log phase, culture supernatants were assayed for the secreted protease, as described in Materials and Methods. No protease activity was detected in the supernatants of the PBAD::eps gfp-epsC strain when grown without arabinose (data not shown). When grown in the presence of 0.001% arabinose, a small amount of extracellular protease activity was detected (Fig. 4, top). In contrast, the protease activity measured in supernatants from cultures grown with 0.01% arabinose was more than seven-fold higher than those grown with 0.001% arabinose (Fig. 4, bottom). Supernatants from cells with gfp-epsC expressed from the native promoter exhibited an intermediate level of protease secretion (Fig. 4, center). The elevation of secreted protease activity upon increased eps gene expression correlated with an increased number of fluorescent foci, suggesting that some or all of the additional fluorescent foci may represent functional T2S complexes.
Native EpsG is localized around the cell periphery in a pattern similar to that of chromosomally expressed GFP-EpsC and GFP-EpsM.
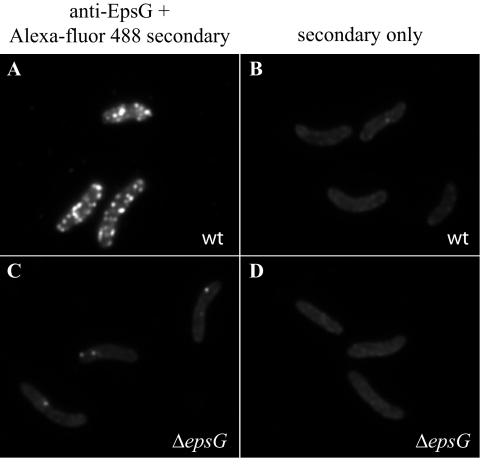
To confirm that the fluorescence patterns observed with the chromosomal gfp fusion strains represent the distribution of native Eps proteins and were not artifacts of GFP fusion to the proteins, we examined the localization of the Eps complex in wild-type V. cholerae cells by immunofluorescence. Unfortunately, there was no signal above background noise apparent with anti-EpsC antibodies, likely due to low antibody recognition of the native protein and/or a relatively low quantity of protein in the cell (data not shown). We were able, however, to determine the spatial distribution of native EpsG, the most abundant protein of the T2S complex (37, 49). In these experiments, we consistently observed bright fluorescent foci distributed along the full length of the bacterial cell (Fig. 5A). No fluorescence above background was observed in the ΔepsG mutant, except for occasional dots (Fig. 5C). The fluorescence obtained with Alexa Fluor 488 antibody that is shown in panels B and D is likely to only be autofluorescence, as fixed cells with no antibody incubation exhibited the same background fluorescence (not shown). The localization with the anti-EpsG antibodies mirrors what was observed with the chromosomally expressed GFP-EpsM and GFP-EpsC fusions, once again suggesting distinctly punctate localization for the Eps complex throughout the cell envelope.
FIG. 5.
Localization of native EpsG by immunofluorescence. Following fixation and treatment with lysozyme and EDTA, V. cholerae TRH7000 wild-type (wt) (A) and ΔepsG mutant cells (C) were incubated with anti-EpsG and Alexa-fluor 488-conjugated goat anti-rabbit IgG and visualized by fluorescence microscopy as described in Materials and Methods. (B and D) Wild-type and ΔepsG mutant cells incubated with Alexa Fluor 488-conjugated IgG only.
The subcellular localization of the Eps complex bears a resemblance to the bacterial cytoskeletal protein MreB, which is hypothesized to form helical filaments along the length of the cell (for recent reviews, see references 6, 8, and 56). We explored the possibility that filamentous MreB might play a role in Eps localization and/or assembly by use of the MreB-perturbing agent A22. Following addition of A22 and subsequent disruption of MreB filaments, monitored by cell rounding and dispersal of GFP-MreB filaments (not shown), GFP-EpsC foci were still apparent along the circumference of the cell (see Fig. S2 in the supplemental material). Although it cannot be concluded that GFP-EpsC is localized to positions equivalent to those in untreated cells, the data suggest that the formation of GFP-EpsC foci occurs independently of filamentous MreB. That the T2S apparatus assembles independently of MreB filaments was confirmed by the finding that toxin secretion was unaffected by A22 (see Fig. S2 in the supplemental material).
Characterization of in-frame gene deletions in the gfp fusion strains.
The gfp-epsC and gfp-epsM strains offer the unique opportunity to observe the dynamics of these fusion proteins in the context of the otherwise wild-type cell, presenting a powerful tool for exploring protein-protein interactions and determining what is required for their spatial distribution. To begin to delineate the roles of other Eps proteins in the establishment and maintenance of the GFP-EpsC and GFP-EpsM foci, we made a series of gene replacements in the gfp-epsC and gfp-epsM strains. For additional controls, we introduced the same mutations in gfp-free wild-type V. cholerae TRH7000 in parallel. Proper in-frame replacement of each gene with a gene cassette conferring antibiotic resistance was confirmed by PCR and sequencing, and expression of the other Eps proteins verified by Western blotting (not shown). We noted that levels of GFP-EpsC, EpsL, and EpsM increased in the gfp-epsC ΔepsD strain, though not due to a polar effect, because expression of genes both upstream and downstream of the insertion appeared to be affected (Fig. 2A). GFP-EpsC production was also higher in log-phase cultures in both the ΔepsL and epsM mutant backgrounds. The increased production of Eps proteins also occurs in the non-gfp strains and in other mutants generated previously and is thought to be a result of upregulation of the eps promoter whenever secretion via the T2S pathway is prevented (not shown). This is likely mediated by the alternative sigma factor σE, which is upregulated in eps mutants (55) and which has been shown, by microarray analysis, to regulate expression of the eps genes (13).
No secreted protease activity was detected in the supernatants of overnight cultures for any of the deletion mutant strains (Table 3). Protease secretion was restored in both the TRH7000 and gfp fusion mutant strains upon introduction of plasmids expressing the missing genes (Table 3). The pEpsD plasmid restored approximately 50% of secreted protease activity to the various ΔepsD strains without induction; however, the secretion defect was fully complemented when expression was increased with 10 μM IPTG.
GFP-EpsC requires EpsD for focal assembly.
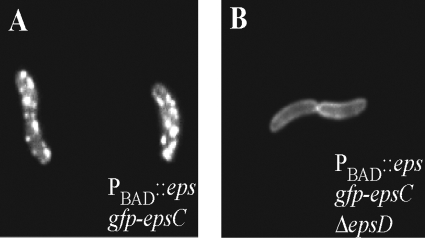
EpsC orthologs have been suggested to interact with orthologs of outer membrane pore protein EpsD (2, 28, 40) and the inner membrane proteins EpsL and EpsM (20, 29, 44, 57). To begin to dissect the roles of these proteins in formation of GFP-EpsC foci and to determine if GFP fusion technology in combination with fluorescence microscopy provides a useful alternative to molecular and biochemical procedures in mapping protein-protein interactions within the T2S complex, we examined the gfp-epsC strains containing deletions of epsD, epsL, and epsM by fluorescence microscopy. Removal of epsD resulted in loss of the fluorescent foci associated with GFP-EpsC and dispersal of the fluorescence along the entire cell membrane, suggesting that EpsC requires EpsD for focal assembly (Fig. 6B). The membrane fluorescence in the gfp-epsC ΔepsD strain was brighter than that in the gfp-epsC strain with an intact epsD gene, consistent with the two- to threefold-increased levels of the fusion detected on Western blots (Fig. 2A; compare lanes 2 and 3). Expression of EpsD from a plasmid restored punctate fluorescence to the gfp-epsC ΔepsD strain, upon induction with 10 μM IPTG (Fig. 6J), the IPTG concentration also required for full complementation of the protease secretion defect (Table 3).
Although the production of all Eps proteins was increased in the ΔepsD strain, we sought to verify that the dispersed fluorescence of GFP-EpsC in the ΔepsD strain was not simply due to upregulation of gfp-epsC by removing epsD in the PBAD::eps gfp-epsC strain. Because the native promoter has been replaced in this strain with the arabinose-inducible promoter, the level of production of Eps proteins, including GFP-EpsC, was unchanged upon deletion of epsD. Similar to what was observed with the native promoter, GFP-EpsC fluorescence in the absence of EpsD was dispersed throughout the membrane and lacking all punctate fluorescence (Fig. 7B). These studies indicate a crucial role for EpsD in the formation of fluorescent GFP-EpsC foci.
FIG. 7.
GFP-EpsC localization in the absence and presence of EpsD following overexpression of the eps operon. GFP-EpsC was expressed at similar levels and displayed nonpunctate membrane localization in the PBAD::eps gfp-epsC ΔepsD strain (B) compared to PBAD::eps gfp-epsC (A) following arabinose-mediated induction of the entire eps operon.
Upon deletion of either epsL or epsM, GFP-EpsC still formed punctate spots of fluorescence in the cell membranes (Fig. 6C and D). Cells from both deletion strains displayed fluorescent foci along their peripheries just like the otherwise wild-type gfp-epsC strain; however, slightly more foci occurred at the tips of the cells from the deletion strains. There was no overall shift in fluorescence to imply a polar bias, but the more frequent appearance of fluorescent spots at the poles was notable. Over 40% of the gfp-epsC ΔepsL cells contained polar foci, compared to 23% of gfp-epsC cells. In both the ΔepsL and ΔepsM mutants, GFP-EpsC was clearly capable of assembling into foci; however, the subtle increase in polarity of the fusion may indicate that it is not being efficiently maintained in the lateral membrane.
EpsL and EpsM have been shown to participate in stabilizing interactions with one another, resulting in mutual protection from proteolysis in both E. coli and V. cholerae (48). Western blot analysis confirmed this mutual stabilization of EpsL and EpsM in the gfp-epsC mutant strain. In log-phase cultures, EpsL and EpsM were found at slightly reduced levels in the absence of the other, an effect that was magnified in stationary-phase culture (Fig. 2B and C, lanes 9 and 10), perhaps due to increased levels of proteases during this growth phase. In stationary-phase cultures, the cells of the gfp-epsC strain examined by fluorescence microscopy were shorter in length but still retained ample fluorescent foci (Fig. 6E). The gfp-epsC ΔepsL strain retained fluorescent foci as well; however, the majority of GFP-EpsC accumulated at the polar membrane (Fig. 6G). Over 80% of these cells contained polar foci. In the epsM mutant background, GFP-EpsC also accumulated at the polar membrane in stationary-phase cultures (Fig. 6H). Under these conditions, the only punctate fluorescence visible was at the poles, with the remainder of the fluorescent signal in the cytoplasm. The effects of the ΔepsL and epsM mutations in the gfp-epsC strain were complemented upon expression of the pEpsL and pEpsM plasmids, respectively, restoring lateral fluorescent foci to the cells in the stationary-phase cultures (Fig. 6K and L). Taken together, our data reveal that GFP-EpsC is sensitive to proteolysis in the absence of EpsL and EpsM and that residual GFP-EpsC that escapes degradation accumulates at the poles. This suggests that the EpsL-EpsM complex stabilizes EpsC in a conformation that is required for its maintenance in the lateral membrane. Similar to the polar accumulation of overexpressed GFP-EpsM, these results show that imbalanced expression of GFP-EpsC in comparison to that of certain Eps proteins can also result in mislocalization to the pole.
GFP-EpsM foci are not generated in the absence of EpsD, EpsC, or EpsL.
As with GFP-EpsC, dispersed fluorescence was observed when GFP-EpsM was examined in the absence of EpsD (Fig. 8C). The fluorescence was evenly distributed in the membrane and the cytoplasm, again suggesting that EpsD is required for formation of GFP-EpsM foci. The gfp-epsM ΔepsC and gfp-epsM ΔepsL strains had similar appearances, indicating that each of these proteins is also required for localization of GFP-EpsM (Fig. 8B and D). Fluorescent foci were restored in each gfp-epsM deletion strain upon expression of the corresponding complementing plasmid (Fig. 8F to H). The lack of GFP-EpsM foci in the absence of EpsD, EpsC, and EpsL is consistent with the model that the focal complex is built upon EpsD and that the assembly of GFP-EpsM into the complex requires EpsC and EpsL.
Colocalization of GFP-EpsM and mCherry-EpsC.
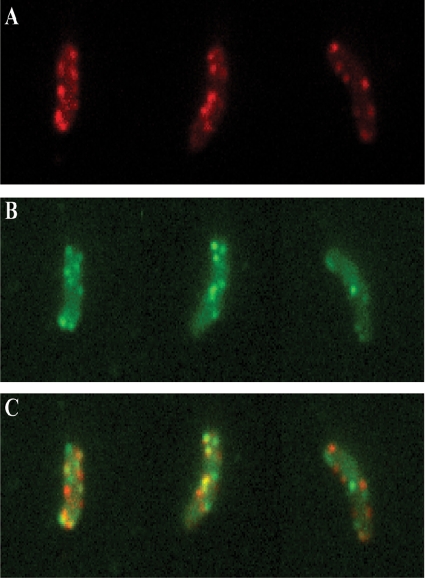
Because orthologs of EpsC have been implicated in direct interactions with orthologs of EpsL and EpsM and our data show that EpsC and EpsM form similar patterns of fluorescence when expressed as GFP fusions, we sought to colocalize EpsC and EpsM by producing mCherry-EpsC and GFP-EpsM in the same cell. Due to the relatively low fluorescence of mCherry, the mcherry-epsC construct was inserted in place of epsC on the chromosome of the PBAD::eps gfp-epsM strain, creating PBAD::eps mcherry-epsC gfp-epsM. This strain carries both tagged genes in addition to chromosomal copies of the unlabeled genes and is capable of increased production of Eps proteins in the presence of arabinose. Measured protease activity in the supernatant of PBAD::eps mcherry-epsC gfp-epsM was comparable to that of wild-type V. cholerae, indicating that these fusions support T2S (Table 3). Fig. 9A, B, and C show the dual-labeled cells when imaged to visualize mCherry, GFP, or both, respectively. Similar to what was seen when MreB was simultaneously labeled with two different fluorescent markers (16), GFP-EpsM and mCherry-EpsC showed partial colocalization. These two tagged proteins were present in sufficient quantities to be simultaneously visualized in 11% of the fluorescent foci.
FIG. 9.
GFP-EpsM and mCherry-EpsC colocalization in V. cholerae cells. Cells of PBAD::eps producing both mCherry-EpsC and GFP-EpsM were imaged with DsRed (A) and GFP (B) filters. (C) Overlays of DsRed and GFP signals are shown, with GFP-EpsM in green, mCherry-EpsC in red, and overlapping signals in yellow.
DISCUSSION
Our new studies of Eps complex localization indicate that accurate representation of intracellular distribution relies upon expression of GFP fusions in stoichiometric ratios with their partner proteins at levels as close to wild type as possible. Here we demonstrate that chromosomal replacement of the epsM gene with a gfp-tagged version produced fluorescent focal points along the cell membranes, whereas GFP-EpsM expression from a plasmid resulted in brighter, more-even membrane fluorescence with occasional patches of higher intensity. It seems likely that the focal points of fluorescence in the gfp-epsM strain represent fusion assembly into complexes with other Eps proteins and that the continuous fluorescence throughout the membrane upon pGFP-EpsM plasmid expression consists of unincorporated fusion protein. Perhaps there are more complexes forming than are readily apparent, but they may be obscured by the abundance of unincorporated GFP-EpsM dispersed in the membranes. When pGFP-EpsM plasmid expression was further increased with 10 μM IPTG, the pattern shifted dramatically to a predominantly polar localization. The intensely bright polar GFP-EpsM spots do not likely represent inclusion bodies in the traditional sense, as they are Triton X-100 soluble, similar to native EpsM (26), and clearly, sufficient GFP is correctly folded to form the fluorophore and emit fluorescence.
Similar findings have been reported with the K. oxytoca homolog PulM, which also accumulates at the polar membrane upon overexpression (4). For these localization studies, gfp-pulM was introduced onto the E. coli chromosome at the λ attachment site and induced with IPTG, with the rest of the pul operon expressed from a plasmid. The localization of GFP-PulM in this context looks very similar to our images of uninduced plasmid-encoded GFP-EpsM expressed in the V. cholerae epsM mutant, with an even signal along the circumference of the cell and subtle patches of brighter fluorescence (Fig. 1B). It may be, as with our studies, that more-discrete fluorescent foci could emerge upon more balanced expression of the other Pul proteins with GFP-PulM.
Although overexpression of GFP-EpsM alone results in subcellular localization patterns radically different from that of the chromosomally expressed GFP-EpsM, simultaneous overexpression of GFP-EpsM with the other Eps components from the pBAD promoter results in many distinct fluorescent foci unchanged from those seen when the fusion protein was expressed from the native promoter (Fig. 2). The intensity of the fluorescent foci is also increased; however, at this point it is not possible to determine if each focus represents completely assembled and functional T2S complexes or if some of them consist of assembly intermediates. Our data clearly underscore the importance of localizing components of multiprotein complexes in stoichiometric balance with their interacting partners, however, to determine their subcellular locations.
The general pattern of fluorescence in the gfp-epsC and gfp-epsM strains is reminiscent of other GFP fusion localization patterns that have emerged in the literature in recent years, including the Sec apparatus (7, 54) and several proteins involved in bacterial cell wall synthesis (reviewed in reference 52), including penicillin-binding proteins and bacterial cytoskeletal protein MreB, which appears to form a helical structure. There are likely insufficient complexes in a single V. cholerae cell to generate a contiguous helix of T2S machineries; however, we examined a possible role for MreB filaments in directing insertion and assembly of the T2S complex. Treatment with MreB inhibitor A22 did not appear to disrupt the formation of fluorescent foci in the gfp-epsC strain or interrupt T2S. We cannot say with certainty that the fluorescent foci are maintained at the same precise positions in the A22-treated cells, but secreted protease activity in these cultures further suggested that the T2S complexes are assembled and functional.
Using the chromosomal gfp fusion strains that we constructed, we were also able to study the effects of changing the balance between fusion proteins and their interacting partners using gene deletions, rather than overexpression of a single GFP fusion. The fluorescent foci formed in the gfp-epsC and PBAD::eps gfp-epsC strains dispersed upon deletion of epsD, suggesting that EpsD is critical to the localization of EpsC. On the other hand, GFP-EpsC is capable of forming foci in the absence of either EpsL or EpsM, though there appears to be some degradation of GFP-EpsC and mislocalization to the poles, which becomes very pronounced when both EpsL and EpsM are absent, as in the stationary-phase cultures. Thus, EpsL and EpsM are likely involved in keeping EpsC in a conformation that is required for its maintenance in the lateral membrane. EpsC homologs of other organisms have been shown to interact with both EpsL and EpsM homologs (20, 29, 41, 44, 45, 57), so it may be that direct interactions occur between each of the three proteins. Alternatively, EpsC may interact directly only with EpsL, and EpsM assists by stabilizing this interaction. The gfp-epsM ΔepsL strain does not exhibit fluorescent foci, consistent with GFP-EpsM requiring EpsL for localization. EpsC and EpsD are each required for formation of GFP-EpsM foci, as well, again consistent with the model that these proteins are prerequisites for EpsL and EpsM localization. We propose a model in which EpsC and EpsD form a dock on which other components of the complex might assemble, with EpsL and EpsM playing a more passive role, perhaps by keeping EpsC in a conformation that allows for its maintenance in the lateral membrane.
In an attempt to localize EpsC and EpsM to the same visible foci in the cell, we simultaneously labeled EpsC with the red fluorescent protein mCherry and EpsM with GFP. A merged image (Fig. 9C) shows the colocalization pattern of mCherry-EpsC and GFP-EpsM. Although some yellow foci, representing the overlap of red and green fluorescent proteins, are present, many foci are nonoverlapping. It is possible that the overlapping foci are the only locations in the cell where fully assembled T2S complexes exist. The visible red or green foci could therefore represent T2S complexes not yet fully assembled. Considering the relative instability of GFP-EpsM (Fig. 2A and D), it also seems reasonable that these foci may contain a mixture of full-length fluorescent forms of the fusions and those that have been cleaved and are no longer fluorescent.
In many colocalization studies, steric hindrance is often a factor (58), and it is conceivable that there simply is not enough physical space for two oligomeric, fluorescently tagged proteins to bind and interact in the same complex. When Dye and colleagues labeled MreB, a protein known to form long, helical filaments in Caulobacter cresentus, with two different fluorescent markers, they observed only partial colocalization of the two tagged versions (16), suggesting that a high percentage of visible overlap may not be possible. In the case of the T2S complex, although the individual components that make up the system are known, exactly how these proteins come together to create a large multiprotein complex is not well understood. EpsD is believed to form a ring-like assembly of 12 subunits in the outer membrane, while EpsL and EpsM are thought to exist as an unknown number of dimers in the inner membrane (25; also see Fig. S1 in the supplemental material). Neither the number of EpsC proteins needed to link the two membrane substructures together nor the number of fluorescent molecules necessary for visible foci to be detected is known. It is possible that the nonoverlapping foci represent T2S complexes containing both EpsM and EpsC, but not yet a complete oligomer of one or the other fluorescently labeled protein.
Our microscopy experiments indicate that EpsC localization requires EpsD, but it is unclear based on the current data whether both proteins are necessary for formation of a docking subcomplex or whether EpsD initiates placement of EpsC. The very recent finding that the EpsD homolog PulD localizes in a punctate pattern throughout the cell envelope when expressed in E. coli in the absence of PulC-PulN provides support for the latter suggestion (5). Oligomerization of EpsD, for example, may be the driving force behind nucleation of EpsC, and once EpsD pores are formed, their diffusion through the membrane may be constrained by interactions with other cell wall components, such as peptidoglycan and lipopolysaccharides.
We will continue to exploit this cell biological approach of GFP-EpsC localization to elucidate the relationship between the EpsC and EpsD proteins in vivo, to ensure that critical interactions with the cell envelope are maintained and to further dissect the roles of these two proteins in localization of the T2S complex. Studies of other combinations of gfp fusions and deletions of eps genes will also help us continue to refine our model of T2S complex assembly. Similar approaches have been very successful in defining the ordered assembly of other localized multiprotein complexes. For example, the recruitment of Fts cell division components to the septum has been found to occur in a sequential fashion (21, 30). With this approach and others that employ fluorescent proteins as tools for assessing protein-protein interactions in living cells, we expect to identify stages of assembly that may be otherwise difficult to elucidate outside the context of the membrane environment and the complete T2S complex.
Supplementary Material
Acknowledgments
We thank Michael Bagdasarian for anti-EpsG antiserum and Christine Jacobs-Wagner for the pKS mCherry plasmid. We are grateful to Marianna Shvartsbeyn for assistance with construction of pEpsCD, Sean Devine for assistance with construction of pVC1200, and Maria Scott for creation of the pGFP-EpsC plasmid.
This work was supported by grant AI49294 from the National Institutes of Health (to M.S.), and S.R.L. and M.D.G. were supported in part by National Institutes of Health training grants HL007698 and AI007258, respectively.
Footnotes
Published ahead of print on 27 February 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Bitter, W., M. Koster, M. Latijnhouwers, H. de Cock, and J. Tommassen. 1998. Formation of oligomeric rings by XcpQ and PilQ, which are involved in protein transport across the outer membrane of Pseudomonas aeruginosa. Mol. Microbiol. 27209-219. [DOI] [PubMed] [Google Scholar]
- 2.Bleves, S., M. Gerard-Vincent, A. Lazdunski, and A. Filloux. 1999. Structure-function analysis of XcpP, a component involved in general secretory pathway-dependent protein secretion in Pseudomonas aeruginosa. J. Bacteriol. 1814012-4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bouley, J., G. Condemine, and V. E. Shevchik. 2001. The PDZ domain of OutC and the N-terminal region of OutD determine the secretion specificity of the type II out pathway of Erwinia chrysanthemi. J. Mol. Biol. 308205-219. [DOI] [PubMed] [Google Scholar]
- 4.Buddelmeijer, N., O. Francetic, and A. P. Pugsley. 2006. Green fluorescent chimeras indicate nonpolar localization of pullulanase secreton components PulL and PulM. J. Bacteriol. 1882928-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buddelmeijer, N., M. Krehenbrink, F. Pecorari, and A. P. Pugsley. 2009. Type II secretion system secretin PulD localizes in clusters in the Escherichia coli outer membrane. J. Bacteriol. 191161-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cabeen, M. T., and C. Jacobs-Wagner. 2005. Bacterial cell shape. Nat. Rev. Microbiol. 3601-610. [DOI] [PubMed] [Google Scholar]
- 7.Campo, N., H. Tjalsma, G. Buist, D. Stepniak, M. Meijer, M. Veenhuis, M. Westermann, J. P. Muller, S. Bron, J. Kok, O. P. Kuipers, and J. D. Jongbloed. 2004. Subcellular sites for bacterial protein export. Mol. Microbiol. 531583-1599. [DOI] [PubMed] [Google Scholar]
- 8.Carballido-Lopez, R. 2006. The bacterial actin-like cytoskeleton. Microbiol. Mol. Biol. Rev. 70888-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casadaban, M. J., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138179-207. [DOI] [PubMed] [Google Scholar]
- 10.Chami, M., I. Guilvout, M. Gregorini, H. W. Remigy, S. A. Muller, M. Valerio, A. Engel, A. P. Pugsley, and N. Bayan. 2005. Structural insights into the secretin PulD and its trypsin resistant core. J. Biol. Chem. 28037732-37741. [DOI] [PubMed] [Google Scholar]
- 11.Cianciotto, N. P. 2005. Type II secretion: a protein secretion system for all seasons. Trends Microbiol. 13581-588. [DOI] [PubMed] [Google Scholar]
- 12.Connell, T. D., D. J. Metzger, J. Lynch, and J. P. Folster. 1998. Endochitinase is transported to the extracellular milieu by the eps-encoded general secretory pathway of Vibrio cholerae. J. Bacteriol. 1805591-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ding, Y., B. M. Davis, and M. K. Waldor. 2004. Hfq is essential for Vibrio cholerae virulence and downregulates sigma expression. Mol. Microbiol. 53345-354. [DOI] [PubMed] [Google Scholar]
- 14.Donnenberg, M. S., and J. B. Kaper. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 594310-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Douet, V., L. Loiseau, F. Barras, and B. Py. 2004. Systematic analysis, by the yeast two-hybrid, of protein interaction between components of the type II secretory machinery of Erwinia chrysanthemi. Res. Microbiol. 15571-75. [DOI] [PubMed] [Google Scholar]
- 16.Dye, N. A., Z. Pincus, J. A. Theriot, L. Shapiro, and Z. Gitai. 2005. Two independent spiral structures control cell shape in Caulobacter. Proc. Natl. Acad. Sci. USA 10218608-18613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Filloux, A. 2004. The underlying mechanisms of type II protein secretion. Biochim. Biophys. Acta 1694163-179. [DOI] [PubMed] [Google Scholar]
- 18.Fullner, K. J., and J. J. Mekalanos. 1999. Genetic characterization of a new type IV-A pilus gene cluster found in both classical and El Tor biotypes of Vibrio cholerae. Infect. Immun. 671393-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Furste, J. P., W. Pansegrau, R. Frank, H. Blocker, P. Scholz, M. Bagdasarian, and E. Lanka. 1986. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene 48119-131. [DOI] [PubMed] [Google Scholar]
- 20.Gerard-Vincent, M., V. Robert, G. Ball, S. Bleves, G. P. F. Michel, A. Lazdunski, and A. Filloux. 2002. Identification of XcpP domains that confer functionality and specificity to the Pseudomonas aeruginosa type II secretion apparatus. Mol. Microbiol. 441651-1665. [DOI] [PubMed] [Google Scholar]
- 21.Goehring, N. W., and J. Beckwith. 2005. Diverse paths to midcell: assembly of the bacterial cell division machinery. Curr. Biol. 15R514-R526. [DOI] [PubMed] [Google Scholar]
- 22.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1774121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirst, T. R., J. Sanchez, J. B. Kaper, S. J. Hardy, and J. Holmgren. 1984. Mechanism of toxin secretion by Vibrio cholerae investigated in strains harboring plasmids that encode heat-labile enterotoxins of Escherichia coli. Proc. Natl. Acad. Sci. USA 817752-7756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hofstra, H., and B. Witholt. 1984. Kinetics of synthesis, processing, and membrane transport of heat-labile enterotoxin, a periplasmic protein in Escherichia coli. J. Biol. Chem. 25915182-15187. [PubMed] [Google Scholar]
- 25.Johnson, T. L., J. Abendroth, W. G. J. Hol, and M. Sandkvist. 2006. Type II secretion: from structure to function. FEMS Microbiol. Lett. 255175-186. [DOI] [PubMed] [Google Scholar]
- 26.Johnson, T. L., M. E. Scott, and M. Sandkvist. 2007. Mapping critical interactive sites within the periplasmic domain of the Vibrio cholerae type II secretion protein EpsM. J. Bacteriol. 1899082-9089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaper, J. B., J. G. Morris, Jr., and M. M. Levine. 1995. Cholera. Clin. Microbiol. Rev. 848-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Korotkov, K. V., B. Krumm, M. Bagdasarian, and W. G. Hol. 2006. Structural and functional studies of EpsC, a crucial component of the type 2 secretion system from Vibrio cholerae. J. Mol. Biol. 363311-321. [DOI] [PubMed] [Google Scholar]
- 29.Lee, H. M., J. R. Chen, H. L. Lee, W. M. Leu, L. Y. Chen, and N. T. Hu. 2004. Functional dissection of the XpsN (GspC) protein of the Xanthomonas campestris pv. campestris type II secretion machinery. J. Bacteriol. 1862946-2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Margolin, W. 2005. FtsZ and the division of prokaryotic cells and organelles. Nat. Rev. Mol. Cell Biol. 6862-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marsh, J. W., and R. K. Taylor. 1998. Identification of the Vibrio cholerae type 4 prepilin peptidase required for cholera toxin secretion and pilus formation. Mol. Microbiol. 291481-1492. [DOI] [PubMed] [Google Scholar]
- 32.Menard, R., P. J. Sansonetti, and C. Parsot. 1993. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol. 1755899-5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meselson, M., and R. Yuan. 1968. DNA restriction enzyme from E. coli. Nature 2171110-1114. [DOI] [PubMed] [Google Scholar]
- 34.Michel, G., S. Bleves, G. Ball, A. Lazdunski, and A. Filloux. 1998. Mutual stabilization of the XcpZ and XcpY components of the secretory apparatus in Pseudomonas aeruginosa. Microbiology 1443379-3386. [DOI] [PubMed] [Google Scholar]
- 35.Miller, G., and M. Feiss. 1988. The bacteriophage lambda cohesive end site: isolation of spacing/substitution mutations that result in dependence on Escherichia coli integration host factor. Mol. Gen. Genet. 212157-165. [DOI] [PubMed] [Google Scholar]
- 36.Nouwen, N., N. Ranson, H. Saibil, B. Wolpensinger, A. Engel, A. Ghazi, and A. P. Pugsley. 1999. Secretin PulD: association with pilot PulS, structure, and ion-conducting channel formation. Proc. Natl. Acad. Sci. USA 968173-8177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nunn, D. N., and S. Lory. 1993. Cleavage, methylation, and localization of the Pseudomonas aeruginosa export proteins XcpT, XcpU, XcpV, and XcpW. J. Bacteriol. 1754375-4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Opalka, N., R. Beckmann, N. Boisset, M. N. Simon, M. Russel, and S. A. Darst. 2003. Structure of the filamentous phage pIV multimer by cryo-electron microscopy. J. Mol. Biol. 325461-470. [DOI] [PubMed] [Google Scholar]
- 39.Overbye, L. J., M. Sandkvist, and M. Bagdasarian. 1993. Genes required for extracellular secretion of enterotoxin are clustered in Vibrio cholerae. Gene 132101-106. [DOI] [PubMed] [Google Scholar]
- 40.Possot, O. M., M. Gerard-Vincent, and A. P. Pugsley. 1999. Membrane association and multimerization of secreton component PulC. J. Bacteriol. 1814004-4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Possot, O. M., G. Vignon, N. Bomchil, F. Ebel, and A. P. Pugsley. 2000. Multiple interactions between pullulanase secreton components involved in stabilization and cytoplasmic membrane association of PulE. J. Bacteriol. 1822142-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pugsley, A. P., M. G. Kornacker, and I. Poquet. 1991. The general protein-export pathway is directly required for extracellular pullulanase secretion in Escherichia coli K12. Mol. Microbiol. 5343-352. [DOI] [PubMed] [Google Scholar]
- 43.Py, B., L. Loiseau, and F. Barras. 2001. An inner membrane platform in the type II secretion machinery of Gram-negative bacteria. EMBO Rep. 2244-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robert, V., A. Filloux, and G. P. Michel. 2005. Subcomplexes from the Xcp secretion system of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 25243-50. [DOI] [PubMed] [Google Scholar]
- 45.Robert, V., F. Hayes, A. Lazdunski, and G. P. Michel. 2002. Identification of XcpZ domains required for assembly of the secreton of Pseudomonas aeruginosa. J. Bacteriol. 1841779-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sandkvist, M. 2001. Biology of type II secretion. Mol. Microbiol. 40271-283. [DOI] [PubMed] [Google Scholar]
- 47.Sandkvist, M., M. Bagdasarian, S. P. Howard, and V. J. Dirita. 1995. Interaction between the autokinase EpsE and EpsL in the cytoplasmic membrane is required for extracellular secretion in Vibrio cholerae. EMBO J. 141664-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sandkvist, M., L. P. Hough, M. M. Bagdasarian, and M. Bagdasarian. 1999. Direct interaction of the EpsL and EpsM proteins of the general secretion apparatus in Vibrio cholerae. J. Bacteriol. 1813129-3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sandkvist, M., L. O. Michel, L. P. Hough, V. M. Morales, M. Bagdasarian, M. Koomey, V. J. Dirita, and M. Bagdasarian. 1997. General secretion pathway (eps) genes required for toxin secretion and outer membrane biogenesis in Vibrio cholerae. J. Bacteriol. 1796994-7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sandkvist, M., V. Morales, and M. Bagdasarian. 1993. A protein required for secretion of cholera toxin through the outer membrane of Vibrio cholerae. Gene 12381-86. [DOI] [PubMed] [Google Scholar]
- 51.Schafer, A., A. Tauch, W. Jager, J. Kalinowski, G. Thierbach, and A. Puhler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 14569-73. [DOI] [PubMed] [Google Scholar]
- 52.Scheffers, D. J., and M. G. Pinho. 2005. Bacterial cell wall synthesis: new insights from localization studies. Microbiol. Mol. Biol. Rev. 69585-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scott, M. E., Z. Y. Dossani, and M. Sandkvist. 2001. Directed polar secretion of protease from single cells of Vibrio cholerae via the type II secretion pathway. Proc. Natl. Acad. Sci. USA 9813978-13983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shiomi, D., M. Yoshimoto, M. Homma, and I. Kawagishi. 2006. Helical distribution of the bacterial chemoreceptor via colocalization with the Sec protein translocation machinery. Mol. Microbiol. 60894-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sikora, A. E., S. R. Lybarger, and M. Sandkvist. 2007. Compromised outer membrane integrity in Vibrio cholerae type II secretion mutants. J. Bacteriol. 1898484-8495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stewart, G. C. 2005. Taking shape: control of bacterial cell wall biosynthesis. Mol. Microbiol. 571177-1181. [DOI] [PubMed] [Google Scholar]
- 57.Tsai, R. T., W. M. Leu, L. Y. Chen, and N. T. Hu. 2002. A reversibly dissociable ternary complex formed by XpsL, XpsM and XpsN of the Xanthomonas campestris pv. campestris type II secretion apparatus. Biochem. J. 367865-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Voorhout, W. F., J. J. Leunissen-Bijvelt, J. L. Leunissen, and A. J. Verkleij. 1986. Steric hindrance in immunolabelling. J. Microsc. 141303-310. [DOI] [PubMed] [Google Scholar]
- 59.Voulhoux, R., G. Ball, B. Ize, M. L. Vasil, A. Lazdunski, L. F. Wu, and A. Filloux. 2001. Involvement of the twin-arginine translocation system in protein secretion via the type II pathway. EMBO J. 206735-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.