Abstract
The Epstein-Barr virus (EBV) latency III program imposed by EBNA2 and LMP1 is directly responsible for immortalization of B cells in vitro and is thought to mediate most immunodeficiency-related posttransplant lymphoproliferative diseases in vivo. To answer the question whether and how this proliferation program is related to c-Myc, we have established the transcriptome of both c-Myc and EBV latency III proliferation programs using a Lymphochip specialized microarray. In addition to EBV-positive latency I Burkitt lymphoma lines and lymphoblastoid cell lines (LCLs), we used an LCL expressing an estrogen-regulatable EBNA2 fusion protein (EREB2-5) and derivative B-cell lines expressing a constitutively active or tetracycline-regulatable c-myc gene. A total of 897 genes were found to be fourfold or more up- or downregulated in either one or both proliferation programs compared to the expression profile of resting EREB2-5 cells. A total of 661 (74%) of these were regulated similarly in both programs. Numerous repressed genes were known targets of STAT1, and most induced genes were known to be upregulated by c-Myc and to be involved in cell proliferation. In keeping with the gene expression patterns, inactivation of c-Myc by a chemical inhibitor or by conditional expression of dominant-negative c-Myc and Max mutants led to proliferation arrest of LCLs. Most genes differently regulated in both proliferation programs corresponded to genes induced by NF-κB in LCLs, and many of them coded for immunoregulatory and/or antiapoptotic molecules. Thus, c-Myc and NF-κB are the two main transcription factors responsible for the phenotype, growth pattern, and biological properties of cells driven into proliferation by EBV.
Initially isolated from African Burkitt lymphoma (BL) cells, Epstein-Barr virus (EBV) is the first transforming virus described in humans. This gammaherpesvirus, belonging to lymphocryptoviruses, is widely distributed around the world, and over 95% of adults have been infected at some time. Most cases of primary infection are asymptomatic, but some do have clinical symptoms, presenting as a benign self-limiting lymphoproliferative disease, so-called infectious mononucleosis. During primary infection, EBV infects and induces continuous proliferation of resting B cells. In vivo, EBV-infected B cells are actively eliminated by a vigorous cytotoxic immune response, allowing the spontaneous resolution of EBV primary infection. Yet EBV persists silently in the memory B-cell compartment of the organism throughout life and may be reactivated in cases of immunodeficiency (25, 56). The efficient immortalization capacity of EBV is routinely demonstrated by establishment of lymphoblastoid cell lines (LCLs) in vitro. Immunodeficiency-related B-cell lymphomas, including posttransplant lymphoproliferative disorders (PTLDs), are caused directly by EBV (45), reflecting the transforming capacity of the virus and the destruction of the T-cell compartment of the host by the immunodeficiency (27). Thus, it is usually conceded that LCLs represent an in vitro model of PTLDs. EBV is also associated with various cancers, including BL, Hodgkin's lymphomas, T-cell lymphomas, and nasopharyngeal carcinomas.
The EBV genome persists in the host cell in an episomal form. Three latency viral programs have been described for this virus both in vitro and in vivo. Two small nonpolyadenylated RNAs (EBER1 and EBER2) and a large transcription unit called BART (BamHI-A rightward transcript) or CST (complementary strand transcript), giving rise to a number of microRNAs, are expressed in all forms of latency. Latency I is characterized by the expression of the viral protein EBNA1 that is essential for episomal maintenance of the EBV genome and that is found in EBV-associated BL tumors. Latency II corresponds to the expression of EBNA1, latent membrane protein 1 (LMP1), LMP2a, and LMP2b. LMP1 reroutes the TRAF (tumor necrosis factor [TNF] receptor-associated factor) and TRADD (TNF receptor death domain) adaptors of the TNF receptor family and is responsible for continuous activation of NF-κB (19). LMP2a activates the kinases Syk and Lyn of the B-cell receptor (BCR), providing a continuous survival signal to the cell (58). Latency II is characteristic of almost all EBV-associated tumors except BL tumors and PTLDs. EBV of latency III is present in LCLs in vitro and in most PTLDs in vivo. Latency III, also called the viral proliferation program by some authors (54), is characterized by the expression of EBNA2 that secondarily governs the expression of the EBNA3 family as well as of the LMP proteins. A fourth EBV latency state, latency 0, has been described that is characterized by the almost complete silencing of the virus. Latency 0 represents the state of the virus in the G0 resting memory B-cell reservoir in vivo (5, 27, 55).
One of the key issues raised by the transition from a resting B cell to its EBV-infected counterpart is to identify the cellular transcriptional targets of the virus responsible for cell reprogramming, i.e., for induction of continuous proliferation and B-cell immortalization. This question has not been fully addressed experimentally. The transforming properties of the three major EBV latency proteins, EBNA2, LMP1, and LMP2a, have been extensively studied and point to two obvious cellular pathways usurped by EBV: Notch and NF-κB. EBNA2 acts at the transcriptional level through direct interaction with RBP/Jκ (also called CBF1) as a Notch equivalent. In some instances, Notch can partially replace EBNA2 (15, 18). Functional substitution of Notch by EBNA2 is very likely to be responsible for arresting B-cell differentiation (56), but the Notch pathway has not been shown to directly induce proliferation of EBV-immortalized B cells (5). In some experimental models, the active form of Notch1 could cause growth suppression of the cells, accompanied by cell cycle inhibition and apoptosis (37). NF-κB activation is mainly caused by LMP1 and is associated with protection against apoptosis (5). Most so-called “activation markers” expressed in viral latency II or III are targets of LMP1 and have been shown to be regulated by NF-κB. Expression profiling studies confirmed that most genes regulated by LMP1 in B cells are dependent on NF-κB (6). But NF-κB activation by LMP1, first described by Laherty et al. (30), is in fact an ambivalent event. It certainly contributes to protection of EBV-immortalized B cells against apoptosis (7, 12), but it also promotes interaction of B cells and T cells and T-cell-dependent B-cell killing through induction of adhesion molecules, expression of CD95, and upregulation of molecules involved in antigen presentation (19, 32). Overactivation of NF-κB by LMP1 may also lead to apoptosis by ligand-independent activation of CD95 (31). Consistent with a role in protection against apoptosis, LMP2a has been shown to provide B cells lacking a functional BCR with a survival signal in vivo (8) and to increase NF-κB activation (16). LMP2a has also been shown to activate the Notch pathway (2). But LMP2a has not been shown, so far, to drive cell proliferation directly.
Another putative transcriptional target of EBV is c-Myc. Through dimerization with Max, c-Myc is a master transcription factor regulating the cell cycle machinery. EBNA2 acts as a positive regulator of c-myc gene expression in the context of its native P1-P2 promoter after infection of resting B cells (21). Activation of NF-κB by LMP1 is also able to enhance c-myc expression (10). LCLs and PTLDs express increased levels of c-Myc, like most cases of aggressive lymphomas with a high proliferative index (29, 51). Thus, the c-myc gene appears to be a natural cellular target responsible for EBV-driven B-cell proliferation. This concept is supported by a recent report showing growth inhibition of one LCL by the c-Myc chemical inhibitor 10058-F4 (14). But lessons from in vitro models and human BL tumors suggest that c-myc disregulation alone is probably not able to convert normal B cells into tumor cells (34, 60). Importantly, c-Myc is dispensable for cell growth as shown in c-myc knockout fibroblasts (36). c-Myc overexpression is known to induce activation of the serine-threonine kinases ATM (ataxia-telangiectasia mutated) and ATR (ATM-Rad3-related), leading to phosphorylation and activation of the proapoptotic protein p53 and to cell apoptosis (34). c-Myc-induced p53 activation is interpreted as a physiological fail-safe mechanism that may prevent inappropriate proliferation of normal cells. Of note, we along with others have noticed some p53 activation in EBV-immortalized LCLs (10, 32). It is agreed that proliferation of BL tumor cells requires secondary genetic and epigenetic alterations in the p53 tumor suppressor pathway (34). But such genetic alterations are rare in EBV-related PTLDs (43) and nearly absent in LCLs despite years of continuous culture. Reciprocally, acquisition of the growth pattern and phenotype of BL cells by c-Myc overexpression requires that the EBV latency III proliferation program be extinguished in vitro (41, 44) and in vivo (22, 23, 58). This is in line with the finding that c-Myc downregulates NF-κB (47, 52, 57). We have established the expression profiles of both c-Myc-dependent and EBNA2-dependent latency III proliferation programs using BL cell lines, LCLs, and primary B-cell model systems in which the viral proliferation program and an introduced c-myc gene can be switched on and off by estrogen and tetracycline, respectively (41, 44, 48, 49). Studying both the interferon and NF-κB response genes led us to show that c-Myc overexpression in BL tumor cells may directly contribute to immune escape through repression of the interferon response (48). This biological feature of the c-Myc proliferation program would be incompatible with the immunological status of EBV latency III proliferating cells (40). Thus, so far, c-Myc-driven latency I and EBNA2-driven latency III proliferation programs seem to be incompatible, and a direct role of c-Myc in proliferation of EBV-immortalized B cells has not been formerly demonstrated.
Here, we raise the question whether the EBV-driven proliferation program of latency III is related to the c-Myc proliferation program. To address this question, we have now compared the whole expression profile of both programs with that of resting B cells that have been arrested by switching off EBNA2 in conditionally immortalized B cells.
MATERIALS AND METHODS
Cellular models.
The following cellular models have been used for the EBNA2-dependent EBV latency III program: LCLs PRI, TSOC, OUL, RUD, 1602, 1.11, 1.13, and 1.25. The EBV-positive EREB2-5 cell line with an EBNA2-estrogen chimeric receptor (function of EBNA2 inducible by estrogen) was cultured in the absence of estrogen for 48 h (0 h; resting cells) and treated with estrogen for 1, 2, 3, 6, and 8 h and continuously (24).
EBV-negative BL41 and EBV-positive latency I BL cell lines Akata, Mukira, BL29, Elijah, the BL-like A1 cell line (derived from the EREB2-5 cell line transfected with an Igκ-c-myc minilocus construct and deprived of estrogen), and P493-6 cells (an EREB2-5-derivative cell line transfected with a Tet-off-inducible c-myc vector grown continuously in the absence of estrogen and tetracycline) served as cellular models for the c-Myc program (42). P493-6 cells were also included in the c-myc-off state (0 h; cultured in the presence of 1 μg/ml tetracycline [Sigma-Aldrich]) and were used 8 h after tetracycline was washed out (reinduction of c-myc).
All cell lines were cultured in RPMI 1640 medium (Gibco BRL-Life Technologies, Cergy-Pontoise, France) supplemented with 10% decomplemented fetal calf serum (D. Dutscher, Biotech GmbH), 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine (Gibco BRL-Life Technologies) at 37°C in a humidified 5% CO2 atmosphere. EREB2-5 cells were grown in the presence of 1 μM estradiol (Sigma-Aldrich, St Louis, MO). P493-6 cells were grown continuously in the absence of estrogen and tetracycline (Sigma-Aldrich) (24, 42).
RNA isolation and cDNA microarray screening.
RNAs were extracted from each cell line and in each cell condition as well as from a lymphopool of reference using the Trizol reagent (Invitrogen) (1). Hybridizations were performed as described previously (1). Briefly, after reverse transcription, amplification of cDNAs was associated with Cy5 labeling for each cell condition and with Cy3 labeling of the cDNAs from the lymphopool reference. Cy5-labeled cDNAs of each condition were cohybridized with Cy3-labeled cDNAs of the lymphopool on a Lymphochip cDNA array as described previously (1). For each condition and for each gene, the specific signal was defined as the Cy5/Cy3 ratio.
Statistical analysis of DNA microarrays.
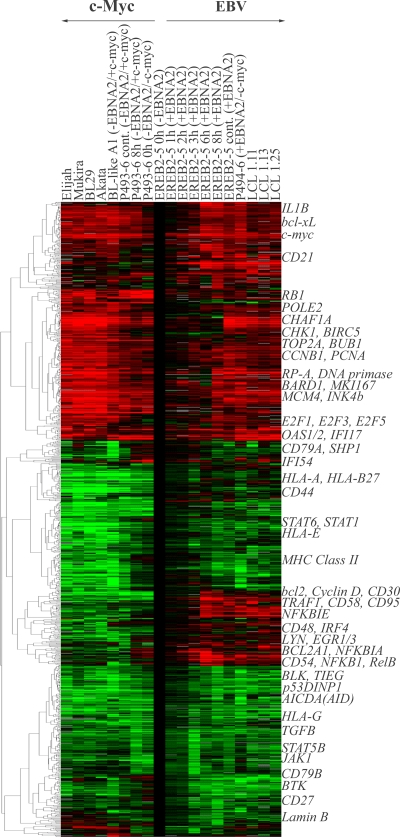
We have previously reported the expression profile of 66 genes (48) out of the 12,069 of the Lymphochip DNA microarray (1). Here, we have analyzed the whole gene expression profile. For each gene, the Cy5/Cy3 ratio of the condition studied and the Cy5/Cy3 ratio of the EREB2-5 cells deprived of estrogen (resting cells) were calculated and corresponded to the level of induction or repression of a given gene from the resting to the proliferative state. Then, a first filtering step was applied, consisting in the selection of the genes for which at least one condition exhibits at least fourfold induction or repression. The second filtering step consisted in eliminating all the inconsistent duplicates. The result was the selection of 897 genes (Fig. 1).
FIG. 1.
Hierarchical clustering of the 897 genes selected. Duplicates were eliminated by selection of the sequence corresponding to the median of all the sequences of the same gene. Then, filtering was performed by selection of genes with at least a fourfold change in at least one of the cell conditions compared to estrogen deprived EREB2-5 cells as a model of resting G1 B cells (EREB2-5 cells at 0 h in the absence of EBNA2). Culture conditions are given at the top of the panel and indicate the cell line, number of hours of treatment (cont, continuously), and the presence (+) or absence (−) of EBNA2 and/or c-Myc. Expression levels are shown in red (induced) and green (repressed) relative to the expression level of arrested EREB2-5 cells. Representative genes of an expression profile cluster are indicated on the right-hand side. MHC, major histocompatibility complex.
Statistical analysis was done by combining the techniques of hierarchical clustering, K-mean clustering, and principal component analysis using the Cluster and Treeview programs after formatting the tables with the Excel software. The 897 genes were partitioned by K-mean clustering so that the number of genes was above 15 in each group. The resulting number of groups obtained was 18 (G1 to G18). All pairs of groups for which Pearson's correlation coefficient was over 0.9 were merged. Then, the closest groups were merged 2 by 2 according to their proximity by hierarchical clustering and principal component analysis of the mean vectors. This was repeated until maximization of the absolute value of χ2 (13). The final resulting number of classes was 10 (C1 to C10). Functional annotation of genes was performed manually after consulting the following sites: http://www.ncbi.nlm.nih.gov/sites/entrez (OMIM section), http://www.genenames.org/cgi-bin/hgnc_search.pl, http://www.myccancergene.org (c-Myc target genes), and http://people.bu.edu/gilmore/nf-kb/index.html (NF-κB target genes).
Nylon filter arrays analysis.
Nylon filter arrays were designed that represent 2,253 human genes. The genes were selected for being involved in tumor processing, cell cycle, and apoptosis. The filters contain oligonucleotides (60 bp) that are identical with a sequence within the 3′ prime end of the corresponding mRNA [approximately 0- to 1,000-bp distance from the poly(A) site]. Each oligonucleotide was spotted twice on the filter array. In addition, 10 Escherichia coli sequences and 10 randomly selected antisense sequences from spotted genes were added as negative controls.
P493-6 cells proliferating in the absence of tetracycline were treated with different concentrations of tetracycline ranging from 0 to 8 ng/ml for 48 h and with 100 ng/ml for full c-Myc repression. RNA and protein were prepared by standard methods (48).
The labeling, hybridization, scanning, and data processing procedures were performed as described earlier (35). In brief, a standard reverse transcription reaction was performed using 4 μg of total RNA in the presence of 50 μCi of [α-33P]dCTP (3,000 Ci/mmol; Perkin Elmer Life Sciences, MA) and an oligo(dT)20 primer. The hybridization temperature was 40°C.
To ensure the highest data reliability, we produced 18 data points in total per gene, and the average was calculated from all 18 data points. Total RNA from three independent experiments was prepared from cells at each c-Myc level. The RNA samples were labeled individually, and each sample was hybridized with three filters. We differentiated between significance of expression, which determines whether a signal on the filter is significantly above the background, and significance of differential expression, which indicates whether expression of a gene is different in two samples. The expression data of two different probes were analyzed by means of a two-tailed t test for independent samples (P ≤ 0.05) (35).
c-Myc-responsive genes were determined by pairwise comparisons of expression values at each c-Myc level with the expression values of the reference sample c-Myc-off (100 ng/ml tetracycline). Genes that were defined as c-Myc-responsive had to fulfill the following criteria: (i) significance of differential expression at least for the comparison of c-Myc-off versus c-Myc-on (without tetracycline), (ii) at least a twofold change in expression between c-Myc-off and c-Myc-on, and (iii) significance of expression at least in the c-Myc-on or c-Myc-off samples for c-Myc-induced and c-Myc-repressed genes, respectively.
Plasmid constructs.
The pRT-1 inducible vector was derived from the previously described CKR516 vector (12) in which the Eμ-Rous sarcoma virus promoter was replaced by an Eμ-chicken-beta-actin promoter. The EBNA1 gene was added to maintain episomal replication of the vector also in EBV-negative cells. The enhanced green fluorescent protein marker was replaced by an inactive truncated version of never growth factor receptor (NGFR) lacking the cytoplasmic domain (NGFR-t). The bidirectional tetracycline-inducible promoter drives the expression of both NGFR-t and the cDNA of interest. A complete description of these vectors is given elsewhere (4). The cDNA coding for hemagglutinin (HA)-tagged dominant-negative mutants of c-Myc and Max described previously (17), named Δc-Myc and MaxΔBR, respectively, were cloned into the SfiI sites.
Transfection and cell sorting.
Stable transfection of LCL PRI and hygromycin selection (Calbiochem) were performed as described previously (3, 12). Induction was stable after 4 weeks of selection. After a 24-h treatment with 1 μg/ml doxycycline (Sigma-Aldrich, Saint-Louis, MO), the cells became NGFR-t positive, with transfection rates varying from 60 to 90%. The NGFR-t-expressing cells were purified following the manufacturer's protocol (MACs microbeads; Miltenyi Biotech, Auburn, CA). Briefly, viable cells were isolated by Ficoll-Paque Plus density gradient centrifugation (Eurobio, Les Ulis, France) and divided into two batches, one treated with 1 μg/ml doxycycline for 24 h and one without doxycycline. Both batches were then submitted in parallel to cell purification. Incubation was performed for 30 min with 1 μg of monoclonal antibody (MAb) anti-NGFR (BD Pharmingen, San Diego, CA) for 106 doxycycline-treated cells or without primary antibody for untreated cells. Cell separation was performed with 20 μl of goat anti-mouse immunoglobulin G (IgG) magnetic beads (Miltenyi Biotech, Bergisch Gladbach, Germany) for 107 cells. For immunoblotting analysis, purified cells (5 × 106 cells) were lysed in 100 μl of blue Laemmli lysis buffer (Bio-Rad, Hercules, CA) and 5% β-mercaptoethanol (Bio-Rad), and then the lysates were sonicated. For flow cytometry analysis, purified cells were resuspended at 106 cells/ml. For proliferation assays, cells were plated in 96-well plates (1.5 × 104/well) in medium containing 10% fetal calf serum. During 4 days, proliferation rates were obtained using a CellTiter 96 AQueous One Solution cell proliferation assay (Promega, France).
Immunoblotting analysis.
Immunoblotting was performed as described elsewhere (32). After the lysate was cleared, proteins were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto nitrocellulose membranes (Schleicher & Schuell, Dassel, Germany) for probing with mouse anti-HA tag (clone 6E2; 1/1,000 dilution; Cell Signaling Technology, CA) and anti-c-Myc (clone 9E10; 1/200 dilution; Santa Cruz Biotechnology, Beverly, MA) MAbs and anti-histone H3 rabbit polyclonal antibody (1/2,000 dilution; Cell Signaling Technology). Specifically bound antibodies were detected with a 1/5,000 dilution of peroxidase-conjugated goat anti-mouse or goat anti-rabbit IgG (Dako, Trapes, France) and visualized with an enhanced chemiluminescence detection system (Amersham, Orsay, France).
Growth inhibition assay.
The LCL PRI, TSOC, OUL, RUD, 1602, 1.25, and EREB2-5 cells were treated with the c-Myc chemical inhibitor 10058-F4 [(Z,E)-5-(4-ethylbenzylidine)-2-thioxothiazolidin-4-one] (Calbiochem, La Jolla, CA) at concentrations ranging between 15 and 120 μM. The concentration of the inhibitor that resulted in 50% growth inhibition (GI50) was calculated by linear regression analysis with a linear correlation coefficient, r2, of >0.9. During time course experiments, cell growth was evaluated each 24 h in 96-well plates (1.5 × 105 cells/ml) using a CellTiter 96 AQueous One Solution cell proliferation assay (Promega).
Apoptosis analysis by flow cytometry.
Cells were doubly stained with annexin V-fluorescein isothiocyanate (FITC) (BD Pharmingen) and propidium iodide (PI; Sigma-Aldrich) in cold phosphate-buffered saline (PBS)-CaCl2-MgCl2 (Invitrogen, Cergy-Pontoise, France) as described previously (3) and analyzed by a FACSCalibur cytometer (BD Pharmingen).
Analysis of DNA synthesis by a BrdU pulse-labeling technique.
Exponentially growing cells (2×106) were pulse labeled with 18 μg/ml bromodeoxyuridine (BrdU; Sigma-Aldrich) for 3 h, washed with PBS, and fixed with 50% ice-cold ethanol for 30 min. Cells were then incubated for 30 additional minutes with 2 N HCl to partially denature the DNA, then washed, and resuspended in PBS-1% bovine serum albumin-0.5% Tween 20 solution. Incorporated BrdU was stained with 20 μl of FITC-conjugated anti-BrdU mouse MAb (BD Pharmingen) for 30 min at room temperature. Samples were then washed in PBS-1% bovine serum albumin-0.5% Tween 20 solution and resuspended in PBS containing 50 μg/ml PI. Bivariate distributions of BrdU amounts (FITC) versus DNA content (PI) were analyzed by a FACSCalibur cytometer (BD Pharmingen). Of note, treatment of cells with the 10058-F4 compound resulted in an autofluorescence increase. Therefore, the regions defining the G0/G1, S, and G2/M phases were set up in the valleys separating the different phases of the cell cycle.
RESULTS
Classification of genes induced in EBV latency III and/or c-Myc-driven proliferation programs.
For this study, we exploited our inducible cellular models to compare the EBV latency III- and the c-Myc-driven proliferation programs. In the EREB2-5 cell line, latency III proliferation is governed by an estrogen-inducible EBNA2-estrogen receptor fusion protein that can be reversibly switched on and off by the addition or withdrawal of estrogen (24). The cell line P493-6 is derived from EREB2-5 cells transfected with a Tet-off- inducible c-myc vector and is thus doubly inducible for c-Myc and EBNA2. In other words, this cellular model can be induced to proliferate either by induction of the c-Myc or the EBNA2 latency III proliferation program (42, 44). Taking advantage of this doubly inducible cell system, we additionally studied P493-6 cells in the c-myc-off/EBNA2-on condition (i.e., in the presence of both tetracycline and estrogen) as an additional model for EBV latency III-driven proliferation. The cell line A1 has been generated by introducing an Igκ-c-myc minilocus into EREB2-5 cells. It proliferates in the absence of estrogen and recapitulates features of a BL cell with t(2;8) translocation. To consolidate the results obtained with the EREB2-5-derived cell model systems, we used three classical LCLs (1.11, 1.13, and 1.25) and four EBV-positive latency I BL cell lines (Elijah, Mukira, BL29, and Akata) as cellular models for constitutive EBV latency III- and c-Myc-driven proliferation (latency I), respectively. As EREB2-5 cells enter a resting state after estrogen withdrawal, the transcriptome of cells proliferating either on an EBV-driven or a c-Myc-driven proliferation program was compared to the one of estrogen-deprived EREB2-5 cells.
A total of 897 genes were at least fourfold induced or repressed in at least one of the conditions, EBV latency III- and/or c-Myc-driven proliferating cells. Hierarchical clustering of the 897 differentially expressed selected genes in proliferating cells, compared to resting EREB2-5 cells, gave rise to four main branches (Fig. 1). Two branches corresponded to genes that were upregulated (337 genes) and downregulated (243 genes) in both proliferation programs, one branch to genes that were repressed predominantly in c-Myc-driven proliferating cells (211 genes), and one branch to genes induced in EBV latency III proliferating cells only (106 genes). Thus, hierarchical clustering confirmed that the majority of differentially expressed genes (580 out of 897 genes) were regulated in a similar manner in both categories of proliferating cells.
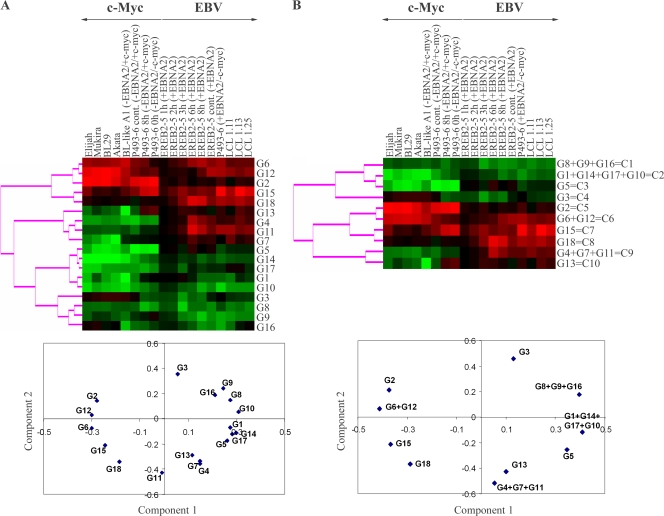
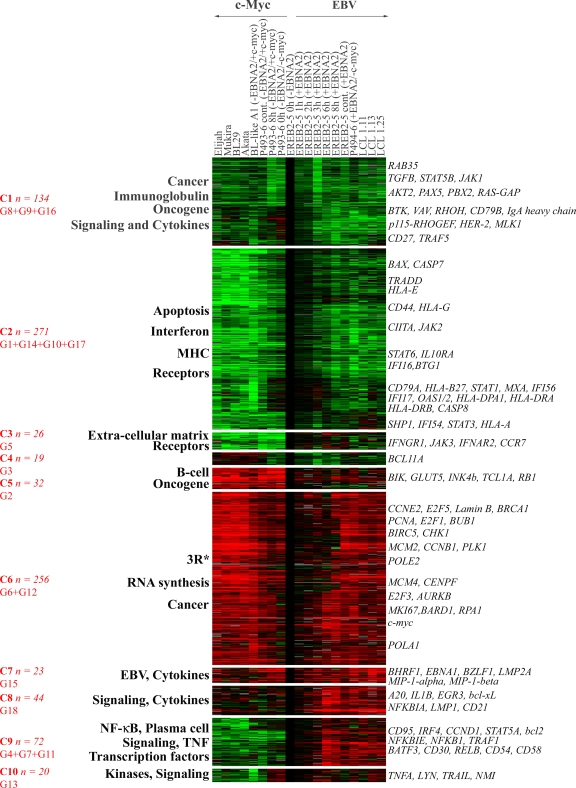
To find out the different classes of coregulated genes in proliferating cells, we combined K-mean clustering, hierarchical clustering, and principal component analysis using cluster analysis software (1). For K-mean clustering, the maximum number of groups was set up in order to get at least 15 genes in each group. With this criterion, K-mean clustering analysis suggested the existence of 18 different groups, named G1 to G18 (Fig. 2A). Each group was then taken as a metagene, corresponding to the mean expression of the genes of the group. Clustering and principal component analysis of these 18 metagenes showed that the distance between some of these metagenes was very small. We then merged step by step and two by two the closest metagenes. In order to select which pair of metagenes to merge, we used the χ2 maximization test as a criterion for maximization of the dispersion (13). This led us to generate 10 different classes, C1 to C10, from the 18 original groups, with each class having a specific expression profile (Fig. 2B). Functional analysis of these classes showed that, except for class C4 (19 genes), all classes of genes were characterized by a specific over- or underrepresentation of genes belonging to specific functional groups (Fig. 3 and Table 1). The detailed list of most significant genes in each class is given in the supplemental material.
FIG. 2.
Results of the first step of K-mean clustering. The 897 selected genes were initially divided in 18 groups, G1 to G18, using the K-mean clustering method. The K-mean groups for which the Pearson correlation coefficient was above 0.9 were aggregated 2 by 2. Then, the closest groups were aggregated 2 by 2 either after hierarchical clustering or principal component analysis. The choice between both methods was given by the maximum increase of absolute value of χ2. This was repeated until the maximum of the χ2 value was obtained. Proximity of the groups was graphically visualized by hierarchical clustering and principal component analysis. (A) Hierarchical clustering (upper panel) and principal component analysis (lower panel) of the 18 groups G1 to G18. (B) Hierarchical clustering (upper panel) and principal component analysis (lower panel) of the 10 merged classes (C1 to C10). Culture conditions are given at the tops of the panels and indicate the cell line, number of hours of treatment (cont, continuously), and the presence (+) or absence (−) of EBNA2 and/or c-Myc.
FIG. 3.
Expression profile of the 10 K-mean classes. Each K-mean class of Fig. 2B was clustered in a hierarchical manner. The expression profile was obtained with the Treeview program. Overrepresented functions are annotated on the left. Some putatively interesting genes are annotated on the right. The merged classes and the number of genes are indicated in red. 3R, genes involved in DNA replication, repair, and/or recombination. Culture conditions are given at the top of the panel and indicate the cell line, number of hours of treatment (cont, continuously), and the presence (+) or absence (−) of EBNA2 and/or c-Myc. MHC, major histocompatibility complex.
TABLE 1.
Mean variation tendency of classes and corresponding functional categories of genes regulated in both c-Myc and EBV latency III proliferating cells compared to resting cells
| Class of genes | Main functional categories | Mean change in expression (n-fold) ina:
|
|
|---|---|---|---|
| c-Myc proliferation program | EBV latency III proliferation program | ||
| C1 | Oncogenes/cancer | ↘ −1.9 | ↘ −2.4 |
| Signaling/cytokines | |||
| Immunoglobulin | |||
| C2 | Apoptosis | ↘↘ −3.3 | ↘ −1.8 |
| Interferon response | |||
| Surface receptors | |||
| MHC | |||
| C3 | Extracellular matrix | ↘↘ −5.1 | → −1.1 |
| Receptors | |||
| C5 | B-cell lineage markers | ↗↗ 5.6 | → 1.3 |
| Oncogenes | |||
| C6 | 3Rb | ↗↗ 2.9 | ↗ 2.2 |
| RNA synthesis | |||
| C7 | Cytokines | ↗ 2.2 | ↗↗ 3.1 |
| EBV | |||
| C8 | Cytokines | → 1.2 | ↗↗ 2.8 |
| NF-κB | |||
| Signaling | |||
| C9 | Plasma cell differentiation | ↘ −2.1 | ↗ 1.9 |
| Extracellular communication | |||
| NF-κB | |||
| Signaling | |||
| Transcription factors | |||
| Apoptosis | |||
| C10 | Interferon targets | ↘ −1.6 | → 1.3 |
| Kinases/signaling | |||
Arrows indicate the tendency in the mean change in expression levels as follows: →, −1.5- to 1.5-fold; ↘, −2.5- to −1.5-fold; ↗, 1.5- to 2.5-fold; ↘↘, less than −2.5-fold; ↗↗, >2.5-fold.
3R, genes involved in DNA replication, repair, and/or recombination.
Commonly regulated genes in both EBV latency III- and c-Myc-driven proliferation programs are shared in three classes corresponding to 74% of differentially expressed genes.
Classes C1 and C2 harbored 134 and 271 genes, respectively (Fig. 3; see also Fig. S1 and S2 in the supplemental material). These two classes identify genes that were downregulated in cells driven by both the c-Myc- and the EBV latency III-driven proliferation programs during the transition from the resting to the proliferating state. Expression of genes of class C1 was moderately repressed in a similar fashion in both proliferation programs (Table 1). Among genes belonging to class C1, there are genes of the Ras pathway (rab35 and ras-gap), tgfb, genes of BCR signaling, and pax5. Expression of class C2 genes was markedly decreased in c-Myc-driven proliferating cells and moderately decreased in EBV latency III-driven proliferating cells. These genes would appear to be overexpressed in EBV latency III-driven proliferating cells compared to c-Myc-driven proliferating cells only and not to arrested cells (Table 1). Among genes belonging to class C2, there are various genes of the interferon response such as ifi16, ifi54, ifi56, stat1, stat3, stat6, mxA, and oas1. Additionally, genes involved in apoptosis as well as in major histocompatibility complex regulation were found in this class.
Class C6 of 256 genes clearly corresponds to the proliferation program of the cells (Fig. 3; see also Fig. S5 in the supplemental material). This class contains genes induced in both the EBV latency III and the c-Myc proliferation programs (Table 1). It contains all of the repair, recombination, and replication genes including pcna, mki67, genes of the cyclins, e2f factors, dna polymerases, the brca1 gene, and the c-myc gene itself, as well as genes involved in RNA synthesis and chromosomal maintenance. In keeping with the high proliferation rate of BL cells compared to LCLs, genes of this class were more highly expressed in c-Myc than in EBV latency III proliferating cells (Table 1).
Altogether, comparison of the transcriptomes of cells driven into proliferation by either c-Myc or EBV shows that classes C1, C2, and C6 comprising 661 genes (74%) were similarly induced or repressed in both c-Myc and EBV latency III proliferation programs. These similarities between both proliferation programs led to the hypothesis that c-Myc could be one of the main active transcription factors not only in BL latency I cells but also in EBV latency III proliferating cells.
Few genes are specifically regulated in c-Myc proliferating cells.
Class C3 of 26 genes corresponds to genes specifically repressed in the c-Myc program (Fig. 3 and Table 1; see also Fig. S3 in the supplemental material). Genes coding for cytokine receptors were overrepresented in this class, including interferon type I and II receptors and extracellular matrix proteins. Class C5 of 32 genes corresponds to genes specifically upregulated in the c-Myc program and includes genes controlling B-cell differentiation and some oncogenes such as tcl1a (Fig. 3; see also Fig. S4 in the supplemental material).
Genes specifically upregulated in proliferating cells in EBV latency III point to the transcription factor NF-κB.
Classes C7, C8, and C9 harbored genes induced in EBV latency III proliferating cells. Class C7 contains 23 genes specifically upregulated in EBV latency III and upregulated to a lesser and variable extent in BL, A1, and P493-6 cells driven by c-Myc. These include the EBV genes bhrf1, ebna1, bzlf1, and lmp2a, as well as the chemokine genes mip-1-a and mip-1-b (Fig. 3; see also Fig. S6 in the supplemental material). Because the Lymphochip was not designed for the analysis of viral gene expression and most viral genes were only sparsely represented on the chip, expression of viral genes was validated by an independent method using immunoblotting and/or quantitative reverse transcription-PCR analysis (see Fig. S10 in the supplemental material).
Class C8 consists of 44 genes, and class C9 consists of 72 genes; these genes are upregulated in the EBV latency III program and are unaffected (class C8) or downregulated in cells driven into proliferation by c-Myc (class C9) (Table 1 and Fig. 3; see also Fig. S7 and S8 in the supplemental material). These classes include the lmp1 gene from EBV; genes coding for NF-κB subunits and IκB proteins such as nfkb1, nfkb2, relB, c-rel, nfkbia, and nfkbie; genes coding for cytokines such as il1b; genes involved in intracellular signal transduction; antiapoptotic genes such as c-iap2, c-flip, bcl2a1, bcl2, bcl-xL, and a20 as well as cd95; genes involved in the intercellular communication such as adhesion molecules like CD48, CD54, CD58, CD82, and H-cadherin; and some transcription factors such as STAT5α, JunB, and GATA3. Class C9 also includes genes involved in plasma cell differentiation such as irf4 and cd138. Class C10 consists of 20 genes that are weakly induced or unaffected by the EBV latency III program and are repressed in BL, A1, and P493-6 cells driven by c-Myc. Some of these appear to be particularly strongly repressed in A1 cells, a cell line characterized by particularly high c-Myc expression levels. These include some interferon response genes and genes coding for tyrosine kinases such as Lyn and Fgr (Fig. 3; see also Fig. S9 in the supplemental material).
Altogether, 159 genes (18%) corresponding to classes C7, C8, C9, and C10 were induced specifically in EBV latency III proliferating cells; many of these code for immunoregulatory and/or antiapoptotic molecules and are targets of NF-κB.
Validation of c-Myc targets genes identified on the Lymphochip.
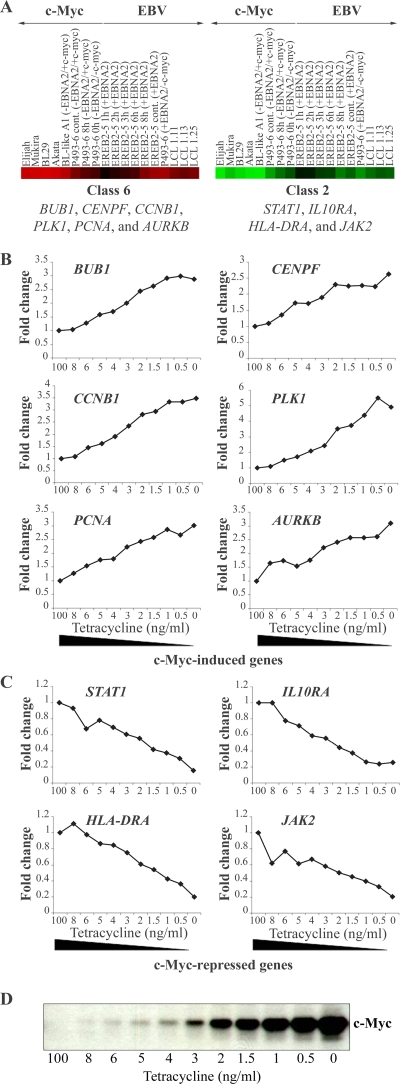
Some of the putative c-Myc targets identified on the Lymphochip were validated by an independent experimental approach as genes transcriptionally regulated by this transcription factor (35). To this end, proliferating P493-6 cells were incubated with increasing doses of tetracycline for 48 h. RNA was isolated, reverse transcribed, radioactively labeled, and subjected to nylon filter hybridization. The filter arrays represented 2,253 human genes related to cancer. As shown in Fig. 4, there was a strict negative or positive correlation between expression of the gene of interest and the tetracycline concentration that correlated inversely with c-Myc expression (48). All the genes in common between the Lymphochip and the filter arrays were indeed identified as positive or negative c-Myc target genes, pointing to the high reliability of the Lymphochip expression data.
FIG. 4.
Validation of c-Myc-regulated target genes identified on the Lymphochip cDNA array (C6 and C2) by nylon filter hybridization. (A) Six individual genes (bub1, cenpf, ccnb1, plk1, pcna, and aurkb) of C6 and four individual genes (stat1, il10ra, hla-dra, and jak2) of C2 were selected as representative genes present on the Lymphochip and on nylon filters. Culture conditions are given at the top of the panel and indicate the cell line, number of hours of treatment (cont, continuously), and the presence (+) or absence (−) of EBNA2 and/or c-Myc. (B and C) RNAs were isolated from P493-6 cells treated with different concentrations of tetracycline (0 to 8 ng/ml and 100 ng/ml), reverse transcribed into 33P-labeled cDNA, and hybridized to the filters. The expression was quantified, and the expression levels of the selected genes at the highest tetracycline concentration (100 ng/ml) were set to 1. The relative change is presented as a function of tetracycline concentration corresponding inversely to the c-Myc concentration. The expression levels of positively regulated and negatively regulated c-Myc targets are shown in panels B and C, respectively. (D) The relative change is presented as a function of tetracycline concentration corresponding inversely to the c-Myc protein concentration as revealed by immunoblotting. A clear dose response of the expression of the selected genes to different tetracycline concentrations indicates that the genes are regulated directly or indirectly by c-Myc.
c-Myc activity is required for proliferation of EBV latency III proliferating cells.
Transcriptome analysis showed that about 92% of induced genes in EBV latency III proliferating cells during the switch from quiescence to continuous proliferation were likely to be related directly or indirectly to c-Myc and NF-κB, suggesting that they act synergistically to impose the EBV-induced proliferation program of latency III. LMP1-mediated constitutive activation of NF-κB is a well-known feature of LCLs (38, 39) and is instrumental in making the cells highly immunogenic and in protecting them from apoptosis (6, 7, 12, 32). On the other hand, c-Myc has been shown to antagonize the action of NF-κB, rendering the cells nonimmunogenic and highly susceptible for induction of apoptosis (48, 50, 52, 57). At least in the pathogenesis of BL, it appears that c-Myc and NF-κB are incompatible (22, 23, 42). Given that c-Myc and NF-κB act antagonistically, that NF-κB may also exert proliferative effects (46), and that c-Myc may be dispensable for proliferation (36), we attempted to formally address the role of c-Myc in proliferation of LCLs.
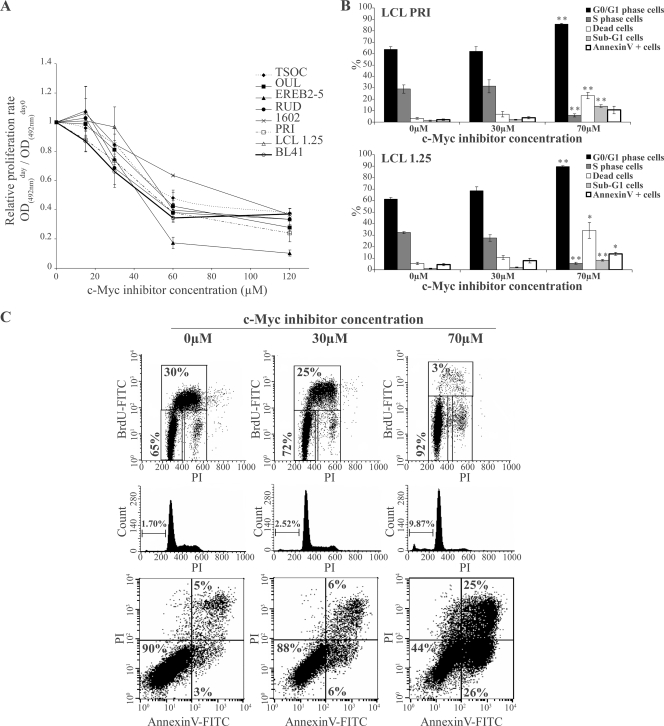
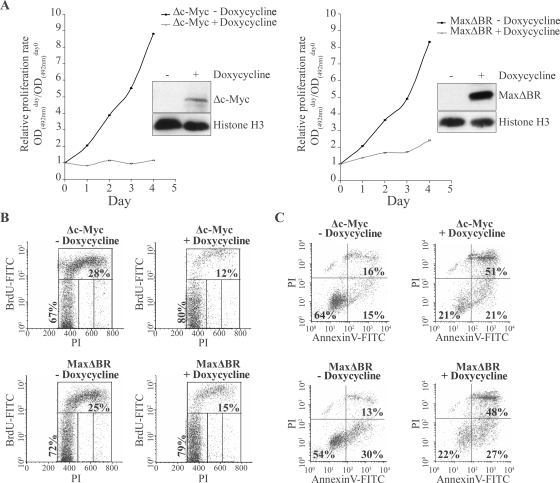
In a first step, we studied the action of the c-Myc chemical inhibitor 10058-F4 compound (36) on cell growth and apoptosis of LCLs. As shown on Fig. 5A, all LCLs tested, including EREB2-5 and 1.25 cells, were similarly sensitive to the 10058-F4 compound in a dose-dependent manner relative to the proliferation rate. At 48 h of treatment, the various GI50 values for LCLs ranged from 43 to 59 μM except for 1602 cells, with a GI50 of 92 μM. As a positive control for cells driven into proliferation by c-Myc, the EBV-negative BL cell line BL41 was used and showed similar growth inhibition, with a GI50 of 45 μM. Treatment with 10058-F4 resulted in a dose-dependent S-phase arrest leading to accumulation of cells in G0/G1 (Fig. 5B and C). Part of the cells died either by apoptosis (annexin V-positive cells) or by necrosis (PI-positive and annexin V-negative cells) (Fig. 5C). To confirm the results obtained with the chemical inhibitor, we cloned two dominant-negative mutants of c-Myc/Max complex, Δc-Myc and MaxΔBr, in the pRT-1 vector (a vector harboring the bidirectional tetracycline-inducible promoter that drives the expression of both NGFR-t and the cDNA of interest) (4) and stably transfected this construct into the LCL PRI. After doxycycline treatment, NGFR-t-positive cells were sorted and compared to NGFR-t-negative control cells with respect to their proliferation rate. Induction of the expression of both Δc-Myc and MaxΔBr proteins resulted in a prolonged arrest of proliferation in cells transfected with Δc-Myc and a significant and prolonged decrease of proliferation rate in cells transfected with MaxΔBr (Fig. 6A and B). This effect was associated with an increase in cell death in both Δc-Myc- and MaxΔBr-transfected PRI LCL cells (Fig. 6C). Therefore, results of both chemical and functional inhibition experiments indicate that c-Myc is necessary for proliferation of cells in EBV latency III. The data indicate that LCLs are very sensitive to c-Myc repression and cell cycle arrest and readily die by apoptosis or necrosis rather than entering a long-lasting resting state.
FIG. 5.
Effect of a synthetic c-Myc inhibitor on proliferation of LCL cells. (A) Both EBV-negative BL41 cells and the LCLs TSOC, OUL, EREB2-5, RUD, 1602, PRI, and 1.25 were treated for 48 h with increasing concentrations (15 to 120 μM) of the synthetic inhibitor 10058-F4. At day 0, 1.5 × 104untreated and treated cells were seeded into wells of 96-wells plates. The relative proliferation rate was assayed using a colorimetric method as described in Materials and Methods. (B) The LCL PRI (upper panel) and 1.25 (lower panel) cells at 1.5 × 104 cells/ml were treated for 48 h with 30 μM and 70 μM 10058-F4 leading to 20% and 80% inhibition of proliferation, respectively. The G0/G1 and S phases of the cell cycle were assayed by flow cytometry after BrdU incorporation (FITC) and PI staining of DNA content. Apoptosis was assayed by measuring percentages of cells with a decreased DNA content (sub-G1 cells) and having externalized phosphatidylserines (annexin V-positive and PI-negative cells). The dead cells represent apoptotic (annexin V-positive and PI-negative cells) plus necrotic cells (PI-positive cells). Similar results were obtained with the EREB2-5 cells (data not shown). Error bars indicate standard error from the mean from three independent sets of experiments. A significant difference with a P value of <0.05 (*) or <0.01 (**) compared to untreated control cells using a Student's t test is indicated. (C) A typical flow cytometry result of the effect of the synthetic c-Myc inhibitor 10058-F4 on proliferation and cell death of LCL 1.25 cells. BrdU-FITC/PI biparametric graphs are shown in the top panels (the different regions were set up in the valleys separating each phase of the cell cycle because of an increase in autofluorescence after treatment of cells with 10058-F4). Middle panels show a sub-G1 peak assay. Lower panels are annexin V-FITC/PI biparametric graphs. Concentrations of 10058-F4 are indicated at the top of the set of graphs, and informative percentages are indicated in each graph. OD, optical density.
FIG. 6.
Effect of functional repression of c-Myc on proliferation of LCL cells. PRI-LCL was stably transfected with the inducible vector pRT-1 which contains cDNA coding for dominant-negative mutants of c-Myc or Max, named Δc-Myc and MaxΔBR, respectively. After 24 h (day 0) of doxycycline induction, the NGFR-t-expressing cells were purified by cell sorting. (A) At day 0, 1.5 × 104untreated cells (−Doxycycline) and doxycycline treated cells (+Doxycycline) were seeded into wells of 96-wells plates. The following days, the number of viable cells was measured using a colorimetric method as described in Materials and Methods. Cells were harvested, and whole-cell extracts were subjected to immunoblotting analysis with anti-HA tag and anti-histone H3 antibodies. (B) At day 2, cells were pulse labeled with BrdU for the last 3 h, fixed, and stained with FITC-conjugated anti-BrdU mouse MAb and PI. BrdU uptake (FITC) versus DNA content (PI) was evaluated by flow cytometry. Representative density plots are shown. PI and FITC fluorescence intensities are plotted on the x and y axes, respectively. (C) At day 2, cells were doubly stained with annexin V-FITC and PI and then analyzed by flow cytometry. FITC and PI fluorescence intensities are plotted on the x and y axes, respectively. Informative percentages are indicated in each graph. Representative density plots are shown. OD, optical density.
DISCUSSION
We have addressed the question whether and how the EBV latency III proliferation program is related to the c-Myc proliferation program. To this end, we have analyzed the expression profiles of EREB2-5 cells, a conditional cell line with an estrogen-regulatable EBNA2-estrogen receptor fusion protein, two derivative cell lines harboring additionally either a constitutively active (A1 cells) or a tetracycline-regulatable (P493-6 cells) c-myc gene and BL and LCL lines. As a reference, we took EREB2-5 cells that were arrested by withdrawal of estrogen and compared the expression profiles of cells in c-Myc-driven and EBV latency III-driven proliferation programs to that of arrested EREB2-5 cells in order to identify genes induced and repressed during the switch from the resting to the proliferating state. Genes were filtered to allow us to select only those that were at least fourfold changed in at least one condition. Using this filter, we found 897 genes putatively regulated at a high level among approximately 12,000 targets, corresponding to 7.5% of tested genes with this cDNA array. It is of note that, using these criteria, we obtained classes of genes with a mean relative change of at least 1.5-fold in at least one of the two proliferating programs (see Results) (Table 1); this ratio of relative change is often considered as the minimal significant threshold ratio. Combining K-mean clustering, hierarchical clustering, and principal component analysis and searching to maximize the dispersion as a criterion to define the optimum number of classes, we identified 10 classes of genes, called C1 to C10.
With the cDNA array technology, expression of numerous genes can be quantified in one step, giving rise to a molecular portrait of a biological system and/or pointing to key transcriptional regulators (11). In our case, the two obvious key regulators were c-Myc and NF-κB. However, as recently noted by Koltai and Weingarten-Baror (28), we have to keep in mind that for some genes discrepancies may exist between cDNA array results and results from other techniques and that not all probes of cDNA arrays are reliably specific, and some cross-reactivity may occur. This means that results obtained from cDNA arrays have to be validated for each individual gene by another technique, especially if the gene belongs to a small class. We found two small classes of genes, C5 (32 genes) and C7 (23 genes). A selected set of positive and negative c-Myc target genes has been independently confirmed by filter hybridization (Fig. 4).
Class C5, was specifically expressed in cells driven into proliferation by c-Myc. This included P493-6 cells with c-Myc in the off state. Apparently, the Tet07 promoter is leaky, which results in a low level of c-Myc expression, even in the off condition (reference 42 and unpublished results). These low levels of c-Myc are sufficient to keep the cells alive and to drive proliferation at a very low rate. Even if these results have to be checked, it is interesting that the TCL1 oncogene, identified as a cofactor in BL tumors (26, 53), was specifically expressed in this class C5. This class also contains the glucose transporter protein 5 and members of the immune receptor translocation-associated protein family located on chromosome 1q21, a region that is frequently translocated in B-cell malignancies and amplified in BL tumors (34).
The major advantage of cDNA arrays is the rapid screening of numerous genes, allowing very quick elaboration of functional hypotheses without any a priori assumptions. Strikingly, we found that 74% of the genes for which induction or repression had been observed in one or both proliferation programs were similarly induced or repressed in cells driven into proliferation either by c-Myc or by the viral latency III gene expression program. In our experimental design, the BL cell lines represent tumor cell lines that may have acquired genetic or epigenetic changes in addition to the Ig/c-Myc translocation that have been selected for in vivo. In contrast, A1 and P493-6 cells represent cells derived from normal B cells and driven into proliferation merely by high c-Myc expression. It cannot, of course, be excluded that these cells have also acquired additional changes, but such changes, if they exist, have not been selected for in vivo. Remarkably, the comparison of the expression profiles of BL, A1, and P493-6 cells proliferating on a c-Myc program did not provide evidence for significant differences (data not shown). This underlines the notion that c-Myc is the master transcription factor responsible for proliferation of BL cell lines. c-Myc target genes were clearly separated into two categories, with induced and repressed genes defined in reference to the expression profile of arrested EREB2-5 cells. c-Myc-induced genes were those involved in proliferation, RNA and DNA synthesis, DNA repair, and chromosome maintenance and segregation. c-Myc-repressed genes were those involved in immune recognition and in the interferon response. c-Myc targets have been reported by various groups including us (9, 20, 33, 49). Combined with functional studies, our expression profiling approach of c-Myc-regulated genes has identified two distinct functions of c-Myc, induction of proliferation and impairment of immune recognition of tumor cells by inhibition of the NF-κB and the interferon responses (48). Indeed, the hallmark of the BL signature compared to diffuse large B-cell lymphomas as revealed by transcriptome analysis of fresh tumor specimens is the c-Myc signature: upregulation of c-Myc target genes and downregulation of genes involved in the NF-κB and interferon responses (9, 20).
The fact that 74% of genes were similarly up- or downregulated in cells driven into proliferation by either c-Myc or the viral latency III expression program is an indication that the transcriptional activity of c-Myc is also a determining factor of the EBV latency III program. Of note, compared to resting cells, the downregulation of interferon response genes was much less pronounced in cells proliferating on an EBV latency III than on a c-Myc program. This is most likely due to the lower level of c-Myc expression in LCL than in BL cells (47) and/or to induction of NF-κB in LCLs. If the expression profiles of only BL and LCL cells are compared, these genes would thus appear to be overexpressed in LCLs relative to BL cells.
We have previously reported that c-Myc is a direct target gene of EBNA2 (21). This has recently been confirmed in LCLs conditionally immortalized by recombinant virus encoding a 4-hydroxytamoxifen-regulatable EBNA2-estrogen receptor fusion protein (59). To our knowledge, the functional role of c-Myc for regulation of proliferation and apoptosis of LCLs has not been fully explored experimentally. An inhibitory effect of the c-Myc chemical inhibitor 10058-F4 on cell proliferation has been reported only on a single LCL (14). We have shown here that functional interference with c-Myc either by a chemical c-Myc inhibitor or by overexpression of dominant-negative versions of c-Myc or Max inhibited proliferation of all LCLs tested. Cells were arrested in S phase and accumulated in G0/G1 phase. Cell cycle arrest induced by inhibiting the function of c-Myc either by a chemical inhibitor or by dominant-negative c-Myc or Max constructs was associated with induction of cell death, with about half of the cells exhibiting features of apoptosis (annexin V positive and PI negative) and the other half showing features of necrosis (annexin V positive and PI positive).
Using the pRT-1 vector and doxycycline treatment, it was not possible to extend the period of analysis to 96 h (12; also data not shown) because long-term doxycycline treatment of both Δc-Myc- and MaxΔBr-transfected cells resulted in selection of doxycycline-insensitive cells, despite the fact that over 90% of the cell had died at day 4 (data not shown). In contrast, long-term culture of LCLs with the 10058-F4 compound led to complete cell death (data not shown). The specificity of the c-Myc inhibitor 10058-F4 is underlined by the fact that it was shown to affect the proliferation of c-Myc-overexpressing but not of c-Myc-deficient rat fibroblasts (14, 36). Altogether, these data provided evidence that c-Myc is absolutely required for proliferation of EBV-immortalized B-cells.
The second group of important genes consists of those specifically induced in cells driven into proliferation by the EBV latency III genes. These comprise 18% of differentially expressed genes and can be classified into two categories. Class C8 consists of 44 genes that are unchanged in cells driven into proliferation by c-Myc and induced in the viral latency III program, the most prominent genes being the viral oncogene EBNA2 and LMP1 that affect the expression of this whole class of genes. Classes C9 and C10 correspond to genes repressed in c-Myc and induced in EBV latency III proliferating cells. Remarkably, most of these genes are LMP1 and NF-κB target genes, and there is a high level of concordance with previously published results (6, 10). These NF-κB target genes include members of the TNF receptor family, TRAF/TRADD molecules, genes protecting the cell from apoptosis, genes coding for inflammatory cytokines and chemokines, and genes involved in cellular adhesion and antigen presentation that impose a highly immunogenic phenotype on EBV-infected cells. Thus, regarding the overall importance of c-Myc and NF-κB for the EBV latency III program, c-Myc as a downstream target of EBNA2 as well as LMP1 (10) is quantitatively dominating the expression profile of EBV latency III, yet LMP1 as the critical activator of NF-κB is responsible for the dramatic difference in the growth pattern and phenotype of cells driven into proliferation by either c-Myc or the EBV latency III program. Detailed analysis of the data indicates that NF-κB activation through LMP1 efficiently counteracts the transcriptional repression of immunoregulatory genes that is imposed by c-Myc alone. In this view, class C3 and the genes of class C2 that are repressed during c-Myc-driven proliferation and are unchanged or repressed to a much lesser extent in cells in EBV latency III (including infgr1, jak3, infar2, and ccr7 in class C3 and the major histocompatibility complex class II and interferon response genes in class C2) may be classified together with class C8 and C9. Genes in classes C2 and C3 are repressed by c-Myc to a similar extent as genes in classes C8 and C9 but are induced to a much lesser extent by the viral latency III genes.
Altogether, these results indicate that the growth program of BL cells is mainly governed by one single master transcriptional system that is c-Myc itself, supported by some other oncogenes such as TCL1 as cofactors. In EBV-immortalized B cells the transition from quiescence to EBV latency III-induced proliferation is governed by two main master transcriptional systems, c-Myc and Rel/NF-κB. During EBV latency III-induced proliferation, induction of c-Myc appears to be predominantly associated with induction of proliferation, whereas induction of NF-κB would be responsible for protection from apoptosis, the inflammatory response, and the immunogenicity of EBV-immortalized cells and for partly counteracting c-Myc's repression of the interferon response.
Supplementary Material
Acknowledgments
This work was supported by the Institut National du Cancer, Cancéropôle Grand-Sud-Ouest, Ligue Nationale contre le Cancer, Conseil Régional du Limousin, and Association pour la Recherche sur le Cancer. UMR-CNRS 6101 is a Laboratoire Labelisé Ligue Nationale contre le Cancer. N.F. was successively supported by the Conseil Régional du Limousin, Cancéropôle GSO, and Comité Orientation Recherche Cancer en Limousin. S.D.P. was successively supported by the Ministère de l'Enseignement Supérieur et de la Recherche and by the Association pour la Recherche contre le Cancer. G.W.B. was supported by a grant from the Wilhelm Sander foundation.
Footnotes
Published ahead of print on 4 March 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Alizadeh, A. A., M. B. Eisen, R. E. Davis, C. Ma, I. S. Lossos, A. Rosenwald, J. C. Boldrick, H. Sabet, T. Tran, X. Yu, J. I. Powell, L. Yang, G. E. Marti, T. Moore, J. Hudson, Jr., L. Lu, D. B. Lewis, R. Tibshirani, G. Sherlock, W. C. Chan, T. C. Greiner, D. D. Weisenburger, J. O. Armitage, R. Warnke, R. Levy, W. Wilson, M. R. Grever, J. C. Byrd, D. Botstein, P. O. Brown, and L. M. Staudt. 2000. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403503-511. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, L. J., and R. Longnecker. 2008. An auto-regulatory loop for EBV LMP2A involves activation of Notch. Virology 371257-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baran-Marszak, F., J. Feuillard, I. Najjar, C. Le Clorennec, J. M. Bechet, I. Dusanter-Fourt, G. W. Bornkamm, M. Raphael, and R. Fagard. 2004. Differential roles of STAT1α and STAT1β in fludarabine-induced cell cycle arrest and apoptosis in human B cells. Blood 1042475-2483. [DOI] [PubMed] [Google Scholar]
- 4.Bornkamm, G. W., C. Berens, C. Kuklik-Roos, J. M. Bechet, G. Laux, J. Bachl, M. Korndoerfer, M. Schlee, M. Holzel, A. Malamoussi, R. D. Chapman, F. Nimmerjahn, J. Mautner, W. Hillen, H. Bujard, and J. Feuillard. 2005. Stringent doxycycline-dependent control of gene activities using an episomal one-vector system. Nucleic Acids Res. 33e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bornkamm, G. W., and W. Hammerschmidt. 2001. Molecular virology of Epstein-Barr virus. Philos. Trans. R. Soc. Lond. B 356437-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cahir-McFarland, E. D., K. Carter, A. Rosenwald, J. M. Giltnane, S. E. Henrickson, L. M. Staudt, and E. Kieff. 2004. Role of NF-κ B in cell survival and transcription of latent membrane protein 1-expressing or Epstein-Barr virus latency III-infected cells. J. Virol. 784108-4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cahir-McFarland, E. D., D. M. Davidson, S. L. Schauer, J. Duong, and E. Kieff. 2000. NF-kappa B inhibition causes spontaneous apoptosis in Epstein-Barr virus-transformed lymphoblastoid cells. Proc. Natl. Acad. Sci. USA 976055-6060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casola, S., K. L. Otipoby, M. Alimzhanov, S. Humme, N. Uyttersprot, J. L. Kutok, M. C. Carroll, and K. Rajewsky. 2004. B cell receptor signal strength determines B cell fate. Nat. Immunol. 5317-327. [DOI] [PubMed] [Google Scholar]
- 9.Dave, S. S., K. Fu, G. W. Wright, L. T. Lam, P. Kluin, E. J. Boerma, T. C. Greiner, D. D. Weisenburger, A. Rosenwald, G. Ott, H. K. Muller-Hermelink, R. D. Gascoyne, J. Delabie, L. M. Rimsza, R. M. Braziel, T. M. Grogan, E. Campo, E. S. Jaffe, B. J. Dave, W. Sanger, M. Bast, J. M. Vose, J. O. Armitage, J. M. Connors, E. B. Smeland, S. Kvaloy, H. Holte, R. I. Fisher, T. P. Miller, E. Montserrat, W. H. Wilson, M. Bahl, H. Zhao, L. Yang, J. Powell, R. Simon, W. C. Chan, and L. M. Staudt. 2006. Molecular diagnosis of Burkitt's lymphoma. N. Engl. J. Med. 3542431-2442. [DOI] [PubMed] [Google Scholar]
- 10.Dirmeier, U., R. Hoffmann, E. Kilger, U. Schultheiss, C. Briseno, O. Gires, A. Kieser, D. Eick, B. Sugden, and W. Hammerschmidt. 2005. Latent membrane protein 1 of Epstein-Barr virus coordinately regulates proliferation with control of apoptosis. Oncogene 241711-1717. [DOI] [PubMed] [Google Scholar]
- 11.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 9514863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feuillard, J., M. Schuhmacher, S. Kohanna, M. Asso-Bonnet, F. Ledeur, R. Joubert-Caron, P. Bissieres, A. Polack, G. W. Bornkamm, and M. Raphael. 2000. Inducible loss of NF-κB activity is associated with apoptosis and Bcl-2 down-regulation in Epstein-Barr virus-transformed B lymphocytes. Blood 952068-2075. [PubMed] [Google Scholar]
- 13.Gasch, A. P., and M. B. Eisen. 2002. Exploring the conditional coregulation of yeast gene expression through fuzzy k-means clustering. Genome Biol. 3RESEARCH0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gomez-Curet, I., R. S. Perkins, R. Bennett, K. L. Feidler, S. P. Dunn, and L. J. Krueger. 2006. c-Myc inhibition negatively impacts lymphoma growth. J. Pediatr. Surg. 41207-211. [DOI] [PubMed] [Google Scholar]
- 15.Gordadze, A. V., R. Peng, J. Tan, G. Liu, R. Sutton, B. Kempkes, G. W. Bornkamm, and P. D. Ling. 2001. Notch1IC partially replaces EBNA2 function in B cells immortalized by Epstein-Barr virus. J. Virol. 755899-5912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guasparri, I., D. Bubman, and E. Cesarman. 2008. EBV LMP2A affects LMP1-mediated NF-κB signaling and survival of lymphoma cells by regulating TRAF2 expression. Blood 1113813-3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hermeking, H., D. A. Wolf, F. Kohlhuber, A. Dickmanns, M. Billaud, E. Fanning, and D. Eick. 1994. Role of c-myc in simian virus 40 large tumor antigen-induced DNA synthesis in quiescent 3T3-L1 mouse fibroblasts. Proc. Natl. Acad. Sci. USA 9110412-10416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hofelmayr, H., L. J. Strobl, G. Marschall, G. W. Bornkamm, and U. Zimber-Strobl. 2001. Activated Notch1 can transiently substitute for EBNA2 in the maintenance of proliferation of LMP1-expressing immortalized B cells. J. Virol. 752033-2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huen, D. S., S. A. Henderson, D. Croom-Carter, and M. Rowe. 1995. The Epstein-Barr virus latent membrane protein-1 (LMP1) mediates activation of NF-κB and cell surface phenotype via two effector regions in its carboxy-terminal cytoplasmic domain. Oncogene 10549-560. [PubMed] [Google Scholar]
- 20.Hummel, M., S. Bentink, H. Berger, W. Klapper, S. Wessendorf, T. F. Barth, H. W. Bernd, S. B. Cogliatti, J. Dierlamm, A. C. Feller, M. L. Hansmann, E. Haralambieva, L. Harder, D. Hasenclever, M. Kuhn, D. Lenze, P. Lichter, J. I. Martin-Subero, P. Moller, H. K. Muller-Hermelink, G. Ott, R. M. Parwaresch, C. Pott, A. Rosenwald, M. Rosolowski, C. Schwaenen, B. Sturzenhofecker, M. Szczepanowski, H. Trautmann, H. H. Wacker, R. Spang, M. Loeffler, L. Trumper, H. Stein, and R. Siebert. 2006. A biologic definition of Burkitt's lymphoma from transcriptional and genomic profiling. N. Engl. J. Med. 3542419-2430. [DOI] [PubMed] [Google Scholar]
- 21.Kaiser, C., G. Laux, D. Eick, N. Jochner, G. W. Bornkamm, and B. Kempkes. 1999. The proto-oncogene c-myc is a direct target gene of Epstein-Barr virus nuclear antigen 2. J. Virol. 734481-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelly, G., A. Bell, and A. Rickinson. 2002. Epstein-Barr virus-associated Burkitt lymphomagenesis selects for downregulation of the nuclear antigen EBNA2. Nat. Med. 81098-1104. [DOI] [PubMed] [Google Scholar]
- 23.Kelly, G. L., A. E. Milner, R. J. Tierney, D. S. Croom-Carter, M. Altmann, W. Hammerschmidt, A. I. Bell, and A. B. Rickinson. 2005. Epstein-Barr virus nuclear antigen 2 (EBNA2) gene deletion is consistently linked with EBNA3A, -3B, and -3C expression in Burkitt's lymphoma cells and with increased resistance to apoptosis. J. Virol. 7910709-10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kempkes, B., D. Spitkovsky, P. Jansen-Durr, J. W. Ellwart, E. Kremmer, H. J. Delecluse, C. Rottenberger, G. W. Bornkamm, and W. Hammerschmidt. 1995. B-cell proliferation and induction of early G1-regulating proteins by Epstein-Barr virus mutants conditional for EBNA2. EMBO J. 1488-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kieff, E., and A. B. Rickinson. 2001. Epstein-Barr virus and its replication, p. 2511-2627. In D. M. Knipe and P. M. Howley (ed.), Virology, 4th ed. Lippincott Williams and Wilkins, Philadelphia, PA.
- 26.Kiss, C., J. Nishikawa, K. Takada, P. Trivedi, G. Klein, and L. Szekely. 2003. T cell leukemia I oncogene expression depends on the presence of Epstein-Barr virus in the virus-carrying Burkitt lymphoma lines. Proc. Natl. Acad. Sci. USA 1004813-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knowles, D. M. 1999. Immunodeficiency-associated lymphoproliferative disorders. Mod. Pathol. 12200-217. [PubMed] [Google Scholar]
- 28.Koltai, H., and C. Weingarten-Baror. 2008. Specificity of DNA microarray hybridization: characterization, effectors and approaches for data correction. Nucleic Acids Res. 362395-2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Korkolopoulou, P., E. Patsouris, G. Pangalis, A. Tsenga, J. Elemenoglou, E. Thomas-Tsangli, D. Spandidos, and C. Kittas. 1993. A comparative assessment of proliferating cell nuclear antigen, c-myc p62, and nucleolar organizer region staining in non-Hodgkin's lymphomas: a histochemical and immunohistochemical study of 200 cases. Hum. Pathol. 24371-377. [DOI] [PubMed] [Google Scholar]
- 30.Laherty, C. D., H. M. Hu, A. W. Opipari, F. Wang, and V. M. Dixit. 1992. The Epstein-Barr virus LMP1 gene product induces A20 zinc finger protein expression by activating nuclear factor kappa B. J. Biol. Chem. 26724157-24160. [PubMed] [Google Scholar]
- 31.Le Clorennec, C., T. S. Ouk, I. Youlyouz-Marfak, S. Panteix, C. C. Martin, J. Rastelli, E. Adriaenssens, U. Zimber-Strobl, J. Coll, J. Feuillard, and C. Jayat-Vignoles. 2008. Molecular basis of cytotoxicity of Epstein-Barr virus (EBV) latent membrane protein 1 (LMP1) in EBV latency III B cells: LMP1 induces type II ligand-independent autoactivation of CD95/Fas with caspase 8-mediated apoptosis. J. Virol. 826721-6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Le Clorennec, C., I. Youlyouz-Marfak, E. Adriaenssens, J. Coll, G. W. Bornkamm, and J. Feuillard. 2006. EBV latency III immortalization program sensitizes B cells to induction of CD95-mediated apoptosis via LMP1: role of NF-κB, STAT1, and p53. Blood 1072070-2078. [DOI] [PubMed] [Google Scholar]
- 33.Li, Z., S. Van Calcar, C. Qu, W. K. Cavenee, M. Q. Zhang, and B. Ren. 2003. A global transcriptional regulatory role for c-Myc in Burkitt's lymphoma cells. Proc. Natl. Acad. Sci. USA 1008164-8169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindstrom, M. S., and K. G. Wiman. 2002. Role of genetic and epigenetic changes in Burkitt lymphoma. Semin. Cancer Biol. 12381-387. [DOI] [PubMed] [Google Scholar]
- 35.Machl, A. W., C. Schaab, and I. Ivanov. 2002. Improving DNA array data quality by minimising “neighbourhood” effects. Nucleic Acids Res. 30e127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mateyak, M. K., A. J. Obaya, S. Adachi, and J. M. Sedivy. 1997. Phenotypes of c-Myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ. 81039-1048. [PubMed] [Google Scholar]
- 37.Morimura, T., R. Goitsuka, Y. Zhang, I. Saito, M. Reth, and D. Kitamura. 2000. Cell cycle arrest and apoptosis induced by Notch1 in B cells. J. Biol. Chem. 27536523-36531. [DOI] [PubMed] [Google Scholar]
- 38.Mosialos, G. 1997. The role of Rel/NF-κB proteins in viral oncogenesis and the regulation of viral transcription. Semin. Cancer Biol. 8121-129. [DOI] [PubMed] [Google Scholar]
- 39.Mosialos, G., M. Birkenbach, R. Yalamanchili, T. VanArsdale, C. Ware, and E. Kieff. 1995. The Epstein-Barr virus transforming protein LMP1 engages signaling proteins for the tumor necrosis factor receptor family. Cell 80389-399. [DOI] [PubMed] [Google Scholar]
- 40.Najjar, I., F. Baran-Marszak, C. Le Clorennec, C. Laguillier, O. Schischmanoff, I. Youlyouz-Marfak, M. Schlee, G. W. Bornkamm, M. Raphael, J. Feuillard, and R. Fagard. 2005. Latent membrane protein 1 regulates STAT1 through NF-κB-dependent interferon secretion in Epstein-Barr virus-immortalized B cells. J. Virol. 794936-4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pajic, A., A. Polack, M. S. Staege, D. Spitkovsky, B. Baier, G. W. Bornkamm, and G. Laux. 2001. Elevated expression of c-myc in lymphoblastoid cells does not support an Epstein-Barr virus latency III-to-I switch. J. Gen. Virol. 823051-3055. [DOI] [PubMed] [Google Scholar]
- 42.Pajic, A., D. Spitkovsky, B. Christoph, B. Kempkes, M. Schuhmacher, M. S. Staege, M. Brielmeier, J. Ellwart, F. Kohlhuber, G. W. Bornkamm, A. Polack, and D. Eick. 2000. Cell cycle activation by c-myc in a Burkitt lymphoma model cell line. Int. J. Cancer 87787-793. [DOI] [PubMed] [Google Scholar]
- 43.Poirel, H. A., A. Bernheim, A. Schneider, M. Meddeb, S. Choquet, V. Leblond, F. Charlotte, F. Davi, D. Canioni, E. Macintyre, M. F. Mamzer-Bruneel, I. Hirsch, O. Hermine, A. Martin, P. Cornillet-Lefebvre, M. Patey, O. Toupance, J. L. Kemeny, P. Deteix, and M. Raphael. 2005. Characteristic pattern of chromosomal imbalances in posttransplantation lymphoproliferative disorders: correlation with histopathological subcategories and EBV status. Transplantation 80176-184. [DOI] [PubMed] [Google Scholar]
- 44.Polack, A., K. Hortnagel, A. Pajic, B. Christoph, B. Baier, M. Falk, J. Mautner, C. Geltinger, G. W. Bornkamm, and B. Kempkes. 1996. c-myc activation renders proliferation of Epstein-Barr virus (EBV)-transformed cells independent of EBV nuclear antigen 2 and latent membrane protein 1. Proc. Natl. Acad. Sci. USA 9310411-10416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rea, D., H. J. Delecluse, S. J. Hamilton-Dutoit, L. Marelle, I. Joab, L. Edelman, J. F. Finet, M. Raphael, et al. 1994. Epstein-Barr virus latent and replicative gene expression in post-transplant lymphoproliferative disorders and AIDS-related non-Hodgkin's lymphomas. Ann. Oncol. 5(Suppl 1)113-116. [DOI] [PubMed] [Google Scholar]
- 46.Sanchez-Beato, M., A. Sanchez-Aguilera, and M. A. Piris. 2003. Cell cycle deregulation in B-cell lymphomas. Blood 1011220-1235. [DOI] [PubMed] [Google Scholar]
- 47.Schlee, M., M. Holzel, S. Bernard, R. Mailhammer, M. Schuhmacher, J. Reschke, D. Eick, D. Marinkovic, T. Wirth, A. Rosenwald, L. M. Staudt, M. Eilers, F. Baran-Marszak, R. Fagard, J. Feuillard, G. Laux, and G. W. Bornkamm. 2007. c-Myc activation impairs the NF-κB and the interferon response: implications for the pathogenesis of Burkitt's lymphoma. Int. J. Cancer. 1201387-1395. [DOI] [PubMed] [Google Scholar]
- 48.Schlee, M., M. Schuhmacher, M. Holzel, G. Laux, and G. W. Bornkamm. 2007. c-MYC impairs immunogenicity of human B cells. Adv. Cancer Res. 97167-188. [DOI] [PubMed] [Google Scholar]
- 49.Schuhmacher, M., F. Kohlhuber, M. Holzel, C. Kaiser, H. Burtscher, M. Jarsch, G. W. Bornkamm, G. Laux, A. Polack, U. H. Weidle, and D. Eick. 2001. The transcriptional program of a human B cell line in response to Myc. Nucleic Acids Res. 29397-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Staege, M. S., S. P. Lee, T. Frisan, J. Mautner, S. Scholz, A. Pajic, A. B. Rickinson, M. G. Masucci, A. Polack, and G. W. Bornkamm. 2002. MYC overexpression imposes a nonimmunogenic phenotype on Epstein-Barr virus-infected B cells. Proc. Natl. Acad. Sci. USA 994550-4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takano, Y., M. Saegusa, M. Ikenaga, and I. Okayasu. 1997. Apoptosis and proliferative activity of non-Hodgkin's lymphomas: comparison with expression of bcl-2, p53 and c-myc proteins. Pathol. Int. 4790-94. [DOI] [PubMed] [Google Scholar]
- 52.Tanaka, H., I. Matsumura, S. Ezoe, Y. Satoh, T. Sakamaki, C. Albanese, T. Machii, R. G. Pestell, and Y. Kanakura. 2002. E2F1 and c-Myc potentiate apoptosis through inhibition of NF-κB activity that facilitates MnSOD-mediated ROS elimination. Mol. Cell 91017-1029. [DOI] [PubMed] [Google Scholar]
- 53.Teitell, M. A., M. A. Lones, S. L. Perkins, W. G. Sanger, M. S. Cairo, and J. W. Said. 2005. TCL1 expression and Epstein-Barr virus status in pediatric Burkitt lymphoma. Am. J. Clin. Pathol. 124569-575. [DOI] [PubMed] [Google Scholar]
- 54.Thorley-Lawson, D. A. 2005. EBV the prototypical human tumor virus—just how bad is it? J. Allergy Clin. Immunol. 116251-261, 262. [DOI] [PubMed] [Google Scholar]
- 55.Thorley-Lawson, D. A. 2001. Epstein-Barr virus: exploiting the immune system. Nat. Rev. Immunol. 175-82. [DOI] [PubMed] [Google Scholar]
- 56.Thorley-Lawson, D. A., and G. J. Babcock. 1999. A model for persistent infection with Epstein-Barr virus: the stealth virus of human B cells. Life Sci. 651433-1453. [DOI] [PubMed] [Google Scholar]
- 57.You, Z., L. V. Madrid, D. Saims, J. Sedivy, and C. Y. Wang. 2002. c-Myc sensitizes cells to tumor necrosis factor-mediated apoptosis by inhibiting nuclear factor kappa B transactivation. J. Biol. Chem. 27736671-36677. [DOI] [PubMed] [Google Scholar]
- 58.Young, L. S., and A. B. Rickinson. 2004. Epstein-Barr virus: 40 years on. Nat. Rev. Cancer 4757-768. [DOI] [PubMed] [Google Scholar]
- 59.Zhao, B., S. Maruo, A. Cooper, M. R. Chase, E. Johannsen, E. Kieff, and E. Cahir-McFarland. 2006. RNAs induced by Epstein-Barr virus nuclear antigen 2 in lymphoblastoid cell lines. Proc. Natl. Acad. Sci. USA 1031900-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zindy, F., C. M. Eischen, D. H. Randle, T. Kamijo, J. L. Cleveland, C. J. Sherr, and M. F. Roussel. 1998. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 122424-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.