Abstract
The Mre11–Rad50–Xrs2 (MRX) complex has an important function in the maintenance of genomic integrity by contributing to the detection and repair of chromosome breaks. Here we show that the complex is recruited to sites of paused forks where it stabilizes the association of essential replisome components. Interestingly, this function is not dependent on the S phase checkpoint or the nuclease activity of Mre11. We find that disruption of the MRX complex leads to a loss of fork recovery and a failure to properly complete DNA replication when cells are exposed to replication stress. Our data suggest that one critical function of the MRX complex during replication is to promote the cohesion of sister chromatids at paused forks, offering an explanation for why MRX deficiency leads to a loss of cell viability and high levels of chromosome rearrangements under conditions of replication stress.
Keywords: DNA replication, MRX complex, replisome stability, sister chromatid cohesion (SCC), stalled forks
Introduction
Accurate replication of DNA is essential for the preservation of genomic integrity and the continuation of life. Sites in the genome susceptible to genomic breaks and rearrangements correspond with pauses in replication fork progression (Cha and Kleckner, 2002; Lambert et al, 2005). Thus, maintaining fork structures during replication stress is paramount for ensuring that DNA breaks do not occur and that synthesis can continue once the stress is overcome.
In Saccharomyces cerevisiae, the Mre11/Rad50/Xrs2 (MRX) complex, similar to its human counterpart Mre11/Rad50/Nbs1 (MRN), preserves genomic integrity. Mre11 interacts with itself and both Xrs2 and Rad50. Mre11 has intrinsic DNA-binding activity, associates with the ends of linear DNA molecules and duplexed DNA and has nuclease activity specified by four phosphoesterase motifs in the amino terminal end of the protein (Furuse et al, 1998; Usui et al, 1998; Paull and Gellert, 1999; de Jager et al, 2001; Hopfner et al, 2002). The overall structure of the Rad50 component is typical of a class of proteins known as the ‘structural maintenance of chromosome' proteins, which are necessary for chromosome condensation and sister chromatid cohesion (SCC) (reviewed in Hirano, 2002). The ends of Rad50 contain Walker A and B nucleotide (NTP)-binding motifs (Alani et al, 1989), which are separated by an extended coiled-coil structure bound in the centre of the protein by a putative hinge region (Alani et al, 1989; Dolganov et al, 1996). The crystal structure of Rad50 indicates that a dimer can form between two Rad50 molecules, allowing a flexible bridge to be generated, which can tether duplexed DNA molecules over long distances of up to 1200 Å (Hopfner et al, 2002). Xrs2 is the most divergent member of the complex and is less well characterized.
Mutations in the genes encoding components of the MRX complex lead to well-characterized defects including telomere shortening, meiotic defects and DNA damage sensitivity (reviewed in D'Amours and Jackson, 2002). Interestingly, the nuclease activity of Mre11 has been shown to be dispensable for some of these processes including homologous recombination (HR) and telomere maintenance, indicating the complex has critical functions distinct from its DNA end-processing ability (Bressan et al, 1998; Moreau et al, 1999; Tsukamoto et al, 2001). Structural characterization of the MRX/MRN complex shows an elongated conformation, with tethering properties important for bridging sister chromatids during HR (Williams et al, 2007). Indeed, mutations in the coiled-coil domain of Rad50 that negatively influence the tethering functions of the complex render cells sensitive to genotoxic stress, underscoring the importance of complex structure preservation (Hopfner et al, 2002). It has been proposed that the MRX complex serves as a ‘critical glue' that is necessary for cohesion establishment between sister chromatids during DNA metabolic activities (Williams et al, 2007).
During repair, the interconnection between the MRX complex and Tel1 kinase is well documented. The MRX complex localizes to DNA double-stranded breaks (DSBs) very rapidly (Lisby et al, 2004; Shroff et al, 2004) and recruits Tel1 (Nakada et al, 2003). The DSB repair function of the MRX complex is both initiated and regulated by Tel1 on a pathway parallel to Mec1, leading to Rad53 activation (Usui et al, 2001). Furthermore, Tel1-dependent phosphorylation of Mre11 and Xrs2 has been shown to directly influence the proper repair of DNA damage (D'Amours and Jackson, 2001; Usui et al, 2001). However, there are phenotypic differences between mre11Δ and tel1Δ cells that are most notable when cells are faced with stress during DNA replication. For example, Mre11-deficient cells show sensitivity to the replication inhibitor hydroxyurea (HU), whereas tel1Δ cells do not (Supplementary Figure 1; D'Amours and Jackson, 2001). Consistent with this, disruption of any component in the MRX complex leads to very high rates of gross chromosomal rearrangements (GCRs), a phenotype attributed to replication specific events (Chen and Kolodner, 1999; Myung et al, 2001). These data suggest a function for the MRX complex during replication that is distinct from Tel1 and its function during repair.
In contrast to the DNA damage repair pathway, the characterization of MRX during replication has remained limited even though there is precedence for the complex having a fundamental role in replication. First, in mammalian cells, the MRN complex colocalizes with proliferating cell nuclear antigen and sites of BrdU incorporation during S phase (Mirzoeva and Petrini, 2003). Second, in a cyclin-dependent kinase-mediated manner, MRN interacts with replication protein A (RPA) for recruitment to replication centres (Robison et al, 2004; Olson et al, 2007). Lastly, work in Xenopus laevis has shown that Mre11 promotes replication fork restart and prevents the formation of DSBs in the newly replicated DNA (Costanzo et al, 2001; Trenz et al, 2006). Currently, however, a mechanism for how Mre11 aids fork recovery, allowing replication to restart remains undefined. In this study, we have investigated the molecular function of the MRX complex during DNA replication. We show that the complex is recruited to forks during HU-induced pausing and that it stabilizes components of the replisome independently of S phase checkpoint activation and Mre11 nuclease activity. Our data suggest that the integrity of the complex is essential for replisome stability during fork pausing and for promoting cohesion between sister chromatids during replication stress. We suggest that the tethering function of the MRX complex (Hopfner et al, 2002; Williams et al, 2007) has a stabilizing influence on paused replisomes, allowing replication recovery. Our model offers one explanation for why MRX deficiency leads to replication stress sensitivity and high levels of chromosome rearrangements.
Results
In the absence of Mre11 fork progression is altered during HU exposure
Cells that lack a functional MRX complex have an unstable genome with high rates of GCRs (Myung et al, 2001). Because such rearrangements have been attributed to events at blocked replication forks (Lambert et al, 2005), we wanted to determine the function of the MRX complex functions during replication. We first visualized replication by DNA combing and compared mre11Δ with wild-type cells as described earlier (Tourriere et al, 2005). We monitored both a ‘normal S phase' using asynchronous early log-phase cultures pulse labelled with BrdU for 40 min or an ‘S phase during replicative stress' where asynchronous early log-phase cultures were pulse labelled with BrdU for 3 h during 0.2 M HU treatment.
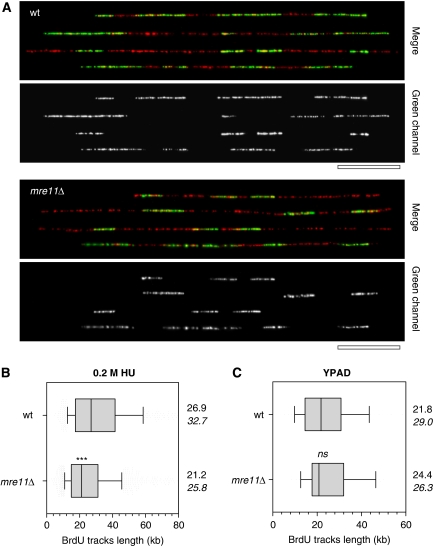
DNA fibres were purified and stretched on silanized coverslips and subsequently revealed after denaturation with an anti-ssDNA antibody (red, Figure 1A). The lengths of newly replicated DNA tracks were detected with anti-BrdU (green, Figure 1A). Wild type and mre11Δ showed identical track lengths after 40 min of BrdU incorporation under normal conditions (Figure 1C). In contrast, we found after 3 h of BrdU incorporation that mre11Δ cells treated with replication stress have 25% shorter tracks (25.8 kb) than wild-type cells (32.7 kb; Figure 1A and B). These data support a role for Mre11 during replication under conditions of HU-induced pausing and suggest that in mre11Δ cells, forks are either more prone to collapse or continue replication but at a suboptimal rate compared with wild type.
Figure 1.
Mre11 is required for normal replication fork progression in the presence of HU. Exponentially growing wild-type (JC604) and mre11Δ (JC616) cells were labelled for 3 h with 200 μg/ml BrdU in the presence of 200 mM HU. Genomic DNA fibres were stretched on silanized coverslips by DNA combing and newly replicated DNA was detected with an anti-BrdU antibody. DNA fibres were counterstained with an anti-ssDNA antibody. (A) Representative fibres of wild type (top panel) and mre11Δ (bottom panel) are shown with DNA in red and BrdU in green; Bar: 50 kb. (B) Distribution of BrdU tracks length in HU-treated cells. Box: 25–75 percentile range. Whiskers: 10–90 percentile range. Median (vertical bars) and mean values (italics) are indicated in kb. ***P<0.0001; Mann–Whitney rank sum test. (C) Distribution of BrdU tracks length in untreated wt and mre11Δ cells. ns: P=0.62; Mann–Whitney rank sum test.
Mre11 is necessary for replisome stability during HU-induced fork stalling
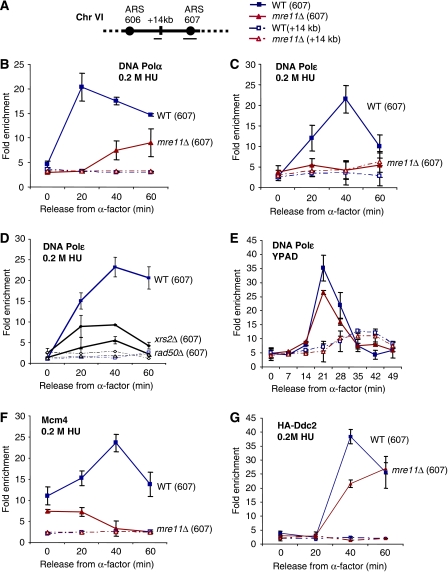
Given that the replication pattern was altered in mre11Δ cells during HU exposure, we wanted to assess the stability of replisome components at stalled forks by performing chromatin immunoprecipitation (ChIP). Either mre11Δ or wild-type cells were synchronized in G1 with α-factor and then released into S phase in the presence of HU. Under these conditions, origins fire as normal, but the rate of fork progression is significantly inhibited because of nucleotide depletion (Santocanale and Diffley, 1998; Alvino et al, 2007). The recovered DNA was quantified by real-time PCR with primer pairs to ARS607 and a negative control region 14 kb from the origin as described earlier (Figure 2A; Cobb et al, 2003; Cobb et al, 2005). First, we determined the association of DNA polymerase α and ɛ with replication forks upon HU treatment. In wild-type cells, HA-tagged DNA Polα was loaded onto forks by 20 min as cells entered into S phase and remained associated with forks for up to 60 min (Figure 2B). In mre11Δ cells, we were able to recover low levels of DNA Polα at both 40 and 60 min; however, there was a severe defect that was most notable at 20 min (Figure 2B). In the absence of Mre11, the defect in DNA Polɛ recovery was even more severe. In wild-type cells, DNA Polɛ associates with ARS607 from 20 to 60 min, with peak enrichment at 40 min (Figure 2C), yet in mre11Δ cells, there was a total loss of DNA Polɛ recovered at all times monitored (Figure 2C, data not shown). A similar loss of DNA Polɛ was observed when other MRX complex members were disrupted, with virtually no recovery observed in rad50Δ cells (Figure 2D). The disruption of Xrs2 also resulted in a significant decrease in DNA Polɛ recovery, however, the effect was slightly less pronounced (Figure 2D). These data show that disruption of any component of the MRX complex results in a considerable alteration in DNA Polɛ recovery at stalled forks compared with wild-type cells. Interestingly, the contribution of Mre11 to DNA Polɛ stabilization was dependent on fork pausing, because when cells were released from α-factor into media without added HU, the profiles were similar with only a small decrease observed in mre11Δ cells (Figure 2E). These data are consistent with our DNA combing data showing little discrepancy between mre11Δ and wild-type cells in the absence of HU.
Figure 2.
Stability of replisome components in mre11Δ cells. (A) Genomic regions amplified for ChIP analysis on Chr VI are as described earlier (Cobb et al, 2003, 2005) and correspond to early firing origin ARS607 (filled symbol) and a nonorigin site, +14 kb (open symbols). ChIP was performed using the following strains with either αMyc (9E10) or αHA (F-7, Santa-Cruz) antibodies on cells released from α-factor into YPAD+0.2 M HU (B) for HA-Polα in wild-type (JC539) and mre11Δ (JC538) cells; (C) Myc-Polɛ was monitored after released into 0.2 M HU comparing wild-type (JC285) with mre11Δ (JC388) and (D) rad50Δ (JC796) and xrs2Δ (JC757) cells. (E) Myc-Polɛ in wild-type (JC285) and mre11Δ (JC388) cells was monitored after released into YPAD without HU at 19°C. (F) Myc-Mcm4 was monitored after released into 0.2 M HU comparing wild type (JC171) with mre11Δ (JC559). (G) HA-Ddc2 in wild-type (JC257) and mre11Δ (JC549) cells was determined after release into 0.2 M HU. Error bars represent the standard deviation in fold enrichment of multiple runs of at least three different ChIP experiments.
We questioned whether a defect at the origin or an alteration in the initiation of replication could account for the decrease in DNA Polα and Polɛ recovered at forks in mre11Δ cells. However, the similar ChIP profiles in YPAD (Figure 2E) suggest that the loss of DNA polymerases recovered in mre11Δ cells during HU treatment is not likely attributed to some major disruption in the initiation of replication in cells lacking Mre11 per se. Indeed, such an intrinsic effect on origin firing would most certainly be visible in the absence of HU as well but is not. Additionally, 2D gel analysis showed only a slight decrease in the efficiency of initiation in mre11Δ cells (Supplementary Figure 2A), which might help explain the small decrease in DNA Polɛ observed in the absence of damage (Figure 2E). Finally, the levels of Orc2 and Mcm4 recovered at replication origins in G1 are almost indistinguishable for wild-type and mre11Δ cells, indicating proper pre-RC assembly (Figures 2F; Supplementary Figure 2B), although we see a dramatic loss of Mcm4 recovery at the fork in mre11Δ cells upon entry into S phase in the presence of replication stress (Figure 2F).
During stalls in replication, the checkpoint protein complex Mec1–Ddc2 is recruited to forks (Osborn and Elledge, 2003; Cobb et al, 2005), and in mec1Δ and checkpoint defective cells, replisome components are destabilized leading to irreversible fork catastrophe (Tercero and Diffley, 2001; Lucca et al, 2004; Cobb et al, 2005). Therefore, to determine whether the loss of polymerase recovery at forks in mre11Δ cells could be attributed to a defect in Mec1–Ddc2 recruitment, we performed ChIP on Ddc2 during HU treatment. Surprisingly, the ChIP profile for Ddc2 looked similar for wild type and cells lacking Mre11 (Figure 2G). Ddc2 is present at stalled forks in mre11Δ cells at the same time points where a loss of Mcm4, DNA Polα and Polɛ are observed (Figures 2B, C and F). This suggests that the function of the MRX complex stabilizes replisome components but does so independently of the presence of Mec1–Ddc2 at stalled forks.
In summary, our ChIP data indicate altered recovery of replisome components when MRX-deficient cells are exposed to replication stress, but there is little or no alterations in either the recruitment of Mec1–Ddc2 under these conditions or in the timing of origin activation in mre11Δ cells.
The MRX complex is recruited to stalled forks and promotes replication recovery after fork pausing
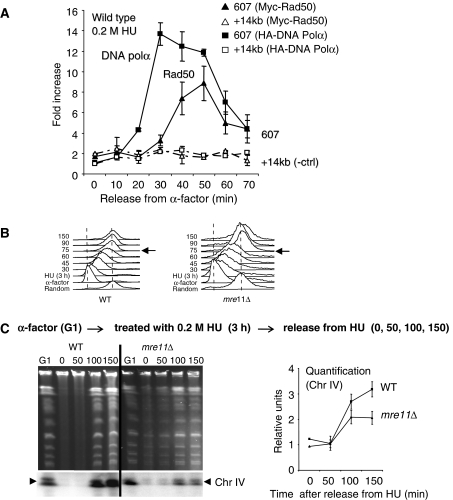
We then determined whether the function of the MRX complex during HU-induced replication stress is a result of direct interaction of the complex with paused forks. We performed ChIP as described in Figure 2 (Cobb et al, 2003, 2005) using a strain containing both an epitope tagged HA-DNA Polα and Myc-Rad50. Cellular extracts were divided and immunoprecipitations were performed with either α-HA to detect DNA Polα as a marker for the location of the replication fork or αMyc to detect Rad50 as a marker for the complex (ChIP on Rad50 gave more reliable results than Mre11). Upon release into S phase, we consistently observed a 5–6-fold enrichment for Rad50 that correlated with the presence of DNA Polα at ARS607 (Figure 3A), indicating that the MRX complex is present at paused forks. The recovery of Rad50 at forks was dependent on replication stress because no preferential recovery was observed in cells released into media without HU (data not shown).
Figure 3.
The MRX complex is recruited to forks and promotes recovery during pauses in replication. ChIP using αMyc (9E10) and αHA (F-7, Santa-Cruz) antibodies as described in Figure 2 was performed on (A) Myc-Rad50 and HA-Polα in a wild-type background (JC528), where enrichment at ARS607 (solid bar) and +14 kb from ARS607 (speckled bar) is compared at the indicated time points after release into 0.2 M HU. (B) FACS analysis was performed as described in Cobb et al (2003). Wild-type (JC467) and mre11Δ (JC371) cells were blocked with α-factor for 90 min (G1) and released into S phase with 0.2 M HU for 3 h before cells were washed and resuspended in YPAD to monitor the progression through S phase. Samples are taken at indicated time points (30, 45, 60, 75, 90, 150) after HU removal. (C) Ethidium bromide stained PFGE for samples arrested in G1 with α-factor for 90 min (G1) before treatment with 0.2 M HU for 180 min (0 min time point). HU was removed and cells were washed and released into YPAD media with samples taken at 50, 100 and 150 min. Southern blot analysis was performed to monitor ChrIV replication (lower panel) with quantification of triplicate experiments for Chr IV replication shown in relative units compared with the signal in G1 using Quantity One (Biorad) comparing wild type (JC467) and mre11Δ (JC371).
We then wanted to determine whether the loss of viability in mre11Δ cells could be attributed specifically to defects in replication upon HU treatment. We monitored replication progression in mre11Δ cells after replication stress by FACS analysis. Cells lacking Mre11 traverse S phase by 75 min after 3 h HU exposure and looked similar to wild-type cells (Figure 3B). However, FACS analysis only measures bulk DNA content, and it cannot determine whether a small percentage of the DNA remains unreplicated or whether the apparently replicated chromosomes are intact. Therefore, to examine the integrity of the chromosomes during replication, we performed pulse-field gel electrophoretic (PFGE) analysis to monitor the recovery of DNA synthesis upon HU exposure as described earlier (Desany et al, 1998).
DNA was prepared in agarose plugs from an equal number of cells that were blocked in α-factor (G1 sample) then released into S phase + 0.2 M HU for 3 h. The 0 time point indicates samples after 3 h HU treatment with subsequent samples taken 50–150 min after HU removal (Figure 3C). The replicative state of the genome was visualized by the appearance of fully replicated chromosomes in the pulse-field gel and by Southern blot analysis with a radio-labelled probe to detect fully replicated chromosome IV (bottom arrows, Figure 3C). The quantification indicates the signal relative to G1 levels (Quantity One (BioRad); Figure 3C). In G1 the chromosomes have not started replication and are visualized in the gel and on the blot (Figure 3C). Upon release into S phase + 3 h HU treatment (0 min), the chromosomes are in the process of replication and do not migrate into the gel. In wild-type cells, replication is complete and chromosome re-entry is observed by 100–150 min post-HU removal (Figure 3C). In contrast, the cells lacking Mre11 always exhibited a reduced pattern of chromosome re-entry after HU treatment. Quantification of three independent experiments show that approximately half the level of replicated Chr IV can be detected by 150 min compared with G1 levels in mre11Δ cells (Figure 3C), suggesting a defect in replication recovery upon HU-induced stress. We note that in mre11Δ mutants, we consistently observed a small subpopulation that never blocked in G1 with α-factor and migrated into the gel at time point 0 (Figure 3C), possibly indicating a defect in the G2-M transition. Taken together, the MRX complex is recruited to forks during replication stress (Figure 3A) and in mrx-deficient cells, the replisome components show altered association with sites of replication (Figure 2). This ultimately leads to defects exhibited by a decrease in replication recovery after fork pausing (Figure 3C).
The integrity of the MRX complex but not Mre11 nuclease activity is required for the maintenance of DNA Polɛ with paused forks
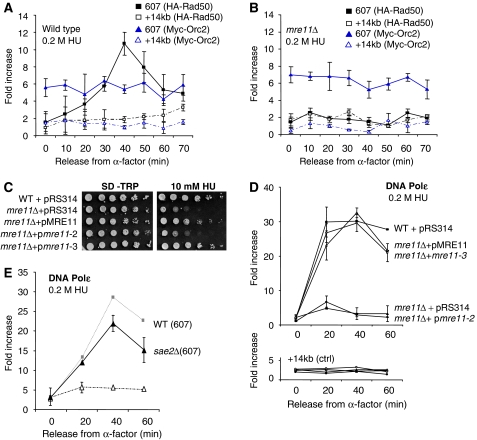
To show that Rad50 recruitment can be used as an indicator for the localization of the entire complex, we performed ChIP on Rad50 in mre11Δ cells (Figures 4A and B). We used Orc2 as a positive control because we had determined that in mre11Δ cells, the levels of Orc2 were indistinguishable from wild type in both G1 and through S phase (Supplementary Figure 2). In the absence of Mre11, there is a complete loss of Rad50 recovery compared with wild-type cells (Figures 4B and C). This strongly suggests that it is the MRX complex that is recruited and not something unique to Rad50.
Figure 4.
The MRX complex functions at the fork independently of Mre11 nuclease activity. ChIP using αMyc (9E10) and αHA (F-7, Santa-Cruz) antibodies as described in Figure 2 was performed on (A) on HA-Rad50 and Myc-Orc2 in wild-type (J764) and (B) mre11Δ (JC743) cells, where enrichment at ARS607 (solid line) and +14 kb from ARS607 (speckled line) is monitored at the indicated time points after release into 0.2 M HU. (C) Drop assays on exponentially growing cells of 1:10 serial dilutions on SC-TRP plates +10 mM HU at 30°C or (D) ChIP on Myc-Polɛ using αMyc (9E10) as described in Figure 2 were performed with wild-type (JC285) cells after transformation with pRS314 vector alone (WT +pRS314, ▪), and in mre11Δ (JC388) cells after transformation with pRS314 vector alone (mre11Δ+pRS314, ▴), or pRS314 carrying either full length MRE11 (mre11Δ+pMRE11, ▵) or mre11-3 (mre11Δ+pmre11-3, ○) and mre11-2 (mre11Δ+pmre11-2, •) constructs from cultures grown in SD-TRP media. (E) ChIP using αMyc (9E10) was performed on Myc-Polɛ in wild-type (JC285) and sae2Δ (JC686) cells as described in Figure 2.
The nuclease activity of Mre11 degrades DNA ends and hairpins by processing misfolded DNA at broken ends (Furuse et al, 1998; Paull and Gellert, 1998; Connelly et al, 1999; Trujillo and Sung, 2001; Lobachev et al, 2002). Because such DNA intermediates likely exist at least transiently at paused or stalled forks, we determined if the nuclease activity of Mre11 and the structural integrity of the MRX complex contributed to polymerases recovery. We introduced into mre11Δ cells mutants of Mre11 were either deficient for complex formation mre11-2 (56–58DLF → FLS) or only nuclease deficient mre11-3 (125–126HD → LV) but still MRX complex assembly proficient (Bressan et al, 1998). We compared isogenic mre11Δ and wild-type strains carrying empty vectors, and mre11Δ cells expressing either plasmid-borne wild-type pMRE11 or one of the mutants pmre11-2 (structurally deficient and nuclease dead) or pmre11-3 (only nuclease dead). Transformation with either pMRE11 or pmre11-3 in mre11Δ cells rescued the phenotype and DNA Polɛ association with stalled forks, with levels returning to that observed in wild-type cells (Figure 4D). As a control, we showed that pmre11-3 cells were sensitive to growth on media containing 0.01% methyl methanesulfonate, which forms DNA lesions and is consistent with nuclease deficiency (Supplementary Figure 3). In stark contrast to pmre11-3, transformation with pmre11-2 did not complement DNA Polɛ recovery and levels remained indistinguishable from those in mre11Δ cells (Figure 4D). This indicates that the integrity of the MRX complex is essential for replisome stability at the fork, but that the nuclease activity of Mre11 is not required either for DNA Polɛ maintenance or cell viability during exposure to HU (Figure 4C and D).
Sae2 is also involved in processing meiotic and mitotic double-strand breaks, and it has been shown to cooperatively function with the MRX complex to process hairpin DNA structures (Lengsfeld et al, 2007). Therefore, we performed ChIP on Myc-Polɛ in sae2Δ cells and observed a similar profile to wild-type cells upon release into HU (Figure 4E). Taken together, these data suggest that while the MRX complex is recruited to stalled forks, replisome maintenance is independent of DNA processing by the complex or its partners.
Genetic interactions between Mre11 and components of the replisome
We have determined that the MRX complex has a role during replication, but it was unclear how it was functioning. To investigate a potential mechanism for the MRX complex, we took a genetic approach and combined the loss of Mre11 with temperature sensitive (ts) replisome alleles. These mutants show defects in replication and cell viability at a nonpermissive temperature (37°C) and include pol2-12 and pol2-16, which compromise the catalytic subunit of DNA Polɛ, as well as mcm10-43 and mcm5-3 (Budd and Campbell, 1993; Kesti et al, 1999; Kawasaki et al, 2000; Dziak et al, 2003). Mcm5 is a component of the MCM2-7 helicase and is important for the loading of Mcm10, which in turn is important for the loading of additional replication factors including RPA and DNA Polα. After the initiation of replication, the Mcm2-7 helicase, Mcm10 and the replicative polymerases including DNA Polɛ, migrate with the fork during elongation.
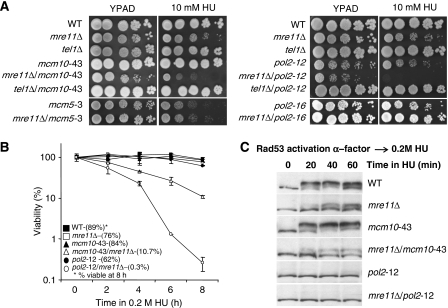
Strains were monitored for cell growth in the presence of the replication inhibitor HU. A slight defect was observed in mre11Δ cells as well as in the mcm5-3 and pol2-12 ts alleles at the semipermissive temperature; 30°C (Figure 5A). Strikingly, a synergistic loss of viability was observed when mre11Δ was coupled with either the mcm10-43 or pol2-12 allele that was most evident in the mre11Δ/pol2-12 double mutant (Figure 5A). Consistent with the drop assays, mre11Δ/mcm10-43 and mre11Δ/pol2-12 cells showed extremely poor recovery after transient exposure to 0.2 M HU, with viability dropping to 10.7 and 0.3%, respectively, after 8 h (Figure 5B).
Figure 5.
Mre11 shows synergistic interactions with replisome components. (A) Drop assays on YPAD±10 mM HU were performed with exponentially growing cultures at permissive temperature (25°C) with 1:5 serial dilutions before 2 days incubation at 30°C (B) Cell viability was monitored as colony outgrowth from asynchronous cultures grown at permissive temperature (25°C) before transient exposure to 0.2 M HU at 30°C for the indicated times with values normalized to survival at time point 0 and (C) the phosphorylation status of Rad53 was monitored by western blot analysis after cells were blocked in G1 with α-factor and released in 0.2 M HU for the indicated time at 30°C using anti-Rad53 antibodies (yC-19, Santa Cruz) for the following strains: wild type (JC467), mre11Δ (JC371) mcm10-43 (JC216), mcm10-43/mre11Δ (JC554), pol2-12 (JC617), pol2-12/mre11Δ (JC553), mcm5-3 (JC227), mcm5-3/mre11Δ (JC564), pol2-16 (JC583) pol2-16/mre11Δ (JC619), tel1Δ (JC65), mcm10-43/tel1Δ (JC591), pol2-12/mre11Δ (JC612).
These data might indicate that the pol2-12 and mcm10-43 mutant alleles generate damage during replication that is then recognized and processed through the canonical DNA damage repair cascade by the MRX complex together with Tel1, a pathway characterized earlier and termed as the TM checkpoint pathway (Usui et al, 2001). However, drop assays showed that in contrast to mre11Δ, no mutants containing the deletion of TEL1 were sensitive to HU; and the double-mutant phenotypes observed with mre11Δ were not recapitulated in tel1Δ/mcm10-43 and tel1Δ/pol2-12 cells (Figure 5A). As well, to determine whether the loss of viability in the mre11Δ/mcm10-43 and mre11Δ/pol2-12 double mutants was due to a failure in the replication checkpoint, we also monitored Rad53 phosphorylation when cells were synchronously released into S phase in the presence of 0.2 M HU. We observed similar kinetics in wild-type and mcm10-43 cells for Rad53 activation peaking at 40 min (Figure 5C). When Mre11 was deleted, a slight defect in full Rad53 activation was observed, which is consistent with earlier reports (D'Amours and Jackson, 2001; Nakada et al, 2004) and the phenotype of the double mutant mre11Δ/mcm10-43 resembled that of mre11Δ. Also in agreement with earlier reports (Navas et al, 1995), pol2-12 cells were deficient in the S phase checkpoint after release into 0.2 M HU (Figure 5C), therefore, we do not interpret the additive loss of viability observed in mre11Δ/mcm10-43 and mre11Δ/pol2-12 cells as solely a deficiency in the Rad53 checkpoint activation. This is particularly clear for mre11Δ/pol2-12 cells where the dramatic loss of viability cannot be explained simply by a defect in checkpoint activation because pol2-12 alone shows complete abrogation of Rad53 phosphorylation (Figure 5C). Taken together, these data suggest a function for Mre11 during HU-induced fork arrest that is distinct from and not correlating with Tel1 or Rad53 checkpoint signalling.
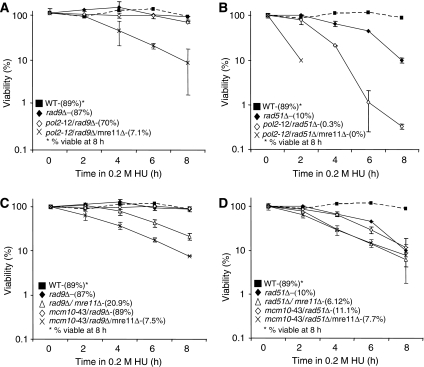
Consistent with an independence of the intra-S checkpoint after HU treatment, no additive loss of recovery was observed when pol2-12 and mcm10-43 were combined with a rad9Δ mutation (Figures 6A and C). The viability after 8 h in the double-mutant cells mcm10-43/rad9Δ (89%) and pol2-12/rad9Δ (70%) was similar to the single mutants mcm10-43 (84%), pol2-12 (62%) and rad9Δ (87%; Figure 5B). As well, the percentage of viable cells after 8 h of HU treatment for the triple-mutant cells mre11Δ/mcm10-43/rad9Δ (7.5%) and mre11Δ/pol2-12/rad9Δ (7.1%) looked similar to the double mutants mre11Δ/mcm10-43 and mre11Δ/pol2-12, with a small alleviation in cellular death observed when Rad9 was disrupted in the mre11Δ/pol2-12 (0.3%) mutant background (compare Figures 5B with 6A and C).
Figure 6.
Epistatic analysis between replisome alleles with mre11Δ and rad9Δ or rad51Δ. (A–D) Cell viability was monitored as colony outgrowth from asynchronous cultures grown at permissive temperature (25°C) before transient exposure to 0.2 M HU at 30°C for the indicated times with values normalized to survival at time point 0. All experiments were performed at the same time as Figure 5B for the following strains: rad9Δ (JC176), rad9Δ/mre11Δ (JC568) mcm10-43/rad9Δ (JC569), mcm10-43/rad9Δ/mre11Δ (JC570), pol2-12/rad9Δ (JC618), pol2-12/rad9Δ/mre11Δ (JC572), rad51Δ (JC445), rad51Δ/mre11Δ (JC633) mcm10-43/rad51Δ (JC634), mcm10-43/rad51Δ/mre11Δ (JC629), pol2-12/rad51Δ (JC630), pol2-12/rad51Δ/mre11Δ (JC683).
In contrast to Rad9-mutant cells, a severe additive loss of viability was observed upon the disruption of Rad51. The double mutants rad51Δ/mcm10-43 (11.1%) and rad51Δ/pol2-12 (0.3%) showed a loss of viability after 8 h of HU exposure similar to double mutants with mre11Δ (compare Figures 5B with 6B and D), and the triple mutant mre11Δ/mcm10-43/rad51Δ (7.5%) showed a similar loss of viability as both double mutants mre11Δ/mcm10-43 (10.7%) and mre11Δ/rad51Δ (6.12%), suggestive of an epistatic relationship. These genetic interactions indicate the importance of Mre11 when DNA replication is compromised and suggest that the function of Mre11 might principally be within the HR pathway for recovery after HU exposure. The triple mutant mre11Δ/pol2-12/rad51Δ did show the most severe phenotype with no survivors after 8 h of HU (Figure 6B), suggesting in the pol2-12 background that Mre11 is not functioning solely within the Rad51-dependent HR pathway. However, we note the identical and severe loss of viability in both double mutants mre11Δ/pol2-12 (0.3%) and rad51Δ/ pol2-12 (0.3%; Figures 5B and 6B).
The MRX complex and SCC factors provide support to forks during stalls in replication
The genetic studies showed that disrupting Mre11 in combination with the alleles pol2-12 and mcm10-43 leads to synergistic HU sensitivity (Figure 5A). Interestingly, common to both Pol2 and Mcm10 are their interactions with factors necessary for SCC. The pol2-12 allele has a 22 amino acid truncation in the C-terminal domain and results in the disruption of SCC (Edwards et al, 2003). Also, Mcm10 has been shown to bind Ctf4, a factor that associates with replication forks and is critical for the establishment of SCC (Wittmeyer and Formosa, 1997; Hanna et al, 2001; Mayer et al, 2001; Bermudez et al, 2003; Gambus et al, 2006; Lengronne et al, 2006; Skibbens et al, 2007; Zhu et al, 2007).
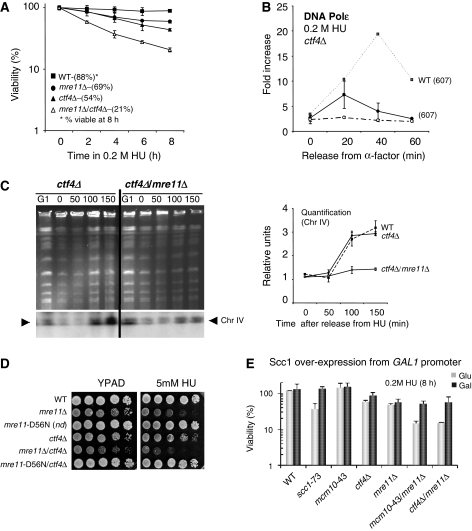
Interestingly, Pan et al (2006) reported that ctf4Δ/mre11Δ double-mutant cells exhibit growth defects. This in combination with the loss of viability we observed in mre11Δ/mcm10-43 cells (Figures 5 and 6) led us to investigate Mre11 and Ctf4 during S phase in the presence of replication stress. We report here an additive decrease in cell viability in mre11Δ/ctf4Δ double mutants after transient exposure to HU, suggesting at least a partial overlap in function for these proteins in response to replication stress (Figure 7A). To determine what effect the deletion of Ctf4 would have on the replisome, we performed ChIP on Myc-Polɛ in ctf4Δ cells and observed a similar profile to the recovery levels observed in mrx-mutant cells (Figure 7B). Because the loss of DNA Polɛ was so pronounced in the single mutants, determining an additive affect for replisome stability was precluded in mre11Δ/ctf4Δ mutant cells.
Figure 7.
The MRX complex functions on a pathway parallel to SCC factors to maintain forks during replication stress. (A) Cell viability was monitored as described earlier in Figure 5B for the following strains: wild type (JC467), mre11Δ (JC371), ctf4Δ (JC648), mre11Δ/ctf4Δ(JC662). (B) ChIP using αMyc (9E10) was performed on Myc-Polɛ in wild-type (JC285) and ctf4Δ (JC670) cells as described in Figure 2. (C) Ethidium bromide stained PFGE for samples arrested in G1 with α-factor for 90 min (G1) before treatment with 0.2 M HU for 180 min (0). HU was removed and cells were washed and released into YPAD media for 50, 100 and 150 min. Southern blot analysis was performed to monitor ChrIV replication (lower panel) with quantification of triplicate experiments for Chr IV replication shown in relative units compared with the signal in G1 using Quantity One (Biorad) strains used are ctf4Δ (JC648) and ctf4Δ/mre11Δ (JC662) with all experiments perform at the same time as Figure 3C. (D) Drop assays on YPAD±5 mM HU were performed from exponentially growing cultures with 1:10 serial dilutions at 30°C comparing wild type (JC467), mre11Δ (JC371), mre11D56N(JC606), ctf4Δ (JC648), mre11Δ/ctf4Δ (JC662) and mre11D56N/ctf4Δ (JC647). (E) Cell viability was monitored as colony outgrowth on SCC1overexpression induced (gal) or repressed (glu) from asynchronous cultures grown at permissive temperature (25°C) before transient exposure to 0.2 M HU at 30°C for 8 h. Galactose-induced SCC1 overexpression and glucose repressed SCC1 started 90 min before HU exposure and was maintained throughout all the experiments. The percentage survival is normalized to the cellular viability at time 0 for the following strains: wild type (JC784), scc1-73 (JC786), mcm10-43 (JC787), ctf4Δ (JC795), mre11Δ (JC801), mre11Δ/mcm10-43 (JC803), mre11Δ/ctf4Δ (JC797).
However, to determine whether mre11Δ/ctf4Δ mutants had an additive defect during replication, we monitored recovery from HU treatment by performing PFGE analysis as in Figure 3C. In contrast to mre11Δ cells, which did not recover by 150 min post-HU treatment, cells lacking Ctf4 resumed replication with kinetics similar to wild-type cells and full re-entry was observed at 100–150 min (Figures 7C). This suggests that the altered association of DNA Polɛ during HU treatment is reversible in ctf4Δ cells and that the fork remains in a competent state to resume replication once the HU is removed. Strikingly, at 150 min post-HU treatment, the mre11Δ/ctf4Δ cells showed a severe recovery defect with the level of fully replicated chromosomes remaining unchanged from 0 to 150 min (Figure 7C). In summary, these data suggest that Mre11 and the SCC establishment factor Ctf4 share a partially redundant function for recovery from HU-induced fork pausing.
Drop assays show growth defects in mre11Δ/ctf4Δ double mutants on YPAD and sensitivity to growth on HU (Figures 7D; Pan et al, 2006). As our data suggest a structural role for the MRX complex to promote cohesion between sister chromatids, we reasoned that a nuclease dead Mre11 would not exhibit the same additive phenotype as mre11Δ when combined with a disruption in Ctf4. Indeed this was the case, and when using the nuclease dead mre11-D56N separation of function allele (Krogh et al, 2005) there was no additive sensitivity in ctf4Δ/mre11-D56N double-mutant cells on 5 mM HU (Figure 7D).
We have interpreted the additive loss of viability after exposure to HU in mre11Δ/ctf4Δ and mre11Δ/mcm10-43 double mutants to be independent of checkpoint activation and a result of disruptions in cohesion between sister chromatids at paused forks. Indeed, it has been proposed that one essential function of the MRX complex is to provide bridging cohesion between two strands of DNA (Hopfner et al, 2002). To address this more directly, we monitored cellular viability in these double mutants during transient exposure to HU while overexpressing the cohesin factor Scc1; which is a cohesin complex member regulated in a cell cycle dependent manner, peaking in S phase (Michaelis et al, 1997). We reasoned that an increase in cohesin complex assembly from Scc1 overexpression would promote SCC and restore cell viability if mre11Δ/ctf4Δ and mre11Δ/mcm10-43 double mutants exhibit cohesion-related defects.
Scc1 was placed under control of the galactose inducible GAL1 promoter and integrated into the genome at the LEU2 locus in wild-type and mutant cells (Sullivan et al, 2004). Cultures were grown overnight in YPLG glycerol before transfer to media containing either 2% galactose or glucose followed by 0.2 M HU treatment. Cultures containing 2% galactose showed robust Scc1 expression (Supplementary Figure 4). As a positive control, we show that overexpression of Scc1 during HU treatment restored viability in cells harbouring the ts allele scc1-73 (Ciosk et al, 2000). Similarly, the overexpression of Scc1 restored viability in both mre11Δ/mcm10-43 (51.3%) and mre11Δ/ctf4Δ (55.8%) double-mutant cells to the level of mre11Δ (55.4%) cells, providing direct evidence that SCC defects are a major contributor to cell death in these double mutants during replication stress (Figure 7E).
Discussion
We describe here a role for the MRX complex during DNA synthesis and show in three different ways by: DNA combing, ChIP and PFGE analysis that in the absence of Mre11, replication is altered upon exposure to replication stress. During pauses in replication, the MRX complex is recruited to forks (Figures 3A) but its role there appears distinct from DNA end processing and checkpoint signalling events. Our work presents in vivo data for the MRX complex that is specific to replication forks and which supports the earlier reported structural function of the complex (Chen et al, 2001; Hopfner et al, 2002; Williams et al, 2008). We suggest that upon its recruitment to paused forks, the MRX complex provides cohesive support to sister chromatids, and in SCC compromised mutant cells, the MRX complex becomes important for efficient fork recovery in the face of replication stress. Through MRX scaffold abilities (Chen et al, 2001; Hopfner et al, 2002; Williams et al, 2008), the complex promotes replication recovery possibly through a Rad51- and HR-dependent mechanism.
The MRX complex is recruited to stalled forks stabilizing the replisome
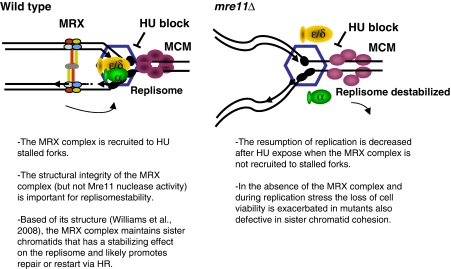
When cells are exposed to HU-induced replication stress, forks do not collapse but remain in a competent conformation to resume high fidelity and rapid replication once the stress is overcome. The MRX complex has a role in this process (compare WT and mre11Δ, Figure 3C). The complex is recruited to stalled forks in wild-type cells (Figure 3A) and functions either during a stall or in aiding fork recovery or both; however, distinguishing between these two events is difficult to demonstrate. Nonetheless, its presence at HU stalled forks in wild-type cells suggests that the complex has a function before and distinct from a function in the repair of DNA DSBs arising from forks that have collapsed. Indeed, support of this is demonstrated by alterations in replisome association and the loss of replication recovery in mre11Δ cells upon HU treatment (Figures 2 and 3C).
We have proposed a function for the MRX complex during replication based on EM structural data, which showed the complex can tether sister chromatids (Hopfner et al, 2002). In contrast to mammals and Xenopus, live cell imaging detected no Mre11 focus formation in wild-type cells upon HU treatment, but only after forks collapse in checkpoint defective cells (Lisby et al, 2004). We do not think our ChIP results showing Rad50 recruitment to forks (Figure 3A) directly contradicts these reports and suggest that its function at HU-paused forks might involve fewer or more dispersed MRX complex molecules when ‘tethering' sister chromatids, which would be difficult to detect by microscopy. One can also speculate that because Mre11 can interact with itself (Nairz and Klein, 1997; Paull and Gellert, 1998), having the complex present at stalled forks in anticipation of collapse might also aid in the rapid recruitment of additional MRX complex molecules to repair/prevent the formation of DNA DSBs upon breakdown.
One question still remaining is what signals MRX recruitment and what does it interact with at stalled forks. We see an interaction between RPA and Mre11 by co-immunoprecipitation that might have functional significance, yet this interaction did not increase upon HU treatment (data not shown). This is different from the scenario in human cells where a physical interaction between components of the MRN complex and RPA has been reported to increase during HU treatment and signal MRN localization to replication centres (Olson et al, 2007). Also, we did not detect a physical interaction between the MRX complex and either DNA Polɛ or Polα (Supplementary Figure 5). This suggests that the influence of MRX on replisome stability is likely indirect in nature, and also argues that the MRX complex is not recruited to forks through direct interaction with these polymerases.
During HU exposure replication is impaired in the absence of Mre11
Fork progression was altered in mre11Δ cells upon exposure to HU replication stress but not in the absence of damage. This alteration in progression did not, however, indicate a complete cessation of replication (Figure 1A and B), and the ChIP data should be interpreted within this context. One explanation for the lack of DNA Polɛ (Figure 2C and D) detected at stalled forks in the absence of MRX complex members is that aberrant DNA conformations are generated that preclude efficient antibody access to replisome components in mrx-deficient cells, resulting is the loss of detectable enrichment. Second, DNA Polɛ is present but with reduced stoichiometry and below the levels of detection, however this is difficult to demonstrate. Finally, DNA Polɛ might very well be lost from forks. This would be consistent with experiments performed in X. laevis egg extracts showing DNA Polɛ association with chromatin during replicational stress dependent on Mre11 (Trenz et al, 2006). Currently, we are trying to determine whether replication continues with a compensating polymerase such as DNA Polδ; however, these experiments have proven to be quite difficult because the inclusion of an HA epitope tag on DNA Polδ in mre11Δ cells (but not wild type) renders cells extremely sick (Supplementary Figure 6). This prevents ChIP but underscores the importance of DNA Polδ in mre11Δ cells.
The nuclease activity of Mre11 is not required for replisome maintenance during pauses in replication
GCRs are attributed to polymerase destabilization and mishaps at replication forks (Kolodner et al, 2002; Cobb et al, 2005; Lambert et al, 2005). The results presented here are consistent with the relatively low rates of chromosomal rearrangement reported for the nuclease dead mre11-3 allele compared with the high rates seen in mre11Δ cells and mrx alleles that disrupt complex formation (Smith et al, 2005), such as mre11-2. Our data support a structural function for the MRX complex that is dependent upon the integrity of the interactions among the Mre11, Rad50 and Xrs2 components. We show that the nuclease activity of Mre11 is dispensable for replisome maintenance at paused forks, but that the structural integrity of the complex is essential. This is evident from DNA Polɛ complementation assays where the ectopic expression of mre11-3 in mre11Δ restored DNA Polɛ recovery to wild-type levels, but with mre11-2 the recovery of DNA Polɛ remained as in mre11Δ cells (Figure 4D). Similar to cells transformed with mre11-3, the endogenous nuclease dead mre11-D56N allele is not sensitive to low levels of HU (Krogh et al, 2005) and showed no additive sensitivity when coupled with a disruption of the SCC establishment factor Ctf4 during replication stress (Figure 7D). Taken together, these data indicate that Mre11 nuclease activity does not contribute in any detectable way to MRX function at stalled forks for recovery from replication stress.
Together SCC factors and the MRX complex promote the resumption of DNA replication
Our data support a model where under conditions of replication stress maintaining newly synthesized daughter strands promotes fork stability (Figure 8). We propose that preserving the architecture behind the fork is important for replication recovery, possibly through an HR-dependent restart mechanism. The model we present here suggests that cohesion between sister chromatids during times of replication stress involves the MRX complex. This model is supported by our observations that the MRX complex is recruited to paused forks (Figures 3A and 4A and B) as well as our genetic data that indicate the MRX complex has overlapping functions with factors important for SCC. Indeed, the loss of Mre11 in combination with SCC-deficient mutants cells results in a loss of viability when cells were confronted with replication stress. This model is also supported by structural data showing that the MRX complex can bridge two strands of duplexed DNA, the distance of newly synthesized sister chromatids (Hopfner et al, 2002). In summary, we present here a novel function for the MRX complex during replication that involves conformational preservation of newly synthesized DNA behind stalled forks. During pauses in replication if sister chromatids are not maintained, the consequence is disruption of the replisome at the fork, and ultimately the loss of chromosome integrity, a hallmark of mrx-deficient cells.
Figure 8.
Model for role of the MRX complex at stalled forks. The MRX complex is present at stalled replication forks and is important for fork recovery. The MRX complex is important for the association of DNA polymerase α and ɛ and possibly other replisome factors (represented by the hexagon) with stalled forks. In the absence of the MRX complex and of properly established SCC, sister chromatids are not held together, and this has a destabilizing influence on the fork resulting in a loss of fork recovery and cell viability.
Materials and methods
Strains and plasmids
All strains are listed in Table I. The mre11Δ∷HIS3 and ctf4Δ∷TRP1 disruptions were constructed using pFA6a PCR-based cassettes (Longtine et al, 1998) and verified by PCR and phenotypic analyses. pRS314-MRE11-FL-TRP-HA, -mre11-2-TRP-HA, and -mre11-3-TRP-HA were kind gifts from the Dr J Petrini at the Sloan-Kettering Institute (Bressan et al, 1998). The plasmid YIplac128 GAL-Scc1-3HA was kindly provided by F Ulhmann at the Cancer Research, UK, and used for integration of SCC1 gene under control of the galactose inducible GAL1 promoter at the LEU2 locus in JC-770 (Sullivan et al, 2004).
Table 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| JC-65 | JC-141 with tel1∷URA3 | This study |
| JC-141 | MATα, ade2-1, trp1-1, his3-11, -15, ura3-1, leu2-3,-112, can1-100 | R Rothstein (W303-1A) |
| JC-171 | JC-467 with CDC54- 13Myc∷KanMX6 | This study |
| JC-176 | MATa can1-100, ade2-1, trp1-1, his3-11, -15, ura3-1, leu2-3,-112, rad9∷LEU2 | |
| JC-216 | MATa, ade2-1, trp1-1, his3-11, -15, ura3-1, leu2-3,-112, can1-100, mcm10-43 | Kawasaki et al, 2000 |
| JC-227 | MATα, leu2-3,-112ura3-52, his4-34,mcm5-3 | Dziak et al, 2003 |
| JC-257 | JC-467 with RPA1-13Myc∷TRP, Ddc2-3HA∷URA3 | This study |
| JC-285 | JC-467 with POL2-13Myc∷KanMX6 | Bjergbaek et al, 2005 |
| JC-371 | JC-467 with mre11∷HIS3 | This study |
| JC-388 | JC-285 with mre11∷HIS3 | This study |
| JC-445 | MATa, ade2-1, trp1-1, his3-11, -15, ura3-1, leu2-3,-112, rad51∷URA3 | This study |
| JC-467 | MATa, ade2-1, trp1-1, his3-11, -15, ura3-1, leu2-3,-112, can1-100, pep4∷LEU2 | R Rothstein (W303-1A) |
| JC-528 | MATa, ade2-1, trp1-1, his3-11, -15, ura3-1, leu2-3,-112, can1-100, RAD50-13Myc∷KanMX6, CDC17-3HA∷TRP1 | This study |
| JC-538 | JC 539 with mre11∷HIS3 | This study |
| JC-539 | JC 467 with CDC17-3HA∷TRP1 | This study |
| JC-549 | JC-257 with mre11∷HIS3 | This study |
| JC-553 | JC-617 with mre11∷HIS3 | This study |
| JC-554 | JC-216 with mre11∷HIS3 | This study |
| JC-559 | JC 171 with mre11∷HIS3 | This study |
| JC-564 | JC-227 with mre11∷ HIS3 | This study |
| JC-568 | JC-371 with rad9∷TRP1 | This study |
| JC-569 | JC-216 with rad9∷LEU2 | This study |
| JC-570 | JC-569 with mre11∷HIS3 | This study |
| JC-572 | JC-618 with mre11∷HIS3 | This study |
| JC-583 | MATa, ade2-1, trp1-1, his3-11, -15, ura3-1, leu2-3,-112, pol2-16 | Kesti et al, 1999 |
| JC-591 | JC-216 with tel1∷URA3 | This study |
| JC-604 | MATa ade2-1,trp1-1, can1-100, leu2-3, 112, his3-11, 15, ura3-1, GAL, psi+,RAD5,URA3∷GPD-TK | Tourriere et al, 2005 |
| JC-606 | MATa ade2-1, trp1-1, his3-11, -15, ura3-1, leu2-3,-112, can1-100 with mre11-D56N | Krogh et al, 2005 |
| JC-612 | JC-617 with tel1∷URA3 | This study |
| JC-616 | JC-604 with mre11∷HIS3 | This study |
| JC-617 | MATa, ade2-1, trp1-1, his3-11, -15, ura3-1, leu2-3,-112, can1-100, pol2-12 | This study derived from Budd and Campbell, 1993 |
| JC-618 | JC-617 with rad9∷LEU2 | This study |
| JC-619 | JC-583 with mre11∷HIS3 | This study |
| JC-629 | JC-633 with mcm10-43 | This study |
| JC-630 | JC-617 with rad51∷URA3 | This study |
| JC-633 | JC-371 with rad51∷URA3 | This study |
| JC-634 | JC-216 with rad51∷URA3 | This study |
| JC-647 | JC-606 with ctf4∷TRP1 | This study |
| JC-648 | JC-467 with ctf4∷TRP1 | This study |
| JC-662 | JC-648 with mre11∷HIS3 | This study |
| JC-670 | JC-285 with ctf4∷TRP1 | This study |
| JC-683 | JC-633 with pol2-12 | This study |
| JC-686 | JC-285 with sae2∷URA3 | This study |
| JC-743 | JC764 with mre11∷HIS3 | This study |
| JC-757 | JC-285 with xrs2∷HIS3 | This study |
| JC-764 | MATa, RAD50-3HA∷TRP1, ORC2-9Myc∷LEU2, ade2-1, trp1-1, his3-11, -15, ura3-1, leu2-3,-112 | This study |
| JC-770 | MATa, ade2-1, trp1-1, his3-11, -15, ura3-1, leu2-3,-112, can1-100 | This study |
| JC-784 | MATa, GAL-SCC1-3HA∷LEU2, ade2-1, trp1-1, his3-11, -15, ura3-1, leu2-3,-112, can1-100, | This study |
| JC-786 | JC-784 with scc1-73 | This study |
| JC-787 | JC-784 with mcm10-43 | This study |
| JC-795 | JC-784 with ctf4∷URA3 | This study |
| JC-796 | JC-285 with rad50∷KanMX6 | This study |
| JC-797 | JC-795 with mre11∷HIS3 | This study |
| JC-801 | JC-784 with mre11∷HIS3 | This study |
| JC-803 | JC-787 with mre11∷HIS3 | This study |
| All strains are W303 background unless otherwise indicated. | ||
Survival and drop assays
Survival assays were performed on cultures grown overnight in YPAD at 25°C before exposure to 0.2M HU at 30°C for the indicated times. In total, 500 cells were plated in triplicate and the percentage of viability was determined from the 0 time point after release from α-factor into HU. Drop assays were performed with 1:5 serial dilutions of cultures grown at 25°C and plated on YPAD±HU and incubated at 30°C. For complementation experiments, cultures were grown on -TRP selective media and drop assays were performed with 1:5 dilutions on SD-TRP +/− HU and incubated at 30°C. Survival assays with SCC1 overexpression were performed on exponentially grown cultures in YPLG glycerol at 25°C before shifting to YPAD or YPA galactose (2%) for 90 min before 8 h HU treatment at 30°C in the same media. In all, 500 cells were plated in duplicate on YPAD or YPA galactose media with the percentage viability determined from 0 time point after release.
Chromatin immunoprecipitation
ChIP was performed as described earlier in Cobb et al (2003, 2005) except that washes buffer contained 0.01% SDS. DNA was quantified by real-time PCR using the ABI 7900 Sequence Detector System in the Southern Alberta Cancer Research Institute (provided by the Alberta Cancer Foundation). Primer sequences are available upon request. α-mouse Dynabeads were coupled with monoclonal antibodies against Myc (9E10) or HA (F-7; SC-7392 Santa-Cruz). Uncoupled Dynabeads were used for background controls.
For complementation experiments, cultures were grown on -TRP selective media before synchronization with α-factor in YPAD pH 5.0 and released in YPAD+0.2 M HU. The subsequent steps of ChIP were performed as described earlier.
PFGE and 2D gel electrophoresis
Yeast cells were embedded in agarose plugs (3 × 107 cells/plug), and genomic DNA was extracted as described (Lengronne et al, 2001). Agarose plugs were used for PFGE. Yeast chromosomes were separated by PFGE (Chef DRII, BioRad) for 18 h at 6V/cm 60′′/60′′, stained with ethidium bromide and transferred to Hybond XL (GE Healthcare). Southern blot analysis for chromosome IV was detected using a probe to the Rad9 gene. All experiments were performed in a least triplicate, and quantification was carried out using Quantity One (BioRad).
DNA combing
DNA combing was performed as described (Michalet et al, 1997). HU was added 30 min before the addition of BrdU. BrdU was detected with a rat monoclonal antibody (BU1/75, AbCys) followed by treatment with a secondary antibody coupled to Alexa 488 (Molecular Probes). DNA molecules were counterstained with an anti-ssDNA antibody (MAB3034, Chemicon) and an anti-mouse IgG coupled to Alexa 546 (Molecular Probes). DNA fibres were analysed on a Leica DM6000B microscope equipped with a CoolSNAP HQ CCD camera (Roper Scientifics) and a 40 × objective. Image acquisition was performed with MetaMorph (Roper Scientifics). Representative images of DNA fibres were assembled from different fields of view with Adobe Photoshop as described (Pasero et al, 2002).
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6
Supplementary Table I
Supplemental Data
Acknowledgments
We thank all members of the Cobb and Pasero Laboratories for helpful discussions. We thank Etienne Schwob and the DNA combing facility of Montpellier for silanized coverslips. We are also grateful to J Campbell, A Sugino, L Symington, C Sjogren for yeast strains and to J Petrini for providing plasmids. We thank F Uhlmann for the Scc1 overexpression vector and helpful suggestions. We thank Susan Lees-Miller and David Shore for reading this manuscript before submission. JC is an Alberta Heritage Foundation for Medical Research Scholar, and work in the JC laboratory is funded by Grants from the Canadian Institutes for Health Research # MOP-82736 and the Alberta Cancer Board # 23575. Research in the PP laboratory is funded by Fondation pour la Recherche Médicale, CNRS and INCa.
References
- Alani E, Subbiah S, Kleckner N (1989) The yeast RAD50 gene encodes a predicted 153-kD protein containing a purine nucleotide-binding domain and two large heptad-repeat regions. Genetics 122: 47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvino GM, Collingwood D, Murphy JM, Delrow J, Brewer BJ, Raghuraman MK (2007) Replication in hydroxyurea: it's a matter of time. Mol Cell Biol 27: 6396–6406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermudez VP, Maniwa Y, Tappin I, Ozato K, Yokomori K, Hurwitz J (2003) The alternative Ctf18-Dcc1-Ctf8-replication factor C complex required for sister chromatid cohesion loads proliferating cell nuclear antigen onto DNA. Proc Natl Acad Sci USA 100: 10237–10242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjergbaek L, Cobb JA, Tsai-Pflugfelder M, Gasser SM (2005) Mechanistically distinct roles for Sgs1p in checkpoint activation and replication fork maintenance. EMBO J 24: 405–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressan DA, Olivares HA, Nelms BE, Petrini JH (1998) Alteration of N-terminal phosphoesterase signature motifs inactivates Saccharomyces cerevisiae Mre11. Genetics 150: 591–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd ME, Campbell JL (1993) DNA polymerases delta and epsilon are required for chromosomal replication in Saccharomyces cerevisiae. Mol Cell Biol 13: 496–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha RS, Kleckner N (2002) ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science 297: 602–606 [DOI] [PubMed] [Google Scholar]
- Chen C, Kolodner RD (1999) Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat Genet 23: 81–85 [DOI] [PubMed] [Google Scholar]
- Chen L, Trujillo K, Ramos W, Sung P, Tomkinson AE (2001) Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol Cell 8: 1105–1115 [DOI] [PubMed] [Google Scholar]
- Ciosk R, Shirayama M, Shevchenko A, Tanaka T, Toth A, Nasmyth K (2000) Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol Cell 5: 243–254 [DOI] [PubMed] [Google Scholar]
- Cobb JA, Bjergbaek L, Shimada K, Frei C, Gasser SM (2003) DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J 22: 4325–4336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb JA, Schleker T, Rojas V, Bjergbaek L, Tercero JA, Gasser SM (2005) Replisome instability, fork collapse, and gross chromosomal rearrangements arise synergistically from Mec1 kinase and RecQ helicase mutations. Genes Dev 19: 3055–3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly JC, de Leau ES, Leach DR (1999) DNA cleavage and degradation by the SbcCD protein complex from Escherichia coli. Nucleic Acids Res 27: 1039–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo V, Robertson K, Bibikova M, Kim E, Grieco D, Gottesman M, Carroll D, Gautier J (2001) Mre11 protein complex prevents double-strand break accumulation during chromosomal DNA replication. Mol Cell 8: 137–147 [DOI] [PubMed] [Google Scholar]
- D'Amours D, Jackson SP (2001) The yeast Xrs2 complex functions in S phase checkpoint regulation. Genes Dev 15: 2238–2249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amours D, Jackson SP (2002) The Mre11 complex: at the crossroads of DNA repair and checkpoint signalling. Nat Rev Mol Cell Biol 3: 317–327 [DOI] [PubMed] [Google Scholar]
- de Jager M, Dronkert ML, Modesti M, Beerens CE, Kanaar R, van Gent DC (2001) DNA-binding and strand-annealing activities of human Mre11: implications for its roles in DNA double-strand break repair pathways. Nucleic Acids Res 29: 1317–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desany BA, Alcasabas AA, Bachant JB, Elledge SJ (1998) Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway [In Process Citation]. Genes Dev 12: 2956–2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolganov GM, Maser RS, Novikov A, Tosto L, Chong S, Bressan DA, Petrini JH (1996) Human Rad50 is physically associated with human Mre11: identification of a conserved multiprotein complex implicated in recombinational DNA repair. Mol Cell Biol 16: 4832–4841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziak R, Leishman D, Radovic M, Tye BK, Yankulov K (2003) Evidence for a role of MCM (mini-chromosome maintenance) 5 in transcriptional repression of sub-telomeric and Ty-proximal genes in Saccharomyces cerevisiae. J Biol Chem 278: 27372–27381 [DOI] [PubMed] [Google Scholar]
- Edwards S, Li CM, Levy DL, Brown J, Snow PM, Campbell JL (2003) Saccharomyces cerevisiae DNA polymerase epsilon and polymerase sigma interact physically and functionally, suggesting a role for polymerase epsilon in sister chromatid cohesion. Mol Cell Biol 23: 2733–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M, Nagase Y, Tsubouchi H, Murakami-Murofushi K, Shibata T, Ohta K (1998) Distinct roles of two separable in vitro activities of yeast Mre11 in mitotic and meiotic recombination. EMBO J 17: 6412–6425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, Edmondson RD, Labib K (2006) GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol 8: 358–366 [DOI] [PubMed] [Google Scholar]
- Hanna JS, Kroll ES, Lundblad V, Spencer FA (2001) Saccharomyces cerevisiae CTF18 and CTF4 are required for sister chromatid cohesion. Mol Cell Biol 21: 3144–3158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T (2002) The ABCs of SMC proteins: two-armed ATPases for chromosome condensation, cohesion, and repair. Genes Dev 16: 399–414 [DOI] [PubMed] [Google Scholar]
- Hopfner KP, Craig L, Moncalian G, Zinkel RA, Usui T, Owen BA, Karcher A, Henderson B, Bodmer JL, McMurray CT, Carney JP, Petrini JH, Tainer JA (2002) The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature 418: 562–566 [DOI] [PubMed] [Google Scholar]
- Kawasaki Y, Hiraga S, Sugino A (2000) Interactions between Mcm10p and other replication factors are required for proper initiation and elongation of chromosomal DNA replication in Saccharomyces cerevisiae. Genes Cells 5: 975–989 [DOI] [PubMed] [Google Scholar]
- Kesti T, Flick K, Keranen S, Syvaoja JE, Wittenberg C (1999) DNA polymerase epsilon catalytic domains are dispensable for DNA replication, DNA repair, and cell viability. Mol Cell 3: 679–685 [DOI] [PubMed] [Google Scholar]
- Kolodner RD, Putnam CD, Myung K (2002) Maintenance of genome stability in Saccharomyces cerevisiae. Science 297: 552–557 [DOI] [PubMed] [Google Scholar]
- Krogh BO, Llorente B, Lam A, Symington LS (2005) Mutations in Mre11 phosphoesterase motif I that impair Saccharomyces cerevisiae Mre11-Rad50-Xrs2 complex stability in addition to nuclease activity. Genetics 171: 1561–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert S, Watson A, Sheedy DM, Martin B, Carr AM (2005) Gross chromosomal rearrangements and elevated recombination at an inducible site-specific replication fork barrier. Cell 121: 689–702 [DOI] [PubMed] [Google Scholar]
- Lengronne A, McIntyre J, Katou Y, Kanoh Y, Hopfner KP, Shirahige K, Uhlmann F (2006) Establishment of sister chromatid cohesion at the S. cerevisiae replication fork. Mol Cell 23: 787–799 [DOI] [PubMed] [Google Scholar]
- Lengronne A, Pasero P, Bensimon A, Schwob E (2001) Monitoring S phase progression globally and locally using BrdU incorporation in TK(+) yeast strains. Nucleic Acids Res 29: 1433–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengsfeld BM, Rattray AJ, Bhaskara V, Ghirlando R, Paull TT (2007) Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol Cell 28: 638–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Barlow JH, Burgess RC, Rothstein R (2004) Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118: 699–713 [DOI] [PubMed] [Google Scholar]
- Lobachev KS, Gordenin DA, Resnick MA (2002) The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell 108: 183–193 [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961 [DOI] [PubMed] [Google Scholar]
- Lucca C, Vanoli F, Cotta-Ramusino C, Pellicioli A, Liberi G, Haber J, Foiani M (2004) Checkpoint-mediated control of replisome-fork association and signalling in response to replication pausing. Oncogene 23: 1206–1213 [DOI] [PubMed] [Google Scholar]
- Mayer ML, Gygi SP, Aebersold R, Hieter P (2001) Identification of RFC(Ctf18p, Ctf8p, Dcc1p): an alternative RFC complex required for sister chromatid cohesion in S. cerevisiae. Mol Cell 7: 959–970 [DOI] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R, Nasmyth K (1997) Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91: 35–45 [DOI] [PubMed] [Google Scholar]
- Michalet X, Ekong R, Fougerousse F, Rousseaux S, Schurra C, Hornigold N, van Slegtenhorst M, Wolfe J, Povey S, Beckmann JS, Bensimon A (1997) Dynamic molecular combing: stretching the whole human genome for high-resolution studies. Science 277: 1518–1523 [DOI] [PubMed] [Google Scholar]
- Mirzoeva OK, Petrini JH (2003) DNA replication-dependent nuclear dynamics of the Mre11 complex. Mol Cancer Res 1: 207–218 [PubMed] [Google Scholar]
- Moreau S, Ferguson JR, Symington LS (1999) The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol Cell Biol 19: 556–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung K, Datta A, Kolodner RD (2001) Suppression of spontaneous chromosomal rearrangements by S phase checkpoint functions in Saccharomyces cerevisiae. Cell 104: 397–408 [DOI] [PubMed] [Google Scholar]
- Nairz K, Klein F (1997) mre11S—a yeast mutation that blocks double-strand-break processing and permits nonhomologous synapsis in meiosis. Genes Dev 11: 2272–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada D, Hirano Y, Sugimoto K (2004) Requirement of the Mre11 complex and exonuclease 1 for activation of the Mec1 signaling pathway. Mol Cell Biol 24: 10016–10025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada D, Matsumoto K, Sugimoto K (2003) ATM-related Tel1 associates with double-strand breaks through an Xrs2-dependent mechanism. Genes Dev 17: 1957–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navas TA, Zhou Z, Elledge SJ (1995) DNA polymerase epsilon links the DNA replication machinery to the S phase checkpoint. Cell 80: 29–39 [DOI] [PubMed] [Google Scholar]
- Olson E, Nievera CJ, Lee AY, Chen L, Wu X (2007) The Mre11-Rad50-Nbs1 complex acts both upstream and downstream of ataxia telangiectasia mutated and Rad3-related protein (ATR) to regulate the S-phase checkpoint following UV treatment. J Biol Chem 282: 22939–22952 [DOI] [PubMed] [Google Scholar]
- Osborn AJ, Elledge SJ (2003) Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes Dev 17: 1755–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Ye P, Yuan DS, Wang X, Bader JS, Boeke JD (2006) A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell 124: 1069–1081 [DOI] [PubMed] [Google Scholar]
- Pasero P, Bensimon A, Schwob E (2002) Single-molecule analysis reveals clustering and epigenetic regulation of replication origins at the yeast rDNA locus. Genes Dev 16: 2479–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull TT, Gellert M (1998) The 3′–5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol Cell 1: 969–979 [DOI] [PubMed] [Google Scholar]
- Paull TT, Gellert M (1999) Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev 13: 1276–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robison JG, Elliott J, Dixon K, Oakley GG (2004) Replication protein A and the Mre11.Rad50.Nbs1 complex co-localize and interact at sites of stalled replication forks. J Biol Chem 279: 34802–34810 [DOI] [PubMed] [Google Scholar]
- Santocanale C, Diffley JF (1998) A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature 395: 615–618 [DOI] [PubMed] [Google Scholar]
- Shroff R, Arbel-Eden A, Pilch D, Ira G, Bonner WM, Petrini JH, Haber JE, Lichten M (2004) Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr Biol 14: 1703–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens RV, Maradeo M, Eastman L (2007) Fork it over: the cohesion establishment factor Ctf7p and DNA replication. J Cell Sci 120(Pt 15): 2471–2477 [DOI] [PubMed] [Google Scholar]
- Smith S, Gupta A, Kolodner RD, Myung K (2005) Suppression of gross chromosomal rearrangements by the multiple functions of the Mre11-Rad50-Xrs2 complex in Saccharomyces cerevisiae. DNA Repair (Amst) 4: 606–617 [DOI] [PubMed] [Google Scholar]
- Sullivan M, Hornig NCD, Porstmann T, Uhlmann F (2004) Studies on substrate recognition by the budding yeast separase. J Biol Chem 279: 1191–1196 [DOI] [PubMed] [Google Scholar]
- Tercero JA, Diffley JF (2001) Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 412: 553–557 [DOI] [PubMed] [Google Scholar]
- Tourriere H, Versini G, Cordon-Preciado V, Alabert C, Pasero P (2005) Mrc1 and Tof1 promote replication fork progression and recovery independently of Rad53. Mol Cell 19: 699–706 [DOI] [PubMed] [Google Scholar]
- Trenz K, Smith E, Smith S, Costanzo V (2006) ATM and ATR promote Mre11 dependent restart of collapsed replication forks and prevent accumulation of DNA breaks. EMBO J 25: 1764–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo KM, Sung P (2001) DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50*Mre11 complex. J Biol Chem 276: 35458–35464 [DOI] [PubMed] [Google Scholar]
- Tsukamoto Y, Taggart AK, Zakian VA (2001) The role of the Mre11-Rad50-Xrs2 complex in telomerase-mediated lengthening of Saccharomyces cerevisiae telomeres. Curr Biol 11: 1328–1335 [DOI] [PubMed] [Google Scholar]
- Usui T, Ogawa H, Petrini JH (2001) A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol Cell 7: 1255–1266 [DOI] [PubMed] [Google Scholar]
- Usui T, Ohta T, Oshiumi H, Tomizawa J, Ogawa H, Ogawa T (1998) Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell 95: 705–716 [DOI] [PubMed] [Google Scholar]
- Williams RS, Williams JS, Tainer JA (2007) Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem Cell Biol 85: 509–520 [DOI] [PubMed] [Google Scholar]
- Williams RS, Moncalian G, Williams JS, Yamada Y, Limbo O, Shin DS, Groocock LM, Cahill D, Hitomi C, Guenther G, Moiani D, Carney J, Russell P, Tainer JA (2008) Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell 135: 97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmeyer J, Formosa T (1997) The Saccharomyces cerevisiae DNA polymerase alpha catalytic subunit interacts with Cdc68/Spt16 and with Pob3, a protein similar to an HMG1-like protein. Mol Cell Biol 17: 4178–4190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Ukomadu C, Jha S, Senga T, Dhar SK, Wohlschlegel JA, Nutt LK, Kornbluth S, Dutta A (2007) Mcm10 and And-1/CTF4 recruit DNA polymerase alpha to chromatin for initiation of DNA replication. Genes Dev 21: 2288–2299 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6
Supplementary Table I
Supplemental Data