Abstract
All of the known self-transmissable plasmids of the Archaea have been found in the genus Sulfolobus. To gain more insight into archaeal conjugative processes, four newly isolated self-transmissable plasmids, pKEF9, pHVE14, pARN3 and pARN4, were sequenced and subjected to a comparative sequence analysis with two earlier sequenced plasmids, pNOB8 and pING1. The analyses revealed three conserved and functionally distinct sections in the genomes. Section A is considered to encode the main components of the conjugative apparatus, where two genes show low but significant sequence similarity to sections of genes encoding bacterial conjugative proteins. A putative origin of replication is located in section B, which is highly conserved in sequence and contains several perfect and imperfect direct and inverted repeats. Further downstream, in section C, an operon encoding six to nine smaller proteins is implicated in the initiation and regulation of replication. Each plasmid carries an integrase gene of the type that does not partition on integration, and there is strong evidence for their integration into host chromosomes, where they may facilitate intercellular exchange of chromosomal genes. Two plasmids contain hexameric short regularly spaced repeats (SRSR), which have been implicated in plasmid maintenance, and each plasmid carries multiple recombination motifs, concentrated in the variable regions, which likely provide sites for genomic rearrangements.
Keywords: pARN3, pARN4, pHVE14, pKEF9, SRSR cluster
Introduction
Bacterial conjugation has been studied extensively in proteobacteria and especially in the F-plasmid system of Escherichia coli K12. Several proteins participate in the processing and transport of single-stranded DNA (Dtr), whereas other proteins, carrying transmembrane helices, facilitate mating pair formation (Mpf), which requires pili synthesis (Lanka and Wilkins 1995). Homologs of these proteins are encoded in conjugative plasmids of other bacterial phyla, which suggests that the core conjugative apparatus is widely conserved among bacteria (Pansegrau and Lanka 1996, Christie 2001). The apparatus is also encoded in a range of conjugative transposons, which have the capacity both to transpose intracellularly and to conjugate intercellularly (Salyers et al. 1995).
More recently, it has become clear that alternative conjugative systems exist in some Gram-positive bacteria, where conjugal transfer is initiated without pili by a simpler transfer process (Errington et al. 2001). For example, some Streptomyces plasmids encode a single Tra protein for which neither DNA-processing activity nor site-specific nicking of the plasmid DNA has been detected (Grohmann et al. 2003). This protein shows sequence similarity to the septal DNA translocators SpoIIIE and FtsK, which can both translocate double-stranded DNA (Errington et al. 2001, Grohmann et al. 2003). The possibility that conjugation also involves transfer of double-stranded DNA via cell-to-cell contact is supported by experiments with Streptomyces plasmid pSAM2 (Possoz et al. 2001).
Archaeal self-transmissable plasmids have been found only in diverse strains of the hyperthermophilic genus Sulfolobus, where they occur in about 3% of isolated strains (Prangishvili et al. 1998). Two have already been characterized—pNOB8 from the Japanese strain Sulfolobus NOB8H2 (Schleper et al. 1995, She et al. 1998) and pING1 from Sulfolobus islandicus strain HEN2P2 (Prangishvili et al. 1998, Stedman et al. 2000). Comparative sequence analyses reveal minimal significant sequence similarity between open reading frames (ORFs) of archaeal and bacterial conjugative plasmids. The exceptions are two large archaeal proteins that show significant sequence similarity to limited sections of the bacterial TraG and TrbE proteins that both carry Walker A and B, and other motifs (Walker et al. 1982, She et al. 1998). In bacteria, both proteins are thought to be involved in coordinating the transport of single-stranded DNA through membrane pores (Schröder et al. 2002, Rabel et al. 2003). There is also evidence from electron microscopic studies of conjugating Sulfolobus cultures that extensive cellular contact occurs, suggesting that DNA is transfered directly through cell membranes (Schleper et al. 1995).
Intercellular transfer of chromosomal genes has been demonstrated in Sulfolobus acidocaldarius and likely occurs via conjugation. An archaeal intron encoding a homing enzyme was shown to transfer between cells, inserting into the single chromosomal 23S rRNA gene (Aagaard et al. 1995), and various marker genes have been shown to exchange intercellularly between chromosomes (Grogan 1996, Reilly and Grogan 2001). These phenomena may be facilitated by proteins encoded in a conjugative plasmid encaptured in the S. acidocaldarius genome (Garrett et al. 2004).
To gain further insight into the archaeal conjugative apparatus and the evolution of conjugative processes in general, we isolated and sequenced four self-transmissable plasmids, pKEF9, pHVE14, pARN3 and pARN4, from different S. islandicus strains and subjected them to comparative genome analyses.
Materials and methods
Plasmid preparation and DNA sequencing
Methods for sampling, enrichment and plating of Sulfolobus strains from Iceland were described previously (Zillig et al. 1996, 1998). The self-transmissable plasmids pKEF9, pHVE14, pARN3 and pARN4 were propagated in another host strain, Sulfolobus solfataricus P2, after mixing at a donor:recipient ratio of 1:10,000 (Prangishvili et al. 1998). Plasmid DNA was isolated on Qiagen columns (QIAfilter, Plasmid Maxi Kit, Qiagen, Westburg, Germany) and digested by EcoRI to confirm the plasmid’s identity. Shotgun libraries were prepared in pUC18 from 2 kb fragments of sonicated plasmid DNA. Cloned DNA was isolated in a Biorobot 8000 (Qiagen) and sequenced in MegaBACE 1000 Sequenators (Amersham Biotech, Amersham, U.K.). Plasmid sequences were assembled by Sequencer 3.1.2., and remaining small gaps or ambiguous sequences were resolved by primer walking.
Comparative sequence analyses
Open reading frames containing more than 40 amino acids with ATG, GTG or TTG start codons were found with the program MUTAGEN (Brügger et al. 2003). Searches for sequence matches were made against public databases with BLAST2 (Altschul et al. 1997). Open reading frame comparisons and isoelectric point (pI) determinations were performed with MUTAGEN. Transmembrane regions were identified with TMHMM (www.cbs.dtu.dk/services), TMpred (www.ch.embnet.org) and Drawhca (Callebaut et al. 1997). Signal peptide sequences were predicted with SignalP (www.cbs.dtu.dk/services). Open reading frame sequences were also checked for conserved motifs and protein family relationships (http://pfam.wustl.edu/ and http://www.ncbi.nlm.nih.gov/COG/new/). Amino acid repeat sequences were identified with RADAR (Heger and Holm 2000), and α-helices were predicted with GOR IV (Garnier et al. 1996). Direct and inverted repeats of nucleotide sequences were detected with DNA Strider (Marck 1988). Sequences were aligned with T-coffee (Notredame et al. 2000).
Results
Genome sequences
The self-transmissable plasmids, pKEF9, pHVE14, pARN3 and pARN4, were derived from single colonies obtained from enrichment cultures sampled from Iceland (Zillig et al. 1996, 1998). Copy numbers in their natural hosts were generally low and, therefore, the plasmids were propagated in S. solfataricus P2 to produce higher yields (Prangishvili et al. 1998, Stedman et al. 2000). Restriction fragment patterns in agarose gels corresponded to those reported earlier for pKEF9, pARN3 and pARN4 (Prangishvili et al. 1998). The plasmid pHVE14 was isolated from the same enrichment culture as pHVE14/5 (Prangishvili et al. 1998), but produced a different restriction fragment pattern when digested by EcoRI (data not shown).
The complete sequence of each plasmid was determined at about a fourfold genome coverage. Sequence ambiguities were resolved by primer walking on plasmid DNA or clones thereof. The plasmid sizes correspond to the approximate sizes predicted from restriction fragment patterns (Prangishvili 1998). General properties of the plasmid genomes and their sequence accession numbers are listed in Table 1.
Table 1.
Sulfolobus self-transmissable plasmids used in this study. Abbreviations: SRSR = short regularly spaced repeat; and ORF = open reading frame.
| Plasmid 1 | G+C content (%) | Size (bp) | HexamerSRSR cluster | Recombination motifs | Predicted ORFs | Accession no |
| pKEF9 | 37.1 | 28,930 | 1 | 12 | 40 | AJ748321 |
| pHVE14 | 37.4 | 35,422 | 0 | 11 | 54 | AJ748324 |
| pARN3 | 36.3 | 26,200 | 0 | 10 | 40 | AJ748322 |
| pARN4 | 37.1 | 26,476 | 0 | 11 | 36 | AJ748323 |
| pING1 | 37.3 | 24,554 | 0 | 8 | 35 | NC004852 |
| pNOB8-33 | 37.3 | 33,345 | 1 | 20 | 41 | AJ010405 |
1 pING1 (Stedmanet al. 2000) and pNOB8-33 (She et al. 1998) were published earlier.
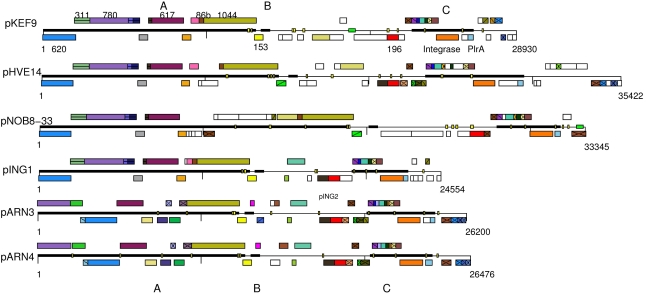
Gene maps of each plasmid were generated with MUTAGEN (Brügger et al. 2003) and each ORF was checked for matches against public sequence databases. The gene maps of pKEF9, pHVE14, pARN3 and pARN4 are aligned with the earlier sequenced pNOB8 and pING1, and homologous genes are color-coded (Figure 1). Plasmids pKEF9 and pHVE14 are closest to pNOB8 and pING1 in gene organization and sequence, whereas pARN3 and pARN4 form a separate group. Conserved ORFs show 50 to 80% amino acid sequence identity within each group, and 15 to 64% sequence identity between the groups. They are defined forthwith as the pKEF and pARN groups.
Figure 1.
Comparative gene map of the Sulfolobus self-transmissable plasmids. Thick, black horizontal lines delineate the genomic sections A, B and C. The main conserved open reading frames (ORFs) in section A, the highly conserved ORF153 and ORF196, the integrase and PlrA are labeled for pKEF9. Homologs in the different plasmids can be identified by color and pattern. Open reading frames shown as an empty box have no homologs in the other plasmids. The short regularly spaced repeat (SRSR) clusters are indicated by green rectangular boxes on the linear genomes of pKEF9 and pNOB8-33. Recombination motifs are represented by yellow boxes on the linear genomes. The location of the deletion in pNOB8-33 is downstream from ORF617. The deletion variant plasmid pING2 extends from downstream of ORF1044 to the center of the operon in section C.
Fifteen homologous ORFs are shared among the six self-transmissable plasmids (Table 2). The locations of the larger ORFs are labeled on the pKEF9 genome in Figure 1 and their homologs in the other plasmids can be identified from the color codes. The conserved ORFs are concentrated within the genomic sections A, B and C, which appear to be functionally distinct (Figure 1). Section A carries genes that are implicated in conjugation, section B contains a putative replication origin, and section C includes an operon with six to nine short genes, some of which are involved in the initiation of plasmid replication, an integrase and the DNA binding protein, PlrA. The latter protein is conserved in all published sequences of Sulfolobus plasmids and may have an important regulatory role (Figure 1). Between sections B and C and downstream from section C, the genomes are more variable in gene content and sequence (Figure 1).
Table 2.
Genes conserved in each of the Sulfolobus self-transmissable plasmids. Abbreviations: ORF = open reading frame; aa = amino acids; pI = isoelectric point; and TMH = transmembrane helix.
| Section | ORF 1 | Size range (aa) | Identity (%) | pI | TMH motifs | Homology |
| A | 620 | 618–624 | 25–89 | 5–7.5 | 2 | TrbE |
| A | 311 | 222–312 | 15–83 | 9–10 | 0–1 | |
| A | 780 | 660–780 | 19–88 | 5–7 | 1 | Signal peptide protein |
| A | 617 | 507–622 | 21–89 | 5–10 | 10–12 | |
| A | 86b/138 | 86/138 | 17–68 | ~9 | 3 | |
| A | 1044 | 1025–1044 | 33–91 | 8–9 | 2 | TraG |
| 153 | 153–166 | 17–100 | ~10 | |||
| 196 | 192–253 | 48–100 | ~10 | |||
| C | 102 | 102–108 | 25–100 | 4.5–7.5 | ||
| C | 106 | 99–165 | 64–99 | ~5 | RepA | |
| C | 62 | 61–72 | 52–96 | ~5 | CopG | |
| C | 87 | 87–92 | 51–89 | ~10 | Leucine zipper | |
| C | 98 | 83–101 | 20–100 | ~10 | ||
| 419 | 419–458 | 41–93 | ~10 | 4 pING1/0 others | Integrase | |
| 77 | 73–99 | 46–96 | 10–11 | plrA | ||
1 ORF numbers are from the pKEF9 genome. ORF153 is 100% conserved in pKEF9, pING1, pARN3 and pARN4.
Conjugative proteins
Section A comprises up to half of each genome (~13.5 kb) and contains six conserved ORFs. The order and sequence of the genes are highly conserved in the pKEF plasmid group, and the genes are arranged in putative operons transcribed in the same direction, except for ORF620, which is a single gene that is transcribed in the opposite direction (Figure 1). In the pARN plasmids, the order and sequences of the genes are more varied. All of the conserved and some less conserved ORFs in section A are predicted to form transmembrane helix (TMH) motifs. For example, ORF617 carries 10 to 12 TMH motifs throughout its length, whereas the others have one to three (Table 2).
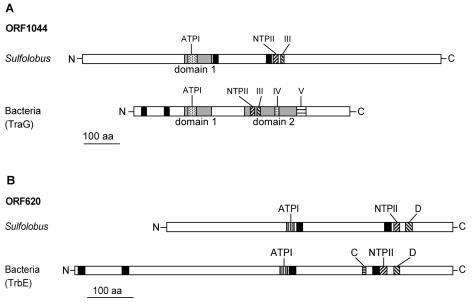
ORF1044 of pKEF9 exhibits sequence motifs and domain structures characteristic of the smaller (500–700 amino acids) bacterial TraG proteins that participate in conjugation and Type IV secretion systems (Balzer et al. 1994). The archaeal protein carries domain 1 with a Walker A motif for ATP binding, and part of domain 2, which includes both the Walker B motif for NTP binding and motif III, but lacks motifs IV and V (Figure 2A). Moreover, two TMHs lie between the two Walker A and B motifs in the archaeal protein, whereas they precede the Walker A and B motifs in bacterial TraG proteins (Schröder et al. 2002). The sequence identity is 20–25% in the domain–motif regions (Figure 2A), whereas the sequence alignments outside of the motif regions show < 10% identity and are considered insignificant. Moreover, the 300 to 400 amino acids in the C-terminal region of ORF1044 is archaea-specific (Figure 2A).
Figure 2.
Comparison of motif patterns in ORF1044 and ORF620 of Sulfolobus and the bacterial TraG and TrbE homologs, respectively. The conserved domains 1 and 2, and the motifs III, IV and V are labeled. ATPI indicates a Walker A motif and NTPII represents a Walker B motif. Black boxes denote putative transmembrane spanning helices. (A) ORF1044. Conserved domains and motifs were identified by alignment of the Sulfolobus ORFs with 19 different bacterial TraG homologs (Schröder et al. 2002). (B) ORF620. Conserved motifs were identified by alignment with 19 different TrbE homologs of the VirB4 family (Rabel et al. 2003).
ORF620 also carries a series of motifs found in the larger (~800 amino acids) bacterial TrbE proteins, including Walker A and B motifs and two membrane-spanning segments (Figure 2B). The TrbE proteins belong to the VirB4 superfamily and, like TraG, are required for DNA transfer and the Type IV secretion apparatus (Rabel et al. 2003). ORF620 shows low sequence similarity to bacterial TrbE proteins outside of the motif regions and lacks the N-terminal region carrying TMHs (Figure 2B).
ORF617 is the only protein that can generate multiple TMHs, 10 in proteins of the pKEF group and 12 in proteins of the pARN group. For pKEF proteins, these putative helices are concentrated in the N-terminal two thirds of the protein, whereas the C-terminal region is quite basic (pI 11.5), suggesting a DNA binding function. In pARN plasmids, the ORF is smaller (507–509 amino acids) and the TMHs occur throughout the protein. The presence of multiple TMHs is consistent with the protein being involved in transmembrane pore formation. In the pKEF plasmid group, the ORF immediately upstream from ORF617 (Figure 1) carries three putative TMHs. Two are located in the 60 amino acids of the C-terminal region and show 32/64% sequence identity/similarity to the N-terminal region of ORF617, suggesting that there is partial gene duplication.
ORF311 is rich in charged amino acids (35 to 41%), is basic (pI 9–10.5) and contains an imperfect direct repeat of 50–70 amino acids that is predicted to be α-helical. Proteins of the pKEF group also contain a putative TMH near the N-terminus.
ORF780 exhibits a putative signal peptide sequence and shows 20–36% sequence identity over approximately 550 amino acids with the signal peptide protein, SSO1053, of S. solfataricus (She et al. 2001, Albers and Driessen 2002). Like other secreted proteins of Sulfolobus, it is rich in asparagine (10%) and tyrosine (11%).
Section A proteins of self-transmissible plasmids that are highly conserved likely generate the core conjugative apparatus in Sulfolobus.
Putative origin of replication
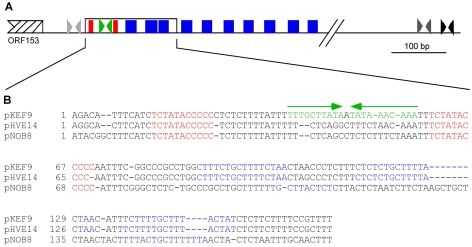
Section B, which follows section A and ORF153, is a highly conserved region (Figure 1). About 170 bp at the beginning of this section exhibits a sequence identity of > 95% for the five Icelandic plasmids and about 60% identity with pNOB8. It contains several direct and inverted repeat sequences (Figure 3A) including the 10(11) bp perfect direct repeat sequence, TCTATACCCC(C), separated by a 34–35 bp A+T-rich region, which corresponds to three helical turns and would position the repeats on the same side of the DNA helix. The region also contains three larger imperfect direct repeats, YYYTY TGCTTT(T)RCTAA (Figure 3B). Additional copies of the large repeat are also located downstream from this region, although they are not all positionally conserved in each plasmid (Figure 3A).
Figure 3.
(A) Putative replication origin. Red boxes = 10–11 bp perfect direct repeats; blue boxes = 16 bp imperfect direct repeats; dark grey arrows = inverted repeats conserved in all plasmids; green arrows = inverted repeats conserved in pKEF9, pING1, pARN3 and pARN4; and light grey arrows = inverted repeats conserved in pHVE14 and pNOB8. Black arrows = plasmid-specific inverted repeats. (B) The 170 bp conserved region. The 10–11 bp repeats are shown in red and 16 bp imperfect repeats are shown in blue. The total number of these imperfect repeats is: pKEF9 = 13, pHVE14 = 10; pNOB8 = 6; pING1 = 12; pARN3 = 9; and pARN4 = 8. Inverted repeats are shown in green.
The inverted repeats in this region include the 10 bp TTTGcTTATA lying within the 34–35 bp A+T-rich region of pKEF9, pING1, pARN3 and pARN4 (Figure 3A). Inverted repeats are also located 39 bp upstream of the 170 bp conserved region in pNOB8 and pHVE14, an 8 bp repeat TATaCTTA in pNOB8 and a 13 bp repeat TTACTAaTAACCT in pHVE14 (Figure 3A), which exhibit no common sequence (positions denoted by lower case letters are non-conserved). Several inverted repeats are located 500 – 800 bp downstream of the conserved 170 bp region (Figure 3A). One of these, TAA(G)GG GC-3–4 bp-GCCC(C)TTA, occurs in each plasmid.
The 170 bp region described is the only one where the nucleotide sequence is highly conserved in all the self-transmissable plasmids, including the small pING1 derivatives, pING2 and pING3, and contains multiple direct and inverted repeat sequences that are characteristic of replication origins of bacterial plasmids (del Solar et al. 1998). Therefore, this region is a strong candidate for a replication origin. This inference is supported by G+C skew analyses (Tillier and Collins 2000), which produce a large trough at this position for each plasmid (data not shown). No similarities were detected with the repeat sequences in the chromosomal replication origins of S. solfataricus.
Plasmid replication and maintenance
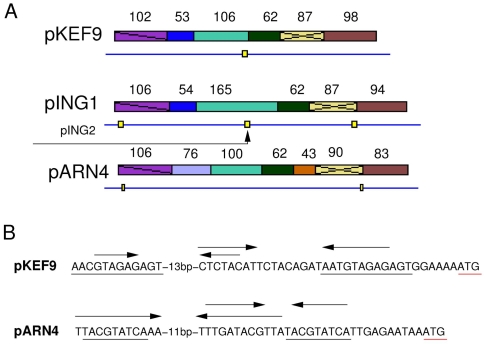
An operon containing six to nine short genes is located in section C (Figure 1). For most adjacent genes, stop and start codons overlap at the sequence ATGA, such that consecutive ORFs change reading frames. Five of the genes, including the first and last two, occur in the same order in each plasmid (Figure 4). The plasmids carry between one and four additional ORFs, some of which are also present in other plasmids (Figures 1 and 4). In general, the ORFs are rich in leucines (11–12.7%) and charged amino acids (36–39%), mainly glutamic acid and lysines. Database searches provide evidence implicating two of the proteins in DNA replication.
Figure 4.
(A) Putative replication operon from pKEF9, pING1 and pARN4. Homologous open reading frames (ORFs) are shown in identical colors and the ORF sizes are given. Positions of the recombination motifs are indicated by yellow boxes on the linear genomes. The region of the operon present in the deletion derivative pING2 is mapped below pING1. (B) Promoter regions for the regulatory site of the putative CopG homolog on pKEF9 and pARN4 are aligned. The start codon of the first gene in the operon is underlined in red. Inverted repeats are marked by arrows. Direct repeats are underlined.
ORF106, the most conserved ORF in the operon, shows the best database match, over positions 18–102 (32/53% identity/similarity), to the replication initiator protein, RepA, of Haemophilus ducrey and a similar match to RepA of plasmid pRUM of Enterococcus faecium. The matching region of ORF106 is bordered by a putative recombination motif and no database matches were found for the downstream part of the ORF (Figure 4A).
The highly conserved ORF87 exhibits a leucine zipper motif from positions 27 to 55, with five leucines each interspaced by six amino acids, that could facilitate protein–protein interactions in a RepA-like protein complex. Such motifs are located near the N-terminus of many bacterial RepA proteins (del Solar et al. 1998).
ORF62 shows a significant sequence match to the CopG protein family and we infer that it may regulate expression of the operon. This is supported by the observation that each operon contains at least one set of inverted repeat sequences in its promoter region, which resembles sequences preceding a RepA homolog encoded in the Sulfolobus cryptic plasmid pRN1 that has been shown to be regulated by a CopG homolog (Lipps et al. 2001a) (Figure 4B). One set of inverted repeats borders the box A promoter motif and another is located between the box A motif and the start codon.
Two genes located downstream from the operon are conserved in each plasmid. One gene encodes an integrase, of the non-partitioning type, which probably facilitates chromosomal integration (She et al. 2002). This is consistent with the observed presence of copies of a pNOB8-like plasmid in the chromosome of Sulfolobus tokodaii and of a pARN-type plasmid in the chromosome of S. acidocaldarius (Garrett et al. 2004).
ORF80, downstream from the integrase gene, encodes a PlrA protein that generates an atypical leucine zipper motif and binds to a conserved sequence within its own promoter (Lipps et al. 2001b). However, the conserved promoter sequence of the pRN family plasmids was absent. It is conserved in all known Sulfolobus plasmids and is thought to have an important regulatory role (Garrett et al. 2004).
Short regularly spaced repeat clusters
pKEF9 contains a cluster of six SRSRs (Figure 5), which is similar to that detected earlier in pNOB8 (She et al. 1998), but is absent from the other plasmids (Table 1, Figure 1). For pNOB8, it was considered to be a possible parS element involved, together with the encoded ParA and ParB-like homologs, in copy number control (She et al. 1998). Although pKEF9 does not carry the partial parA and parB homologs, the SRSR motif may still play a role in plasmid maintenance because, in our study, pKEF9 and pNOB8 were the most stably maintained of the six plasmids in foreign Sulfolobus hosts.
Figure 5.
Alignment of short regularly spaced repeat (SRSR) sequences of pKEF9 (red). Conserved sequence positions are indicated. The genome nucleotide positions of the SRSR clusters are: pKEF9 = 19,067–18,720; and pNOB8-33 = 32,855–33,199.
Variable regions and putative recombination sites
The genomic regions between sections B and C, and downstream from section C, are more variable in size, gene content and sequence (Figure 1). They are also relatively rich in the putative recombination motifs (Table 1), which may contribute to their variability. The latter correspond closely to sequences similar to the interrupted inverted repeat TAAACTGGGGAGTTTA, which can generate perfect hairpin structures. This motif is involved in rearrangements of pING plasmids (Stedman et al. 2000), and similar “hairpin” motifs provide sites for rearrangements in the Sulfolobus pRN family plasmids (Peng et al. 2000).The locations of the motifs are indicated on the genome maps (Figures 1 and 4A), and only a few are conserved in position across different genomes. For example, the position of the motif in ORF165 of pING1 is conserved in pKEF9, pHVE14 and pNOB8, but not in the pARN plasmids (Figure 4A). Moreover, identical motifs are located in the C-terminal part of ORF1044 in each plasmid (Figure 1), bordering amino acid positions 900 to 970. The region between these sites shows little sequence similarity, whereas the region downstream of the second motif is identical in pKEF9, pING1, pARN3 and pARN4.
Discussion
Although we still know little about the mechanism of conjugation in archaea, we have delineated a conserved plasmid region considered to encode the main proteins involved with conjugation (section A). It is striking how few proteins are involved, in marked contrast to the conjugative systems of the proteobacteria, which require large multicomponent protein complexes. No proteins are encoded that are homologous to the bacterial relaxosome complex that generates a single-stranded copy of plasmid DNA prior to transfer, nor were homologous genes detected in the chromosome of the conjugating host S. solfataricus (She et al. 2001). This is consistent with experimental evidence showing that the bacterial relaxosome interacts with the C-terminal region of the TraG protein (Gomis-Rüth et al. 2001), which is absent from the partial Sulfolobus homolog (Figure 2). Therefore, we infer that a simpler conjugative mechanism operates in Sulfolobus, probably involving transfer of double-stranded DNA.
No proteins were detected that are homologous to proteins of the bacterial Dtr and Mpf apparatus other than TrbE and TraG. The latter belong to a large protein family of the Type IV secretion apparatus, which is involved in intercellular transfer of proteins (Christie 2001, Grohmann et al. 2003). They are also membrane-associated and carry domain structures including Walker A and B motifs. Moreover, they facilitate single-stranded DNA transport during proteobacterial conjugation by linking the Dtr complex with the Mpf apparatus (Schröder et al. 2002, Rabel et al. 2003). Because significant sequence similarity between Sulfolobus and the proteobacterial TrbE and TraG proteins is limited to the regions around the Walker A and B motifs (Figure 2), it is likely that the two Sulfolobus proteins act as energizers in a novel DNA transfer process. Electron micrographs suggest that this occurs after extensive cell-to-cell contact and not through pili interactions (Schleper et al. 1995).
ORF617, the only protein carrying several transmembrane helical segments (10–12 TMH), may contribute to a membrane pore through the ether-linked lipid membrane, possibly by mimicking the single membrane spanning protein that facilitates DNA transfer in some smaller self-transmissable plasmids of Gram-positive bacteria (Possoz et al. 2001), although there is no significant sequence similarity.
pING2, a deletion derivative of pING4, is not self-transmissable, but can replicate and be mobilized in the presence of pING1 (Stedman et al. 2000). (The possibility that pING2 integrates into the larger plasmid prior to conjugation, and is excised afterwards, has not been excluded (Prangishvili et al. 1998).) After transfer to another cell, it replicates initially at much higher copy numbers than pING1, but is then gradually lost from the cell culture during continuous growth (Stedman et al. 2000). These properties are consistent with pING2 lacking the conjugative apparatus encoded in section A, but containing the origin of replication in section B, and the putative repA homolog (ORF165) in section C (Figures 1 and 4A). Moreover, the initial high copy number of the pING2 derivative is consistent with the deletion of the gene encoding CopG which is predicted to regulate expression of the repA homolog (Figures 4A and 4B). Moreover, the impaired maintenance of pING2, relative to pING1, may reflect the absence of other ORFs from the operon in site C (Figure 4A).
The SRSR clusters found in pKEF9 and pNOB8 (Figure 1) are also present in archaeal chromosomes where, generally, they exist in multiple clusters and are larger, often containing over 100 repeats (She et al. 2001). The working hypothesis for their function is that they are involved in chromosomal segregation by analogy to eukaryotic centromere elements (Charlebois et al. 1998). To date, a protein has been isolated from S. solfataricus P2 that binds specifically both to the pNOB8 SRSR repeats and to the closely similar SRSR clusters in the S. solfataricus P2 chromosome, producing an opening of the DNA structure at the center of each repeat (Peng et al. 2003). It has also been demonstrated that the chromosomal SRSR clusters can generate long RNA transcripts (Tang et al. 2002). However, although the clusters are likely to be involved in plasmid maintenance and chromosome segregation, the mechanism by which this occurs remains to be determined.
The putative recombination motifs that exhibit the consensus sequence TAAACTGGGGAGTTTA and can generate perfect hairpin structures are likely involved with plasmid rearrangements. Two of these motifs flank a region of pING4 and recombine in vivo to yield the small derivative plasmid pING2 (Stedman et al. 2000) (Figure 4A). Similar sequence motifs also occur in the cryptic Sulfolobus plasmids of the pRN family, where they flank a region of variable length and are likely to provide recombination sites for plasmid changes (Peng et al. 2000). Motifs are also conserved in pNOB8 and in the pNOB8-type plasmid integrated into the S. tokodaii genome, suggesting they may also facilitate rearrangements in Sulfolobus chromosomes (Garrett et al. 2004).
These results provide an overview of self-transmissable plasmids of Sulfolobus and of the protein components essential for conjugation and plasmid replication. The archaeal conjugative apparatus appears to be much simpler than that of most known Gram-negative bacterial systems. There is a superficial similarity to systems operating in some Gram-positive bacteria with respect to the occurrence of direct cell-to-cell contact prior to DNA transfer and the possibility of double-stranded DNA transfer, as has been proposed for some self-transmissable plasmids of Streptomyces (Possoz et al. 2001, Errington et al. 2001). Expression and functional analyses of the key protein components identified in this work should lead to a more detailed understanding of this important process in the archaeal domain of life.
Acknowledgments
This work was supported by a grant to B.G. from the Danish Technical Science Research Council and by an Archaeal Centre Grant to R.A.G. from the Danish Natural Science Research Council. K.B. received a Ph.D. grant from Copenhagen University.
References
- R1.Aagaard C., Dalgaard J.Z., Garrett R.A. Inter-cellular mobility and homing of an archaeal rDNA intron confers selective advantage over intron-cells of Sulfolobus acidocaldarius . Proc. Natl. Acad. Sci. USA. 1995;92:12285–12289. doi: 10.1073/pnas.92.26.12285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R2.Albers S.V., Driessen A.M. Signal peptides of secreted proteins of the archaeon Sulfolobus solfataricus: a genomic survey. Arch. Microbiol. 2002;177:209–216. doi: 10.1007/s00203-001-0386-y. [DOI] [PubMed] [Google Scholar]
- R3.Altschul S., Madden T., Schaffer A., Zhang J., Zhang Z., Miller W., Lipman D. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;2:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R4.Balzer D., Pansegrau W., Lanka E. Essential motifs of relaxase (TraI) and TraG proteins involved in conjugative transfer of plasmid RP4. J. Bacteriol. . 1994;176:4285–4295. doi: 10.1128/jb.176.14.4285-4295.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R5.Brügger K., Redder P., Skovgaard M. MUTAGEN: Multi-user tool for annotating genomes. Bioinformatics. 2003;19:2480–2481. doi: 10.1093/bioinformatics/btg336. [DOI] [PubMed] [Google Scholar]
- R6.Callebaut I., Labesse G., Durand P., Poupon A., Canard L., Chomilier J., Mornon J.P. Deciphering protein sequence information through hydrophobic cluster analysis (HCA): current status and perspectives. Cell. Mol. Life Sci. 1997;53:621–645. doi: 10.1007/s000180050082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R7.Charlebois R.L., She Q., Sprott D.P., Sensen C.W. , Garrett R.A. Sulfolobus genome: from genomics to biology. Curr. Opin. Microbiol. 1998;1:584–588. doi: 10.1016/s1369-5274(98)80093-3. [DOI] [PubMed] [Google Scholar]
- R8.Christie P.J. Type IV secretion: intercellular transfer of macromoleules by systems ancestrally related to conjugation machines. Mol. Microbiol. 2001;40:294–305. doi: 10.1046/j.1365-2958.2001.02302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R9.del Solar G., Giraldo R., Ruiz-Echevarria M.J., Espinosa M., Diaz-Orejas R. Replication and control of circular bacterial plasmids. Microbiol. Mol. Biol. Rev. 1998;62:434–464. doi: 10.1128/mmbr.62.2.434-464.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R10.Errington J., Bath J., Wu L.J. DNA transport in bacteria. Nat. Rev. Mol. Cell Biol. 2001;2:538–545. doi: 10.1038/35080005. [DOI] [PubMed] [Google Scholar]
- R11.Garnier J., Gibrat J.F., Robson B. GOR method for predicting protein secondary structure from amino acid sequence. Methods Enzymol. 1996;266:540–553. doi: 10.1016/s0076-6879(96)66034-0. [DOI] [PubMed] [Google Scholar]
- R12.Garrett R.A., Redder P., Greve B., Brügger K., Chen L., She Q. Archaeal plasmids. In: Funnell B.E., Phillips G.J., editors. Plasmid Biology. Washington: A.S.M. Press; 2004. pp. 377–392. [Google Scholar]
- R13.Gomis-Rüth F.X., Moncalian G., Perez-Luque R., Gonzalez A., Cabezon E., De La Cruz F., Coll M. The bacterial conjugation protein TrwB resembles ring helicases and F1-ATPase. Nature. 2001;409:637–641. doi: 10.1038/35054586. [DOI] [PubMed] [Google Scholar]
- R14.Grogan D.W. Exchange of genetic markers at extremely high temperatures in the archaeon Sulfolobus acidocaldarius . J. Bacteriol. 1996;178:3207–3211. doi: 10.1128/jb.178.11.3207-3211.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R15.Grohmann E., Muth G., Espinosa M. Conjugative plasmid transfer in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 2003;67:277–301. doi: 10.1128/MMBR.67.2.277-301.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R16.Heger A., Holm L. Rapid automatic detection and alignment of repeats in protein sequences. Proteins. 2000;41:224–237. doi: 10.1002/1097-0134(20001101)41:2<224::aid-prot70>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- R17.Lanka E., Wilkins B. DNA processing reactions in bacterial conjugation. Annu. Rev. Biochem. 1995;64:141–169. doi: 10.1146/annurev.bi.64.070195.001041. [DOI] [PubMed] [Google Scholar]
- R18.Lipps G., Stegert M., Krauss G. Thermostable and site-specific DNA binding of the gene product ORF56 from the Sulfolobus islandicus plasmid pRN1, a putative archaeal plasmid copy control protein. Nucleic Acids Res. 2001;29:904–913. doi: 10.1093/nar/29.4.904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R19.Lipps G., Ibanez P., Stroessenreuther T., Hekimian K., Krauss G. The protein ORF80 from the acidophilic and thermophilic archaeon Sulfolobus islandicus binds highly site-specifically to double-stranded DNA and represents a novel type of basic leucine zipper protein. Nucleic Acids Res. 2001;29:4973–4982. doi: 10.1093/nar/29.24.4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R20.Marck C. ‘DNA Strider’: a ‘C’ program for the fast analysis of DNA and protein sequences on the Apple Macintosh family of computers. Nucleic Acids Res. 1988;16:1829–1836. doi: 10.1093/nar/16.5.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R21.Notredame C., Higgins D.G., Heringa J. T-coffee: a novel method for fast and accurate multiple sequence alignments. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- R22.Pansegrau W., Lanka E. Enzymology of DNA transfer by conjugative mechanisms. Prog. Nucleic Acid Res. Mol. Biol. 1996;54:197–251. doi: 10.1016/s0079-6603(08)60364-5. [DOI] [PubMed] [Google Scholar]
- R23.Peng X., Holz I., Zillig W., Garrett R.A., She Q. Evolution of the family of pRN plasmids and their integrase-mediated insertion into the chromosome of the crenarchaeon Sulfolobus solfataricus . J. Mol. Biol. 2000;303:449–454. doi: 10.1006/jmbi.2000.4160. [DOI] [PubMed] [Google Scholar]
- R24.Peng X., Brügger K., Shen B., Chen L., She Q., Garrett R.A. Genus-specific protein binding to the large clusters of DNA repeats (short regularly spaced repeats) present in Sulfolobus genomes. J. Bacteriol. 2003;185:2410–2417. doi: 10.1128/JB.185.8.2410-2417.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R25.Possoz C., Ribard C., Cagnat J., Guéreau M. The integrative element pSAM2 from Streptomyces: kinetics and mode of conjugal transfer. Mol. Microbiol. 2001;42:159–166. doi: 10.1046/j.1365-2958.2001.02618.x. [DOI] [PubMed] [Google Scholar]
- R26.Prangishvili D., Albers S.-V., Holz I., et al. Conjugation in archaea: frequent occurrence of conjugative plasmids in Sulfolobus . Plasmid. 1998;40:190–202. doi: 10.1006/plas.1998.1363. [DOI] [PubMed] [Google Scholar]
- R27.Rabel C., Grahn A.M., Lurz R., Lanka E. The VirB4 family of proposed traffic nucleoside triphosphatases: common motifs in plasmid RP4 TrbE are essential for conjugation and phage adsorption. J. Bacteriol. 2003;185:1045–1058. doi: 10.1128/JB.185.3.1045-1058.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R28.Reilly M.S., Grogan D.W. Characterization of intragenic recombination in a hyperthermophilic archaeon via conjugational DNA exchange. J. Bacteriol. 2001;183:2943–2946. doi: 10.1128/JB.183.9.2943-2946.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R29.Salyers A.A., Shoemaker N.B., Stevens A.M., Li L.Y. Conjugative transposons: an unusual and diverse set of integrated gene transfer elements. Microbiol. Rev. 1995;59:579–590. doi: 10.1128/mr.59.4.579-590.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R30.Schleper C., Holz I., Janekovic D., Murphy J., Zillig W. A multicopy plasmid of the extremely thermophilic archaeonSulfolobus effects its transfer to recipients by mating. J. Bacteriol. 1995;177:4417–4426. doi: 10.1128/jb.177.15.4417-4426.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R31.Schröder G., Krause S., Zechner E.L., Traxler B., Yeo H.-J., Lurz R., Waksman G., Lanka E. TraG-like proteins of DNA transfer systems and of Helicobacter pylori type IV secretion system: inner membrane gate for exported substrates? J. Bacteriol. 2002;184:2767–2779. doi: 10.1128/JB.184.10.2767-2779.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R32.She Q., Phan H., Garrett R.A., Albers S.V., Stedman K.M., Zillig W. Genetic profile of pNOB8 from Sulfolobus: the first conjugative plasmid from an archaeon. Extremophiles. 1998;2:417–425. doi: 10.1007/s007920050087. [DOI] [PubMed] [Google Scholar]
- R33.She Q., Singh R.K., Confalonieri F., et al. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. USA. 2001;98:7835–7840. doi: 10.1073/pnas.141222098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R34.She Q., Brügger K., Chen L. Archaeal integrative genetic elements and their impact on genome evolution. Res. Microbiol. 2002;153:325–332. doi: 10.1016/s0923-2508(02)01331-1. [DOI] [PubMed] [Google Scholar]
- R35.Stedman K.M., She Q., Phan H., Holz I., Singh H., Prangishvili D., Garrett R.A., Zillig W. The pING family of conjugative plasmids from the extremely thermophilic archaeon Sulfolobus islandicus: insights into recombination and conjugation in crenarchaeota. J. Bacteriol. 2000;182:7014–7020. doi: 10.1128/jb.182.24.7014-7020.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R36.Tang T.-H., Bachellerie J.-P., Rozhdestvensky T., et al. Identification of 86 candidates for small non-messenger RNAs from the archaeonArchaeoglobus fulgidus . Proc. Natl. Acad. Sci. USA. 2002;99:7536–7541. doi: 10.1073/pnas.112047299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R37.Tillier E.R., Collins R.A. The contributions of replication orientation, gene direction, and signal sequences to base-composition asymmetries in bacterial genomes. J. Mol. Evol. . 2000;50:249–257. doi: 10.1007/s002399910029. [DOI] [PubMed] [Google Scholar]
- R38.Walker J.E., Saraste M., Runswick M.J., Gay N.J. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R39.Zillig W., Prangishvili D., Schleper C., Elferink M., Holz I., Albers S., Janekovic D., Gotz D. Viruses, plasmids and other genetic elements of thermophilic and hyperthermophilic archaea. FEMS Microbiol. Rev. 1996;18:225–236. doi: 10.1111/j.1574-6976.1996.tb00239.x. [DOI] [PubMed] [Google Scholar]
- R40.Zillig W., Arnold H.P., Holz I., et al. Genetic elements in the extremely thermophilic archaeon Sulfolobus . Extremophiles. 1998;2:131–140. doi: 10.1007/s007920050052. [DOI] [PubMed] [Google Scholar]