Abstract
Classification and regression tree (CART) analysis was applied to genome-wide tetranucleotide frequencies (genomic signatures) of 195 archaea and bacteria. Although genomic signatures have typically been used to classify evolutionary divergence, in this study, convergent evolution was the focus. Temperature optima for most of the organisms examined could be distinguished by CART analyses of tetranucleotide frequencies. This suggests that pervasive (nonlinear) qualities of genomes may reflect certain environmental conditions (such as temperature) in which those genomes evolved. The predominant use of GAGA and AGGA as the discriminating tetramers in CART models suggests that purine-loading and codon biases of thermophiles may explain some of the results.
Keywords: bioinformatics, convergence, decision tree, extremophile, genomic signature, hyperthermophile, purine-loading, tetranucleotide frequencies, thermophile, virtual coding strand
Introduction
It was Erwin Chargaff who first noticed the similarity in the ratios of adenine and thymine and of cytosine and guanine in DNA (Chargaff et al. 1949). Chargaff sustained a lifelong interest in the complexities of genomes, in many cases speculating far ahead of the development of methods and instrumentation necessary to test his ideas. For example, in his Essays on Nucleic Acids, Chargaff (1963) predicted the significance of frequencies of oligonucleotide motifs to perform linguistic analyses of DNA “texts,” noting that “a great deal can be learned about an unknown language through a study of its phonemes, their frequency, distribution density and allophonic relationships.”
A paucity of DNA sequences and constraints on computational methods limited the pace of research on motif frequencies through the 1980s. Microbiologists were aware of the importance of GC ratios in the characterization of archaea and bacteria, but Chargaff’s initiative was not followed up until the 1990s when Samuel Karlin and others began to apply computational analyses to a growing suite of completely sequenced genomes (Karlin et al. 1994, Fertil et al. 2005, Paz et al. 2006, Vasilevskaya et al. 2006).
In 1992, with only partially sequenced bacterial, archaeal and eukaryotic genomes, as well as some completed virus genomes, Karlin and colleagues explored the relative abundances of di-, tri- and tetra-nucleotides (Burge et al. 1992). Karlin et al. (1994) focused on dinucleotide relative abundances, which, they considered, constituted a “robust” genome signature. In Karlin and Ladunga (1994) and Karlin and Burge (1995), “genomic signature” was more formally defined as a basis for discriminating among genomes.
Seven complete microbial genomes and a number of partial genomes were available in 1997 for genomic signature analysis using short oligonucleotides, which, as noted by Karlin et al. (1997), may overcome some of the problems of inter-genome comparisons by alignment techniques. Because of the close correlation that Karlin et al. found between di-, tri and tetranucleotides, most subsequent studies have been confined to the study of dinucleotide frequencies only. However, some authors (Pride et al. 2003, Teeling et al. 2004, Fertil et al. 2005) have investigated tetramer frequencies on the assumption that their study yields an advantageous trade-off between accuracy and speed of analysis.
Since the initial work of Karlin and colleagues, numerous studies have confirmed the efficacy of a non-alignment, genomic signature approach to genome analysis. For example, van Passel et al. (2006a) examined the dinucleotides of 334 bacterial and archaeal genomes and noted a congruency between genomic signatures and 16S RNA sequences. Confirming previous results (e.g., Karlin and Burge 1995, Pride et al. 2003), these authors concluded that the genomic signature constitutes a distinctive phylogenetic signal and may better reflect evolutionary relationships than single gene comparisons. Confidence in the technique resulted in the suggestion by van Passel et al. (2006a) that, based on genomic signatures, certain enteric gamma proteobacterial species be combined, whereas other species, such as Buchnera aphidocola, could be split.
On a large scale, genomic signature analyses are relatively unaffected by the heterogeneities that result from horizontal gene transfers. Yet, the genome-wide pervasiveness of genomic signatures (Jernigan and Baran 2002) allows intragenomic analysis in search of local heterogeneities, thus giving rise to the possibility of identifying horizontal transfers (Karlin 2001, Lio 2002, Dufraigne et al. 2005). The property of pervasiveness may be the reason that studies of genomic frequencies based only on partial genomes are so promising as it allows for the analysis and classification of plasmids (van Passel et al. 2006b) and short sequences, such as genome fragments of 30,000–40,000 bp (Teeling et al. 2004), 5000 bp (Woyke et al. 2006), and even as small as 400 bp (Sandberg et al. 2001).
Genomic signatures as potential indicators of convergent evolution of sequences
Genomic signature analysis seems applicable in classifying instances of both divergent and convergent evolution. Several genomic signature analyses, such as that of Foerstner et al. (2005), have addressed convergent evolution of some sequences, resulting from selection driven by the same environmental conditions, independent of phylogeny. Such selective pressures include physical constraints on the functions of the DNA molecule, as well as the structures of the proteins encoded. Thermophilic archaea and bacteria have been investigated for evidence of genome convergence under the influence of extreme temperatures. Consideration of this question dates back to biochemical studies of AT/CG ratios and dinucleotide compositions in the 1960–1970s (reviewed in Karlin and Burge 1995).
Doolittle (1994) noted that sequence convergence (a sort of “molecular mimicry”) can be difficult to distinguish from horizontal transfers and from statistically insignificant short matches. However, whole-genome signature analyses may identify horizontal transfers as heterogeneities within the genome (Karlin 2001, Lio 2002, Dufraigne et al. 2005).
Campbell et al. (1999) compared signatures of five thermophilic archaea with signatures of 22 bacteria (mostly proteobacteria and Gram positives). In contrast to other studies, they concluded that thermophily in archaeal species was not evident from the genomic signatures. However, subsequent studies, including this one, suggest that, on the contrary, there are genome-wide signatures for thermophilic and hyperthermophilic microorganisms.
Trends in amino acid compositions of proteins have some genome-wide influence on codons and, therefore, some effect on a genomic signature. Amino acid use by thermophiles includes the replacement of polar non-charged amino acids with charged amino acids such as lysine, arginine, aspartic acid and glutamic acid (Cambillau and Claverie 2000, Suhre and Claverie 2003). A similar analysis of two psychrophilic archaea (Saunders et al. 2003) showed a bias for polar non-charged amino acids. Carbone et al. (2005) (and others reviewed in Carbone et al. 2005) determined that a “codon bias signature” separated thermophiles from mesophiles in a set of 16 archaea and 80 bacteria.
Karlin et al. (1994) and Karlin and Burge (1995) speculated that environmental influences such as pH, temperature and salinity might influence dinucleotide genomic signatures. In a study of seven complete and several partial genomes, Karlin et al. (1997) noted that the three thermophiles had significantly lower proportions of the dinucleotide CG. Kawashima et al. (2000) and Suhre and Claverie (2003) concluded that the “dinucleotide statistical index,” computed from dinucleotide frequencies, showed more pure pyrimidine dinucleotides (TC combinations) and more pure purine dinucleotides (AG combinations) in hyperthermophiles. An in vitro investigation by Xia et al. (2002) of Pasteurella multocida cultivated for approximately 14,400 generations at 45 °C (i.e., above the optimum temperature of 37 °C) resulted in a decrease of GC% and an increase of TA, TT and AA dimers. That both coding and non-coding sequences show genomic signatures (e.g., Karlin and Burge 1995, Karlin and Mrazek 1996, Campbell et al. 1999) supports hypotheses of sequence convergence. Convergent sequence signatures in non-coding regions may reflect similar environmental pressures on fundamental DNA activities such as replication and repair.
Archaea versus bacteria and hyperthermophily versus mesophily?
Currently, most hyperthermophiles for which the genome has been completely sequenced are archaeons. Therefore, it is important to consider whether divergence between thermophiles and mesophiles reflects phylogeny or temperature preference. For example, Carbone et al. (2005) found signature differences in hyperthermophiles and thermophiles, although the initial goal of the study had been to separate archaea (all of which, in the dataset they used, were hyperthermophiles) from bacteria. Graham et al. (2000) used coding sequences of nine archaea to find signature sets of genes, some pertaining to unique archaeal functions and metabolisms such as methanogenesis. However, all of the organisms in the study were hyperthermophiles, a factor which might account, at least in part, for the genes found to be in common. Fadiel et al. (2003) probed for repeats of at least 25 bp in archaea and found “remarkable” signatures in non-coding regions. However, the investigated set of seven archaea consisted entirely of hyperthermophiles, whereas the comparison set of six bacteria consisted solely of mesophiles. It is uncertain, therefore, whether the 25 bp signatures identified are characteristic solely of archaea or hyperthermophiles, or both.
Classification and regression tree analyses indexing pervasive genomic signatures
We investigated genomic signatures by classification and regression tree (CART) analysis (Breiman et al. 1984), a powerful method for developing a classification scheme, categorizing an organismic characteristic on the basis of any number of classifying (predictor) variables. Some studies have used linear discriminant analysis (Carbone et al. 2005) to classify microbes based on codon biases. Other techniques have been used to classify fragments of DNA according to nucleotide composition. These include machine learning methods such as the self-organizing map (SOM) (e.g., Kohonen 1990, Abe et al. 2003, 2005, 2006) and the support-vector-machine (SVM) (e.g., Tsirigos and Rigoutsos 2005, McHardy et al. 2007). In other studies, e.g., Lin et al. 2003, CART has allowed efficient use of large collections of classifying variables to identify nonlinear relationships among the classifiers, yielding a simple sequential set of classification rules.
Tree-based methods such as CART have been applied in mining large datasets, such as microarrays, to detect discriminating factors for classification (Boulesteix et al. 2003). The CART method (Hermanek and Guggenmoos-Holzmann 1994, Masic et al. 1998) has also been used to winnow phenotypes, symptoms and prognoses for diagnostic characteristics with which to create decision trees to aid in medical diagnoses. As such, CART is a method of classifying organisms based on DNA sequences.
Genomic signature analysis is based on a significantly different paradigm (Karlin et al. 1997) to that of alignment methods, including synteny, and it therefore requires different methods of comparison. It is the pervasive, linguistic quality of genomic signatures (unlike sequence alignment) that lends itself to classification schemes such as CART. The versatility of genomic signatures, their ability to classify either convergent groups or divergent groups, to be unaffected by horizontal transfers on a global scale and yet to identify horizontal transfers on a local level, is the reason that signatures are effectively indexed by CART analysis.
Genomic sequences have most oftern been catalogued phylogenetically, and the datasets are typically of genes, excluding non-coding regions. Much work remains to be done with other sorts of cataloguing systems. Genomic signatures as analyzed by CART afford such an alternative. In this study, we found CART to be useful in building classification schemes relating tetramer (or other oligomer) frequencies to characteristics of interest. The method is successful in revealing short lists of discriminating tetramers, the frequencies of which distinguished three temperature ranges, hyperthermophily, thermophily and mesophily. Short decision trees were generated and shown to be effective predictors of the temperature preferences of known organisms external to the training data used to generate the trees.
Materials and methods
Sequence collection
One hundred and ninety-five fully sequenced microbial genomes (24 archaeal and 171 bacterial species), protein tables and associated annotations were downloaded from NCBI GenBank (http://www.ncbi.nlm.nih.gov/genomes/lproks.cgi on April 1, 2006). Only one strain of any organism was included in the set, and organisms not identified to species level were excluded. Of the 195 genomes meeting our criteria, 16 are classified by NCBI as hyperthermophiles, of which 12 are archaeons, 14 are thermophiles, of which five are archaeons, and 165 are mesophiles, of which seven are archaeons (Supplementary Table S1).
Producing a virtual coding strand for each genome
A “virtual coding strand” for each genome was produced from the positive strand. A virtual coding strand is comprised of the entire positive strand, including all coding and non-coding regions. It allows a more precisely controlled experimental system by eliminating the extra variable of gene orientation. To produce the virtual coding strand each incidence of a gene positioned on the complementary strand (as indicated by the associated protein table) was placed in the “correct” (sense) 5′ to 3′ orientation by replacing the “reversed” (antisense) gene with its reverse complement. Coding regions with overlapping start-stop locations were handled independently, and each region was concatenated onto the virtual coding strand. Any “corrected” genes (reversed or overlapping) were flanked by delimiting Ns at both ends to eliminate tetramer counts across these virtual intergenic boundaries. In cases where organisms have more than one chromosome, the chromosomes were concatenated, separated by an ‘N’. Thus a complete “sense” strand, including all coding and non-coding regions with all genes in 5′ to 3′ order, was constructed and used for subsequent steps in the analysis.
Collecting overlapping tetramers (and other oligomers)
Counts of tetramers across each virtual coding strand were recorded by moving a four-base window through the strand one base at a time. Only tetramers containing A, C, G and T were counted. Dimers, trimers and pentamers were also collected and analyzed to determine the optimal size for the subsequent CART analyses.
Randomized control sequences
Two variations of random sequences were constructed and used as controls for testing the external validity of each decision tree. First, for each of the 195 organisms, independent draws from a multinomial distribution, with probabilities set to the empirical proportions of A, C, G, T and N within that organism’s sequence, were made at each nucleotide site to generate a simulated genome of the same length as the original genome. This is analogous to rolling a five-sided die for each nucleotide to produce a pseudorandom strand with approximately the same nucleotide ratio as the original organism. Second, a first-order Markov chain model was used. Here, conditional on the value of the previous nucleotide, one of five different multinomial distributions were used to generate the simulated strand. Figuratively, we have five different five-sided dice, one each for the A, C, G, T or N that occupies the position of the previous nucleotide. If the previous nucleotide is an A, then the computer selects the five-sided die corresponding to A and “rolls it” to generate the current nucleotide, where the proportions of A, C, G, T and N on A’s die are the empirical proportions of AA, AC, AG, AT, and AN on the original genome. In this way, a pseudorandom strand is created with a first-order Markov model using the original genome’s empirical, one-step transition probabilities (e.g., Ewens and Grant 2001).
Classification and regression trees
Classification and regression trees are decision trees for determining a set of logical conditions that provide classification of cases based on the values of a set of classifier variables. One advantage of classification trees is that they are non-parametric, assuming neither a linear, nor even a continuous, nonlinear relationship between the classifiers and the dependent variable. An important advantage is their simplicity. Classification trees often yield simpler models than their parametric counterparts, as well as straightforward classification of new cases. In this paper, archaeal and bacterial genomes are classified on the basis of the intra-genomic relative frequencies of tetramers. We use the freely available rpart implementation of CART (Breiman et al. 1984) in the R statistical package Development Core Team (http://www.R-project.org). All trees were generated so that any node with nine or fewer observations was left as a leaf (i.e., not split) and each leaf contained at least three observations.
To begin, the intra-genomic tetranucleotide or tetramer frequencies of all 195 genomes were used to build a CART model classifying the genomes according to temperature range: hyperthermophile versus non-hyperthermophile (thermophile and mesophile) and mesophile versus non-mesophile (hyperthermophile and thermophile). The validity of these CART models for predicting temperature ranges of genomes omitted from the model was then tested by leaving out one genome at a time and building a decision tree using the other 194 genomes. The omitted genome was then classified with the resulting model. The process of predicting the omitted genome, hereafter referred to as “leave-one-out,” was repeated for all of the 195 genomes. As a third test of the external predictive validity of the CART models, 50 randomly selected genomes were set aside and a classification tree was built using the remaining 145 genomes. The temperature ranges for each of the 50 omitted genomes was then predicted. Building a model with 145 genomes and predicting 50 randomly selected genomes (hereafter referred to as "50-randomly-selected-genomes") was repeated over 40 iterations.
Additional controls
Four additional controls were included to highlight the limits and versatility of CART analyses. First, the intra-genomic tetramer frequencies of all 195 original genomes were used to build a CART model classifying genomes according to temperature range, which was used to classify the temperature range of the randomly generated genomes (both multinomial distribution and first-order Markov). Second, the temperature ranges for all 195 original genomes were randomly permuted and reassigned to all 195 organisms. Then using all 195 organisms with randomly reassigned temperature ranges, a full leave-one-out analysis was performed counting the number of temperature range misclassifications. This process of randomly permuting the temperature ranges and performing a full leave-one-out analysis was carried out 100 times, computing the average number of temperature range misclassifications over all 100 iterations. Third, we used the GC ratios of each genome (Table 1) as the sole classifier in a CART analysis to classify microbial temperature regimes. Finally, the intra-genomic relative frequencies of tetramers were used to discriminate archaea from bacteria to demonstrate the versatility of this technique as applied to a pervasive genomic signature.
Table 1.
The NCBI-reported temperature optima and GC% of 16 hyperthermophilic (12 archaea and 4 bacteria) and 14 thermophilic (5 archaea and 9 bacteria) species. An additional 165 mesophilic (7 archaea, 158 bacteria) species were also included in our analysis (data not shown). Leave-one-out mispredictions shows the type of misprediction when classifying the temperature optimum of an organism (hyperthermphile versus non-hyperthermophile (hyper) or mesophile versus non-mesophile (meso)) based on a model from the genomes of the other 194 organisms, using tetramer frequencies as the sole classifier.
| Temperature optima (°C) | Kingdom | Species | GC% | Leave-one-out mispredictions |
| Hyperthermophilic | ||||
| 103 | Archaea | Pyrococcus abyssi | 42.0 | |
| 100 | Archaea | Pyrobaculum aerophilum | 52.0 | |
| 100 | Archaea | Pyrococcus furiosus | 42.0 | |
| 98 | Archaea | Methanopyrus kandleri | 60.0 | Hyper |
| 98 | Archaea | Pyrococcus horikoshii | 42.0 | |
| 96 | Archaea | Aquifex aeolicus | 43.0 | |
| 93 | Archaea | Aeropyrum pernix | 67.0 | |
| 90 | Archaea | Nanoarchaeum equitans | 31.6 | Hyper and meso |
| 85 | Archaea | Methanococcus jannaschii | 31.3 | |
| 85 | Archaea | Sulfolobus solfataricus | 35.8 | |
| 85 | Archaea | Thermococcus kodakaraensis KOD1 | 52.0 | |
| 83 | Archaea | Archaeoglobus fulgidus | 46.0 | |
| 80 | Archaea | Sulfolobus tokodaii | 32.8 | Hyper |
| 80 | Bacteria | Thermotoga maritima | 45.0 | Hyper |
| 78 | Bacteria | Carboxydothermus hydrogenoformans | 42.0 | Hyper and meso |
| 75 | Bacteria | Thermoanaerobacter tengcongensis | 37.6 | Meso |
| Thermophilic | ||||
| 72 | Archaea | Sulfolobus acidocaldarius DSM 639 | 36.7 | Hyper |
| 68 | Bacteria | Thermus thermophilus HB27 | 69.4 | |
| 65 | Archaea | Methanobacterium thermoautotrophicum | 49.5 | |
| 60 | Bacteria | Geobacillus kaustophilus HTA426 | 52.0 | Meso |
| 60 | Archaea | Picrophilus torridus DSM 9790 | 36.0 | Meso |
| 60 | Bacteria | Symbiobacterium thermophilum IAM14863 | 68.7 | |
| 60 | Archaea | Thermoplasma volcanium | 50.0 | |
| 59 | Archaea | Thermoplasma acidophilum | 50.0 | |
| 58 | Bacteria | Moorella thermoacetica ATCC 39073 | 55.8 | |
| 55 | Bacteria | Thermosynechococcus elongatus | 53.9 | Meso |
| 52 | Bacteria | Thermobifida fusca YX | 67.5 | |
| 48 | Bacteria | Chlorobium tepidum TLS | 56.0 | Meso |
| 45 | Bacteria | Methylococcus capsulatus Bath | 63.6 | Meso |
| 45 | Bacteria | Streptococcus thermophilus CNRZ1066 | 40.0 | Meso |
Results and discussion
CART with genomic signatures: an accurate discrimination of temperature classifications
There were relatively few misclassifications from the CART models and from tests of those models. As discussed below, many misclassifications may represent inconsistent reporting of temperature optima rather than errors in the CART analyses. This parallels the experience of van Passel et al. (2006a), who speculated that some bacteria apparently misclassified in their study may actually have been correctly classified by their method, but incorrectly classified in the literature. Here, the criteria for classifying organisms by temperature optima are those of the latest edition of The Prokaryotes (Dworkin 1999): hyperthermophile > 80 °C, thermophile 60–80 °C, mesophile 15–60 °C and psychrophile < 15 °C.
The classification tree of hyperthermophiles versus non-hyperthermophiles (the set of thermophiles and mesophiles) was almost error-free. Two of 16 hyperthermophiles, Carboxydothermus hydrogenoformans and Thermotoga maritima, appeared at first to be misclassified by a tree designed to distinguish them from non-hyperthermophiles (Figure 1); however, these species may not be true hyperthermophiles. According to the NCBI database, C. hydrogenoformans with a temperature optimum of 78 °C is a hyperthermophile; whereas, by Dworkin’s (1999) definition, it should be considered a thermophile, as confirmed in this CART analysis. Thermotoga maritima is listed with a temperature optimum of 80 °C, which places it on the boundary between temperature classifications. All 179 non-hyperthermophiles were correctly classified (Figure 1).
Figure 1.
Classification tree of hyperthermophile (hyper) versus non-hyperthermophile (non-hyper) based on tetramer relative frequencies of 195 genomes. Each split shows the tetramer selected by CART for each level of classification and the relative frequencies for each direction. Nodes show the classified temperature range and number of organisms classified in each temperature category according to NCBI (non-hyper/hyper). Organisms misclassified in this model are listed under the appropriate nodes.
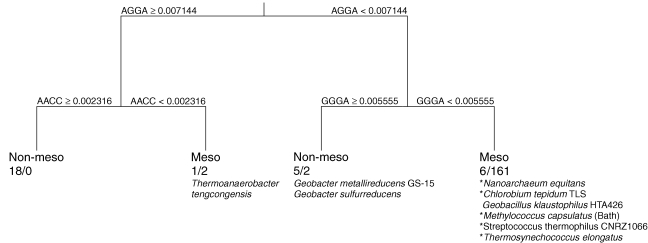
The classification tree based on the tetramer frequencies over the virtual coding strands of all 195 genomes for mesophiles versus non-mesophiles (the set of hyperthermophiles and thermophiles) resulted in a total of nine misclassifications: only two out of 165 mesophiles, Geobacter metallireducens and G. sulfurreducens were misclassified (Figure 2). Seven out of 30 non-mesophiles were misclassified. This included the hyperthermophile Nanoarchaeum equitans, an exceptionally small organism with a tiny genome and an extreme AT bias of 32%. Neoarchaeum equitans is the only known parasitic or symbiotic archaeon with another archaeon (Ignicoccus) as a host and is the sole member of its own phylum.
Figure 2.
Classification tree of mesophile (meso) versus non-mesophile (non-meso) based on tetramer relative frequencies of 195 genomes. Each split shows the tetramer selected by CART for each level of classification and the relative frequencies for each direction. Nodes show the classified temperature range and number of organisms classified in each temperature category according to NCBI (non-meso/meso). Organisms misclassified in this model are listed under the appropriate nodes. Asterisks (*) indicate organisms that may not be misclassified given current understanding of temperature optima (see Discussion).
Five of the organisms classified in the NCBI database as hyperthermophiles or thermophiles were classified as mesophiles on the mesophile versus non-mesophile tree (see asterisks (*) in Figure 2). However, with the exception of Geobacillus kaustophilus, their reported temperature optima of 45 °C (Methylococcus capsulatus and Streptococcus thermophilus), 48 °C (Chlorobium tepidum) and 55 °C (Thermosynechococcus elongatus) fall well below the temperature range for thermophily.
A leave-one-out analysis was performed to test the external validity of these models. Misclassified organisms and their temperature optima are indicated in Table 1. In many cases, the CART analysis revealed temperature optima that are inconsistent with the NCBI-reported temperature classifications. In the 195 tests of models for hyperthermophiles versus non-hyperthermophiles, only eight organisms were misclassified, including three incorrectly classified in the original models discussed above, namely, Carboxydothermus hydrogenoformans and Thermatoga maritima, which may not be true hyperthermophiles, and Nanoarchaeum equitans, an atypical archaeon. The other five apparently misclassified organisms included the hyperthermophile Sulfolobus tokadaii with a borderline temperature optimum of 80°C. It also included Sulfolobus acidocaldarius with a temperature optimum of 70–75 °C according to the NCBI database, but of 80 °C according to Chen et al. (2005), indicating that its status remains uncertain (Table 1).
In the leave-one-out testing of 195 mesophile versus non-mesophile models, there were 14 misclassifications. However, on close scrutiny, several more ambiguities in temperature classifications became apparent including N. equitans, which may be a true outlier in any dataset. In addition, Picrophilus torridus, with a borderline optimum of 60 °C, may be misclassified as a thermophile.
When testing the predictive ability of the CART 50-randomly-selected-genomes model, an average of 2.6 out of 50 genomes (5.2%) were misclassified on the hyperthermophile versus non-hyperthermophile trees. An average of 5.1 out of 50 genomes (10.2%) were misclassified on the mesophile versus non-mesophile trees.
The total number of misclassifications when building models using all the genomes (hereafter referred to as the “model-with-all”) and the total number of missed predictions in the leave-one-out and 50-randomly-selected-genomes analyses are shown in Table 2. For the leave-one-out analysis, there were eight misclassifications in the hyperthermophiles versus non-hyperthermophile grouping (five hyperthermophiles, one thermophile and two mesophiles) and 14 misclassifications in the mesophile versus non-mesophile grouping (three hyperthermophiles, six thermophiles and five mesophiles).
Table 2.
Number of misclassifications for two temperature optimum ranges, hyperthermophiles versus non-hyperthermophiles (hyper vs. non-hyper) and mesophiles versus non-mesophiles (meso vs. non-meso), using all tetramer frequencies over the entire virtual coding strand. Model-with-all is the number of misclassifications when building a model using all 195 genomes. Leave-one-out is the number of mispredictions when classifying an organism against a model built using the other 194 genomes. 50-randomly-selected-genomes is the average number (over 40 iterations) of misclassifications when building a model on 145 genomes and attempting to classify the other 50 randomly selected genomes.
| Temperature range | Model-with-all | Leave-one-out | 50-randomly-selected-genomes |
| Hyper vs. non-hyper | 2 (1%) | 8 (4.1%) | 2.6 (5.2%) |
| Meso vs. non-meso | 9 (4.6%) | 14 (7.2%) | 5.1 (10.2%) |
When genome sequences were randomized by a single multinomial at each nucleotide, the classifications had at least twice the number of misclassifications as the original experiments (Table 3). Misclassifications in genomes generated by a first order Markov method were much closer to the original experiments. In the hyperthermophile versus non-hyperthermophile model, the error rate was identical, although different organisms were misclassified. This suggests that a considerable amount of information exists in the dimer frequencies that comprise the tetramers, a hypothesis that was confirmed by experiments with dimers, which yielded slightly less accurate predictions than tetramers (see Supplementary Table S2).
Table 3.
Number of leave-one-out mispredictions when classifying an organism into two temperature optimum ranges, hyperthermophile versus non-hyperthermophile (hyper vs. non-hyper) and mesophile versus non-mesophile (meso vs. non-meso) using all tetramers frequencies over the entire vitrual coding strand, based on a model from the genomes of the other 194 organisms. The leave-one-out classifications were made for the 195 original genomes (original) and two types of randomly generated control sequences, multinomial (multi) and Markov.
| Temperature range | Original | Random control | |
| Multi | Markov | ||
| Hyper vs. non-hyper | 8 (4.1%) | 16 (8.2%) | 8 (4.1%) |
| Meso vs. non-meso | 14 (7.2%) | 59 (30.3%) | 21 (10.8%) |
When temperature ranges for the 195 genomes were permuted and randomly reassigned to the 195 organisms, the resulting models in the leave-one-out analyses yielded a substantial increase in the misclassification rate. When differentiating between hyperthermophiles and non-hyperthermophiles, the average misclassification rate was 14.4% (28.1 of 195 over 100 iterations). This yields about the same misclassification rate one would expect when ignoring all concomitant information and blindly classifying organisms with probabilities equal to their relative frequency in the sample. In this case, randomly classifying an organism as non-hyperthermophile with probability 0.918 (179/195) or a hyperthermophile with probability 0.082 (16/195) yields an expected misclassification rate of 16.4%, 8.2% in the two categories, respectively. Similarly, permuting the temperature ranges and randomly reassigning them to the 195 organisms yields an average misclassification rate of 27% (52.8 of 195 over 100 iterations), whereas random classification based solely on relative frequency yields an expected misclassification rate of 30.8%. In both cases, the misclassification rates are substantially higher than achieved by the original leave-one-out CART analysis. For hyperthermophile versus non-hyperthermophile, the misclassification rate more than tripled (4.1% using the classification tree, 14.4% using random assignment) and for mesophile versus non-mesophile, the misclassification rate nearly quadrupled (7.2% using the classification tree, 27% using random assignment). The GC% was not an accurate predictor of temperature optimum. This is confirmed by the poor performance (similar to “Multinomial” in Table 3) of CART in differentiating temperature optima (Table 4). However, genomic signatures have proved useful in showing divergent relationships. As expected, a CART analysis of tetramers performed well in separating archaea from bacteria. The model-with-all had 5 (2.6%) misclassifications all of which were archaea. Whereas the leave-one-out prediction missed 15 (7.7%): 9 archaea and 6 bacteria. Using the CART analysis, about 21% (5/24) of archaea were misclassified and no bacteria (0/171) were misclassified. Using a random classification scheme based on the relative frequencies of archaea and bacteria in the sample, we would expect about 87.7% (21/24) of archaea to be misclassified and about 12.3% (21/171) of bacteria to be misclassified.
Table 4.
Number of organisms misclassified using GC% to model and predict temperature optimum range. Model-with-all is the number of misclassifications when building a model using all 195 genomes. Leave-one-out is the number of mispredictions when classifying an organism against a model built using the other 194 genomes. Models predicted the temperature optimum range of organisms: hyperthermophile versus non-hyperthermophile (hyper vs. non-hyper), and mesophile versus non-mesophile (meso vs. non-meso).
| Temperature range | Model-with-all | Leave-one-out |
| Hyper vs. non-hyper | 15 (7.7%) | 23 (11.8%) |
| Meso vs. non-meso | 27 (13.8%) | 42 (21.5%) |
Tetramers were chosen as the classifier after parallel sets of experiments run with dimers, trimers and pentamers confirmed a slightly higher correct overall classification using tetramers (Table S2). In addition, for each motif size of length L = {2, 3, 4, 5}, each genome was cross-validated using a model built with the other 194 genomes. The number of tested organisms that were incorrectly classified was recorded for each experimental classification (hyperthermophile versus non-hyperthermophile and mesophile versus non-mesophile; Table S2). Further, a tree was built based on both trimers and tetramers, simultaneously, as potential classifiers, yielding results similar to those from a tree fitted with tetramers alone. In particular, GAGA continued to be chosen as the best first-decision classifier, even when trimers were considered with tetramers. Based on these experimental trials, we found that tetramers provided the most effective classification scheme.
There were just a few discriminating tetramers in nearly every one of the 195 CART models generated for each of the temperature comparisons. The predominant tetramer in the models of hyperthermophile versus non-hyperthermophile was GAGA, used as the initial split parameter in 192 out of 195 trees; the remaining three models used the tetramer AGAG. In all cases, a higher proportion of these tetramers was associated with the hyperthermophilic classification. In the models of mesophiles versus non-mesophiles, AGGA was the primary split parameter in 193 cases; the remaining two were split on GAGA. In this case, a higher proportion of these tetramers was associated with the non-mesophilic classification. The most commonly used tetramers in secondary splits were ATCA (for the hyperthermophile versus non-hyperthermophile) and GGGA and AACC (for mesophile versus non-mesophile). As noted by other authors (e.g., Lao and Forsdyke 2000, Lambros et al. 2003, and Paz et al. 2004), the coding regions of many thermophilic genomes are purine-loaded and possess codon biases that reflect that loading. Our method of using a “virtual coding strand” eliminated the variable of gene orientation. This may accentuate the signal from such codon biases in nucleotide composition by insuring that all genic regions (and therefore all codons) are counted in the sense direction.
Conclusion
Genomic signatures, as represented by intra-genomic relative frequencies of tetramers, yielded effective discriminators of temperature optima in archaea and bacteria when analyzed by CART. The implication of these results extends beyond the examples examined in this study. CART analysis may be used to explore a variety of hypotheses about genomic sequences. There may be practical applications including the identifications of temperature optima of partial genomes such as in some metagenomic analyses of communities. Furthermore, this approach may yield information about the phylogenies of high temperature organisms and may suggest explorations of other pressures on genomic convergences such as high salt conditions. Since CART uses a few signature oligonucleotides to construct trees, it may lead to identification of particular traits associated with those nucleotides. The method may be seen as an in silico complement to genomic signature tags (Dunn et al. 2002) by which genomes are sampled for a subset of fragments reflecting the whole. CART queries may also reveal overrepresented sequences of some functional significance as in binding sites of non-coding regions.
Supplementary material
Supplementary Table S1. A list of the 195 organisms of which the genomes were used in this study (24 archaea and 171 bacteria). The complete genome sequences were downloaded from GenBank (http://www.ncbi.nlm.nih.gov/genomes/lproks.cgi) on April 1, 2006.
Supplementary Table S2. Classification and regression tree analysis results using four oligonucleotide frequency lengths over the entire virtual coding strand to classify organisms into two temperature optimum ranges, hyperthermophile versus non-hyperthermophile (hyper vs. non-hyper) and mesophile versus non-mesophile (meso vs. non-meso). Tetramers provide the most effective classification scheme considering both models built. Model-with-all is the number of misclassifications when building a model using all 195 genomes. Leave-one-out is the number of mispredictions when classifying an organism against a model built using the other 194 genomes.
Acknowledgments
We thank Robert Obar, Harvard Medical School, and Shawn McCafferty, Biology Department, Wheaton College, for their insights on portions of this work and Greg Williams for his database expertise. We thank the reviewers for their valuable input to the manuscript. Partial support was provided by the National Science Foundation’s Course, Curriculum and Laboratory Improvement program (CCLI-EMD) under Grant No. 0340761 and Susanne Woods and Molly Easo Smith of the Wheaton College Provost Office.
References
- R1.Abe T., Kanaya S., Kinouchi M., Ichiba Y., Kozuki T., Ikemura T. Genome Res. Vol. 13. Cold Spring Harbor Lab; 2003. Informatics for unveiling hidden genome signatures; pp. 693–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R2.Abe T., Sugawara H., Kinouchi S., Kanaya S., Ikemura T. Novel phylogenetic studies of genomic sequence fragments derived from uncultured microbe mixtures in environmental and clinical samples. DNA Res. 2005;12:281–290. doi: 10.1093/dnares/dsi015. [DOI] [PubMed] [Google Scholar]
- R3.Abe T., Sugawara H., Kinouchi M., Ikemura T. Self-Organizing Map (SOM) unveils and visualizes hidden sequence characteristics of a wide range of eukaryote genomes. Gene. 2006;365:27–34. doi: 10.1016/j.gene.2005.09.040. [DOI] [PubMed] [Google Scholar]
- R4.Boulesteix A.L., Tutz G., Strimmer K. Bioinformatics. 18. Vol. 19. Oxford Univ Press; 2003. A CART-based approach to discover emerging patterns in microarray data; pp. 2465–2472. [DOI] [PubMed] [Google Scholar]
- R5.Breiman L., Friedman J.H., Olshen R.A., Stone C.J. New York: Chapman and Hall; 1984. Classification and regression trees.368 [Google Scholar]
- R6.Burge C., Campbell A.M., Karlin S. Proc. Natl. Acad. Sci. USA. 4. Vol. 89. National Acad Sciences; 1992. Over- and under-representation of short oligonucleotides in DNA sequences; pp. 1358–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R7.Cambillau C., Claverie J.M. J. Biol. Chem. Vol. 275. ASBMB; 2000. Structural and genomic correlates of hyperthermostability; pp. 32383–32386. [DOI] [PubMed] [Google Scholar]
- R8.Campbell A., Mrazek J., Karlin S. Proc. Natl. Acad. Sci. USA. Vol. 96. USA: 1999. Genome signature comparisons among prokaryote, plasmid and mitochondrial DNA; pp. 9184–9189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R9.Carbone A., Kepes F., Zinovyev A. Codon bias signatures, organization of microorganisms in codon space and lifestyle. Mol. Biol. Evol. 2005;22:547–561. doi: 10.1093/molbev/msi040. [DOI] [PubMed] [Google Scholar]
- R10.Chargaff E. New York: Elsevier; 1963. Essays on Nucleic Acids.211 [Google Scholar]
- R11.Chargaff E., Vischer E., Doniger R., Green C., Misani F. The composition of desoxypentose nucleic acids of thymus and spleen. J. Biol. Chem. 1949;177:405–416. [PubMed] [Google Scholar]
- R12.Chen L., Brugger K., Skovgaard M., et al. The Genome of Sulfolobus acidocaldarius, a model organism of the Crenarchaeota. J. Bacteriol. 2005;187:4992–4999. doi: 10.1128/JB.187.14.4992-4999.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R13.Doolittle R.F. Convergent evolution: the need to be explicit. Trends Biochem. Sci. 1994;19:15–18. doi: 10.1016/0968-0004(94)90167-8. [DOI] [PubMed] [Google Scholar]
- R14.Dufraigne C., Fertil B., Lespinats S., Giron A., Deschavanne P. Detection and characterization of horizontal transfers in prokaryotes using genomic signature. Nucleic Acids Res. 2005;33:1–14. doi: 10.1093/nar/gni004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R15.Dunn J., McCorkle S., Praissman L., Hind G., van der Lelie D., Bahou W., Gnatenko D., Krause M. Genomic signature tags (GSTs): a system for profiling genomic DNA. Genome Res. 2002;12:1756–1765. doi: 10.1101/gr.306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R16.Dworkin M. New York: Springer-Verlag; 1999. The Prokaryotes: an evolving electronic resource for the microbiological community.4770 [Google Scholar]
- R17.Ewens W.J., Grant G.R. New York: Springer-Verlag; 2001. Statistical methods in bioinformatics: an introduction; pp. 303–310. [Google Scholar]
- R18.Fadiel A., Lithwick S., Ganji G., Scherer S.W. Remarkable sequence signatures in archaeal genome. Archaea. 2003;1:185–190. doi: 10.1155/2003/458235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R19.Fertil B., Massin M., Lespinats S., Devic C., Dumee P., Giron A. GENSTYLE: exploration and analysis of DNA sequences with genomic signature. Nucleic Acids Res. 2005;33:W512–W515. doi: 10.1093/nar/gki489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R20.Foerstner K.U., von Mering C., Hooper S.D., Bork P. Environments shape the nucleotide composition of genomes. EMBO Rep. 2005;6:1208–1213. doi: 10.1038/sj.embor.7400538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R21.Graham D.E., Overbeek R., Olsen G.J., Woese C.R. An archaeal genome signature. Proc. Natl. Acad. Sci. USA. 2000;97:3304–3308. doi: 10.1073/pnas.050564797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R22.Hermanek P., Guggenmoos-Holzmann I. Classification and regression trees (CART) for estimation of prognosis in patients with gastric carcinoma. J. Cancer Res. Clin. Oncol. 1994;120:309–313. doi: 10.1007/BF01236389. [DOI] [PubMed] [Google Scholar]
- R23.Jernigan R.W., Baran R.H. Pervasive properties of the genomic signature. BMC Bioinformatics. 2002;3:1–12. doi: 10.1186/1471-2164-3-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R24.Karlin S. Detecting anomalous gene clusters and pathogenicity islands in diverse bacterial genomes. Trends Microbiol. 2001;9:335–343. doi: 10.1016/s0966-842x(01)02079-0. [DOI] [PubMed] [Google Scholar]
- R25.Karlin S., Burge C. Dinucleotide relative abundance extremes: a genomic signature. Trends Genet. 1995;11:283–290. doi: 10.1016/s0168-9525(00)89076-9. [DOI] [PubMed] [Google Scholar]
- R26.Karlin S., Ladunga I. Comparisons of eukaryotic genomic sequences. Proc. Natl. Acad. Sci. USA. 1994;262:459–472. doi: 10.1073/pnas.91.26.12832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R27.Karlin S., Mrazek J. What drives codon choices in human genes? J. Mol. Biol. 1996;262:459–472. doi: 10.1006/jmbi.1996.0528. [DOI] [PubMed] [Google Scholar]
- R28.Karlin S., Ladunga I., Blaisdell B.E. Heterogeneity of genomes: measures and values. Proc. Natl. Acad. Sci. USA. 1994;91:12837–12841. doi: 10.1073/pnas.91.26.12837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R29.Karlin S., Mrazek J., Campbell A. Compositional biases of bacterial genomes and evolutionary implications. J. Bacteriol. 1997;179:3899–3913. doi: 10.1128/jb.179.12.3899-3913.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R30.Kawashima T., Amano N., Koike H., et al. Archaeal adaptation to higher temperatures revealed by genomic sequence of Thermoplasma volcanium . Proc. Natl. Acad. Sci. USA. 2000;97:14257–14262. doi: 10.1073/pnas.97.26.14257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R31.Kohonen T. The self-organizing map. Proc. IEEE. 1990;78:1464–1480. [Google Scholar]
- R32.Lambros R.J., Mortimer J., Forsdyke D. Optimum growth temperature and base composition of open reading frames in prokaryotes. Extremophiles. 2003;7:443–450. doi: 10.1007/s00792-003-0353-4. [DOI] [PubMed] [Google Scholar]
- R33.Lao P., Forsdyke D. Thermophilic bacteria strictly obey Szybalski’s Transcription direction rule and politely purine-load RNAs with both adenine and guanine. Genome Res. 2000;10:228–236. doi: 10.1101/gr.10.2.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R34.Lin S., Patel S., Duncan A., Goodwin L. Using decision trees and support vector machines to classify genes by names. In: Scheffer T., Lesser U., editors. Proc. European Workshop on Data Mining and Text Mining for Bioinformatics (<a href="http://www.mpi-inf.mpg.de/~scheffer/publications/ws03proc/">http://www.mpi-inf.mpg.de/~scheffer/publications/ws03proc/</a>); 2003. pp. 35–41. [Google Scholar]
- R35.Lio P. Investigating the relationship between genome structure, composition and ecology in prokaryotes. Mol. Biol. Evol. 2000;19:789–800. doi: 10.1093/oxfordjournals.molbev.a004136. [DOI] [PubMed] [Google Scholar]
- R36.Masic N., Gagro A., Rabati S., Sabioncello A., Dai G., Jaki B., Vitale B. Decision-tree approach to the immunophenotype-based prognosis of the B-cell chronic lymphocytic leukemia. Am. J. Hematol. 1998;59:143–148. doi: 10.1002/(sici)1096-8652(199810)59:2<143::aid-ajh7>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- R37.McHardy A.C., Martin H.G., Tsirigos A., Hugenholtz P., Rigoutsos I. Accurate phylogenetic classification of variable-length DNA fragments. Nat. Methods. 2007;4:63–72. doi: 10.1038/nmeth976. [DOI] [PubMed] [Google Scholar]
- R38.Paz A., Mester D., Baca I., Nevo E., Korol A. Adaptive role of increased frequency of polypurine tracts in mRNA sequencs of thermophilic prokaryotes. Proc. Natl. Acad. Sci. USA. 2004;101:2951–2956. doi: 10.1073/pnas.0308594100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R39.Paz A., Kirzhner V., Nevo E., Korol A. Coevolution of DNA-interacting proteins and genome “dialect”. Mol. Biol. Evol. 2006;23:56–64. doi: 10.1093/molbev/msj007. [DOI] [PubMed] [Google Scholar]
- R40.Pride D.T., Meinersmann R.J., Wassenaar T.M., Blaser M.J. Evolutionary implications of microbial genome tetranucleotide frequency biases. Genome Res. 2003;13:145–158. doi: 10.1101/gr.335003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R41.Sandberg R., Winberg G., Branden C.-I., Kaske A., Ernberg I., Cöster J. Capturing whole-genome characteristics in short sequences using a naïve Bayesian classifier. Genome Res. 2001;11:1404–1409. doi: 10.1101/gr.186401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R42.Saunders N.F.W., Thomas T., Curmi P.M.G., et al. Mechanisms of thermal adaptation revealed from the genomes of the antarctic archaea Methanogenium frigidum and Methanococcoides burtonii . Genome Res. 2003;13:1580–1588. doi: 10.1101/gr.1180903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R43.Suhre K., Claverie J.M. Genomic correlates of hyperthermostability, an update. J. Biol. Chem. 2003;278:17198–17202. doi: 10.1074/jbc.M301327200. [DOI] [PubMed] [Google Scholar]
- R44.Teeling H., Meyerdierks A., Bauer M., Amann R., Glockner F.O. Application of tetranucleotide frequencies for the assignment of genomic fragments. Environ. Microbiol. 2004;6:938–947. doi: 10.1111/j.1462-2920.2004.00624.x. [DOI] [PubMed] [Google Scholar]
- R45.Tsirigos A., Rigoutsos I. A sensitive, support-vector-machine method for the detection of horizontal gene transfers in viral, archaeal and bacterial genomes. Nucleic Acids Res. 2005;33:3699–3707. doi: 10.1093/nar/gki660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R46.van Passel M.W.J., Kuramae E.E., Luyf A.C.M., Bart A., Boekhout T. The reach of the genome signature in prokaryotes. BMC Evol. Biol. 2006;6:1–27. doi: 10.1186/1471-2148-6-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R47.van Passel M.W.J., Bart A., Luyf A.C.M., van Kampen A.H.C., van der Ende A. Compositional discordance between prokaryotic plasmids and host chromosomes. BMC Genomics. 2006;7:26. doi: 10.1186/1471-2164-7-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R48.Vasilevskaya V.V., Gusev L.V., Khokhlov A.R. Protein sequences as literature text. Macromol. Theory Simul. 2006;15:425–431. [Google Scholar]
- R49.Woyke T., Teeling H., Ivanova N.N., et al. Symbiosis insights through metagenomic analysis of a microbial consortium. Nature. 2006;443:950–955. doi: 10.1038/nature05192. [DOI] [PubMed] [Google Scholar]
- R50.Xia X., Wei T., Xie Z., Danchin A. Genomic changes in nucleotide and dinucleotide frequencies in Pasteurella multocida cultured under high temperature. Genetics. 2002;161:1385–1394. doi: 10.1093/genetics/161.4.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1. A list of the 195 organisms of which the genomes were used in this study (24 archaea and 171 bacteria). The complete genome sequences were downloaded from GenBank (http://www.ncbi.nlm.nih.gov/genomes/lproks.cgi) on April 1, 2006.
Supplementary Table S2. Classification and regression tree analysis results using four oligonucleotide frequency lengths over the entire virtual coding strand to classify organisms into two temperature optimum ranges, hyperthermophile versus non-hyperthermophile (hyper vs. non-hyper) and mesophile versus non-mesophile (meso vs. non-meso). Tetramers provide the most effective classification scheme considering both models built. Model-with-all is the number of misclassifications when building a model using all 195 genomes. Leave-one-out is the number of mispredictions when classifying an organism against a model built using the other 194 genomes.