Abstract
The human motor neuron degenerative disease spinal muscular atrophy with respiratory distress type 1 (SMARD1) is caused by loss of function mutations of immunoglobulin µ-binding protein 2 (IGHMBP2), a protein of unknown function that contains DNA/RNA helicase and nucleic acid-binding domains. Reduced IGHMBP2 protein levels in neuromuscular degeneration (nmd) mice, the mouse model of SMARD1, lead to motor neuron degeneration. We report the biochemical characterization of IGHMBP2 and the isolation of a modifier locus that rescues the phenotype and motor neuron degeneration of nmd mice. We find that a 166 kb BAC transgene derived from CAST/EiJ mice and containing tRNA genes and activator of basal transcription 1 (Abt1), a protein-coding gene that is required for ribosome biogenesis, contains the genetic modifier responsible for motor neuron rescue. Our biochemical investigations show that IGHMBP2 associates physically with tRNAs and in particular with tRNATyr, which are present in the modifier and with the ABT1 protein. We find that transcription factor IIIC-220 kDa (TFIIIC220), an essential factor required for tRNA transcription, and the helicases Reptin and Pontin, which function in transcription and in ribosome biogenesis, are also part of IGHMBP2-containing complexes. Our findings strongly suggest that IGHMBP2 is a component of the translational machinery and that these components can be manipulated genetically to suppress motor neuron degeneration.
INTRODUCTION
Spinal muscular atrophy with respiratory distress type 1 (SMARD1) is an autosomal recessive neurodegenerative disease (1,2). The disease is also known as distal spinal muscular atrophy 1 (DSMA1, MIM no. 604320). The primary pathology of SMARD1 consists of degeneration of spinal cord motor neurons and axonal loss leading to muscular atrophy (1,2). Diaphragmatic paralysis is one of the earliest and most severe manifestations of the disease and typically leads to early infant death (1,2). SMARD1 results from mutations in the immunoglobulin µ-binding protein 2 (IGHMBP2) (1,2). The human IGHMBP2 is a 993 amino acid protein that contains an RNA/DNA helicase domain (found in the superfamily I helicases), an R3H, single-stranded nucleic acid-binding domain and a zinc finger domain (1–3). IGHMBP2 mRNA is ubiquitously expressed (4). On the basis of in vitro binding assays, IGHMBP2 has been proposed to bind DNA and was given several names in the literature: glial factor 1 [GF1 (5)], immunoglobulin S-µ-binding protein-2 [Smbp2 or IGHMBP2 (4,6)], rat insulin-enhancer-binding protein [Rip1 (7)] and cardiac transcription factor 1 [Catf1 (8)]. Proposed functions, based on in vitro data, include transcriptional activation, immunoglobulin class switching and pre-mRNA splicing (4–9). In neurons, IGHMBP2 is predominantly found in the cytoplasm, including dendrites and axons (3), arguing that IGHMBP2 has a major function outside the nucleus. The presence of an RNA helicase domain and a single-stranded nucleic acid-binding domain suggests that IGHMBP2 may function in RNA processing, regulation or metabolism.
Neuromuscular degeneration (nmd) mice that are homozygous for mutations of the Ighmbp2 gene develop a phenotype that is very similar to SMARD1 (10). The pathology of nmd mice consists of motor neuron degeneration with axonal loss leading to neurogenic muscle atrophy and death by ∼8–12 weeks (10–12). We previously reported that the nmd mutation is a single A-to-G transition 23 bp into intron 4 of the mouse Ighmbp2 gene that creates a new favorable splice donor, resulting in a frameshift and premature stop codon (10). In nmd homozygous mice, low levels (∼20%) of correctly spliced mRNA are evident in all nmd tissues, indicating that this is a hypomorphic allele and not a complete null mutation (10). We reported that the motor neuron degeneration of nmd mice was completely corrected by transgenic expression of wild-type IGHMBP2 driven by a neuron-specific promoter (11). This finding demonstrates that motor neuron degeneration in nmd mice is cell autonomous and is a direct consequence of reduced IGHMBP2 protein levels in motor neurons. Interestingly, these transgenic mice later developed a primary myopathy affecting both skeletal muscles and the myocardium, suggesting that IGHMBP2 is also essential for myocyte maintenance (11). Such primary myopathy has not been described in human patients with SMARD1, presumably because it is superceded by the severe motor neuronopathy that leads to early death, before manifestation of the primary myopathy (11).
Mice contain 418 tRNA genes and humans contain 505 tRNA genes, corresponding to tRNAs that decode the standard 20 amino acids (13). Approximately 6% of these genes contain introns (13). tRNAs are transcribed in the nucleus by RNA polymerase III as longer pre-tRNA transcripts (14,15). tRNA genes contain intragenic RNA polymerase III promoters whose core elements consist of the A and B boxes (whose sequences roughly correspond to the D- and T-loops of the mature tRNA) (16,17). The A and B boxes are recognized by a multisubunit complex known as general transcription factor IIIC (TFIIIC). Mammalian TFIIIC220, one of the subunits of TFIIIC, binds directly to box B of RNA polymerase III promoters and is essential for tRNA transcription (16,17). Pre-tRNA transcripts contain 5′-leader and 3′-trailer sequences (14,15). Several tRNAs also contain a single, short (14–60 nt) intron which is found in the anticodon loop, typically one nucleotide 3′ to the anticodon (13–15). Processing of pre-tRNAs involves removal of the 5′-leader by RNAse P, removal of the 3′-trailer by various endo- and exo-nucleases, splicing of the intron, addition of the CCA trinucleotide to the 3′-end and numerous tRNA modifications (14,15,18,19). Addition of the CCA trinucleotide typically occurs prior to tRNA splicing and is followed by aminoacylation and export of the mature tRNA to the cytoplasm (15).
The biogenesis of ribosomes in eukaryotes is a complicated, step-wise process that involves more than 200 proteins and an equal number of small nucleolar RNAs (snoRNAs) (20). RNA polymerase I generates a large pre-rRNA precursor (termed 47S/45S) in mammals that contains the mature 18S, 28S and 5.8S rRNAs. A series of endonucleolytic cleavages and exonucleolytic processing steps leads to generation of the mature rRNAs (21). snoRNAs are involved in the processing and modification of ribosomal RNA (rRNA) and are found in cells complexed with proteins (snoRNPs). U3 snoRNP is a large ribonucloprotein composed of a 217 nucleotide U3 snoRNA and multiple proteins (22) and directs the first cleavage of the pre-rRNA precursor (22,23).
Spinal muscular atrophy (SMA), although distinct from SMARD1, shares many clinical and pathological features with SMARD1. SMA is caused by deletions or loss of function mutations of the survival of motor neurons (SMN) gene (24). The pathology of SMA consists of degeneration of spinal cord motor neurons, leading to muscular atrophy (25). The SMN protein associates with seven additional proteins, known as Gemins2–8, into a large complex termed the SMN complex (26–28). The SMN complex plays critical roles in the biogenesis of spliceosomal small nuclear RNPs (snRNPs) and possibly other RNPs (26,29–32).
Here, we show that IGHMBP2 is not a component of the SMN complex, but rather it interacts with a unique set of small RNAs that includes tRNATyr, and with proteins, including helicases, TFIIIC220 and activator of basal transcription 1 (ABT1), that are required for tRNA transcription and ribosome biogenesis and function. These biochemical observations are corroborated in mutant nmd mice by the analysis of a genetic suppressor of motor neuron disease encoding multiple tRNATyr genes and ABT1 that we have shown to associate physically with IGHMBP2. Our findings strongly suggest that IGHMBP2 is a component of the translational machinery and that these components can be manipulated genetically to suppress motor neuron degeneration.
RESULTS
IGHMBP2 is not a component of the SMN complex
Because SMARD1 and SMA have similar phenotypes, caused by degeneration of motor neurons, we first wanted to investigate whether IGHMBP2 could interact with SMN or with other components of the SMN complex. We cloned the human IGHMBP2 cDNA in a eukaryotic expression vector in frame with a FLAG epitope tag and generated a stable human 293T cell line, expressing FLAG-IGHMBP2 and another 293T cell line expressing the empty FLAG vector as a negative control. We used anti-FLAG beads to immunoprecipitate FLAG-IGHMBP2 and we probed the immunoprecipitates on western blots using antibodies against components of the SMN complex: SMN, Gemin2, Gemin3, Gemin5 and Gemin6. As shown in Supplementary Material, Figure S1, none of these proteins co-immunoprecipitate with IGHMBP2, indicating that IGHMBP2 does not associate with the SMN complex.
Generation of a monoclonal antibody against IGHMBP2
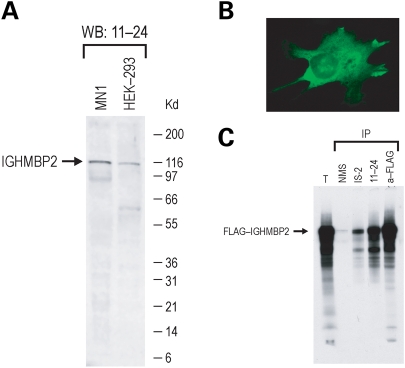
Next, we generated a monoclonal antibody (mAb11-24) against IGHMBP2 by immunizing mice with an amino-terminal portion of human IGHMBP2 that was generated as recombinant protein in bacteria. As shown in Figure 1, mAb11-24 recognized IGHMBP2 in western blots, in immunoprecipitations (IPs) and by immunofluorescence. mAb11-24 recognizes a protein band of ∼110 kDa corresponding to IGHMBP2 and also a faster migrating band at ∼60 kDa, which is inconsistently seen, and likely represents a proteolytic fragment of IGHMBP2. The predominant localization of IGHMBP2 in human NT2 cells and in mouse MN-1 cells is cytoplasmic but there is also a fraction of IGHMBP2 that is localized in the nucleus. This is consistent with prior reports using a rabbit polyclonal anti-IGHMBP2 antibody (3).
Figure 1.
Properties of the monoclonal antibody against IGHMBP2. (A) Total cell lysates from mouse MN1 cells or human 293T cells were resolved on 4–12% NuPAGE gel, blotted and probed with 11-24 mAb. Molecular mass markers (in kilodaltons) are shown on the right. (B) Indirect immunofluorescence of IGHMBP2 on NT2 cell line using 11-24 mAb analyzed by epifluorescence microscopy. (C) FLAG-IGHMBP2 was in vitro-translated in the presence of 35S-methionine, and radiolabeled FLAG-IGHMBP2 was immunoprecipitated with non-immune mouse serum (NMS), immune serum from a mouse immunized with recombinant IGHMBP2 (IS-2), the 11-24 mAb, or anti-FLAG (M2) mAb. Immunoprecipitated proteins were resolved on 4–12% NuPAGE gel and detected by autoradiography. Total (T) contains 20% of input used for IPs.
Identification of proteins that associate with IGHMBP2
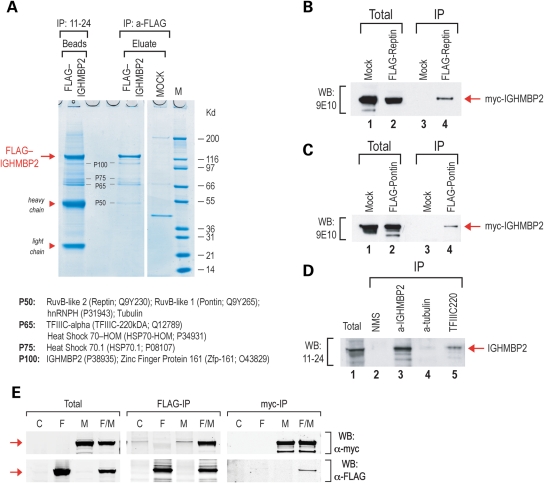
To identify possible protein partners of IGHMBP2, we used 293T cells stably expressing either FLAG-IGHMBP2 or the FLAG empty vector, in IP experiments. We chose this approach because we can perform IPs by using two different monoclonal antibodies, anti-FLAG and also mAb11-24 against IGHMBP2. Proteins that co-immunoprecipitate with both antibodies are likely to represent authentic IGHMBP2-associated proteins. Furthermore, competitive elution with 3XFLAG peptide from the anti-FLAG immunoprecipitates results in much cleaner co-IP patterns since the antibody chains do not contaminate the samples. As shown in Figure 2, other proteins co-immunoprecipitated with FLAG-IGHMBP2 and were found in both anti-FLAG and mAb11-24 immunoprecipitates. We identified several of these proteins by mass spectrometry and the results are shown in Figure 2A. Interestingly, we identified two closely related ATPase/helicases, Reptin (Tip48, RuvB-like2) and Pontin (Tip49, RuvB-like1) co-immunoprecipitating with IGHMBP2. To confirm the association of IGHMBP2 with Reptin and Pontin, we performed reverse co-IP experiments. None of the antibodies that we have tested was able to efficiently immunoprecipitate Reptin or Pontin, although many of them worked well on western blots. To overcome the lack of immunoprecipitating antibodies for Reptin and Pontin, we used FLAG-tagged constructs for Reptin and Pontin and co-transfected these plasmids along with a plasmid expressing myc-IGHMBP2 in 293T cells. Cells transfected with myc-IGHMBP2 and FLAG-empty vector served as negative controls. We then performed IPs with anti-FLAG antibodies and probed the immunoprecipitates with anti-myc (9E10) antibody. As shown in Figure 2B and C, myc-IGHMBP2 co-immunoprecipitated with FLAG-Reptin and FLAG Pontin. Peptides corresponding to transcription factor IIIC, 220 KDa subunit (TFIIIC220; also known as TFIIICa) were obtained from the P65 band, indicating that TFIIIC220, a large 220 kDa protein, fragmented during cell lysis and IP. To confirm the IGHMBP2–TFIIIC220 association, we performed IPs in 293 cells using an antibody against the endogenous TFIIIC220 and probed the immunoprecipitates with mAb11-24 against IGHMBP2. As shown in Figure 2D (lane 5), IGHMBP2 co-immunoprecipitated with TFIIIC220. Peptides corresponding to tubulin chains were also identified in IGHMBP2 immunoprecipitates. To further assess these findings, we performed IPs using anti-α-tubulin antibody from 293T cells and probed the immunoprecipitates with mAb11-24 against IGHMBP2 but detected only a faint interaction (Fig. 2D, lane 4). These results indicate that IGHMBP2 associates with the helicases Reptin and Pontin and with TFIIIC220. Co-IP experiments showed that IGHMBP2 did not interact with hnRNP H, HSP70 or Zfp161 (Supplementary Material, Fig. S2). Since many RNA or DNA helicases form dimers or homomers, we tested whether IGHMBP2 self-associates. For this, we expressed either FLAG-IGHMBP2, or myc-IGHMBP2, or both FLAG- and myc-IGHMBP2 in 293T cells along with an empty vector as negative control (Fig. 2E). Co-IPs were then performed with either anti-FLAG or anti-myc antibodies, and the immunoprecipitates were probed on western blots with anti-myc (Fig. 2E, top panel) or anti-FLAG (Fig. 2E, bottom panel) antibodies, respectively. The presence of myc-IGHMBP2 in the FLAG-IGHMBP2 immunoprecipitates and the presence of FLAG-IGHMBP2 in the myc-IP immunoprecipitates (Fig. 2E) indicate that IGHMBP2 is able to self-associate in vivo.
Figure 2.
IGHMBP2 self-associates and associates with TFIIIC220, and the helicases Reptin and Pontin. (A) Identification of IGHMBP2-associated proteins. Immunoprecipitations (IPs) were performed with anti-FLAG or 11-24 (anti-IGHMBP2) antibodies from 293 cells expressing FLAG-IGHMBP2 or from mock-transfected 293 cells. Immunoprecipitates from the anti-FLAG resins were eluted with 3× FLAG peptide. Immunoprecipitates from the 11-24 IP (beads) and eluates from the anti-FLAG IPs were resolved on a 4–12% NuPAGE gel and visualized by Coomassie blue staining. Arrow indicates the FLAG-IGHMBP2 protein. Proteins that co-precipitate with both antibodies (band P50 is obscured from the 11-24 IPs from the antibody chain) are indicated. Molecular mass markers (in kilodaltons) are shown on the right. The heavy and light chains of the 11-24 antibody are indicated. Protein bands P100, P75, P65 and P50 were excised from the gel (FLAG-IGHMBP2 eluate) and identified by mass spectrometry. The corresponding proteins (and Genbank protein IDs) from these bands are listed below. (B–D) Validation of IGHMBP2-interacting proteins by reverse co-IP experiments. (B) Two hundred and ninety-three cells expressing myc-IGHMBP2 were transfected with FLAG-Reptin (lane 2) or with empty vector (mock, lane 1). IPs were performed with anti-FLAG antibody from the mock-transfected (lane 3) or FLAG-Reptin-transfected cells (lane 4), and the immunoprecipitates were probed with anti-myc (9E10) antibody on western blot. (C) Two hundred and ninety-three cells expressing myc-IGHMBP2 were transfected with FLAG-Pontin (lane 2) or with empty vector (mock, lane 1). IPs were performed with anti-FLAG antibody from the mock-transfected (lane 3) or FLAG-Pontin-transfected cells (lane 4), and the immunoprecipitates were probed with anti-myc (9E10) antibody on western blot. (D) IPs were performed from 293 cells using antibodies against: TFIIIC220 (lane 5), α-tubulin (lane 4), IGHMBP2 (11-24 mAb, lane 3), NMS (non-immune mouse serum; negative control, lane 2). The immunoprecipitates were probed with 11-24 (anti-IGHMBP2 mAb) on western blot. (E) IGHMBP2 self-associates. 293T cells were transfected with FLAG-IGHMBP2 (F), or myc-IGHMBP2 (M), or both FLAG-IGHMBP2 and myc-IGHMBP2 (F/M), or were mock-transfected (C). IPs were performed with either anti-FLAG or anti-myc antibodies, and the immunoprecipitates were probed on western blots with anti-myc (top panel) or anti-FLAG (bottom panel) antibodies. Total represents ∼10% of lysate prior to IPs. Arrow indicates epitope-tagged IGHMBP2 protein.
Identification of small RNAs that associate with IGHMBP2
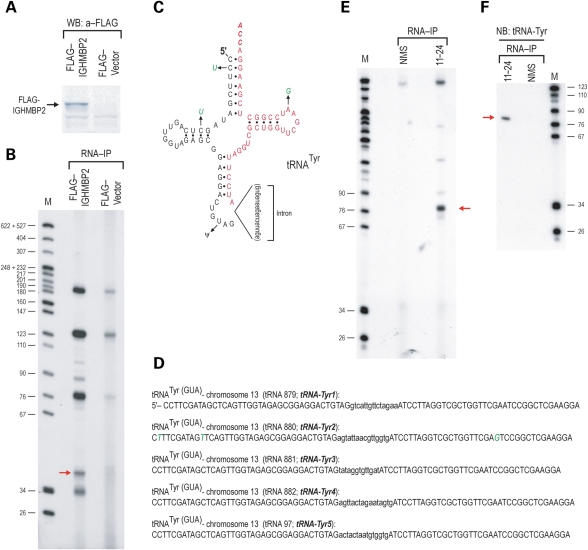
We next asked whether IGHMBP2 associates with small RNAs. We generated a stable MN1 cell line, expressing FLAG-IGHMBP2 and another MN1 cell line expressing the empty FLAG vector as a negative control (Fig. 3A). MN1 is a well-characterized hybrid mouse motor neuronal cell line (33–35). We then performed IPs with anti-FLAG antibody from MN1-FLAG-IGHMBP2 and from MN1-FLAG cells. RNA was then isolated from the immunoprecipitates, 3′-end-labeled with [32P]Cp and T4 RNA ligase and resolved on a 10% UREA–PAGE. Two discrete RNA bands (∼34 and ∼40 nt) were present in the FLAG-IGHMBP2 immunoprecipitates only (Fig. 3B). We then directionally cloned the ∼40 nt RNA band from the FLAG-IGHMBP2 immunoprecipitates by employing the same strategy that we used to clone miRNAs and the (mt)tRNAMet from Ago2 immunoprecipitates (36,37) (detailed in 38). Of the 30 clones sequenced, three clones corresponded to the 3′-end or tRNATyr (Fig. 3C and D), one clone corresponded to the 3′-end of tRNAMet (CAU anticodon), one clone to the 3′-end of tRNAArg (UCU anticodon) and the remainder represented clones with adapters only. These results indicated that IGHMBP2 associated with tRNAs and, in particular, tRNATyr. We were unable to clone the smaller, ∼34 nt band, but it likely corresponds to 5′-ends of tRNAs. Because the RNAs from the FLAG-IGHMBP2 immunoprecipitates were 3′-end-labeled with a single [32P] pCp molecule, the stoichiometry of the RNAs may be extrapolated from band intensity. As shown in Figure 3B, the ratio of the ∼40 nt to the ∼34 nt band is 1:1, suggesting that they may be derived from processing of a tRNA molecule. In experiments described in what follows, we detect association of IGHMBP2 with intact tRNAs (Fig. 3E and F). Cleavage of tRNAs in the anticodon loop has been shown to occur as a conserved response to oxidative stress in yeast, plants and mammalian cells (39) and it is likely that such fragmentation occurred prior to or during cell lysis and RNA IP. The RNA bands at ∼75, ∼120 and ∼180 nt correspond in size to tRNAs, 5S rRNA and 5.8S rRNAs. The 5S and 5.8S rRNAs (which we have identified by northern blots; our unpublished data) are likely non-specific as we often observe them in small RNA IPs using various antibodies [such as anti-Ago antibodies (36)], and in the anti-FLAG vector (negative control) RNA immunoprecipitate (Fig. 3B).
Figure 3.
IGHMBP2 associates with small RNAs and in particular with tRNA-Tyr. (A) Lysates from mouse MN1 cells stably expressing FLAG-IGHMBP2 or vector only (FLAG-vector) were probed on a western blot with a monoclonal antibody against the FLAG epitope. (B) RNA immunoprecipitations (RNA-IP) were performed with anti-FLAG antibody from FLAG–IGHMBP2-expressing or mock-transfected (vector only) MN1 cells. RNA was isolated from the immunoprecipitates, 3′-end-labeled with [5′-32P]-pCp and T4 RNA ligase and resolved by electrophoresis on a 10% denaturing PAGE. Nucleotide sizes of the radiolabeled marker (M) are shown on the left. Red arrow at ∼40 nt indicates small RNAs that were directionally cloned and sequenced. (C) Secondary structure of mouse tRNA-Tyr (tRNA-882; numbering is adapted from 13) with cloned sequences shown in red, including the post-transcriptionally added CCA trinucleotide at the 3′-end. Nucleotide differences of tRNA 880 (tRNA-Tyr2) are indicated in green; all other nucleotides between the various tRNAs-Tyr listed above are identical. The pseudo-uridine modification of uridine present in the center of the anticodon of all tRNA-Tyr is shown with the symbol ψ. (D) Sequences of mouse tRNA-Tyr genes contained in the modifier locus (see Fig. 4). Introns are in lower case; note that intronic sequences differ between the various tRNA-Tyr genes. (E) RNA-IP was performed with 11-24 mAb against endogenous IGHMBP2 from human HeLa cells or with non-immune mouse serum (NMS, negative control). RNA was isolated from the immunoprecipitates, 3′-end-labeled with [5′-32P]-pCp and T4 RNA ligase and resolved by electrophoresis on a 10% denaturing PAGE. Nucleotide sizes of the radiolabeled marker (M) are shown on the left. Red arrow indicates tRNAs. (F) RNA-IP was performed from HeLa cells as in (E), and the isolated RNA was transferred to a nylon membrane and probed with a 5′-end-labeled DNA oligo, antisense to tRNA-Tyr.
Next, we isolated RNAs that co-immunoprecipitated with endogenous IGHMBP2 from mAb11-24 immunoprecipitates from HeLa cells; non-immune mouse serum (NMS) served as a negative control. We 3′-end-labeled the RNA with pCp and T4 RNA ligase and analyzed the small RNAs on a 10% UREA–PAGE. As shown in Figure 3E, a strong band at ∼75 nt, corresponding in size to tRNAs, was present in mAb11-24 immunoprecipitates but not in the negative (NMS) control. To confirm the association of tRNATyr with endogenous IGHMBP2, we probed RNAs that were isolated from mAb11-24 immunoprecipitates from HeLa cells with an oligonucleotide probe against tRNATyr on a northern blot. As shown in Figure 3F, tRNATyr was detected specifically in mAb11-24 immunoprecipitates. These findings indicate that IGHMBP2 associates with small RNAs and, in particular, with tRNATyr.
Positional complementation cloning of the Mnmc modifier using BAC transgenic rescue
During the course of our original fine-mapping of the nmd mouse mutation, we observed that ∼25% of the (B6-nmd2J × CAST/EiJ)F2 nmd2J/nmd2J mice were very mildly affected with regard to the extent of neurogenic atrophy and paralysis (10,11). We mapped the modifier region to mouse Chr 13 and completed construction of a congenic B6.CAST-MnmC line of mice that are homozygous B6 at all loci except around the region containing the MnmC gene. Just as in the original F2 intercross, this single Chr 13 locus suppresses the nmd motor neuron disease in a semidominant fashion but fails to rescue the onset or progression of dilated cardiomyopathy (11). Using the B6.CAST-Mnm congenic strain in a mapping cross, we have obtained additional recombinants to reduce the genetic interval to 0.03 cM between markers CA1 and CAGA. We have constructed a contig across most of the 430 kb Mnm critical interval from CAST-derived (CHORI-26 library, http://bacpac.chori.org) BAC clones (Fig. 4A).
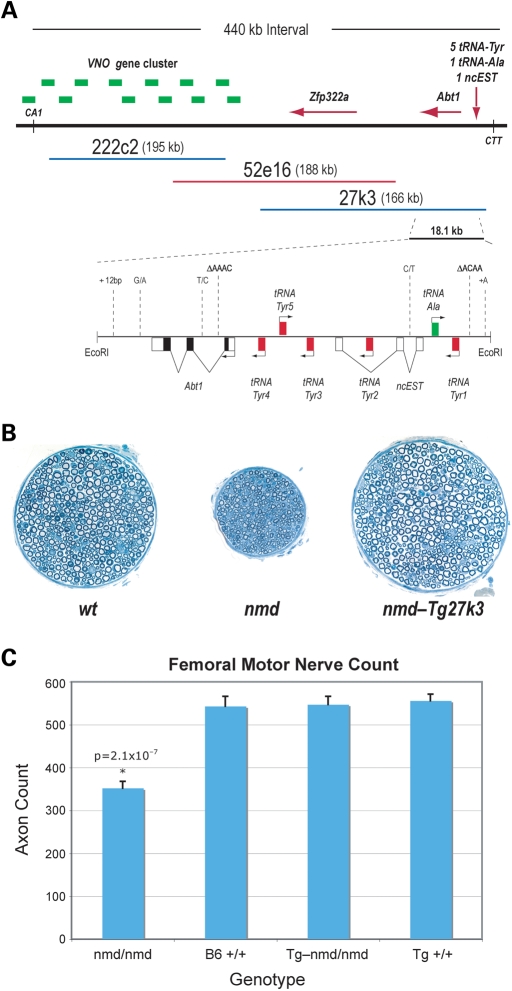
Figure 4.
MnM modifier locus and BAC-27K3 rescuing transgene. (A) Transgenic positional complementation cloning the Mnm modifier. Schematic of the 430 kb Mnm modifier locus and of the BAC 27k3 (blue) that suppresses nmd motor neuron degeneration and contains the sequences necessary to confer the Mnm effect. The 18.1 kb EcoRI fragment from BAC-27k3 that contains the five tRNA-Tyr and Abt1 genes is shown magnified below. The polymorphisms between B6 and CAST are also shown. Boxed genes and exons above the line are transcribed from left to right and those below the line are transcribed from right to left. Coding genes and functional tRNAs are depicted as is a non-coding EST (ncEST). (B) Cross-sections of the motor branch of the femoral nerve from a 35-day-old B6-+/+ mouse, a 41-day-old B6-nmd/nmd mouse and a 37-day-old B6-Tg27k3 nmd/nmd rescued mouse. There is a significant reduction in the total number of myelinated axons in the nmd femoral motor nerves that is completely rescued by the 27k3 BAC transgene. (C) Axon counts from femoral motor nerves from homozygous B6 nmd mice (nmd/nmd, n = 13), wild-type B6 mice (+/+, n = 6), B6-Tg27k3 nmd/nmd rescued mice (Tg nmd/nmd, n = 8) and B6-Tg27k3 (Tg +/+, n = 4).
To further localize the Mnm modifier within our genetic interval, we undertook a transgenic rescue strategy using the ∼166 kb CAST-derived BAC clone (CH26-27K3). As shown in Figure 4A, BAC-27k3 contains two protein-coding genes, Abt1 and Zfp322a. In addition, 1 non-coding EST (ENSMUSESTG00000018860) and 24 tRNA genes (10 tRNAVal, 6 tRNAAla, 5 tRNATyr, 2 tRNAIle and 1 tRNAMet) have been annotated to the region between 23 380 802 and 23 527 157 bp on mouse Chr 13 (NCBI build 37). We have identified eight independent B6-Tg(BAC27k3) transgenic founders and crossed each of them with B6-+/nmd mice to generate B6-Tg(BAC27k3) nmd/nmd mutant mice. Seven of eight 27k3 founder lines completely recapitulate the MnmC modifier rescue phenotype seen in our B6.CAST-Mnm congenic line though one founder line (line 60) displays a generally mild, yet variable rescue phenotype among littermates. Thus, we have been able to limit the Mnm modifier effect to a single 166 kb CAST-derived BAC clone. We quantitated the extent of the motor neuron rescue in nmd mice carrying the BAC-27k3 transgene by counting the number of myelinated axons derived from the motor branch of the femoral nerve. As shown in Figure 4B and C, the 27k3 modifier rescued completely the motor neuron degeneration of nmd mice.
Shotgun sequencing of BAC-27k3 to identify CAST/EiJ sequence polymorphisms was compared with the public mouse C57BL/6J genome sequence. No coding polymorphisms, aberrant alternative splicing events or real-time reverse transcriptase (RT)–polymerase chain reaction (PCR) expression differences have been detected in the CAST-derived Abt1 or Zfp322a sequences that might point to the cause of the genetic modifier effect (data not shown). Apart from the triplication of an ∼10 kb repetitive element in CAST detected by DNA fingerprint analysis of BAC-27k3, we have been surprised by the low rate of polymorphisms between C57BL/6J and CAST/EiJ in our interval (∼1 SNP/4.1 kb) given that the two subspecies are thought to have diverged approximately 0.5–1 million years ago. However, this is consistent with the mosaic nature of the genome of common laboratory strains with intervals of high SNP density and low SNP density reflecting the recent contributions of musculus, domesticus and castaneus-derived chromosomal segments during inbreeding (40). The ∼10 kb triplication is relatively common among several inbred strains based on Southern blotting (data not shown) and does not appear to be associated with the modifier effect though the sequence contains three tRNA genes (tRNAAla and two tRNAVal genes). Complete sequencing of an 18.1 kb EcoRI fragment of BAC-27k3 containing the Abt1 gene, six tRNA genes and one noncoding EST revealed only seven polymorphisms in intergenic or intronic regions (summarized in Fig. 4A, GenBank accession number EU880213) that do not immediately suggest a mechanism for the modifier effect of the CAST/EiJ allele versus the C57BL/6J allele in suppressing motor neuron disease. The syntenic genomic area in humans is found on chromosome 6 and contains a cluster of four tRNATyr genes and the ABT1 gene.
The identification of five tRNATyr genes in the 27k3 modifier and the physical association of tRNATyr with IGHMBP2 are particularly intriguing and indicate that IGHMBP2 may function in tRNA pathways or in translation. This is further supported by the association of IGHMBP2 with TFIIIC220, a general transcription factor for RNA polymerase III, which is essential for transcription of tRNA genes.
The genetic modifier rescues the motor neuron degeneration of nmd mice without restoring the protein levels of IGHMBP2
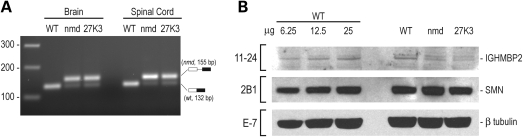
To test whether the genetic modifier functions by correcting the splicing defect of nmd mice, we assayed the levels of aberrantly spliced Ighmbp2 pre-mRNA in nmd and in B6-Tg(BAC27k3) nmd mice that express the genetic modifier. RT–PCR with primers spanning the point mutation in intron 4 of Ighmbp2 gene of nmd mice leads to the inclusion of 23 nucleotides from intron 4 in the aberrant Ighmbp2 pre-mRNA from nmd mice and the generation of a 155 base pair (bp) PCR product. The same set of primers generates a 132 bp PCR product for the correctly spliced, wild-type Ighmbp2 transcript. In nmd mice, the predominant Ighmbp2 transcript is the aberrant one with much lower levels of the wild-type transcript (10). As shown in Figure 5A, the genetic modifier does not restore the splicing defect of the Ighmbp2 gene. We also assayed the IGHMBP2 protein levels of brain lysates from wild-type, nmd and B6-Tg(BAC27k3) nmd mice on western blots and found that the genetic modifier does not restore the protein levels of IGHMBP2 (Fig. 5B). These findings indicate that the modifier rescues the motor neuron degeneration of nmd mice by a mechanism that does not affect IGHMBP2 protein levels.
Figure 5.
The genetic modifier rescues the motor neuron degeneration of nmd mice without restoring the protein levels of IGHMBP2. (A) RT–PCR was performed with primers spanning the point mutation in intron 4 of Ighmbp2 gene of nmd mice that leads to the inclusion of 23 nucleotides in the aberrant Ighmbp2 pre-mRNA from nmd mice. The aberrant Ighmbp2 transcript is the major transcript generated in nmd mice and in the B6-Tg(BAC27k3) nmd mice. (B) Indicated amounts of brain tissue lysates from wild-type (wt), nmd or B6-Tg(BAC27k3) nmd mice were probed with the indicated antibodies on western blots.
IGHMBP2 associates with the ABT1 protein, whose gene is present in the 27k3 modifier
The presence of the Abt1 gene in the 27k3 modifier is particularly interesting because ABT1 is an essential factor required for pre-ribosomal RNA (pre-rRNA) processing and maturation of ribosomes (41). In the initial immunopurification of FLAG-IGHMBP2, we did not detect a clear protein band at ∼31 kDa, corresponding to ABT1, in the Coomassie-blue stained FLAG-IGHMBP2 eluates (Fig. 2A). However, since ABT1 is a small protein and the sensitivity of Coomasie blue is low, we performed co-IP experiments to test whether there is an association of IGHMBP2 with ABT1. We found that there are no anti-ABT1 antibodies suitable for IP. We thus generated a myc-tagged ABT1 construct and co-transfected it along with a plasmid expressing FLAG-IGHMBP2 in 293T cells. We then performed IPs with anti-FLAG or anti-myc antibodies and probed the immunoprecipitates with anti-myc (9E10) or anti-FLAG antibodies to detect co-precipitating ABT1 and IGHMBP2, respectively. As shown in Figure 6, myc-ABT1 co-immunoprecipitated with FLAG-IGHMBP2, indicating that there is a physical association between IGHMBP2 and ABT1. However, co-IP experiments showed that IGHMBP2 did not interact with ZNF322a, the other protein-coding gene found in the 27k3 modifier (Supplementary Material, Fig. S2).
Figure 6.
IGHMBP2 associates with ABT1. 293T cells were co-transfected with FLAG-IGHMBP2 and myc-ABT1. IPs were performed with anti-myc antibody (left panel) or with anti-FLAG antibody (right panel), and the immunoprecipitates were probed on western blots with anti-FLAG antibody (to detect FLAG-IGHMBP2) and with 9E10 anti-myc antibody (to detect myc-ABT1). Total represents 10% of lysate prior to IP.
DISCUSSION
Dysregulation of pre-mRNA, rRNA or tRNA pathways is emerging as an important feature of neurodegenerative disorders and in particular in degeneration of motor neurons or their axons.
SMA is caused by reduced SMN protein levels, and in the SMA mouse, this reduction results in generalized pre-mRNA splicing defects (42). Most cells, except motor neurons, apparently tolerate such splicing defects caused by reduced SMN levels. It is also possible that in motor neurons, SMN may have additional, as yet unidentified functions that promote axonal and neuronal survival.
Charcot–Marie–Tooth (CMT) neuropathies are common diseases of the peripheral nervous system that are caused by degeneration of axons (either axonal or demyelinating) of motor neurons or sensory neurons. Mutations of the tyrosyl-tRNA synthetase (also known as YARS), the enzyme that aminoacylates tRNATyr, cause dominant intermediate CMT neuropathy type C (DI-CMTC) (43). Mutations of glycyl-tRNA synthetase (also known as GARS), the enzyme that aminoacylates tRNAGly, cause CMT neuropathy type 2D (CMTD2) and distal spinal muscular atrophy type V (dSMA-V) (44,45). Recent studies indicate that dSMA-V and CMTD2 are best characterized as primary motor neuronopathies (45). These findings strongly indicate that disruption of tRNA function may underlie neurodegeneration.
Amyotrophic lateral sclerosis type 4 (ALS4) is caused by mutations of senataxin (46). Senataxin has an RNA helicase domain that is similar to that of IGHMBP2 and shows significant similarity to the yeast helicase Sen1p, which is involved in the processing and maturation of tRNAs and other small noncoding RNAs (47). Senataxin is localized to the cytoplasm and in the nucleolus of neurons, suggesting a function in ribosomal RNA biogenesis and/or translation (48).
Two RNA-binding proteins are also major players in the etiopathogenesis of sporadic and familial ALS. Cytoplasmic aggregation of the TARDBP RNA-binding protein [TAR-DNA-binding protein 43 (TDP 43)] is found in ALS motor neurons (49), and mutations of TARDBP cause familial ALS (50–53). Mutations of FUS (fused in sarcoma), another RNA-binding protein, cause familial ALS, and like TARDBP, FUS aggregates in the cytoplasm of affected motor neurons (54,55).
Our study adds SMARD1 as another motor neuron degenerative disease that is likely caused by dysregulation of tRNA and rRNA pathways. Mice contain 10 tRNATyr genes, all of which contain introns (13). Humans contain 15 tRNATyr genes, 14 of which contain introns (13). The tRNATyr genes are unique among tRNA genes because in virtually all organisms, from yeast to humans, they contain introns (13). Another exceptional feature of tRNATyr is the presence of a pseudo-uridine modification of the uridine that is found in the center of the GUA anticodon in all tRNATyr from yeast to humans (Fig. 3C) and which is required for proper codon recognition (56). Furthermore, this modification is dependent on the presence of the intron and leads to the hypothesis that the strong evolutionary pressure to maintain the intron in virtually all tRNATyr genes from yeast to humans may be to direct this pseudo-uridine modification (56,57). In mice, half of all tRNATyr genes are located in an ∼8 kb region of chromosome 13 (13). Intriguingly, we found that this cluster of the five tRNATyr genes is present in the BAC-27k3 transgene that rescues motor neuron degeneration of nmd mice, and tRNATyr physically associates with IGHMBP2. The association of IGHMBP2 with TFIIIC220 is also very interesting because TFIIIC220 binds to tRNA promoters and is an essential factor for tRNA transcription.
The finding that the ABT1, Reptin and Pontin proteins associate with IGHMBP2 is also interesting. Both Reptin and Pontin are evolutionary conserved and ubiquitously expressed ∼50 kDa proteins that contain ATPase/helicase domains and are found in multiple complexes involved in chromatin remodeling and gene transcription (58–67). Reptin and Pontin also associate with U3 snoRNA and are required for U3 snoRNP biogenesis. ABT1 is an ∼31 kDa protein that is evolutionary conserved from yeast (Saccharomyces cerevisiae) to humans. Mouse Abt1 was initially implicated in general transcription on the basis of its ability to associate with TATA-binding protein and to stimulate weakly the activity of reporter genes (68). However, subsequent protein–protein interaction studies and localization studies indicated that ABT1, like its yeast homolog, functions in ribosome biogenesis (41). The yeast homolog of ABT1 is called Esf2p (for eighteen S factor 2 protein), is an essential protein and biochemical and genetic studies have unequivocally demonstrated that it functions in ribosome biogenesis (41). Specifically, Esf2p associates with U3 snoRNP (small nucleolar ribonucleoprotein) and the 5′ ETS (external transcribed spacer) of pre-ribosomal RNA (pre-rRNA) and is essential for early pre-rRNA processing (41). U3 snoRNP contains four proteins that are common to most snoRNPs and also many additional proteins, which are required for its assembly and function. Reptin and Pontin are two of the proteins that associate with U3 snoRNP and are required for its assembly in human cells (22). The yeast homolog of Reptin is also required for the assembly of yeast snoRNPs including yeast U3 snoRNP (69). Our finding that IGHMBP2 also associates with Reptin and Pontin and the fact that the Abt1 gene, whose protein product is required for ribosome biogenesis, is contained in the 27k3 genetic modifier suggest that IGHMBP2 plays a role in ribosome biogenesis or function.
While our manuscript was under revision, Guenther et al. (70) reported that IGHMBP2 associates with ribosomes. Our findings are in support of a role of IGHMBP2 in mRNA translation, and our biochemical and genetic investigations have led to a remarkable convergence of the pathways that IGHMBP2 is likely to function, which include ribosome and tRNA function, both of which are essential for mRNA translation. Future work will focus on delineating the molecular mechanism(s) of IGHMBP2 function and its specific requirements for motor neuron maintenance.
MATERIALS AND METHODS
Mice and genotypic selection
Mice were maintained in humidity and temperature-controlled rooms with a 12:12 dark:light cycle. They were given an NIH-mouse/rat diet with 6% fat, except the ALR/LtJ strain, which received 4% diet (5K54, PMI Feeds, Inc., St Louis, MO, USA) ad libitum with free access to water (HCl-acidified, pH 2.8–3.2). All procedures performed on the animals were reviewed and approved by our institutional animal care and use committee. The C57BL/6J–MnmC congenic strain [B6.CAST-(D13Mit17-D13Mit14)/Cx] was constructed by 10 backcross generations of a (B6.BKS-+/nmd2J × CAST/EiJ)F2 mouse to C57BL/6J as described previously (10). For construction of a high-resolution genetic map of the MnmC locus, 3528 (B6.CAST-+/nmd MnmB/MnmC) F2 mice were phenotyped and genotyped to identify recombinants using the flanking SSLP markers CA1 (27k3CA1F CCAGCAAGGAAGGGGAAAAC and 27k3CA1R TGGAGCTTGTTTCATCCACC) and CAGA (CAGA-F2 ATTTTGTCACTCCCTCCCTGCC and CAGA-R2 CTAAAACCTTCAAAGCAAACTGGG). F2 mice containing recombinant MnmB/MnmC genotypes in the absence of a homozygous nmd/nmd mutation (+/+ or +/nmd) were mated to known +/nmd heterozygotes until mutant nmd progeny appeared to determine the status of the Chr 13 Mnm modifier allele.
Generation and characterization of transgenic BAC-27K3 mice
Screening of CHORI-26 CAST BAC library by Southern blot hybridization was performed by TJL Molecular Biology Service using five probes derived from B6 sequences across the genetic interval (primer sequences available on request). CHORI-26 BAC 27K3 was microinjected into fertilized C57BL/6J embryos to generate eight independent transgenic C57BL/6J-Tg(BAC-27K3) founder lines that were identified with primers flanking polymorphic SSLP repeats: F49 TCTGCTCTGAATCCAACTGG and R51 AACATACCCATAGCCTCTAC (112 bp in B6 and 91 bp in CAST); F54 TGTTCCCAGCTGACCTTATC and R56 GCAGAGAAAGACAGATGGAC (116 bp in B6 and 134 bp in CAST). Transgenic-positive mice (+/+ for nmd) were bred to heterozygous B6.BKS-+/nmd mice and the F1 transgenic-positive heterozygous (+/nmd) offspring were backcrossed to B6.BKS-+/nmd mice to generate transgenic-positive mutant nmd/nmd mice (nmd-Tg27K3) for phenotypic analysis.
Histology and axon counts
Tissue was fixed by transcardial perfusion with 2% glutaraldehyde/ 2% paraformaldehyde in 0.1 m cacodylate buffer. Femoral nerves were dissected free and postfixed overnight in the same fixative and then processed for plastic embedding. For counts of axons, left and right nerves were taken whenever possible, and counts were averaged so that each n represents one mouse and the average count of the left and right nerve. The total number of myelinated axons in each nerve was counted using light microscopy on 0.5 µm sections stained with Toluidine blue. Images were taken on a Nikon E600 microscope using a Leica DFC300 FX digital camera.
DNA sequencing
EcoRI fragments of CHORI-26 CAST BAC clone 27K3 were subcloned into Invitrogen pCR 4-TOPO vector and were end-sequenced with vector primers T3 and T7 on an Applied Biosystems 3700 DNA sequencer. Complete sequencing of an 18.1 kb EcoRI fragment containing the Abt1 gene was accomplished using a transposon insertion kit, EZ::Tn-Tn5 TET-1 and EZ::TN KAN-2 according to the manufacturers instructions (Epicentre Technologies), and the sequence information was deposited into GenBank (Accession no. EU880213).
DNA constructs
The open-reading frames of IGHMBP2, ABT1, Zfp161 were cloned downstream of the CMV promoter into modified pcDNA3 vectors (Invitrogen) containing either the myc-tag or the FLAG-tag sequences at the amino terminus. All of the constructs were analyzed by DNA sequencing.
Antibodies
The anti-IGHMBP2 monoclonal antibody mAb11-24 was produced by immunization of BALB/c mice and fusion of spleen cells with Sp2/0 myeloma cells. The mouse sera and the hybridoma culture supernatants were both screened by ELISA, western blot, IP and immunofluorescence. Hybridoma cell lines were cloned to homogeneity by limiting dilution. Ig subclass was determined using an isotyping kit (Roche). Antibodies used were as follows: 11-24 mouse IgG1 monoclonal anti-IGHMBP2; 2B1, mouse IgG1 monoclonal anti-SMN; 2E17, mouse IgG1 monoclonal anti-Gemin2; 12H12, mouse IgG1 monoclonal anti-Gemin3; 17D10, mouse IgG1 monoclonal anti-Gemin4; 9E10, mouse IgG1 monoclonal anti-myc (Developmental Studies Hybridoma Bank); A-14, affinity-purified rabbit polyserum anti-myc 9E10 epitope (Santa Cruz Biotechnologies); M2, mouse IgG1 monoclonal anti-Flag (Sigma); anti-TFIIIC220 antibody (sc-23110, Santa Cruz, Inc.); anti-α-tubulin antibody (Ab4074, Abcam); anti-β-tubulin antibody (Ab6046, Abcam or E-7 from Developmental Studies Hybridoma Bank); anti-hnRNP H (Bethyl); anti-HSP70 (Abcam); anti-ZNF322a (Aviva).
Cell culture, IP, small RNA cloning
Cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen). Cells were transfected by the standard calcium phosphate method. Following overnight incubation with DNA, cells were washed and fresh medium was added. Stable clones of MN1 and 293T were obtained by selection in the presence of G418 (100 µg/ml). IP experiments were performed from total cell extracts followed by western blotting as described previously (36). For small RNA cloning, RNA was isolated from FLAG-IGHMBP2 immunoprecipitates; 3′-end labeling, directional adapter ligation, cloning and sequencing were performed as described previously (38).
In vitro translation and IP
FLAG-Zfp161, myc-IGHMBP2 and FLAG-IGHMBP2 proteins were generated by in vitro translation using the TNT T7 Quick-Coupled Transcription/Translation System in the presence of 35S-methionine (Promega) following the manufacturing instructions. The in vitro translated proteins were mixed and allowed to interact for 1 h at 4°C. IPs were carried out using either anti-myc antibody (9E10) or anti-FLAG M2 beads (Sigma). After binding, the beads were washed five times with RSB-100 buffer (20 mm Tris–HCL, pH 7.5, 1200 m NaCl, 2.5 mm MgCl2, 0.5% Triton X100 and complete EDTA-free protease inhibitors) and boiled in SDS–PAGE loading buffer. The IPs were analyzed by NuPAGE and exposed to a Biomax XAR film (Kodak).
Affinity purification of IGHMBP2 protein complex
FLAG-IGHMBP2 and control cell lines were grown in the presence of G418 (100 µg/ml). Total cell extracts were prepared by resuspending cell pellets in RSB-100 (10 mm Tris–HCl, pH 7.4, 2.5 mm MgCl2, 100 mm NaCl) buffer containing 0.1% Nonidet P-40 and protease inhibitors. Cells were sonicated briefly. Following centrifugation at 10 000 rpm for 15 min, supernatants were added to anti-FLAG beads (Sigma) pre-washed with the same buffer. Extracts were incubated with anti-FLAG beads for 2 h at 4°C. Supernatants were discarded, and beads were extensively washed with RSB-100 containing 0.02% Nonidet P-40. Three high-salt washes were performed with 10 bed volumes of RSB-300 (300 mm NaCl) buffer containing 0.02% Nonidet P-40 for 15 min at 4°C. Following three washes with RSB-100 containing 0.02% Nonidet P-40, bound complexes were eluted with 10 bed volumes of the same buffer containing 0.25–0.5 mg/ml 3X-FLAG peptide (Sigma) for 1 h at 4°C.
Mass spectrometry
Bands were excised from Coomassie-stained NuPAGE gel, in gel-digested with trypsin and peptides were identified by tandem MS-MS. The peptide sequences obtained from mass spectrometry for selected proteins that co-immunoprecipitate with FLAG-IGHMBP2 (Fig. 2) were RuvB-like 2 (Reptin/Tip48; Q9Y230): AVLIAGQPGTGK, VYSLFLDESR, QASQGMVGQLAAR; RuvB-like 1 (Pontin/Tip49; Q9Y265): LDPSIFESLQK, TALALAIAQELGSK, ALESSIAPIVIFASNR; TFIIIC220 (TFIIICa, TFIIIC-220Kda; Q12789): MGITPLR, KNITNDIR; hnRNPH (P31943): YIEIFK, THYDPPR, IQNGAQGIR, VHIEIGPDGR, HTGPNSPDTANDGFVR; β-tubulin (P05218): YLTVAAVFR, FPGQLNADLR, LAVNMVPFPR, ISEQFTAMFR, ISVYYNEATGGK; α-tubulin (P05209): FDLMYAK, LSVDYGKK, EDMAALEK, DVNAAIATIK, EIIDLVLDR, AVFVDLEPTVIDEVR; HSP70 (P08107): SAVEDEGLK, LLQDFFNGR, VEIIANDQGNR, TTPSYVAFTDTER; Zfp-161 (O43829): HENNMHSER.
Reverse transcriptase–PCR
Total RNA was extracted from forebrain or spinal cord of wild-type, nmd or nmd-Tg27K3 mice using TRIzol reagent (Invitrogen). RNA was treated with DNAse I (Roche) and reverse-transcribed with oligo-dT using Advantage RT-for-PCR kit (Clontech). The cDNA was used as a template for PCR with primers E4F (5′-CGCCTGAAAAAAGCACTGATGAC) and E5R (5′-TGTGTTGTAGAAAGAGAGTGGGG), and the PCR products were analyzed on 2% Agarose electrophoresis gel.
Immunofluorescence
NT2 cells plated on glass coverslips were briefly washed with PBS, fixed in 2% formaldehyde-PBS for 30 min at room temperature and permeabilized in 0.5% Triton X-100-PBS for 5 min at room temperature. Cells were blocked in 3% bovine serum albumin in PBS for 30 min at room temperature. Double-label immunofluorescence experiments were performed by separate sequential incubations of each primary antibody diluted in PBS containing 3% bovine serum albumin followed by the specific secondary antibody coupled to fluorescein isothiocyanate or TRITC. All incubations were at room temperature for 1 h.
SUPPLEMENTARY MATERIAL
FUNDING
Supported by NIH grant NS056070 to Z.M. and G.A.C.
Supplementary Material
ACKNOWLEDGEMENTS
G.A.C. and Z.M. are equal senior contributors and guided the genetic and biochemical experiments, respectively. We thank the members of our laboratories and Francesco Lotti for discussions and many helpful suggestions. We are grateful to G. Dreyfuss for anti-SMN and anti-Gemin antibodies.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Grohmann K., Schuelke M., Diers A., Hoffmann K., Lucke B., Adams C., Bertini E., Leonhardt-Horti H., Muntoni F., Ouvrier R., et al. Mutations in the gene encoding immunoglobulin mu-binding protein 2 cause spinal muscular atrophy with respiratory distress type 1. Nat. Genet. 2001;29:75–77. doi: 10.1038/ng703. [DOI] [PubMed] [Google Scholar]
- 2.Grohmann K., Varon R., Stolz P., Schuelke M., Janetzki C., Bertini E., Bushby K., Muntoni F., Ouvrier R., Van Maldergem L., et al. Infantile spinal muscular atrophy with respiratory distress type 1 (SMARD1) Ann. Neurol. 2003;54:719–724. doi: 10.1002/ana.10755. [DOI] [PubMed] [Google Scholar]
- 3.Grohmann K., Rossoll W., Kobsar I., Holtmann B., Jablonka S., Wessig C., Stoltenburg-Didinger G., Fischer U., Hubner C., Martini R., et al. Characterization of Ighmbp2 in motor neurons and implications for the pathomechanism in a mouse model of human spinal muscular atrophy with respiratory distress type 1 (SMARD1) Hum. Mol. Genet. 2004;13:2031–2042. doi: 10.1093/hmg/ddh222. [DOI] [PubMed] [Google Scholar]
- 4.Fukita Y., Mizuta T.R., Shirozu M., Ozawa K., Shimizu A., Honjo T. The human S mu bp-2, a DNA-binding protein specific to the single-stranded guanine-rich sequence related to the immunoglobulin mu chain switch region. J. Biol. Chem. 1993;268:17463–17470. [PubMed] [Google Scholar]
- 5.Kerr D., Khalili K. A recombinant cDNA derived from human brain encodes a DNA binding protein that stimulates transcription of the human neurotropic virus JCV. J. Biol. Chem. 1991;266:15876–15881. [PubMed] [Google Scholar]
- 6.Mizuta T.R., Fukita Y., Miyoshi T., Shimizu A., Honjo T. Isolation of cDNA encoding a binding protein specific to 5′-phosphorylated single-stranded DNA with G-rich sequences. Nucleic Acids Res. 1993;21:1761–1766. doi: 10.1093/nar/21.8.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shieh S.Y., Stellrecht C.M., Tsai M.J. Molecular characterization of the rat insulin enhancer-binding complex 3b2. Cloning of a binding factor with putative helicase motifs. J. Biol. Chem. 1995;270:21503–21508. doi: 10.1074/jbc.270.37.21503. [DOI] [PubMed] [Google Scholar]
- 8.Sebastiani G., Durocher D., Gros P., Nemer M., Malo D. Localization of the Catf1 transcription factor gene to mouse chromosome 19. Mamm. Genome. 1995;6:147–148. doi: 10.1007/BF00303264. [DOI] [PubMed] [Google Scholar]
- 9.Molnar G.M., Crozat A., Kraeft S.K., Dou Q.P., Chen L.B., Pardee A.B. Association of the mammalian helicase MAH with the pre-mRNA splicing complex. Proc. Natl Acad. Sci. USA. 1997;94:7831–7836. doi: 10.1073/pnas.94.15.7831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cox G.A., Mahaffey C.L., Frankel W.N. Identification of the mouse neuromuscular degeneration gene and mapping of a second site suppressor allele. Neuron. 1998;21:1327–1337. doi: 10.1016/s0896-6273(00)80652-2. [DOI] [PubMed] [Google Scholar]
- 11.Maddatu T.P., Garvey S.M., Schroeder D.G., Hampton T.G., Cox G.A. Transgenic rescue of neurogenic atrophy in the nmd mouse reveals a role for Ighmbp2 in dilated cardiomyopathy. Hum. Mol. Genet. 2004;13:1105–1115. doi: 10.1093/hmg/ddh129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maddatu T.P., Garvey S.M., Schroeder D.G., Zhang W., Kim S.-Y., Nicholson A.I., Davis C.J., Cox G.A. Dilated cardiomyopathy (DCM) in the nmd mouse: transgenic rescue and QTLs that improve cardiac function and survival. Hum. Mol. Genet. 2005;14:3179–3189. doi: 10.1093/hmg/ddi349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schattner P., Brooks A.N., Lowe T.M. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005;33:W686–W689. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hopper A.K., Phizicky E.M. tRNA transfers to the limelight. Genes Dev. 2003;17:162–180. doi: 10.1101/gad.1049103. [DOI] [PubMed] [Google Scholar]
- 15.Wolin S.L., Matera A.G. The trials and travels of tRNA. Genes Dev. 1999;13:1–10. doi: 10.1101/gad.13.1.1. [DOI] [PubMed] [Google Scholar]
- 16.Geiduschek E.P., Kassavetis G.A. The RNA polymerase III transcription apparatus. J. Mol. Biol. 2001;310:1–26. doi: 10.1006/jmbi.2001.4732. [DOI] [PubMed] [Google Scholar]
- 17.Schramm L., Hernandez N. Recruitment of RNA polymerase III to its target promoters. Genes. Dev. 2002;16:2593–2620. doi: 10.1101/gad.1018902. [DOI] [PubMed] [Google Scholar]
- 18.Paushkin S.V., Patel M., Furia B.S., Peltz S.W., Trotta C.R. Identification of a human endonuclease complex reveals a link between tRNA splicing and pre-mRNA 3′ end formation. Cell. 2004;117:311–321. doi: 10.1016/s0092-8674(04)00342-3. [DOI] [PubMed] [Google Scholar]
- 19.Nakanishi K., Nureki O. Recent progress of structural biology of tRNA processing and modification. Mol. Cells. 2005;19:157–166. [PubMed] [Google Scholar]
- 20.Kiss T. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell. 2002;109:145–148. doi: 10.1016/s0092-8674(02)00718-3. [DOI] [PubMed] [Google Scholar]
- 21.Rouquette J., Choesmel V., Gleizes P.E. Nuclear export and cytoplasmic processing of precursors to the 40S ribosomal subunits in mammalian cells. EMBO J. 2005;24:2862–2872. doi: 10.1038/sj.emboj.7600752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watkins N.J., Lemm I., Ingelfinger D., Schneider C., Hossbach M., Urlaub H., Luhrmann R. Assembly and maturation of the U3 snoRNP in the nucleoplasm in a large dynamic multiprotein complex. Mol. Cell. 2004;16:789–798. doi: 10.1016/j.molcel.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 23.Kass S., Tyc K., Steitz J.A., Sollner-Webb B. The U3 small nucleolar ribonucleoprotein functions in the first step of preribosomal RNA processing. Cell. 1990;60:897–908. doi: 10.1016/0092-8674(90)90338-f. [DOI] [PubMed] [Google Scholar]
- 24.Lefebvre S., Burglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M., et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 25.Iannaccone S.T., Burghes A. Spinal muscular atrophies. Adv. Neurol. 2002;88:83–98. [PubMed] [Google Scholar]
- 26.Gubitz A.K., Feng W., Dreyfuss G. The SMN complex. Exp. Cell Res. 2004;296:51–56. doi: 10.1016/j.yexcr.2004.03.022. [DOI] [PubMed] [Google Scholar]
- 27.Paushkin S., Gubitz A.K., Massenet S., Dreyfuss G. The SMN complex, an assemblyosome of ribonucleoproteins. Curr. Opin. Cell Biol. 2002;14:305–312. doi: 10.1016/s0955-0674(02)00332-0. [DOI] [PubMed] [Google Scholar]
- 28.Meister G., Eggert C., Fischer U. SMN-mediated assembly of RNPs: a complex story. Trends Cell Biol. 2002;12:472–478. doi: 10.1016/s0962-8924(02)02371-1. [DOI] [PubMed] [Google Scholar]
- 29.Fischer U., Liu Q., Dreyfuss G. The SMN-SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell. 1997;90:1023–1029. doi: 10.1016/s0092-8674(00)80368-2. [DOI] [PubMed] [Google Scholar]
- 30.Meister G., Fischer U. Assisted RNP assembly: SMN and PRMT5 complexes cooperate in the formation of spliceosomal UsnRNPs. EMBO J. 2002;21:5853–5863. doi: 10.1093/emboj/cdf585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pellizzoni L., Yong J., Dreyfuss G. Essential role for the SMN complex in the specificity of snRNP assembly. Science. 2002;298:1775–1779. doi: 10.1126/science.1074962. [DOI] [PubMed] [Google Scholar]
- 32.Narayanan U., Achsel T., Luhrmann R., Matera A.G. Coupled in vitro import of U snRNPs and SMN, the spinal muscular atrophy protein. Mol. Cell. 2004;16:223–234. doi: 10.1016/j.molcel.2004.09.024. [DOI] [PubMed] [Google Scholar]
- 33.Salazar-Grueso E.F., Kim S., Kim H. Embryonic mouse spinal cord motor neuron hybrid cells. Neuroreport. 1991;2:505–508. doi: 10.1097/00001756-199109000-00002. [DOI] [PubMed] [Google Scholar]
- 34.Brooks B.P., Merry D.E., Paulson H.L., Lieberman A.P., Kolson D.L., Fischbeck K.H. A cell culture model for androgen effects in motor neurons. J. Neurochem. 1998;70:1054–1060. doi: 10.1046/j.1471-4159.1998.70031054.x. [DOI] [PubMed] [Google Scholar]
- 35.Lieberman A.P., Harmison G., Strand A.D., Olson J.M., Fischbeck K.H. Altered transcriptional regulation in cells expressing the expanded polyglutamine androgen receptor. Hum. Mol. Genet. 2002;11:1967–1976. doi: 10.1093/hmg/11.17.1967. [DOI] [PubMed] [Google Scholar]
- 36.Mourelatos Z., Dostie J., Paushkin S., Sharma A., Charroux B., Abel L., Rappsilber J., Mann M., Dreyfuss G. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 2002;16:720–728. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maniataki E., Mourelatos Z. Human mitochondrial tRNAMet is exported to the cytoplasm and associates with the Argonaute 2 protein. RNA. 2005;11:849–852. doi: 10.1261/rna.2210805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maniataki E., De Planell Saguer M.D., Mourelatos Z. Immunoprecipitation of microRNPs and directional cloning of microRNAs. Methods Mol. Biol. 2005;309:283–294. doi: 10.1385/1-59259-935-4:283. [DOI] [PubMed] [Google Scholar]
- 39.Thompson D.M., Lu C., Green P.J., Parker R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA. 2008;14:2095–2103. doi: 10.1261/rna.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wade C.M., Kulbokas E.J., III, Kirby A.W., Zody M.C., Mullikin J.C., Lander E.S., Lindblad-Toh K., Daly M.J. The mosaic structure of variation in the laboratory mouse genome. Nature. 2002;420:574–578. doi: 10.1038/nature01252. [DOI] [PubMed] [Google Scholar]
- 41.Hoang T., Peng W.T., Vanrobays E., Krogan N., Hiley S., Beyer A.L., Osheim Y.N., Greenblatt J., Hughes T.R., Lafontaine D.L. Esf2p, a U3-associated factor required for small-subunit processome assembly and compaction. Mol. Cell. Biol. 2005;25:5523–5534. doi: 10.1128/MCB.25.13.5523-5534.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Z., Lotti F., Dittmar K., Younis I., Wan L., Kasim M., Dreyfuss G. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell. 2008;133:585–600. doi: 10.1016/j.cell.2008.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jordanova A., Irobi J., Thomas F.P., Van Dijck P., Meerschaert K., Dewil M., Dierick I., Jacobs A., De Vriendt E., Guergueltcheva V., et al. Disrupted function and axonal distribution of mutant tyrosyl-tRNA synthetase in dominant intermediate Charcot–Marie–Tooth neuropathy. Nat. Genet. 2006;38:197–202. doi: 10.1038/ng1727. [DOI] [PubMed] [Google Scholar]
- 44.Antonellis A., Ellsworth R.E., Sambuughin N., Puls I., Abel A., Lee-Lin S.Q., Jordanova A., Kremensky I., Christodoulou K., Middleton L.T., et al. Glycyl tRNA synthetase mutations in Charcot–Marie–Tooth disease type 2D and distal spinal muscular atrophy type V. Am. J. Hum. Genet. 2003;72:1293–1299. doi: 10.1086/375039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sivakumar K., Kyriakides T., Puls I., Nicholson G.A., Funalot B., Antonellis A., Sambuughin N., Christodoulou K., Beggs J.L., Zamba-Papanicolaou E., et al. Phenotypic spectrum of disorders associated with glycyl-tRNA synthetase mutations. Brain. 2005;128:2304–2314. doi: 10.1093/brain/awh590. [DOI] [PubMed] [Google Scholar]
- 46.Chen Y.Z., Bennett C.L., Huynh H.M., Blair I.P., Puls I., Irobi J., Dierick I., Abel A., Kennerson M.L., Rabin B.A., et al. DNA/RNA helicase gene mutations in a form of juvenile amyotrophic lateral sclerosis (ALS4) Am. J. Hum. Genet. 2004;74:1128–1135. doi: 10.1086/421054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ursic D., Himmel K.L., Gurley K.A., Webb F., Culbertson M.R. The yeast SEN1 gene is required for the processing of diverse RNA classes. Nucleic Acids Res. 1997;25:4778–4785. doi: 10.1093/nar/25.23.4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen Y.Z., Hashemi S.H., Anderson S.K., Huang Y., Moreira M.C., Lynch D.R., Glass I.A., Chance P.F., Bennett C.L. Senataxin, the yeast Sen1p orthologue: characterization of a unique protein in which recessive mutations cause ataxia and dominant mutations cause motor neuron disease. Neurobiol. Dis. 2006;23:97–108. doi: 10.1016/j.nbd.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 49.Neumann M., Sampathu D.M., Kwong L.K., Truax A.C., Micsenyi M.C., Chou T.T., Bruce J., Schuck T., Grossman M., Clark C.M., et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 50.Van Deerlin V.M., Leverenz J.B., Bekris L.M., Bird T.D., Yuan W., Elman L.B., Clay D., Wood E.M., Chen-Plotkin A.S., Martinez-Lage M., et al. TARDBP mutations in amyotrophic lateral sclerosis with TDP-43 neuropathology: a genetic and histopathological analysis. Lancet Neurol. 2008;7:409–416. doi: 10.1016/S1474-4422(08)70071-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kabashi E., Valdmanis P.N., Dion P., Spiegelman D., McConkey B.J., Vande Velde C., Bouchard J.P., Lacomblez L., Pochigaeva K., Salachas F., et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat. Genet. 2008;40:572–574. doi: 10.1038/ng.132. [DOI] [PubMed] [Google Scholar]
- 52.Sreedharan J., Blair I.P., Tripathi V.B., Hu X., Vance C., Rogelj B., Ackerley S., Durnall J.C., Williams K.L., Buratti E., et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gitcho M.A., Baloh R.H., Chakraverty S., Mayo K., Norton J.B., Levitch D., Hatanpaa K.J., White C.L., III, Bigio E.H., Caselli R., et al. TDP-43 A315T mutation in familial motor neuron disease. Ann. Neurol. 2008;63:535–538. doi: 10.1002/ana.21344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kwiatkowski T.J., Jr, Bosco D.A., LeClerc A.L., Tamrazian E., Vanderburg C.R., Russ C., Davis A., Gilchrist J., Kasarskis E.J., Munsat T., et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 55.Vance C., Rogelj B., Hortobágyi T., De Vos K.J., Nishimura A.G., Sreedharan J., Hu J., Smith B., Ruddy D., Wright P., et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johnson P.F., Abelson J. The yeast tRNATyr gene intron is essential for correct modification of its tRNA product. Nature. 1983;302:681–687. doi: 10.1038/302681a0. [DOI] [PubMed] [Google Scholar]
- 57.van Tol H., Beier H. All human tRNATyr genes contain introns as a prerequisite for pseudouridine biosynthesis in the anticodon. Nucleic Acids Res. 1988;16:1951–1966. doi: 10.1093/nar/16.5.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kanemaki M., Makino Y., Yoshida T., Kishimoto T., Koga A., Yamamoto K., Yamamoto M., Moncollin V., Egly J.M., Muramatsu M., et al. Molecular cloning of a rat 49-kDa TBP-interacting protein (TIP49) that is highly homologous to the bacterial RuvB. Biochem. Biophys. Res. Commun. 1997;235:64–68. doi: 10.1006/bbrc.1997.6729. [DOI] [PubMed] [Google Scholar]
- 59.Bauer A., Huber O., Kemler R. Pontin52, an interaction partner of beta-catenin, binds to the TATA box binding protein. Proc. Natl Acad. Sci. USA. 1998;95:14787–14792. doi: 10.1073/pnas.95.25.14787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kanemaki M., Kurokawa Y., Matsu-ura T., Makino Y., Masani A., Okazaki K., Morishita T., Tamura T.A. TIP49b, a new RuvB-like DNA helicase, is included in a complex together with another RuvB-like DNA helicase, TIP49a. J. Biol. Chem. 1999;274:22437–22444. doi: 10.1074/jbc.274.32.22437. [DOI] [PubMed] [Google Scholar]
- 61.Bauer A., Chauvet S., Huber O., Usseglio F., Rothbacher U., Aragnol D., Kemler R., Pradel J. Pontin52 and reptin52 function as antagonistic regulators of beta-catenin signalling activity. EMBO J. 2000;19:6121–6130. doi: 10.1093/emboj/19.22.6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ikura T., Ogryzko V.V., Grigoriev M., Groisman R., Wang J., Horikoshi M., Scully R., Qin J., Nakatani Y. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- 63.Shen X., Mizuguchi G., Hamiche A., Wu C. A chromatin remodelling complex involved in transcription and DNA processing. Nature. 2000;406:541–544. doi: 10.1038/35020123. [DOI] [PubMed] [Google Scholar]
- 64.Saurin A.J., Shao Z., Erdjument-Bromage H., Tempst P., Kingston R.E. A Drosophila Polycomb group complex includes Zeste and dTAFII proteins. Nature. 2001;412:655–660. doi: 10.1038/35088096. [DOI] [PubMed] [Google Scholar]
- 65.Ohdate H., Lim C.R., Kokubo T., Matsubara K., Kimata Y., Kohno K. Impairment of the DNA binding activity of the TATA-binding protein renders the transcriptional function of Rvb2p/Tih2p, the yeast RuvB-like protein, essential for cell growth. J. Biol. Chem. 2003;278:14647–14656. doi: 10.1074/jbc.M213220200. [DOI] [PubMed] [Google Scholar]
- 66.Gstaiger M., Luke B., Hess D., Oakeley E.J., Wirbelauer C., Blondel M., Vigneron M., Peter M., Krek W. Control of nutrient-sensitive transcription programs by the unconventional prefoldin URI. Science. 2003;302:1208–1212. doi: 10.1126/science.1088401. [DOI] [PubMed] [Google Scholar]
- 67.Jonsson Z.O., Jha S., Wohlschlegel J.A., Dutta A. Rvb1p/Rvb2p recruit Arp5p and assemble a functional Ino80 chromatin remodeling complex. Mol. Cell. 2004;16:465–477. doi: 10.1016/j.molcel.2004.09.033. [DOI] [PubMed] [Google Scholar]
- 68.Oda T., Kayukawa K., Hagiwara H., Yudate H.T., Masuho Y., Murakami Y., Tamura T.A., Muramatsu M.A. A novel TATA-binding protein-binding protein, ABT1, activates basal transcription and has a yeast homolog that is essential for growth. Mol. Cell. Biol. 2000;20:1407–1418. doi: 10.1128/mcb.20.4.1407-1418.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.King T.H., Decatur W.A., Bertrand E., Maxwell E.S., Fournier M.J. A well-connected and conserved nucleoplasmic helicase is required for production of box C/D and H/ACA snoRNAs and localization of snoRNP proteins. Mol. Cell. Biol. 2001;21:7731–7746. doi: 10.1128/MCB.21.22.7731-7746.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guenther U.P., Handoko L., Laggerbauer B., Jablonka S., Chari A., Alzheimer M., Ohmer J., Plottner O., Gehring N., Sickmann A., et al. IGHMBP2 is a ribosome-associated helicase inactive in the neuromuscular disorder distal SMA type 1 (DSMA1) Hum. Mol. Genet. 2009;18:1288–1300. doi: 10.1093/hmg/ddp028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.