Abstract
Despite many advances in membrane proteomics during the last decade the fundamental problem of accessing the transmembrane regions itself has only been addressed to some extent. The present study establishes a method for the nano-LC-based analysis of complex membrane proteomes on the basis of a methanolic porcine pancreatic elastase digest to increase transmembrane coverage. Halobacterium salinarium purple and Corynebacterium glutamicum membranes were successfully analyzed by using the new protocol. We demonstrated that elastase digests yield a large proportion of transmembrane peptides, facilitating membrane protein identification. The potential for characterization of a membrane protein through full sequence coverage using elastase is there but is restricted to the higher abundance protein components. Compatibility of the work flow with the two most common mass spectrometric ionization techniques, ESI and MALDI, was shown. Currently better results are obtained using ESI mainly because of the low response of MALDI for strictly neutral peptides. New findings concerning elastase specificity in complex protein mixtures reveal a new prospect beyond the application in shotgun experiments. Furthermore peptide mass fingerprinting with less specific enzymes might be done in the near future but requires an adaptation of current search algorithms to the new proteases.
Upon the introduction of modern mass spectrometric ionization techniques, such as MALDI (1) and ESI (2), extremely powerful and valuable tools were given to researchers for the identification and characterization of proteins. Nevertheless the intricate analysis of membrane subproteomes still represents one of the major challenges despite the amount of “success” reported in the literature. Until now, there was no general protocol available to address membrane proteomes as a whole, demonstrating their degree of difficulty and complexity.
During the past years, two different strategies in proteome analysis have evolved: the more widespread bottom-up and top-down proteomics. The latter approach has been shown to provide access to the transmembranal regions of membrane proteins and has the power to characterize complete protein primary structures including labile covalent modifications (3, 4). Nevertheless there are limitations due to sample complexity, emphasizing the need for a liquid chromatographic separation on the protein level, which is comparable to LC peptide separation. Therefore, the bottom-up variant is the most commonly used method for the proteomics analysis of complex membrane samples. Improvements in the bottom-up work flow were achieved by adapting sample cleanup and prefractionation processes (5–10) and subsequently by the development of modified and optimized separation techniques. The two main work flows, the gel-based approach mainly carried out with 2D1 SDS-PAGE (11, 12) and the shotgun identification of proteolytically digested protein mixtures and their multidimensional separation via liquid chromatography (13), had to be adapted. The separation of proteins via classical 2D SDS-PAGE is only possible up to a GRAVY score of ∼0.4 (14, 15). Despite lacking the separation power of the IEF-SDS-PAGE system, derived techniques like the doubled SDS- (16) and 16-benzyldimethyl-n-hexadecylammonium chloride/SDS-PAGE (17) represent an improvement for hydrophobic proteins. Advances in the nLC separation of hydrophobic peptides, e.g. the separation at elevated temperatures (18) and the use of LC-compatible detergents (19), yielded significant success. When combining both prominent separation techniques, the one-dimensional SDS-LC analysis proved to be more effective than 2D PAGE as well (20, 21).
One of the remaining steps still offering room for improvements is the proteolytic procedure. The original multidimensional protein identification technique using trypsin has been successfully improved by the application of proteinase K under high pH conditions, yielding an increased number of membrane protein identifications (22). Further modification of this method by recleaving the isolated membranous parts with cyanogen bromide increased the accessibility to membrane-spanning peptides (18).
Despite the suitability of proteinase K for the shotgun analysis of membrane proteomes, trypsin is the prevalent enzyme choice in most current proteomics approaches because of its very specific cleavage behavior. Calculations have implied that alternative proteases are preferentially suited for the analysis of membrane proteomes (23). Proteases other than trypsin have been utilized only to a small degree and mostly for the targeted analysis of protein complexes. Pepsin, for example, was used for the characterization of an aquaporin (24), and elastase and subtilisin were used as proteases in a triple digest approach of protein complexes and lens tissue (25). Further improvements in the accessibility of the membrane proteome have been shown for tryptic (26, 27) and tryptic/chymotryptic (28) digests by performing the proteolytic treatment in the presence of methanol.
During membrane proteomics method development, purple membranes from Halobacterium sp. NRC-1, consisting to a large extent of the H+-ATPase bacteriorhodopsin, have widely been used as reference in a variety of cases. Full BR sequence coverage has been reported by several publications using different techniques (27, 29). Such exorbitant sequence coverage amounts are only achievable for the most abundant proteins within a complex mixture. The identification of less expressed proteins, which certainly are more in the researcher's interest, occurs via lower peptide numbers (27).
Generally the basic tryptic cleavage sites are predominantly located in the loops facing the cytoplasm and to a lesser extent in the extracellular or periplasmic loops but are very scarce within the transmembrane helix stretches. This mostly limits the potential tryptic fragments to loop components and larger peptides containing at least one TM helix. The use of another protease, preferentially cleaving after neutral aliphatic residues, should not succumb to this lack of cleavage sites.
One of the previously mentioned proteases, porcine pancreatic elastase, was reported to possess potential cleavage specificity at the carboxyl-terminal side of small neutral amino acids (30). It has been used previously for the examination of single protein phosphorylations (31) but was described not to be suited for the cleavage of complex membrane-containing samples because of a supposedly limited activity when applied to them (22). Despite this finding, elastase has been successfully utilized in a mass spectrometric analysis as early as 1974 when it was regarded as an ideal protease for future mass spectrometric studies (32). At the same time, most of the experiments to characterize elastase by analyzing its S1 pocket (for nomenclature, see Ref. 33) binding capabilities (34) as well as cleavage preferences of ester and protein substrates (35–39) were performed.
Based on previous research concerning elastase, we set up an nLC-based membrane proteomics analysis. This large data set was used to characterize the protease behavior of elastase and the physicochemical properties of the detected peptides. The PM model was used to establish a method to analyze more complex Corynebacterium glutamicum membranes, which have been analyzed previously using different 2D techniques (7, 28). We demonstrated that a combination of elastase and methanol is particularly suitable for an nLC-based membrane proteome analysis and results in a significantly increased number of TM peptides. Additionally the promising PMF application of elastase digests is discussed.
EXPERIMENTAL PROCEDURES
Bacterial Cells and Chemicals—
C. glutamicum strain DM 1698 (derived from ATCC 21527) membranes were purified as described before (7). Purple membranes from Halobacterium salinarium were a generous gift from Norbert Dencher (Physical Biochemistry, Technische Universität, Darmstadt, Germany). Tosylphenylalanyl chloromethyl ketone-treated dimethylated trypsin was purchased from Sigma-Aldrich. Porcine elastase (Grade II) was from Roche Diagnostics. All other chemicals and solvents were obtained from Carl Roth (Karlsruhe, Germany) if not otherwise noted. Only HPLC-grade or better solvents were used throughout the experiments in this study.
Proteolytic Digest—
The proteolytic treatment represents a modified version of the specific integral membrane peptide level enrichment as described previously (28). One hundred micrograms of PM were washed using 25 mm NH4HCO3 buffer (pH 8.0). After a 2-min centrifugation at 10,000 × g the supernatant was removed, and the pellet was gently resuspended in methanol. Following a 20-min period of sonication, the methanol was diluted to 60% using 25 mm NH4HCO3 buffer. The proteolytic digest was started by adding either 2 μg of trypsin or 10 μg of elastase and incubated for 24 h at 30 °C. To avoid any adhesion of the membrane slurry to the tubes, the whole digest process was performed without any external shaking. Prior to storing the peptide-containing supernatant at −20 °C, the sample was mixed and centrifuged at 10,000 × g for 2 min.
In the case of the more complex CB, for which protein content was determined after the method of Lowry (40), an additional digestive step before the actual proteolytic treatment was introduced to remove the loosely attached cytoplasmic contaminants. Two hundred micrograms of washed membranes were digested with the appropriate enzyme in 25 mm NH4HCO3 for 12 h at 30 °C. Then the supernatant was discarded after centrifugation, and the pellet underwent the same procedure as described above, starting with the methanol addition.
MALDI MS—
Mass spectra of in-solution digests were acquired using either an Applied Biosystems/MDS Sciex 4800 TOF/TOF™ Analyzer or a Bruker Daltonics Ultraflex™ TOF/TOF. Appropriate dilutions of the digest solutions were spotted on a target via the standard dried droplet preparation protocol. The matrix solution containing 2 mg/ml α-cyano-4-hydroxycinnamic acid and all dilutions were prepared with 50% ACN, 50% H2O, 0.5% TFA solvent. Reduction of salt contaminations was achieved by washing each preparation with ice-cold 5% formic acid.
nLC-MALDI MS/MS—
Nanoflow LC separation was carried out using an Agilent Technologies 1100 series HPLC system consisting of a nanocapillary pump, solvent degasser, a fraction collector, and a thermostatic autosampler. Samples containing 25 μg of PM or 200 μg of CB proteins were vacuum-centrifuged to a volume of about 8 μl. Solvent A (92% H2O, 8% ACN, 0.1% TFA) was added to obtain a total volume of 18 μl. This peptide mixture was injected onto the precolumn (Agilent Zorbax™ 300 SB C18, 75 μm × 3 mm). Preconcentration and desalting were carried out by flushing with Solvent A at a flow rate of 20 μl/min. The separation of the peptide mixture was performed on a 75-μm × 150-mm, C18 3.5-μm, 100-Å LC Packings Dionex PepMap™ 100 column at a flow rate of 300 nl/min with increasing ACN concentration. A step gradient mobile phase system was used: 3–55% Solvent B (95% ACN, 5% H2O, 0.1% TFA) in 110 min, 55–100% Solvent B in 10 min, 100% Solvent B for 10 min, 100–3% Solvent B in 3 min, and 2 min at 3% Solvent B. The separated peptides were mixed on a tee (Upchurch Scientific) with matrix solution supplied by an auxiliary pump (flow rate, 1.2 μl/min). This solution contained 3 mg/ml α-CHCA (Bruker Daltonics, Bremen, Germany) dissolved in 70% ACN, 30% H2O, 0.1% TFA and spiked with 20 fmol (final amount) of [Glu1]fibrinopeptide B (Bachem, Weil, Germany) per spot for internal calibration. The final mixture was directly spotted every 15 s on a blank 123 × 81-mm Opti-TOF™ LC/MALDI Insert metal target.
Subsequent MALDI-TOF/TOF measurements were carried out using the 4800 TOF/TOF Analyzer. All peptides used for calibration were taken from the Sequazyme™ Peptide Mass Standards kit by Applied Biosystems. Spectra were acquired in the positive reflector mode between 700 and 6000 m/z with fixed laser intensity. A total of 750 laser shots per spot were accumulated. An eight-point plate model default calibration followed by internal recalibration to the spiked [Glu1]fibrinopeptide B mass was performed to ensure proper MS/MS precursor accuracy within 30 ppm. The precursor selection for MS/MS was carried out via the software job-wide interpretation feature of the instrument to avoid unnecessary multiple selections of identical precursor peptides. Up to six precursors per spot were selected for fragmentation, each requiring a minimum signal-to-noise ratio of 50. The fragmentation of the selected precursors was performed at a collision energy of 1 kV using air as collision gas at a pressure of 1 × 10−6 torr. Depending on the spectral quality, 1250 up to 2500 laser shots were recorded. Potential matrix cluster signals were removed from precursor selection by excluding all masses in the range from 700 to 1400 m/z having values of 0.030 ± 0.1 m/z as well as the internal calibrant mass.
nLC-ESI MS/MS—
Twenty-five micrograms of PM or 200 μg of CB protein digest were prepurified with C18 syringe tips (C18-AR, Varian), and the peptides were eluted with Solvent B (80% ACN, 0.1% formic acid), vacuum-concentrated, and reconstituted in 20 μl of Solvent A (2% ACN, 0.1% formic acid). The nLC-ESI-MS/MS system was operated generally as described previously (28, 41) except that mass spectra were recorded on an LTQ-Orbitrap instead of an LTQ. The linear ion trap and orbitrap were operated in parallel, i.e. during a full MS scan on the orbitrap at a resolution of 60,000, MS/MS spectra of the four most intense precursors were detected in the ion trap. Further instrument settings were used as described by Fischer et al. (28). The solvent gradient was from 0 to 50% Solvent B in 120 min followed by 10 min of Solvent B and 10 min of Solvent A with a flow rate of ∼250 nl/min on the capillary column.
Database Searches—
Mascot generic format (mgf) files were retrieved from each nLC-MALDI MS/MS run using the built-in Peaks2Mascot feature, exporting up to 65 peaks per MS/MS spectrum, each requiring a minimum signal-to-noise of 5. The corresponding ESI files were generated by first extracting all scans from the Xcalibur binary files using the export_msn tool from Thermo Electron. All settings were at default except for the number of grouped scans, which was reduced to 1. The generated .dta files containing the MS/MS data were then merged into a single .mgf file using the merge.pl script hosted on the Mascot™ site (Matrix Science Ltd.). MS/MS peak lists from three to six replicate runs were concatenated into one single file for the database search.
MS/MS queries were processed using the Mascot database search engine v2.2.03 (Matrix Science Ltd.; Ref. 42). Data were analyzed using the following settings: <30-ppm MS precursor mass tolerance and <0.5-Da MS/MS mass tolerance for MALDI-TOF/TOF and <5-ppm MS precursor mass tolerance and <0.8-Da MS/MS mass tolerance for ESI-LTQ-Orbitrap. When tryptic searches were performed, up to four missed cleavages were taken into consideration. In all elastase searches, the number of missed cleavages was set to the maximum value of 9 if any enzyme specificity was defined. Amino-terminal pyroglutamate formation was considered as variable modification in all PM database searches in addition to methionine oxidation into the resulting sulfoxide, which was considered for both samples. For this purpose a custom Halobacterium database was generated from Swiss-Prot/TrEMBL containing 2490 entries as of November 30, 2007. With respect to database searches involving CB samples, a C. glutamicum ATCC 13032 Bielefeld database containing 3058 sequences was provided by Jörn Kalinowski (43). False discovery rates given are those originating from the internal Mascot decoy database search function. A decoy database consisting of same length random protein sequences is automatically generated and searched. False discovery rate is calculated by dividing the absolute number of decoy hits by the sum of decoy and true hits against the actual database.
Result Interpretation and Data Analysis—
All statistical analyses in this study are exclusively based on peptides having Mascot MS/MS ions scores exceeding the “identity or extensive homology threshold” (p < 0.05). The required threshold value for each individual peptide is stated in supplemental Table S1. Neither general PMF data nor low scoring peptides were included even if after manual inspection a true hit was obtained. In the case of multiple fragmentations of identical precursors, because of recurrence in repetitive runs, only data from the highest scoring peptide were kept. PMF and protein sequence coverages were calculated using Biotools™ 2.2 from Bruker Daltonics. Peptide pI values were iterated based on amino acid pI values given in Ref. 44. Hydrophobicity was estimated via GRAVY score calculation (14). Protein transmembrane stretches were predicted using the TMHMM algorithm (45) and displayed in the TMRPres2D viewer (46).
RESULTS
Protease Characterization—
Important protease characteristics in proteomics include stability, specificity, reproducibility, and autolysis. With the results obtained from multiple elastase digests, we observed no stability problems under the given conditions. To confirm these observations, elastase-digested C. glutamicum membranes were analyzed via SDS-PAGE after different time points (supplemental Fig. S1). Activity was present for at least 12 h, and apparently no enzyme degradation could be observed up to 48 h.
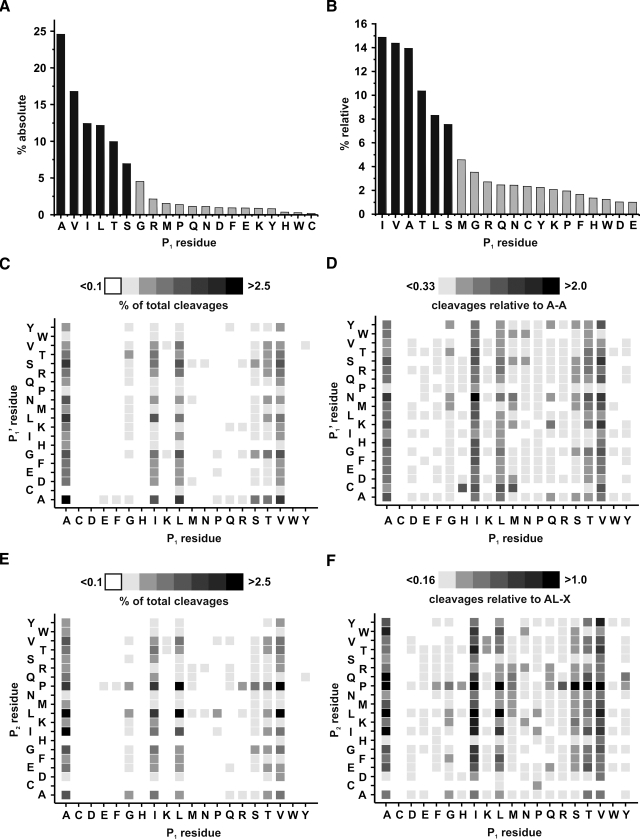
MS/MS data evaluation of 6577 significantly identified Halobacterium and Corynebacterium peptides revealed an absolute specificity of over 82% for cleavage of Ala, Val, Leu, Ile, Ser, and Thr in the P1 position, i.e. the residue before the cleaved peptide bond (Fig. 1A). If the statistical distribution of the amino acid was taken into consideration, this number decreased to 69% because of the frequent occurrence of these residues in the protein sequences (Fig. 1B). Nevertheless elastase did exhibit certain specific qualities and can be treated as a semispecific protease. With respect to the obtained information, reproducibility definitely is no concern at all. When comparing different elastase digests of crude non-LC-separated purple membranes, MS spectra only varied within the regular biological variability (supplemental Fig. S2). Minor differences among digests occurred by additional or missing peptides especially if a large number of potential cleavage sites were clustered in a certain sequence region. The resulting outcome, namely the sequence coverage, is mostly comparable because clustered cleavage sites usually lead to the formation of peptide clusters. Autolysis was prominent when exerting the power of nLC separation but could be easily circumvented by blacklisting known autolytic signals for MS/MS precursor selection (for a complete list of all identified peptides see supplemental Table S1). Extending the specificity analysis to the following P1′ (Fig. 1, C and D) and the second-to-last P2 (Fig. 1, E and F) residues yielded additional insight in elastase proteolysis. Similar to most other proteases, no peptide was liberated if a proline residue occupied the S1′ position of elastase following the bond to be hydrolyzed. However, the rigid proline imino acid structure seemed to be advantageous for cleavage if it was localized two positions ahead of the cleaved bond. Histidine, aspartate, and cysteine exhibited a contrary behavior when present in the P2 position with cysteine being the most interfering amino acid in any of the three examined positions.
Fig. 1.
Carboxyl-terminal P1 residue cleavage specificity of porcine elastase based on MS/MS data of 6577 peptides. A, absolute percentage of occurred cleavages after the stated amino acid. B, relative percentage of cleavages considering amino acid distribution in the sequences of identified protein hits (totalling 648,423 residues). C, cleavage dependence of P1 and P1′ residues based on absolute number of occurred cleavages. D, relative frequency of P1-P1′ cleavages, based on amino acid distribution in the protein sequences, normalized to the most frequently observed cleavage Ala-Ala. E, cleavage dependence of P1 and P2 residues based on absolute number of occurred cleavages. F, relative frequency of P2P1-X cleavages, based on amino acid distribution in the protein sequences, normalized to the most frequently observed cleavage Ala-Leu-X.
MS/MS Search Specificity—
MALDI-TOF/TOF data of digested PM, mainly consisting of the proton pump bacteriorhodopsin, was used to determine the optimal search settings for “elastatic” digests. Regarding the total number of identified peptides and proteins, searches without any enzyme definition yielded the best results, although higher MS/MS ion scores were required to surpass the Mascot identity threshold. We identified 673 PM peptides, 120 of which were matched to BR. This was the top hit using the above search parameters (Table I). Defining a cleavage specificity of AVLIST reduced these numbers to 501 and 86, respectively. Roughly one-quarter of peptides were lost because of cleavages occurring outside the AVLIST definition. Analogous restriction of the specificity to the four most prominent residues (AVLI) resulted in a further decrease of identified peptides (328 and 50). Extending the definition by including glycine as a potential cleavage site (AVLISTG) did not improve identified peptide numbers any further (486 and 86). BR sequence coverage was highest (88%) using the AVLIST definition. Low scoring peptides, just passing the reduced AVLIST identity threshold, cover the additional sequence stretches. However, no defined enzyme specificity was maintained as the parameter of choice for this MS/MS evaluation because the main focus of LC-MS/MS analyses is on peptide and protein identification.
Table I.
Search specificity overview of PM nLC-MALDI-MS/MS experiments performed throughout this work
Shown are the number of repetitive runs, search engine enzyme definitions used, identified peptides, false discovery rates against decoy database, identified proteins, BR protein hit number, matched BR peptides, and BR sequence coverage.
| Sample
|
||||||
|---|---|---|---|---|---|---|
| PM trypsin MALDI | PM trypsin MALDI | PM elastase MALDI | PM elastase MALDI | PM elastase MALDI | PM elastase MALDI | |
| No. of runs | 4 | 4 | 6 | 6 | 6 | 6 |
| Search specificity | KR | None | AVLISTG | AVLIST | AVLI | None |
| Identified peptides | 264 | 310 | 486 | 501 | 328 | 673 |
| False positive (%) | 10.6 | 6.2 | 5.1 | 5.7 | 9.4 | 3.3 |
| Identified proteins | 160/52 | 119/49 | 160/48 | 160/47 | 137/30 | 165/56 |
| BR protein hit no. | 20 | 1 | 1 | 1 | 1 | 1 |
| BR peptides | 4 | 39 | 86 | 86 | 50 | 120 |
| BR sequence coverage (%) | 18.5 | 40.6 | 81.8 | 88 | 73.1 | 75.1 |
Enzymatic Comparison—
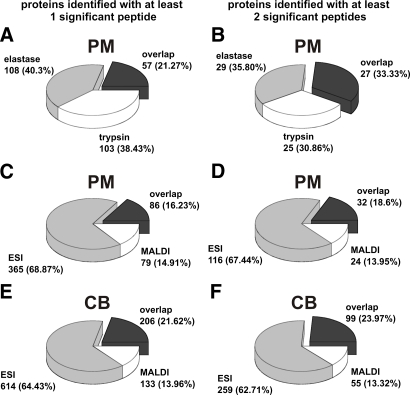
For a more detailed comparison with the standard protease trypsin, we simply substituted elastase for trypsin in our PM nLC-MALDI-MS/MS work flow. The resulting number of identified proteins was more or less comparable to the elastatic alternative (Table II). Proteins identified by only one significant peptide, the so-called “one-hit wonders,” were not significantly increased using either protease. So elastase does not generate more protein hits based solely on one significant peptide but has an increased average peptide-per-protein ratio. It has to be taken into consideration that slightly more elastase runs were necessary to achieve this equality. This is because of the need for extensive fragmentation of the increased number of generated peptides in the elastase digests. A comparison of single run results supports this observation, showing increased mean protein but lower mean peptide values per run in the tryptically digested PM samples. Furthermore when comparing these two enzymes on an nLC background, three differences could be observed. First of all, the false positive rate of trypsin-digested samples was much higher under our chosen quality criteria. This can be attributed to the very low required ion score to exceed the identity threshold, sometimes as low as 12. Adjusting the Mascot significance threshold to p < 0.02 (required ion score, 16) would halve the trypsin false positive rate but only marginally reduce the number of peptide hits by ∼10% (data not shown). To avoid significant loss of true positive peptides by using this stringent threshold, the standard significance threshold of p < 0.05 was used in all subsequent searches. In studies with protein identification emphasis, the now increased number of false positives mostly consisted of low scoring or low mass peptides, which anyway would be rejected or require manual evaluation. Second, the identified proteins were basically two subsets with only limited overlap. This is also true for the Mascot protein hit number. Of the first 30 protein hits of the elastase digest only half of the proteins appear among the top 30 tryptic hits (supplemental Table S2). Seven of the elastase top 30 proteins are completely absent from the trypsin list. Six proteins are absent vice versa, pointing out complementary characteristics of both proteases. Overall only 57 of the 268 proteins are present in both result lists (Fig. 2A). The remaining protease-exclusive hits are more or less equally distributed between elastase and trypsin. If only peptides with two or more significant peptide matches are considered, the proportion of overlap increases to roughly one-third, leaving each protease with another third of identified proteins (Fig. 2B). This demonstrates that equal proportions of one-peptide protein hits are generated, and thus no drawback for any protease exists. The third and most striking observation was the low BR ranking (Mascot hit number 20) with only four matched peptides in the tryptic digest even though this protein basically represents more than 90% of the total protein content of the sample (Table I). This clearly demonstrates the existence of proteins that simply are not suited for the standard protease trypsin. Surprisingly BR reappeared as the top hit if no specificity was defined in the tryptic search. This is due to 35 additional non- or semitryptic peptides found under these settings that are extensively prominent at the carboxyl terminus and to a lesser extent in the second loop region (supplemental Fig. S2). The truncation seems also to be exclusive to the BR protein as only 11 additional non-tryptic non-BR peptides were found (Table I).
Table II.
General overview of PM/CB nLC-ESI-MS/MS and nLC-MALDI-MS/MS experiments performed
Shown are the number of repetitive runs, search engine enzyme definitions used, mean peptides identified per single nLC run, total identified peptides, identified TM peptides, false discovery rates against decoy database, mean proteins identified per single nLC run, total identified proteins, number of one-hit wonders, one-hit wonders identified solely by one small <700-Da peptide, and decoy database hits of <700-Da peptides. Data from all single run experiments are given in supplemental Table S5. Combined peptide and protein data are given in supplemental Tables S1 and S2.
| Sample
|
|||||
|---|---|---|---|---|---|
| PM trypsin MALDI | PM elastase MALDI | PM elastase ESI | CB elastase MALDI | CB elastase ESI | |
| No. of runs | 4 | 6 | 4 | 4 | 3 |
| Search specificity | KR | None | None | None | None |
| Peptides (mean ± S.D.) | 145 ± 40 | 229 ± 49 | 808 ± 182 | 521 ± 100 | 1205 ± 143 |
| Identified peptides | 264 | 673 | 1675 | 1443 | 2786 |
| TM peptides | 20 | 174 | 604 | 149 | 749 |
| False positive (%) | 10.6 | 3.3 | 13.8 | 2.1 | 11.9 |
| Proteins (mean ± S.D.) | 98 ± 19 | 58 ± 13 | 225 ± 38 | 155 ± 40 | 449 ± 57 |
| Identified proteins | 160/52 | 165/56 | 451/148 | 339/154 | 820/358 |
| One-hit wonders | 108 | 109 | 303 | 185 | 462 |
| One-hit wonders <700 Da | 0 | 0 | 146 | 0 | 190 |
| Decoy hits <700 Da (%) | 0 | 0 | 33.9 | 0 | 34.4 |
Fig. 2.
Pie charts representing overlap as well as enzyme- and instrument-exclusive protein hits, respectively. A and B display the comparison of elastase and tryptic digests of PM analyzed by nLC-MALDI-TOF/TOF. The remaining diagrams demonstrate the protein distribution among the MALDI and ESI instruments: C and D for PM digests; E and F for CB digests. Left-hand charts A, C, and E include one-hit wonders; right-hand charts B, D, and F only factor in proteins with at least two significant peptides.
PMF Application—
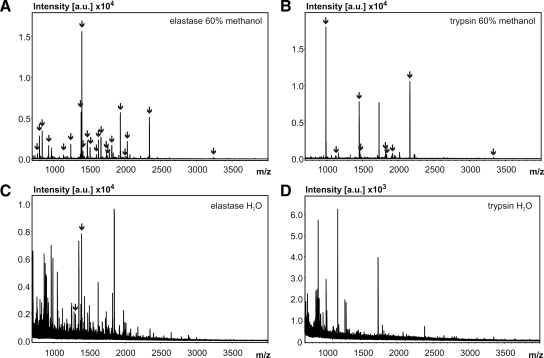
Although these results imply that nLC separation is a prerequisite, improvement was also visible in PMF applications without any nLC separation step (Fig. 3). Both protease change and methanol addition increased the quality of MALDI BR peptide mass fingerprints of PM digests. Aqueous digests massively suffered from interfering, probably lipid-generated signals, which were reduced upon digestion in methanol. Given an adequate mass accuracy, elastase was capable of drastically improving PMF results of tough candidate proteins for trypsin (Fig. 3, A and B). BR sequence coverage increased from 35.9 to 83.9% just by replacing the protease. In the standard non-organic buffer system, elastase and trypsin yielded poor results for BR (Fig. 3, C and D).
Fig. 3.
PMF spectra of 500-ng PM digests under different conditions. BR peaks were matched within 10 ppm after internal calibration. Minimum peak signal-to-noise ratio requirement was 6. Arrows depict peaks matched to BR. A, elastase digest in 60% methanol. B, trypsin digest in 60% methanol. C, elastase digest in aqueous solution. D, trypsin digest in aqueous solution. a.u., arbitrary units.
Theoretical matching of 81 nLC-MALDI-MS/MS-verified BR AVLIST peptides against its sequence and two “false” protein sequences suggests that a mass accuracy below 5 ppm is required for optimal peak matching using the AVLIST definition (supplemental Fig. S4A). Multiple matches to one mass are mostly accounted to isobaric peptides usually spanning the same sequence stretch. The false positive rate rapidly grows beyond that point, impeding protein identification with mass accuracies above 50 ppm. Completely nonspecific proteases like proteinase K require a substantially higher accuracy for a comparable rate of false positive hits (supplemental Fig. S3B) that drastically limits its PMF capabilities.
Instrument Comparison—
Besides protease comparison, an interesting aspect was to run our protocol on two commonly used instruments. Considering that an ESI instrument always generates an excess of MS/MS spectra compared with MALDI, it can therefore be regarded as the more “efficient” technology in terms of useful spectra. The percentage of queries yielding significant peptide identifications was substantially lower in ESI (12.8%) than in MALDI (27.9%) (data not shown). Despite having included one run less in our evaluation, the absolute number of successfully evaluated peptides was still higher in ESI (Table II). Consequently ESI clearly surpassed MALDI by ∼2-fold with regard to identified peptides and the correlated protein number. Hence the sequence coverage of the model protein BR was marginally higher when using ESI. Worth mentioning is the noticeable cluster formation; some were present in both, and others were present solely on the ESI instrument (Fig. 4). The only part exclusively covered by MALDI was seven amino acids near the carboxyl terminus.
Fig. 4.
BR sequence coverage display. Identified peptide locations are indicated by thin bars underneath the protein sequence. TM stretches are indicated by gray shading. A, significant peptides identified in the nLC-MALDI MS/MS work flow. B, significant peptides identified in the nLC-ESI MS/MS work flow.
Regarding whole proteomes, an overlapping proportion of proteins was identified by both instruments. Approximately 20% of the CB proteins were identified using both instruments, but the number of instrument-exclusive hits was unequally distributed, favoring ESI to a large extent regardless of the inclusion of one-hit wonders into the calculations (Fig. 2, E and F). This dominance of electrospray ionization has to be treated cautiously because depending on the sample used the fraction of proteins identified solely by a single small ESI peptide (<700 Da) was up to 50% (Table II). These small peptides are responsible for approximately one-third of decoy database hits in the ESI searches. This demonstrates that the advantage of the extended mass range of ESI instruments has to be treated very cautiously when data relying on small peptides are to be interpreted.
The PM protein distribution was in the same range on both instruments (Fig. 2, C and D). A more detailed view of the top 75 CB protein list shows 65 shared identifications and only six (ESI) or four (MALDI) proteins missing on the entire list for the other instrument (Table III). Apparently the coverage of the high abundance proteins is more or less equal using any instrument.
Table III.
Top 75 protein list of CB nLC-ESI-MS/MS and nLC-MALDI-MS/MS experiments
Shown are the number of matched ESI and MALDI peptides, accession number, mass, length, pI, and Mascot hit numbers. “None” indicates that the protein was absent from the respective experiment. Proteins with predicted TM stretches are highlighted by bold type. For a complete list of all proteins see supplemental Table S2. ABC, ATP-binding cassette; PTS, phosphotransferase system.
| No. | Peptides found
|
Accession no. | Protein description | Mascot hits
|
Protein mass (Da) | Protein length | Protein pI | ||
|---|---|---|---|---|---|---|---|---|---|
| ESI | MALDI | ESI | MALDI | ||||||
| 1 | 97 | 166 | P00772 | Elastase 1 | 2 | 1 | 28,803 | 266 | 8.42 |
| 2 | 144 | 61 | Cg1537 | ptsG; glucose-specific enzyme II BC component of PTS | 1 | 3 | 72,525 | 683 | 5.13 |
| 3 | 74 | 72 | Cg0446 | sdhA; succinate dehydrogenase A | 3 | 2 | 74,632 | 673 | 5.37 |
| 4 | 71 | 46 | Cg2705 | amyE; maltose-binding protein precursor | 4 | 4 | 49,417 | 443 | 4.36 |
| 5 | 65 | 22 | Cg3009 | Hypothetical protein predicted by Glimmer/Critica | 5 | 9 | 6,158 | 57 | 3.47 |
| 6 | 62 | 10 | Cg2181 | ABC-type peptide transport system, secreted component | 6 | 23 | 57,653 | 534 | 3.9 |
| 7 | 47 | 3 | Cg2120 | ptsF; sugar specific PTS system, fructose mannitol-specific transport protein | 7 | 63 | 70,457 | 688 | 5.31 |
| 8 | 39 | 47 | Cg2409 | ctaC; cytochrome c oxidase chain II | 10 | 5 | 39,607 | 359 | 4.74 |
| 9 | 8 | 46 | Cg0587 | tuf; elongation factor Tu | 47 | 6 | 43,824 | 396 | 4.92 |
| 10 | 44 | 22 | Cg2911 | ABC-type manganese/zinc transport system, secreted manganese/zinc-binding (lipo)protein | 8 | 8 | 33,986 | 314 | 4.09 |
| 11 | 41 | 22 | Cg0447 | sdhB; succinate dehydrogenase B | 11 | 10 | 26,631 | 249 | 5.4 |
| 12 | 39 | 2 | Cg2925 | ptsS; enzyme II sucrose protein | 9 | 181 | 69,104 | 661 | 4.96 |
| 13 | 30 | 17 | Cg2404 | qcrA1; Rieske iron-sulfur protein | 16 | 17 | 45,156 | 408 | 5.29 |
| 14 | 29 | 9 | Cg1366 | atpA; probable ATP synthase α chain protein | 14 | 38 | 58,715 | 547 | 4.86 |
| 15 | 28 | 26 | Cg3138 | Membrane protease subunit, stomatin/prohibitin homolog | 13 | 7 | 34,765 | 325 | 4.77 |
| 16 | 27 | 21 | Cg2780 | ctaD; probable cytochrome c oxidase polypeptide subunit | 15 | 12 | 65,059 | 584 | 6.91 |
| 17 | 27 | 19 | Cg2403 | qcrB; cytochrome b, membrane protein | 19 | 15 | 59,770 | 539 | 6.38 |
| 18 | 26 | 13 | Cg2708 | msiK1; ABC-type sugar transport system, ATPase component | 12 | 18 | 40,290 | 376 | 5.53 |
| 19 | 25 | 12 | Cg2875 | Hypothetical protein predicted by Glimmer | 17 | 22 | 3,872 | 38 | 3.92 |
| 20 | 23 | 0 | Cg1332 | Putative secreted hydrolase | 21 | None | 55,920 | 527 | 4.4 |
| 21 | 22 | 1 | Cg3182 | cop1; trehalose corynomycoyl transferase | 18 | 148 | 70,616 | 657 | 4.85 |
| 22 | 22 | 5 | Cg1363 | atpE; ATP synthase C chain | 23 | 76 | 8,117 | 80 | 4.42 |
| 23 | 22 | 1 | Cg0413 | cmt1; trehalose corynomycoyl transferase | 27 | 426 | 39,481 | 365 | 4.56 |
| 24 | 19 | 2 | Cg0683 | Permease | 24 | 153 | 55,630 | 533 | 5.78 |
| 25 | 18 | 6 | Cg0953 | Na+/proline, Na+/pantothenate symporter | 20 | 47 | 57,232 | 551 | 5.98 |
| 26 | 18 | 5 | Cg0737 | ABC-type transport system, secreted lipoprotein component | 30 | 49 | 31,722 | 299 | 4.05 |
| 27 | 16 | 18 | Cg1656 | ndh; NADH dehydrogenase | 22 | 13 | 50,926 | 467 | 5.68 |
| 28 | 17 | 11 | Cg1368 | atpD; ATP synthase α subunit | 31 | 20 | 52,519 | 483 | 4.74 |
| 29 | 12 | 17 | Cg2843 | pstB; ABC-type phosphate transport system, ATPase component | 33 | 16 | 28,196 | 257 | 5.71 |
| 30 | 4 | 17 | Cg0583 | fusA; elongation factor G | 105 | 11 | 77,842 | 709 | 4.81 |
| 31 | 16 | 5 | Cg3008 | porA; porin | 38 | 94 | 4,677 | 45 | 3.77 |
| 32 | 15 | 2 | Cg1603 | Conserved membrane protein | 25 | 88 | 41,873 | 397 | 4.91 |
| 33 | 15 | 9 | Cg3186 | cmt2; trehalose corynomycoyl transferase | 26 | 26 | 37,163 | 341 | 5.41 |
| 34 | 15 | 6 | Cg3365 | rmpC; putative ribitol transport membrane protein | 28 | 62 | 52,700 | 513 | 8.78 |
| 35 | 15 | 9 | Cg3186 | cmt2; trehalose corynomycoyl transferase | 26 | 26 | 37,163 | 341 | 5.41 |
| 36 | 14 | 1 | Cg1314 | putP; proline transport system | 35 | 103 | 56,610 | 524 | 4.91 |
| 37 | 14 | 4 | Cg0834 | Bacterial extracellular solute-binding protein | 39 | 81 | 45,049 | 424 | 3.85 |
| 38 | 2 | 14 | Cg0957 | fas-IB; fatty-acid synthase | 32 | 14 | 314,960 | 2969 | 4.74 |
| 39 | 13 | 7 | Cg1081 | ABC-type multidrug transport system, ATPase component | 29 | 24 | 33,285 | 308 | 4.77 |
| 40 | 13 | 5 | Cg1556 | Conserved hypothetical protein | 44 | 65 | 33,325 | 304 | 9.68 |
| 41 | 13 | 4 | Cg1362 | atpB; ATP synthase F0 subunit 6 | 78 | 92 | 36,385 | 322 | 9.51 |
| 42 | 12 | 10 | Cg0924 | ABC-type cobalamin/Fe3+-siderophores transport system | 34 | 28 | 35,668 | 338 | 4 |
| 43 | 12 | 9 | Cg0756 | cstA; putative carbon starvation protein A | 37 | 33 | 81,038 | 759 | 6.06 |
| 44 | 12 | 1 | Cg1082 | Putative membrane protein | 41 | 182 | 30,196 | 277 | 9.26 |
| 45 | 12 | 1 | Cg0359 | Putative membrane protein | 64 | 1057 | 21,555 | 201 | 9.62 |
| 46 | 1 | 12 | Cg1787 | ppc; phosphoenolpyruvate carboxylase | 295 | 19 | 103,134 | 919 | 4.92 |
| 47 | 0 | 12 | Cg0791 | pyc; pyruvate carboxylase | None | 21 | 123,027 | 1140 | 5.39 |
| 48 | 11 | 1 | Cg2262 | ftsY; signal recognition particle GTPase | 52 | 173 | 60,607 | 576 | 4.33 |
| 49 | 11 | 1 | Cg2361 | Cell division initiation protein, antigen 84 homolog | 61 | 348 | 38,662 | 365 | 4.87 |
| 50 | 10 | 11 | Cg0445 | sdhCD; succinate dehydrogenase CD | 85 | 27 | 28,354 | 257 | 9.77 |
| 51 | 10 | 0 | Cg2810 | Na+/H+-Dicarboxylate symporter | 36 | None | 47,955 | 465 | 7.71 |
| 52 | 10 | 4 | Cg2195 | Putative secreted or membrane protein | 43 | 68 | 11,897 | 115 | 4.1 |
| 53 | 10 | 0 | Cg3368 | ABC transporter permease protein | 53 | None | 57,131 | 528 | 9.02 |
| 54 | 10 | 0 | Cg2912 | ABC-type cobalamin/Fe3+-siderophores transport system | 54 | None | 24,475 | 230 | 9.34 |
| 55 | 6 | 10 | Cg2196 | Putative secreted or membrane protein | 153 | 29 | 12,721 | 119 | 4.21 |
| 56 | 9 | 5 | Cg2845 | pstC; ABC-type phosphate transport system, permease component | 42 | 59 | 37,569 | 355 | 9.66 |
| 57 | 9 | 4 | Cg2704 | ABC-type sugar transport system, permease component | 63 | 75 | 31,528 | 281 | 9.46 |
| 58 | 0 | 9 | Cg1437 | ilvC; ketol-acid reductoisomerase | None | 31 | 36,136 | 338 | 4.55 |
| 59 | 1 | 9 | Cg1790 | pgk; phosphoglycerate kinase | 136 | 37 | 42,671 | 405 | 4.55 |
| 60 | 5 | 9 | Cg2958 | butA; l-2,3-butanediol dehydrogenase/actetoin reductase | 164 | 40 | 27,047 | 258 | 4.62 |
| 61 | 9 | 8 | Cg2211 | Putative membrane protein | 70 | 45 | 16,356 | 147 | 6.36 |
| 62 | 3 | 8 | Cg3227 | lldA; putative l-lactate dehydrogenase | 452 | 32 | 45,686 | 420 | 5.72 |
| 63 | 7 | 8 | Cg1429 | Putative membrane protein | 88 | 42 | 36,855 | 352 | 4.44 |
| 64 | 3 | 8 | Cg1685 | tatX; SEC-independent protein secretion pathway component | 754 | 50 | 12,036 | 105 | 5.28 |
| 65 | 0 | 8 | Cg2444 | Hypothetical protein predicted by Glimmer/Critica | None | 51 | 19,830 | 181 | 4.33 |
| 66 | 8 | 7 | Cg2405 | qcrC; cytochrome c1 | 79 | 30 | 31,134 | 295 | 6 |
| 67 | 8 | 6 | Cg1791 | gap; glyceraldehyde-3-phosphate dehydrogenase | 55 | 43 | 36,023 | 334 | 5.16 |
| 68 | 8 | 6 | Cg1001 | mscL; large conductance mechanosensitive channel | 59 | 44 | 14,526 | 135 | 4.84 |
| 69 | 3 | 7 | Cg1111 | eno; enolase | 144 | 25 | 44,921 | 425 | 4.65 |
| 70 | 1 | 7 | Cg0766 | icd; isocitrate dehydrogenase | 227 | 34 | 80,032 | 738 | 4.72 |
| 71 | 0 | 7 | Cg1290 | metE; homocysteine methyltransferase | None | 36 | 81,263 | 745 | 4.78 |
| 72 | 2 | 7 | Cg1880 | thrS; threonyl-tRNA synthetase | 386 | 54 | 78,124 | 700 | 5.04 |
| 73 | 0 | 6 | Cg2523 | malQ; 4-α-glucanotransferase | None | 39 | 78,475 | 706 | 4.76 |
| 74 | 0 | 6 | Cg2499 | glyS; glycyl-tRNA synthetase (glycine-tRNA ligase) | None | 41 | 53,011 | 461 | 5.28 |
| 75 | 1 | 6 | Cg1737 | acn; aconitase | 449 | 46 | 102,104 | 943 | 4.53 |
Peptide Physicochemical Properties—
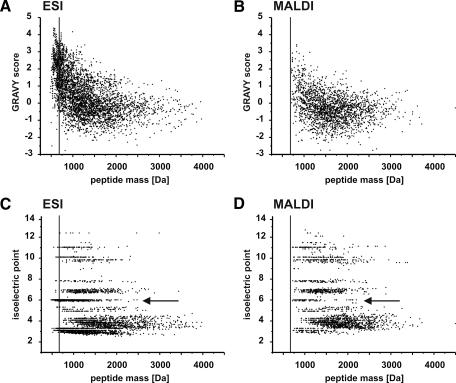
Peptide pI values and GRAVY scores represent approved values for the characterization of the physicochemical properties of a peptide. However, if instruments based on such different physical processes are compared, plain peptide mass and charge have to be factored in as well. Under the normal operating conditions, small peptides with masses below 700 Da were only accessible to the ESI instrument (Fig. 5A). To some extent MALDI can overcome this deficit by having a 2-fold increase in relative peptide numbers exceeding 2000 Da. Regarding hydrophobicity, the biggest difference between both techniques was the accumulated occurrence of high positive GRAVY score peptides on the ESI instrument (Fig. 6, A and B). Around 20% of all identified CB and PM peptides possessed GRAVY scores above 2.0; this fraction did not surpass the 5% mark using MALDI (Fig. 5B). Needless to say, a significant proportion of these peptides was below the 700 m/z cutoff of the MALDI-TOF/TOF. Peptide pI distributions (Fig. 6, C and D) displayed further differences partly linked to the above discussed observations. First of all, the percentage of acidic peptides with pI values of 5 and below was similar (ranging from 51.6 to 56.0%) in all elastase digests regardless of the instrument used (Fig. 5C). So no discrimination of acidic peptides among the instruments was observed. Second, basic peptides having pI values beyond 7 were 2–3-fold more prominent on the MALDI instrument. This effect was more significant for His- and Arg-containing peptides. With MALDI being equal to ESI regarding acidic peptides and superior in terms of identifying basic peptides, its biggest drawback is its low performance for neutral peptides. These can be divided in peptides where neutrality is achieved by equal amounts of acidic and basic side chains (subsequently termed “balanced charges”) or by the complete lack of any dissociable side-chain moieties (subsequently termed “aliphatic”). The pI of the aliphatic group is only influenced by the peptide termini functions and is therefore always 6.02. MALDI experiences extreme difficulties in identifying these aliphatic neutral peptides. Although ∼25% of peptides detected by ESI belong to this group, their MALDI percentage is below 5%. It has to be considered, once more, that about half (580 of 1119) of the ESI aliphatic neutral peptides possessed molecular masses below 700 Da. Still those surpassing the 700-Da threshold exceeded the number of MALDI-identified aliphatic neutral peptides by a large margin. Although the total number of identified ESI peptides was more than twice the number of MALDI peptides (4461 versus 2116 peptides), this proportion dropped to 1.5-fold (3009 versus 2039) if peptides below 700 Da and aliphatic neutral peptides were omitted (Fig. 5D). The absence of these peptides in MALDI is the reason for the lack of peptides exceeding a GRAVY score of 2. As a matter of fact, data obtained from tryptic digests yielded no hint to this observation because the mandatory basic residue prohibited the necessary physicochemical states of pI 6.02 and the resultant extremely high GRAVY values (Fig. 5, B and C).
Fig. 5.
Peptide physicochemical properties statistics of nLC-MS/MS experiments. A, relative distributions of peptide mass ranges. B, relative distributions of peptide GRAVY scores. C, relative distributions of peptide pI values (acidic, pI < 5; basic, pI > 7; neutral (aliphatic), pI = 6.02; neutral (balanced charges), 7 > pI > 5, pI ≠ 6.02). D, fractions of neutral (aliphatic) peptides among transmembrane peptides. E, fractions of neutral (aliphatic) and low mass peptides among all identified peptides. F, fractions of low mass peptides within TM peptides. elas., elastase; tryp., trypsin.
Fig. 6.
Instrument comparison of peptide GRAVY and pI values. ESI (A) and MALDI (B) peptide GRAVY scores plotted against calculated masses are shown. ESI (C) and MALDI (D) peptide pI values plotted against calculated masses are shown. Arrows indicate clusters of neutral aliphatic peptides, which are significantly reduced on the MALDI instrument. The small peptide (<700 Da) threshold is indicated by a vertical line.
Transmembrane Coverage—
The proportion of peptides spanning protein TM regions is an important indication of the effectiveness of any membrane proteomics strategy. The tryptic digest again was inferior when it comes to the relative proportion of transmembrane peptides (Fig. 5, E and F). Elastase simply generated a higher number of membrane-spanning peptides, whereas most tryptic identifications of membrane proteins are achieved over loop coverage. This observation is extremely prominent in BR; just one tryptic peptide partially had membrane localization. The corresponding elastase experiment covered transmembrane parts with 89 peptides (supplemental Tables S3 and S4). Several membrane protein identifications in the CB sample are in line with this observation. The top 75 protein list of nLC-ESI and nLC-MALDI data (Table III) depicts a variety of membrane-localized proteins, including their respective transmembrane peptides (supplemental Table S4). They include prominent respiratory chain, transport, and ion channel proteins with challenging candidates like cytochrome b, ATP synthase subunits a and c, and cytochrome c oxidase I. Another membrane protein, the II BC component of the CB phosphotransferase system (Cg1537), exhibited a BR-comparable Arg/Lys distribution in its middle transmembrane part and was well covered by elastatic peptides. The same goes for membrane proteins of smaller size; e.g. eight peptides were liberated from the single helix of the CB porin (Cg3008).
In parallel to the high GRAVY peptides, the data obtained from the ESI instrument include more elastatic peptides covering TM helices. Concerning the absolute number of TM peptides, ESI clearly surpassed MALDI. However, it has to be mentioned that more than half of the TM peptides of the ESI results were exactly those aliphatic neutral peptides with no dissociable side-chain functions as highlighted in the previous section. This fraction of TM peptides in MALDI represents only 17%. Again peptides below 700 Da represent a significant component of the TM fractions among the ESI peptides (Fig. 5F). The minimal TM helical length of ∼17 amino acids and its resulting minimal peptide mass clearly demonstrate the accessibility of elastase to the membrane-spanning regions.
DISCUSSION
The intent of this study was to establish a new method for the preferential identification of complex membrane proteomes on the basis of porcine pancreatic elastase as the digestive enzyme. We showed that elastase applied on two membrane samples exhibits ∼70% P1 residue specificity for the six amino acids Ala, Val, Leu, Ile, Ser, and Thr. The remaining 30% of cleavages occurring outside this specificity window are no surprise if S1 pocket geometry (47) and deduced possible interactions of the inserting side chain are considered. In our opinion, size- and hydrophobic interaction-based exclusion of certain side chains simply cannot be as efficient as having additional ionic repulsion like trypsin does (48, 49). It has to be considered as well that the elastase used in this study still may contain trace amounts of other pancreatic proteases like trypsin and chymotrypsin, which influence the observed specificity. The specificity of elastase can be supportive in elastatic proteome studies to increase identification significance.
In our PMF experiments, the methanol addition dramatically increased the efficiency of both tryptic and elastatic BR peptide mass fingerprints, most likely due to the solvation of the hydrophobic products and/or increasing BR accessibility in the membrane. Despite elastase being superior to trypsin in this particular case, this is only caused by the inherent lack of tryptic cleavage sites in BR as well as in many other membrane proteins. Still the peptides generated in the elastase digest are surrounding basic amino acids within the BR sequence due to the typical MALDI discrimination effects (50). Elastase PMF applications will most likely be even more successful as soon as search engine algorithms are adapted to wider specificities of three to six potential cleavage sites. If another characteristic of less specific proteases, i.e. the formation of peptide clusters, is implemented in the significance evaluation by the search engine, its results would improve remarkably. The required instrument mass accuracy is already attainable when using current mass analyzers like the Orbitrap. In our opinion, the gain of information through the observed elastase specificity is absolutely sufficient for PMF applications. The PMF capability of proteinase K, despite its usefulness regarding membrane samples, is very limited because of its lack of specificity.
Our nLC-MALDI approach of methanolic trypsin-digested purple membranes yielded four significantly identified peptides, resulting in poor sequence coverage of 18.5%. An in silico tryptic digest of BR contains six fragments without missed cleavages in the mass range of a modern MALDI mass spectrometer, so our result of four peptides can be interpreted as a realistic outcome. For a 27-kDa membrane protein, the low number of tryptic cleavage sites does not give rise to higher expectations. As a consequence such proteins trail other less abundant proteins in the protein hit ranking, as seen in the BR example, or might even be absent. The most abundant protein identified as hit number 20 in a database search is unacceptable for any method (Table II and supplemental Table S2). When tryptic specificity was omitted, 35 additional non- or semitryptic peptides were found that return BR to the top position in the protein identification list. This observation was surprising as a less specific cleavage behavior of trypsin in methanol was not mentioned by Blonder et al. (27), who established the 60% methanol/trypsin protocol. Another study published by Strader et al. (51) even demonstrated that trypsin specificity is higher in the presence of 60% methanol for a variety of sample amounts. Contrary to these statements, this work might indicate a certain loss of tryptic specificity in the presence of methanol. This finding has to be handled with care because it was derived from a 310-peptide MS/MS analysis of a sample whose content is dominated by one single protein. Only 11 additional peptides that do not originate from the abundant BR protein were identified in the nonspecific tryptic search. Furthermore the observed specificity loss seems to be marginal because the non- and semispecific peptides are only detectable after nano-LC separation.
The simple change of protease to elastase dramatically boosted BR peptide number and sequence coverage. The nLC-MALDI experiment significantly identified 120 BR peptides (75.1% sequence coverage), and the nLC-ESI variant was even more successful, delivering 295 positive identifications (93.2% sequence coverage); both setups delivered BR as the top search hits. This high sequence coverage is comparable to BR coverage reported in the literature by Blonder et al. (27) but with a drastically increased peptide count. Using a tryptic digest in 60% methanol, it was demonstrated that an achievement of almost 100% coverage is possible. However, it has to be considered that this analysis was performed on an electrospray ionization instrument, which is important for result comparison. The authors concede a low spectral quality for three large peptides of the 13 BR peptides that highly contributes to the reported sequence coverage value. Other publications achieving similar results also depend on these large proteolytic fragments (29), which carry charges too high (3+) for efficient ESI-ion trap fragmentation or require the lower accuracy MALDI-TOF linear mode. The top-down BR characterization offered excellent mass accuracy and sequence coverage; similar results have been achieved for other membrane proteins as well (3, 4). However, top-down experiments generally have higher requirements concerning sample amount and purity than peptide-based approaches. In addition, the lack of comparable separation techniques for membrane proteins restricts these analyses to less complex sample mixtures.
The 295 BR peptides of our approach are less prone to false positive identification caused by single low quality MS/MS data as it easily could happen with the 13 tryptic peptides from the cited experiment. The increased number of potential cleavage sites in comparison with trypsin virtually eliminates the dependence on large peptides, simplifying any protein identification, and bears noteworthy potential for a complete protein characterization. Regarding the non-BR protein identifications in the PM sample, both ESI and MALDI elastase data sets contain the same membrane proteins as the reference table in the literature (27). We do not want to point at explicit differences between both analyses on the protein level because of potential variability in sample origin and purification but have to stress that we identified most membrane proteins with an increased number of peptides (supplemental Table S2).
The application of our protocol to the CB membrane sample clearly indicates that the identification of membrane proteins via their transmembrane stretches is possible. We identified a large number of TM peptides, especially in the ESI experiment (26.9%). Nevertheless very high sequence coverages were only attainable for the more abundant proteins within the sample (e.g. Cg1537; supplemental Table S4). If adequate protein amounts are present, there is potential for full coverage. For the identification of lower abundance proteins, which will typically rely on few peptides, the higher peptide-per-protein ratio of the elastase digest definitely is beneficial. However, some drawbacks have to be considered when dealing with the combination of a high complexity sample and a broad specificity protease like elastase. First the higher number of occurring cleavages leads to a reduction of the effective sample amount due to a dilution effect. This could be overcome by better separation strategies to handle the increased peptide diversity. Established techniques like separation of even less specific proteinase K-digested samples by the multidimensional protein identification technique (18, 22) could be transferred to our method. However it is not obvious whether the 2D SCX-C18-nLC setup offers the required orthogonality to fractionate the inhomogeneous peptide population generated in an elastase digest. The physicochemical properties of elastatic peptides discussed in this study should encourage the development of an appropriate separation process by LC specialists.
When comparing nLC-coupled MALDI and ESI outcomes of the elastase digest protocol, a fair comparison is generally impossible. The well established on-line coupling of nLC to an ESI instrument is more straightforward and thus better understood in regard to its problems than the MALDI spotting procedure (52). The loss of separation power through the MALDI spotting process can hardly be compensated despite the advantage of the off-line measurement. Additionally differences are caused by the mass analyzers used, the TOF and the linear ion trap-Orbitrap, that cannot be separated from those originating from the ionization technique or coupling. Despite this, certain effects can be attributed exclusively to the ionization method. Part of the superiority of ESI regarding the number of identified peptides could be attributed to the extended mass range of the ESI instrument below 700 m/z and the improved nLC coupling. The reason that this cannot be compensated by the improved ability of MALDI to detect peptides above 2000 Da might be the poor recovery rates of higher mass hydrophobic peptides from the C18-nLC column. We have to stress that the informational value of larger peptides is substantially higher than that of smaller ones. Additionally false positive matching of these peptides generally is less likely because of the reduced sequence redundancy within the database. Thus large peptides contribute more to successful protein identification and are therefore preferred. A large proportion of the disadvantage of MALDI compared with ESI in transmembrane peptide identification, which is the relevant criterion for the success of any membrane proteomics study, originates from the absence of small peptides and of peptides containing only uncharged residues. The low detection efficiency when using the standard matrix α-CHCA can be attributed to two effects: first a less efficient incorporation into the matrix crystals due to less strong ion pair formation and second, and presumably more important, the less efficient protonation of the lower basicity terminal amino group (50, 53). Even if such peptides are detected, their low signal intensity prohibits the acquisition of informative laser-induced fragmentation MS/MS spectra. Although the preferred detection of Arg-containing peptides is well known from proteomics studies using MALDI and α-CHCA as the matrix (50), the discrimination of neutral peptides in MALDI has not been relevant to most proteomics studies because tryptic peptides contain either a basic lysine or arginine side chain. As shown also by the data presented here, electrospray ionization is much less prone to this discrimination effect; on the contrary the ESI response was shown to be promoted by hydrophobic characteristics of peptides or by hydrophobic alkyl tags introduced by derivatization (54, 55).
To summarize, our results indicate that elastase is a well suited enzyme for proteolyzing complex membrane samples. Even PMF applications should be possible because of its biased specificity toward six amino acids. Some inherent problems of trypsin, like the lack of tryptic cleavage sites in membrane proteins, can be circumvented by elastase usage. At present ESI instruments appear to be the best suited for this purpose, although MALDI developments might overcome this drawback in the near future. Then the complementary advantages from both proteases could be optimally used.
Supplementary Material
Acknowledgments
We thank Norbert Dencher and Holger Seelert for the ready supply of H. salinarium purple membrane samples, Jörn Kalinowski for giving us access to an established C. glutamicum database, and Dimitrios Papasotiriou and Thorsten Jaskolla for carefully reading the manuscript and helpful discussion.
Footnotes
Published, MCP Papers in Press, December 30, 2008, DOI 10.1074/mcp.M800223-MCP200
The abbreviations used are: 2D, two-dimensional; α-CHCA, α-cyano-4-hydroxycinnamic acid; BR, bacteriorhodopsin; CB, C. glutamicum; PM, purple membrane; PMF, peptide mass fingerprint; TM, transmembrane; nLC, nanoflow LC.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Karas, M., and Hillenkamp, F. ( 1988) Laser desorption ionization of proteins with molecular masses exceeding 10,000 daltons. Anal. Chem. 60, 2299–2301 [DOI] [PubMed] [Google Scholar]
- 2.Fenn, J. B., Mann, M., Meng, C. K., Wong, S. F., and Whitehouse, C. M. ( 1989) Electrospray ionization for mass spectrometry of large biomolecules. Science 246, 64–71 [DOI] [PubMed] [Google Scholar]
- 3.Whitelegge, J. P., Halgand, F., Souda, P., and Zabrouskov, V. ( 2006) Top-down mass spectrometry of integral membrane proteins. Exp. Rev. Proteomics 3, 585–596 [DOI] [PubMed] [Google Scholar]
- 4.Zabrouskov, V., and Whitelegge, J. P. ( 2007) Increased coverage in the transmembrane domain with activated-ion electron capture dissociation for top-down Fourier-transform mass spectrometry of integral membrane proteins. J. Proteome Res. 6, 2205–2210 [DOI] [PubMed] [Google Scholar]
- 5.Lehner, I., Niehof, M., and Borlak, J. ( 2003) An optimized method for the isolation and identification of membrane proteins. Electrophoresis 24, 1795–1808 [DOI] [PubMed] [Google Scholar]
- 6.Goshe, M. B., Blonder, J., and Smith, R. D. ( 2003) Affinity labeling of highly hydrophobic integral membrane proteins for proteome-wide analysis. J. Proteome Res. 2, 153–161 [DOI] [PubMed] [Google Scholar]
- 7.Schluesener, D., Fischer, F., Kruip, J., Rögner, M., and Poetsch, A. ( 2005) Mapping the membrane proteome of Corynebacterium glutamicum. Proteomics 5, 1317–1330 [DOI] [PubMed] [Google Scholar]
- 8.Zhang, H., Lin, Q., Ponnusamy, S., Kothandaraman, N., Lim, T. K., Zhao, C., Kit, H. S., Arijit, B., Rauff, M., Hew, C. L., Chung, M. C., Joshi, S. B., and Choolani, M. ( 2007) Differential recovery of membrane proteins after extraction by aqueous methanol and trifluoroethanol. Proteomics 7, 1654–1663 [DOI] [PubMed] [Google Scholar]
- 9.Wittig, I., Braun, H. P., and Schägger, H. ( 2006) Blue native PAGE. Nat. Protoc. 1, 418–428 [DOI] [PubMed] [Google Scholar]
- 10.Wittig, I., Karas, M., and Schägger, H. ( 2007) High-resolution clear-native electrophoresis for in-gel functional assays an fluorescence studies of membrane protein complexes. Mol. Cell. Proteomics 6, 1215–1225 [DOI] [PubMed] [Google Scholar]
- 11.Görg, A., Weiss, W., and Dunn, M. J. ( 2004) Current two-dimensional electrophoresis technology for proteomics. Proteomics 4, 3665–3685 [DOI] [PubMed] [Google Scholar]
- 12.O'Farrell, P. H. ( 1975) High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 250, 4007–4021 [PMC free article] [PubMed] [Google Scholar]
- 13.Washburn, M. P., Wolters, D., and Yates, J. R., III ( 2001) Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19, 242–247 [DOI] [PubMed] [Google Scholar]
- 14.Kyte, J., and Doolittle, R. F. ( 1982) A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132 [DOI] [PubMed] [Google Scholar]
- 15.Pedersen, S. K., Harry, J. L., Sebastian, L., Baker, J., Traini, M. D., McCarthy, J. T., Manoharan, A., Wilkins, M. R., Gooley, A. A., Righetti, P. G., Packer, N. H., Williams, K. L., and Herbert, B. R. ( 2003) Unseen proteome: mining below the tip of the iceberg to find low abundance and membrane proteins. J. Proteome Res. 2, 303–311 [DOI] [PubMed] [Google Scholar]
- 16.Rais, I., Karas, M., and Schägger, H. ( 2004) Two-dimensional electrophoresis for the isolation of integral membrane proteins and mass spectrometric identification. Proteomics 4, 2567–2571 [DOI] [PubMed] [Google Scholar]
- 17.Hartinger, J., Stenius, K., Hogemann, D., and Jahn, R. ( 1996) 16-BAC/SDS-PAGE: a two-dimensional gel electrophoresis system suitable fort the separation of integral membrane proteins. Anal. Biochem. 240, 126–133 [DOI] [PubMed] [Google Scholar]
- 18.Speers, A. E., Blackler, A. R., and Wu, C. C. ( 2007) Shotgun analysis of integral membrane proteins facilitated by elevated temperature. Anal. Chem. 79, 4613–4620 [DOI] [PubMed] [Google Scholar]
- 19.Chen, E. I., Cociorva, D., Norris, C. L., and Yates, J. R., III ( 2007) Optimization of mass spectrometry-compatible surfactants for shotgun proteomics. J. Proteome Res. 6, 2529–2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simpson, R. J., Conolly, L. M., Eddes, J. S., Pareira, J. J., Moritz, R. L., and Reid, G. E. ( 2000) Proteomic analysis of the human colon carcinoma cell line LIM 1215: development of a membrane protein database. Electrophoresis 21, 1707–1732 [DOI] [PubMed] [Google Scholar]
- 21.Reinders, J., Zahedi, R. P., Pfanner, N., Meisinger, C., and Sickmann, A. ( 2006) Toward the complete yeast mitochondrial proteome: multidimensional separation techniques for mitochondrial proteomics. J. Proteome Res. 5, 1543–1554 [DOI] [PubMed] [Google Scholar]
- 22.Wu, C. C., MacCoss, M. J., Howell, K. E., and Yates, J. R., III ( 2003) A method for the comprehensive proteomic analysis of membrane proteins. Nat. Biotechnol. 21, 508–51012721575 [Google Scholar]
- 23.Fischer, F., and Poetsch, A. ( 2006) Protein cleavage strategies for an improved analysis of the membrane proteome. Proteome Sci. 4, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han, J., and Schley, K. L. ( 2004) Proteolysis and mass spectrometric analysis of an integral membrane: aquaporin 0. J. Proteome Res. 3, 807–812 [DOI] [PubMed] [Google Scholar]
- 25.MacCoss, M. J., McDonald, W. H., Saraf, A., Sadygov, R., Clark, J. M., Tasto, J. J., Gould, K. L., Wolters, D., Washburn, M., Weiss, A., Clark, J. I., and Yates, J. R., III ( 2002) Shotgun identification of protein modifications from protein complexes and lens tissue. Proc. Natl. Acad. Sci. U. S. A. 99, 7900–7905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blonder, J., Goshe, M. B., Moore, R. J., Pasa-Tolic, L., Masselon, C. D., Lipton, M. S., and Smith, R. D. ( 2002) Enrichment of integral membrane proteins for proteomic analysis using liquid chromatography-tandem mass spectrometry. J. Proteome Res. 1, 351–360 [DOI] [PubMed] [Google Scholar]
- 27.Blonder, J., Conrads, T. P., Yu, L. R., Terunuma, A., Janini, G. M., Issaq, H. J., Vogel, J. C., and Veenstra, T. D. ( 2004) A detergent- and cyanogen bromide-free method for integral membrane proteomics: Application to Halobacterium purple membranes and the human epidermal membrane proteome. Proteomics 4, 31–45 [DOI] [PubMed] [Google Scholar]
- 28.Fischer, F., Wolters, D., Rögner, M., and Poetsch, A. ( 2006) Toward the complete membrane proteome: high coverage of integral membrane proteins through transmembrane peptide detection. Mol. Cell. Proteomics 5, 444–453 [DOI] [PubMed] [Google Scholar]
- 29.Yu, Y. Q., Gilar, M., and Gebler, J. C. ( 2004) A complete peptide mapping of membrane proteins: a novel surfactant aiding the enzymatic digestion of bacteriorhodopsin. Rapid Commun. Mass Spectrom. 18, 711–715 [DOI] [PubMed] [Google Scholar]
- 30.Bieth, J. G. ( 1998) Pancreatic elastase, in Handbook of Proteolytic Enzymes, pp. 42–46, Academic Press, London
- 31.Schlosser, A., Pipkorn, R., Bossemeyer, D., and Lehmann, W. D. ( 2001) Analysis of protein phosphorylation by a combination of elastase digestion and neutral loss tandem mass spectrometry. Anal. Chem. 73, 170–176 [DOI] [PubMed] [Google Scholar]
- 32.Morris, H. R., Batley, K. E., Harding, N. C., Bjur, R. A., Dann, J. G., and King, R. W. ( 1974) Dihydrofolate reductase: low-resolution mass-spectrometric analysis of an elastase digest as a sequencing tool. Biochem. J. 137, 409–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schechter, I., and Berger, A. ( 1967) On the size of the active site in proteases. I. Papain. Biochem. Biophys. Res. Commun. 27, 157–162 [DOI] [PubMed] [Google Scholar]
- 34.Lu, W., Apostol, I., Qasim, M. A., Warne, N., Wynn, R., Zhang, W. L., Anderson, S., Chiang, Y. W., Ogin, E., Rothberg, I., Ryan, K., and Laskowski, M., Jr. ( 1997) Binding of amino acid side-chains to S1 cavities of serine proteinases. J. Mol. Biol. 266, 441–461 [DOI] [PubMed] [Google Scholar]
- 35.Thomson, A., and Kapadia, S. B. ( 1979) The specificity of the S1 and S2 subsites of elastase. Eur. J. Biochem. 102, 111–116 [DOI] [PubMed] [Google Scholar]
- 36.Narayanan, A. S., and Anwar, R. A. ( 1969) The specificity of purified porcine pancreatic elastase. Biochem. J. 114, 11–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naughton, M. A., and Sanger, F. ( 1961) Purification and specificity of pancreatic elastase. Biochem. J. 78, 156–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vered, M., Burstein, Y., and Gertler, A. ( 1985) Digestion of elastin by porcine pancreatic elastase I and elastase II. Int. J. Pept. Protein Res. 25, 76–84 [DOI] [PubMed] [Google Scholar]
- 39.Jinsmaa, Y., and Yoshikawa, M. ( 1999) Enzymatic release of neocasomorphin and β-casomorphin from bovine β-casein. Peptides 20, 957–962 [DOI] [PubMed] [Google Scholar]
- 40.Lowry, O. H., Rosebrough, N. J., Farr, A. L., and Randall, R. J. ( 1951) Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193, 265–275 [PubMed] [Google Scholar]
- 41.Wolters, D. A., Washburn, M. P., and Yates, J. R., III ( 2001) An automated multidimensional protein identification technology for shotgun proteomics. Anal. Chem. 73, 5683–5690 [DOI] [PubMed] [Google Scholar]
- 42.Perkins, D. N., Pappin, D. J., Creasy, D. M., and Cottrell, J. S. ( 1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 43.Kalinowski, J., Bathe, B., Bartels, D., Bischoff, N., Bott, M., Burkovski, A., Dusch, N., Eggeling, L., Eikmanns, B. J., Gaigalat, L., Goesmann, A., Hartmann, M., Huthmacher, K., Krämer, R., Linke, B., McHardy, A. C., Meyer, F., Möckel, B., Pfefferle, W., Pühler, A., Rey, D. A., Rückert, C., Rupp, O., Sahm, H., Wendisch, V. F., Wiegräbe, I., and Tauch, A. ( 2003) The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of l-aspartate-derived amino acids and vitamins. J. Biotechnol. 104, 5–25 [DOI] [PubMed] [Google Scholar]
- 44.Nelson, D. L., and Cox, M. M. ( 2000) Lehninger Principles of Biochemistry, 3rd Ed., p. 125, W. H. Freeman and Co., Bedford, NY
- 45.Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E. L. ( 2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580 [DOI] [PubMed] [Google Scholar]
- 46.Spyropoulos, I. C., Liakopoulos, T. D., Bagos, P. G., and Hamodrakas, S. J. ( 2004) TMRPres2D: high quality visual representation of transmembrane protein models. Bioinformatics 20, 3258–3260 [DOI] [PubMed] [Google Scholar]
- 47.Meyer, E., Cole, G., Radhakrishnan, R., and Epp, O. ( 1988) Structure of native porcine pancreatic elastase at 1.65 Å resolutions. Acta Crystallogr. Sect. B Struct. Sci. 44, 28–38 [DOI] [PubMed] [Google Scholar]
- 48.Vajda, T., and Szabó, T. ( 1976) Specificity of trypsin and alpha-chymotrypsin towards neutral substrates. Acta Biochim. Biophys. Acad. Sci. Hung. 11, 287–294 [PubMed] [Google Scholar]
- 49.Gráf, L., Jancsó, A., Szilágyi, L., Hegyi, G., Pintér, K., Náray-Szabó, G., Hepp, J., Medzihradszky, K., and Rutter, W. J. ( 1988) Electrostatic complementarity within the substrate-binding pocket of trypsin. Proc. Natl. Acad. Sci. U. S. A. 85, 4961–4965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krause, E., Wenschuh, H., and Jungblut, P. R. ( 1999) The dominance of arginine-containing peptides in MALDI-derived tryptic mass fingerprints of proteins. Anal. Chem. 71, 4160–4165 [DOI] [PubMed] [Google Scholar]
- 51.Strader, M. B., Tabb, D. L., Hervey, W. J., Pan, C., and Hurst, G. B. ( 2006) Efficient and specific trypsin digestion of microgram to nanogram quantities of proteins in organic-aqueous solvent systems. Anal. Chem. 78, 125–134 [DOI] [PubMed] [Google Scholar]
- 52.Mirgorodskaya, E., Braeuer, C., Fucini, P., Lehrach, H., and Gobom, J. ( 2006) Nanoflow liquid chromatography coupled to matrix-assisted laser desorption/ionization mass spectrometry: sample preparation, data analysis, and application to the analysis of complex peptide mixtures. Proteomics 5, 299–408 [DOI] [PubMed] [Google Scholar]
- 53.Jaskolla, T. W., Lehmann, W. D., and Karas, M. ( 2008) 4-Chloro-α-cyanocinnamic acid is an advanced, rationally designed MALDI matrix. Proc. Natl. Acad. Sci. U. S. A. 105, 12200–12205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cech, N. B., and Enke, C. G. ( 2000) Relating electrospray ionization response to nonpolar character of small peptides. Anal. Chem. 72, 2717–2723 [DOI] [PubMed] [Google Scholar]
- 55.Frahm, J. L., Bori, I. D., Comins, D. L., Hawkridge, A. M., and Muddiman, D. C. ( 2007) Achieving augmented limits of detection for peptides with hydrophobic alkyl tags. Anal. Chem. 79, 3989–3995 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.