Abstract
Gene families with multiple members are predicted to have individuals with overlapping functions. We examined all of the Arabidopsis (Arabidopsis thaliana) myosin family members for their involvement in Golgi and other organelle motility. Truncated fragments of all 17 annotated Arabidopsis myosins containing either the IQ tail or tail domains only were fused to fluorescent markers and coexpressed with a Golgi marker in two different plants. We tracked and calculated Golgi body displacement rate in the presence of all myosin truncations and found that tail fragments of myosins MYA1, MYA2, XI-C, XI-E, XI-I, and XI-K were the best inhibitors of Golgi body movement in the two plants. Tail fragments of myosins XI-B, XI-F, XI-H, and ATM1 had an inhibitory effect on Golgi bodies only in Nicotiana tabacum, while tail fragments of myosins XI-G and ATM2 had a slight effect on Golgi body motility only in Nicotiana benthamiana. The best myosin inhibitors of Golgi body motility were able to arrest mitochondrial movement too. No exclusive colocalization was found between these myosins and Golgi bodies in our system, although the excess of cytosolic signal observed could mask myosin molecules bound to the surface of the organelle. From the preserved actin filaments found in the presence of enhanced green fluorescent protein fusions of truncated myosins and the motility of myosin punctae, we conclude that global arrest of actomyosin-derived cytoplasmic streaming had not occurred. Taken together, our data suggest that the above myosins are involved, directly or indirectly, in the movement of Golgi and mitochondria in plant cells.
The Arabidopsis (Arabidopsis thaliana) myosin gene family contains 17 members: myosin group XI, which includes 13 members (myosins XI-A, -B, -C, -D, -E, -F, -G, -H, -I, -J, and -K, MYA1, and MYA2), and myosin group VIII, which includes four members (ATM1, ATM2, myosin VIIIA, and myosin VIIIB). Both groups are related to unconventional myosin V (Berg et al., 2001; Foth et al., 2006). The Arabidopsis myosins contain a conserved motor domain with ATPase and actin-binding activities, a number of IQ domains that bind myosin light chains, a coiled-coil domain for dimerization, and a specific tail that binds different cargo (Kinkema and Schiefelbein, 1994; Tominaga et al., 2003). Using these functional domains, myosins convert chemical energy from ATP hydrolysis into physical movement along actin fibers, carrying with their tails membrane-bound organelles or RNA/protein complexes (Li and Nebenführ, 2008b).
Plant myosins have been implicated in various cellular activities, such as cytoplasmic streaming (Shimmen and Yokota, 2004; Esseling-Ozdoba et al., 2008), plasmodesmata function (Baluska et al., 2001; Volkmann et al., 2003), organelle movement (Nebenführ et al., 1999; Jedd and Chua, 2002), cytokinesis (Molchan et al., 2002; Collings et al., 2003; Volkmann et al., 2003), endocytosis (Volkmann et al., 2003; Baluska et al., 2004; Samaj et al., 2005), and targeted RNA transport (Hamada et al., 2003). Actomyosin mediated cytoplasmic streaming found in various algae cells reach velocities of up to 100 μm s−1, which is the fastest known myosin-mediated movement (Shimmen and Yokota, 1994).
The information that exists regarding specific roles of each plant myosin is rather limited. Immunolocalization studies indicated that myosin XIs are associated with various particles in lily (Lilium longiflorum) and tobacco (Nicotiana tabacum) pollen tubes (Yokota et al., 1995), with mitochondria, plastids, and low-density membranes in maize (Zea mays) root cells (Liu et al., 2001; Wang and Pesacreta, 2004), and with endoplasmic reticulum (ER) in tobacco BY2 cells (Yokota et al., 2008). Specific antibodies against MYA2 showed that it is associated with peroxisomes in epidermal and guard cells of Arabidopsis leaves (Hashimoto et al., 2005). More recent studies using recombinant DNA fusions to fluorescent proteins showed localization of the tails of MYA2, MYA1, XI-K, and XI-I to peroxisomes (Li and Nebenführ, 2007; Reisen and Hanson, 2007) and MYA1 partially localized to Golgi (Li and Nebenführ, 2007). Furthermore, it was shown that peroxisomes, Golgi, and mitochondrial motility were arrested by dominant negative mutants of myosin XI-K and myosin XI-E (Avisar et al., 2008b; Sparkes et al., 2008). Arrest of organelle motility was also found in Arabidopsis knockout plants xi-k and mya2 (Peremyslov et al., 2008) and double mutants xi-k/mya1, xi-k/mya2, and mya2/xi-b (Prokhnevsky et al., 2008). In contrast, the association of the single globular tail domain of MYA1 or MYA2 with peroxisomes did not arrest their motility (Li and Nebenführ, 2007). Knockout plants for myosin xi-k and mya2 had root hair phenotypes (Ojangu et al., 2007; Peremyslov et al., 2008); however, all other 11 myosin XI single knockouts looked normal under regular growth conditions (Peremyslov et al., 2008). Reciprocal stimulation between dimerization via the coiled-coil domains of MYA1 and organelle binding was suggested (Li and Nebenführ, 2008a). As for myosin VIII, immunostaining studies showed that it localized to the cell periphery at plant-specific structures such as plasmodesmata and cytokinetic cell plates (Reichelt et al., 1999; Baluska et al., 2001). Recent data from our laboratory and from others confirmed the presence of myosin VIII in plasmodesmata (Golomb et al., 2008) and the cell plate (Van Damme et al., 2004) and further provided evidence for its involvement with endocytosis (Golomb et al., 2008; Sattarzadeh et al., 2008) and its colocalization with the ER (Golomb et al., 2008). In addition, it was shown that myosin VIII is involved in the plasmodesmata targeting of the beet yellows virus protein Hsp70h (Avisar et al., 2008a).
We have determined the role of all 17 genes through transient overexpression of dominant negative forms in leaf epidermal cells. Fluorescent dominant negative fusions not only provide data on the subcellular location but also provide a relatively easy way of determining expression. Additionally, overexpression of dominant negative forms can expose a role of an individual member, which might be masked by redundant activity, if it was silenced. In order to undertake such a large-scale study, we needed to choose an efficient, fast, and reproducible expression system. Therefore, Agrobacterium tumefaciens-mediated transient expression in Nicotiana leaves was suitable.
RESULTS
Isolation and Generation of eGFP and mRFP Fusions to the 17 Arabidopsis Myosins
In order to compare the involvement of all annotated Arabidopsis myosins with Golgi movement, we isolated, sequenced, and subcloned a fragment containing the IQ, coiled-coil, and tail domains of all of them downstream to enhanced green fluorescent protein (eGFP) or containing the coiled-coil and tail domains downstream of mono red fluorescent protein (mRFP; Fig. 1). The idea was to create dominant negative clones of each myosin family member that lacks the head actin-binding domain but should still be able to bind cargo. This should saturate the binding sites and compete out the function of endogenous wild-type myosin molecules. Sequence analysis of several independent clones in both laboratories revealed the following differences in translation compared with the annotated database sequences. In myosin XI-D, instead of 1,256-KSLDLFVFMYLFQ-1,268, we found 1,256-VSFTRPP-1,262, which probably results from a different splicing site and an A-to-V change in position 1,082. In myosin XI-G, nucleotides 3,217 to 3,261 are missing; again, this seems to be a different splicing site prediction. In myosin VIIIB, we found that the predicted 932-VVFLPDVC-939 is 932-ELLSEQFE-939; again, this is probably a result of different splicing. The clone of ATM1 in our hands contains an 865G-to-865R change (Knight and Kendrick-Jones, 1993). Genevestigator analysis of expression patterns of Arabidopsis myosins revealed that myosins ATM2 and VIIIB from the group of myosin VIII and myosins XI-A, XI-B, XI-C, XI-D, XI-E, and XI-J are expressed at high levels in pollen. These myosins are expressed also in the mature leaf but at levels two to five times lower than other myosin family members (Zimmermann et al., 2004; Supplemental Fig. S1). Previously, it was shown that myosins from Nicotiana benthamiana possess a high degree of sequence and functional homology to their Arabidopsis counterparts (Avisar et al., 2008b). In addition, the functions of Arabidopsis myosins XI-K and XI-E were proven in N. tabacum (Sparkes et al., 2008). Therefore, we decided to compare different fragments of all 17 Arabidopsis myosins in N. tabacum (N.t.) and N. benthamiana (N.b.) and to study their subcellular localization and ability to arrest the motility of Golgi and other different organelles.
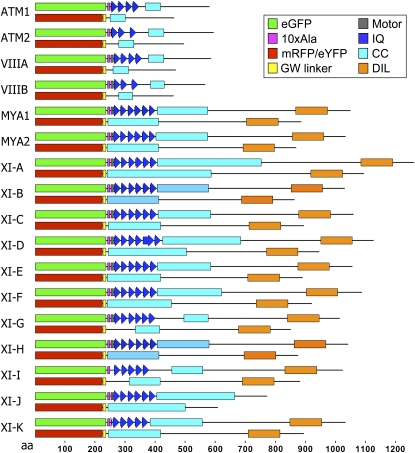
Figure 1.
Schematic representation of the eGFP/mRFP-myosin fusions used in this work. A fragment containing the IQ tail domain of each of the 17 Arabidopsis annotated myosin cDNAs was fused to the C terminus of eGFP with a linker of 10×Ala. In some of the constructs, a few amino acids from the motor domain were included. The tail fusions did not contain the IQ domain and were fused using Gateway (GW) to either mRFP or eYFP. CC, Coiled coil; DIL, Dilute.
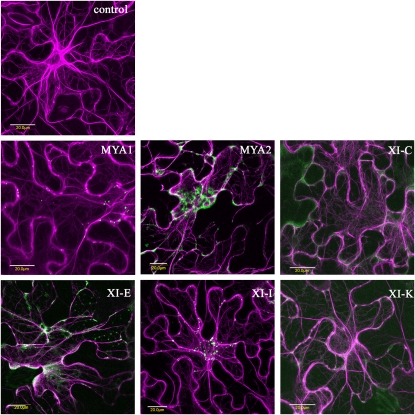
Myosin Tail and IQ Tail Fusions Are Located to Various Subcellular Structures in Tobacco Leaf Epidermal Cells
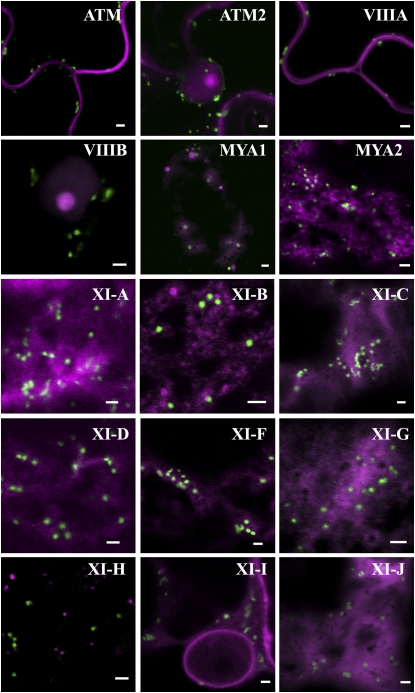
First, the subcellular localizations of tail and IQ tail myosin truncations were compared with each other and with Golgi bodies. Myosin tails were coexpressed with a fluorescent Golgi body marker (sialyl transferase [ST]-XFP) in N.t. epidermal cells (Fig. 2); in addition, the localization of the two different myosin fusions, mRFP-tail and eGFP-IQ tail, were compared by coexpression in N.t. (Supplemental Fig. S3) and the eGFP-IQ tail fusions were expressed in N.b. (Supplemental Fig. S2). A general observation was that while class VIII fusions mainly located to the plasma membrane (ATM1, ATM2, VIIIA, and VIIIB; Fig. 2; Supplemental Fig. S2) and/or the nucleolus (ATM2 and VIIIB; Fig. 2), class XI fusions tended to be in more motile punctate structures (MYA1, MYA2, XI-E, XI-H, and XI-I) or diffuse (XI-C, XI-D, XI-F, XI-J, and XI-K) throughout the cytoplasm (Fig. 2; Supplemental Fig. S2). XI-A, XI-B, and XI-G tail/IQ tail expression tended to vary from motile puncta to diffuse. Interestingly, both XI-I truncations collocated to punctate structures and also collocated to the nuclear envelope in both plant species (Fig. 2; Supplemental Figs. S2, S3, and S5).
Figure 2.
The subcellular localization patterns of Golgi and Arabidopsis myosin tail fragments in N.t. Myosin tail fragments fused to mRFP (magenta) were coexpressed with a Golgi marker (ST-GFP; green) in N.t. leaf epidermal cells. See Sparkes et al. (2008) for images of XI-E and XI-K tail fusions coexpressed with a Golgi marker. Bars = 2 μm.
None of the myosin fusions solely colocated with Golgi bodies, but the surrounding cytosolic location could represent a small proportion of bound myosin fusions (Fig. 2). Upon coexpression, most of the IQ tail and tail fusions of the same myosin gene colocated to the same structures in N.t. with few exceptions: the ATM2 IQ tail fusion did not colocate to the nucleolus unlike the shorter tail fusion, XI-A and XI-C tail and IQ tail fusions were diffuse throughout the cytosol, and the IQ tail fusion was also in the nucleoplasm (Supplemental Fig. S3). Thus, it is suggested that subcellular localization is similar whether or not the IQ domain is present.
The Effect of Each of the Arabidopsis Myosin Fragments on Golgi Body Motility
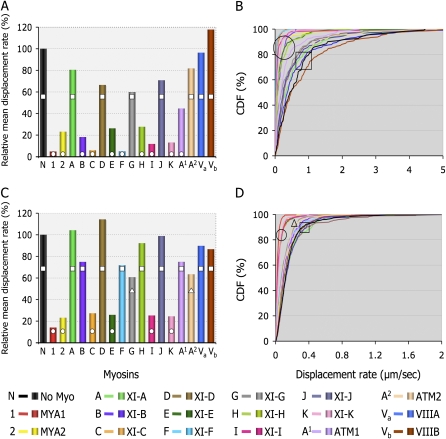
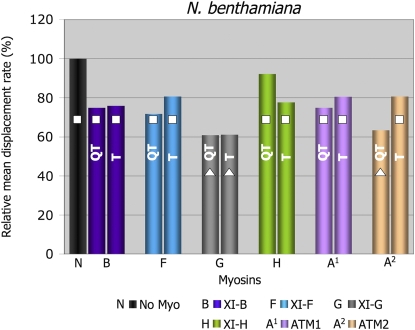
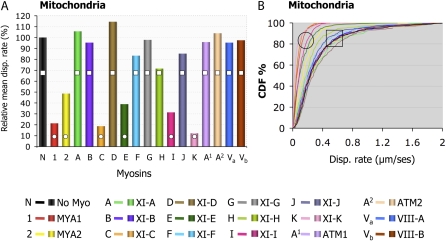
In order to study the effect of each myosin on the motility of Golgi, we coexpressed each myosin fragment with a Golgi marker in two different Nicotiana species. Tail fusions were expressed in N.t. and IQ tail fusions were expressed in N.b. Myosin fusions that behaved differently in the two Nicotiana species were expressed and compared in N.b. This comparison allowed us to verify variations derived from either the plant or the length of the myosin fusion. Owing to the variability of organelle movement within a cell (i.e. fast, slow, and stationary), large samples of organelles were analyzed from at least 10 movies from three to five independent experiments. Volocity software was used to track organelles and calculate velocity and displacement rates of each tracked organelle over time. Displacement rate is the final distance an organelle moves between the first and last frame of the time-lapse sequence over time. We found that organelles that were undergoing short-range oscillations in one spot, but appeared by eye to have stopped moving (Supplemental Movies S1–S4), obtained somewhat unexpectedly high values of velocity calculated by the software. The two data sets from N.t. and N.b. were acquired using different rates of image acquisition, which had a different influence on velocity values determined for such oscillatory movements (i.e. a higher frame rate measured more oscillations, resulting in higher velocities than those captured using a lower frame rate). However, this did not affect displacement rate measurements, since this is a measure of movement away from the starting point and is not significantly affected by short-range oscillatory motion. The two data sets generated from N.b. and N.t., therefore, were compared using the displacement rate values, as they are a better reflection of the perturbation of directional organelle movement and allow comparisons to be drawn between data sets from the two species. To confirm that the different rates of image acquisition did not affect the velocity or displacement rate calculated for organelles in similar plant species, high and low rates of image capture were compared in N.b., but no significant differences were found (Supplemental Table S2.). Figure 3 shows the mean displacement rate (normalized against the respective controls) and cumulative distribution frequency graphs that describe the distribution of displacement rates within a Golgi population in the presence of each truncated myosin fusion. Comparisons between the two data sets, from N.t. (Fig. 3A) and from N.b. (Fig. 3C), reveal that both tail and IQ tail fusions of myosins MYA1, MYA2, XI-C, XI-E, XI-I, and XI-K significantly inhibited Golgi displacement rates in both plants (see Supplemental Table S1 for statistical analysis; Supplemental Movies S1 and S2). Figure 3 also shows that myosins XI-B, XI-F, XI-H, and ATM1 significantly interfered with Golgi movement only as tail fusions in N.t. (Fig. 3, A and B) but not as IQ tail fusions in N.b. (Fig. 3, C and D). In addition, myosins XI-G and ATM2 partially interfered with Golgi movement only as IQ tail fusions in N.b. plants (Fig. 3, C and D). In order to understand the origin of the differences obtained in the two systems, the tail fusions of myosins XI-G, ATM2, XI-B, XI-F, XI-H, and ATM1 were analyzed in N.b. Figure 4 shows that except for the ATM2 tail fusion, all other tail fusions had a similar effect on Golgi body motility to the IQ tail fusions in N.b. The tail fusion of ATM2 did not affect Golgi movement, although its IQ tail fragment did to some extent (Supplemental Table S1). Importantly, when IQ tail mutants of XIB, XIF, and ATM1 were expressed in N.t., they exhibited an inhibitory effect on Golgi body movement similar to their tail fusion counterparts in this plant (data not shown). Thus, it seems that the source of variability between the two expression systems is the plant species itself and not an intrinsic difference in the myosin fragment fused to a fluorescent marker protein.
Figure 3.
Displacement rates of Golgi bodies in the presence of 17 Arabidopsis myosin tail fusions in N.t. (A and B) and myosin IQ tail fusions in N.b. (C and D). Using Volocity software, displacement rates were calculated for Golgi bodies in the presence of myosin tail fusions (A and B) or myosin IQ tail fusions (C and D). Values are expressed as percentages of the control displacement rate of Golgi bodies in each system. A and C, Plots of mean displacement. B and D, Cumulative distribution frequency (CDF) plots. Columns or curves overlaid with different shapes are statistically different at P < 0.05. (Data for XI-E and XI-K tail fusions in N.t. are from Sparkes et al. [2008].)
Figure 4.
Displacement rates of Golgi in the presence of tail or IQ tail fusions in N.b. The myosin tail fusions that gave different results in N.t. compared with their IQ tail counterparts in N.b. were expressed in N.b., and the mean displacement rates were calculated. Columns overlaid with different shapes are statistically different at P < 0.05.
Myosin Inhibitors of Golgi Motility Affect Mitochondrial Movement as Well
Next, we addressed the question of whether myosin truncations specifically perturbed Golgi movement only or whether they affected other organelle classes. Assessing the displacement rates of mitochondria in the presence of myosin IQ tail mutants in N.b. showed that the same myosins that inhibited Golgi movement in the two systems were able to arrest mitochondrial motility as well (Fig. 5; Supplemental Movie S3; Supplemental Table S1). The rest of the putative dominant negative mutants of myosins XI-A, XI-B, XI-D, XI-F, XI-G, XI-H, XI-J, ATM1, ATM2, VIIIA, and VIIIB had no significant effect on the movement of mitochondria in N.b. leaves (Fig. 5; Supplemental Table S1). The inhibitory IQ tail myosin fusions did not specifically colocalize to Golgi or mitochondria in N.b. (Supplemental Fig. S5).
Figure 5.
Displacement rates for mitochondria in the presence of IQ tail truncations of all 17 Arabidopsis myosins in N.b. leaves. All 17 myosin IQ tail fusions were coexpressed with mitochondria markers in N.b. leaves. Abaxial epidermal cells were analyzed by confocal microscopy 48 h after infiltration. Time-lapse image acquisition was performed, and organelle tracking was performed by Volocity. A, The mean displacement (disp.) rates were normalized to control organelle displacement in the absence of myosin mutant and plotted here as percentage of control (100%). B, Cumulative distribution frequency (CDF) plots showing the behavior of organelle populations in the presence of the indicated myosins.
Myosin-Truncated Fragments Do Not Indiscriminately Arrest Cytoplasmic Streaming
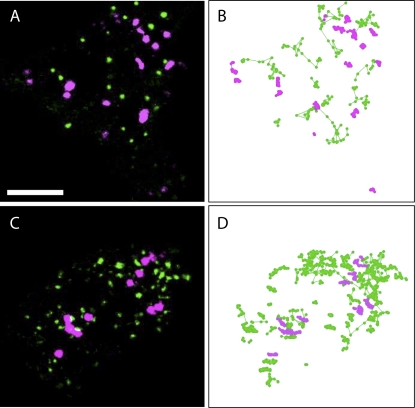
To ensure that myosin-truncated fragments are not nonspecifically interfering with actomyosin-derived cytoplasmic streaming, actin was labeled by the DsRed FABD2 marker (Voigt et al., 2005) and examined in the presence of each eGFP-IQ tail myosin fusion in N.b. Figure 6 shows no detectable actin disruption in the presence of the six myosin mutants of MYA1, MYA2, myosin XI-C, XI-E, XI-I, and XI-K. Furthermore, we compared the motility of myosin punctae of MYA1 and MYA2 with Golgi body movement in the same time-lapse movie acquired from N.b. While Golgi bodies are motionless in the presence of eGFP-IQ tail, the MYA1 and MYA2 punctae are motile (with mean displacement rate 10 times higher than Golgi in these clips). Some of the MYA1 and MYA2 punctae moved in linear routes that may correspond to streaming in cytoplasmic strands (Fig. 7; Supplemental Movie S4). eGFP-MYA1 and eGFP-MYA2 spots can move without the motor domain, because they are either bound to organelles or vesicles that move by other wild-type active motor proteins or they are aggregates of the GFP chimera itself that are dragged along by cytoplasmic streaming.
Figure 6.
Actin fibers are unaffected in the presence of the inhibitory eGFP-myosin mutants. An actin marker, DsRED-FABD2, was coexpressed with eGFP myosin fusions in N.b. leaves. Confocal images of abaxial epidermal cells are shown. Actin is not disrupted in the presence of the myosin mutants. Bars = 20 μm.
Figure 7.
Tracking of both MYA1 and MYA2 punctae as well as Golgi stacks in the same cell. To demonstrate that arresting organelle motility was not accompanied by a general arrest of cytoplasmic streaming, both MYA1 or MYA2 and Golgi stacks were tracked in the same leaf. eGFP-IQ tail and the Golgi marker, ST-mRFP, were coexpressed in leaf abaxial epidermal cells of N.b. and observed by confocal microscopy 48 h after infiltration. A and B, MYA1 (green) and Golgi (magenta). C and D, MYA2 (green) and Golgi (magenta). A and C, The first frames of the time-lapse sequences. B and D, Tracks of Golgi (magenta) and eGFP-IQ tail puncta (green). While Golgi stacks are almost completely motionless, puncta of MYA1 and MYA2 are motile. Bar = 8 μm.
These data provide strong evidence that the myosin mutants are not arresting all cytoplasmic motion in the cells but are specifically inhibiting the motility of the tested Golgi and mitochondria.
DISCUSSION
Using dominant negative myosin mutants lacking the head motor domain, we found six myosins (MYA1, MYA2, and myosins XI-C, XI-E, XI-I, and XI-K) that were more efficient in inhibiting the motility of Golgi bodies in two different Nicotiana species either as IQ tail or tail truncations. Additionally, IQ tail fragments of these myosins also inhibited mitochondrial motility in N.b. This suggests that these myosins are involved in transporting these organelles in plant cells either in a collaborative, synergistic manner or simply a redundant manner. Although transient expression of Arabidopsis genes in other plant systems is an accepted, widely used method, the data presented here still have to be reconfirmed, in the future, in Arabidopsis plants. The calculated organelle velocity obtained in N.b. was smaller than that obtained in N.t. and smaller than reported previously (Nebenführ et al., 1999; Avisar et al., 2008b; Sparkes et al., 2008). The lower image frame rate used in N.b. movies in contrast to the higher frame rate image acquisition used in N.t. raised the possibility of biased analysis toward slow-moving organelles in N.b. However, when fast and slow frame rates of image acquisition were compared in N.b., no differences in measured organelle motility were found. This suggested that the differences found in organelle velocity between the two plants are the result of different growth conditions of N.b. and N.t. in the two laboratories. Interestingly, mutants of ATM1, ATM2, XI-B, XI-G, XI-H, and XI-F exhibited conflicting results in the two species of Nicotiana plants: while myosins XI-B, XI-F, XI-H, and ATM1 were inhibitory only as tail fusions in N.t., myosins XI-G and ATM2 slightly inhibited Golgi movement only as IQ tail fusions in N.b. However, when all of these myosin constructs were expressed side by side in N.b., they affected Golgi movement in a similar way (except for the slight effect observed with IQ tail of ATM2). Thus, it may be concluded that species-specific differences rather than intrinsic differences in the myosin gene fusions expressed resulted in the different rates observed in the different plants (i.e. N.t. versus N.b.). Again, this also emphasizes the conclusion that the major players in Golgi movement are myosins MYA1, MYA2, XI-C, XI-E, XI-I, and XI-K.
The slight effect of ATM1 (tail in N.t.) and ATM2 (IQ tail in N.b.) with Golgi movement is in agreement with our previous observation that ATM1 colocalized with the ER (Golomb et al., 2008) and raises the possibility of an indirect regulation of Golgi motility via ER motion (Boevink et al., 1998; Runions et al., 2006; Hawes et al., 2008) that is driven by myosins (Yokota et al., 2008). Interestingly, we have recently shown that Golgi bodies are physically attached to the ER (Sparkes et al., 2009), which could potentially be via a myosin scaffold at the ER-Golgi interface, and we have evidence that ER remodeling still occurs in the absence of the Golgi bodies (I.A. Sparkes, unpublished data).
Our findings regarding IQ tail truncations of Arabidopsis myosins XI-K and XI-E in N.b. as inhibitors of Golgi and mitochondrial movement are in agreement with previous publications (Avisar et al., 2008b; Sparkes et al., 2008). Taken together, the new findings presented here are the strong involvement of myosin XI-C in the motility of Golgi and mitochondria, the involvement of myosin XI-I in the movement of Golgi and mitochondria, its localization to the nuclear envelope, and the different combinations of myosins that are involved in the movement of these two different organelles.
The subcellular location of tail and IQ tail fusions vary from plasma membrane (ATM, ATM2, VIIIA, and VIIIB), diffuse in the cytosol (XI-A, MYA2, XI-B, XI-C, XI-D, XI-E, XI-F, XI-G, XI-I, XI-J, and XI-K), nuclear envelope (XI-I), nucleolus (VIIIB and ATM2 tail only), to motile puncta (MYA1, MYA2, XI-A, XI-B, XI-E, XI-G, XI-H, XI-I, and XI-K). Previous expression studies of fluorescent fusions to class VIII (Golomb et al., 2008) and class XI (Reisen and Hanson, 2007) myosins corroborate these subcellular locations. However, Li and Nebenführ (2007) found that MYA1 fusions occasionally colocated to Golgi bodies, and Reisen and Hanson (2007) did not observe XI-I tail on the nuclear envelope. These discrepancies could be due to differences in the constructs under test and the experimental systems used Different patterns of subcellular localization were found among the best myosin mutants inhibiting Golgi movement: while MYA1, MYA2, and myosins XI-E, XI-K, and XI-I locate to punctate structures of varying size, only myosin XI-I encircles the nucleus and myosin XI-C is completely diffuse in the cytoplasm. This could indicate that the effect or lack of an effect is not due to a specific pattern of localization.
Interference of a dominant negative myosin mutant with the function of other myosins could be executed in several putative ways: (1) competition for myosin-binding sites, (2) competition among adaptor proteins that mediate myosin-cargo interactions, (3) competition for a limited pool of light chains, (4) dimerization of the mutant headless myosin with wild-type myosin, and/or (5) nonspecific inhibition of total cytoplasmic streaming by the toxic dominant negative mutant.
Taken together, the data presented here and in the literature suggest that option 2 is more likely; however, more data are required to draw discerning conclusions. It was shown that a fragment of N.b. myosin XI-K including the IQ domain, coiled coil, and tail was able to arrest the motility of Golgi, mitochondria, and peroxisomes in N.b. leaves (Avisar et al., 2008b). A shorter fragment of N.b. myosin XI-K containing only the globular tail domain was equally efficient in arresting motility of Golgi stacks in the same assay system (Avisar et al., 2008b). Furthermore, motility inhibition of all three organelles was shown by transient expression of RNA interference of N.b. myosin XI-K in N.b. leaves (Avisar et al., 2008b). Similarly, inhibition of the motility of Golgi, mitochondria, and peroxisomes was obtained by transiently expressing tail fragments containing the coiled-coil and globular tail domains of Arabidopsis myosins XI-K and XI-E in N.t. (Sparkes et al., 2008). At the level of the whole plant, it was shown that organelle movement was inhibited in root hairs and leaves of both myosin XI-K knockout plants and in plants overexpressing the globular tail domain of myosin XI-K as a dominant negative mutant (Peremyslov et al., 2008). Thus, competition for light chains is not likely to play a role in the inhibition of organelle motility by dominant negative myosin mutants, because it was manifested by fragments lacking the IQ domains, by RNA interference, and in knockout plants. Since none of the inhibitory myosin fragments solely colocalized to the inhibited organelles, it seems that saturating binding sites on the cargo itself may not be the inhibitory mechanism in our system. However, since a more diffuse cytoplasmic location could reflect low-level or partial binding to the organelle surface, this possibility cannot be formally discounted. Of note, colocalization between eGFP/enhanced yellow fluorescent protein (eYFP) fusions of myosin fragments and organelles was observed in other systems. Fragments from MYA1, MYA2, and myosins XI-K and XI-I containing the globular tail domains 1 and 2 colocalized to peroxisomes, as shown by bimolecular fluorescence complementation in Arabidopsis leaves (Li and Nebenführ, 2007). The second domain of MYA1 globular tail, GT2, was partially localized to Golgi stacks (Li and Nebenführ, 2007). A fragment containing the tail domain without the coiled coil from MYA2 colocalized to peroxisomes (Reisen and Hanson, 2007). On the other hand, no such colocalization was observed when myosin fragments from XI-K and XI-E including the coiled-coil and tail domains were expressed in N.t. (Sparkes et al., 2008). From these observations, we conclude that localization of myosin fragments depends on correct protein folding of the myosin fragment expressed and, perhaps, on the action of other genes expressed in the specific plant system.
Gross inhibition of cytoplasmic streaming by dominant negative myosin mutants is unlikely, because continuous movement of myosin puncta is presented here. Significant motility of eGFP-myosin fusions was also shown previously (Reisen and Hanson, 2007; Sparkes et al., 2008). Heterodimers of myosins were detected in low percentage in different systems; however, it was shown that homodimers are more thermodynamically stable (Dechesne et al., 1987; Kerwin and Bandman, 1991; Singh and Bandman, 2006), and heterodimerization of plant myosins is still to be tested. Interestingly, heterologous interaction between two subdomains of the globular tails GT1 from MYA1 and GT2 from MYA2, XI-C, or XI-K was observed in yeast two-hybrid analysis (Li and Nebenführ, 2007). However, no such interaction was observed between two intact tails in yeast two-hybrid analysis (Li and Nebenführ, 2007) or in vivo. Therefore, our assumption is that motility inhibition of organelles by myosin dominant negative mutants is the result of competition for relevant ligands. Candidates for tail ligands might be members of the small GTPase Rab family, as shown recently (Hashimoto et al., 2008), or other molecules similar to those interacting with myosin V (Li and Nebenführ, 2008b) that are still to be characterized in plants.
CONCLUSION
Apart from mutants of myosin XI-K and MYA2, all other family members of myosin XI had no observable phenotypes, excluding a moderate decrease in peroxisome movement observed in mya1 knockout plants and in plants expressing the MYA1 dominant negative globular tail domain (Peremyslov et al., 2008). Therefore, we conclude that myosins XI-K, MYA1, and MYA2 are major players in organelle motility in leaves and root hairs (Peremyslov et al., 2008; Prokhnevsky et al., 2008). Myosins XI-C and XI-E may be major players in pollen tubes, where they are highly expressed, but they are only minor players in leaves and root hairs. This might be the reason that no phenotypes were observed in the published knockout plants (Peremyslov et al., 2008). In addition, we discovered that the myosin XI-I mutant is very potent in inhibiting Golgi and mitochondrial movement. Despite the fact that its level of expression in all tissues is similar to or higher than that of MYA1 and MYA2 and myosin XI-K, no phenotype was observed in its knockout plant. One explanation may be that its dominant negative mutant interferes directly or indirectly with myosin XI-K, MYA2, and MYA1 activities.
MATERIALS AND METHODS
Generation of IQ Tail and Tail Constructs
IQ tail fusions were amplified by PCR from first-strand cDNA synthesized using oligo(dT) and reverse transcriptase from Arabidopsis (Arabidopsis thaliana) whole plant (including flowers) RNA. The ATM1 clone was kindly provided by Dieter Volkmann from the University of Bonn (Reichelt et al., 1999). The fragments were cloned into the plasmid pART7 downstream to eGFP, with a linker of 10 Ala amino acids between the restriction sites KpnI and BamHI, beside myosin fragments XI-A, XI-D, and XI-G, which were cloned between KpnI and XbaI, and XI-H, which was cloned between KpnI and ClaI. The expression cassettes were cloned into pART27 binary vector using NotI cleavage. All binary expression vectors were transformed into Agrobacterium tumefaciens strain GV3101.
Myosin tail fusions were amplified using IQ tail fusions as templates or by reverse transcription-PCR using the SuperScript III one-step reverse transcription-PCR platinum Taq HiFi kit (Invitrogen) from mRNA extracted from various Arabidopsis tissues using the Nucleospin RNA II kit (Macherey-Nagel). The ATM tail clone was amplified from the clone received from J.K. Jones (Knight and Kendrick-Jones, 1993). All amplified products were cloned into pDONOR 207 using Gateway technology according to Invitrogen and subsequently recombined into binary vectors resulting in eYFP (using pCAMBIA 1300 backbone vector; Sparkes et al., 2005) or mRFP (using pB7WGR2 vector) fusions. The binary expression vectors were transformed into A. tumefaciens strain GV3101.
Plants and Transient Expression
Nicotiana benthamiana plants were grown in peat in a controlled growth room at 25°C with optimum light of 16 h daily. Nicotiana tabacum plants were grown according to Sparkes et al. (2005).
N.b. leaf epidermal cells were infiltrated as follows. A. tumefaciens strain GV3101 was transformed with the plasmid and grown at 28°C for 24 h. The bacteria were precipitated and resuspended to a final optical density at 600 nm of 0.5 in the following buffer: 10 mm MES, pH 5.6, 10 mm MgCl2, and 100 μm acetosyringone (Sigma Aldrich). Leaves of 3-week-old N.b. plants were infiltrated with the bacterial culture using a 1-mL syringe as described previously (Golomb et al., 2008). Expression of the fluorescent chimera in the leaf cells was analyzed after 48 h.
N.t. leaf epidermal cells were infiltrated as outlined by Sparkes et al. (2006), with an optical density at 600 nm of 0.1 for myosin constructs and an optical density at 600 nm of 0.04 for Golgi marker (α-2,6-ST-cyan fluorescent protein [CFP]/GFP). Images were taken 48 to 72 h after infiltration.
Microscopy
For N.b., an Olympus IX81/FV500 laser-scanning microscope was used to observe fluorescently labeled cells with the following filter sets. To observe eGFP, we used the eGFP channel, 488-nm excitation, and BA505-525; to observe mRFP, we used 543-nm excitation and BA610. The objective used was PlanApo 60X1.00 WLSM ∞/0.17. When eGFP and mRFP were detected in the same sample, we used dichroic mirror 488/543. In all cases, where more than one color was monitored, sequential acquisition was performed.
For N.t., a Zeiss LSM META 510 confocal microscope was used with the following settings: dual imaging of eYFP and CFP were captured using multitracking in-line switching mode. CFP was excited with 458 nm and eYFP with 514 nm using a 458/514 dichroic mirror, and the subsequent emission was detected using 470- to 500-nm and 560- to 615-nm band-pass filters, respectively. Similarly, eGFP and mRFP dual imaging was captured using multitracking in-line switching mode. eGFP was excited with 488 nm and mRFP with 543 nm using the 488/543 dichroic mirror, and the subsequent emission was detected using 505- to 530-nm and 570- to 650-nm band-pass filters, respectively.
Organelle Tracking
For N.b., ST-mRFP was used as a Golgi marker (Saint-Jore-Dupas et al., 2006). The mitochondrial marker was made by Nelson et al. (2007) and obtained from The Arabidopsis Information Resource. To monitor both organelle and myosin motilities, time-lapse image acquisition every 2.6 s was performed exclusively from cells expressing both myosin and the organelle-specific marker. Control samples (with no myosin interference) were also captured at a high rate of image acquisition (every 0.4 s), and velocity measurements were compared with the low-frame-rate recordings. Organelle tracking was done using the Volocity 4.21 module of Improvision, and velocity was calculated from 100 to 580 organelles in the presence of each myosin. Size, shape, and intensity parameters were specified so that most organelles will be defined by the software. Tracks combining 12 to 20 sequential images were calculated by the shortest path model, and mean velocity or displacement rate were calculated for each track. Statistical tests were done using KaleidaGraph 4.03 (Synergy Software). General linear model analysis was carried out followed by Scheffe's multiple comparison test. This method was selected due to the uneven number of replicates between the treatments. Statistically significant differences between the treatments were determined using P < 0.05.
For N.t., Golgi bodies were tracked and analyzed according to Sparkes et al. (2008), except that 100 frames were captured over 14.29 s (7 frames s−1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Transcription profile of Arabidopsis myosin family members according to Genevestigator in arbitrary units.
Supplemental Figure S2. The subcellular localization of eGFP-IQ tail fusions of all Arabidopsis myosins in N.b. leaf epidermal cells.
Supplemental Figure S3. Comparative subcellular localization of mRFP-tail and eGFP-IQ tail myosin fusions in N.t.
Supplemental Figure S4. Subcellular localization of IQ tail inhibitory myosin truncations compared with Golgi and mitochondria localization.
Supplemental Table S1. Statistical analysis (Scheffe's test) of the data obtained from displacement rate analysis for Golgi bodies and mitochondria in the presence of all Arabidopsis myosins in the two plants, N.t. and N.b.
Supplemental Movie S1. Motility of Golgi bodies in the presence of Arabidopsis myosin tail fusions in N.t. (bar = 2 μm).
Supplemental Movie S2. Motility of Golgi bodies in the presence of Arabidopsis myosin IQ tail fusions in N.b. (bar = 10 μm).
Supplemental Movie S3. Motility of mitochondria in the presence of Arabidopsis myosin IQ tail fusions in N.b. (bar = 6 μm).
Supplemental Movie S4. Motionless Golgi bodies in the presence of moving Arabidopsis MYA1 and MYA2 eGFP-IQ tail particles in N.b. (bar = 8 μm).
Supplementary Material
Acknowledgments
We thank J.K. Jones and D. Volkmann for providing us with the ATM construct, Nicholas Teanby for help with the cumulative distribution frequency analysis, and Janet Evins for technical support at Oxford Brookes University.
The work was supported by the Israeli Science Foundation (grant no. 752/05), by BARD, the United States-Israel Binational Agricultural Research and Development Fund (grant no. IS–4038–07 to E.S.), and by the Leverhulme Trust (grant no. F/00 382/G to I.A.S.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Einat Sadot (vhesadot@agri.gov.il).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Avisar D, Prokhnevsky AI, Dolja VV (2008. a) Class VIII myosins are required for plasmodesmatal localization of a closterovirus Hsp70 homolog. J Virol 82 2836–2843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avisar D, Prokhnevsky AI, Makarova KS, Koonin EV, Dolja VV (2008. b) Myosin XI-K is required for rapid trafficking of Golgi stacks, peroxisomes, and mitochondria in leaf cells of Nicotiana benthamiana. Plant Physiol 146 1098–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baluska F, Cvrckova F, Kendrick-Jones J, Volkmann D (2001) Sink plasmodesmata as gateways for phloem unloading: myosin VIII and calreticulin as molecular determinants of sink strength? Plant Physiol 126 39–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baluska F, Samaj J, Hlavacka A, Kendrick-Jones J, Volkmann D (2004) Actin-dependent fluid-phase endocytosis in inner cortex cells of maize root apices. J Exp Bot 55 463–473 [DOI] [PubMed] [Google Scholar]
- Berg JS, Powell BC, Cheney RE (2001) A millennial myosin census. Mol Biol Cell 12 780–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boevink P, Oparka K, Santa Cruz S, Martin B, Betteridge A, Hawes C (1998) Stacks on tracks: the plant Golgi apparatus traffics on an actin/ER network. Plant J 15 441–447 [DOI] [PubMed] [Google Scholar]
- Collings DA, Harper JD, Vaughn KC (2003) The association of peroxisomes with the developing cell plate in dividing onion root cells depends on actin microfilaments and myosin. Planta 218 204–216 [DOI] [PubMed] [Google Scholar]
- Dechesne CA, Bouvagnet P, Walzthony D, Leger JJ (1987) Visualization of cardiac ventricular myosin heavy chain homodimers and heterodimers by monoclonal antibody epitope mapping. J Cell Biol 105 3031–3037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esseling-Ozdoba A, Houtman D, Van Lammeren AA, Eiser E, Emons AM (2008) Hydrodynamic flow in the cytoplasm of plant cells. J Microsc 231 274–283 [DOI] [PubMed] [Google Scholar]
- Foth BJ, Goedecke MC, Soldati D (2006) New insights into myosin evolution and classification. Proc Natl Acad Sci USA 103 3681–3686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golomb L, Abu-Abied M, Belausov E, Sadot E (2008) Different subcellular localizations and functions of Arabidopsis myosin VIII. BMC Plant Biol 8 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada S, Ishiyama K, Choi SB, Wang C, Singh S, Kawai N, Franceschi VR, Okita TW (2003) The transport of prolamine RNAs to prolamine protein bodies in living rice endosperm cells. Plant Cell 15 2253–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto K, Igarashi H, Mano S, Nishimura M, Shimmen T, Yokota E (2005) Peroxisomal localization of a myosin XI isoform in Arabidopsis thaliana. Plant Cell Physiol 46 782–789 [DOI] [PubMed] [Google Scholar]
- Hashimoto K, Igarashi H, Mano S, Takenaka C, Shiina T, Yamaguchi M, Demura T, Nishimura M, Shimmen T, Yokota E (2008) An isoform of Arabidopsis myosin XI interacts with small GTPases in its C-terminal tail region. J Exp Bot 59 3523–3531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawes C, Osterrieder A, Hummel E, Sparkes I (2008) The plant ER-Golgi interface. Traffic 9 1571–1580 [DOI] [PubMed] [Google Scholar]
- Jedd G, Chua NH (2002) Visualization of peroxisomes in living plant cells reveals acto-myosin-dependent cytoplasmic streaming and peroxisome budding. Plant Cell Physiol 43 384–392 [DOI] [PubMed] [Google Scholar]
- Kerwin B, Bandman E (1991) Assembly of avian skeletal muscle myosins: evidence that homodimers of the heavy chain subunit are the thermodynamically stable form. J Cell Biol 113 311–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinkema M, Schiefelbein J (1994) A myosin from a higher plant has structural similarities to class V myosins. J Mol Biol 239 591–597 [DOI] [PubMed] [Google Scholar]
- Knight AE, Kendrick-Jones J (1993) A myosin -like protein from a higher plant. J Mol Biol 231 148–154 [DOI] [PubMed] [Google Scholar]
- Li JF, Nebenführ A (2007) Organelle targeting of myosin XI is mediated by two globular tail subdomains with separate cargo binding sites. J Biol Chem 282 20593–20602 [DOI] [PubMed] [Google Scholar]
- Li JF, Nebenführ A (2008. a) Inter-dependence of dimerization and organelle binding in myosin XI. Plant J 55 478–490 [DOI] [PubMed] [Google Scholar]
- Li JF, Nebenführ A (2008. b) The tail that wags the dog: the globular tail domain defines the function of myosin V/XI. Traffic 9 290–298 [DOI] [PubMed] [Google Scholar]
- Liu L, Zhou J, Pesacreta TC (2001) Maize myosins: diversity, localization, and function. Cell Motil Cytoskeleton 48 130–148 [DOI] [PubMed] [Google Scholar]
- Molchan TM, Valster AH, Hepler PK (2002) Actomyosin promotes cell plate alignment and late lateral expansion in Tradescantia stamen hair cells. Planta 214 683–693 [DOI] [PubMed] [Google Scholar]
- Nebenführ A, Gallagher LA, Dunahay TG, Frohlick JA, Mazurkiewicz AM, Meehl JB, Staehelin LA (1999) Stop-and-go movements of plant Golgi stacks are mediated by the acto-myosin system. Plant Physiol 121 1127–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51 1126–1136 [DOI] [PubMed] [Google Scholar]
- Ojangu EL, Jarve K, Paves H, Truve E (2007) Arabidopsis thaliana myosin XIK is involved in root hair as well as trichome morphogenesis on stems and leaves. Protoplasma 230 193–202 [DOI] [PubMed] [Google Scholar]
- Peremyslov VV, Prokhnevsky AI, Avisar D, Dolja VV (2008) Two class XI myosins function in organelle trafficking and root hair development in Arabidopsis. Plant Physiol 146 1109–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokhnevsky AI, Peremyslov VV, Dolja VV (2008) Overlapping functions of the four class XI myosins in Arabidopsis growth, root hair elongation, and organelle motility. Proc Natl Acad Sci USA 105 19744–19749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichelt S, Knight AE, Hodge TP, Baluska F, Samaj J, Volkmann D, Kendrick-Jones J (1999) Characterization of the unconventional myosin VIII in plant cells and its localization at the post-cytokinetic cell wall. Plant J 19 555–567 [DOI] [PubMed] [Google Scholar]
- Reisen D, Hanson MR (2007) Association of six YFP-myosin XI-tail fusions with mobile plant cell organelles. BMC Plant Biol 7 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runions J, Brach T, Kuhner S, Hawes C (2006) Photoactivation of GFP reveals protein dynamics within the endoplasmic reticulum membrane. J Exp Bot 57 43–50 [DOI] [PubMed] [Google Scholar]
- Saint-Jore-Dupas C, Nebenführ A, Boulaflous A, Follet-Gueye ML, Plasson C, Hawes C, Driouich A, Faye L, Gomord V (2006) Plant N-glycan processing enzymes employ different targeting mechanisms for their spatial arrangement along the secretory pathway. Plant Cell 18 3182–3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaj J, Read ND, Volkmann D, Menzel D, Baluska F (2005) The endocytic network in plants. Trends Cell Biol 15 425–433 [DOI] [PubMed] [Google Scholar]
- Sattarzadeh A, Franzen R, Schmelzer E (2008) The Arabidopsis class VIII myosin ATM2 is involved in endocytosis. Cell Motil Cytoskeleton 65 457–468 [DOI] [PubMed] [Google Scholar]
- Shimmen T, Yokota E (1994) Physiological and biochemical aspects of cytoplasmic streaming. Int Rev Cytol 155 97–139 [Google Scholar]
- Shimmen T, Yokota E (2004) Cytoplasmic streaming in plants. Curr Opin Cell Biol 16 68–72 [DOI] [PubMed] [Google Scholar]
- Singh S, Bandman E (2006) Dimerization specificity of adult and neonatal chicken skeletal muscle myosin heavy chain rods. Biochemistry 45 4927–4935 [DOI] [PubMed] [Google Scholar]
- Sparkes IA, Hawes C, Baker A (2005) AtPEX2 and AtPEX10 are targeted to peroxisomes independently of known endoplasmic reticulum trafficking routes. Plant Physiol 139 690–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes IA, Ketelaar T, de Ruijter NCA, Hawes C (2009) Grab a Golgi: laser trapping of Golgi bodies reveals in vivo interactions with the endoplasmic reticulum. Traffic 10 567–571 [DOI] [PubMed] [Google Scholar]
- Sparkes IA, Runions J, Kearns A, Hawes C (2006) Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat Protocols 1 2019–2025 [DOI] [PubMed] [Google Scholar]
- Sparkes IA, Teanby NA, Hawes C (2008) Truncated myosin XI tail fusions inhibit peroxisome, Golgi, and mitochondrial movement in tobacco leaf epidermal cells: a genetic tool for the next generation. J Exp Bot 59 2499–2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga M, Kojima H, Yokota E, Orii H, Nakamori R, Katayama E, Anson M, Shimmen T, Oiwa K (2003) Higher plant myosin XI moves processively on actin with 35 nm steps at high velocity. EMBO J 22 1263–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme D, Bouget FY, Van Poucke K, Inze D, Geelen D (2004) Molecular dissection of plant cytokinesis and phragmoplast structure: a survey of GFP-tagged proteins. Plant J 40 386–398 [DOI] [PubMed] [Google Scholar]
- Voigt B, Timmers AC, Samaj J, Muller J, Baluska F, Menzel D (2005) GFP-FABD2 fusion construct allows in vivo visualization of the dynamic actin cytoskeleton in all cells of Arabidopsis seedlings. Eur J Cell Biol 84 595–608 [DOI] [PubMed] [Google Scholar]
- Volkmann D, Mori T, Tirlapur UK, Konig K, Fujiwara T, Kendrick-Jones J, Baluska F (2003) Unconventional myosins of the plant-specific class VIII: endocytosis, cytokinesis, plasmodesmata/pit-fields, and cell-to-cell coupling. Cell Biol Int 27 289–291 [DOI] [PubMed] [Google Scholar]
- Wang Z, Pesacreta TC (2004) A subclass of myosin XI is associated with mitochondria, plastids, and the molecular chaperone subunit TCP-1alpha in maize. Cell Motil Cytoskeleton 57 218–232 [DOI] [PubMed] [Google Scholar]
- Yokota E, McDonald AR, Liu B, Shimmen T, Palevitz BA (1995) Localization of a 170kDa myosin heavy chain in plant cells. Protoplasma 185 178–187 [Google Scholar]
- Yokota E, Ueda S, Tamura K, Orii H, Uchi S, Sonobe S, Hara-Nishimura I, Shimmen T (2008) An isoform of myosin XI is responsible for the translocation of endoplasmic reticulum in tobacco cultured BY-2 cells. J Exp Bot 60 197–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.