Abstract
Helicobacter pylori strains display remarkable genetic diversity, and the presence of strains bearing the toxigenic vacA s1 allele, a complete cag pathogenicity island (PAI), cagA alleles containing multiple EPIYA phosphorylation sites, and expressing the BabA adhesin correlates with development of gastroduodenal disease in adults. To better understand the genetic variability present among pediatric strains and its relationship to disease, we characterized H. pylori strains infecting 47 pediatric North American patients. Prevalence of mixed infection was assessed by random amplified polymorphic DNA analysis of multiple H. pylori clones from each patient. Microarray-based comparative genomic hybridization was used to examine the genomic content of the pediatric strains. The cagA and vacA alleles were further characterized by allele-specific PCR. A range of EPIYA motif configurations were observed for the cagA gene, which was present in strains from 22 patients (47%), but only 19 (41%) patients contained a complete cag PAI. Thirty patients (64%) were infected with a strain having the vacA s1 allele, and 28 patients (60%) had the babA gene. The presence of a functional cag PAI was correlated with ulcer disease (P = 0.0095). In spite of declining rates of H. pylori infection in North America, at least 11% of patients had mixed infection. Pediatric strains differ in their spectrum of strain-variable genes and percentage of absent genes in comparison to adult strains. Most children were infected with H. pylori strains lacking the cag PAI, but the presence of a complete cag PAI, in contrast to other virulence markers, was associated with more severe gastroduodenal disease.
It is estimated that >50% of the world's population is colonized with Helicobacter pylori in the stomach, making it one of the most common bacterial pathogens of humans. H. pylori infection is generally acquired in childhood (24, 33) and can persist for life. Gastritis (inflammation of the gastric mucosa) results in all who are colonized with H. pylori, but some hosts remain asymptomatic, while others develop peptic ulcers, gastric adenocarcinomas, and mucosa-associated lymphoid tissue lymphoma. Gastric cancer is the second leading cause of cancer death worldwide, and 63% of gastric cancer cases in 2002 were attributable to H. pylori infection (38, 49). While severe disease most often presents in adulthood, children display H. pylori-associated gastritis and the incidence of ulcer disease among infected children was 6.8% in a European pediatric population (31). Many studies have examined bacterial, host, and environmental risk factors associated with development of H. pylori-associated diseases in adults, but similar studies in children have been limited.
Genetic differences among H. pylori strains contribute to differences in disease outcome among infected individuals in adult populations. The gene encoding VacA, which induces vacuolation of host cells, is present in nearly all H. pylori strains, but a number of allele types have been defined. Strains having the type s1 vacA signal sequence and the m1 vacA middle region allele (vacA s1/m1) are associated with ulcer disease (9). The cag pathogenicity island (PAI) encodes a type IV secretion system (T4SS) (1, 15) that translocates the CagA protein effector, also encoded in the island, into host cells. Presence of the cag PAI is associated with increased inflammation, promoting host cell interleukin-8 (IL-8) production, and cagA-positive strains are associated with peptic ulcers (50) as well as gastric cancer (13). Inside the host cell, CagA protein becomes tyrosine phosphorylated at C-terminal EPIYA (Glu-Pro-Ile-Tyr-Ala) sites by src family kinases, deregulates SHP-2, and induces the hummingbird phenotype (26, 45). Strains having more C-type EPIYA motifs, the major phosphorylation site, induce stronger effects on host cells and are associated with gastric cancer (7, 12, 35). The presence of a functional allele of babA, a gene encoding an adhesin that mediates binding to Lewis B antigens expressed on gastric epithelial cells, is associated with duodenal ulcer and gastric adenocarcinoma (21).
While these H. pylori genes and alleles have been associated with disease outcome in adults, studies in children have provided mixed results. A recent study identified two genes (jhp0562, coding for a putative glycosyltransferase, and jhp0870, coding for an outer membrane protein) associated with peptic ulcer disease in children, but not adults, suggesting a different spectrum of genetic risk factors in adults and children (37). Studies using a whole-genome microarray-based approach have been done to investigate the variability in genomic content of H. pylori strains, but these studies have included mostly strains from adult patients (25, 29, 41, 42). Studies of the genetic variability of pediatric H. pylori strains have largely been limited to genes previously associated with virulence in adult populations. To better understand the genetic variability present among pediatric strains, we used whole-genome microarray-based comparative genomic hybridization to examine the genomic content of H. pylori strains isolated from symptomatic North American children and compared the pediatric isolate genetic variability to that observed in adult strains. We then examined the frequency of known virulence genes and virulence alleles among the pediatric H. pylori strains and the associations of strain genotype with the clinical and histological characteristics of the patients.
MATERIALS AND METHODS
Patient population and endoscopic evaluation.
Patients were selected randomly from subjects for whom a H. pylori culture was available for genotyping from three primary centers (Miami Children's Hospital, Miami, FL; Rainbow Babies & Children's Hospital, Cleveland, OH; and Children's Healthcare of Atlanta at Egleston, Atlanta, GA) and subjects referred to the primary centers from additional U.S. and Canadian Centers. Gastric biopsies were obtained during a diagnostic fiber-optic upper endoscopy performed at the discretion of the pediatric gastroenterologist because of the subjects' persistent gastrointestinal symptoms. The study cohort was accrued over a 3-year period, and patients were selected for analysis using a random numbering scheme. All patients were treated with eradication therapy. Disease diagnoses were defined as follows: normal gross appearance, erosions, ulcers, and nodularity. The study protocols and procedures for the protection of human subjects were approved by the Institutional Review Board of Emory University.
H. pylori isolation and histopathologic evaluation.
One biopsy from the antrum was collected and frozen for H. pylori isolation, and separate biopsies from the antrum and/or fundus were fixed in formalin and processed for histology at each clinical center. Histologic preparations and frozen biopsies were shipped to Emory University for histopathologic review and processing, respectively. For histopathologic evaluation, hematoxylin-and-eosin-stained slides were graded using the visual analog scale of the Sydney classification, which guided analysis of the density of H. pylori, and the amounts of neutrophils, mononuclear inflammatory cells, and intestinal metaplasia (19). Biopsy samples were cultured as described previously (22). The primary growth of each biopsy site was pooled and frozen in brain heart infusion broth containing 10% fetal bovine serum, 20% glycerol, and 0.2% β-cyclodextrin for storage at −80°C. For single-colony clone isolation, dilutions of the frozen primary growth were plated to obtain well-separated colonies. Individual colonies were then amplified by growth on horse blood medium (43) and frozen as individual clone stocks, and genomic DNA was prepared (Wizard Prep; Promega).
RAPD-PCR fingerprinting.
Random amplified polymorphic DNA (RAPD) fingerprinting was performed as previously described (2) with primers 1254 and 14216. PCR products were electrophoresed in 1% agarose gels for comparison of banding patterns.
aCGH.
Each strain was examined by a two-color array competitive genomic hybridization (aCGH) with a reference sample containing an equal molar mixture of the two sequenced strains 26695 and J99 used to design the probes on the microarray as described previously (41). Each isolate was analyzed on at least two microarrays. Data extraction and processing were performed as previously described (42), and data were simplified into a binary score for gene presence (score of 1) and absence (score of 0). Gene calls are given in Table S2 in the supplemental material.
PCR genotyping of vacA and cagA alleles.
vacA signal sequence and mid-region PCR typing was performed as described by Atherton et al. (9, 10). The numbers and types of EPIYA motifs present in cagA were determined as described previously (8).
Coculture experiments.
The human gastric adenocarcinoma cell line AGS (ATCC CRL-1739) was cocultured with H. pylori strains for analysis of IL-8 release and CagA translocation at 24 h as described previously (42).
Reference strains.
The control strains were G27 (cag PAI+ vacA s1a/m1) (18), Tx30a (ATCC 51932) (cag PAI− vacA s2/m2), 26695 (cagA ABC EPIYA) (48), and J99 (cagA BC EPIYA) (4).
Statistical analysis.
Tests of association between different clinical characteristics, patient demographics, and strain genotypes were performed using either the Fisher's exact test or the Spearman rank correlation, as appropriate. The genomic content of the pediatric strains, based on the aCGH, was compared to that of 71 H. pylori strains isolated from adult patients for which aCGH had been previously performed and data reported (25, 41). Differences in the mean percentages of genes absent between the pediatric strains and the adult strains were assessed by t test. Because the arrays include all the genes present in strains 26695 and J99, data from these 2 of the 15 strains reported by Salama et al. (41) were removed from the analysis of the difference in mean percentages of genes absent. The relative proportions of strain-variable genes in each functional class that were variable in only the adult strains were analyzed by chi-square or the Fisher's exact test, as appropriate. Because the comparison of the pediatric array data and the adult array data could be potentially biased by including globally representative adult strains and pediatric strains from only North American patients, the analyses were repeated, comparing the pediatric strains to only the hpEurope and/or hpAfrica1 strains included in the publication by Gressmann et al. (25). Because the arrays include genes present in an hpEurope strain (26695) and an hpAfrica1 strain (J99), other hpEurope and hpAfrica1 strains have significantly fewer genes absent based on array CGH compared to other populations of H. pylori (25). To avoid bias in the comparison, the test for difference in mean percentages of genes absent was repeated, restricting the adult strains to only hpEurope and hpAfrica1 strains. The analysis of disproportionate distribution of genes variable only in adult strains among different functional classes was repeated, restricting the adult strains to only hpEurope strains, the group to which most of the North American pediatric strains likely belong (20, 25). Statistical analyses were performed using SAS version 9.1 (SAS Institute, Cary, NC).
Microarray data accession number.
Raw microarray data from this study are available in GEO (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE13317.
RESULTS
A pediatric cohort of H. pylori-associated gastric symptoms.
The patients' demographic characteristics, endoscopic presentation, and histological scores are presented in Table 1. Histology slides of biopsy material from the antrum were obtained from 35 of the 47 patients and from the fundus from 22 patients. There were 15 patients with biopsies available for both sites and 5 patients with no histology samples. All but five children had neutrophils and mononuclear cells in at least one of the gastric sites. Of these five children, two showed no inflammation, and one of these two had marked complete intestinal metaplasia. The remaining three children had only mononuclear cells corresponding to chronic gastritis. Of the 15 patients with biopsies from the antrum and the fundus, 12 had different quantitation of parameters for the two sites. Six had discordant scores for H. pylori colonization density (two had higher scores for antrum, and four had higher scores for fundus), seven had discordant scores for neutrophil infiltration (five had higher scores for antrum, and two had higher scores for fundus), and eight had discordant scores for monocyte infiltration (six had higher scores for antrum, and two had higher scores for fundus). While level of H. pylori density was positively correlated with level of neutrophil infiltration in this population (P = 0.007; Spearman's rank correlation), the discordant scores for inflammation between the two biopsy sites were not related to discordant H. pylori density scores.
TABLE 1.
Patient characteristics
| Parameter | Characteristic | Resulta |
|---|---|---|
| Age (yr) | 12 (3-21)b | |
| Gender | Male | 27 (57) |
| Female | 16 (34) | |
| Unknown | 4 (9) | |
| Race | White | 24 (51) |
| Black | 15 (32) | |
| Other | 5 (11) | |
| Unknown | 3 (6) | |
| Endoscopic presentation | Normal | 22 (47) |
| Nodularity | 16 (34) | |
| Gastritis | 2 (4) | |
| Ulcer | 5 (11) | |
| Unknown | 2 (4) | |
| Antrum | ||
| H. pylori density | Normal | 2 (4) |
| Mild | 12 (26) | |
| Moderate | 10 (21) | |
| Marked | 11 (23) | |
| Unknown | 12 (26) | |
| Neutrophil infiltration | Normal | 3 (6) |
| Mild | 7 (15) | |
| Moderate | 13 (28) | |
| Marked | 12 (26) | |
| Unknown | 12 (26) | |
| Monocyte infiltration | Normal | 2 (4) |
| Mild | 3 (6) | |
| Moderate | 5 (11) | |
| Marked | 25 (53) | |
| Unknown | 12 (26) | |
| Fundus | ||
| H. pylori density | Normal | 4 (9) |
| Mild | 5 (11) | |
| Moderate | 7 (15) | |
| Marked | 6 (13) | |
| Unknown | 25 (53) | |
| Neutrophil infiltration | Normal | 6 (13) |
| Mild | 3 (6) | |
| Moderate | 8 (17) | |
| Marked | 5 (11) | |
| Unknown | 25 (53) | |
| Monocyte infiltration | Normal | 2 (4) |
| Mild | 5 (11) | |
| Moderate | 8 (17) | |
| Marked | 7 (15) | |
| Unknown | 25 (53) |
Except as noted for patient age, values represent the number (percentage) of patients with the result (n = 47).
Median and range.
RAPD typing shows evidence of mixed infection in a subset of patients.
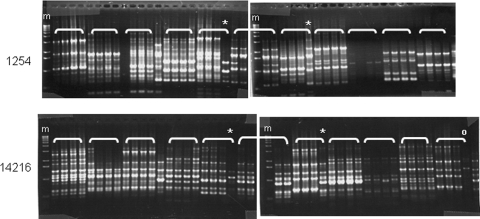
We performed RAPD typing of four single-colony isolates from each patient using two different primers to assess the relatedness of strains. As expected, clones from different patients yielded different RAPD patterns while clones from the same patient showed nearly identical DNA fragment patterns regardless of the primer used (Fig. 1). In four patients, we observed one clone with a distinct RAPD pattern using both primers, suggesting infection with two distinct strain types. Two additional patients displayed differences with a single primer. Heterogeneous colony morphology observed in some patients did not correlate with observed differences in RAPD patterns. Thus, possibly six patients positive for H. pylori had infection with multiple strain types.
FIG. 1.
Ethidium bromide-stained agarose gel showing RAPD profiles of four single-colony isolates per patient obtained with primers 1254 (top) and 14216 (bottom). Brackets identify clones from the same patient. m, marker; *, clone showing different RAPD patterns from the other three patient clones with both primers. °, clone showing a different RAPD pattern with a single primer (14216 in this example).
aCGH shows variability in genomic content.
To further analyze the genetic differences among H. pylori isolates within and between patients we performed microarray-based CGH (aCGH) on one representative isolate of each RAPD type present in each patient. For one patient with apparently distinct clones by RAPD analysis, the second clone type did not grow well enough to obtain sufficient DNA for aCGH. We analyzed an additional clone obtained from a patient (Em38) (see below). Thus, we analyzed a total of 53 isolates from 47 patients.
We obtained data for 1,675 genes, of which 1,324 were present in all strains and 351 were absent in at least one of the strains (see Table S2 in the supplemental material). None of the genes was absent in all strains. Of the 351 genes that were variably present, 334 genes were also variably present among the 71 adult strains for which the results of aCGH were previously reported (25, 41) and 17 genes were variably present among only the pediatric strains. However, only seven of these genes (HP0105, HP0498, HP0843-0845, HP0888, and HP1265) have not been previously identified as variably present among H. pylori strains isolated from other populations (25, 29, 41, 42). HP0888 was absent in two strains and the other six genes were absent in only one strain. Of the 1,324 genes that were present in all of the pediatric strains, 197 were variably present among the adult strains. Two functional classes were overrepresented among the genes that were variably present in only the adult strains: transport and binding proteins (P = 0.0006; chi-square) and cell envelope proteins (P < 0.0001; chi-square). These two functional classes were also overrepresented when the analysis was restricted to the 15 hpEurope adult strains (P = 0.003 for transport and binding proteins and P = 0.02 for cell envelope proteins).
The percentage of genes absent for each pediatric strain ranged from 2.3% to 10.6%. In comparison, the percentage of genes absent in a global collection of 69 adult strains ranged from 6.6% to 17.7%. The mean percentage of genes absent from the pediatric strains (6.4%; standard deviation, 2.3%) was significantly lower than that of the 69 adult strains (10.7%; standard deviation, 2.4%) and the subset of 23 hpEurope and hpAfrica1 adult strains (9.4%; standard deviation, 1.8%), which may more accurately reflect strain variation representative of North America (20, 25) (P < 0.0001 for both comparisons; t test).
aCGH reveals variation in the presence of virulence genes.
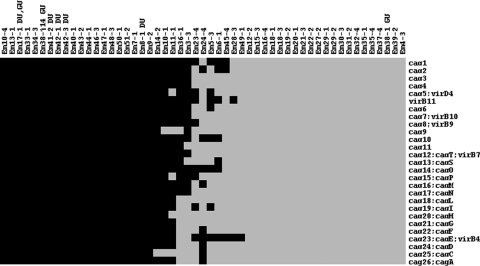
Among the variable genes were those whose presence has been associated with virulence, including the 27 cag PAI genes and the Lewis B antigen binding protein (babA) gene. Twenty-eight patients (60%) were infected with a strain having the babA gene. In addition, jhp0562, a gene associated with ulcer development in children but not adults (37), was variably present among the strains and 43 patients (81%) were infected with a strain having jhp0562. We were particularly interested in the cag PAI because its presence has been associated with severe disease in adults, but it was absent in the majority of strains in this population (Fig. 2). Interestingly, four of five patients with ulcer diagnosis contained a strain with a complete PAI versus 14 of 40 patients without ulcer. Since our initial RAPD typing analysis had revealed that infection with distinct strain types can occur in this patient population, we wondered whether the fifth ulcer patient (Em38) did indeed harbor a cag PAI-containing strain. To explore this possibility, we replated the initial frozen culture sweep, harvested 21 clones for analysis, and found 1 clone showing a distinct RAPD pattern (Em38-j14). Microarray analysis showed this clone contains all the genes in the cag PAI. Thus, in this cohort the cagA gene was present in 22 patients (47%), but only 19 (41%) bore a strain containing a complete PAI and therefore encoding a T4SS that could deliver CagA to host cells.
FIG. 2.
Presence (black) or absence (gray) of the 27 cag PAI genes in 53 H. pylori clones isolated from 47 pediatric patients. Each column represents a different clone and each row the indicated gene. Clones isolated from patients with duodenal (DU) and gastric (GU) ulcers are indicated.
aCGH establishes coinfection with genetically distinct strains.
Our RAPD analysis indicated seven patients might be infected with genetically distinct strains. We used the aCGH data to further explore the relatedness of clones from six of these patients (we were unable to obtain sufficient DNA from one clone). Pairwise comparison of gene content between clones showed that clones from the four patients with distinct RAPD patterns using both primers differed at 54 to 100 loci (Table 2). In contrast, the two patients with clones showing an altered RAPD pattern with only one primer differed at 13 and 15 loci (Table 2). Previous analyses of strains from children and adults revealed frequent infection with highly related clones that differ in 0 to 22 loci and rare cases in which much larger numbers of loci differ between clones that constitute distinct strains (42). Using these criteria, five of the seven patients with possible mixed infection had infection with distinct strain types, while two patients were infected with a single strain type. Thus at least 11% of patients had a mixed infection.
TABLE 2.
Genetic content of strains from patients with two clones analyzed
| Patient | Presence of RAPD differencesa | No. of loci differentb | babA status | cag PAI status |
|---|---|---|---|---|
| Em10 | Both | 92 | 10-1 yes, 10-4 no | 10-1 partial, 10-4 full |
| Em18 | Both | 64 | Both no | Both no |
| Em29 | Only 14216 | 13 | Both no | Both no |
| Em35 | Only 1254 | 15 | Both no | Both no |
| Em38 | Both | 100 | Both yes | 38-1 no, 38-j14 full |
| Em42 | Both | 54 | Both yes | Both yes |
The RAPD primers were 1254 and 14216.
The number of gene loci showing differential presence or absence between paired isolates as measured by aCGH.
For two patients, the genes that varied between the distinct infecting strain types may impart a different pathogenic potential (Table 2). In patient Em10, clone Em10-1 contained the babA gene but lacked cag9 and cag25, while clone Em10-4 lacked babA but contained a complete cag PAI. In patient, Em38 both clones contained babA, but clone Em38-1 lacked the entire cag PAI, while clone Em38-j14 had a complete cag PAI. While the cag PAI has been shown to be unstable during human infection (30), clones Em38-1 and Em38-j14 differed at an additional 73 gene loci, making it unlikely that Em38-1 was a cag PAI− derivative of Em38-j14. In the remaining two patients containing distinct strain types, both either lacked (patient Em18) or contained (patient Em42) the full cag PAI and babA.
Analysis of bacterial virulence gene alleles.
In addition to variation in presence and absence of virulence genes among strains, allelic variation of the secreted cytotoxins VacA and CagA correlate with disease severity in some populations. We used established PCR assays (8, 10) to analyze the VacA and CagA types present in this population. We determined the vacA allelic types at both the signal sequence region and the mid-region for all strains (see Table S1 in the supplemental material). Thirty patients (64%) were infected with a strain having the toxigenic vacA s1 allele, and 17 patients (36%) were exclusively infected with a strain having a vacA s2 allele. Of the 30 patients infected with a strain having a vacA s1 allele, 22 (73%) had the vacA s1/m1 allele, 5 (17%) had a vacA s1/m2 allele, 2 (7%) were positive for both the m1 and the m2 middle regions by PCR assay, and 1 (3%) had a band that was smaller than the m1 product by PCR assay.
For strains containing the cagA gene, we explored the number and type of CagA EPIYA phosphorylation site motifs present (Table 3). Among the 24 cagA-positive strains (from 22 patients), different CagA EPIYA motif configurations were observed. Seventeen (71%) had cagA alleles with the ABC EPIYA configuration, 1 (4%) had the BC EPIYA configuration, 2 (8%) had the BCC EPIYA configuration, and 1 (4%) had the ABCC EPIYA configuration. Three strains did not produce any product in the EPIYA PCR assay. However, one of the patients infected with a strain that did not produce any EPIYA PCR product had a mixed infection and the other strain had an ABC EPIYA motif configuration. Three of the patients infected with a strain having an ABC EPIYA motif configuration and one patient infected with a strain that did not produce any product in the EPIYA PCR assay did not have a full cag PAI.
TABLE 3.
cag PAI genetic and phenotypic virulence characteristics of patient strains
| Straina | Presence of gene(s) by arrayb
|
cagA EPIYAc | Presence of CagA proteind | Translocation of CagAe
|
IL-8 releasef | Ulcerg | ||
|---|---|---|---|---|---|---|---|---|
| cag PAI | cagA | CagA delivery | Hummingbird phenotype | |||||
| Em44-1 | Full | + | ABC | Yes | Yes | Yes | ++ | |
| Em10-4* | Full | + | ABC | Yes | No | No | − | |
| Em42-1* | Full | + | ABC | Yes | Yes | Yes | ++ | D |
| Em42-3* | Full | + | ABC | Yes | Yes | Yes | ++ | D |
| Em43-2 | Full | + | ABC | Yes | Yes | Yes | ++ | |
| Em38-j14* | Full | + | ABCC | No | No | Yes | − | G |
| Em8-1 | Full | + | BC | Yes | No | Yes | (+/−) | D |
| Em17-1 | Full | + | BCC | Yes | Yes | Yes | + | G, D |
| Em41-2 | Full | + | No bands | Yes | Yes | Yes | ++ | D |
| Em10-1* | Partial | + | No bands | No | No | No | − | |
| Em31-2 | None | − | No | No | No | − | ||
| Em38-1* | None | − | No | No | No | − | G | |
| Em15-3 | None | − | No | No | No | − | ||
Asterisks indicate patients with two distinct strains.
Gene presence assessed by aCGH.
Measured by PCR typing.
Assessed by immunoblotting.
Translocation of CagA was determined by immunoblot detection of tyrosine-phosphorylated CagA or the hummingbird phenotype.
IL-8 release was detected by ELISA during coculture of AGS cells with H. pylori strains.
D, duodenal; G, gastric.
Association of bacterial virulence genotypes with pathology.
Of the 40 patients who did not have an ulcer, 14 (35%) were infected with a strain that had a complete cag PAI. In contrast, all five of the patients that had an ulcer were infected with a strain having a complete cag PAI. Infection with at least one strain containing a complete cag PAI was significantly associated with development of ulcer disease (by Fisher's exact test, P = 0.0095 when Em38-j14 [ulcer, PAI+] is included and P = 0.14 when Em38-j14 is excluded). All 5 ulcer patients were also infected with a strain having the vacA s1 allele and the jhp0562 gene, compared to 16 (40%) and 35 (88%), respectively, of the 40 patients who did not have an ulcer. Four (80%) of the 5 ulcer patients were infected with a strain having a babA gene, compared to 24 (60%) of the 40 patients who did not have an ulcer. However, the presence of neither the vacA s1 allele, the jhp0562 gene, nor the babA gene was significantly associated with ulcer disease (by Fisher's exact test, P = 0.14 for vacA s1, P = 1 for jhp0562, and P = 0.64 for babA). Of the 19 patients who were infected with at least one strain having a complete cag PAI, 16 were also infected with at least one strain having a vacA s1 allele and a babA gene. Having an infection that was positive for a complete cag PAI, a vacA s1 allele, and a babA gene was associated with having an ulcer (P = 0.060; Fisher's exact test). No significant associations were found between being infected with a strain having a complete cag PAI, a vacA s1 allele, or a babA gene and H. pylori colonization density, neutrophil infiltration, or monocyte infiltration.
cag PAI is functional in strains from patients with and without ulcer.
We saw a strong association for infection with a strain containing a complete set of cag PAI genes and development of ulcer disease. This led us to explore whether the T4SS system was functional in the strains from ulcer patients as well as within a subset of strains, including two with a complete set of cag PAI genes, one with a partial set, and two having none of the cag PAI genes, from patients without ulcer. To measure T4SS function, we measured translocation of the CagA effector into host cells and induction of proinflammatory cytokine release during coculture of each clinical isolate with a human gastric epithelial cell line (AGS) (Table 3; and see Fig. S1 in the supplemental material). To measure CagA translocation into host cells, we employed two assays. We used light microscopy to observe CagA-dependent host cell morphology changes, termed the “hummingbird” phenotype (45). We also monitored CagA entry into host cells by its acquisition of phosphorylation on tyrosine residues in the EPIYA motifs. As shown in Fig. S1A in the supplemental material, most but not all strains tested (6/8) containing a full complement of cag PAI genes induced IL-8 production above background, although there was considerable variation in the levels of IL-8 secreted in response to different isolates. All strains that induced IL-8 secretion induced the hummingbird phenotype, while strains lacking some or all of the cag PAI genes did not (Table 3; and see Fig. S1B in the supplemental material).
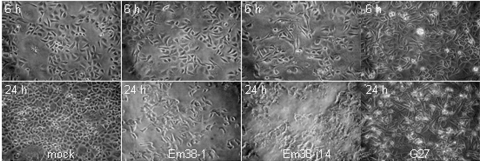
Interestingly, two strains (Em8-1 and Em38-j14) that induced little or no IL-8 secretion showed the hummingbird phenotype. Strain Em8-1 had detectable CagA protein, but no tyrosine-phosphorylated CagA could be detected even at 24 h (see Fig. S1C in the supplemental material). In the IL-8 assay, there was low, but detectable IL-8 secretion resulting in a +/− call (Table 3). These results suggest that the T4SS of this strain was in fact functional. Strain Em38-j14 showed no detectable IL-8 secretion in the coculture experiment, and neither CagA protein nor phospho-CagA could be detected with the antibodies used. Interestingly, light microscopy revealed a higher percentage of cells showing the hummingbird morphology at 6 h for strain Em38-j14 than for positive control strain G27 with a functional PAI (Fig. 3), suggesting an active cag PAI T4SS. At 24 h, when the G27 strain showed a high percentage of cells with the hummingbird morphology, cells incubated with Em38-j14 appeared to have lost membrane integrity and many detached from the substrate (Fig. 3). This cytotoxicity may account for the lack of detectable IL-8 production. In conclusion, the strains containing all cag PAI genes showed evidence by at least one of three assays that the T4SS and effectors were active in strains from 5/5 ulcer patients and 2/3 patients without ulcer. Conversely, 4/4 strains lacking all or part of the cag PAI showed no activity in any of the three assays.
FIG. 3.
Light micrograph of AGS cells cocultured with the indicated H. pylori strains for 6 h (top) or 24 h (bottom). Strain Em38-1 (PAI−) does not display the hummingbird phenotype at either time point, and strain Em38-j14 (PAI+) shows many elongated cells at 6 h and cytotoxicity at 24 h (compared to the mock and positive control strain G27 [PAI+]).
DISCUSSION
This study investigated the genetic variability of H. pylori strains isolated from North American children. By comparing the genetic variability of the pediatric strains to what has been reported for adult strains, we observed that pediatric strains differ in their spectrum of strain-variable genes and the percentage of absent genes. Variability in the presence of virulence genes and virulence alleles was also observed among the pediatric strains. A relatively low proportion of the strains contained a complete cag PAI, but the presence of a complete cag PAI was associated with ulcer.
H. pylori strains show considerable genetic diversity between individuals, and individuals can be coincidentally infected with genetically distinct strain types (47). In order to correlate disease outcome with bacterial genotypes, it is necessary to assess all genotypes present in the infected person. Using RAPD followed by aCGH to quantify the number of loci different among isolates, we found evidence that 11% of patients had infection with multiple genetically distinct strain types in a population with a low overall incidence of infection (14 to 15%) (Table 2). Previous work has suggested rare (0.8 to 7%) mixed infections among children mainly by observation of multiple vacA genotypes measured using PCR assays (17, 22, 39). Depending on the relative proportion of clones and the efficiency of PCR priming among different strains, the vacA PCR assay may underestimate the frequency of mixed infection. On the other hand, heterogeneity of vacA genotypes can be observed in an otherwise similar strain background (11), thus overestimating the frequency of mixed infection.
For one patient (Em38), analysis of 4 clones did not provide evidence of mixed infection, but screening of 25 clones revealed the presence of a strain with a distinct genotype from the rest of the population. Thus, our observation that 11% of patients harbor multiple strain types likely represents a minimal estimate of the true amount of mixed infection. In all cases of mixed infection, there appeared to be a dominant strain (75% or 96% of clones). While we were careful to allow sufficient time for growth of all clones during the initial plating and picked both large and small colony types when present, we cannot exclude the possibility that the observed strain prevalence was selected by in vitro growth.
We were motivated to search for an additional strain type in patient Em38 because of the gastric ulcer diagnosis and the absence of the cag PAI in the first strain isolated. Subsequent experiments confirmed the presence of an active cag PAI in the second strain type. If we hypothesize that this second strain was responsible for the pathological outcome, this suggests that a minority strain can still contribute to disease development. Alternatively, this clone's distribution may have been higher at the disease site (body) than the biopsy site (antrum) or selected against during growth in vitro.
The pediatric strains had a significantly lower percentage of genes absent compared to adult strains. It is possible that gene loss, including loss of genes that have a role in transmission and early infection, accumulates during the course of persistent infection. Because H. pylori infection is generally acquired in childhood, the pediatric strains may have a lower percentage of genes missing than adult strains because of their carriage for a shorter length of time from transmission. Strains that have a smaller amount of gene loss would be more likely to retain genes important for transmission and would be better able to infect a new host. Interestingly, while early molecular fingerprinting studies suggested that children acquire infection from their parents, and predominantly their mother, recent work has suggested that other household members contribute to transmission, including children (44). If recently transmitted strains in the pediatric population have a lower percentage of genes missing, they may represent a more robust source of new infections. Furthermore, examination of genes that are variably present in adult strains but always present in pediatric strains could help to identify genes that have a role in H. pylori transmission.
The genes that were lost in this pediatric population showed a different spectrum of functional annotation from that observed in adult isolate variable genes. Two functional classes, transport and binding proteins and cell envelope proteins, were overrepresented among the genes that were variably present in only the adult strains (and thus uniformly present in pediatric isolates). The cell envelope proteins that were variably present only in adult strains included many of the over 60 outer membrane proteins (OMPs) encoded in the H. pylori genome. Some OMPs have been shown to be adhesins (27, 32, 36). Because these proteins are expressed on the surface of the bacterial cell and interact with the host, having a larger repertoire of OMPs that can be expressed could be an advantage when H. pylori is adapting to a new host. Similarly, the H. pylori genome encodes many apparently redundant transport proteins, including multiple putative iron-siderophore uptake receptors (3). Whether these genes are truly redundant or transport a range of compounds found in different hosts remains to be determined. Certain omp or transporter genes that are important only during transmission and/or early infection or for which there is no receptor or substrate in a particular host may be more likely to be lost due to the absence of selection to maintain their presence during chronic infection. Interestingly, this gene loss is likely in just a subset of the strain population. Individual isolates from the same patient of the same strain differed in the carriage of 13 to 15 genes in this study (Table 2), and other studies using adults have observed up to 67 genes variably present within one person's strain population(28, 42). Possible mechanisms for this gene loss include the existence of direct sequence repeats in the genome that can mediate deletions (6) and observed gene conversion events between related outer membrane proteins (5, 46). The retention of a more complete spectrum of these cell envelope and transport and binding protein genes by pediatric strains suggests roles for these genes during transmission and/or early infection. Future studies that compare the genomic contents of pediatric strains and adult strains from the same population will help to further identify H. pylori genes involved in transmission. Additionally, pediatric strains may represent a better resource for studying the functional activity of these genes.
Using both microarray and PCR approaches, we were able to describe the frequency of adult bacterial virulence gene markers in our population. Interestingly, a complete and functional cag PAI showed the most significant association with ulcer disease development and addition of other adult bacterial virulence markers did not improve the association. Previous studies of pediatric populations using serology or PCR targeting CagA found an association in one case (40), but not others (17, 22), including North American children. Our phenotypic analysis showed that strains containing CagA, but lacking other genes of the PAI, do not induce proinflammatory and cell signaling phenotypes. On the other hand, most strains with a genetically complete PAI (8/9) displayed the expected cellular phenotypes and may better predict strain virulence. While one study suggested an association of CagA-positive strains with increased inflammation (34), we did not observe such an association. This may result from a lower control-to-case ratio when testing histologic parameters versus clinical diagnosis.
The prevalence of the cag PAI was lower (41%) than might be expected from the prevalence in North American adults (62%) predicted by CagA serology from a population-based survey(16). Other studies have observed a lower prevalence of CagA positivity in children compared to adults and an increase in CagA positivity with age (17, 40). Interestingly, in the two cases where mixed infection was observed with strains differing in their cag PAI status, the cag PAI+ strain appeared to be the minority strain. Since the immune responses of children to H. pylori infection appear to differ from those of adults(14, 23, 34), it is intriguing to speculate that a functional cag PAI may be detrimental to bacterial colonization or persistence in children while enhancing bacterial survival as the immune response matures. A better understanding of the interaction of cag PAI+ strains with the pediatric immune system may provide clues as to why a complete cag PAI uniquely predicted development of pediatric ulcer disease compared to other adult H. pylori virulence gene markers.
Supplementary Material
Acknowledgments
This work was supported by a grant from The Pew Charitable Trusts awarded to N.R.S. and by RO1 DK53708 from the NIH.
The contents of this study are solely the responsibility of the authors and do not necessarily represent the official views of the NIDDK.
No conflict of interest exists for the authors of this study.
Footnotes
Published ahead of print on 22 April 2009.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Akopyants, N. S., S. W. Clifton, D. Kersulyte, J. E. Crabtree, B. E. Youree, C. A. Reece, N. O. Bukanov, E. S. Drazek, B. A. Roe, and D. E. Berg. 1998. Analyses of the cag pathogenicity island of Helicobacter pylori. Mol. Microbiol. 2837-53. [DOI] [PubMed] [Google Scholar]
- 2.Akopyanz, N., N. O. Bukanov, T. U. Westblom, S. Kresovich, and D. E. Berg. 1992. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 205137-5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alm, R. A., J. Bina, B. M. Andrews, P. Doig, R. E. W. Hancock, and T. J. Trust. 2000. Comparative genomics of Helicobacter pylori: analysis of the outer membrane protein families. Infect. Immun. 684155-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397176-180. [DOI] [PubMed] [Google Scholar]
- 5.Amundsen, S. K., J. Fero, L. M. Hansen, G. A. Cromie, J. V. Solnick, G. R. Smith, and N. R. Salama. 2008. Helicobacter pylori AddAB helicase-nuclease and RecA promote recombination-related DNA repair and survival during stomach colonization. Mol. Microbiol. 69994-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aras, R. A., J. Kang, A. I. Tschumi, Y. Harasaki, and M. J. Blaser. 2003. Extensive repetitive DNA facilitates prokaryotic genome plasticity. Proc. Natl. Acad. Sci. USA 10013579-13584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Argent, R. H., M. Kidd, R. J. Owen, R. J. Thomas, M. C. Limb, and J. C. Atherton. 2004. Determinants and consequences of different levels of CagA phosphorylation for clinical isolates of Helicobacter pylori. Gastroenterology 127514-523. [DOI] [PubMed] [Google Scholar]
- 8.Argent, R. H., Y. Zhang, and J. C. Atherton. 2005. Simple method for determination of the number of Helicobacter pylori CagA variable-region EPIYA tyrosine phosphorylation motifs by PCR. J. Clin. Microbiol. 43791-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Atherton, J. C., P. Cao, R. M. Peek, Jr., M. K. Tummuru, M. J. Blaser, and T. L. Cover. 1995. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J. Biol. Chem. 27017771-17777. [DOI] [PubMed] [Google Scholar]
- 10.Atherton, J. C., T. L. Cover, R. J. Twells, M. R. Morales, C. J. Hawkey, and M. J. Blaser. 1999. Simple and accurate PCR-based system for typing vacuolating cytotoxin alleles of Helicobacter pylori. J. Clin. Microbiol. 372979-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aviles-Jimenez, F., D. P. Letley, G. Gonzalez-Valencia, N. Salama, J. Torres, and J. C. Atherton. 2004. Evolution of the Helicobacter pylori vacuolating cytotoxin in a human stomach. J. Bacteriol. 1865182-5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Basso, D., C. F. Zambon, D. P. Letley, A. Stranges, A. Marchet, J. L. Rhead, S. Schiavon, G. Guariso, M. Ceroti, D. Nitti, M. Rugge, M. Plebani, and J. C. Atherton. 2008. Clinical relevance of Helicobacter pylori cagA and vacA gene polymorphisms. Gastroenterology 13591-99. [DOI] [PubMed] [Google Scholar]
- 13.Blaser, M. J., G. I. Perez-Perez, H. Kleanthous, T. L. Cover, R. M. Peek, P. H. Chyou, G. N. Stemmermann, and A. Nomura. 1995. Infection with Helicobacter pylori strains possessing cagA is associated with an increased risk of developing adenocarcinoma of the stomach. Cancer Res. 552111-2115. [PubMed] [Google Scholar]
- 14.Camorlinga-Ponce, M., F. Aviles-Jimenez, L. Cabrera, R. Hernandez-Pando, O. Munoz, J. Soza, and J. Torres. 2003. Intensity of inflammation, density of colonization and interleukin-8 response in the gastric mucosa of children infected with Helicobacter pylori. Helicobacter 8554-560. [DOI] [PubMed] [Google Scholar]
- 15.Censini, S., C. Lange, Z. Xiang, J. E. Crabtree, P. Ghiara, M. Borodovsky, R. Rappuoli, and A. Covacci. 1996. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc. Natl. Acad. Sci. USA 9314648-14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho, I., M. J. Blaser, F. Francois, J. P. Mathew, X. Y. Ye, J. D. Goldberg, and E. J. Bini. 2005. Helicobacter pylori and overweight status in the United States: data from the Third National Health and Nutrition Examination Survey. Am. J. Epidemiol. 162579-584. [DOI] [PubMed] [Google Scholar]
- 17.Costa Lopes, A. I., A. Palha, L. Monteiro, M. Olcastro, A. Pelerito, and A. Fernandes. 2006. Helicobacter pylori genotypes in children from a population at high gastric cancer risk: no association with gastroduodenal histopathology. Am. J. Gastroenterol. 1012113-2122. [DOI] [PubMed] [Google Scholar]
- 18.Covacci, A., S. Censini, M. Bugnoli, R. Petracca, D. Burroni, G. Macchia, A. Massone, E. Papini, Z. Xiang, N. Figura, et al. 1993. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc. Natl. Acad. Sci. USA 905791-5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dixon, M. F., R. M. Genta, J. H. Yardley, and P. Correa. 1996. Classification and grading of gastritis. The updated Sydney System. International Workshop on the Histopathology of Gastritis, Houston 1994. Am. J. Surg. Pathol. 201161-1181. [DOI] [PubMed] [Google Scholar]
- 20.Falush, D., T. Wirth, B. Linz, J. K. Pritchard, M. Stephens, M. Kidd, M. J. Blaser, D. Y. Graham, S. Vacher, G. I. Perez-Perez, Y. Yamaoka, F. Megraud, K. Otto, U. Reichard, E. Katzowitsch, X. Wang, M. Achtman, and S. Suerbaum. 2003. Traces of human migrations in Helicobacter pylori populations. Science 2991582-1585. [DOI] [PubMed] [Google Scholar]
- 21.Gerhard, M., N. Lehn, N. Neumayer, T. Boren, R. Rad, W. Schepp, S. Miehlke, M. Classen, and C. Prinz. 1999. Clinical relevance of the Helicobacter pylori gene for blood-group antigen-binding adhesin. Proc. Natl. Acad. Sci. USA 9612778-12783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gold, B. D., L. J. van Doorn, J. Guarner, M. Owens, D. Pierce-Smith, Q. Song, L. Hutwagner, P. M. Sherman, O. L. de Mola, and S. J. Czinn. 2001. Genotypic, clinical, and demographic characteristics of children infected with Helicobacter pylori. J. Clin. Microbiol. 391348-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gottrand, F., F. Cullu, D. Turck, P. Vincent, L. Michaud, M. O. Husson, E. Martin-Delasalle, and J. P. Farriaux. 1997. Normal gastric histology in Helicobacter pylori-infected children. J. Pediatr. Gastroenterol. Nutr. 2574-78. [DOI] [PubMed] [Google Scholar]
- 24.Granstrom, M., Y. Tindberg, and M. Blennow. 1997. Seroepidemiology of Helicobacter pylori infection in a cohort of children monitored from 6 months to 11 years of age. J. Clin. Microbiol. 35468-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gressmann, H., B. Linz, R. Ghai, K. P. Pleissner, R. Schlapbach, Y. Yamaoka, C. Kraft, S. Suerbaum, T. F. Meyer, and M. Achtman. 2005. Gain and loss of multiple genes during the evolution of Helicobacter pylori. PLoS Genet. 1e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Higashi, H., R. Tsutsumi, A. Fujita, S. Yamazaki, M. Asaka, T. Azuma, and M. Hatakeyama. 2002. Biological activity of the Helicobacter pylori virulence factor CagA is determined by variation in the tyrosine phosphorylation sites. Proc. Natl. Acad. Sci. USA 9914428-14433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ilver, D., A. Arnqvist, J. Ogren, I. M. Frick, D. Kersulyte, E. T. Incecik, D. E. Berg, A. Covacci, L. Engstrand, and T. Borén. 1998. Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science 279373-377. [DOI] [PubMed] [Google Scholar]
- 28.Israel, D. A., N. Salama, U. Krishna, U. M. Rieger, J. C. Atherton, S. Falkow, and R. M. Peek, Jr. 2001. Helicobacter pylori genetic diversity within the gastric niche of a single human host. Proc. Natl. Acad. Sci. USA 9814625-14630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joyce, E. A., K. Chan, N. R. Salama, and S. Falkow. 2002. Redefining bacterial populations: a post-genomic reformation. Nat. Rev. Genet. 3462-473. [DOI] [PubMed] [Google Scholar]
- 30.Kersulyte, D., H. Chalkauskas, and D. E. Berg. 1999. Emergence of recombinant strains of Helicobacter pylori during human infection. Mol. Microbiol. 3131-43. [DOI] [PubMed] [Google Scholar]
- 31.Koletzko, S., F. Richy, P. Bontems, J. Crone, N. Kalach, M. L. Monteiro, F. Gottrand, D. Celinska-Cedro, E. Roma-Giannikou, G. Orderda, S. Kolacek, P. Urruzuno, M. J. Martinez-Gomez, T. Casswall, M. Ashorn, H. Bodanszky, and F. Megraud. 2006. Prospective multicentre study on antibiotic resistance of Helicobacter pylori strains obtained from children living in Europe. Gut 551711-1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mahdavi, J., B. Sonden, M. Hurtig, F. O. Olfat, L. Forsberg, N. Roche, J. Angstrom, T. Larsson, S. Teneberg, K. A. Karlsson, S. Altraja, T. Wadstrom, D. Kersulyte, D. E. Berg, A. Dubois, C. Petersson, K. E. Magnusson, T. Norberg, F. Lindh, B. B. Lundskog, A. Arnqvist, L. Hammarstrom, and T. Boren. 2002. Helicobacter pylori SabA adhesin in persistent infection and chronic inflammation. Science 297573-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitchell, H. M., Y. Y. Li, P. J. Hu, Q. Liu, M. Chen, G. G. Du, Z. J. Wang, A. Lee, and S. L. Hazell. 1992. Epidemiology of Helicobacter pylori in southern China: identification of early childhood as the critical period for acquisition. J. Infect. Dis. 166149-153. [DOI] [PubMed] [Google Scholar]
- 34.Munoz, L., M. Camorlinga, R. Hernandez, S. Giono, G. Ramon, O. Munoz, and J. Torres. 2007. Immune and proliferative cellular responses to Helicobacter pylori infection in the gastric mucosa of Mexican children. Helicobacter 12224-230. [DOI] [PubMed] [Google Scholar]
- 35.Naito, M., T. Yamazaki, R. Tsutsumi, H. Higashi, K. Onoe, S. Yamazaki, T. Azuma, and M. Hatakeyama. 2006. Influence of EPIYA-repeat polymorphism on the phosphorylation-dependent biological activity of Helicobacter pylori CagA. Gastroenterology 1301181-1190. [DOI] [PubMed] [Google Scholar]
- 36.Odenbreit, S., M. Till, D. Hofreuter, G. Faller, and R. Haas. 1999. Genetic and functional characterization of the alpAB gene locus essential for the adhesion of Helicobacter pylori to human gastric tissue. Mol. Microbiol. 311537-1548. [DOI] [PubMed] [Google Scholar]
- 37.Oleastro, M., L. Monteiro, P. Lehours, F. Megraud, and A. Menard. 2006. Identification of markers for Helicobacter pylori strains isolated from children with peptic ulcer disease by suppressive subtractive hybridization. Infect. Immun. 744064-4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parkin, D. M. 2006. The global health burden of infection-associated cancers in the year 2002. Int. J. Cancer 1183030-3044. [DOI] [PubMed] [Google Scholar]
- 39.Podzorski, R. P., D. S. Podzorski, A. Wuerth, and V. Tolia. 2003. Analysis of the vacA, cagA, cagE, iceA, and babA2 genes in Helicobacter pylori from sixty-one pediatric patients from the Midwestern United States. Diagn. Microbiol. Infect. Dis. 4683-88. [DOI] [PubMed] [Google Scholar]
- 40.Queiroz, D. M., E. N. Mendes, A. S. Carvalho, G. A. Rocha, A. M. Oliveira, T. F. Soares, A. Santos, M. M. Cabral, and A. M. Nogueira. 2000. Factors associated with Helicobacter pylori infection by a cagA-positive strain in children. J. Infect. Dis. 181626-630. [DOI] [PubMed] [Google Scholar]
- 41.Salama, N., K. Guillemin, T. K. McDaniel, G. Sherlock, L. Tompkins, and S. Falkow. 2000. A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc. Natl. Acad. Sci. USA 9714668-14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salama, N. R., G. Gonzalez-Valencia, B. Deatherage, F. Aviles-Jimenez, J. C. Atherton, D. Y. Graham, and J. Torres. 2007. Genetic analysis of Helicobacter pylori strain populations colonizing the stomach at different times postinfection. J. Bacteriol. 1893834-3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salama, N. R., G. Otto, L. Tompkins, and S. Falkow. 2001. Vacuolating cytotoxin of Helicobacter pylori plays a role during colonization in a mouse model of infection. Infect. Immun. 69730-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwarz, S., G. Morelli, B. Kusecek, A. Manica, F. Balloux, R. J. Owen, D. Y. Graham, S. van der Merwe, M. Achtman, and S. Suerbaum. 2008. Horizontal versus familial transmission of Helicobacter pylori. PLoS Pathog 4e1000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Segal, E. D., J. Cha, J. Lo, S. Falkow, and L. S. Tompkins. 1999. Altered states: involvement of phosphorylated CagA in the induction of host cellular growth changes by Helicobacter pylori. Proc. Natl. Acad. Sci. USA 9614559-14564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Solnick, J. V., L. M. Hansen, N. R. Salama, J. K. Boonjakuakul, and M. Syvanen. 2004. Modification of Helicobacter pylori outer membrane protein expression during experimental infection of rhesus macaques. Proc. Natl. Acad. Sci. USA 1012106-2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suerbaum, S., and C. Josenhans. 2007. Helicobacter pylori evolution and phenotypic diversification in a changing host. Nat. Rev. Microbiol. 5441-452. [DOI] [PubMed] [Google Scholar]
- 48.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, J. C. Venter, et al. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388539-547. [DOI] [PubMed] [Google Scholar]
- 49.WHO. 2006. Fact sheet no. 297. Cancer. World Health Organization, Geneva, Switzerland.
- 50.Xiang, Z., S. Censini, P. F. Bayeli, J. L. Telford, N. Figura, R. Rappuoli, and A. Covacci. 1995. Analysis of expression of CagA and VacA virulence factors in 43 strains of Helicobacter pylori reveals that clinical isolates can be divided into two major types and that CagA is not necessary for expression of the vacuolating cytotoxin. Infect. Immun. 6394-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.