Abstract
The Jun-Fos heterodimeric transcription factor is the terminal link between the transfer of extracellular information in the form of growth factors and cytokines to the site of DNA transcription within the nucleus in a wide variety of cellular processes central to health and disease. Here, using isothermal titration calorimetry, we report detailed thermodynamics of the binding of bZIP domains of Jun-Fos heterodimer to synthetic dsDNA oligos containing the TGACTCA cis-element and all possible single nucleotide variants thereof encountered widely within the promoters of a diverse array of genes. Our data show that Jun-Fos heterodimer tolerates single nucleotide substitutions and binds to TGACTCA variants with affinities in the physiologically relevant micromolar-submicromolar range. The energetics of binding are richly favored by enthalpic forces and opposed by entropic changes across the entire spectrum of TGACTCA variants in agreement with the notion that protein-DNA interactions are largely driven by electrostatic interactions and intermolecular hydrogen bonding. Of particular interest is the observation that the Jun-Fos heterodimer binds to specific TGACTCA variants in a preferred orientation. Our 3D atomic models reveal that such orientational preference results from asymmetric binding and may in part be attributable to chemically distinct but structurally equivalent residues R263 and K148 located within the basic regions of Jun and Fos, respectively. Taken together, our data suggest that the single nucleotide variants of the TGACTCA motif modulate energetics and orientation of binding of the Jun-Fos heterodimer and that such behavior may be a critical determinant of differential regulation of specific genes under the control of this transcription factor. Our study also bears important consequences for the occurrence of single nucleotide polymorphisms within the TGACTCA cis-element at specific gene promoters between different individuals.
Keywords: AP1-DNA thermodynamics, Jun-Fos heterodimer, bZIP domain, Single nucleotide variants
The transcription factor AP1 (activator protein 1), comprised largely of constituent proteins Jun and Fos, executes the terminal stage of many critical signaling cascades that initiate at the cell surface and reach their climax in the nucleus (1, 2). Upon activation by MAP kinases, AP1 binds to the TGACTCA consensus motif and many closely related sequences within the promoters of a multitude of genes as Jun-Jun homodimer or Jun-Fos heterodimer. In so doing, Jun and Fos recruit the transcriptional machinery to the site of DNA and switch on expression of genes involved in a diverse array of cellular processes such as cell growth and proliferation, cell cycle regulation, embryonic development and cancer (3-5).
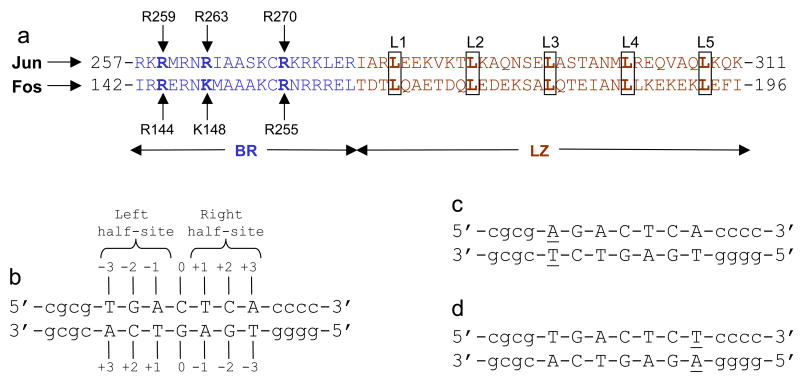
Jun and Fos recognize the TGACTCA and related sequences at the promoters of specific genes through their so-called basic zipper (bZIP) domains (Figure 1a). The bZIP domain can be further dissected into two well-defined functional subdomains termed the basic region (BR) at the N-terminus followed by the leucine zipper (LZ) at the C-terminus. The leucine zipper is a highly conserved protein module found in a wide variety of cellular proteins and usually contains a signature leucine at every seventh position within the five successive heptads of amino acid residues. The leucine zippers adapt continuous α-helices in the context of Jun-Jun homodimer or Jun-Fos heterodimer by virtue of their ability to wrap around each other in a coiled coil dimer (6-8). Such intermolecular arrangement brings the basic regions at the N-termini of bZIP domains into close proximity and thereby enables them to insert into the major grooves of DNA at the promoter regions in an optimal fashion in a manner akin to a pair of forceps (8). The basic regions are believed to be unstructured and fold into α-helices only upon association with DNA in a coupled folding-binding manner (9-13). While the α-helices are held together by numerous inter-helical hydrophobic contacts and salt bridges, hydrogen bonding between the sidechains of basic residues in the basic regions and the DNA bases accounts for high affinity binding of bZIP domains to DNA.
Figure 1.
Protein and DNA sequences. (a) Subdivision of bZIP domain into its respective N-terminal basic region (BR) and the C-terminal leucine zipper (LZ) for Jun and Fos transcription factors. The BR and LZ subdomains are colored blue and brown, respectively. The five signature leucines (L1-L5) characteristic of LZ subdomains are boxed and bold faced. The basic residues within the BR subdomains that hydrogen bond with specific DNA bases are labeled by vertical arrows. (b) Nucleotide sequence of 15-mer dsDNA oligo containing the TGACTCA motif. The TGACTCA motif is capitalized whilst the flanking nucleotides are shown in small letters. The numbering of various nucleotides relative to the central C/G base pair (assumed to be at zero position) in both strands is indicated. The TGA and TCA half-sites within this motif are also marked. (c) Nucleotide sequence of dsDNA oligo containing the T→A mutation at −3 position and herein referred to as the A-3 oligo. The variant nucleotides in both strands are underlined. (d) Nucleotide sequence of dsDNA oligo containing the A→T mutation at +3 position and herein referred to as the T+3 oligo. The variant nucleotides in both strands are underlined. Note that the A-3 and T+3 oligos shown in (c) and (d) are examples of a pair of symmetrically related dsDNA oligos in that they contain identical half-sites in opposite directions— these half-sites are indistinguishable upon the rotation of the variant motif by 180° in the plane of the paper (two-fold symmetry).
X-ray crystal structure analysis shows that the bZIP domains of Jun-Fos heterodimer bind to the TGACTCA sequence in a non-preferred orientation (8), implying that this promoter element does not dictate the orientation of the Jun-Fos heterodimer. Given that the AP1 transcription factor cooperates and acts in concert with a diverse array of other transcription factors, including many steroid hormone receptors, within the transcription initiation complex in regulating gene expression, the orientational binding of Jun-Fos heterodimer to gene promoters could be a key determinant of its transcriptional potency. But how would this be achieved? Since the discovery of TGACTCA motif as an essential cis-element for recognizing the AP1 transcription factor within the promoter of metallothionein 2A gene over two decades ago (14), there has been a growing number of studies demonstrating the key role of single nucleotide variants (SNVs) of this element within a diverse spectrum of genes for recruiting AP1 to the site of transcriptional machinery (15-27). This exciting episode of spicing up the TGACTCA motif with genetic variation across a diverse array of gene promoters took yet another twist recently upon the demonstration that this element may not only be subject to SNVs but that single nucleotide polymorphisms (SNPs) may also feature heavily within this element across different individuals, especially those from distinct ethnic groups (28). Thus, while SNVs may allow differential regulation of specific AP1-responsive genes, SNPs may determine the phenotypic makeup of an individual by virtue of their ability to differentially modulate the binding of transcription factors to identical promoter regions between different individuals and thus could account for differential response of individuals to specific diseases. A SNP within the promoter of MDM2 ubiquitin ligase gene, a negative regulator of p53 tumor suppressor, has indeed been implicated in cancer (29).
Although the binding of AP1 to promoter elements containing the TGACTCA sequence has been extensively explored in biophysical terms over the past two decades or so, the effect of genetic variations within this sequence upon protein-DNA interaction remains hitherto poorly understood. In an attempt to analyze the effect of such genetic variations within the TGACTCA sequence on AP1-DNA interactions and whether such variations could dictate their relative orientation, we have employed here isothermal titration calorimetry (ITC) to study detailed thermodynamics of the binding of bZIP domains of Jun-Fos heterodimer to synthetic dsDNA oligos containing the TGACTCA cis-element and all possible SNVs thereof encountered widely within the promoters of a diverse array of genes. Our data suggest that TGACTCA variants modulate energetics and orientation of binding of the Jun-Fos heterodimer and that such behavior may be a critical determinant of differential regulation of specific genes under the control of this transcription factor. Our study also bears important consequences for the occurrence of SNPs within the TGACTCA cis-element at the promoters of identical genes in different individuals.
Materials and Methods
Protein preparation
bZIP domains of human Jun and Fos were cloned and expressed as described previously (30). Briefly, the proteins were cloned into pET102 bacterial expression vector, with an N-terminal thioredoxin (Trx)-tag and a C-terminal polyhistidine (His)-tag, using Invitrogen TOPO technology. Additionally, thrombin protease sites were introduced at both the N- and C-termini of the proteins to aid in the removal of tags after purification. Proteins were subsequently expressed in Escherichia coli Rosetta2(DE3) bacterial strain (Novagen) and purified on Ni-NTA affinity column using standard procedures. Further treatment of bZIP domains of Jun and Fos on MonoQ ion-exchange column coupled to GE Akta FPLC system led to purification of recombinant domains to apparent homogeneity as judged by SDS-PAGE analysis. The identity of recombinant proteins was confirmed by MALDI-TOF mass spectrometry analysis. Final yields were typically between 10-20mg protein of apparent homogeneity per liter of bacterial culture. As noted previously (30), the treatment of recombinant proteins with thrombin protease significantly destabilized the bZIP domains of both Jun and Fos and both domains appeared to be proteolytically unstable. For this reason, all experiments reported herein were carried out on recombinant fusion bZIP domains of Jun and Fos containing a Trx-tag at the N-terminus and a His-tag at the C-terminus. The tags were found to have no effect on the binding of these domains to DNA under all conditions used here. Protein concentrations were determined as described earlier (30). Jun-Fos bZIP heterodimers were generated by mixing equimolar amounts of the purified bZIP domains of Jun and Fos. The efficiency of bZIP heterodimerization was close to 100% as judged by Native-PAGE and size exclusion chromatography (SEC) analysis using a Hiload Superdex 200 column.
DNA synthesis
HPLC-grade 15-mer DNA oligos containing the TGACTCA consensus motif and all possible single nucleotide variants thereof were commercially obtained from Sigma Genosys. The flanking nucleotides were appropriately chosen to prevent self-annealing of sense and antisense strands. The design of such oligos and the numbering of various nucleotides relative to the central C/G base pair are depicted in Figures 1b-d. Oligo concentrations were determined spectrophotometrically on the basis of their extinction co-efficients derived from their nucleotide sequences using the online software OligoAnalyzer 3.0 (Integrated DNA Technologies) based on the nearest-neighbor model (31). Double-stranded DNA (dsDNA) oligos were generated as described earlier (30).
ITC measurements
Isothermal titration calorimetry (ITC) experiments were performed on Microcal VP-ITC instrument and data were acquired and processed using Microcal ORIGIN software. All measurements were repeated 2-3 times. Briefly, the bZIP domains of Jun-Fos heterodimer and dsDNA oligos were prepared in 50mM Tris, 200mM NaCl, 1mM EDTA and 5mM β-mercaptoethanol at pH 8.0. The experiments were initiated by injecting 20 × 10μl aliquots of 50-100μM of dsDNA oligo from the syringe into the calorimetric cell containing 1.8ml of 5-10μM of the bZIP domains of Jun-Fos heterodimer at 25°C. The data were fit to a 1-site model derived from the binding of a ligand to a macromolecule using the law of mass action to extract the various thermodynamic parameters as described previously (30). Although Tris buffer is not ideally suited for ITC analysis due to its high ionization enthalpy, enthalpy of binding of Jun-Fos heterodimer to dsDNA oligos was identical in both Tris and phosphate buffers, implying that the values of enthalpy being reported here solely arise from the binding process with no contributions from coupled equilibria due to protonation/deprotonation.
Structural modeling
3D structures of bZIP domains of Jun-Fos heterodimer in complex with dsDNA oligos containing the TGACGCA variant motif in two possible orientations I and II were modeled using the crystal structure of bZIP domains of Jun-Fos heterodimer in complex with a dsDNA oligo containing the TGACTCA sequence as a template (with PDB code of 1FOS) in MODELLER (32). Additionally, hydrogen bonding restraints were introduced. For orientation I, hydrogen bonds were added between the NH1 atom of R263 in Jun and O6 atom of G+1 in the sense strand of TGACGCA motif, between the NH2 atom of R263 in Jun and O6 atom of G+1 in the sense strand of TGACGCA motif, and between the NZ atom of K148 in Fos and O4 atom of T+1 in the antisense strand of TGACGCA motif. For orientation II, hydrogen bonds were added between the NH1 atom of R263 in Jun and O4 atom of T+1 in the antisense strand of TGACGCA motif, between the NH2 atom of R263 in Jun and O4 atom of T+1 in the antisense strand of TGACGCA motif, and between the NZ atom of K148 in Fos and O6 atom of G+1 in the sense strand of TGACGCA motif. In each case, a total of 100 structural models were calculated and the structure with the lowest energy, as judged by the MODELLER Objective Function, was selected for further energy minimization in MODELLER prior to analysis. The structures were rendered using RIBBONS (33).
Results and Discussion
Jun-Fos heterodimer tolerates single nucleotide substitutions at all positions within the TGACTCA motif
A previous study based on an analysis of highly qualitative nature demonstrated that single nucleotide substitutions at specific positions within the TGACTCA motif completely abrogated binding by the bZIP domains of Jun-Fos heterodimer (34). This conclusion is highly surprising given that all possible single nucleotide variants of the TGACTCA motif are encountered within the promoter regions of a diverse array of genes whereby they act in concert with other promoter elements to regulate the action of AP1 transcription factor in a differential manner (15-27). In light of this argument coupled with the fact that the overall binding energy results from the summation of all interactions between protein residues and DNA bases in a cooperative manner, we reasoned that single substitutions within the TGACTCA motif may result in the reduction of free energy associated with such protein-DNA interactions but are unlikely to lead to complete abrogation within the physiological context. To test our hypothesis, we introduced all possible single substitutions within the TGACTCA motif and measured their effect on the binding of bZIP domains of Jun-Fos heterodimer using the powerful and highly quantitative technique of ITC. Figure 2 shows representative isotherms obtained from such experiments, while detailed analysis of all the associated thermodynamic parameters is presented in Table 1. Our data indeed support our hypothesis that no single nucleotide substitution at any position within the TGACTCA motif is sufficient per se to completely abolish the binding of Jun-Fos heterodimer. The binding affinities observed are in the physiologically relevant range and vary by up to 60-fold from a value of 0.14μM to 8.10μM, implying that genetic variations within the TGACTCA motif at distinct gene promoters may be critical determinants of tightly modulating the transcriptional potency of the Jun-Fos heterodimer.
Figure 2.
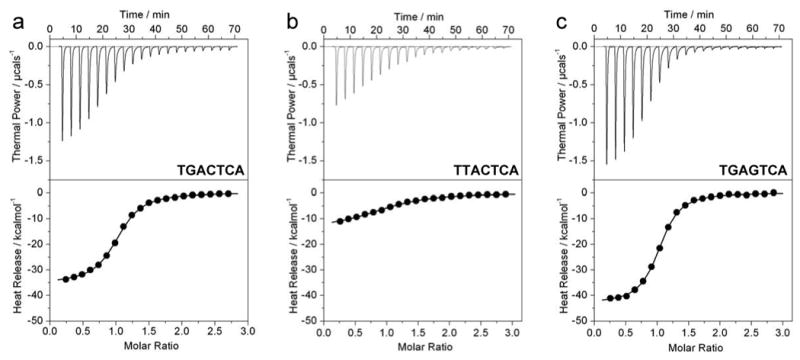
Representative ITC isotherms for the binding of bZIP domains of Jun-Fos heterodimer to dsDNA oligos containing the promoter sites TGACTCA (a), TTACTCA (b) and TGAGTCA (c). Note that the variant motif TGAGTCA is related to the wildtype TGACTCA motif by 2-fold symmetry. The position of the variant nucleotide in each of the sites relative to the consensus sequence TGACTCA is underlined. The solid lines represent the fit of the data points in the lower panels to a function based on the binding of a ligand to a macromolecule using the Microcal ORIGIN software (41).
Table 1.
Experimentally determined thermodynamic parameters for the binding of bZIP domains of Jun-Fos heterodimer to dsDNA oligos containing the wild-type (WT) consensus motif TGACTCA and all possible single nucleotide variants thereof obtained from ITC measurements at 25°C and pH 8.0
| Motif | Sequence | n | Kd/μM | ΔH/kcalmol-1 | TΔS/kcalmol-1 | ΔG/kcalmol-1 | Gene Promoter |
|---|---|---|---|---|---|---|---|
| WT | TGACTCA | 1.02 ± 0.01 | 0.21 ± 0.01 | -35.08 ± 0.22 | -25.94 ± 0.19 | -9.12 ± 0.03 | Metallothionein 2A |
| A-3 | AGACTCA | 0.98 ± 0.09 | 3.55 ± 0.76 | -31.02 ± 0.59 | -23.56 ± 0.48 | -7.45 ± 0.13 | Aflatoxin B1 aldehyde reductase AKR7 |
| C-3 | CGACTCA | 0.97 ± 0.04 | 5.96 ± 2.28 | -19.70 ± 0.56 | -12.53 ± 0.32 | -7.16 ± 0.23 | Transcriptional activator ORF50 |
| G-3 | GGACTCA | 1.03 ± 0.06 | 2.82 ± 0.17 | -31.36 ± 0.36 | -23.80 ± 0.36 | -7.58 ± 0.04 | Mu-opioid receptor MOR1 |
| A-2 | TAACTCA | 0.98 ± 0.01 | 6.31 ± 0.08 | -26.48 ± 0.28 | -19.37 ± 0.30 | -7.10 ± 0.01 | Cytochrome P450 enzyme CYP27B1 |
| C-2 | TCACTCA | 1.05 ± 0.04 | 8.10 ± 1.06 | -30.74 ± 0.20 | -23.77 ± 0.27 | -6.95 ± 0.08 | Monocyte Chemotactic Protein MCP1 |
| T-2 | TTACTCA | 1.00 ± 0.02 | 2.08 ± 0.05 | -13.98 ± 0.14 | -6.23 ± 0.17 | -7.76 ± 0.01 | Transcriptional regulator JE/MCAF |
| C-1 | TGCCTCA | 0.92 ± 0.03 | 4.13 ± 0.59 | -29.89 ± 0.21 | -22.53 ± 0.30 | -7.35 ± 0.08 | Pro-opiomelanocortin hormone POMC |
| G-1 | TGGCTCA | 1.06 ± 0.01 | 3.90 ± 0.50 | -33.61 ± 0.13 | -26.21 ± 0.06 | -7.39 ± 0.08 | Bone protein osteocalcin |
| T-1 | TGTCTCA | 0.98 ± 0.01 | 3.62 ± 0.30 | -36.89 ± 0.06 | -29.46 ± 0.11 | -7.43 ± 0.05 | Transforming growth factor TGFB1 |
| A0 | TGAATCA | 0.95 ± 0.04 | 0.58 ± 0.04 | -39.88 ± 0.10 | -31.36 ± 0.06 | -8.52 ± 0.04 | Mitochondrial proton carrier UCP1 |
| G0 | TGAGTCA | 1.05 ± 0.04 | 0.14 ± 0.01 | -42.87 ± 0.16 | -33.52 ± 0.21 | -9.34 ± 0.03 | Cell cycle regulator cyclin D1 |
| T0 | TGATTCA | 1.04 ± 0.04 | 0.50 ± 0.02 | -38.85 ± 0.21 | -30.25 ± 0.21 | -8.60 ± 0.02 | Tyrosine hydroxylase TH |
| A+1 | TGACACA | 1.00 ± 0.03 | 0.92 ± 0.03 | -37.06 ± 0.14 | -28.82 ± 0.17 | -8.24 ± 0.02 | Glucocorticoid receptor GR |
| C+1 | TGACCCA | 0.95 ± 0.06 | 4.45 ± 0.40 | -31.95 ± 0.09 | -24.63 ± 0.02 | -7.31 ± 0.05 | Interferon IFN-γ |
| G+1 | TGACGCA | 0.95 ± 0.01 | 0.64 ± 0.01 | -32.78 ± 0.13 | -24.33 ± 0.15 | -8.46 ± 0.01 | ECM glycoprotein fibronectin 1 |
| A+2 | TGACTAA | 1.04 ± 0.04 | 2.00 ± 0.05 | -20.90 ± 0.09 | -13.11 ± 0.13 | -7.78 ± 0.01 | MAP kinase phosphatase MKP3 |
| G+2 | TGACTGA | 0.92 ± 0.03 | 5.92 ± 1.05 | -28.16 ± 0.19 | -21.01 ± 0.08 | -7.14 ± 0.11 | Steroidogenic acute regulator STAR |
| T+2 | TGACTTA | 1.07 ± 0.03 | 5.16 ± 0.15 | -27.88 ± 0.11 | -20.65 ± 0.08 | -7.22 ± 0.02 | Nerve growth Factor NGF |
| C+3 | TGACTCC | 1.04 ± 0.02 | 3.07 ± 0.29 | -31.90 ± 0.11 | -24.35 ± 0.04 | -7.53 ± 0.06 | Chloride channel CLC5 |
| G+3 | TGACTCG | 0.97 ± 0.02 | 1.80 ± 0.17 | -33.33 ± 0.06 | -25.46 ± 0.02 | -7.85 ± 0.06 | Amyloid precursor protein β-APP |
| T+3 | TGACTCT | 1.02 ± 0.03 | 3.30 ± 0.15 | -42.46 ± 0.13 | -34.97 ± 0.10 | -7.49 ± 0.03 | Tumor suppressor p53 |
Note that the DNA sequence shown for the TGACTCA motif and its single nucleotide variants corresponds to the sense strand only and nucleotides flanking these motifs have been omitted for clarity (see Figures 1b-d). The substituted nucleotide relative to the TGACTCA motif is underlined. One example of a gene promoter that contains a particular TGACTCA variant for recruiting the AP1 transcription factor is provided for physiological relevance (15-27). The values for the stoichiometry (n), affinity (Kd) and enthalpy change (ΔH) accompanying the binding of bZIP domains of Jun-Fos heterodimer to dsDNA oligos were obtained from the fit of a function, based on the binding of a ligand to a macromolecule using the law of mass action (41), to the ITC isotherms. Free energy of binding (ΔG) was calculated from the relationship ΔG=RTlnKd, where R is the universal molar gas constant (1.99 cal/mol/K) and T is the absolute temperature (K). Entropic contribution (TΔS) to binding was calculated from the relationship TΔS=ΔH-ΔG. Errors were calculated from 2-3 independent measurements. All errors are given to one standard deviation.
To further rationalize the effect of such genetic variations on the transcriptional output of Jun-Fos heterodimer, we categorized the spectrum of binding affinities observed between bZIP domains and variants of TGACTCA motif into three major classes: those substitutions that bind with an affinity similar to the wildtype (WT) motif containing the TGACTCA sequence, those that bind with submicromolar affinity and those that bind with micromolar affinity. The only motif that binds to bZIP domains with an affinity similar to the WT motif is the one containing G at 0 position (G0) within the TGACTCA sequence. That this is so is expected in light of the fact that the TGAGTCA motif is related to the TGACTCA motif by a two-fold symmetry with identical TGA and TCA half-sites in both the sense and antisense strands but in opposite orientations. There are four single nucleotide substitutions within the TGACTCA sequence that reduce the binding affinity of bZIP domains only moderately by up to about four-fold to submicromolar levels. These include the substitutions A at 0 position (A0), T at 0 position (T0), A at +1 position (A+1) and G at +1 position (G+1). All other single nucleotide substitutions within the TGACTCA motif reduce the binding affinity of bZIP domains by at least an order of magnitude to micromolar levels. In summary, we report here for the first time that no single nucleotide substitutions at any given position within the TGACTCA motif abrogate the binding of bZIP domains of Jun-Fos heterodimer. Our new findings suggest that single nucleotide substitutions within the cis-acting promoter elements may have evolved as a subtle mechanism to differentially regulate transcriptional activity of AP1 at distinct promoters through differential binding. It is likely that the effect of such single nucleotide substitutions on the energetics of binding directly correlates with the transcriptional activity of AP1 under physiological context. This hypothesis will be tested in our future studies.
Enthalpy-entropy compensation buffers the binding of Jun-Fos heterodimer to single nucleotide variants of the TGACTCA motif
It is evident from our data that the binding of bZIP domains to DNA is largely driven by favorable enthalpic contributions accompanied by entropic penalty at physiological temperatures regardless of the position of nucleotide substitution within the TGACTCA motif (Table 1). This is not at all surprising given that an extensive network of hydrogen bonding, electrostatic interactions and hydrophobic contacts between residues in the basic regions of bZIP domains and DNA have to be established. On the other side, the entropic penalty largely results from the overall difference between two major opposing entropic forces. The favorable entropic force is the increase in the degrees of freedom of water molecules while the unfavorable entropic force is the decrease in the degrees of freedom of backbone and sidechain atoms within the protein and DNA molecules upon complexation. The substantial entropic penalty observed here thus suggests that the protein and DNA molecules experience a greater loss of entropy than that gained by water molecules upon intermolecular association.
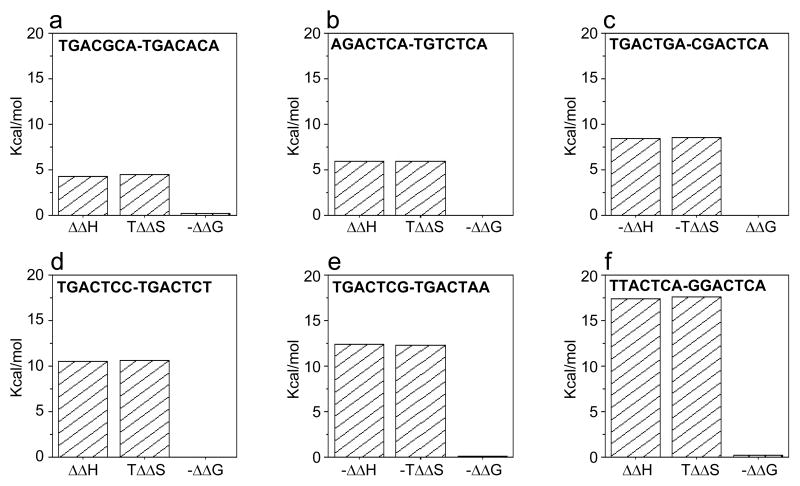
It should be particularly noted here that enthalpy and entropy are not necessarily opposing forces but they often act in an antagonistic manner in biological systems to maintain more or less constant energetics of binding in response to external factors such as temperature and internal changes such as mutations. Such thermodynamic homeostasis, or thermostasis as a portmanteau, also appears to be a hallmark of the binding of bZIP domains of Jun-Fos heterodimer to TGACTCA motif containing various single nucleotide substitutions. As exquisitely illustrated in Figure 3, various pairs of TGACTCA variants undergo enthalpy-entropy compensation with little or negligible effect on the overall binding energetics. These data thus underscore how subtle genetic variations within cis-elements may not necessarily translate into loss of binding energetics and that such a feat may be accomplished through an underlying enthalpy-entropy compensatory mechanism intrinsic to biological systems.
Figure 3.
Differential thermodynamic signatures for the binding of bZIP domains of Jun-Fos heterodimer to various pairs of dsDNA oligos containing TGACTCA variants with similar affinities: (a) TGACGCA relative to TGACACA; (b) AGACTCA relative to TGTCTCA; (c) TGACTGA relative to CGACTCA; (d) TGACTCC relative to TGACTCT; (e) TGACTCG relative to TGACTAA; and (f) TTACTCA relative to GGACTCA. The position of the variant nucleotide in each of the sites relative to the consensus sequence TGACTCA is underlined. ΔΔH, TΔΔS and ΔΔG were calculated from the relationships ΔΔH=ΔHx-ΔHy, TΔΔS=TΔSx-TΔSy and ΔΔG=ΔGx-ΔGy, where the subscripts x and y denote the corresponding thermodynamic parameters for the binding of bZIP domains of Jun-Fos heterodimer to dsDNA oligo x relative to oligo y, respectively (Table 1).
Jun-Fos heterodimer binds to specific variants of the TGACTCA motif in a preferred orientation
The TGA and TCA half-sites within the TGACTCA motif are related by a two-fold symmetry — 180° rotation of either half-site about the central C/G base pair within the context of dsDNA generates the other (Figures 1b-d). However, the TGACTCA motif is not a perfect palindrome and therefore Jun and Fos may have a preference for one half-site over the other due to non-identical contacts with the central C/G base pair. Such a scenario may blossom into the binding of Jun and Fos to TGACTCA motif with a preferred orientation. If this is indeed true, one would expect differential energetics of binding of Jun and Fos to dsDNA oligos containing symmetrically related TGACTCA variants since non-identical contacts with TGA and TCA half-sites would almost certainly be expected to result in varying binding affinities. On the other hand, if Jun and Fos engage in identical contacts with TGA and TCA half-sites, this would not be expected to result in differential energetics of binding to dsDNA oligos containing symmetrically related TGACTCA variants. This latter scenario would be indicative of non-preferred orientation of Jun-Fos heterodimer in association with DNA due to the fact that equivalent energetics of binding would allow the two monomers to freely exchange with each half-site. In an attempt to analyze the extent to which Jun and Fos may exhibit such orientational preference and the extent to which it may be modulated by genetic variations, we plotted relative binding affinities of symmetrically related pairs of dsDNA oligos containing TGACTCA variants to bZIP domains of Jun-Fos heterodimer (Figure 4).
Figure 4.

Analysis of relative binding affinities of symmetrically related pairs of dsDNA oligos containing TGACTCA variants to bZIP domains of Jun-Fos heterodimer. Relative binding affinity is defined as the ratio of the binding affinity of one dsDNA oligo to Jun-Fos heterodimer over that of the other. The nomenclature for the various dsDNA oligos shown is the same as that indicated in Table 1.
It is clearly evident from our data that the vast majority of symmetrically related pairs of dsDNA oligos bind to Jun-Fos heterodimer with relative binding affinities of close to unity, implying that they most likely bind without a preferred orientation due to little or negligible differences in the energetics of binding. Among these symmetrically related pairs of dsDNA oligos that appear to assume non-preferred orientation are the TGACTCA (WT) and TGAGTCA (G0) motifs — an observation that is consistent with X-ray structural analysis of Jun-Fos heterodimer in complex with dsDNA oligo containing the TGACTCA motif and a number of other studies reported previously (8, 35-37). Although the finding that Jun and Fos bind to TGACTCA motif in a non-preferred orientation was expected, the fact that they employ quite a distinct interplay between underlying enthalpic and entropic forces is being reported here for the first time (Table 1). Such enthalpy-entropy compensation to smooth out any differentiation between the overall binding energetics to symmetrically related WT and G0 dsDNA oligos cannot be accounted for by structural data and points to the need for further understanding of protein-DNA interactions in biophysical terms. We believe that such differences in the underlying thermodynamic parameters for the binding of Jun-Fos heterodimer to symmetrically related WT and G0 dsDNA oligos are most likely due to the differences in the flanking nucleotides. In order to avoid self-annealing of sense and antisense strands, it has not been possible at this stage to completely eliminate the contributions of flanking nucleotides on our thermodynamic measurements reported herein. However, it should be noted that such contributions should be minimal given that the flanking nucleotides make no discernable contact with Jun-Fos heterodimer (8). We will fully explore the effect of flanking nucleotides on the thermodynamics of binding of Jun-Fos heterodimer to DNA in our future studies. Interestingly, our analysis also reveals that Jun-Fos heterodimer binds in a non-preferred orientation to symmetrically related dsDNA oligos containing the TGAATCA (A0) and TGATTCA (T0) motifs, indicating that the central C/G base pair can be substituted by A/T base pair without any effect on the orientation of Jun-Fos heterodimer. However, unlike the central C/G base pair, the underlying interplay between entropic and enthalpic forces appears to be very similar in the case of A/T base pair (Table 1). Perhaps even more striking is the observation that the Jun-Fos heterodimer binds without preferred orientation to symmetrically related motifs A-3/T+3, C+3/G-3, A-2/T+2, C-2/G+2, T-2/A+2 and C+1/G+1, which all contain a non-central single nucleotide substitution.
The preference for orientation however does not completely escape the Jun-Fos heterodimer in its quest to bind to single nucleotide variants of the TGACTCA motif. The symmetrically related motifs C-3/G+3, T-1/A+1 and C-1/G+1 clearly exhibit differential energetics of binding to Jun-Fos heterodimer as demonstrated through their relative binding affinities of over 3-fold, 4-fold and 6-fold, respectively. That this is so implies strongly that the Jun-Fos heterodimer binds to these pairs of symmetrically related motifs in a preferred orientation due to formation of non-identical contacts with DNA in the two possible orientations allowed.
3D atomic models provide structural basis of the binding of Jun-Fos heterodimer to the TGACGCA variant in a preferred orientation
X-ray crystallography analysis provided the structural basis for the non-preferred orientation of Jun-Fos heterodimer in complex with dsDNA oligo containing the TGACTCA motif (8). This seminal work revealed that the lack of such preference for orientation was not due to the symmetric binding of Jun and Fos to the central C/G base pair but, on the contrary, asymmetric interactions were observed between R270 in Jun and R155 in Fos with DNA bases. Thus, while R270 sidechain was observed to hydrogen bond with the central C (C0) in one strand, R155 sidechain engaged in hydrogen bonding with the central G (G0) in the other strand. However, such binding asymmetry should not be expected to translate into preferred orientation for Jun-Fos heterodimer due to the fact that R270 and R155 respectively occupy structurally equivalent positions within the α-helical basic regions of Jun and Fos, and are therefore able to exchange freely with each other as exquisitely demonstrated in the crystal structure (8). In other words, the R270-C0 and R155-G0 contacts are energetically equivalent due to the involvement of an identical hydrogen bonding partner, guanidino moiety, in each case. Thus, although the lack of orientational preference upon the interaction of Jun-Fos heterodimer with TGACTCA motif can be rationalized in structural terms, the question as to how certain single nucleotide substitutions within this motif might confer oriented binding raises further curiosity.
Our data presented here suggest strongly that the Jun-Fos heterodimer binds to CGACTCA, TGACTCG, TGTCTCA, TGACACA, TGCCTCA and TGACGCA variants of the TGACTCA motif in a preferred orientation (Figure 4). In an effort to gain insights into the structural basis of such preference for oriented binding, we modeled 3D structures of the bZIP domains of Jun-Fos heterodimer in complex with dsDNA oligos containing the TGACGCA motif in two possible orientations, hereinafter referred to as orientations I and II (Figure 5). Given that the T→G substitution at +1 position within the TGACTCA motif to generate the TGACGCA motif would destroy the symmetric location of T at +1 position (T+1) within both strands, it might be reasonable to suspect that the oriented binding could result from the differential protein-DNA contacts at G+1 in the sense strand and its topologically equivalent counterpart T+1 in the antisense strand within the TGACGCA motif. Our structural analysis indeed reveals that G+1 in the sense strand and its topological equivalent T+1 in the antisense strand within the TGACGCA motif make non-equivalent contacts with Jun-Fos heterodimer. Thus, while G+1 in the sense strand hydrogen bonds with the guanidino moiety of R263 in Jun, T+1 in the antisense strand is involved in hydrogen bonding with the ε-amino group of K148 in orientation I (Figure 5a). These contacts are exquisitely mirrored by R263 and K148 in orientation II through the free exchange of Jun and Fos due to occupation of structurally equivalent positions within the basic regions (Figure 5b). Given the unique chemistry of guanidino moiety relative to ε-amino group, R263-G+1 and K148-T+1 contacts are likely to be energetically non-equivalent and thereby such energetic difference could favor the binding of Jun-Fos heterodimer to TGACGCA motif in only one of two possible orientations with important consequences on its transcriptional role. It is of worthy note that the TGACGCA element is found within the promoters of genes such as fibronectin 1 and sodium/iodide symporter (22, 23). In the latter case, it is immediately flanked between the cis-elements for the TTF-1 and Pax-8 transcription factors (38-40). Given such topological arrangement, it is conceivable that the precise orientation of Jun-Fos heterodimer in association with the TGACGCA element within the promoter of sodium/iodide symporter gene may be a critical determinant of the nature of other interacting cellular partners and hence gene expression.
Figure 5.
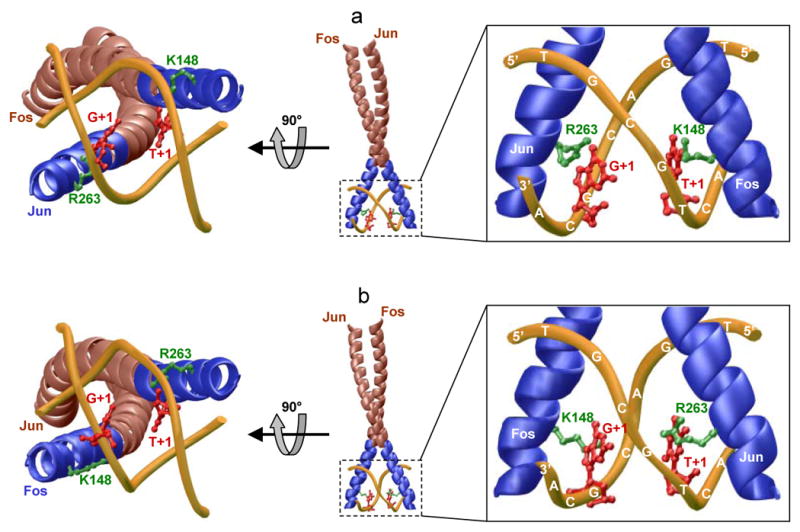
3D structural models of bZIP domains of Jun-Fos heterodimer in complex with dsDNA oligos containing the TGACGCA motif in two possible orientations I (a) and II (b). The two orientations are related by a 180°-rotation of the Jun-Fos heterodimer about the dyad axis of symmetry. The backbones of LZ and BR subdomains of the bZIP heterodimer are colored brown and blue, respectively. The backbone of dsDNA is shown in yellow. The sidechains of R263 in Jun and K148 in Fos are colored green. The guanine at position +1 (G+1) in the sense strand TGACGCA and thymine at position +1 (T+1) in the antisense strand TGCGTCA are colored red. Insets show close-up views of contacts between specific bZIP residues with DNA bases.
Although our 3D structural models provide a compelling rationale for the binding of Jun-Fos heterodimer to the TGACGCA motif with a preferred orientation, it should be borne in mind that only structural analysis through experimental means can confirm the accuracy of such models and which one of the two possible orientations may actually be the preferred one. We also emphasize that for the binding of Jun-Fos heterodimer to other variants of the TGACTCA motif with a preferred orientation, R263 and K148 may not necessarily be responsible for differential protein-DNA contacts but rather the role of additional residues within the basic regions may have to be invoked. It has indeed been previously shown that amino acid substitutions within the basic regions of bZIP domains can result in oriented binding of Jun-Fos heterodimer to DNA (37). In short, our study warrants extensive X-ray crystallographic analysis of Jun-Fos heterodimer in complex with specific TGACTCA variants to fully unravel the structural basis of the preference for oriented binding and, needless to say, our future efforts will be directed along these lines of curiosity.
Conclusions
Transcription factors do not act alone but rather in concert in a cooperative manner within the transcriptional initiation complex responsible for switching on gene expression. The architecture of such cooperative machinery parallels the design of composite sites within gene promoters for the recognition of a diverse array of transcription factors that often arrive in droves and in association with each other. The orientation of transcription factors relative to each other as well as DNA coupled with the sequence of specific cis-elements thus play a key role in gauging the transcriptional output in a spatial and temporal manner. Because of such inter-dependence and added versatility, the cis-elements within a composite site may not necessarily contain an optimal sequence for binding to a corresponding transcription factor. Understanding how subtle nucleotide changes within such cis-elements may affect their binding energetics to trans-factors is thus of paramount importance to not only unraveling the regulatory mechanisms of transcriptional machinery but may also offer insights into the role of genetic variations, such as single nucleotide polymorphisms, in specific promoter regions between different individuals. It is this curiosity that led to the design of our current study.
Here, we report how single nucleotide variants of the TGACTCA consensus motif modulate energetics and orientation of binding of the AP1 transcription factor. The major conclusions of our study are that the single nucleotide substitutions within the TGACTCA motif do not abrogate the binding of Jun-Fos heterodimer. On the contrary, the Jun-Fos heterodimer binds to the TGACTCA variants with affinities in the physiologically relevant micromolar-submicromolar range. Given that TGACTCA variants are widely encountered within the gene promoters (15-27), we believe that such differential energetics of binding may have emerged as an evolutionary mechanism for the differential regulation of AP1-responsive genes. Our data also indicate that certain single nucleotide variants of the TGACTCA motif may also dictate the orientation of the Jun-Fos heterodimer in complex with DNA and that such a feat may be attributable to chemically distinct amino acid residues located in structurally equivalent positions within the basic regions of Jun and Fos. Supporting this corollary is the salient observation that amino acid substitutions within the basic regions of bZIP domains can result in oriented binding of Jun-Fos heterodimer to DNA (37). Our present study thus mirrors this previous finding in that specific nucleotide substitutions within the TGACTCA motif can also accomplish a similar fate on the Jun-Fos heterodimer.
In concluding, the demonstration that single nucleotide variants within the TGACTCA motif can modulate energetics and orientation of binding of Jun-Fos heterodimer suggests that it may be a general feature of other cis-elements acting within the gene promoters. Our study thus clearly provides a precedent for guiding the design of future experiments to expand our understanding of the regulatory mechanisms underlying the operation of the transcriptional machinery.
Abbreviations
- AP1
Activator protein 1
- BR
Basic region
- His
Polyhistidine tag
- ITC
Isothermal titration calorimetry
- LZ
Leucine zipper
- MAP
Mitogen-activated protein
- SNP
Single nucleotide polymorphism
- SNV
Single nucleotide variant
- Trx
Thioredoxin
- bZIP
Basic zipper
Footnotes
This work was supported by funds from the National Institutes of Health (Grant# R01-GM083897), the American Heart Association (Grant# 0655087B) and the UM/Sylvester Braman Family Breast Cancer Institute to AF.
References
- 1.Chinenov Y, Kerppola TK. Close encounters of many kinds: Fos-Jun interactions that mediate transcription regulatory specificity. Oncogene. 2001;20:2438–2452. doi: 10.1038/sj.onc.1204385. [DOI] [PubMed] [Google Scholar]
- 2.Angel P, Karin M. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim Biophys Acta. 1991;1072:129–157. doi: 10.1016/0304-419x(91)90011-9. [DOI] [PubMed] [Google Scholar]
- 3.Baxevanis AD, Vinson CR. Interactions of coiled coils in transcription factors: where is the specificity? Curr Opin Genet Dev. 1993;3:278–285. doi: 10.1016/0959-437x(93)90035-n. [DOI] [PubMed] [Google Scholar]
- 4.Raivich G, Behrens A. Role of the AP-1 transcription factor c-Jun in developing, adult and injured brain. Prog Neurobiol. 2006;78:347–363. doi: 10.1016/j.pneurobio.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 5.Milde-Langosch K. The Fos family of transcription factors and their role in tumourigenesis. Eur J Cancer. 2005;41:2449–2461. doi: 10.1016/j.ejca.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 6.Curran T, Franza BR., Jr Fos and Jun: the AP-1 connection. Cell. 1988;55:395–397. doi: 10.1016/0092-8674(88)90024-4. [DOI] [PubMed] [Google Scholar]
- 7.Halazonetis TD, Georgopoulos K, Greenberg ME, Leder P. c-Jun dimerizes with itself and with c-Fos, forming complexes of different DNA binding affinities. Cell. 1988;55:917–924. doi: 10.1016/0092-8674(88)90147-x. [DOI] [PubMed] [Google Scholar]
- 8.Glover JN, Harrison SC. Crystal structure of the heterodimeric bZIP transcription factor c-Fos-c-Jun bound to DNA. Nature. 1995;373:257–261. doi: 10.1038/373257a0. [DOI] [PubMed] [Google Scholar]
- 9.Weiss MA, Ellenberger T, Wobbe CR, Lee JP, Harrison SC, Struhl K. Folding transition in the DNA-binding domain of GCN4 on specific binding to DNA. Nature. 1990;347:575–578. doi: 10.1038/347575a0. [DOI] [PubMed] [Google Scholar]
- 10.Weiss MA. Thermal unfolding studies of a leucine zipper domain and its specific DNA complex: implications for scissor's grip recognition. Biochemistry. 1990;29:8020–8024. doi: 10.1021/bi00487a004. [DOI] [PubMed] [Google Scholar]
- 11.Thompson KS, Vinson CR, Freire E. Thermodynamic characterization of the structural stability of the coiled-coil region of the bZIP transcription factor GCN4. Biochemistry. 1993;32:5491–5496. doi: 10.1021/bi00072a001. [DOI] [PubMed] [Google Scholar]
- 12.Patel L, Abate C, Curran T. Altered protein conformation on DNA binding by Fos and Jun. Nature. 1990;347:572–575. doi: 10.1038/347572a0. [DOI] [PubMed] [Google Scholar]
- 13.Saudek V, Pastore A, Castiglione Morelli MA, Frank R, Gausepohl H, Gibson T, Weih F, Roesch P. Solution structure of the DNA-binding domain of the yeast transcriptional activator protein GCN4. Protein Eng. 1990;4:3–10. doi: 10.1093/protein/4.1.3. [DOI] [PubMed] [Google Scholar]
- 14.Lee W, Haslinger A, Karin M, Tjian R. Activation of transcription by two factors that bind promoter and enhancer sequences of the human metallothionein gene and SV40. Nature. 1987;325:368–372. doi: 10.1038/325368a0. [DOI] [PubMed] [Google Scholar]
- 15.Hengerer B, Lindholm D, Heumann R, Ruther U, Wagner EF, Thoenen H. Lesion-induced increase in nerve growth factor mRNA is mediated by c-fos. Proc Natl Acad Sci U S A. 1990;87:3899–3903. doi: 10.1073/pnas.87.10.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Therrien M, Drouin J. Pituitary pro-opiomelanocortin gene expression requires synergistic interactions of several regulatory elements. Mol Cell Biol. 1991;11:3492–3503. doi: 10.1128/mcb.11.7.3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koike M, Kuroki T, Nose K. Common target for 12-O-tetradecanoylphorbol-13-acetate and ras in the transcriptional enhancer of the growth factor-inducible JE gene. Mol Carcinog. 1993;8:105–111. doi: 10.1002/mc.2940080207. [DOI] [PubMed] [Google Scholar]
- 18.Fabre S, Manin M, Pailhoux E, Veyssiere G, Jean C. Identification of a functional androgen response element in the promoter of the gene for the androgen-regulated aldose reductase-like protein specific to the mouse vas deferens. J Biol Chem. 1994;269:5857–5864. [PubMed] [Google Scholar]
- 19.Shyy JY, Lin MC, Han J, Lu Y, Petrime M, Chien S. The cis-acting phorbol ester “12-O-tetradecanoylphorbol 13-acetate”-responsive element is involved in shear stress-induced monocyte chemotactic protein 1 gene expression. Proc Natl Acad Sci U S A. 1995;92:8069–8073. doi: 10.1073/pnas.92.17.8069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larose M, Cassard-Doulcier AM, Fleury C, Serra F, Champigny O, Bouillaud F, Ricquier D. Essential cis-acting elements in rat uncoupling protein gene are in an enhancer containing a complex retinoic acid response domain. J Biol Chem. 1996;271:31533–31542. doi: 10.1074/jbc.271.49.31533. [DOI] [PubMed] [Google Scholar]
- 21.Goldberg D, Polly P, Eisman JA, Morrison NA. Identification of an osteocalcin gene promoter sequence that binds AP1. J Cell Biochem. 1996;60:447–457. doi: 10.1002/(SICI)1097-4644(19960315)60:4%3C447::AID-JCB2%3E3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 22.Moriguchi Y, Matsubara H, Mori Y, Murasawa S, Masaki H, Maruyama K, Tsutsumi Y, Shibasaki Y, Tanaka Y, Nakajima T, Oda K, Iwasaka T. Angiotensin II-induced transactivation of epidermal growth factor receptor regulates fibronectin and transforming growth factor-beta synthesis via transcriptional and posttranscriptional mechanisms. Circ Res. 1999;84:1073–1084. doi: 10.1161/01.res.84.9.1073. [DOI] [PubMed] [Google Scholar]
- 23.Chun JT, Di Dato V, D'Andrea B, Zannini M, Di Lauro R. The CRE-like element inside the 5′-upstream region of the rat sodium/iodide symporter gene interacts with diverse classes of b-Zip molecules that regulate transcriptional activities through strong synergy with Pax-8. Mol Endocrinol. 2004;18:2817–2829. doi: 10.1210/me.2004-0020. [DOI] [PubMed] [Google Scholar]
- 24.Hayama A, Uchida S, Sasaki S, Marumo F. Isolation and characterization of the human CLC-5 chloride channel gene promoter. Gene. 2000;261:355–364. doi: 10.1016/s0378-1119(00)00493-5. [DOI] [PubMed] [Google Scholar]
- 25.Borner C, Hollt V, Kraus J. Involvement of activator protein-1 in transcriptional regulation of the human mu-opioid receptor gene. Mol Pharmacol. 2002;61:800–805. doi: 10.1124/mol.61.4.800. [DOI] [PubMed] [Google Scholar]
- 26.Wang SE, Wu FY, Chen H, Shamay M, Zheng Q, Hayward GS. Early activation of the Kaposi's sarcoma-associated herpesvirus RTA, RAP, and MTA promoters by the tetradecanoyl phorbol acetate-induced AP1 pathway. J Virol. 2004;78:4248–4267. doi: 10.1128/JVI.78.8.4248-4267.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dwivedi PP, Anderson PH, Omdahl JL, Grimes HL, Morris HA, May BK. Identification of growth factor independent-1 (GFI1) as a repressor of 25-hydroxyvitamin D 1-alpha hydroxylase (CYP27B1) gene expression in human prostate cancer cells. Endocr Relat Cancer. 2005;12:351–365. doi: 10.1677/erc.1.00920. [DOI] [PubMed] [Google Scholar]
- 28.Bream JH, Ping A, Zhang X, Winkler C, Young HA. A single nucleotide polymorphism in the proximal IFN-gamma promoter alters control of gene transcription. Genes Immun. 2002;3:165–169. doi: 10.1038/sj.gene.6363870. [DOI] [PubMed] [Google Scholar]
- 29.Bond GL, Hu W, Bond EE, Robins H, Lutzker SG, Arva NC, Bargonetti J, Bartel F, Taubert H, Wuerl P, Onel K, Yip L, Hwang SJ, Strong LC, Lozano G, Levine AJ. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell. 2004;119:591–602. doi: 10.1016/j.cell.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 30.Seldeen KL, McDonald CB, Deegan BJ, Farooq A. Coupling of folding and DNA-binding in the bZIP domains of Jun-Fos heterodimeric transcription factor. Arch Biochem Biophys. 2008;473:48–60. doi: 10.1016/j.abb.2008.02.024. [DOI] [PubMed] [Google Scholar]
- 31.Cantor CR, Warshaw MM, Shapiro H. Oligonucleotide interactions. 3. Circular dichroism studies of the conformation of deoxyoligonucleotides. Biopolymers. 1970;9:1059–1077. doi: 10.1002/bip.1970.360090909. [DOI] [PubMed] [Google Scholar]
- 32.Marti-Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, Sali A. Comparative Protein Structure Modeling of Genes and Genomes. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 33.Carson M. Ribbons 2.0. J Appl Crystallogr. 1991;24:958–961. [Google Scholar]
- 34.Risse G, Jooss K, Neuberg M, Bruller HJ, Muller R. Asymmetrical recognition of the palindromic AP1 binding site (TRE) by Fos protein complexes. Embo J. 1989;8:3825–3832. doi: 10.1002/j.1460-2075.1989.tb08560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chytil M, Peterson BR, Erlanson DA, Verdine GL. The orientation of the AP-1 heterodimer on DNA strongly affects transcriptional potency. Proc Natl Acad Sci U S A. 1998;95:14076–14081. doi: 10.1073/pnas.95.24.14076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leonard DA, Kerppola TK. DNA bending determines Fos-Jun heterodimer orientation. Nat Struct Biol. 1998;5:877–881. doi: 10.1038/2316. [DOI] [PubMed] [Google Scholar]
- 37.Leonard DA, Rajaram N, Kerppola TK. Structural basis of DNA bending and oriented heterodimer binding by the basic leucine zipper domains of Fos and Jun. Proc Natl Acad Sci U S A. 1997;94:4913–4918. doi: 10.1073/pnas.94.10.4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taki K, Kogai T, Kanamoto Y, Hershman JM, Brent GA. A thyroid-specific far-upstream enhancer in the human sodium/iodide symporter gene requires Pax-8 binding and cyclic adenosine 3′,5′-monophosphate response element-like sequence binding proteins for full activity and is differentially regulated in normal and thyroid cancer cells. Mol Endocrinol. 2002;16:2266–2282. doi: 10.1210/me.2002-0109. [DOI] [PubMed] [Google Scholar]
- 39.Schmitt TL, Espinoza CR, Loos U. Characterization of a thyroid-specific and cyclic adenosine monophosphate-responsive enhancer far upstream from the human sodium iodide symporter gene. Thyroid. 2002;12:273–279. doi: 10.1089/10507250252949388. [DOI] [PubMed] [Google Scholar]
- 40.Ohno M, Zannini M, Levy O, Carrasco N, di Lauro R. The paired-domain transcription factor Pax8 binds to the upstream enhancer of the rat sodium/iodide symporter gene and participates in both thyroid-specific and cyclic-AMP-dependent transcription. Mol Cell Biol. 1999;19:2051–2060. doi: 10.1128/mcb.19.3.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wiseman T, Williston S, Brandts JF, Lin LN. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]