Abstract
Frequent gene duplications in the genome incessantly supply new genetic materials for functional innovation presumably driven by positive Darwinian selection. This mechanism in the desaturase gene family has been proposed to be important in triggering the pheromonal diversification in insects. With the recent completion of a dozen Drosophila genomes, a genome-wide perspective is possible. In this study, we first identified homologs of desaturase genes in 12 Drosophila species and noted that while gene duplication events are relatively frequent, gene losses are not scarce, especially in the desat1–desat2–desatF clade. By reconciling the gene tree with species phylogeny and the chromosomal synteny of the sequenced Drosophila genomes, at least one gene loss in desat2 and a minimum of six gene gains (resulting in seven desatF homologs, α-η), three gene losses and one relocation in desatF were inferred. Upon branching off the ancestral desat1 lineage, both desat2 and desatF gained novel functions through accelerating protein evolution. The amino acid residues under positive selection located near the catalytic sites and the C-terminal region might be responsible for altered substrate selectivity between closely related species. The association between the expression pattern of desatF-α and the chemical composition of cuticular hydrocarbons implies that the ancestral function of desatF-α is the second desaturation at the four carbons after the first double bond in diene synthesis, and the shift from bisexual to female-specific expression in desatF-α occurred in the ancestral lineage of Drosophila melanogaster subgroup. A relationship between the number of expressed desatF homologs and the diene diversification has also been observed. These results suggest that the molecular diversification of fatty acid desaturases after recurrent gene duplication plays an important role in pheromonal diversity in Drosophila.
Keywords: cuticular hydrocarbon, fatty acid desaturase, gene duplication, pheromonal diversity, positive selection
Introduction
Fatty acid desaturases are enzymes that catalyze the introduction of double bonds at specific positions of fatty acids. Desaturases play essential roles in both lipid metabolism and the maintenance of proper structure and function of biological membranes in living organisms. Studies on desaturases in insects have shown that their roles in lipid biosynthesis also contribute to the precursor diversity of cuticular hydrocarbons and sex pheromones (Roelofs and Rooney 2003). The cuticular surface of insects bears a lipid layer that functions primarily to limit water loss (Howard and Blomquist 1982). In some insects, cuticular hydrocarbons present a rich reservoir of chemicals that are important in species and gender recognition, dominance and fertility cues, task-specific cues, and chemical mimicry (reviewed in Howard and Blomquist 2005). In Drosophila, the existence of fatty-acid–derived cuticular hydrocarbons which act in females as important attractive cues for males has been known for decades. Differences in hydrocarbon profiles between species have been proposed to contribute to sexual isolation, but only two desaturase genes have been identified to be responsible for pheromonal differences between sibling species or geographical races of Drosophila (Coyne et al. 1999; Dallerac et al. 2000; Takahashi et al. 2001; Fang et al. 2002; Chertemps et al. 2006; Legendre et al. 2008). In fact, both genes originated by gene duplication, suggesting that the increase of gene number of the desaturase gene family enlarges the pheromone diversification between closely related species in insects (Knipple et al. 2002; Roelofs and Rooney 2003; Greenberg et al. 2006; Xue et al. 2007).
Eight fatty acid desaturase genes have been identified in Drosophila melanogaster (http://www.flybase.org/blast/; Crosby et al. 2007). Of which, infertile crescent, encoding an enzyme with Δ4-desaturase activity, is the most distantly related member belonging to the Sphingolipid subfamily. All the other 7 desaturase genes are grouped in the First Desaturase subfamily that introduces the first double bond into the saturated acyl chain at the Δ9 position (Hashimoto et al. 2008). We shall focus on the seven members of the First Desaturase subfamily because at least three of them are involved in the biosynthesis of pheromonal hydrocarbons which are diversified chemicals for mate recognition in Drosophila (Dallerac et al. 2000; Takahashi et al. 2001; Labeur et al. 2002; Chertemps et al. 2006). The two tandemly duplicated Δ9 desaturase genes, desat1 and desat2, are responsible for adding the first double bond into unsaturated fatty acid precursors leading to monoenes in both sexes (Dallerac et al. 2000; Labeur et al. 2002). The roles that desat2 plays in the differential adaptation to ecological changes and the behavioral isolation between Z and M races of D. melanogaster are well documented (Takahashi et al. 2001; Fang et al. 2002; Greenberg et al. 2003). Recently, the third desaturase gene, desatF (aka Fad2), has been functionally characterized in the production of the female dienes (Chertemps et al. 2006, 2007; Legendre et al. 2008). This female-specific expression might be acquired after desatF originated by a retrotransposition event because desatF is the only intronless member, whereas all the other desaturase genes have multiple exons (Bai et al. 2007).
The importance of gene duplication has long been appreciated (Ohno 1970). Yet, gene losses have only recently attracted attention through comparative genomic studies (Hahn et al. 2007). A general notion is that frequent gene gains and losses through duplication and pseudogenization increase genetic variation and thereby contribute to species divergence. The diversity of moth sex pheromones is such an example which suggests that multiple birth-and-death processes of desaturases are subject to sexual selection between closely related species (Knipple et al. 2002; Roelofs and Rooney 2003). In addition, natural selection could act as an effective sieve to increase beneficial gene duplicates, whereas elimination of duplicated and/or existing genes might also provide changes that otherwise could not have occurred (Wang et al. 2006). Because desaturases possess functions in both ecological adaptation and mate recognition in Drosophila, one would expect that natural and/or sexual selection may act on this gene family as in the cases of accessory gland proteins (reviewed in Clark et al. 2006). To test this idea, we first identified all homologs of desaturases from the 12 sequenced Drosophila genomes to understand the birth-and-death processes of the desaturase gene family. Next, we asked if any signature of positive selection could be detected, especially in the lineage after gene duplication. If there are, the next questions would be which amino acid residues are under positive selection and whether they are located at the sites with implication in functional adaptation. Finally, we addressed how duplication events of desaturases lead to the pheromonal diversification by analyzing the gene expression patterns and cuticular hydrocarbon profiles.
Materials and Methods
Sequence Data
Coding sequences of seven fatty acid desaturase genes, desat1, desat2, Fad2, CG8630, CG9743, CG9747, and CG15531 of D. melanogaster, were used to Blast against the 12 Drosophila genomes at FlyBase (http://www.flybase.org/blast/; Crosby et al. 2007). The orthologs of each gene were identified by reciprocal Blast and conserved synteny. The results were further confirmed with GBrowser at Flybase (Wilson et al. 2008) and annotation tracts at the University of California–San Cruz Genome Browser (http://genome.ucsc.edu/; Karolchik et al. 2003).
Phylogeny Reconstruction
Phylogeny of three fatty acid desaturase genes, desat1, desat2, and desatF, was reconstructed by maximum parsimony (PAUP* 4.0b10; Swofford 2002) and Bayesian inference (MrBayes 3.1.2; Huelsenbeck and Ronquist 2001). Sites with ambiguous alignment were not included. No weighting was assigned for maximum parsimony analyses, and gaps were treated as missing data. Branch support was obtained from bootstrapping with 1,000 replicates. Sites with gaps were excluded from Bayesian inference. In Bayesian analyses, 2 independent tests, each with one cold and seven heated Markov chains, were run for 2 million generations. Trees were sampled every 1,000 generations, and 500 of the sampled trees were described as burn-in while summarizing the result. In the summarized tree, posterior probabilities were indicated on each branch, and maximum likelihood method was applied to estimate the branch length (Yang 2006).
Tests for Positive Selection
To detect selection, sequences were analyzed with maximum likelihood–based methods implemented in CODEML of PAML 4 (Yang 2007). CODEML estimates the ratio of nonsynonymous to synonymous substitution rate (ω) under models allowing ω vary among sites (site models), branches (branch models), and a combination of both (branch site models). In all tests, the likelihood ratio test (LRT) was performed in comparing the null model against the alternative model. The test statistic 2Δℓ = 2(ℓ1 − ℓ0), where ℓ0 and ℓ1 are the log likelihood values under the null and alternative hypotheses, respectively, was compared with the chi-square distribution, with the degree of freedom to be the difference in the number of parameters between the two hypotheses. All sites with ambiguous alignment and gaps were excluded from the analysis. For better convergence and to avoid too many parameters to be estimated at the same time, branch lengths were estimated under model M0 (one-ratio), which assigns 1 ω value across the whole tree and sequence, and then is fixed in subsequent analyses.
To test if ω values vary among desat1, desat2, and desatF, branch models, where ω values are allowed to vary between lineages, were performed. We set the joint corresponding ω values of desat1, desat2, desatF, and all other branches as ω1, ω2, ωF, and ω0. Test scheme is listed in table 1. If the null hypothesis is rejected, it means that different ω values exist between two target clades.
Table 1.
LRTs among desat1, desat2, and desatF Clades under Branch Models
| H0 | H1 | df | 2Δl | P Value | ω1 | ω2 | ωF | |
| desat1 versus desat2 | ω1 = ω2, ωF = ω0 | ω1 ≠ ω2, ωF = ω0 | 1 | 12.46 | 4.1 × 10−4 | 0.0493 | 0.0753 | — |
| desat1 versus desatF | ω1 = ωF, ω2 = ω0 | ω1 ≠ ωF, ω2 = ω0 | 1 | 30.29 | 3.7 × 10−8 | 0.0494 | — | 0.0904 |
| desat2 versus desatF | ω2 = ωF, ω1 = ω0 | ω2 ≠ ωF, ω1 = ω0 | 1 | 4.70 | 0.03 | — | 0.0748 | 0.0906 |
NOTE.—df, degree of freedom. ω1, ω2, and ωF denote the ω ratios of desat1, desat2, and desatF clades, respectively.
To detect positive selection acting only on certain codons in certain branches, branch site models were performed. The alternative hypothesis assigns some sites in the target branch to be under positive selection, whereas the null hypothesis does not. All branches except the root branch on the tree were tested. For branches with significant P values after Bonferroni correction (Anisimova and Yang 2007), sites under positive selection with posterior probability higher than 0.95 in “Bayes empirical Bayes” analysis were listed.
Flanking Sequence Analysis
Because retrotransposition, DNA-mediated transposition, and inversions occurring at flanking regions could all result in nontandem gene duplication events, we further identified the break point of each copy by DotPlot, compared the similarity between duplicate genes with their flanking sequences, and searched traces of repetitive sequences and transposons. To determine the boundary of each transposition and its flanking sequences, syntenic genomic regions of each desatF locus between species were compared using DotPlot in the GCG Wisconsin Package (Version 10.3, Accelrys Inc.). Repetitive sequences, including poly-A, of the flanking regions were identified by RepeatMasker (http://www.repeatmasker.org/). Transposable elements were recognized by Blast against D. melanogaster transposable element database at Flybase (http://www.flybase.org).
Gene Expression by Reverse Transcriptase–Polymerase Chain Reaction
Adult desatF expression was performed by reverse transcriptase–polymerase chain reaction (RT-PCR) from total RNA. Total RNA was extracted from 3- to 5-day-old adults by TRIzol reagent (Invitrogen, Carlsbad, CA) and then treated by DNase using DNA-free (Ambion, Foster City, CA) according to the manufacturer's instructions. Reverse transcription was carried out with SuperScript III First-Strand Synthesis System (Invitrogen) using oligo(dT)20 primer. Gene-specific primers were used for further polymerase chain reaction (PCR) amplification. The primer sequences and detailed PCR conditions are available upon request. An internal control for the reverse transcription reaction was conducted with primers specific to Act5C mRNA. In addition, we also performed controls with RNA samples amplified without reverse transcriptase to verify the absence of genomic DNA contamination.
Results
Identifying Fatty Acid Desaturase Genes
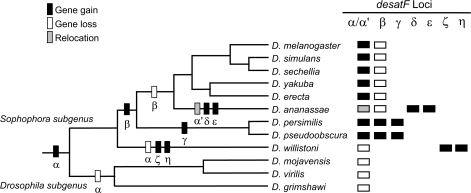
We systematically identified the fatty acid desaturase homologs from the sequenced Drosophila genomes by reciprocal Blast and conserved chromosomal synteny with seven members of the desaturase gene family in D. melanogaster. The annotation and chromosomal location for each gene are listed in supplementary table S1 (Supplementary Material online). Based on a large-scale analysis on a wide range of eukaryotic genomes, the 3 genes, desat1, desat2, and desatF, involved in the biosynthesis of cuticular hydrocarbons in D. melanogaster formed a single cluster, and the other four genes are more distantly related to this cluster (Hashimoto et al. 2008). Of which, desat1 and desat2 are tandemly duplicated copies, whereas desatF originated by a single retrotransposition. Based on phylogenetic analyses of these three desaturase genes (supplementary fig. S1, Supplementary Material online), the retrotransposition event took place before the tandem duplication of desat1 and desat2. Because desat1 and desat2 exist in all 12 Drosophila genomes, we inferred that the retrotransposition event predates the split of the Drosophila and Sophophora subgenera, and the absence of desatF in the three species of Drosophila subgenus, that is, Drosophila grimshawi, Drosophila mojavensis, and Drosophila virilis, is the secondary loss after the retrotransposition. As expected, all desatF homologs are intronless except that in Drosophila yakuba, GE21776, in which a putative 32-bp intron together with a 10-bp deletion was predicted to result in a truncated protein with 14 amino acids short. If we consider GE21776, a single-exon gene as all other desatF homologs do, then the D. yakuba reference genome might contain a nonfunctional allele due to the 10-bp deletion in the coding region that gave rise to a premature stop codon. A notable feature in desatF clade is that three homologous copies were found in each Drosophila ananassae, Drosophila pseudoobscura, and Drosophila persimilis, respectively. In D. ananassae, three annotated desatF genes, namely GF24026, GF18504, and GF16174, occurred in the regions that are not homologous to the desatF-α location in D. melanogaster, that is, they are located at nonsyntenic regions. In D. pseudoobscura genome (R2.2), one syntenically conserved desatF was annotated as dpse_GA20691 (on XR, aka Muller element D, homologous to 3L of D. melanogaster), and two additional annotated single-exon desaturase genes, GA27148 and GA27452, were identified on the second chromosome (Muller element E, homologous to 3R of D. melanogaster). The fact that homologs of these two genes have also been found in D. persimilis (GL23117 and GL22317) suggests that these two genes are additional members of desatF in D. obscura group. In addition, two desatF homologs in Drosophila willistoni, GK17186 and GK11373, are also resident in nonsyntenic regions. Moreover, only six desaturase genes are annotated in the Drosophila erecta genome (CAF1), and the decrease in gene number is due to the loss of desat2 in this reference genome.
Assigning Gene Gains and Losses of desatF
To understand the evolutionary history of desatF in Drosophila, we reconciled the gene tree of desatF (supplementary fig. S1, Supplementary Material online) with the species tree to estimate the minimum number of duplication events (fig. 1). After the birth of desatF (locus α) through retrotransposition from the common ancestor of desat1 and desat2, at least six gene gains and three gene losses gave rise to seven paralogous desatF loci (α-η) in the Drosophila lineage. The desatF-α is absent from all three species of Drosophila subgenus, suggesting that either the gene was lost in the common ancestor of these species or independent losses occurred in each branch. Based on the phylogeny (supplementary fig. S1, Supplementary Material online), the second duplication, which gave rise to locus desatF-β, took place in the common ancestor of D. melanogaster and D. obscura species groups. This locus was subsequently lost in the lineage leading to D. melanogaster species group. The third duplication created an obscura lineage–specific locus (desatF-γ). In D. ananassae, none of the desatF copies is located at the syntenic region of desatF-α. We assigned ana_GF24026 as desatF-α′ because this locus is on the same chromosome arm (Muller element D) as other desatF-α. Relocation of this locus could occur by multiple inversion events. The other two desatF copies of D. ananassae are not orthologous either to desatF-β or to desatF-γ of D. pseudoobscura (supplementary fig. S1, Supplementary Material online), so we proposed that at least two rounds of lineage-specific duplications generating desatF-δ and desatF-ϵ in the lineage of D. ananassae. Similarly, two additional rounds of duplication took place in the lineage of D. willistoni to result in another two desatF copies, desatF-ζ and desatF-η.
FIG. 1.—
Gene gains and losses of desatF genes in Drosophila. Using the well-defined phylogeny of the 12 sequenced Drosophila species (Drosophila 12 Genomes Consortium 2007), the distribution of seven paralogous loci α-η of desatF was assigned on the right based on the gene tree (supplementary fig. S1, Supplementary Material online).
Because multiple nontandem desatF loci were found in D. ananassae, D. pseudoobscura, D. persimilis, and D. willistoni, it is of great interest to investigate the underlying mechanisms of their duplication. The fact that all the desatF-α orthologs contain poly-A tracts within 500 bp downstream of the stop codon is consistent with their retrogene nature. Among these desatF paralogs, sequence similarities between desatF-β and desatF-γ in both D. pseudoobscura and D. persimilis extend to ∼150 bp upstream and ∼590 bp downstream of the coding regions. In addition, desatF-γ is flanked by two Helitron transposable elements, one on each side, indicating that desatF-γ is duplicated by DNA-mediated transposition. Similarly, desatF-η of D. willistoni might also be duplicated by DNA-mediated transposition based on Helitron elements located at flanking regions. Based on the sequence divergence between the two copies (desatF-ζ and desatF-η, fig. 2), the duplication could occur in an early branch of the D. willistoni group (66.2 Ma, Tamura et al. 2004) rather than in D. willistoni per se although the sequence information from other species of D. willistoni group is not available. When comparing flanking sequences of the two desatF loci in D. willistoni, only desatF-ζ contains the poly-A tract. On the other hand, desatF-η has no poly-A tract and is flanked by inverted Helitron sequences. It is not clear that desatF-η is duplicated either from the ancestral desatF-α or from desatF-ζ by DNA-mediated mechanism. Because desatF-α has been completely degenerated from D. willistoni genome, it is not possible to check the similarity of flanking sequences. On the other hand, desatF-ζ and desatF-η do not share any sequence similarity beyond the coding region. Even if desatF-ζ is the parental copy of desatF-η, this duplication event must have occurred long time ago. Similarly, the divergence among three paralogs in D. ananassae (desatF-α′, desatF-δ, and desatF-ϵ) is also high, so the two duplication events might have taken place after the split of D. melanogaster subgroup and D. ananassae subgroup (44.2 Ma, Tamura et al. 2004). The little homology (about 40% identity) shared by the 5′ regions of the three loci provides little evidence on the duplication mechanism except for desatF-α′ which contains the poly-A tract. Both desatF-δ and desatF-ϵ might arise from independent retrotranspositions or DNA-mediated transpositions because neither poly-A tracts in the 3′ regions nor DNA transposons in the flanking sequences were detected. Alternatively, only one copy was duplicated from desatF-α′ and these two genes were generated by tandem duplication and were subsequently separated by chromosomal rearrangements.
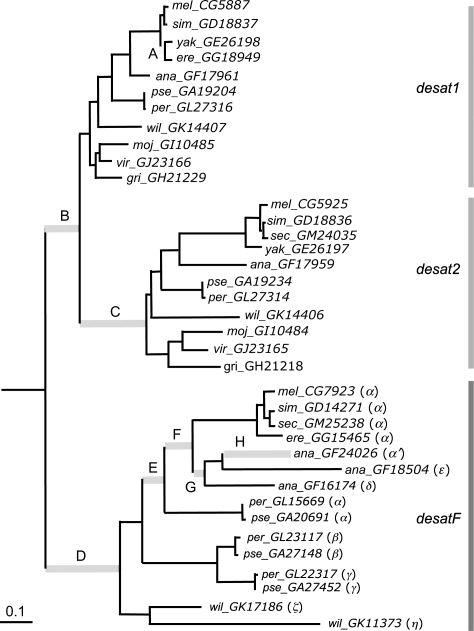
FIG. 2.—
Branches with positive selection under branch site models were labeled on the phylogeny of the desat1–desat2–desatF clade. The phylogeny is reconstructed based on the gene tree (supplementary fig. S1, Supplementary Material online) with minor adjustments in desat1 and desat2 according to species tree (FlyBase). Branch lengths were estimated using maximum likelihood method with general time reversible model and gamma distribution.
Identify Sites under Positive Selection
As desat1, desat2, and desatF have evolved different functions in pheromone biosynthesis, we are interested in how selection, if there is any, governed the functional diversification of the three genes. To address this question, we first compared the estimated ω under the branch models with a known phylogeny. The phylogeny of all homologs of desat1, desat2, and desatF (fig. 2) was reconstructed based on the gene tree (supplementary fig. S1, Supplementary Material online) with minor adjustments in the desat1 and desat2 subclades according to the species tree (Drosophila 12 Genomes Consortium 2007). The results showed that the ω ratios of the three clades are significantly different from one another (table 1). Among them, the ω value of desatF (ωF) is the highest and the ω of the parental copy, desat1, (ω1) is the lowest. Despite the differences in ω values among three genes, the fact that all the estimated ω do not exceed 1 implies that most codons are under purifying selection. The higher ω in both desat2 and desatF could result from either positive selection or relaxation of functional constraint in a small portion of codons on some branches.
Because gene gains and losses occurred frequently in the desat1–desat2–desatF clade, it is likely that ω ratios also vary within each subclade. We therefore performed the branch site models to identify candidate sites that are subject to positive selection for each branch, especially on the branches after duplication events. Of 72 branches, eight were detected under positive selection (branches A–H in fig. 2). The putative positive sites with posterior probabilities higher than 0.95 are given in table 2. Only one or two sites were suggested on branches A, F, and G, whereas multiple positive changes were assigned on the other five branches (B–E and H). Interestingly, these five branches are the ones right after gene duplication. Among them, 11 and 12 positive sites inferred on the branches leading to desat2 (C) and desatF (D), respectively, are the highest. About half of these sites (6/11 for Desat2 and 7/12 for DesatF) are located in the C-terminal regions (after residue 270 of supplementary fig. S2, Supplementary Material online), including sites around the third histidine box.
Table 2.
Putative Positively Selected Sites Inferred by Branch Site Models
| Branch | 2Δl | P Value | Number of Sites | Sites under Positive Selection |
| A | 12.142 | 4.93 × 10−04 | 1 | L106T |
| B | 12.269 | 4.61 × 10−04 | 7 | M135W, L147I, Q177T, I227L, C228A, K276G, T379V |
| C | 28.970 | 7.35 × 10−08 | 11 | W93Y, S98Q, S104G, V236I, I260Q, F316W, S342S, A361E, T362L, I375A, T381V |
| D | 17.067 | 3.61 × 10−05 | 12 | A88S, T119S, F180W, L189C, F250H, A270S, N288E, S292I, T295R, W300Y, T313S, K333R |
| E | 11.981 | 5.37 × 10−04 | 4 | Y83I, T107F, C228I, P252M |
| F | 11.493 | 6.99 × 10−04 | 1 | R209E |
| G | 12.885 | 3.31 × 10−04 | 2 | Y155I, Y274F |
| H | 12.694 | 3.67 × 10−04 | 4 | T100D, L106A, L111G, N282S |
NOTE.—Branches with significant P values (degree of freedom = 1) after Bonferroni correction under branch site models and putative adaptive sites with posterior probability higher than 0.95 in Bayes empirical Bayes analysis are listed. Numbers labeled on these sites indicate the sequence positions in the consensus sequence. Capital letters flanking the sites indicate the amino acid states before and after the change, respectively. S342S is the site involved in two nonsynonymous changes according to the model.
Expression Differences of desatF Homologs
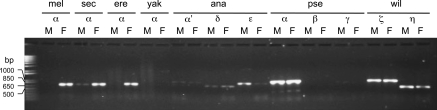
To investigate whether all the desatF homologs are functional loci, the expression patterns of these desatF genes were performed in both sexes for these Drosophila species (fig. 3). In D. melanogaster species subgroup, desatF-α was only expressed in females of D. melanogaster and D. erecta, predominantly expressed in females of Drosophila sechellia, and no expression in both sexes of D. simulans (Chertemps et al. 2006) and D. yakuba. In D. obscura species group, the desatF-α orthologs, GA20691 in D. pseudoobscura and GL15669 in D. persimilis, were expressed in both sexes. For all the paralogs of desatF-α, most of them were expressed in both sexes except desatF-β and desatF-γ. The expression of desatF-β was not detected in either sex, and desatF-γ was only expressed in males in both D. pseudoobscura and D. persimilis.
FIG. 3.—
RNA expression of desatF homologs (α, α′, β, γ, δ, ϵ, ζ, and η) in adult males (M) and females (F) of Drosophila melanogaster (mel), Drosophila sechellia (sec), Drosophila erecta (ere), Drosophila yakuba (yak), Drosophila pseudoobscura (pse), and Drosophila willistoni (wil) by RT-PCR with gene-specific primers.
Discussion
Comparative genomic studies revealing frequent gene gains and losses provide an opportunity to examine how genetic diversity shapes functional divergence. In this study, multiple gene gains and losses were identified in the desat1–desat2–desatF clade, and both desat2 and desatF lineages exhibit an accelerated rate of evolution as indicated by their overall ω values. More interestingly, most of the positively selected sites inferred from our analyses occurred on the branches right after duplication events. These putatively selected sites might be responsible for the functional divergence among desaturase genes. In desatF, multiple gene duplication events occurred independently in several Drosophila lineages. The changes of desatF expression patterns, most likely by recruiting new cis-regulatory elements, are concordant with duplication events. Because DesatF is responsible for diene production in D. melanogaster, the relationship between the gain and loss of desatF genes and diversification of the hydrocarbon profile in Drosophila is discussed below.
Birth-and-Death Process of Desaturase Genes
The frequent duplication and pseudogenization in the desat1–desat2–desatF clade follows the birth-and-death model (Ohta and Nei 1994). A conservative estimate suggests that recurrent gene duplication events occurred independently in the Sophophora lineage (fig. 1). As indicated by both theoretical models and empirical data, the most common fate of a duplicated gene is pseudogenization as most of the mutations are deleterious (Lynch and Conery 2000). In desaturase gene family, several desat2 and desatF alleles have undergone a pseudogenization or eliminated from the genome after duplication. A signature of degeneration, the 16-bp deletion in the promoter region of desat2, has been reported in D. melanogaster populations (Dallerac et al. 2000; Takahashi et al. 2001). In this study, we uncovered another degenerated allele of desat2 in D. erecta. A large portion of desat2 coding sequence has been deleted, and only the last exon and part of the 3′ untranslated region can be recognized in the genome. In desatF clade, we have assigned several gene losses after gene duplication, especially twice in desatF-α, one in the lineage leading to Sophophora and the other one in D. willistoni (fig. 1). It is clear that the pseudogenization level indicated by deletion, loss-of-function, and elimination from the genome in desatF (at least three losses, fig. 1) is higher than that in desat2 (one loss). This phenomenon is congruent with the genealogy of desat1–desat2–desatF clade (fig. 2 and supplementary fig. S1 [Supplementary Material online]), that is, desatF is duplicated from the ancestral branch earlier than desat2. Another indication of nonfunction is the absence of expression. No desatF expression could be detected in both sexes of D. simulans and D. yakuba (fig. 3). For D. simulans, nonsense mutation was not found in the current genome release, suggesting that this gene is either expressed in other developmental stages or at the early stage of pseudogenization. The desatF-α allele in D. yakuba reference genome is more likely to be a degenerated one with a frameshift mutation (10-bp deletion) causing a premature stop codon rather than a functional one with a de novo intron as annotated in the current genome release (CAF1).
The other possible fates of a duplicated gene are subfunctionalization or neofunctionalization (Force et al. 1999). In both cases, cis-regulatory evolution would be detected by gene expression changes. The relocation of desatF, like other retrogenes, would have a better chance to recruit new regulatory elements (reviewed in Long et al. 2003). All the insertion sites of desatF homologs are located in the intergenic regions. It is not clear if they had recruited some of the existing promoters or enhancers of the neighboring genes because most of the functional regulatory sequences are largely unknown. The only identified regulatory sequence, binding motif of Doublesex female protein (DSXF), is 9 bp in length that could also evolve de novo by mutation mechanism. The expression patterns of desatF-α are bisexual in D. pseudoobscura and D. persimilis but shift to be female biased in D. erecta, D. sechellia, and D. melanogaster. This derived female-biased desatF-α expression in these three species may be evolved by the recruitment of a DSXF motif in the 5′-flanking region of desatF-α on the branch leading to D. melanogaster species subgroup (Legendre et al. 2008). On the other hand, the fact that desatF-γ in D. pseudoobscura and D. persimilis switches to be male specific indicates that desatF-γ acquired the male-specific cis-regulatory element after duplicated from either desatF-α or desatF-β. If desatF-γ was directly duplicated from desatF-α, the male-specific expression of desatF-γ evolved after the relocation from X chromosome (XR, Muller element D) to autosome (2, Muller element E). This observation fits in the widely known pattern of dominant male expression of new retrogenes (Betrán et al. 2002; Dai et al. 2006).
Accelerated Protein Evolution and Functional Diversification of the Desaturase Gene Family
In addition to cis-regulatory evolution, functional differentiation of duplicate copies through accelerated protein evolution is extremely important for genetic novelty. In the desaturase gene family, upon branching off desat1, both desat2 and desatF gained novel functions through accelerated protein evolution. Genome-wide analyses on the substitution rates of paralogous genes revealed that accelerated protein evolution resulting in asymmetric divergence often observed in duplicated gene pairs. The asymmetric evolution rate can be contributed by relaxation of selective constrains, especially for young duplicates, and positive selection acting on beneficial mutations (Conant and Wagner 2003; Zhang et al. 2003). The rate asymmetry is greater in gene pairs duplicated through retrotransposition than in tandemly duplicated pairs (Cusack and Wolfe 2007). In the case of desat1–desat2–desatF evolution, the asymmetric evolution rate is no exception. The ancestral copy, desat1, evolves at the slowest rate. The tandemly duplicated copy, desat2, diverges at the moderate rate. The retrogene, desatF, evolves at the highest rate. Based on our phylogenetic analyses (fig. 2 and supplementary fig. S1 [Supplementary Material online]), both desat2 and desatF duplicated from the ancestral desat1 before the two subgenera, Drosophila and Sophophora, split around 62.9 Ma (Tamura et al. 2004). Given the long divergence time between these duplicated genes, the protein changes due to relaxation of selection only contribute to a small fraction of the total increase in protein evolution. The fact that functions of Desat1, Desat2, and DesatF have been demonstrated to be different in substrate selectivity in D. melanogaster (Dallerac et al. 2000; Chertemps et al. 2006, 2007) implies that the acquisition of new function is the key to the preservation of functional copies of desat2 and desatF.
In D. melanogaster, the ancestral Desat1 is an Δ9 desaturase which introduces the first double bond at the Δ9 position (Δ position is relative to the carboxyl end) of either the palmitic acid (C16:0) to produce ω7 fatty acids (ω position is relative to the methyl end) or the less preferred stearic acid (C18:0) to yield ω9 fatty acids (Wicker-Thomas et al. 1997; Dallerac et al. 2000). The tandemly duplicated gene, desat2, also encodes a Δ9 desaturase but switches its substrate preference to myristic acid (C14:0) to produce ω5 fatty acids (Dallerac et al. 2000). The intronless desatF in D. melanogaster, that is, desatF-α in this study, is possibly a Δ11–Δ15 desaturase, which performs the second desaturation at four carbons after the first double bonds in monosaturated ω5 and ω7 fatty acids with C22–C26 carbon length, subsequently leading to n,(n + 4)-Cm:2 dienes (cuticular hydrocarbons of m carbon atoms in length with two double bonds at the nth and (n + 4)th carbon positions), for example, heptacosadiene (7,11-C27:2 and 5,9-C27:2) and nonacosadiene (7,11-C29:2), in females of D. melanogaster (Chertemps et al. 2006, 2007; Legendre et al. 2008). Desat1 and Desat2 act on unsaturated fatty acids with different substrate preferences, whereas DesatF chooses longer monosaturated fatty acids as substrates. Amino acid changes accumulated around the regions of the catalytic sites on the desat2 and desatF branches might be responsible for such functional divergences. In desaturases, amino acid changes around the three conserved histidine box motifs and the C-terminal region might contribute to the regioselectivity and stereoselectivity (Fox et al. 1993; Libisch et al. 2000; Hoffmann et al. 2007; Meesapyodsuk et al. 2007). In our study, about 50% of the putative positively selected sites inferred on the branches leading to desat2 and desatF clades are concentrated. Nevertheless, the roles of these amino acid changes on the novel substrate selectivity of Desat2 and DesatF remain to be verified by functional assays.
What would be the major driving force shaping the functional diversification of desaturase gene family along Drosophila lineage? In D. melanogaster, loss-of-function allele of desat2 is responsible for cold tolerance that could be an adaptive trait when ancestral D. melanogaster population migrated out of Africa (Greenberg et al. 2003, 2006). On the other hand, because desatF is involved in the biosynthesis of cuticular dienes which are major female sex pheromones in some Drosophila species, it is possible that sexual selection is the main driving force, at least in some lineages, during desatF evolution. In D. melanogaster species subgroup, 7,11-C27:2 diene is the major female hydrocarbon in sexually dimorphic species including D. melanogaster and D. sechellia, whereas 7-C23:1 monoene is the major female hydrocarbon in monomorphic species including D. simulans and Drosophila mauritiana. The 7,11-C27:2 stimulates courtship of the two sexually dimorphic species but inhibits the two monomorphic species. Similarly, 7-C23:1 is also recognized by males as a chemical cue for sexual isolation between closely related species (Antony et al. 1985; Coyne et al. 1994; Ferveur and Sureau 1996). Accordingly, both natural and sexual selection might play important roles in shaping the accelerated evolution of desaturase genes.
Because desatF-α is responsible for the major cuticular diene in D. melanogaster, the potential roles of desatF-α homologs leading to diene diversification in Drosophila would be the next question to ask. To address this question, we first summarized the expression patters of desatF and the major cuticular dienes (table 3). In D. melanogaster, desatF-α is only expressed in females, resulting in 7,11-dienes (e.g., 7,11-C27:2 and 7,11-C29:2) to be the major hydrocarbons in females, whereas males have only monoenes (Chertemps et al. 2006; Legendre et al. 2008). In addition to D. melanogaster, the female-specific desatF-α expression in D. sechellia and D. erecta is also strongly associated with sexual dimorphic dienes. On the other hand, the lack of dienes in D. simulans and D. yakuba is correlated with lack of desatF-α expression. Also, no n,(n + 4)-Cm:2 dienes were detected in species lacking desatF-α, including D. ananassae, D. willistoni, D. mojavensis, and D. virilis. Outside the D. melanogaster species subgroup, desatF-α locus was independently lost in several lineages but remains in the two species of D. obscura species group, D. pseudoobscura and D. persimilis. The syntenic desatF-α orthologs are expressed bisexually in D. pseudoobscura and D. persimilis which show high abundant 5,9-dienes (e.g., 5,9-C25:2 and 5,9-C27:2) in both sexes. These new observations suggest that the ancestral function of DesatF-α is to produce n,(n + 4)-Cm:2 dienes. This ancestral function of desatF-α is conserved in all these Drosophila species but D. erecta. The major female-specific diene in D. erecta is tritriacontadiene (9,23-C33:2) in which the carbon length between two double bonds is 14. It is possible that DesatF-α of D. erecta has acquired a novel substrate regioselectivity to produce n,(n + 14)-Cm:2 dienes after leaving the D. yakuba–D. melanogaster lineage.
Table 3.
Summary of Major Cuticular Dienes and desatF-α Expression in Drosophila
| Major Cuticular Dienes |
desatF-α |
||||
| Species | Male | Female | References | Present | Expression |
| D. melanogaster | None | n,(n + 4)-C27:2; n,(n + 4)-C29:2 | Antony and Jallon (1982); Jallon (1984) | Yes | Female specific |
| 7,11-heptacosadiene; 5,9-heptacosadiene; 7,11-nonacosadiene | |||||
| D. simulans | None | None | Pechine et al. (1985); Jallon and David (1987) | Yes | No |
| D. sechellia | n,(n + 4)-C27:2 | n,(n + 4)-C27:2 | Jallon and David (1987); Cobb et al. (1989) | Yes | Female biased |
| 7,11-heptacosadiene (only ∼1.4% of total cuticular hydrocarbons) | 7,11-heptacosadiene | ||||
| D. yakuba | None | None | Mas and Jallon (2005) | Yes | No |
| D. erecta | None | n,(n + 14)-C33:2 | Pechine et al. (1988) | Yes | Female specific |
| 9,23-tritriacontadiene | |||||
| D. ananassae | n,(n + 20)-C31:2; n (n + 22)-C31:2 | n,(n + 20)-C31:2; n,(n + 22)-C31:2 | Doi et al. 1997 | No | — |
| 5,25-hentriacontadiene; 4,26-hentriacontadiene | 5,25-hentriacontadiene; 4,26-hentriacontadiene | ||||
| D. pseudoobscura | n,(n + 4)-C25:2; n (n + 4)-C27:2 | n,(n + 4)-C25:2; n,(n + 4)-C27:2 | Blomquist et al. (1985) | Yes | Bisexual |
| 5,9-pentacosadiene; 5,9-heptacosadiene | 5,9-pentacosadiene; 5,9-heptacosadiene | ||||
| D. persimilis | n,(n + 4)-C25:2 | n,(n + 4)-C25:2 | Noor and Coyne 1996 | Yes | Bisexual |
| 5,9-pentacosadiene | 5,9-pentacosadiene | ||||
| D. willistoni | n,(n + (16∼22))-Cm:2 | n,(n + (16∼22))-Cm:2 | Wang CC, Fang S, unpublished data | No | — |
A relationship between the number of expressed desatF homologs and the diene diversification in these species has also been observed. The expression of desatF-α, the only desatF homolog in D. melanogaster subgroup, contributes to the diene production of either n,(n + 4)-Cm:2 or n,(n + 14)-Cm:2. Outside the D. melanogaster subgroup, there is more than one desatF homolog. In D. ananassae, the various cuticular dienes, for example n,(n + 20)-Cm:2 and n,(n + 22)-Cm:2, are correlated with the expression of multiple desatF homologs, that is, desatF-α′, desatF-δ, and desatF-ϵ (fig. 3). Similarly, the lineage-specific desatF-ζ and desatF-η in D. willistoni might contribute to the complicated C33 and C35 dienes, including n,(n + 16)-Cm:2, n,(n + 18)-Cm:2, n,(n + 20)-Cm:2, and n,(n + 22)-Cm:2. In D. pseudoobscura, the desatF-γ, which is lowly expressed in males, might be involved in synthesizing the low quantity of the unusual n,(n + 9)-Cm:2 or n,(n + 11)-Cm:2 diene. As these unusual dienes appear in very low quantities, whether any of these dienes is sexual dimorphic remains unknown. Based on our observation on the number of desatF homologs and the diene complexity, we hypothesize that different DesatF exhibit different regioselectivity of the second desaturation of long chain fatty acid. Nevertheless, further functional assay is necessary to test the hypothetic roles of these desatF homologs on the diene diversity. It would not be surprised if functional divergence at the desat1–desat2–desatF clade drives the cuticular hydrocarbon diversification among Drosophila species as studies on the desaturase gene family and the pheromone diversification between closely related species in moths have been well documented (Knipple et al. 2002; Roelofs and Rooney 2003; Xue et al. 2007).
Supplementary Material
Supplementary table S1 and figures S1 and S2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
We thank Ziheng Yang for his advices and comments on an early draft; Esther Betrán, Y. Henry Sun, J. J. Emerson, Yu-Ping Poh, Alex Hon-Tsen Yu, Hurng-Yi Wang, Chi-Chun Chen, Hwei-yu Chang, and Ya-Jen Cheng for their comments and helpful discussion; Wan-Ju Shen for helping in the RNA expression; and Hsiao-Yung Ho for consulting in cuticular hydrocarbon analyses. We also thank the anonymous reviewers for their insightful comments. The genome sequences of the 12 Drosophila species provided by Fly community are greatly appreciated. This work was funded by the National Science Council (Taiwan, Republic of China) grants to S.F., C.T.T., and S.C.T.
References
- Anisimova A, Yang Z. Multiple hypothesis testing to detect adaptive protein evolution affecting individual branches and sites. Mol Biol Evol. 2007;24:1219–1228. doi: 10.1093/molbev/msm042. [DOI] [PubMed] [Google Scholar]
- Antony C, Davis TL, Carlson DA, Pechiné JM, Jallon JM. Compared behavioral responses of male Drosophila melanogaster (Canton-S) to natural and synthetic aphrodisiacs. J Chem Ecol. 1985;11:1617–1629. doi: 10.1007/BF01012116. [DOI] [PubMed] [Google Scholar]
- Antony C, Jallon JM. The chemical basis for sex recognition in Drosophila melanogaster. J Insect Physiol. 1982;28:873–880. [Google Scholar]
- Bai Y, Casola C, Feschotte C, Betrán E. Comparative genomics reveals a constant rate of origination and convergent acquisition of functional retrogenes in Drosophila. Genome Biol. 2007;8:R11. doi: 10.1186/gb-2007-8-1-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betrán E, Thornton K, Long M. Retroposed new genes out of the X in Drosophila. Genome Res. 2002;12:1854–1859. doi: 10.1101/gr.604902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomquist GJ, Toolson EC, Nelson DR. Epicuticular hydrocarbons of Drosophila pseudoobscura (Diptera; Drosophilidae): identification of unusual alkadiene and alkatriene positional isomers. Insect Biochem. 1985;15:25–34. [Google Scholar]
- Chertemps T, Duportets L, Labeur C, Ueda R, Takahashi K, Saigo K, Wicker-Thomas C. A female-biased expressed elongase involved in long-chain hydrocarbon biosynthesis and courtship behavior in Drosophila melanogaster. Proc Natl Acad Sci USA. 2007;104:4273–4278. doi: 10.1073/pnas.0608142104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chertemps T, Duportets L, Labeur C, Ueyama M, Wicker-Thomas C. A female-specific desaturase gene responsible for diene hydrocarbon biosynthesis and courtship behaviour in Drosophila melanogaster. Insect Mol Biol. 2006;15:465–473. doi: 10.1111/j.1365-2583.2006.00658.x. [DOI] [PubMed] [Google Scholar]
- Clark NL, Aagaard JE, Swanson WJ. Evolution of reproductive proteins from animals and plants. Reproduction. 2006;131:11–22. doi: 10.1530/rep.1.00357. [DOI] [PubMed] [Google Scholar]
- Cobb M, Burnet B, Blizard R, Jallon JM. Courtship in Drosophila sechellia: its structure, functional aspects, and relationship to those of other members of the Drosophila melanogaster species group. J Insect Behav. 1989;2:63–89. [Google Scholar]
- Conant GC, Wagner A. Asymmetric sequence divergence of duplicate genes. Genome Res. 2003;13:2052–2058. doi: 10.1101/gr.1252603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne JA, Crittenden AP, Mah K. Genetics of a pheromonal difference contributing to reproductive isolation in Drosophila. Science. 1994;265:1461–1464. doi: 10.1126/science.8073292. [DOI] [PubMed] [Google Scholar]
- Coyne JA, Wicker-Thomas C, Jallon JM. A gene responsible for a cuticular hydrocarbon polymorphism in Drosophila melanogaster. Genet Res. 1999;73:189–203. doi: 10.1017/s0016672398003723. [DOI] [PubMed] [Google Scholar]
- Crosby MA, Goodman JL, Strelets VB, Zhang P, Gelbart WM the FlyBase Consortium. FlyBase: genomes by the dozen. Nucleic Acids Res. 2007;35:D486–D491. doi: 10.1093/nar/gkl827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusack BP, Wolfe KH. Not born equal: increased rate asymmetry in relocated and retrotransposed rodent gene duplicates. Mol Biol Evol. 2007;24:679–686. doi: 10.1093/molbev/msl199. [DOI] [PubMed] [Google Scholar]
- Dai H, Yoshimatsu TF, Long M. Retrogene movement within- and between-chromosomes in the evolution of Drosophila genomes. Gene. 2006;385:96–102. doi: 10.1016/j.gene.2006.04.033. [DOI] [PubMed] [Google Scholar]
- Dallerac R, Labeur C, Jallon JM, Knipple DC, Roelofs WL, Wicker-Thomas C. A Δ9 desaturase gene with a different substrate specificity is responsible for the cuticular diene hydrocarbon polymorphism in Drosophila melanogaster. Proc Natl Acad Sci USA. 2000;97:9449–9454. doi: 10.1073/pnas.150243997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi M, Nemoto T, Nakanishi H, Kuwahara Y, Oguma Y. Behavioral response of males to major sex pheromone component, (Z,Z)-5,25-hentriacontadiene, of Drosophila ananassae females. J Chem Eco. 1997;23:2067–2078. [Google Scholar]
- Drosophila 12 Genomes Consortium. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Fang S, Takahashi A, Wu CI. A mutation in the promoter of desaturase 2 is correlated with sexual isolation between Drosophila behavioral races. Genetics. 2002;162:781–784. doi: 10.1093/genetics/162.2.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferveur JF, Sureau G. Simultaneous influence on male courtship of stimulatory and inhibitory pheromones produced by live sex-mosaic Drosophila melanogaster. Proc R Soc Lond B Biol Sci. 1996;263:967–973. doi: 10.1098/rspb.1996.0143. [DOI] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox BG, Shanklin J, Somerville C, Munck E. Stearoyl-acyl carrier protein Δ9 desaturase from Ricinus communis is a diiron-oxo protein. Proc Natl Acad Sci USA. 1993;90:2486–2490. doi: 10.1073/pnas.90.6.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg AJ, Moran JR, Coyne JA, Wu CI. Ecological adaptation during incipient speciation revealed by precise gene replacement. Science. 2003;302:1754–1757. doi: 10.1126/science.1090432. [DOI] [PubMed] [Google Scholar]
- Greenberg AJ, Moran JR, Fang S, Wu CI. Adaptive loss of an old duplicated gene during incipient speciation. Mol Biol Evol. 2006;23:401–410. doi: 10.1093/molbev/msj045. [DOI] [PubMed] [Google Scholar]
- Hahn MW, Han MV, Han SG. Gene family evolution across 12 Drosophila genomes. PLoS Genet. 2007;3:2135–2146. doi: 10.1371/journal.pgen.0030197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto K, Yoshizawa AC, Okuda S, Kuma K, Goto S, Kanehisa M. The repertoire of desaturases and elongases reveals fatty acid variations in 56 eukaryotic genomes. J Lipid Res. 2008;49:183–191. doi: 10.1194/jlr.M700377-JLR200. [DOI] [PubMed] [Google Scholar]
- Hoffmann M, Hornung E, Bushch S, Kassner N, Ternes P, Braus GH, Feussner I. A small membrane-peripheral region close to the active center determines regioselectivity of membrane-bound fatty acid desaturases from Aspergillus nidulans. J Biol Chem. 2007;282:26666–26674. doi: 10.1074/jbc.M705068200. [DOI] [PubMed] [Google Scholar]
- Howard RW, Blomquist GJ. Chemical ecology and biochemistry of insect hydrocarbons. Ann Rev Entomol. 1982;27:149–172. doi: 10.1146/annurev-ento-031620-071754. [DOI] [PubMed] [Google Scholar]
- Howard RW, Blomquist GJ. Ecological, behavioral, and biochemical aspects of insect hydrocarbons. Ann Rev Entomol. 2005;50:371–393. doi: 10.1146/annurev.ento.50.071803.130359. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Jallon JM. A few chemical words exchanged by Drosophila during courtship and mating. Behav Genet. 1984;14:441–478. doi: 10.1007/BF01065444. [DOI] [PubMed] [Google Scholar]
- Jallon JM, David JR. Variations in cuticular hydrocarbons among the eight species of the Drosophila melanogaster subgroup. Evolution. 1987;41:294–302. doi: 10.1111/j.1558-5646.1987.tb05798.x. [DOI] [PubMed] [Google Scholar]
- Karolchik D, Baertsch R, Diekhans M, et al. (13 co-authors) The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipple DC, Rosenfield CL, You MM, Jeong SE. Evolution of the integral membrane desaturase gene family in moths and flies. Genetics. 2002;162:1737–1752. doi: 10.1093/genetics/162.4.1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labeur C, Dallerac R, Wicker-Thomas C. Involvement of desat1 gene in the control of Drosophila melanogaster pheromone biosynthesis. Genetica. 2002;114:269–274. doi: 10.1023/a:1016223000650. [DOI] [PubMed] [Google Scholar]
- Legendre A, Miao XX, Da Lage JL, Wicker-Thomas C. Evolution of a desaturase involved in female pheromonal cuticular hydrocarbon biosynthesis and courtship behavior in Drosophila. Insect Biochem Mol Biol. 2008;38:244–255. doi: 10.1016/j.ibmb.2007.11.005. [DOI] [PubMed] [Google Scholar]
- Libisch B, Michaelson LV, Lewis MJ, Shewry PR, Napier JA. Chimeras of Δ6-fatty acid and Δ8-shingolipid desaturases. Biochem Biophys Res Commun. 2000;279:779–785. doi: 10.1006/bbrc.2000.4023. [DOI] [PubMed] [Google Scholar]
- Long M, Betrán E, Thornton K, Wang W. The origin of new genes: glimpses from the young and old. Nat Rev Genet. 2003;4:865–875. doi: 10.1038/nrg1204. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Mas F, Jallon JM. Sexual isolation and cuticular hydrocarbon differences between Drosophila santomea and Drosophila yakuba. J Chem Ecol. 2005;31:2747–2752. doi: 10.1007/s10886-005-7570-5. [DOI] [PubMed] [Google Scholar]
- Meesapyodsuk D, Reed DW, Covello PS, Qiu X. Primary structure, regioselectivity, and evolution of the membrane-bound fatty acid desaturases of Claviceps purpurea. J Biol Chem. 2007;282:20191–20199. doi: 10.1074/jbc.M702196200. [DOI] [PubMed] [Google Scholar]
- Noor MA, Coyne JA. Genetics of a difference in cuticular hydrocarbons between Drosophila pseudoobscura and D. persimilis. Genet Res. 1996;68:117–123. doi: 10.1017/s0016672300034005. [DOI] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. Berlin (Germany): Springer-Verlag; 1970. [Google Scholar]
- Ohta T, Nei M. Divergent evolution and evolution by the birth-and-death process in the immunoglobulin VH gene family. Mol Biol Evol. 1994;11:469–482. doi: 10.1093/oxfordjournals.molbev.a040127. [DOI] [PubMed] [Google Scholar]
- Pechine JM, Antony C, Jallon JM. Precise characterization of cuticular compounds in young Drosophila by mass spectrometry. J Chem Ecol. 1988;14:1071–1085. doi: 10.1007/BF01019336. [DOI] [PubMed] [Google Scholar]
- Pechine JM, Pereza F, Antony C, Jallon JM. A further characterization of Drosophila cuticular monoenes using a mass spectrometry method to localize double bonds in complex mixtures. Anal Biochem. 1985;145:177–182. doi: 10.1016/0003-2697(85)90344-6. [DOI] [PubMed] [Google Scholar]
- Roelofs WL, Rooney AP. Molecular genetics and evolution of pheromone biosynthesis in Lepidoptera. Proc Natl Acad Sci USA. 2003;100:9179–9184. doi: 10.1073/pnas.1233767100a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. PAUP*. Phylogenetic analysis using parsimony (*and other methods) Sunderland (MA): Sinauer Associates; 2002. [Google Scholar]
- Takahashi A, Tsaur SC, Coyne JA, Wu CI. The nucleotide changes governing cuticular hydrocarbon variation and their evolution in Drosophila melanogaster. Proc Natl Acad Sci USA. 2001;98:3920–3925. doi: 10.1073/pnas.061465098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Subramanian S, Kumar S. Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol Biol Evol. 2004;21:36–44. doi: 10.1093/molbev/msg236. [DOI] [PubMed] [Google Scholar]
- Wang X, Grus WE, Zhang J. Gene losses during human origins. PLoS Biol. 2006;4:366–377. doi: 10.1371/journal.pbio.0040052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker-Thomas C, Henriet C, Dallerac R. Partial characterization of a fatty acid desaturase gene in Drosophila melanogaster. Insect Biochem Mol Biol. 1997;27:963–972. doi: 10.1016/s0965-1748(97)00077-5. [DOI] [PubMed] [Google Scholar]
- Wilson RJ, Goodman JL, Strelets VB, the FlyBase Consortium FlyBase: integration and improvements to query tools. Nucleic Acids Res. 2008;36:D588–D593. doi: 10.1093/nar/gkm930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue B, Rooney AP, Kajikawa M, Okada N, Roelofs WL. Novel sex pheromone desaturases in the genomes of corn borers generated through gene duplication and retroposon fusion. Proc Natl Acad Sci USA. 2007;104:4467–4472. doi: 10.1073/pnas.0700422104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. Computational molecular evolution. Oxford: Oxford University Press; 2006. pp. 176–177. [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Zhang P, Gu Z, Li WH. Different evolutionary patterns between young duplicate genes in the human genome. Genome Biol. 2003;4:R56. doi: 10.1186/gb-2003-4-9-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.