Abstract
The p53tumour suppressor is regulated mainly by Mdm2, an E3 ubiquitin ligase that promotes the ubiquitylation and proteasome-mediated degradation of p53. Many agents that induce p53 are inhibitors of transcription, suggesting that the p53 pathway can detect a signal(s) arising from transcriptional malfunction. Mdm2 associates with TAFII250, a component of the general transcription factor TFIID. Inactivation of TAFII250 in ts13 cells, which express a temperature-sensitive mutant of TAFII250, leads to the induction of p53 and cell cycle arrest. In the present study, we show that TAFII250 stimulates the ubiquitylation and degradation of p53 in a manner that is dependent upon Mdm2 and requires its acidic domain. Mechanistically, TAFII250 downregulates Mdm2 auto-ubiquitylation, leading to Mdm2 stabilization, and promotes p53-Mdm2 association through a recently defined second binding site in the acidic domain of Mdm2. These data provide a novel route through which TAFII250 can directly influence p53 levels and are consistent with the idea that the maintenance of p53 turnover is coupled to the integrity of RNA polymerase II transcription.
Keywords: p53, Mdm2, TAFII250, transcription factor, ubiquitylation
Introduction
The p53 tumour suppressor protein is a short-lived transcription factor that is stabilized and activated, mainly through post-translational mechanisms, in response to a wide variety of cellular stresses, including DNA damage and hyper-proliferation (reviewed by Vousden and Lu (2002) and Yee and Vousden (2005)). Activated p53 orchestrates the transactivation or transrepression of many target genes leading to growth arrest, senescence or apoptosis.
The stability of p53 is regulated mainly by the oncoprotein Mdm2, an E3 ubiquitin ligase that mediates the ubiquitylation and rapid proteasome-dependent degradation of p53 and Mdm2 itself (Fang et al., 2000; Honda and Yasuda, 2000); reviewed by Michael and Oren (2003). p53 and Mdm2 form an autoregulatory loop in which p53 positively regulates mdm2 expression while Mdm2 negatively regulates p53 levels and activity (Wu et al., 1993). Cellular stresses that induce p53 disrupt the p53–Mdm2 interaction with the effect of attenuating p53 degradation (Ashcroft et al., 2000).
Mdm2 is a nuclear phosphoprotein that contains several conserved regions including an N-terminal p53-binding domain, a C-terminal RING finger domain required to mediate ubiquitylation of p53 and auto-ubiquitylation (Fang et al., 2000; Honda and Yasuda, 2000), and a central acidic region that is important for p53 degradation (Argentini et al., 2001; Zhu et al., 2001; Blattner et al., 2002; Kawai et al., 2003; Meulmeester et al., 2003). Several proteins interact with the acidic domain including p14ARF (Midgley et al., 2000), the TATA-binding protein (TBP) (Leveillard and Wasylyk, 1997) and a number of other transcriptional proteins and ribosomal proteins; reviewed by Ganguli and Wasylyk (2003). Importantly, the acidic domain has recently been identified as a second point of contact between Mdm2 and p53 that interacts with a critical ubiquitylation signal in the core domain of p53 (Shimizu et al., 2002; Kulikov et al., 2006; Wallace et al., 2006; Yu et al., 2006).
The TBP-associated factor (TAF), TAFII250 (also designated CCG1 or TAF1 in humans), is the largest component of the general transcription factor TFIID and acts as a scaffold for the assembly of TFIID from TBP and a number of other TAFs (reviewed by Wasserman and Sauer (2001). Functionally, TAFII250 mediates promoter recognition and DNA binding by TFIID, alteration of chromatin structure, and posttranslational modification of general transcription factors, thereby facilitating transcriptional initiation. TAFII250 is an essential protein and is required for progression of the cell cycle and repression of apoptosis in mammalian cells (Wasserman and Sauer, 2001). Interestingly, TAFII250 interacts directly with the RING finger domain of Mdm2 (Leveillard and Wasylyk, 1997).
Blockage of RNA polymerase II transcription is thought to be a key factor in the induction of p53 by agents that elicit DNA damage (Ljungman and Lane, 2004). However, the mechanisms through which failed transcription activates p53 are only partly understood. The BHK-derived cell lines, ts13 and tsBN462, encode a temperature sensitive mutant of TAFII250 (G690D) that is active at 33°C but dysfunctional when shifted to 39°C (Sekiguchi et al., 1988). At the non-permissive temperature, ts13 cells undergo a G1 phase cell cycle arrest in a manner that shows striking similarity to a DNA damage response, and is dependent upon ATR-mediated phosphorylation and activation of p53 (Buchmann et al., 2004). Additionally, induction of the p53 pathway under these conditions is likely to involve decreased TAFII250-dependent expression of Mdm2 (Wasylyk and Wasylyk, 2000) as well as direct phosphorylation of p53 itself by TAFII250 (Li et al., 2004). Failure to express key TAFII250-dependent cell cycle-specific genes, such as cyclins A, D1 and D3, is also like to contribute to growth arrest at the nonpermissive temperature (Suzuki-Yagawa et al., 1997).
In the present study, we confirm that p53 is induced in ts13 cells grown at restrictive temperature, independently of a heat shock response. We also show that TAFII250 interacts with Mdm2 in vivo and promotes Mdm2-mediated degradation of p53. Using an in vivo ubiquitylation assay, we show that TAFII250 stimulates p53 ubiquitylation in a manner that is dependent upon Mdm2 and its acidic domain. The mechanism of this effect involves TAFII250-mediated inhibition of Mdm2 auto-ubiquitylation and stabilization of Mdm2, as well as stimulation of p53-Mdm2 association through the acidic domain of Mdm2.
Results
TAFII250 and Mdm2 endogenous proteins interact in cultured cells
To confirm the previously reported interaction between TAFII250 and Mdm2 in cultured cells (Leveillard and Wasylyk, 1997), immunoprecipitation analysis was performed using extracts of the osteosarcoma cell line SJSA (which expresses elevated levels of endogenous Mdm2). Proteins were immunoprecipitated with anti-TAFII250 (clone 6B3) and anti-Mdm2 (SMP14 and 4B2) antibodies. As shown in Figure 1 TAFII250 was observed to immunoprecipitate with an anti-TAFII250 antibody and to co-immunoprecipitate Mdm2, but was absent when an irrelevant antibody was used (upper panel). Similarly, in the reciprocal analysis, Mdm2 was found to co-immunoprecipitate when the TAFII250 antibody was used. These data confirm that Mdm2 and TAFII250 associate in vivo (Leveillard and Wasylyk, 1997).
Figure 1.
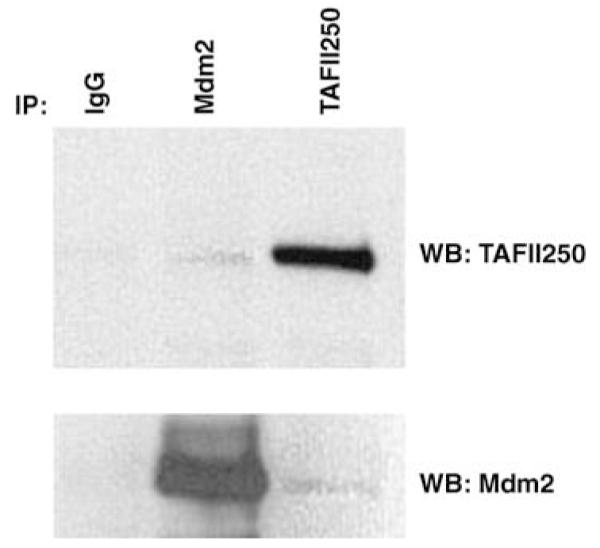
Mdm2 interacts with the general transcription factor TFIID in cultured cells. SJSA cells (6 × 107) were harvested and proteins in the extracts were immunoprecipitated with anti-TAFII250 monoclonal antibody (clone 6B3), anti-Mdm2 monoclonal antibodies (SMP14 and 4B2) and IgG mouse as a control. Immunoprecipitates were analysed by Western blotting with anti-TAFII250 antibody or anti-Mdm2 antibodies as indicated.
TAFII250 is implicated in p53-Mdm2 feedback loop
To investigate the role of TAFII250 in the p53-Mdm2 pathway ts13 cells (Sekiguchi et al., 1988) were maintained at 33°C then shifted to 39°C for different periods of time. Cell extracts were analysed by Western blotting for p53 and p21 (as p21 is expressed from a p53-responsive promoter). As shown in Figure 2a the p53 levels increased reaching a maximum between 6 and 8 h and was accompanied by increases in p21 levels. Lack of any significant p53 or p21 induction in BHK21 cells (the parental line for ts13) (Figure 2b), or in ts13 cells in which wild-type TAFII250 was expressed ectopically (Figure 2c), indicated that p53 induction did not occur simply as a result of heat shock. These data show that reduction of TAFII250 activity leads to p53 activation and are in keeping with the conclusions of others that TAFII250 regulates p53 levels and the p53 response (Wasylyk and Wasylyk, 2000; Buchmann et al., 2004; Li et al., 2004).
Figure 2.
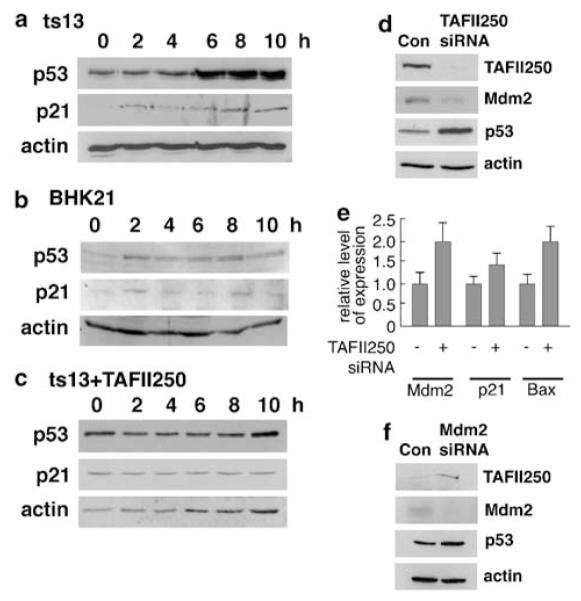
Mdm2 and TAFII250 are reciprocally regulated. ts13 cells (a) or the parental line BHK21 (b) were maintained at 33°C then shifted to 39°C and subsequently harvested and lysed in 2 × SDS loading buffer at different time intervals post-shift (0, 2, 4, 6, 8, 10 h). The cell extracts were analysed by Western blotting using DO-1 antibody for p53, the C-19-G antibody (Santa Cruz) for p21 and 20–33 antibody (Sigma-Aldrich Company Ltd., Dorset, UK) for actin.(c) ts13 cells were transfected with 10 μg of TAFII250 expression vector per 10 cm plate. The next day, cells were shifted to 39°C, harvested at different time points and analysed by western blotting for p53 and actin protein levels. Transfection efficiencies were generally in the range of 50–70% in this experiment. (d–f) U2OS cells were transfected with siRNA duplex for TAFII250 (d and e) or Mdm2 (f). As controls, the cells were transfected with siRNA duplexes of random sequence (d–f). Cells transfected with siRNA were harvested after 24 h (e and f) or 48 h (d). Levels of TAFII250, Mdm2 (monoclonal antibody 4B2), p53 and actin proteins were analysed by Western blotting (d and f) as above. (e) The relative levels Mdm2, p21 and Bax mRNAs in the TAFII250 siRNA-treated as compared with the scrambled siRNA-treated cells were measured by Taqman.
The relationship between TAFII250 and the p53-Mdm2 pathway was confirmed following knock down of endogenous wild-type TAFII250 in U2OS cells by siRNA. Consistent with the results obtained following inactivation of temperature-sensitive TAFII250, decreased levels of wild-type TAFII250 resulted in an increase in p53 levels (Figure 2d). This was accompanied by a stimulation of expression of the p21, Bax and Mdm2 genes (Figure 2e) consistent with the activation of p53. Notably, however, Mdm2 protein levels were reduced following TAFII250 knock down (Figure 2d), contrasting with the increased (presumably p53-dependent) expression of the Mdm2 gene. These observations, together with the association of TAFII250 and Mdm2 (Figure 1), suggested the possibility that TAFII250 might regulate the level of Mdm2 protein (see below). The induction of Mdm2 expression following depletion of TAFII250 levels is also in sharp contrast to previously published findings that TAFII250 stimulates Mdm2 expression (Wasylyk and Wasylyk, 2000). Interestingly, knock down of Mdm2 in these cells resulted in a stimulation of the levels of both p53 and TAFII250 (Figure 2f), suggesting that there is a reciprocal regulation between Mdm2 and TAFII250.
TAFII250 promotes the ubiquitylation and turnover of p53
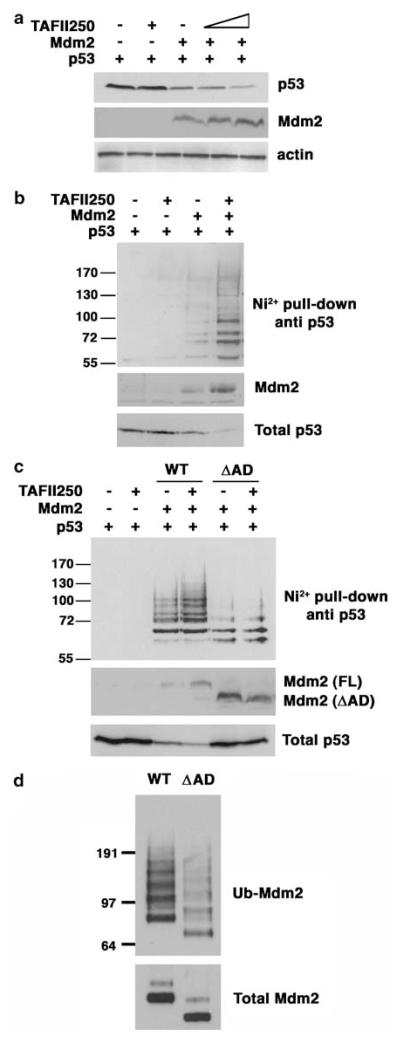
To explore the possibility that TAFII250 might regulate Mdm2 by protein–protein interaction, H1299 cells (p53-null cells) were co-transfected with plasmids encoding Mdm2 and p53 together with different amounts of a plasmid expressing TAFII250. Western blot analysis of the extracts show that p53 levels decreased when Mdm2 was expressed, whereas expression of TAFII250 alone had no effect on p53 levels (Figure 3a). Significantly, however, co-expression of TAFII250 stimulated the Mdm2-mediated reduction in p53 levels, consistent with the idea that TAFII250 can mediate its effect on p53 through modulating the activity of Mdm2.
Figure 3.
TAFII250 promotes ubiquitylation and degradation of p53. (a) H1299 cells were cotransfected with 1 μg of p53, 5 μg of Mdm2 and 5 and 10 μg of TAFII250 expression vectors. Cells were harvested 36 h after transfection and lysed with 2 × SDS sample buffer. The cells extracts were analysed for p53 protein levels. (b) Ni2+ pull-down was performed using extracts of H1299 cells that had been transfected with 1 μg of p53, 1 μg of Mdm2, 1 μg of TAFII250 and 2 μg of His-Ubiquitin expression vectors. The His-tagged ubiquitylated proteins were purified with Ni2+ agarose beads and analysed by western blotting using anti-p53 antibody DO-1. (c) p53 ubiquitylation was measured in H1299 cells transfected with plasmids encoding p53, TAFII250 and Mdm2 (wild-type and ΔAD mutant) as indicated. (d) Mdm2 (wild-type and ΔAD mutant) auto-ubiquitylation was measured in H1299 cells transfected with plasmids encoding wild type or ΔAD mutant Mdm2.
To investigate whether TAFII250 interferes with p53 ubiquitylation, an in vivo ubiquitylation assay was used in which H1299 cells were co-transfected with plasmids expressing wild-type human p53, Mdm2 and His-tagged ubiquitin. At 36 h post-transfection, the cells were lysed in 6 M guanidinium buffer, conditions that prevent deubiquitylation of proteins and disrupt any non-covalent protein–protein interactions. The His-ubiquitylated proteins were purified by affinity chromatography using Ni2+-agarose beads, then subsequently eluted and analysed by Western blotting with the anti-p53 antibody, DO-1. Figure 3b shows that p53 is ubiquitylated by Mdm2 but not by TAFII250. Significantly, p53 ubiquitylation increased when TAFII250 was co-expressed with Mdm2 and this was accompanied by a concomitant increase in the degradation of p53. Notably, the Mdm2 levels were higher in the presence of TAFII250. These data indicate that TAFII250 can promote a reduction in p53 levels occurs through stimulation of Mdm2-mediated ubiquitylation of p53, and that this may be brought about through a TAFII250-dependent increase in Mdm2 levels.
Deletion of the Mdm2 acidic domain inhibits Mdm2-mediated ubiquitylation of p53 (Kawai et al., 2003; Meulmeester et al., 2003). Consistent with these findings, an Mdm2 ΔAD mutant lacking the acidic domain (amino acids 222–270) generated only a low level of ubiquitylation of p53 and was refractory to the presence of TAFII250 (Figure 3c). Significantly, the levels of the Mdm2 ΔAD mutant were not stimulated by TAFII250. This again suggests that the mechanism may involve a TAFII250-dependent increase in Mdm2 levels. (Note, however, that the levels of the ΔAD mutant were significantly higher than those of wild-type Mdm2 (Figure 3c, bottom panel). This reflects impaired autoubiquitylation activity in the ΔAD mutant (as measured in the presence of MG132 to stabilize the proteins; Figure 3d)).
TAFII250 increases Mdm2 levels and downregulates Mdm2 auto-ubiquitylation
To confirm whether TAFII250 can stimulate Mdm2 levels, H1299 cells were transfected with a plasmid expressing Mdm2 together with increasing amounts of the plasmid expressing TAFII250. Western analysis of the cell extracts revealed a TAFII250-dose-dependent increase in the levels of Mdm2 (Figure 4a) consistent with the idea that TAFII250 might stabilize Mdm2.
Figure 4.
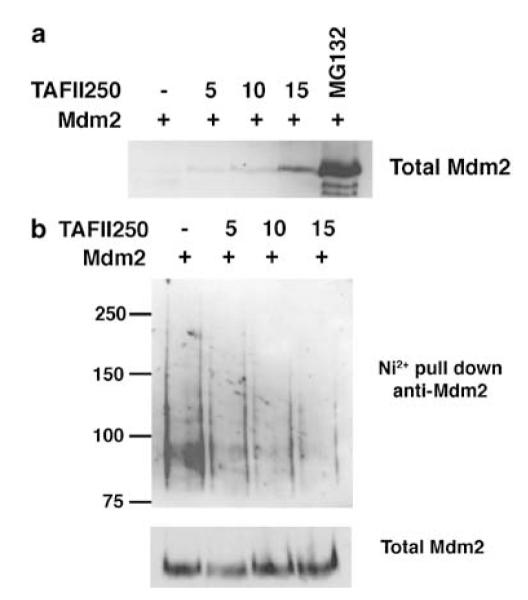
TAFII250 increases Mdm2 levels and downregulates Mdm2 auto-ubiquitylation. (a) H1299 cells were transfected with 5 μg of Mdm2 together with increasing amounts of TAFII250 expression vector, as indicated in the figure. 20 μM of MG132 was added to one of the plates for 4 h. Total cell extracts were analysed by Western blotting using the anti-Mdm2 antibodies SMP14 and D12. (b) The transfections were performed as above but in the presence of 2 μg of the His-Ubiquitin expression vector. Additionally, the cells were treated with 20 μM MG132 for 4 h before harvest.
In addition to controlling the levels of p53, Mdm2 can also promote its own ubiquitylation and proteasome-dependent degradation (Fang et al., 2000; Honda and Yasuda, 2000). To determine whether TAFII250 could influence Mdm2 auto-ubiquitylation, H1299 cells were transfected with plasmids expressing Mdm2 and His-tagged ubiquitin, together with increasing amounts of the TAFII250 expression plasmid. This experiment was carried out in presence of 20 μM of the proteasome inhibitor, MG132, for 4 h before harvesting to block Mdm2 degradation, thereby permitting comparison of ubiquitylation at equal levels of Mdm2. As before, His-tagged proteins were purified on Ni2+ resin and ubiquitylated Mdm2 was detected by Western blotting. The data (Figure 4b) show that TAFII250 stimulates a dose-dependent reduction in Mdm2 auto-ubiquitylation. Interestingly, a similar effect has been observed following the phosphorylation of Mdm2 by AKT (Feng et al., 2004). These data suggest that TAFII250 can regulate the ability of Mdm2 to undergo auto-ubiquitylation leading to the stabilization of Mdm2.
TAFII250 increases the half-life of Mdm2
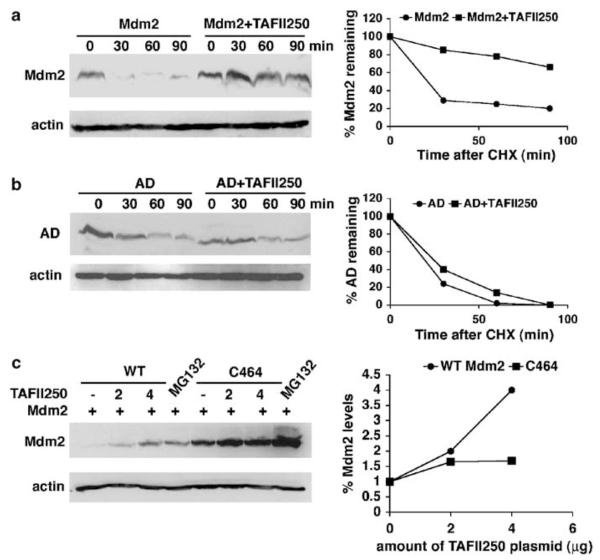
To determine whether TAFII250-mediated inhibition of auto-ubiquitylation affected the half-life of Mdm2, H1299 cells were co-transfected with Mdm2 and TAFII250 expression vectors, and at 36 h post-transfection, the cells were treated with 10 μg/ml of the translation inhibitor, cycloheximide. Subsequently, at different time points, cell extracts were prepared and the remaining Mdm2 protein levels determined by Western blotting. As shown in Figure 5a, the half-life of Mdm2 increased from 37 to 133 min upon TAFII250 expression, consistent with the idea that TAFII250 protects Mdm2 from degradation. In contrast to wild-type Mdm2, TAFII250 had no effect on the turnover of the acidic domain mutant (Figure 5b), indicating that the acidic domain plays an important role in the stabilization mechanism.
Figure 5.
TAFII250 stabilizes Mdm2. (a) Mdm2 half-life was measured in the presence and absence of TAFII250. H1299 cells were transfected with 5 μg of Mdm2 expression vector, with 5 μg of TAFII250 expression vector or empty vector. At 36 h post-transfection the cells were treated with 10 μg/ml of cycloheximide and harvested at different time points as indicated. Extracts were analysed by Western blotting for the expression levels of Mdm2 using SMP14 and D12 antibodies. The intensities of the signals were quantified and represented in a graph (right). (b) The acidic domain is important for the stabilization of Mdm2 by TAFII250. H1299 cells were transfected with 5 μg ‘AD’ encoding Mdm2 that contains a deletion in the acidic domain (222–270 aa), together with TAFII250 (5 μg) expression plasmid or with empty vector (5 μg). The densities of the Mdm2 signals were quantified and represented in the graph on the right. (c) H1299 cells were transfected with plasmids expressing wild-type Mdm2 or the C464 mutant (2 μg) together with increasing amounts of TAFII250 expression vector (0, 2, 4 μg). One plate of cells transfected with Mdm2 and one transfected with C464 were treated with 20 μM of MG132 for 4 h. Cells were harvested and lysates were analysed by Western blotting with anti-Mdm2 antibody (SMP14 and D12).
The RING finger domain of Mdm2 is necessary for efficient ubiquitylation and degradation of p53 and Mdm2 itself. The cysteine 464 residue is essential for the E3 ligase activity of Mdm2 and substitution of this residue increases Mdm2 stability (Honda and Yasuda, 2000). Our data suggest that the increase of Mdm2 protein levels by TAFII250 was due to downregulation of Mdm2 auto-ubiquitylation activity, leading to inhibition of Mdm2 degradation, and not to any transcriptional effect (Figures 4 and 5a). This model predicts that the levels of C464 mutant should not be affected by TAFII250. To test this idea, H1299 cells were co-transfected with wild-type Mdm2 or the C464 mutant of Mdm2 together with increasing amounts of TAFII250. As shown in Figure 5c, TAFII250 clearly increased the protein levels of wild type Mdm2. In contrast, C464 protein levels, already elevated due to lack of auto-ubiquitylation, did not increase greatly; (the small increase in C464A levels observed following treatment with MG132 probably arises because H1299 cells retain a low level of active endogenous Mdm2 that may mediate some turnover of the C464A mutant). These data indicate that TAFII250 regulates the degradation of Mdm2 through stabilization and, given that H1299 cells lack p53, this occurs in a manner that is independent of p53.
TAFII250 promotes the association of p53 and Mdm2
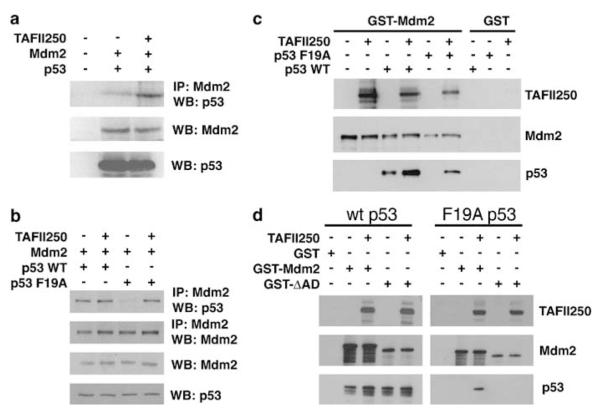
To explore further the mechanism by which TAFII250 can stimulate p53 turnover, the influence of TAFII250 on the p53-Mdm2 interaction was initially examined using extracts prepared from H1299 cells that had been transfected with expression plasmids for Mdm2 alone or Mdm2 together with TAFII250. The volumes of these extracts were adjusted empirically such that the concentration of Mdm2 protein was the same in each case. These extracts were then mixed with extracts of H1299 cells that had been transfected with a p53-expressing plasmid. Subsequently, the ability of the Mdm2 proteins to associate with p53 was determined following immunoprecipitation of Mdm2 and Western blotting to detect p53. The data from this analysis (Figure 6a) revealed that Mdm2 that had been expressed in the presence of TAFII250 bound two to three times more p53 than the Mdm2 that had been expressed alone.
Figure 6.
TAFII250 promotes the interaction of Mdm2 and p53. (a) H1299 cells were transfected with plasmids expressing Mdm2 alone or Mdm2 together with TAFII250. Cell extracts were prepared and the volumes adjusted empirically to yield equal concentrations of Mdm2 in each extract. H1299 cells were also transfected, separately, with a plasmid expressing wild-type human p53. Subsequently, extracts were prepared and equal volumes mixed with the extracts from the Mdm2- or Mdm2/TAFII250-expressing cells. Mdm2 was subsequently immunoprecipitated and the amount of co-immunoprecipitating p53 was measured by Western blotting using the anti-p53 polyclonal antibody, CM-1. (b) H1299 cells were transfected with plasmids encoding TAFII250, Mdm2 and wild type or a F19A mutant of p53. Cells were treated with the proteasome inhibitor, MG132, 6 h before harvesting. Mdm2 and p53 were detected by Western blotting following immunoprecipitation of Mdm2 (upper panels). The total levels of Mdm2 and p53 are also shown (lower panels). (c and d) GST-Mdm2, GST-Mdm2-ΔAD (d) or GST alone were bound to glutathione-sepharose beads and subsequently incubated with p53 WT or a F19A mutant of p53 in the presence or absence of TAFII250. Immobilized proteins were detected by Western blotting.
To determine whether TAFII250 could affect the p53–Mdm2 interaction in cultured cells, H1299 cells were transfected with plasmids encoding TAFII250, Mdm2 and either wild-type p53 or a F19A mutant of p53 that does not bind through its N-terminus to Mdm2 (Bottger et al., 1997). Co-immunoprecipitation assays confirmed that, while Mdm2 associates tightly with wild-type p53, the interaction between Mdm2 and the F19A mutant was very weak (Figure 6b: compared lanes 1 and 3). Expression of TAFII250 did not appear to affect the level of Mdm2-wild-type p53 association, suggesting that the interaction between p53 and Mdm2 (at least through their respective N-termini (see below)) occurs efficiently in vivo. Strikingly, however, the interaction between Mdm2 and the F19A mutant p53 was restored by the presence of TAFII250 (Figure 6b: compare lanes 3 and 4). These data raised the possibility that TAFII250 might promote interaction of p53 with the second p53-binding site in the acidic domain of Mdm2.
The effect of TAFII250 on p53-Mdm2 association was also analysed in vitro using purified proteins. As was observed using the cell extracts (Figure 6a), TAFII250 stimulated the interaction between wild-type p53 and GST-Mdm2 in pull-down experiments (Figure 6c). The data confirmed the lack of association between Mdm2 and the F19A mutant p53 protein (Figure 6b). Notably, however, as with the co-immunoprecipitation experiments (Figure 6b), TAFII250 was able to promote interaction between the F19A p53 mutant and Mdm2 (Figure 6c). To determine whether the acidic domain was required for TAFII250-mediated p53-Mdm2 association, the pull-downs were carried out using GST-ΔAD-Mdm2. In this case TAFII250 failed to promote association between F19A p53 and the ΔAD mutant (Figure 6d). These data are consistent with the idea that TAFII250 can stimulate p53-Mdm2 association through the second p53-binding site in the acidic domain of Mdm2, independently of interaction between the N-terminal binding domains of p53 and Mdm2.
Discussion
In this study, we demonstrate that Mdm2 and TAFII250 associate in cultured cells (Figure 1) and that a robust induction of p53 occurs following temperature-sensitive inactivation of TAFII250 in the ts13 cells (Figure 2). These data are in close agreement with the findings of other groups who have studied the interaction between TAFII250 and the p53 pathway (Leveillard and Wasylyk, 1997; Wasylyk and Wasylyk, 2000; Buchmann et al., 2004; Li et al., 2004). In order to understand the underlying mechanism(s) that leads to the induction of p53, we investigated the influence of TAFII250 on p53 levels and ubiquitylation. We find that TAFII250 promotes the ubiquitylation and turnover of p53 in an Mdm2-dependent manner (Figure 3). The mechanism of this effect involves inhibition of Mdm2 auto-ubiquitylation, leading to the stabilization of Mdm2 (Figures 4 and 5). Additionally, we find that TAFII250 promotes the association of p53 with Mdm2, both in cultured cells and in vitro, in a manner that requires the acidic domain of Mdm2 and is independent of the Mdm2 binding function in the N-terminus of p53 (Figure 6). Taken together, these findings suggest that Mdm2 ubiquitylation function and interaction with p53 can be regulated physiologically by TAFII250. However, while we are confident that the link between TAFII250 and the p53 response is well established (Leveillard and Wasylyk, 1997; Wasylyk and Wasylyk, 2000; Buchmann et al., 2004; Li et al., 2004), and that our interpretation of the data is the most plausible one, we cannot rule out the possibility that varying the levels of proteins such as TAFII250 that are involved in fundamental cellular processes may lead to p53 activation through alterations in numerous indirectly linked cellular pathways. On the other hand, our demonstration that purified TAFII250 can stimulate the interaction between purified p53 and Mdm2 proteins in vitro strongly favours the idea that TAFII250 can directly interact with and regulate Mdm2 physiologically.
A model describing the influence of TAFII250 on the p53–Mdm2 interaction is shown in Figure 7. p53-Mdm2 association occurs principally between the N-termini of the two proteins. Recently, however, a second point of contact has been identified that involves the core domain (DNA-binding domain) of p53 and the acidic domain of Mdm2 (Kulikov et al., 2006; Shimizu et al., 2002; Wallace et al., 2006; Yu et al., 2006). The model arising from these studies proposes that association of the N-terminus of p53 with the N-terminus of Mdm2 triggers a conformational change in Mdm2 that permits the acidic domain to associate with a ubiquitylation signal in the core domain of p53 (Wallace et al., 2006). This second point of p53-Mdm2 contact is critical for p53 ubiquitylation to occur (Kulikov et al., 2006; Wallace et al., 2006). In the present study, we confirm that the acidic domain is required for efficient Mdm2-mediated ubiqutylation of p53 and for Mdm2 auto-ubiquitylation (Figure 3). Additionally, we find TAFII250-dependent stimulation of p53 ubiquitylation also requires the acidic domain of Mdm2 (Figure 3). Strikingly, TAFII250 can promote the interaction between Mdm2 and a mutant of p53 (F19A; (Bottger et al., 1997)) that cannot bind to Mdm2 through its N-terminal Mdm2-binding domain (Figure 6). Since TAFII250 does not appear to affect association of p53 and Mdm2 through their N-terminal interacting domains (Figure 6d), these data fit with the idea that TAFII250 can facilitate the interaction of the core domain of p53 with the acidic domain of Mdm2. Additionally, the ability of TAFII250 to promote interaction between p53 and Mdm2 may contribute to the reduction in Mdm2 auto-ubiquitylation given that the E3 ligase function of Mdm2 would now be directed selectively towards p53. The ability of TAFII250 to bring p53 and Mdm2 together is reminiscent of the ability of YY1 to promote the p53-Mdm2 interaction (Gronroos et al., 2004; Sui et al., 2004) and suggests that YY1 and TAFII250 may share a common mechanism for this effect, such as acting in the capacity of a scaffold.
Figure 7.
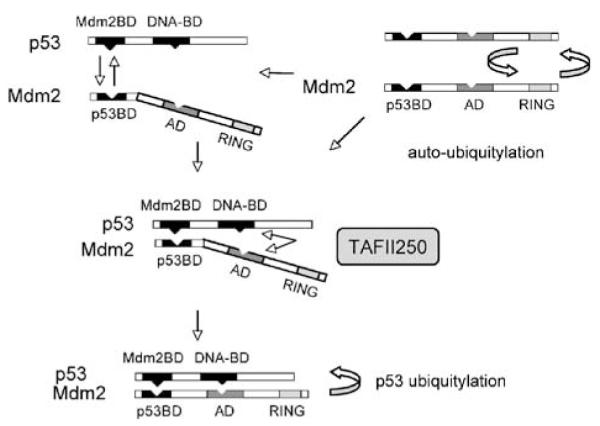
A model describing the ability of TAFII250 to promote ubiquitylation and turnover of p53. For a description of the events depicted in this model, see the Discussion.
The temperature-sensitive defect in the transcription factor TAFII250 gives rise to a G1 phase arrest that is characteristic of the DNA damage response and is mediated by p53 (Wasylyk and Wasylyk, 2000; Buchmann et al., 2004). Altered TAFII250-dependent expression of Mdm2 (Wasylyk and Wasylyk, 2000), loss of the inhibitory phosphorylation of p53 at Thr55 (Li et al., 2004), and ATR-mediated Ser15 phosphorylation and activation of p53 (Buchmann et al., 2004) are likely to be key mechanistic events in this process. Our data demonstrate that stabilization of Mdm2 by TAFII250 promotes Mdm2-mediated ubiquitylation and turnover of p53 (Figures 3–5). We propose that loss of the p53 ubiquitylation function of Mdm2 is likely to be a major contributory factor, together with these other mechanisms, in the induction of p53 following TAFII250 inactivation.
In addition to the effect of TAFII250 on p53 turnover, we also find that knock down of Mdm2 leads to the induction of TAFII250 (Figure 2e). We have not yet explored the mechanism of this effect. However, it is an interesting observation that suggests the intriguing possibility of a negative feedback loop in which Mdm2 can downregulate the levels of its activator p53. Elucidation of this mechanism will be an important issue that will require further significant in-depth exploration.
Ljungman and Lane (2004) have put forward a strong case proposing that the RNA polymerase II transcription machinery may play a vital role in DNA damage surveillance and in triggering DNA damage responses to suppress mutagenesis. They highlight various lines of evidence that underpin the close association between RNA polymerase II transcription and the p53 response, including the observations that agents that block transcription lead to the induction of p53, that the DNA repair-associated transcription factor complex TFIIH phosphorylates and regulates p53, and that p53 degradation normally requires the participation of the p300 co-activator protein in the role of a scaffold protein. When viewed from this perspective, the data in the present study not only support this theme, but suggest plausible molecular mechanisms through which failure of the core transcription apparatus can be signalled directly to the p53 induction machinery. For example, recruitment and activation of Mdm2 by TAFII250 at specific promoters could help maintain low levels of p53. Thus, if transcriptional integrity was compromised, the ability to stabilize Mdm2 and promote its association with p53 would fail thereby contributing to elevated p53 levels. However, a model incorporating this mechanism is likely to be complex and will have to take into account the involvement and interplay of other mechanisms discussed above, such as the activation of ATR and its knock-on effects on p53 and Chk1 (Buchmann et al., 2004). This model also raises the more philosophical question of why a core transcription factor such as TAFII250 should maintain such a close relationship with Mdm2. Perhaps the answer lies in the knowledge that TAFII250 is tightly linked with the cell cycle and is responsible for promoting the expression of a range of cell cycle genes (Suzuki-Yagawa et al., 1997; O’Brien and Tjian, 2000). TAFII250 would be ideally suited to keep p53 under tight control as cells traverse the cycle. Mdm2 itself has been linked to the expression of cell cycle genes such as cyclin A (Leveillard and Wasylyk, 1997). Therefore the maintenance of Mdm2 function would make biological sense under these circumstances.
Materials and methods
Plasmids
Plasmids expressing human Mdm2, C464 Mdm2, p53, F19A mutant p53 and His6-ubiquitin were provided by Dr Dimitris Xirodimas (University of Dundee). The human wild-type p53-expressing plasmid has been previously described (Dumaz and Meek, 1999). The Mdm2 acidic domain mutant (ΔAD) was constructed by polymerase chain reaction (PCR) amplification using the primers ΔAD_for 5′-attcgatggcgtccctgtaga-3′ and ΔAD_rev 5′-gatgaagatgatgaggtatatcaagtt-3′. The sequence was confirmed to ensure that only the desired mutation had been incorporated. pCMV-TAFII250 vector was provided by Dr Neil Perkins (University of Dundee). Synthetic siRNA duplex for Mdm2 and TAFII250 was from Dharmacon.
Cell culture and transfection
H1299 (p53-null lung carcinoma-derived), U2OS and OSA (SJSA-1, both wild-type p53-positive osteocarcinoma-derived) cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% foetal calf serum (FCS) and 100 U/ml penicillin/streptomycin at 37°C and 5% CO2 in a humidified atmosphere. Cells were transiently transfected using either standard calcium phosphate precipitation or lipofectamine (Invitrogen Ltd., Fountain Drive, Paisley, UK) according to the manufacturer’s instructions.
Immunoprecipitations and antibodies
Cells were lysed in NP-40 lysis buffer (10 mM Tris-HCl pH 7.5, 2mM EDTA, 150 mM NaCl, 1% (v/v) Ipegal, Sigma-Aldrich Company Ltd., Poole, Dorset, UK) supplemented with protease inhibitors. Equal amounts of lysate were incubated with 2 μg of anti-TAFII250 (clone 6B3, Upstate) or anti-Mdm2 (SMP14, 4B2 and/or D12, Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA), following which protein G Sepharose beads were added and rocked gently for 2 h at 4°C. The beads were washed three times with NP-40 buffer, and 25 μl of 2 × sodium dodecyl sulphate (SDS) loading buffer was added. Immunoprecipitated proteins were analysed by Western blotting using TAFII250- or Mdm2-specific antibodies.
Western blotting
Cell extracts were subjected to SDS–polyacrylamide gel electrophoresis and Western blotting using standard conditions. Detection of Mdm2 and TAFII250 was carried out using the antibodies described in the previous section. p53 was detected using the monoclonal antibody, DO1 unless otherwise indicated. Horse-radish peroxidase-coupled goat anti-mouse and rabbit anti-goat IgG (DakoCytomation, Cambridgeshire, UK) were used as secondary antibodies. Detection was carried out by chemiluminescence and the bands were quantitated by densitometry.
In vivo ubiquitylation assay
The ubiquitylation assay relies on the transfection of cells with a plasmid encoding His-tagged ubiquitin followed by the capture and purification of ubiquitylated proteins from cell extracts using nickel-agarose beads. This assay has been described in significant detail elsewhere (Xirodimas et al., 2001). The purified ubiquitylated proteins were analysed by Western blotting.
Protein purification and protein–protein interaction
Sf9 insect cells were infected with recombinant baculovirus expressing HA-TAFII250. The recombinant protein was purified as described with anti-HA antibody (12CA5). GST-Mdm2, GST-ΔAD-Mdm2 or GST (500 ng) were immobilized on glutathione-Sepharose (Amersham plc, Little Chalfont, Buckinghamshire, UK) and incubated with 100 ng of wild-type p53, or the F19A mutant p53 (Bottger et al., 1997) (both purified from bacterial extracts), and 100 ng of TAFII250 recombinant proteins, at 4°C for 2 h. Bound proteins were analysed by Western blotting.
RNA preparation and real-time RT–PCR-total
RNA was extracted using RNeasy columns (Qiagen Ltd., Crawley, West Sussex, UK). RNA (300–600 ng) was incubated with random primers (Promega, Southampton, UK) and Superscript II reverse transcriptase (Invitrogen) to generate cDNA. Real-time PCR was carried out with an ABI Prism 7700 sequence detector using the following protocol: 50°C for 2 min, 95°C for 10 min and 40 cycles of 95°C for 15 s and 60°C for 1 min. Mdm2, p21 and Bax primers and 6-FAM/TAMRA labelled probes were used as previously described (Saville et al., 2004).
Acknowledgements
We are very grateful to Dimitris Xirodimas and Frances Fuller-Pace for advice and critical review of the manuscript; and Frank Sauer and Tobias Maile for providing human HA-TAFII250 baculovirus. This work was supported by the Association for International Cancer Research and Cancer Research UK.
References
- Argentini M, Barboule N, Wasylyk B. The contribution of the acidic domain of MDM2 to p53 and MDM2 stability. Oncogene. 2001;20:1267–1275. doi: 10.1038/sj.onc.1204241. [DOI] [PubMed] [Google Scholar]
- Ashcroft M, Taya Y, Vousden KH. Stress signals utilize multiple pathways to stabilize p53. Mol Cell Biol. 2000;20:3224–3233. doi: 10.1128/mcb.20.9.3224-3233.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner C, Hay TJ, Meek DW, Lane DP. Hypophosphorylation of Mdm2 augments p53 stability. Mol Cell Biol. 2002;22:6170–6182. doi: 10.1128/MCB.22.17.6170-6182.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottger A, Bottger V, Sparks A, Liu WL, Howard SF, Lane DP. Design of a synthetic Mdm2-binding mini protein that activates the p53 response in vivo. Curr Biol. 1997;7:860–869. doi: 10.1016/s0960-9822(06)00374-5. [DOI] [PubMed] [Google Scholar]
- Buchmann AM, Skaar JR, DeCaprio JA. Activation of a DNA damage checkpoint response in a TAF1-defective cell line. Mol Cell Biol. 2004;24:5332–5339. doi: 10.1128/MCB.24.12.5332-5339.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumaz N, Meek DW. p53-serine15 phosphorylation stimulates transactivation function but does not directly influence interaction with HDM2. EMBO J. 1999;18:7002–7010. doi: 10.1093/emboj/18.24.7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S, Jensen JP, Ludwig RL, Vousden KH, Weissman AM. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J Biol Chem. 2000;275:8945–8951. doi: 10.1074/jbc.275.12.8945. [DOI] [PubMed] [Google Scholar]
- Feng J, Tamaskovic R, Yang Z, Brazil DP, Merlo A, Hess D, et al. Stabilization of Mdm2 via decreased ubiquitination is mediated by protein kinase B/Akt-dependent phosphorylation. J Biol Chem. 2004;279:35510–35517. doi: 10.1074/jbc.M404936200. [DOI] [PubMed] [Google Scholar]
- Ganguli G, Wasylyk B. p53-independent functions of Mdm2. Mol Cancer Res. 2003;1:1027–1035. [PubMed] [Google Scholar]
- Gronroos E, Terentiev AA, Punga T, Ericsson J. YY1 inhibits the activation of the p53 tumor suppressor in response to genotoxic stress. Proc Natl Acad Sci USA. 2004;101:12165–12170. doi: 10.1073/pnas.0402283101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda R, Yasuda H. Activity of MDM2, a ubiquitin ligase, toward p53 or itself is dependent on the RING finger domain of the ligase. Oncogene. 2000;19:1473–1476. doi: 10.1038/sj.onc.1203464. [DOI] [PubMed] [Google Scholar]
- Kawai H, Wiederschain D, Yuan Z-M. Critical contribution of the MDM2 acidic domain to p53 ubiquitination. Mol Cell Biol. 2003;23:4939–4947. doi: 10.1128/MCB.23.14.4939-4947.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulikov R, Winter M, Blattner C. Binding of p53 to the central domain of Mdm2 is regulated by phosphorylation. J Biol Chem. 2006;281:28575–28583. doi: 10.1074/jbc.M513311200. [DOI] [PubMed] [Google Scholar]
- Leveillard T, Wasylyk B. The Mdm2 C-terminal region binds TAFII250 and is required for Mdm2 regulation of the cyclin A promoter. J Biol Chem. 1997;272:30651–30661. doi: 10.1074/jbc.272.49.30651. [DOI] [PubMed] [Google Scholar]
- Li HH, Li AG, Sheppard HM, Liu X. Phosphorylation on Thr-55 by TAF1 mediates degradation of p53: a role for TAF1 in cell G1 progression. Mol Cell. 2004;13:867–878. doi: 10.1016/s1097-2765(04)00123-6. [DOI] [PubMed] [Google Scholar]
- Ljungman M, Lane DP. Transcription – guarding the genome by sensing DNA damage. Nat Rev Cancer. 2004;4:727–737. doi: 10.1038/nrc1435. [DOI] [PubMed] [Google Scholar]
- Meulmeester E, Frenk R, Stad R, de Graaf P, Marine J-C, Vousden KH, et al. Critical role for a central part of Mdm2 in the ubiquitylation of p53. Mol Cell Biol. 2003;23:4929–4938. doi: 10.1128/MCB.23.14.4929-4938.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael D, Oren M. The p53-Mdm2 module and the ubiquitin system. Semin Cancer Biol. 2003;13:49–58. doi: 10.1016/s1044-579x(02)00099-8. [DOI] [PubMed] [Google Scholar]
- Midgley CA, Desterro JM, Saville MK, Howard S, Sparks A, Hay RT, et al. An N-terminal p14ARF peptide blocks Mdm2-dependent ubiquitination in vitro and can activate p53 in vivo. Oncogene. 2000;19:2312–2323. doi: 10.1038/sj.onc.1203593. [DOI] [PubMed] [Google Scholar]
- O’Brien T, Tjian R. Different functional domains of TAFII250 modulate expression of distinct subsets of mammalian genes. Proc Natl Acad Sci USA. 2000;97:2456–2461. doi: 10.1073/pnas.97.6.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saville MK, Sparks A, Xirodimas DP, Wardrop J, Stevenson LF, Bourdon JC, et al. Regulation of p53 by the ubiquitin-conjugating enzymes UbcH5B/C in vivo. J Biol Chem. 2004;279:42169–42181. doi: 10.1074/jbc.M403362200. [DOI] [PubMed] [Google Scholar]
- Sekiguchi T, Miyata T, Nishimoto T. Molecular cloning of the cDNA of human X chromosomal gene (CCG1) which complements the temperature-sensitive G1 mutants, tsBN462 and ts13, of the BHK cell line. EMBO J. 1988;7:1683–1687. doi: 10.1002/j.1460-2075.1988.tb02996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu H, Burch LR, Smith AJ, Dornan D, Wallace M, Ball KL, et al. The conformationally flexible S9–S10 linker region in the core domain of p53 contains a novel MDM2 binding site whose mutation increases ubiquitination of p53 in vivo. J Biol Chem. 2002;277:28446–28458. doi: 10.1074/jbc.M202296200. [DOI] [PubMed] [Google Scholar]
- Sui G, Affarel B, Shi Y, Brignone C, Wall NR, Yin P, et al. Yin Yang 1 is a negative regulator of p53. Cell. 2004;117:859–872. doi: 10.1016/j.cell.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Suzuki-Yagawa Y, Guermah M, Roeder RG. The ts13 mutation in the TAF(II)250 subunit (CCG1) of TFIID directly affects transcription of D-type cyclin genes in cells arrested in G1 at the nonpermissive temperature. Mol Cell Biol. 1997;17:3284–3294. doi: 10.1128/mcb.17.6.3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden KH, Lu X. Live or let die: the cell’s response to p53. Nature Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- Wallace M, Worrall E, Pettersson S, Hupp TR, Ball KL. Dual-site regulation of MDM2 E3-ubiquitin ligase activity. Mol Cell. 2006;23:251–263. doi: 10.1016/j.molcel.2006.05.029. [DOI] [PubMed] [Google Scholar]
- Wasserman DA, Sauer F. TAFII250: a transcriptional toolbox. J Cell Sci. 2001;114:2895–2902. doi: 10.1242/jcs.114.16.2895. [DOI] [PubMed] [Google Scholar]
- Wasylyk C, Wasylyk B. Defect in the p53-Mdm2 autoregulatory loop resulting from inactivation of TA-F(II)250 in cell cycle mutant tsBN462 cells. Mol Cell Biol. 2000;20:5554–5570. doi: 10.1128/mcb.20.15.5554-5570.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Bayle JH, Olson D, Levine AJ. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- Xirodimas D, Saville MK, Edling C, Lane DP, Lain S. Different effects of p14ARF on the levels of ubiquitinated p53 and Mdm2 in vivo. Oncogene. 2001;20:4972–4983. doi: 10.1038/sj.onc.1204656. [DOI] [PubMed] [Google Scholar]
- Yee KS, Vousden KH. Complicating the complexity of p53. Carcinogenesis. 2005;28:1317–1322. doi: 10.1093/carcin/bgi122. [DOI] [PubMed] [Google Scholar]
- Yu GW, Rudiger S, Veprintsev D, Freund S, Fernandez-Fernandez MR, Fersht AR. The central region of HDM2 provides a second binding site for p53. Proc Natl Acad Sci USA. 2006;103:1227–1232. doi: 10.1073/pnas.0510343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Yao J, Wani G, Wani MA, Wani AA. Mdm2 mutant defective in binding p300 promotes ubiquitination but not degradation of p53: evidence for the role of p300 in integrating ubiquitination and proteolysis. J Biol Chem. 2001;276:29695–29701. doi: 10.1074/jbc.M102634200. [DOI] [PubMed] [Google Scholar]