Abstract
We describe a protocol in which dimethyl sulfate (DMS) modification of the base-pairing faces of unpaired adenosine and cytidine nucleotides is used for structural analysis of RNAs and RNA-protein complexes (RNPs). The protocol is optimized for RNAs of small to moderate size (≤500 nucleotides). The RNA or RNP is first exposed to DMS under conditions that promote formation of the folded structure or complex, as well as ‘control’ conditions that do not allow folding or complex formation. The positions and extents of modification are then determined by primer extension, polyacrylamide gel electrophoresis (PAGE), and quantitative analysis. From changes in the extent of modification upon folding or protein binding (appearance of a ‘footprint’), it is possible to detect local changes in RNA secondary and tertiary structure, as well as the formation of RNA-protein contacts. This protocol takes 1.5–3 days to complete, depending on the type of analysis used.
Keywords: structure mapping, ribozyme, RNP, chemical footprinting, dimethyl sulfate
Introduction
Methods for studying RNA and RNP structure
All organisms encode a host of RNAs that fold to specific three-dimensional structures, many of which associate functionally with proteins. These RNAs and RNA-protein (RNP) complexes perform a diverse range of essential functions, from pre-mRNA splicing and translation to maintenance of chromosome ends. Critical to understanding the functional roles and properties of these complexes is an understanding of their structures and dynamics. Thus, a correspondingly large array of methods has been fashioned to probe RNA and RNP structure.
Experimental methods to probe RNA and RNP structures are also diverse, and can be (imperfectly) divided into three main classes: spectroscopic, physical, and chemical. Spectroscopic methods use electromagnetic radiation and its interactions with matter, in tremendously varied ways, to report on biomolecular structure. Commonly used methods in this category include x-ray crystallography1 and NMR2, as well as fluorescence methods, including those at the single-molecule level3, small angle x-ray scattering4, electron paramagnetic resonance5, and atomic absorption and emission spectroscopy6. Physical methods, most prominently sedimentation velocity7 and native polyacrylamide gel mobility8, are used to obtain information about the size and shape of a molecule or complex by measuring its movement in solution or through a polymer matrix.
There is also a powerful and diverse collection of chemical methods. The main group is ‘footprinting’, the subject of the current protocol. The general theme of this type of method is that the reactivities to a chemical probe of individual nucleotides within an RNA are measured, and from the relative reactivities, features of the structure of the RNA can be deduced. A particularly powerful method is hydroxyl radical footprinting, in which modification of the RNA backbone leads to strand scission9,10. This method gives valuable information on tertiary structure and can be performed with millisecond time resolution11. Further, the hydroxyl radicals can be generated in solution or site-specifically within the molecule12. However, hydroxyl radical methods do not probe the bases and is therefore not useful for detecting changes in secondary structure. Thus, several alternative chemical probes are used to monitor the accessibility of the bases. Dimethyl sulfate (DMS), the subject of this protocol, is one of the most commonly used probes for this purpose, as discussed further below. Other probes of bases are 1-cyclohexyl-3-(2-morpholinoethyl) carbodiimide metho-p-toluene sulfonate (CMCT, which reacts primarily with N3 of U and N1 of G), diethyl pyrocarbonate (DEPC, which reacts primarily with N7 of A), and kethoxal (which reacts with N1 and N2 of G). An analogous ‘chemical’ method is probing with complementary oligonucleotides, which allows measurement of accessibility for stretches of ~10 nucleotides within a folded or folding RNA13,14. All of the chemical methods above can also be applied as ‘modification-interference’ approaches, in which the RNA is first modified under denaturing conditions so that each reactive nucleotide is equally modified, and then the experimenter determines which modifications inhibit folding or function. A battery of additional chemical reagents has also been established for this method (NAIM15). Additionally, methods have recently been developed that monitor the conformation and/or flexibility of each 2′-OH group within the ribose backbone by measuring the ability to cleave at the adjacent phosphodiester linkage (in-line probing16) or to attack an exogenously added electrophile (SHAPE17,18). The latter method appears to be exceptionally useful for monitoring secondary structure and is being adapted for high-throughput use19. Finally, for catalytic RNAs, the enzymatic activity of the RNA can be useful as a ‘chemical’ probe for formation of the native state20.
DMS footprinting: history and theory
Of the chemical reagents used to probe RNA structure, dimethyl sulfate (DMS) is one of the oldest and most versatile. It was introduced for RNA structure mapping in 198021, when Peattie and Gilbert adapted methods that had been used for sequencing DNA and RNA22,23. This early method allowed detection of methylation by DMS at N7 of guanosine and N3 of cytidine nucleotides, because these modifications facilitate cleavage of the chain upon reduction with sodium borohydride and treatment with aniline (for guanosine) or upon treatment with hydrazine and then aniline (for cytidine). Thus, the sites of methylation were determined by polyacrylamide gel electrophoresis (PAGE) analysis of end-labeled RNA. An analogous method was used to ‘footprint’ the binding of proteins to DNA24–26.
A major innovation came a few years later, when it was shown that two positions of methylation by DMS, N1 of adenosine and N3 of cytidine27, could be detected by extending a primer with reverse transcriptase (RT), which is blocked opposite the modified sites28,29 (Fig. 1a and b). Methylation at N7 of guanosine was also detected by cleaving the RNA21 prior to reverse transcription. This new method was powerful for two reasons. First, the ability to detect modifications on the base-pairing faces of two different nucleotides in the same experiment gave a robust and sensitive probe for RNA secondary structure. Second, because the primer to be extended could be complementary to any portion of the modified RNA, it allowed much larger RNAs to be studied conveniently than was possible with methods that required end-labeled RNA.
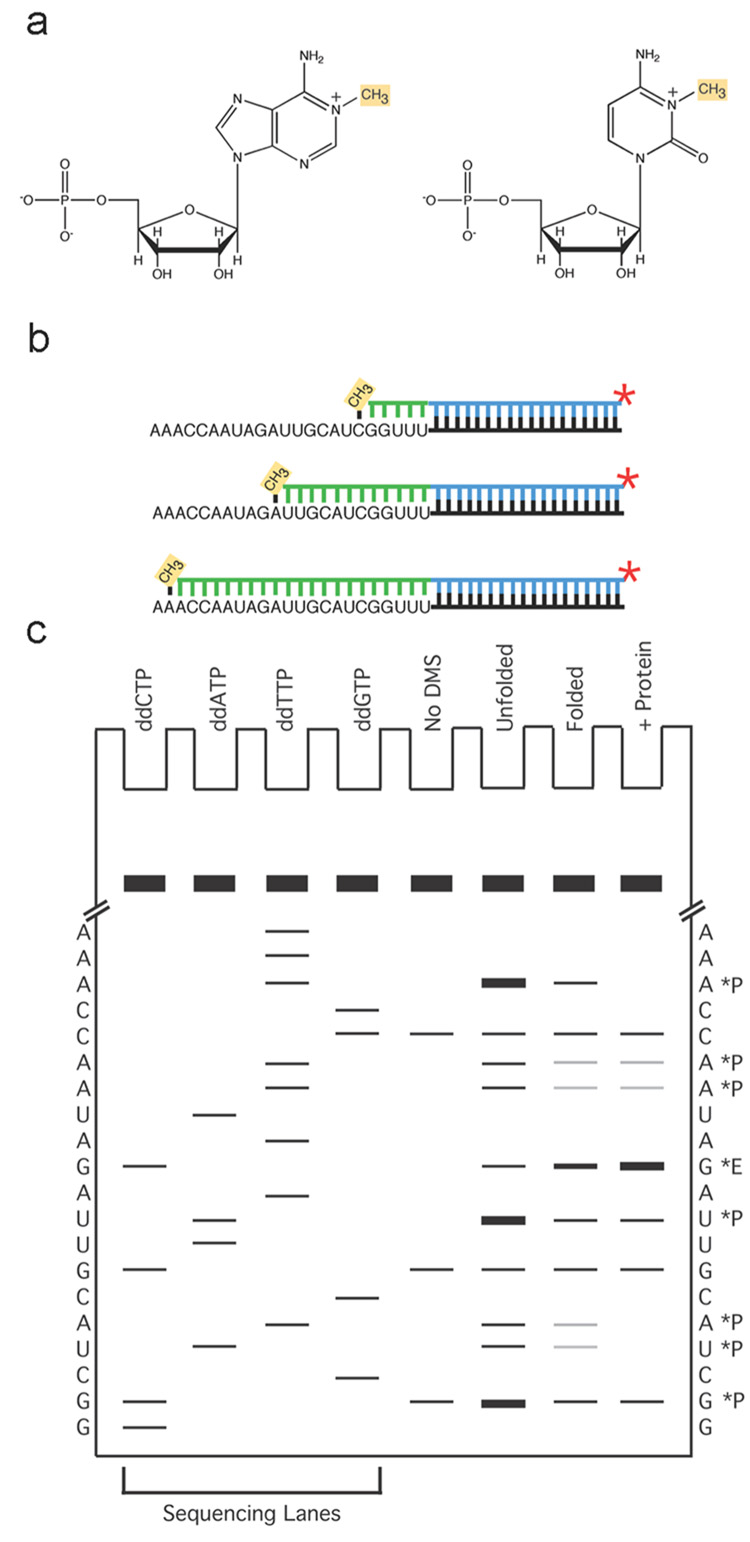
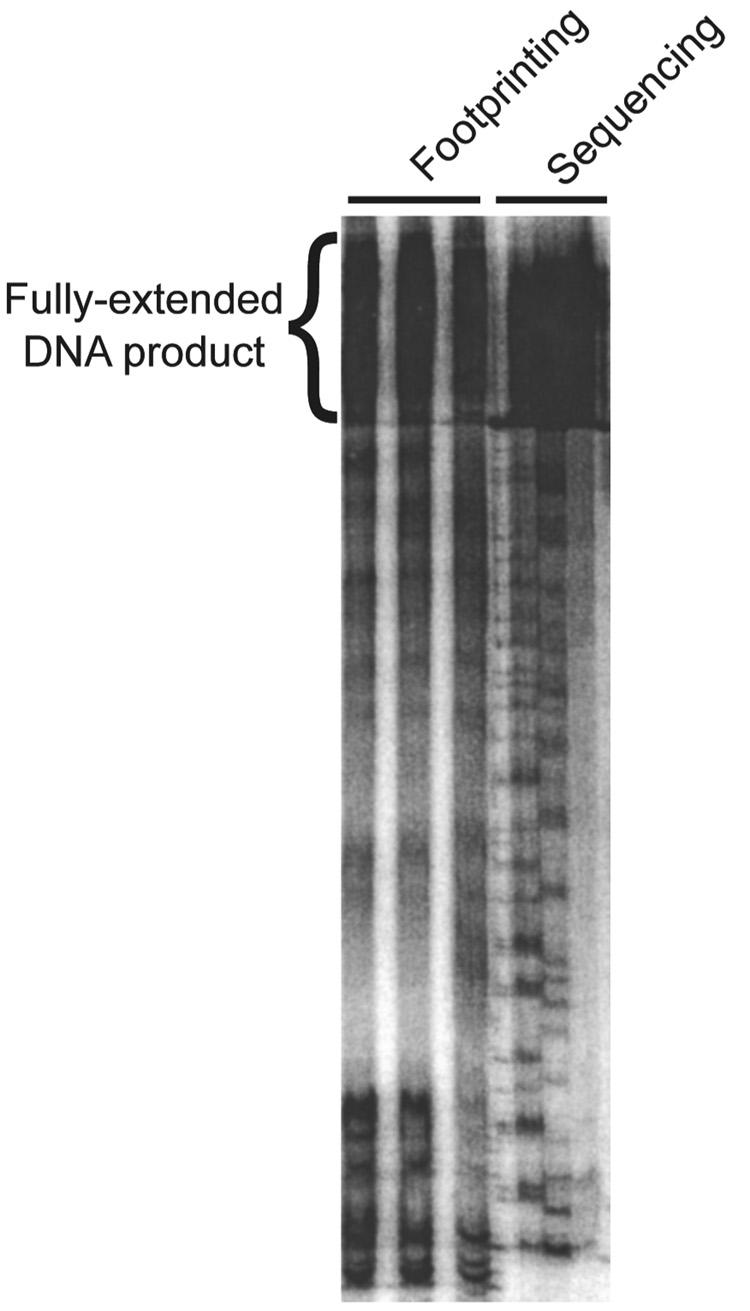
Figure 1. General schematic diagram of DMS footprinting.
a, The modifications detected by this method are methylation at N1 of adenine and N3 of cytosine, as indicated in yellow. b, Primer extension. The DMS modification reaction is carried out under limiting conditions so that each molecule has no more than an average of one detectable modification. Reverse transcription is illustrated (product in green) proceeding from a radiolabeled primer (blue line) until it is blocked at a position one nucleotide upstream from a methylated A or C nucleotide. This reaction on a population of modified RNAs generates a family of radiolabeled products whose lengths are determined by the positions of modifications, and the proportions of these different products reflect the extents of modification at each position. c, PAGE analysis of reverse transcription products. Sequencing lanes at the left are used to determine the position of modification for each experimental band. To the left and right of the gel is shown the sequence of the RNA, as determined from the sequencing reactions. The four lanes at the right show expected results from the experimental lanes. The lane illustrating a reaction that was not treated with DMS (the ‘No DMS’ lane) displays several bands, which is a typical result. These are DMS-independent positions of reverse transcriptase stopping or pausing. In contrast, the bands that are enriched in the DMS treated samples (‘Unfolded’, ‘Folded’, and ‘+ protein’) reflect DMS modification. Note that these bands are one position lower on the gel than A and C nucleotides as determined in the sequencing lanes. This is because reverse transcriptase is blocked one position upstream from the modified position. Nucleotides that give changes in extent of modification upon folding or protein binding are indicated with asterisks and ‘P’ for protection or ‘E’ for enhancement.
Applications of DMS footprinting
DMS footprinting has proven to be a tremendously versatile method and has been applied to a large fraction of known structured RNAs. Initial applications of the method centered on the ribosomal RNAs and RNA-protein complexes (RNPs)30–44. More recently, as the diversity of known structured RNAs has increased dramatically, so has the range of RNAs and RNPs that have been characterized by DMS footprinting, including group I45–54 and group II introns55,56, RNase P RNA57, natural and artificial small catalytic RNAs58–60, structured elements in non-coding regions of mRNAs61–64, the signal recognition particle RNA65,66, and spliceosomal snRNAs13,67–69.
There are several strategies for interpretation of the data from DMS footprinting experiments. One strategy is to analyze the DMS modification pattern in the context of a high-resolution structure to establish the correctness of the structure or to identify regions that do not conform to predictions from the structure, which may suggest that these regions are dynamic or can exist in multiple conformations. It is also used in the absence of a crystal structure to establish or refute the existence of specific structural elements, especially secondary structures30,59,70. Footprinting can be a valuable constraint for structure prediction, and has even been incorporated in an algorithm for structure predication based mainly on free energy calculation71. However, a limitation of DMS footprinting pattern for structure prediction or comparison with known structures is that the degree of reactivity is not a simple reflection of solvent accessibility, but rather depends on more complicated factors such as the local electrostatic environment72.
Other valuable strategies involve comparisons of footprinting experiments under different conditions or in the presence and absence of a bound ligand as a means of identifying regions of the structure that change or to localize the binding region of the ligand. The methylation reaction rate is relatively insensitive to changes in solution conditions such as pH73,74 and monovalent ion concentration51,74, and increases only mildly with increased temperature (<2-fold for a 15 °C change, ref. 45), allowing changes in reactivity to be attributed to structural changes rather than changes in conditions. Thus, DMS footprinting has been used to monitor a pH-dependent conformational change in the ribosome73–75, RNA structural changes in response to changes in ionic conditions or mutation51,76,77, and it has been used extensively to monitor protein binding to ribosomal RNAs31,36–40,78–81, Ro protein binding to Y RNAs 82, as well as RNA-RNA associations in the ribosome32,41,42,44, and binding of a small RNA regulator to an mRNA83. It has also been used to monitor binding of small ligands to RNAs, notably the binding of antibiotics to ribosomal complexes84–87.
DMS footprinting is also used to follow conformational or folding transitions within RNA. Here, the footprinting patterns, and therefore features of the structures, are compared for two different structures or intermediates. Some folding intermediates are so long-lived at moderate or low temperature that they are easily trapped and probed with DMS53,88–90. It is also possible to block folding or conformational changes at defined points in a reaction sequence; e.g. by withholding protein factors that are required for a conformational transition to proceed42. These methods have also been applied to follow conformational transitions mediated by RNA chaperones52,91. Recently, rapid quench techniques have been applied to DMS footprinting, and it has been reported that RNA can be modified to detectable levels on the timescale of milliseconds92,93.
DMS is also useful as a reagent for the related but distinct method of modification interference94–97. In a modification interference experiment, the RNA is first treated with DMS under denaturing conditions so that all reactive nucleotides are modified. Then, functional RNA molecules are separated from non-functional ones, and the positions of modification are determined for both pools. Positions where modifications are absent or underrepresented in the pool of functional molecules are interpreted as making important contacts, so that methylation at these positions blocks is inhibitory.
A final application of DMS footprinting is to perform it in vivo. DMS is readily absorbed into cells and has been used to footprint RNAs in a variety of organisms and cell types98–105. A protocol for DMS footprinting in vivo has recently been published106.
Overview of the DMS footprinting procedure
Here we present a simple protocol used routinely in our labs for quantitative DMS footprinting of structured RNAs and RNA-protein complexes (Fig. 1). We have used the protocol to footprint group I intron RNAs and ribozyme derivatives ranging in length from 300 to 500 nucleotides51–54. The protocol is optimized for RNAs of this size or smaller that are prepared by in vitro transcription or are naturally abundant, and it is expected to be readily adaptable for use with longer RNAs (see Variations of the method with respect to RNA length). The basic steps of the DMS footprinting method are unchanged since the mid-1980s, and there are several published methods107–111. The RNA or RNP is treated with a low concentration of DMS for a time sufficient to produce, at most, a small number of modifications per molecule. The methylated RNA is then deproteinized and isolated and a radiolabeled primer is annealed to the RNA (Fig. 1b). A reverse transcription reaction is performed to extend the primer to the sites of modification, and after treatment to destroy the RNA while leaving intact the newly-formed cDNA, positions of modification are determined by denaturing PAGE (Fig. 1c). Sequencing reactions using the standard dideoxynucleotide method112 are run in parallel to map the sites of methylation to the sequence of the RNA. The sites of modification and the extents of modification are then compared between the experimental conditions and a reference condition, typically the RNA in the absence of Mg2+ or in the absence of protein. A control reaction that is treated identically except for the absence of DMS (abbreviated herein as ‘non-DMS’ control) is also necessary to establish whether a reverse transcription ‘stop’ is caused by DMS methylation or some DMS-independent source, such as truncated RNA or a sequence that blocks reverse transcription. These comparisons can be done by eye or, as emphasized here, by quantitatively determining the intensity of the band reflecting modification at each position and for each experimental condition.
The next step is to interpret the changes in modification in terms of their molecular origins. Protections from modification, relative to an unfolded control, can arise from secondary structure formation, and this is often the simplest interpretation. However, protections can also arise from formation of tertiary contacts involving the base-pairing face. A ubiquitous example is the type I A-minor tertiary contact, which includes hydrogen bonding between N1 of a single-stranded A and the 2′-OH of a base-paired nucleotide113,114 and, as expected, gives protection from DMS modification46,53. When a comparison is made in the presence and absence of a protein, protein-dependent protections are expected to arise from direct contacts as well as from changes in RNA secondary or tertiary structure induced by protein binding.
Advantages and limitations of DMS footprinting
Strengths of DMS footprinting are:
It is very likely to detect changes in secondary structure because it provides signals on the base-pairing faces of adenosine and cytidine nucleotides.
It is also capable of detecting changes in tertiary structure.
The reaction is fast, allowing high time resolution.
The reaction rate is not strongly dependent on solution conditions, allowing probing of structural changes with changes in conditions.
Relative to enzymatic probes (i.e. nucleases), the small size of DMS allows high spatial resolution.
Limitations of DMS footprinting are:
The sensitivity of the method to changes in both secondary and tertiary structure can make it difficult to deduce the origin of a change. A common way to address this problem is to test candidate secondary structure changes by constructing and testing mutations that are expected strengthen or weaken one of the proposed secondary structures76. A second strategy is to combine DMS with other chemical probes, such as CMCT, kethoxal, and DEPC107,110, which modify other positions (see above), and can therefore provide corroborating evidence for a secondary structure change. An additional strategy is to combine DMS footprinting with enzymatic footprinting methods, which can be highly specific for secondary structure formation but have lower spatial resolution than the smaller chemical probes107.
DMS does not provide a signal for accessibility of the phosphodiester backbone of the RNA. Therefore, DMS footprinting is often performed in conjunction with hydroxyl radical footprinting46,49, which is sensitive to the changes in solvent accessibility that often arise from tertiary structure formation or RNA-protein contacts, but does not depend strongly on the formation of secondary structure115,116. This combination is particularly effective for time-resolved studies, because both methods can provide data in the regime of milliseconds11,92. Other probes that have been used to assess backbone accessibility are ethylnitrosourea107,111 and peroxynitrous acid117–119. Further, as noted above, SHAPE17,18 and in-line probing16 can provide information on the conformation and/or flexibility of the ribose sugars, and these may also be useful in conjunction with DMS.
DMS footprinting, like other footprinting methods, provides local information on accessibility, but it does not provide a signal for changes in global properties of the RNA. For this reason, footprinting of RNA folding has recently been performed in conjunction with sedimentation velocity7 or small angle x-ray scattering120, which give complementary information on global size and shape.
Experimental Design
RNA Handling
There are several precautions that one should take whenever working with RNA. Unintentional degradation of RNA caused by RNase contamination is a ubiquitous problem, and the consequences are particularly severe for footprinting experiments. Some useful precautions for dealing with RNase contamination are as follows:
Keep sterile any solution or buffer that will come into contact with RNA. Also make smaller aliquots of solutions and buffers for reactions with RNA and store at −20 °C. Discard any solution or buffer suspected of RNase contamination and prepare it again.
Wear gloves at all times during your experiment and change to a fresh pair when the gloves you are wearing have been compromised (e.g. after touching a surface that is not known to be free of RNases).
Autoclaving does not inactivate most RNases, so use of plasticware such as disposable tips and microcentrifuge tubes (RNase-free) is recommended. If glassware must be used, bake the glassware at 200 °C110.
If necessary, use aerosol barrier pipet tips to prevent contamination.
If necessary, use RNase inhibitors (e.g., if working with protein preparations known to have RNase contamination). We have found RNasin Plus (Promega) to be effective. There are also RNase inhibitor solutions designed to clean surfaces, such as RNase Zap (Ambion).
Formation of the desired RNA or RNP structure
For DMS footprinting experiments using in vitro transcribed RNA, the first step is to fold the RNA to the desired structure. It is common to begin by heating the RNA in the absence of Mg2+ to ~90 °C and cooling it slowly to ensure that each experiment begins with the same ‘unfolded’ structure or ensemble of structures. Then Mg2+ is added to initiate tertiary folding. Because RNAs are susceptible to misfolding, it is often necessary to incubate the RNA in the presence of Mg2+ for an extended time or at elevated temperature to allow formation of the native structure20,121–132. Before embarking on footprinting experiments, it is critical to establish by using functional assays the conditions and incubation time necessary to achieve the desired RNA structure.
Next, protein is added if an RNP is to be formed. There are three general requirements for this step. First, the protein or proteins must be in stoichiometric excess over the RNA (of course, the binding stoichiometry must be considered in making this calculation). Because DMS provides structural information about the RNA, it is critical that essentially all of the RNA be in complex with the protein, whereas the presence of excess protein that is not in complex with the RNA does not affect the results. Second, the concentration of protein must be above the equilibrium constant (Kd) for formation of the complex. If the protein concentration is not high enough, a substantial fraction of the RNA will remain free in solution at equilibrium. It is not necessary that the RNA concentration be above the Kd because it will be driven into the complex by the high protein concentration. Third, an incubation of RNA and protein must be performed for sufficient time to allow the complex to form. Under protein concentrations typically employed (nM to µM), complex formation is usually fast enough that this is not a problem. As above for folding of the RNA, conditions that give complete binding of the RNA to the protein should be established in separate experiments (functional assays or direct binding assays) prior to performing DMS footprinting reactions. In practice, it is useful to vary the protein concentration and incubation time to confirm experimentally that the protein is saturating and therefore that it is the complexed RNA that is being probed by DMS.
DMS Handling
DMS is a highly toxic chemical. Read the Material Safety Data Sheet (MSDS available at www.sigmaaldrich.com) before use. DMS is carcinogenic and easily absorbed through the skin, so wear gloves at all times when working with DMS. It is also advisable to wear eye protection and a lab coat. DMS is also very toxic by inhalation, so you should work in a fume hood when handling DMS.
Consult your institution’s environmental health and safety department (EHS) regarding proper waste disposal of DMS solid waste. Decompose liquid DMS waste by adding 10 N NaOH and mixing, followed by neutralization with 10 N HCl (check with pH paper). Once decomposed and neutralized, dispose of the solution in the sink. DMS spills should be handled by trained personnel from EHS.
Chemical modification by DMS
The footprinting experiment should be performed under conditions that give only a low level of modification per molecule. In principle, it is desirable to have less than one modification per molecule because, for molecules that are modified in multiple positions, it is possible that the first modification will influence the DMS accessibility of other positions. Further, the extension products that reflect modification at long distances from the primer will be underrepresented because reverse transcription will stop at the modification closest to the primer. On the other hand, there is greater signal with a higher overall level of modification. It should also be noted that the methylation by DMS of guanosine at N-7, typically silent in DMS footprinting experiments because it does not block RT, occurs approximately an order of magnitude faster than the observed modifications27, so that even if the reaction is performed with only one adenosine or cytidine modification, the reaction is not truly ‘single hit kinetics’ because there will likely be several guanosine nucleotides modified per molecule. In practice, a balance is struck between having a low enough level of modification to minimize any problems of the types described above, yet a high enough level to produce readily-detectable signals. A reasonable starting point is to vary the DMS concentration over a 5–10-fold range (0.1 – 1% by volume) and vary the time of incubation with DMS (1–10 min for room temperature incubations, 20–25 °C). Longer incubations do not give linear increases in modification because DMS is hydrolyzed in aqueous solution45.
DMS reaction quench
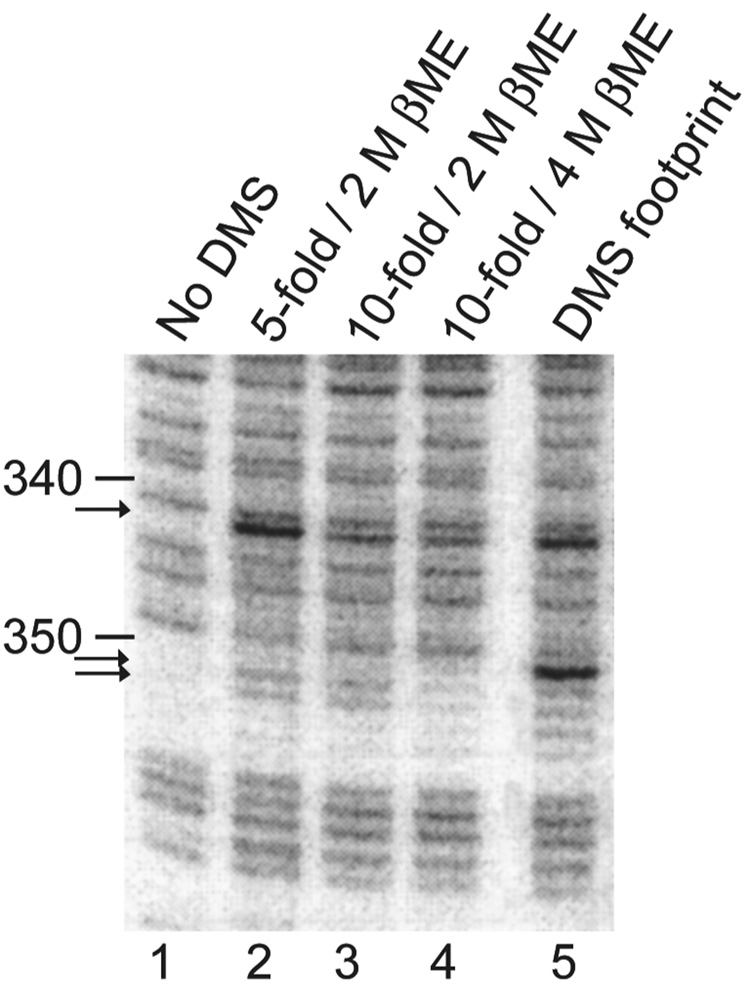
For time-resolved measurements, it is critical that the DMS modification reaction be quenched effectively at the desired time. Even for experiments in which precise time resolution is not necessary, it is important to have an effective quench because the solution conditions are changed upon proceeding to the next step of the procedure, so if the DMS modification reaction continues, some of the observed modification will have arisen under conditions other than the desired experimental conditions. Published methods that we examined either did not include a quench step107–111, instead relying on termination of the DMS reaction upon ethanol precipitation, or included a quenching step of β-mercaptoethanol addition that we found to be insufficient to fully protect the RNA from further reaction with DMS (Fig. 2). Therefore, we modified existing protocols by including substantially more β-mercaptoethanol and a greater dilution of the footprinting reaction (see Procedure), and we confirmed the effectiveness of this quench solution by mixing DMS with RNA to which quench solution had already been added (Fig. 2).
Figure 2. Establishment of a solution quench of DMS modification reaction.
Samples in lanes 2–4 were diluted as indicated, from 50 µl, into a solution containing β-mercaptoethanol at the indicated concentrations (200 µl or 450 µl). DMS was then added (1 µl of DMS diluted 1:3 with EtOH) and samples were incubated at room temperature for 5 min before precipitating with EtOH. Lane 1 was treated identically to lane 3 except that DMS was not added, and lane 5 was treated identically to lane 3 except that DMS (1 µl, diluted 1:3 with EtOH) was added first. The sample (50 µl) was incubated at room temperature for 1 min with DMS, followed by addition of quench solution. Arrows indicate nucleotides that are modified detectably in lanes 2 and 3, indicating that these quench solutions were not sufficient to protect the RNA from modification. Lane 4 shows nearly perfect quenching (although modification at position 342 is still visible), with final concentrations of 3.6 M β-mercaptoethanol and 0.067% DMS. The quench conditions that we describe in the Procedure are 3.8 M β-mercaptoethanol and 0.033% (3.5 mM) DMS, which gave no detectable modification in an equivalent experiment (not shown).
Primer extension
After deproteinizing and isolating the RNA, a reverse transcription reaction is performed to determine the positions and extents of modification (Fig. 1b). For longer RNAs (>100–150 nucleotides), multiple primers are necessary. To ensure strong and specific annealing, primers should be designed such that they are 18–25 nucleotides long, and fortuitous complementarity with other regions of the RNA should be minimized (< 4 or 5 consecutive base pairs is desired). Primers should be spaced so that they are complementary to regions in the target RNA no more than 200 nucleotides apart111, and for quantitative footprinting we recommend that they be no more than 100 nucleotides apart. This spacing will ensure that the footprinting pattern from each primer can be quantitatively read, with single nucleotide resolution, to the positions that can be analyzed by the next primer.
Two strategies have been commonly employed to label the products with 32P; either the primer is labeled in a separate step prior to the extension reaction107,108,111 or the extension reaction is performed in the presence of labeled nucleotides107–110. The former method is used here. Labeling of the primer is strongly recommended for small RNAs (≤100 nucleotides) because it is critical that all products of primer extension be labeled sufficiently as to be visible. We recommend labeling of the primer for all quantitative footprinting analysis because this method produces products that have the same specific activity regardless of length. If labeled nucleotides are used instead, a correction must be made to account for the increased specific activity of the longer products. Further, the primer labeling method results in a greater fraction of the label being present in the short products, which are more easily resolved and quantitated. The principal disadvantage of this method is that an extra step is required, as the primers must first be end-labeled (see REAGENT SETUP below).
The reverse transcriptase enzyme (RT) that is most commonly used is from avian myeloblastosis virus (AMV, see Reagents), because its long half-life at higher temperatures (42–50°C) allows the reverse transcription reaction to be performed under conditions that destabilize RNA secondary structures and therefore increase the efficiency of extension. More recently, the reverse transcriptase from the Moloney strain of murine leukemia virus (MMLV) has been engineered so that it is also active up to 50°C (e.g. the SuperScript family of RTs by Invitrogen), allowing this enzyme to be used effectively for primer extension18,133. Thus, both enzymes are listed as options in our protocol.
Analysis
Traditionally, much of the analysis has consisted of inspecting gel images and comparing band intensities by eye. Quantitative methods, such as manual boxing of bands using ImageQuant software (see Step 39 and Fig. 4), have historically been difficult and much more time-consuming, but with new software quantitative methods are becoming increasingly accessible. A semi-automated program for gel analysis has recently been developed (SAFA), which is freely available and substantially reduces the time required134 (see Step 39 and Fig. 3). Although this software was developed for analysis of hydroxyl radical footprinting, which typically gives peaks whose intensities differ from each other less than those from DMS footprinting, a side-by-side analysis of DMS footprinting data with SAFA and manual boxing of individual bands using ImageQuant gave comparable results53. Recent versions of ImageQuant TL (Amersham, GE Healthcare) are also very efficient and versatile.
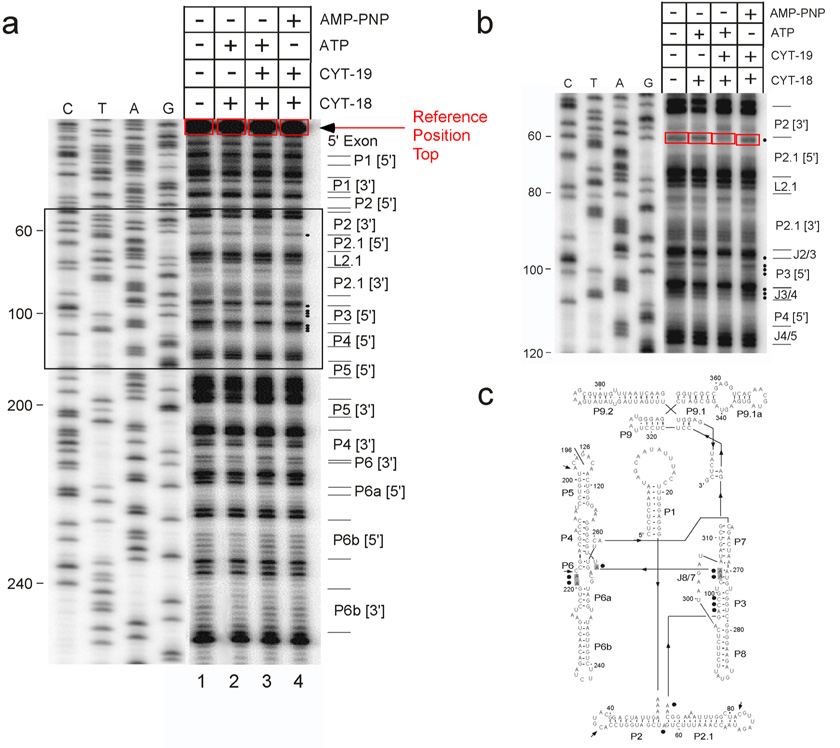
Figure 4. DMS footprinting of an RNA-protein complex and quantitative analysis using manual boxing.
This experiment monitored formation of a complex between a variant of the Tetrahymena LSU intron (ΔP5abc) and the specific-binding protein CYT-18, and the effect of the DEAD-box chaperone protein CYT-19 on this complex52. a, Footprinting gel. 20 nM RNA was pre-incubated with 100 nM CYT-18 alone or also with 100 nM CYT-19 plus 1 mM ATP or AMP-PNP as indicated above each lane, and then modified with 0.2% DMS. Modifications were mapped by primer extension with MMLV RT, using a 5′-labeled primer complementary to intron positions 274–298. Results were analyzed by manual boxing. The band at the top of the gel, representing fully extended product, is quantitated as a reference. Bands to be analyzed (labeled with black dots) differ in intensity between the four experimental lanes. They are boxed and quantitated and the counts are expressed as a percentage of the reference counts in Table 2. Lanes C, T, A and G are dideoxy sequencing ladders of the plasmid DNA using the same 5′-labeled primer. b, Enlargement of the boxed region, which contains the area of interest. Intron positions are indicated to the left, and group I intron regions are indicated to the right. Boxes are shown for one of the analyzed nucleotides (A57). c, Secondary structure showing nucleotides whose modification level was dependent on the added proteins, as indicated by the this and other experiments (not shown). Shading indicates nucleotides that were protected upon CYT-18 addition (differences of at least 0.01, see Table 2 for quantiation of A103-A105), and closed circles indicate nucleotides that were protected upon addition of CYT-19 and ATP. Arrows indicate nucleotides that were enhanced for reactivity by addition of CYT-19 and ATP. All of the nucleotides that were protected upon CYT-18 addition were protected further upon addition of CYT-19 (Table 2), presumably because CYT-18 does not bind stably to the misfolded ribozyme that accumulates in the absence of CYT-19 (A. Chadee and R. Russell, unpublished results). All of the highlighted nucleotides gave bands in reactions with DMS that were substantially more intense than the corresponding reactions without DMS (not shown).
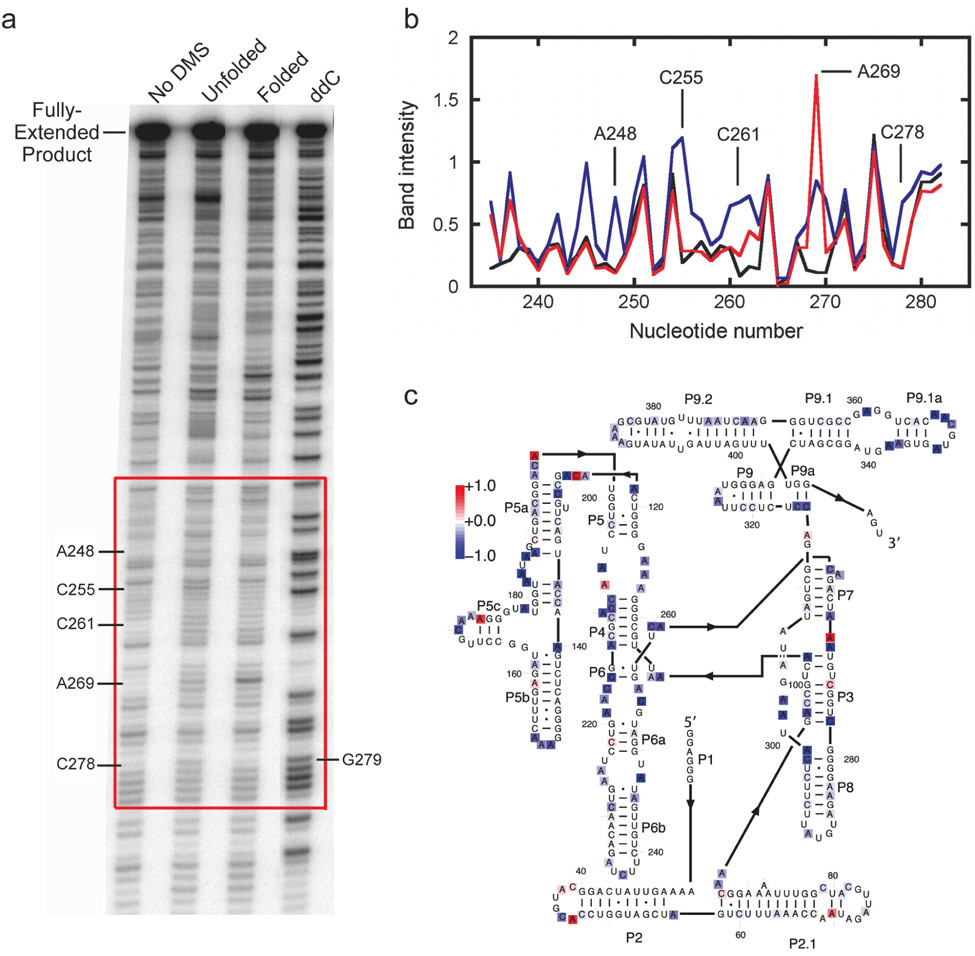
Figure 3. Quantitative analysis of DMS footprinting to monitor RNA folding.
This experiment monitors Mg2+ dependent folding of the Tetrahymena group I ribozyme53. a, DMS footprinting gel. The DMS exposure time for this experiment was relatively short, 1 min at 25 °C with 0.67% DMS, such that the background from RNase degradation and/or pauses by RT is significant relative to the DMS-dependent signal (bands in lane marked ‘No DMS’). However, DMS-dependent bands are readily observed, as indicated by nucleotide labels. The label at the right, G279, is included to illustrate that fragments indicating DMS modification migrate more rapidly by one nucleotide than the corresponding fragments in the sequencing lanes. b, Analysis using SAFA of the region of panel a indicated by the red square. Band intensity was determined by dividing the integrated intensity of each band by the corresponding intensity for the fully-extended primer (determined by boxing this band using Image Quant), and multiplying this by 1000. Thus, an intensity value of 1 means that the indicated band was 0.1% as intense as the fully-extended product, or for the DMS-dependent bands, that roughly 0.1% of the RNA was methylated at this position. c, Comparison of the DMS protection patterns for the folded and unfolded RNAs. Values were obtained by subtracting the intensity values in the unfolded form from those representing the folded form, so that a negative value (colored blue as indicated by the scale bar) represents a nucleotide that is protected in the folded RNA relative to the unfolded RNA, whereas a nucleotide colored red represents a nucleotide that displays enhanced reactivity in the folded RNA.
Quantitation has two advantages over qualitative analysis. First, it is systematic, eliminating the possibility of selectively interpreting changes where they are expected. Second, it allows the data to be normalized to correct for differences in loading between lanes. Two methods of normalization are to divide the intensity of each band by the average of all of the measured bands in a given lane134 or to divide the intensity of each band by the intensity of the fully-extended product at the top of each lane53.
To interpret changes in band intensity between conditions, it is necessary to establish that the intensity arises from DMS modification and not from a DMS-independent effect causing RT to stop at that position (see Fig. 1c). Thus, it is essential to compare a reaction that has been treated identically but not exposed to DMS. Bands should only be interpreted if there is not a strong band at the corresponding position in this control. Identities of the bands are established by comparisons with dideoxynucleotide sequencing reactions using the same primer and run on the same gel. Because DMS modification causes RT to stop one nucleotide before the damaged site, a band resulting from modification at a given nucleotide runs one position lower on the gel than the corresponding band in sequencing reactions.
Variations of the method with respect to RNA length
We have used the protocol here for analysis of RNAs ranging in length from 300–500 nucleotides51–54. Other published protocols are optimized for smaller111 or larger37,108,110 RNAs, and therefore have some differences from this one. It should be noted that most of these differences, listed below, depend on the sequence and structural properties of the RNA being probed, not on its length per se, so it is important to survey conditions to find those optimized to a given RNA.
For larger RNAs, there is increased likelihood that primers will have some affinity for non-target sites within the RNA, so it may be critical to cool the solution slowly during the primer annealing step (step 17)110. It is also important to design primers such that fortuitous complementarity with other regions is minimized.
For larger RNAs, it is necessary to use more primers to completely footprint a larger RNA than a smaller one, as each primer will give quantitative footprinting information on only a limited number of nucleotides downstream from the primer (typically <200 nucleotides).
For larger RNAs, it will be necessary to achieve a higher level of modification per RNA to obtain an interpretable signal; a general suggestion is one modification for every 300 nucleotides110. This requirement does not mean that larger RNAs require longer incubations and/or higher concentrations of DMS, because a given DMS incubation produces a certain frequency of modification on a per nucleotide basis, such that a larger RNA will have more modifications per RNA.
For larger RNAs that have very stable structures, it may be useful to perform the primer extension reaction at a higher temperature than the 42 °C used here, which is possible using thermostable RT enzymes that were not available or in common use when earlier protocols were published107–110.
For larger RNAs, it is also possible to label the products of reverse transcription by using labeled nucleotides rather than end-labeling the primers. This method has the advantage of ease, with fewer experimental steps, and it has been suggested that this method is preferable when the yield of annealed primer is low because excess primer can be used without obscuring the shortest elongation products107. However, we recommend labeling the primer for all quantitative applications (see Primer Extension above).
The necessity of degrading the RNA prior to gel electrophoresis (steps 25–26)18,111 may depend on RNA length. This step may not be necessary for very small RNAs, as the heteroduplex will be less stable. Some protocols have omited this step and instead included a heating step (at neutral pH) prior to gel loading109,110, which presumably disrupts structure within the DNA products as well as the heteroduplex. We have found that this heating step is unnecessary if the RNA is degraded and extension products are separated on a gel run at 55–60 °C (see Fig. 3 and Fig 4).
Materials
REAGENTS
Dimethyl sulfate (Aldrich D186309)
 DMS is highly toxic and carcinogenic. See DMS Handling in Experimental Design section.
DMS is highly toxic and carcinogenic. See DMS Handling in Experimental Design section.Reverse Transcriptase enzyme (RT): Avian Myeloblastosis Virus (AMV, Amersham) or Moloney Murine Leukemia Virus (MMLV, SuperScript, Invitrogen)
T4 polynucleotide kinase (New England Biolabs M0201L)
DNA primers (Integrated DNA Technologies): DNA primers are user-specific and should be designed as described above in the Introduction.
β-Mercaptoethanol (EMD Chemicals EM6010)
Phenol/chloroform/isoamyl alcohol (25:24:1) from EMD Chemicals EM6805
 Chloroform is believed to be a carcinogen; avoid inhalation
Chloroform is believed to be a carcinogen; avoid inhalationEthanol
 Ethanol is highly flammable
Ethanol is highly flammable[γ-32P]ATP, 160 µCi/µl (>3000 Ci/mmol; Perkin Elmer NEG035C)
 You should complete a course in radiological safety or receive equivalent training before working with radioactive materials. Use an acrylic shield that blocks beta emissions when working with 32P. Keep a Geiger counter nearby and periodically monitor your work area for contamination.
You should complete a course in radiological safety or receive equivalent training before working with radioactive materials. Use an acrylic shield that blocks beta emissions when working with 32P. Keep a Geiger counter nearby and periodically monitor your work area for contamination.dNTP mix (10 mM each; New England Biolabs N0447S)
ddNTPs (set of 4 at 5 mM each; Amersham 27-2045-01)
3 M sodium acetate, pH 5.0
Acrylamide, 40% (w/v) volume, 29:1 acrylamide:bisacrylamide (EMD Chemicals, EM1700)
 Acrylamide is toxic and may be carcinogenic. Wear gloves and safety glasses.
Acrylamide is toxic and may be carcinogenic. Wear gloves and safety glasses.TEMED (N,N,N',N'-Tetramethylethylenediamine; EMD Chemicals EM8920)
Wonder wedge separation tool (Hoefer SE1514)
Whatman 3MM paper (35 × 45 cm)
Quench solution (β-mercaptoethanol (30% v/v), 0.3M sodium acetate)
25X TTE Buffer (1 L: 216 g Tris base, 72 g Taurine, 4 g EDTA)
10X annealing buffer: 500 mM Tris pH 8.3, 600 mM NaCl, 100 mM DTT
Primer extension mix (see REAGENT SETUP below)
Sequencing extension mixes (see REAGENT SETUP below)
Loading dye (90% (v/v) formamide, 20 mM EDTA, 0.04% (w/v) bromophenol blue, 0.04% (w/v) xylene cyanol)
EQUIPMENT
Sequencing electrophoresis apparatus (Owl)
Phosphorimager (Amersham, GE Healthcare)
Heat Block, Water Bath, or Thermocycler
Gel Dryer (Thermo Savant or BioRad)
Acrylic shield to block beta emissions
Geiger counter to monitor work area for radiological contamination
REAGENT SETUP
32P-labeling of DNA primers
Each oligonucleotide primer should be 5′-end-labeled before the DMS footprinting experiment is performed. Mix 1 µl of each primer (~20 µM final concentration) with 1 µl (10 U) of T4 polynucleotide kinase (PNK), 1 µl of [γ-32P]ATP, and the recommended buffer for the PNK enzyme, and incubate for 1 hour at 37 °C. Use [γ-32P]ATP of high specific activity and concentration (>3000 Ci/mmol, 160 µCi/µl). Purify the labeled primer using 20% (w/v) denaturing PAGE and visualize the primer by radiography. While exposing the gel, carefully poke holes with a syringe needle through both the film and the gel to mark the relative positions, so that the gel and film can be properly aligned later. After developing the film, re-align the gel and film and cut out the gel slices containing the labeled primers. Elute overnight at 4 °C by soaking each slice in 100 µl of 10 mM Tris, pH 8.0, 1 mM EDTA buffer. Quantitate the specific activity of the labeled primer by measuring 1 µl in a scintillation counter. We omit scintillation fluid for simplicity, counting Cerenkov radiation and multiplying by a factor of 3 to correct for the low counting efficiency. The level of activity should be at least 50–100k dpm/µl for best results. Store the labeled primer at −20 °C and use within 2–4 weeks (the half-life of 32P is 2 weeks). A similar method for labeling oligonucleotides has recently been published18.
 It is possible to substitute a gel filtration step for purification of the labeled primer, but we do not recommend this unless the primer is known to be homogeneous in length, because any heterogeneity at the 5’ end will lead to multiple bands arising from a given position of DMS modification.
It is possible to substitute a gel filtration step for purification of the labeled primer, but we do not recommend this unless the primer is known to be homogeneous in length, because any heterogeneity at the 5’ end will lead to multiple bands arising from a given position of DMS modification.
Primer extension mix
Mix 1 µl of 4X annealing buffer containing 50 mM MgCl2, 1 µl of 4 mM dNTP (4 mM of each dNTP), 0.2 µl of RT (10 U/µl), and 1.3 µl of H2O. Scale up preparation of this solution according to the number of primer extension reactions to be performed (3.5 µl will be added to each primer extension reaction). CRITICAL: Prepare this solution shortly before it will be used and store it on ice.
Sequencing extension mixes
Prepare four solutions identically to the Primer extension mix above, except instead of adding 1.3 µl of H2O, add 0.3 µl of H2O and 1 µl of a 1 mM solution of one of the four ddNTPs. Scale up volumes according to the number of primers to be used in the experiment (3.5 µl will be added to each sequencing reaction). For each primer, there will be four sequencing reactions, one for each ddNTP. Hint: after performing initial experiments with a given primer and RNA, it is not necessary to perform all four sequencing reactions; one or two reactions are sufficient to unambiguously assign all of the bands in the experimental lanes. CRITICAL: Prepare these solutions shortly before they will be used and store them on ice.
Preparation of an 8% (w/v) polyacrylamide gel
Assemble two plates (20 cm × 43 cm) using 0.35 mm spacers. The plates must be sealed on the sides and at the bottom to prevent leaking. We use electrical tape and large binding clamps, but clamps supplied with electrophoresis apparatus may also be used. Dissolve 48 g of urea in 50 ml of H2O, 20 ml of 40% (w/v) acrylamide (29:1 acrylamide:bisacrylamide), and 4 ml of 25X TTE buffer in a 250 ml flask; mix well and bring the total volume to 100 ml with H2O. Add 1 ml of 10% (w/v) ammonium persulfate and mix well. Add 100 µl of TEMED, mix well, and pour immediately and carefully between the plates. Insert an appropriate comb (0.35 mm) between the plates and allow 30–40 min for polymerization. Monitor the gel solution remaining in the flask to determine when the gel has polymerized. When the gel is polymerized, carefully pull out the comb, remove the tape and clamps, mount the gel on the gel apparatus, fill the top chamber with 1X TTE buffer, and forcefully rinse out the wells with 1X TTE buffer. We use a 50 ml syringe with an 18 gauge needle to rinse out the wells.
 Rinse out the wells again, immediately prior to loading the samples, because urea tends to accumulate in the wells over time.
Rinse out the wells again, immediately prior to loading the samples, because urea tends to accumulate in the wells over time.
PROCEDURE
Folding of RNA
1. Fold RNA by adding Mg2+ ion. We provide the specific conditions that we use for folding of the Tetrahymena group I ribozyme. Appropriate conditions for native folding of other RNAs may be similar but must be determined on a case-by-case basis. To a 1.5 ml microcentrifuge tube add:
| Component | Amount | Final concentration |
|---|---|---|
| 10X Buffer | 2.5 µl | 50 mM Na-MOPS, pH 7.0 |
| 10X MgCl2 | 2.5 µl | 10 mM MgCl2 |
| 10X RNA | 2.5 µl | 20 nM – 2 µM |
| H2O | 17.5 µl (less if adding protein; see step 3) |
 Also set up a control reaction (non-DMS control) that will be treated identically except for DMS treatment (steps 5–6). If the experiment includes protein binding, perform two control reactions without DMS: one with protein and one without protein.
Also set up a control reaction (non-DMS control) that will be treated identically except for DMS treatment (steps 5–6). If the experiment includes protein binding, perform two control reactions without DMS: one with protein and one without protein.
 We typically use 2 µM RNA (final concentration) in our DMS footprinting experiments. At this concentration, the RNA precipitates quickly and efficiently at room temperature (20–25 °C) in ethanol (step 8 and step 12). At lower RNA concentrations, incubation at −20 °C for at least an hour may be necessary, as well as the addition of carrier RNA (addressed below).
We typically use 2 µM RNA (final concentration) in our DMS footprinting experiments. At this concentration, the RNA precipitates quickly and efficiently at room temperature (20–25 °C) in ethanol (step 8 and step 12). At lower RNA concentrations, incubation at −20 °C for at least an hour may be necessary, as well as the addition of carrier RNA (addressed below).
2. Incubate RNA under conditions known to give the desired structure: 50 °C for 30 min to generate native Tetrahymena group I ribozyme.
 Incubation conditions for folding RNA can be important for forming native structure. In the case of the Tetrahymena ribozyme, incubation for the equivalent time at 25 °C results in an inactive misfolded ribozyme.
Incubation conditions for folding RNA can be important for forming native structure. In the case of the Tetrahymena ribozyme, incubation for the equivalent time at 25 °C results in an inactive misfolded ribozyme.
Formation of RNA-protein complex (optional)
3. If studying the structure of an RNP, form RNA-Protein Complex. To the folded RNA from step 2, add protein to achieve a final volume of 25 µl.
4. Incubate to allow complete complex formation between the RNA and protein.
 To ensure that the RNA binds quantitatively to the protein, the protein should be present at a concentration well above the Kd and should be in excess of the RNA.
To ensure that the RNA binds quantitatively to the protein, the protein should be present at a concentration well above the Kd and should be in excess of the RNA.
 Remember to include two control reactions without DMS (steps 5–6): one with protein and one without protein.
Remember to include two control reactions without DMS (steps 5–6): one with protein and one without protein.
Chemical modification by DMS
5. Carefully dilute DMS (see REAGENTS) 6-fold in ethanol.
 DMS is extremely toxic and carcinogenic. Wear gloves and safety glasses and work with DMS in a fume hood.
DMS is extremely toxic and carcinogenic. Wear gloves and safety glasses and work with DMS in a fume hood.
6. Add 1 µl of DMS:ethanol solution to RNA (0.65% (v/v) final concentration of DMS) and incubate at 25 °C for 1 minute. For the non-DMS control(s), do not add DMS, but do perform an identical incubation and continue to step 7.
 In carrying out an initial experiment, vary the DMS concentration and incubation time (e.g., 1, 5, and 10 min) to achieve an optimal level of DMS modification (see Experimental Design).
In carrying out an initial experiment, vary the DMS concentration and incubation time (e.g., 1, 5, and 10 min) to achieve an optimal level of DMS modification (see Experimental Design).
Quench reaction and RNA purification
7. Add 475 µl of Quench solution (see REAGENTS). Add the quench solution to the non-DMS control samples as well as the experimental samples
8. Add 2 volumes of ethanol (1 ml) and pellet RNA by centrifugation (16100 × g, 20 minutes at room temperature: 20 – 25 °C).
 If the concentration of RNA is less than 10 µg/ml, add 10 µg of tRNA (or other RNA) as a carrier for RNA precipitation prior to adding the ethanol and incubate at −20 °C for at least an hour. Under the conditions stated in this protocol (a final concentration of 2 µM of a ≥300 nucleotide RNA at step 1), a −20 °C incubation is not necessary because the RNA precipitates quickly and efficiently at room temperature.
If the concentration of RNA is less than 10 µg/ml, add 10 µg of tRNA (or other RNA) as a carrier for RNA precipitation prior to adding the ethanol and incubate at −20 °C for at least an hour. Under the conditions stated in this protocol (a final concentration of 2 µM of a ≥300 nucleotide RNA at step 1), a −20 °C incubation is not necessary because the RNA precipitates quickly and efficiently at room temperature.
9. Remove the supernatant and resuspend the RNA pellet in 0.3 M sodium acetate (100 µl).
 Be very careful not to disturb RNA pellet when removing the supernatant. Disturbing the RNA pellet may result in the loss of the RNA.
Be very careful not to disturb RNA pellet when removing the supernatant. Disturbing the RNA pellet may result in the loss of the RNA.
10. Add an equal volume of Phenol/Chloroform/Isoamyl alcohol and mix well manually or by vortexing. Centrifuge briefly to separate the layers.
11. Carefully remove the top aqueous phase, which contains the RNA, and transfer to a fresh 1.5 ml microcentrifuge tube.
 Avoid carryover of phenol because it can inhibit the subsequent reverse transcriptase reactions (steps 15–24). As the phenol extraction (steps 10 – 11) is mainly to remove proteins, the phenol extraction step is optional for RNA only reactions, but absolutely necessary for RNA-protein reactions.
Avoid carryover of phenol because it can inhibit the subsequent reverse transcriptase reactions (steps 15–24). As the phenol extraction (steps 10 – 11) is mainly to remove proteins, the phenol extraction step is optional for RNA only reactions, but absolutely necessary for RNA-protein reactions.
12. Add 250 µl of ethanol to precipitate the RNA and pellet the RNA by centrifugation (16,100 × g, 20 minutes at room temperature).
 As noted above, if the concentration of RNA is below 10 µg/ml, an incubation at −20 °C for at least an hour may be necessary to ensure efficient precipitation.
As noted above, if the concentration of RNA is below 10 µg/ml, an incubation at −20 °C for at least an hour may be necessary to ensure efficient precipitation.
13. Remove the supernatant, wash the RNA pellet with 100 µl of ice-cold 70% (v/v) ethanol and centrifuge (16100 × g, 20 minutes at room temperature).
14. Carefully and thoroughly remove the 70% (v/v) ethanol, allow the RNA pellet to air dry for 5–10 minutes, and resuspend the RNA pellet in 20 µl of 1X Annealing Buffer.
 It is important to remove residual ethanol because it can interfere with reverse transcription. We recommend removing the last visible traces of ethanol with a small (≤200 µl) pipet tip, followed by leaving the tube sitting on the bench to air dry. An alternative to air drying is to use a vacuum chamber (Speedvac) to dry the pellet. We do not recommend this procedure as it can lead to loss of the pellet and makes resuspension of the pellet difficult, and in our hands the procedure including air drying is sufficient.
It is important to remove residual ethanol because it can interfere with reverse transcription. We recommend removing the last visible traces of ethanol with a small (≤200 µl) pipet tip, followed by leaving the tube sitting on the bench to air dry. An alternative to air drying is to use a vacuum chamber (Speedvac) to dry the pellet. We do not recommend this procedure as it can lead to loss of the pellet and makes resuspension of the pellet difficult, and in our hands the procedure including air drying is sufficient.
 The RNA sample may be stored at −20°C overnight.
The RNA sample may be stored at −20°C overnight.
Reverse Transcription Reaction (Primer extension)
15. Transfer 2 µl of each experimental (and non-DMS control) RNA solution (0.05 – 5 pmol) from step 14 to separate microcentrifuge tubes (or appropriate PCR tubes if using a thermocycler for heating steps) and add 0.5 µl of 32P-labeled primer (50–200k dpm/µl, typically 0.01–0.05 pmol using the protocol above, 32P-labeling of DNA primers).  Always use a acrylic shield when working with 32P. Keep a Geiger counter nearby and periodically monitor your work area for contamination. If the primer is less than 50k dpm/µl, add a larger volume of primer. We have added up to 0.8 µl with no further adjustments to the protocol and without adverse effects on primer extension.
Always use a acrylic shield when working with 32P. Keep a Geiger counter nearby and periodically monitor your work area for contamination. If the primer is less than 50k dpm/µl, add a larger volume of primer. We have added up to 0.8 µl with no further adjustments to the protocol and without adverse effects on primer extension.
16. Incubate at 85 °C for 1 min. This can be done using a heat block or a thermocycler.
17. Quickly cool to room temperature to allow the primer to anneal to the RNA. For RNAs of moderate size, this can be done by simply removing the tube from the heat block or thermocycler. For larger RNAs with more potential binding sites for the primers, it may be necessary to cool slowly using a thermocycler (we have used a program that changes the temperature from 85 °C to 25 °C at 1.0 °C/sec) or by removing your samples in a small tray containing water at 85 °C to allow the solution to cool over ~10 min109.
18. Add 3.5 µl of Primer extension mix (see Reagent Setup) and incubate at 42 °C for 45 min.
Dideoxynucleotide sequencing reactions
19. Prepare sequencing reactions at the same time as the primer extension reactions. Dilute original 10X stock of RNA (not treated with DMS and not the non-DMS control, 10X RNA from step 1 or the stock solution that was used to prepare the RNA in step 1) in 1X annealing buffer (20 nM – 2 µM final RNA concentration). Make 10 µl per primer to be used.
20. To 10 µl of RNA solution, add 2.5 µl of 32P-labeled primer (50–200k dpm/µl)
21. Incubate at 85 °C for 1 min, then cool to room temperature to anneal primer and RNA. (See step 17.)
22. Add 2.5 µl of RNA-primer solution to each of four microcentrifuge tubes.
23. To each tube, add 3.5 µl of a different Sequencing extension mix (see REAGENTS); each tube should therefore contain a different ddNTP.
Hint: It may also be useful to perform a fifth reaction in which Primer extension mix is added instead of Sequencing extension mix as a control reaction to assess the efficiency of the primer extension reaction. If it is working well, this reaction will produce only low levels of incompletely-extended products, with the vast majority of the material present as fully-extended product.
24. Incubate at 42 °C for 45 min. Sequencing reactions should be treated identically to the experimental (primer extension) samples for all subsequent steps
Degradation of RNA
25. Degrade RNA to leave only the 32P-labeled, extended single stranded ssDNA. Add 1µl of 2 N NaOH, mix well, and incubate at 95°C for 3 min; then cool to room temperature by placing tubes on the bench.
26. Add 1µl of 2 N HCl to neutralize reaction, mix well, and then add 1.5 µl of 3 M sodium acetate.
 Check pH of one reaction with pH paper. After establishing that the additions of HCl and sodium acetate give a pH near neutrality (5–8), these two additions can be combined into a single 2.5 µl addition to reduce the number of pipetting steps.
Check pH of one reaction with pH paper. After establishing that the additions of HCl and sodium acetate give a pH near neutrality (5–8), these two additions can be combined into a single 2.5 µl addition to reduce the number of pipetting steps.
27. Add 20 µl of ethanol to precipitate the ssDNA.
28. Centrifuge (16100 × g, 20 minutes at room temperaure) to pellet the ssDNA.
29. Remove supernatant and resuspend the ssDNA pellet in 6 µl of Loading Dye (see Reagents)
 To ensure that the precipitation was successful, determine the relative levels of radioactive material in the supernatant and the pellet with a Geiger counter. If the DNA has precipitated efficiently, the counts should be in the pellet. If significant radioactive material is in the supernatant, add unlabeled carrier RNA or DNA (1 mg/ml), incubate the supernatant at −20 °C for at least an hour and repeat the centrifugation. Again, check the success of the precipitation with a Geiger counter. For the protocol as written, an incubation at −20 °C is not necessary.
To ensure that the precipitation was successful, determine the relative levels of radioactive material in the supernatant and the pellet with a Geiger counter. If the DNA has precipitated efficiently, the counts should be in the pellet. If significant radioactive material is in the supernatant, add unlabeled carrier RNA or DNA (1 mg/ml), incubate the supernatant at −20 °C for at least an hour and repeat the centrifugation. Again, check the success of the precipitation with a Geiger counter. For the protocol as written, an incubation at −20 °C is not necessary.
Denaturing polyacrylamide gel electrophoresis (PAGE)
30. Prepare an 8% (w/v) sequencing-length denaturing polyacrylamide gel (see REAGENT SETUP) with 1X TTE Running Buffer.
 Tris-taurine-EDTA (TTE) buffer is used instead of Tris-borate-EDTA buffer (TBE) to enhance resolution. Taurine is typically recommended as a glycerol-tolerant buffer, and we find that taurine gives sharper bands than borate for this protocol.
Tris-taurine-EDTA (TTE) buffer is used instead of Tris-borate-EDTA buffer (TBE) to enhance resolution. Taurine is typically recommended as a glycerol-tolerant buffer, and we find that taurine gives sharper bands than borate for this protocol.
31. Pre-run the gel for 30–60 min (50–75 W) or until it reaches a temperature of 50–60 °C. (Measure with a thermometer or by touching the plate with the power off. If the gel is warm enough, this will produce discomfort within a few seconds.) Heating the gel is important to prevent the labeled DNA fragments from forming secondary structures during gel migration. We recommend attaching an aluminum plate to the gel plate to ensure a uniform temperature.  High voltage. Never touch the gel apparatus while the power is on.
High voltage. Never touch the gel apparatus while the power is on.
32. Load and run sequencing reactions on the gel alongside DMS experimental lanes. All lanes must be loaded before the run is begun so that all of the lanes will be properly aligned.
 To ensure that each lane on the gel contains approximately the same level of radioactive material, count 1 µl of each sample in a scintillation counter and normalize the loading volumes accordingly.
To ensure that each lane on the gel contains approximately the same level of radioactive material, count 1 µl of each sample in a scintillation counter and normalize the loading volumes accordingly.
 To increase the read length, load each reaction twice (~2–3 µl per load, assuming all samples are equivalent in specific activity). Run the first load for approximately one hour at 50–75 W (until the leading dye, bromophenol blue, is near the bottom of the gel). Then load the second set of samples, in a different set of wells, and run for an additional hour. It is also necessary to load the sequencing reactions again in this second set.
To increase the read length, load each reaction twice (~2–3 µl per load, assuming all samples are equivalent in specific activity). Run the first load for approximately one hour at 50–75 W (until the leading dye, bromophenol blue, is near the bottom of the gel). Then load the second set of samples, in a different set of wells, and run for an additional hour. It is also necessary to load the sequencing reactions again in this second set.
33. After run, remove the gel from the electrophoresis apparatus and carefully separate the plates with a plate-separating tool (a spatula or a plastic wedge. Spatulas tend to break the plates more frequently; we recommend the Wonder Wedge, see Materials). Be very careful to separate the plates with the entire gel remaining only on one of the plates, so that the gel does not tear.
34. With the plate lying on the bench, place a piece of Whatman 3MM paper (35 × 45 cm) on top of the gel and press down all along the gel. The gel should stick to the paper. Carefully pull the gel off the plate and cover with plastic wrap. Cut off the extra paper and plastic wrap around the gel.
35. Place the gel in a gel dryer for ≥30 minutes at 85 °C under a vacuum.
 It is necessary to dry the gel completely, because if it is not dry it may crack extensively when removed from the dryer, which can make analysis impossible. The amount of time necessary for complete drying varies with gel dryer. For most, 30 min is sufficient, but it is a good idea to let it dry a bit longer to ensure that it is completely dry. We have dried gels for up to 90 min without adverse effects.
It is necessary to dry the gel completely, because if it is not dry it may crack extensively when removed from the dryer, which can make analysis impossible. The amount of time necessary for complete drying varies with gel dryer. For most, 30 min is sufficient, but it is a good idea to let it dry a bit longer to ensure that it is completely dry. We have dried gels for up to 90 min without adverse effects.
36. Expose the gel (typically 1 – 16 hours, depending on level of radioactivity used and length of RNA) using a phosphorimager screen.
 Longer exposures (up to 48 hours) are recommended when the labeled primer is below 50k dpm/µl.
Longer exposures (up to 48 hours) are recommended when the labeled primer is below 50k dpm/µl.
37. Scan the image using a phosphorimager.
Analysis
38. Look at the results of the sequencing reactions to determine which bands in the experimental lanes with DMS correspond to modification of which nucleotides.
 Remember that methylation of A or C nucleotides blocks RT one position upstream, so methylation produces a fragment that runs faster by one nucleotide than the corresponding position in the sequencing lanes (see Fig. 1c and Fig. 3a).
Remember that methylation of A or C nucleotides blocks RT one position upstream, so methylation produces a fragment that runs faster by one nucleotide than the corresponding position in the sequencing lanes (see Fig. 1c and Fig. 3a).
39. Determine integrated intensities for each band. Use software such as SAFA134 or ImageQuant TL (Amersham, GE Healthcare) to analyze each position (Fig. 3), or use ImageQuant to manually draw boxes around selected bands for further analysis (Fig. 4).
40. Determine the overall level of DMS modification for each experimental condition. Regardless of the method used in step 39, manually box both the fully-extended product and the entire lane including this product. After subtracting the gel background by drawing boxes of equal size at a location that does not show any signal, compare the intensities of the fully-extended product and the entire lane to determine the fraction of extensions that gave full-length product. If the overall level of DMS modification is one hit per molecule or less and the primer extension is efficient, the intensity from the fully-extended primer should be significant relative to the entire lane. For the example shown in Fig. 3a, the fully-extended bands represent 45–55% of the total intensity. The lane corresponding to the reaction run without DMS should give a higher value than the other lanes and is a direct measure of the efficiency of primer extension and the level of RNase degradation of the RNA.
41. Compare integrated intensity values for the same nucleotide under different experimental conditions (i.e. in the presence and absence of protein or under different solution conditions, Fig. 3b). Quantitative comparison can be made by subtracting the intensity values under one set of conditions from those under a second set of conditions.
 This comparison does not explicitly involve the corresponding intensity value from the reaction run without DMS, but it is important to evaluate this band and not to interpret changes if there is significant intensity in this lane, indicating a background caused by RNA degradation or RT pausing. A good general rule is not to interpret a position unless the intensity from least one of the experimental conditions is at least 2-fold larger than the corresponding band in the absence of DMS53.
This comparison does not explicitly involve the corresponding intensity value from the reaction run without DMS, but it is important to evaluate this band and not to interpret changes if there is significant intensity in this lane, indicating a background caused by RNA degradation or RT pausing. A good general rule is not to interpret a position unless the intensity from least one of the experimental conditions is at least 2-fold larger than the corresponding band in the absence of DMS53.
42. Consider the changes in protection pattern in the context of the secondary and/or tertiary structure of the RNA, if available. The program SAFA can generate a color-coded representation of the difference data if the corresponding secondary structure is entered (Fig. 3c).
Steps 1–14 take 3–4 hours.
Steps 15–35 take 5–7 hours.
Step 36 takes 1–48 hours, depending on the level of radioactivity of the labeled primer.
Steps 37–42 take 2 hours to 1 day, depending on the method of analysis.
Troubleshooting
Problems with the experiment will only become apparent after PAGE (steps 30–37). See Anticipated Results below and Table 1 for troubleshooting advice.
Table 1.
Troubleshooting
| Problem | Reason | Solution |
|---|---|---|
| Smeary gel | RNA is not completely degraded after reverse transcription (see Fig. 5). |
Use pH paper to make sure the pH is 13–14 after NaOH addition (step 25). |
| Sodium acetate from the final precipitation step is present in the ssDNA sample. |
Wash pellet with 70% ethanol (v/v) before resuspending in gel loading dye (step 28). |
|
| Low signal in lanes with DMS | Insufficient reaction of DMS with RNA |
Increase DMS concentration and/or incubation time (steps 5 & 6). |
| Weak full-length bands and strong product bands in lanes with DMS |
Over-reaction of DMS with RNA | Decrease DMS concentration and/or reaction time (steps 5 & 6). |
| Extensive banding in the absence of DMS treatment; good extension in sequencing lanes |
Poor extension by RT caused by problems with DMS treatment step |
Make sure that phenol is not being carried over into the primer extension reaction. If necessary, perform a chloroform extraction after step 11. If footprinting an RNP, check whether bands are present in the ‘no DMS’ lanes, both with and without protein. If the bands are only present with the protein, the protein preparation is likely contaminated with RNase activity. Use an RNase inhibitor or purify the protein further to remove the contaminating RNase. |
| Extensive banding in the absence of DMS treatment and in sequencing lanes |
RNA stock is degraded | Check by end-labeling the RNA and analyzing the length distribution of the prep by PAGE. |
| Poor extension by RT caused by reaction conditions |
RT (step 18) is sensitive to Mg2+ concentration. Make sure the final Mg2+ concentration is 3–4 mM in excess of dNTPs (which bind stoichiometrically to one Mg2+ ion). Try increasing the temperature of the primer extension reaction (up to 50 °C), as the RNA may form secondary structure that inhibits RT, and higher temperature may destabilize this structure. |
|
| Poor extension by RT caused by inactive enzyme |
Replace RT with new enzyme stock (step 18). |
|
| Poor choice of RT enzyme | Switch to a different RT (step 18). While traditionally primer extension is performed using AMV-RT, engineered versions of MMLV such as Superscript (Invitrogen) are also used and may be more effective for a given RNA. |
|
| Primer binds in more than one position on the RNA. (If using more than one primer, this problem will be apparent because the problem will be primer -specific.) |
Construct a new primer (see Primer Extension). Make sure that there are not additional complementary regions within the RNA. |
|
| Additional bands preset because primer is heterogeneous at its 5′-end |
Purify the primer carefully by PAGE (see “32P-labeling of DNA primers” in Reagent Setup). |
|
| No extension in experimental lanes; good extension in sequencing lanes |
RNA lost in precipitation after DMS treatment. |
Add carrier (like RNA or linear acrylamide) for precipitation (step 14) or increase RNA concentration in footprinting reactions. |
| RNA did not resuspend after the pellet was dried |
We recommend air drying rather than using a Speedvac (step 14) to prevent this problem. |
|
| Strong inhibition of RT from phenol carryover |
Remove the aqueous phase carefully (step 11) to ensure that phenol is not carried over. If necessary, perform a chloroform extraction to ensure complete removal of phenol. |
|
| Bands absent or very faint in all lanes, including the band representing the primer |
Radioactive material was lost in final precipitation |
Check pellet and supernatant with a Geiger counter. If necessary, incubate at −20 °C for 1 hr after adding ethanol (step 29). |
| Insufficient label was used in primer extension |
Make sure solution of primer is at least 50k dpm/µl. |
Anticipated Results
There are several features of the data to check to make sure that the experiment has been performed successfully and has given reliable results.
Discrete bands should be visible as a ladder, as shown in Fig. 3a and Fig. 4a. A gel that does not give this result is shown in Fig. 5 (see Smeary gels in Table 1).
If the experiment includes formation of an RNA-protein complex, compare the reactions without DMS that were performed in the presence and absence of protein. If the bands corresponding to incomplete extension are more intense in the reaction that included protein, this suggests RNase contamination of the protein stock. Consider using an RNase inhibitor in a future experiment or further purify the protein (see RNA Handling above).
Bands corresponding to A and C nucleotides should be more intense in the lanes from reactions that included DMS than from the control reaction without DMS. If footprinting an RNP, it is critical to include and check the reaction that included protein but did not include DMS to rule out the possibility that the increased banding arises from RNase contamination of the protein preparation.
Figure 5.
DMS footprinting gel from an experiment in which the RNA degradation steps were omitted (steps 25–26). See Table 1, Smeary gel.
If it is clear that there are DMS-dependent bands and that the intensities differ between experimental conditions, these differences can be interpreted in terms of structure changes upon protein binding or changes in solution conditions. It is anticipated that tertiary folding of the RNA or protein binding will result in the formation of additional contacts, some of which will appear as protection from DMS modification (protections) relative to an unfolded or unbound control reaction. Note, however, that even ‘unfolded’ RNA, in the absence of Mg2+, typically includes regions that remain base-paired135,136 and are therefore protected from DMS modification51. It is also likely that folding or protein binding will result in enhanced DMS reactivity of certain nucleotides or segments, because the formation of contacts may cause certain segments to be exposed to solvent or constrained in an orientation that is strongly favorable to reaction of DMS.
In the case of protein binding, it is not straightforward to determine which protections result from direct contacts with the protein and which result from additional contacts within the RNA induced by protein binding. In certain cases, insight into this question may be gained by performing footprinting in the absence of the protein but in the presence of high concentrations of Mg2+, which may induce the RNA to fold to a similar structure as protein binding at lower Mg2+ concentration. Note that functional assays are critical for establishing whether high Mg2+ concentration does actually give a structure similar to that induced by protein binding. Provided that these structures are similar, protections that appear in the presence of high Mg2+ concentration result from contacts within the RNA, while protections that only appear in the presence of bound protein are strong candidates to reflect direct contacts with the protein137.
Table 2.
Manual boxing analysis from experiment shown in Fig. 4
| No protein | CYT-18 | CYT-18, CYT-19 & ATP | CYT-18, CYT-19 & AMP-PNP | |||||
|---|---|---|---|---|---|---|---|---|
| Intensity* | Fraction | Intensity* | Fraction | Intensity* | Fraction | Intensity* | Fraction | |
| Reference | 11,000 | (1) | 13,000 | (1) | 9,000 | (1) | 11,000 | (1) |
| A57 | 250 | 0.023 | 290 | 0.022 | 120 | 0.013 | 260 | 0.024 |
| A95 | 450 | 0.041 | 610 | 0.047 | 45 | 0.005 | 430 | 0.039 |
| A97 | 340 | 0.031 | 460 | 0.035 | 54 | 0.006 | 350 | 0.032 |
| C98 | 350 | 0.032 | 400 | 0.031 | 36 | 0.004 | 340 | 0.031 |
| C99 | 330 | 0.030 | 380 | 0.029 | 36 | 0.004 | 340 | 0.031 |
| A103 | 560 | 0.051 | 310 | 0.024 | 120 | 0.013 | 530 | 0.048 |
| A104 | 550 | 0.050 | 300 | 0.023 | 110 | 0.012 | 520 | 0.047 |
| A105 | 560 | 0.051 | 310 | 0.024 | 81 | 0.009 | 540 | 0.049 |
Intensity values were obtained by manually boxing each band using ImageQuant. Units are arbitrary.
Acknowledgments
We thank Yaqi Wan for assistance in preparing Fig. 3, and Liz Doherty and Jennifer Doudna for early advice on footprinting and for sharing their protocol. Research in our labs is supported by grants from the NIH (R01-GM070456 to R.R. and R01-GM037951 to Alan M. Lambowitz) and the Welch Foundation (F-1563 to R.R.).
Footnotes
Competing Interests Statement
The authors declare no competing financial interests.
Contributor Information
Pilar Tijerina, Email: pilar@icmb.utexas.edu.
Sabine Mohr, Email: sabine@mail.utexas.edu.
References
- 1.Golden BL. Preparation and crystallization of RNA. Methods Mol. Biol. 363;2007:239–257. doi: 10.1007/978-1-59745-209-0_12. [DOI] [PubMed] [Google Scholar]
- 2.Wu H, Finger LD, Feigon J. Structure determination of protein/RNA complexes by NMR. Methods Enzymol. 2005;394:525–545. doi: 10.1016/S0076-6879(05)94022-6. [DOI] [PubMed] [Google Scholar]
- 3.Myong S, Stevens BC, Ha T. Bridging conformational dynamics and function using single-molecule spectroscopy. Structure. 2006;14:633–643. doi: 10.1016/j.str.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 4.Lipfert J, Doniach S. Small-angle X-ray scattering from RNA, proteins, and protein complexes. Annu. Rev. Biophys. Biomol. Struct. 2007;36:307–327. doi: 10.1146/annurev.biophys.36.040306.132655. [DOI] [PubMed] [Google Scholar]
- 5.Kim NK, Murali A, DeRose VJ. A distance ruler for RNA using EPR and site-directed spin labeling. Chem. Biol. 2004;11:939–948. doi: 10.1016/j.chembiol.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 6.Das R, Travers KJ, Bai Y, Herschlag D. Determining the Mg2+ stoichiometry for folding an RNA metal ion core. J. Am. Chem. Soc. 2005;127:8272–8273. doi: 10.1021/ja051422h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Su LJ, Brenowitz M, Pyle AM. An alternative route for the folding of large RNAs: apparent two-state folding by a group II intron ribozyme. J. Mol. Biol. 2003;334:639–652. doi: 10.1016/j.jmb.2003.09.071. [DOI] [PubMed] [Google Scholar]
- 8.Lilley DM. Analysis of global conformational transitions in ribozymes. Methods Mol. Biol. 2004;252:77–108. doi: 10.1385/1-59259-746-7:077. [DOI] [PubMed] [Google Scholar]
- 9.Tullius TD, Greenbaum JA. Mapping nucleic acid structure by hydroxyl radical cleavage. Curr. Opin. Chem. Biol. 2005;9:127–134. doi: 10.1016/j.cbpa.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 10.Brenowitz M, Chance MR, Dhavan G, Takamoto K. Probing the structural dynamics of nucleic acids by quantitative time-resolved and equilibrium hydroxyl radical "footprinting". Curr. Opin. Struct. Biol. 2002;12:648–653. doi: 10.1016/s0959-440x(02)00366-4. [DOI] [PubMed] [Google Scholar]
- 11.Shcherbakova I, Mitra S, Beer RH, Brenowitz M. Fast Fenton footprinting: a laboratory-based method for the time-resolved analysis of DNA, RNA and proteins. Nucleic Acids Res. 2006;34:e48. doi: 10.1093/nar/gkl055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Culver GM, Noller HF. Directed hydroxyl radical probing of RNA from iron(II) tethered to proteins in ribonucleoprotein complexes. Methods Enzymol. 2000;318:461–475. doi: 10.1016/s0076-6879(00)18070-x. [DOI] [PubMed] [Google Scholar]
- 13.Black DL, Pinto AL. U5 small nuclear ribonucleoprotein: RNA structure analysis and ATP-dependent interaction with U4/U6. Mol. Cell. Biol. 1989;9:3350–3359. doi: 10.1128/mcb.9.8.3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zarrinkar PP, Williamson JR. Kinetic intermediates in RNA folding. Science. 1994;265:918–924. doi: 10.1126/science.8052848. [DOI] [PubMed] [Google Scholar]
- 15.Ryder SP, Ortoleva-Donnelly L, Kosek AB, Strobel SA. Chemical probing of RNA by nucleotide analog interference mapping. Methods Enzymol. 2000;317:92–109. doi: 10.1016/s0076-6879(00)17008-9. [DOI] [PubMed] [Google Scholar]
- 16.Soukup GA, Breaker RR. Relationship between internucleotide linkage geometry and the stability of RNA. RNA. 1999;5:1308–1325. doi: 10.1017/s1355838299990891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE) J Am Chem Soc. 2005;127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 18.Wilkinson KA, Merino EJ, Weeks KM. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 19.Mortimer SA, Weeks KM. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J. Am. Chem. Soc. 2007;129:4144–4145. doi: 10.1021/ja0704028. [DOI] [PubMed] [Google Scholar]
- 20.Russell R, Herschlag D. New pathways in folding of the Tetrahymena group I RNA enzyme. J. Mol. Biol. 1999;291:1155–1167. doi: 10.1006/jmbi.1999.3026. [DOI] [PubMed] [Google Scholar]
- 21.Peattie DA, Gilbert W. Chemical probes for higher-order structure in RNA. Proc. Natl. Acad. Sci. U. S. A. 1980;77:4679–4682. doi: 10.1073/pnas.77.8.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maxam AM, Gilbert W. A new method for sequencing DNA. Proc. Natl. Acad. Sci. U. S. A. 1977;74:560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peattie DA. Direct chemical method for sequencing RNA. Proc. Natl. Acad. Sci. U. S. A. 1979;76:1760–1764. doi: 10.1073/pnas.76.4.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilbert W, Maxam AM, Mirzabekov A. In: Control of Ribosome Synthesis. Kjeldgaard NO, Malaloe O, editors. New York: Academic; 1976. pp. 139–148. [Google Scholar]
- 25.Johnsrud L. Contacts between Escherichia coli RNA polymerase and a lac operon promoter. Proc. Natl. Acad. Sci. U. S. A. 1978;75:5314–5318. doi: 10.1073/pnas.75.11.5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siebenlist U, Gilbert W. Contacts between Escherichia coli RNA polymerase and an early promoter of phage T7. Proc. Natl. Acad. Sci. U. S. A. 1980;77:122–126. doi: 10.1073/pnas.77.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lawley PD, Brookes P. Further studies on the alkylation of nucleic acids and their constituent nucleotides. Biochem. J. 1963;89:127–138. doi: 10.1042/bj0890127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Inoue T, Cech T. Secondary structure of the circular form of the Tetrahymena rRNA intervening sequence: a technique for RNA structure analysis using chemical probes and reverse transcriptase. Proc. Natl. Acad. Sci. USA. 1985;82:648–652. doi: 10.1073/pnas.82.3.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lempereur L, et al. Conformation of yeast 18S rRNA. Direct chemical probing of the 5′ domain in ribosomal subunits and in deproteinized RNA by reverse transcriptase mapping of dimethyl sulfate-accessible sites. Nucleic Acids Res. 1985;13:8339–8357. doi: 10.1093/nar/13.23.8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moazed D, Stern S, Noller HF. Rapid chemical probing of conformation in 16 S ribosomal RNA and 30 S ribosomal subunits using primer extension. J. Mol. Biol. 1986;187:399–416. doi: 10.1016/0022-2836(86)90441-9. [DOI] [PubMed] [Google Scholar]
- 31.Stern S, Wilson RC, Noller HF. Localization of the binding site for protein S4 on 16 S ribosomal RNA by chemical and enzymatic probing and primer extension. J. Mol. Biol. 1986;192:101–110. doi: 10.1016/0022-2836(86)90467-5. [DOI] [PubMed] [Google Scholar]
- 32.Moazed D, Noller HF. Transfer RNA shields specific nucleotides in 16S ribosomal RNA from attack by chemical probes. Cell. 1986;47:985–994. doi: 10.1016/0092-8674(86)90813-5. [DOI] [PubMed] [Google Scholar]
- 33.Egebjerg J, Leffers H, Christensen A, Andersen H, Garrett RA. Structure and accessibility of domain I of Escherichia coli 23 S RNA in free RNA, in the L24-RNA complex and in 50 S subunits. Implications for ribosomal assembly. J. Mol. Biol. 1987;196:125–136. doi: 10.1016/0022-2836(87)90515-8. [DOI] [PubMed] [Google Scholar]
- 34.Baudin F, et al. Higher-order structure of domain III in Escherichia coli 16S ribosomal RNA, 30S subunit and 70S ribosome. Biochimie. 1987;69:1081–1096. doi: 10.1016/0300-9084(87)90008-3. [DOI] [PubMed] [Google Scholar]
- 35.Mougel M, Philippe C, Ebel JP, Ehresmann B, Ehresmann C. The E. coli 16S rRNA binding site of ribosomal protein S15: higher-order structure in the absence and in the presence of the protein. Nucleic Acids Res. 1988;16:2825–2839. doi: 10.1093/nar/16.7.2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Svensson P, Changchien LM, Craven GR, Noller HF. Interaction of ribosomal proteins, S6, S8, S15 and S18 with the central domain of 16 S ribosomal RNA. J. Mol. Biol. 1988;200:301–308. doi: 10.1016/0022-2836(88)90242-2. [DOI] [PubMed] [Google Scholar]
- 37.Stern S, Changchien LM, Craven GR, Noller HF. Interaction of proteins S16, S17 and S20 with 16 S ribosomal RNA. J. Mol. Biol. 1988;200:291–299. doi: 10.1016/0022-2836(88)90241-0. [DOI] [PubMed] [Google Scholar]
- 38.Powers T, Changchien LM, Craven GR, Noller HF. Probing the assembly of the 3′ major domain of 16 S ribosomal RNA. Quaternary interactions involving ribosomal proteins S7, S9 and S19. J. Mol. Biol. 1988;200:309–319. doi: 10.1016/0022-2836(88)90243-4. [DOI] [PubMed] [Google Scholar]
- 39.Powers T, Stern S, Changchien LM, Noller HF. Probing the assembly of the 3′ major domain of 16 S rRNA. Interactions involving ribosomal proteins S2, S3, S10, S13 and S14. J. Mol. Biol. 1988;201:697–716. doi: 10.1016/0022-2836(88)90468-8. [DOI] [PubMed] [Google Scholar]
- 40.Moazed D, Robertson JM, Noller HF. Interaction of elongation factors EF-G and EF-Tu with a conserved loop in 23S RNA. Nature. 1988;334:362–364. doi: 10.1038/334362a0. [DOI] [PubMed] [Google Scholar]
- 41.Moazed D, Noller HF. Interaction of tRNA with 23S rRNA in the ribosomal A, P, and E sites. Cell. 1989;57:585–597. doi: 10.1016/0092-8674(89)90128-1. [DOI] [PubMed] [Google Scholar]
- 42.Moazed D, Noller HF. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 43.Stern S, Powers T, Changchien LM, Noller HF. RNA-protein interactions in 30S ribosomal subunits: folding and function of 16S rRNA. Science. 1989;244:783–790. doi: 10.1126/science.2658053. [DOI] [PubMed] [Google Scholar]
- 44.Moazed D, Noller HF. Sites of interaction of the CCA end of peptidyl-tRNA with 23S rRNA. Proc. Natl. Acad. Sci. U. S. A. 1991;88:3725–3728. doi: 10.1073/pnas.88.9.3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Banerjee AR, Jaeger JA, Turner DH. Thermal unfolding of a group I ribozyme: the low-temperature transition is primarily disruption of tertiary structure. Biochemistry. 1993;32:153–163. doi: 10.1021/bi00052a021. [DOI] [PubMed] [Google Scholar]
- 46.Murphy FL, Cech TR. An independently folding domain of RNA tertiary structure within the Tetrahymena ribozyme. Biochemistry. 1993;32:5291–5300. doi: 10.1021/bi00071a003. [DOI] [PubMed] [Google Scholar]
- 47.Mohr G, Caprara MG, Guo Q, Lambowitz AM. A tyrosyl-tRNA synthetase can function similarly to an RNA structure in the Tetrahymena ribozyme. Nature. 1994;370:147–150. doi: 10.1038/370147a0. [DOI] [PubMed] [Google Scholar]
- 48.Emerick VL, Woodson SA. Fingerprinting the folding of a group I precursor RNA. Proc. Natl. Acad. Sci. U.S.A. 1994;91:9675–9679. doi: 10.1073/pnas.91.21.9675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doherty EA, Doudna JA. The P4-P6 domain directs higher order folding of the Tetrahymena ribozyme core. Biochemistry. 1997;36:3159–3169. doi: 10.1021/bi962428+. [DOI] [PubMed] [Google Scholar]
- 50.Caprara MG, Myers CA, Lambowitz AM. Interaction of the Neurospora crassa mitochondrial tyrosyl-tRNA synthetase (CYT-18 protein) with the group I intron P4-P6 domain. Thermodynamic analysis and the role of metal ions. J. Mol. Biol. 2001;308:165–190. doi: 10.1006/jmbi.2001.4581. [DOI] [PubMed] [Google Scholar]
- 51.Russell R, et al. Exploring the folding landscape of a structured RNA. Proc. Natl. Acad. Sci. U.S.A. 2002;99:155–160. doi: 10.1073/pnas.221593598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mohr S, Stryker JM, Lambowitz AM. A DEAD-box protein functions as an ATP-dependent RNA chaperone in group I intron splicing. Cell. 2002;109:769–779. doi: 10.1016/s0092-8674(02)00771-7. [DOI] [PubMed] [Google Scholar]
- 53.Russell R, et al. The paradoxical behavior of a highly structured misfolded intermediate in RNA folding. J. Mol. Biol. 2006;363:531–544. doi: 10.1016/j.jmb.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 54.Russell R, Tijerina P, Chadee AB, Bhaskaran H. Deletion of the P5abc peripheral element accelerates early and late folding steps of the Tetrahymena group I ribozyme. Biochemistry. 2007;46:4951–4961. doi: 10.1021/bi0620149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Costa M, Christian EL, Michel F. Differential chemical probing of a group II self-splicing intron identifies bases involved in tertiary interactions and supports an alternative secondary structure model of domain V. RNA. 1998;4:1055–1068. doi: 10.1017/s1355838298980670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Konforti BB, Liu Q, Pyle AM. A map of the binding site for catalytic domain 5 in the core of a group II intron ribozyme. EMBO J. 1998;17:7105–7117. doi: 10.1093/emboj/17.23.7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tranguch AJ, Kindelberger DW, Rohlman CE, Lee JY, Engelke DR. Structure-sensitive RNA footprinting of yeast nuclear ribonuclease P. Biochemistry. 1994;33:1778–1787. doi: 10.1021/bi00173a022. [DOI] [PubMed] [Google Scholar]
- 58.Kumar PK, Taira K, Nishikawa S. Chemical probing studies of variants of the genomic hepatitis delta virus ribozyme by primer extension analysis. Biochemistry. 1994;33:583–592. doi: 10.1021/bi00168a025. [DOI] [PubMed] [Google Scholar]
- 59.Butcher SE, Burke JM. Structure-mapping of the hairpin ribozyme. Magnesium-dependent folding and evidence for tertiary interactions within the ribozyme-substrate complex. J. Mol. Biol. 1994;244:52–63. doi: 10.1006/jmbi.1994.1703. [DOI] [PubMed] [Google Scholar]
- 60.Keiper S, Bebenroth D, Seelig B, Westhof E, Jaschke A. Architecture of a Diels-Alderase ribozyme with a preformed catalytic pocket. Chem. Biol. 2004;11:1217–1227. doi: 10.1016/j.chembiol.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 61.Wang YH, Sczekan SR, Theil EC. Structure of the 5′ untranslated regulatory region of ferritin mRNA studied in solution. Nucleic Acids Res. 1990;18:4463–4468. doi: 10.1093/nar/18.15.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rettberg CC, Prere MF, Gesteland RF, Atkins JF, Fayet O. A three-way junction and constituent stem-loops as the stimulator for programmed -1 frameshifting in bacterial insertion sequence IS911. J. Mol. Biol. 1999;286:1365–1378. doi: 10.1006/jmbi.1999.2546. [DOI] [PubMed] [Google Scholar]
- 63.Fernandez-Miragall O, Martinez-Salas E. Structural organization of a viral IRES depends on the integrity of the GNRA motif. RNA. 2003;9:1333–1344. doi: 10.1261/rna.5950603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ruschak A, et al. Secondary structure models of the 3′ untranslated regions of diverse R2 RNAs. RNA. 2004;10:978–987. doi: 10.1261/rna.5216204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rose MA, Weeks KM. Visualizing induced fit in early assembly of the human signal recognition particle. Nat. Struct. Biol. 2001;8:515–520. doi: 10.1038/88577. [DOI] [PubMed] [Google Scholar]
- 66.Menichelli E, Isel C, Oubridge C, Nagai K. Protein-induced conformational changes of RNA during the assembly of human signal recognition particle. J. Mol. Biol. 2007;367:187–203. doi: 10.1016/j.jmb.2006.12.056. [DOI] [PubMed] [Google Scholar]
- 67.Krol A, et al. Solution structure of human U1 snRNA. Derivation of a possible three-dimensional model. Nucleic Acids Res. 1990;18:3803–3811. doi: 10.1093/nar/18.13.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hartmuth K, Raker VA, Huber J, Branlant C, Luhrmann R. An unusual chemical reactivity of Sm site adenosines strongly correlates with proper assembly of core U snRNP particles. J. Mol. Biol. 1999;285:133–147. doi: 10.1006/jmbi.1998.2300. [DOI] [PubMed] [Google Scholar]
- 69.Ast G, Pavelitz T, Weiner AM. Sequences upstream of the branch site are required to form helix II between U2 and U6 snRNA in a trans-splicing reaction. Nucleic Acids Res. 2001;29:1741–1749. doi: 10.1093/nar/29.8.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brunel C, Romby P, Westhof E, Ehresmann C, Ehresmann B. Three-dimensional model of Escherichia coli ribosomal 5 S RNA as deduced from structure probing in solution and computer modeling. J. Mol. Biol. 1991;221:293–308. doi: 10.1016/0022-2836(91)80220-o. [DOI] [PubMed] [Google Scholar]
- 71.Mathews DH, et al. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl. Acad. Sci. U. S. A. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lavery R, Pullman A. A new theoretical index of biochemical reactivity combining steric and electrostatic factors An application to yeast tRNAPhe. Biophys. Chem. 1984;19:171–181. doi: 10.1016/0301-4622(84)85017-6. [DOI] [PubMed] [Google Scholar]
- 73.Muth GW, Ortoleva-Donnelly L, Strobel SA. A single adenosine with a neutral pKa in the ribosomal peptidyl transferase center. Science. 2000;289:947–950. doi: 10.1126/science.289.5481.947. [DOI] [PubMed] [Google Scholar]
- 74.Bayfield MA, Dahlberg AE, Schulmeister U, Dorner S, Barta A. A conformational change in the ribosomal peptidyl transferase center upon active/inactive transition. Proc. Natl. Acad. Sci. U. S. A. 2001;98:10096–10101. doi: 10.1073/pnas.171319598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Muth GW, Chen L, Kosek AB, Strobel SA. pH-dependent conformational flexibility within the ribosomal peptidyl transferase center. RNA. 2001;7:1403–1415. [PMC free article] [PubMed] [Google Scholar]
- 76.Altuvia S, Kornitzer D, Teff D, Oppenheim AB. Alternative mRNA structures of the cIII gene of bacteriophage lambda determine the rate of its translation initiation. J. Mol. Biol. 1989;210:265–280. doi: 10.1016/0022-2836(89)90329-x. [DOI] [PubMed] [Google Scholar]
- 77.Matsumura S, Ikawa Y, Inoue T. Biochemical characterization of the kink-turn RNA motif. Nucleic Acids Res. 2003;31:5544–5551. doi: 10.1093/nar/gkg760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Egebjerg J, Christiansen J, Brown RS, Larsen N, Garrett RA. Protein L18 binds primarily at the junctions of helix II and internal loops A and B in Escherichia coli 5 S RNA. Implications for 5 S RNA structure. J. Mol. Biol. 1989;206:651–668. doi: 10.1016/0022-2836(89)90573-1. [DOI] [PubMed] [Google Scholar]
- 79.Moazed D, Samaha RR, Gualerzi C, Noller HF. Specific protection of 16 S rRNA by translational initiation factors. J. Mol. Biol. 1995;248:207–210. doi: 10.1016/s0022-2836(95)80042-5. [DOI] [PubMed] [Google Scholar]
- 80.Merryman C, Moazed D, McWhirter J, Noller HF. Nucleotides in 16S rRNA protected by the association of 30S and 50S ribosomal subunits. J. Mol. Biol. 1999;285:97–105. doi: 10.1006/jmbi.1998.2242. [DOI] [PubMed] [Google Scholar]
- 81.Dahlquist KD, Puglisi JD. Interaction of translation initiation factor IF1 with the E. coli ribosomal A site. J. Mol. Biol. 2000;299:1–15. doi: 10.1006/jmbi.2000.3672. [DOI] [PubMed] [Google Scholar]
- 82.Green CD, Long KS, Shi H, Wolin SL. Binding of the 60-kDa Ro autoantigen to Y RNAs: evidence for recognition in the major groove of a conserved helix. RNA. 1998;4:750–765. doi: 10.1017/s1355838298971667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Argaman L, Altuvia S. fhlA repression by OxyS RNA: kissing complex formation at two sites results in a stable antisense-target RNA complex. J. Mol. Biol. 2000;300:1101–1112. doi: 10.1006/jmbi.2000.3942. [DOI] [PubMed] [Google Scholar]
- 84.Moazed D, Noller HF. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature. 1987;327:389–394. doi: 10.1038/327389a0. [DOI] [PubMed] [Google Scholar]
- 85.Woodcock J, Moazed D, Cannon M, Davies J, Noller HF. Interaction of antibiotics with A- and P-site-specific bases in 16S ribosomal RNA. EMBO J. 1991;10:3099–3103. doi: 10.1002/j.1460-2075.1991.tb07863.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Spickler C, Brunelle MN, Brakier-Gingras L. Streptomycin binds to the decoding center of 16 S ribosomal RNA. J. Mol. Biol. 1997;273:586–599. doi: 10.1006/jmbi.1997.1323. [DOI] [PubMed] [Google Scholar]
- 87.Purohit P, Stern S. Interactions of a small RNA with antibiotic and RNA ligands of the 30S subunit. Nature. 1994;370:659–662. doi: 10.1038/370659a0. [DOI] [PubMed] [Google Scholar]
- 88.Moazed D, Van Stolk BJ, Douthwaite S, Noller HF. Interconversion of active and inactive 30 S ribosomal subunits is accompanied by a conformational change in the decoding region of 16 S rRNA. J. Mol. Biol. 1986;191:483–493. doi: 10.1016/0022-2836(86)90143-9. [DOI] [PubMed] [Google Scholar]
- 89.Powers T, Daubresse G, Noller HF. Dynamics of in vitro assembly of 16 S rRNA into 30 S ribosomal subunits. J. Mol. Biol. 1993;232:362–374. doi: 10.1006/jmbi.1993.1396. [DOI] [PubMed] [Google Scholar]
- 90.Powers T, Noller HF. A temperature-dependent conformational rearrangement in the ribosomal protein S4.16 S rRNA complex. J. Biol. Chem. 1995;270:1238–1242. doi: 10.1074/jbc.270.3.1238. [DOI] [PubMed] [Google Scholar]
- 91.Karginov FV, Uhlenbeck OC. Interaction of Escherichia coli DbpA with 23S rRNA in different functional states of the enzyme. Nucleic Acids Res. 2004;32:3028–3032. doi: 10.1093/nar/gkh640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hennelly SP, et al. A time-resolved investigation of ribosomal subunit association. J. Mol. Biol. 2005;346:1243–1258. doi: 10.1016/j.jmb.2004.12.054. [DOI] [PubMed] [Google Scholar]
- 93.Fabbretti A, et al. The real-time path of translation factor IF3 onto and off the ribosome. Mol Cell. 2007;25:285–296. doi: 10.1016/j.molcel.2006.12.011. [DOI] [PubMed] [Google Scholar]
- 94.Herr W, Chapman NM, Noller HF. Mechanism of ribosomal subunit association: discrimination of specific sites in 16 S RNA essential for association activity. J. Mol. Biol. 1979;130:433–449. doi: 10.1016/0022-2836(79)90433-9. [DOI] [PubMed] [Google Scholar]
- 95.Peattie DA, Herr W. Chemical probing of the tRNA--ribosome complex. Proc. Natl. Acad. Sci. U. S. A. 1981;78:2273–2277. doi: 10.1073/pnas.78.4.2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.von Ahsen U, Noller HF. Identification of bases in 16S rRNA essential for tRNA binding at the 30S ribosomal P site. Science. 1995;267:234–237. doi: 10.1126/science.7528943. [DOI] [PubMed] [Google Scholar]
- 97.Pan J, Woodson SA. Folding intermediates of a self-splicing RNA: mispairing of the catalytic core. J. Mol. Biol. 1998;280:597–609. doi: 10.1006/jmbi.1998.1901. [DOI] [PubMed] [Google Scholar]
- 98.Zaug AJ, Cech TR. Analysis of the structure of Tetrahymena nuclear RNAs in vivo: telomerase RNA, the self-splicing rRNA intron, and U2 snRNA. RNA. 1995;1:363–374. [PMC free article] [PubMed] [Google Scholar]
- 99.Charpentier B, Stutz F, Rosbash M. A dynamic in vivo view of the HIV-I Rev-RRE interaction. J. Mol. Biol. 1997;266:950–962. doi: 10.1006/jmbi.1996.0858. [DOI] [PubMed] [Google Scholar]
- 100.Iseni F, Baudin F, Blondel D, Ruigrok RW. Structure of the RNA inside the vesicular stomatitis virus nucleocapsid. RNA. 2000;6:270–281. doi: 10.1017/s135583820099109x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bricker AL, Belasco JG. Importance of a 5′ stem-loop for longevity of papA mRNA in Escherichia coli. J. Bacteriol. 1999;181:3587–3590. doi: 10.1128/jb.181.11.3587-3590.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Doktycz MJ, Larimer FW, Pastrnak M, Stevens A. Comparative analyses of the secondary structures of synthetic and intracellular yeast MFA2 mRNAs. Proc. Natl. Acad. Sci. U. S. A. 1998;95:14614–14621. doi: 10.1073/pnas.95.25.14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Luo D, Condon C, Grunberg-Manago M, Putzer H. In vitro and in vivo secondary structure probing of the thrS leader in Bacillus subtilis. Nucleic Acids Res. 1998;26:5379–5387. doi: 10.1093/nar/26.23.5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Waldsich C, Grossberger R, Schroeder R. RNA chaperone StpA loosens interactions of the tertiary structure in the td group I intron in vivo. Genes Dev. 2002;16:2300–2312. doi: 10.1101/gad.231302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Disney MD, Haidaris CG, Turner DH. Uptake and antifungal activity of oligonucleotides in Candida albicans. Proc. Natl. Acad. Sci. U. S. A. 2003;100:1530–1534. doi: 10.1073/pnas.0337462100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wells SE, Hughes JM, Igel AH, Ares M., Jr Use of dimethyl sulfate to probe RNA structure in vivo. Methods Enzymol. 2000;318:479–493. doi: 10.1016/s0076-6879(00)18071-1. [DOI] [PubMed] [Google Scholar]
- 107.Ehresmann C, et al. Probing the structure of RNAs in solution. Nucleic Acids Res. 1987;15:9109–9128. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Christiansen J, Garrett R. Enzymatic and chemical probing of ribosomal RNA-protein interactions. Methods Enzymol. 1988;164:456–468. doi: 10.1016/s0076-6879(88)64061-4. [DOI] [PubMed] [Google Scholar]
- 109.Stern S, Moazed D, Noller HF. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 1988;164:481–489. doi: 10.1016/s0076-6879(88)64064-x. [DOI] [PubMed] [Google Scholar]
- 110.Merryman C, Noller HF. In: RNA:Protein Interactions, A Practical Approach. Smith CWJ, editor. Oxford: Oxford University Press; 1998. pp. 237–253. [Google Scholar]
- 111.Brunel C, Romby P. Probing RNA structure and RNA-ligand complexes with chemical probes. Methods Enzymol. 2000;318:3–21. doi: 10.1016/s0076-6879(00)18040-1. [DOI] [PubMed] [Google Scholar]
- 112.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U. S. A. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cate JH, et al. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 114.Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc. Natl. Acad. Sci. U.S.A. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Balasubramanian B, Pogozelski WK, Tullius TD. DNA strand breaking by the hydroxyl radical is governed by the accessible surface areas of the hydrogen atoms of the DNA backbone. Proc. Natl. Acad. Sci. U.S.A. 1998;95:9738–9743. doi: 10.1073/pnas.95.17.9738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Latham JA, Cech TR. Defining the inside and outside of a catalytic RNA molecule. Science. 1989;245:276–282. doi: 10.1126/science.2501870. [DOI] [PubMed] [Google Scholar]
- 117.King PA, Jamison E, Strahs D, Anderson VE, Brenowitz M. 'Footprinting' proteins on DNA with peroxonitrous acid. Nucleic Acids Res. 1993;21:2473–2478. doi: 10.1093/nar/21.10.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chaulk SG, MacMillan AM. Characterization of the Tetrahymena ribozyme folding pathway using the kinetic footprinting reagent peroxynitrous acid. Biochemistry. 2000;39:2–8. doi: 10.1021/bi992167e. [DOI] [PubMed] [Google Scholar]
- 119.Kent O, Chaulk SG, MacMillan AM. Kinetic analysis of the M1 RNA folding pathway. J. Mol. Biol. 2000;304:699–705. doi: 10.1006/jmbi.2000.4263. [DOI] [PubMed] [Google Scholar]
- 120.Kwok LW, et al. Concordant exploration of the kinetics of RNA folding from global and local perspectives. J. Mol. Biol. 2006;355:282–293. doi: 10.1016/j.jmb.2005.10.070. [DOI] [PubMed] [Google Scholar]
- 121.Lindahl T, Adams A, Fresco JR. Renaturation of transfer ribonucleic acids through site binding of magnesium. Proc. Natl. Acad. Sci. U.S.A. 1966;55:941–948. doi: 10.1073/pnas.55.4.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Weidner H, Crothers DM. Pathway-dependent refolding of E. coli 5S RNA. Nucleic Acids Res. 1977;4:3401–3414. doi: 10.1093/nar/4.10.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sugimoto N, Kierzek R, Turner DH. Kinetics for reaction of a circularized intervening sequence with CU, UCU, CUCU, and CUCUCU: mechanistic implications from the dependence on temperature and on oligomer and Mg2+ concentrations. Biochemistry. 1988;27:6384–6392. doi: 10.1021/bi00417a029. [DOI] [PubMed] [Google Scholar]
- 124.Walstrum SA, Uhlenbeck OC. The self-splicing RNA of Tetrahymena is trapped in a less active conformation by gel purification. Biochemistry. 1990;29:10573–10576. doi: 10.1021/bi00498a022. [DOI] [PubMed] [Google Scholar]
- 125.Herschlag D, Cech TR. Catalysis of RNA cleavage by the Tetrahymena thermophila ribozyme. 1. Kinetic description of the reaction of an RNA substrate complementary to the active site. Biochemistry. 1990;29:10159–10171. doi: 10.1021/bi00496a003. [DOI] [PubMed] [Google Scholar]
- 126.Zarrinkar PP, Wang J, Williamson JR. Slow folding kinetics of RNase P RNA. RNA. 1996;2:564–573. [PMC free article] [PubMed] [Google Scholar]
- 127.Pan J, Thirumalai D, Woodson SA. Folding of RNA involves parallel pathways. J. Mol. Biol. 1997;273:7–13. doi: 10.1006/jmbi.1997.1311. [DOI] [PubMed] [Google Scholar]
- 128.Esteban JA, Banerjee AR, Burke JM. Kinetic mechanism of the hairpin ribozyme. Identification and characterization of two nonexchangeable conformations. J. Biol. Chem. 1997;272:13629–13639. doi: 10.1074/jbc.272.21.13629. [DOI] [PubMed] [Google Scholar]
- 129.Pan T, Sosnick TR. Intermediates and kinetic traps in the folding of a large ribozyme revealed by circular dichroism and UV absorbance spectroscopies and catalytic activity. Nat. Struct. Biol. 1997;4:931–938. doi: 10.1038/nsb1197-931. [DOI] [PubMed] [Google Scholar]
- 130.Russell R, Herschlag D. Probing the folding landscape of the Tetrahymena ribozyme: Commitment to form the native conformation is late in the folding pathway. J. Mol. Biol. 2001;308:839–851. doi: 10.1006/jmbi.2001.4751. [DOI] [PubMed] [Google Scholar]
- 131.Pichler A, Schroeder R. Folding problems of the 5′ splice site containing the P1 stem of the group I thymidylate synthase intron: substrate binding inhibition in vitro and mis-splicing in vivo. J. Biol. Chem. 2002;277:17987–17993. doi: 10.1074/jbc.M111798200. [DOI] [PubMed] [Google Scholar]
- 132.Chadalavada DM, Senchak SE, Bevilacqua PC. The folding pathway of the genomic hepatitis delta virus ribozyme is dominated by slow folding of the pseudoknots. J. Mol. Biol. 2002;317:559–575. doi: 10.1006/jmbi.2002.5434. [DOI] [PubMed] [Google Scholar]
- 133.Gerard GF, Fox DK, Nathan M, D'Alessio JM. Reverse transcriptase. The use of cloned Moloney murine leukemia virus reverse transcriptase to synthesize DNA from RNA. Mol. Biotechnol. 1997;8:61–77. doi: 10.1007/BF02762340. [DOI] [PubMed] [Google Scholar]
- 134.Das R, Laederach A, Pearlman SM, Herschlag D, Altman RB. SAFA: Semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA. 2005;11:344–354. doi: 10.1261/rna.7214405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Stein A, Crothers DM. Conformational changes of transfer RNA. The role of magnesium(II) Biochemistry. 1976;15:160–168. doi: 10.1021/bi00646a025. [DOI] [PubMed] [Google Scholar]
- 136.Grilley D, Soto AM, Draper DE. Mg2+-RNA interaction free energies and their relationship to the folding of RNA tertiary structures. Proc. Natl. Acad. Sci. U. S. A. 2006;103:14003–14008. doi: 10.1073/pnas.0606409103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Matsuura M, Noah JW, Lambowitz AM. Mechanism of maturase-promoted group II intron splicing. EMBO J. 2001;20:7259–7270. doi: 10.1093/emboj/20.24.7259. [DOI] [PMC free article] [PubMed] [Google Scholar]