Abstract
Solvent molecules play key roles in the conformational dynamics of proteins. Here we use single molecule force-clamp spectroscopy to probe the role played by the stabilizing osmolyte glycerol on the conformational ensembles visited by a single ubiquitin protein folding after mechanical extension. Using a variety of force-pulse protocols, we find that glycerol stabilizes the native state of ubiquitin, making it more resistant to mechanical unfolding. We also find that although glycerol enhanced the hydrophobic collapse of unfolded and highly extended ubiquitins, it had no effect on the resulting collapsed states that are essential precursors of the folded state. These disparate effects of glycerol may be the result of distinct structural roles played by solvent molecules at the transition state of each folding ensemble. Our results open the way for a detailed analysis of the transition state structures that form along the folding trajectory of a mechanically extended protein.
Keywords: force-clamp, glycerol, single molecule, solvent
Understanding the mechanisms by which proteins acquire their native topology remains an important question in biology. The acquisition of the native state involves a delicate interplay between the structural search for the native state and desolvation of the solvent exposed protein backbone. Water molecules are inextricably involved in this dynamic conversion between the different protein conformations encountered along the folding pathway (1). Furthermore, water molecules mediate the hydrogen bonding network that determines the final structure of the native state of the protein and the dynamics of contraction of the unfolded polypeptide (2, 3). A comprehensive description of the (un)folding reaction must therefore incorporate the solvating environment that envelopes the protein and determines its structure, dynamics, and often function (1, 4). Studies using bulk experimental techniques have provided a wealth of information on the thermodynamic properties of proteins upon modification of the solvent environment (5–8) and molecular mechanisms for osmolyte induced protein stability have been proposed (9). However, direct insight into the role of osmolyte molecules on the structure of the transition state of a protein, which is the main determinant of protein dynamics, can only be gained at the single molecule level.
Single molecule force spectroscopy is a valuable tool to uncover the structure of the transition state of a protein under the application of a stretching force along a well-defined coordinate, the end-to-end length (10, 11). While earlier pioneering experiments allowed location of the mechanical transition state of several proteins (12–14), molecular dynamics simulations complemented those experimental findings by providing atomic-level insight into the key events that determine protein unfolding, namely the dynamic rupture of interstrand hydrogen bonds (15). Most recently, the combination of solvent substitution with force-clamp spectroscopy has proved successful in experimentally providing a molecular perspective on the role of solvent molecules in protein unfolding (16, 17) and in understanding the driving forces involved in the collapse of an extended polypeptide (18). Specifically, the use of solvent substitution provides a useful tool to uncover the molecular architecture of the mechanical transition state of proteins in detail. These experiments demonstrated that solvent bridging plays a pivotal role in the mechanical stability of the native state of the I27 protein under force (16). However, despite this significant progress, these studies have been mostly limited to the study of thermodynamically stable states, namely the unfolded and the folded structures (16, 18), and little is known about the transition state structures of the evolving conformations acquired by the polypeptide as it folds.
A significant advantage of single molecule techniques is their ability to individually monitor the full folding trajectory of a single molecule (19, 20). In the case of mechanical experiments, applying a stretching force to a protein slows down the folding mechanism exponentially, which permits capturing the conformational dynamics followed by a single protein from a totally extended conformation to its natively refolded form (21–23). Recent measurements conducted in constant force mode have allowed us to separate the different stages encompassing the folding reaction of the small protein ubiquitin (22, 24). These experiments permitted identification of 3 distinct conformational ensembles visited during the folding process by probing the mechanical stability of each protein conformation as its structural fingerprint. This ensemble of conformations comprise the folded and unfolded protein forms, along with a recently discovered ensemble of minimum energy collapsed structures (MECS) that are obligate precursors to the native form (25). Such structurally heterogeneous and mechanically labile MECS originate after the protein's hydrophobic collapse, and interconvert into the native state following a 2-state, rate limiting reaction (26–28). Interestingly, this set of mechanically labile structures has been also identified in analogous experiments using ubiquitin monomers, suggesting that they are not a particular feature of polyproteins (25).
The capability of force-clamp spectroscopy to drive a single protein to different regions of the energy landscape provides the unprecedented opportunity to probe the effect of solvents on each distinct conformational ensemble visited by a single ubiquitin protein along its folding pathway (18, 22, 25). Using a variety of force-pulse protocols, here we show that glycerol, a protecting osmolyte (7), readily separates the distinct phases of the mechanical folding of ubiquitin by stabilizing the native state. In contrast to the glycerol sensitivity of the extended and native state of ubiquitin, we measure that the mechanical resistance of an ensemble of collapsed states is largely insensitive to the solvent environment. Our experiments reveal the role of glycerol in the different phases of the folding reaction while providing plausible molecular interpretations of the mechanical transition state of each particular protein conformation.
Results
Broad Separation of Folding Phases in Glycerol Solutions.
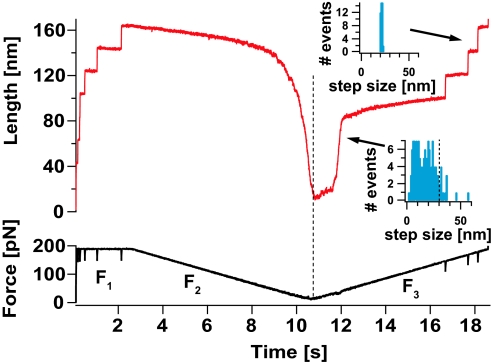
To capture and fully separate the conformational dynamics sampled by a single protein during its folding trajectory, we first apply a force-ramp protocol. This protocol permits the observation of the full force-length relationship of an extended polyprotein during protein collapse and folding. Fig. 1 shows a single ubiquitin polyprotein in 30% vol/vol aqueous glycerol being first unfolded under a constant high force of 190 pN to a highly extended conformation where all of the residues are exposed to the solvent. Under force-clamp conditions, stretching a ubiquitin polyprotein results in a well-defined series of step increases in length of approximately 20 nm, marking the unfolding and extension of the individual modules in the chain (F1 regime). The force is then relaxed linearly down to 10 pN, while monitoring the end-to-end length of the protein as it collapses (F2). Upon reaching 10 pN the force is ramped back up again to 190 pN (F3). As the force begins to rise, the collapsed polypeptide immediately shows resistance to extension. When the force reaches approximately 50–60 pN, we observe a rapid stepwise extension of the collapsed conformations. The step size distribution for the unraveling of such collapsed structures in glycerol is broad (Bottom Inset) and similar to that measured in PBS solution (25). Interestingly, the majority (≈90%) of the measured step sizes lies below 29 nm (dashed line in the histogram Inset), indicating that the distribution of step sizes corresponding to the collapsed conformations is largely contained within the contour length of a single ubiquitin monomer. This result suggests that just after hydrophobic collapse, the polyprotein chain rapidly segregates into its individual ubiquitin monomers. A small number of larger steps (≈10%) were also observed, because of either lack of sufficient time resolution or conformations that involved interactions between neighboring ubiquitin domains (29–31). Although multiple domains in a polyubiquitin chain are known to interact with each other forming ubiquitin dimers (31) and tetramers (29), such inherently flexible complexes are likely to be the result of lattice interactions rather than stable conformations (29). Thus, it is not likely that intra-chain binding plays a role in the collapsing dynamics that we observe. At a much higher force, in excess of 150 pN, we begin to observe the unfolding of the stabilized native ubiquitin proteins that have had time to refold within the short time that the protein was held in its collapsed length, exhibiting a narrow distribution of step sizes centered at ≈20 nm (Top Inset). This experiment clearly demonstrates the 3 distinct phases of a folding polypeptide separated by the different effects of glycerol, namely the native folded protein, the extended protein and the collapsed protein. It is important to note that the 3 phases are observed in the same polypeptide molecule as it progresses toward the native state. This presents a unique opportunity to separate the timescales of the distinct folding ensembles within a single molecule experiment. Independently studying the effect of force and solvent substitution on the native, unfolded, and collapsed states of ubiquitin provides an ideal experimental platform to compare their distinct properties.
Fig. 1.
Wide separation of the folding ensembles in glycerol solutions. We use a force-ramp protocol in 30% glycerol solution to completely separate the 3 conformational ensembles of a folding polypeptide, demonstrating that they are structurally distinct. The ubiquitin polyprotein is first unfolded under a constant force of 190 pN (F1 regime) during 2.5 s, resulting in a well-defined series of step increases in length of 20 nm that mark the unfolding and extension of the individual modules in the chain. The force is then relaxed linearly down to 10 pN, while monitoring the contraction of the end-to-end length of the highly extended ubiquitin polyprotein as it undergoes hydrophobic collapse (F2). Upon reaching 10 pN, the force is linearly ramped back up again to 190 pN (F3). When the force value reaches approximately 50–60 pN, the protein undergoes a heterogeneous stepwise extension (Bottom Inset) marking the unraveling of the ensemble of collapsed conformations. At a much higher force of approximately 150 pN we observe the appearance of the 20 nm steps (Top Inset) that mark the native ensemble.
Probing the Physicochemical Properties of the Native Conformations.
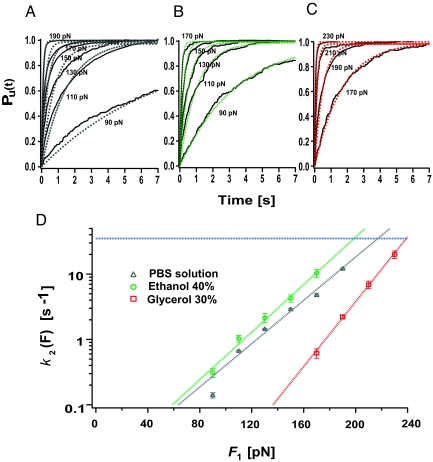
We begin by measuring the properties of the native conformations of the ubiquitin protein by measuring their unfolding force dependency under different solvent environments. In our experiments, we probe the mechanical properties of the native state of mechanically stable proteins by studying their unfolding kinetics under the effect of a constant stretching force (Fig. 2A) (10, 32). To gain insight into the molecular structure of the mechanical transition state of unfolding, we measure the force dependency of the unfolding rate of single polyproteins using the Bell model (10, 16, 24, 33), which allows us to estimate the distance to the transition state, Δx, and the height of the energy barrier, ΔG, that define the unfolding reaction as described in the supporting information (SI) Text. In combination with solvent substitution, these experiments have revealed the molecular architecture of the unfolding transition state of the protein I27, demonstrating that solvent bridging in the transition state region determines Δx. In particular, recent experiments showed that upon replacement of water by increasing amounts of the larger molecule glycerol, the distance to the unfolding transition state increases from Δx = 2.5 Å, the size of a water molecule, up to Δx = 4.4 Å, consistent with the size of a glycerol molecule. Furthermore, the replacement of water with glycerol greatly increases ΔG, thereby mechanically stabilizing the native state (16). Whether this trend represents a general mechanism for the transition state architecture of other mechanically stable proteins such as ubiquitin remains experimentally unexplored.
Fig. 2.
The mechanical stability of the native state of ubiquitin is strongly dependent on the solvent environment. Applying a constant stretching force to a ubiquitin polyprotein results in length versus time trajectories showing the characteristic staircase of unfolding events, where each step of 20 nm marks the individual unfolding of a ubiquitin module in the chain. The average time-course of unfolding was obtained by summation and normalization of n > 50 recordings. (A–C) Multiple trace averages of unfolding events for ubiquitin in (A) PBS aqueous solution, data from (25), (B) ethanol solution (40% vol/vol), and (C) glycerol solution (30% vol/vol) at different constant pulling forces. We obtain the average rate of unfolding of ubiquitin by fitting a single exponential to the average time-course of unfolding at each particular force. (D) Semilogarithmic plot of the rate of unfolding of ubiquitin, k2, as a function of the pulling force in; PBS aqueous solution, gray triangles; 40% ethanol, green circles; 30% glycerol, red squares, obtained from the data shown in (A–C). The dashed line in each case represents the fit of the Arrhenius/Bell term, k2(F) = k0,2exp(FΔx2/kT), to the experimental data. From this fit we obtain the values of the rate constant in the absence of force, k0,2, and the distance to the transition state, Δx2, for each different solvent environment. Assuming a prefactor A = 106 s−1 we can estimate the height of the energy barrier of unfolding, ΔG2, by using the relationship k0,2 = Aexp(−ΔG2/kT). The obtained values are shown in Table S1. These results show that both ΔG2 and Δx2 are strongly dependent on the solvent environment. Blue horizontal lines in D represent the limit in the rate resolution for experiments conducted with cantilevers with a spring constant of 15 pN/nm.
Fitting the Bell equation to the data Fig. 2D (gray dashed line) corresponding to the force-dependent unfolding rate of ubiquitin in standard PBS buffered aqueous solution (Fig. 2C, gray triangles) resulted in k0,2 = 8.8 × 10−3 ± 5.0 × 10−4 s−1 and Δx2 = 1.6 ± 0.1 Å, in close agreement with earlier experiments (10). From the measured value of k0,2 and assuming a prefactor A = 106 s−1 (34) we readily estimated the value of ΔG2 = 11.0 kcal mol−1 (25). To investigate the effect of solvent substitution on the mechanical stability of the native state of ubiquitin, we have now extended our measurements to study its unfolding force-dependency in solutions containing 40% vol/vol ethanol and 30% vol/vol glycerol. Fig. 2 B and C show the averaged unfolding trajectories and their corresponding exponential fits obtained at different pulling forces in 40% vol/vol ethanol and 30% vol/vol glycerol, respectively. The unfolding force dependency obtained from these data are shown in Fig. 2D (40% vol/vol ethanol, green circles; 30% vol/vol glycerol, red squares). From the data, it is apparent that glycerol, a stabilizing osmolyte (9), makes the native state of ubiquitin significantly more resistant to mechanical unfolding. This effect is not solely because of viscosity, but rather reflects an increase in the activation barrier height (35). Arrhenius fits to the data (Fig. 2D, dashed lines) show that glycerol causes an increase in the activation energy barrier to unfold by ΔΔG2 ≈3.2 kcal/mol. By contrast, substitution with a 40% vol/vol ethanol solution has only a minor effect (ΔΔG2 ≈ 0.05 kcal/mol). Furthermore, while the measured distance to the transition state increases when replacing PBS solution with 30% vol/vol glycerol from Δx2, water = 1.6 ± 0.1 Å to Δx2,gly = 2.3 ± 0.1 Å, the value remains almost unaltered upon the addition of 40% vol/vol ethanol, Δx2,ethanol = 1.7 ± 0.1 Å. The values of the parameters defining the energy landscape under all tested chemical environments, namely the height of the activation energy barrier, ΔG2, and the position of the transition state, Δx2, are shown in Table S1.
The Extent of Protein Collapse from the Unfolded State Is Significantly Enhanced by Glycerol.
In our single-molecule assay, the complete unfolding reaction leads the protein to a totally extended conformation, close to its full contour length, where all amino acids are exposed to the solvent environment and native contact formation is rare (18, 36). Studying the collapse of such extended proteins at the single molecule level permits a direct determination of the major driving forces governing the process. In particular, the extent of protein collapse was notably altered by changing the polarity of the solvent environment, which modifies the strength of hydrophobic interactions between the amino acids in the extended polyprotein chain. These experiments strongly supported the view that hydrophobic forces play a major role in the collapse of an extended protein (18). To further understand the mechanical and physiochemical properties of the unfolded state of ubiquitin, we use the force-ramp protocol depicted in Fig. 1. The protein is first unfolded under constant force to a highly extended conformation. Then, the force is linearly decreased down to 10 pN, triggering the contraction of the protein. This protocol permits the observation of the full force-length relationship of the extended protein as it collapses. Fig. 3A shows the collapsing trajectories of 136 different fully extended polyubiquitin molecules such as the one observed in Fig. 1 in a 30% vol/vol glycerol solution. Remarkably, we observe a high degree of heterogeneity in the responses. Such heterogeneity can be explained in terms of the different strength of the hydrophobic driving forces that act on each particular collapse trajectory, which depends on the initial combination of dihedral angles sampled by the protein once it is unfolded and stretched at 190 pN (18). In some cases, the protein collapses very little during the ramp down of the force down to 10 pN (F2 region in Fig. 1). In others, a large contraction of the protein was observed. Proteins were often observed to fold during the force-ramp protocol, as in the example shown in Fig. 1. These folding events were detected as an increase in the length by multiples of approximately 20 nm after restoration of the force to 190 pN. To compare all of the recordings, we normalized their length by the value measured in the initial extended conformation at 190 pN. The normalized length, L(F)/L (190 pN), is shown in Fig. 3A as a function of the force during the ramp down in force to 10 pN. Proteins that failed to fold during the ramp down process (blue traces in Fig. 3A, n = 65) showed large variations in their extent of collapse. Proteins that folded (red traces in Fig. 3A, n = 71) collapsed much further than the failures. This is apparent from the histogram of L(F)/L (190 pN) measured at 12 pN in Fig. 3D where proteins which failed to fold are shown in blue and proteins that successfully folded are shown in red. In the presence of glycerol the extent of collapse is greatly enhanced, causing a much larger number of molecules to fold (52%) during the ramp-down of the force (red traces in Fig. 3A). Strikingly, the large hydrophobic collapse measured in 30% vol/vol glycerol solutions contrasts with the much reduced collapse observed in PBS solution and 40% vol/vol ethanol solution. In particular, 40% vol/vol ethanol greatly reduces the extent of collapse and prevents folding during the force ramp down (18). This is readily apparent when comparing the extent of collapse measured at 12 pN. Fig. 3 B–D show L(F)/L (190) at 12 pN for proteins exposed to 40% vol/vol ethanol (Fig. 3B), PBS solution (Fig. 3C), and 30% vol/vol glycerol (Fig. 3D).
Fig. 3.
The extent of protein collapse from highly extended states is enhanced in glycerol solutions. (A) Plot of 136 individual collapse trajectories in a 30% vol/vol glycerol solution such as the one shown in Fig. 1 (F2). To compare all of the recordings, we normalized their length, L(F), by the value measured in the initial extended conformation at 190 pN, L(190 pN). The normalized length, L(F)/L (190 pN), is recorded as the pulling force is linearly decreased from 190 pN down to 10 pN. Proteins that failed to fold during the ramp down process (n = 65) are shown in blue. Proteins that folded (which is confirmed during the ramp-up section of the recording as depicted in Fig. 1; F3 region) are shown in red and collapsed much further than the failures (n = 71). The large hydrophobic collapse measured in 30% glycerol solutions (red bars, folders; blue bars, failures) contrasts with the much reduced collapse observed in a 40% ethanol solution. The extent of the collapse in each solvent environment can be compared by measuring the normalized protein length, L(F)/L(190), at a force of 12 pN in (A) 40% ethanol, (B) PBS solution, and (C) 30% glycerol.
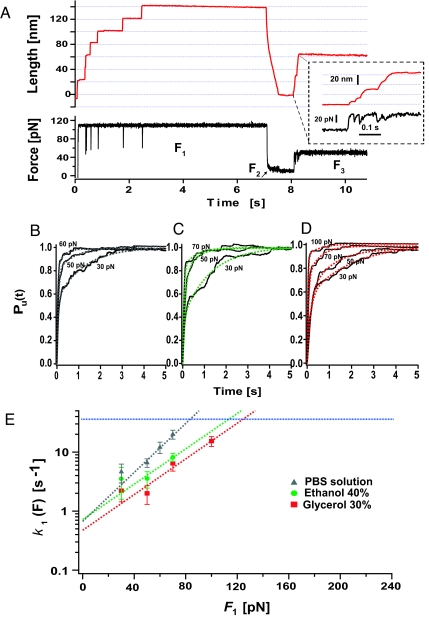
Probing the Physicochemical Properties of the Ensemble of Collapsed Conformations.
The combination of force clamp and solvent substitution has revealed the important and distinct effect that the solvent environment has on the mechanical stability of both the folded and unfolded states of ubiquitin. It is therefore appealing to investigate the response of the recently identified ensemble of collapsed states (25) upon the application of a stretching force under different solvent environments. To study the force-dependent unraveling rate of this ensemble of conformations in standard PBS solution, we devised a force quench protocol (Fig. 4A) described in detail in the SI Text section that specifically probes the mechanical resistance of these structures under force. Fig. 4E shows a logarithmic plot of the rate of unraveling of the collapsed conformations, k1, as a function of the pulling force (gray triangles) in standard PBS aqueous solution (25). We fitted the force-dependency of k1 with a simple Arrhenius term, obtaining k0,1 = 0.7 s−1 and <Δx1> = 2.0 Å. Assuming the same prefactor value used to describe the unfolding energy barrier of the native conformations (34) of A = 106 s−1, we calculate an average activation energy of <ΔG1> approximately 8.4 kcal/mol (25). It is remarkable that the distance to the transition state for the collapsed structures is similar to that of the native state Δx1 approximately 2 Å. We now extend these results by studying the effect of solvent substitution on the force dependency of the unraveling rate of the collapsed conformations (Fig. 4E). Fig. 4 C and D show averaged traces and their corresponding double exponential fits obtained at different forces in solutions containing 40% vol/vol ethanol and 30% vol/vol glycerol. Fig. 4E shows the force dependency of the unraveling rate of the collapsed conformations in 40% vol/vol ethanol (green circles) and 30% vol/vol glycerol (red squares). These results demonstrate that both ethanol and glycerol have a minor effect on the force-dependency of the collapsed states. Bell fits to the data (solid lines) give activation energies and distances to the transition state similar to those found in PBS solution (Δxethanol = 1.4 Å, Δxgly = 1.4 Å). Extrapolated to zero force, the unraveling rates converge to the same value of ≈0.5–0.7 s−1, respectively (Fig. 4D), which implies a vanishingly small change in the height of the unraveling barrier (ΔΔG1 = 0.2 kcal/mol).
Fig. 4.
The collapsed conformations are solvent-insensitive. (A) We use the 3-pulse protocol (F1 to F3) to measure the force-dependent extension of the collapsed ensemble. After a standard force-quench sequence (F1 and F2), the force-dependent rate of extension of the collapsed conformations k1 is measured by varying F3 within the range 30–70 pN in (B) PBS aqueous solution, data from (25), (C) ethanol solution (40% vol/vol), and (D) glycerol solution (30% vol/vol). We measured the unfolding rate from the weighted average of biexponential fits to the unraveling time course of 25–50 recordings at each force. (E) We fit an Arrhenius term to k1 (F) to estimate the average size of the activation energy barrier, <ΔG1>, and distance to the transition state, <Δx1> of the collapsed conformations in PBS solution and in solutions containing 40% ethanol and 30% glycerol. The data shows that the Arrhenius fits for all 3 solvent environments converge to the same rate at 0 force, indicating that <ΔG1> is solvent-independent.
Discussion
Protective and denaturing osmolytes such as ethanol and glycerol are known to destabilize or stabilize the native state of proteins against a wide variety of environmental stresses such as high pressure (37) or temperature (38). Here, we have discovered that ethanol and glycerol affect the activation energy barrier to mechanical unfolding of the protein ubiquitin, expanding the repertoire of known function of these osmolytes. Specifically, our results show that although ethanol slightly destabilizes the native state of ubiquitin (ΔΔG = 0.05 kcal/mol), glycerol significantly enhances it by ΔΔG = 3.8 kcal/mol. Although the effects of both ethanol and glycerol are in qualitative agreement with the results obtained from transfer free energy calculations (39), the molecular origin of these changes to the free energy remain elusive. Indeed, despite extensive studies it has proved difficult to provide a universal molecular mechanism of solvent effects on protein destabilization/stabilization. Although a ‘direct mechanism’ suggests that the cosolvent directly and favorably interacts with the protein backbone (40), an ‘indirect mechanism’ suggests that the cosolvent disrupts the water structure, thereby modifying its solvation properties (41). Our experiments at the single molecule level provide a different perspective to this important area by directly capturing the role of solvent molecules in the structure of the mechanical unfolding transition state of a protein. Crucially, this level of experimental control provides a unique test of either a direct or indirect solvent stabilization mechanism in the context of mechanical stability (35). In our experiments probing mechanical unfolding of ubiquitin in 40% vol/vol ethanol show that Δx remains unchanged in this aqueous ethanol solution as compared with water (Δxethanol = 1.7 Å, Δxwater = 1.6 Å), suggesting that ethanol molecules are not actively involved in the unfolding transition state structure of this protein. These results therefore suggest that ethanol slightly destabilizes ubiquitin through an indirect mechanism, disrupting the water structure. Indeed, previous structural studies of aqueous alcohol solutions have shown that the extended hydrogen bond network of water is significantly altered in the presence of an alcohol (42). In contrast to ethanol, the distance to the transition state is measured to increase from Δxwater = 1.6 Å in water to Δxgly = 2.3 Å upon addition of 30% vol/vol glycerol, suggesting that the larger molecule glycerol plays an active role in the transition state structure, which is consistent with a direct mechanism of protein stabilization. The involvement of glycerol molecules in the mechanical transition state of unfolding observed for ubiquitin, resulting in an increase in Δx, is consistent with the results obtained for the protein I27 (35). Interestingly, while the increase in Δx for the protein I27 in glycerol was approximately 1.6 Å the increase in Δx for ubiquitin is only 0.7 Å. This smaller increase in Δx suggests that the mechanical transition state structure in ubiquitin, thought to be placed between the β1–β5 sheets (43, 44), is less sensitive to the size of the solvent molecules.
In addition to the native state of ubiquitin, the extended state is also found to be highly sensitive to the solvent environment. While protein collapse is significantly hampered in an aqueous ethanol environment, it is readily enhanced in aqueous glycerol. Protein stability is known to be strongly determined by its solvation properties with the surrounding solvent environment. The observation of an enhancement in the hydrophobic collapse of ubiquitin in glycerol is not inconsistent with a proposed backbone based theory of protein folding, in which unfavourable interactions between the protein backbone and glycerol's polar surface area drive the collapse of the unfolded protein (9, 45). However, the heterogeneity observed in the collapsing trajectories can only be explained in terms of the different initial solvation properties of side chains in the unfolded protein. These initial conformations result from the different combination of dihedral angles explored by the extended polyprotein (46). The heterogeneity in the collapsing trajectories, mainly driven by hydrophobic interactions, is likely to result in the conformational disorder observed in the ensemble of collapsed states (Fig. 1, Bottom Inset).
Strikingly, the behavior of both the native and extended proteins contrasts with the measured solvent insensitivity of the collapsed conformations. It is interesting to consider what structural properties of the collapsed conformations could result in this solvent insensitivity. One plausible explanation is that solvent molecules are excluded from the internal structure of the collapsed conformations. Thus solvent molecules would not play an integral role in their transition state structure. Indeed, this scenario would account for the similar Δx values measured for the different solvents (Fig. 4 and Table S1), suggesting that solvent bridging is not an important mechanism for these structures. Instead, each collapsed conformation may be held together by interactions that are mostly hydrophobic in origin. The mechanical resistance exhibited by the collapsed conformations would thus reflect the strength of this hydrophobic packing. Interestingly, such a hydrophobic packing scenario has been observed for the protein ankyrin, which consists of smaller ankyrin repeats which stack into a helical supramolecular structure via hydrophobic forces (47). Similarly to our single molecule experiments, such ankyrin structures are largely insensitive to osmolytes (48), yet exhibit mechanical resistance which is similar in scale to that measured for the collapsed conformations (49). Alternatively, the solvent insensitivity of the collapsed conformations may reflect structures which are preferentially solvated by water molecules from the ethanol/glycerol aqueous mixtures. Thus the transition state structures of these structures will only involve water molecules resulting in an unchanging Δx and ΔG. In this scenario, the collapsed conformations would require final expulsion of water molecules to reach the native fold. Indeed, this is not inconsistent with previous computational studies which suggest that the final step of the folding process from a near-native collapsed structure to the final folded conformation involves a desolvation process that squeezes out water molecules in the vicinity of the partially hydrated core (1). It is feasible that in our experiments, such a folding step is represented at the end of the collapsing trajectories (Figs. 1 and 3), which are largely affected by the solvent environment. Indeed, once the protein reaches the folded length, it remains a puzzle how the native contacts are formed to regain its mechanical stability. We hypothesize that this transition encompasses the correct rearrangement of side-chains and contact pairs in each individual collapsed structure to reach the final native structure, and that this last step is not crucially determined by the solvent environment.
Here, we demonstrate the success of using solvent substitution combined with single molecule force-clamp spectroscopy as an independent probe of the evolving structures of a polypeptide as it folds. These experiments open the way for a detailed examination of the transition state structures that populate the folding trajectory of a protein.
Materials and Methods
Protein Engineering.
WT-Ubiquitin polyprotein was subcloned using the BamHI and BglII restriction sites (50). The nine-domain ubiquitin was cloned into the pQE80L (Qiagen) expression vector, and transformed into the BLR(DE3) Escherichia coli expression strain and purified by histidine metal-affinity chromatography with Talon resin (Clontech) and by gel-filtration using a Superdex 200 HR column (GE Biosciences).
Force Spectroscopy.
Force-clamp atomic force microscopy experiments were conducted at room temperature using a home-made set-up under force-clamp conditions described elsewhere (10). Experiments were carried out in a sodium phosphate buffer solution, specifically, 50 mM sodium phosphate (Na2HPO4 and NaH2PO4), and 150 mM NaCl, pH = 7.4. Samples of glycerol (≥ 99%) and ethanol (≥ 99%) were obtained from Sigma-Aldrich and used without additional purification. Solvent mixtures were prepared to obtain the desired volume fraction, vol/vol, ratio of the co-solvent and viscosity.
Supplementary Material
Acknowledgments.
We thank Jasna Brujić and members of the J.M.F. laboratory for discussions. S.G-M. thanks the Generalitat de Catalunya for a postdoctoral fellowship through the NANO and Beatriu de Pinós programs, and also the Fundación Caja Madrid for financial support. This work was supported by National Institutes of Health Grants HL66030 and HL61228 (to J.M.F).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0902090106/DCSupplemental.
References
- 1.Cheung MS, Garcia AE, Onuchic JN. Protein folding mediated by solvation: Water expulsion and formation of the hydrophobic core occur after the structural collapse. Proc Natl Acad Sci USA. 2002;99:685–690. doi: 10.1073/pnas.022387699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tarek M, Tobias DJ. Role of protein-water hydrogen bond dynamics in the protein dynamical transition. Phys Rev Lett. 2002;88:138101. doi: 10.1103/PhysRevLett.88.138101. [DOI] [PubMed] [Google Scholar]
- 3.Qu YX, Bolen CL, Bolen DW. Osmolyte-driven contraction of a random coil protein. Proc Natl Acad Sci USA. 1998;95:9268–9273. doi: 10.1073/pnas.95.16.9268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pal SK, Peon J, Zewail AH. Biological water at the protein surface: Dynamical solvation probed directly with femtosecond resolution. Proc Natl Acad Sci USA. 2002;99:1763–1768. doi: 10.1073/pnas.042697899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sorenson JM, Hura G, Soper AK, Pertsemlidis A, Head-Gordon T. Determining the role of hydration forces in protein folding. J Phys Chem B. 1999;103:5413–5426. [Google Scholar]
- 6.Rariy RV, Klibanov AM. Correct protein folding in glycerol. Proc Natl Acad Sci USA. 1997;94:13520–13523. doi: 10.1073/pnas.94.25.13520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gekko K, Timasheff SN. Mechanism of protein stabilization by glycerol: Preferential hydration in glycerol-water mixtures. Biochemistry. 1981;20:4667–4676. doi: 10.1021/bi00519a023. [DOI] [PubMed] [Google Scholar]
- 8.O'Brien EP, Ziv G, Haran G, Brooks BR, Thirumalai D. Effects of denaturants and osmolytes on proteins are accurately predicted by the molecular transfer model. Proc Natl Acad Sci USA. 2008;105:13403–13408. doi: 10.1073/pnas.0802113105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Street TO, Bolen DW, Rose GD. A molecular mechanism for osmolyte-induced protein stability. P Natl Acad Sci USA. 2006;103:13997–14002. doi: 10.1073/pnas.0606236103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schlierf M, Li H, Fernandez JM. The unfolding kinetics of ubiquitin captured with single-molecule force-clamp techniques. Proc Natl Acad Sci USA. 2004;101:7299–7304. doi: 10.1073/pnas.0400033101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wiita AP, Ainavarapu SRK, Huang HH, Fernandez JM. Force-dependent chemical kinetics of disulfide bond reduction observed with single-molecule techniques. Proc Natl Acad Sci USA. 2006;103:7222–7227. doi: 10.1073/pnas.0511035103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H, Carrion-Vazquez M, Oberhauser AF, Marszalek PE, Fernandez JM. Point mutations alter the mechanical stability of immunoglobulin modules. Nat Struct Biol. 2000;7:1117–1120. doi: 10.1038/81964. [DOI] [PubMed] [Google Scholar]
- 13.Brockwell DJ, et al. Mechanically unfolding the small, topologically simple protein L. Biophys J. 2005;89:506–519. doi: 10.1529/biophysj.105.061465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perez-Jimenez R, Garcia-Manyes S, Ainavarapu SR, Fernandez JM. Mechanical unfolding pathways of the enhanced yellow fluorescent protein revealed by single molecule force spectroscopy. J Biol Chem. 2006;281:40010–40014. doi: 10.1074/jbc.M609890200. [DOI] [PubMed] [Google Scholar]
- 15.Lu H, Schulten K. The key event in force-induced unfolding of titin's immunoglobulin domains. Biophys J. 2000;79:51–65. doi: 10.1016/S0006-3495(00)76273-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dougan L, Feng G, Lu H, Fernandez JM. Solvent molecules bridge the mechanical unfolding transition state of a protein. Proc Natl Acad Sci USA. 2008;105:3185–3190. doi: 10.1073/pnas.0706075105. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Dougan L, Koti AS, Genchev G, Lu H, Fernandez JM. A single-molecule perspective on the role of solvent hydrogen bonds in protein folding and chemical reactions. Chemphyschem. 2008;22:2836–2847. doi: 10.1002/cphc.200800572. [DOI] [PubMed] [Google Scholar]
- 18.Walther KA, Grater F, Dougan L, Badilla CL, Berne BJ, Fernandez JM. Signatures of hydrophobic collapse in extended proteins captured with force spectroscopy. Proc Natl Acad Sci USA. 2007;104:7916–7621. doi: 10.1073/pnas.0702179104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lipman EA, Schuler B, Bakajin O, Eaton WA. Single-molecule measurement of protein folding kinetics. Science. 2003;301:1233–1235. doi: 10.1126/science.1085399. [DOI] [PubMed] [Google Scholar]
- 20.Hoffmann A, et al. Mapping protein collapse with single-molecule fluorescence and kinetic synchrotron radiation circular dichroism spectroscopy. Proc Natl Acad Sci USA. 2007;104:105–110. doi: 10.1073/pnas.0604353104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cecconi C, Shank EA, Bustamante C, Marqusee S. Direct observation of the three-state folding of a single protein molecule. Science. 2005;309:2057–2060. doi: 10.1126/science.1116702. [DOI] [PubMed] [Google Scholar]
- 22.Fernandez JM, Li H. Force-clamp spectroscopy monitors the folding trajectory of a single protein. Science. 2004;303:1674–1678. doi: 10.1126/science.1092497. [DOI] [PubMed] [Google Scholar]
- 23.Schlierf M, Berkemeier F, Rief M. Direct observation of active protein folding using lock-in force spectroscopy. Biophys J. 2007;93:3989–3998. doi: 10.1529/biophysj.107.114397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia-Manyes S, Brujic J, Badilla CL, Fernandez JM. Force-clamp spectroscopy of single-protein monomers reveals the individual unfolding and folding pathways of I27 and ubiquitin. Biophys J. 2007;93:2436–2446. doi: 10.1529/biophysj.107.104422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garcia-Manyes S, Dougan L, Badilla CL, Brujic J, Fernandez JM. Direct observation of an ensemble of stable collapsed states in the mechanical folding of ubiquitin. Proc Natl Acad Sci USA. 2009 doi: 10.1073/pnas.0901213106. doi/ 10.1073/pnas.0901213106. [DOI] [PMC free article] [PubMed]
- 26.Honeycutt JD, Thirumalai D. Metastability of the folded states of globular proteins. Proc Natl Acad Sci USA. 1990;87:3526–3529. doi: 10.1073/pnas.87.9.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Camacho CJ, Thirumalai D. Minimum energy compact structures of random sequences of heteropolymers. Phys Rev Lett. 1993;71:2505–2508. doi: 10.1103/PhysRevLett.71.2505. [DOI] [PubMed] [Google Scholar]
- 28.Camacho CJ, Thirumalai D. Kinetics and thermodynamics of folding in model proteins. P Natl Acad Sci USA. 1993;90:6369–6372. doi: 10.1073/pnas.90.13.6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phillips CL, Thrower J, Pickart CM, Hill CP. Structure of a new crystal form of tetraubiquitin. Acta Crystallogr D Biol Crystallogr. 2001;57:341–344. doi: 10.1107/s090744490001800x. [DOI] [PubMed] [Google Scholar]
- 30.Cook WJ, Jeffrey LC, Kasperek E, Pickart CM. Structure of tetraubiquitin shows how multiubiquitin chains can be formed. J Mol Biol. 1994;236:601–609. doi: 10.1006/jmbi.1994.1169. [DOI] [PubMed] [Google Scholar]
- 31.Cook WJ, Jeffrey LC, Carson M, Chen Z, Pickart CM. Structure of a diubiquitin conjugate and a model for interaction with ubiquitin conjugating enzyme (E2) J Biol Chem. 1992;267:16467–16471. doi: 10.2210/pdb1aar/pdb. [DOI] [PubMed] [Google Scholar]
- 32.Brujic J, Hermans RI, Walther KA, Fernandez JM. Single-molecule force spectroscopy reveals signatures of glassy dynamics in the energy landscape of ubiquitin. Nat Phys. 2006;2:282–286. [Google Scholar]
- 33.Bell GI. Models for the specific adhesion of cells to cells. Science. 1978;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 34.Li MS, Klimov DK, Thirumalai D. Thermal denaturation and folding rates of single domain proteins: Size matters. Polymer. 2004;45:573–579. [Google Scholar]
- 35.Dougan L, Fang G, Lu H, Fernandez J.M. Solvent molecules bridge the unfolding transition state of a protein. Proc Natl Acad Sci USA. 2008:105. doi: 10.1073/pnas.0706075105. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Li MS, Hu CK, Klimov DK, Thirumalai D. Multiple stepwise refolding of immunoglobulin domain I27 upon force quench depends on initial conditions. Proc Natl Acad Sci USA. 2006;103:93–98. doi: 10.1073/pnas.0503758103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruan K, Xu C, Li T, Li J, Lange R, Balny C. The thermodynamic analysis of protein stabilization by sucrose and glycerol against pressure-induced unfolding. Eur J Biochem. 2003;270:1654–1661. doi: 10.1046/j.1432-1033.2003.03485.x. [DOI] [PubMed] [Google Scholar]
- 38.Gekko K, Timasheff SN. Thermodynamic and kinetic examination of protein stabilization by glycerol. Biochemistry. 1981;20:4677–4686. doi: 10.1021/bi00519a024. [DOI] [PubMed] [Google Scholar]
- 39.Tanford C. Contribution of hydrophobic interactions to stability of globular conformation of proteins. J Am Chem Soc. 1962;84:4240. [Google Scholar]
- 40.Hua L, Zhou RH, Thirumalai D, Berne BJ. Urea denaturation by stronger dispersion interactions with proteins than water implies a 2-stage unfolding. Proc Natl Acad Sci USA. 2008;105:16928–16933. doi: 10.1073/pnas.0808427105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bennion BJ, Daggett V. The molecular basis for the chemical denaturation of proteins by urea. Proc Natl Acad Sci USA. 2003;100:5142–5147. doi: 10.1073/pnas.0930122100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dougan L, et al. Methanol-water solutions: A bi-percolating liquid mixture. J Chem Phys. 2004;121:6456–6462. doi: 10.1063/1.1789951. [DOI] [PubMed] [Google Scholar]
- 43.Irback A, Mitternacht S, Mohanty S. Dissecting the mechanical unfolding of ubiquitin. Proc Natl Acad Sci USA. 2005;102:13427–13432. doi: 10.1073/pnas.0501581102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kleiner A, Shakhnovich E. The mechanical unfolding of ubiquitin through all-atom Monte Carlo simulation with a Go-type potential. Biophys J. 2007;92:2054–2061. doi: 10.1529/biophysj.106.081257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rose GD, Fleming PJ, Banavar JR, Maritan A. A backbone-based theory of protein folding. Proc Natl Acad Sci USA. 2006;103:16623–16633. doi: 10.1073/pnas.0606843103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Auton M, Bolen DW. Predicting the energetics of osmolyte-induced protein folding/unfolding. Proc Natl Acad Sci USA. 2005;102:15065–15068. doi: 10.1073/pnas.0507053102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee G, Abdi K, Jiang Y, Michaely P, Bennett V, Marszalek PE. Nanospring behaviour of ankyrin repeats. Nature. 2006;440:246–249. doi: 10.1038/nature04437. [DOI] [PubMed] [Google Scholar]
- 48.Wetzel SK, Settanni G, Kenig M, Binz HK, Pluckthun A. Folding and unfolding mechanism of highly stable full-consensus ankyrin repeat proteins. J Mol Biol. 2008;376:241–257. doi: 10.1016/j.jmb.2007.11.046. [DOI] [PubMed] [Google Scholar]
- 49.Li L, Wetzel S, Pluckthun A, Fernandez JM. Stepwise unfolding of ankyrin repeats in a single protein revealed by atomic force microscopy. Biophys J. 2006;90:L30–32. doi: 10.1529/biophysj.105.078436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carrion-Vazquez M, et al. Mechanical and chemical unfolding of a single protein: a comparison. Proc Natl Acad Sci USA. 1999;96:3694–3699. doi: 10.1073/pnas.96.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.