Abstract
Prion diseases are transmissible neurodegenerative disorders of prion protein (PrP) conformation. Prion replication by conversion of benign PrPC isoforms into disease-specific PrPSc isoforms is intimately involved in prion disease pathogenesis and may be initiated in cholesterol-rich caveolae-like domains (CLD). Concentrations of the cholesterol transporter ATP-binding cassette A1 protein (ABCA1) are elevated in pre-clinical scrapie prion infected mice and in prion infected cells in vitro. Elevation of ABCA1 in prion infected brain is not a direct consequence of local PrPSc accumulation; levels of ABCA1 are comparable in brain regions that differ dramatically in the amount of PrPSc. Similarly, ABCA1 concentrations are identical in normal mice, transgenic mice overexpressing PrP, and PrP knockout mice. In contrast, PrPC and PrPSc levels, but not Prnp mRNA, were increased by overexpression of ABCA1 in N2a neuroblastoma cells and scrapie prion infected N2a cells (ScN2a). Conversely, RNAi-mediated knock down of Abca1 expression decreased the concentrations of PrPC in N2a cells and of PrPSc in ScN2a cells. These results suggest that ABCA1’s effects on PrPC levels are posttranslational and may reflect an increase in of PrPC stability, mediated either indirectly by increasing membrane cholesterol and CLD formation or by other functions of ABCA1. The increased supply of PrPC available for conversion would lead to increased PrPSc formation.
Creutzfeldt-Jakob disease and Kuru in humans, bovine spongiform encephalopathy, scrapie in sheep, and chronic wasting disease in cervids are members of the group of neurodegenerative protein folding disorders caused by prions (Prusiner, 1998). Prion replication involves conversion of benign cellular isoforms, designated PrPC, into disease-specific isoforms designated PrPSc. The cellular site for prion replication is not well defined; in vitro and in vivo studies suggest that it takes place at plasma membrane, where the initial interaction between endogenous GPI-anchored PrPC and exogenous PrPSc occurs, and in endosomal compartments (Campana et al., 2005, Harris, 1999). Substantial evidence suggests that PrPSc generation takes place in cholesterol-rich, detergent insoluble membrane domains that are identified by various names including caveolae-like domains (CLD), detergent resistant membranes, or lipid rafts (Caughey & Raymond, 1991, Kaneko et al., 1997, Sarnataro et al., 2004, Vey et al., 1996). Cholesterol is required for PrPC cell surface expression and stability (Caughey & Raymond, 1991, Gilch et al., 2006, Kaneko et al., 1997, Sarnataro et al., 2004) and pharmacological treatments that lower cellular cholesterol reduce PrPSc generation and prolong prion incubation time (Bate et al., 2004, Gilch et al., 2006, Marella et al., 2002, Taraboulos et al., 1995).
Genes involved in cholesterol metabolism and transport are consistently found among differentially expressed genes (DEGs) in prion infected mice and cell lines. Among them is Abca1, whose mRNA levels were increased by 2–3 fold in the brains of prion infected C57Bl/6J and FVB/NCr mice (Riemer et al., 2004, Xiang et al., 2007, Xiang et al., 2004). We have recently extended these observations and found that increased Abca1 expression is detectable in whole brain beginning at approximately two thirds of the incubation period, before the onset of clinical signs, in multiple prion strain-mouse strain combinations (D. Hwang, I. Lee, H, Yoo, N. Gehlenborg, N. Price, B. Ogata, D. Baxter, R. Pitstick, R.Y., D. Spicer, J. Hohmann, S. Dearmond, G.A.C., L. Hood, in preparation). This lack of mouse strain or prion strain specificity suggests that the increased expression of ABCA1 is a common feature of prion disease. However, as is the case for many DEGs, the significance of Abca1 mRNA elevation in prion disease is not clear.
ABCA1 is a membrane-associated protein that belongs to the ATP-binding cassette protein superfamily of multicellular organisms and is localized in membranous cellular compartments including the plasma membrane, the Golgi stack and endosomes. In brain, ABCA1 is expressed in neurons, astrocytes and glial cells (Fukumoto et al., 2002, Koldamova et al., 2003). ABCA1 is involved in the transport of intracellular cholesterol and caveolae from the trans-Golgi to the plasma membrane and ABCA1 mutations cause Tangier disease (Lawn et al., 1999, Orso et al., 2000). The potential importance of cholesterol in prion disease has been established by the inhibitory effect of pharmacological depletion of cellular cholesterol (Bate et al., 2004, Gilch et al., 2006, Marella et al., 2002, Taraboulos et al., 1995). The involvement of cholesterol in prion disease and the role of ABCA1 in cholesterol transport provided the impetus to examine interactions among ABCA1, PrP, and PrPSc formation.
Methods
Cells, Antibodies, and Mice
N2a and ScN2a cells were obtained from Dr. Stanley B. Prusiner, University of California San Francisco, San Francisco, CA and maintained at 37° C in 5% CO2 atmosphere in Dulbecco’s modified eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS). 8-Br-cAMP (Sigma, St. Louis, MO) treatment was used to enhance expression of ABCA1. Cells were incubated for 24 hours with 8-Br-cAMP in complete medium at the concentrations indicated followed by washing and immunoblotting. Primary antibodies were obtained from the following sources: anti-PrP recombinant human Fab D18 from Dr. Stanley B. Prusiner (Peretz et al., 2001), anti-ABCA1 and β-actin from Novus Biologicals, Inc; Littleton, CO, anti-caveolin-1 from Sigma, St. Louis, MO. FVB Tg(MoPrP-A)B4053 mice overexpressing mouse PrP-A (Carlson et al., 1994), FVB.129-Prnptm1Zrch null mice (Bueler et al., 1992), and FVB/NCr mice were maintained in the Animal Resource Center at the McLaughlin Research Institute. All protocols were reviewed and approved the Institutional Animal Care and Use Committee.
Scrapie Inoculation and Brain Harvest
Three week old FVB/NCr mice were anesthetized with isoflurane and inoculated intracerebrally with 20 μl of a 1:10 dilution of brain homogenate in phosphate buffered saline from either clinically ill RML scrapie inoculated mice (RML) or from normal mice (clean) and observed for clinical signs as previously described (Carlson et al., 1994). Brains were harvested from euthanized mice and snap frozen at −80° C for storage. For some experiments, brains were dissected into fore and hindbrain. Forebrain consisted of olfactory bulb, cortex, hippocampus, thalamus and hypothalamus and hindbrain consisted of cerebellum, medulla, pons and the majority of the midbrain.
Western Blot Analysis
Brain lysates were prepared in ice-cold lysis buffer (10mM Tris-HCl pH 7.5, 150mM NaCl, 0.5% TritonX-100, 0.5% Na-deoxycholate) by repeatedly passing through successively smaller gauge needles. Lysates were cleared by centrifuging at 4400 × g for 5 min at 4° C and were stored at −80° C with or without complete protease inhibitor (Roche). N2a or ScN2a cells were washed three times with chilled PBS (calcium and magnesium free) and lysed in chilled lysis buffer and prepared as described above. Total protein content was determined using the detergent compatible BCA Protein assay (Pierce, Rockford, IL) with BSA as a standard. For PrPSc detection, 40 μg of protein lysate was digested with PK at 1:50 (w/w) for 30 minutes at 37° C. Digestion was stopped using Pefablock (Roche) at a final concentration of 1mM. All samples were boiled with sample buffer at 100° C for 10 minutes, except samples designated for ABCA1 detection. ABCA1 specific samples were incubated with sample buffer at room temperature for 30 minutes. Unless otherwise noted, samples containing 40 μg of protein were run on 12% SDS-PAGE Tris/Glycine gels (Invitrogen, Carlsbad, CA). Proteins were transferred onto nitrocellulose membranes (1h, 25volts) that were then blocked in 5% milk in TBST (10mM Tris-HCl, pH 7.8, 150mM NaCl, 0.05% Tween-20) for 1 hr at room temperature. Blots were incubated overnight at 4° C with primary antibodies (1μg/ml anti-PrP; 1:1000 anti-ABCA1; 1:1000 anti-cav-1) followed by incubation with corresponding secondary antibodies conjugated with HRPO for 1 hr at room temperature. Protein bands were developed with SuperSignal West pico chemiluminescence substrate (Pierce, Rockford, IL) for 5 min, and exposed to CL-XPosure Film (Pierce, Rockford, IL). Band intensities were quantified using Quantity One software using a VersaDoc Imaging System 3000 (Bio-Rad). To control for loading differences, immunoblots were stripped in stripping buffer (50mM Tris-HCl, pH 6.8, 2% SDS and 100 mM β-mercaptoethanol) for 30 minutes at 50° C followed by washing with TBST, and reblocking. Blots were then probed with anti-β actin antibody (1:2000) and HRPO-conjugated secondary antibody, prior to development. The different sample preparation methods for PrPC and ABCA1 detection necessitated their detection on separate filters; therefore PrPC and ABCA1 signals were normalized to β-actin on the corresponding filter. PK-digestion precluded β-actin detection on PrPSc filters.
RT-PCR
Total RNAs were isolated using RNeasy mini kit (Qiagen) and equal amounts of RNA were reverse-transcribed into cDNAs using Superscript III first-strand synthesis system for RT-PCR (Invitrogen, CA) into cDNA using oligo-dT primer. The cDNAs were used for gene specific PCR mediated amplification. For specific PCR amplification of Prnp cDNA the primers were (5′-3′): AAAAAGCGGCCAAAGCCTGG and CTTGTTCCACTGATTAT, which yield a 229 base pair product. The Gpdh primers were (5′-3′): ATGGGTGTGAACCACGAGAA and AGGCATGGACTGTGGTCAT and yield a 144 base pair product. The Abca1 primers, which yield a 82 base pair product, were (5′-3′): CATTAAGGACATGCACAAGGTCC and AGGATTTTCTGGTGGACAATGAAA. For Prnp and Gpdh, PCR was performed for 40 cycles of 95° C for 30 s, 52° C for 15 s, and 72° C for 45 s. For Abca1, the annealing temperature was 54° C. The amplified products were analyzed by agarose gel electrophoresis.
RNA Interference
All duplex siRNA were synthesized by Dharmacon Inc., Lafayette, CO. Control siRNA was directed against a luciferase gene. N2a and ScN2a cells were transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) with a non-specific siRNA (sense sequence 5′-UAAGGCUAUGAAGAGAUACUU, antisense sequence 5′-GUAUCUCUUCAUAGCCUUAUU), or Abca1 siRNA probes targeting different positions of Abca1 (NM_013454) mRNA. Abca1 siRNA probes, all 5′-3′ with sense sequence followed by antisense sequence, are: ABCA1 #1, GAAGAAAUAUUCCUCAAAGUU and CUUUGAGGAAUAUUUCUUCUU; ABCA1 #2, CCAAAUGGCUCUGUGUAUAUU and UAUACACAGAGCCAUUUGGUU; ABCA1 #3, GGAGAGAACUAAUAAGAUCUU and GAUCUUAUUAGUUCUCUCCUU; and ABCA1 #4, GGAGAGAAGCUUUCAAUGAUU and UCAUUGAAAGCUUCUCUCCUU-3. 72 hr after transfection, cells were lysed for Western blot analysis with anti-ABCA1, anti-caveolin-1 and anti-PrP antibodies.
Immunocytochemistry and Microscopy
Neuroblastoma cells were transfected with GFP-ABCA1 (Fitzgerald et al., 2001) or GFP (Clonetech, Mountain View, CA) constructs using Lipofectamine 2000 (Invitrogen; Carlsbad, CA) and 48 hr after transfection, cells were seeded to glass cover slips for 24 hr. The cover slips were washed with PBS and fixed in 4% paraformaldehyde (PFA)-PBS for 30 minutes. Cross-linking was terminated by 0.1M glycine-PBS treatment for 30 minutes. Cells were washed with PBS and non-specific binding sites were blocked by incubating with 5% BSA and 5% normal goat serum in PBS for 1 hr. After blocking, the cells were washed with PBS and incubated with primary antibody (anti-PrP, Fab D18 5 μg/ml) in 1% BSA and 1% normal goat serum in PBS overnight at 4° C. Following overnight incubation, cells were incubated with diluted (1:1000) nuclear DNA stain (DAPI) (Molecular Probes-Invitrogen, Carlsbad, CA) for 2 min and washed twice with PBS for 10 minutes. Cells were incubated with the fluorophore-conjugated secondary antibody (rhodamine- labeled goat anti-human Ig, Pierce, Rockford, IL) for 2 hr. Cells were washed three times with PBS and cover slips were mounted with anti-fade mounting medium (Molecular Probes-Invitrogen, Carlsbad, CA). All incubations and solutions were at 4° C. Fluorescence microscopy was performed using a Nikon TE2000 photomicroscope and fluorescence intensity was quantified using METAMORPH software (Molecular Devices, Sunnyvale, CA). Comparison of the PrP fluorescence intensity in GFP-ABCA1 and GFP transfected cells was based on the result that the intensity distribution of the signal in non-transfected cells (GFP negative) in the two samples was identical.
Results
Prion Infection Elevates ABCA1 Protein Levels
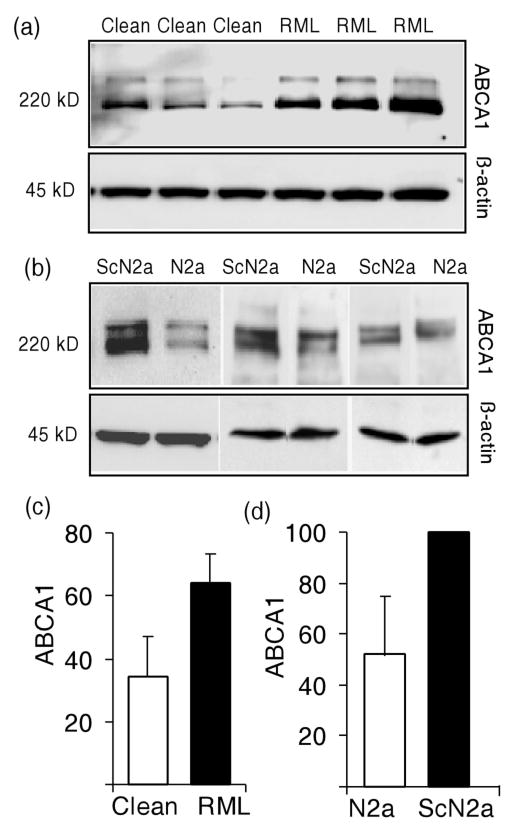
To test whether the increased Abca1 mRNA expression that is seen prion disease translates into higher protein levels, Western blot analyses were performed. Brain were harvested from mice inoculated either with normal or RML brain homogenate 20 weeks previously and lysates for immunoblotting were prepared. Two bands that may be due to phosphorylation or other post-translational processes are detected by the anti-ABCA1 antibody (Bortnick et al., 2000, Koldamova et al., 2003). Both bands were included in our quantitation and levels normalized to β-actin staining on the same blot. ABCA1 levels were approximately 2-fold higher (P ≤ 0.05) in the brains of clinically ill mice infected with the Rocky Mountain Laboratory (RML) mouse-adapted scrapie prion strain than in age-matched mice given normal brain homogenate (Fig. 1 a and c). Similar results were obtained in cell culture. Since each pair of cell cultures was analyzed on a separate blot, β-actin normalized intensity level of ABCA1 in infected ScN2a cells was assigned a value of 100 and the normalized ABCA1 in N2a cells expressed as a percentage. ABCA1 protein levels were approximately 2-fold higher (P ≤ 0.05) in three independent cultures of persistently scrapie-infected N2a cells (ScN2a) than in three cultures of uninfected N2a cells (Fig. 1 b and d).
Fig. 1.
Scrapie prion infection increases ABCA1 levels. (a) Lysates of brain homogenates from FVB/NCr mice inoculated either with normal brain homogenate (Clean) or with RML prion-infected brain homogenate (RML) were prepared 20 weeks after inoculation. Infected mice showed clinical signs of disease. Lysates from six individual mice were analyzed by immunoblotting using anti-ABCA1 and anti-β actin antibodies. (b) Western blots of lysates from independent cultures of N2a and ScN2a cells using anti-ABCA1 and anti-β actin antibodies. (c) Densitometric quantitation of ABCA1 relative to β-actin in uninfected (‘clean’, open bar) and scrapie prion infected (‘RML’, black bar) mice and N2a or ScN2A cells is shown. The two-fold difference between uninfected and infected mice was statistically significant (P ≤ 0.05). (d) Quantitation of ABCA1 in N2a and ScN2a cells. Since each pair was run on its own blot, following normalization to β-actin, ABCA1 intensity in ScN2a was assigned a value of ‘100’ and ABCA1 level expressed as a percentage. The twofold difference between N2a and ScN2a cells was statistically significant (P ≤ 0.05). Error bars indicate standard deviation from the mean.
Changes in PrPC or PrPSc Concentration Do Not Alter ABCA1 Expression Levels
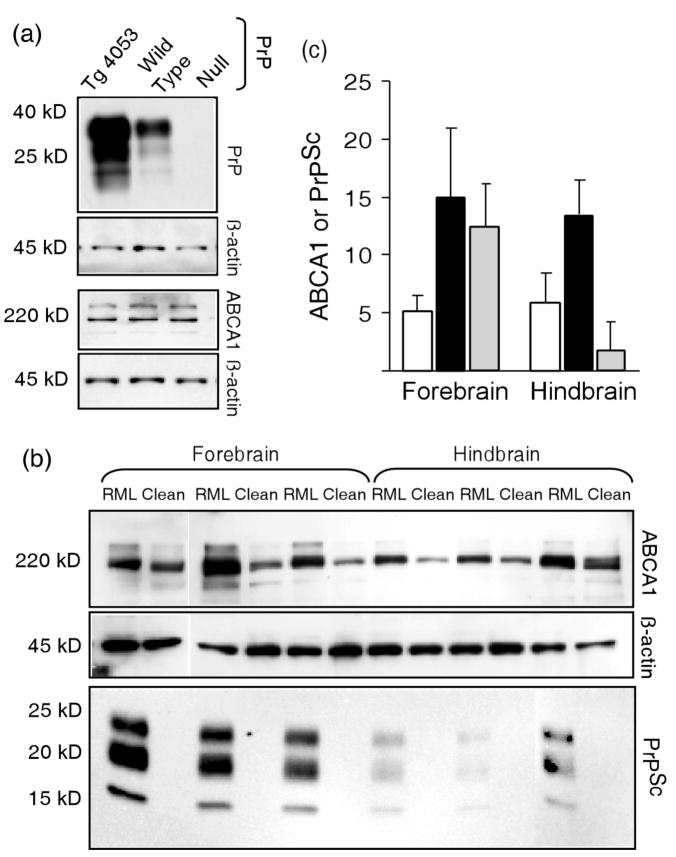
ABCA1 levels were independent of PrPC concentration since ABCA1 levels were similar in Prnp null mice, wild type mice, and transgenic mice (Tg4053) that overexpress PrPC (Fig. 2a). We then tested the possibility that elevation of ABCA1 in prion-infected mice was a local effect of accumulation of PrPSc. PrPSc does not accumulate uniformly throughout the brain and varies depending on the particular prion strain and host. We exploited the regional differences in PrPSc distribution in mice infected with RML scrapie prions to assess the effect of PrPSc concentration on ABCA1 levels. In these mice, more PrPSc is found in hippocampus, thalamus, and cerebral cortex than in hindbrain structures such as the brain stem and cerebellum. Significantly more (~8-fold, P ≤ 0.01) PK-resistant PrPSc is found in the forebrain than in the hindbrain of clinically ill mice (Fig. 2 b and c). In contrast, levels of ABCA1, which were ~2-fold higher in prion infected mice than in uninfected mice, were similar in forebrain and hindbrain (Fig. 2 b and c). Mice inoculated with normal brain homogenate also had similar ABCA1 levels in forebrain and hindbrain (Fig. 2 b and c). The two-fold increase in forebrain and hindbrain was consistent with the increase in ABCA1 seen in whole brain homogenates from infected mice (Fig 1 a and c). These results suggest that the increase in ABCA1 levels is not a direct response to PrPSc accumulation. The independence of ABCA1 levels and regional PrPC or PrPSc concentrations suggests that the elevation of ABCA1 in prion infected cells or mice is a consequence of infection, not PrPSc deposition. Although direct involvement of ABCA1 in conversion seems unlikely, the consequences of alteration of ABCA1 concentration were explored.
Fig. 2.
ABCA1 expression levels are independent of PrPC and PrPSc concentrations. (a) Equal amounts of brain lysates from age matched Prnp null mice, wild type, or transgenic mice (Tg4053) overexpressing PrPC were subjected to Western blot analysis using anti-PrP, anti-ABCA1 and anti-β actin antibodies. The three lines of mice did not differ in ABCA1 concentration. (b) Lysates of homogenates of forebrains and hindbrains from mice inoculated 20 weeks previously with either with scrapie brain homogenate (RML) or normal brain homogenate (Clean) were analyzed by Western blot using anti-ABCA1 and anti-β actin antibodies. Lysates for detection of PrPSc were digested with PK prior to Western blot analysis with anti-PrP Fab. (c) Forebrain and hindbrain concentrations of ABCA1 in uninfected mice (open bars) or scrapie prion-infected mice (black bars), and of PK-resistant PrPSc (gray bars). Each bar is the mean of three mice ± the standard deviation. The concentrations of β-actin-normalized ABCA1 from prion infected mice were approximately two-fold greater (P ≤ 0.05) in both forebrains and hindbrains from scrapie infected mice than from the uninfected mice, but forebrain and hindbrain ABCA1 concentrations did not differ. There was approximately 8-fold more PrPSc in forebrain than in hindbrain (P < .01).
Increased Expression of ABCA1 Elevates PrPC and PrPSc Levels
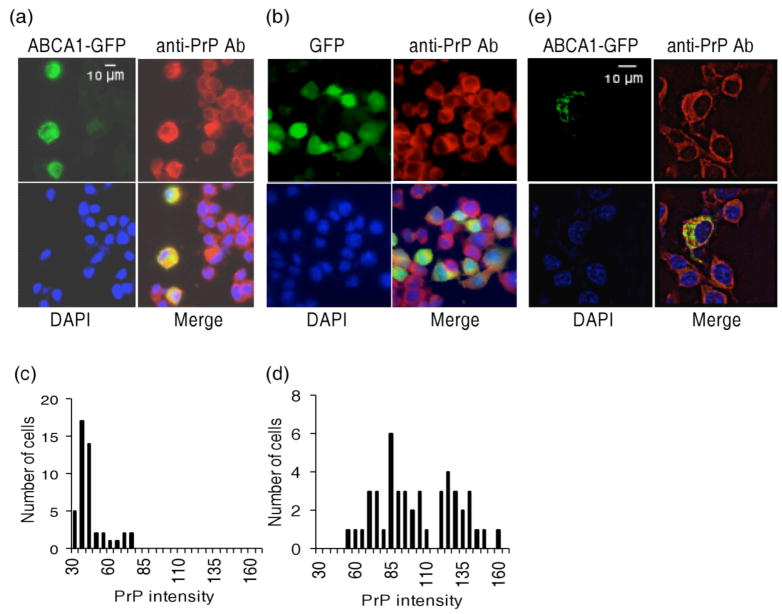
The impact of increasing ABCA1 expression on endogenous PrPC concentration was tested by transiently overexpressing a GFP-ABCA1 chimeric construct in N2a cells. Previous studies with this GFP-ABCA1 construct have shown that it is fully competent for cholesterol transport and co-localizes with endogenous ABCA1 (Fitzgerald et al., 2001). Consistent with previous observations in N2a and 293 cells (Fitzgerald et al., 2001, Fukumoto et al., 2002), GFP-ABCA1 decorated perinuclear compartments and cytoplasmic vesicular structures, while GFP-transfected control cells showed a uniform distribution of fluorescence throughout the cytosol (Fig. 3 a, b and e). Quantification of PrP-specific fluorescence intensity in GFP expressing cells showed significantly (P < .0001, Students T test) higher levels of PrP in GFP-ABCA1 expressing cells than in cells transfected with GFP alone or in non-transfected cells (Fig. 3 c and d). The PrP intensity distribution cells transfected with GFP alone was not different from that of non-transfected cells in populations containing either GFP-ABCA1 expressing cells or GFP expressing cells. PrPC and GFP-ABCA1 did not co-localize (Fig. 3e).
Fig. 3.
Overexpression of GFP-ABCA1 in N2a cells increases PrPC concentration. (a) GFP-ABCA1 and (b) GFP transfected neuroblastoma cells were analyzed by immunofluorescence 72 hr after transfection using anti-PrP Fab and DAPI staining. Green, red and blue pseudocolors show localization of GFP-ABCA1, PrP, and the nuclei; ‘merge’ shows their overlap. PrP fluorescence intensity in arbitrary units was quantified and expressed as a frequency distribution for (c) GFP-ABCA1 transfected cells and (d) GFP transfected cells. (e) Z-section images showing lack of co-localization of GFP-ABCA1 and PrP.
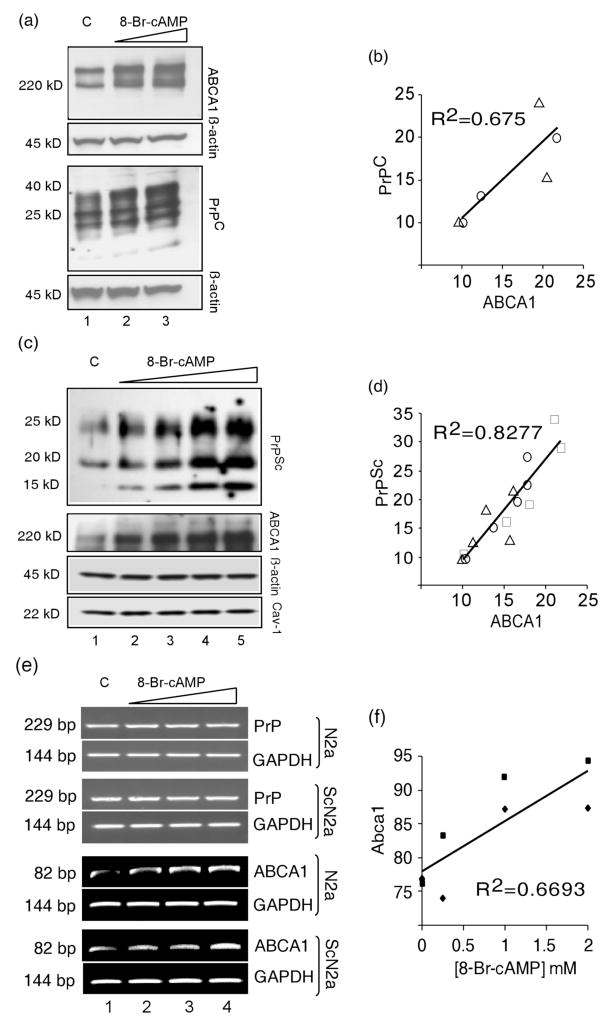
Enhanced expression of ABCA1 from its endogenous gene also leads to increased levels of PrP. The murine Abca1 gene possesses a strong cAMP responsive element (CRE) in its first intron and expression of Abca1 can be enhanced by cAMP analogues (Le Goff et al., 2006). The Prnp gene lacks consensus CRE sites and PrP mRNA levels are not increased by cAMP analogue treatment (Cabral et al., 2002). cAMP analogues have been often used to induce ABCA1 expression for studying its role in cholesterol transport (Lawn et al., 1999, Le Goff et al., 2006) and are used here to induce endogenous ABCA1 expression and study its impact on the concentration of PrPC or PrPSc. Treatment of N2a cells with the cAMP analogue 8-Br-cAMP at 1 or 2 mM for 24 hr caused a dose-dependent increase of both ABCA1 and PrP in N2a cells (Fig. 4 a, and b). Densitometric analysis of Western blots, one of which is shown in Fig. 4 a, with the β-actin normalized PrPC and ABCA1 levels in untreated cultures produced a positive correlation (R2 = 0.674, P = .022) between the levels of ABCA1 and PrPC in response to 8-Br-cAMP (Fig. 4b). A Western blot from a second experiment is shown in Supplementary Fig. S1.
Fig. 4.
PrPC and PrPSc concentrations increase in proportion to 8-Br-c AMP-induced expression of ABCA1. (a) N2a cells were untreated (C, lane 1) or treated with the cAMP analogue 8-Br-cAMP at 1mM (lane 2), or 2 mM (lane 3). Cell lysates were analyzed by immunoblotting with anti-ABCA1, anti-PrP, and anti-β-actin antibodies as indicated. (b) Concentrations of ABCA1 and PrPC are correlated in 8-Br-cAMP treated N2a cells (R2 = 0.675, P = 0.02). PrPC and ABCA1 levels were determined by densitometry from the immunoblot shown in Fig 4a and from a second experiment shown in Supplementary Figure S1. Circles and triangles distinguish the two experiments. (c) Immunoblots of lysates from untreated ScN2a cells (C, lane 1) or treated with cAMP analogue 8-Br-cAMP at 0.25 mM (lane 2), 0.5 mM (lane 3), 1 mM (lane 4), 2 mM (lane 5). In addition to anti-ABCA1, anti-PrP, and Anti-β-actin antibodies, anti-Caveolin-1 (Cav-1) was used for immunoblotting. (d) Concentrations of ABCA1 and PrPSc are correlated in 8-Br-cAMP treated ScN2a cells (R2 = 0.8277, P < 0.001). PrPSc and ABCA1 levels were determined by densitometry from the immunoblot shown in Fig 4b and from two additional experiments shown in Supplementary Figure S2. Circles, triangles, and squares distinguish the three experiments. (e) Effect of 8-Br-cAMP treatment on expression of Prnp (PrP), Gapdh (GAPDH), and Abca1 (ABCA1) mRNA assessed by RT-PCR. Ethidium bromide stained gels are shown. Untreated controls (C) are in lane 1, 8-Br-cAMP concentrations used for treatment were 0.5 mM (lane 2), 1 mM (lane 3), and 2 mM (lane 4). (f) Abca1 expression increases with concentration of 8-Br-cAMP. Squares indicate values from N2a cells and diamonds indicate values from ScN2a cells. Prnp and Gapdh mRNA levels did not change with 8-Br-cAMP treatment.
PrPSc levels show more variability among individual ScN2a cultures than do PrPC levels in N2a cultures. In three independent experiments, one of which is shown in Fig. 4c, ScN2a cultures were treated with 0.25, 0.5, 1, or 2 mM 8-Br-cAMP and levels of PK-resistant PrPSc and ABCA1 were determined by immunoblotting; the additional two experiments are shown in Supplementary Fig. S2. Since ABCA1 has been reported to function in transport of caveolae (Orso et al., 2000), we also monitored level of caveolin-1 (CAV1) in some experiments; no effect of 8-Br-cAMP treatment on CAV1 levels was found. The 8-Br-cAMP-induced elevation ABCA1 and PrPSc concentrations were highly correlated (R2 = 0.8277, P < .001) as shown in Fig. 4d. The increase of PrPSc in ScN2a cells (Fig. 4 c and d) likely reflects the ABCA1-induced increase in PrPC concentration (Fig. 4 a and b). Prnp mRNA levels in N2a and ScN2a appeared to be unchanged by 8Br-cAMP treatment, while there was a clear increase in ABCA1 mRNA levels detectable by RT-PCR (Fig. 4e and f). This is consistent with the previous observations made in PC-12 and C6 cells and the presence of CRE sequence in Abca1 but its absence in Prnp. Although effects of many other cAMP inducible genes such as solute carrier family 15 member 3 (Slc15a3), which is also increased in prion disease, on PrPC concentration cannot be ruled out, our results are consistent with an effect of ABCA1 as shown in Fig. 3 where transient overexpression of an ABCA1-GFP construct increased PrPC. Although there is a highly significant correlation of ABCA1 and PrP, this is only a correlation, and, on its own, is only consistent with an effect of ABCA1 and does not demonstrate a direct link.
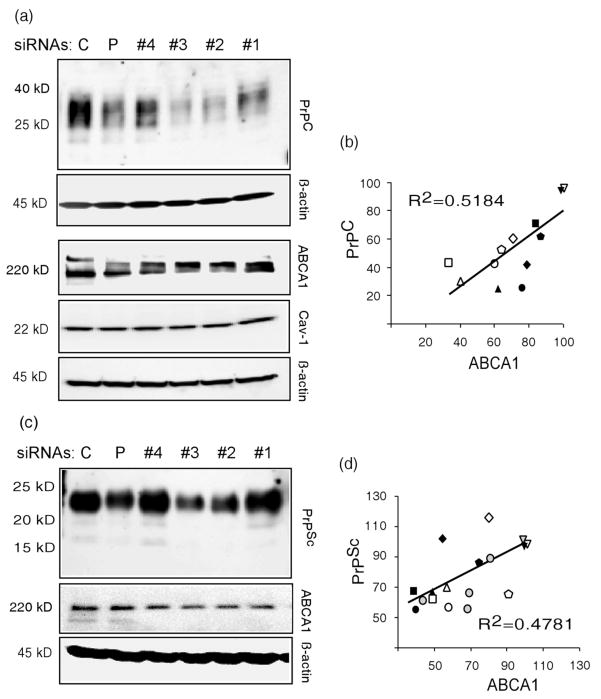
Knockdown of Abca1 Expression Reduces PrPC Levels and Production of PrPSc
RNAi mediated knockdown of ABCA1 expression was used to further assess the effects of ABCA1 concentration on PrP levels. In two independent experiment, four RNAi probes targeting different regions of ABCA1 mRNA and a pool of the four Abca1-specific probes, were used to reduce expression of Abca1 in N2a and ScN2a cells; and a non-specific RNAi was used as a control. Western blots for N2a and ScN2a cells are shown in Fig. 5a and c, and in Supplementary Fig. S3 and S4. Among the RNAi probes siRNA #2 and #3 were the most effective in reducing ABCA1 levels. Due to the variability in PrPSc among ScN2a cultures, four additional ScN2a cultures were treated with non-specific RNAi or ABCA1 #3; western blots from these cultures are shown in Supplementary Fig. S5. The RNAi probes had no effect on CAV1 levels. The four siRNA probes individually and pooled varied in their efficiency in reducing ABCA1 protein levels in N2a and ScN2a cells, which allowed us to assess the concentration dependent correlation between target (ABCA1) and effector (PrP) gene products (Fig. 5b and d). Intensity of immmunostaining, normalized to β-actin, of control cultures in each experiment was assigned a value of 100. As shown in Fig. 5 b and d, the degree of PrPC and PrPSc reduction corresponded to the efficiency of Abca1 knockdown, and ABCA1 and PrPC or PrPSc concentrations were directly correlated (PrPC, R2 = 0.5184, P = .0032; PrPSc, R2 = 0.4781, P < .0005). These results demonstrate that PrPC concentrations in the cell are affected by and proportional to ABCA1 levels and that reduction of ABCA1 inhibits PrPSc production.
Fig. 5.
Levels of PrPC and PrPSc are reduced in proportion to ABCA1 knockdown by RNAi. (a) N2a cells were transfected with a non-specific siRNA (C), a pool of Abca1-specific siRNAs (P), or four individual siRNA probes targeting different regions in the Abca1 mRNA (#1–4, see Materials and Methods). After 72 hr, cell lysates were assayed in Western blots using anti-ABCA1, anti-Cav-1, and anti-β-actin antibodies. Immunoblots from a second experiment are shown in Supplementary Fig. S3. (b) A plot of ABCA1 concentration vs. PrPC shows a correlation of PrPC and ABCA1 concentrations (R2 = 0.5184, P = .0032). Abca1 specific siRNAs 1, 2, 3, 4, the siRNA pool, and non specific siRNA are indicated by squares, triangles, circles, diamonds, pentagons and inverted triangles. Filled and open symbols differentiate the two experiments. (c) ScN2a cells were transfected with a non-specific siRNA (C), or four siRNAs individually (#1–4) or pooled (P). Lysates were PK digested and analyzed in Western blots using anti-PrP antibody. Lysates not treated with PK were used for anti-ABCA1 and anti-β actin antibodies. A repeat of this experiment is shown in Supplementary Figure S4. Four separate cultures of ScN2a cells were treated with siRNA #3 and lysates immunoblotted with PrP, ABCA1, and β-actin; the results are shown in Supplementary Figure S5. (d) PrPSc and ABCA1 concentrations are correlated (R2 = 0.4781, P < 0.0005). Abca1 specific siRNAs 1, 2, 3, 4, the siRNA pool, and non-specific siRNA are annotated by squares, triangles, circles, diamonds, pentagons and inverted triangles. Filled, open, and shaded symbols differentiate the three sets of experiments.
Discussion
Analysis of differential gene expression in prion infected mice suggests many perturbations in cholesterol homeostasis in addition to upregulation of Abca1 (Riemer et al., 2004, Xiang et al., 2007, Xiang et al., 2004) D. Hwang, et al., in preparation). To mention only a few examples, the sterol-O-acyltransferase 1 gene (Soat1) that encodes acyl-coenzyme A:cholesterol acyltransferase (ACAT) is overexpressed in prion-infected mice. Expression of the apolipoprotein genes Apod and Apoe also increase in parallel with Abca1 during the course of infection. LDL receptor mRNA is reduced in scrapie infected mouse brain (Riemer et al., 2004). The mechanism by which prion infection alters cholesterol homeostasis is not known, but may be related to alterations in re-cycling of PrP, synaptic membrane degeneration or to cellular stress responses.
Although the cause for increased Abca1 expression in prion disease has not been determined it is unlikely to be a direct effect of increased levels of extracellular PrPSc since the levels of ABCA1 were reduced to the same extent in regions of the brain that differ in the amount of PrPSc that has accumulated. In the case of FVB/NCr mice infected with RML scrapie prions shown in Fig. 2b and c, much less PK-resistant PrPSc is found in hindbrain structures than in more anterior regions while ABCA1 in forebrain and hindbrain showed similar increases. Other combinations of prion strains, mouse strains, or species show entirely different patterns of PrPSc deposition (DeArmond & Ironside, 1999, Kuczius & Groschup, 1999). There is no evidence for direct interactions among PrP isoforms and ABCA1; we noted no effect of PrPC concentrations on ABCA1 levels (Fig. 2a) and GFP-ABCA1 and PrP did not colocalize (Fig. 3e). It is likely that ABCA1 is not directly involved in prion replication but increases as a secondary consequence of prion infection.
Although Abca1 is only one of several genes involved in cholesterol homeostasis that is differentially expressed in prion-infected mice, altering its concentration is sufficient to modulate levels of PrP in cultured cells. Our studies did not address the mechanism by which ABCA1 affects PrPC and PrPSc in cultured cells. ABCA1 is a multifunctional protein and, in addition to its role in cholesterol transport, has reported functions in apoptosis, lipoprotein metabolism, and flipping of the bilipid membrane. As noted above, we did not observe co-localization of PrPC with GFP-ABCA1 as expected since ABCA1 is not found in CLD while PrPC is (Mendez et al., 2001, Vey et al., 1996), so a direct effect of ABCA1 on PrP seems unlikely. CAV1 is expressed in N2a and ScN2a cells, but its levels do no change in proportion to changes in ABCA1 concentration (Fig. 4c and Supplementary Fig. S2 and S5). Therefore, it seems unlikely that the role of ABCA1 on PrP concentration is mediated by an effect on CAV1 or caveolae. Given the well-documented effects of changes in cholesterol levels on PrPSc formation and prion incubation time in culture, an effect of ABCA1 on cholesterol is an attractive possibility. Reduction of membrane cholesterol either by inhibition of its biosynthesis with, lovastatin or squalestatin or by removal from the membrane with filipin dramatically reduces production of PrPSc in cultured cells (Bate et al., 2004, Gilch et al., 2006, Marella et al., 2002, Taraboulos et al., 1995). Simvastatin treatment also prolonged prion incubation time in mice (Mok et al., 2006). Although ABCA1 transports cholesterol to lipoprotein acceptors, increased expression of ABCA1 also can cause a general increase in membrane cholesterol. Once cholesterol is in membrane, it is distributed non-uniformly by lateral diffusion and condensation into cholesterol-rich CLD, which do not provide cholesterol for efflux to apolipoproteins (Mendez et al., 2001). Under this scenario, our demonstration that increased ABCA1 enhances PrPSc formation might be explained by an increased supply of PrPC due to more or larger CLD that serve as sites for the GPI-anchored protein. Since Prnp gene expression was not affected by changes in ABCA1 levels, increased membrane cholesterol and CLD may enhance cell surface localization and stability of PrPC. Additional work is needed to determine the mechanism by which changes in ABCA1 concentration affect levels of PrP.
The effects of ABCA1 on prion incubation time in mice are unknown at present. Preliminary results from analysis of a newly identified Abca1 mutant mouse with low serum cholesterol levels indicate reduced concentrations of PrPC in brain (unpublished results), which is consistent with the in vitro results presented here. PrP transgenic and Prnp gene ablated mice were used to demonstrate that prion incubation time is proportional to the concentration of PrPC and even a modest change in PrPC levels can substantially alter the rate of PrPSc formation and disease progression (Bueler et al., 1994, Carlson et al., 1994, Prusiner et al., 1990). Our studies in N2a and ScN2a cells lead us to hypothesize that reduced in ABCA1 concentration in mice may lead to longer incubation times. Whether the elevation of ABCA1 in prion infected mice has any consequences on prion replication will require additional studies.
Supplementary Material
Acknowledgments
We thank Michael L. Fitzgerald for providing the GFP-ABCA1 construct, D. William Provance for microscopy and image analysis, and Drs. Bermingham, Cabin, and Mercer for helpful comments and discussion. This work was supported by grants DAMD17-03-1-0321and DAMD17-03-1-0450 from the National Prion Research Program, U.S. Army Medical Research and Materiel Command and by program project grant NS41997 from the National Institute for Neurologic Disorders and Stroke, U. S. Public Health Service.
Abbreviations
- ABCA1
ATP-binding cassette transporter A1
- CLD
caveolae-like domain
- PK
Proteinase K
- Prnp
prion protein gene
- PrPC
cellular isoform of prion protein
- PrPSc
disease-specific isoform of the prion protein
- Cav-1
caveolin-1
- N2a
mouse neuroblastoma cell line
- ScN2a
scrapie-infected N2a cells
- siRNA
small interfering RNA
References
- Bate C, Salmona M, Diomede L, Williams A. Squalestatin cures prion-infected neurons and protects against prion neurotoxicity. J Biol Chem. 2004;279:14983–90. doi: 10.1074/jbc.M313061200. [DOI] [PubMed] [Google Scholar]
- Bortnick AE, Rothblat GH, Stoudt G, Hoppe KL, Royer LJ, McNeish J, Francone OL. The correlation of ATP-binding cassette 1 mRNA levels with cholesterol efflux from various cell lines. J Biol Chem. 2000;275:28634–40. doi: 10.1074/jbc.M003407200. [DOI] [PubMed] [Google Scholar]
- Bueler H, Fischer M, Lang Y, Bluethmann H, Lipp HP, DeArmond SJ, Prusiner SB, Aguet M, Weissmann C. Normal development and behaviour of mice lacking the neuronal cell-surface PrP protein. Nature. 1992;356:577–82. doi: 10.1038/356577a0. [DOI] [PubMed] [Google Scholar]
- Bueler H, Raeber A, Sailer A, Fischer M, Aguzzi A, Weissmann C. High prion and PrPSc levels but delayed onset of disease in scrapie-inoculated mice heterozygous for a disrupted PrP gene. Mol Med. 1994;1:19–30. [PMC free article] [PubMed] [Google Scholar]
- Cabral AL, Lee KS, Martins VR. Regulation of the cellular prion protein gene expression depends on chromatin conformation. J Biol Chem. 2002;277:5675–82. doi: 10.1074/jbc.M104815200. [DOI] [PubMed] [Google Scholar]
- Campana V, Sarnataro D, Zurzolo C. The highways and byways of prion protein trafficking. Trends Cell Biol. 2005;15:102–11. doi: 10.1016/j.tcb.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Carlson GA, Ebeling C, Yang SL, Telling G, Torchia M, Groth D, Westaway D, DeArmond SJ, Prusiner SB. Prion isolate specified allotypic interactions between the cellular and scrapie prion proteins in congenic and transgenic mice. Proc Natl Acad Sci U S A. 1994;91:5690–4. doi: 10.1073/pnas.91.12.5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caughey B, Raymond GJ. The scrapie-associated form of PrP is made from a cell surface precursor that is both protease- and phospholipase-sensitive. J Biol Chem. 1991;266:18217–23. [PubMed] [Google Scholar]
- DeArmond SJ, Ironside JW. Neuropathology of Prion Diseases. In: Prusiner SB, editor. Prion Biology and Diseases. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1999. pp. 585–652. [Google Scholar]
- Fitzgerald ML, Mendez AJ, Moore KJ, Andersson LP, Panjeton HA, Freeman MW. ATP-binding cassette transporter A1 contains an NH2-terminal signal anchor sequence that translocates the protein’s first hydrophilic domain to the exoplasmic space. J Biol Chem. 2001;276:15137–45. doi: 10.1074/jbc.m100474200. [DOI] [PubMed] [Google Scholar]
- Fukumoto H, Deng A, Irizarry MC, Fitzgerald ML, Rebeck GW. Induction of the cholesterol transporter ABCA1 in central nervous system cells by liver X receptor agonists increases secreted Abeta levels. J Biol Chem. 2002;277:48508–13. doi: 10.1074/jbc.M209085200. [DOI] [PubMed] [Google Scholar]
- Gilch S, Kehler C, Schatzl HM. The prion protein requires cholesterol for cell surface localization. Mol Cell Neurosci. 2006;31:346–53. doi: 10.1016/j.mcn.2005.10.008. [DOI] [PubMed] [Google Scholar]
- Harris DA. Cellular biology of prion diseases. Clin Microbiol Rev. 1999;12:429–44. doi: 10.1128/cmr.12.3.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko K, Vey M, Scott M, Pilkuhn S, Cohen FE, Prusiner SB. COOH-terminal sequence of the cellular prion protein directs subcellular trafficking and controls conversion into the scrapie isoform. Proc Natl Acad Sci U S A. 1997;94:2333–8. doi: 10.1073/pnas.94.6.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koldamova RP, Lefterov IM, Ikonomovic MD, Skoko J, Lefterov PI, Isanski BA, DeKosky ST, Lazo JS. 22R-hydroxycholesterol and 9-cis-retinoic acid induce ATP-binding cassette transporter A1 expression and cholesterol efflux in brain cells and decrease amyloid beta secretion. J Biol Chem. 2003;278:13244–56. doi: 10.1074/jbc.M300044200. [DOI] [PubMed] [Google Scholar]
- Kuczius T, Groschup MH. Differences in proteinase K resistance and neuronal deposition of abnormal prion proteins characterize bovine spongiform encephalopathy (BSE) and scrapie strains. Mol Med. 1999;5:406–18. [PMC free article] [PubMed] [Google Scholar]
- Lawn RM, Wade DP, Garvin MR, Wang X, Schwartz K, Porter JG, Seilhamer JJ, Vaughan AM, Oram JF. The Tangier disease gene product ABC1 controls the cellular apolipoprotein-mediated lipid removal pathway. J Clin Invest. 1999;104:R25–31. doi: 10.1172/JCI8119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Goff W, Zheng P, Brubaker G, Smith JD. Identification of the cAMP-responsive enhancer of the murine ABCA1 gene: requirement for CREB1 and STAT3/4 elements. Arterioscler Thromb Vasc Biol. 2006;26:527–33. doi: 10.1161/01.ATV.0000201042.00725.84. [DOI] [PubMed] [Google Scholar]
- Marella M, Lehmann S, Grassi J, Chabry J. Filipin prevents pathological prion protein accumulation by reducing endocytosis and inducing cellular PrP release. J Biol Chem. 2002;277:25457–64. doi: 10.1074/jbc.M203248200. [DOI] [PubMed] [Google Scholar]
- Mendez AJ, Lin G, Wade DP, Lawn RM, Oram JF. Membrane lipid domains distinct from cholesterol/sphingomyelin-rich rafts are involved in the ABCA1-mediated lipid secretory pathway. J Biol Chem. 2001;276:3158–66. doi: 10.1074/jbc.M007717200. [DOI] [PubMed] [Google Scholar]
- Mok SW, Thelen KM, Riemer C, Bamme T, Gultner S, Lutjohann D, Baier M. Simvastatin prolongs survival times in prion infections of the central nervous system. Biochem Biophys Res Commun. 2006;348:697–702. doi: 10.1016/j.bbrc.2006.07.123. [DOI] [PubMed] [Google Scholar]
- Orso E, Broccardo C, Kaminski WE, Bottcher A, Liebisch G, Drobnik W, Gotz A, Chambenoit O, Diederich W, Langmann T, Spruss T, Luciani MF, Rothe G, Lackner KJ, Chimini G, Schmitz G. Transport of lipids from golgi to plasma membrane is defective in tangier disease patients and Abc1-deficient mice. Nat Genet. 2000;24:192–6. doi: 10.1038/72869. [DOI] [PubMed] [Google Scholar]
- Peretz D, Williamson RA, Kaneko K, Vergara J, Leclerc E, Schmitt-Ulms G, Mehlhorn IR, Legname G, Wormald MR, Rudd PM, Dwek RA, Burton DR, Prusiner SB. Antibodies inhibit prion propagation and clear cell cultures of prion infectivity. Nature. 2001;412:739–43. doi: 10.1038/35089090. [DOI] [PubMed] [Google Scholar]
- Prusiner SB. Prions. Proc Natl Acad Sci U S A. 1998;95:13363–83. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner SB, Scott M, Foster D, Pan KM, Groth D, Mirenda C, Torchia M, Yang SL, Serban D, Carlson GA, et al. Transgenetic studies implicate interactions between homologous PrP isoforms in scrapie prion replication. Cell. 1990;63:673–86. doi: 10.1016/0092-8674(90)90134-z. [DOI] [PubMed] [Google Scholar]
- Riemer C, Neidhold S, Burwinkel M, Schwarz A, Schultz J, Kratzschmar J, Monning U, Baier M. Gene expression profiling of scrapie-infected brain tissue. Biochem Biophys Res Commun. 2004;323:556–64. doi: 10.1016/j.bbrc.2004.08.124. [DOI] [PubMed] [Google Scholar]
- Sarnataro D, Campana V, Paladino S, Stornaiuolo M, Nitsch L, Zurzolo C. PrP(C) association with lipid rafts in the early secretory pathway stabilizes its cellular conformation. Mol Biol Cell. 2004;15:4031–42. doi: 10.1091/mbc.E03-05-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taraboulos A, Scott M, Semenov A, Avrahami D, Laszlo L, Prusiner SB. Cholesterol depletion and modification of COOH-terminal targeting sequence of the prion protein inhibit formation of the scrapie isoform. J Cell Biol. 1995;129:121–32. doi: 10.1083/jcb.129.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vey M, Pilkuhn S, Wille H, Nixon R, DeArmond SJ, Smart EJ, Anderson RG, Taraboulos A, Prusiner SB. Subcellular colocalization of the cellular and scrapie prion proteins in caveolae-like membranous domains. Proc Natl Acad Sci U S A. 1996;93:14945–9. doi: 10.1073/pnas.93.25.14945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang W, Hummel M, Mitteregger G, Pace C, Windl O, Mansmann U, Kretzschmar HA. Transcriptome analysis reveals altered cholesterol metabolism during the neurodegeneration in mouse scrapie model. J Neurochem. 2007 doi: 10.1111/j.1471-4159.2007.04566.x. [DOI] [PubMed] [Google Scholar]
- Xiang W, Windl O, Wunsch G, Dugas M, Kohlmann A, Dierkes N, Westner IM, Kretzschmar HA. Identification of differentially expressed genes in scrapie-infected mouse brains by using global gene expression technology. J Virol. 2004;78:11051–60. doi: 10.1128/JVI.78.20.11051-11060.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.