Abstract
Background
Microtubules (MTs) are assembled by heterodimers of α- and β-tubulins, which provide tracks for directional transport and frameworks for the spindle apparatus and the phragmoplast. MT nucleation and dynamics are regulated by components such as the γ-tubulin complex which are conserved among eukaryotes, and other components which are unique to plants. Following remarkable progress made in the model plant Arabidopsis thaliana toward revealing key components regulating MT activities, the completed rice (Oryza sativa) genome has prompted a survey of the MT cytoskeleton in this important crop as a model for monocots.
Scope
The rice genome contains three α-tubulin genes, eight β-tubulin genes and a single γ-tubulin gene. A functional γ-tubulin ring complex is expected to form in rice as genes encoding all components of the complex are present. Among proteins that interact with MTs, compared with A. thaliana, rice has more genes encoding some members such as the MAP65/Ase1p/PRC1 family, but fewer for the motor kinesins, the end-binding protein EB1 and the mitotic kinase Aurora. Although most known MT-interacting factors have apparent orthologues in rice, no orthologues of arabidopsis RIC1 and MAP18 have been identified in rice. Among all proteins surveyed here, only a few have had their functions characterized by genetic means in rice. Elucidating functions of proteins of the rice MT cytoskeleton, aided by recent technical advances made in this model monocot, will greatly advance our knowledge of how monocots employ their MTs to regulate their growth and form.
Key words: Cytoskeleton, kinesins, microtubules (MTs), microtubule-associated proteins (MAPs), motors, rice, Oryza sativa
INTRODUCTION
The 25-nm tubular structure of microtubules (MTs) was first described in the monocot Phleum pratense (Timothy-grass) over four decades ago (Ledbetter and Porter, 1963). Since then, MTs have been implicated in almost every single aspect of intracellular activities, from cell division to cell morphogenesis in plants, as well as among other eukaryotic organisms. Assembled by the heterodimers of α- and β-tubulin GTPases in a head-to-tail fashion, MTs exhibit their characteristic dynamic instability phenomenon in vitro and in vivo (Baskin, 2000). The polarity of MTs is defined with a more dynamic (faster growing and shortening) plus end and less dynamic minus end. The terminal sub-unit at the minus end is α-tubulin, and at the plus end is β-tubulin. MTs often exhibit a rapid catastrophic process resulting from forming a cap of GDP-β-tubulin following GTP hydrolysis, which precedes a slower rescue process to recruit tubulin dimers containing GTP-bound β-tubulin. This phenomenon is known as dynamic instability, which is observed more frequently at the plus end than at the minus end.
In plants, one of the most noticeable features of the dynamic MT network is reflected by the four MT arrays exhibited by mitotic cells in somatic tissues. They are the parallel cortical MTs, the pre-prophase band (PPB), the barrel-shaped mitotic apparatus and the phragmoplast. The rapid turnover of these arrays was first observed in live cells by microinjection of fluorescently labelled tubulins, and later by green fluorescent protein (GFP) fusion proteins with α-tubulin or an MT-binding motif (Zhang et al., 1990; Marc et al., 1998; Ueda et al., 1999; Granger and Cyr, 2000; Kumagai et al., 2001). Besides the dynamic instability property intrinsic to MTs, their rapid turnover in plant cells must be modulated by MT-associated proteins (MAPs) whose activities may be regulated in a cell cycle-dependent manner. Progress made in the past few years has resulted in the identification of proteins which decorate all arrays or a particular array, mainly in A. thaliana. A number of these proteins are either remotely related to MAPs in organisms of other kingdoms, or specific to flowering plants. Unfortunately, very little progress has been made in characterizing proteins which are associated with MTs in rice and other monocots.
In the past years, fascinating discoveries have been made to allow MTs to be connected with various aspects of plant growth and development. One of the remaining questions is how plant cells remodel their MTs in response to hormones such as auxins and brassinosteroids. Recently, a previously identified auxin-inducible gene has turned out to encode a MAP which may participate in spatial regulation of cell division (Buschmann et al., 2006). Another quest for plant MAPs has resulted in a phospholipase D isoform as a potential linker between cortical MTs and the plasma membrane (Gardiner et al., 2001). In addition, external cues such as salt stress also trigger responses of MT reorganization (Shoji et al., 2006; Wang et al., 2007a). Now, there is no doubt that plant cells need MAPs for MT-based activities (Lloyd and Hussey, 2001). It is foreseeable that mining plant genomes and proteomes by various approaches will soon result in the identification of more plant MAPs that mediate various growth responses.
The excitement generated by functional studies of plant MTs and MAPs has been expressed in several review articles (Hussey and Hawkins, 2001; Sedbrook, 2004; Hamada, 2007; Kaloriti et al., 2007; Mineyuki, 2007). However, most if not all published studies have been carried out in dicot systems such as A. thaliana and tobacco BY-2 cells. A proteomic attempt showed promise in identifying proteins interacting with tubulin in rice (Chuong et al., 2004). Following in the footsteps of the initial description of MTs in monocots, the sequenced rice genome has prompted a survey of the inventory of the MT cytoskeleton and its interacting proteins in this model species of Graminaceae. The sequence analysis reported here was largely carried out by referring to two annotation systems: the Rice Genome Annotation Project (RGAP, http://rice.plantbiology.msu.edu/) with calls noted as LOC_Osxxgxxxxx, and the Rice Annotation Project (RAP, http://rapdb.dna.affrc.go.jp/) with calls noted as Osxxgxxxxxxx. Proteins discussed in this article are listed in Table 1. By summarizing the rice MT cytoskeleton based on the best current understanding of plant MTs, it is anticipated that in coming years additions of novel rice proteins will be made to the inventory or rice MAPs. It is hoped that this survey will help integrate functions of the proteins of the rice MT cytoskeleton with historical observations on MT-based cellular processes, e.g. cell division, in monocot systems such as onion and spiderwort.
Table 1.
Proteins discussed in this review
| Protein family | Members in rice |
|---|---|
| Tubulin | OsTUA (3), OsTUB (8), OsTUG (1) |
| Tubulin-folding factors | Tubulin-specific chaperones A–E (1 each), OsARL2 (1) |
| γ-Tubulin ring complex | OsGCP2–4 (1 each), OsGCP5 (2), OsGCP6 (1), OsNEDD1/GCP-WD (1) |
| Aurora kinase | OsAUR1 (1), OsAUR2 (1) |
| TPX2/WVD2 | OsWDL (6) |
| MAP65/Ase1p | OsMAP65 (11) |
| XMAP215/TOG | OsMOR1 (1) |
| MAP70 | OsMAP70 (4) |
| AIR9 | OsAIR9 (1) |
| TOR1/SPR2 | OsTOR1/SPR2 (4) |
| EB1 | OsEB1a (1), OsEB1b (1) |
| SPR1 | OsSPR1 (4) |
| CLASP | OsCLASP (3) |
| TAN | OsTAN (1) |
| TON1 | OsTON1 (2) |
| TON2 | OsTON2 (1) |
| Katanin | OsKSS1/SGL1 (p60) (1), OsKLS (p80) (3) |
| PLDδ | OsPLDδ (1) |
| MAP190 | OsMAP190 (1) |
| Kinesin | OsKinesin (52) |
Numbers in parentheses represent how many similar proteins are encoded by the rice genome.
THE TUBULIN FAMILY PROTEINS
Early persistent investigations have resulted in the conclusion that plant tubulins are encoded by multigene families, e.g. there are six α-tubulin, nine β-tubulin and two γ-tubulin genes in the model organism A. thaliana (Kopczak et al., 1992; Snustad et al., 1992; Liu et al., 1994). Although it is always tempting to postulate that different isoforms of tubulins may take on different tasks, to date experimental data have not been able to prove this assumption despite ample evidence of tissue-specific expression patterns of tubulin genes (Eun and Wick, 1998; Goddard et al., 1998). When polymerization-competitive bovine brain α- and β-tubulins were delivered by microinjection into stamen hair cells of the spiderwort Tradescantia virginiana, they entered polymerization/depolymerization cycles and were incorporated into various arrays of MTs (Zhang et al., 1990). Hence, different isoforms of tubulins are believed to be functionally equivalent, and when expressed in the same cells they exhibit functional redundancy, as shown by the two γ-tubulin genes in A. thaliana (Pastuglia et al., 2006).
Prior to the completion of the rice genome, the quest for rice tubulin genes had already uncovered all α- and β-tubulins (Kang et al., 1994; Qin et al., 1997; Yoshikawa et al., 2003). There are three α-tubulin genes (OsTUA) and eight β-tubulin genes (OsTUB) in rice. Expression analysis indicated that tissue-specific expression was found among individual members of these tubulins (Qin et al., 1997; Yoshikawa et al., 2003). For example, OsTUB8 was specifically expressed in anthers (Yang et al., 2007). This fact mirrors the finding that AtTUA1 and AtTUB9 are preferentially expressed in anthers in A. thaliana (Carpenter et al., 1992; Cheng et al., 2001).
Plant γ-tubulin was first detected in all MT arrays and found to be pronounced toward minus ends of MTs in the onion Allium cepa 15 years ago (Liu et al., 1993). Plant γ-tubulin and its associated proteins are not restricted to the MT minus end, and often appear on the wall of extant MTs to initiate the formation of MT branches (Murata et al., 2005). Genetic approaches have confirmed that γ-tubulin is essential for MT organization in A. thaliana (Binarova et al., 2006; Pastuglia et al., 2006). Surprisingly, there is only a single γ-tubulin gene (OsTUG) in the rice genome. The protein encoded by this LOC_Os05g06450/Os05g0156600 locus is expected to be essential for MT organization as seen for the redundant role of two γ-tubulin genes in A. thaliana (Pastuglia et al., 2006).
Despite the significantly larger genome size of rice compared with that of A. thaliana, the fact that there are fewer tubulin genes in rice makes it an attractive system for future genetic manipulation of functions of individual tubulins.
PREPARING TUBULINS FOR POLYMERIZATION: TUBULIN-SPECIFIC CHAPERONES
Six tubulin-specific chaperones (A–E) and ARL2 are conserved proteins which are solely devoted to folding tubulins to make heterodimers competent for polymerization. Studies in A. thaliana have indicated that their functions are indispensable for MT-dependent cellular activities, and mutations often lead to embryo lethal phenotypes (Kirik et al., 2002a, b; Steinborn et al., 2002; Tzafrir et al., 2002).
In the rice genome, single genes encode tubulin-specific chaperones A–E and ARL2: LOC_Os02g57150/Os02g0816500 (113 amino acids), LOC_Os04g59560/Os04g0692100 (267 amino acids), LOC_Os02g35110/Os02g0557000 (353 amino acids), LOC_Os10g36490/Os10g0508500 (1140 amino acids), LOC_Os05g01500/Os05g0105300 (433 amino acids) and LOC_Os02g22140/Os02g0327100 (186 amino acids), respectively. Similarly to their counterparts in other organisms, chaperones B and E contain a CAP-Gly domain toward their N-termini, which is the key element of the MT-binding domain in proteins such as the dynactin component p150Glued and CLIP170. Whether the CAP-Gly motif of these two proteins binds MTs in vivo awaits further examination. The ARL2 protein is a small GTPase, and is known as an ADP-ribosylation factor.
AT THE MT MINUS END: THE γ-TUBULIN RING COMPLEX
Unlike α- and β-tubulins whose heterodimers act as bricks of the MT cylinder, γ-tubulin forms a ring complex (γTuRC) with five other accessory proteins in metazoans (Wiese and Zheng, 2006). The proteins of the complex are often abbreviated as GCPs (γ-tubulin complex proteins) or Grips (γ-tubulin ring proteins), which typically decorate the MT minus end. The accessory proteins are structurally related, and all contain five homologous regions or two grip motifs, with regions I and II for motif 1 and regions III–V for motif 2 (Gunawardane et al., 2000; Murphy et al., 2001). The most critical players of the γTuRC are γ-tubulin plus Spc97p/GCP2 and Spc98p/GCP3, which are conserved among all eukaryotes and form the γ-tubulin small complex (γTuSC), which is the core of the large γTuRC (Wiese and Zheng, 2006). The budding yeast Saccharomyces cerevisiae only produces γTuSC (Wiese and Zheng, 2006). The γ-tubulin complex plays an essential role in MT nucleation. Recently, it has been found that a WD40 repeat-containing protein NEDD1 or GCP-WD interacts with γTuRC, and is essential for spindle assembly, probably due to its role in recruiting γTuRC to the centrosome, the MT-organizing centre (MTOC) in animal cells (Gunawardane et al., 2003; Haren et al., 2006; Lüders et al., 2006; Manning and Kumar, 2007).
Homologues of GCP2-6 and NEDD1/GCP-WD have been identified among land plants by mining the available plant genomes (Murata et al., 2007). Among them, the GCP2/Spc97p and GCP3/Spc98p homologues have been proved to form a soluble complex with γ-tubulin, and decorate the nuclear envelope (Erhardt et al., 2002; Seltzer et al., 2007).
The rice genome contains single genes for homologues of GCP2/Spc97p, GCP3/Spc98p, GCP4 and GCP6: LOC_Os04g42330/Os04g0501700, LOC_Os09g27370/Os09g0446200, LOC_Os05g06260/Os05g0154500 and LOC_Os04g47906/Os04g0566800, respectively. However, two genes encode homologues of GCP5, i.e. LOC_Os12g41290/Os12g0606100 and LOC_Os02g32340/Os02g0523300, with homology as high as 90 % identity at the amino acid level. This scenario is almost identical to that in A. thaliana as a model dicot also containing two GCP5-encoding genes: At1g80260 and At1g20570. In addition, homologues of GCP2-4 and GCP6, with >80 % amino acid sequence identity with those rice counterparts, are encoded by single genes in A. thaliana (Murata et al., 2007).
A single putative NEDD1/GCP-WD homologue was identified by analysing the rice genome. This hypothetic protein encoded by the LOC_Os09g09470/Os09g0267500 locus is 65 % identical to the protein encoded by the At5g05970 locus in A. thaliana. The rice protein has the highest homology with human NEDD1 toward the N-terminal WD40 repeats. It is unclear whether the plant homologues function like the mammalian NEDD1/GCP-WD in the recruitment of the γ-tubulin complex. Given the fact that the spindle pole body functions as a centrosome-like primary MTOC, to date no such WD40 repeat-containing proteins have been identified as a NEDD1/GCP-WD homologue in fungi. It is rather intriguing whether the plant counterpart functions in the same way as the animal protein. More specifically, it would be critical to examine whether it is involved in MT nucleation on the wall of extant MTs.
THE MITOTIC KINASE AURORA
The Aurora family enzymes are serine/threonine kinases with molecular weights of approx. 30–40 kDa. In animals and fungi, Aurora kinases act in discrete regions of the spindle apparatus, such as the centrosome, kinetochores and the spindle midzone (Carmena and Earnshaw, 2003). The frog Aurora A isoform Eg2 directly binds to MTs in vitro, implying a direct interaction between the kinase and proteins associated with MTs in vivo (Roghi et al., 1998). The animal Aurora A appears at the centrosome and MTs toward spindle poles, and activates MAPs and kinesin motors essential for MT nucleation and organization (Marumoto et al., 2005). Because beads coated with Aurora A are able to promote spindle assembly in the absence of the centrosome or chromosomes, Aurora A has earned the nickname of ‘guardian of poles’ (Tsai and Zheng, 2005). The animal Aurora B forms a complex with the inner centromere protein (INCENP), survivin and borealin, and the complex appears at the centromere before the onset of anaphase and in the spindle midzone and the midbody after anaphase (Ruchaud et al., 2007). They are referred to as components of the chromosomal passenger complex (CPC) because of their dynamic behaviour during mitosis. Aurora B substrates also include MAPs and kinesins, but they are different from those of Aurora A because of different intracellular localizations.
Three Aurora kinases of two classes are encoded by the A. thaliana genome. AtAUR1 and AtAUR2 form the first class, and AtAUR3 the second, and all of them were predominantly expressed in tissues enriched with dividing cells (Demidov et al., 2005). Driven by the 35S promoter, their cDNAs were expressed in fusions with the GFP-coding sequence in cultured tobacco BY-2 cells (Demidov et al., 2005; Kawabe et al., 2005). Both AtAUR1–GFP and AtAUR2–GFP fusions decorated the nuclear envelope and structures resembling mitotic spindles, and the AtAUR1–GFP signal also conspicuously appeared at the site of the forming cell plate during cytokinesis. AtAUR3–GFP, however, exhibited a dot-like pattern on chromosomes, probably at the centromere/kinetochore region. Overexpression of AtAUR3 in BY-2 cells has profound effects, causing alteration of the distribution of the MT nucleation factor γ-tubulin, partial disassembly of spindle MTs and misorientation of the cell division plane (Kawabe et al., 2005).
Rice expresses only two distinct Aurora kinases, OsAUR1 (LOC_Os01g09580/Os01g0191800) of 292 amino acids and OsAUR2 (LOC_Os03g55620/Os03g0765000) of 279 amino acids, belonging to the first and second classes mentioned above, respectively. Similarly to AtAURs, sequence comparison shows no preference of homology for OsAURs to either animal Aurora A or Aurora B. Thus, novel activities of plant Aurora kinases are expected.
THE TPX2 HOMOLOGUE AND RELATED WVD2 PROTEINS
In mammals, Aurora A is targeted to the centrosome and spindle MTs by the MAP TPX2 (Kufer et al., 2002). TPX2 also plays a critical role in directing the kinesin XKLP2 to the centrosome in the frog Xenopus (Wittmann et al., 2000). Among TPX2 proteins identified in different organisms, a signature motif in the MT-binding site has been identified as pfam06886 with the KLEEK penta-peptide (http://pfam.sanger.ac.uk/).
In A. thaliana, an expressed protein of 758 amino acids encoded by the At1g03780 locus has been identified as a putative TPX2 homologue, which bears a region resembling the signature motif (Perrin et al., 2007). Several other expressed proteins related to this homologue also share this feature, but with significantly smaller sizes of approx. 200–300 amino acids. One of them is the WVD2 (WAVE-DAMPENED 2) protein which acts as an MT-bundling and stabilizing factor (Yuen et al., 2003). WVD2 and seven related WDL1–7 proteins all contain the pfam06886 motif (Yuen et al., 2003; Kaloriti et al., 2007; Perrin et al., 2007). When WVD2 or its homologues are expressed constitutively at elevated levels, cortical MTs are often bundled and stabilized, resulting in reduced anisotropic cell elongation, and consequently short and thick stems and roots (Perrin et al., 2007). Based on sequence comparison, it was determined that at least 14 proteins bear the TPX2 pfam06886 motif in A. thaliana (data not shown). Among them, WVD2 and WDL1 are able to bind directly to MTs in vitro and in vivo, indicating that they are authentic MAPs (Perrin et al., 2007). It would not be surprising if other proteins in this group of 14 turn out to be true MAPs as well because of the presence of the KLEEK penta-peptide inside the signature motif.
In the rice genome, the LOC_Os07g32390/Os07g0507200 locus encodes a protein sharing 45 % amino acid sequence identity with that of At1g03780. While there is little if any doubt that rice and arabidopsis TPX2 homologues are indeed MAPs, it would be intriguing to determine whether either or both of them function in targeting any Aurora kinase or kinesin to the spindle or spindle poles. Based on the criterion of the presence of the penta-peptide KLEEK as in WVD2 and WDLs, six putative proteins related to WVD2 were identified. They are LOC_Os02g10690/Os02g0200800, LOC_Os03g58480/Os03g0799100, LOC_Os06g40450/Os06g0606800, LOC_Os07g08790/Os07g0185500, LOC_Os11g36340/Os11g0571900 and LOC_Os11g38010/Os11g0592600, which could be considered to encode OsWDL proteins. Two additional loci, LOC_Os03g11400/Os03g0212600 and LOC_Os09g13650/Os09g0307300, may also encode MAPs because the deduced sequences contain the pfam06886 motif with an incompletely conserved KLEEK region.
In contrast to the TPX2 homologue which may be specifically involved in MT organization in spindles, WVD2 and WDL proteins may play roles in modulating the cortical MT array to facilitate anisotropic cell expansion. We are eager to learn the function of the rice TPX2 homologue, and the physiological significance of having so many WDL isoforms in rice and A. thaliana.
MEDIATING MT–MT INTERACTIONS: SPLENDID MEMBERS OF THE MAP65/ASE1P/PRC FAMILY
Early heroic endeavours in isolating plant MAPs resulted in clear candidates purified from cultured carrot cells (Cyr and Palevitz, 1989). Whether plant MAPs could be purified to homogeneity was no longer questioned after MAP65 was purified from tobacco BY-2 cells (Jiang and Sonobe, 1993). It was not realized at the time that similar proteins could be found in organisms across eukaryotic kingdoms. The yeast homologue of MAP65 was identified as a protein critical for anaphase B spindle elongation, thus named as Ase1p for anaphase spindle elongation 1 protein (Pellman et al., 1995). The mammalian homologue PRC1 (protein regulating cytokinesis 1) was identified as an important MAP playing a regulatory role in cytokinesis (Jiang et al., 1998).
The initial cloning of tobacco MAP65 cDNA was followed by revealing nine genes encoding MAP65-like proteins in A. thaliana (Smertenko et al., 2000; Hussey et al., 2002). Surprisingly, different A. thaliana MAP65 isoforms exhibit dramatically different localization patterns when expressed in fusions with GFP in BY-2 cells (Van Damme et al., 2004b). While MAP65-1 and -5 fusions decorate cortical MTs, MAP65-4 only appears on MTs in the spindle pole region during mitosis (Van Damme et al., 2004a, b). MAP65 isoforms also differentially decorate phragmoplast MTs, with MAP65-3 appearing conspicuously near the MT plus end (Van Damme et al., 2004a). Most surprisingly, endogenous MAP65-6 is associated with mitochondria, probably implying its role in mediating the interaction between the organelle and MTs (T. Mao et al., 2005). Biochemical studies conclude that MAP65-1 and probably other isoforms form homodimers of approx. 25 nm to cross-link MTs, suggesting a role in bundling antiparallel MTs in the phragmoplast mid-zone (Smertenko et al., 2004; Wicker-Planquart et al., 2004). Activities of MAP65 are regulated by phosphorylation, especially by cyclin-dependent kinase (Cdk) during the cell cycle (G. Mao et al., 2005; Sasabe et al., 2006; Smertenko et al., 2006). It has also been found that the MT-bundling activity of tobacco MAP65-1 is downregulated when phosphorylated by a cytokinesis-specific mitogen-activated protein kinase cascade to allow phragmoplast MTs to be disassembled (Sasabe and Machida, 2006; Sasabe et al., 2006). Genetic evidence pinpoints AtMAP65-3/PLE for its critical role in cytokinesis (Müller et al., 2004; Caillaud et al., 2008). Because multiple endogenous MAP65 proteins are present at the plus end of phragmoplast MTs, as revealed by GFP fusions and immunolocalization, it remains to be understood how their activities are coordinated spatially and temporally.
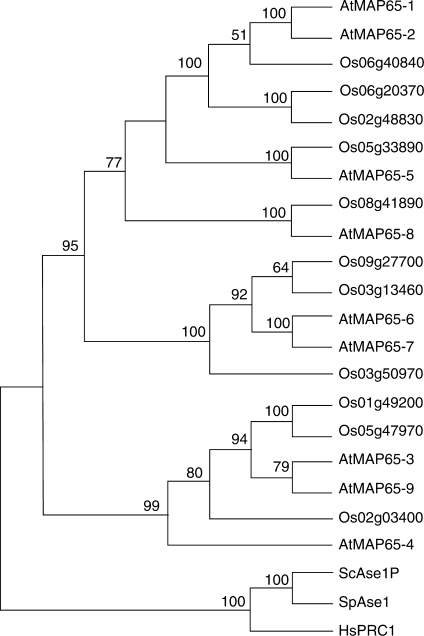
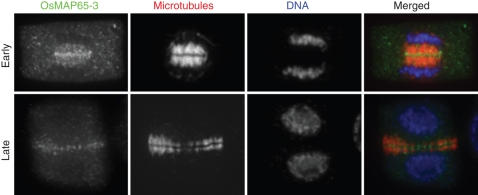
Eleven rice genes were identified as encoding MAP65 homologues (Table 2). It appears that an additional member has been added to each of the two clades containing AtMAP65-1/AtMAP65-2 and AtMAP65-6/AtMAP65-7 (Fig. 1). The homology between the hypothetical OsMAP65-3 (LOC_Os01g49200/Os01g0685900) and AtMAP65-3 is 48 % identity/64 % similarity at the amino acid level. Because of the aforementioned role of AtMAP65-3 in cytokinesis, it was important to test whether OsMAP65-3 behaved similarly during mitotic cell division. When the endogenous OsMAP65-3 protein was probed with isoform-specific antibodies, it appeared near the plus end of the phragmoplast MTs during cytokinesis (Fig. 2). Hence, the result has suggested that OsMAP65-3 may play a similar role to that of AtMAP65-3. A recent genetic analysis indicated that insertional mutations at the OsMAP65-3 locus caused dwarf and low-fertility phenotypes (Miyao et al., 2007). Whether the phenotype is a result of a cytokinesis defect awaits further examination of the mutants. It would be rather interesting to dissect distinct developmental roles of these 11 MAP65 isoforms in rice.
Table 2.
List of MAP65 family proteins in rice
| RGAP IDs | RAP IDs |
|---|---|
| LOC_Os01g49200 | Os01g0685900* |
| LOC_Os02g03400 | Os02g0126300 |
| LOC_Os02g48830 | Os02g0720200 |
| LOC_Os03g13460 | Os03g0237600 |
| LOC_Os03g50970 | Os03g0719000 |
| LOC_Os05g33890 | Os05g0409400 |
| LOC_Os05g47970 | Os05g0552900 |
| LOC_Os06g20370 | Os06g0308300 |
| LOC_Os06g40840 | Os06g0611000 |
| LOC_Os08g41890 | Os08g0531100 |
| LOC_Os09g27700 | Os09g0450300 |
IDs were assigned by RGAP and RAP, respectively.
* Known as MAP65-3.
Fig. 1.
Neighbor–Joining tree of MAP65 family proteins. Phylogenetic relationships among the MAP65 members of rice (Os), A. thaliana (At), the budding yeast Saccharomyces cerevisiae (ScAse1p), the fission yeast Schizosaccharomyces pombe (SpAse1) and human (HsPRC1) are shown. For rice proteins, shown in this figure were IDs annotated by RGAP omitting LOC_. The AtMAP65 members were assigned according to published nomenclature (Hussey and Hawkins, 2001). The GenBank accession numbers for the others are: NP_116582 for ScAse1p, CAC21482 for SpAse1 and AAC02688 for HsPRC1. Bootstrap values at the branches represent the percentages obtained in 1000 replications. Only values >50 % are presented.
Fig. 2.
Immunolocalization of OsMAP65-3 in developing phragmoplasts. OsMAP65-3 was detected by an isoform specific antibody, MTs by an anti-α-tubulin antibody, and DNA by DAPI (4′,6-diamidino-2-phenylindole). Cells at an early and late stage of cytokinesis are shown. OsMAP65-3 appeared in a broader distribution pattern in the early phragmoplast, and then became more concentrated in the phragmoplast midline in the late phragmoplast. In the merged image, OsMAP65-3 was pseudocoloured in green, MTs in red and DNA in blue.
The yeast protein Ase1p is a substrate of the Aurora B-type kinase Ipl1p, and its phosphorylation is critical for spindle assembly (Kotwaliwale et al., 2007). It is intriguing whether a similar phosphorylation event regulates the activity of OsMAP65-3 and/or other isoforms.
PROMOTING MT ASSEMBLY AT THE PLUS END: XMAP215/TOG FAMILY PROTEINS
The founding member of this family, XMAP215, was first identified more than two decades ago as a MAP which could promote centrosome-nucleated MT assembly (Gard and Kirschner, 1987a, b). A decade later, a genetic screen recovered the homologous Stu2p as an essential MAP required for spindle assembly in budding yeast (Wang and Huffaker, 1997). The outstanding feature of this family of proteins is the presence of HEAT (huntingtin, elongation factor-3, A sub-unit of PR65, Tor-kinase) repeats scattered across the polypeptides (Gard et al., 2004). The protein functions as an MT polymerase by adding new tubulin dimers to the plus end while moving with the growing plus end (Howard and Hyman, 2007).
The plant gene homologous to XMAP215 was first isolated when examining the mor1 mutant defective in MT organization in A. thaliana (Whittington et al., 2001). Allelic mutations also cause defects in cytokinesis during microgametophyte formation (Twell et al., 2002). Detailed examinations of the mor1 mutant suggest that the MOR1 protein decorates all MT arrays during mitosis, and is essential for various aspects of MT organization during cell division, in addition to ensuring the integrity of cortical MTs (Eleftheriou et al., 2005; Kawamura et al., 2006).
Unlike in A. thaliana, the O. sativa genome contains two linked genes, LOC_Os01g60040/Os01g0816400 and LOC_Os01g60050/Os01g0816500, which are expressed for MOR1/XMAP215-related proteins of 1998 and 326 amino acids, respectively. It seems that the protein encoded by the first gene is an apparent orthologue of MOR1 due to its similarity in size to AtMOR1 of 1978 amino acids. The protein encoded by the latter shows >60 % sequence identity with the N-terminal approx. 300 amino acid region of OsMOR1. Hence, it is rather mysterious whether this smaller protein acts as a MAP in vivo.
In the frog Xenopus, XMAP215 is a substrate of Aurora A, and its phosphorylation is critical for its function in MT nucleation (Giet et al., 2002). Again, possible Aurora phosphorylation events might play critical roles in regulating functions of XMAP215 homologues in rice and other plants.
A PLANT-SPECIFIC MAP FAMILY: THE MAP70 PROTEINS
The aforementioned penta-peptide KLEEK can also be found in a family of plant-specific MAPs known as MAP70s (Korolev et al., 2007). However, this penta-peptide does not fall in the determined MT-binding site of AtMAP70 (Korolev et al., 2005). It is possible that the N-terminal regions of AtMAP70s may constitute another MT-binding pocket which cannot be simply revealed by a truncated fusion. It has been shown by both overexpression and RNA interference (RNAi)-mediated gene silencing that the most abundantly expressed isoform AtMAP70-5 plays a role in anisotropic cell expansion in stems (Korolev et al., 2007).
While there are five MAP70 isoforms in A. thaliana, four were detected in rice by sequence comparison (Korolev et al., 2005). Among them, proteins encoded by LOC_Os02g50320/Os02g0736100, LOC_Os06g14080/Os06g0251700 and LOC_Os12g44340/Os12g0640900 are closely related to each other and to AtMAP70-1–4. However, the fourth isoform, encoded by LOC_Os03g11650/Os03g0215700, is more divergent.
WHEN AUXIN MEETS MTS: AIR9
AIR9, previously known as an auxin-induced gene in root cultures, was rediscovered as a gene encoding a 187 kDa MAP (Buschmann et al., 2006). Its N-terminal basic and serine-rich domain forms an MT-binding site. Besides decorating cortical MTs, the PPB and the phragmoplast when AIR9 was expressed in a GFP fusion, the fusion protein was inserted at the cortical site previously occupied by the PPB during cytokinesis followed by an inward expansion (Buschmann et al., 2006).
A single rice gene LOC_Os07g05470/Os07g0148800 encodes an AIR9-homologous protein which shares 70 % amino acid sequence identity only within the A9 repeats. However, this predicted rice protein lacks the short basic and serine-rich region, and the following leucine-rich repeats found toward the N-terminus of AIR9. Therefore, it is questionable whether this much smaller rice protein indeed acts as a MAP, and whether it plays an essential role in plant cell division and development.
ANOTHER PLANT-SPECIFIC MAP FAMILY: TORTIFORLIA 1 (TOR1)/SPIRAL 2 (SPR2) AND RELATED PROTEINS
Two independent quests for genes whose mutations were responsible for the helical growth phenotype resulted in the identification of the TOR1/SPR2 protein as another plant-specific MAP of 864 amino acids (Buschmann et al., 2004; Shoji et al., 2004). TOR1/SPR2 and five homologous proteins in A. thaliana contain multiple HEAT repeats. Three lines of evidence support TOR1/SPR2 being an authentic MAP (Buschmann et al., 2004; Shoji et al., 2004). First, GFP fusion proteins decorate MT arrays during mitosis when ectopically expressed. Secondly, bacterially expressed and purified proteins co-sediment with MTs in vitro. Thirdly, a tobacco TOR1/SPR2 homologue was detected in the MAP fraction from BY-2 cells (Shoji et al., 2004). The tor1 and spr2 mutations cause cortical MTs to form left-handed helices, leading to defects in directional cell elongation and right-handed helical growth in longitudinally expanding organs (Buschmann et al., 2004; Shoji et al., 2004).
The closest rice TOR1/SPR2 homologue is encoded by the LOC_Os07g33630/Os07g0520400 and LOC_Os09g38710/Os09g0560000 loci, with >36 % amino acid sequence identity. Two other genes, LOC_Os02g50640/Os02g0739900 and LOC_Os06g13600/Os06g0244700, encode proteins with 27 and 26 % identity to TOR1/SPR2, respectively. Remaining questions include how the endogenous protein acts on cortical MTs to regulate their organization and underlying cellulose microfibril deposition in rice.
MT PLUS END-TRACKING PROTEINS: +TIPS
Among proteins decorating MTs, the most spectacular ones are those which specifically accumulate at the plus end of growing MTs. Collectively, they are named as MT plus end-tracking proteins, commonly known as +TIPs. When labelled with fluorescent proteins such as GFP, +TIPs often exhibit a comet-like dynamic localization pattern. The +TIPs include diverse proteins with no sequence homology to each other, and range from proteins as small as 30 kDa to as large as the cytoplamic dynein complex of >1000 kDa (Akhmanova and Hoogenraad, 2005). Besides dynein, several MT motor kinesins have also been determined to be +TIPs (Wu et al., 2006). The tip-tracking mechanisms include treadmilling for some +TIPs and surfing for others (Asbury, 2008).
Two classes of plant +TIPs, EB1 proteins and SPIRAL 1 (SPR1) proteins, are discussed here.
EB1
As the founding member of +TIPs, EB1 (end-binding protein 1) was discovered as a binding partner of the tumour suppressor protein APC (adenomatous polyposis coli) (Su et al., 1995). EB1 is present among all examined eukaryotic organisms, and acts as the heart for mediating other +TIPs to interact with MT plus ends (Lansbergen and Akhmanova, 2006). This small protein of approx. 300 amino acids contains a CH (calponin homology) domain at the N-terminus required for MT binding and an EB1 signature motif toward the C-terminus. In animal cells, the function of EB1 for promoting MT polymerization and stability is absolutely essential for MT organization during interphase and for spindle assembly (Rogers et al., 2002).
In A. thaliana, three EB1 isoforms in two classes have been identified (Chan et al., 2003; Mathur et al., 2003; Dixit et al., 2006; Bisgrove et al., 2008). AtEB1a and AtEB1b are 78 % identical to each other, and 49 % identical to AtEB1c (Bisgrove et al., 2008). The endogenous AtEB1 proteins preferentially decorate MT plus ends in various MT arrays during mitosis, and GFP fusions of AtEB1a and AtEB1b clearly reveal comet-like localization (Chan et al., 2003; Mathur et al., 2003; Dixit et al., 2006; Bisgrove et al., 2008). AtEB1a–GFP also appears at the MT-nucleating sites in spindles and phragmoplasts (Chan et al., 2005). AtEB1c resides in the nucleus during interphase, and associates with spindle and phragmoplast MTs, preferentially towards the plus end, during mitosis (Dixit et al., 2006; Bisgrove et al., 2008). A T-DNA insertional mutation at the EB1c locus confers hypersensitivity to the MT-poisoning agent oryzalin, and the sensitivity is enhanced when additional EB1 genes are compromised, indicating some overlapping functions among isoforms (Bisgrove et al., 2008). Although the AtEB1 proteins play a role in cellular activities required for responses to touch and gravity, they do not seem to be essential, which is different from their animal counterparts. If plant EB1 plays a role in mitosis, such a role is probably redundantly covered by other proteins acting on MTs.
Only two rice genes are found to encode EB1: LOC_Os04g54940/Os04g0642100 (OsEB1a) and LOC_Os10g35580/Os10g0498900 (OsEB1b). OsEB1a and AtEB1a are closely related, and have 65 % sequence identity; while OsEB1b and AtEB1c are closely related, and have 59 % identity. Thus, rice produces one EB1 protein for each class. However, it is unclear how the rice EB1 proteins behave in living cells.
SPR1
In analysing A. thaliana mutants exhibiting altered cell expansion resulting in twisted growth as described above for spr2, two independent efforts were converged at the SPR1 gene encoding a 119-amino acid protein (Nakajima et al., 2004; Sedbrook et al., 2004). The reports also concluded that endogenous as well as a functional SPR1–GFP fusion protein localized to cortical MTs and mitotic arrays. SPR1–GFP often appeared in a comet-like pattern, reminiscent of a +TIP (Sedbrook et al., 2004). Five other proteins are encoded by the SPR1-like SP1L1–5 genes, and their more conserved N- and C-terminal regions are connected by less conserved central regions (Nakajima et al., 2006). The MT association is conferred by the combination of both conserved regions (Nakajima et al., 2004). The twisted growth pattern caused by the spr1 mutation can be greatly enhanced by mutations in SP1L genes, indicating redundant functions among these homologous proteins (Nakajima et al., 2006).
An intriguing sequence pattern among SPR1 proteins is the presence of two almost identical peptide stretches at the N- and the C-terminus, ‘GGGQSSLGYLF’ and ‘GGGSSLDYLF’, respectively. Such a feature is repeated among four small proteins of 102–128 amino acids encoded by the LOC_Os03g30430/Os03g0417800, LOC_Os04g48870 (not annotated by RAP), LOC_Os11g41150/Os11g0629400 and LOC_Os12g31780/Os12g0502000 loci in the rice genome. These proteins probably play a constitutive role in regulating MT dynamics in rice based on the conserved sequence. Further genetic studies would allow a conclusion to be drawn as to whether the loss of rice SPR1-like proteins could cause a twisted growth phenotype.
The CLASP homologue: not a CLIP170-binding protein
Another often talked about +TIP in animal cell is CLASP (CLIP170-associated protein). CLIP170 (cytoplasmic linker protein 170) is a MAP which plays a role in cell migration and axon guidance (Galjart, 2005). CLASP proteins share limited similarity with XMAP215/TOG by having multiple HEAT repeats and an N-terminal TOG domain. The A. thaliana genome contains the single At2g20190 gene encoding a CLASP homologue (Ambrose et al., 2007; Kirik et al., 2007). Although AtCLASP appears preferentially toward the MT plus end, its localization pattern is clearly different from that of EB1 or SPR1 as AtCLASP also decorates cortical MTs in a punctate manner besides a comet-like appearance (Kirik et al., 2007). A mitotic function of AtCLASP is expected as: (a) AtCLASP is associated with the PPB and the phragmoplast; and (b) T-DNA insertions at the corresponding locus cause drastic reductions in the mitotic index and mitotic zone in the root (Ambrose et al., 2007; Kirik et al., 2007). Consequently, a significant reduction of overall vertical growth is demonstrated by the mutant plants, which also exhibit cell swelling when anisotropic expansion is required.
Surprisingly, the rice genome contains three genes encoding proteins highly homologous to AtCLASP: LOC_Os04g42840/Os04g0507500, LOC_Os02g40430/Os02g0617300 and LOC_Os02g57120/Os02g0816300. The outstanding questions about these rice CLASPs are whether they are authentic MAPs, whether they interact with EB1 as their animal counterparts and whether they are involved in responding to internal and external cues to modulate MT behaviour.
Because there is no CLIP170 homologue found in plants according to sequence homology, an obvious question is what partner(s) the plant CLASP may have, if any.
TANGLED 1: A CELL DIVISION SITE MARKER
Four decades ago, the PPB was discovered in wheat as an MT-based band present at the cell cortex prior to nuclear envelope breakdown, which forecasts the future division plane (Pickett-Heaps and Northcote, 1966a, b). Since then, this remarkable landmark has been inspiring plant cell biologists to search for the footprint left by the PPB at the cortical site. Besides the aforementioned AIR9 protein, the TANGLED (TAN) protein has also generated great enthusiasm as an indicator marking the site once occupied by the PPB (Walker et al., 2007). An earlier study indicated that the maize TAN protein acted as a MAP in vitro (Smith et al., 2001). A fluorescent fusion protein of AtTAN decorated the PPB, and remained at the cell division site as discontinuous dots encircling the cell cortex throughout the rest of mitosis (Walker et al., 2007). The rice genome contains a single locus encoding an apparent TAN orthologue: LOC_Os02g26140/Os02g0459300. The hypothetically deduced polypeptide of 415 amino acids has 68 and 42 % sequence identities with the maize and arabidopsis orthologues, respectively. How TAN interacts with MTs and influences cell division plane determination remains to be determined.
TON1 AND TON2: PROTEINS REQUIRED FOR PPB FORMATION
Earlier genetic screens recovered ton and fass mutants bearing a phenotype of an extremely compressed apical–basal axis reflected by a maximum plant height of a few centimetres (Traas et al., 1995; McClinton and Sung, 1997). When mutant cells undergo mitosis, MTs fail to be organized into the PPB array, but the spindle and phragmoplast arrays can still be formed. The TON1 locus contains two tandem genes encoding redundant TON1a and TON1b with 85 % sequence identity, which decorate cortical MTs and PPB (Azimzadeh et al., 2008). TON1 is related the animal centrosomal protein FOP, and interacts with the calcium-binding protein centrin. It was proposed that TON1 is an MT-organizing factor at the cell cortex, reminiscent of FOP at the centrosome (Azimzadeh et al., 2008). The loci of LOC_Os11g01170/Os11g0102600 and LOC_Os12g01170/Os12g0102200 encode identical TON1 proteins in rice.
The TON2/FASS protein is a member of the B′′ family of protein phosphatase 2A (PP2A) regulatory sub-units, and interacts with the AtAα regulatory sub-unit (McClinton and Sung, 1997; Camilleri et al., 2002). The involvement of PP2A suggests that the phosphorylation status of cytoskeletal proteins at the cell cortex is critical for MT organization into the PPB array. The rice genome contains a single TON2 gene LOC_Os05g05710/Os05g0149800. It would be rather interesting to identify the substrate of PP2A at the cell cortex at early stages of mitosis.
MT-SEVERING AND -DESTABILIZING FACTORS: KATANIN AND BEYOND
In animal cells, the rapid shortening of cytoplasmic MTs when cells enter mitosis is brought about by the katanin complex as an MT-severing ATPase (McNally and Vale, 1993). The complex contains a 60 kDa catalytic sub-unit and an 80 kDa accessory sub-unit. The plant homologue of the katanin catalytic sub-unit, AtKTN1/KSS1 (katanin or katanin small sub-unit), was first identified as a component responsible for ordered deposition of cellulose microfibrils, while it was also detected by direct sequence comparison (Burk et al., 2001; McClinton et al., 2001). Its MT-severing activity has been verified in vitro and in vivo (Burk and Ye, 2002; Stoppin-Mellet et al., 2002, 2006). Surprisingly, overexpression of the native AtKTN1 protein randomized cortical MTs but did not fragment them, and consequently altered the deposition of cellulose microfibrils (Burk et al., 2007). Other genetic analyses on various aspects of plant development have agreed that katanin plays a critical role in regulating the organization of cortical MTs, which is essential for anisotropic cell expansion in both arabidopsis and rice (Bichet et al., 2001; Webb et al., 2002; Bouquin et al., 2003; Komorisono et al., 2005). It has also been found that the loss of katanin caused upregulation of the expression of gibberellin (GA) biosynthesis genes in arabidopsis and rice (Bouquin et al., 2003; Komorisono et al., 2005). Because GA plays a critical role in regulating plant height, this phenomenon of altered gene expression could be interpreted as a feedback regulation resulting from the dwarf phenotype exhibited by the katanin mutants. The OsKSS1 encoded by the SGL1 gene is located at LOC_Os01g49000/Os01g0683100.
The 80 kDa accessory sub-unit (KLS for katanin large sub-unit) contains a WD40 domain, and plays a role in targeting the catalytic sub-unit to the centrosome in animal cells (McNally et al., 2000). There are at least four p80 homologues encoded by the A. thaliana genome, but their physiological roles remain to be characterized (Bouquin et al., 2003). Based on the presence of the N-terminal WD40 repeats and the C-terminal AtKTN1/KSS1-interacting domain, three loci in the rice genome encode homologues of this sub-unit: LOC_Os04g58130/Os04g0677700, LOC_Os01g57210/Os01g0780400 and LOC_Os10g35200/Os10g0494800.
Besides katanin, a plant-specific 18 kDa protein known as MAP18 has been documented as a destabilizing factor on cortical MTs (Wang et al., 2007b). This protein may be another regulator of the dynamic MT network at the cell cortex during rapid cell expansion in A. thaliana. However, there is no obvious homologue of MAP18 in rice. It remains unclear whether other monocot species have MAP18 homologues.
In animal cells, members of the Kinesin-13 sub-family are critical MT depolymerases regulating dynamics of MT ends (Howard and Hyman, 2007). However, similar functions have not been observed in plant cells (Lu et al., 2005).
EMERGING NOVEL PLANT MAPS: PLD/P90, MAP190 AND RIC1
A few novel MAPs have been isolated by various approaches. They have been implicated in regulating MT organization at various stages of cell growth.
PLDδ
In a quest for MAPs that were associated with the plasma membrane in tobacco BY-2 cells, a 90 kDa protein was isolated and turned out to be the phospholipase PLDδ (Marc et al., 1996; Gardiner et al., 2001). Endogenous PLDδ indeed decorates cortical MTs and MT arrays during mitosis in tobacco cells. In the rice genome, there are at least 13 genes encoding forms of PLDα, PLDβ, PLDγ and PLDδ. Among them, LOC_Os09g37100/Os09g0543100 encodes PLDδ.
MAP190
MAP190 was isolated from BY-2 cells as a putative MAP which also interacted with actin microfilaments in vitro (Igarashi et al., 2000). The protein contains a calcium-binding EF-hand motif toward the C-terminus. Besides decorating mitotic MT arrays, it also appeared in the nucleus. The rice LOC_Os06g36710/Os06g0562700 locus encodes a protein related to tobacco MAP190.
RIC1
RIC (ROP-interactive CRIB motif-containing protein) proteins contain the CRIB (Cdc42/Rac-interactive binding) motif, and specifically interact with the GTP-bound form of the small GTPase ROP (Wu et al., 2001). Among RICs identified in A. thaliana, RIC1 is associated with cortical MTs in leaf pavement cells, and acts as an MT-stabilizing factor (Fu et al., 2005). While in rice there are a number of genes encoding proteins containing the CRIB motif, none of them seems to be an apparent RIC1 orthologue. Because RIC1 plays a critical role in morphogenesis of leaf epidermal cells in A. thaliana, the absence of a rice orthologue may suggest that RIC1 probably is associated with a signalling route specific to dicots. Specific signalling routes may be part of the molecular mechanisms specifying different appearances in leaf epidermal cells in dicots and monocots.
THE KINESIN SUPERFAMILY
Among all cytoskeletal protein families, kinesins constitute the largest family, especially in flowering plants (Lee and Liu, 2004). Kinesins belong to one of the three classes of cytoskeleton-based motor proteins, and use the energy of ATP hydrolysis to move along MT tracks. Members of the kinesin superfamily contain a highly conserved approx. 350 amino acid catalytic core bearing ATPase activity and a nucleotide-dependent MT-binding domain. Kinesins often exhibit a tripartite structure, with the motor domain of the catalytic core plus the neck generating directional forces, a coiled-coil stalk for dimerization, and diverse tail domains for recognizing their cargoes. The ‘neck’ domain, a 34 amino acid peptide for plus end-directed kinesins or a 14 amino acid peptide for minus end-directed kinesins, determines the directionality of the motors (Endow, 1999). Kinesins are classified according to the sequence homology in their motor domains, and have 14 designated sub-families and some orphans (Lawrence et al., 2004). Except for members of Kinesin-5 and a few members of Kinesin-14 which have conserved non-motor sequences and mitotic functions, most plant kinesins are distinctly divergent from their animal and fungal counterparts when their non-motor domains are examined (Lee and Liu, 2007). Based on earlier drafts of the rice genomes of indica and japonica, >40 genes have been shown to encode kinesins in rice (Richardson et al., 2006). Rice kinesins distribute across most of the 14 sub-families, except for Kinesin-2, -3, -9 and -11 (Richardson et al., 2006). According to the genome annotation of the cultivar japonica, there are at least 52 kinesins in rice (Table 3). While a few kinesins have been experimentally investigated in A. thaliana and other dicot species such as tobacco and cotton, very limited information has been obtained about rice kinesins.
Table 3.
List of kinesin family proteins in rice
| RGAP | RAP | Sub-family | Known as |
|---|---|---|---|
| LOC_Os01g14090 | Os01g0243100 | 14 | |
| LOC_Os01g15540 | Os01g0260100 | 14 | |
| LOC_Os01g33040 | Os01g0513900 | 7 | OsNACK1 |
| LOC_Os01g42070 | Os01g0605500 | 8 | |
| LOC_Os01g43580 | Os01g0625200 | 13 | |
| LOC_Os01g54080 | Os01g0744000 | 14 | |
| LOC_Os02g01180 | Os02g0101800 | 6 | |
| LOC_Os02g13570 | Os02g0229500+ | ||
| LOC_Os02g13580 | Os02g0229600 | 14 | |
| LOC_Os02g28850 | Os02g0489800 | 12 | Kinesin-12B |
| LOC_Os02g43050 | Os02g0644400 | 7 | |
| LOC_Os02g43130 | Os02g0645100 | 7 | |
| LOC_Os02g50910 | Os02g0742800 | 4 | |
| LOC_Os02g53520 | Os02g0775400 | 7 | K16 |
| LOC_Os02g56540 | Os02g0810200 | UG | PAKRP2 |
| LOC_Os03g02290 | Os03g0114000 | 14 | |
| LOC_Os03g05820 | Os03g0152900 | UG | |
| LOC_Os03g17164 | Not annotated | 5 | |
| LOC_Os03g18980 | Os03g0301800 | 14 | |
| LOC_Os03g39020 | Os03g0587200 | 12 | |
| LOC_Os03g53920 | Os03g0750200 | 12 | |
| LOC_Os03g56260 | Os03g0773600 | 8 | |
| LOC_Os03g64415 | Os03g0862200 | 14 | |
| LOC_Os04g28260 | Os04g0350300 | 12 | PAKRP1, Kinesin-12A |
| LOC_Os04g30720 | Os04g0375900 | 10 | |
| LOC_Os04g45580 | Os04g0538800 | 7 | |
| LOC_Os04g53760 | Os04g0629700 | 14 | |
| LOC_Os04g57140 | Os04g0666900 | 14 | KCBP |
| Not annotated | Not annotated | 14 | OsKCH1 |
| LOC_Os05g02670 | Os05g0117798 | 5 | |
| LOC_Os05g06280 | Os05g0154700 | 13 | Kinesin-13A |
| LOC_Os05g33030 | Os05g0397900 | 14 | |
| LOC_Os05g38480 | Os05g0459400 | 10 | |
| LOC_Os05g44560 | Os05g0521300 | 14 | |
| LOC_Os06g04560 | Os06g0137100 | UG | |
| LOC_Os06g11380 | Os06g0217600 | 14 | |
| LOC_Os06g36080 | Os06g0554700 | 14 | |
| LOC_Os07g01490 | Os07g0105700 | 14 | |
| LOC_Os07g44400 | Os07g0638000 | 12 | |
| LOC_Os08g02380 | Os08g0117000 | 1 | |
| LOC_Os08g43400 | Os08g0547500 | 7 | |
| LOC_Os08g44420 | Os08g0558400 | 5 | |
| LOC_Os09g02650 | Os09g0114500 | 4 | |
| LOC_Os09g25380 | Os09g0421200 | 7 | |
| LOC_Os09g35890 | Os09g0528000 | 7 | |
| LOC_Os10g36880 | Os10g0512800 | 7 | |
| LOC_Os11g35090 | Os11g0552600 | 7 | |
| LOC_Os11g37140 | Os11g0581000 | 12 | |
| LOC_Os11g42800 | Os11g0648100 | 14 | |
| LOC_Os11g44880 | Os11g0672400 | 14 | |
| LOC_Os12g36100 | Os12g0547500 | 14 | |
| LOC_Os12g39980 | Os12g0590500 | 12 | |
| LOC_Os12g42160 | Os12g0616000 | 14 |
IDs of these proteins were assigned by RGAP and RAP, respectively. Documented members are listed with their known names. Note that one of the Kinesin-14 members has been annotated into two loci by RGAP and RAP, and OsKCH1 on chromosome IV was not annotated by both. Orphan kinesins were noted as UG for ungrouped.
A previous sequence analysis has pinpointed distinct domains found among the rice kinesins, suggesting their distinct functions (Richardson et al., 2006). Here some interesting features of rice kinesins in three expanded sub-families are summarized. Others are listed in Table 3.
Kinesin-7
The founding member of this sub-family is CENP-E, a kinesin which appears at the kinetochore region during mitosis in mammals, and interacts with mitotic checkpoint proteins for its role in mitotic progression (Yen et al., 1992; Vos et al., 2006). Kinesin-7 is expanded in flowering plants. There are at least ten members in rice (Table 3). Among them, two have been studied by genetic and biochemical means.
The dwarf bamboo shoot 1 (dbs1) mutation at the OsNACK1 (LOC_Os01g33040/Os01g0513900) locus encoding a Kinesin-7 member causes severe dwarfism in rice (Sazuka et al., 2005). OsNACK1 is an apparent orthologue of NACK1 in arabidopsis and tobacco. NACK1 governs the localization of a mitogen-activated protein kinase cascade to the phragmoplast midzone, and plays a critical role in MT turnover in the phragmoplast (Nishihama et al., 2002; Sasabe and Machida, 2006). The dbs1 mutant exhibits cell wall stubs in rapidly dividing cells, reflecting defects in cytokinesis, a phenotype similar to that in A. thaliana (Nishihama et al., 2002; Strompen et al., 2002; Sazuka et al., 2005).
The kinesin K16 encoded by the LOC_Os02g53520/Os02g0775400 locus, another rice Kinesin-7 member, has been studied biochemically for its motor activity compared with that of a conventional kinesin from brain (Umeki et al., 2006). It exhibits a weaker interaction with MTs and a lower affinity for nucleotides. How this motor operates in vivo is unclear.
Kinesin-12
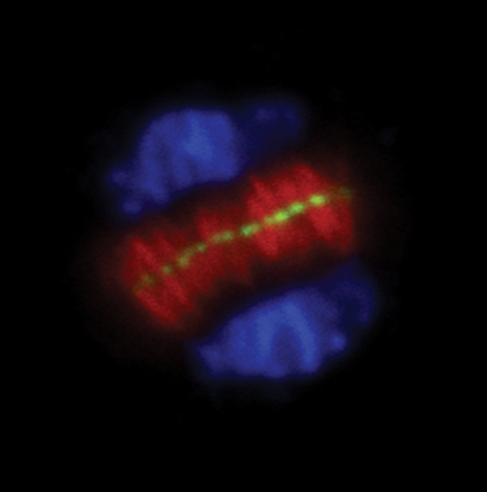
While there are six members of the Kinesin-12 sub-family in A. thaliana, seven have been identified in rice (Table 3). According to the neck sequences present in these proteins, they are most probably plus end-directed motors. OsKinesin-12A/PAKRP1 encoded by the LOC_Os04g28260/Os04g0350300 locus was detected in rapidly dividing cells, and exclusively appeared at the plus end of phragmoplast MTs (Fig. 3). Three closely related kinesins are encoded by the rice genome at the loci of LOC_Os02g28850/Os02g0489800, LOC_Os03g39020/Os03g0587200 and LOC_Os11g37140/Os11g0581000. Their apparent orthologues in A. thaliana exhibit redundant functions as only the absence of both AtKinesin-12A and AtKinesin-12B leads to the loss of the bipolar organization pattern of phragmoplast MTs in developing pollen grains (Pan et al., 2004; Lee et al., 2007). Similar functional redundancy is expected for the rice orthologues.
Fig. 3.
Immunolocalization of OsKinesin-12A in the phragmoplast. OsKinesin-12A was labelled with an antibody raised against a truncated version of the protein, MTs by an anti-α-tubulin antibody, and DNA by DAPI (4′,6-diamidino-2-phenylindole). OsKinesin-12A was pseudocolored in green, MTs in red, and DNA in blue. OsKinesin-12A exclusively decorated the plus end of phragmoplast MTs.
Two members of the Kinesin-12 sub-family have been implicated in orienting the phragmoplast during cytokinesis in arabidopsis, and thus were named POKs (phragmoplast orienting kinesins) (Müller et al., 2006). Similarly, two apparent POK orthologues are encoded by the LOC_Os07g44400/Os07g0638000 and LOC_Os12g39980/Os12g0590500 loci.
An additional Kinesin-12 member is encoded by the LOC_Os03g53920/Os03g0750200 locus, which is most closely related to the kinesin encoded by At3g44050 in A. thaliana. Neither has been reported in the literature.
Kinesin-14
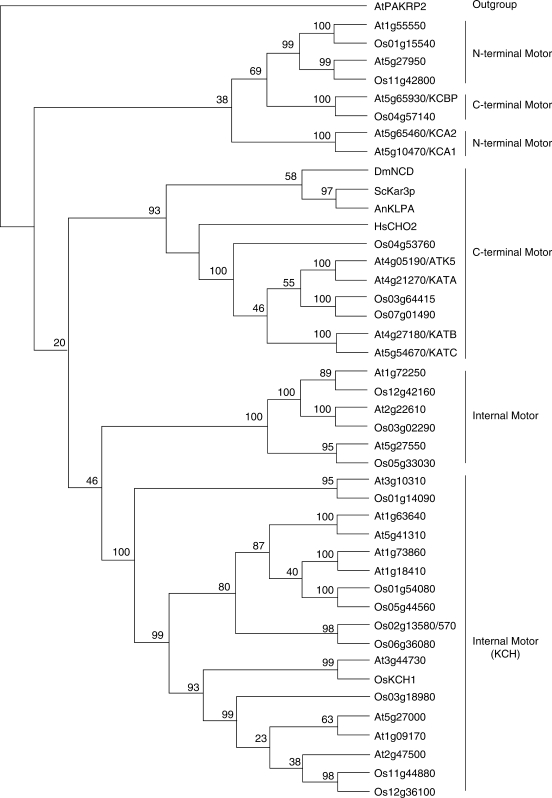
All Kinesin-14 members are minus end-directed motors as they all contain the conserved neck domain determining such directionality. This sub-family has been expanded the most compared with other sub-families of plant kinesins. This may reflect the fact that flowering plants do not make the minus end-directed motor dynein. Several distinct sub-groups can be distinguished within this subfamily (Fig. 4). Among them, apparent orthologues of the yeast Kar3p decorate the MT-overlapping region in mitotic spindles and the phragmoplast, and play an evolutionarily conserved role in organizing MTs in spindles and the phragmoplast during mitosis and meiosis (Liu et al., 1996; Chen et al., 2002; Marcus et al., 2003; Ambrose et al., 2005). Four Kar3p-like kinesins, namely KATA/ATK1, KATB, KATC and ATK5, are made in A. thaliana. The rice genome has only two genes, LOC_Os04g53760/Os04g0629700 and LOC_Os07g01490/Os07g0105700, encoding proteins which resemble these four kinesins. These two rice kinesins are more closely related to KATA/ATK1 and its paralogue ATK5 than to the KATB and KATC paralogues. Interestingly, rice does not have apparent orthologues of KATB and KATC.
Fig. 4.
Neighbor–joining tree of members of the Kinesin-14 subfamily. Phylogenetic relationships among Kinesin-14 members of rice (Os), A. thaliana (At), and representatives from the budding yeast S. cerevisiae (ScKar3p), the filamentous fungus Aspergillus nidulans (AnKLPA), the fly Drosophila melanogaster (DmNCD) and human (HsCHO2) are presented. AtPAKRP2 was used as an outgroup kinesin. For rice proteins, shown in this figure were IDs annotated by RGAP omitting LOC_. Documented plant kinesins are listed with their abbreviated names following the locus calls. Among Kinesin-14 members, those having their motor domains located in the N-terminus, the C-terminus, and the middle are highlighted as N-terminal motor, C-terminal motor, internal motor, and KCHs as specialized internal motor kinesins. Bootstrap values at the branches represent the percentages obtained in 1000 replications.
A calmodulin-binding kinesin has been added to the inventory of Kinesin-14 in photosynthetic organisms (Reddy et al., 1996; Richardson et al., 2006). This so-called KCBP contains a Ca2+/calmodulin-binding peptide toward its very C-terminus, and an extended coiled-coil domain between the N-terminal nucleotide-independent MT-binding domain and the motor domain. As in A. thaliana, a single KCBP is encoded by the LOC_Os04g57140/Os04g0666900 locus in the rice genome. Detailed studies in A. thaliana and other organisms indicate that KCBP functions in MT organization/stability in interphase and mitotic cells, and consequently in cell morphogenesis (Oppenheimer et al., 1997; Reddy and Day, 2000; Vos et al., 2000; Preuss et al., 2003).
Minus end-directed kinesins in fungi and animals in the Kinesin-14 sub-family typically have the motor domain located toward their C-termini. One surprise is that the plant Kinesin-14 sub-family contains members having their motor domains located toward the N-terminus or in the middle (Day et al., 2000). Among those with N-terminal motors, arabidopsis At1g55550 and At5g27950 have apparent rice orthologues of LOC_Os01g15540/Os01g0260100 and LOC_Os11g42800/Os11g0648100, respectively. Overexpression of the tobacco orthologue of At5g27950 causes cortical MTs to be converted into a radial array emanating from a single perinuclear focus (Goto and Asada, 2007). Unfortunately, in vivo functions of these kinesins are unknown. Two other closely related arabidopsis N-terminal motor Kinesin-14s are KCA1 and KCA2. As targets of Cdk, they appear close to the plasma membrane, demarcating the division site in a cell cycle-dependent manner (Vanstraelen et al., 2004, 2006). However, rice does not have an apparent KCA1/KCA2 orthologue.
Another unique feature is that a plant-specific group of Kinesin-14 distinguish themselves from other members of Kinesin-14 for having a CH domain towards the N-terminus of the polypeptide. These distinctive kinesins have been named KCH, for kinesin with CH domain, which confers binding to F-actin in vitro (Preuss et al., 2004). The motor domain is located in the middle of the KCH kinesins. At least eight rice genes encode KCHs. Because of its unique actin-binding capability, they could probably be the long sought after dynamic linkers between MTs and F-actin in flowering plants. Unfortunately, this potential function for the founding member KATD and other KCHs has not been tested by genetic means in A. thaliana (Tamura et al., 1999). It is foreseeable that arrays such as the PPB and the phragmoplast, where MTs closely interact with F-actin in a cell cycle-dependent manner, would need KCHs for reorganization of F-actin in response to rapid remodelling of MTs.
PERSPECTIVES
Very little has been learnt about the MT cytoskeleton in rice. The completed rice genome has presented an inventory of MT-interacting proteins whose functions are largely unknown. Functional characterization of these proteins will reveal MT-based molecular mechanisms that regulate critical cellular events that may be common to plants or unique to monocots. Rapidly growing resources such as the pools of T-DNA and transposon insertion mutants, and ample collections of cDNA clones are making rice an unmatchable model crop for studies aimed at improving Graminaceous crops (Jung et al., 2008). The recent introduction of another monocot model, Brachypodium distachyon, will help make great leaps toward understanding the growth and form of monocots (Opanowicz et al., 2008). By writing this review, we would like to invite colleagues to investigate the rice MT system for its role in growth and development in this remarkable crop, and to extend the studies to other monocot species such as B. distachyon.
ACKNOWLEDGEMENTS
Our investigation of the MT cytoskeleton has been graciously supported by grants from the Chemical Sciences, Geosciences and Biosciences Division, Office of Science, US Department of Energy (contract #: DE-FG02-04ER15554), and the National Research Initiative (NRI) of the US Department of Agriculture (grant #: 2005-35304-16031).
LITERATURE CITED
- Akhmanova A, Hoogenraad CC. Microtubule plus-end-tracking proteins: mechanisms and functions. Current Opinion in Cell Biology. 2005;17:47–54. doi: 10.1016/j.ceb.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Ambrose JC, Li W, Marcus A, Ma H, Cyr R. A minus-end-directed kinesin with plus-end tracking protein activity is involved in spindle morphogenesis. Molecular Biology of the Cell. 2005;16:1584–1592. doi: 10.1091/mbc.E04-10-0935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose JC, Shoji T, Kotzer AM, Pighin JA, Wasteneys GO. The Arabidopsis CLASP gene encodes a microtubule-associated protein involved in cell expansion and division. The Plant Cell. 2007;19:2763–2775. doi: 10.1105/tpc.107.053777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asbury CL. XMAP215: a tip tracker that really moves. Cell. 2008;132:19–20. doi: 10.1016/j.cell.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimzadeh J, Nacry P, Christodoulidou A, et al. Arabidopsis TONNEAU1 proteins are essential for preprophase band formation and interact with centrin. The Plant Cell. 2008;20:2146–2159. doi: 10.1105/tpc.107.056812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin TI. The cytoskeleton. In: Buchanan BB, Gruissem W, Jones RL, editors. Biochemistry and molecular biology of plants. San Diego: American Society of Plant Biologists; 2000. pp. 202–258. [Google Scholar]
- Bichet A, Desnos T, Turner S, Grandjean O, Höfte H. BOTERO1 is required for normal orientation of cortical microtubules and anisotropic cell expansion in Arabidopsis. The Plant Journal. 2001;25:137–148. doi: 10.1046/j.1365-313x.2001.00946.x. [DOI] [PubMed] [Google Scholar]
- Binarova P, Cenklova V, Prochazkova J, et al. γ-Tubulin is essential for acentrosomal microtubule nucleation and coordination of late mitotic events in Arabidopsis. The Plant Cell. 2006;18:1199–1212. doi: 10.1105/tpc.105.038364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisgrove SR, Lee Y-RJ, Liu B, Peters N, Kropf DL. The microtubule plus-end binding protein EB1 functions in root responses to touch and gravity signals in Arabidopsis. The Plant Cell. 2008;20:396–410. doi: 10.1105/tpc.107.056846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouquin T, Mattsson O, Naested H, Foster R, Mundy J. The Arabidopsis lue1 mutant defines a katanin p60 ortholog involved in hormonal control of microtubule orientation during cell growth. Journal of Cell Science. 2003;116:791–801. doi: 10.1242/jcs.00274. [DOI] [PubMed] [Google Scholar]
- Burk D, Ye Z. Alteration of oriented deposition of cellulose microfibrils by mutation of a katanin-like microtubule-severing protein. The Plant Cell. 2002;14:2145–2160. doi: 10.1105/tpc.003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burk DH, Liu B, Zhong R, Morrison WH, Ye Z-H. A katanin-like protein regulates normal cell wall biosynthesis and cell elongation. The Plant Cell. 2001;13:807–827. [PMC free article] [PubMed] [Google Scholar]
- Burk DH, Zhong R, Ye ZH. The katanin microtubule severing protein in plants. Journal of Integrated Plant Biology. 2007;49:1174–1182. [Google Scholar]
- Buschmann H, Chan J, Sanchez-Pulido L, Andrade-Navarro MA, Doonan JH, Lloyd CW. Microtubule-associated AIR9 recognizes the cortical division site at preprophase and cell-plate insertion. Current Biology. 2006;16:1938–1943. doi: 10.1016/j.cub.2006.08.028. [DOI] [PubMed] [Google Scholar]
- Buschmann H, Fabri CO, Hauptmann M, et al. Helical growth of the Arabidopsis mutant tortifolia1 reveals a plant-specific microtubule-associated protein. Current Biology. 2004;14:1515–1521. doi: 10.1016/j.cub.2004.08.033. [DOI] [PubMed] [Google Scholar]
- Caillaud M-C, Lecomte P, Jammes F, et al. MAP65-3 microtubule-associated protein is essential for nematode-induced giant cell ontogenesis in Arabidopsis. The Plant Cell. 2008;19:423–437. doi: 10.1105/tpc.107.057422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camilleri C, Azimzadeh J, Pastuglia M, Bellini C, Grandjean O, Bouchez D. The Arabidopsis TONNEAU2 gene encodes a putative novel protein phosphatase 2A regulatory subunit essential for the control of the cortical cytoskeleton. The Plant Cell. 2002;14:833–845. doi: 10.1105/tpc.010402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmena M, Earnshaw WC. The cellular geography of aurora kinases. Nature Reviews Molecular Cell Biology. 2003;4:842–854. doi: 10.1038/nrm1245. [DOI] [PubMed] [Google Scholar]
- Carpenter JL, Ploense SE, Snustad DP, Silflow CD. Preferential expression of an α-tubulin gene of Arabidopsis in pollen. The Plant Cell. 1992;4:557–571. doi: 10.1105/tpc.4.5.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J, Calder G, Fox S, Lloyd C. Localization of the microtubule end binding protein EB1 reveals alternative pathways of spindle development in Arabidopsis suspension cells. The Plant Cell. 2005;17:1737–1748. doi: 10.1105/tpc.105.032615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J, Calder GM, Doonan JH, Lloyd CW. EB1 reveals mobile microtubule nucleation sites in Arabidopsis. Nature Cell Biology. 2003;5:967–971. doi: 10.1038/ncb1057. [DOI] [PubMed] [Google Scholar]
- Chen C, Marcus A, Li W, et al. The Arabidopsis ATK1 gene is required for spindle morphogenesis in male meiosis. Development. 2002;129:2401–2409. doi: 10.1242/dev.129.10.2401. [DOI] [PubMed] [Google Scholar]
- Cheng Z, Snustad DP, Carter JV. Temporal and spatial expression patterns of TUB9, a β-tubulin gene of Arabidopsis thaliana. Plant Molecular Biology. 2001;47:389–398. doi: 10.1023/a:1011628024798. [DOI] [PubMed] [Google Scholar]
- Chuong SD, Good AG, Taylor GJ, Freeman MC, Moorhead GB, Muench DG. Large-scale identification of tubulin-binding proteins provides insight on subcellular trafficking, metabolic channeling, and signaling in plant cells. Molecular and Cellular Proteomics. 2004;3:970–983. doi: 10.1074/mcp.M400053-MCP200. [DOI] [PubMed] [Google Scholar]
- Cyr RJ, Palevitz BA. Microtubule-binding proteins from carrot.1. Initial characterization and microtubule bundling. Planta. 1989;177:245–260. doi: 10.1007/BF00392813. [DOI] [PubMed] [Google Scholar]
- Day IS, Miller C, Golovkin M, Reddy AS. Interaction of a kinesin-like calmodulin-binding protein with a protein kinase. Journal of Biological Chemistry. 2000;275:13737–45. doi: 10.1074/jbc.275.18.13737. [DOI] [PubMed] [Google Scholar]
- Demidov D, Van Damme D, Geelen D, Blattner FR, Houben A. Identification and dynamics of two classes of Aurora-like kinases in arabidopsis and other plants. The Plant Cell. 2005;17:836–848. doi: 10.1105/tpc.104.029710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit R, Chang E, Cyr R. Establishment of polarity during organization of the acentrosomal plant cortical microtubule array. Molecular Biology of the Cell. 2006;17:1298–1305. doi: 10.1091/mbc.E05-09-0864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eleftheriou EP, Baskin TI, Hepler PK. Aberrant cell plate formation in the Arabidopsis thaliana microtubule organization 1 mutant. Plant and Cell Physiology. 2005;46:671–675. doi: 10.1093/pcp/pci068. [DOI] [PubMed] [Google Scholar]
- Endow SA. Determinants of molecular motor directionality. Nature Cell Biology. 1999;1:E163–E167. doi: 10.1038/14113. [DOI] [PubMed] [Google Scholar]
- Erhardt M, Stoppin-Mellet V, Campagne S, et al. The plant Spc98p homologue colocalizes with γ-tubulin at microtubule nucleation sites and is required for microtubule nucleation. Journal of Cell Science. 2002;115:2423–2431. doi: 10.1242/jcs.115.11.2423. [DOI] [PubMed] [Google Scholar]
- Eun SO, Wick SM. Tubulin isoform usage in maize microtubules. Protoplasma. 1998;204:235–244. [Google Scholar]
- Fu Y, Gu Y, Zheng Z, Wasteneys G, Yang Z. Arabidopsis interdigitating cell growth requires two antagonistic pathways with opposing action on cell morphogenesis. Cell. 2005;120:687–700. doi: 10.1016/j.cell.2004.12.026. [DOI] [PubMed] [Google Scholar]
- Galjart N. CLIPs and CLASPs and cellular dynamics. Nature Reviews Molecular Cell Biology. 2005;6:487–498. doi: 10.1038/nrm1664. [DOI] [PubMed] [Google Scholar]
- Gard DL, Kirschner MW. A microtubule-associated protein from Xenopus eggs that specifically promotes assembly at the plus-end. Journal of Cell Biology. 1987;105:2203–2215. doi: 10.1083/jcb.105.5.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gard DL, Kirschner MW. Microtubule assembly in cytoplasmic extracts of Xenopus oocytes and eggs. Journal of Cell Biology. 1987;105:2191–2201. doi: 10.1083/jcb.105.5.2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gard DL, Becker BE, Josh Romney S. MAPping the eukaryotic tree of life: structure, function, and evolution of the MAP215/Dis1 family of microtubule-associated proteins. International Review of Cytology. 2004;239:179–272. doi: 10.1016/S0074-7696(04)39004-2. [DOI] [PubMed] [Google Scholar]
- Gardiner JC, Harper JDI, Weerakoon ND, et al. A 90-kD phospholipase D from tobacco binds to microtubules and the plasma membrane. The Plant Cell. 2001;13:2143–2158. doi: 10.1105/TPC.010114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giet R, McLean D, Descamps S, et al. Drosophila Aurora A kinase is required to localize D-TACC to centrosomes and to regulate astral microtubules. Journal of Cell Biology. 2002;156:437–451. doi: 10.1083/jcb.200108135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard RH, Villemur R, Silflow CD, Wick SM. Generation of chicken polyclonal antibodies against distinct maize isotubulins. Protoplasma. 1998;204:226–234. [Google Scholar]
- Goto Y, Asada T. Excessive expression of the plant kinesin TBK5 converts cortical and perinuclear microtubules into a radial array emanating from a single focus. Plant and Cell Physiology. 2007;48:753–761. doi: 10.1093/pcp/pcm045. [DOI] [PubMed] [Google Scholar]
- Granger CL, Cyr JL. Microtubule reorganization in tobacco BY-2 cells stably expressing GFP–MBD. Planta. 2000;210:502–509. doi: 10.1007/s004250050037. [DOI] [PubMed] [Google Scholar]
- Gunawardane RN, Martin OC, Cao K, et al. Characterization and reconstitution of Drosophila γ-tubulin ring complex subunits. Journal of Cell Biology. 2000;151:1513–1523. doi: 10.1083/jcb.151.7.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane RN, Martin OC, Zheng YX. Characterization of a new γTuRC subunit with WD repeats. Molecular Biology of the Cell. 2003;14:1017–1026. doi: 10.1091/mbc.E02-01-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada T. Microtubule-associated proteins in higher plants. Journal of Plant Research. 2007;120:79–98. doi: 10.1007/s10265-006-0057-9. [DOI] [PubMed] [Google Scholar]
- Haren L, Remy MH, Bazin I, Callebaut I, Wright M, Merdes A. NEDD1-dependent recruitment of the γ-tubulin ring complex to the centrosome is necessary for centriole duplication and spindle assembly. Journal of Cell Biology. 2006;172:505–515. doi: 10.1083/jcb.200510028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard J, Hyman AA. Microtubule polymerases and depolymerases. Current Opinion in Cell Biology. 2007;19:31–35. doi: 10.1016/j.ceb.2006.12.009. [DOI] [PubMed] [Google Scholar]
- Hussey PJ, Hawkins TJ. Plant microtubule-associated proteins: the HEAT is off in temperature-sensitive mor1. Trends in Plant Science. 2001;6:389–392. doi: 10.1016/s1360-1385(01)02090-8. [DOI] [PubMed] [Google Scholar]
- Hussey PJ, Hawkins TJ, Igarashi H, Kaloriti D, Smertenko A. The plant cytoskeleton: recent advances in the study of the plant microtubule-associated proteins MAP-65, MAP-190 and the Xenopus MAP215-like protein, MOR1. Plant Molecular Biology. 2002;50:915–924. doi: 10.1023/a:1021236307508. [DOI] [PubMed] [Google Scholar]
- Igarashi H, Orii H, Mori H, Shimmen T, Sonobe S. Isolation of a novel 190 kDa protein from tobacco BY-2 cells: possible involvement in the interaction between actin filaments and microtubules. Plant and Cell Physiology. 2000;41:920–931. doi: 10.1093/pcp/pcd015. [DOI] [PubMed] [Google Scholar]
- Jiang CJ, Sonobe S. Identification and preliminary characterization of a 65-kDa higher-plant microtubule-associated protein. Journal of Cell Science. 1993;105:891–901. doi: 10.1242/jcs.105.4.891. [DOI] [PubMed] [Google Scholar]
- Jiang W, Jimenez G, Wells NJ, et al. PRC1: a human mitotic spindle-associated CDK substrate protein required for cytokinesis. Molecular Cell. 1998;2:877–885. doi: 10.1016/s1097-2765(00)80302-0. [DOI] [PubMed] [Google Scholar]
- Jung KH, An G, Ronald PC. Towards a better bowl of rice: assigning function to tens of thousands of rice genes. Nature Reviews Genetics. 2008;9:91–101. doi: 10.1038/nrg2286. [DOI] [PubMed] [Google Scholar]
- Kaloriti D, Galva C, Parupalli C, Khalifa N, Galvin M, Sedbrook JC. Microtubule associated proteins in plants and the processes they manage. Journal of Integrated Plant Biology. 2007;49:1164–1173. [Google Scholar]
- Kang MS, Choi YJ, Kim MC, Lim CO, Hwang I, Cho MJ. Isolation and characterization of two β-tubulin cDNA clones from rice. Plant Molecular Biology. 1994;26:1975–1979. doi: 10.1007/BF00019507. [DOI] [PubMed] [Google Scholar]
- Kawabe A, Matsunaga S, Nakagawa K, et al. Characterization of plant Aurora kinases during mitosis. Plant Molecular Biology. 2005;58:1–13. doi: 10.1007/s11103-005-3454-x. [DOI] [PubMed] [Google Scholar]
- Kawamura E, Himmelspach R, Rashbrooke MC, et al. MICROTUBULE ORGANIZATION 1 regulates structure and function of microtubule arrays during mitosis and cytokinesis in the Arabidopsis root. Plant Physiology. 2006;140:102–114. doi: 10.1104/pp.105.069989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik V, Grini PE, Mathur J, et al. The Arabidopsis tubulin-folding cofactor A gene is involved in the control of the α/β-tubulin monomer balance. The Plant Cell. 2002;a 14:2265–2276. doi: 10.1105/tpc.003020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik V, Mathur J, Grini PE, et al. Functional analysis of the tubulin-folding cofactor C in Arabidopsis thaliana. Current Biology. 2002;b 12:1519–1523. doi: 10.1016/s0960-9822(02)01109-0. [DOI] [PubMed] [Google Scholar]
- Kirik V, Herrmann U, Parupalli C, Sedbrook JC, Ehrhardt DW, Hülskamp M. CLASP localizes in two discrete patterns on cortical microtubules and is required for cell morphogenesis and cell division in Arabidopsis. Journal of Cell Science. 2007;120:4416–4425. doi: 10.1242/jcs.024950. [DOI] [PubMed] [Google Scholar]
- Komorisono M, Ueguchi-Tanaka M, Aichi I, et al. Analysis of the rice mutant dwarf and gladius leaf 1. Aberrant katanin-mediated microtubule organization causes up-regulation of gibberellin biosynthetic genes independently of gibberellin signaling. Plant Physiology. 2005;138:1982–1993. doi: 10.1104/pp.105.062968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopczak SD, Haas NA, Hussey PJ, Silflow CD, Snustad DP. The small genome of Arabidopsis contains at least six expressed α-tubulin genes. The Plant Cell. 1992;4:539–547. doi: 10.1105/tpc.4.5.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolev AV, Chan J, Naldrett MJ, Doonan JH, Lloyd CW. Identification of a novel family of 70 kDa microtubule-associated proteins in Arabidopsis cells. The Plant Journal. 2005;42:547–555. doi: 10.1111/j.1365-313X.2005.02393.x. [DOI] [PubMed] [Google Scholar]
- Korolev AV, Buschmann H, Doonan JH, Lloyd CW. AtMAP70-5, a divergent member of the MAP70 family of microtubule-associated proteins, is required for anisotropic cell growth in Arabidopsis. Journal of Cell Science. 2007;120:2241–2247. doi: 10.1242/jcs.007393. [DOI] [PubMed] [Google Scholar]
- Kotwaliwale CV, Frei SB, Stern BM, Biggins S. A pathway containing the Ipl1/aurora protein kinase and the spindle midzone protein Ase1 regulates yeast spindle assembly. Developmental Cell. 2007;13:433–445. doi: 10.1016/j.devcel.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufer TA, Silljé HH, Körner R, Gruss OJ, Meraldi P, Nigg EA. Human TPX2 is required for targeting Aurora-A kinase to the spindle. Journal of Cell Biology. 2002;158:617–623. doi: 10.1083/jcb.200204155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai F, Yoneda A, Tomida T, Sano T, Nagata T, Hasezawa S. Fate of nascent microtubules organized at the M/G1 interface, as visualized by synchronized tobacco BY-2 cells stably expressing GFP–tubulin: time-sequence observations of the reorganization of cortical microtubules in living plant cells. Plant and Cell Physiology. 2001;42:723–732. doi: 10.1093/pcp/pce091. [DOI] [PubMed] [Google Scholar]
- Lansbergen G, Akhmanova A. Microtubule plus end: a hub of cellular activities. Traffic. 2006;7:499–507. doi: 10.1111/j.1600-0854.2006.00400.x. [DOI] [PubMed] [Google Scholar]
- Lawrence CJ, Dawe RK, Christie KR, et al. A standardized kinesin nomenclature. Journal of Cell Biology. 2004;167:19–22. doi: 10.1083/jcb.200408113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledbetter MC, Porter KR. A ‘microtubule’ in plant cell fine structure. Journal of Cell Biology. 1963;19:239–250. doi: 10.1083/jcb.19.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YRJ, Liu B. Cytoskeletal motors in Arabidopsis. Sixty-one kinesins and seventeen myosins. Plant Physiology. 2004;136:3877–3883. doi: 10.1104/pp.104.052621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y-RJ, Liu B. Cytoskeletal motor proteins in plant cell division. In: Verma DPS, Hong Z, editors. Plant cell monographs: cell division control in plants. Berlin: Springer; 2007. pp. 169–193. [Google Scholar]
- Lee YRJ, Li Y, Liu B. Two homologous phragmoplast-associated kinesins play a critical role in cytokinesis during male gametogenesis in Arabidopsis. The Plant Cell. 2007;19:2595–2605. doi: 10.1105/tpc.107.050716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Marc J, Joshi HC, Palevitz BA. A γ-tubulin-related protein associated with the microtubule arrays of higher plants in a cell cycle-dependent manner. Journal of Cell Science. 1993;104:1217–1228. doi: 10.1242/jcs.104.4.1217. [DOI] [PubMed] [Google Scholar]
- Liu B, Joshi HC, Wilson TJ, Silflow CD, Palevitz BA, Snustad DP. γ-Tubulin in Arabidopsis-gene sequence, immunoblot, and immunofluorescence studies. The Plant Cell. 1994;6:303–314. doi: 10.1105/tpc.6.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Cyr RJ, Palevitz BA. A kinesin-like protein, KatAp, in the cells of Arabidopsis and other plants. The Plant Cell. 1996;8:119–132. doi: 10.1105/tpc.8.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd C, Hussey P. Microtubule-associated proteins in plants – why we need a MAP. Nature Reviews Molecular Cell Biology. 2001;2:40–7. doi: 10.1038/35048005. [DOI] [PubMed] [Google Scholar]
- Lu L, Lee Y-RJ, Pan R, Liu B. An internal motor kinesin is associated with the Golgi apparatus and plays a role in trichome morphogenesis in Arabidopsis. Molecular Biology of the Cell. 2005;16:811–823. doi: 10.1091/mbc.E04-05-0400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüders J, Patel UK, Stearns T. GCP-WD is a γ-tubulin targeting factor required for centrosomal and chromatin-mediated microtubule nucleation. Nature Cell Biology. 2006;8:137–147. doi: 10.1038/ncb1349. [DOI] [PubMed] [Google Scholar]
- Manning J, Kumar S. NEDD1: function in microtubule nucleation, spindle assembly and beyond. International Journal of Biochemistry and Cell Biology. 2007;39:7–11. doi: 10.1016/j.biocel.2006.08.012. [DOI] [PubMed] [Google Scholar]
- Mao G, Chan J, Calder G, Doonan JH, Lloyd CW. Modulated targeting of GFP–AtMAP65-1 to central spindle microtubules during division. The Plant Journal. 2005;43:469–478. doi: 10.1111/j.1365-313X.2005.02464.x. [DOI] [PubMed] [Google Scholar]
- Mao T, Jin L, Li H, Liu B, Yuan M. Two microtubule-associated proteins of the Arabidopsis MAP65 family function differently on microtubules. Plant Physiology. 2005;138:654–662. doi: 10.1104/pp.104.052456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marc J, Sharkey DE, Durso NA, Zhang M, Cyr RJ. Isolation of a 90-kD microtubule-associated protein from tobacco membranes. The Plant Cell. 1996;8:2127–2138. doi: 10.1105/tpc.8.11.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marc J, Granger CL, Brincat J, et al. A GFP–MAP4 reporter gene for visualizing cortical microtubule rearrangements in living epidermal cells. The Plant Cell. 1998;10:1927–1940. doi: 10.1105/tpc.10.11.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus AI, Li W, Ma H, Cyr RJ. A kinesin mutant with an atypical bipolar spindle undergoes normal mitosis. Molecular Biology of the Cell. 2003;14:1717–1726. doi: 10.1091/mbc.E02-09-0586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marumoto T, Zhang D, Saya H. Aurora-A – a guardian of poles. Nature Reviews Cancer. 2005;5:42–50. doi: 10.1038/nrc1526. [DOI] [PubMed] [Google Scholar]
- Mathur J, Mathur N, Kernebeck B, Srinivas BP, Hulskamp M. A novel localization pattern for an EB1-like protein links microtubule dynamics to endomembrane organization. Current Biology. 2003;13:1991–1997. doi: 10.1016/j.cub.2003.10.033. [DOI] [PubMed] [Google Scholar]
- McClinton RS, Sung ZR. Organization of cortical microtubules at the plasma membrane in Arabidopsis. Planta. 1997;201:252–260. doi: 10.1007/s004250050064. [DOI] [PubMed] [Google Scholar]
- McClinton RS, Chandler JS, Callis J. cDNA isolation, characterization, and protein intracellular localization of a katanin-like p60 subunit from Arabidopsis thaliana. Protoplasma. 2001;216:181–190. doi: 10.1007/BF02673870. [DOI] [PubMed] [Google Scholar]
- McNally FJ, Vale RD. Identification of katanin, an ATPase that severs and disassembles stable microtubules. Cell. 1993;75:419–429. doi: 10.1016/0092-8674(93)90377-3. [DOI] [PubMed] [Google Scholar]
- McNally K, Bazirgan O, McNally F. Two domains of p80 katanin regulate microtubule severing and spindle pole targeting by p60 katanin. Journal of Cell Science. 2000;113:1623–1633. doi: 10.1242/jcs.113.9.1623. [DOI] [PubMed] [Google Scholar]
- Mineyuki Y. Plant microtubule studies: past and present. Journal of Plant Research. 2007;120:45–51. doi: 10.1007/s10265-006-0063-y. [DOI] [PubMed] [Google Scholar]
- Miyao A, Iwasaki Y, Kitano H, et al. A large-scale collection of phenotypic data describing an insertional mutant population to facilitate functional analysis of rice genes. Plant Molecular Biology. 2007;63:625–635. doi: 10.1007/s11103-006-9118-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller S, Smertenko A, Wagner V, Heinrich M, Hussey P, Hauser M. The plant microtubule-associated protein AtMAP65-3/PLE is essential for cytokinetic phragmoplast function. Current Biology. 2004;14:412–417. doi: 10.1016/j.cub.2004.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller S, Han S, Smith LG. Two kinesins are involved in the spatial control of cytokinesis in Arabidopsis thaliana. Current Biology. 2006;16:888–894. doi: 10.1016/j.cub.2006.03.034. [DOI] [PubMed] [Google Scholar]
- Murata T, Sonobe S, Baskin TI, et al. Microtubule-dependent microtubule nucleation based on recruitment of γ-tubulin in higher plants. Nature Cell Biology. 2005;7:961–968. doi: 10.1038/ncb1306. [DOI] [PubMed] [Google Scholar]
- Murata T, Tanahashi T, Nishiyama T, Yamaguchi K, Hasebe M. How do plants organize microtubules without a centrosome? Journal of Integrated Plant Biology. 2007;49:1154–1163. [Google Scholar]
- Murphy SM, Preble AM, Patel UK, et al. GCP5 and GCP6: two new members of the human γ-tubulin complex. Molecular Biology of the Cell. 2001;12:3340–3352. doi: 10.1091/mbc.12.11.3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K, Furutani I, Tachimoto H, Matsubara H, Hashimoto T. SPIRAL1 encodes a plant-specific microtubule-localized protein required for directional control of rapidly expanding Arabidopsis cells. The Plant Cell. 2004;16:1178–1190. doi: 10.1105/tpc.017830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K, Kawamura T, Hashimoto T. Role of the SPIRAL1 gene family in anisotropic growth of Arabidopsis thaliana. Plant and Cell Physiology. 2006;47:513–522. doi: 10.1093/pcp/pcj020. [DOI] [PubMed] [Google Scholar]
- Nishihama R, Soyano T, Ishikawa M, et al. Expansion of the cell plate in plant cytokinesis requires a kinesin-like protein/MAPKKK complex. Cell. 2002;109:87–99. doi: 10.1016/s0092-8674(02)00691-8. [DOI] [PubMed] [Google Scholar]
- Opanowicz M, Vain P, Draper J, Parker D, Doonan JH. Brachypodium distachyon: making hay with a wild grass. Trends in Plant Science. 2008;13:172–177. doi: 10.1016/j.tplants.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Oppenheimer DG, Pollock MA, Vacik J, et al. Essential role of a kinesin-like protein in Arabidopsis trichome morphogenesis. Proceedings of the National Academy of Sciences, USA. 1997;94:6261–6266. doi: 10.1073/pnas.94.12.6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan R, Lee YRJ, Liu B. Localization of two homologous Arabidopsis kinesin-related proteins in the phragmoplast. Planta. 2004;220:156–164. doi: 10.1007/s00425-004-1324-4. [DOI] [PubMed] [Google Scholar]
- Pastuglia M, Azimzadeh J, Goussot M, et al. γ-Tubulin is essential for microtubule organization and development in Arabidopsis. The Plant Cell. 2006;18:1412–1425. doi: 10.1105/tpc.105.039644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellman D, Bagget M, Tu YH, Fink GR, Tu H. Two microtubule-associated proteins required for anaphase spindle movement in Saccharomyces cerevisiae. Journal of Cell Biology. 1995;130:1373–1385. doi: 10.1083/jcb.130.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin RM, Wang Y, Yuen CY, Will J, Masson PH. WVD2 is a novel microtubule-associated protein in Arabidopsis thaliana. The Plant Journal. 2007;49:961–971. doi: 10.1111/j.1365-313X.2006.03015.x. [DOI] [PubMed] [Google Scholar]
- Pickett-Heaps JD, Northcote DH. Organization of microtubules and endoplasmic reticulum during mitosis and cytokinesis in wheat meristems. Journal of Cell Science. 1966;a 1:109–120. doi: 10.1242/jcs.1.1.109. [DOI] [PubMed] [Google Scholar]
- Pickett-Heaps JD, Northcote DH. Cell division in the formation of the stomatal complex of the young leaves of wheat. Journal of Cell Science. 1966;b 1:121–128. doi: 10.1242/jcs.1.1.121. [DOI] [PubMed] [Google Scholar]
- Preuss ML, Delmer DP, Liu B. The cotton kinesin-like calmodulin-binding protein associates with cortical microtubules in cotton fibers. Plant Physiology. 2003;132:154–160. doi: 10.1104/pp.103.020339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss ML, Kovar DR, Lee Y-RJ, Staiger CJ, Delmer DP, Liu B. A plant-specific kinesin binds to actin microfilaments and interacts with cortical microtubules in cotton fibers. Plant Physiology. 2004;136:3945–3955. doi: 10.1104/pp.104.052340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin XQ, Giani S, Breviario D. Molecular cloning of three rice alpha-tubulin isotypes: differential expression in tissues and during flower development. Biochimica et Biophysica Acta. 1997;1354:19–23. doi: 10.1016/s0167-4781(97)00110-3. [DOI] [PubMed] [Google Scholar]
- Reddy A, Day I. The role of the cytoskeleton and a molecular motor in trichome morphogenesis. Trends in Plant Science. 2000;5:503–505. doi: 10.1016/s1360-1385(00)01792-1. [DOI] [PubMed] [Google Scholar]
- Reddy ASN, Safadi F, Narasimhulu SB, Golovkin M, Hu X. A novel plant calmodulin-binding protein with a kinesin heavy chain motor domain. Journal of Biological Chemistry. 1996;271:7052–7060. doi: 10.1074/jbc.271.12.7052. [DOI] [PubMed] [Google Scholar]
- Richardson DN, Simmons MP, Reddy AS. Comprehensive comparative analysis of kinesins in photosynthetic eukaryotes. BMC Genomics. 2006;7:18. doi: 10.1186/1471-2164-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers SL, Rogers GC, Sharp DJ, Vale RD. Drosophila EB1 is important for proper assembly, dynamics, and positioning of the mitotic spindle. Journal of Cell Biology. 2002;158:873–884. doi: 10.1083/jcb.200202032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roghi C, Giet R, Uzbekov R, et al. The Xenopus protein kinase pEg2 associates with the centrosome in a cell cycle-dependent manner, binds to the spindle microtubules and is involved in bipolar mitotic spindle assembly. Journal of Cell Science. 1998;111:557–572. doi: 10.1242/jcs.111.5.557. [DOI] [PubMed] [Google Scholar]
- Ruchaud S, Carmena M, Earnshaw WC. Chromosomal passengers: conducting cell division. Nature Reviews Molecular Cell Biology. 2007;8:798–812. doi: 10.1038/nrm2257. [DOI] [PubMed] [Google Scholar]
- Sasabe M, Machida Y. MAP65: a bridge linking a MAP kinase to microtubule turnover. Current Opinion in Plant Biology. 2006;9:563–570. doi: 10.1016/j.pbi.2006.09.010. [DOI] [PubMed] [Google Scholar]
- Sasabe M, Soyano T, Takahashi Y, et al. Phosphorylation of NtMAP65-1 by a MAP kinase down-regulates its activity of microtubule bundling and stimulates progression of cytokinesis of tobacco cells. Genes and Development. 2006;20:1004–1014. doi: 10.1101/gad.1408106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sazuka T, Aichi I, Kawai T, Matsuo N, Kitano H, Matsuoka M. The rice mutant dwarf bamboo shoot 1: a leaky mutant of the NACK-type kinesin-like gene can initiate organ primordia but not organ development. Plant and Cell Physiology. 2005;46:1934–1943. doi: 10.1093/pcp/pci206. [DOI] [PubMed] [Google Scholar]
- Sedbrook JC. MAPs in plant cells: delineating microtubule growth dynamics and organization. Current Opinion in Plant Biology. 2004;7:632–640. doi: 10.1016/j.pbi.2004.09.017. [DOI] [PubMed] [Google Scholar]
- Sedbrook JC, Ehrhardt DW, Fisher SE, Scheible WR, Somerville CR. The Arabidopsis SKU6/SPIRAL1 gene encodes a plus end-localized microtubule-interacting protein involved in directional cell expansion. The Plant Cell. 2004;16:1506–1520. doi: 10.1105/tpc.020644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seltzer V, Janski N, Canaday J, et al. Arabidopsis GCP2 and GCP3 are part of a soluble gamma-tubulin complex and have nuclear envelope targeting domains. The Plant Journal. 2007;52:322–331. doi: 10.1111/j.1365-313X.2007.03240.x. [DOI] [PubMed] [Google Scholar]
- Shoji T, Narita NN, Hayashi K, et al. Plant-specific microtubule-associated protein SPIRAL2 is required for anisotropic growth in arabidopsis. Plant Physiology. 2004;136:3933–3944. doi: 10.1104/pp.104.051748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji T, Suzuki K, Abe T, et al. Salt stress affects cortical microtubule organization and helical growth in Arabidopsis. Plant and Cell Physiology. 2006;47:1158–1168. doi: 10.1093/pcp/pcj090. [DOI] [PubMed] [Google Scholar]
- Smertenko A, Saleh N, Igarashi H, et al. A new class of microtubule-associated proteins in plants. Nature Cell Biology. 2000;2:750–753. doi: 10.1038/35036390. [DOI] [PubMed] [Google Scholar]
- Smertenko AP, Chang HY, Wagner V, Kaloriti D, Fenyk S, Sonobe S, et al. The Arabidopsis microtubule-associated protein AtMAP65-1: molecular analysis of its microtubule bundling activity. The Plant Cell. 2004;16:2035–2047. doi: 10.1105/tpc.104.023937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smertenko AP, Chang HY, Sonobe S, et al. Control of the AtMAP65-1 interaction with microtubules through the cell cycle. Journal of Cell Science. 2006;119:3227–3237. doi: 10.1242/jcs.03051. [DOI] [PubMed] [Google Scholar]
- Smith LG, Gerttula SM, Han S, Levy J. TANGLED1: a microtubule binding protein required for the spatial control of cytokinesis in maize. Journal of Cell Biology. 2001;152:231–236. doi: 10.1083/jcb.152.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snustad DP, Haas NA, Kopczak SD, Silflow CD. The small genome of Arabidopsis contains at least nine expressed β-tubulin genes. The Plant Cell. 1992;4:549–556. doi: 10.1105/tpc.4.5.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinborn K, Maulbetsch C, Priester B, et al. The Arabidopsis PILZ group genes encode tubulin-folding cofactor orthologs required for cell division but not cell growth. Genes and Development. 2002;16:959–971. doi: 10.1101/gad.221702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoppin-Mellet V, Gaillard J, Vantard M. Functional evidence for in vitro microtubule severing by the plant katanin homologue. Biochemical Journal. 2002;365:337–342. doi: 10.1042/BJ20020689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoppin-Mellet V, Gaillard J, Vantard M. Katanin's severing activity favors bundling of cortical microtubules in plants. The Plant Journal. 2006;46:1009–1017. doi: 10.1111/j.1365-313X.2006.02761.x. [DOI] [PubMed] [Google Scholar]
- Strompen G, El Kasmi F, Richter S, et al. The Arabidopsis HINKEL gene encodes a kinesin-related protein involved in cytokinesis and is expressed in a cell cycle-dependent manner. Current Biology. 2002;12:153–158. doi: 10.1016/s0960-9822(01)00655-8. [DOI] [PubMed] [Google Scholar]
- Su LK, Burrell M, Hill DE, et al. APC binds to the novel protein EB1. Cancer Research. 1995;55:2972–2977. [PubMed] [Google Scholar]
- Tamura K, Nakatani K, Mitsui H, Ohashi Y, Takahashi H. Characterization of katD, a kinesin-like protein gene specifically expressed in floral tissues of Arabidopsis thaliana. Gene. 1999;230:23–32. doi: 10.1016/s0378-1119(99)00070-0. [DOI] [PubMed] [Google Scholar]
- Traas J, Bellini C, Nacry P, Kronenberger J, Bouchez D, Caboche M. Normal differentiation patterns in plants lacking microtubular preprophase bands. Nature. 1995;375:676–677. [Google Scholar]
- Tsai MY, Zheng Y. Aurora A kinase-coated beads function as microtubule-organizing centers and enhance RanGTP-induced spindle assembly. Current Biology. 2005;15:2156–2163. doi: 10.1016/j.cub.2005.10.054. [DOI] [PubMed] [Google Scholar]
- Twell D, Park SK, Hawkins TJ, et al. MOR1/GEM1 has an essential role in the plant-specific cytokinetic phragmoplast. Nature Cell Biology. 2002;4:711–714. doi: 10.1038/ncb844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzafrir I, McElver JA, Liu CM, et al. Diversity of TITAN functions in Arabidopsis seed development. Plant Physiology. 2002;128:38–51. [PMC free article] [PubMed] [Google Scholar]
- Ueda K, Matsuyama T, Hashimoto T. Visualization of microtubules in living cells of transgenic Arabidopsis thaliana. Protoplasma. 1999;206:201–206. [Google Scholar]
- Umeki N, Mitsui T, Umezu N, Kondo K, Maruta S. Preparation and characterization of a novel rice plant-specific kinesin. Journal of Biochemistry. 2006;139:645–654. doi: 10.1093/jb/mvj074. [DOI] [PubMed] [Google Scholar]
- Van Damme D, Bouget F, Van Poucke K, Inze D, Geelen D. Molecular dissection of plant cytokinesis and phragmoplast structure: a survey of GFP-tagged proteins. The Plant Journal. 2004;a 40:386–398. doi: 10.1111/j.1365-313X.2004.02222.x. [DOI] [PubMed] [Google Scholar]
- Van Damme D, Van Poucke K, Boutant E, Ritzenthaler C, Inze D, Geelen D. In vivo dynamics and differential microtubule-binding activities of MAP65 proteins. Plant Physiology. 2004;b 136:3956–3967. doi: 10.1104/pp.104.051623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanstraelen M, Torres Acosta J, De Veylder L, Inze D, Geelen D. A plant-specific subclass of C-terminal kinesins contains a conserved a-type cyclin-dependent kinase site implicated in folding and dimerization. Plant Physiology. 2004;135:1417–1429. doi: 10.1104/pp.104.044818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanstraelen M, Van Damme D, De Rycke R, Mylle E, Inze D, Geelen D. Cell cycle-dependent targeting of a kinesin at the plasma membrane demarcates the division site in plant cells. Current Biology. 2006;16:308–314. doi: 10.1016/j.cub.2005.12.035. [DOI] [PubMed] [Google Scholar]
- Vos JW, Safadi F, Reddy ASN, Hepler PK. The kinesin-like calmodulin binding protein is differentially involved in cell division. The Plant Cell. 2000;12:979–990. doi: 10.1105/tpc.12.6.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos LJ, Famulski JK, Chan GK. How to build a centromere: from centromeric and pericentromeric chromatin to kinetochore assembly. Biochemistry and Cell Biology. 2006;84:619–639. doi: 10.1139/o06-078. [DOI] [PubMed] [Google Scholar]
- Walker KL, Müller S, Moss D, Ehrhardt DW, Smith LG. Arabidopsis TANGLED identifies the division plane throughout mitosis and cytokinesis. Current Biology. 2007;17:1827–1836. doi: 10.1016/j.cub.2007.09.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang PJJ, Huffaker TC. Stu2p: a microtubule-binding protein that is an essential component of the yeast spindle pole body. Journal of Cell Biology. 1997;139:1271–1280. doi: 10.1083/jcb.139.5.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Li JJ, Yuan M. Salt tolerance requires cortical microtubule reorganization in Arabidopsis. Plant and Cell Physiology. 2007;a 48:1534–1547. doi: 10.1093/pcp/pcm123. [DOI] [PubMed] [Google Scholar]
- Wang X, Zhu L, Liu B, et al. Arabidopsis MICROTUBULE-ASSOCIATED PROTEIN18 functions in directional cell growth by destabilizing cortical microtubules. The Plant Cell. 2007;b 19:877–889. doi: 10.1105/tpc.106.048579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb M, Jouannic S, Foreman J, Linstead P, Dolan L. Cell specification in the Arabidopsis root epidermis requires the activity of ECTOPIC ROOT HAIR 3 – a katanin-p60 protein. Development. 2002;129:123–131. doi: 10.1242/dev.129.1.123. [DOI] [PubMed] [Google Scholar]
- Whittington AT, Vugrek O, Wei KJ, et al. MOR1 is essential for organizing cortical microtubules in plants. Nature. 2001;411:610–613. doi: 10.1038/35079128. [DOI] [PubMed] [Google Scholar]
- Wicker-Planquart C, Stoppin-Mellet V, Blanchoin L, Vantard M. Interactions of tobacco microtubule-associated protein MAP65-1b with microtubules. The Plant Journal. 2004;39:126–134. doi: 10.1111/j.1365-313X.2004.02115.x. [DOI] [PubMed] [Google Scholar]
- Wiese C, Zheng Y. Microtubule nucleation: γ-tubulin and beyond. Journal of Cell Science. 2006;119:4143–4153. doi: 10.1242/jcs.03226. [DOI] [PubMed] [Google Scholar]
- Wittmann T, Wilm M, Karsenti E, Vernos I. TPX2, a novel Xenopus MAP involved in spindle pole organization. Journal of Cell Biology. 2000;149:1405–1418. doi: 10.1083/jcb.149.7.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Gu Y, Li S, Yang Z. A genome-wide analysis of Arabidopsis Rop-interactive CRIB motif-containing proteins that act as Rop GTPase targets. The Plant Cell. 2001;13:2841–2856. doi: 10.1105/tpc.010218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Xiang X, Hammer JAR. Motor proteins at the microtubule plus-end. Trends in Cell Biology. 2006;16:135–143. doi: 10.1016/j.tcb.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Yang GX, Jan A, Komatsu S. Functional analysis of OsTUB8, an anther-specific beta-tubulin in rice. Plant Science. 2007;172:832–838. [Google Scholar]
- Yen TJ, Li G, Schaar BT, Szilak I, Cleveland DW. CENP-E is a putative kinetochore motor that accumulates just before mitosis. Nature. 1992;359:536–539. doi: 10.1038/359536a0. [DOI] [PubMed] [Google Scholar]
- Yoshikawa M, Yang GX, Kawaguchi K, Komatsu S. Expression analyses of beta-tubulin isotype genes in rice. Plant and Cell Physiology. 2003;44:1202–1207. doi: 10.1093/pcp/pcg150. [DOI] [PubMed] [Google Scholar]
- Yuen CY, Pearlman RS, Silo-Suh L, Hilson P, Carroll KL, Masson PH. WVD2 and WDL1 modulate helical organ growth and anisotropic cell expansion in Arabidopsis. Plant Physiology. 2003;131:493–506. doi: 10.1104/pp.015966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DH, Wadsworth P, Hepler PK. Microtubule dynamics in living dividing plant cells – confocal imaging of microinjected fluorescent brain tubulin. Proceedings of the National Academy of Sciences, USA. 1990;87:8820–8824. doi: 10.1073/pnas.87.22.8820. [DOI] [PMC free article] [PubMed] [Google Scholar]