Abstract
The nearly-neutral theory represents a development of Kimura’s Neutral Theory of Molecular Evolution that makes testable predictions that go beyond a mere null model. Recent evidence has strongly supported several of these predictions, including the prediction that slightly deleterious variants will accumulate in a species that has undergone a severe bottleneck or in cases where recombination is reduced or absent. Because bottlenecks often occur in speciation and slightly deleterious mutations in coding regions will usually be nonsynonymous, we should expect that the ratio of nonsynonymous to synonymous fixed differences between species should often exceed the ratio of nonsynonymous to synonymous polymorphisms within species. Numerous data support this prediction, although they have often been wrongly interpreted as evidence for positive Darwinian selection. The use of conceptually flawed tests for positive selection has become widespread in recent years, seriously harming the quest for an understanding of genome evolution. When properly analyzed, many (probably most) claimed cases of positive selection will turn out to involve the fixation of slightly deleterious mutations by genetic drift in bottlenecked populations. Slightly deleterious variants are a transient feature of evolution in the long term, but they have had substantial impact on contemporary species, including our own.
Introduction
Motoo Kimura’s Neutral Theory of Molecular Evolution provides the central organizing concepts for modern evolutionary biology. The so-called “nearly-neutral” theory is a corollary of Kimura’s theory that has attracted considerable attention recently. My purpose here is to review the concept of “near-neutrality” in relation to the Neutral Theory and to review some recent evidence relating to the importance of slightly deleterious mutations in evolution. Many biologists tend to avoid population genetics because of its mathematical complexities. Nonetheless, the Neutral Theory and its “nearly neutral” corollary make straightforward predictions that can be expressed verbally or with a minimum of mathematics. In this review I concentrate on such simple predictions rather than on the mathematical complexity of their derivation.
In its inception, population genetics was a field characterized by a wealth of theory and a paucity of data. For example, phenomena such as heterozygote advantage were modeled theoretically before any actual cases were known. The situation has reversed dramatically with the advent of rapid sequencing and genotyping technologies. Now population genetics is a field awash with data, whereas the theoretical framework used to interpret this data is often inadequate (Hughes et al. 2006; Hughes 2007a). In such a situation, it is important that studies be designed so as to constitute critical tests among competing hypotheses.
I will argue that the nearly-neutral theory, because it makes a number of testable predictions, provides the opportunity for testing the predictions of the Neutral Theory against those of selectionist alternatives. Moreover, a recent resurgence of selectionist thinking in evolutionary has created a kind of crisis in evolutionary biology. This crisis is both unnecessary and self-inflicted, resulting from the widespread use of ill-conceived statistical methods (Hughes 2007a; Hughes et al. 2006: Sabeti et al. 2006). I will show how the nearly-neutral theory, by providing a more plausible explanation of the observed results than does the hypothesis of widespread positive selection, has the potential to resolve this crisis in favor of Kimura’s theory.
In the view presented here, the nearly-neutral theory is best understood as a corollary or development of Kimura’s Neutral Theory of Molecular evolution, rather than as an independent theory competing with the Neutral Theory. Still less should the nearly-neutral theory be viewed as a kind of “selectionism lite”. Rather, the nearly-neutral theory makes testable predictions regarding the dynamics of a certain class of mutation – those that are slightly deleterious in their fitness effect – under realistic conditions. Critics of Kimura have sometimes dismissed the Neutral Theory as a mere null hypothesis – a theory of “no effect.” But near-neutrality describes conditions under which the Neutral Theory is not a mere null hypothesis; indeed, conditions under which the Neutral Theory makes bold predictions which are based on rigorous reasoning and yet seem counter-intuitive to biologists schooled on decades of selectionist story-telling. Near-neutrality thus represents the leading edge of the Neutral Theory, where that theory as a whole stands or falls.
Evolution as a Population Process
Evolutionary biologists are accustomed to assert that their discipline provides a central unifying theory for all branches of biology theory behind biology (Mayr 1982). There were two major theoretical advances in evolutionary biology in the Twentieth Century. The first was the synthesis of Mendelian genetics and Darwin’s hypothesis of natural selection, which gave rise to “Neo-Darwinism” and gave birth to population genetics as a distinct discipline within the biological sciences. The second was Motoo Kimura’s reformulation of population genetics taking into account the fact that natural populations are finite, which culminated in his formulation of the Neutral Theory of Molecular Evolution.
When Mendel’s work was rediscovered early in the 20th Century, the Neo-Darwinians reinterpreted evolution as change in gene frequencies within populations and developed simple models of natural selection in which fitness differences among alleles lead to gene frequency change. Perhaps the most important aspect of Neo-Darwinism was its emphasis on evolutionary change as a population process, leading to an appreciation of the population as a level of biological reality beyond the molecular, the cellular, and the organismal. However, there were other aspects of the Neo-Darwinian synthesis that were not constructive. One was a tendency to elevate assumptions into dogmas. For example, simple models of natural selection made the obviously unrealistic, but mathematically convenient, assumption of an infinite population size. When Sewall Wright examined population genetics in finite populations, thereby discovering the important process known as genetic drift, his results were dismissed as a mere curiosity of limited real-world value, since most natural populations were assumed to be very large (Fisher and Ford 1950).
As a result of the over-literal application of simplistic models, evolutionary biologists began to view natural selection as a virtually omnipotent force, shaping every aspect of the phenotype so as to optimize the Darwinian fitness of the organism. Gould and Lewontin (1979) named this viewpoint the “Panglossian paradigm” and charged Neo-Darwinism with a propensity for “adaptive story-telling”; that is, the concoction of plausible but untested stories regarding the adaptive significance of phenotypic traits.
The Neutral Theory of Molecular Evolution
Beginning in the mid-1950’s, Kimura (1955, 1957, 1964) examined the consequences of finite population size not only for genetic drift but also for natural selection. Perhaps because of the advanced mathematics used in this work, it attracted relatively little attention among evolutionary biologists until Kimura (1968) proposed the radically new hypothesis that genetic drift is the dominant process in evolution both within populations and over evolutionary time (the “Neutral Theory of Molecular Evolution”). Kimura did not deny the importance of natural selection. Rather, he emphasized the distinction between two types of selection: (1) purifying (or conservative natural selection) which acts to eliminate deleterious mutations; and (2) positive (Darwinian) selection, favor advantageous mutants. Because deleterious mutations are very common, the Neutral Theory predicts that purifying selection is ubiquitous; but, since advantageous mutations are predicted to be very rare, positive selection is predicted to be a rare phenomenon.
Kimura’s proposal gave rise to the so-called selectionist-neutralist controversy in the 1970’s. At the time, the available data for testing the theory consisted of a limited number of amino acid sequences and data on population frequency of protein variants using the allozyme technique. Even with these limited data, several important predictions of the Neutral Theory were supported.
One important premise of the Neutral Theory is that most mutations within coding regions are deleterious, because a majority of them are nonsynonymous (amino acid-altering) and thus have a disruptive effect on protein structure. Furthermore, the more important a domain is to the function of the protein, the more disruptive amino acid changes are predicted to be. As a consequence, the Neutral Theory predicts that the most functionally important regions of proteins (such as the active sites of enzymes) will generally evolve more slowly than functionally important regions. By contrast, on the selectionist view, the most important regions of proteins would be expected to evolve the fastest, as adaptation is constantly fine-tuned. Of course, the neutralist view turned out to be correct. Indeed it is by now routine for biologists to infer that a given protein domain or genomic region must have an important function because it is evolutionarily “conserved”; i.e., because it evolves slowly.
When rapid nucleotide sequencing became possible at the end of the 1970’s, Kimura used the neutral theory to make a prediction regarding the pattern of nucleotide substitution in protein coding genes (Kimura 1977). On the premise that most nonsynonymous mutations are selectively deleterious and thus tend to be eliminated by purifying selection, one might predict that the number of synonymous substitutions per synonymous site (dS) will exceed the number of nonsynonymous substitutions per nonsynonymous site (dN) in most protein-coding genes. Kimura (1977) himself initially tested this prediction in a rather roundabout way, because he did not develop a method of estimating dS and dN directly. Subsequently, when methods of estimating these quantities become available, this prediction has been supported overwhelmingly in comparative sequence analyses, demonstrating the prevalence of purifying selection on protein-coding regions and the comparative rarity of positive selection.
At the same time, advances in molecular biology revealed a reality very different from what might be expected under the Panglossian Paradigm. The genomes of eukaryotes were found to consist largely of non-coding DNA of no discernible function, while many processes at the molecular level (for example, intron splicing and RNA editing, to name just two) seemed to have an ad hoc, Rube Goldberg-esque quality inconsistent with the Neo-Darwinist vision of phenotypic optimality.
The Neutral Theory of Molecular Evolution provided the theoretical foundation for the newly emerging discipline of bioinformatics. Homology search and alignment – two staples of the bioinformaticist’s arsenal – depend on the fact that functionally important regions of sequences evolve slowly. Likewise, the overwhelmingly conservative nature of protein evolution, predicted by the Neutral Theory, opens to structural biologists the possibility of using evolutionary relationships as a source of information in inferring tertiary structures, thereby reducing the problem of solving the millions of protein structures found in nature to one of solving perhaps as few as 1,000 structural families (Chothia 1992; Chen and Kurgan 2007).
Given the importance of his contribution, it is not an exaggeration to say that Kimura was the most important evolutionary biologist since Darwin. What is perhaps surprising in Kimura’s case, given the impact of his work on the biological sciences, is that its significance has so far been little appreciated by the general educated public or by philosophers and historians of ideas. To take one example, a recent book entitled Evolution: the History of an Idea (Larson 2004) includes not a single mention of Kimura. To my mind, this is rather like writing a history of physics without mentioning Einstein.
The philosopher Daniel Dennett, in his book Darwin’s Dangerous Idea, proposes that Darwin’s key (and “dangerous”) insight was that evolution is an “algorithmic process” (Dennett 1995). By “algorithmic,” one gathers that Dennett means essentially deterministic. But determinism was hardly a bold or “dangerous” idea in Darwin’s time, having been a familiar concept in Western thought since at least the Stoics. Rather, one might suggest that the truly new idea in evolutionary biology is that of Kimura (building on the work of Sewall Wright), which along with Heisenberg’s Uncertainty Principle and Gödel’s proof of the incompleteness of mathematics, formed part of a Twentieth Century revolution in thought that for the first time revealed the universe as non-algorithmic.
The Neutral Theory and the Nearly-Neutral Theory
One prediction of the Neutral Theory is that the effectiveness of natural selection depends on the effective population size. Kimura (1983) suggested that the behavior of an allele is controlled mainly by genetic drift when its relative advantage or disadvantage, measured by the selection coefficient (s), is less than twice the reciprocal of the effective population size (Ne); i.e. |s| < 1/(2Ne). Such an allele is referred to as “almost neutral” or “nearly neutral.” Li (1978) proposed a more relaxed definition of near-neutrality; namely, when |s| < 1/Ne. Nei (2005), taking into account the random variation among individuals with respect to numbers of offspring, proposed |s| < 1/√2Ne as a statistical definition of neutrality. Whichever of these criteria is more appropriate, the theory predicts that, in a very small population, genetic drift becomes such a powerful force that natural selection (whether positive or purifying) cannot overcome it unless the selection is very strong.
These theoretical considerations led to the recognition that there exists a certain category of mutations that can be described as “nearly neutral.” Kimura’s associate, Tomoko Ohta, developed the theoretical understanding of this type of mutation, giving rise to the so-called “nearly-neutral theory” (Ohta 1973, 2002). The nearly-neutral theory represents the application of key concepts of the Neutral Theory to the study of variants with small selection coefficients, particularly slightly deleterious mutations. Unlike strictly neutral alleles, the fate of nearly neutral alleles depends on effective population size. Thus, when the effective population size becomes small enough (on the order of the reciprocal of the selection coefficient), a slightly deleterious mutation will act as if it is neutral and can drift to a high frequency or even to fixation. However, if the effective population size is large for a long time, selection will decrease the frequency of slightly deleterious variants in the population and eventually eliminate them.
The fate of slightly advantageous mutants will likewise depend on effective population size, but there are reasons for believing that slightly advantageous mutants will be very rare. In recent years there has been increased attention to the question of slightly advantageous mutations, and some authors have suggested that these may occur as frequently as slightly deleterious mutations. Sawyer et al. (2007) revived an argument of Fisher (1930), who suggested that advantageous and disadvantageous mutations should occur roughly in equal frequencies by analogy with the functioning of a mechanical device such as a microscope. Fisher likened mutations to adjustments of the focus of the microscope up or down, thereby perturbing it from the precise focus that provides a distinct image. We can forgive Fisher this fanciful image on the grounds that he was writing before there existed any knowledge of the genetic code or of protein chemistry. But it is interesting to note that the roots of his analogy lie in a kind of teleological thinking, characteristic of Neo-Darwinism, that saw natural selection as a kind of “designer,” fine-tuning phenotypes.
It may be true that, as generally assumed by the classic models of quantitative genetics, the number of alleles in a population that slightly increase the value of some quantitative trait is likely to be approximately equal to the number of alleles that slightly decrease its value. But if there is stabilizing selection on the trait, any mutation that causes the trait value to depart from the optimal value – whether by increasing it or by decreasing it – will be at least slightly deleterious. Moreover, the facts of protein chemistry argue strongly against the hypothesis that advantageous mutations are anywhere near as frequent as deleterious mutations. First of all, the majority of mutations in coding regions are nonsynonymous. Furthermore, there is evidence that the deleterious effect of a given nonsynonymous mutation depends on the chemical distance between the new amino acid and that which it replaced (Yampolsky et al. 2005). And, although the nature of the genetic code provides some degree of buffering against radical amino acid changes, a higher proportion of the possible replacements for any given amino acid will introduce a chemically dissimilar amino acid than a chemically similar amino acid (Miyata et al. 1979).
Testing Predictions of the Nearly Neutral Theory
In recent years, the availability of substantial data on sequence polymorphism within natural populations, as well as sequences of orthologous genes from closely related species, has created a situation where several predictions of the nearly-neutral theory can be tested. In some cases, the authors who have analyzed these data have provided explicit tests of these predictions. In other cases, the authors have presented results that support the nearly-neutral theory but have provided selectionist explanations for the results, often based on the use of flawed tests for positive selection. Here I discuss three predictions of the nearly-neutral theory and some data that support them.
(1) The nearly-neutral theory predicts that there will be evidence of ongoing purifying selection against slightly deleterious variants
Such slightly deleterious variants will be particularly common in a species that has undergone a prolonged bottleneck. In protein-coding regions, slightly deleterious variants will be overwhelmingly nonsynonymous. This leads to the prediction that, especially in populations that have undergone a prolonged bottleneck, we should see an excess of rare nonsynonymous polymorphisms.
As mentioned previously, comparison of the number of synonymous nucleotide substitutions per site (dS) and the number of nonsynonymous substitutions per nonsynonymous site (dN) has provided evidence of the prevalence of purifying selection. Within a population, the mean of all pairwise dS values among a set of allelic sequences is known as the synonymous nucleotide diversity (πS), while the mean of all pairwise dN values is known as the nonsynonymous nucleotide diversity (πN). At most loci, πS is greater than πN, reflecting the effect of purifying selection in eliminating or decreasing the frequency of deleterious nonsynonymous variants.
However, the comparison of πS and πN does not capture all of the information available from data on sequence polymorphism regarding the action of purifying selection. Strongly deleterious mutations will often be eliminated very quickly even in a very small population; for instance, a mutation with a dominant lethal effect will be eliminated instantly. Past events of elimination of strongly deleterious mutations play some role in lowering πN relative to πS. But there may also be slightly deleterious nonsynonymous variants in a population, which are subject to ongoing purifying selection that decreases their frequency in comparison to that of synonymous variants in the same genes but has not yet eliminated them from the population. Other types of statistical analysis are required to detect the presence of such relatively rare nonsynonymous variants.
In humans, much of the data on polymorphism is in the form of single-nucleotide polymorphisms (SNP). One approach to analysis of SNP data is to compare gene diversity at SNP sites categorized by their location within genes (Hughes et al. 2003). Where xi is the frequency of the ith allele (nucleotide) at a given locus (site), the gene diversity (“heterozygosity”) is 1- Σxi2 (Nei 1987). Since human SNP sites are almost always bi-allelic, another frequently used measure is “minor allele frequency” (MAF), the frequency of the less common allele, which shows similar patterns to those revealed by analysis of gene diversity. Even though nonsynonymous SNPs are over-represented in most human SNP data sets, a comparison of gene diversity at nonsynonymous SNPs with that at other SNPs is unaffected by this ascertainment bias.
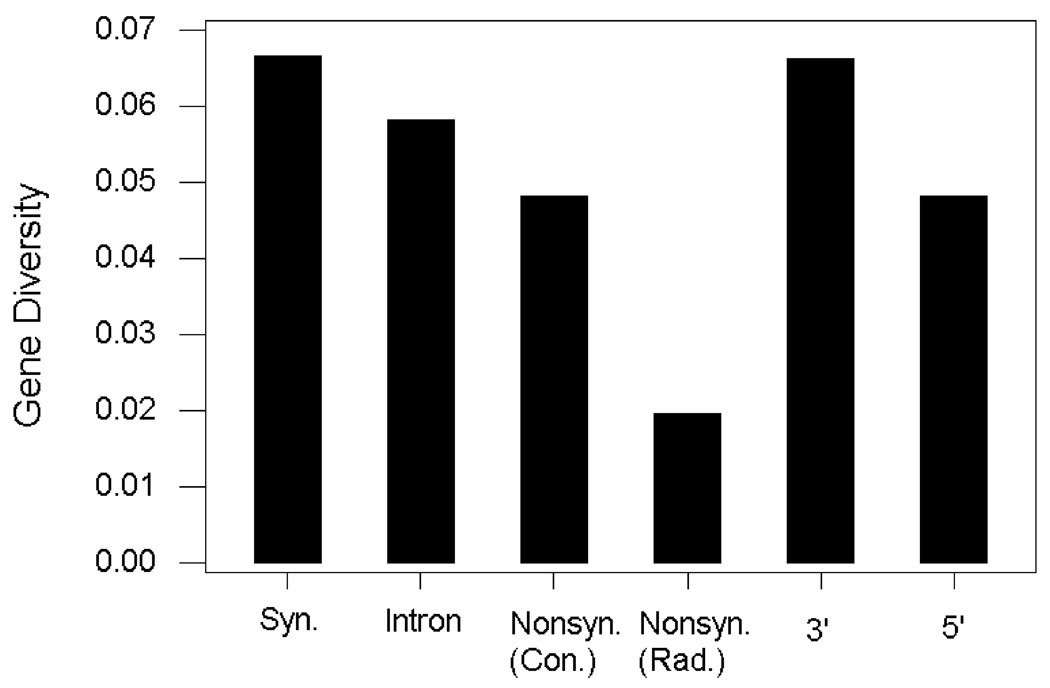
Gene diversity at nonsynonymous SNP sites in the human genome tends to be lower than that at silent SNPs, particularly if the nonsynonymous SNP causes a radical change (Hughes et al. 2003). Figure 1 illustrates this trend with data from 4119 SNPs, categorized follows: synonymous; intronic; 5’ (including both the UTR and other 5’ SNPs outside the UTR); 3’; conservative nonsynonymous; and radical nonsynonymous (Hughes et al. 2005). Interestingly, there is evidence of somewhat reduced gene diversity in the 5’ region, which may include sites involved in the regulation of expression.
Figure 1.
Median gene diversity in a sample of 102 humans representing worldwide human genetic diversity at 4119 SNP sites classified by location and coding effect (Hughes et al. 2005). Median gene diversity differed significantly among categories (Kruskal-Wallis test; P < 0.001).
Results such as these suggest that the human population includes a large number of functionally important polymorphic sites at which gene diversity is reduced because of ongoing purifying selection (Hughes et al. 2003, 2005; Freudenberg-Hua et al. 2003; Sunyaev et al. 2001; Zhao et al. 2003). Because the MAF at these sites is in the range of 1–10%, these are very different from classic Mendelian disease genes, which have frequencies of one in a thousand or lower. Thus, the selectively disfavored allele at these SNP loci is likely to be only slightly deleterious. The abundance of slightly deleterious variants subject to ongoing purifying selection is precisely what the nearly-neutral theory predicts we should find in species such as humans which has undergone a prolonged bottleneck in its history, followed by a rapid expansion (Harpending et al. 1998; Li and Sadler 1991; Liu et al. 2006; Tenesa et al. 2007).
Another approach to the detection of rare nonsynonymous mutations is based on Tajima’s (1989) observation that useful inferences regarding population processes can be derived from a comparison of the average number of pairwise nucleotide differences with the number of segregating (polymorphic) sites, the latter corrected for the number of sequences compared. The difference between these two quantities divided by its standard error is Tajima’s D statistic. Under ideal conditions, a positive value of D indicates an excess of polymorphic variants at intermediate frequencies, suggestive of balancing selection, while a negative value of D indicates an excess of rare variants, indicating ongoing purifying selection.
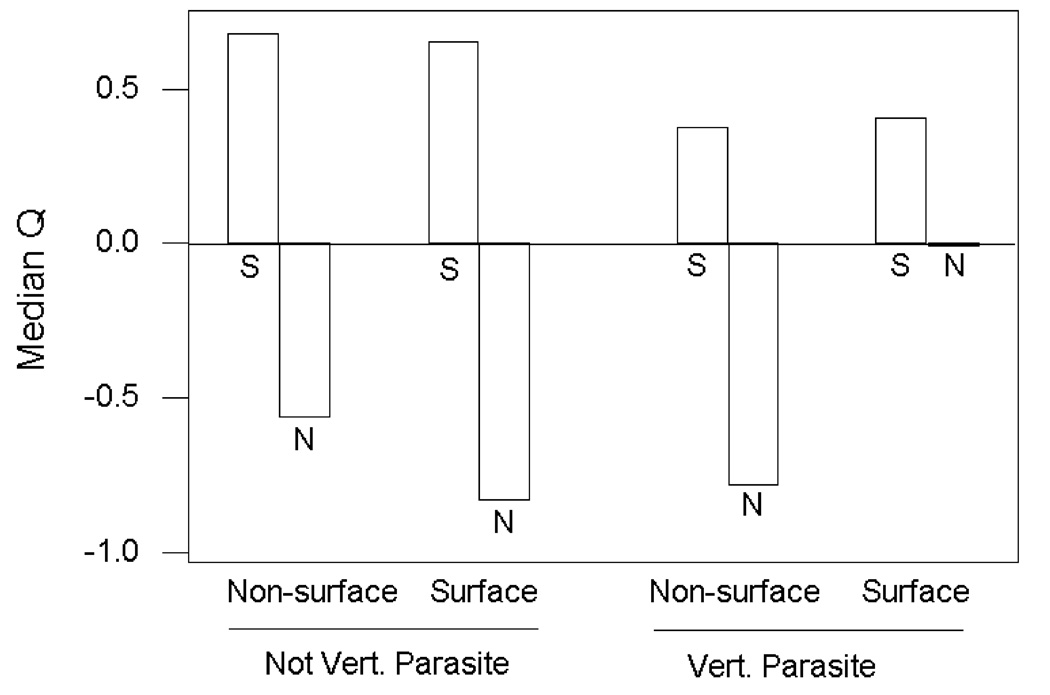
The excess of rare nonsynonymous variants in particular suggests the presence of slightly deleterious variants in the process of being reduced in frequency by purifying selection. A number of studies have applied Tajima’s D or related statistics separately to synonymous sites and nonsynonymous. When data sets from a number of different species or populations are compared, it is preferable to use the ratio (Q) of Tajima’s D to the absolute value of its theoretical minimum, since Tajima’s D is not independent of sample size (Schaeffer 2002; Hughes 2005; Hughes and Hughes 2007a). Figure 2 illustrates median values of this statistic computed separately for synonymous polymorphisms (designated Qsyn) and nonsynonymous polymorphisms (Qnon) 149 datasets of polymorphism at bacterial loci categorized as to whether the bacterial species is a parasite of vertebrates and as to whether or not the gene encodes a surface protein (Hughes 2005).
Figure 2.
Median Q (measure of skewness toward rare polymorphic variants) at synonymous (S) and nonsynonymous (N) sites in149 protein-coding genes of bacteria, categorized as to whether or not the bacterial species is parasitic on vertebrates and whether or not the protein in expressed on the bacterial cell surface (Hughes 2005). In the case of synonymous sites, median Q did not differ significantly among categories (Kruskal-Wallis test); in the case of nonsynonymous sites, there was a significant difference among categories (Kruskal-Wallis test; P < 0.001).
Median Qsyn was positive in each category, while median Qnon was always negative (Figure 2). Within each category median Qsyn was always significantly greater than Qnon (Wilcoxon signed rank test; P < 0.001 in each case). This result shows that there was an excess of rare nonsynonymous polymorphisms but not of synonymous polymorphisms. There was no significant difference among the categories with respect to median Qsyn, but there was a significant difference with respect to median Qnon (Figure 2). Median Qnon was much higher in the case of genes encoding surface proteins of bacteria parasitic on vertebrates (Figure 2).
Even in the case of genes encoding surface proteins of bacteria parasitic on vertebrates, there was more of a skew toward rare polymorphisms at nonsynonymous than at nonsynonymous sites. The latter genes are particularly important because they may include some loci at which balancing selection, driven by host immune recognition, is acting to maintain polymorphisms (Hughes 2005). Thus, even in a data set that includes some credible examples of positive selection, the results show a predominance of slightly deleterious variants at nonsynonymous polymorphic sites.
(2) In the absence of back-mutation, recombination plays an essential role in purging slightly deleterious variants
If most nonsynonymous polymorphisms are slightly deleterious, regions of low recombination are expected to show both a higher level of nonsynonymous polymorphism and a greater rate of accumulation of nonsynonymous substitutions over evolutionary time. Note that this is the opposite of what would be predicted if a substantial number of nonsynonymous variants are selectively advantageous. In the latter case, the rate of nonsynonymous substitution should be highest is regions of high recombination because recombination allows advantageous variants to become fixed.
There have been a number of recent tests of the prediction that nonsynonymous substitutions will accumulate at a more rapid rate in non-recombining regions. For example, it has been shown that the dN/dS ratio in between-species comparisons is five times or more as high for genes on non-recombining sex chromosomes (the mammalian Y or avian W) as on recombining sex chromosomes (like the mammalian X or the avian Z; Berlin and Ellgren 2006; Wyckoff et al. 2002). Moreover, Haddrill and colleagues (2007) showed that dN /dS across the Drosophila melanogaster and D. yakuba genomes was negatively correlated with the recombination rate. Recombination occurs rarely if at all between mitochondrial genomes of animals, and mitochondrial protein coding genes have been extensively sequenced. However, relatively few studies have compared mitochondrial and nuclear polymorphism within the same populations. On the basis of data summarized by Eyre Walker (2006), the ratio of the number of polymorphic nonsynonymous sites to polymorphic synonymous sites in nuclear genes of D. melanogaster is about 0.25. However, for mitochondrial genes, this ratio seems to be much higher (about 0.41; Rand et al. 1994). Likewise, Weinreich and Rand (2000) surveying published data from a number of animal species, found overall higher levels of nonsynonymous polymorphism in mitochondrial than in nuclear genes.
The previously mentioned excess of rare nonsynonymous variants in bacteria (Hughes 2005) is also consistent with the expectation that slightly deleterious mutations will accumulate when recombination is reduced, since bacteria have limited recombination. Similar reasoning can also explain an excess of nonsynonymous polymorphisms in Arabidopsis thaliana, where a mating system based on partial self-fertilization leads to reduced recombination and thus inefficient purifying selection (Bustamante et al. 2002).
Mamirova et al. (2007) studied this question further by examining seven orthologous genes of proteobacteria and the mitochondria of mammals. As might be predicted, they found that purifying selection is less efficient in obligately intracellular proteobacteria than in their free-living relatives, since obligately intracellular species have low effective population sizes and reduced opportunities for recombination (Mamirova et al. 2007). Surprisingly, however, these authors found that purifying selection is more efficient in the mitochondria than in proteobacteria. The latter result is probably due to the fact that mammalian mitochondrial genomes have a high mutation rate and an extremely high transition:transversion ratio (Vigilant et al. 1991; Belle et al. 2005). The high rate of transitional mutations leads to an enhanced possibility of back-mutation, which serves to compensate to some extent for the lack of recombination.
(3) Because speciation often involves a population bottleneck, fixation of slightly deleterious mutations will often be seen in comparisons between species
There is a great deal of evidence consistent with the above prediction, but the picture is complicated by widespread use of an inappropriate test for positive selection known as the McDonald-Kreitman (MK) test. McDonald and Kreitman (1991) pointed out that comparison of the numbers of fixed synonymous (Fs) and nonsynonymous (Fn) between two species with the numbers of segregating synonymous (Ss) and nonsynonymous (Sn) sites within one (or both) species can be informative regarding the action of natural selection. The typical MK test arranges these four quantities in the form of a contingency table. On the hypothesis that under strict neutrality Fn:Fs should equal Sn:Ss, it is argued that a pattern of Fn:Fs > Sn:Ss is evidence of positive selection favoring adaptive divergence between two species. Conversely, a pattern of Sn:Ss > Fn:Fs might be taken of as evidence of balancing selection maintaining polymorphism at the amino acid level within the species. In practice, the latter type of inference is rarely made; and the MK test is usually used to test for adaptive divergence between species.
However, the nearly-neutral theory offers an alternative explanation of a pattern where Fn:Fs > Sn:Ss; namely, fixation of slightly deleterious variants during a bottleneck accompanying speciation (Hughes 2007a; Hughes et al. 2006; Eyre-Walker et al. 2002; Ohta 1993). This pattern might be expected to be very common if bottlenecks frequently accompany speciation (e.g., DeSalle et al. 1988; Li et al. 1999; Dusfour et al. 2007). In most uses of the MK test, no effort is made to decide between these two alternative interpretations; and the pattern Fn:Fs > Sn:Ss is erroneously taken as a certain indicator of positive selection.
McDonald and Kreitman’s (1991) original paper applied the MK test to the alcohol dehydrogenase (Adh) gene of Drosophila melanogaster and found greater Fn:Fs than Sn:Ss. They mentioned that this result might be explained by the fixation of slightly deleterious alleles during a bottleneck, but they argued that the hypothesis of adaptive evolution is “simpler.” However, Ohta (1993) showed that Fn:Fs is much greater in Adh of Hawaiian Drosophila than in either of the melanogaster or obscura species groups. Because the Hawaiian species are known to have undergone population bottlenecks in speciation (Desalle and Templeton 1988), this observation strongly supports the “nearly neutral” hypothesis of fixation of slightly deleterious alleles during population bottlenecks, rather than the hypothesis of positive selection (Ohta 1993).
Aside from the failure to consider alternative hypotheses, there are numerous other problems with the MK test. As mentioned previously, there are two aspects of sequence polymorphism: (1) the average number of pairwise nucleotide differences; and (2) the number of segregating sites. In treating polymorphism, the MK test deals with the latter only, meaning that it gives excessive weight to rare polymorphisms. Moreover, several authors have developed statistics based on the MK test that are even more problematic than the test itself. For example, Rand and Kann (1996) defined a “neutrality index” (NI) as the measure (Sn/Fn)/(Ss/Fs). Adaptive divergence between species is alleged to occur when NI is less than 1. Such a measure, being a ratio of ratios, compounds the statistically undesirable properties of ratio data. Ratios are very sensitive to stochastic error, particularly in the denominator. Moreover, NI is undefined if Fn, Ss, or Fs is zero.
Data from Wise and colleagues (1998) on polymorphism in the mitochondrial NADH2 gene of human and chimpanzee illustrate the dilemmas of the MK test and NI very nicely. Since each species is used as the outgroup to the other, Fn is 10 for both species, and Fs is 82. For human, Sn is 10, while Ss is 11. For chimpanzee, Sn is 7, while Ss is 32. Thus, in the human data, NI is 7.45, while for chimpanzee NI is 1.79. For human, the MK test is highly significant (P < 0.001); but this means that Sn:Ss is significantly greater than Fn:Fs (Wise et al. 1998).
Such results are hard to explain on selectionist grounds, but are easily explained by the nearly-neutral theory. Whatever bottlenecks may have occurred at the time of the separation of human and chimpanzee lineages, the effects are not detectable in this case because each species is used as the basis of comparison for the other. However, it is well known that the chimpanzee has a larger effective population size than human, since it did not suffer the prolonged bottleneck that occurred in early modern Homo sapiens (Yu et al. 2003). Because of its larger effective population size, the chimpanzee has been able to purge slightly deleterious mutations in the NADH2 gene, resulting in low Sn:Ss (0.219). Because of its much smaller long-term effective population size, the human has not been so successful in purging slightly deleterious variants, resulting in an excess of nonsynonymous polymorphisms in the NADH2 gene, resulting in a much higher Sn:Ss (0.909).
Bazin and colleagues (2006) applied an approach based on the MK test to a data set of mitochondrial and nuclear gene sequences from a variety of animal species, yielding some provocative results. These authors also made use of a neutrality index, but it was defined somewhat differently than that of Rand and Kann (1996). Instead of counts of the numbers of segregating synonymous and nonsynonymous sites within a species, Bazin et al. (2006) estimated the synonymous nucleotide diversity (πS) and nonsynonymous nucleotide diversity (πN). Bazin et al. (2006) computed their neutrality index (NI) as (πN / πN)/(dN/dS), where dN and dS are, respectively, the numbers of synonymous substitutions per synonymous site and the number of nonsynonymous substitutions per nonsynonymous site between the species studied and a related species. Again, a value of NI less than 1 was taken as evidence of adaptive evolution between species. Using πS and πN provides a method less influenced by rare polymorphisms than are Ss and Sn, as used in the traditional MK test; but this formulation of NI is equally bedeviled by the inherent problems of ratio data.
Bazin et al. (2006) reported NI values lower than 1 for mitochondrial genes of both vertebrates and invertebrates, but greater than 1 in nuclear genes in both vertebrates and invertebrates. The observation that NI is lower for mitochondrial genes than for nuclear genes is easily explained by the nearly-neutral theory given that low NI reflects mainly the fixation of slightly deleterious mutations during speciation. Because of the difficulty of removing deleterious mutations from the non-recombining mitochondrial genome, it is consistent with the predictions of the nearly-neutral theory that dN is higher relative to dS in mitochondrial than in nuclear genes (Weinreich and Rand 2000). Thus, there is no need to invoke positive selection to explain NI values less than 1 in the case of mitochondrial genes.
A less easily explained observation was that median NI for mitochondrial genes of invertebrates was significantly lower than that of vertebrates (Bazin et al. 2006). Part of this difference may have been due to relatively low πN in mitochondrial genes of invertebrates, reflecting more efficient ongoing purifying selection in invertebrate populations than in vertebrate populations, resulting from larger effective population sizes in the former. However, Bazin et al. (2006) reported that, in between-species comparisons, mean dN/dS was significantly higher in mitochondrial genes of invertebrates than in those of invertebrates. This seems to go against what would be expected based on the assumption that effective population sizes are larger in vertebrates than invertebrates.
One possible explanation relates to the fact that effective population sizes of nuclear and mitochondrial genomes, though probably correlated, are not the same. Lynch (2007) summarized estimates of the ratio of mitochondrial effective population size to nuclear effective population size for 12 species of vertebrates (including 8 species of mammals) and 7 species of invertebrates (including 5 species of insects). The mean ratio for vertebrates (1.22) was nearly four times that for invertebrates (0.31). One possible explanation for such a difference is that in vertebrates, particularly mammals, there may be on average a greater variance in mating success among males than is seen in invertebrates. Because variance in mating success among mammals lowers the nuclear but not the mitochondrial effective population size, it increases the ratio of mitochondrial to nuclear effective population sizes (Lynch 2007). Consistent with this hypothesis is the observation that systems of female-defense polygyny (which lead to high variation in reproductive success among males) are rare in insects but common in mammals (Thornhill and Alcock 1983). Certainly, none of the invertebrates in Lynch’s (2007) data have such a mating system, while several of the mammals do. On the other hand, Lynch’s (2007) estimates of the ratio of mitochondrial effective population size to nuclear effective population size are based on the assumption of neutrality and thus might be questioned by those who take a selectionist position.
Other factors that may cause differences in the ratio of mitochondrial to nuclear effective population sizes are the mutation rate and transitional bias. As mentioned previously, a high mutation rate and a strong transitional bias help to eliminate slightly deleterious mutations, thus increasing the mitochondrial effective population size. Lynch (2007) summarizes estimates of the ratio of mitochondrial to nuclear mutation rates; these data show a mean ratio of 19.1 for vertebrates (15 taxa) as compared to 7.7 for bilaterian invertebrates (8 taxa).
Estimating dN/dS in mitochondrial genomes is further complicated by the transitional bias seen in most mitochondrial genomes (Vigilant et al. 1991; Belle et al. 2005). Transitional variants will be particularly common at synonymous sites, where most of them will probably not be subject to purifying selection. There is evidence of a saturation phenomenon, whereby the observed transition:transversion ratio declines steeply in comparisons of mitochondrial genomes from distantly related species, indicating that dS between more distantly related mitochondrial genes is likely to be substantially underestimated (Blouin et al. 1998; Tamura 1992). Thus, it is important that approximately equal distant outgroups be used for computation of dN/dS in mitochondrial genomes of different species.
Interestingly, comparison of taxa within the vertebrates and within the invertebrates in Bazin et al.’s (2006) data revealed patterns consistent with differences in effective population size. For example, because aquatic environments are buffered to some extent against such factors as climatic change that may strongly impact populations of terrestrial animals, teleost fishes might be expected to have larger effective population sizes than tetrapods; and mean dN/dS was lower for teleosts than for tetrapods. Similarly, among arthropods, crustaceans showed lower mean dN/dS than insects (Bazin et al. 2005).
It is also worth noting that numerous studies have found evidence that, contrary to the conclusion of Bazin et al. (2005), polymorphism in mitochondrial genomes reflects what we would expect based on long-term effective population size. For example, Hughes and Hughes (2007a) surveyed mitochondrial sequence data from birds and found the highest diversities in tropical mainland species, while diversities were lower in tropical island species and in temperate zone species. This result is consistent with the expectation of the highest effective population sizes in the tropical mainland species, which have generally larger ranges than island species and have not experienced the bottlenecks undergone by North Temperate Zone species due to recent glaciation. The latter, particularly Nearctic migrant species, showed evidence of recent population bottlenecks in the form of an excess of rare nonsynonymous polymorphisms (Hughes and Hughes 2007a).
Any credible hypothesis extensive between-species positive selection on mitochondrial genes requires a biological mechanism behind the alleged selection; but it is uncertain what that mechanism might be. Xenomitochondrial mice, in which mitochondria have been transplanted between one species of Mus and another, show decreasing viability as a function of increasing evolutionary distance, evidently as a result of mitochondrial-nuclear mismatch in the oxidative phosphorylation complexes (Trounce 2004); but drift alone can explain such divergences. Blier and colleagues (2006) found no differences in enzyme activity, thermal sensitivity or thermostability of mitochondrial enzymes between two fish species that evolved in very different thermal environments. These results are important because they suggest that adaptive diversification of mitochondrial genes between species has not occurred even when one might expect it on ecological grounds. On the other hand, the evidence for purifying selection on mitochondrial genes is immense; one need only mention the fact that more than 250 mitochondrially linked genetic diseases have been described in humans (Mancuso et al. 2007).
Several studies using variants of the MK test show a high level of “positive selection” on between-species amino acid differences in comparisons between Drosophila melanogaster and related Drosophila species (Sawyer et al. 2007; Shapiro et al. 2007; Smith and Eyre-Walker 2002), whereas in human-chimpanzee comparisons a much lower proportion of genes show evidence of “positive selection” (Bustamante 2005; Gojobori 2007). This is a rather surprising difference, given the pronounced morphological and behavioral differences between human and chimpanzee and the much less obvious phenotypic differences among species of the melanogaster species complex. However, this result can be explained by the way the MK test handles within-species polymorphism.
The high rate of “positive selection” detected by the MK test in the ancestry of D. melanogaster can be explained by fixation of slightly deleterious mutations during a bottleneck in the process of speciation. D. melanogaster has a very large long-term effective population size, as indicated by a high level of genetic diversity (Li and Sadler 1991). With an origin in Sub-Saharan Africa, this species was largely unaffected by Pleistocene glaciation, a major cause of bottlenecks in animal species of the North Temperate zones (Hughes and Hughes 2007a). Given a large effective population size for a long time, the nearly-neutral theory predicts that slightly deleterious mutations will have a good chance of being purged by purifying selection. Thus, the highly effective purifying selection within D. melanogaster, by lowering Sn, causes Fn to appear large by comparison.
The human species, by contrast, underwent a recent bottleneck of long duration early in the origin of modern humans (Harpending et al. 1998). As mentioned previously, the human population shows evidence of an excess of rare nonsynonymous polymorphisms, as expected if many of these polymorphisms represent slightly deleterious mutations that increased in frequency during the bottleneck and now are in the process of being eliminated by purifying selection (Hughes et al. 2003). Consistent with this interpretation, Bustamante et al. (2005) reported a genome-wide Fn:Fs ratio of 0.60 in human-chimpanzee comparisons, but an Sn:Ss ratio of 0.91 within the human species. By contrast, a recent analysis of data from D. melanogaster showed an Fn:Fs ratio of 0.37 and a Sn:Ss ratio of 0.31 (Gojobori et al. 2007). The simplest explanation for this difference is that numerous slightly deleterious polymorphisms serve to increase the relative value of Sn in humans.
Positive Selection at the Molecular Level: a Host of Unjustified Claims
Kimura never denied the importance of positive Darwinian selection in adaptive evolution. Rather, he predicted that, although genetic drift and purifying selection predominate at the molecular level, positive selection does occur, although relatively rarely. One development since the publication of Kimura’s (1983) summary of the status of the neutral theory has been an intense interest in testing for positive selection at the molecular level. Some of these tests have provided unquestionable evidence of positive selection. However, because several widely used methods of testing for positive selection are both biologically and statistically problematic, there have been a vast number of poorly justified claims of positive selection in recent years.
In the previous section, I discussed some of the problems with one widely used method of detecting positive selection, the MK test. Here I will briefly describe the problems with another widely used approach, the so-called codon-based methods. Moreover, I argue that, even aside from statistical problems, the recent quest for positive selection has been extraordinarily misguided because it has sought a “signature “ of positive selection that is in fact unlikely to occur in most cases of positive selection. I conclude that many – in fact probably almost all – claims of positive selection in the literature in fact represent cases where purifying selection is relaxed or is inefficient.
The reasoning behind both the MK test and codon based methods can be traced back to the prediction that, if purifying selection is acting to remove a substantial fraction of nonsynonymous mutations whereas synonymous mutations are neutral or nearly so, dS will exceed dN, the pattern that is in fact observed in most genes. On the other hand, if positive Darwinian selection has acted to favor repeated changes at the amino acid sequence level, one might expect a reversal of the usual pattern (Hughes and Nei 1988). The paradigmatic example of such a pattern involves the genes of the vertebrate major histocompatibility complex (MHC); and it is worthwhile to mention a few unique features of the MHC case that have often not been appreciated by evolutionary biologists (Hughes 2007a).
The MHC genes encode molecules that present peptides to T cells, and several loci are highly polymorphic. Doherty and Zinkernagel (1975) proposed that this polymorphism is maintained by overdominant selection relating to the wider immune surveillance of an individual heterozygous at the MHC loci. Because of the availability of a crystal structure of a class I MHC molecule, Hughes and Nei (1988) were able to test Doherty and Zinkernagel’s (1975) hypothesis that selection on the MHC molecules is related to their peptide-binding function. Specifically, Hughes and Nei (1988) tested the prediction that positive selection should act on the codons encoding peptide-binding region (PBR) of the MHC molecule, while purifying selection should predominate elsewhere in the coding sequence. As predicted, they found dN > dS in the codons encoding the PBR, but dS > dN in the remainder of the gene (Hughes and Nei 1988).
Several characteristics of the MHC case distinguish it from many other cases where positive selection on coding sequences has been alleged. First, Hughes and Nei (1988) were testing an a priori hypothesis based on the biological reasoning of Doherty and Zinkernagel (1975). Moreover, the type of selection acting on the PBR of the MHC molecules is such that numerous amino acid changes have been favored over time in a limited set of codons (Hughes and Hughes 1995). This unusual pattern occurs because the vertebrate MHC is involved in a coevolutionary process involving pathogen detection, but many other cases of positive selection at the molecular level are unlikely to have this property.
By contrast to the MHC case, so-called “codon-based” methods of testing for positive selection are typically applied in cases where there is no a priori hypothesis regarding the target of positive selection. Codon-based methods have been designed on the premise that the MHC example provides a “signature of positive selection”; namely, one or more codons in which dN > dS across a phylogeny. Extending this approach, so-called “branch-site” methods identify as “positively selected” a codon with dN > dS even in just one branch of a phylogeny. There are two major problems with these approaches: (1) they fail to rule out alternative hypotheses because they are based on a false premise; and (2) they target a kind of selection that is likely to be confined to only a few rare cases.
The major problem with these methods is the assumption that the existence of one or more codons with dN > dS implies the presence of positive selection. But this assumption is not true. Given the stochastic nature of the mutational process, it is to be expected that such codons will occur by chance in the absence of positive selection (Hughes and Friedman 2005). In fact, computer simulations show that even under purifying selection, individual codons with dN > dS are found at quite a high frequency. In the simulations of Zhang et al. (2005), this “signature” was found on an individual branch in a given simulated tree somewhat less than 5% of the time; but this means that it was found on at least one branch of the tree 50% of the time or more. Thus, although the “codon-based” and “branch-site” methods may be perfectly valid as tests of the hypothesis that there exist one or more codons with dN > dS, that is not the same thing as testing the hypothesis of positive selection.
Note that I am not simply arguing that these methods have a high rate of “false positives.” Rather, I am arguing that these methods are inherently invalid because they are based on a false premise. As a consequence, the vast majority of cases of “positive selection” inferred by these methods are likely to be erroneous.
Secondly, these methods focus on only one type of positive selection – selection favoring repeated amino acid changes at a limited number of sites – that is likely to be rare (Hughes 2007a). A recent survey of published cases where the molecular basis of a phenotypic change is known found that a substantial number involved single amino acid changes and/or loss of function mutations (Hoekstra and Coyne 2007). None of these cases would be detected by methods that look for repeated amino acid changes in a set of codons. Unless there is some known reason why a given gene is expected to be involved in a coevolutionary process involving protein-protein recognition, the search for a set of codons with the property dN > dS as a “signature of positive selection” represents an egregious example of barking up the wrong tree (Hughes 2007a).
When applied without a biologically meaningful a priori hypothesis, methods of detecting positive selection that look for repeated amino acid changes as a “signature” of positive selection probably mainly identify codons that are poorly aligned (Wong et al. 2008) and/or subject to little functional constraint. Thus, these methods have the dangerously misleading property that they identify as functionally important the residues that are in fact the most functionally unimportant. Unfortunately, these methods are often applied to cases – such as the evolution of major human pathogens – where an erroneous assessment of the role of selection may have serious public health consequences; for instance, in influencing the development of vaccines or other therapeutic strategies.
The nearly-neutral theory predicts that the efficiency of purifying selection changes over evolutionary time in response to changes in effective population size. When comparing species whose last common ancestor was in the distant past, it is possible that many bottlenecks and recoveries have occurred since the lineages diverged. Statistical methods may often identify as “positively selected” changes that in fact represent the fixation of slightly deleterious mutations.
Sometimes of course, when a slightly deleterious mutation is fixed during a bottleneck, afterwards an advantageous mutation that compensates for the deleterious mutation may occur and may be fixed by positive selection. Because they focus on sets of codons with multiple nonsynonymous substitutions, both MK and codon-based tests may sometimes identify as “positive selection” a mixture of slightly deleterious mutations fixed during a bottleneck and subsequently fixed “compensatory” mutations. Of course, the latter are cases of positive selection; but, if such compensatory changes provide only a return to the status quo ante-bottleneck, they are not the basis for evolutionary novelties. Moreover, these statistical methods provide no way of distinguishing which nonsynonymous changes were the deleterious ones fixed by chance and which were the compensatory ones fixed by positive selection. Thus, identifying such a mixture of deleterious and compensatory changes as “positive selection” contributes little to our understanding of the molecular basis of adaptive phenotypes.
Even if the MK test or codon-based methods identify cases where positive selection is actually occurring, this information contributes little to progress in biology because no function-based hypothesis is tested. Rather, a set of possibly positively selected amino acid changes are identified, with no information regarding the phenotypic effects of these changes or the factors that might favor them. In the pre-molecular era, evolutionary biology was handicapped by the fact that only phenotypes could be studied, with no knowledge of their genetic basis; the result was a conceptual divide between the mechanism of natural selection, acting at the nucleotide sequence level, and the phenotypic adaptations alleged to result from it. The techniques of molecular biology held the promise of bridging that gap (Hughes 1999), but that promise is threatened by the use of inappropriate statistical methods. Methods that claim to present evidence for positive selection at the sequence level – but with no evidence regarding the phenotypic effects of the allegedly selected substitutions – serve only to reintroduce the conceptual divide between natural selection and adaptation. As a consequence, the widespread use of such inappropriate methods represents a gigantic step backwards for evolutionary biology as an empirical science.
Conclusions
It took biologists 50–60 years (from The Origin of Species to the Neo-Darwinian synthesis in the 1920’s and ‘30’s) to work out the implications of the hypothesis of natural selection. During the intervening period, Darwin’s ideas went in and out of fashion but remained misunderstood and highly controversial. It seems likely that a similar process is now taking place with Kimura’s Neutral Theory of Molecular Evolution. And, if we can estimate the likely duration of this process based on the history of Darwin’s ideas, it seems likely that this process will continue for another decade or two.
An encouraging sign of progress is the recent publication of Michael Lynch’s (2007) book The Origins of Genome Architecture. Lynch (2007) develops the theme that “although small population size promotes the accumulation of mutations that are mildly deleterious in the short term, the resultant alterations to gene and genomic architecture can provide a potential setting for secondary adaptive changes” (p. 70). The originality of Lynch’s vision is that he illustrates how nonadaptive forces may have given rise to raw materials that could later be exploited by natural selection. An example is provided by introns, which have an obviously deleterious effect on the efficiency of protein synthesis but can be exploited for purposes both of regulating protein synthesis and of enhancing protein diversity. One need not agree with every scenario Lynch (2007) proposes to appreciate his conclusion that “a strong belief in Darwin’s principle of natural selection is not a sufficient condition for understanding evolution” (p. 370).
Another important development has been a renewed appreciation of the importance of mutation in evolution (Nei 2005, 2007). Molecular biology has made us aware of categories of mutation unknown to the Neo-Darwinists, including mutations in regulatory regions; complete or partial gene duplication and deletion; recombination that brings together portions of different genes (“exon shuffling”); and the restructuring of the genome by transposable elements and retroviruses. According to the hypothesis of Nei (2007), mutations fixed by genetic drift rather than natural selection play a major role in phenotypic evolution as well as in molecular evolution. Several recent studies have demonstrated that mutations in transcription factor binding sites (almost certainly not positively selected) can cause phenotypic differences among species (Borneman et al. 2007; McGregor et al. 2007).
The nearly-neutral theory plays an important role in deepening our understanding of evolution because it incorporates into evolutionary theory the important biological phenomena of slightly deleterious mutation and changes in population size. Of course, given sufficiently large effective population size, slightly deleterious mutations will eventually be eliminated or neutralized by positively selected compensatory changes. Yet slightly deleterious mutations can contribute to subsequent adaptive evolution by creating the conditions that make possible a future adaptation.
Moreover, in spite of their transient nature, slightly deleterious mutations have great interest for understanding the biology of our own species and of many other species with which we share the earth. First, because of the bottlenecked history of the human species, slightly deleterious variants appear to be especially abundant in our own species, where they may play key roles in complex disease (Hughes et al. 2003). Second, because many emerging infectious agents of humans and their domestic animals have bottlenecked population histories, slightly deleterious variants may play important roles in the evolution of infectious disease agents (Hughes 2007b; Hughes and Hughes 2007b; Pybus et al. 2007). For both of these reasons, understanding the evolutionary role of slightly deleterious mutations may have important medical applications. Finally, particularly for species of the North Temperate Zone, the recent glaciation history of the earth has had a significant impact on population history, leading to bottlenecks and the accumulation of slightly deleterious variants (Hughes and Hughes 2007a). Thus, in spite of their transient nature, the contemporary biotic world is profoundly marked by slightly deleterious mutations.
Acknowledgment
This research was supported by grant GM43940 from the National Institutes of Health.
References
- Bazin E, Glémin S, Galtier N. Population size does not influence mitochondrial genetic diversity in animals. Science. 2006;312:570–572. doi: 10.1126/science.1122033. [DOI] [PubMed] [Google Scholar]
- Belle EM, Piganeau G, Garnder M, Eyre-Walker A. An investigation of the variation in the transitional bias among various animal mitochondrial DNA. Gene. 2005;355:58–66. doi: 10.1016/j.gene.2005.05.019. [DOI] [PubMed] [Google Scholar]
- Berlin S, Ellegren H. Fast accumulation of nonsynonymous mutations on the female-specific W chromosome in birds. J. Mol. Evol. 2006;62:66–72. doi: 10.1007/s00239-005-0067-6. [DOI] [PubMed] [Google Scholar]
- Blier PU, Breton S, Desrosiers V, Lemieux H. Functional conservatism in mitochondrial evolution: insight from hybridization of arctic and brook charrs. J. Exp. Zool. (Mol. Dev. Evol.) 2006;306B:425–432. doi: 10.1002/jez.b.21089. [DOI] [PubMed] [Google Scholar]
- Blouin MS, Yowell CA, Courtney CH, Dame JB. Substitution bias, rapid saturation, and the use of mtDNA for nematode systematics. Mol. Biol. Evol. 1998;15:1719–1727. doi: 10.1093/oxfordjournals.molbev.a025898. [DOI] [PubMed] [Google Scholar]
- Borneman AR, Gianoulis TA, Zhang ZD, Yu H, Rozowsky J, Seringhaus MR, Wang LY, Gerstein M, Snyder M. Divergence of transcription factor binding sites across related yeast species. Science. 2007;317:815–819. doi: 10.1126/science.1140748. [DOI] [PubMed] [Google Scholar]
- Bustamante CD, Nielsen R, Sawyer SA, Olsen KM, Purugganan MD, Hartl DL. The cost of inbreeding in Arabidopsis. Nature. 2002;416:531–534. doi: 10.1038/416531a. [DOI] [PubMed] [Google Scholar]
- Bustamante CD, Fledel-Alon A, Williamson S, Nielsen R, Hubisz MT, Glanowski S, Tanenbaum DM, White TJ, Sninsky JJ, Hernandez RD, Civello D, Adams MD, Cargill M, Clark AG. Natural selection on protein-coding genes in the human genome. Nature. 2005;437:1153–1157. doi: 10.1038/nature04240. [DOI] [PubMed] [Google Scholar]
- Chen K, Kurgan L. PFRES: protein fold classification by using evolutionary information and predicted secondary structure. Bioinformatics. 2007;23:2843–2850. doi: 10.1093/bioinformatics/btm475. [DOI] [PubMed] [Google Scholar]
- Chothia C. Proteins. One thousand families for the molecular biologist. Nature. 1992;357:543–544. doi: 10.1038/357543a0. [DOI] [PubMed] [Google Scholar]
- Dennett DC. Darwins’s Dangerous Idea. New York: Simon and Schuster; 1995. [Google Scholar]
- Desalle R, Templeton AR. Founder effects and the rate of mitochondrial DNA evolution in Hawaiian Drosophila. Evolution. 1988;42:1076–1084. doi: 10.1111/j.1558-5646.1988.tb02525.x. [DOI] [PubMed] [Google Scholar]
- Doherty PC, Zinkernagel RM. Enhanced immunologic surveillance in mice heterozygous at the H-2 complex. Nature. 1975;256:50–52. doi: 10.1038/256050a0. [DOI] [PubMed] [Google Scholar]
- Dusfour I, Michaux J, Harbach RE, Manguin S. Speciation and phylogeography of the Southeast Asian Anopheles sundiacus complex. Infect Genet Evol. 2007;7:484–493. doi: 10.1016/j.meegid.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A. Changing effective population size and the McDonald-Kreitman test. Genetics. 2002;162:2017–2024. doi: 10.1093/genetics/162.4.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A. The genomic rate of adaptive evolution. Trends Ecol. Evol. 2006;21:569–575. doi: 10.1016/j.tree.2006.06.015. [DOI] [PubMed] [Google Scholar]
- Fisher RA. The Genetical Theory of Natural Selection. Oxford: Oxford University Press; 1930. [Google Scholar]
- Fisher RA, Ford EB. The “Sewall Wright” effect. Heredity. 1950;4:117–119. doi: 10.1038/hdy.1950.8. [DOI] [PubMed] [Google Scholar]
- Friedman R, Hughes AL. Likelihood-ratio tests for positive selection of human and mouse duplicate genes reveal non-conservative and anomalous properties of widely used methods. Mol. Phyl. Evol. 2007;42:388–397. doi: 10.1016/j.ympev.2006.07.015. [DOI] [PubMed] [Google Scholar]
- Freudenberg-Hua Y, Freudenberg J, Kluck N, Cichon S, Propping P, Nöthen MM. Single nucleotide variation analysis in 65 candidate genes for CNS disorders in a representative sample of the European population. Genome Res. 2003;13:2271–2276. doi: 10.1101/gr.1299703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gojobori J, Tang H, Akey JM, Wu C-I. Adaptive evolution in humans revealed by the negative correlation between the polymorphism and fixation phases of evolution. Proc. Natl. Acad. Sci. USA. 2007;104:3907–3912. doi: 10.1073/pnas.0605565104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ, Lewontin RC. The spandrels of San Marco and the Panglossian paradigm: a critique of the adaptationist programme. Proc. R. Soc. Lond. B. 1979;205:581–598. doi: 10.1098/rspb.1979.0086. [DOI] [PubMed] [Google Scholar]
- Haddrill PR, Halligan DL, Tomaras D, Charlesworth B. Reduced efficacy of selection in regions of the Drosophila genome that lack crossing over. Genome Biology. 2007;8:R18. doi: 10.1186/gb-2007-8-2-r18. 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harpending HC, Batzer MA, Gurven M, Jorde LB, Rogers AR, Sherry ST. Genetic traces of ancient demography. Proc. Natl. Acad. Sci. USA. 1998;95:1961–1967. doi: 10.1073/pnas.95.4.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra HE, Coyne JA. The locus of evolution: evo devo and the genetics of adaptation. Evolution. 2007;61:995–1016. doi: 10.1111/j.1558-5646.2007.00105.x. [DOI] [PubMed] [Google Scholar]
- Hughes AL. Adaptive Evolution of Genes and Genomes. New York: Oxford University Press; 1999. [Google Scholar]
- Hughes AL. Evidence for abundant slightly deleterious polymorphisms in bacterial populations. Genetics. 2005;169:533–538. doi: 10.1534/genetics.104.036939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL. Looking for Darwin in all the wrong places: the misguided quest for positive selection at the nucleotide sequence level. Heredity. 2007a;99:364–373. doi: 10.1038/sj.hdy.6801031. [DOI] [PubMed] [Google Scholar]
- Hughes AL. Micro-scale signature of purifying selection in Marburg virus genomes. Gene. 2007b;392:266–272. doi: 10.1016/j.gene.2006.12.038. [DOI] [PubMed] [Google Scholar]
- Hughes PC, Friedman R. Variation in the pattern of synonymous and nonsynonymous difference between two fungal genomes. Mol. Biol. Evol. 2005;22:1320–1324. doi: 10.1093/molbev/msi120. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Hughes MK. Natural selection on the peptide-binding regions of major histocompatibility complex molecules. Immunogenetics. 1995;42:233–243. doi: 10.1007/BF00176440. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Hughes MA. Coding sequence polymorphism in avian mitochondrial genomes reflects population histories. Mol. Ecol. 2007a;16:1369–1376. doi: 10.1111/j.1365-294X.2007.03242.x. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Hughes MA. More effective purifying selection in RNA viruses than in DNA viruses. Gene. 2007b;404:117–125. doi: 10.1016/j.gene.2007.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Nei M. Pattern of nucleotide substitution at MHC class I loci reveals overdominant selection. Nature. 1988;335:167–170. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Packer B, Welch R, Bergen AW, Chanock SJ, Yeager M. Widespread purifying selection at polymorphic sites in human protein-coding loci. Proc. Natl. Acad. Sci. USA. 2003;100:15754–15757. doi: 10.1073/pnas.2536718100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Packer B, Welsch R, Chanock SJ, Yeager M. High level of functional polymorphism indicates a unique role of natural selection at human immune system loci. Immunogenetics. 2005;57:821–827. doi: 10.1007/s00251-005-0052-7. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Friedman R, Glenn NL. The future of data analysis in evolutionary genomics. Curr. Genomics. 2006;7:227–234. [Google Scholar]
- Kimura M. Solution of a process of random genetic drift with a continuous model. Proc. Natl. Acad. Sci. USA. 1955;41:144–150. doi: 10.1073/pnas.41.3.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. Some problems of stochastic processes in genetics. Ann. Math. Stat. 1957;28:882–891. [Google Scholar]
- Kimura M. Diffusion models in population genetics. J. Appl. Prob. 1964;1:177–232. [Google Scholar]
- Kimura M. Evolutionary rate at the molecular level. Nature. 1968;217:624–626. doi: 10.1038/217624a0. [DOI] [PubMed] [Google Scholar]
- Kimura M. Preponderance of synonymous changes as evidence for the neutral theory of molecular evolution. Nature. 1977;267:275–276. doi: 10.1038/267275a0. [DOI] [PubMed] [Google Scholar]
- Kimura M. The Neutral Theory of Molecular Evolution. Cambridge: Cambridge University Press; 1983. [Google Scholar]
- Larson EJ. Evolution: The Remarkable History of a Scientific Theory. New York: Modern Library; 2004. [Google Scholar]
- Li WH. Maintenance of genetic variability under the joint effect of mutation, selection and random drift. Genetics. 1978;90:349–382. doi: 10.1093/genetics/90.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W-H, Sadler LA. Low nucleotide diversity in man. Genetics. 1991;129:513–523. doi: 10.1093/genetics/129.2.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YJ, Satta Y, Takahata N. Paleo-demography of the Drosophilamelanogaster subgroup: application of the maximum likelihood method. Genes Genet Syst. 1999;74:117–127. doi: 10.1266/ggs.74.117. [DOI] [PubMed] [Google Scholar]
- Liu H, Prugnolle F, Manica A, Balloux F. A geographically explicit genetic model of worldwide human settlement history. Am. J. Hum. Genet. 2006;79:230–237. doi: 10.1086/505436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. The Origins of Genomic Architecture. Sunderland MA: Sinauer; 2007. [Google Scholar]
- McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:114–116. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- McGregor A, Orgogozo V, Delon I, Zanet J, Srinivasan DG, Payre F, Stern DL. Morphological evolution through multiple cis-regulatory mutations at a single. Nature. 2007;448:587–590. doi: 10.1038/nature05988. [DOI] [PubMed] [Google Scholar]
- Mamirova L, Popadin K, Gelfand MS. Purifying selection in mitochondria, free-living and obligate intracellular proteobacteria. BMC Evol Biol. 2007;7:17. doi: 10.1186/1471-2148-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso M, Filosto M, Choub A, Tentorio M, Broglio L, Padovani A, Siciliano G. Mitochondrial DNA-related disorders. Disci. Rep. 2007;27:31–37. doi: 10.1007/s10540-007-9035-2. [DOI] [PubMed] [Google Scholar]
- Mayr E. The Growth of Biological Thought. Cambridge, MA: Belknap Press; 1982. [Google Scholar]
- Miyata T, Miyazawa S, Yasunaga T. Two types of amino acid substitution in protein evolution. J. Mol. Evol. 1979;12:219–236. doi: 10.1007/BF01732340. [DOI] [PubMed] [Google Scholar]
- Nei M. Molecular Evolutionary Genetics. New York: Columbia University Press; 1987. [Google Scholar]
- Nei M. Selectionism and neutralism in molecular evolution. Mol. Biol. Evol. 2005;22:2318–2342. doi: 10.1093/molbev/msi242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. The new mutation theory of phenotypic evolution. Proc. Natl. Acad. Sci. USA. 2007;104:12235–12242. doi: 10.1073/pnas.0703349104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T. Slightly deleterious mutant substitutions in evolution. Nature. 1973;246:96–98. doi: 10.1038/246096a0. [DOI] [PubMed] [Google Scholar]
- Ohta T. Amino acid substitution at the Adh locus of Drosophila is facilitated by small population size. Proc. Natl. Acad. Sci. USA. 1993;90:4548–4551. doi: 10.1073/pnas.90.10.4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T. Near-neutrality in evolution of genes and gene regulation. Proc. Natl. Acad. Sci. USA. 2002;99:16134–16137. doi: 10.1073/pnas.252626899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pybus OG, Rambaut A, Belshaw R, Freckleton RP, Drummond AJ, Holmes EC. Phylogenetic evidence for deleterious mutation load in RNA viruses and its contribution to viral evolution. Mol. Biol. Evol. 2007;24:845–852. doi: 10.1093/molbev/msm001. [DOI] [PubMed] [Google Scholar]
- Rand DM, Kann LM. Excess amino acid polymorphism in mitochondrial DNA: contrasts among genes from Drosophila, mice, and humans. Mol. Biol. Evol. 1996;13:735–748. doi: 10.1093/oxfordjournals.molbev.a025634. [DOI] [PubMed] [Google Scholar]
- Rand DM, Dorfsman M, Kann LM. Neutral and non-neutral evolution of Drosophila mitochondrial DNA. Genetics. 1994;138:741–756. doi: 10.1093/genetics/138.3.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeti PC, Schaffner SK, Fry B, Lohmueller J, Varilly P, Shamovsky O, Palma A, Mikkelsen TS, Altshuler D, Lander ES. Positive natural selection in the human lineage. Science. 2006;312:1614–1620. doi: 10.1126/science.1124309. [DOI] [PubMed] [Google Scholar]
- Sawyer SA, Parsch J, Zhang Z, Hartl DL. Prevalence of positive selection among nearly neutral amino acid replacements in Drosophila. Proc. Natl. Acad. Sci. USA. 2007;104:6504–6510. doi: 10.1073/pnas.0701572104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer SW. Molecular population genetics of sequence length diversity in the Adh region of Drosophila pseudoobscura. Genet. Res. 2002;80:163–175. doi: 10.1017/s0016672302005955. [DOI] [PubMed] [Google Scholar]
- Shapiro JA, Huang W, Zhang C, Hubisz MJ, Lu J, Turissini DA, Fang S, Wang H-Y, Hudson RR, Nielsen R, Chen Z, Wu C-I. Adaptive evolution in the Drosophila genome. Proc. Natl. Acad. Sci. USA. 2007;104:2271–2276. doi: 10.1073/pnas.0610385104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith NG, Eyre-Walker A. Adaptive evolution in Drosophila. Nature. 2002;415:1022–1024. doi: 10.1038/4151022a. [DOI] [PubMed] [Google Scholar]
- Sunyaev S, Ramensky V, Koch I, Lathe W, III, Kondrashov AS, Bork P. Prediction of deleterious human alleles. Human Mol. Genet. 2001;10:591–597. doi: 10.1093/hmg/10.6.591. [DOI] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K. The rate and pattern of nucleotide substitution in Drosophila mitochondrial DNA. Mol. Biol. Evol. 1992;9:814–825. doi: 10.1093/oxfordjournals.molbev.a040763. [DOI] [PubMed] [Google Scholar]
- Tenesa A, Navarro P, Hayes BJ, Duffy DL, Clarke GM, Goddard ME, Visscher PM. Recent human effective population size estimated from linkage disequilibrium. Genome Res. 2007;17:520–526. doi: 10.1101/gr.6023607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornhill R, Alcock M. The Evolution of Insect Mating Systems. Cambridge MA: Harvard University Press; 1983. [Google Scholar]
- Trounce IA, McKenzie M, Cassar CA, Ingraham CA, Lerner CA, Dunn DA, Donegan CL, Takeda K, Pogozelski WK, Howell RL, Pinkert CA. Development and initial characterization of xenomitochondrial mice. J. Bionergetics Biomembranes. 2004;36:421–427. doi: 10.1023/B:JOBB.0000041778.84464.16. [DOI] [PubMed] [Google Scholar]
- Vigilant L, Stoneking M, Harpending H, Hawkes K, Wilson AC. African populations and the evolution of human mitochondrial DNA. Science. 1991;253:1503–1507. doi: 10.1126/science.1840702. [DOI] [PubMed] [Google Scholar]
- Weinreich DM, Rand DM. Contrasting patterns of nonneutral evolution in proteins encoded in neuclear and mitochondrial genomes. Genetics. 2000;156:385–399. doi: 10.1093/genetics/156.1.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise CA, Sram M, Easteal S. Departure from neutrality at the mitochondrial NADH dehydrogenase subunit 2 gene in humans, but not in chimpanzees. Genetics. 1998;148:409–421. doi: 10.1093/genetics/148.1.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong KM, Suchard MA, Huelsenbeck JP. Alignment uncertainty and genomic analysis. Science. 2008;319:473–476. doi: 10.1126/science.1151532. [DOI] [PubMed] [Google Scholar]
- Wyckoff GJ, Joyce L, Wu C-I. Molecular evolution of functional genes on the mammalian Y chromosome. Mol. Biol. Evol. 2002;19:1633–1636. doi: 10.1093/oxfordjournals.molbev.a004226. [DOI] [PubMed] [Google Scholar]
- Yampolsky LY, Kondrashov FA, Kondrashov AS. Distribution of the strength of selection against amino acid replacements in human proteins. Hum. Mol. Genet. 2005;14:3191–3201. doi: 10.1093/hmg/ddi350. [DOI] [PubMed] [Google Scholar]
- Yu N, Jensen-Seaman MI, Chemnick L, Kidd JR, Deinard AS, Ryder O, Kidd KK, Li W-H. Low nucleotide diversity in chimpanzees and bonobos. Genetics. 2003;164:1511–1518. doi: 10.1093/genetics/164.4.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Nielsen R, Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol. Biol. Evol. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Fu Y-X, Hewett-Emmett D, Boerwinkle E. Investigating single nucleotide polymorphism (SNP) density in the human genome and its implications for molecular evolution. Gene. 2003;312:207–213. doi: 10.1016/s0378-1119(03)00670-x. [DOI] [PubMed] [Google Scholar]