Abstract
The flavoenzyme proline dehydrogenase catalyzes the first step of proline catabolism, the oxidation of proline to pyrroline-5-carboxylate. Here we report the first crystal structure of an irreversibly inactivated proline dehydrogenase. The 1.9 Å resolution structure of Thermus thermophilus proline dehydrogenase inactivated by the mechanism-based inhibitor N-propargylglycine shows that N5 of the flavin cofactor is covalently connected to the ε-amino group of Lys99 via a 3-carbon linkage, consistent with the mass spectral analysis of the inactivated enzyme. The isoalloxazine ring has a butterfly angle of 25°, which suggests that the flavin cofactor is reduced. Two mechanisms can account for these observations. In both, N-propargylglycine is oxidized to N-propargyliminoglycine. In one mechanism, this α,β-unsaturated iminium compound is attacked by the N5 atom of the now reduced flavin to produce a 1,4-addition product. Schiff base formation between Lys99 and the imine of the 1,4-addition product releases glycine and links the enzyme to the modified flavin. In the second mechanism, hydrolysis of N-propargyliminoglycine yields propynal and glycine. A 1,4-addition reaction with propynal coupled with Schiff base formation between Lys99 and the carbonyl group tethers the enzyme to the flavin via a 3-carbon chain. The presumed non-enzymatic hydrolysis of N-propargyliminoglycine and the subsequent rebinding of propynal to the enzyme make the latter mechanism less likely.
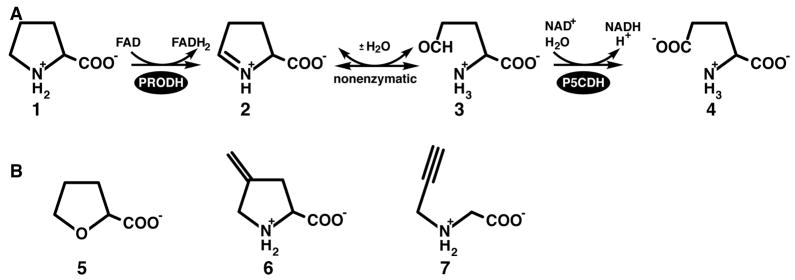
The catabolic pathway for proline involves a 4-electron oxidation to glutamate, and consists of two enzyme-catalyzed reactions coupled by a nonenzymatic equilibrium (Scheme 1A). The flavoenzyme proline dehydrogenase (PRODH) catalyzes the first reaction of the pathway, the oxidation of proline to Δ1-pyrroline-5-carboxylate (P5C) (1 to 2 of Scheme 1A). P5C is hydrolyzed nonenzymatically to glutamate semialdehyde (3). P5C dehydrogenase (P5CDH) completes the transformation of proline to glutamate by catalyzing the oxidation of glutamate semialdehyde (3 to 4). PRODH and P5CDH are separate enzymes in eukaryotes and in some bacteria, such as Thermus thermophilus, but they are fused into the bifunctional enzyme proline utilization A (PutA) in other bacteria (1–4). Human PRODH is of medical interest because it contributes to p53-mediated apoptosis by serving as a superoxide generator (5–10). Also, mutations in the PRODH gene have been linked to increased susceptibility to schizophrenia (11–13).
Scheme 1.
Reversible inhibition of PRODH by proline analogs has been well studied. Kowaloff and coworkers first reported in 1977 that L-lactate is a competitive inhibitor of PRODH activity isolated from rat liver mitochondria (14). Since then, inhibition of highly purified bacterial PRODHs and PutAs by L-tetrahydro-2-furoic acid (THFA, 5 of Scheme 1B), L-lactate and acetate has been studied in detail using structural and kinetic methods (15–17). The structures show that these inhibitors bind in the active site at the si face of the FAD isoalloxazine ring, forming electrostatic and nonpolar noncovalent interactions with conserved residues. In particular, the inhibitor carboxylate group forms ionic interactions with two conserved arginine residues and a conserved lysine. Accordingly, kinetic experiments show these compounds to be classic competitive inhibitors with inhibition constant values (Ki) ranging from sub-millimolar for THFA to 30 mM for acetate.
In contrast, there is only one report of irreversible inhibition of PRODH by a mechanism-based inactivator. Tritsch et al. showed that 4-methylene-L-proline (6, Scheme 1B) is a mechanism-based inactivator of PRODH activity from rat liver mitochondrial suspensions (18). Inactivation occurred in a pseudo-first order process (KI = 5 mM and kinact = 0.117 min−1) and PRODH activity was protected from inactivation by the addition of L-proline or L-lactate. The proposed mechanism of inactivation involves the oxidation of 6 to Δ1-pyrroline-3-methylene-5-carboxylate, followed by the nucleophilic attack of the enzyme on the electrophilic methylene carbon atom, yielding a covalently modified, inactive enzyme. The identity of the enzyme nucleophile and structure of the inactivation complex have not been reported. Motivated by the observation that propargyl-containing compounds are mechanism-based inactivators of other flavoenzymes, we have investigated the inhibition of Thermus thermophilus PRODH (TtPRODH) by N-propargylglycine (7, Scheme 1B). Results of this work are reported herein, including a crystal structure of the inactivated enzyme as well as characterizations of inactivation kinetics and spectral changes caused by inactivation.
EXPERIMENTAL PROCEDURES
Synthesis
The synthesis of N-propargylglycine was based on literature procedures (19, 20), modified as follows. Iodoacetic acid (2.5 g, 13.4 mmol) was combined with an ~8-fold excess of propargylamine (6 g, 109 mmol) and yielded 0.65 g of product (43% yield) after crystallization (3×) from ethanol. The 1H nuclear magnetic resonance (NMR) spectrum corresponded to the previously reported one (19), but was recorded in a different solvent (100 mM Na2HPO4 buffer, pH ~ 9.5) and at higher field (500 MHz). The spectrum was recorded in 100% H2O on a Varian Unity INOVA-500 spectrometer using selective pre-saturation of the water signal with a 2-s pre-saturation interval. The lock signal was dimethyl-d6 sulfoxide. Chemical shifts were standardized to the dimethyl-d6 sulfoxide signal at 2.49 ppm. 1H NMR (H2O, 500 MHz) δ 2.68 (1H, t, J = 2.53 Hz), 3.36 (2H, s), 3.57 (2H, d, J = 2.53 Hz).
Crystallization
TtPRODH was expressed and purified as previously reported (21). Purified TtPRODH (3 mg/mL) was incubated with N-propargylglycine for 30 min at a ratio of 1 mg of protein per 1 mg of inhibitor. Based on a protein monomer mass of 37,983 Da, the molar ratio of inactivator to enzyme was approximately 300. Crystals were grown using conditions similar to those used to crystallize oxidized TtPRODH (21). The inactivated enzyme was pipetted (2 μL) into sitting drop crystallization trays, mixed with an equal volume of reservoir solution, sealed with clear tape and placed in an incubator at room temperature. The optimal reservoir consisted of 100 mM imidazole buffer at pH 7, 100 mM MgCl2, 14 % 2-methyl-2,4-pentanediol and 5 mM fresh dithiothreitol. Oval-shaped crystals formed overnight and had dimensions of 0.4 mm ×0.2 mm × 0.1 mm. The crystals were pale yellow compared to oxidized TtPRODH crystals, which exhibit an intense yellow color (21). Loss of yellow color indicates that the FAD is reduced. Crystals were cryo-protected with reservoir buffer containing 25 % 2-methyl-2,4-pentanediol and inhibitor, picked up with mounting loops, and plunged into liquid nitrogen.
X-ray Diffraction Data Collection, Processing, and Refinement
A 1.9 Å resolution data set was collected at beamline 4.2.2 of the Advanced Light Source using a NOIR-1 detector. The data set used for structure determination consisted of 144 images with detector distance of 130 mm, oscillation range of 1°/frame and exposure time of 20 s/frame. Integration and scaling were performed with d*TREK (22). The space group is P212121 with unit cell dimensions a = 82.3 Å, b = 90.2 Å and c = 94.9 Å. There are two protein molecules in the asymmetric unit with 47 % solvent. The previously reported structure of oxidized TtPRODH was also determined from crystals having space group P212121 and similar unit cell lengths (a = 82.1 Å, b = 89.6 Å, c = 94.3 Å (17)). Data processing statistics are listed in Table 1.
Table 1.
Data Collection and Refinement Statisticsa
| Wavelength (Å) | 1.00 |
| Diffraction resolution (Å) | 65 – 1.9 (1.95 – 1.90) |
| No. of observations | 325777 |
| No. of unique reflections | 55959 |
| Redundancy | 5.8 (5.8) |
| Completeness (%) | 99.3 (100) |
| Rmerge | 0.059 (0.356) |
| Average I/σ | 12.5 (3.3) |
| Wilson B-factor (Å2) | 33 |
| No. of protein chains | 2 |
| No. of protein residues | 588 |
| No. of water molecules | 334 |
| No. of non-hydrogen atoms | 5222 |
| Rcryst | 0.195 (0.239) |
| Rfreeb | 0.222 (0.318) |
| RMSDc | |
| Bond lengths (Å) | 0.011 |
| Bond angles (deg.) | 1.31 |
| Ramachandran plotd | |
| Most favored (%) | 94.9 |
| Additional allowed (%) | 4.9 |
| Generously allowed (%) | 0.2 |
| Average B-factors (Å2) | |
| Protein | 32 |
| FAD | 36 |
| Lys99 side chain (CB-NZ) | 35 |
| 3-carbon link | 36 |
| Water | 37 |
| MPD | 45 |
| PDB entry | 2EKG |
The structure of inactivated TtPRODH was determined using molecular replacement as implemented in MOLREP (23). The search model consisted of the (βα)8 core (residues 40–281) of oxidized TtPRODH (PDB entry 2G37). The flavin and active site residues, including Lys99, were omitted from the search model. A clear solution was identified with 2 molecules in the asymmetric unit, correlation coefficient of 0.64 and R-factor of 0.372. The model from molecular replacement was extended and improved with several rounds of model building in COOT (24) and restrained refinement with TLS in REFMAC5 (25). TLS domains were identified with TLSMD (26). The test set used for Rfree calculations was based on the set used previously for refinement of oxidized TtPRODH due to the similarity in the crystal forms.
The final model includes residues 5 – 293 of chain A and residues −5 to 293 of chain B. (Residues numbered from −5 to zero of chain B are part of the histidine affinity tag.) Each chain contains a covalently modified flavin as described in Results. The solvent model includes 334 water molecules and four 2-methyl-2,4-pentanediol molecules. The two protein chains have similar conformations. The root mean square deviation between the two chains is 0.76 Å for 279 equivalent Cα atoms. Within the core of the enzyme (residues 40 – 281), the deviation is only 0.17 Å. The active sites of the two proteins in the asymmetric unit are also nearly identical. Thus, the discussion below will focus on only one of the chains.
Kinetic and Spectroscopic Characterization of the Inactivation of TtPRODH
Kinetic data were obtained on a Cary 100 spectrophotometer. A fresh 10 mM stock solution of N-propargylglycine (7, Scheme 1B) dissolved in 20 mM potassium phosphate buffer, pH = 7.4 was prepared. TtPRODH (10 μM) was incubated at 25 °C with N-propargylglycine at concentrations of 0.5 mM, 0.75 mM, 1.0 mM and 2.5 mM. Aliquots were removed every 3 min for 18 min and assayed at 25 °C using a previously described PRODH assay based on reduction of dichlorophenolindophenol (DCPIP) (17, 27) with L-proline (50 mM) as the substrate. Inactivation parameters were estimated using the analysis of Meloche (28) and Kitz and Wilson (29), except that a single global fitting calculation was performed rather than the standard method (30) of extracting half-lives from plots of the natural logarithm of activity versus time and making a Kitz and Wilson replot. In the analysis used here, the percentage of activity remaining was plotted versus time and the data for all four inactivator concentrations were fit globally using Origin 7.0 to Equation 1:
| (1) |
In Equation 1, A(t) is the percent activity remaining after incubating the enzyme with the inactivator at concentration I for time t. The parameters A and y0 are fitting parameters, which are determined for each inactivator concentration simultaneously. KI and kinact are the desired inactivation parameters. Note that Equation 1 combines the equation for the Kitz and Wilson replot (Equation 4 of Silverman (30)),
| (2) |
with an expression describing pseudo-first-order kinetics,
Absorbance spectra of the bound FAD cofactor were acquired at 25 °C with a Cary 100 spectrophotometer using the wavelength range 300 – 600 nm. To measure the absorbance spectrum of the oxidized enzyme, TtPRODH was added to a quartz cuvette at 5 μM in 20 mM potassium phosphate buffer at pH 7.4. A quartz cuvette with buffer served as the reference. To monitor spectral changes caused by inactivation, the inhibitor (i.e., 7) was pipetted into the cuvette containing the oxidized enzyme to a final concentration of 0.5 mM. The solution was mixed by inverting the cuvette and spectra were collected immediately after returning the cuvette to the cuvette holder and at subsequent 46-s intervals.
Mass Spectrometry
TtPRODH was dialyzed overnight against 100 mM ammonium acetate. The enzyme was then incubated with a 300-molar excess of inactivator (based on enzyme monomer mass) for 30 min and the sample was submitted to the University of Missouri Proteomics Core for analysis using nanospray QqTOF mass spectrometry.
RESULTS
Mass Spectrometry of TtPRODH Treated with N-propargylglycine
Activity assays indicated that N-propargylglycine (7) inactivated TtPRODH. To determine whether inactivation resulted in the covalent modification of enzyme, TtPRODH was incubated with N-propargylglycine and analyzed by nanospray mass spectrometry. The mass spectrum indicated that the sample contained two major components having masses of 37981 Da and 38805 Da. The 37981 Da component corresponds to the native enzyme without the flavin, which has a predicted mass of 37983 Da. Release of the flavin during nanospray ionization is consistent with the noncovalent binding of the flavin to the enzyme. The 38805 Da component corresponds to the inactivated enzyme. The difference in mass between the two components is 824 Da, which exceeds the mass of FAD (784 Da) by 40 Da. These results are consistent with inactivation resulting in covalent attachment of the flavin to the enzyme via a species derived from N-propargylglycine having three C atoms.
Structure of TtPRODH Treated with N-propargylglycine
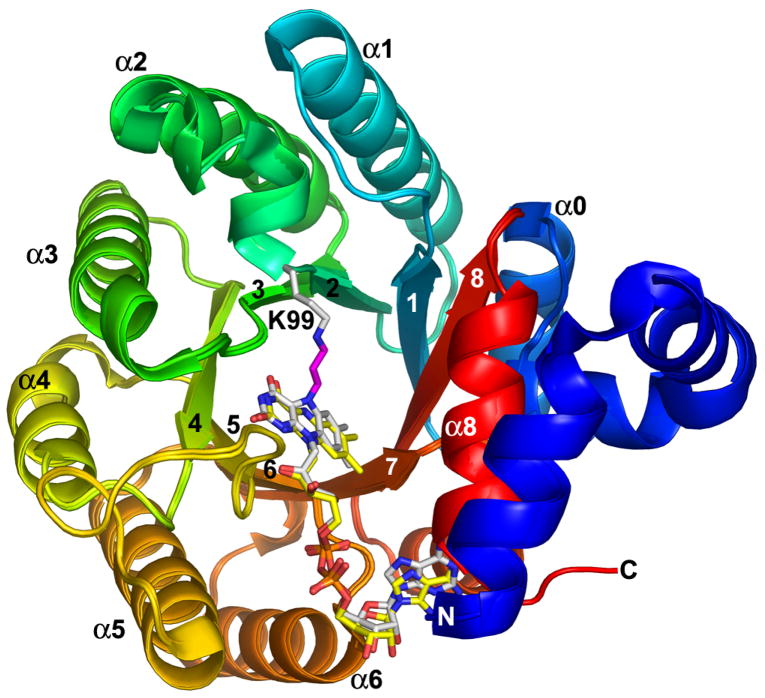
The 1.9 Å resolution crystal structure of TtPRODH inactivated by a species derived from N-propargylglycine was solved using molecular replacement (Table 1). As expected, the inactivated enzyme has the same overall fold as the oxidized enzyme (Figure 1). In fact, the root mean square deviation between the oxidized and inactivated enzymes is only 0.2 Å for Cα atoms. As shown in Figure 1, ribbon representations of the two enzymes superimpose almost perfectly.
Figure 1.
Superposition of N-propargylglycine-inactivated TtPRODH and oxidized TtPRODH. The proteins are colored using a rainbow scheme, with blue at the N-terminus and red at the C-terminus. Strands and helices of the (βα)8 barrel are labeled. The FAD cofactors are colored white in the inactivated enzyme and yellow in the oxidized enzyme. Lys99 of the inactivated enzyme is represented in white sticks. The three-carbon link between the Lys99 ε-amino group and the FAD N5 is shown in magenta. This figure and others were prepared with PyMOL (W. L. DeLano (2002) The PyMOL Molecular Graphics System).
The catalytic core of oxidized TtPRODH is a distorted (βα)8 barrel. The major distortion from the classic triosephosphate isomerase (βα)8 barrel is that the last helix of the barrel (α8) packs above the C-terminal ends of the strands of the barrel rather than alongside β8 (Figure 1). The FAD binding site is at the C-terminal ends of the strands of the barrel with the re face packed tightly against strands 4–6 and the si face available for hydride transfer from substrates (Figure 1). The distorted (βα)8 barrel fold and location of the FAD binding site are conserved in bacterial monofunctional PRODHs and PutAs (31).
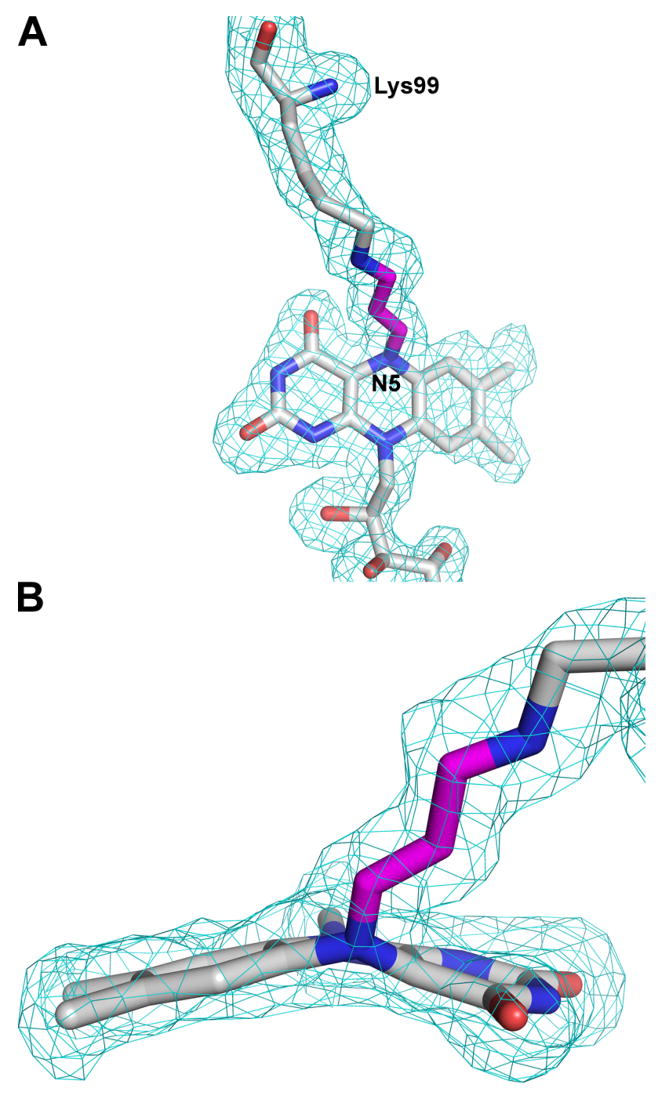
Electron density maps clearly indicated that the flavin was covalently attached to the inactivated enzyme, in contrast to the native enzyme to which the flavin binds noncovalently. A strong tube-shaped electron density feature connecting the ε-amino of Lys99 and the N5 atom of the isoalloxazine ring was observed (Figure 2A). Moreover, electron density maps indicated that the flavin isoalloxazine ring was highly bent (Figure 2B), which contrasts with the planar isoalloxazine ring of the oxidized enzyme. These features were present in both TtPRODH molecules in the asymmetric unit. A model consistent with these electron density features was built and refined.
Figure 2.
Two views of the lysine-FAD adduct along with electron density. The map is a simulated annealing σA-weighted Fo - Fc omit map contoured at 2.0 σ. Prior to map calculation, the lysine-flavin adduct was omitted and simulated annealing refinement was performed using PHENIX (47). The model shown is the final refined model. The three-carbon link between the Lys99 and the flavin N5 atom is shown in magenta.
The tubular electron density feature between Lys99 and the flavin N5 was modeled as a 3-carbon covalent linkage (Figures 2, 3). The covalent link is consistent with the mass spectral data and the proposed mechanism of inactivation (vide infra). Further support for the proposed mechanism comes from the observation that that the cofactor shifts 0.8 Å in the active site toward Lys99 (Figure 1).
Figure 3.
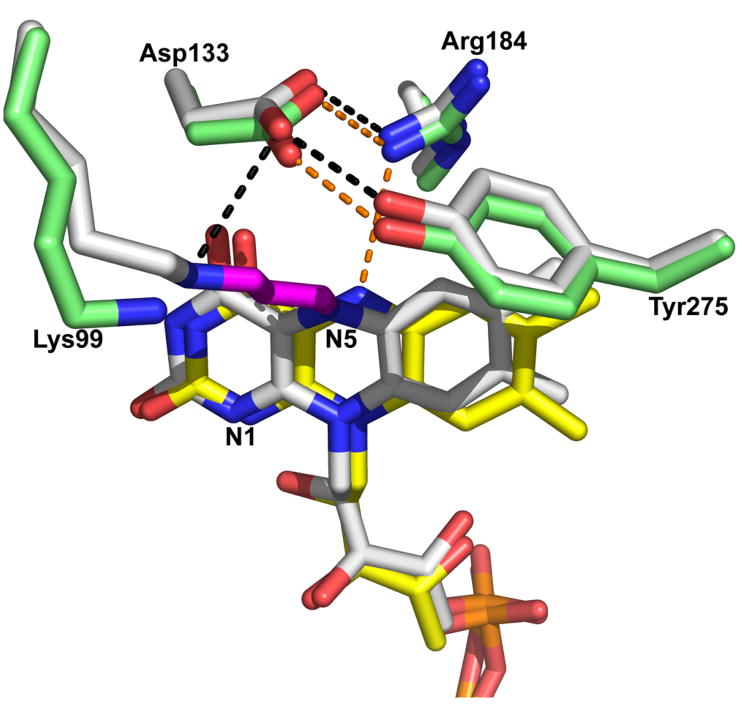
Comparison of the active sites of N-propargylglycine-inactivated TtPRODH and oxidized TtPRODH. The inactivated enzyme is colored white with hydrogen bonds represented by black dashed lines. The oxidized enzyme has yellow FAD, green protein side chains and hydrogen bonds represented by orange dashed lines.
The final refined model shows that the flavin isoalloxazine ring is bent along the N5–N10 axis such that the si face is convex (Figures 2B, 3). This type of deviation from planarity is known as butterfly bending (32). The butterfly angle of inactivated TtPRODH is approximately 25°. The planar isoalloxazine conformation is typically associated with the oxidized state of the flavin, whereas highly non-planar isoalloxazine conformations indicate reduction of the flavin. The butterfly angle of 25° observed here is rather large for flavoenzymes (32), and suggests that the FAD of inactivated TtPRODH is reduced. Aside from the butterfly bending of the isoalloxazine ring and covalent modification of N5, the conformation of the cofactor is identical to that of the oxidized enzyme (Figure 3).
Inactivation also results in small shifts (< 1 Å) of the side chains of active site residues Arg184, Asp133 and Tyr275 (Figure 3). In the oxidized enzyme, Arg184 donates a hydrogen bond to the flavin N5 (3.2 Å), while Asp133 forms an ion pair with Arg184 and a hydrogen bond with Tyr275. The hydrogen-bonding pair Asp133-Tyr275 forms the ceiling of the active site by contacting methylene groups of the proline ring (15). Inactivation of TtPRODH causes Arg184 and the flavin N5 to move apart by 0.7 Å. The hydrogen-bonding residues Asp133 and Tyr275 shift in concert 0.5 Å away from the flavin isoalloxazine to avoid steric clash with the dimethylbenzene ring (Figure 3). Despite the movements described above, the electrostatic interactions among Arg184, Asp133 and Tyr275 are maintained in the inactivated enzyme.
Spectral Changes Associated with the Inactivation of TtPRODH
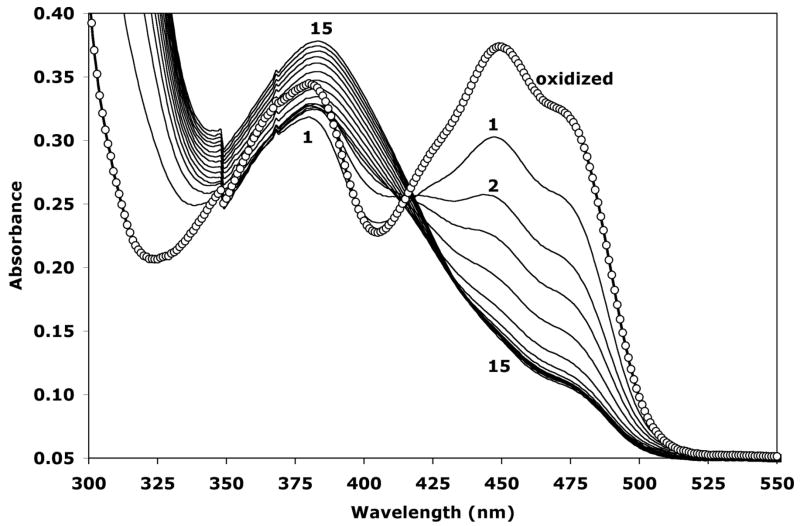
Oxidized TtPRODH shows a characteristic flavoenzyme absorption spectrum with λmax at 449 nm and 380 nm (Figure 4, curve with circles). The spectrum changed significantly upon addition of N-propargylglycine. The λmax at 449 nm disappeared over time. The λmax at 380 nm initially decreased and then gradually increased (Figure 4). Moreover, the peak at 380 nm not only increased but also shifted to a longer wavelength of 383 nm (Figure 4). The consistent decrease of A449 suggests that the cofactor is reduced during inactivation of the enzyme. In contrast, the increase in absorbance in the 380–386 nm range observed here was not observed during potentiometric and proline titrations of TtPRODH (17). Hence, these data suggest that the spectral feature in this region arises from a new flavin species that is formed during inactivation. This new species is presumably the covalently modified FAD described in the previous section.
Figure 4.
Spectral changes caused by inactivation of TtPRODH by N-propargylglycine (7). The spectrum of oxidized TtPRODH is represented by the curve with circles and has maxima at λ = 449 nm and 380 nm. Spectra 1–15 were acquired after the addition of N-propargylglycine. Curve 1 corresponds to the first spectrum obtained immediately after adding the inhibitor and mixing. Curves 2–15 were collected at 92-s intervals. Note that the maximum at λ = 449 nm disappears and a new maximum centered at λ = 383 nm appears as time advances.
Kinetic Characterization of TtPRODH Inactivation
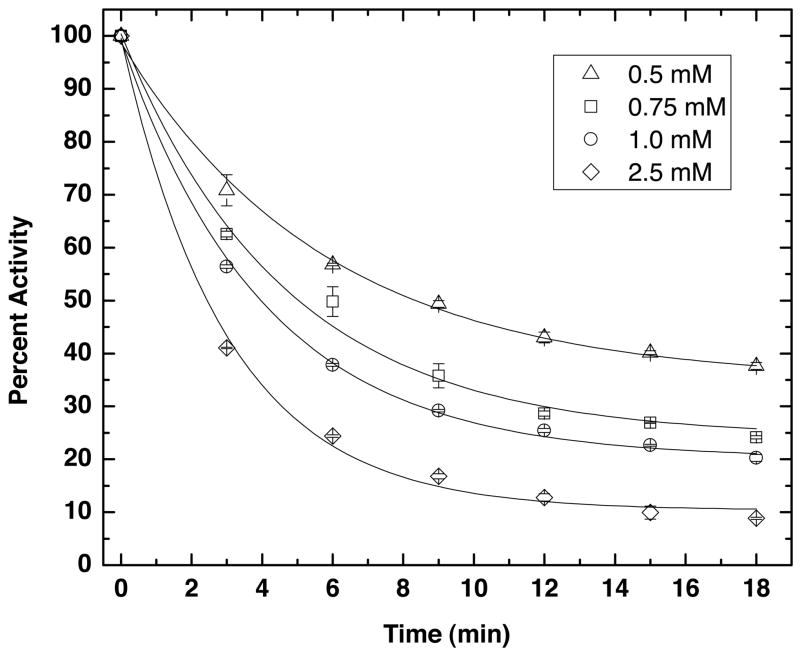
N-propargylglycine inactivated TtPRODH in a time- and concentration-dependent manner, as shown in Figure 5. Inactivation parameters were estimated by global fitting of kinetic data measured at four different inactivator concentrations to Equation 1, and resulted in values of kinact = 0.43 ± 0.05 min−1 and KI = 0.8 ± 0.2 mM. The value of 0.43 min−1 for kinact indicates that the Kitz and Wilson replot (t1/2 versus 1/I, Equation 2) intersects the ordinate above the origin at 1.6 min. This result is consistent with the formation of a dissociable complex between the enzyme and N-propargylglycine before inactivation and covalent attachment.
Figure 5.
Kinetics of TtPRODH inactivation by N-propargylglycine at 25 °C using the DCPIP activity assay. The percent activity remaining after incubation with N-propargylglycine is plotted as a function of time for four inactivator concentrations. The curves represent the best least-squares global fit to equation 1. The coefficient of determination of the fit is R2 = 0.997. The inactivation parameters obtained from fitting are kinact = 0.43 ± 0.05 min−1 and KI = 0.8 ± 0.2 mM.
DISCUSSION
Mechanism of Inactivation of TtPRODH by N-propargylglycine
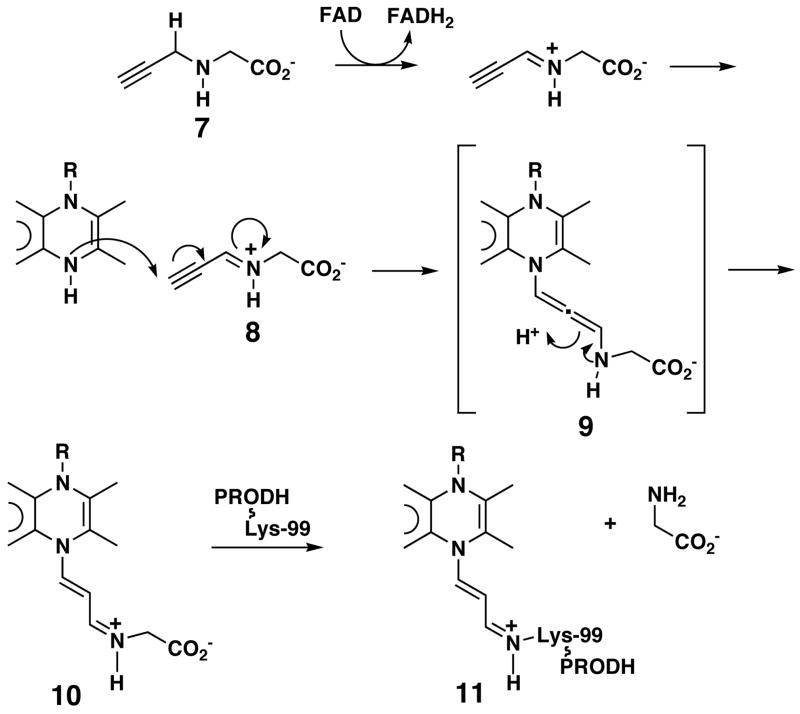
The structural, spectroscopic and mass spectral data presented here show that the FAD of N-propargylglycine-inactivated TtPRODH is reduced and that the flavin N5 is covalently attached to the ε-amino group of Lys99 by a 3-carbon chain. This active site event is also consistent with the kinetic analysis, suggesting the formation of a dissociable active site complex. These observations are consistent with two inactivation mechanisms (Schemes 2 and 3). In both mechanisms, N-propargylglycine is initially oxidized to N-propargyliminoglycine (8) with concomitant reduction of FAD to FADH2 (Schemes 2 and 3), analogous to the oxidation of proline to P5C (Scheme 1A). In the first mechanism (Scheme 2), the flavin N5 attacks the terminal carbon of 8 to form the allenic species 9, which rearranges to produce 10, and thereby completes the 1,4-addition. The ε-amino group of Lys99 can then attack the iminium ion in 10, which displaces glycine and results in the formation of an imine linkage between Lys99 and the modified flavin (11).
Scheme 2.
Scheme 3.
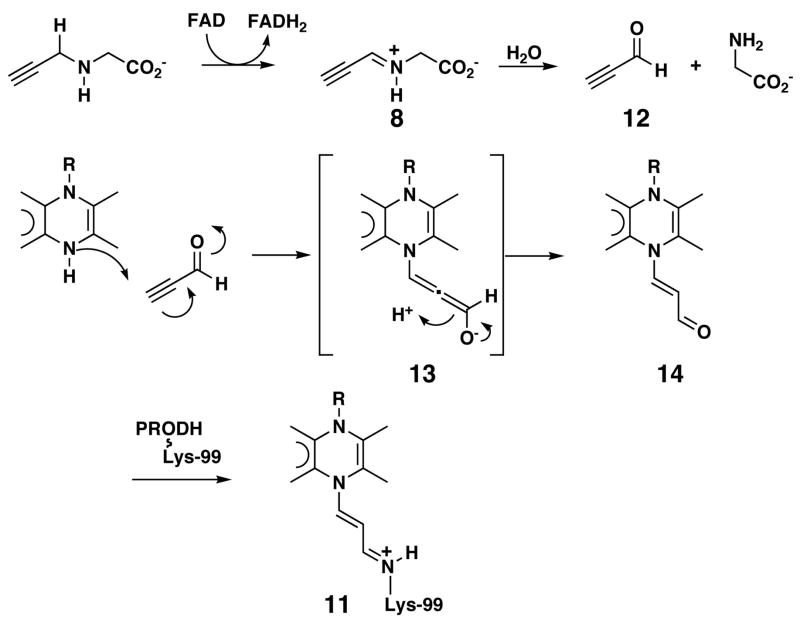
In the second mechanism, N-propargyliminoglycine is hydrolyzed to propynal (12) and glycine (Scheme 3). The hydrolysis step is analogous to the hydrolysis of P5C (2) to glutamate semialdehyde (3). Modification of the flavin N5 will then occur by the route described above where propynal is the electrophile. In this mechanism, species 8 must undergo hydrolysis either on the enzyme or in solution. Because the hydrolysis of P5C is thought to be a non-enzymatic process, it can be reasonably argued that 8 will also be hydrolyzed in solution. This step and the necessary rebinding of 12 to the enzyme make this mechanism less appealing than the one shown in Scheme 2. However, it cannot be ruled out as a possibility.
The ε-amino group of Lys99 needs to adopt the neutral ionization state to carry out nucleophilic attack on the iminium ion in 10 or to form a Schiff base with the aldehyde group of 14. If the pKa of Lys99 is unperturbed from its solution value of ~10.0, the neutral form represents about 1 % of the population of Lys99 at the pH used for inactivation (8.0). Examination of the crystal structures of the native and inactivated enzymes does not suggest an obvious mechanism to lower the pKa of Lys99. It may be that the small amount of the uncharged species present at pH 8 reacts with 10 or 14. This reaction would shift the ionization equilibrium of Lys99 and place an additional amount of the lysine in the reactive state. In this manner, all of the lysine could eventually react with either 10 or 14.
The nucleophilic attack of the flavin N5 at the acetylene group of either 8 or 12 is analogous to that proposed by Maycock and colleagues for inactivation of monoamine oxidase by dimethylpropargylglycine (33). Several observations suggest that the FAD is reduced, including the faint color of the crystals, decrease of absorbance near 450 nm during inactivation, and large butterfly angle of the isoalloxazine ring (15, 17, 34). The isoalloxazine ring is planar in structures of oxidized TtPRODH and the PutA PRODH domain. The recent structure of the E. coli PutA PRODH domain reduced by dithionite shows that reduction of the flavin induces a butterfly angle of 22° (35), which is comparable to the butterfly angle of inactivated TtPRODH.
Comparison to Structures of Other Inactivated Flavoenzymes
Mechanism-based inactivation has been studied for several flavoenzymes (33, 36–38). The inactivators used include N-propargylglycine, dimethylpropargylamine, vinylglycine, 2-hydroxy-3-butynoate, rasagiline and a propargylamine-derivatized peptide. Crystal structures of some of these irreversibly inactivated flavoenzymes have been determined. For example, Binda, et al. reported structures of inactivated monoamine oxidase B with propargyl-based inactivators, such as rasagiline (36, 37). In the rasagiline-inactivated enzyme, the flavin is modified at the N5 atom as in N-propargylglycine-inactivated TtPRODH. The structure reported here is different from rasagiline-inactivated monoamine oxidase in that the FAD in TtPRODH is covalently connected to a protein side chain. Our structure is similar to that of a histone lysine–specific demethylase inactivated by a peptide containing a propargylamine-derivatized lysine residue (38). In both TtPRODH and the demethylase, the flavin N5 is covalently tethered to a lysine side chain by a 3-carbon linker and the isoalloxazine is bent. The TtPRODH structure described here is unique in that the enzyme itself contributes the reactive lysine whereas the inactivator provides the lysine for the demethylase.
Potential Applications of Mechanism-Based Inactivators of PRODH
The discovery that N-propargylglycine is a mechanism-based inactivator of T. thermophilus PRODH provides a new avenue for developing specific inhibitors of PRODHs and PutAs. Mechanism-based PRODH inhibitors could find application as pesticides because proline, via proline catabolism, is a major fuel source for insects during flight (39, 40). Of particular interest is Glossina morsitans, a major vector for transmission of parasitic African trypanosomes, which cause trypanosomiasis (African Sleeping Sickness) and Chagas disease. In addition to the use of proline by the vector, insect stages of trypanosomatids utilize proline catabolism for energy (41). PRODHs from these organisms are thus potential targets for pesticide and anti-trypanosome development. It is notable that the only other reported mechanism-based inactivator of PRODH, 4-methylene-L-proline, was shown to be lethal to Glossina (18).
PRODH inactivators also offer a new way to modulate proline catabolism in eukaryotic cells and thus they could be useful for studying the role of PRODH in cancer and apoptosis. Human PRODH (proline-specific isoform encoded on chromosome 22q11) is a pro-apoptotic protein that helps reduce carcinogenesis in humans by serving as a superoxide generator. The gene for human PRODH is induced by the cell proliferation regulator p53. Overexpression of PRODH in colorectal cancer cell lines and various carcinomas induces p53-mediated apoptosis (5, 42–44). Further research has shown that PRODH induces apoptosis in a proline-dependent manner by superoxide generation, even without utilizing p53-mediated pathways (5). Mechanism-based inactivators of human PRODH represent a possible alternative to gene inactivation and RNAi for studying the connection between proline metabolism and apoptosis in the setting of cancer cells.
Acknowledgments
We thank Prof. Peter Tipton for insightful discussions, Prof. Michael Henzl for advice on fitting kinetic data, Dr. Jay Nix of ALS beamline 4.2.2 for help with data collection and processing, and Beverly Dague for performing mass spectrometry experiments. The ALS is supported by the Director, Office of Science, Office of Basic Energy Sciences, Materials Sciences Division, of the U. S. Department of Energy under Contract No. DE-AC03-76SF00098 at Lawrence Berkeley National Laboratory.
Footnotes
This research was supported by the National Institutes of Health Grants GM-65546 (to JJT) and GM-41239 (to CPW).
Coordinates and structure factors have been deposited in the Protein Data Bank under Accession Number 2EKG
Abbreviations: DCPIP, dichlorophenolindophenol; NMR, nuclear magnetic resonance; PRODH, proline dehydrogenase; PDB, Protein Data Bank; P5C, Δ1-pyrroline-5-carboxylate; P5CDH, Δ1-pyrroline-5-carboxylate dehydrogenase; PutA, Proline utilization A; THFA, L-tetrahydro-2-furoic acid; TtPRODH, Thermus thermophilus proline dehydrogenase.
References
- 1.Menzel R, Roth J. Enzymatic properties of the purified putA protein from Salmonella typhimurium. J Biol Chem. 1981;256:9762–9766. [PubMed] [Google Scholar]
- 2.Brown ED, Wood JM. Redesigned purification yields a fully functional PutA protein dimer from Escherichia coli. J Biol Chem. 1992;267:13086–13092. [PubMed] [Google Scholar]
- 3.Ostrovsky de Spicer P, Maloy S. PutA protein, a membrane-associated flavin dehydrogenase, acts as a redox-dependent transcriptional regulator. Proc Natl Acad Sci USA. 1993;90:4295–4298. doi: 10.1073/pnas.90.9.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker DF, Thomas EA. Redox properties of the PutA protein from Escherichia coli and the influence of the flavin redox state on PutA-DNA interactions. Biochemistry. 2001;40:4714–4721. doi: 10.1021/bi0019491. [DOI] [PubMed] [Google Scholar]
- 5.Donald SP, Sun XY, Hu CA, Yu J, Mei JM, Valle D, Phang JM. Proline oxidase, encoded by p53-induced gene-6, catalyzes the generation of proline-dependent reactive oxygen species. Cancer Res. 2001;61:1810–1815. [PubMed] [Google Scholar]
- 6.Hu CA, Donald SP, Yu J, Lin WW, Liu Z, Steel G, Obie C, Valle D, Phang JM. Overexpression of proline oxidase induces proline-dependent and mitochondria-mediated apoptosis. Mol Cell Biochem. 2006 doi: 10.1007/s11010-006-9276-6. [DOI] [PubMed] [Google Scholar]
- 7.Liu Y, Borchert GL, Donald SP, Surazynski A, Hu CA, Weydert CJ, Oberley LW, Phang JM. MnSOD inhibits proline oxidase-induced apoptosis in colorectal cancer cells. Carcinogenesis. 2005;26:1335–1342. doi: 10.1093/carcin/bgi083. [DOI] [PubMed] [Google Scholar]
- 8.Polyak K, Xia Y, Zweier JL, Kinzler KW, Vogelstein B. A model for p53-induced apoptosis. Nature. 1997;389:300–305. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 9.Pandhare J, Cooper SK, Phang JM. Proline oxidase, a proapoptotic gene, is induced by troglitazone: evidence for both peroxisome proliferator-activated receptor gamma-dependent and -independent mechanisms. J Biol Chem. 2006;281:2044–2052. doi: 10.1074/jbc.M507867200. [DOI] [PubMed] [Google Scholar]
- 10.Liu Y, Borchert GL, Surazynski A, Hu CA, Phang JM. Proline oxidase activates both intrinsic and extrinsic pathways for apoptosis: the role of ROS/superoxides, NFAT and MEK/ERK signaling. Oncogene. 2006;25:5640–5647. doi: 10.1038/sj.onc.1209564. [DOI] [PubMed] [Google Scholar]
- 11.Gogos JA, Gerber DJ. Schizophrenia susceptibility genes: emergence of positional candidates and future directions. Trends Pharmacol Sci. 2006;27:226–233. doi: 10.1016/j.tips.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 12.Liu H, Abecasis GR, Heath SC, Knowles A, Demars S, Chen YJ, Roos JL, Rapoport JL, Gogos JA, Karayiorgou M. Genetic variation in the 22q11 locus and susceptibility to schizophrenia. Proc Natl Acad Sci U S A. 2002;99:16859–16864. doi: 10.1073/pnas.232186099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu H, Heath SC, Sobin C, Roos JL, Galke BL, Blundell ML, Lenane M, Robertson B, Wijsman EM, Rapoport JL, Gogos JA, Karayiorgou M. Genetic variation at the 22q11 PRODH2/DGCR6 locus presents an unusual pattern and increases susceptibility to schizophrenia. Proc Natl Acad Sci USA. 2002;99:3717–3722. doi: 10.1073/pnas.042700699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kowaloff EM, Phang JM, Granger AS, Downing SJ. Regulation of proline oxidase activity by lactate. Proc Natl Acad Sci U S A. 1977;74:5368–5371. doi: 10.1073/pnas.74.12.5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang M, White TA, Schuermann JP, Baban BA, Becker DF, Tanner JJ. Structures of the Escherichia coli PutA proline dehydrogenase domain in complex with competitive inhibitors. Biochemistry. 2004;43:12539–12548. doi: 10.1021/bi048737e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu W, Gincherman Y, Docherty P, Spilling CD, Becker DF. Effects of proline analog binding on the spectroscopic and redox properties of PutA. Arch Biochem Biophys. 2002;408:131–136. doi: 10.1016/s0003-9861(02)00535-0. [DOI] [PubMed] [Google Scholar]
- 17.White TA, Krishnan N, Becker DF, Tanner JJ. Structure and kinetics of monofunctional proline dehydrogenase from Thermus thermophilus. J Biol Chem. 2007;282:14316–14327. doi: 10.1074/jbc.M700912200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tritsch D, Mawlawi H, Biellmann JF. Mechanism-based inhibition of proline dehydrogenase by proline analogues. Biochim Biophys Acta. 1993;1202:77–81. doi: 10.1016/0167-4838(93)90065-y. [DOI] [PubMed] [Google Scholar]
- 19.Dietrich RF, Marletta MA, Kenyon GL. Carbon-13 nuclear magnetic resonance studies of creatine, creatinine and some of their analogs. Org Magn Reson. 1980;13:79–88. [Google Scholar]
- 20.Rowley GL, Greenleaf AL, Kenyon GL. On the specificity of creatine kinase. New glycocyamines and glycocyamine analogs related to creatine. J Am Chem Soc. 1971;93:5542–5551. doi: 10.1021/ja00750a038. [DOI] [PubMed] [Google Scholar]
- 21.White TA, Tanner JJ. Cloning, purification and crystallization of Thermus thermophilus proline dehydrogenase. Acta Crystallogr. 2005;F61:737–739. doi: 10.1107/S1744309105019779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pflugrath JW. The finer things in X-ray diffraction data collection. Acta Crystallogr. 1999;55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 23.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J Appl Cryst. 1997;30:1022–1025. [Google Scholar]
- 24.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 25.Winn MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003;374:300–321. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
- 26.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. 2006;D62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 27.Abrahamson JL, Baker LG, Stephenson JT, Wood JM. Proline dehydrogenase from Escherichia coli K12. Properties of the membrane-associated enzyme. Eur J Biochem. 1983;134:77–82. doi: 10.1111/j.1432-1033.1983.tb07533.x. [DOI] [PubMed] [Google Scholar]
- 28.Meloche HP. Bromopyruvate inactivation of 2-keto-3-deoxy-6-phosphogluconic aldolase. I Kinetic evidence for active site specificity. Biochemistry. 1967;6:2273–2280. doi: 10.1021/bi00860a002. [DOI] [PubMed] [Google Scholar]
- 29.Kitz R, Wilson IB. Esters of methanesulfonic acid as irreversible inhibitors of acetylcholinesterase. J Biol Chem. 1962;237:3245–3249. [PubMed] [Google Scholar]
- 30.Silverman RB. Mechanism-based enzyme inactivators. Methods Enzymol. 1995;249:240–283. doi: 10.1016/0076-6879(95)49038-8. [DOI] [PubMed] [Google Scholar]
- 31.Tanner JJ. Structural biology of proline catabolism. Amino Acids. 2008 doi: 10.1007/s00726-008-0062-5. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lennon BW, Williams CH, Jr, Ludwig ML. Crystal structure of reduced thioredoxin reductase from Escherichia coli: structural flexibility in the isoalloxazine ring of the flavin adenine dinucleotide cofactor. Protein Sci. 1999;8:2366–2379. doi: 10.1110/ps.8.11.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maycock AL. Flavin suicide inhibitor adducts. Methods Enzymol. 1980;66:294–302. doi: 10.1016/0076-6879(80)66472-6. [DOI] [PubMed] [Google Scholar]
- 34.Lee YH, Nadaraia S, Gu D, Becker DF, Tanner JJ. Structure of the proline dehydrogenase domain of the multifunctional PutA flavoprotein. Nat Struct Biol. 2003;10:109–114. doi: 10.1038/nsb885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang W, Zhang M, Zhu W, Zhou Y, Wanduragala S, Rewinkel D, Tanner JJ, Becker DF. Redox-induced changes in flavin structure and roles of flavin N(5) and the ribityl 2′-OH group in regulating PutA--membrane binding. Biochemistry. 2007;46:483–491. doi: 10.1021/bi061935g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Binda C, Hubalek F, Li M, Herzig Y, Sterling J, Edmondson DE, Mattevi A. Crystal structures of monoamine oxidase B in complex with four inhibitors of the N-propargylaminoindan class. J Med Chem. 2004;47:1767–1774. doi: 10.1021/jm031087c. [DOI] [PubMed] [Google Scholar]
- 37.Binda C, Hubalek F, Li M, Herzig Y, Sterling J, Edmondson DE, Mattevi A. Binding of rasagiline-related inhibitors to human monoamine oxidases: a kinetic and crystallographic analysis. J Med Chem. 2005;48:8148–8154. doi: 10.1021/jm0506266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang M, Culhane JC, Szewczuk LM, Gocke CB, Brautigam CA, Tomchick DR, Machius M, Cole PA, Yu H. Structural basis of histone demethylation by LSD1 revealed by suicide inactivation. Nat Struct Mol Biol. 2007;14:535–539. doi: 10.1038/nsmb1255. [DOI] [PubMed] [Google Scholar]
- 39.Saktor B. Biochemical adaptations for flight in the insect. Biochem Soc Symp. 1976;41:111–131. [PubMed] [Google Scholar]
- 40.Custer AV. Stoichiometric estimates of the biochemical conversion efficiencies in tsetse metabolism. BMC Ecol. 2005;5:6. doi: 10.1186/1472-6785-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bringaud F, Riviere L, Coustou V. Energy metabolism of trypanosomatids: adaptation to available carbon sources. Mol Biochem Parasitol. 2006;149:1–9. doi: 10.1016/j.molbiopara.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 42.Maxwell SA, Davis GE. Differential gene expression in p53-mediated apoptosis-resistant vs. apoptosis-sensitive tumor cell lines. Proc Natl Acad Sci USA. 2000;97:13009–13014. doi: 10.1073/pnas.230445997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maxwell SA, Rivera A. Proline oxidase induces apoptosis in tumor cells, and its expression is frequently absent or reduced in renal carcinomas. J Biol Chem. 2003;278:9784–9789. doi: 10.1074/jbc.M210012200. [DOI] [PubMed] [Google Scholar]
- 44.Rivera A, Maxwell SA. The p53-induced gene-6 (proline oxidase) mediates apoptosis through a calcineurin-dependent pathway. J Biol Chem. 2005;280:29346–29354. doi: 10.1074/jbc.M504852200. [DOI] [PubMed] [Google Scholar]
- 45.Engh RA, Huber R. Accurate bond and angle parameters for x-ray protein structure refinement. Acta Crystallogr. 1991;A47:392–400. [Google Scholar]
- 46.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 47.Adams PD, Gopal K, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Pai RK, Read RJ, Romo TD, Sacchettini JC, Sauter NK, Storoni LC, Terwilliger TC. Recent developments in the PHENIX software for automated crystallographic structure determination. J Synchrotron Rad. 2004;11:53–55. doi: 10.1107/s0909049503024130. [DOI] [PubMed] [Google Scholar]