Abstract
Mitochondrial DNA is thought to be especially prone to oxidative damage by reactive oxygen species generated through electron transport during cellular respiration. This damage is mitigated primarily by the base excision repair (BER) pathway, one of the few DNA repair pathways with confirmed activity on mitochondrial DNA. Through genetic epistasis analysis of the yeast Saccharomyces cerevisiae, we examined the genetic interaction between each of the BER proteins previously shown to localize to the mitochondria. In addition, we describe a series of genetic interactions between BER components and the MutS homolog MSH1, a respiration-essential gene. We show that, in addition to their variable effects on mitochondrial function, mutant msh1 alleles conferring partial function interact genetically at different points in mitochondrial BER. In addition to this separation of function, we also found that the role of Msh1p in BER is unlikely to be involved in the avoidance of large-scale deletions and rearrangements.
DEPLETION of mitochondrial function has been implicated in the human aging process as well as in several inherited and aging-related disorders (Wallace 2005; Weissman et al. 2007). Much of this dysfunction may be attributed to mitochondrial genome instability, as the respiratory capacity of the mitochondria is dependent on an intact genome. Since respiration is essential for the survival of eukaryotic obligate aerobes, the facultative anaerobe Saccharomyces cerevisiae is an ideal model system for mitochondrial studies. Despite the difference in size between the mitochondrial genomes of yeast and humans, the encoded components are required for the same process, cellular energy production (Foury et al. 1998). Therefore, studying how S. cerevisiae maintain mitochondrial DNA (mtDNA) could lend valuable insight into mitochondrial genome maintenance in higher eukaryotes (Perocchi et al. 2008).
The necessary process of electron transport during respiration can cause damage to proteins, lipids, and nucleic acids through the formation of reactive oxygen species (ROS) (Longo et al. 1996). Because mtDNA exists in this harsh environment, it is thought that it is especially prone to oxidative damage (Bohr 2002). Damaged bases can be mutagenic by misincorporation opposite the damage by the replicative polymerase or by translesion synthesis beyond the damaged base. Therefore, the repair of oxidative lesions is essential for the stability of the mitochondrial genome.
An important mechanism for repair of oxidative DNA damage is the base excision repair (BER) pathway (Croteau and Bohr 1997; Nilsen and Krokan 2001; Bohr 2002). This pathway is well studied in the nucleus of many organisms, and isoforms of several key components have been shown to localize to the mitochondrial compartment (Rosenquist et al. 1997; You et al. 1999; Vongsamphanh et al. 2001). However, despite their extensive nuclear and biochemical characterization, the role of these isoforms in the repair of mtDNA is poorly understood.
BER is initiated when an N-glycosylase recognizes a damaged base and cleaves the glycosidic bond between it and the sugar-phosphate backbone, creating an apurinic/apyrimidinic (AP) site that can be repaired by one of two BER pathways. In short patch BER, the AP site is processed by an AP endonuclease on the 5′ side of the damaged base and by the AP lyase activity of a glycosylase, or polymerase β, on the 3′ side of the damage, to create a single-strand gap (Wilson et al. 1998). This gap is filled by a DNA polymerase and then ligated to complete the repair. In the alternative method of long-patch BER, the DNA is again cleaved by an AP endonuclease to generate an available 3′-end for synthesis by a DNA polymerase at the nick, displacing the existing sequence containing the abasic site and creating a 5′ flap. This flap is cleaved by a flap endonuclease, and the resulting nick is sealed by DNA ligase, completing the repair. Biochemical studies suggest that both short-patch and long-patch pathways are active in mitochondria (Akbari et al. 2008; Liu et al. 2008; Szczesny et al. 2008).
In this study, we examine the mitochondrial roles of Apn1p, Ntg1p, and Ogg1p, three well-studied BER components. The N-glycosylase Ogg1p is important for the repair of oxidatively damaged DNA, and studies of ogg1-Δ strains have found an increase in point mutations in both nuclear and mitochondrial DNA (Thomas et al. 1997; Singh et al. 2001). In yeast, it was previously demonstrated that a deletion of the N-glycosylase NTG1, or the AP endonuclease APN1, leads to a decrease in mitochondrial mutations as measured by rates of erythromycin resistance, suggesting that the actions of Ntg1p and Apn1p create mutagenic intermediates in mtDNA during repair (Phadnis et al. 2006). This stands in contrast to the increases seen for nuclear DNA mutation rates in the presence of these deletion alleles, indicating that it is not always possible to extrapolate the mitochondrial function of BER proteins on the basis of their nuclear functions, thus making mitochondrial-specific studies necessary (Ramotar et al. 1991, 1993; Alseth et al. 1999; Bennett 1999). In addition, there are likely to be mitochondrial-specific players in the pathway. Here we show that the mismatch repair homolog Msh1p plays multiple roles in mitochondrial BER.
Msh1p is the only one of six yeast homologs of MutS, the bacterial mismatch repair protein, which has been found localized to the mitochondria (Reenan and Kolodner 1992; Chi and Kolodner 1994). Msh1p is essential for mitochondrial function and maintenance of mtDNA, necessitating the use of partial function mutants to study the role of Msh1p in mtDNA maintenance (Mookerjee et al. 2005). Although the effects of its disruption have been examined in multiple studies, the mechanism by which Msh1p acts to carry out its essential functions remains unclear (Reenan and Kolodner 1992; Koprowski et al. 2002; Mookerjee et al. 2005; Mookerjee and Sia 2006). Its role as a mitochondrial mismatch repair protein has been disputed, particularly since there are no other mismatch repair proteins that localize to the mitochondria. However, since mtDNA has such a high potential requirement for BER, it is possible that this pathway in the mitochondria may utilize Msh1p. Previous studies have shown genetic interactions between Msh1p and the BER proteins Ogg1p, Apn1p, and Ntg1p (Dzierzbicki et al. 2004; Kaniak et al. 2009). Using msh1 alleles disrupted in conserved DNA binding and ATPase domains, we have examined the frequency and spectrum of mutations responsible for the different mutation rates seen with each allele.
MATERIALS AND METHODS
Strains and media:
Rich dextrose medium (YPD) contains 1% yeast extract, 2% Bacto peptone, and 2% dextrose. Rich glycerol medium (YPG) contains 1% yeast extract, 2% Bacto peptone, and 2% glycerol. YPD geneticin medium is YPD supplemented with 200 mg/liter geneticin (Invitrogen). YG erythromycin medium contains 1% yeast extract and 2% glycerol supplemented with 50 mm sodium phosphate buffer (pH 6.5) and 4 g/liter of erythromycin (MP Biomedicals). Synthetic dextrose media contain 0.17% yeast nitrogen base, 0.5% ammonium sulfate, 2% dextrose, and necessary amino acids. Synthetic glycerol (SGly) media contain 0.17% yeast nitrogen base, 0.5% ammonium sulfate, 2% glycerol, and necessary amino acids.
All S. cerevisiae strains used in this study are isogenic to the D273-10B-derived DFS188 strain (MATa ura3-52 leu2-3, 112 lys2 his3 arg8∷hisG; ρ+) (Sia et al. 2000) (Table 1). Primers located ∼200 bp upstream and downstream of the gene of interest were used to amplify the kanMX cassette from the S. cerevisiae deletion collection (Open Biosystems). The ogg1-Δ strain was created using primers 5′-AGGCATTTGAAGCGTCCTGATTCATAATTGCGATTTTTATTTATCAACCAGCAGATTCTAACTGAGAGTGCACC-3′ and 5′-CGCCTTTTCGGTCGCGTGCTTTTATC GTGGTATTTACTATGACTTTTTAACCTGTGCGGTATTTTCACACCGC-3′ to amplify the HIS3 gene from the pRS413 plasmid (Stratagene, La Jolla, CA). Because msh1 mutant strains are difficult to manipulate while maintaining respiratory competence, multiple methods of strain construction were utilized. Strains containing multiple deletions of BER genes or deletion of a BER gene in an msh1 mutant background were constructed by mating otherwise isogenic single mutants of opposite mating type and then sporulating and dissecting the resulting diploid strains. The strains msh1-F105A ntg1-Δ, msh1-R813W apn1-Δ, and msh1-R813W ntg1-Δ were constructed by deleting APN1 or NTG1 in the appropriate msh1 mutant background. The resulting strains were nonrespiring, so functional mtDNA was reintroduced by cytoduction. Briefly, the cells were mated with NPY3 (DFS160 ρ+), which contains a kar1-1 mutation preventing nuclear fusion, and haploid respiring cytoductants with the DFS188 nuclear background were screened for a Lys− and respiration-proficient phenotype. To construct the msh1-F105A ogg1-Δ strain, the msh1-F105A strain was transformed with the Ura-selectable pRS416-MSH1 plasmid (Mookerjee et al. 2005) to maintain a wild-type copy of Msh1p. The resulting strain was mated to DFS188 ogg1-Δ, dissected to isolate the double mutant, and then grown on 5-FOA to select for loss of the plasmid. The resulting strain was nonrespiring and was cytoduced with wild-type mtDNA as described above.
TABLE 1.
Strains used in this study
| Strain | Relevant genotype | Reference |
|---|---|---|
| DFS188 | MATaura3-52 leu2-3, 112 lys2 his3 arg8∷hisG | Sia et al. (2000) |
| NRY127 | DFS188 apn1-Δ∷kanMX | Phadnis et al. (2006) |
| RCY029 | DFS188 ntg1-Δ∷kanMX | Phadnis et al. (2006) |
| YSM130 | DFS188 ogg1-Δ∷HIS3 | This study |
| LJY148 | DFS188 apn1-Δ∷kanMX ntg1-Δ∷kanMX | This study |
| LJY400 | DFS188 apn1-Δ∷kanMX ntg1-Δ∷kanMX ogg1-Δ∷HIS3 | This study |
| LJY165 | DFS188 apn1-Δ∷kanMX ogg1-Δ∷HIS3 | This study |
| LJY134 | DFS188 ntg1-Δ∷kanMX ogg1-Δ∷HIS3 | This study |
| YSM054 | DFS188 msh1-F105A | Mookerjee and Sia (2006) |
| LJY173 | DFS188 msh1-F105A apn1-Δ∷kanMX | This study |
| LJY137 | DFS188 msh1-F105A ntg1-Δ∷kanMX | This study |
| LJY218 | DFS188 msh1-F105A ogg1-Δ∷HIS3 | This study |
| YSM084 | DFS188 msh1-G776D | Mookerjee and Sia (2006) |
| LJY156 | DFS188 msh1-G776D apn1-Δ∷kanMX | This study |
| LJY208 | DFS188 msh1-G776D ntg1-Δ∷kanMX | This study |
| YSM148 | DFS188 msh1-G776D ogg1-Δ∷HIS3 | This study |
| LJY399 | DFS188 msh1-G776D apn1-Δ∷kanMX ogg1-Δ∷HIS3 | This study |
| NPY108 | DFS188 + pRK2 | Phadnis et al. (2006) |
| NPY109 | DFS188 + pRK2 APN1 | Phadnis et al. (2006) |
| LJY382 | DFS188 msh1-G776D + pRK2 | This study |
| LJY383 | DFS188 msh1-G776D + pRK2 APN1 | This study |
| YSM105 | DFS188 msh1-R813W | Mookerjee and Sia (2006) |
| LJY183 | DFS188 msh1-R813W apn1-Δ∷kanMX | This study |
| LJY180 | DFS188 msh1-R813W ntg1-Δ∷kanMX | This study |
| EAS789 | DFS188 msh1-R813W ogg1-Δ∷HIS3 | This study |
For construction of the msh1-G776D + pRK2 and msh1-G776D + pRK2 APN1 strains, YSM84 was transformed with the appropriate plasmid. The resulting strains were nonrespiring, so functional mtDNA was reintroduced by cytoduction as described above.
Mitochondrial point mutation rates:
Point mutation rates (mutations/cell division) were estimated by performing fluctuation analysis using spontaneous resistance to the drug erythromycin (EryR) as a reporter. Independent colonies were isolated on YPG plates and grown at 30° for 3 days. Twenty independent colonies were used to inoculate 5 ml each of YPG, which were grown at 30° with agitation for 2 days. For the Apn1p overexpression studies, independent colonies were isolated on SGly–Leu media, and singles were used to inoculate SGly–Leu media and grown for 3 days. Appropriate dilutions were plated on YPG, allowing growth of all respiring cells in a culture and YG plates supplemented with 4 g/liter erythromycin to select EryR colonies. Plates were incubated at 30° for 7 days. Rates of erythromycin resistance per cell division were determined using the method of the median (Lea and Coulson 1949). Each rate was determined from at least two independent assays.
Determining the mutational spectra:
The spectra of mutations causing erythromycin resistance were determined in mutant strains by amplifying the region of the 21S rRNA gene previously shown to give rise to EryR strains from independently derived EryR colonies (Sor and Fukuhara 1982). This region, which is located at 1900–2000 on the mitochondrial genome, was amplified using primers 5′-GGTAAATAGCAGCCTTATTATG-3′ and 5′-CGATCTATCTAATTACAGTAAAGC-3′. The amplified product was sequenced at the Cornell University sequencing facility.
Respiration loss assay:
To determine the proportion of nonrespiring cells in each strain, cultures were grown overnight in YPG medium and colonies were isolated on YPG plates. Ten to 20 independent YPD cultures were inoculated with a single colony each and grown overnight, and appropriate dilutions were plated on YPG + 0.1% dextrose medium. All tests were performed in duplicate, and the average median values were used for analysis.
Statistical analysis:
Fluctuation analyses were performed two or more times. Statistical analysis was performed using Instat 3 for Macintosh (GraphPad Software, San Diego). Error is represented as ±SEM. Two-tailed P-values were calculated using unpaired t-tests to compare average rates and frequencies. Mutational spectra were compared by chi-square analysis.
RESULTS
Differential functions of BER on mitochondrial and nuclear DNA:
In the nucleus, various effects have been reported for deletion of NTG1 and APN1. Deletion of APN1 has been reported to increase nuclear mutation rates 2- to 12-fold, and reported effects of NTG1 deletion range from insignificant to 40-fold over wild-type levels (Ramotar et al. 1991, 1993; Bennett 1999; Hanna et al. 2004; Doudican et al. 2005). To determine how mtDNA is affected by these deletions, we assayed for spontaneous resistance to EryR. In agreement with previously published findings (Phadnis et al. 2006), we found that the mitochondrial point mutation rates in the ntg1-Δ strain (0.13 × 10−7 mutations/cell division) and the apn1-Δ strain (0.41 × 10−7 mutations/cell division) were significantly lower than the wild-type rate of 1.2 × 10−7 mutations/cell division (P < 0.001 in both cases) (Table 2). These findings suggest that the context in which these BER enzymes function in the mitochondria may be substantially different from that in the nucleus, despite identical biochemistry. Furthermore, we show that the rates of mitochondrial point mutations in an apn1-Δ ntg1-Δ strain (0.75 × 10−7 mutations/cell division) are not significantly different from rates of mutation in a strain containing a deletion of APN1 or NTG1 alone (P = 0.25 and 0.18, respectively), which is consistent with their action in a single genetic pathway for prevention of EryR mutations.
TABLE 2.
Rates of erythromycin resistance
| Relevant genotype | Rate of erythromycin resistance (×10−7) (standard error of the mean) |
|---|---|
| Wild type | 1.2 (0.01) |
| msh1-F105A | 6.4 (0.12) |
| msh1-F105A apn1-Δ | 25 (0.70) |
| msh1-F105A ntg1-Δ | 28 (4.29) |
| msh1-F105A ogg1-Δ | 5.2 (1.6) |
| msh1-G776D | 32 (6.0) |
| msh1-G776D apn1-Δ | 1.2 (0.18) |
| msh1-G776D ntg1-Δ | 5.0 (1.92) |
| msh1-G776D ogg1-Δ | 20 (3.98) |
| Wild type + pRK2 | 3.9 (0.58) |
| Wild type + pRK2APN1 | 11 (1.35) |
| msh1-G776D + pRK2 | 19 (4.6) |
| msh1-G776D + pRK2APN1 | 14 (3.36) |
| msh1-R813W | 4.2 (0.83) |
| msh1-R813W apn1-Δ | 9.0 (1.0) |
| msh1-R813W ntg1-Δ | 7.6 (0.80) |
| msh1-R813W ogg1-Δ | 41 (2.07) |
| apn1-Δ | 0.13 (0.01) |
| ntg1-Δ | 0.32 (0.05) |
| ogg1-Δ | 4.9 (0.6) |
| apn1-Δ ntg1-Δ | 0.75 (0.32) |
| apn1-Δ ogg1-Δ | 2.1 (0.39) |
| ntg1-Δ ogg1-Δ | 2.6 (0.65) |
| apn1-Δ ntg1-Δ ogg1-Δ | 3.5 (1.4) |
In contrast to the decreased rates in apn1-Δ and ntg1-Δ, we show that deletion of OGG1 causes a significant, almost 5-fold increase in mitochondrial point mutations (4.9 × 10−7 mutations/cell division P = 0.025), which is consistent with previous results (Singh et al. 2001). Additional deletion of APN1 or NTG1 in an ogg1-Δ strain increases the median point mutation rate by 16- and 8.2-fold relative to the rate of apn1-Δ and ntg1-Δ strains alone (P = 0.032 and 0.009, respectively). With a mitochondrial point mutation rate of 3.5 × 10−7, the apn1-Δ ntg1-Δ ogg1-Δ strain is not significantly different from either the apn1-Δ ogg1-Δ or the ntg1-Δ ogg1-Δ strain (P = 0.33 and 0.62, respectively). In contrast to their effects on mitochondrial point mutations, we have found that deletion of APN1, NTG1, or OGG1 does not significantly affect the frequency of respiration loss as compared to the wild-type strain (P = 0.40, 0.60, and 0.37, respectively) (Table 3).
TABLE 3.
Respiration loss
| Relevant genotype | % nonrespiring cells |
|---|---|
| Wild type | 0.87 |
| msh1-F105A | 88 |
| msh1-F105A apn1-Δ | 90 |
| msh1-F105A ntg1-Δ | 84 |
| msh1-F105A ogg1-Δ | 97 |
| msh1-G776D | 99 |
| msh1-G776D apn1-Δ | 98 |
| msh1-G776D ntg1-Δ | 99 |
| msh1-G776D ogg1-Δ | 99 |
| msh1-G776D apn1-Δ ogg1-Δ | >99 |
| Wild type + pRK2 | 1.1 |
| Wild type + pRK2APN1 | 0.78 |
| msh1-G776D + pRK2 | 99 |
| msh1-G776D + pRK2APN1 | 99 |
| msh1-R813W | 25 |
| msh1-R813W apn1-Δ | 19 |
| msh1-R813W ntg1-Δ | 20 |
| msh1-R813W ogg1-Δ | 12 |
| apn1-Δ | 0.26 |
| ntg1-Δ | 0.53 |
| ogg1-Δ | 1.5 |
| apn1-Δ ntg1-Δ | 0.75 |
| apn1-Δ ogg1-Δ | 2.1 |
| ntg1-Δ ogg1-Δ | 2.6 |
| apn1-Δ ntg1-Δ ogg1-Δ | 3.4 |
The difference between mitochondrial and nuclear phenotypes conferred by disruption of the BER pathway suggests a role for unique mitochondrial components. As demonstrated by epistasis analysis, we reveal specific interactions between MSH1 and the BER genes using mitochondrial point mutation accumulation as an output parameter, consistent with a role for Msh1p in BER (Table 2). Moreover, these genetic relationships are dependent on defined mutations to the conserved functional domains of MSH1, suggesting that specific activities of Msh1p contribute differently to BER as described below.
DNA binding by Msh1p occurs early in mitochondrial BER:
Mutation at a conserved phenylalanine residue within the MutS homolog mismatch recognition domain disrupts both the mismatch recognition and the DNA-binding capacities of MutS homologs (Bowers et al. 1999; Dufner et al. 2000; Yamamoto et al. 2000; Drotschmann et al. 2001; Schofield et al. 2001). In MSH1, we find that this mutation, msh1-F105A, confers a point mutation rate of 6.3 × 10−7 mutations/cell division, about fivefold over wild type (P < 0.001). This relatively mild defect is contrasted by a respiration competence that is only 10% of the wild-type strain (Tables 2 and 3). The levels of Msh1p-F105A are comparable to steady-state levels of Msh1p, demonstrating that the associated phenotypes are specific to disruption of the DNA-binding domain (Mookerjee et al. 2005). Additional deletion of OGG1 does not significantly alter the mitochondrial point mutation phenotype (P = 0.08), consistent with Msh1p-binding activity functioning upstream of Ogg1p. This finding suggests that Msh1p may bind to damaged bases or mismatches and interact with or even preferentially bind particular downstream components of BER.
In contrast to the interaction observed with the OGG1 deletion, msh1-F105A, in conjunction with the APN1 or NTG1 deletion, synergistically increases point mutation accumulation over levels observed with msh1-F105A alone (25 × 10−7 and 29 × 10−7 mutations/cell division; P < 0.001 and P = 0.026, respectively), suggesting that the repair steps following Msh1p binding are at least partially independent of pathways involving Apn1p and Ntg1p. Unlike point mutation rates, deletion of APN1, NTG1, or OGG1 does not alter the respiration loss seen in the msh1-F105A strain, indicating that the mutator phenotype conferred by the msh1-F105A allele is specifically due to a defect in BER, while respiration loss occurs through gross instability that is likely a separate feature of MSH1 disruption (Table 3).
ATP-bound Msh1p functions in downstream BER events:
The msh1-G776D substitution within the conserved ATPase domain is analogous to mutations that disrupt the ability of MutS homologs to bind ATP (Alani et al. 1997; Studamire et al. 1998). The steady-state levels of Msh1p-G776D are comparable to levels of Msh1p, demonstrating that the associated phenotypes do not result from altered protein stability (Mookerjee et al. 2005). It is the most deleterious of the three mutations studied here, conferring a 27-fold increase in point mutation rates relative to wild-type rates (32 × 10−7 mutations/cell division; P = 0.036), as well as an almost complete loss of respiratory competence (Table 2, Table 3). However, in conjunction with deletion of APN1 or NTG1, we find a complete rescue of the msh1-G776D EryR phenotype to wild-type levels (P = 0.83 and 0.19, respectively). In contrast, loss of Ogg1p, which, like Ntg1p, can initiate BER by glycosidic bond cleavage, does not significantly alter the rate conferred by msh1-G776D alone (P = 0.17). Since the activities of Apn1p and Ntg1p appear to be important for the msh1-G776D phenotype, these data are consistent with a model in which Msh1p bound to ATP functions downstream of endonucleolytic cleavage. One prediction of this model is that an excess of Apn1p would create more damage and increase the mitochondrial point mutation rate. Under these experimental conditions, we find that an extra copy of APN1 under a copper-inducible promoter increases the mutation rate relative to the wild-type rate (P = 0.04), but does not affect the mutation rate relative to msh1-G776D alone (P = 0.35).
As with msh1-F105A strains, respiration loss in msh1-G776D strains is not affected by deletion of the BER genes or overexpression of Apn1p (Table 3). However, the msh1-G776D apn1-Δ ogg1-Δ strain has an even more dramatic respiration loss than msh1-G776D alone, and it is difficult to maintain respiring cells, indicating that these proteins do play a partially redundant role in maintaining respiratory competence. With this level of mitochondrial impairment, it is impossible to accurately measure mitochondrial point mutation rates.
Msh1p ATPase activity is epistatic to APN1 or NTG1 deletion:
In contrast to the msh1-G776D allele, an R813W substitution in Msh1p presumably allows ATP binding and a binding-dependent conformational change, as in other MutS homologs, but disrupts ATP hydrolysis (Studamire et al. 1999). Therefore, comparing these mutants should allow us to differentiate between those functions that require ATPase activity and those that require only ATP binding. The msh1-R813W allele confers a 3.5-fold increase in point mutation accumulation compared to wild type, which under the conditions of these experiments is not quite significant (4.2 × 10−7 mutations/cell division; P = 0.067) (Table 2), and an ∼25% loss of respiration (Table 3). Unlike the msh1-G776D allele, the rate of EryR in msh1-R813W strains is not significantly altered by additional deletion of APN1 or NTG1 (P = 0.14 and 0.24, respectively), while deletion of OGG1 results in a 9.7-fold increase over the rate of msh1-R813W alone (41 × 10−7 mutations/cell division; P < 0.001). As with msh1-F105A and msh1-G776D, respiration loss conferred by the msh1-R813W allele is unaffected by the additional deletion of APN1, NTG1, or OGG1 individually.
Disruption of BER shifts the mutational spectra of mtDNA:
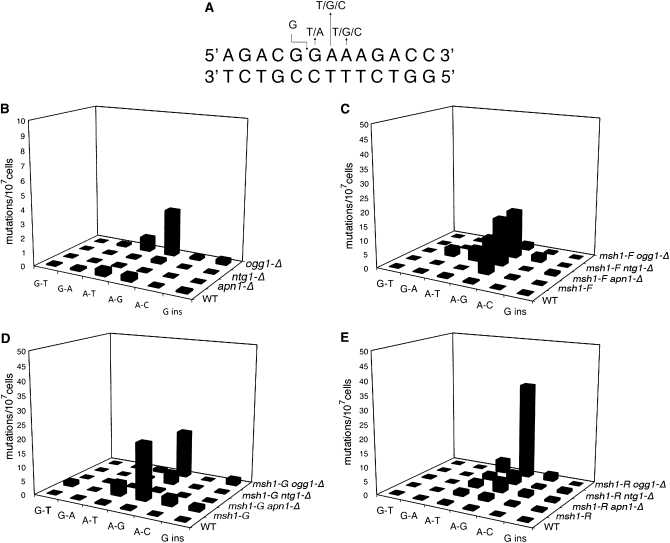
An important consideration of these studies is not only the rate of mutation accumulation, but also the mutational spectrum, which can indicate a specific mutational mechanism. Because a different biochemical activity is disrupted in each of the msh1-F105A, msh1-G776D, and msh1-R813W mutants, they may display different mutator effects. To determine the frequencies of mutational events responsible for EryR with each of the msh1 alleles, the region from 1900 to 2000 of the mitochondrial 21S rRNA gene was sequenced from independently isolated EryR colonies to determine the types of substitutions that give rise to EryR in individual colonies. Contained within this 100-bp region is a stretch of four nucleotides whose mutation commonly confers EryR (Figure 1A) (Vanderstraeten et al. 1998; Kalifa and Sia 2007). All sequenced EryR clones in this study resulted from mutations in this region. In wild-type EryR strains, the majority of mutations found are A-to-G transitions and A-to-T transversions at frequencies that are not affected by deletion of APN1 (P = 0.61 and P = 1.0). However, deletion of NTG1 causes a significant shift toward A-to-G transitions as compared to the wild-type strain (P = 0.047) (Table 4).
Figure 1.—
(A) The sequence from 1945 to 1957 of the 21S rRNA gene encoded in the mitochondrial genome, the most common site of nucleotide substitutions conferring erythromycin resistance. (B–E) Frequency of base changes in EryR mutant strains. The spectrum of mutations for each strain was determined by sequencing from independent erythromycin-resistant strains, as described in materials and methods. Graphs represent data from 15 to 50 independent colonies for each strain. The frequency of each type of substitution was estimated by multiplying the fraction of each type by the average frequency of EryR for each strain.
TABLE 4.
Spectra of mutations conferring erythromycin resistance
| % with nucleotide substitutions
|
|||||||
|---|---|---|---|---|---|---|---|
| Relevant genotype | A to C | A to G | A to T | G to A | G to T | G insertion | Total sequenced |
| Wild type | 3 | 34 | 13 | 14 | 6 | 9 | 35 |
| msh1-F105A | 5 | 95 | 0 | 0 | 0 | 0 | 19 |
| msh1-F105A apn1-Δ | 0 | 67 | 18 | 12 | 0 | 3 | 33 |
| msh1-F105A ntg1-Δ | 11 | 79 | 26 | 0 | 0 | 0 | 19 |
| msh1-F105A ogg1-Δ | 12 | 76 | 12 | 0 | 0 | 0 | 17 |
| msh1-G776D | 10 | 65 | 15 | 0 | 5 | 5 | 20 |
| msh1-G776D apn1-Δ | 21 | 42 | 11 | 21 | 0 | 5 | 19 |
| msh1-G776D ntg1-Δ | 5 | 82 | 9 | 0 | 0 | 5 | 22 |
| msh1-G776D ogg1-Δ | 0 | 84 | 5 | 0 | 0 | 11 | 19 |
| Wild type + pRK2 | 14 | 47 | 21 | 7 | 0 | 0 | 14 |
| Wild type + pRK2APN1 | 23 | 46 | 23 | 8 | 0 | 0 | 13 |
| msh1-G776D + pRK2 | 3 | 69 | 22 | 3 | 0 | 3 | 32 |
| msh1-G776D + pRK2APN1 | 3 | 79 | 9 | 3 | 0 | 6 | 34 |
| msh1-R813W | 15 | 40 | 35 | 0 | 0 | 10 | 20 |
| msh1-R813W apn1-Δ | 18 | 49 | 23 | 0 | 3 | 8 | 39 |
| msh1-R813W ntg1-Δ | 8 | 54 | 26 | 0 | 0 | 13 | 39 |
| msh1-R813W ogg1-Δ | 6 | 83 | 11 | 0 | 0 | 0 | 18 |
| apn1-Δ | 16 | 28 | 38 | 16 | 3 | 0 | 32 |
| ntg1-Δ | 6 | 61 | 19 | 6 | 0 | 6 | 31 |
| ogg1-Δ | 3 | 67 | 20 | 3 | 0 | 7 | 30 |
Ogg1p recognizes and excises multiple types of damaged bases from DNA. Its most well-studied substrate is 8-oxo-7,8-dihydroguanine (8-oxo-G), but it acts with similar efficiencies on 7,8-dihydro-8-oxoadenine, 2,6-diamino-4-hydroxy-5-formamidopyrimidine, and 4,6-diamino-5-formamidopyrimidine (FapyA) (van der Kemp et al. 1996; Klungland et al. 1999; Krishnamurthy et al. 2008). It has been shown that 8-oxo-G mispairs with adenine, and in subsequent rounds of replication, causes a G-to-T transversion (Grollman and Moriya 1993). We observed a 6% frequency of G to T in our wild-type strains, indicating that, while detectable, it is not a highly represented mutation. We expected that deletion of OGG1 would increase G-to-T events through failure of 8-oxo-G repair. Surprisingly, we found that, in an ogg1-Δ strain, none of 30 EryR colonies sequenced contained this mutation. However, 67% of these contained an A-to-G transition, almost double the 34% A-to-G frequency of EryR colonies in wild-type cells (P = 0.013). These data suggest that G-to-T substitutions do occur and yet are not increased by loss of Ogg1p. Instead, it suggests that, in the mitochondria, Ogg1p preferentially prevents mutations caused by damaged adenine or thymine. On the basis of studies of mtDNA in mammalian systems (Tanaka and Ozawa 1994; Reyes et al. 1998), we predict that adenine is more likely to be the damaged base. Both FapyA and hypoxanthine lesions have been shown to mispair with cytosine, resulting in A-to-G mutations in subsequent rounds of replication, and these represent significant lesions in mtDNA (Reyes et al. 1998; Tudek 2003).
Erythromycin resistance in msh1-F105A strains results from A-to-G substitutions in 95% of the mutants sequenced (Table 4). This mutational spectrum is also significantly shifted toward A-to-G transitions in the msh1-G776D strain (P = 0.048), although not in the msh1-R813W strain (P = 0.77). Using the data presented in Tables 2 and 4, we can estimate the frequency of each type of base substitution observed. As shown in Figure 1, B–E, the frequency of mutational changes at adenine residues increases in ogg1-Δ and msh1 mutant strains. These data indicate that each msh1 allele affects both the rate and the spectrum of mutation differently. In addition, we find that, when the mutation rates are reduced to wild-type levels in the msh1-G776D apn1-Δ and msh1-G776D ntg1-Δ strains, the mutational spectra are indistinguishable from apn1-Δ and ntg1-Δ strains with respect to the number of A-to-G substitutions (P = 0.37 and 0.14, respectively).
DISCUSSION
The importance of maintaining the mitochondrial genome for respiratory function cannot be overstated, as is demonstrated by the variety and severity of inherited mitochondrial diseases, as well as the correlation between mtDNA mutation accumulation, ROS production, and aging (Loeb et al. 2005; Kai et al. 2006; Vermulst et al. 2007). To understand how accumulating mtDNA mutations affect cellular functions over time, we need to elucidate the mechanisms that contribute to mtDNA maintenance. Base excision repair was the earliest confirmed pathway of mtDNA repair, and its presence is particularly intriguing, given the prevalent assumption that mtDNA is highly susceptible to oxidative damage, the main substrate for BER activity.
In the nuclear model of both short-patch and long-patch BER, AP endonuclease cleavage is a required intermediate step following base removal. The similarities between apn1-Δ and ntg1-Δ, and their disparity to ogg1-Δ with respect to point mutation accumulation, suggest that, in the mitochondria, Apn1p and Ntg1p are connected in function while Ogg1p function is at least partially independent. The finding that accumulation of mitochondrial point mutations in the apn1-Δ ntg1-Δ ogg1-Δ strain is similar to that in both the apn1-Δ ogg1-Δ and ntg1-Δ ogg1-Δ strains further supports the connection between Apn1p and Ntg1p. We hypothesize that Apn1p and Ntg1p form a functional complex in mitochondria. Such an Apn1p–Ntg1p complex may function in a nucleotide incision repair (NIR) role where glycosylase activity is absent and endonucleolytic cleavage initiates repair, while Ogg1p plays no role in this type of repair. The strong mutational shift toward A-to-G transitions in both ogg1-Δ and ntg1-Δ strains, but not in apn1-Δ, however, may suggest that NIR activity is not primarily acting on damaged adenine nucleotides and that Apn1p and Ntg1p activities are not completely interdependent. Unfortunately, the protein components of short-patch BER, long-patch BER, and NIR overlap, making it difficult to separate the contribution of each pathway by genetic analysis.
Msh1p appears to constitute an additional component of the mitochondrial BER pathways. Each of the mutant alleles results in a unique set of phenotypes consistent with multiple roles for Msh1p in BER, as well as with additional roles in genome stability. Previously, we attributed the mutator phenotypes resulting from the msh1-G776D, msh1-R813W, and msh1-F105A alleles to defects in the same Msh1p mechanism (Mookerjee et al. 2005). However, we observe here that the deleterious effects of these alleles on point mutation are differentially dependent on the activities of BER enzymes. For example, the increased point mutation rates in msh1-G776D mutant strains seem to require functional Apn1p and Ntg1p, presumably due to the normal role of Msh1p in an Apn1p- and Ntg1p-dependent base excision repair pathway. In contrast, point mutation accumulation in msh1-R813W strains is unaffected by deletion of APN1 or NTG1, and the msh1-F105A allele appears defective independently from an Apn1p- and Ntg1p-containing pathway.
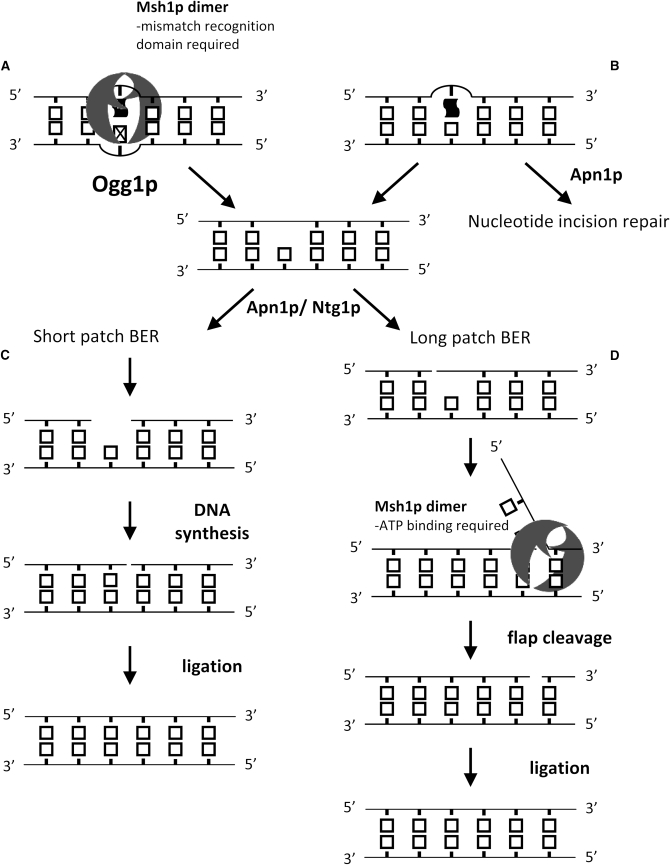
These data support a model for Msh1p function in mitochondrial BER in which at least two partially overlapping pathways exist (Figure 2). Because disruption of the mismatch recognition/DNA-binding domain in the msh1-F105A allele confers similar phenotypes with regards to point mutation rates and mutational spectra as the deletion of OGG1 does, we propose that Msh1p may aid in the function of Ogg1p through its DNA-binding activity (Figure 2A). It is possible that Msh1p recognizes a specific DNA aberration and directs Ogg1p to the damaged site or stabilizes an existing association. In Figure 2A, we have depicted the Msh1p substrate as an oxidative lesion opposite a mispair. Such binding is consistent with the known binding specificity of the human MutSα complex (Mazurek et al. 2002; Larson et al. 2003), but for Msh1p we do not exclude recognition of other lesions. Because deletion of APN1 and NTG1 results in similar phenotypes, we propose that they act in a complex that does not require Msh1p binding to initiate repair (Figure 2B) but does require that Msh1p bind ATP to complete the repair (Figure 2D). In our model, we have depicted Msh1p bound to a flap intermediate since Msh2p–Msh6p complexes have been demonstrated to bind such structures (Surtees and Alani 2006). Studamire et al. have shown that the nuclear Msh2p–Msh3p complex is important for single-strand annealing, presumably by binding flapped DNA structures. This activity is disrupted by a mutation in MSH2 analogous to msh1-G776D but not by the mutation analogous to msh1-R813W. These data suggest that the ability of the Msh2p–Msh3p complex to bind ATP, but not necessarily to hydrolyze it, is necessary for this structure recognition (Studamire et al. 1998). Our data suggest a similar separation of function for Msh1p, where the mutator phenotype of msh1-R813W is only slightly enhanced while it is dramatically increased in msh1-G776D.
Figure 2.—
Proposed model for Msh1p in mitochondrial BER. Distorted solid boxes (A and B) represent damaged bases, while the box with an “X” (A) represents a mismatch across from a damaged base after replication by a translesion polymerase. The double half-moon structures represent Msh1p dimers.
The disparities between the point mutation rates and respiration loss in the msh1 mutants suggest that the participation of Msh1p in BER is likely unrelated to its role in maintaining larger-scale mtDNA stability. Respiration loss is a common marker for the overall stability of the mitochondrial genome. Cells can lose respiration both through accumulation of point mutations in the mitochondrial genome and through large-scale rearrangements and deletions of mtDNA. In fact, such rearrangements, resulting in ρ− petites, are the primary mechanisms that generate nonrespiring cells in laboratory yeast cultures (Dujon 1981). The frequency at which these events occur is high and may result in respiration loss in several percentages of cells in a culture after overnight growth. Point mutation rates, which occur on the order of one in 107 cells for EryR, would have to increase to phenomenal levels before they would begin to be observed as increases in respiration loss over this background of ρ− cells. Consequently, increases in point mutations do not necessarily result in a correlative effect on respiration loss. So, while the mitochondrial point mutations in the msh1-G776D mutant strain can be suppressed by deletion of APN1 or NTG1, the high rate of respiration loss is not similarly reduced. Therefore, the essential function of Msh1p is to stabilize the mitochondrial genome through a broader mechanism than point mutation prevention.
Most of the proteins functioning within the mitochondria are encoded in the nucleus, and many have additional, well-studied roles throughout the cell. This study exemplifies the necessity for examination of mitochondrial-specific roles of proteins that also function elsewhere. We have shown that Apn1p, Ntg1p, and Ogg1p have unexpected mitochondrial phenotypes and that their function is closely tied to the mitochondrial specific protein, Msh1p. We have also demonstrated that Msh1p plays multiple roles in mitochondrial base excision repair but that this function is separate from a broader role in maintaining respiratory competence of the mitochondria.
Acknowledgments
We thank Linnell Randall and Shweta Krishnan for help with the fluctuation analysis. We also thank Lidza Kalifa for critical reading of the manuscript. This work was supported by grants from the National Science Foundation (MCB0543084) and the National Institutes of Health (GM63626).
References
- Akbari, M., T. Visnes, H. E. Krokan and M. Otterlei, 2008. Mitochondrial base excision repair of uracil and AP sites takes place by single-nucleotide insertion and long-patch DNA synthesis. DNA Repair (Amst.) 7 605–616. [DOI] [PubMed] [Google Scholar]
- Alani, E., T. Sokolsky, B. Studamire, J. J. Miret and R. S. Lahue, 1997. Genetic and biochemical analysis of Msh2p-Msh6p: role of ATP hydrolysis and Msh2p-Msh6p subunit interactions in mismatch base pair recognition. Mol. Cell. Biol. 17 2436–2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alseth, I., L. Eide, M. Pirovano, T. Rognes, E. Seeberg et al., 1999. The Saccharomyces cerevisiae homologues of endonuclease III from Escherichia coli Ntg1 and Ntg2 are both required for efficient repair of spontaneous and induced oxidative DNA damage in yeast. Mol. Cell. Biol. 19 3779–3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, R. A., 1999. The Saccharomyces cerevisiae ETH1 gene, an inducible homolog of exonuclease III that provides resistance to DNA-damaging agents and limits spontaneous mutagenesis. Mol. Cell. Biol. 19 1800–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohr, V. A., 2002. Repair of oxidative DNA damage in nuclear and mitochondrial DNA, and some changes with aging in mammalian cells. Free Radic. Biol. Med. 32 804–812. [DOI] [PubMed] [Google Scholar]
- Bowers, J., T. Sokolsky, T. Quach and E. Alani, 1999. A mutation in the MSH6 subunit of the Saccharomyces cerevisiae MSH2–MSH6 complex disrupts mismatch recognition. J. Biol. Chem. 274 16115–16125. [DOI] [PubMed] [Google Scholar]
- Chi, N.-W., and R. D. Kolodner, 1994. Purification and characterization of MSH1, a yeast mitochondrial protein that binds to DNA mismatches. J. Biol. Chem. 269 29984–29992. [PubMed] [Google Scholar]
- Croteau, D. L., and V. A. Bohr, 1997. Repair of oxidative damage to nuclear and mitochondrial DNA in mammalian cells. J. Biol. Chem. 272 25409–25412. [DOI] [PubMed] [Google Scholar]
- Doudican, N. A., B. Song, G. S. Shadel and P. W. Doetsch, 2005. Oxidative DNA damage causes mitochondrial genomic instability in Saccharomyces cerevisiae. Mol. Cell. Biol. 25 5196–5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drotschmann, K., W. Yang, F. E. Brownewell, E. T. Kool and T. A. Kunkel, 2001. Asymmetric recognition of DNA local distortion. Structure-based functional studies of eukaryotic Msh2-Msh6. J. Biol. Chem. 276 46225–46229. [DOI] [PubMed] [Google Scholar]
- Dufner, P., G. Marra, M. Raschle and J. Jiricny, 2000. Mismatch recognition and DNA-dependent stimulation of the ATPase activity of hMutS-alpha is abolished by a single mutation in the hMSH6 subunit. J. Biol. Chem. 275 36550–36555. [DOI] [PubMed] [Google Scholar]
- Dujon, B., 1981. Mitochondrial genetics and functions, pp. 505–635 in The Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Dzierzbicki, P., P. Koprowski, M. U. Fikus, E. Malc and Z. Ciesla, 2004. Repair of oxidative damage in mitochondrial DNA of Saccharomyces cerevisiae: involvement of the MSH1-dependent pathway. DNA Repair (Amst) 3 403–411. [DOI] [PubMed] [Google Scholar]
- Foury, F., T. Roganti, N. Lecrenier and B. Purnell, 1998. The complete sequence of the mitochondrial genome of Saccharomyces cerevisiae. FEBS Lett. 440 325–331. [DOI] [PubMed] [Google Scholar]
- Grollman, A. P., and M. Moriya, 1993. Mutagenesis by 8-oxoguanine: an enemy within. Trends Genet. 9 246–249. [DOI] [PubMed] [Google Scholar]
- Hanna, M., B. L. Chow, N. J. Morey, S. Jinks-Robertson, P. W. Doetsch et al., 2004. Involvement of two endonuclease III homologs in the base excision repair pathway for the processing of DNA alkylation damage in Saccharomyces cerevisiae. DNA Repair (Amst.) 3 51–59. [DOI] [PubMed] [Google Scholar]
- Kai, Y., C. Takamatsu, K. Tokuda, M. Okamoto, K. Irita et al., 2006. Rapid and random turnover of mitochondrial DNA in rat hepatocytes of primary culture. Mitochondrion 6 299–304. [DOI] [PubMed] [Google Scholar]
- Kalifa, L., and E. A. Sia, 2007. Analysis of Rev1p and Polζ in mitochondrial mutagenesis suggests an alternative pathway of damage tolerance. DNA Repair (Amst.) 6 1732–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaniak, A., P. Dzierzbicki, A. T. Rogowska, E. Malc, M. Fikus et al., 2009. Msh1p counteracts oxidative lesion-induced instability of mtDNA and stimulates mitochondrial recombination in Saccharomyces cerevisiae. DNA Repair (Amst.) 8 318–329. [DOI] [PubMed] [Google Scholar]
- Klungland, A., I. Rosewell, S. Hollenbach, E. Larsen, G. Daly et al., 1999. Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc. Natl. Acad. Sci. USA 96 13300–13305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koprowski, P., M. U. Fikus, P. Mieczkowski and Z. Ciesla, 2002. A dominant mitochondrial mutator phenotype of Saccharomyces cerevisiae conferred by msh1 alleles in the sequence encoding the ATP-binding domain. Mol. Genet. Genomics 266 988–994. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy, N., K. Haraguchi, M. M. Greenberg and S. S. David, 2008. Efficient removal of formamidopyrimidines by 8-oxoguanine glycosylases. Biochemistry 47 1043–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson, E. D., K. Iams and J. T. Drummond, 2003. Strand-specific processing of 8-oxoguanine by the human mismatch repair pathway: inefficient removal of 8-oxoguanine paired with adenine or cytosine. DNA Repair (Amst.) 2 1199–1210. [DOI] [PubMed] [Google Scholar]
- Lea, D. E., and C. A. Coulson, 1949. The distribution of the numbers of mutants in bacterial populations. J. Genet. 49 264–285. [DOI] [PubMed] [Google Scholar]
- Liu, P., L. Qian, J. S. Sung, N. C. de Souza-Pinto, L. Zheng et al., 2008. Removal of oxidative DNA damage via FEN1-dependent long-patch base excision repair in human cell mitochondria. Mol. Cell. Biol. 28 4975–4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb, L. A., D. C. Wallace and G. M. Martin, 2005. The mitochondrial theory of aging and its relationship to reactive oxygen species damage and somatic mtDNA mutations. Proc. Natl. Acad. Sci. USA 102 18769–18770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo, V. D., E. B. Gralla and J. S. Valentine, 1996. Superoxide dismutase activity is essential for stationary phase survival in Saccharomyces cerevisiae. Mitochondrial production of toxic oxygen species in vivo. J. Biol. Chem. 271 12275–12280. [DOI] [PubMed] [Google Scholar]
- Mazurek, A., M. Berardini and R. Fishel, 2002. Activation of human MutS homologs by 8-oxo-guanine DNA damage. J. Biol. Chem. 277 8260–8266. [DOI] [PubMed] [Google Scholar]
- Mookerjee, S. A., and E. A. Sia, 2006. Overlapping contributions of Msh1p and putative recombination proteins Cce1p, Din7p, and Mhr1p in large-scale recombination and genome sorting events in the mitochondrial genome of Saccharomyces cerevisiae. Mutat. Res. 595 91–106. [DOI] [PubMed] [Google Scholar]
- Mookerjee, S. A., H. D. Lyon and E. A. Sia, 2005. Analysis of the functional domains of the mismatch repair homologue Msh1p and its role in mitochondrial genome maintenance. Curr. Genet. 47 84–99. [DOI] [PubMed] [Google Scholar]
- Nilsen, H., and H. E. Krokan, 2001. Base excision repair in a network of defence and tolerance. Carcinogenesis 22 987–998. [DOI] [PubMed] [Google Scholar]
- Perocchi, F., E. Mancera and L. M. Steinmetz, 2008. Systematic screens for human disease genes, from yeast to human and back. Mol. Biosyst. 4 18–29. [DOI] [PubMed] [Google Scholar]
- Phadnis, N., R. Mehta, N. Meednu and E. A. Sia, 2006. Ntg1p, the base excision repair protein, generates mutagenic intermediates in yeast mitochondrial DNA. DNA Repair (Amst.) 5 829–839. [DOI] [PubMed] [Google Scholar]
- Ramotar, D., S. C. Popoff, E. B. Gralla and B. Demple, 1991. Cellular role of yeast Apn1 apurinic endonuclease/3′diesterase: repair of oxidative and alkylation DNA damage and control of spontaneous mutation. Mol. Cell. Biol. 11 4537–4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramotar, D., C. Kim, R. Lillis and B. Demple, 1993. Intracellular localization of the Apn1 DNA repair enzyme of Saccharomyces cerevisiae. Nuclear transport signals and biological role. J. Biol. Chem. 268 20533–20539. [PubMed] [Google Scholar]
- Reenan, R. A. G., and R. D. Kolodner, 1992. Characterization of insertion mutations in the Saccharomyces cerevisiae MSH1 and MSH2 genes: evidence for separate mitochondrial and nuclear functions. Genetics 132 975–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes, A., C. Gissi, G. Pesole and C. Saccone, 1998. Asymmetrical directional mutation pressure in the mitochondrial genome of mammals. Mol. Biol. Evol. 15 957–966. [DOI] [PubMed] [Google Scholar]
- Rosenquist, T. A., D. O. Zharkov and A. P. Grollman, 1997. Cloning and characterization of a mammalian 8-oxoguanine DNA glycosylase. Proc. Natl. Acad. Sci. USA 94 7429–7434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield, M. J., F. E. Brownewell, S. Nayak, C. Du, E. T. Kool et al., 2001. The Phe-X-Glu DNA binding motif of MutS. The role of hydrogen bonding in mismatch recognition. J. Biol. Chem. 276 45505–45508. [DOI] [PubMed] [Google Scholar]
- Sia, E. A., C. A. Butler, M. Dominska, P. Greenwell, T. D. Fox et al., 2000. Analysis of microsatellite mutations in the mitochondrial DNA of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97 250–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, K. D., B. Sigala, H. A. Sikder and C. Schwimmer, 2001. Inactivation of Saccharomyces cerevisiae OGG1 DNA repair gene leads to an increased frequency of mitochondrial mutants. Nucleic Acids Res. 29 1381–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sor, F., and H. Fukuhara, 1982. Identification of two erythromycin resistance mutations in the mitochondrial gene coding for the large ribosomal RNA in yeast. Nucleic Acids Res. 10 6571–6577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studamire, B., T. Quach and E. Alani, 1998. Saccharomyces cerevisiae Msh2p and Msh6p ATPase activities are both required during mismatch repair. Mol. Cell. Biol. 18 7590–7601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studamire, B., G. Price, N. Sugawara, J. E. Haber and E. Alani, 1999. Separation-of-function mutations in Saccharomyces cerevisiae MSH2 that confer mismatch repair defects but do not affect nonhomologous-tail removal during recombination. Mol. Cell. Biol. 19 7558–7567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surtees, J. A., and E. Alani, 2006. Mismatch repair factor MSH2–MSH3 binds and alters the conformation of branched DNA structures predicted to form during genetic recombination. J. Mol. Biol. 360 523–536. [DOI] [PubMed] [Google Scholar]
- Szczesny, B., A. W. Tann, M. J. Longley, W. C. Copeland and S. Mitra, 2008. Long patch base excision repair in mammalian mitochondrial genomes. J. Biol. Chem. 283 26349–26356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, M., and T. Ozawa, 1994. Strand asymmetry in human mitochondrial DNA mutations. Genomics 22 327–335. [DOI] [PubMed] [Google Scholar]
- Thomas, D., A. D. Scot, R. Barbey, M. Padula and S. Boiteux, 1997. Inactivation of OGG1 increases the incidence of G · C → T · A transversions in Saccharomyces cerevisiae: evidence for endogenous oxidative damage to DNA in eukaryotic cells. Mol. Gen. Genet. 254 171–178. [DOI] [PubMed] [Google Scholar]
- Tudek, B., 2003. Imidazole ring-opened DNA purines and their biological significance. J. Biochem. Mol. Biol. 36 12–19. [DOI] [PubMed] [Google Scholar]
- van der Kemp, P. A., D. Thomas, R. Barbey, R. de Oliveira and S. Boiteux, 1996. Cloning and expression in Escherichia coli of the OGG1 gene of Saccharomyces cerevisiae, which codes for a DNA glycosylase that excises 7,8-dihydro-8-oxoguanine and 2,6-diamino-4-hydroxy-5-N-methylformamidopyrimidine. Proc. Natl. Acad. Sci. USA 93 5197–5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderstraeten, S., S. Van den Brule, J. Hu and F. Foury, 1998. The role of 3′-5′ exonucleolytic proofreading and mismatch repair in yeast mitochondrial DNA error avoidance. J. Biol. Chem. 273 23690–23697. [DOI] [PubMed] [Google Scholar]
- Vermulst, M., J. H. Bielas, G. C. Kujoth, W. C. Ladiges, P. S. Rabinovitch et al., 2007. Mitochondrial point mutations do not limit the natural lifespan of mice. Nat. Genet. 39 540–543. [DOI] [PubMed] [Google Scholar]
- Vongsamphanh, R., P.-K. Fortier and D. Ramotar, 2001. Pir1p mediates translocation of the yeast Apn1p endonuclease into the mitochondria to maintain genomic stability. Mol. Cell. Biol. 21 1647–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace, D. C., 2005. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu. Rev. Genet. 39 359–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman, L., N. C. de Souza-Pinto, T. Stevnsner and V. A. Bohr, 2007. DNA repair, mitochondria, and neurodegeneration. Neuroscience 145 1318–1329. [DOI] [PubMed] [Google Scholar]
- Wilson, D. M., B. P. Engelward and L. Samson, 1998. Prokaryotic base excision repair, pp. 29–64 in DNA Damage and Repair. Vol. I: DNA Repair in Prokaryotes and Lower Eukaryotes, edited by J. A. Nickoloff and M. F. Hoekstra. Humana Press, Totowa, NJ.
- Yamamoto, A., M. J. Schofield, I. Biswas and P. Hsieh, 2000. Requirement for Phe36 for DNA binding and mismatch repair by Escherichia coli MutS protein. Nucleic Acids Res. 28 3564–3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You, H. J., R. L. Swanson, C. Harrington, A. H. Corbett, S. Jinks-Robertson et al., 1999. Saccharomyces cerevisiae Ntg1p and Ntg2p: broad specificity N-glycosylases for the repair of oxidative DNA damage in the nucleus and mitochondria. Biochemistry 38 11298–11306. [DOI] [PubMed] [Google Scholar]