Summary
DIM-5 is a SUV39-type histone H3 Lys9 methyltransferase that is essential for DNA methylation in N. crassa. We report the structure of a ternary complex including DIM-5, S-adenosyl-l-homocysteine, and a substrate H3 peptide. The histone tail inserts as a parallel strand between two DIM-5 strands, completing a hybrid sheet. Three post-SET cysteines coordinate a zinc atom together with Cys242 from the SET signature motif (NHXCXPN) near the active site. Consequently, a narrow channel is formed to accommodate the target Lys9 side chain. The sulfur atom of S-adenosyl-l-homocysteine, where the transferable methyl group is to be attached in S-adenosyl-l-methionine, lies at the opposite end of the channel,~4Å away from the target Lys9 nitrogen. Structural comparison of the active sites of DIM-5, an H3 Lys9 trimethyltransferase, and SET7/9, an H3 Lys4 monomethyltransferase, allowed us to design substitutions in both enzymes that profoundly alter their product specificities without affecting their catalytic activities.
Introduction
Histone lysine methylation is part of the histone code that regulates chromatin function (Jenuwein and Allis, 2001; Strahl and Allis, 2000). Histone lysine (K) methyl-transferases (HKMT) differ both in their substrate specificity for the various acceptor lysines as well as in their product specificity for the number of methyl groups (one, two, or three) they transfer. Known targets for HKMT include Lys4, 9, 27, 36, and 79 in histone H3 and Lys20 in histone H4 (reviewed in Marmorstein, 2003). The extent of methylation at these residues is not fully defined, however. The S. cerevisiae SET1 protein can catalyze di-and trimethylation of H3 Lys4, and trimethylation of Lys4 is thought to be present exclusively in active genes (Santos-Rosa et al., 2002). Human SET7/9 protein, on the other hand, generates exclusively mono-methyl Lys4 of H3 (Kwon et al., 2003; Xiao et al., 2003). Furthermore, DIM-5 of N. crassa generates primarily tri-methyl Lys9, which marks chromatin regions for DNA methylation (Tamaru et al., 2003).
The SET domain, which is approximately 130 amino acids in length, is found in all but one known HKMT. HKMTs can be classified according to the presence or absence, and nature of, sequences surrounding the SET domain (Baumbusch et al., 2001; Kouzarides, 2002). Representatives of the major families include SUV39, SET1, SET2, EZ, and RIZ. The SET7/9 and SET8 proteins do not fit into these families (Figure 1). The SUV39 family includes the greatest number of HKMTs. Crystal structures have recently been determined for several SET domain proteins. These include two SUV39 family proteins, DIM-5 and Clr4 (Zhang et al., 2002; Min et al., 2002), a Rubisco MTase (Trievel et al., 2002), four SET7/9 structures in various configurations (Wilson et al., 2002; Jacobs et al., 2002; Kwon et al., 2003; Xiao et al., 2003), and a viral protein that contains only the SET domain (Manzur et al., 2003). These structures revealed that the highly conserved residues of the SET domain (magenta in Figure 1) form a knot-like structure that constitutes the active site of the enzymes. Most recently, the structure of SET7/9 complexed with a peptide revealed its substrate binding site (Xiao et al., 2003).
Figure 1. Domain Structure of SET HKMT Families.
The DIM-5 protein (the smallest known member of the Suv39 family) contains four segments: a weakly conserved amino-terminal region (light blue), a pre-SET domain (yellow) containing nine invariant cysteines, the SET region (green) containing signature motifs NHXCXPN and ELXFDY (magenta), and the post-SET domain (gray) containing three invariant cysteines.
DIM-5 is a SUV39-type histone H3 Lys9 MTase from N. crassa that is essential for DNA methyllation in vivo (Tamaru and Selker, 2001). The structure of DIM-5 com-plexed with AdoHcy revealed that the pre-SET domain forms a Zn3Cys9 triangular zinc cluster (Zhang et al., 2002). However, the three-Cys post-SET domain was disordered in that structure, as well as in the structure of cofactor-free Clr4 (Min et al., 2002). We proposed that the post-SET domain may form another zinc binding site that is essential for catalytic activity and results in sensitivity to metal chelators (Zhang et al., 2002). In order to define the role of this domain and to establish the interactions between the enzyme and substrate, we determined the structure of a ternary complex of DIM-5, methyl-donor product AdoHcy, and a histone peptide.
Results and Discussion
We tested the activity of DIM-5 on three synthetic peptides corresponding to the histone H3 N-terminal residues 1–15, 1–13, and 5–15 (Figure 2A). Trimethylated lysine was the main product for all three substrates. Among them, H3 (1–15) was by far the best substrate, synthesizing trimethylated product 15 to 20 times faster than the other two peptides. Crystals of DIM-5 complexed with H3 (1–15) peptide and AdoHcy were obtained around pH 8.5, where the enzyme is active (Zhang et al., 2002). The structure was solved at 2.6Å resolution by molecular replacement using the coordinates of DIM-5 in the absence of substrate (Zhang et al., 2002). Analysis of the difference Fourier maps clearly indicated electron densities for AdoHcy, the post-SET amino acids of DIM-5, and the structured portion of the H3 peptide (residues 7–12).
Figure 2. DIM-5 Kinetics and Ternary Structure.
(A) Mass spectrometry analysis of different H3 peptides as DIM-5 substrates. The relative amount of each peptide species, expressed as a percentage of the sum of intensity of all related peaks, was plotted over the full time courses of the reactions.
(B) GRASP (Nicholls et al., 1991) surface charge distribution (blue for positive, red for negative, white for neutral). The H3 peptide and AdoHcy are shown as stick models.
(C) Ribbon (Carson, 1997) diagram colored as in Figure 1. The pre-SET residues (yellow) form a Zn3Cys9 triangular zinc cluster. The SET residues (green) and the N-terminal region are folded into six β sheets surrounding a knot-like structure (magenta). The post-SET residues (gray) bind the fourth zinc atom, adjacent to the substrate H3 peptide (red) and AdoHcy (blue).
(D) The substrate H3 peptide (red), superimposed on an omit electron density contoured at 4.0 σ (orange), is inserted as a parallel β strand (red in Figure 2C) between two DIM-5 strands, β 10 (green) and β 18 (magenta). The side chain density for H3 Arg-8 is complete at lower contour levels (2.5 σ in Fobs-Fcal and 0.8 σ in 2Fobs-Fcal).
Peptide Recognition
The histone tail peptide binds in a surface groove (Figure 2B), inserted as a parallel β strand (red in Figure 2C) between two DIM-5 strands, β10 (green) and β18 (magenta), and completes a six-stranded hybrid β sheet (3↑, 9↑, 11↓, 10↑, H3↑, 18↑). The insertion of the target H3 subpeptide as a β strand is reminiscent of the interactions seen in the heterochromatin protein HP1 with a methyllacet histone H3 peptide (Jacobs and Khorasanizadeh, 2002; Nielsen et al., 2002), though in that case, the H3 peptide is inserted as an antiparallel strand between two HP1 strands. The binding of H3 peptide as a β strand may also explain why acetylated peptides are poor substrates for HKMTs (Nakayama et al., 2001; Rea et al., 2000; Schultz et al., 2002). There is evidence that acetylation of histone N termini increases their helical content (Wang et al., 2000), and a helical H3 tail is not expected to fit in the HKMT binding groove.
Recognition of the target Lys9 is achieved through a variety of interactions between DIM-5 and the surrounding H3 sequence, including backbone-backbone, backbone-side chain, and side chain-side chain: (1) The main chain (N-H and C=O) of H3 Lys9 hydrogen bonds with DIM-5 residues L205 (C=O) and A207 (N-H). (2) While the main chain N-H of H3 Ser10 hydrogen bonds the backbone carbonyl of Y283, its side chain hydroxyl oxygen forms a hydrogen bond with the side chain of DIM-5 D209 (Figure 2D). This interaction appears to be critical for peptide recognition. Phosphorylation of Ser10 prevents H3 Lys9 methylation by SUV39H1, Clr4, SETDB1 (Nakayama et al., 2001; Rea et al., 2000; Schultz al., 2002), as well as by DIM-5 (unpublished data). It not surprising that a negatively charged phosphate group on Ser10 would disrupt its interaction with D209. Replacing the highly conserved D209 with Lys, Glu, or Gln also abolished or reduced DIM-5 activity, without affecting AdoMet crosslinking (see below), suggesting that both side chain length and charge are important for the interaction with Ser10. (3) The main chain carbonyl of H3 Thr11 hydrogen bonds the side chain of Q285, while side chain fits into the space between the side chain F206 and the hydrophobic portion of K210. Thr11 may provide critical information for substrate recognition: the sequence around Lys9 and Lys27 (QARK9STorAARK27SA) differs at this position, and DIM-5 does not methylate Lys27 (Tamaru et al., 2003).
The ends of the H3 (1–15) peptide, residues 13–15 and 1–6, which are important for the methylation reaction (Figure 2A), appear disordered in the current model. These residues could not be unambiguously identified because they colocalize to the general area of the beginning or the end of disordered protein residues 286–304, respectively (Figures 2C and 2D; see Experimental Procedures). Residues 286–304 correspond to a region highly variable in length and sequence among HKMT proteins, suggesting that the disordered region may contribute to the substrate specificity of different HKMTs. Further biochemical and structural analysis aimed toward a thermodynamic understanding will be required to resolve this interesting aspect of DIM-5 substrate recognition and allow direct observation of the interaction between this part of the enzyme and peptide substrate.
The Post-SET
Besides the bound peptide, the most prominent difference between the DIM-5 structures with and without peptide is the post-SET structure. The post-SET region was unstructured in substrate-free DIM-5 (Zhang et al., 2002). There are three conserved cysteine residues in this region that are essential for HKMT activity (Rea et al., 2000; Schultz et al., 2002; Zhang et al., 2002). Based on biochemical and genomic analyses, we suggested that these three cysteines form a metal binding site when coupled with a fourth cysteine near the active site (C244 in the signature motif N241HXCXPN247 of DIM-5) (Zhang et al., 2002). This is indeed what we observed in the current ternary structure (Figure 3A)—a zinc ion is tetrahedrally coordinated by C244, C306, C308, and C313. Interestingly, the position of the imdidazole ring the highly conserved H242 suggests that its N∊2 atom could provide a fifth coordination to the zinc atom (Figure 3A), while its Nδ1 atom hydrogen bonds the back-rounding bone N-H of Y283 and further stabilizes the interaction between the active site and the post-SET metal center.
Figure 3. The Methylation Mechanism.
(A) The post-SET zinc ion and the AdoHcy binding site. The zinc ion is presented as a red ball, coordinated by four cysteines, C244 (magenta) and C306XC308X4C313 (gray). AdoHcy is superimposed onto a difference electron density map contoured at 4.0 σ (orange). Dashed lines indicate the hydrogen bonds. One face of the AdoHcy adenine base lies against the aliphatic portion of R159, whose guanidino group forms a bifurcated salt bridge with the two carboxyl oxygen atoms of E278. The polar edge of the adenine base forms three hydrogen bonds to the DIM-5 backbone: the exocyclic amino group N6 with the carbonyl oxygen of H242, the ring N1 with the amide of L307, and the ring N7 with the amide of H242. The adenine ring carbon C8 makes van der Waals contacts to the Y283 hydroxyl and with the side chain carbonyl Oδ1 of N241; this explains the complete loss of AdoMet crosslinking in N241Q and Y283F mutants (Zhang et al., 2002). The two ribose hydroxyls interact with the main chain amide of V203 and the side chain carboxyl of D202. The amino group of the homocysteine moiety hydrogen bonds the side chain Oδ1 of N241, while its side chain amino group forms two hydrogen bonds with the backbone carbonyl of W161 and with the side chain of E278. The carboxylate group of the homocysteine moiety interacts with Y204 and the backbone amide of W161.
(B) Close-up view of the H3 peptide binding site with Lys9 inserted into a channel.
(C) The target Lys binding site (stereo). The arrow indicates the movement of the methyl group transferred from the AdoMet methylsulfonium group to the target amino group.
(D) DIM-5 activity (LogCPM) as a function of pH.
(E) AdoHcy bound in a large surface pocket, allowing for processive methylation. The green ellipse indicates the location where the AdoHcy homocysteine moiety binds in the peptide-free structure (Zhang et al., 2002).
The post-SET zinc binding site is close to the active site (Figure 3A). The structured post-SET region brings in C-terminal residues that participate in both AdoHcy and peptide binding: (1) The main chain amide nitrogen of L307 hydrogen bonds with the ring nitrogen N1 of the AdoHcy adenine base (Figure 3A). (2) The side chain of L317 packs against the AdoHcy adenine ring. (3) W318 forms part of the channel that accommodates the target Lys (see below) and provides van der Waals contacts to Ala7 of H3 and to one of the hydroxyls of the AdoHcy ribose. (4) R314 forms a salt bridge with D282. These interactions are consistent with the observation that simultaneous replacement of the three post-SET cysteines with serines abolished both DIM-5 AdoMet cross-linking and MTase activity (Zhang et al., 2002). In addition, replacement of either of the last two C-terminal residues, L317 orW318, with alanine significantly reduced, but did not abolish, AdoMet crosslinking and MTase activity (Figure 4A). Close examination of the post-SET region of many SET proteins, including SUV39, SET1, and SET2 families, suggests that these interactions between the post-SET domain and the active site are highly conserved; multiple hydrophobic residues are typically present after the post-SET cysteines, and there is usually a positively charged residue (R314 in DIM-5) follow1ing the last post-SET cysteine whenever an Asp (D282 in DIM-5) is present in the active site (Figure 4B). We suggest that the metal center we observed in DIM-5 is universal among all SET proteins with the Cys-rich post-SET. As this metal center is absolutely required for enzymatic activity, it represents a good target to design inhibitors that disrupt metal coordination, as successful for numerous metalloenzymes such as matrix metallo-proteinases (reviewed in Bode et al. 1999; Coussens et al. 2002).
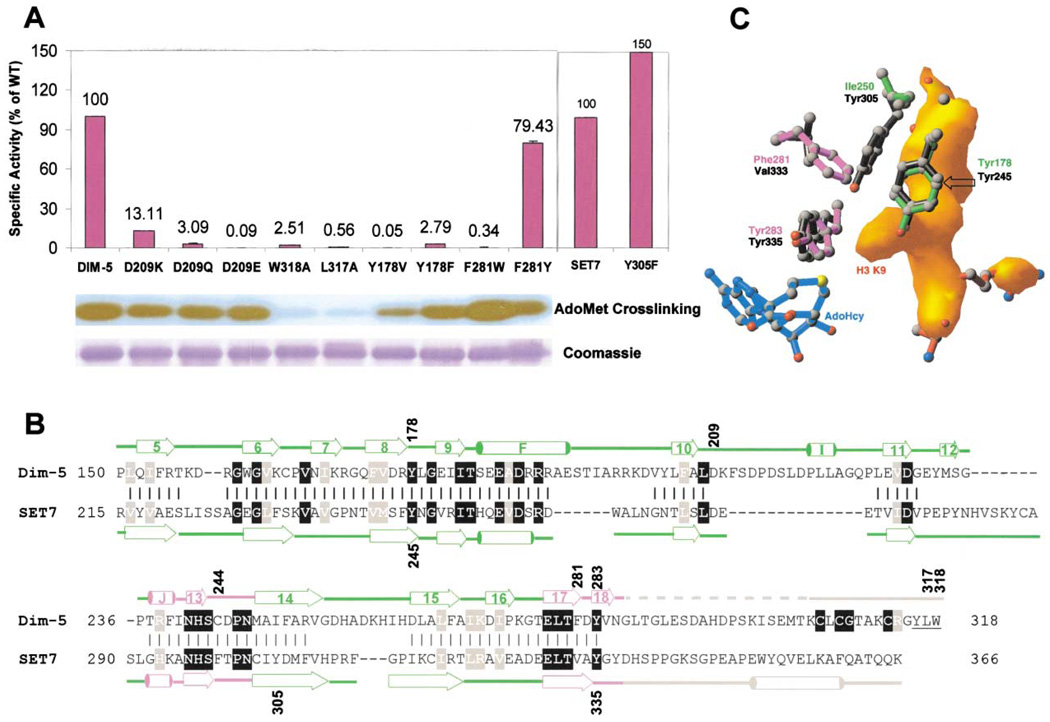
Figure 4. Enzymatic Properties of Recombinant DIM-5 and SET7/9 Mutants.
(A) Activities of DIM-5 and SET7/9 mutants using histone substrate (top). AdoMet crosslinking experiments of DIM-5 showing fluorograph (middle) and Coomassie stain (bottom).
(B) Structure-based sequence alignment of DIM-5 and SET7/9. Secondary structures shown are based on Wilson et al. (2002) and Zhang et al. (2002). Vertical bars indicate residues that align spatially. Residues identical (black background) or similar (gray background) between the two enzymes, as well as the post-SET region of DIM-5, are highlighted. Numbered residues are described in the text. C-terminal hydrophobic residues of DIM-5 are underlined.
(C) Structural comparison of active sties in the ternary DIM-5 (in color) and binary SET7/9-AdoHcy (in black) (PDB 1MT6; Jacobs et al., 2002). The bound peptide in DIM-5 is represented as a solid electron density (orange), with the target Lys surrounded by either two Tyr and one Phe (DIM-5) or three Tyr (SET7/9).
Comparison of DIM-5 with SET7/9 (Kwon et al., 2003; Xiao et al., 2003) and the Rubisco MTase (Trievel et al., 2002), two SET proteins that do not have a Cys-rich post-SET domain, reveals a remarkable example of convergent evolution. In particular, like DIM-5, these two enzymes rely on residues C-terminal of the SET domain for the formation of lysine channel, but do so by packing of an α helix, rather than a metal center, onto the active site.
Target Lysine and Catalytic Mechanism
The side chain of the target Lys9 is deposited into a narrow channel (Figure 3B), seen only when the post-SET region becomes structured. However, the corresponding channel in SET7/9 and Rubisco MTase can be preformed (Kwon et al., 2003; Trievel et al., 2002). The aromatic side chains of F206, F281, Y283, and the carboxyl-terminal residue W318 form the channel wall and make van der Waals contacts to the methylene part of the Lys9 side chain (Figure 3C). The Y283 hydroxyl group is hydrogen bonded to the backbone carbonyl oxygen of I240. This interaction is also observed in the absence of the peptide substrate (Zhang et al., 2002), suggesting that the Tyr ring is in a relatively fixed position to guide the side chain of the target lysine into the channel. At the bottom of the channel, the terminal amino group of the substrate lysine hydrogen bonds the Y178 hydroxyl and is ~4 Å from the AdoHcy sulfur atom, where the transferable methyl group will be attached in AdoMet. The AdoHcy sulfur is also ~4 Å away from the Y283 hydroxyl oxygen.
We previously showed that DIM-5 has an unusually sevhigh pH optimum (~10) and is extremely sensitive to salt (Zhang et al., 2002). At pH 10, the ∊ amino group of the target lysine and the hydroxyl groups of Y178 and Y283 (which would all have typical pKa values of ~10) should be partially deprotonated. The observed interactions suggest that deprotonated Y178 (O−) interacts with the terminal amino group of the target Lys and thereby facilitates its deprotonation, while deprotonated Y283 (O−) stabilizes the positive charge on the AdoMet methylsulfonium group (CH3-S+). As a result of these interactions, the deprotonated amino group (NH2) of the target lysine is able to nucleophilically attack the positively charged AdoMet methylsulfonium without any general base.
The interactions described above readily explain several experimental observations: (1) The involvement of three potential deprotonation events (the target lysine, Y178 and Y283) is consistent with the pH profile of DIM-5; note that in a log plot of activity against pH, the slope intercepts at approximately 3 pH units (Figure 3D). (2) A Y283F mutation abolished both AdoMet crosslinking and MTase activity (Zhang et al., 2002), consistent with a critical role for the affected hydroxyl group in binding AdoMet. (3) A Y178V mutation caused complete loss of MTase activity and reduced crosslinking with AdoMet (Figure 4A). Similarly, a Y178F mutation also dramatically reduced DIM-5 activity but had little effect on AdoMet crosslinking (Figure 4A). This is consistent with the Y178 hydroxyl being in direct contact with the target nitrogen atom and playing an essential role in catalysis.
AdoHcy Binding and Processivity
The methyl-donor product, AdoHcy, is located in an open concave pocket (Figure 3E) in a folded conformation. A similar cofactor conformation was observed in Rubisco MTase (Trievel et al., 2002) and SET7/9 (Jacobs et al., 2002; Kwon et al., 2003; Xiao et al., 2003). The AdoHcy is kinked in a manner similar to that of AdoHcy bound to the class III MTase CbiF (Schubert et al., 2003), a MTase that acts on the ring carbons of precorrin substrates (Schubert et al., 1998), but is significantly different from the extended conformation most frequently observed in the widespread class I MTases such as the DNA MTases (Cheng and Roberts, 2001). DIM-5 interacts with all three moieties of AdoHcy, the adenine base, the ribose, and the homocysteine, through van der Waals contacts and hydrogen bonds (Figure 3A).
Interestingly, the concave pocket is larger than necessary to accommodate just one AdoHcy. In particular, there is an open space next to the bound AdoHcy in the orientation shown in Figure 3E, where a less-ordered cofactor, surrounded by the highly conserved residues R155, W161, Y204, and R238, was observed in the binary structure of DIM-5-AdoHcy (Zhang et al., 2002). This suggests that the bound AdoHcy moves toward the active site upon peptide binding and accounts for the reduced, but not abolished, AdoMet binding ability of the R155H, W161F, Y204F, and R238H mutants (Zhang et al., 2002). This movement path would permit the exchange of the reaction product AdoHcy with AdoMet without releasing the peptide substrate and therefore should allow methyl transfers to proceed processively. Indeed, DIM-5 forms trimethyl-lysine with little accumulation of mono- and di- intermediates (Figure 5A; Tamaru et al., 2003). Considering that different methylation products might have different signaling properties (Santos-Rosa et al., 2002; Czermin et al., 2002; Tamaru et al., 2003), it is important to understand the structural basis for this apparent processivity.
Figure 5. Mass Spectrometry Analysis of Methylation Kinetics.
(A) Representative spectra at various time points for WT DIM-5, its F281Y variant, WT SET7/9, and its Y305F variant are shown. The peaks for unmodified (Um) substrate and mono-, di-, and trimethylated products are labeled. Unlabeled minor peaks correspond to the sodium adducts of the major peaks (+23 Da).
(B) The relative amount of each peptide species over the full time courses of the reactions, expressed as a percentage of the sum of intensity of all related peaks.
(C) Spectra for three DIM-5 mutants having severely impaired catalytic activity but with normal product specificity.
Basis for Product Specificity
DIM-5 and SET7/9 generate distinct products: DIM-5 forms trimethyl-lysine (Figure 5A, first column; and Tamaru et al., 2003) and SET7/9 forms only monomethyl-lysine (Figure 5A, third column; and Xiao et al., 2003). A likely explanation for their different product specificities is that residues in the lysine binding channel of SET7/9 sterically exclude the target lysine side chain with methyl group(s). To identify any such residue(s), we superimposed the residues surrounding the target lysine in the DIM-5 ternary structure with those in the binary structure of SET7/9 complexed with AdoHcy (Jacobs et al., 2002) (Figure 4C). As expected from the primary sequence alignment (Figure 4B), Y178 and Y283 of DIM-5 superimpose well with Y245 and Y335 of SET7/9.We also discovered that the edge of the F281 phenyl ring in DIM-5 points to the same position as the Y305 hydroxyl in SET7/9, both in close proximity to the terminal amino group of target lysine (Figure 4C). Although these two residues are not aligned at the primary sequence level, we hypothesized that the Y305 hydroxyl in SET7/9 may be the source of steric hindrance limiting methylation.
To test this hypothesis, we replaced F281 with a Tyr in DIM-5 (F281Y) and replaced Y305 with a Phe in SET7/9 (Y305F). We found that the F281Y mutation did not significantly impact the total activity of DIM-5 on histones, while the Y305F mutation resulted in an increase in activity of SET7/9 on histones (Figure 4A). We then monitored the kinetics of product formation with the H3 peptide (residues 1–15) as substrate, using MALDI-TOF mass spectrometry. Figure 5 shows representative spectra and the time course for each enzyme. The wild-type (WT) DIM-5 produces trimethyl-lysine as the predominant product even while a significant amount of unmodified substrate is still present (5 min), consistent with the idea that the enzyme is processive. Interestingly, DIM-5 F281Y initially converted unmodified substrate faster than WT (5 min), but the reaction stalled at the mono-methyl stage (compare 5 and 30 min) and then very slowly converted mono- to dimethylated product (compare 30 min and 3 hr). A trace amount of trimethylated product was observed only after prolonged incubation (3 hr). These results sharply contrast with those obtained with other DIM-5 variants with reduced overall catalytic activity, such as Y178F, R238H, and W318A (Figure 5C). After overnight incubation of these feeble enzymes, a substantial amount of unmodified substrate remains, but the trimethyl product is much more prominent than with F281Y. We conclude that the F281Y mutation changed the product specificity of DIM-5 from a tri-MTase to a mono- and di-MTase without affecting over-all catalytic activity.
In the case of SET7/9, the WT enzyme produced only monomethyl lysine plus a trace amount of dimethyl lysine after overnight incubation. Mutant Y305F, however, produced dimethyl lysine at an accelerated rate, and even traces of trimethylated product were seen after an overnight incubation (Figure 5A, fourth column). The specific activity of the Y305F mutant is higher than that of WT (Figure 4A), perhaps due its ability to add a second methyl group.
It was recently reported that a Y245A substitution in SET7/9 has little activity with unmodified substrate but allows SET7/9 to utilize mono- or dimethylated peptide as substrate to form di- or trimethylated product, respectively (Xiao et al., 2003). An analogous mutation in DIM-5, Y178V, abolished enzymatic activity on both histones and unmodified peptide substrates. In contrast, the residual activity of the Y178F mutant generated trimethyl-lysine (Figure 5C). The fact that Y178 of DIM-5 (Y245 of SET7/9) is highly conserved across enzymes with mono-, di-, and tri-specificities (Figure 4B) is consistent with the idea that this residue is primarily concerned with general catalysis rather than product specificity. Conversely, mutations at F281 in DIM-5 and Y305 in SET7/9 alter specificity without significantly affecting catalytic potential.
Conclusions
We determined the crystal structure of a histone H3 lysine 9 MTase, DIM-5 from N. crassa, in complex with an H3 peptide substrate and the methyl-donor product AdoHcy and carried out mutational and biochemical studies to elucidate the catalytic mechanism and the product specificity of this enzyme. We found that the SET domain has a surface groove that binds the methylatable tail of histone H3. The post-SET region contributes to cofactor and peptide binding by forming a zinc binding site in conjunction with a conserved cysteine in the knot-like structure near the active site. Our findings on this three-Cys domain should be relevant to the large number of SET proteins sporting the post-SET domain, including members of the SUV39, SET1, and SET2 families.
We also determined the structural basis of product specificity by engineering variants of DIM-5 and SET7/9 that have altered specificity. Variants that differ in production of mono-, di- or trimethyl lysine provide a resource to investigate the possibility that different methylation states on a given lysine may signal differently. For example, the F281Y mutant of DIM-5 can be used to test whether trimethyl Lys9 is essential in signaling DNA methylation. The predominantly euchromatic H3 Lys9 MTase G9a (Tachibana et al., 2002) is a strong di-MTase and a much weaker tri-MTase (data not shown; and Xiao et al. 2003). It would be interesting to examine the effect of converting G9a to either a mono- or a tri-MTase.
Experimental Procedures
Protein Expression and Crystallography
N. crassa DIM-5 protein was expressed and purified as described (Zhang et al., 2002). For cocrystallization, an H3 peptide (residues 1–15) was added at a final concentration of 2 mM to purified DIM-5 protein (12 mg/ml in 20 mM glycine [pH 9.8], 150 mM NaCl, 5 mM DTT, 5% glycerol, and 600 µM AdoHcy). Crystals were obtained using the hanging drop method at 16°C, with mother liquor containing 0.1 M Tris (pH 8.4–8.6), 20%–25% polyethylene glycol 2000 monomethyl ether, 0.2 M trimethylamine, and 5 mM DTT.
X-ray data from a single frozen crystal were collected on an ADSC Q315 CCD detector at beamline X25 at the National Synchrotron Light Source, Brookhaven National Laboratory. The exposure time for a 1° rotation was 120 s at 1.0Å wavelength with 400 mm detector-to-sample distance. Data acquisition and processing for a total of 135° rotation used the HKL2000 software package (Otwinowski and Minor, 1997). Crystallographic data statistics are shown in Table 1. Data from 10.0–4.0 Å were used in the structure solution by molecular replacement. All data to 2.6 Å were used for refinement.
Table 1.
Summary of X-Ray Diffraction Data
| Space group | P212121 |
| Cell dimensions (Å) | 68.26 × 94.17 × 114.69 |
| Synchrotron beamline | NSLS X25 |
| Resolution range (Å) | 35–2.59/2.68–2.59 |
| Completeness (%) | 91.6/67.6 |
| R linear (%) | 0.088/0.228 |
| <I/σ(I)> | 23.3/4.5 |
| Unique reflections | 21,803/1,578 |
| R factor | 0.22/0.30 |
| R free (5% data) | 0.32/0.38 |
| Observed total reflections | 98,025 |
| Number of atoms | |
| Protein | 3,954 |
| Peptide | 98 |
| AdoHcy | 52 |
| Zinc | 8 |
| Estimated coordinate error (Å) | |
| From Luzzati plot | 0.35 |
| From Sigmaa | 0.46 |
| Rms deviation from ideal values | |
| Bond lengths (Å) | 0.009 |
| Bond angles (°) | 1.6 |
The coordinates of substrate-free DIM-5 (PDB 1ML9) were used to search the molecular replacement solution using the program AMoRe (Navaza, 2001). With reference to the search model, two solutions were found: the orientation of the DIM-5 molecule in space group P212121 corresponds to Eulerian rotations of (103.07°, 80.86°, 0.15°) and (95.48°, 45.64°, 109.30°), with translations along a, b, and c axes of (0.384, 0.0547, 0.0630) and (0.0966, 0.6179, 0.1581) in fractional coordinates, respectively. The solutions, with the correlation coefficient of 0.488 and R factor of 0.44, indicated each asymmetric unit contains two complexes. The contact between the two DIM-5 molecules is mediated through N-terminal residues 30–45.
The resulting model, optimized by rigid-body refinement of X-PLOR (Brünger, 1992), provided an initial phase that was further improved by an overall anisotropic B factor optimization (B11 = 20.7, B22 = 17.5, and B33 = 19.9) and a bulk solvent correction (X-PLOR), resulting in a R factor of 0.33 and R free of 0.37. The difference Fourier maps (2Fobs-Fcal, αcal and Fobs-Fcal, αcal), phased from the protein model, were then calculated at 3.0, 2.8, and 2.6 Å , respectively, and inspected using the graphic program O (Jones and Kjeldgard, 1997). Electron densities, without 2-fold noncrys-tallographic symmetry averaging, were clearly visible in both molecules corresponding to the AdoHcy, the zinc coordinated by post-SET Cys residues, and the structured portion of the H3 peptide. These segments were positioned manually to fit the electron density. One cycle (100 steps) of least-squares positional refinement using meaX- PLOR gave an R factor of 0.26 and R free of 0.33. Several cycles of least-squares refinement of positional and individual B factors, followed by manual model building using O, were carried out. The noncrystallographic symmetry restraints were imposed on the two complexes during the refinement (with NCS weight of 300). A series of simulated annealing omit maps were used to guide the manual model fitting.
At this stage of refinement, we realized that the disordered ends of the peptide, residues 13–15 or 1–6, colocalize to the general area of the beginning or the end of disordered protein residues 286–304, respectively (Figures 2C and 2D). Discontinued densities do exist, but we were not able to unambiguously distinguish between the peptide and protein densities. The assignment of solvent molecules to these densities would reduce the difference between values of R factor and R free; but we took a conservative approach without including such solvent molecules in the final model (with R factor of 0.22 and R free of 0.32).
Besides residues 286–304 between the SET and post-SET regions, two other segments of DIM-5 were not modeled in the final structure: the N-terminal residues 17–25 and residues 90–96 of the pre-SET domain. In addition, a few stretches of residues (52–61, 85–89, and 97–98 of pre-SET, and 190–202 and 212–224 of SET) are flexible, as indicated by disordered side chains and relatively higher crystallographic thermal factors of >75 Å2 (2–3 times higher than the rest of the protein). As a result, many side chains of residues within or near the flexible stretches were modeled only as alanine (pre-SET residues: K53, N54, Q60, V64, S70, E72, E73, and D83; SET residues: E181, S185, E186, E194, S195, T196, R199, R200, D215, S216, L217, L221, and E227). The flexible stretches are clustered together in the folded structure: for example, the loop after strand β3 (K53, N54) is next to strand β9 (E181) and helix αF (S185 and E186); two 310 helices, αA (Q60) and αI (L221), are packed together.
As noted, the pre-SET domain contains nine invariant cysteine residues that are grouped into two segments of five and four cysteines separated by a disordered region (residues 90–96). We noticed that the first Cys segment (residues 50–99) is more mobile (with an average thermal value of 70 Å2) than the second segment (residues 100–150) with an average thermal value of 40 Å2. This observation suggests an intriguing possibility that the zinc can be transferred from pre-SET triangular cluster to the post-SET domain, analogous to methallothiomeins containing two metal clusters (Jacob et al., 1998). The dynamic nature of the pre-SET domain is confirmed by a second data set, from a different crystal, collected at beamline 17-ID of the Advanced Photon Source, Argonne National Laboratory. This time we refined the structure using tighter restraints on NCS (weight = 700, instead of 300 used in the previous refinement). The tighter NCS restraints resulted in a smaller difference between R factor (0.26) and R free (0.32) (again, no water molecules were included) at resolution range of 10–2.8 Å (28,713 reflections). However, the resulting structure is nearly identical to the previous one, particularly in the local regions around the active site.
Mutagenesis and Methyltransferase Assays
Amino acid replacements were obtained using the QuikChange site-directed mutagenesis protocol (Stratagene). All mutants were sequenced to verify the presence of the intended mutation and the absence of additional mutations. Purification of the DIM-5 mutant proteins, enzymatic assays using calf thymus histones (Sigma H4524) as substrate, and AdoMet crosslinking analyses were carried out as described (Zhang et al., 2002). Full-length SET7/9 (366 residues) and mutant proteins were expressed and purified in similar ways as the DIM-5 proteins.
Mass Spectrometry Analysis of the Kinetic Progression of Methylation Reaction
Methylation reactions were carried out in 50 mM glycine (pH 9.5), 4 mM DTT, 250 µM AdoMet, 20 µM peptide, and 0.05 mg/ml enzyme (~1.2 µM) at 23°C for DIM-5 and 37°C for SET7/9. Reactions were stopped by addition of TFA to 0.5%. For mass measurement, 1 µl of reaction mixture with TFA was added directly to 5 µl of CHCA (a-cyano-4-hydroxycinnamic acid) matrix, and 1 µl was spotted on a stainless steel sample plate and rapidly air-dried. Mass was measured by MALDI-TOF on an Applied Biosystems Voyager System 4258 machine (Chemistry Department, Emory University) operated in linear mode using reaction mix without enzyme for calibration. Each measurement was the average of ten spectra collected at ten different positions with 200 shots per position.
Acknowledgments
We thank Dr. Yi Zhang for the cDNA clone of SET7/9, Ms. Chen Qiu for X-ray data collection, Drs. Michael Becker (X25 of NSLS) and Lisa J. Keefe (17ID of APS) for access to beamlines, Dr. John M. Denu for the suggestion of a log plot of activity against pH, and Drs. Robert M. Blumenthal and Paul Wade for critical comments on the manuscript. These studies were supported in part by U.S. Public Health Services grants GM49245 and GM61355 (to X.Z. and X.C.) and GM35690 (to E.U.S).
Footnotes
Accession Numbers
The coordinates of the structure have been deposited in the Protein Data Bank (ID code 1PEG).
References
- Baumbusch LO, Thorstensen T, Krauss V, Fischer A, Naumann K, Assalkhou R, Schulz I, Reuter G, Aalen RB. The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res. 2001;29:4319–4333. doi: 10.1093/nar/29.21.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode W, Fernandez-Catalan C, Tschesche H, Grams F, Nagase H, Maskos K. Structural properties of matrix metallo-proteinases. Cell. Mol. Life Sci. 1999;55:639–652. doi: 10.1007/s000180050320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger AT. X-PLOR. A System For X-Ray Crystallography and NMR. 3.1 Edition. New Haven, CT: Yale University; 1992. [Google Scholar]
- Carson M. Ribbons. Methods Enzymol. 1997;227:493–505. [PubMed] [Google Scholar]
- Cheng X, Roberts RJ. AdoMet-dependent methylation, DNA methyltransferases and base flipping. Nucleic Acids Res. 2001;29:3784–3795. doi: 10.1093/nar/29.18.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coussens LM, Fingleton B, Matrisian LM. Matrix metalloproteinase inhibitors and cancer: trials and tribulations. Science. 2002;295:2387–2392. doi: 10.1126/science.1067100. [DOI] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- Jacob C, Maret W, Vallee BL. Control of zinc transfer between thionein, metallothionein, and zinc proteins. Proc. Natl. Acad. Sci. USA. 1998;95:3489–3494. doi: 10.1073/pnas.95.7.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs SA, Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- Jacobs SA, Harp JM, Devarakonda S, Kim Y, Rastinejad F, Khorasanizadeh S. The active site of the SET domain is constructed on a knot. Nat. Struct. Biol. 2002;9:833–838. doi: 10.1038/nsb861. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Jones TA, Kjeldgard M. Electron-density map interpretation. Methods Enzymol. 1997;277:173–208. doi: 10.1016/s0076-6879(97)77012-5. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Histone methylation in transcriptional control. Curr. Opin. Genet. Dev. 2002;12:198–209. doi: 10.1016/s0959-437x(02)00287-3. [DOI] [PubMed] [Google Scholar]
- Kwon T, Chang JH, Kwak E, Joachimiak A, Kim YC, Lee J, Cho Y. Mechanism of histone lysine methyl transfer revealed by the structure of SET7/9-AdoMet. EMBO J. 2003;22:292–303. doi: 10.1093/emboj/cdg025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzur KL, Farooq A, Zeng L, Plotnikova O, Koch AW, Sachchidanand, Zhou MM. A dimeric viral SET domain meth-yltransferase specific to Lys27 of histone H3. Nat. Struct. Biol. 2003;10:187–196. doi: 10.1038/nsb898. [DOI] [PubMed] [Google Scholar]
- Marmorstein R. Structure of SET domain proteins: a new twist on histone methylation. Trends Biochem. Sci. 2003;28:59–62. doi: 10.1016/S0968-0004(03)00007-0. [DOI] [PubMed] [Google Scholar]
- Min J, Zhang X, Cheng X, Grewal SI, Xu RM. Structure of the SET domain histone lysine methyltransferase Clr4. Nat. Struct. Biol. 2002;9:828–832. doi: 10.1038/nsb860. [DOI] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- Navaza J. Implementation of molecular replacement in AMoRe. Acta Crystallogr. D Biol. Crystallogr. 2001;57:1367–1372. doi: 10.1107/s0907444901012422. [DOI] [PubMed] [Google Scholar]
- Nicholls A, Sharp KA, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- Nielsen PR, Nietlispach D, Mott HR, Callaghan J, Bannister A, Kouzarides T, Murzin AG, Murzina NV, Laue ED. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature. 2002;416:103–107. doi: 10.1038/nature722. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- Schubert HL, Wilson KS, Raux E, Woodcock SC, Warren MJ. The X-ray structure of a cobalamin biosynthetic enzyme, cobalt-precorrin-4 methyltransferase. Nat. Struct. Biol. 1998;5:585–592. doi: 10.1038/846. [DOI] [PubMed] [Google Scholar]
- Schubert HL, Blumenthal RM, Cheng X. Many paths to methyltransfer: a chronicle of convergence. Trends Biochem. Sci. 2003;28:329–335. doi: 10.1016/S0968-0004(03)00090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., 3rd SETDB1: a novel KAP-1-associated his-tone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, Fukuda M, Takeda N, Niida H, Kato H, Shinkai Y. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16:1779–1791. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaru H, Selker EU. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature. 2001;414:277–283. doi: 10.1038/35104508. [DOI] [PubMed] [Google Scholar]
- Tamaru H, Zhang X, McMillen D, Singh P, Nakayama J, Grewal SI, Allis CD, Cheng X, Selker EU. Trimethylated lysine9 of histone H3 is a mark for DNA methylation in Neurosporacrassa. Nat. Genet. 2003;34:75–79. doi: 10.1038/ng1143. [DOI] [PubMed] [Google Scholar]
- Trievel RC, Beach BM, Dirk LM, Houtz RL, Hurley JH. Structure and catalytic mechanism of a SET domain protein methyltransferase. Cell. 2002;111:91–103. doi: 10.1016/s0092-8674(02)01000-0. [DOI] [PubMed] [Google Scholar]
- Wang X, Moore SC, Laszckzak M, Ausio J. Acetylation increases the alpha-helical content of the histone tails of the nucleosome. J. Biol. Chem. 2000;275:35013–35020. doi: 10.1074/jbc.M004998200. [DOI] [PubMed] [Google Scholar]
- Wilson JR, Jing C, Walker PA, Martin SR, Howell SA, Black-burn GM, Gamblin SJ, Xiao B. Crystal structure and functional analysis of the histone methyltransferase SET7/9. Cell. 2002;111:105–115. doi: 10.1016/s0092-8674(02)00964-9. [DOI] [PubMed] [Google Scholar]
- Xiao B, Jing C, Wilson JR, Walker PA, Vasisht N, Kelly G, Howell S, Taylor IA, Blackburn GM, Gamblin SJ. Structure and catalytic mechanism of the human histone methylManzur, transferase SET7/9. Nature. 2003;421:652–656. doi: 10.1038/nature01378. [DOI] [PubMed] [Google Scholar]
- Zhang X, Tamaru H, Khan SI, Horton JR, Keefe LJ, Selker EU, Cheng X. Structure of the Neurospora SET domain protein DIM-5, a histone H3 lysine methyltransferase. Cell. 2002;111:117–127. doi: 10.1016/s0092-8674(02)00999-6. [DOI] [PMC free article] [PubMed] [Google Scholar]