Abstract
Transcriptome analyses have been performed on mature trichomes isolated from wild-type Arabidopsis leaves and on leaf trichomes isolated from the gl3–sst sim double mutant, which exhibit many attributes of immature trichomes. The mature trichome profile contained many highly expressed genes involved in cell wall synthesis, protein turnover, and abiotic stress response. The most highly expressed genes in the gl3–sst sim profile encoded ribosomal proteins and other proteins involved in translation. Comparative analyses showed that all but one of the genes encoding transcription factors previously found to be important for trichome formation, and many other trichome-important genes, were preferentially expressed in gl3–sst sim trichomes. The analysis of genes preferentially expressed in gl3–sst sim led to the identification of four additional genes required for normal trichome development. One of these was the HDG2 gene, which is a member of the HD–ZIP IV transcription factor gene family. Mutations in this gene did not alter trichome expansion, but did alter mature trichome cell walls. Mutations in BLT resulted in a loss of trichome branch formation. The relationship between blt and the phenotypically identical mutant, sti, was explored. Mutations in PEL3, which was previously shown to be required for development of the leaf cuticle, resulted in the occasional tangling of expanding trichomes. Mutations in another gene encoding a protein with an unknown function altered trichome branch formation.
Keywords: Cell wall, cell differentiation, homeodomain, wax, trichome
INTRODUCTION
Arabidopsis leaf trichome development has served as a model for addressing basic biological questions concerning the control of cell fate and cell differentiation (Marks et al., 1991; Szymanski et al., 2000; Larkin et al., 2003; Hulskamp, 2004). Mutations in over 40 different genes result in a loss-of-function trichome phenotype (see Table 1 for gene names and leaf trichome phenotype). In some of the mutants, the only phenotype is an alteration in trichome development; however, most mutants exhibit other developmental defects. These latter mutants reinforce the generality of the trichome model for the study of plant development.
Table 1.
Gene Names and Associated Loss-of-Function Trichome Phenotypes for Many Known Mutants (Only Trichome Phenotypes Are Listed).
| Gene | Name | Loss-of-function phenotype | Reference |
| GL1 | GLABROUS1 | No trichomes | (Oppenheimer et al., 1991) |
| GL2 | GLABRA2 | Aborted trichomes | (Rerie et al., 1994) |
| GL3 | GLABRA3 | Less branched | (Payne et al., 2000) |
| EGL3 | ENHANCER OF GLABRA3 | Less branched | (Zhang et al., 2003) |
| GL3 EGL3 | No trichomes | (Zhang et al., 2003) | |
| TTG1 | TRANSPARENT TESTA GLABRA1 | No trichomes | (Walker et al., 1999) |
| TTG2 | TRANSPARENT TESTA GLABRA2 | Less branched | (Johnson et al., 2002) |
| MYB23 | ATMYB23 | Less branched | (Kirik et al., 2005) |
| TRY | TRIPTYCHON | Extra branched/clusters | (Schellmann et al., 2002) |
| CPC | CAPRICE | More trichomes | (Wada et al., 1997) |
| TRY/CPC | Larger clusters | (Schellmann et al., 2002) | |
| ETC1 | ENHANCER OF TRY AND CPC1 | Larger TRY/CPC clusters | (Kirik et al., 2004a) |
| ETC2 | ENHANCER OF TRY AND CPC2 | Larger TRY/CPC clusters | (Kirik et al., 2004b) |
| HDG11 | HOMEODOMAIN GLABROUS11 | Extra branched | (Nakamura et al., 2006) |
| HDG12 | HOMEODOMAIN GLABROUS12 | Enhances hdg11 | (Nakamura et al., 2006) |
| SPK1 | SPIKE1 | Unbranched | (Qiu et al., 2002) |
| CPR5 | CONSTITUTIVE PATHOGENE RESPONSE5 | Smaller | (Kirik et al., 2001) |
| YRE | YORE-YORE | Smaller | (Kurata et al., 2003) |
| ADL1 | ARABIDOPSIS DYNAMIN-LIKE1 | Less branched | (Kang et al., 2003) |
| SIM | SIAMESE | Multi-cellular | (Churchman et al., 2006) |
| PYM | POLYCHOME | Extra branched | (Hase et al., 2006) |
| KAK | KAKTUS | Extra branched | (Downes et al., 2003) |
| SPY | SPINDLY | Extra branched | (Perazza et al., 1998) |
| AN | ANGUSTIFOLIA | Less branched | (Folkers et al., 2002) |
| SCD1 | STOMATAL CYTOKINESIS-DEFECTIVE 1–1 | Unbranched | (Falbel et al., 2003) |
| CYCA2;3 CYCLINA2;3 | Less endoreduplication | (Imai et al., 2006) | |
| SPI | SPIRRIG | Distorted | (Saedler, 2005) |
| SAC1 | SUPPRESSOR OF ACTIN1 | Smaller | (Zhong et al., 2005) |
| MUR2 | MURUS2 | Reduced papillae | (Vanzin et al., 2002) |
| STI | STICHEL | Unbranched | (Ilgenfritz et al., 2003) |
| CSP1 | CELL SHAPE PHENOTYPE | Less branched | (Chary et al., 2008) |
| SAD2 | SENSITIVE TO ABA AND DROUGHT2 | Fewer trichomes | (Gao et al., 2008) |
| HYP6 | HYPOCOTYL6 | Smaller | (Sugimoto-Shirasu et al., 2002) |
| RHL2 | ROOT HAIRLESS2 | Smaller | (Sugimoto-Shirasu et al., 2002) |
| TBR | TRICHOME BIREFRINGENCE1 | No birefringence | (NIta, 2005) |
| BRT1 | BRIGHT TRICHOME1 | Highly fluorescent | (Sinlapadech et al., 2007) |
| SHV3/SVL1SHAVEN3/SHV3-LIKE1 | Collapsed | (Hayashi et al., 2008) | |
| LEFTY1 LEFTY1 | Less branched | (Abe et al., 2004) | |
| ZWI | ZWICHEL | Less branched (swollen) | (Oppenheimer et al., 1997) |
| LEFTY2 | LEFTY2 | Less branched | (Abe et al., 2004) |
| KIC | KCBP-INTERACTING CA2+ BINDING PROTEIN | Less branched | (Reddy et al., 2004) |
| FRA2 | FRAGILE FIBER2 | Less branched | (Burk et al., 2001) |
| KIS | KIESEL | Less developed | (Kirik et al., 2002) |
| MYO XIK MYOSIN XIK | Smaller | (Ojangu et al., 2007) | |
| PIR | PIROGI | Distorted | (Basu et al., 2004) |
| CRK | CROOKED | Distorted | (Mathur et al., 2003b) |
| WRM | WURN | Distorted | (Mathur et al., 2003a) |
| DIS1 | DISTORTED1 | Distorted | (Mathur et al., 2003a) |
| DIS2 | DISTORTED2 | Distorted | (El-Din El-Assal et al., 2004) |
| GRL | GNARLED | Distorted | (El-Assal Sel et al., 2004) |
| DIS3 | DISTORTED3 | Distorted | (Basu et al., 2005) |
| MYB106 | MYB106 (NOEK: NOK) | Extra branched | (Jakoby et al., 2008) |
| HDG2 | HOMEODOMAIN GLABR2 | Less developed cell wall | This study and Nakamura et al. (2006) |
| PEL3 | PERMEABLE LEAVES3 | Some tangled | This study and Tanaka et al. (2004) |
| BLT | BRANCHLESS TRICHOME | Unbranched | This study |
| SVB | SMALLER WITH VARIABLE BRANCHES | Less developed | This study |
The mature Arabidopsis leaf trichome consists of a unicellular structure with a stalk and three to four branches. The development of a trichome can be broken down into stages, beginning with stage one, the cessation of cell division of a protodermal cell (Hulskamp et al., 1994; Szymanski et al., 1998, 1999). Once a nascent trichome stops dividing, it swells to a diameter of ∼20 μm, and stage two begins with expansion out of the leaf surface via a process resembling tip growth. During stage three, secondary sites of expansion at the trichome tip are initiated to produce trichome branches. Branches continue to expand via tip growth during stage four, characterized by branches with blunt tips and a lack of increase in girth. The vast majority of cell expansion occurs during stage five, when trichome tips sharpen and diffuse expansion increases the total trichome length and girth. During stage six, trichome expansion ceases (see Supplemental Figure 1 for representative images of many of these stages). Other cellular events associated with the progression through the stages include endoreduplication of the nuclear DNA to an average of 32–64C during stages one through four, vacuolization during the transition from stage four to five, and the development of surface papillae during stages five and six (Marks, unpublished data; Hulskamp et al., 1994; Marks et al., 2007).
Genes identified by mutational analyses typically affect one or just a couple of the defined stages. For example, mutations in several genes encoding proteins important for controlling F-actin, such as dis1, result in normal trichome development until stage five. During stage five, the dis1 mutant trichomes undergo uneven diffuse expansion causing the trichomes to assume a distorted appearance (Szymanski et al., 1999). Mutations in STI eliminate branch formation and mutations in MUR2 alter papillae formation (Hulskamp et al., 1994; Vanzin et al., 2002).
Mutations in several genes encoding interacting transcription factors affect the stage one cell fate decision. These include the R2R3 MYB gene GL1, two redundant (with respect to trichome formation) bHLH genes, GL3 and EGL3, and TTG1 encoding a WD repeat containing protein (Oppenheimer et al., 1991; Walker et al., 1999; Payne et al., 2000; Zhang et al., 2003). Mutations in GL1, TTG1, or both GL3 and EGL3 result in a loss of trichomes. However, this complex of factors also plays a role in later trichome development, as partial loss-of-function mutations in any of these genes results in smaller, less branched trichomes (Esch et al., 1994; Larkin et al., 1999; Payne et al., 2000; Zhang et al., 2003). Loss-of-function mutations in the R3 MYBs TRY and CPC, whose encoded proteins are thought to competitively limit the interaction between GL1 and the bHLH proteins, mimic some of the phenotypes associated with the overexpression of bHLH genes (Wada et al., 1997; Schellmann et al., 2002; Esch et al., 2003). The GL1–TTG1–GL3/EGL3 complex has been posited to regulate genes required for both positive and negative regulation of trichome outgrowth (Szymanski et al., 2000; Marks and Esch, 2003). The positive regulators include such transcription factors as GL2 and TTG2 and negative regulators such as CPC and TRY (Rerie et al., 1994; Wada et al., 1997; Johnson et al., 2002; Schellmann et al., 2002). Recently, Zhao et al. (2008) obtained in planta evidence that these genes are directly regulated by the complex.
The focus of this report is on the transcriptome profiles of mature wild-type leaf trichomes and those of the glabra3–shapeshifter (gl3–sst) siamese (sim) double mutant (Marks et al., 2007). The gl3–sst mutation results in a reduced interaction between GL1 and the altered GL3 bHLH protein (Esch et al., 2003). This, in turn, results in a loss of coordinated progression through the stages associated with normal trichome development. The trichomes over-expand during stages two and three and rarely fully mature. Most trichomes have branches with blunt tips and generally have walls that lack papillae, suggesting that their development is arrested during stage four. However, unlike stage four trichomes, which appear to expand via tip growth, the mutant trichomes undergo prolonged diffuse expansion, resulting in varied trichome morphologies. SIM encodes a likely inhibitor of cyclin D function (Churchman et al., 2006). Interestingly, sim trichomes continue to divide after initiation, but then develop into fairly normal trichomes (Walker et al., 2000). As described in Marks et al. (2007), gl3–sst sim trichomes are composed of large clusters of cells that rarely advance beyond stage two. These trichomes also exhibit greatly enhanced GL1 expression, which normally decreases as trichomes mature. Thus, gl3–sst sim trichomes are predicted to have many of the attributes associated with the early stages of trichome development.
In a previous report, we described a procedure that allowed large quantities of trichomes to be quickly isolated from both wild-type and gl3–sst sim plants (Marks et al., 2008). The isolated wild-type trichomes were used for a variety of analyses, including a preliminary transcriptome analysis using the Affymetrix ATH1 GeneChip. In this report, we have expanded the transcriptome analyses, and differential gene expression has been used as a tool to identify four additional genes required for normal trichome development. These include genes encoding a transcription factor, an acyl-transferase, and two proteins with unknown function.
RESULTS
Transcriptome Analysis of Wild-Type Trichomes
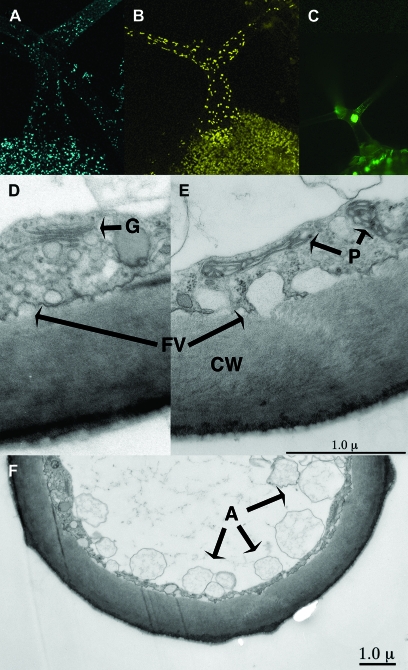
Mature Arabidopsis trichomes are biochemically active, possessing intact glyoxysome, plastid, and nuclear compartments (Figure 1A–1C). TEM analysis of a mature trichome branch revealed an intact cytoplasmic and vacuole system (Figure 1D–1F). Structures likely corresponding to Golgi, plastids, and putative autophagosomes are discernable. In addition, secretion via vesicles into the extra-cellular spaces appears to be ongoing in a mature trichome. This notion of metabolic activity is further supported by the observation of abundant cytoplasmic streaming in mature trichomes (Marks, unpublished data; Spitzer et al., 2006). To begin to study the events taking place in mature trichomes, a study of the mature trichome transcriptome was undertaken.
Figure 1.
Image Analysis of Mature Wild-Type Arabidopsis Trichomes.
(A–C) Maximal projections of confocal imaged trichomes expressing peroxisomal targeted CFP fused to peroxin 5 fusion (AT5G56290), plastid targeted YFP fused to wound-responsive family protein (AT1G19660), and nuclear-targeted GFP fused to ankyrin repeat family protein (AT4G19150), respectively.
(D–F) TEM images of cross-sections through a mature trichome branch. A, possible vacuolar autophagosomes; CW, cell wall; G, Golgi apparatus; FV, plasma membrane fused vesicles; P, plastids.
We previously described a method for isolating sufficient quantities of RNA from mature Arabidopsis trichomes for performing Affymetrix hybridization analyses (Marks et al., 2008). To extend that study, transcriptome profiles for trichomes isolated from five flats of independently grown plants were obtained. As described in Methods, the hybridization data were analyzed using Expressionist software. For these experiments, the Affymetrix MAS5 statistical program was used to filter out probesets with readings not significantly different from background (a P-value of 0.04 was used as the cut-off for calling a particular probeset present or absent). To be considered for further analysis, a probeset needed to be called present in three of the five experiments. Genes meeting this criterion were considered expressed. Genes not meeting this criterion were called not detected. Further, only probesets corresponding to one or more AGI gene IDs were used in our analyses. Overall, 13 328 probsets with an AGI identifier were called present in at least three of the five hybridizations and, of these, over 12 000 were called present in all experiments (Supplemental Table 1).
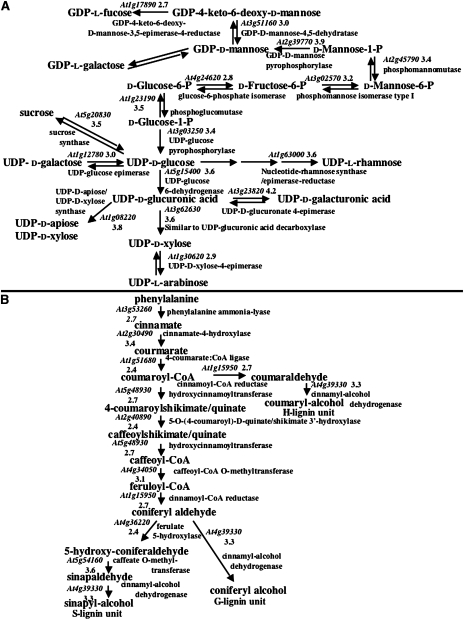
The most highly expressed genes encoded proteins with potential roles in three main biological functions. These genes include MET1A (1); STZ (84); ERD 10 (15), 14 (2) and 15 (24); RCI2A (4); RD19 (26); HSC70-1 (40); LOS1 (48); and COR47 (37), all of which likely play roles in responding to various types of abiotic stresses such as dehydration (the number in parentheses beside the gene name indicates the rank according to the expression level in Supplemental Table 1. The table also contains the full names of these genes). Other genes such as GAMMA-VPE (5); UBC 9 (29) and 28 (35); and UBQ 10 (66), 11 (62) and 14 (55) are likely involved in some aspect of protein turnover. A large number of highly expressed genes, such as GRP-3 (19); AGP4 (78) and 15 (6); FLA7 (28); CES3A (70); and GAE6 (39) likely play roles in cell wall function, biosynthesis or structure. A more extensive analysis of genes involved in cell wall biosynthesis is shown in Figure 2. Genes encoding all the enzymes required to produce cell wall monosaccharides were detected, and nearly every gene was expressed at a level above the normalized mean of 1000 (Figure 2A). In addition, the sucrose transporter AtSUT2/SUC3, which could import the sucrose starter molecule, was highly expressed in trichomes (Supplemental Table 1). Previous analyses showed that Arabidopsis trichomes contain lignin (Marks et al., 2008). As shown in Figure 2B, all the genes required for the synthesis of monolignol subunits were expressed. Along with the TEM images, these results suggest that cell wall synthesis is ongoing in mature trichomes. Given that these cells are not expanding, the new wall is likely contributing to an increase in wall thickness.
Figure 2.
Cell Wall-Related Biochemical Pathways Active in Mature Trichomes.
(A) Synthesis of monosaccharides required for cell wall biosynthesis. Pathway was derived from a Mapman analysis (Usadel et al., 2005).
(B) Synthesis of monolignols required for lignin biosynthesis. Pathway was derived from an AraCyc 4.5 analysis (Mueller et al., 2003). At all steps in both pathways, only the most highly expressed genes for each step are shown. Expression values (shown as a log10 derivative) are shown associated with predicted AGI gene identifiers.
Comparison of the Transcriptomes of Wild-Type Trichomes and Processed Shoots
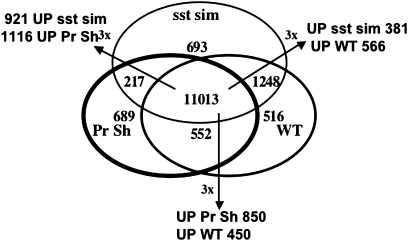
During the trichome isolation, processed shoots were separated from the isolated trichomes. Because the processed shoot tissue experienced the same growth conditions and agitation protocol as the trichomes, it represents the best control tissue for comparison to the isolated trichomes. The bulk of the shoot tissue was derived from expanded leaves and much less from attached meristems. For transcriptome analysis, four processed shoot replicates were generated. For a gene corresponding to a particular probeset to be considered expressed, the probesets corresponding to that gene in three of the four replicates needed to exhibit a signal above background. Using this criterion, 12 470 probesets with AGI identifiers were considered expressed in the processed shoots (Supplemental Table 1). 11 565 genes were co-expressed in both trichomes and processed shoots, and 1764 and 906 were specific to trichomes or processed shoots, respectively (Figure 3). The shared dataset could be subdivided. Using a three-fold difference as a cut-off and passage of a Student's t-test at P ≤ 0.05, 450, and 850 genes were up-regulated in trichomes and processed shoots, respectively. As an initial screen to identify differences between shoots and trichomes, a GO analysis was conducted (data not shown). Not surprisingly, the biggest difference in the 5000 highest expressed genes from each source was an increase in the number of genes encoding products localized to the chloroplast/plastids in processed shoots compared to trichomes. Indeed, 17 of 25 of the most highly expressed genes in processed shoots encoded proteins with functions in chloroplasts, whereas only two such genes were found in the 25 genes most highly expressed in mature trichomes (Supplemental Table 1).
Figure 3.
Composite Venn Diagram Showing Three-Way Comparisons between Transcriptome Profiles for Mature Wild-Type Trichomes, Processed Shoots (Pr Sh), and gl3–sst sim Mutant Trichomes.
Also shown are the numbers of genes that showed a three-fold difference in expression with an associated student's t-test P ≤ 0.05.
A main goal of this analysis was to use the transcriptome profiles to screen for new genes with roles in trichome development. To test the feasibility of this goal, a comparative analysis was undertaken of genes already known to be important for trichome development. This comparison was most fruitful in the analysis of transcription factors known to be involved in regulating various aspects of trichome development (Table 2). Through a variety of studies, loss-of-function mutations in 14 genes encoding transcription factors have been shown to either enhance, eliminate, or alter trichome development (Table 1). Probesets corresponding to 12 of these genes are on the Affymetrix ATH1 GeneChip (Table 2; all expression values were normalized to a mean of 1000). The expression of 10 of these genes was detected in the mature trichome transcriptome, and only TTG1 showed similar levels of expression in both trichomes and leaves, as previously noted (Baudry et al., 2004). The expression of seven of the genes was only detected in the isolated trichomes and not in the processed shoots. The remaining three genes were expressed at a significantly higher level in trichomes vs. processed shoots (160–3.25-fold higher). These results are not surprising, as previous promoter GUS and in situ studies have shown, with the exception of TTG1, that these genes are preferentially expressed in trichomes (see references in Table 1). Mutations in another 37 genes also led to abnormal trichome development and, in many cases, altered shoot development. Expression of 34 of these genes was detected in mature trichomes (Table 2). Of these genes, SIM stood out as showing the greatest level of differential expression (16.3-fold higher in trichomes) followed by TBR (5.11-fold higher). The expression of two genes, HYP6 and PYM, was detected in mature trichomes but not processed shoots. Several other genes, such as WAX2, SAC1, SCD1, ADL1, ZWI, KIS, and SPI, showed moderate but significantly higher expression in trichomes. Differences in expression were not seen in the remaining 23 genes. The fact that many genes did not show preferential trichome expression was not surprising, as mutations in many of these genes alter other aspects of plant development besides that of trichomes.
Table 2.
Comparison of Expression in Wild-Type Trichomes, Processed Shoots, and gl3–sst sim Trichomes of Genes Required for Trichome Development.
| AGI ID | Symbola | WT trib | Shc | Pval1d | Wt/She | gl3–sst simf | Pval2g | sstsim/WTh | Pval3i | sstsim/Shj |
| Transcription factors | ||||||||||
| AT3G27920 | GL1 | 99 ± 41 | NPk | NCl | NC | 3558 ± 706 | 0.00001 | 38.1 | NC | NC |
| AT1G79840 | GL2 | 6756 ± 2372 | NP | NC | NC | 4386 ± 950 | 0.179 | 0.68 | NC | NC |
| AT1G63650 | EGL3 | NP | NP | NC | NC | 110 ± 65 | NC | NC | NC | NC |
| AT5G24520 | TTG1 | 805 ± 12 | 864 ± 77 | 0.141 | 0.94 | 749 ± 320 | 0.474 | 0.88 | 0.373 | 0.87 |
| AT2G37260 | TTG2 | 1388 ± 525 | NP | NC | NC | 1988 ± 427 | 0.154 | 1.50 | NC | NC |
| AT5G40330 | MYB23 | 8602 ± 2466 | NP | NC | NC | 11500 ± 2291 | 0.173 | 1.37 | NC | NC |
| AT5G53200 | TRY | 634 ± 204 | 201 ± 101 | 0.009 | 3.2 | 1220 ± 251 | 0.032 | 2.00 | 0.004 | 6.49 |
| AT2G46410 | CPC | 2743 ± 1187 | NP | NC | NC | 1637 ± 589 | 0.148 | 0.61 | NC | NC |
| AT1G01380 | ETC1 | 2503 ± 1126 | NP | NC | NC | 7323 ± 1354 | 0.011 | 3.19 | NC | NC |
| AT1G73360 | HDG11 | 494 ± 318 | NP | NC | NC | 332 ± 52 | 0.412 | 0.76 | NC | NC |
| AT1G17920 | HDG12 | NP | NP | NC | NC | 69 ± 3.8 | NC | NC | NC | NC |
| AT3G01140 | NOK | 167 ± 40 | NP | NC | NC | 1884 ± 299 | 0.0001 | 11.39 | NC | NC |
| Miscellaneous | ||||||||||
| AT4G16340 | SPK1 | 224 ± 44 | 278 ± 62 | 0.163 | 0.81 | 438 ± 49 | 0.002 | 1.98 | 0.019 | 1.60 |
| AT5G64930 | CPR5 | 1115 ± 228 | 857 ± 138 | 0.068 | 1.29 | 1082 ± 80 | 0.901 | 0.98 | 0.054 | 1.27 |
| AT5G57800 | WAX2 | 2002 ± 367 | 674 ± 324 | 0.003 | 3.24 | 4052 ± 993 | 0.004 | 2.01 | 0.003 | 6.53 |
| AT5G42080 | ADL1 | 1157 ± 221 | 689 ± 169 | 0.008 | 1.69 | 1044 ± 187 | 0.488 | 0.91 | 0.050 | 1.54 |
| AT5G04470 | SIM | 4477 ± 2083 | 275 ± 137 | 0.00007 | 16.3 | 7357 ± 913 | 0.097 | 1.79 | 0.0001 | 29.3 |
| AT3G57860 | PYM | 94 ± 55 | NP | NC | NC | 95 ± 19 | 0.757 | 1.11 | NC | NC |
| AT4G38600 | KAK | 1069 ± 257 | 1038 ± 268 | 0.831 | 1.04 | 1343 ± 101 | 0.110 | 1.28 | 0.155 | 1.33 |
| AT3G11540 | SPY | 493 ± 93 | 448 ± 100 | 0.489 | 1.11 | 649 ± 45 | 0.046 | 1.33 | 0.041 | 1.48 |
| AT1G01510 | AN | 493 ± 131 | 425 ± 98 | 0.431 | 1.15 | 761 ± 313 | 0.149 | 1.49 | 0.085 | 1.72 |
| AT1G49040 | SCD1 | 784 ± 71 | 527 ± 226 | 0.034 | 1.57 | 832 ± 97 | 0.442 | 2.26 | 0.078 | 1.67 |
| AT1G15570 | CYCA2;3 | 155 ± 38 | 107 ± 30 | 0.072 | 1.47 | 199 ± 66 | 0.287 | 1.27 | 0.054 | 1.86 |
| AT1G03060 | SPI | 325 ± 83 | 232 ± 17 | 0.035 | 1.37 | 241 ± 72 | 0.166 | 0.74 | 0.982 | 1.00 |
| AT1G22620 | SAC1 | 470 ± 66 | 306 ± 80 | 0.014 | 1.56 | 344 ± 61 | 0.033 | 0.73 | 0.483 | 1.14 |
| AT2G03220 | MUR2 | 313 ± 160 | 154 ± 52 | 0.051 | 1.95 | 894 ± 114 | 0.007 | 2.80 | 0.0005 | 6.06 |
| AT2G02480 | STI | NP | 106 ± 20 | NC | NC | 353 ± 52 | NC | NC | 0.0009 | 3.32 |
| AT1G68020 | CPS1 | 178 ± 34 | 174 ± 58 | 0.800 | 1.05 | 253 ± 43 | 0.041 | 1.43 | 0.114 | 1.50 |
| AT2G31660 | SAD2 | 350 ± 123 | 245 ± 61 | 0.176 | 1.40 | 304 ± 93 | 0.646 | 0.87 | 0.417 | 1.23 |
| AT3G20780 | HYP6 | 102 ± 46 | NP | NC | NC | 173 ± 16 | 0.087 | 1.69 | NC | NC |
| AT5G02820 | RHL2 | NP | NP | NC | NC | 347 ± 107 | NC | NC | NC | NC |
| AT5G06700 | TBR | 4714 ± 2109 | 859 ± 88 | 0.0002 | 5.11 | 1825 ± 402 | 0.016 | 2.44 | 0.002 | 2.10 |
| AT3G21560 | BRT | 365 ± 117 | 496 ± 246 | 0.339 | 1.33 | 696 ± 233 | 0.062 | 1.90 | 0.292 | 1.45 |
| AT4G26690 | SHV3 | 786 ± 329 | 1190 ± 569 | 0.332 | 0.69 | 298 ± 170 | 0.032 | 0.37 | 0.030 | 0.25 |
| AT5G55480 | SVL1 | 212 ± 85 | 544 ± 74 | 0.186 | 0.83 | 321 ± 100 | 0.012 | 2.77 | 0.048 | 0.58 |
| Microtubule-related | ||||||||||
| AT4G14960 | Lefty1 | 3656 ± 2043 | 2877 ± 1092 | 0.515 | 1.22 | 2540 ± 334 | 0.367 | 0.69 | 0.773 | 0.93 |
| AT5G65930 | ZWI | 307 ± 89 | 174 ± 26 | 0.016 | 1.71 | 286 ± 55 | 0.834 | 0.96 | 0.011 | 1.64 |
| AT1G04820 | Lefty2 | 2937 ± 1663 | 1406 ± 922 | 0.091 | 2.31 | 2130 ± 283 | 0.477 | 0.73 | 0.253 | 1.85 |
| AT2G46600 | KIC | 6224 ± 1206 | 10800 ± 4389 | 0.118 | 0.63 | 3118 ± 760 | 0.005 | 0.50 | 0.021 | 0.29 |
| AT2G34560 | FRA2 | 793 ± 1434 | 1000 ± 533 | 0.863 | 0.94 | 879 ± 494 | 0.932 | 1.02 | 0.939 | 0.96 |
| AT2G30410 | KIS | 2275 ± 549 | 1469 ± 422 | 0.0343 | 1.56 | 2228 ± 250 | 0.983 | 1.00 | 0.051 | 1.52 |
| Actin-related | ||||||||||
| AT5G20490 | MYO XIK | 463 ± 110 | 456 ± 82 | 0.959 | 1.01 | 257 ± 77 | 0.019 | 0.56 | 0.028 | 0.55 |
| AT5G18410 | PIR | 462 ± 161 | 733±355 | 0.170 | 0.65 | 625 ± 114 | 0.196 | 1.41 | 0.782 | 0.92 |
| AT4G01710 | CRK | 540 ± 81 | 427 ± 133 | 0.168 | 1.31 | 517 ± 70 | 0.711 | 0.96 | 0.351 | 1.259 |
| AT3G27000 | WURM | NP | NP | NC | NC | NP | NC | NC | NC | NC |
| AT1G13180 | DIS1 | 257 ± 98 | 198 ± 92 | 0.341 | 1.34 | 310 ± 105 | 0.463 | 1.23 | 0.192 | 1.645 |
| AT1G30825 | DIS2 | 298 ± 71 | 373 ± 73 | 0.164 | 0.79 | 292 ± 47 | 0.961 | 0.99 | 0.152 | 0.79 |
| AT2G35110 | GRL | 476 ± 65 | 521 ± 153 | 0.636 | 0.93 | 530 ± 44 | 0.248 | 1.12 | 0.796 | 1.046 |
| AT2G38440 | DIS3 | 512 ± 166 | 469 ± 92 | 0.792 | 1.06 | 667 ± 241 | 0.335 | 1.31 | 0.162 | 1.385 |
| New mutants | ||||||||||
| AT1G05230 | HDG2 | 1461 ± 256 | NP | NC | NC | 1626 ± 205 | 0.38 | 1.12 | NC | NC |
| AT1G56580 | SVB | 6306 ± 1890 | 1846 ± 958 | 0.006 | 3.89 | 28000 ± 2713 | 0.00009 | 4.56 | 0.001 | 17.74 |
| AT1G64690 | BLT | NP | NP | NC | NC | 1178 ± 364 | NC | NC | NC | NC |
| AT5G23940 | PEL3 | 381 ± 231 | NP | NC | NC | 1145 ± 363 | 0.023 | 3.30 | NC | NC |
Symbol—see Table 1 for full name.
WT tri—mean ± standard deviation of isolated Col wild-type mature trichomes (all values were normalized to 1000).
SH—mean ± standard deviation of processed shoots.
Pval1—P-value obtained from student's t-test between wild-type trichomes and processed shoots.
Wt/Sh—ratio of wild-type trichome values over processed shoot.
gl3–sst sim—mean ± standard deviation of isolated gl3–sst sim trichome clusters.
Pval2—P-value obtained from student's t-test between wild-type and gl3–sst sim trichomes.
sstsim/Wt—ratio of gl3–sst sim over wild-type trichomes.
Pval3—P-value obtained from student's t-test between gl3–sst sim trichomes and processed shoots.
sstsim/Sh—ratio of gl3–sst sim over processed leaf.
NP—not present.
NC—not calculated.
Analysis of the Transcriptome of gl3–sst sim Trichomes
The analysis of mature trichomes only captures the expression profile of genes expressed during the final stage of trichome development. It is likely that the expression profile of earlier staged trichomes would help identify genes required during early trichome development. An example of such a gene is GL1, which is more highly expressed in young developing trichomes than in mature trichomes (Larkin et al., 1993). We have previously shown that the clusters of cells that compose gl3–sst sim trichomes have many attributes of early-stage trichomes, including elevated GL1 expression (Marks et al., 2007). Thus, we wished to determine whether other genes needed for early trichome development were more highly expressed in the double mutant trichomes.
The same procedure used to isolate wild-type mature trichomes was previously shown to be effective for the isolation of gl3–sst sim trichomes (Marks et al., 2008). The double mutant trichomes isolated from three independently grown flats were used to generate probes for hybridization to Affymetrix ATH1 GeneChips. For this analysis, probesets needed to be called present for at least two of the three trials to be considered for study. Using this criterion, 13 170 probesets with AGI IDs were identified (Supplemental Table 1). Of note, the second highest expressed gene was GASA4, which was expressed over seven times more highly in the double mutant than in wild-type trichomes. Previous studies on immature wild-type trichomes also found that this gene was highly expressed (Kryvych et al., 2008). The highest expressed gene has an unknown function, but we show below that this gene is required for trichome differentiation. The third highest expressed gene encodes a member of the translation elongation factor 1-alpha gene family. Either the same or a closely related protein previously was found to be highly abundant in trichomes (Wienkoop et al., 2004). Many of the highest expressed genes encoded either components of ribosomes or proteins with functions related to translation (Supplemental Table 1). Overall, 22 of the 55 most highly expressed genes were found to have functions related to translation. This finding is reflected in the TEM image of a gl3–sst sim trichome shown in Supplemental Figure 2. The cytoplasmic compartment appears to be packed with structures similar in size (20 nM) to ribosomes. Similar to mature trichomes, genes responding to abiotic stress also were highly expressed (Supplemental Table 1).
A comparison of the expression profiles of WT trichomes, gl3–sst sim trichomes, and processed shoot is shown in Figure 3. As shown, 11 013 genes were expressed in all three, whereas 693, 516, and 689 were unique to gl3–sst sim, WT, and processed shoot, respectively. For genes commonly expressed only in gl3–sst sim and WT trichomes, 381 and 566 were expressed three-fold or higher in each type, respectively. Within the population of genes common to gl3–sst sim and processed shoots, 921 and 1116 were expressed three-fold or higher, respectively.
Many of the genes that were more highly expressed in gl3–sst sim trichomes than in either wild-type trichomes or processed shoots encode proteins predicted to be involved in lipid metabolism. In this regard, it is of interest to highlight the R2R3 MYB transcription factor AtMYB30. The expression of AtMYB30 was not detected in either processed shoots or wild-type mature trichomes, but was high in the double mutant trichomes. Raffaele et al. (2008) identified AtMYB30 as having a major role in regulating genes involved in lipid biosynthesis. Using transcriptional profiles from plants that either over or underexpressed AtMYB30, they identified a core set of 18 genes predicted to be regulated by AtMYB30. Table 3 shows the expression levels of these 18 genes in gl3–sst sim trichomes compared to that in mature trichomes and processed shoots. Like AtMYB30, three of the genes were only expressed in gl3–sst sim trichomes. Of the remaining 15 genes, 14 were more highly expressed in gl3–sst sim trichomes, and thereby showed co-regulation with ATMYB30.
Table 3.
T-test Comparing Expression of Genes Predicted to be Regulated by ATMYB30 and Involved in Fatty Acid Biosynthesis in gl3–sst sim Trichomes, Wild-Type Trichomes, and Processed Shoots.
| Pval | Pval | sstsimb | sstsimc | |
| Genea | sstsim vs. WT TRI | sstsim vs. Pro Sh | WT TRI | Pro Sh |
| AT1G01120 KCS1 | 4e–4 | 0.016 | 5.1 | 4.7 |
| AT1G01610 GPAT4 | 1e–4 | 5e–4 | 8.3 | 9.0 |
| AT1G07720 KCS4 | 0.26 | 0.48 | 1.7 | 1.2 |
| AT1G27950 LTP | 0.003 | 0.014 | 8.4 | 12.3 |
| AT1G67730 GL8 | 3e–4 | 0.002 | 2.1 | 3.8 |
| AT2G26250 FDH | 5e–4 | 6e–4 | 12.7 | 7.6 |
| AT2G38530 LTP2 | 0.14 | NPd | 5.1 | 4408 ± 279e |
| AT3G55360 CER10 | 7e–4 | 0.015 | 2.7 | 3.0 |
| AT4G00360 ATT1 | 0.91 | 0.49 | 1.0 | 1.3 |
| AT4G14440 HCD1 | 0.013 | 0.009 | 5.7 | 6.9 |
| AT4G24510 CER2 | 0.01 | 0.14 | 4.5 | 2.5 |
| AT5G47330 PPT1 | NPf | NPd | 214 ± 23g | 214 ± 23g |
| AT5G10480 PAS2 | 2e–4 | 0.003 | 2.6 | 5.4 |
| AT5G57800 WAX2 | 0.004 | 0.003 | 2.0 | 6.5 |
Gene list from Raffaele et al. (2008).
Ratio of means for trichomes from gl3–sst sim to Col wild-type trichomes.
Ratio of means for trichomes from gl3–sst sim to processed shoots.
Processed shoot values called not present.
Mean value for gl3–sst sim trichomes for LTP2.
Col wild-type trichome value called not present.
Mean value for gl3–sst sim trichomes for PPT1.
As stated above, 20 of the 49 genes genetically shown to be important for trichome formation showed significantly higher expression in mature trichomes. To search for more differences, the expression of the 49 genes in gl3–sst sim trichomes was compared to that in both mature trichomes and processed shoots (Table 2). In comparing gl3–sst sim to processed shoots, all of the transcription factors that showed significantly higher expression in mature trichomes also were more highly expressed in the double mutant. The expression of two other transcription factors, EGL3 and HDG12, which was not detected in mature trichomes or processed shoots, was detected in the double mutant. Several other genes encoding transcription factors were more highly expressed in gl3–sst sim compared to mature trichomes. For example, expression levels of GL1 and NOK were 38- and 11-fold higher in gl3–sst sim. In addition, the expression levels of the R3 MYBs TRY and ETC1 were two- and three-fold higher in gl3–sst sim. Within the group of 37 other important genes, the expression of all but one was detected in the double mutant trichomes. Compared to processed shoots, all but three of the genes that were more highly expressed in mature trichomes were also more highly expressed in the double mutant, the exceptions being SAC1, SPI, and KIS. An additional seven genes were more highly expressed in the double mutant than in processed shoots. These included MUR2 and STI, which were expressed 6.1- and 3.3-fold higher in gl3–sst sim compared to processed shoot; the others showed smaller increases. Overall, combining the results for mature and gl3–sst sim trichomes, 28 of 48 genes important for trichome formation were more highly expressed in trichomes than in processed shoots.
Use of Enhanced Expression in gl3–sst sim to Screen for New Mutants
The results above indicate that comparative analyses should be useful for identifying new trichome mutants. Given that 11 of 12 transcription factors required for trichome formation showed enhanced expression in gl3–sst sim trichomes compared to processed shoots, we chose to look for new mutants by identifying additional transcription factors up-regulated in the mutant. Transcription factor genes expressed in the double mutant trichomes but not in processed shoots were ranked by expression level (Table 4). Within the top 10 genes, seven of the transcription factors important for trichome formation were re-identified. To hunt for additional trichome genes, T-DNA insertion lines were obtained from the Arabidopsis stock center corresponding to other genes in the list. Stocks obtained for AGL14 (CS841281) and AGL19 (SALK_000234) did not reveal the presence of trichome mutants; however, insertions in HDG2 resulted in trichome abnormalities. This mutant is shown in Figure 4, and will be discussed in more detail below. An expanded mutant search was conducted by ranking all genes detected in the double mutant but not in processed shoot. Seeds with T-DNA inserts within the most highly ranked genes were screened for trichome defects. This analysis identified two additional new mutants as shown in Figures 5B and 6, and studied in more detail below. Finally, as noted above, the ranking of genes most highly expressed in gl3–sst sim revealed that genes ranked two and three were known to be associated with trichomes. Therefore, we obtained T-DNA insertional lines for the most highly expressed gene. Indeed, these lines also contained mutants with altered trichomes, as shown in Figure 5C.
Table 4.
Ranking of Most Highly Expressed Transcription Factors (TFs) that Are Called Present in gl3–sst sim Mutant Trichomes but Not in Processed Shoot Compared to Ranking for Col Wild-Type (WT) Trichomes.
| Gene | Trichome | Ranking gl3–sst sim |
Ranking WT trichomes |
||
| TFs | All genes | TFs | All genes | ||
| ATMYB23a | 11500 ± 2291 | 1 | 2 | 1 | 3 |
| ETC1a | 7323 ± 1354 | 2 | 7 | 6 | 31 |
| GL2a | 4386 ± 950 | 3 | 15 | 2 | 5 |
| GL1a | 3558 ± 706 | 4 | 18 | 137 | 1481 |
| AGL14 | 2796 ± 662 | 5 | 26 | NP | NP |
| TTG2a | 1988 ± 427 | 6 | 36 | 13 | 68 |
| NOKa | 1884 ± 299 | 7 | 39 | 95 | 1043 |
| AGL19 | 1839 ± 569 | 8 | 41 | 46 | 487 |
| CPCa | 1637 ± 589 | 9 | 48 | 5 | 26 |
| HDG2b | 1626 ± 205 | 10 | 49 | 12 | 65 |
| ATMYB5b | 1488 ± 366 | 11 | 55 | 9 | 48 |
| ATMYB30 | 1295 ± 430 | 12 | 62 | NP | NP |
Encodes protein controlling trichome formation (see Table 1).
Known to be expressed in trichomes (Li et al., 1996).
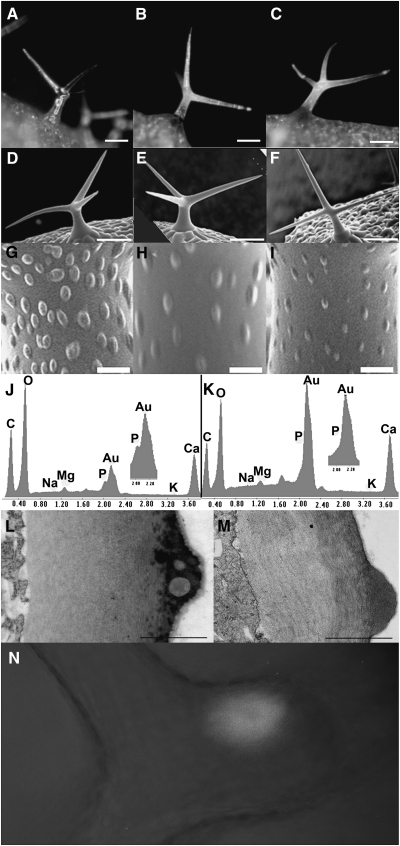
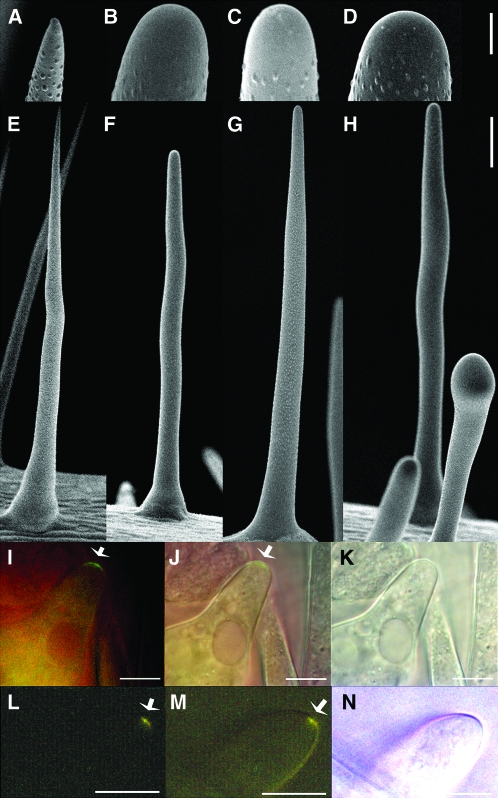
Figure 4.
Analysis of hdg2 Trichome Cell Wall Mutant.
(A–C) Stereomicrographs of trichomes from hdg2-2 mutant, wild-type, and rescued hdg2-2, respectively.
(D–F) SEM of wild-type, hdg2-2, and wild-type transformed with pEGAD MYB5:GFP–cHDG2 transgene showing co-suppressed hdg2-like mutant papillae phenotype, respectively.
(G–I) Higher magnification of trichomes shown in (D–F).
(J, K) Elemental analysis of wild-type and hdg2-2 trichome papillae.
(L, M) TEM of cross-section through branch of wild-type and hdg2-2 mutant trichomes.
(N) Localization of GFP–HDG2 fusion protein in wild-type plant transformed with the pEGAD MYB5:GFP–cHDG2 transgene.
Bars in (A–F) = 100 μm; (G–J) = 1 μm; (L, M) = 0.5 μm.
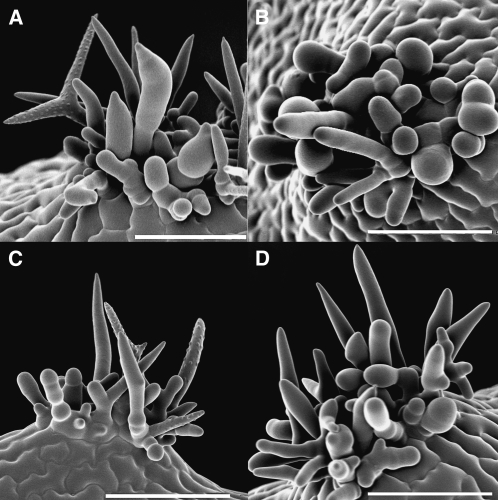
Figure 5.
SEM Images of Wild-Type and Mutant Trichomes.
(A) Wild-type trichome.
(B) blt-1 branchless trichome.
(C) svb-1.
All bars = 100 μm.
Figure 6.
Characterization of the pel3 Mutant.
(A) Wrinkled leaf phenotype of pel3-11 seedling.
(B) Co-grown wild-type Col seedling.
(C, D) Higher magnification of wrinkled leaf showing tangled trichomes in (D).
(E) Wild-type Col inflorescence.
(F) pel 3–11 inflorescence showing lack of expanded petals.
(G) Dissected pel3-11 flower showing trapped petals, anthers, and pollen grains.
(H, I) DIC and fluorescent images of pel3-11 trichomes expressing gfp-PEL3 fusion protein. N highlights position of the nucleus in (H) and (I).
Characterization of New Trichome Mutants
HDG2
Two T-DNA-induced mutant alleles for HDG2 were isolated and characterized as described in Methods. Both of these have T-DNA insertions in either intronic or exonic regions of the gene. The HDG2 mutants have trichomes that appear glass-like under a dissecting microscope as compared to wild-type trichomes (compare Figure 4A to 4B). The mutations do not affect trichome growth or branch number (data not shown and compare Figure 4D (wild-type) to 4E or 4F (mutants)). As shown in Figure 4C, the mutant phenotype was reverted in mutants expressing the genomic coding region of HDG2 under the control of the TRY promoter. The similar phenotype of two independently isolated mutants and the rescue of the phenotype by expression of the HDG2 gene indicate that the glassy trichome phenotype was caused by mutations in the HDG2 gene. Furthermore, within the population of transformants, several presumably co-suppressed T1 plants exhibited the mutant phenotype (Figure 4F). As expected for a transcription factor, a GFP–HDG2 gene construct directed the expression of nuclear localized GFP tagged protein (Figure 4N). Several analyses were performed to study the walls of the mutant. As Shown in Figure 4G–4I, SEM analysis of the trichome surface showed that the mutant trichomes have less developed papillae. Previous elemental analysis showed that the papillae on Arabidopsis trichomes contained phosphorous (P) (Esch et al., 2003). As shown in the comparison of the elemental profiles of wild-type and mutant papillae (Figure 4J and 4K), the mutant papillae lacked P. To explore this aspect of the phenotype further, TEM analysis of wild-type and mutant was conducted. This showed that the mutant papillae lacked the occlusions seen in the wild-type papillae (Figure 4L and 4M). Given the correlation between the lack of both occlusions and P in the mutant, it is likely that the occlusions seen in the wild-type are the sites of accumulation of P-containing compounds. This analysis also shows that the cell walls of the mutant were able to thicken. However, two other features were obviously different. The cuticle layer of the mutant trichomes was much reduced compared to that of wild-type trichomes, and the walls of the mutant stained less.
To begin to correlate the phenotype of the mutant with gene expression, and to identify mis-regulated genes, probes for Affymetrix analysis were generated using RNA isolated from the trichomes of three batches of hdg2 mutants. Genes were considered expressed if the corresponding probesets for two of the three hybridizations showed a signal above background (see Supplemental Table 1 for list of expressed genes). Overall, the wild-type and hdg2 trichome datasets contain 12 999 genes in common, and 330 and 1307 were uniquely expressed in wild-type and hdg2, respectively. Within the common dataset, 82 and 29 passed a t-test with at least a 2.5-fold higher level in wild-type and hdg2 trichomes, respectively. There were two intriguing differences in the differentially expressed genes. First, GL1 expression was significantly higher in the mutant. Second, the expression of the gene encoding CYP94C was greatly reduced in the mutant. The significance of these results will be discussed below.
PEL3
Insertions in At5g23940, which encodes an acyl-transferase, also led to altered trichome phenotype. The expression of this gene was 3.3-fold higher in gl3–sst sim compared to wild-type trichomes, and was not detected in processed shoots. Two different insertion lines were identified that exhibited mutants with similar phenotypes (see Methods). The mutant was characterized as having trichomes that become tangled during leaf expansion (Figure 6D). This resulted in the crinkling of the expanding leaves (Figure 6A and 6C compared to wild-type 6B). In a separate study by others, At5g23940 was identified in a mutant screen for plants exhibiting an altered cuticle layer (Tanaka et al., 2004). The mutants were identified in a screen for plants with enhanced leaf staining by toluidine blue. The authors called the At5g23940-associated mutant PERMEABLE LEAVES3 (PEL3), which is how it will be referred to in this study. However, a trichome phenotype was not noted. The entangling of trichomes with subsequent crinkling of leaves was most pronounced when the plants were grown under low humidity conditions (below 50% humidity). However, even when grown at a higher humidity, differences in trichome expansion could be observed. In the wild-type, the trichomes cleanly slid across one another, but, in the mutant, trichomes displayed a more abrupt sliding pattern (see Supplemental Movies 1 and 2). Developing pel3 trichomes proceed through normal stages of trichome development, but tangling occasionally can be observed during stage 5 (Supplemental Figure 3 of SEM analysis of developing pel3 trichomes). The final trichome size, branch number, and papillae appear normal. As previously noted, pel3 mutants also exhibited reduced fertility, which appeared to be due to the trapping of petals and anthers inside the mutant flowers (Figure 6F and 6G). These phenotypes were largely suppressed in plants expressing a GFP-tagged version of PEL3. Analysis of the GFP–PEL3 plants showed a strong GFP signal in the cytoplasmic and nuclear regions of developing trichomes (Figure 6H and 6I). Confocal analysis suggests that the intense signal in the nuclear regions either reflects fluorescence in the cytoplasm surrounding nuclei or that PEL3 localizes to the nuclear envelope region, as the interior of the trichome nucleus shown in Supplemental Movie 3 displays diminished GFP fluorescence. The nature of the likely biochemical alteration in the cuticle and an understanding of the significance of the localization pattern will require additional experimentation.
BLT
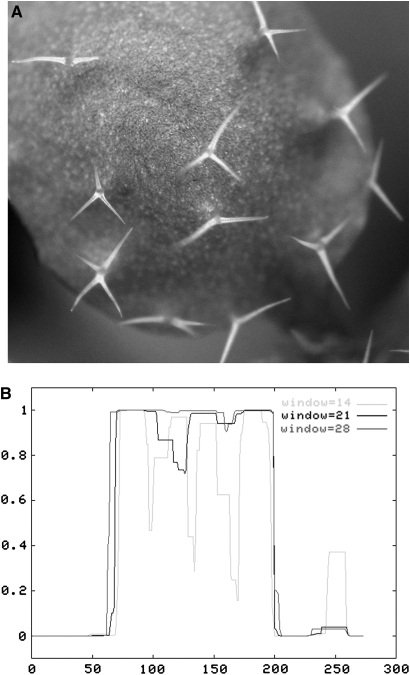
The expression of At1g64690 was not detected in wild-type mature trichomes or processed shoots, but was highly expressed in gl3–sst sim trichomes. In the Col and Ler wild-type backgrounds, leaf trichomes typically have three to four branches. Insertions in At1g64690 resulted in unbranched trichomes (Figure 5B). For this reason, we name the gene BRANCHLESS TRICHOME (BLT). Two mutant alleles have been identified for BLT. One, blt-1, contains a T-DNA insertion in the Col background (stock number CS827202) and the other, blt-2, contains a DS element in the Ler background (stock number CS164367). The phenotype of the mutant alleles has been rescued using a GFP-tagged BLT cDNA (Figure 7A). As shown in Figure 7B, a large portion of the internal amino acid sequence of BLT is predicted to form a coiled-coil domain.
Figure 7.
Characterization of BLT.
(A) Rescue of trichome branches in blt-2 expressing pEGAD TRY:gfp–BLT fusion construct.
(B) Coils analysis of BLT peptide showing that the interior domain of BLT protein has a very high probability of assuming a coiled-coil secondary structure.
x-axis, amino acid residue number; y-axis, probability of forming coiled-coil secondary structure.
The phenotype of blt is very similar to that of sti (Ilgenfritz et al., 2003). To explore the relationship between the two mutants, additional phenotypic and genetic studies were performed. The development of trichomes on both mutants is similar, lacking stage three branch formation (see Supplemental Figure 4A–4C). The unbranched leaf and stem trichomes on these mutants resemble the unbranched trichomes found on the stems of wild-type plants. However, SEM analysis of the tips of the mutant trichomes showed a phenotypic difference. The wild-type unbranched stem trichomes have a sharp point (Figure 8A and 8E), whereas the unbranched trichomes on the leaves and stems of both mutants have blunt tips (compare Figure 5A and 5B, and see Figure 8B, 8C, 8F, and 8G). This indicates that both BLT and STI are required for the initiation of branches, and play a role in branch tip maturation. Double mutants were generated to study the genetic interaction between the two genes (see Methods). As shown in Figure 8D and 8H, the vast majority of the double mutant trichomes resembled those of either of the single mutants. However, an occasional trichome (fewer than 10%) exhibited a more extreme phenotype with stunted growth and a more bloated tip (Figure 8H). Given that the majority of the trichomes on the double mutants had phenotypes no more extreme than those of either of the single mutants, it is likely that the two genes encode products that function in the same developmental pathway. To begin to test for co-localization, GFP-BLT and Tdimer2 RED–STI fusion constructs were moved into the corresponding mutant and wild-type plants (tdimer2 RED is described in more detail in Methods and in Campbell et al., 2002). Both constructs completely rescued the trichome tip phenotype of the corresponding mutants (data not shown), and, as shown in Figure 8I–8N, both fusion proteins localized to the branch tips of stage four trichomes. Images of non-transformed negative control plants for comparison are shown in Supplemental Figure 5.
Figure 8.
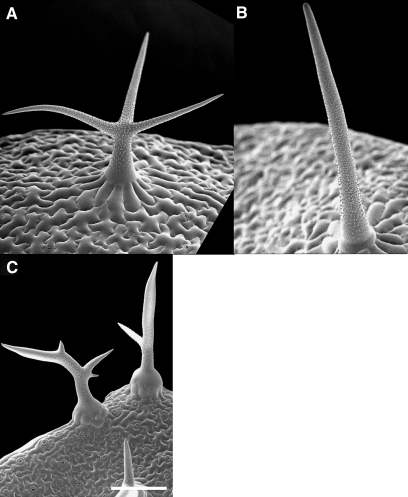
Comparison of blt-1 and sti-ab Mutants.
Higher and lower magnification of stem trichomes of (A, E) wild-type, (B, F) blt-1, (C, G) sti-ab, and (D, H) blt-1 sti-ab double mutant.
(I–K) Fluorescent, merged and DIC images of Col stage three trichome expressing gfp–BLT fusion protein.
(L–N) Fluorescent, merged, and DIC images of sti mutant stage four trichome expressing TdimerRed–cSTI fusion protein. Arrows highlight regions of enhanced fluorescence.
Bars in (A–D) and (I–N) represent 10 μm and in (E–H) 100 μm.
SVB
One final mutant was discovered by analyzing the phenotype of plants with T-DNA inserts in the most highly expressed gene (At1g56580) in the gl3–sst sim transcriptome. This gene encodes a protein with a conserved domain of unknown function (DUF538). The gene was expressed 3.9- and 15.1-fold higher in the double mutant than in either wild-type trichomes or processed shoot tissue, respectively. Two different T-DNA insertions in this gene resulted in plants with similar phenotypes. The trichomes of the mutants were smaller and exhibited branches of variable length and number (Figure 5C). For this reason, we call the gene SMALLER TRICHOME with VARIABLE BRANCHES (SVB). Because attempts to complement the mutant with either GFP-fused or unfused constructs using the GL2 promoter failed, further studies will be required to begin to understand the molecular function of the protein encoded by SVB.
Triple Mutant Analysis
The new mutants were found because of enhanced gene expression in the gl3–sst sim background. To determine whether the expression of these genes plays a role in the gl3–sst sim phenotype, triple mutants were generated for three of the new mutant genes. Figure 9 shows the phenotype of the gl3–sst sim double mutant compared to those of the gl3–sst sim-1 blt-1, gl3–sst sim-1 pel3-1, and gl3–sst sim-1 svb-1 triple mutants (see Methods). The differences between the double and triple mutants were subtle. The most obvious difference was between the double and blt triple (Figure 9A and 9B). All the trichomes in the triple mutants lacked branches and exhibited blunt tips, whereas some members of the cluster found on the double were branched and/or exhibited pointed tips. In the case of the svb triple, trichomes with obvious papillae were not observed (Figure 9D). In the double, a couple of spikes in each mature cluster typically contain papillae. For the pel3 triple, the trichomes may have been more slender (Figure 9C). However, the plants used in this study were grown at a higher humidity where pel3 trichomes appear more normal. In all, the triple mutant phenotypes were not as severe as the single mutant phenotype. This was likely because most of the trichomes in the double mutant trichome clusters never reached a developmental stage at which the functions of the newly discovered genes would have had their maximal developmental effects.
Figure 9.
SEM Analysis of Triple Mutant Leaf Trichome Clusters.
(A) gl3–sst sim doulble mutant.
(B) gl3–sst sim-1 blt-1.
(C) gl3–sst sim-1 pel3-11.
(D) gl3–sst sim-1 svb-1.
All bars = 100 μm.
DISCUSSION
Validation of Datasets
The transcriptome profiles of wild-type mature trichomes and gl3–sst sim and hdg2 mutant trichomes have been analyzed. The plants used in all these studies, except for those used to isolate the hdg2 trichomes, were grown during different times of the year in which the humidity varied, and, in the case of gl3–sst sim, under different lighting regimens (continuous or 16 h light, 8 h dark). Further, the hybridizations for all but hdg2 were performed at two different facilities. All of these parameters are known to generate experimentally induced variance. Given these circumstances, the probesets showing similar levels of expression should reflect genes that are truly expressed in trichomes under most conditions and not genes inadvertently expressed due to specific environmental or experimental factors.
Given that a large degree of experimental variance was expected in these datasets, it was important to find some measures with which to validate the data. In a previous report, qPCR was used to verify a small subset of arbitrarily chosen genes from a preliminary Affymetrix analysis involving one batch of isolated trichomes and processed shoots (Marks et al., 2008). In these datasets, AT3G61260 and AT5G02500 were expressed at similar levels, and AT3G59010 expression was higher in trichomes and AT3G19710 expression was higher in the processed shoot. The qPCR results using cDNA derived from processed shoot and isolated trichomes reflected these findings. In the much larger datasets presented in this report, consisting of five replications of isolated mature wild-type trichomes and four replications of processed shoots, these same trends were maintained. Additional validation comes from a composite of previous reports describing genes that are more highly expressed in mature trichomes. As shown in Table 1, previous mutational analyses identified 12 transcription factors that play a role in trichome development. Eleven of these were previously shown to be preferentially up-regulated in trichomes, whereas TTG1 was shown to be expressed in both leaf trichome and non-trichome cells (see references in Table 1 and Baudry et al., 2004). With the exception of HDG11 and EGL3, the mature trichome and processed leaf datasets showed the same trends. Other studies have shown that genes such as YRE, SIM, and TBR are also preferentially expressed in trichomes, and this also is reflected in the mature trichome vs. processed shoot datasets. Additional validation comes from the expectation that genes encoding proteins involved in photosynthesis would be more highly expressed in processed shoots, and, indeed, this was found to be the case. The known expression patterns of genes could also be used to validate the gl3–sst sim and hdg2 datasets. Most of the same trends seen in the comparison between wild-type and processed shoot profiles were found for similar comparisons between the datasets for the mutants and the processed shoots. The exception was that the known expression patterns for all 12 transcription factors were reflected in the comparison between the gl3–sst sim trichome and processed shoot profiles. Also, for gl3–sst sim, the co-expression of MYB30 with genes previously shown to be co-expressed with MYB30 serves as validation. An additional source of validation for the hdg2 mutant profile was that the hdg2 mutant used for Affymetrix analysis contained an insertion towards the 5’ end of the HDG2 coding sequence, and the expression of the HDG2 gene was not detected in the hdg2 mutant profile. All of these findings support the validity of the datasets.
Search for New Mutants
A key goal of this research was to determine whether comparative analyses between trichome and shoot transcriptional profiles could aid in the identification of new trichome mutants. In this regard, the gl3–sst sim trichomes were thought to offer a good resource for a transcriptional profile that mimics that of immature trichomes. They are composed of large clusters of cells, and have many attributes associated with immature trichomes. For example, many cells in the clusters are small, have thin cell walls, and exhibit low levels of endoreduplication (Marks et al., 2007). Most importantly, GL1 expression was elevated in these cells compared to the expression level in mature trichomes. This latter finding was confirmed in the analysis of the double mutant transcriptional profile. A previously published trichome dataset was obtained by using microcapillary pipettes to withdraw small quantities of cytoplasm from developing trichomes (Kryvych et al., 2008). That study identified GASA4 as a gene highly expressed in immature trichomes. This gene was found to be the third highest expressed gene in the double mutant profile presented in this report. Together, these findings support the notion that the cells within the double mutant trichome clusters share attributes with immature trichomes.
The main strategy used to search for new mutants was to rank genes based on differential gene expression. Several types of gene lists were generated by either comparing the profiles of selected genes such those encoding transcription factors or the total GeneChip profiles. Insertion lines were obtained from ABRC for genes showing the highest level of differential expression. Several hundred lines were ordered, corresponding to a comparable number of differentially expressed genes. For this search, the primary screen was to determine whether plants with altered trichome morphology were present in the original populations obtained from the stock center. Only lines containing plants with altered trichome morphology were further characterized. It is likely that this initial analysis missed some mutants, as not all lines contain seeds homozygous for the selected insertions.
One comparison that proved productive was that generated by ranking the expression levels of transcription factor genes expressed in gl3–sst sim trichomes but not in processed shoots. Seven of 12 transcription factors known to be important for trichome formation were in the top 10 ranked genes. HDG2, an additional transcription factor in the list, was shown to be important for trichome development. Recently, MYB5, the 11th ranked gene, also was shown to have a minor redundant role in trichome development (Gonzalez et al., 2009). A role for MYB30, the 12th ranked gene, has yet to be identified. But it is possible that MYB30 mutant trichomes have chemical alterations that are not detected by visual inspection. Mutants blt and pel3 were identified in the ranked list of all genes expressed in gl3–sst sim trichomes but not in processed shoots. In the gl3–sst sim gene list, BLT and PEL3 were ranked 72 and 76, whereas, in mature trichomes, BLT expression was not detected and PEL3 ranked 333. The final gene identified in this study was SVB. This represented the highest expressed gene in the gl3–sst sim profile. SVB expression was 3.9 and 17.7-fold higher in gl3–sst sim trichomes than in either mature trichomes or processed shoots, respectively.
New Mutants
HDG2
The identification of hdg2 as a trichome mutant is important because it represents the first transcription factor mutant that is only affected in cell wall maturation without changing overall trichome morphology. The key phenotypic difference was that the mutant trichome cell walls appeared more transparent when observed under a Nikon fiber optics ring light attached to a stereomicroscope. Nakamura et al. (2006) previously characterized lines containing T-DNA insertions in HDG2. In their study, they reported on the trichome-specific expression pattern of HDG2, but, under their growth conditions, no differences between hdg2 mutants and wild-type were noted.
Closer examination of the mutant trichomes revealed that the papillae on the outer trichome surfaces were reduced. This was associated with a loss of phosphorous that was previously shown to be associated with wild-type trichome papillae. The lack of the phosphorous-containing compounds was associated with the loss of uncharacterized occlusions in the papillae that were seen in wild-type papillae via TEM. The TEM analysis also showed that the mutant trichomes had a reduced outer cuticle layer. Preliminary results of histochemical staining of the hdg2 trichomes suggest that, compared to wild-type, the cell walls of the branches contain fewer pectins (Ruthenium red staining) and cellulosic compounds (Tinopal LPW staining) and similar levels of lignin (personal communication, Dr Haigler, North Carolina State University). These differences could be responsible for the difference in staining shown in the TEM. However, more work on the chemistry of the mutant trichome cell walls will need to be completed to achieve a better understanding mutant cell wall phenotype.
The papillae are first visible during middle to late stage five of trichome development, when the trichomes are expanding most rapidly. Thus, the many genes regulated by HDG2 and required for papillae development are likely expressed during this stage. Given that HDG2 is highly expressed in mature trichomes, it is possible that some of the HDG2-regulated genes are required for the final maturation of trichomes. HDG2 was expressed at similar levels in mature and gl3–sst sim mutant trichomes. Because the gl3–sst sim trichomes are predicted to be blocked at a stage earlier than stage five, HDG2 may also have a function in controlling earlier stages of trichome development. However, phenotypes associated with abnormal pre-stage five trichome development, such as altered branch number, were not seen. This could indicate: (1) that there were subtle phenotypes associated with a block in early hdg2 trichome development that were not detected, (2) that redundant factors compensated for the loss of HDG2 function during early trichome development, or (3) that HDG2 is up-regulated during early trichome development in anticipation of stage five. There is some support for option (2). HDG2 is most closely related to the HDG family genes ATML1 and PDF2 (Nakamura et al., 2006). These two genes are required for protodermal tissue cell fate (Abe et al., 2003), and, as found in this study, were highly expressed in gl3–sst sim trichomes (see Supplemental File 1).
HDG2 is the fourth member of the HDG family found to have a role in trichome development. Interestingly, each member has a distinct role in trichome development. GL2 was the first HDG gene identified (Rerie et al., 1994). Mutations in GL2 result in a loss of aerial trichome expansion. Instead, the incipient gl2 mutant trichomes expand in the plane of the leaf surface and the mutant trichome cell walls maintain a glassy appearance, lacking papillae (Koornneef et al., 1982). Like HDG2, GL2 is highly expressed in both gl3–sst sim and mature trichomes. Weak mutants of gl2 exhibit trichomes that expand aerially and form branches, but the trichomes on these weak mutants maintain the glassy appearance of immature trichomes. This suggests that GL2 also regulates genes required for cell wall maturation. The products of the HDG11 and 12 genes promote proper branch formation. Loss-of-function hdg11 mutants exhibit extra branched trichomes and, concomitantly, hdg12 mutations enhance the extra-branched phenotype of hdg11 (Nakamura et al., 2006). These trichomes do not show obvious cell wall abnormalities. HDG11 was expressed at a relatively low level in both gl3–sst sim and wild-type trichomes, whereas HDG12 was only detected at a very low level in gl3–sst sim trichomes. Thus, the expression of HDG11 and 12 may be restricted to stage three during branch formation and only be needed at low levels.
To begin to identify the targets of HDG2, the transcriptome profile of mature hdg2 mutant trichomes was generated. The analysis of this profile is ongoing. However, two features of the profile stood out. The first was that GL1 was more highly expressed in the mutant. The prolonged expression of GL1 could account for the juvenile appearance of the hdg2 trichomes. Thus, HDG2 may be a negative regulator of GL1. As HDG2 expression is high during early trichome development when GL1 expression declines, such negative regulation could be an early function of HDG2 in trichome development. The other feature of the hdg2 profile was the reduced expression of CYP94C. There is only one member of this P450 subclass in Arabidopsis, which encodes an enzyme that mediates the omega oxidation of fatty acids and is required for the synthesis of the dicarboxylic acids (Kandel et al., 2007). In Arabidopsis, the main monomer used to create cutin is a dicarboxylic acid (Pollard et al., 2008). Thus, the reduced cuticle layer of hdg2 could be caused in part by the reduced expression of CYP94C.
PEL3
PEL3 was originally identified in a forward genetic screen for cuticle mutants (Tanaka et al., 2004). The mutants were identified in a screen for plants with enhanced leaf staining by toluidine blue. The pel3 mutant showed patchy epidermal staining, but no trichome defect was noted. Here, we show that when plants are subjected to low humidity, some pel3 trichomes become entangled, causing expanding leaves to crinkle. This was the same phenotype noted for wax2 mutants; however, the tangling was attributed to trichome fusion. In the present report, we did not distinguish between tangling and fusion. Given the cuticle phenotype of hdg2, PEL3 represents a candidate for regulation by HDG2. However, comparable levels of PEL3 expression were observed in both wild-type and hdg2 mutant trichomes; this does not preclude the possibility that HDG2 may play a role in controlling PEL3 expression during early trichome development, as the hdg2 profile was obtained from mature trichomes.
PEL3 is a member of the BADH acyl-transferase gene family. Members of this family are responsible for the synthesis of a wide range of biomolecules (D'Auria, 2006). PEL3 protein shows possible overlap in localization with the related CER2 protein. CER2 is required for the formation of the C30 component of epidermal waxes (Lai et al., 2007), and CER2 protein was localized to both nuclear and non-nuclear cellular compartments (Xia et al., 1997). In addition, CER2 previously was shown to be preferentially expressed in developing trichomes, which is in agreement with the present study in which CER2 is expressed at a 4.5-fold higher level in gl3–sst sim trichomes compared to wild-type mature trichomes (see Supplemental Table 1). Given that large quantities of trichomes can be isolated and that the biochemistry of trichomes can be manipulated, future analyses with trichomes should be useful for studying the functions of genes involved in wax and cuticle formation.
BLT
The blt mutant strongly resembles sti, with a phenotype characterized by unbranched trichomes. In addition, a new phenotype of blunt trichome tips for sti was noted that is shared with blt. The genetic and localization studies suggest that the encoded STI and BLT proteins may function in the same pathway. Consistent images from multiple transformants for each construct showed that enhanced fluorescence was detected at the tips of stage three and four expanding trichome branches. These are the stages at which there is a transition from blunt branch tips to sharpened tips, which defines the beginning of stage four. The expression of both BLT and STI was detected in the gl3–sst sim trichomes, but not in mature trichomes, which is consistent with roles of these genes in early trichome development. Interestingly, leaf trichomes on either mutants occasionally develop a bulge towards the trichome base. This phenotype is very similar to that of zwi, suggesting that ZWI may play a role in the same pathway (Hulskamp, 2004). ZWI encodes a kinesin microtubule motor protein that contains a long coiled-coil domain (Oppenheimer et al., 1997; Reddy et al., 1997). Thus, it is possible that the coiled-coil domains of ZWI and BLT proteins could interact. Future studies of BLT may help shed light on the molecular function of STI, which encodes a protein related to prokaryotic replication factors, yet appears not to play a role in trichome DNA replication (Ilgenfritz et al., 2003).
In conclusion, transcriptome profiles have been generated for mature wild-type Arabidopsis trichomes, and for trichomes from two mutants. The data were mined to identify new trichome mutants. This search has not been exhaustive and new comparative analyses will likely result in the identification of additional new mutants. The data also show that many genes expressed in trichomes are likely active in the production of new cell wall material. The manipulation of these genes should aid in our understanding of how plant cell walls develop and how cell shape is controlled.
METHODS
Affymetrix Analysis
Wild-type plants were grown under 24 h light during different times of the year in a growth room controlled for temperature (23°C) but not humidity at the University of Minnesota. Two replications of gl3–sst sim-1 plants were similarly grown at the University of Minnesota, and a third replication was grown at the Samuel Roberts Noble Foundation under a 16/8 h light/dark regime. All three replications of the hdg2-1 mutant were co-grown at the University of Minnesota. The isolation of trichomes, of RNA, and the generation of Affymetrix probes were preformed as described previously (Marks et al., 2008). Three of the wild-type, three of the processed shoot, and two of gl3–sst sim probes were hybridized to the ATH1 GeneChip by the University of Minnesota hybridization facility. The remaining probes were processed at the Samuel Roberts Noble Foundation. The Expressionist Refiner Array software package was used to correct for gradient distortions in the chips and to make present/absent calls. Expressionist Analyst was used to normalize all data to an arithmetic mean of 1000 (each chip independently), perform standard Student's t-tests, and to group the data. Data were subsequently transferred to Excel spreadsheets and ranked according to mean values or fold-differences and P-values. All cell files containing the Affymetrix data are available from ArrayExpress (www.ebi.ac.uk/microarray-as/ae/). The ID numbers of the raw uncorrected non-normalized cell files for gl3–sst sim leaf trichomes, Columbia leaf trichomes, Columbia shoot tissue, and hdg2-2 leaf trichomes are E-MEXP-2013, E-MEXP-2008, E-MEXP-2014, and E-MEXP-2022, respectively.
Mutant Alleles
The first HDG2 mutant allele, hdg2-1, was identified in 1998 during a screen for trichome mutants in an activation tagged population of transformants (Weigel et al., 2000). The characterization of the mutant via TAIL PCR analysis showed that the mutant contained a T-DNA insertion in the second intron of HDG2 (Liu et al., 1995). The second allele, hdg2-2, was derived from homozygous line SALK_127828C (insert in the seventh exon). The hdg2-2 allele was used for the Affymetrix analysis. The two PEL3 mutant alleles, –11 and –12, were derived from SALK_062580 and SALK_036624, respectively (both insertions in the single intron). Of note, the original SALK_036624 seed stock contained a second mutant with long hypocotyls that segregated independently of the PEL3 locus. The two SVB mutant alleles, –1 and –2, were derived from the homozygous lines SALK_073071C and SALK_015997C, respectively (no introns). The two BLT mutant alleles, –1 and –2, were derived from SAIL 632_G06 (CS827202) and GT_5_100529 (CS164367), respectively (no introns). All seed stocks were obtained from ABRC (www.Arabidopsis.org).
Gene Constructs
For in planta expression of BLT, PEL3, HDG2, and STI, modified pEGAD vectors were used (Cutler et al., 2000). As previously described, the CaMV 35S RNA promoter of pEGAD was replaced with the promoter from either MYB5 or TRY, both of which drive expression in trichomes, to generate pEGAD MYB5:gfp and pEGAD TRY:gfp (Esch et al., 2003). To create pEGAD MYB:5TDimer2red, which was used for expressing STI, the plasmid pRSETB-TDimer2 was obtained from Dr R.Y. Tsien (University of California at San Diego). This plasmid contains a dimerized version of dsRED called TDimer2 (Campbell et al., 2002). Primers flanking TDimer2, with added 5’ AgeI and 3’ BsrGI restriction enzyme sites, were used to amplify the TDimer2 coding region. The GFP coding sequence was removed from pEGAD MYB5:gfp by digestion Age/BsrGI and replaced with the corresponding TDimer2 fragment to produce pEGAD MYB5:Tdimer2red. The vectors were further modified by insertion of the Gateway RFA fragment into the SmaI site located downstream of either the GFP or TDimer2 coding regions (Invitrogen; www.invitrogen.com). The coding regions of BLT, PEL3, HDG2, and STI were amplified via PCR, and cloned into the Gateway vector pCR8 (Invitrogen). Clones containing the coding regions in the proper orientation were identified by DNA sequencing, and were used in recombination reactions with the Gateway-modified pEGAD vectors. The template for the BLT coding sequence was pUNI151. At1g64690 was obtained from ABRC (C63754). The templates for HDG2 and PEL3 were cDNAs synthesized using total RNA isolated from Col and gl3–sst shoot apexes, respectively. The template for the STI coding region was a cDNA clone isolated from the CD16 Arabidopsis cDNA library deposited in ABRC by Dr Joe Keeber (University of North Carolina).
Image Analysis (Including Supplementary Movies)
Fluorescent microscopy was performed using either Leica TCS SP2 AOBS confocal or Nikon Diaphot 200 microscopes (Leica Microsystems, www.leica-microsystems.com; Nikon Instruments Inc., www.nikoninstruments.com/). For detection of Tdimer12 red fluorescence, a filter set with emission captured at 585 ± 10 nm was used (Chroma Technology Corp., www.chroma.com). The resulting signal using this filter was yellow, as shown in Figure 8L and 8M.
SEM analyses were performed as previously described (Ahlstrand, 1996; Esch et al., 2004) using an Emitech K1150 Cyro-preparation system and a Hitachi S3500N Scanning Electron Microscope (Emitech Technologies Ltd, www.emitech.co.uk; Hitachi High Technogies, Inc., www.hitachi-hhta.com).
Movies were generated by taking sequential still images with a Canon G5 camera attached to a Nikon SMZ1500 stereomicroscope at 5-min intervals. Images were processed using iPhoto and iMovie software (Nikon Inc., www.nikonusa.com; Apple Computer, www.apple.com). The Supplementary pel3 and Col wild-type movies consisted of approximately 500 images displayed at 10 images per second, depicting trichome development over approximately 40 h.
For TEM analyses, all samples were processed and examined as previously described (Marks et al., 2008).
Isolation of Double and Triple Mutants
To generate triple mutants, pel3-11, svb-2, and blt-1 were each individually crossed to gl3–sst sim-1 double mutants. Within the gl3–sst sim x pel3 and x svb F2 populations, plants displaying the pel3 and svb phenotypes were selected. From the blt x gl3–sst sim F2 population, plants displaying an obvious gl3–sst blt phenotype were selected. F3 populations derived from the selected plants were screened for the presence of plants with clusters of multicellular trichomes, which is the hallmark of the gl3–sst sim genotype. These plants with multicellular trichomes were considered the triple mutants, as no wild-type plants were seen within these segregating populations. This latter finding confirmed the homozygosity of the original selected F2 plants. Seeds were collected from the triple mutants and the resulting F4 generation plants were used for the SEM analysis shown in Figure 9.
The sti mutant used in this study was a gift from Drs Abby Telfer and Scott Poethig (University of Pennsylvania). Previous crosses of this EMS-induced mutant with sti-146 (gift from Dr Martin Hülskamp, University of Köln) confirmed allelism, and the new allele was named sti-ab. To generate the double mutant, sti-ab was crossed with blt-1, which was induced by the insertion of a T-DNA containing a bar gene. F2 plants with branched trichomes were present in the resulting F2 population, which showed that the two mutants were non-allelic. Basta-resistant F2 plants showing the unbranched trichome phenotype were selected. F3 populations derived from the selected F2s were tested for basta resistance. F3 populations with all plants exhibiting unbranched trichomes yet segregating for basta resistance were considered to be derived from F2 plants homozygous for sti and hetereozygous for blt. The F4 seeds were collected from the resistant plants and the resulting F4 populations were tested for basta resistance. F4 populations containing all basta-resistant plants were considered to be derived from F3 sti blt double mutants.
SUPPLEMENTARY DATA
Supplementary Data are available at Molecular Plant Online.
FUNDING
This work was funded by National Science Foundation awards 0343982 and 0605033, a University of Minnesota College of Biological Sciences sabbatical leave award, and the Samuel Roberts Noble Foundation.
Acknowledgments
We thank Gilbert Ahlstrand and Gail Celio of the University of Minnesota CBS Imaging Center for help with the TEM analysis. We thank Tracy Anderson and Mark Sanders (UMN CBS Imaging Center) and Elison Blancaflor (Samuel Roberts Noble Foundation) for help with the confocal microscopy. We thank Candace Haigler (North Carolina State University) for sharing unpublished results concerning the trichome cell walls of the hdg2 mutant. No conflict of interest declared.
References
- Abe M, Katsumata H, Komeda Y, Takahashi T. Regulation of shoot epidermal cell differentiation by a pair of homeodomain proteins in Arabidopsis. Development. 2003;130:635–643. doi: 10.1242/dev.00292. [DOI] [PubMed] [Google Scholar]
- Abe T, Thitamadee S, Hashimoto T. Microtubule defects and cell morphogenesis in the lefty1lefty2 tubulin mutant of Arabidopsis thaliana. Plant Cell Physiol. 2004;45:211–220. doi: 10.1093/pcp/pch026. [DOI] [PubMed] [Google Scholar]
- Ahlstrand G. Low-temperature low-voltage scanning microscopy (LTLVSEM) of uncoated frozen biological materials: a simple alternative. In: Bailey G, Corbett J, Dimlich R, Michael J, Zaluzec N, editors. Proceedings of Microscopy Microanalysis. San Francisco: San Francisco Press; 1996. p. 918. [Google Scholar]
- Basu D, El-Assal SED, Le J, Mallery EL, Szymanski DB. Interchangeable functions of Arabidopsis PIROGI and the human WAVE complex subunit SRA1 during leaf epidermal development. Development. 2004;131:4345–4355. doi: 10.1242/dev.01307. [DOI] [PubMed] [Google Scholar]
- Basu D, Le J, El-Essal SED, Huang S, Zhang CH, Mallery EL, Koliantz G, Staiger CJ, Szymanski DB. DISTORTED3/SCAR2 is a putative Arabidopsis WAVE complex subunit that activates the Arp2/3 complex and is required for epidermal morphogenesis. Plant Cell. 2005;17:502–524. doi: 10.1105/tpc.104.027987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 2004;39:366–380. doi: 10.1111/j.1365-313X.2004.02138.x. [DOI] [PubMed] [Google Scholar]
- Burk DH, Liu B, Zhong R, Morrison WH, Ye ZH. A katanin-like protein regulates normal cell wall biosynthesis and cell elongation. Plant Cell. 2001;13:807–828. [PMC free article] [PubMed] [Google Scholar]
- Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY. A monomeric red fluorescent protein. Proc. Natl Acad. Sci. U S A. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chary SN, Hicks GR, Choi YG, Carter D, Raikhel NV. Trehalose-6-phosphate synthase/phosphatase regulates cell shape and plant architecture in Arabidopsis. Plant Physiol. 2008;146:97–107. doi: 10.1104/pp.107.107441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman ML, et al. SIAMESE, a plant-specific cell cycle regulator, controls endoreplication onset in Arabidopsis thaliana. Plant Cell. 2006;18:3145–3157. doi: 10.1105/tpc.106.044834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler SR, Ehrhardt DW, Griffitts JS, Somerville CR. Random GFP::cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc. Natl Acad. Sci. U S A. 2000;97:3718–3723. doi: 10.1073/pnas.97.7.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Auria JC. Acyltransferases in plants: a good time to be BAHD. Curr. Opin. Plant Biol. 2006;9:331–340. doi: 10.1016/j.pbi.2006.03.016. [DOI] [PubMed] [Google Scholar]
- Downes BP, Stupar RM, Gingerich DJ, Vierstra RD. The HECT ubiquitin-protein ligase (UPL) family in Arabidopsis: UPL3 has a specific role in trichome development. Plant J. 2003;35:729–742. doi: 10.1046/j.1365-313x.2003.01844.x. [DOI] [PubMed] [Google Scholar]
- El-Assal Sel D, Le J, Basu D, Mallery EL, Szymanski DB. Arabidopsis GNARLED encodes a NAP125 homolog that positively regulates ARP2/3. Curr. Biol. 2004;14:1405–1409. doi: 10.1016/j.cub.2004.06.062. [DOI] [PubMed] [Google Scholar]
- El-Din El-Assal S, Le J, Basu D, Mallery EL, Szymanski DB. DISTORTED2 encodes an ARPC2 subunit of the putative Arabidopsis ARP2/3 complex. Plant J. 2004;38:526–538. doi: 10.1111/j.1365-313X.2004.02065.x. [DOI] [PubMed] [Google Scholar]
- Esch JJ, Chen MA, Hillestad M, Marks MD. Comparison of TRY and the closely related At1g01380 gene in controlling Arabidopsis trichome patterning. Plant J. 2004;40:860–869. doi: 10.1111/j.1365-313X.2004.02259.x. [DOI] [PubMed] [Google Scholar]
- Esch JJ, Chen M, Sanders M, Hillestad M, Ndkium S, Idelkope B, Neizer J, Marks MD. A contradictory GLABRA3 allele helps define gene interactions controlling trichome development in Arabidopsis. Development. 2003;130:5885–5894. doi: 10.1242/dev.00812. [DOI] [PubMed] [Google Scholar]
- Esch JJ, Oppenheimer DG, Marks MD. Characterization of a weak allele of the GL1 gene of Arabidopsis thaliana. Plant Mol. Biol. 1994;24:203–207. doi: 10.1007/BF00040586. [DOI] [PubMed] [Google Scholar]
- Falbel TG, Koch LM, Nadeau JA, Segui-Simarro JM, Sack FD, Bednarek SY. SCD1 is required for cytokinesis and polarized cell expansion in Arabidopsis thaliana [corrected] Development. 2003;130:4011–4024. doi: 10.1242/dev.00619. [DOI] [PubMed] [Google Scholar]
- Folkers U, et al. The cell morphogenesis gene ANGUSTIFOLIA encodes a CtBP/BARS-like protein and is involved in the control of the microtubule cytoskeleton. EMBO J. 2002;21:1280–1288. doi: 10.1093/emboj/21.6.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Gong XM, Cao WH, Zhao JF, Fu LQ, Wang XC, Schumaker KS, Guo Y. SAD2 in Arabidopsis functions in trichome initiation through mediating GL3 function and regulating GL1, TTG1 and GL2 expression. J. Integr. Plant Biol. 2008;50:906–917. doi: 10.1111/j.1744-7909.2008.00695.x. [DOI] [PubMed] [Google Scholar]
- Gonzalez A, Mendenhall J, Huo Y, Lloyd A. TTG1 complex MYBs, MYB5 and TT2, control outer seed coat differentiation. Dev. Biol. 2009;325:412–421. doi: 10.1016/j.ydbio.2008.10.005. [DOI] [PubMed] [Google Scholar]
- Hase Y, Trung KH, Matsunaga T, Tanaka A. A mutation in the uvi4 gene promotes progression of endo-reduplication and confers increased tolerance towards ultraviolet B light. Plant J. 2006;46:317–326. doi: 10.1111/j.1365-313X.2006.02696.x. [DOI] [PubMed] [Google Scholar]
- Hayashi S, Ishii T, Matsunaga T, Tominaga R, Kuromori T, Wada T, Shinozaki K, Hirayama T. The glycerophosphoryl diester phosphodiesterase-like proteins SHV3 and its homologs play important roles in cell wall organization. Plant Cell Physiol. 2008;49:1522–1535. doi: 10.1093/pcp/pcn120. [DOI] [PubMed] [Google Scholar]
- Hulskamp M. Plant trichomes: a model for cell differentiation. Nat. Rev. Mol. Cell Biol. 2004;5:471–480. doi: 10.1038/nrm1404. [DOI] [PubMed] [Google Scholar]
- Hulskamp M, Misra S, Jurgens G. Genetic dissection of trichome cell development in Arabidopsis. Cell. 1994;76:555–566. doi: 10.1016/0092-8674(94)90118-x. [DOI] [PubMed] [Google Scholar]
- Ilgenfritz H, Bouyer D, Schnittger A, Mathur J, Kirik V, Schwab B, Chua NH, Jurgens G, Hulskamp M. The Arabidopsis STICHEL gene is a regulator of trichome branch number and encodes a novel protein. Plant Physiol. 2003;131:643–655. doi: 10.1104/pp.014209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai KK, Ohashi Y, Tsuge T, Yoshizumi T, Matsui M, Oka A, Aoyama T. The A-type cyclin CYCA2;3 is a key regulator of ploidy levels in Arabidopsis endoreduplication. Plant Cell. 2006;18:382–396. doi: 10.1105/tpc.105.037309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby MJ, Falkenhan D, Mader MT, Brininstool G, Wischnitzki E, Platz N, Hudson A, Hulskamp M, Larkin J, Schnittger A. Transcriptional profiling of mature Arabidopsis trichomes reveals that NOECK encodes the MIXTA-like transcriptional regulator MYB106. Plant Physiol. 2008;148:1583–1602. doi: 10.1104/pp.108.126979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CS, Kolevski B, Smyth DR. TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell. 2002;14:1359–1375. doi: 10.1105/tpc.001404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel S, Sauveplane V, Compagnon V, Franke R, Millet Y, Schreiber L, Werck-Reichhart D, Pinot F. Characterization of a methyl jasmonate and wounding-responsive cytochrome P450 of Arabidopsis thaliana catalyzing dicarboxylic fatty acid formation in vitro. Febs J. 2007;274:5116–5127. doi: 10.1111/j.1742-4658.2007.06032.x. [DOI] [PubMed] [Google Scholar]
- Kang BH, Busse JS, Bednarek SY. Members of the Arabidopsis dynamin-like gene family, ADL1, are essential for plant cytokinesis and polarized cell growth. Plant Cell. 2003;15:899–913. doi: 10.1105/tpc.009670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik V, Bouyer D, Schobinger U, Bechtold N, Herzog M, Bonneville JM, Hulskamp M. CPR5 is involved in cell proliferation and cell death control and encodes a novel transmembrane protein. Curr. Biol. 2001;11:1891–1895. doi: 10.1016/s0960-9822(01)00590-5. [DOI] [PubMed] [Google Scholar]
- Kirik V, Lee MM, Wester K, Herrmann U, Zheng Z, Oppenheimer D, Schiefelbein J, Hulskamp M. Functional diversification of MYB23 and GL1 genes in trichome morphogenesis and initiation. Development. 2005;132:1477–1485. doi: 10.1242/dev.01708. [DOI] [PubMed] [Google Scholar]
- Kirik V, Mathur J, Grini PE, Klinkhammer I, Adler K, Bechtold N, Herzog M, Bonneville JM, Hulskamp M. Functional analysis of the tubulin-folding cofactor C in Arabidopsis thaliana. Curr. Biol. 2002;12:1519–1523. doi: 10.1016/s0960-9822(02)01109-0. [DOI] [PubMed] [Google Scholar]
- Kirik V, Simon M, Huelskamp M, Schiefelbein J. The ENHANCER OF TRY AND CPC1 gene acts redundantly with TRIPTYCHON and CAPRICE in trichome and root hair cell patterning in Arabidopsis. Dev. Biol. 2004a;268:506–513. doi: 10.1016/j.ydbio.2003.12.037. [DOI] [PubMed] [Google Scholar]
- Kirik V, Simon M, Wester K, Schiefelbein J, Hulskamp M. ENHANCER of TRY and CPC 2 (ETC2) reveals redundancy in the region-specific control of trichome development of Arabidopsis. Plant Mol. Biol. 2004b;55:389–398. doi: 10.1007/s11103-004-0893-8. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Dellaert SWM, van der Veen JH. EMS- and radiation-induced mutation frequencies at individual loci in Arabidopsis thaliana (L) Heynh. Mutation Research. 1982;93:109–123. doi: 10.1016/0027-5107(82)90129-4. [DOI] [PubMed] [Google Scholar]
- Kryvych S, Nikiforova V, Herzog M, Perazza D, Fisahn J. Gene expression profiling of the different stages of Arabidopsis thaliana trichome development on the single cell level. Plant Physiol. Bioch. 2008;46:160–173. doi: 10.1016/j.plaphy.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Kurata T, Kawabata-Awai C, Sakuradani E, Shimizu S, Okada K, Wada T. The YORE-YORE gene regulates multiple aspects of epidermal cell differentiation in Arabidopsis. Plant J. 2003;36:55–66. doi: 10.1046/j.1365-313x.2003.01854.x. [DOI] [PubMed] [Google Scholar]
- Lai C, Kunst L, Jetter R. Composition of alkyl esters in the cuticular wax on inflorescence stems of Arabidopsis thaliana cer mutants. Plant J. 2007;50:189–196. doi: 10.1111/j.1365-313X.2007.03054.x. [DOI] [PubMed] [Google Scholar]
- Larkin JC, Brown ML, Schiefelbein J. How do cells know what they want to be when they grow up? Lessons from epidermal patterning in Arabidopsis. Annu. Rev. Plant Biol. 2003;54:403–430. doi: 10.1146/annurev.arplant.54.031902.134823. [DOI] [PubMed] [Google Scholar]
- Larkin JC, Oppenheimer DG, Pollock S, Marks MD. Arabidopsis GLABROUS1 gene requires downstream sequences for function. Plant Cell. 1993;5:1739–1748. doi: 10.1105/tpc.5.12.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin JC, Walker JD, Bolognesi-Winfield AC, Gray JC, Walker AR. Allele-specific interactions between ttg and gl1 during trichome development in Arabidopsis thaliana. Genetics. 1999;151:1591–1604. doi: 10.1093/genetics/151.4.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SF, Santini JM, Nicolaou O, Parish RW. A novel myb-related gene from Arabidopsis thaliana. FEBS Lett. 1996;379:117–121. doi: 10.1016/0014-5793(95)01461-6. [DOI] [PubMed] [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 1995;8:457–463. doi: 10.1046/j.1365-313x.1995.08030457.x. [DOI] [PubMed] [Google Scholar]
- Marks MD, Esch JJ. Initiating inhibition: control of epidermal cell patterning in plants. EMBO Rep. 2003;4:24–25. doi: 10.1038/sj.embor.embor705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks MD, Esch J, Herman P, Sivakumaran S, Oppenheimer D. A model for cell-type determination and differentiation in plants. Symp. Soc. Exp. Biol. 1991;45:77–87. [PubMed] [Google Scholar]
- Marks MD, Gilding E, Wenger JP. Genetic interaction between glabra3-shapeshifter and siamese in Arabidopsis thaliana converts trichome precursors into cells with meristematic activity. Plant J. 2007;52:352–361. doi: 10.1111/j.1365-313X.2007.03243.x. [DOI] [PubMed] [Google Scholar]
- Marks MD, Gilding E, Betancur L, Chen F, Bauer S, Wenger JP, Dixon RA, Haigler CH. A new method for isolating large quantities of Arabidopsis trichomes for transcriptome, cell wall and other types of analyses. Plant J. 2008;56:483–492. doi: 10.1111/j.1365-313X.2008.03611.x. [DOI] [PubMed] [Google Scholar]
- Mathur J, Mathur N, Kernebeck B, Hulskamp M. Mutations in actin-related proteins 2 and 3 affect cell shape development in Arabidopsis. Plant Cell. 2003a;15:1632–1645. doi: 10.1105/tpc.011676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J, Mathur N, Kirik V, Kernebeck B, Srinivas BP, Hulskamp M. Arabidopsis CROOKED encodes for the smallest subunit of the ARP2/3 complex and controls cell shape by region specific fine F-actin formation. Development. 2003b;130:3137–3146. doi: 10.1242/dev.00549. [DOI] [PubMed] [Google Scholar]
- Mueller LA, Zhang P, Rhee SY. AraCyc: a biochemical pathway database for Arabidopsis. Plant Physiol. 2003;132:453–460. doi: 10.1104/pp.102.017236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Katsumata H, Abe M, Yabe N, Komeda Y, Yamamoto KT, Takahashi T. Characterization of the class IV homeodomain-leucine zipper gene family in Arabidopsis. Plant Physiol. 2006;141:1363–1375. doi: 10.1104/pp.106.077388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nita A. Genetic mapping and molecular characterization of tbr1 mutant in Arabidopsis thaliana. Ph.D. Thesis. University of Potsdam. 2005 [Google Scholar]
- Ojangu EL, Jarve K, Paves H, Truve E. Arabidopsis thaliana myosin XIK is involved in root hair as well as trichome morphogenesis on stems and leaves. Protoplasma. 2007;230:193–202. doi: 10.1007/s00709-006-0233-8. [DOI] [PubMed] [Google Scholar]
- Oppenheimer DG, Herman PL, Sivakumaran S, Esch J, Marks MD. A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell. 1991;67:483–493. doi: 10.1016/0092-8674(91)90523-2. [DOI] [PubMed] [Google Scholar]
- Oppenheimer DG, Pollock MA, Vacik J, Szymanski DB, Ericson B, Feldmann K, Marks MD. Essential role of a kinesin-like protein in Arabidopsis trichome morphogenesis. Proc. Natl Acad. Sci. U S A. 1997;94:6261–6266. doi: 10.1073/pnas.94.12.6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne CT, Zhang F, Lloyd AM. GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics. 2000;156:1349–1362. doi: 10.1093/genetics/156.3.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perazza D, Vachon G, Herzog M. Gibberellins promote trichome formation by uU-regulating GLABROUS1 in Arabidopsis. Plant Physiol. 1998;117:375–383. doi: 10.1104/pp.117.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard M, Beisson F, Li YH, Ohlrogge JB. Building lipid barriers: biosynthesis of cutin and suberin. Trends in Plant Science. 2008;13:236–246. doi: 10.1016/j.tplants.2008.03.003. [DOI] [PubMed] [Google Scholar]
- Qiu JL, Jilk R, Marks MD, Szymanski DB. The Arabidopsis SPIKE1 gene is required for normal cell shape control and tissue development. Plant Cell. 2002;14:101–118. doi: 10.1105/tpc.010346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaele S, Vailleau F, Leger A, Joubes J, Miersch O, Huard C, Blee E, Mongrand S, Domergue F, Roby D. A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. Plant Cell. 2008;20:752–767. doi: 10.1105/tpc.107.054858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy AS, Narasimhulu SB, Day IS. Structural organization of a gene encoding a novel calmodulin-binding kinesin-like protein from Arabidopsis. Gene. 1997;204:195–200. doi: 10.1016/s0378-1119(97)00546-5. [DOI] [PubMed] [Google Scholar]
- Reddy VS, Day IS, Thomas T, Reddy AS. KIC, a novel Ca2+ binding protein with one EF-hand motif, interacts with a microtubule motor protein and regulates trichome morphogenesis. Plant Cell. 2004;16:185–200. doi: 10.1105/tpc.016600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rerie WG, Feldmann KA, Marks MD. The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev. 1994;8:1388–1399. doi: 10.1101/gad.8.12.1388. [DOI] [PubMed] [Google Scholar]
- Saedler R. Role of DISTORTED2, GNARLED AND SPIRRIG in cell morphogenesis of Arabidopsis thaliana. Ph.D. Thesis. Universität zu Köln. 2005 [Google Scholar]
- Schellmann S, Schnittger A, Kirik V, Wada T, Okada K, Beermann A, Thumfahrt J, Jurgens G, Hulskamp M. TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J. 2002;21:5036–5046. doi: 10.1093/emboj/cdf524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinlapadech T, Stout J, Ruegger MO, Deak M, Chapple C. The hyper-fluorescent trichome phenotype of the brt1 mutant of Arabidopsis is the result of a defect in a sinapic acid: UDPG glucosyltransferase. Plant J. 2007;49:655–668. doi: 10.1111/j.1365-313X.2006.02984.x. [DOI] [PubMed] [Google Scholar]
- Spitzer C, Schellmann S, Sabovljevic A, Shahriari M, Keshavaiah C, Bechtold N, Herzog M, Muller S, Hanisch FG, Hulskamp M. The Arabidopsis elch mutant reveals functions of an ESCRT component in cytokinesis. Development. 2006;133:4679–4689. doi: 10.1242/dev.02654. [DOI] [PubMed] [Google Scholar]
- Sugimoto-Shirasu K, Stacey NJ, Corsar J, Roberts K, McCann MC. DNA topoisomerase VI is essential for endoreduplication in Arabidopsis. Curr. Biol. 2002;12:1782–1786. doi: 10.1016/s0960-9822(02)01198-3. [DOI] [PubMed] [Google Scholar]
- Szymanski DB, Jilk RA, Pollock SM, Marks MD. Control of GL2 expression in Arabidopsis leaves and trichomes. Development. 1998;125:1161–1171. doi: 10.1242/dev.125.7.1161. [DOI] [PubMed] [Google Scholar]
- Szymanski DB, Lloyd AM, Marks MD. Progress in the molecular genetic analysis of trichome initiation and morphogenesis in Arabidopsis. Trends Plant Sci. 2000;5:214–219. doi: 10.1016/s1360-1385(00)01597-1. [DOI] [PubMed] [Google Scholar]
- Szymanski DB, Marks MD, Wick SM. Organized F-actin is essential for normal trichome morphogenesis in Arabidopsis. Plant Cell. 1999;11:2331–2347. doi: 10.1105/tpc.11.12.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Tanaka H, Machida C, Watanabe M, Machida Y. A new method for rapid visualization of defects in leaf cuticle reveals five intrinsic patterns of surface defects in Arabidopsis. Plant J. 2004;37:139–146. doi: 10.1046/j.1365-313x.2003.01946.x. [DOI] [PubMed] [Google Scholar]
- Usadel B, et al. Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes, and comparison with known responses. Plant Physiol. 2005;138:1195–1204. doi: 10.1104/pp.105.060459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanzin GF, Madson M, Carpita NC, Raikhel NV, Keegstra K, Reiter WD. The mur2 mutant of Arabidopsis thaliana lacks fucosylated xyloglucan because of a lesion in fucosyltransferase AtFUT1. Proc. Natl Acad. Sci. U S A. 2002;99:3340–3345. doi: 10.1073/pnas.052450699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Tachibana T, Shimura Y, Okada K. Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science. 1997;277:1113–1116. doi: 10.1126/science.277.5329.1113. [DOI] [PubMed] [Google Scholar]
- Walker AR, Davison PA, Bolognesi-Winfield AC, James CM, Srinivasan N, Blundell TL, Esch JJ, Marks MD, Gray JC. The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell. 1999;11:1337–1350. doi: 10.1105/tpc.11.7.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JD, Oppenheimer DG, Concienne J, Larkin JC. SIAMESE, a gene controlling the endoreduplication cell cycle in Arabidopsis thaliana trichomes. Development. 2000;127:3931–3940. doi: 10.1242/dev.127.18.3931. [DOI] [PubMed] [Google Scholar]
- Weigel D, et al. Activation tagging in Arabidopsis. Plant Physiol. 2000;122:1003–1013. doi: 10.1104/pp.122.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienkoop S, Zoeller D, Ebert B, Simon-Rosin U, Fisahn J, Glinski M, Weckwerth W. Cell-specific protein profiling in Arabidopsis thaliana trichomes: identification of trichome-located proteins involved in sulfur metabolism and detoxification. Phytochemistry. 2004;65:1641–1649. doi: 10.1016/j.phytochem.2004.03.026. [DOI] [PubMed] [Google Scholar]
- Xia Y, Nikolau BJ, Schnable PS. Developmental and hormonal regulation of the Arabidopsis CER2 gene that codes for a nuclear-localized protein required for the normal accumulation of cuticular waxes. Plant Physiol. 1997;115:925–937. doi: 10.1104/pp.115.3.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Gonzalez A, Zhao M, Payne CT, Lloyd A. A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development. 2003;130:4859–4869. doi: 10.1242/dev.00681. [DOI] [PubMed] [Google Scholar]
- Zhao M, Morohashi K, Hatlestad G, Grotewold E, Lloyd A. The TTG1-bHLH-MYB complex controls trichome cell fate and patterning through direct targeting of regulatory loci. Development. 2008;135:1991–1999. doi: 10.1242/dev.016873. [DOI] [PubMed] [Google Scholar]
- Zhong R, Burk DH, Nairn CJ, Wood-Jones A, Morrison WH, 3rd, Ye ZH. Mutation of SAC1, an Arabidopsis SAC domain phosphoinositide phosphatase, causes alterations in cell morphogenesis, cell wall synthesis, and actin organization. Plant Cell. 2005;17:1449–1466. doi: 10.1105/tpc.105.031377. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.