Abstract
Rice, the primary source of dietary calories for half of humanity, is the first crop plant for which a high-quality reference genome sequence from a single variety was produced. We used resequencing microarrays to interrogate 100 Mb of the unique fraction of the reference genome for 20 diverse varieties and landraces that capture the impressive genotypic and phenotypic diversity of domesticated rice. Here, we report the distribution of 160,000 nonredundant SNPs. Introgression patterns of shared SNPs revealed the breeding history and relationships among the 20 varieties; some introgressed regions are associated with agronomic traits that mark major milestones in rice improvement. These comprehensive SNP data provide a foundation for deep exploration of rice diversity and gene–trait relationships and their use for future rice improvement.
Keywords: introgression, Oryza sativa, resequencing, SNP discovery
The genomes of domesticated rice, Oryza sativa, contain a wealth of information that can explain the large morphological, physiological, and ecological variation observed in the many varieties cultivated for food. To meet population demands by 2025, rice production must increase by 24% (1). The innovative use of genetic diversity will play a key role in reaching this ambitious goal.
The availability of complete genome sequences provides a starting point to understanding the tremendous diversity of the rice gene pool at a fine scale. Among the organisms with a high-quality genome sequence from at least one individual or strain, such as human, mouse, and Arabidopsis, genomewide surveys of SNP variation in small or moderately sized samples have captured significant portions of within-species variation. In human and mouse, for example, a sampling of 71 and 15 individuals captured 80% and 43% of the genotypic variation, respectively (2, 3). In the model plant, Arabidopsis, 20 diverse varieties captured >90% of the common genotypic variation in the species (4).
We initiated the OryzaSNP project (www.OryzaSNP.org) to discover genetic variation within 20 rice varieties and landraces. These varieties, the OryzaSNPset collection (Table S1), are genetically diverse and actively used in international breeding programs because of their wide range of agronomic attributes (5). Most varieties belong to the 2 main groups, indica and japonica, including tropical and temperate japonica, whereas others represent the aus, deep water, and aromatic rice groups. Adapting a hybridization approach previously used for human, mouse, and Arabidopsis (3, 6, 7), we determined SNP variation in 100 Mb of the rice genome, representing ≈80% of the nonrepetitive portion of the 390-Mb Nipponbare reference genome (8). Here, we describe the discovery of 159,478 high-quality, nonredundant SNPs distributed across the entire genomes of the OryzaSNPset. Relative to the model dicotyledenous plant Arabidopsis (4), typical haplotype blocks in indica rice varieties are longer (≈200 kb). Observed patterns of shared SNPs among groups indicate introgression caused by recent breeding or historical out-crossing events.
Results and Discussion
SNP Prediction and Coverage.
The nonrepetitive sequence (100.1 Mb) used to design 6 ultra-high-density tiling arrays was selected from the high-quality reference genome of O. sativa variety Nipponbare (temperate japonica) (8). The arrays interrogated 26.2% of the genome with low repeat, and therefore high genic content for SNP discovery, and targeted full or partial sequences corresponding to 57% of the 41,042 nontransposable-element-related gene models in The Institute for Genomic Research (TIGR) r5 database (http://rice.plantbiology.msu.edu/).
We randomly selected regions represented on the arrays for dideoxy sequencing to generate 3.6 Mb of double-stranded sequence across all 20 varieties, corresponding to 1.8 Mb of nonredundant sequence in the reference genome. From these sequences, a gold-standard set of curated polymorphisms from unambiguous alignments was compiled for quality assessment and training of our SNP predictors.
Two different computational methods were used for SNP discovery. A model-based (MB) approach, which considered the hybridization signature of a feature (corresponding to a position in the reference genome) and its tiling neighbors (corresponding to sequence bases in the immediate vicinity of that position) (2, 3, 6, 7), identified 242,196 nonredundant SNPs at nonrepetitive sites (Table 1 and Table S2). We also applied a support vector machine (SVM) machine learning (ML) approach that had been used previously for Arabidopsis array data (7). Training sets for the modified ML method included the SNPs in the gold-standard dataset, the experimental hybridization data, information about repetitive oligonucleotides tiled on the arrays, and known polymorphisms in the indica genome (SI Appendix and Table S3). The latter were identified by comparing the Nipponbare genome with a second reference genome from the indica cultivar 93-11 (9) and their use improved the performance of the ML method for SNP detection in indica varieties.
Table 1.
SNP predictions at nonrepetitive sites for varieties by variety group and prediction method
| Group, n | Mean SNPs per variety [recall (%):FDR (%)]* |
|||||||
|---|---|---|---|---|---|---|---|---|
| MBML-union | MBML-intersect | MB only | ML only | |||||
| Temperate japonica, 4 | 14,882 | [NR:NR] | 2,028 | [NR:NR] | 11,044 | [NR:NR] | 1,810 | [NR:NR] |
| Tropical japonica, 3 | 50,221 | [18.4:14.6] | 20,012 | [7.3:8.5] | 12,543 | [2.1:34.4] | 17,666 | [9.0:16.0] |
| Aromatic, 1 | 51,817 | [NR:NR] | 2,022 | [NR:NR] | 48,747 | [NR:NR] | 1,048 | [NR:NR] |
| Aus, 4 | 137,114 | [24.9:11.6] | 63,054 | [12.4:2.1] | 28,195 | [2.7:25.4] | 45,865 | [9.8:17.3] |
| Indica, 8 | 126,702 | [25.3:10.4] | 54,903 | [11.0:2.5] | 29,684 | [3.8:25.5] | 42,115 | [10.5:11.7] |
| All varieties, 20† | 91,203 | [27.8:12.3] | 38,080 | [9.7:3.2] | 24,040 | [10.2:24.7] | 29,083 | [7.9:14.0] |
*Recall and FDR not reported (NR) (in brackets) where <50 SNPs were available for evaluation, because of very low statistical power.
†Numbers are means over all varieties.
A set of 316,373 SNPs at nonrepetitive sites were predicted by the ML method (Table 1 and Table S2). Assessed on the gold-standard SNP dataset, a false discovery rate (FDR) of 8.3% and a recall of 20.9% were observed for ML at nonrepetitive sites across all varieties, compared with 9.1% and 14.4%, respectively, for the MB-detected SNPs. Several SNPs or insertion/deletion polymorphisms in close proximity can suppress hybridization and reduce SNP detection and could account for low recall rates (7, 10). Together, the two datasets (MBML-union dataset) included 397,348 SNPs. Of these, 159,879 SNPs were predicted by both methods (MBML-intersect dataset; Table S2) and constitute a high-quality subset (FDR 2.9%; recall 11.0%) used in subsequent analyses. Approximately one-fourth of the high-quality MBML SNPs were validated at 97% accuracy (S.R. McCouch, personal communication). The genomewide average of SNPs per kb using the MBML-intersect data was 1.6, and the transition/transversion rate (2.1) is similar to other species (11).
Allele frequencies for two-thirds of all sites were ≥0.15, and approximately one-third of SNPs were present in 7–12 varieties (Fig. 1B). The frequency distribution in rice differs markedly from that observed in Arabidopsis, where 50% of nonsynonymous and 40% of synonymous SNPs occurred in only one accession (12, 13), likely because the A. thaliana set included strains chosen for maximal genetic diversity (14), and thus had less population structure than the OryzaSNPset.
Fig. 1.
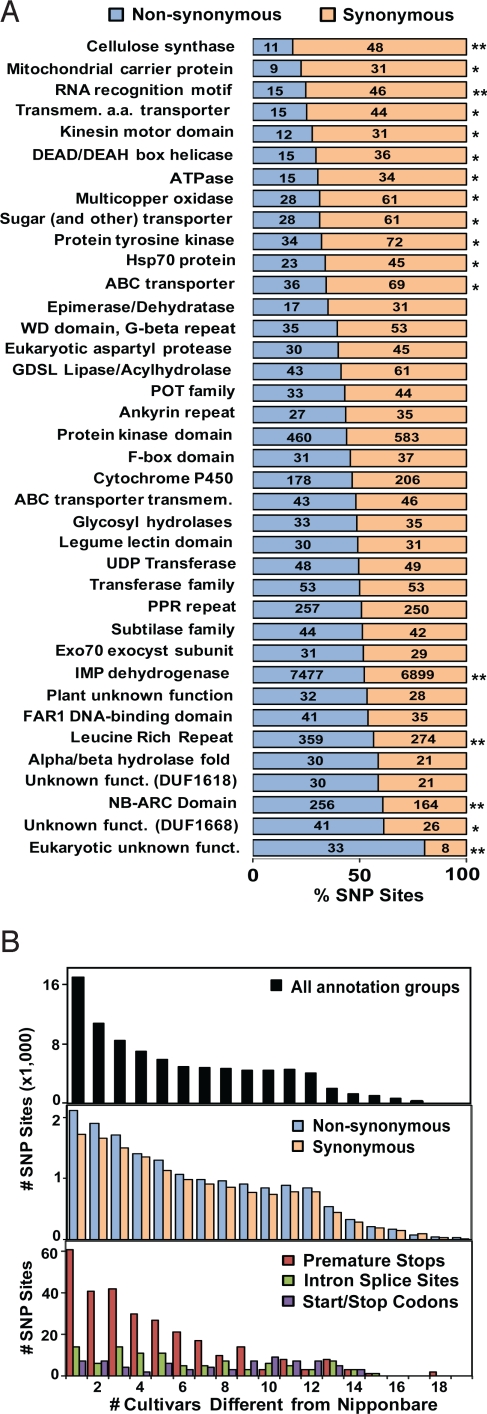
Annotation and distribution of SNPs. (A) Nonsynonymous (12,788) and synonymous (13,698) SNPs predicted from the MBML intersect found in select Pfam domains of rice genes with 30 or more SNPs. χ2 significance of the observed nonsynonymous and synonymous SNP distributions for each Pfam group is shown. *, P < 0.05; **, P < 0.001. (B) Allele frequencies at nonrepetitive MBML-intersect sites for SNPs in all annotation groups (Top), genic SNPs (Middle), and large-effect SNPs (Bottom). Only SNP sites from the MBML-intersect with complete data for >15 varieties were considered. The numbers of varieties with alleles (SNPs) different from the reference variety, Nipponbare, are indicated on the x axis.
Most OrzyaSNPset varieties are donors of agronomic traits, with mapping populations available, enabling rapid application of the discovered SNPs in mapping experiments. As expected, many more coding region SNPs occurred in indica/aus (86.4%) than in japonica varieties, which include the Nipponbare reference from which the array was designed (13.6%). The highest number of SNPs occurred in the aus varieties (www.OryzaSNP.org). Overall, between 26,700 and 57,700 SNPs were detected in japonica × indica pairs in the MBML-intersect dataset. More than 33,000 SNPs distinguish IR64 and Azucena, the parents of widely used doubled haploids and recombinant inbred lines (refs. 15 and 16 and references therein). Within the indica group, >17,000 SNPs are predicted between Zhenshan 97B and Minghui 63, parents of Shanyou 63, the most popular hybrid rice in China from 1985 to 2000.
SNP Annotations and Large-Effect SNPs.
Most high-quality MBML intersect SNPs (91,150/159,879) were located within gene models (Table 2). The proportions of genic SNPs identified as coding, intronic, or UTR (43.5%, 41.6%, and 15.7%, respectively) were different from the proportions identified in Arabidopsis [64.1%, 26.8% and 9.1%, respectively (7)]. The larger rice introns (397 bp) contained more SNPs than Arabidopsis introns [168 bp (17)].
Table 2.
Annotations of nonrepetitive SNPs relative to the TIGR rice gene models
| SNP | MB | ML | MBML-union | MBML-intersect |
|---|---|---|---|---|
| Genic | 135,119 | 171,750 | 215,032 | 91,150 |
| Coding | 57,935 | 74,007 | 91,992 | 39,652 |
| Intronic | 56,693 | 71,779 | 90,301 | 37,883 |
| 5′ UTR | 7,374 | 10,116 | 12,660 | 4,794 |
| 3′ UTR | 14,195 | 17,179 | 21,751 | 9,551 |
| Intergenic | 105,306 | 142,764 | 179,646 | 67,778 |
| Total SNPs | 240,425 | 314,514 | 394,678 | 158,928 |
SNPs on the TIGR rice pseudomolecules were classified as genic or intergenic, and locations within gene models were annotated. The sums of coding, intronic, 5′ UTR, and 3′ UTR SNPs within a column are more than total genic sums because they are given for all overlapping gene models.
Although the ratio of nonsynonymous-to-synonymous SNPs in the array data was 1.2 across all gene models, the ratio dropped to 1.0 for sites in Pfam domains, regions expected to have fewer amino acid substitutions because of domain conservation (Fig. 1A and www.OryzaSNP.org). Genes coding for cellulose synthase, mitochondrial carrier, and amino acid transporter domains, all of which have transmembrane domains, had the lowest nonsynonymous-to-synonymous substitution ratios. In contrast, sequences encoding leucine-rich repeat and NB-ARC domains had a significantly higher ratio of nonsynonymous-to-synonymous SNPs than average (Fig. 1A). Because these domains are common in plant disease- resistance proteins, this finding is consistent with these proteins being particularly diverse because of pathogen pressure (7, 18–20).
Approximately 2.7% of the rice genes contained large-effect SNPs that are expected to affect the integrity of encoded proteins. These include changes predicted to disrupt intron splicing (73 donor-site SNPs, 66 acceptor-site SNPs), introduce premature termination codons (388 SNPs), eliminate translation initiation sites (41 SNPs), and replace nonsense with sense codons (71 SNPs). Dideoxy sequencing was used to validate a subset of 209 large-effect SNPs across the 20 varieties (Table S4). The 16.3% FDR in this set was higher than the MBML average of 2.9% (Table S2), in agreement with what was observed in Arabidopsis (21). The FDR was evenly distributed across rice varieties and allele frequencies and, thus, was not biased against particular SNP calls.
Fifty percent of SNPs resulting in premature stops were rare, occurring in <3 varieties (Fig. 1B). Although this low-frequency skew suggests that premature stops are detrimental, such loss-of-function changes are known to underlie many traits selected during domestication (22). The frequency distributions of SNPs changing intron splice sites and start/stop codons may reflect more neutral selection as compared with SNPs within coding regions. Alternatively, these gene structure features may not be shared with Nipponbare (Fig. 1B).
Phylogenetic Relationships, Population Structure, and Decay of Linkage Disequilibrium.
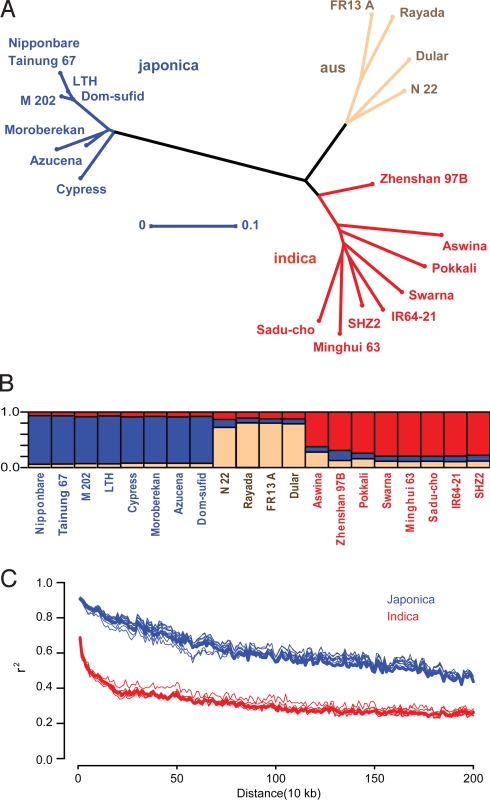
The phylogenetic tree produced using the MBML-intersect dataset (Fig. 2A) revealed 3 distinct groups, with temperate and tropical japonicas closely allied in one group and the other groups correlating with aus and indica types, consistent with other analyses (12, 23). Dom-sufid (aromatic) grouped among the temperate japonica, a discrepancy from the ancestral placement relative to tropical japonica (23). This discrepancy was not observed when using the MB data only and may result from the low prediction rate of SNPs for Dom-sufid by ML. Analysis of population structure by the Bayesian clustering program InStruct (24) also revealed the 3 groups (Fig. 2B).
Fig. 2.
Phylogenetic relationships, population structure, and decay of LD in the OryzaSNPset. (A) Unweighted neighbor-joining dendrogram for nonrepetitive SNPs in the MBML-intersect data (159,879 sites). Horizontal bar indicates distance by simple matching coefficient. (B) Population structure as determined by MB inference using InStruct (24). The 3 groups correspond to indica (red), aus (brown), and japonica (blue). (C) Decay of LD, expressed as r2 as a function of inter-SNP distance for filtered MBML-intersect SNPs, in the indica and japonica varieties, for each chromosome (light) and overall (bold). Limited numbers of japonica SNPs bias LD estimates.
The extent of linkage disequilibrium (LD) impacts both the genotyping effort required for whole-genome association scans and the resolution with which causal regions can be localized. LD reflects the strong population structure of the OryzaSNPset (Fig. 2C). For the MBML intersect dataset, LD extends to ≈200 kb for the indica group, a higher estimate than reported (12–14). The limited number of SNPs among the japonica varieties renders the LD estimation for that group unreliable. When we focused on the regions used by Mather et al. (14), LD levels similar to our genomewide LD decay pattern were observed.
Extended Haplotype Sharing Identifies Large-Scale Introgressions.
Modern rice germplasm is to a large extent shaped by directed breeding, and the OryzaSNP dataset affords an opportunity to assess the degree of introgression and relationships among varietal groups. Most OryzaSNPset varieties are widely used in breeding programs and are under strong selection; thus, important aspects of breeding history can be inferred from introgression patterns among the 3 groups, indica, japonica, and aus. Using a haplotype sharing ratio method in a comparison of all group pairs, we identified patterns of introgression among the 3 varietal groups along the 12 chromosomes (Fig. 3 and Fig. S1).
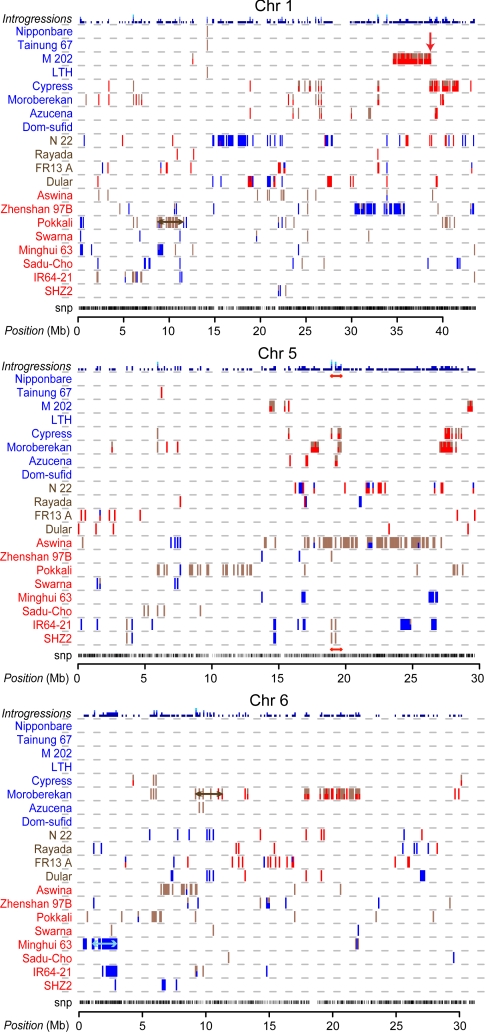
Fig. 3.
Introgressed regions detected in rice chromosomes 1, 5, and 6. The origin of an introgression is indicated by the color of the varietal group (red from indica, blue from japonica, and brown from aus). Each vertical line corresponds to a window of 100 kb. If the source of an introgression is ambiguous, each potential donor is indicated with half of the line. The maximum frequency of introgressions of the same type at the same position is plotted at the top of each chromosome. Red arrow indicates Sd1 (chromosome 1), brown bars indicate saltol (chromosome 1) and NB-ARC regions (chromosome 6), a red bar shows shared introgressions (chromosome 5), and light blue bar indicates introgressed region in Minghui 63 (chromosome 6). See Fig. S1 for introgressions in all chromosomes.
Large introgressions revealed by the SNP data reflect the breeding history of some rice varieties. For example, the japonica varieties Cypress and M202 show large regions on chromosome 1 introgressed from indica or aus (Fig. 3). These modern American semidwarf varieties were previously known to harbor introgressions from the indica variety IR8, the donor of the semidwarf gene Sd1 important in the Green Revolution. The Sd1 locus (25) is located at ≈38.7 Mb on chromosome 1, corresponding to the overlapping introgression regions we observed in Cypress and M202.
The OryzaSNP data also confirmed introgressions from the aus group, a pool of traditional varieties commonly used as donors for abiotic stress tolerance traits into cultivated varieties. On chromosome 1, the indica variety Pokkali contains aus introgression regions that correspond to flanking markers and candidate genes underlying a salt tolerance quantitative trait loci (QTL) (Saltol) between 10.7 and 12.3 Mb (Fig. 3) (26). Moroberekan, a temperate japonica traditional variety from Africa and a popular donor for disease resistance and drought tolerance (27), contains several regions on chromosome 6 introgressed from indica or aus (Fig. 3), one of which colocalizes to a large cluster of NB-ARC-type resistance genes between 9.2 and 11.1 Mb (28). These intriguing introgression patterns suggest that Pokkali and Moroberekan, landraces indigenous to India and Africa, respectively, were involved in cross-breeding with exotic germplasm by early rice farmers.
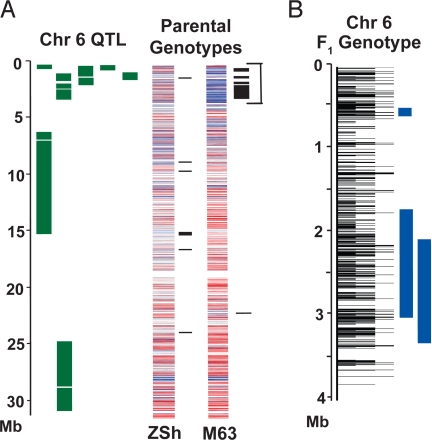
Two indica varieties from China, Minghui 63 and Zhenshan 97B, parents of a popular hybrid rice, Shanyou 63, are well characterized by QTL mapping for various traits and for gene action controlling heterosis (29–31). Minghui 63 shows an introgression on chromosome 6 (0–4 Mb) from japonica that colocalizes with QTL reported in several studies for traits including leaf area, vascular bundle number, root features, plant height, cooking quality, and amylose content (Fig. 4A and Table S5) (29–31). The introgression also contains heterotic loci associated with high yield performance in heterozygous vs. homozygous individuals (Fig. 4B and Table S5) (32). Of particular interest in this region are SNPs in the waxy locus, a starch synthase gene known to affect amylose content in rice grain (14). We detected large-effect SNPs in 4 genes within the region. Although the contribution of these genes to heterosis in Shanyou 63 is unknown, their identification demonstrates the power of the OryzaSNP data for identifying candidates at a high resolution (1 SNP per 2.6 kb in this 4-Mb region).
Fig. 4.
Relationships among SNP genotypes of parental varieties, Zhenshan 97B (ZSh) and Minghui 63 (M63), QTL regions, and F1 heterotic loci on chromosome 6. (A) Molecular marker data from 5 QTL studies (29–31, 38, 39) were assembled and flanking markers were physically mapped to the rice genome (green bars). Parental genotypes at 17,317 SNP sites are shown as red (A allele), blue (B allele), or gray (missing data) lines and introgressions are shown as black bars. QTL for traits including leaf area, vascular bundle number, plant height, root number, cooking quality, amylose content, gel consistency, and shoot weight colocalize with a large introgression in M63. (B) F1 hybrid genotypes at 1,564 SNP sites in the overlapping QTL region at 0–4 Mb. Heterozygous SNP sites are black lines; small = intergenic, medium = UTR/intron/synonymous, and large = nonsynonymous. Blue bars show heterotic loci associated with high-yield performance in heterozygous vs. homozygous individuals (32).
Common patterns of introgression could indicate other selection events. Across the 20 genomes, 295, 66, 12, and 1 positions were found in 2, 3, 4, to 5 varieties, respectively, that carried introgressions of the same type (see plots at top of chromosomes in Fig. 3 and Fig. S1). These patterns of coincidence were significantly different from random by a permutation test using 1,000 resamples (P < 0.025). A majority (70%) of the coincident introgressions occur within 300 kb of one another. For example, in the region from 18.9 to 19.7 on chromosome 5, there are 3 positions where introgressions occur in 5, 4, and 3 of the same or other varieties. Using QTL data extracted from the Gramene database (http://gramene.org), this region was found to be associated with grain quality, carbohydrate content, and panicle traits based on an enrichment test (P < 7.24 E-42 by Fisher's exact test) (33). These blocks of colocalized, shared introgressions cover ≈9% of the genome. The nonrandom distribution and coincidence of introgressions may indicate regions of intense selection, such as those related to desirable traits under domestication.
Haplotype sharing between pairs of accessions identified extended blocks where 90% or more of the SNPs are in common (Fig. S2). Pairwise sharing revealed even more potential introgressions than observed by haplotype sharing across group-pairs. Large regions of aus introgressions in Pokkali occur on chromosome 5 from 8.3 to 13 Mb (Fig. 3 and Fig. S1) and are part of an area that appears highly conserved across all other indica and japonica.
To further examine the history of varietal selection, we tabulated the number and length of introgressions occurring from other varietal groups (Table S6). On average, the modern indica and japonica varieties have 8.4 Mb of introgressions from other groups, whereas the aus type has approximately twice the length of introgressions (17.5 Mb). The average length of introgressions, however, appears to be similar across varietal groups (average 0.15 ± 0.04 Mb per introgression). One possibility is that modern varieties have been under strong selection for specific production environments, thus constraining the introgression of chromosomal fragments. However, the aus types, representing traditional varieties, could be subjected to a lesser degree of selection. Although a larger sample size will be needed to test this hypothesis, this analysis demonstrates the potential of genomewide SNP datasets for probing the history of varietal selection in rice.
Conclusions
Our study provides comprehensive SNP data from a set of rice varieties that captures the impressive genotypic and phenotypic diversity of this important crop plant. An immediate outcome of our work is the detection of chromosomal segments introgressed from one varietal group into another shedding light on the breeding history of rice. Some introgressions correlate with known genomic regions responsible for traits transferred between varietal groups, whereas others represent candidates for additional events of potential significance for breeding. Furthermore, the much-improved knowledge of shared breeding history and genetic relationships enhances traditional methods (e.g., coefficient of parentage; Table S7) for the selection of parents for crossing programs.
The SNP coverage of the rice genome available from our study is more than sufficient to obtain genomewide tag SNPs, especially for regions highly conserved across varietal types, despite the lower estimates of LD (75–150 kb) in previous studies (12–14). Sequencing of additional rice types including Oryza rufipogon, the progenitor of domesticated rice, is an obvious next step to provide more SNPs across all groups.
Last, and perhaps most importantly, the OryzaSNP resource provides the foundation for high-resolution genotyping of hundreds to thousands of additional varieties. Compared with studies of other model plants such as Arabidopsis, a major advantage of rice is the much more extensive information available for a diverse set of known agronomically important traits from thousands of varieties across many different environments. Detailed knowledge of phenotypes, coupled with a deep genotype database, will create a powerful platform for association genetics and discovery of alleles that can be combined to achieve the much-needed increase in rice yield in the coming years.
Materials and Methods
Plant Varieties, Reference Genome Masking, and Target Selection.
Each rice variety (Table S1) was purified by 1 round of single seed descent. Rice genome sequence (Build 4; ref. 8) was masked for repeats (8, 34, 35). Those sequences with no or a single hit (91.6 Mb) and with 2–10 hits (77.6 Mb) were chosen for long-range PCR (LR-PCR) primer design (see SI Appendix).
Array Design, Sample Preparation, and Hybridization.
The 13,586 selected LR-PCR amplicons span 11,343 nonoverlapping fragments and cover 117.8 Mb of unmasked genomic sequence. This genomic fraction was used to design 6 high-density oligonucleotide (25-mer) resequencing arrays that queried 100.1 Mb of the Nipponbare genome by using a tiling strategy (3, 6, 7). The LR-PCR products for each of the 20 rice strains were combined (at ≈8 Mb complexity), fragmented, and labeled (3). Each array, synthesized by Affymetrix, contained ≈20 Mb of tiled sequence and was segmented into 3 chambers. Each chamber was hybridized with a different DNA/hybridization mixture containing labeled target DNAs of 2 strains. Hybridized targets were detected by using confocal scanners.
Base-Calling, SNP Detection, and Normalization.
We used the pattern recognition (MB) algorithms for analysis as described (3, 6) using criteria and quality scoring algorithms specified in SI Appendix (3). To correct for between-array variation and obtain comparability of the data generated by multiple array experiments, hybridization data were quantile-normalized on the level of amplicon pools across all varieties (36).
Repetitive Probe Annotation and Quality Assessment SNPs.
Repetitive probes in the reference genome were annotated by identifying oligomers that match at least one other 25-mer in the target DNA, allowing for some degree of degeneracy. The mismatch criteria distinguished between the 3 match types (exact, inexact, and short 25-mer matches) and bulged 25-mer matches that were restricted to a 1-base bulge located only on 1 strand (ref. 7 and SI Appendix). We used dideoxy sequencing of randomly selected fragments from a subset of the tiled regions to compile a set of true (curated) SNP and non-SNP positions for quality assessment (SI Appendix). A 2-layered approach based on SVMs was applied to predict SNPs from the hybridization data (7).
SNP Annotation.
All SNP locations and tiled regions were mapped relative to the International Rice Genome Sequencing Project (IRGSP) (8) and TIGR (37) pseudomolecules by using the program Vmatch (www.vmatch.de), and SNPs were annotated relative to the IRGSP and TIGR pseudomolecules and to the Rice Annotation Project (RAP) and TIGR gene models (SI Appendix). For the MBML-intersect SNP set, a total of 158,928 of 159,879 IRGSP localized SNPs were mapped to the TIGR pseudomolecules. SNP sites annotated as nonsynonymous, synonymous, or as large-effect changes were extracted from the MBML-intersect dataset, and only sites with high confidence base calls for at least 15/20 cultivars were included in calculations of allele frequencies. For each SNP site, the number of varieties with bases different from the reference were plotted by frequency and annotation category. SNP distribution and annotation processes (www.OryzaSNP.org) are detailed in SI Appendix. Sixty loci containing large-effect SNPs were randomly selected for validation by PCR amplification and dideoxy sequencing at 2× coverage for all amplicons (Table S4 and www.OryzaSNP.org).
Dendrogram Construction and Population Structure.
A pairwise distance matrix using the simple matching coefficient for SNPs at nonrepetitive sites was calculated, and an unweighted neighbor-joining tree was constructed by using DARwin 5 (http://darwin.cirad.fr/darwin) (Fig. 2A). Population structure was determined by MB inference using InStruct (24) on a random subset of 5,000 MBML-intersect SNPs (Fig. 2B).
LD and Introgression Analyses.
Only biallelic nonsingleton SNPs in the MBML-intersect dataset were used to calculate LD as the correlation coefficient r2 between SNP pairs. The mean r2 value was calculated for 10-kb bins based on all pairs of nonsingleton SNPs. Because of the extensive population structure in the sample of 20 varieties, we examined LD decay in each subpopulation separately. Because of the small sample size in the aus group (4 varieties), only indica and japonica groups, with 8 varieties each, were analyzed. Only SNP pairs with no missing data at both loci in at least 6 chromosomes of the 8 varieties were included in the calculations (Fig. 2C).
To study the ancestral contribution of groups to the genome of each variety, we applied a likelihood ratio test method. All putative introgressions between pairs of groups (indica, aus, and japonica) were examined. For every window of 100 Kb with at least 10 SNPs, the ratio of the average sharing of each variety to its own and another group was calculated when at least 3 pairs of comparison occurred in each group. Regions with an average sharing ratio of <0.5 were defined as introgressions (Fig. 3 and Fig. S1). Frequencies of introgressions shared across varieties were plotted. The length, number of introgressions in each variety, and shared introgressions across varieties were tabulated (Table S6). Regions of extensive haplotype sharing, with 90% or more shared SNPs, were determined for each pair of varieties (Fig. S2).
QTL and genetic data for Pokkali, Moroberekan, Minghui 63, and Zhenshan 97B were from published studies (26–31). Physical locations for flanking markers were acquired from Gramene (http://gramene.org) and Michigan State University Rice Genome Annotation (http://rice.plantbiology.msu.edu) databases, or they were inferred by blastn searches of marker-associated sequences and/or marker primers against the reference genome (Figs. 3 and 4 and Table S5).
Data Release.
Processed resequencing data are at www.ncbi.nlm.nih.gov/Traces, SNP annotations, the full dataset, and descriptive information on basic queries are at www.OryzaSNP.org. The dideoxy sequence data generated for data quality and training purposes and large-effect validation are in GenBank (accession nos. FI321710–FI329971 and FI494729–FI495095, respectively).
Supplementary Material
Acknowledgments.
We thank G. Schweikert, H. Huang, G. Nilsen, M. Morenzoni, J. Sheehan, L. Stuve, J. Montgomery, H. Tao, and C. Chen for technical assistance, R. Mauleon for enrichment analyses, and Y.-I. Hsing (Academia Sinica, Taiwan) and Q. Zhang (Huazhong Agricultural University, China) for providing seeds. This work was supported by the International Rice Research Institute, U.S. Department of Agriculture–Cooperative State Research, Education, and Extension Service Grant 2006-35604-16628, and Generation Challenge Program Grant 2005-35. D.W. and G.R. are funded by the Max Planck Society. D.W. is also supported by a Gottfried Wilhelm Leibniz Award of the Deutsche Forschungsgemeinschaft and the Bundesministerium Für Wirtschaftliche Zusammenarbeit. R.M.D. is supported by a Ford Foundation Diversity Fellowship and U.S. Department of Agriculture–Cooperative State Research, Education, and Extension Service—National Research Initiative-Rice–Conservation Assessment Program Grant 2004-35317-14867. K.Z., B.P., and C.D.B. were funded by National Science Foundation Grants 0606461 and 0701382.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. FI321710–FI329971 and FI494729–FI495095).
This article contains supporting information online at www.pnas.org/cgi/content/full/0900992106/DCSupplemental.
References
- 1.International Rice Research Institute. Bringing Hope, Improving Lives: Strategic Plan, 2007–2015. Manila, Philippines: IRRI Press; 2006. [Google Scholar]
- 2.Hinds DA, et al. Whole-genome patterns of common DNA variation in three human populations. Science. 2005;307:1072–1079. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- 3.Frazer KA, et al. A sequence-based variation map of 8.27 million SNPs in inbred mouse strains. Nature. 2007;448:1050–1053. doi: 10.1038/nature06067. [DOI] [PubMed] [Google Scholar]
- 4.Kim S, et al. Recombination and linkage disequilibrium in Arabidopsis thaliana. Nat Genet. 2007;39:1151–1155. doi: 10.1038/ng2115. [DOI] [PubMed] [Google Scholar]
- 5.McNally KL, et al. Sequencing multiple and diverse rice varieties. Connecting whole-genome variation with phenotypes. Plant Physiol. 2006;141:26–31. doi: 10.1104/pp.106.077313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patil N, et al. Blocks of limited haplotype diversity revealed by high-resolution scanning of human chromosome 21. Science. 2001;294:1719–1723. doi: 10.1126/science.1065573. [DOI] [PubMed] [Google Scholar]
- 7.Clark RM, et al. Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science. 2007;317:338–342. doi: 10.1126/science.1138632. [DOI] [PubMed] [Google Scholar]
- 8.International Rice Genome Sequencing Project. The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- 9.Yu J, et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- 10.Zeller G, et al. Detecting polymorphic regions in Arabidopsis thaliana with resequencing microarrays. Genome Res. 2008;18:918–929. doi: 10.1101/gr.070169.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wakeley J. Substitution-rate variation among sites and the estimation of transition bias. Mol Biol Evol. 1994;11:436–442. doi: 10.1093/oxfordjournals.molbev.a040124. [DOI] [PubMed] [Google Scholar]
- 12.Garris AJ, McCouch SR, Kresovich S. Population structure and its effect on haplotype diversity and linkage disequilibrium surrounding the xa5 locus of rice (Oryza sativa L.) Genetics. 2003;165:759–769. doi: 10.1093/genetics/165.2.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olsen KM, et al. Selection under domestication: evidence for a sweep in the rice waxy genomic region. Genetics. 2006;173:975–983. doi: 10.1534/genetics.106.056473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mather KA, et al. The extent of linkage disequilibrium in rice (Oryza sativa L.) Genetics. 2007;177:2223–2232. doi: 10.1534/genetics.107.079616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hittalmani S, et al. Molecular mapping of quantitative trait loci for plant growth, yield, and yield-related traits across three diverse locations in a doubled haploid rice population. Euphytica. 2002;125:207–214. [Google Scholar]
- 16.Ramalingam J, et al. Candidate defense genes from rice, barley, and maize and their association with qualitative and quantitative resistance in rice. Mol Plant Microbe Interact. 2003;16:14–24. doi: 10.1094/MPMI.2003.16.1.14. [DOI] [PubMed] [Google Scholar]
- 17.The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 18.Grant MR, et al. Independent deletions of a pathogen-resistance gene in Brassica and Arabidopsis. Proc Natl Acad Sci USA. 1998;95:15843–15848. doi: 10.1073/pnas.95.26.15843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bakker EG, Toomajian C, Kreitman M, Bergelson J. A genomewide survey of R gene polymorphisms in Arabidopsis. Plant Cell. 2006;18:1803–1818. doi: 10.1105/tpc.106.042614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen J, Araki H, Chen L, Chen J, Tian D. Unique evolutionary mechanism in R genes under the presence/absence polymorphism in Arabidopsis thaliana. Genetics. 2006;172:1243–1250. doi: 10.1534/genetics.105.047290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nordborg M, et al. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 2005;3:e196. doi: 10.1371/journal.pbio.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doebley JF, Gaut BS, Smith BD. The molecular genetics of crop domestication. Cell. 2006;127:1309–1321. doi: 10.1016/j.cell.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 23.Garris AJ, Tai TH, Coburn J, Kresovich S, McCouch S. Genetic structure and diversity in Oryza sativa L. Genetics. 2005;169:1631–1638. doi: 10.1534/genetics.104.035642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao H, Williamson S, Bustamante CD. A Markov chain Monte Carlo approach for joint inference of population structure and inbreeding rates from multilocus genotype data. Genetics. 2007;176:1635–1651. doi: 10.1534/genetics.107.072371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sasaki A, et al. Green revolution: A mutant gibberellin-synthesis gene in rice. Nature. 2002;416:701–702. doi: 10.1038/416701a. [DOI] [PubMed] [Google Scholar]
- 26.Walia H, et al. Comparative transcriptional profiling of two contrasting rice genotypes under salinity stress during the vegetative growth stage. Plant Physiol. 2005;139:822–835. doi: 10.1104/pp.105.065961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Botwright-Acuna TL, Lafitte HR, Wade LJ. Genotype × environment interactions for grain yield of upland rice backcross lines in diverse hydrological environments. Field Crops Res. 2008;108:117–125. [Google Scholar]
- 28.Jeung JU, et al. A novel gene, Pi40(t), linked to the DNA markers derived from NBS-LRR motifs confers broad spectrum of blast resistance in rice. Theor Appl Genet. 2007;115:1163–1177. doi: 10.1007/s00122-007-0642-x. [DOI] [PubMed] [Google Scholar]
- 29.Cui KH, et al. Mapping QTLs for seedling characteristics under different water supply conditions in rice (Oryza sativa) Physiol Plant. 2008;132:53–68. doi: 10.1111/j.1399-3054.2007.00991.x. [DOI] [PubMed] [Google Scholar]
- 30.Ge XJ, Xing YZ, Xu CG, He YQ. QTL analysis of cooked rice grain elongation, volume expansion, and water absorption using a recombinant inbred population. Plant Breeding. 2005;124:121–126. [Google Scholar]
- 31.Zheng X, Wu JG, Lou XY, Xu HM, Shi CH. The QTL analysis on maternal and endosperm genome and their environmental interactions for characters of cooking quality in rice (Oryza sativa L.) Theor Appl Genet. 2008;116:335–342. doi: 10.1007/s00122-007-0671-5. [DOI] [PubMed] [Google Scholar]
- 32.Hua JP, et al. Single-locus heterotic effects and dominance by dominance interactions can adequately explain the genetic basis of heterosis in an elite rice hybrid. Proc Natl Acad Sci USA. 2003;100:2574–2579. doi: 10.1073/pnas.0437907100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hosack DA, Dennis G, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70–R70.78. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Juretic N, Bureau TE, Bruskiewich RM. Transposable element annotation of the rice genome. Bioinformatics. 2004;20:155–160. doi: 10.1093/bioinformatics/bth019. [DOI] [PubMed] [Google Scholar]
- 35.Yuan Q, et al. The TIGR rice genome annotation resource: Annotating the rice genome and creating resources for plant biologists. Nucleic Acids Res. 2003;31:229–233. doi: 10.1093/nar/gkg059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high-density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 37.Ouyang S, et al. The TIGR Rice Genome Annotation Resource: Improvements and new features. Nucleic Acids Res. 2007;35:D883–D887. doi: 10.1093/nar/gkl976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cui KH, et al. Molecular dissection of the genetic relationships of source, sink, and transport tissue with yield traits in rice. Theor Appl Genet. 2003;106:649–658. doi: 10.1007/s00122-002-1113-z. [DOI] [PubMed] [Google Scholar]
- 39.Lian XM, et al. QTLs for low nitrogen tolerance at seedling stage identified using a recombinant inbred line population derived from an elite rice hybrid. Theor Appl Genet. 2005;112:85–96. doi: 10.1007/s00122-005-0108-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.