Abstract
Eukaryotic centromeres and telomeres are specialized chromosomal regions that share one common characteristic: their underlying DNA sequences are assembled into heritably repressed chromatin. Silent chromatin in budding and fission yeast is composed of fundamentally divergent proteins tat assemble very different chromatin structures. However, the ultimate behaviour of silent chromatin and the pathways that assemble it seem strikingly similar among Saccharomyces cerevisiae (S. cerevisiae), Schizosaccharomyces pombe (S. pombe) and other eukaryotes. Thus, studies in both yeasts have been instrumental in dissecting the mechanisms that establish and maintain silent chromatin in eukaryotes, contributing substantially to our understanding of epigenetic processes. In this review, we discuss current models for the generation of heterochromatic domains at centromeres and telomeres in the two yeast species.
Keywords: heterochromatin, RNA, silencing
Introduction
Centromeres
The centromere is essential for proper segregation of chromosomes in mitosis and meiosis and is therefore of vital importance for genetic stability. It is the DNA region in which the kinetochore is formed, a structure that allows chromosomes to associate with spindle microtubules. Centromere function and its many associated proteins are conserved, yet centromere specification is not always hard-wired to the DNA sequence and displays dramatic plasticity (reviewed in Sullivan et al, 2001; Allshire and Karpen, 2008). Centromeres can have different structures depending on their size, the number of kinetochore microtubules they interact with and whether or not they are surrounded by pericentric heterochromatin.
Both Schizosaccharomyces pombe (S. pombe) and Saccharomyces cerevisiae (S. cerevisiae) are monocentric eukaryotes with localized centromeres, in contrast to holocentric organisms such as Caenorhabditis elegans, in which kinetochores form along the entire chromosome. A conserved feature of all centromeres is the special histone H3 variant, called Cnp1 in S. pombe and Cse4 in S. cerevisiae, which is found exclusively within the core centromeric region (Smith, 2002). In most other aspects, budding and fission yeast centromeres are quite different. In S. cerevisiae, complete centromere function is specified by only 125 bp of DNA comprising three distinct centromeric DNA elements (CDE I, II and III). The 15 bp of CDE III is most important as it attracts a complex containing sequence-specific DNA-binding proteins (Ndc10, Cep3, Ctf13 and Skp1). This complex dictates the assembly of the single Cse4-containing nucleosome, which spans the middle AT-rich CDEII element (Meluh et al, 1998; Furuyama and Biggins, 2007). Directly analogous elements are absent in S. pombe. Rather, centromere structure comprises a central core domain (cnt) bearing Cnp1 nucleosomes surrounded by a long inverted repeat. Each centromeric flank can be divided into two regions: the inner repeats (imr), which are specific to each of the three centromeres, and the outer repeats (otr), which are composed of elements known as dg and dh (Bjerling and Ekwall, 2002). The arrangement of dg and dh repeats with respect to each other and to the central core differs at each of the three fission yeast centromeres. Notably, the otr regions in S. pombe are assembled into silent heterochromatin, which is important for proper centromere function (see also accompanying Focus Review by Torras-Llort et al).
Telomeres
The telomere assumes a ‘cap' structure that maintains and protects the ends of eukaryotic linear chromosomes (Zakian, 1996). Telomeres impede chromosomal fusion (end-to-end joining) by blocking activation of the DNA damage checkpoint response and locally impairing double-strand break repair. Most importantly, telomeres and the RNA-directed enzyme telomerase ensure the addition of TG repeats that are otherwise eroded with each successive round of cell division. Collectively, these functions stabilize chromosome ends and contribute to genomic stability. Importantly, the telomeres of both budding and fission yeasts are assembled into silent chromatin structures (Huang, 2002; see also accompanying Focus Review by Luke and Lingner).
Telomeric DNA consists of three main parts: a short single-stranded (ss) 3′ overhang, double-stranded (ds) telomeric repeats and the subtelomeric region. The ss overhang and double-stranded stretch in S. cerevisiae comprise ∼300 bp of an irregular TG1−3 repeat that lies terminal to subtelomeric sequences. The subtelomeric regions include up to four tandem copies of Y′ elements, short internal TG1−3 repeats and an X element composed of imperfect repeats and a conserved 437 bp core (Zakian, 1996).
The telomeric repeats of S. pombe are also 300 bp long, but are somewhat more degenerate. They consist mainly of TTACA(G)n (where n=1–8), and contain interspersed repeats of TTACGG and TTACACGG, each with two Gs. The repeats at both ends of chromosome III are immediately flanked by repeats of ribosomal RNA genes, whereas chromosomes I and II share similar subtelomeric sequences that contain open reading frames (ORFs). Telomere-linked helicases (tlh) are encoded by the most distal ORFs in the subtelomeric regions of chromosomes I (tlh1+) and II (tlh2+). These putative helicases are members of the recQ family and display extensive sequence homology with the dh and dg repeats found at centromeres (cenH like)(Wood et al, 2002; Mandell et al, 2005). Interestingly, the S. cerevisiae Y′ elements also encode a DNA helicase, which is expressed primarily in meiosis (Louis and Haber, 1992; Yamada et al, 1998). In S. pombe, there is conservation among neighbouring ORFs in addition to the homology shared by tlh1+ and tlh2+, indicating that the two subtelomeric regions resulted from a duplication.
Although the terminal telomere sequence is associated with non-histone proteins forming a ‘telosome', subtelomeric regions in both fission and budding yeast are nucleosomal (Vega-Palas et al, 1998; Wiren et al, 2005). Important is the presence, or absence, of post-translational modifications on the histone tails of subtelomeric nucleosomes. In S. cerevisiae, lysines at positions 9, 14, 18, 23 and 27 on H3, at positions 5, 8, 12 and 16 on H4, at position 7 on H2A, and at positions 11 and 16 on H2B are hypoacetylated in subtelomeric chromatin (Thompson et al, 1994; Braunstein et al, 1996; Suka et al, 2001). Moreover, two specific and universally conserved marks of active or open chromatin, H3K4me and H4K16ac, are absent from telomeres in both yeasts. Although S. cerevisiae has no H3K9me at all, this modification is characteristically present throughout fission yeast heterochromatin, including pericentric DNA, subtelomeric domains and at silent mating-type loci (Nakayama et al, 2001). Nucleosomes bearing H3K9me are also typically hypoacetylated on H4K16 and H3K14.
In budding yeast, the hypoacetylated status of histone tails seems to be sufficient to favour the binding of the silent information regulatory (SIR) complex, Sir2-3-4, which in turn ensures a heritable downregulation of transcription of subtelomeric genes. This is called telomeric position effect, or TPE (see below). Sir2, a conserved NAD-dependent histone deacetylase, can act on all lysines of the H3 and H4 tails, but particularly targets H4K16ac (Blander and Guarente, 2004), as well as H3K9ac in S. pombe (Shankaranarayana et al, 2003). Other markers of active chromatin, notably di- and tri-methylated forms of histone H3K79, antagonize the binding of the SIR complex and impair repression of subtelomeric genes (van Leeuwen et al, 2002). Thus, the predominant pattern of histone modification at budding yeast telomeres is an absence of active marks, whereas S. pombe requires the positive signal provided by H3K9me. Intriguingly, in fission yeast Sir2 cooperates with Clr3 to eliminate acetylation marks on both H4K16, H3K9 and K14, which allows for subsequent methylation of H3K9 (Wiren et al, 2005).
In addition to subtelomeric histones, a sequence-specific factor binds the TG-rich telomeric repeats. In almost every species these repeat-binding factors share a myb-like DNA-binding domain (Konig and Rhodes, 1997). In budding yeast, the terminal repeats are bound by the repressor activator protein 1 (Rap1), whereas in S. pombe the analogous protein is called Taz1. In addition, S. pombe, similar to man, has a Rap1 homologue that lacks the DNA-binding domain. Fission yeast Rap1 associates with telomeric repeats through Taz1, again analogous to the association of human Rap1 with Trf1. The additional telomere-associated proteins can be divided into two classes: those that mediate end maintenance by controlling telomerase accessibility, and those that promote silent chromatin. Ku, a heterodimer that binds all DNA ends regardless of sequence, has a special role at telomeres: it contributes both to controlling telomerase and to promoting silent chromatin. In addition, budding yeast Ku has a crucial role in anchoring telomeres to the nuclear envelope (NE), which further facilitates the nucleation and spread of chromatin-mediated gene silencing (see below) (Hediger et al, 2002; Taddei et al, 2004, 2009). In the absence of yKu, TG repeats in yeast shorten, subtelomeric repression is lost and strains become temperature sensitive (Fisher and Zakian, 2005).
The epigenetic nature of centromeres and telomeres
Epigenetics is the study of heritable changes in gene function that occur without a change in the sequence of the DNA. Centromere assembly and propagation provide a unique example of an epigenetic process as protein structures are assembled onto DNA and then stably propagated through numerous cell divisions in a DNA sequence independent manner. Despite their variation in size and sequence composition, the epigenetic aspect of centromeres is highly conserved.
The epigenetic nature of centromeres is manifest in the fact that—although there are different requirements for centromere establishment—a functional centromere is transmitted epigenetically to daughter cells. Even the S. cerevisiae centromere shows epigenetic behaviour. Specifically, mutations in certain kinetochore proteins were shown to abolish de novo establishment of the S. cerevisiae centromere, although functional centromeres could be stably propagated for over 25 generations in this background (Mythreye and Bloom, 2003). Moreover, mutations in the core CDE element reduced the association of cohesin with naïve centromeres, but had little effect on established centromeres (Tanaka et al, 1999). In S. pombe, plasmids with minimal centromeric DNA establish functional centromeres stochastically, but once the functional state is attained it is propagated faithfully (Steiner and Clarke, 1994). Finally, a recent study also showed that heterochromatin and RNA interference (RNAi) are required to establish, but not to maintain, CENP-ACnp1 chromatin at fission yeast centromeres (Folco et al, 2008).
Whether the telomeric functions of capping and end-replication behave epigenetically is unclear, yet telomere-associated gene silencing is one of the classic examples of semi-stable, yet heritable, transcriptional repression (Figure 1) (Gottschling et al, 1990). Both native subtelomeric genes and reporters integrated into telomere proximal zones succumb to transcriptional silencing through chromatin-mediated mechanisms. Despite the fact that the subtelomeric repression of transcription in budding and fission yeast share many heterochromatin-like features, the molecular mechanisms of repression differ significantly, as explained below.
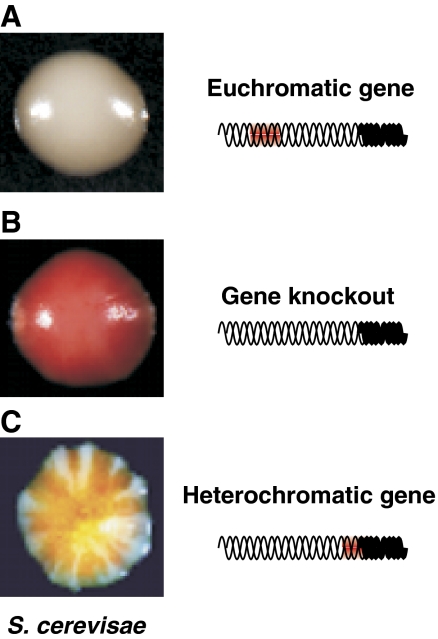
Figure 1.
Variegated expression of a gene on packaging into a heterochromatic structure. (A) Cells expressing the wild-type ADE2 gene from its endogenous, euchromatic locus produce colonies that are white, (B) whereas those lacking the ADE2 gene appear red. (C) Juxtaposition of ADE2 to heterochromatin results in its silencing without changing the underlying coding sequence. Although inherited, the packaging state of ADE2 (euchromatic versus heterochromatic) can switch at a low frequency. This results in a variegating phenotype in a clonal population of cells. An example of a telomeric position effect (TPE) (Gottschling et al, 1990) in S. cerevisiae is shown here.
Position effect variegation
Position effect variegation (PEV) is a universally conserved epigenetic phenomenon through which inserted or translocated genes are influenced by nearby heterochromatin. Thereafter, the ensuing expression status of the gene is clonally inherited. Importantly, centromeric heterochromatin and PEV are only observed in organisms that have extensive domains of repetitive DNA at their centromeres. Thus, the 125-bp centromere of S. cerevisiae is not heterochromatic, and does not silence genes. On the other hand, in fission yeast reporter genes inserted in the centromeric regions cnt, imr and otr are subject to PEV (Allshire et al, 1994, 1995). Depending on the centromeric region, repression of the reporter is more or less pronounced. In the outer repeats, marker genes are tightly repressed, whereas marker genes inserted within the central core or the inner most repeats display a more variegated pattern of expression. The impact of local context on gene silencing is a conserved feature of PEV, which ensures domain- rather than promoter-specific repression. Although budding yeast lacks centromeric PEV, similar events are observed at the silent mating-type loci and near telomeres.
In both budding and fission yeast, epigenetic gene silencing is crucial for mating-type determination, as it guarantees that these single-celled organisms can switch mating-type (Rusche et al, 2003). In each species the haploid genome contains the information needed to form at least two different cell types. In budding yeast, one of the two sets of mating-type information must be kept transcriptionally silent in haploid cells, or else the haploid behaves as a diploid and is unable to mate. In other words, the cell assumes a ‘pseudo diploid' character, suppressing the information needed to form a zygote, undergo meiosis and sporulate. Thus, the robustness of the species requires heritable repression of at least one set of mating-type determining genes.
The mechanisms that ensure mating-type repression in budding yeast, also serve to mediate position-dependent repression at telomeres (Aparicio et al, 1991; reviewed in Huang, 2002; Rusche et al, 2003). In an analogous manner, mechanisms that repress recombination and transcription at fission yeast centromeres contribute to silencing at the mating-type locus and TPE. The repression of mating-type information in both species is robust and extremely stable, whereas TPE is strongly variegating. This variegation is manifest as an ability to switch at a low frequency between ‘on' and ‘off' states and then propagate either state for many generations (Figure 1).
The other criteria that define epigenetic repression and which are fulfilled by flies, S. cerevisiae and S. pombe are as follows:
correlation with an altered chromatin structure that spreads outwards from a site of nucleation, silencing independently of the promoter concerned;
reduced accessibility for large molecules or complexes;
presence of hypoacetylated histones and/or specific marks that bind structural chromatin components;
an involvement of nucleosome-binding non-histone complexes that are limiting in abundance and show sensitivity to gene dosage; and
heritability through either mitotic or meiotic division.
Screens in flies, S. pombe, and S. cerevisiae have identified mutations that enhance or suppress heterochromatin-induced silencing, classically called E(var)s and Su(var)s (Muller, 1930; Wakimoto, 1998). Hundreds of suppressors of PEV have been identified to date, and these have proven to be useful tools to study heritable repression, as well as centromere and telomere biology (Allshire et al, 1995; Pidoux and Allshire, 2004). Some of the mutated genes encode for histone modifying enzymes, heterochromatin proteins (HPs) or histone variants (reviewed in Huang, 2002; Rusche et al, 2003). Notably, genetic approaches such as these have allowed the field to create a general definition of heterochromatin, although the molecular mechanisms may be clearly distinct in different organisms.
Silent chromatin assembly in budding yeast
The assembly of silent chromatin is a multistep process, starting with the nucleation of a nucleosome-binding repression complex at specific regulatory sequences and its subsequent spread into neighbouring sequences. Pioneering studies on the ordered assembly of silent chromatin have been carried out in S. cerevisiae and have provided a foundation for understanding epigenetic repression (reviewed in Rusche et al, 2003). In brief, the formation of silent chromatin in budding yeast requires the association of a heterotrimeric nucleosome-binding SIR complex that contains Sir2, Sir3 and Sir4 proteins in 1:1:1 stoichiometry (Cubizolles et al, 2006). The complex is recruited to DNA by interactions with proteins that bind to chromosome ends or to specific regulatory sites called silencers. At budding yeast telomeres, the SIR complex is recruited by Rap1 and the yKu heterodimer. The Rap1 protein binds once every 18 bp within the TG repeat, and each Rap1 molecule provides a binding site for Sir4 (Luo et al, 2002). Sir4 recruitment is further catalysed by the yKu70/80 heterodimer, which is associated with the telomere through its DNA-end-binding function independently of Sir4 (Gravel et al, 1998; Martin et al, 1999). Importantly, Sir4 binding to Rap1 is antagonized by Rif1/Rif2 (Mishra and Shore, 1999).
Sir4 is necessary for the recruitment of the entire SIR complex, although once nucleated, excess Sir3 can propagate along nucleosomes without Sir4 (Hecht et al, 1996). Sir2's NAD-dependent histone deacetylase activity keeps telomeric nucleosomes in a hypoacetylated state (Imai et al, 2000). Sir2 binds neither DNA nor histones with high affinity, but once recruited by Sir4, Sir2-mediated deacetylation can create a high-affinity binding site for Sir3. Sir3 has dimerization capacity and in complex with Sir2-4, results in the spread of the SIR complex outward from the nucleation site (Hecht et al, 1996; Liaw and Lustig, 2006). Sir3 contributes to the specificity for deacetylated histone tails, whereas Sir4 enhances the affinity of the complex through its ability to bind DNA (Martino et al, 2009) (Figure 2).
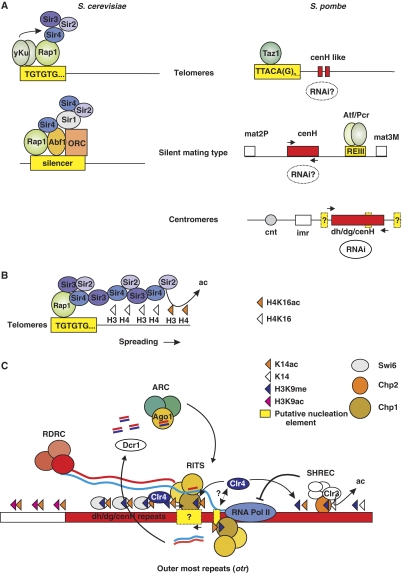
Figure 2.
Silent chromatin assembly in budding and fission yeast. (A) Cis-acting DNA sequences (nucleation sites, yellow boxes) are necessary to nucleate assembly of silent chromatin. Trans-acting proteins that directly bind the nucleation sites are indicated. Nucleation sites at fission yeast centromeres are likely to exist, although they have not been identified to date (yellow boxes). Bidirectional transcription (indicated by black arrows) of cendg/dh/H-like sequences (red boxes) is thought to produce dsRNA, which is processed into siRNAs by the RNAi machinery in S. pombe. siRNAs are required at least for the initiation of heterochromatin assembly at the silent mating-type locus and in addition for the maintenance of heterochromatin at centromeres. (B) Sir3 and Sir4 have dimerization capacity that results in the spread of the SIR complex outward from the nucleation site. Sir3 contributes to the specificity for deacetylated histone tails, whereas Sir4 enhances the affinity of the complex through its ability to bind DNA. Sir2-mediated deacetylation keeps telomeric nucleosomes hypoacetylated creating a high-affinity binding site for Sir3. (C) In S. pombe, the RITS complex promotes Clr4-mediated H3K9 methylation by associating with nascent transcripts through siRNA base pairing, and with methylated H3K9 through the chromodomain of its Chp1 subunit. Low levels of H3K9 methylation are maintained in RNAi mutant cells by a yet to be identified alternative pathway (putative nucleation element, yellow box). Primary siRNAs originating from dsRNA formed by bidirectional transcription of a centromeric sequence could prime further dsRNA synthesis and secondary siRNA generation by recruiting the RDRC complex to the nascent transcript. This would allow the spreading of H3K9me away from the nucleation site. H3K9me is bound by the chromodomain proteins Chp1, Chp2 and Swi6. The binding of Chp2 to H3K9me results in the recruitment of the SHREC complex, which in turn deacetylates H3K14. For unknown reasons this reduces RNA Pol II occupancy.
During the deacetylation reaction catalysed by Sir2, NAD is hydrolysed and generates a by-product called O-acetyl-ADP-ribose (O-AADPR; Tanny et al, 1999; Tanner et al, 2000). This by-product can enhance the stability of the SIR–nucleosomal complex and may provoke a conformational change of the SIR-bound nucleosomal fibre (Tanny et al, 1999; Tanner et al, 2000; Tanny and Moazed, 2001; Liou et al, 2005; Martino et al, 2009). Although these in vitro results are compelling, questions remain as how this works in vivo, because Sir2 deacetylation activity could be replaced in modified yeast by a class I catalytic domain that does not generate O-AADPR, with only minor loss of transcriptional repression (Chou et al, 2008).
Transcriptional silencing itself is thought to arise from sterical hindrance of positive regulators of transcription, by the interaction of the SIR complex with nucleosomes (Hecht et al, 1995). SIR complex association also leads to the sequestration of the silent chromatin at the NE through association with Esc1 (Gartenberg et al, 2004; Taddei et al, 2004). Both the binding of the SIR complex to nucleosomes and the recruitment of silent chromatin to the NE, have been shown to render silent chromatin less accessible to the recombination machinery and to the action of enzymatic probes, such as a bacterial DNA methyltransferase or restriction endonucleases (Gottschling, 1992; Singh and Klar, 1992; Loo and Rine, 1994).
Despite this sequestration, certain classes of DNA-binding proteins seem able to access silent chromatin. For example, recognition sites for the FLP and Cre recombinases located within budding yeast silent chromatin domains are accessible to these enzymes when expressed at high levels (Holmes and Broach, 1996; Cheng et al, 1998). Moreover, promoters within a silenced domain can remain accessible to proteins of the transcription machinery, although the factors that stimulate elongation seem to be excluded (Sekinger and Gross, 2001; Gao and Gross, 2008). Fission yeast heterochromatin may also be accessible to the transcription machinery, because heterochromatin defects have been attributed to specific RNA pol II mutants (Djupedal et al, 2005; Kato et al, 2005). In addition, small interfering RNAs (siRNAs) have been identified, which match pericentromeric heterochromatin (Reinhart and Bartel, 2002; Cam et al, 2005; Buhler et al, 2008). Consistently, it was shown that though transcription of the ‘forward' strand of pericentric DNA repeats was inhibited by heterochromatin formation, the ‘reverse' strand seemed to be transcribed equally in both wild-type and heterochromatin-deficient strains (Volpe et al, 2002). This might suggest that transcription can cooperate with RNA decay mechanisms to keep heterochromatic regions repressed. The implications of this are discussed in more detail below.
Heterochromatin assembly in fission yeast
The assembly of heterochromatin in fission yeast, similar to that in budding yeast, involves orchestrated changes in chromatin modifications. After deacetylation of the histone H3 N-terminus by the class I and II histone deacetylases Clr3 and Clr6 (homologs of the HDACs Hda1 and Rpd3, respectively), and the class III NAD-dependent deacetylase Sir2, the methyltransferase, Clr4, methylates histone H3 at lysine 9, creating a binding site for the Swi6, Chp1 and Chp2 chromodomain proteins (Grewal et al, 1998; Partridge et al, 2000; Nakayama et al, 2001; Bjerling et al, 2002; Shankaranarayana et al, 2003; Motamedi et al, 2008). Swi6 and Chp2 are homologous to HP1 proteins, a conserved family of chromatin factors that recognizes methylated H3K9 in all species (Jacobs et al, 2001; Jacobs and Khorasanizadeh, 2002). Similar to the SIR complex, sequential cycles of Swi6 binding and Clr4 recruitment have been proposed to mediate the spreading of H3K9 methylation along the chromatin fibre (Nakayama et al, 2001; Grewal and Moazed, 2003).
Recent studies have begun to elucidate mechanistic details of assembly and maintenance of these heterochromatic structures. Specifically, it has been shown that the fission yeast chromodomain proteins Swi6, Chp1 and Chp2, although found at both centromeric and telomeric heterochromatin, contribute in distinct ways to heterochromatin assembly at these loci (Thon and Verhein-Hansen, 2000; Partridge et al, 2000, 2002). First, Chp1 contributes to de novo assembly at all sites of heterochromatin, yet contributes to the maintenance of repressed chromatin exclusively at centromeres (Sadaie et al, 2004). This may stem from the fact that different heterochromatic regions are more or less dynamic; centromeric domains seem to be less stable and more in need of establishment events.
Much similar to the situation in S. cerevisiae, the nucleation of heterochromatin in fission yeast requires cis-acting recruitment events (Figure 2), such as the recruitment of S. pombe Rap1 by Taz1 (Kanoh and Ishikawa, 2001; Zhang et al, 2008). Again similar to S. cerevisiae, recruitment pathways are partially redundant: the Taz1–Rap1 interaction is compensated by a second Taz1-dependent pathway that nucleates methylation of H3K9 by Clr4 (Kanoh et al, 2005). At the mating-type locus, an element called REIII recruits ATF/CREB family proteins and helps to nucleate heterochromatin (Jia et al, 2004), whereas two further elements, REII and cenH elements (similar to dg and dh repeats found at the centromere) function cooperatively to enhance heterochromatin formation at the mating-type locus (Ayoub et al, 2000). The cis-acting nucleation sites at centromeres seem to be less well defined. Indeed, recent evidence suggests that transcription of pericentromeric dg and dh repeats has a critical function in heterochromatin assembly (Figure 2). It seems that, in addition to specific DNA sequences, transcription and/or non-coding RNAs (ncRNA) can provide an initial scaffold for the formation of heterochromatin (Cam et al, 2009). This observation, coupled with the fact that strains defective in RNA processing mechanisms compromise PEV (Buhler et al, 2007; Houseley et al, 2007; Murakami et al, 2007; Vasiljeva et al, 2008; Wang et al, 2008), have challenged the paradigm that heterochromatin excludes transcription.
Transcriptional scaffolds for the assembly of silent chromatin
Although it seems paradoxical, transcription may well be a prerequisite for the assembly and maintenance of some forms of silent chromatin. Although we know little about the underlying mechanisms that link RNA to chromatin, there is growing evidence that ncRNAs can contribute to epigenetic inheritance (Bernstein and Allis, 2005). One of the most prominent examples is the ncRNA Xist that is involved in X chromosome inactivation in mammalian females (Leeb et al, 2009; Senner and Brockdorff, 2009). Xist nucleates a repressive chromatin state in cis for almost an entire chromosome. ncRNAs have also been linked to certain forms of gene repression in budding yeast. For instance, a non-coding antisense RNA has been implicated in transcriptional silencing of Ty1 retrotransposons (Berretta et al, 2008), and antisense transcription has been shown to regulate chromatin-dependent silencing of the PHO84 gene in an aging yeast culture (Camblong et al, 2007). The PHO84 antisense RNA is normally kept at a low level by the nuclear exosome, an RNAse complex with 3′–5′ exonucleolytic activity. When this antisense RNA is degraded, PHO84 sense mRNA is present in maximal amounts, yet under stress conditions the antisense ncRNA accumulates and recruits the exosome to the PHO84 gene, reducing the sense message. The PHO84 ncRNA then seems to recruit a histone deacetylase to the locus to further inhibit sense transcription (Camblong et al, 2007). Although this is neither an SIR-dependent nor a heritable state of repression, it does underscore the role of RNA in the suppression of mRNA accumulation.
Finally, ncRNA has recently been detected to bind chromosome ends in which it contributes to the regulation of telomerase. The non-coding telomeric repeat-containing RNA (TERRA) is transcribed towards the chromosomal end in humans, mouse, hamster, zebrafish and in both budding and fission yeasts (Azzalin et al, 2007; Luke et al, 2008; Schoeftner and Blasco, 2008). Its role in gene repression is unclear, and the disruption of TPE through loss of SIR factors actually increased the level of TERRA, meaning that its presence is inversely correlated with repression (see also accompanying focus review by Luke and Lingner, in press). In contrast to this, it was found that fission yeast centromeric dg and dh transcripts are positively correlated with the assembly of heterochromatin in an RNA-dependent manner (Buhler and Moazed, 2007; Grewal and Elgin, 2007; Zaratiegui et al, 2007).
One major difficulty in assigning function to ncRNAs is to distinguish between the effects of transcription per se and a function of the transcript itself. It is possible that RNA Pol II transcribes non-coding DNA to remodel chromatin and that the resulting ncRNA is a non-functional by-product. Indeed, genes can be activated by transcription through promoter regions making DNA sequences more accessible to the transcription machinery (Hirota et al, 2008). It is also possible that genes become silenced as a consequence of transcription interference (Martens et al, 2004; Hongay et al, 2006). Finally, RNA could also actively recruit modifying enzymes that help assemble a higher-order chromatin structure. Interestingly, some HP1 proteins themselves have affinity for RNA (Muchardt et al, 2002), and recent work on the fission yeast Swi6 showed that it specifically interacts with heterochromatic transcripts. This led to the proposal that its RNA-binding activity serves the general function of retaining heterochromatic RNAs on chromatin (Motamedi et al, 2008).
RNAi-mediated heterochromatin assembly
In fission yeast several lines of evidence argue that RNA serves as a scaffold for the assembly of heterochromatin. In particular, the RNAi pathway contributes to repression at fission yeast centromeres, in which siRNAs, together with long ncRNAs, are essential for the formation of heterochromatin at pericentric dg/dh repeats. RNAi is a conserved silencing mechanism that is triggered by double-stranded RNA (dsRNA) (Bartel, 2004; Hannon, 2002). The mechanism of silencing involves the generation of small RNA molecules of ∼22 nucleotides from the longer dsRNAs by an RNase III-like enzyme called Dicer (Bernstein et al, 2001). These siRNAs then load onto an effector complex called RNA-induced silencing complex (RISC). RISC complexes contain Argonaute, which is a member of the conserved Argonaute/PIWI family of proteins that are required for RNAi in a variety of systems (Caudy et al, 2002; Hammond et al, 2001; Hutvagner and Zamore, 2002; Mourelatos et al, 2002; Zamore, 2001). siRNA-programmed RISC targets cognate mRNAs for degradation (Caudy et al, 2003). In a related process, small RNAs, called miRNAs, are produced from hairpin RNA transcripts by Dicer enzymes and programme RISC for translational repression of target mRNAs (Pillai, 2005). In some organisms, the RNAi response also requires an RNA-directed RNA polymerase (RdRp) that may be involved in amplifying dsRNA using siRNAs as primers (Dalmay et al, 2000; Sijen et al, 2001).
Although absent in S. cerevisiae, the key components of RNAi, Dicer and Argonaute, and an RNA-dependent RNA polymerase, are found in S. pombe. Deletion of the genes encoding any of these proteins (Dcr1, Ago1 and Rdp1, respectively) results in loss of H3K9 methylation and Swi6 localization at centromeres (Volpe et al, 2002). Moreover, siRNAs corresponding to centromeric repeats have been identified (Reinhart and Bartel, 2002; Cam et al, 2005; Buhler et al, 2008). Importantly, RNAi turned out to contribute to the initiation of heterochromatin assembly at all heterochromatic loci, but it is only required for maintenance at centromeres (Figure 2) (Jia et al, 2004; Sadaie et al, 2004; Kanoh et al, 2005). Thus, Chp1 and the RNAi machinery seem to be functionally linked. Strikingly, Chp1 resides in a complex together with Ago1, a newly identified factor Tas3, and centromeric siRNAs. This complex has been termed RNA-induced transcriptional silencing (RITS) complex and is required for silencing and high H3K9 methylation levels at centromeric dg and dh repeats (Verdel et al, 2004).
Further biochemical analysis of the S. pombe RNAi proteins resulted in the identification of two additional RNAi effector complexes that are important for centromeric heterochromatin formation: the Argonaute siRNA chaperone (ARC) complex and the RNA-directed RNA polymerase complex (RDRC) (Motamedi et al, 2004; Buker et al, 2007). RDRC has RNA-dependent RNA polymerase activity and is thought to amplify siRNAs (Motamedi et al, 2004). As in the RITS complex, ARC contains siRNAs bound to Argonaute. However, the siRNAs found in ARC are mostly double stranded, suggesting that ARC is a precursor complex involved in siRNA maturation. The RITS complex contains single-stranded siRNAs (Buker et al, 2007), which have been proposed to act as specificity factors for association with chromatin. In principle, siRNAs could target specific chromatin regions by base pairing with either DNA or nascent RNAs (Grewal and Moazed, 2003).
Studies over the past years have provided support for a model in which siRNAs act as guide molecules to target histone modifying enzymes to chromatin through base pairing between siRNA and pre-mRNA, during RNA elongation by RNA pol II. This would allow nascent RNA to serve as a scaffold for the recruitment of histone modifying enzymes (Figure 2). Support for this model comes from artificial tethering of RITS to the transcript of a normally euchromatic gene. Tethering of the RITS complex to ura4+ RNA through a site-specific RNA-binding protein (N protein of phage λ) results in heterochromatin assembly and silencing of the cognate ura4+ gene. This tethering also results in the generation of ura4+-specific siRNAs, and silencing requires proteins associated with both RNAi and heterochromatin (Buhler et al, 2006).
Downregulation of RNA Pol II in fission yeast
Recent work in fission yeast has shed new light on the mechanism of H3K14 deacetylation by the Clr3 HDAC and its relative contribution to chromatin-mediated gene repression. Affinity purification of Clr3 showed a complex termed SHREC (Snf2/Hdac-containing repressor complex) (Sugiyama et al, 2007). Core components of SHREC include Clr1, Clr2, the Clr3 histone deacetylase and the Mit1 chromatin-remodelling protein. SHREC seems to act downstream of heterochromatin assembly (H3K9 methylation) to catalyse the deacetylation of H3K14. This in turn seems to limit transcription by impairing access of DNA Pol II to heterochromatin (Figure 2) (Bjerling et al, 2002; Sugiyama et al, 2007). SHREC itself is targeted to heterochromatic loci by different ligands, two of which are Chp2 or Ccq1. Chp2 seems to be particularly important at centromeres, whereas Ccq1 functions only at the telomere, together with Taz1 (Kanoh et al, 2005; Sugiyama et al, 2007; Motamedi et al, 2008). Briefly, at centromeres the recognition of H3K9me by Chp2 recruits SHREC to facilitate histone H3 deacetylation, which in turn coincides with reduced presence of RNA pol II. On the other hand, Ccq1 circumvents the requirement for H3K9 methylation. Similarly, Atf1/Pcr1 are required for Clr3 targeting to a nucleation site at the mating-type locus, although it is not clear that this acts through physical recruitment of SHREC (Yamada et al, 2005).
The observation that RNA Pol II occupancy at heterochromatic loci increases on deletion of SHREC components implicates SHREC in the restriction of promoter access. However, deletion of all the three chromodomain proteins (Swi6, Chp1 and Chp2) is needed to reach the level of derepression achieved in a clr4Δ strain. This argues that RNA Pol II restriction is only part of the silencing mechanism (Motamedi et al, 2008). The rest may involve RNA decay mechanisms that operate in cis. Importantly, the RNA degradation seems to be different from classical post-transcriptional gene silencing, because it depends on the status of the chromatin from which the RNA is transcribed (Buhler et al, 2006, 2007). Therefore, this is referred to as co-transcriptional gene silencing (CTGS, Figure 3).
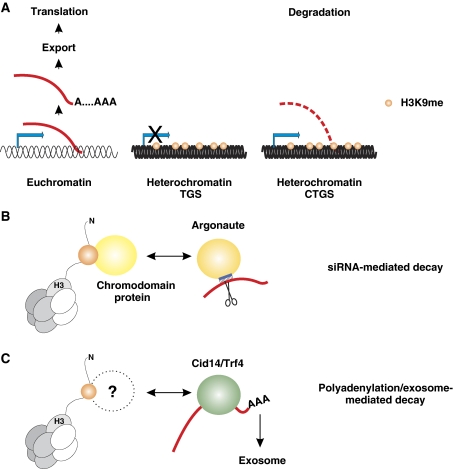
Figure 3.
Chromatin-dependent gene silencing mechanisms operate at a transcriptional and/or post-transcriptional level. (A) Silencing of heterochromatin can be achieved by either shutting off transcription (TGS) or by degradation of heterochromatic RNAs (CTGS). In contrast to classic post-transcriptional gene silencing (PTGS), CTGS depends on the status of chromatin from which the gene is transcribed and is therefore referred to as ‘co-transcriptional'. (B) RNAi-mediated degradation of heterochromatic RNAs. Argonaute-containing complexes can be physically linked to heterochromatin through chromodomain proteins. One histone-octamer is shown in grey. The chromodomain protein binds to methylated K9 (orange) of the unstructured N-terminal tail of histone H3. The siRNA (blue) guides Argonaute to the heterochromatic RNA through base-pairing interaction and induces ‘slicing'. (C) Heterochromatic gene silencing mediated by a non-canonical polyA-polymerase and the exosome. RNAs transcribed from heterochromatic regions are identified by Cid14/Trf4 and marked as aberrant with a short polyA tail. This serves as a signal for the exosome to degrade the RNA.
Chromatin-dependent RNA degradation
At heterochromatic loci in which RNAi is essential for silencing, RNA degradation could theoretically be mediated by the RNAi machinery (Noma et al, 2004; Buhler et al, 2007). Consistent with this idea, recombinant fission yeast Ago1 has siRNA-guided endonucleolytic activity (‘slicer' activity), and siRNAs originating from centromeric RNAs as well as centromeric reporter gene insertions have been detected (Irvine et al, 2006; Buhler et al, 2007; Buker et al, 2007). Intriguingly, heterochromatic siRNA levels increase upon deletion of SHREC components, suggesting that RNAi is compensating for the loss of TGS in these mutant strains (Sugiyama et al, 2007). Furthermore, silencing of heterochromatin has also been shown to require the TRAMP polyadenylation complex and exosome-mediated RNA degradation (Buhler et al, 2007; Murakami et al, 2007; Wang et al, 2008). Importantly, exosome and TRAMP mutant yeast strains show loss of heterochromatic gene silencing without any obvious defects in heterochromatin formation. Furthermore, highly unstable ncRNAs from silent chromatin regions can be detected in S. cerevisiae, which has entirely lost the RNAi pathway (Wyers et al, 2005; Houseley et al, 2007; Vasiljeva et al, 2008). This suggests that CTGS is likely to be a conserved RNA-turnover mechanism that can also function independently of the RNAi pathway to keep heterochromatin silent and further corroborates the concept of CTGS as a heterochromatic gene silencing pathway acting downstream of heterochromatin assembly (Figure 3; Buhler, 2009).
Heterochromatic microenvironments in the interphase nucleus
A striking feature of repetitive DNA and the silent chromatin it engenders is the propensity to stick together to form foci within the nucleus. These are called telomere clusters, or in the case of centromeres, chromocenters. Both can be found around the nucleolus or along the inner face of the NE in yeast and other organisms (reviewed in Akhtar and Gasser, 2007; de Laat and Grosveld, 2007). The result of this spatial arrangement is that the heterochromatin associated with simple repeat DNA creates a subnuclear compartment that sequesters silencing factors and silenced chromatin from the rest of the genome. The relevance of this phenomenon for both TPE and regulation of the rest of the genome were elegantly shown in budding yeast, in which the components that anchor heterochromatin could be identified and ablated by genetic techniques (Taddei et al, 2004, 2009).
Recent work has elaborated a function for these subcompartments. First, with respect to silencing, subcompartments seem to favour repression by overcoming natural restrictions on TPE that are imposed by the limiting abundance of silencing factors (Taddei et al, 2009). Indeed, overexpression of Sir2, Sir3 and Sir4 group wise, or Sir3 or Sir2 alone, enhances repression of reporter genes at telomeres or the HM loci (Maillet et al, 1996). Consistently, native HMR and HML silencers were shown to out-compete telomeres for limiting pools of SIR factors, because of their strong and redundant silencers (Buck and Shore, 1995; Cockell et al, 1998). In an important study, the tethering of a silencer-flanked reporter construct near telomeric foci was found to improve repression in an SIR-dependent manner (Andrulis et al, 1998). Finally, repression at silencer-proximal genes far from telomeres was facilitated by SIR factor overexpression, as well as by compromising telomere anchorage (Maillet et al, 2001; Gartenberg et al, 2004; Taddei et al, 2009). Thus, although a perinuclear anchoring is not absolutely necessary for SIR-mediated repression, it clearly contributes to its efficiency and propagation (Figure 4).
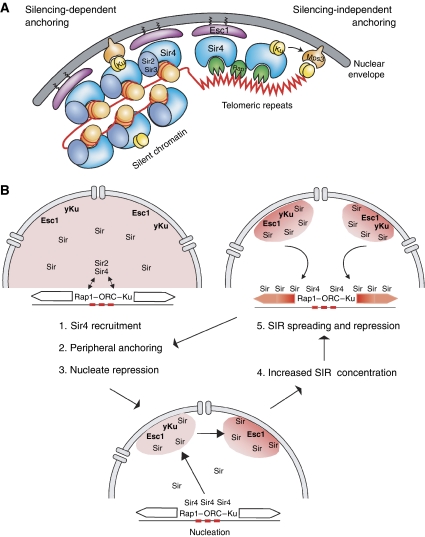
Figure 4.
Telomere anchoring and the promotion of TPE in yeast. (A) We show schematically both the silencing-dependent and the silencing- independent pathways of telomere anchoring in S. cerevisiae. The silencing-dependent pathway primarily exploits the integral SIR complex protein, Sir4, and its high-affinity interaction with Enhancer of Silent chromatin 1, which is peripherally associated with the nuclear envelope (NE). Sir4 can also bind yKu, which in turn mediates interaction with telomerase (Schober et al, 2009). Telomerase then binds Mps3, a SUN domain protein that is an integral component of the NE. At telomeres in S phase the yKu–telomerase–Mps3 pathway is sufficient to anchor telomeres in the absence of silent chromatin or Sir4 (Hediger et al, 2002). (B) We show a sequential model for how the binding of telomeres and their sequestration of SIR factors in foci can seed and the establishment of silencing at budding yeast telomeres. We propose that Sir4 is first recruited at the nucleation centre by DNA-binding proteins that can bind Sir4. These include Rap1, ORC, Abf1 and/or yKu. The presence of Sir4 at the locus will then bring it to the nuclear periphery through one of the two Sir4 anchoring pathways (yKu or Esc1) in which the high local concentrations of Sir proteins will help silencing complexes assemble and spread. Once silenced, the repressed telomere can associate with the NE through Esc1, which also increases the local concentration of Sir proteins and reinforces repression with this zone. Importantly, yKu can bind chromosome ends and link them to the nuclear envelope protein, Mps3, in the absence of repression.
How is attachment at the NE achieved? There are two pathways of anchoring in budding yeast, one of which is enhanced by formation of silent chromatin (Sir4-Esc1, Figure 4A), whereas the other is efficient in its absence (yKu–Mps3). For the generation of telomeric subcompartments this latter pathway is very important, as it allows telomere juxtaposition before heterochromatin formation. The pathway that requires yKu tethers telomeres to the NE through telomerase RNA, and the telomerase subunits, Est2 and Est1 in S-phase cells (Schober et al, 2009), thanks to the ability of the telomerase cofactor Est1 to bind the integral NE protein Mps3 (Uetz et al, 2000; Antoniacci et al, 2007). Mps3 is a member of the conserved SUN domain family, which contains inner NE proteins that interact with both chromatin and the cytoskeleton in many species. In budding yeast it docks yKu70/80-Tlc1-Est1 and Sir4 at the NE (Bupp et al, 2007), whereas it binds other proteins of the nuclear lumen in other species (reviewed in Fridkin et al, 2008). Interfering with this pathway perturbs telomere position and yKu-mediated anchoring in S-phase cells, but not in G1, because Est1 is stable only in S phase (Larose et al, 2007).
Intriguingly, the perturbation of telomere anchoring has more effects than simply the loss of TPE. Cells lacking both functional yeast Ku and Esc1 are viable, yet the dispersed SIR complexes have promiscuous effects on the transcriptome, and most notably at promoters implicated in ribosome biogenesis (Taddei et al, 2009; Zhu et al, 2009). At the same time the endogenous subtelomeric genes are derepressed. Thus, the sequestration of silencing factors in perinuclear foci has functional consequences for genome-wide gene regulation.
Much less is known about the mechanisms that tether heterochromatic regions of mitotically dividing fission yeast to the NE, although all three heterochromatic domains show perinuclear localization (Funabiki et al, 1993). The three centromeres and the MAT locus localize at the nuclear periphery by attaching to the spindle pole body (SPB). The telomeres are also found at the nuclear periphery but on the opposite side of the SPB in the proximity of the nucleolus in two to four clusters (Funabiki et al, 1993; Alfredsson-Timmins et al, 2007). This organization depends on heterochromatin (Ekwall et al, 1996; Alfredsson-Timmins et al, 2007) and can also be affected by mutations in the key factors of RNAi (ago1, dcr1 and rdp1) (Hall et al, 2003). However, despite the loss of centromeric repression, centromere clustering was unaffected in these RNAi mutants, and telomere clustering was lost without affecting telomeric silencing. Moreover, despite a loss of clustering, telomeres remained associated with the NE in RNAi mutants (Hall et al, 2003). Thus, in S. pombe, telomere–telomere interactions, but not centromere–centromere interactions depend on RNAi. Although RNAi is essential for telomere clustering, other pathways—possibly a redundant anchorage pathway such as the Ku pathway in budding yeast—position telomeres at the NE.
Apart from heterochromatin, other genomic elements are able to organize chromatin spatially. Notably, genomic regions designated as chromosome-organizing clamps (COC) are tethered to the nuclear periphery in a heterochromatin-independent manner in S. pombe. This is mediated by the TFIIIC transcription factor complex that normally recruits RNA polymerase III (Noma et al, 2006), yet these sites are not occupied by RNA polymerase III. The functional consequences of tethering COCs to the nuclear periphery are less clear, but they may have a boundary function that could impact complex chromosomal processes such as gene regulation, DNA replication and recombination (Noma et al, 2006). Interestingly, TFIIIC is also known to bind to several sites across the S. cerevisiae genome, called ETC loci, which are similarly independent of RNA polymerase III localization (Moqtaderi and Struhl, 2004).
Meiosis entails major re-arrangements of the nuclear organization of fission yeast chromatin. Chromocenters are detached from the SPB and change places with telomeres, in preparation for the horsetail movements when the meiotic recombination takes place (Chikashige et al, 1997). In meiotic prophase, Taz1 is required for stable association between telomeres and SPB, and loss of the association leads to strong negative phenotypes (Cooper et al, 1998). Indeed, meiotic recombination is reduced, and both spore viability and the ability of zygotes to re-enter mitosis are impaired. Finally, mutations in the RNAi machinery provoke a mild but consistent disruption of meiotic telomere clustering and SPB integrity (Hall et al, 2003). To date it is unclear exactly to which extent meiotic and mitotic elements of nuclear organization are conserved in S. pombe. This awaits a careful genetic dissection of the localization machinery.
Flagging up damage and telomeres
Additional elements and pathways contribute to the perinuclear localization of budding yeast telomeres and centromeres, many being incompletely explained. Some pathways of positioning seem to be linked to the cellular response to a double-strand break, which raises the issue of whether the nuclear periphery influences recombinational repair or telomerase elongation, or both. Data are still scarce on this issue, but it seems that Mps3 may help suppress or regulate certain forms of recombination, such as break-induced repair (Gartenberg, 2009). This could be relevant for telomeres and damage, for instance, when there is no donor sequence for repair by homologous recombination. It remains to be seen exactly how and why telomerase wins over recombination pathways when chromosomal breaks bear TG repeats. This is an important question to solve if we are to understand the molecular structure of a chromosomal end.
Conclusions
Much of our understanding of chromatin-mediated repression comes from the study of model organisms. The two yeasts discussed here have widely different mechanisms of silencing, yet both contribute important principles of action, which have and will continue to guide studies in more biomedically relevant organisms. Budding yeast provides important paradigms for nucleation, propagation and questions of dosage dependence for heterochromatin, whereas S. pombe has contributed many of the models currently explored on how RNA contributes to transcriptional repression in the nucleus. Even principles of nuclear organization are likely to have parallels in higher eukaryotic cells, although other internal nuclear structures may replace the NE as an organizing principle. The power of genetics and population-wide statistics, which are so easy in yeast, will ensure that these organisms remain at the forefront of epigenetic research.
References
- Akhtar A, Gasser SM (2007) The nuclear envelope and transcriptional control. Nat Rev Genet 8: 507–517 [DOI] [PubMed] [Google Scholar]
- Alfredsson-Timmins J, Henningson F, Bjerling P (2007) The Clr4 methyltransferase determines the subnuclear localization of the mating-type region in fission yeast. J Cell Sci 120 (Pt 11): 1935–1943 [DOI] [PubMed] [Google Scholar]
- Allshire RC, Javerzat JP, Redhead NJ, Cranston G (1994) Position effect variegation at fission yeast centromeres. Cell 76: 157–169 [DOI] [PubMed] [Google Scholar]
- Allshire RC, Karpen GH (2008) Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat Rev Genet 9: 923–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allshire RC, Nimmo ER, Ekwall K, Javerzat JP, Cranston G (1995) Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev 9: 218–233 [DOI] [PubMed] [Google Scholar]
- Andrulis ED, Neiman AM, Zappulla DC, Sternglanz R (1998) Perinuclear localization of chromatin facilitates transcriptional silencing. Nature 394: 592–595 [DOI] [PubMed] [Google Scholar]
- Antoniacci LM, Kenna MA, Skibbens RV (2007) The nuclear envelope and spindle pole body-associated Mps3 protein bind telomere regulators and function in telomere clustering. Cell Cycle 6: 75–79 [DOI] [PubMed] [Google Scholar]
- Aparicio OM, Billington BL, Gottschling DE (1991) Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell 66: 1279–1287 [DOI] [PubMed] [Google Scholar]
- Ayoub N, Goldshmidt I, Lyakhovetsky R, Cohen A (2000) A fission yeast repression element cooperates with centromere-like sequences and defines a mat silent domain boundary. Genetics 156: 983–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J (2007) Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318: 798–801 [DOI] [PubMed] [Google Scholar]
- Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Bernstein E, Allis CD (2005) RNA meets chromatin. Genes Dev 19: 1635–1655 [DOI] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366 [DOI] [PubMed] [Google Scholar]
- Berretta J, Pinskaya M, Morillon A (2008) A cryptic unstable transcript mediates transcriptional trans-silencing of the Ty1 retrotransposon in S cerevisiae. Genes Dev 22: 615–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjerling P, Ekwall K (2002) Centromere domain organization and histone modifications. Braz J Med Biol Res 35: 499–507 [DOI] [PubMed] [Google Scholar]
- Bjerling P, Silverstein RA, Thon G, Caudy A, Grewal S, Ekwall K (2002) Functional divergence between histone deacetylases in fission yeast by distinct cellular localization and in vivo specificity. Mol Cell Biol 22: 2170–2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blander G, Guarente L (2004) The Sir2 family of protein deacetylases. Annu Rev Biochem 73: 417–435 [DOI] [PubMed] [Google Scholar]
- Braunstein M, Sobel RE, Allis CD, Turner BM, Broach JR (1996) Efficient transcriptional silencing in Saccharomyces cerevisiae requires a heterochromatin histone acetylation pattern. Mol Cell Biol 16: 4349–4356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck SW, Shore D (1995) Action of a RAP1 carboxy-terminal silencing domain reveals an underlying competition between HMR and telomeres in yeast. Genes Dev 9: 370–384 [DOI] [PubMed] [Google Scholar]
- Buhler M (2009) RNA turnover and chromatin-dependent gene silencing. Chromosoma 118: 141–151 [DOI] [PubMed] [Google Scholar]
- Buhler M, Haas W, Gygi SP, Moazed D (2007) RNAi-dependent and -independent RNA turnover mechanisms contribute to heterochromatic gene silencing. Cell 129: 707–721 [DOI] [PubMed] [Google Scholar]
- Buhler M, Moazed D (2007) Transcription and RNAi in heterochromatic gene silencing. Nat Struct Mol Biol 14: 1041–1048 [DOI] [PubMed] [Google Scholar]
- Buhler M, Spies N, Bartel DP, Moazed D (2008) TRAMP-mediated RNA surveillance prevents spurious entry of RNAs into the Schizosaccharomyces pombe siRNA pathway. Nat Struct Mol Biol 15: 1015–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler M, Verdel A, Moazed D (2006) Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell 125: 873–886 [DOI] [PubMed] [Google Scholar]
- Buker SM, Iida T, Buhler M, Villen J, Gygi SP, Nakayama J, Moazed D (2007) Two different Argonaute complexes are required for siRNA generation and heterochromatin assembly in fission yeast. Nat Struct Mol Biol 14: 200–207 [DOI] [PubMed] [Google Scholar]
- Bupp J, Martin A, Stensrud E, Jaspersen S (2007) Telomere anchoring at the nuclear periphery requires the budding yeast Sad1-UNC-84 domain protein Mps3. J Cell Biol 179: 845–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cam HP, Chen ES, Grewal SI (2009) Transcriptional scaffolds for heterochromatin assembly. Cell 136: 610–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cam HP, Sugiyama T, Chen ES, Chen X, FitzGerald PC, Grewal SI (2005) Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat Genet 37: 809–819 [DOI] [PubMed] [Google Scholar]
- Camblong J, Iglesias N, Fickentscher C, Dieppois G, Stutz F (2007) Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S Cerevisiae. Cell 131: 706–717 [DOI] [PubMed] [Google Scholar]
- Caudy AA, Ketting RF, Hammond SM, Denli AM, Bathoorn AM, Tops BB, Silva JM, Myers MM, Hannon GJ, Plasterk RH (2003) A micrococcal nuclease homologue in RNAi effector complexes. Nature 425: 411–414 [DOI] [PubMed] [Google Scholar]
- Caudy AA, Myers M, Hannon GJ, Hammond SM (2002) Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev 16: 2491–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng TH, Li YC, Gartenberg MR (1998) Persistence of an alternate chromatin structure at silenced loci in the absence of silencers. Proc Natl Acad Sci USA 95: 5521–5526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikashige Y, Ding DQ, Imai Y, Yamamoto M, Haraguchi T, Hiraoka Y (1997) Meiotic nuclear reorganization: switching the position of centromeres and telomeres in the fission yeast Schizosaccharomyces pombe. EMBO J 16: 193–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou CC, Li YC, Gartenberg MR (2008) Bypassing Sir2 and O-acetyl-ADP-ribose in transcriptional silencing. Mol Cell 31: 650–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockell M, Gotta M, Palladino F, Martin SG, Gasser SM (1998) Targeting Sir proteins to sites of action: a general mechanism for regulated repression. Cold Spring Harb Symp Quant Biol 63: 401–412 [DOI] [PubMed] [Google Scholar]
- Cooper JP, Watanabe Y, Nurse P (1998) Fission yeast Taz1 protein is required for meiotic telomere clustering and recombination. Nature 392: 828–831 [DOI] [PubMed] [Google Scholar]
- Cubizolles F, Martino F, Perrod S, Gasser SM (2006) A homotrimer-heterotrimer switch in Sir2 structure differentiates rDNA and telomeric silencing. Mol Cell 21: 825–836 [DOI] [PubMed] [Google Scholar]
- Dalmay T, Hamilton A, Rudd S, Angell S, Baulcombe DC (2000) An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101: 543–553 [DOI] [PubMed] [Google Scholar]
- de Laat W, Grosveld F (2007) Inter-chromosomal gene regulation in the mammalian cell nucleus. Curr Opin Genet Dev 17: 456–464 [DOI] [PubMed] [Google Scholar]
- Djupedal I, Portoso M, Spahr H, Bonilla C, Gustafsson CM, Allshire RC, Ekwall K (2005) RNA Pol II subunit Rpb7 promotes centromeric transcription and RNAi-directed chromatin silencing. Genes Dev 19: 2301–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekwall K, Nimmo ER, Javerzat JP, Borgstrom B, Egel R, Cranston G, Allshire R (1996) Mutations in the fission yeast silencing factors clr4+ and rik1+ disrupt the localisation of the chromo domain protein Swi6p and impair centromere function. J Cell Sci 109 (Pt 11): 2637–2648 [DOI] [PubMed] [Google Scholar]
- Fisher TS, Zakian VA (2005) Ku: a multifunctional protein involved in telomere maintenance. DNA Repair (Amst) 4: 1215–1226 [DOI] [PubMed] [Google Scholar]
- Folco HD, Pidoux AL, Urano T, Allshire RC (2008) Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres. Science 319: 94–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridkin A, Penkner A, Jantsch V, Gruenbaum Y (2008) SUN-domain and KASH-domain proteins during development, meiosis and disease. Cell Mol Life Sci (E-pub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki H, Hagan I, Uzawa S, Yanagida M (1993) Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J Cell Biol 121: 961–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama S, Biggins S (2007) Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc Natl Acad Sci USA 104: 14706–14711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Gross DS (2008) Sir2 silences gene transcription by targeting the transition between RNA polymerase II initiation and elongation. Mol Cell Biol 28: 3979–3994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartenberg MR (2009) Life on the edge: telomeres and persistent DNA breaks converge at the nuclear periphery. Genes Dev 23: 1027–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartenberg MR, Neumann FR, Laroche T, Blaszczyk M, Gasser SM (2004) Sir-mediated repression can occur independently of chromosomal and subnuclear contexts. Cell 119: 955–967 [DOI] [PubMed] [Google Scholar]
- Gottschling DE (1992) Telomere-proximal DNA in Saccharomyces cerevisiae is refractory to methyltransferase activity in vivo. Proc Natl Acad Sci USA 89: 4062–4065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschling DE, Aparicio OM, Billington BL, Zakian VA (1990) Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell 63: 751–762 [DOI] [PubMed] [Google Scholar]
- Gravel S, Larrivee M, Labrecque P, Wellinger RJ (1998) Yeast Ku as a regulator of chromosomal DNA end structure. Science 280: 741–744 [DOI] [PubMed] [Google Scholar]
- Grewal SI, Bonaduce MJ, Klar AJ (1998) Histone deacetylase homologs regulate epigenetic inheritance of transcriptional silencing and chromosome segregation in fission yeast. Genetics 150: 563–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Elgin SC (2007) Transcription and RNA interference in the formation of heterochromatin. Nature 447: 399–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Moazed D (2003) Heterochromatin and epigenetic control of gene expression. Science 301: 798–802 [DOI] [PubMed] [Google Scholar]
- Hall IM, Noma K, Grewal SI (2003) RNA interference machinery regulates chromosome dynamics during mitosis and meiosis in fission yeast. Proc Natl Acad Sci USA 100: 193–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ (2001) Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293: 1146–1150 [DOI] [PubMed] [Google Scholar]
- Hannon GJ (2002) RNA interference. Nature 418: 244–251 [DOI] [PubMed] [Google Scholar]
- Hecht A, Laroche T, Strahl-Bolsinger S, Gasser SM, Grunstein M (1995) Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell 80: 583–592 [DOI] [PubMed] [Google Scholar]
- Hecht A, Strahl-Bolsinger S, Grunstein M (1996) Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature 383: 92–96 [DOI] [PubMed] [Google Scholar]
- Hediger F, Neumann FR, Van Houwe G, Dubrana K, Gasser SM (2002) Live Imaging of telomeres. yKu and Sir proteins define redundant telomere-anchoring pathways in yeast. Curr Biol 12: 2076–2089 [DOI] [PubMed] [Google Scholar]
- Hirota K, Miyoshi T, Kugou K, Hoffman CS, Shibata T, Ohta K (2008) Stepwise chromatin remodelling by a cascade of transcription initiation of non-coding RNAs. Nature 456: 130–134 [DOI] [PubMed] [Google Scholar]
- Holmes SG, Broach JR (1996) Silencers are required for inheritance of the repressed state in yeast. Genes Dev 10: 1021–1032 [DOI] [PubMed] [Google Scholar]
- Hongay CF, Grisafi PL, Galitski T, Fink GR (2006) Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell 127: 735–745 [DOI] [PubMed] [Google Scholar]
- Houseley J, Kotovic K, El Hage A, Tollervey D (2007) Trf4 targets ncRNAs from telomeric and rDNA spacer regions and functions in rDNA copy number control. EMBO J 26: 4996–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y (2002) Transcriptional silencing in Saccharomyces cerevisiae and Schizosaccharomyces pombe. Nucleic Acids Res 30: 1465–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, Zamore PD (2002) A microRNA in a multiple-turnover RNAi enzyme complex. Science 297: 2056–2060 [DOI] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L (2000) Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403: 795–800 [DOI] [PubMed] [Google Scholar]
- Irvine DV, Zaratiegui M, Tolia NH, Goto DB, Chitwood DH, Vaughn MW, Joshua-Tor L, Martienssen RA (2006) Argonaute slicing is required for heterochromatic silencing and spreading. Science 313: 1134–1137 [DOI] [PubMed] [Google Scholar]
- Jacobs SA, Khorasanizadeh S (2002) Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science 295: 2080–2083 [DOI] [PubMed] [Google Scholar]
- Jacobs SA, Taverna SD, Zhang Y, Briggs SD, Li J, Eissenberg JC, Allis CD, Khorasanizadeh S (2001) Specificity of the HP1 chromo domain for the methylated N-terminus of histone H3. EMBO J 20: 5232–5241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia S, Noma K, Grewal SI (2004) RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins. Science 304: 1971–1976 [DOI] [PubMed] [Google Scholar]
- Kanoh J, Ishikawa F (2001) spRap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr Biol 11: 1624–1630 [DOI] [PubMed] [Google Scholar]
- Kanoh J, Sadaie M, Urano T, Ishikawa F (2005) Telomere binding protein Taz1 establishes Swi6 heterochromatin independently of RNAi at telomeres. Curr Biol 15: 1808–1819 [DOI] [PubMed] [Google Scholar]
- Kato H, Goto DB, Martienssen RA, Urano T, Furukawa K, Murakami Y (2005) RNA polymerase II is required for RNAi-dependent heterochromatin assembly. Science 309: 467–469 [DOI] [PubMed] [Google Scholar]
- Konig P, Rhodes D (1997) Recognition of telomeric DNA. Trends Biochem Sci 22: 43–47 [DOI] [PubMed] [Google Scholar]
- Larose S, Laterreur N, Ghazal G, Gagnon J, Wellinger RJ, Elela SA (2007) RNase III-dependent regulation of yeast telomerase. J Biol Chem 282: 4373–4381 [DOI] [PubMed] [Google Scholar]
- Leeb M, Steffen PA, Wutz A (2009) X chromosome inactivation sparked by non-coding RNAs. RNA Biol 6 (E-pub ahead of print) [DOI] [PubMed] [Google Scholar]
- Liaw H, Lustig AJ (2006) Sir3 C-terminal domain involvement in the initiation and spreading of heterochromatin. Mol Cell Biol 26: 7616–7631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou GG, Tanny JC, Kruger RG, Walz T, Moazed D (2005) Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell 121: 515–527 [DOI] [PubMed] [Google Scholar]
- Loo S, Rine J (1994) Silencers and domains of generalized repression. Science 264: 1768–1771 [DOI] [PubMed] [Google Scholar]
- Louis EJ, Haber JE (1992) The structure and evolution of subtelomeric Y' repeats in Saccharomyces cerevisiae. Genetics 131: 559–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke B, Lingner J (in press) TERRA-Telomeric Repeat Containing RNA. EMBO J [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke B, Panza A, Redon S, Iglesias N, Li Z, Lingner J (2008) The Rat1p 5′ to 3′ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol Cell 32: 465–477 [DOI] [PubMed] [Google Scholar]
- Luo K, Vega-Palas MA, Grunstein M (2002) Rap1-Sir4 binding independent of other Sir, yKu, or histone interactions initiates the assembly of telomeric heterochromatin in yeast. Genes Dev 16: 1528–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillet L, Boscheron C, Gotta M, Marcand S, Gilson E, Gasser SM (1996) Evidence for silencing compartments within the yeast nucleus: a role for telomere proximity and Sir protein concentration in silencer-mediated repression. Genes Dev 10: 1796–1811 [DOI] [PubMed] [Google Scholar]
- Maillet L, Gaden F, Brevet V, Fourel G, Martin SG, Dubrana K, Gasser SM, Gilson E (2001) Ku-deficient yeast strains exhibit alternative states of silencing competence. EMBO Rep 2: 203–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandell JG, Bahler J, Volpe TA, Martienssen RA, Cech TR (2005) Global expression changes resulting from loss of telomeric DNA in fission yeast. Genome Biol 6: R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens JA, Laprade L, Winston F (2004) Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature 429: 571–574 [DOI] [PubMed] [Google Scholar]
- Martin SG, Laroche T, Suka N, Grunstein M, Gasser SM (1999) Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell 97: 621–633 [DOI] [PubMed] [Google Scholar]
- Martino F, Kueng S, Robinson P, Tsai-Pflugfelder M, van Leeuwen F, Ziegler M, Cubizolles F, Cockell MM, Rhodes D, Gasser SM (2009) Reconstitution of yeast silent chromatin: multiple contact sites and O-AADPR binding load SIR complexes onto nucleosomes in vitro. Mol Cell 33: 323–334 [DOI] [PubMed] [Google Scholar]
- Meluh PB, Yang P, Glowczewski L, Koshland D, Smith MM (1998) Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell 94: 607–613 [DOI] [PubMed] [Google Scholar]
- Mishra K, Shore D (1999) Yeast Ku protein plays a direct role in telomeric silencing and counteracts inhibition by Rif proteins. Curr Biol 9: 1123–1126 [DOI] [PubMed] [Google Scholar]
- Moqtaderi Z, Struhl K (2004) Genome-wide occupancy profile of the RNA polymerase III machinery in Saccharomyces cerevisiae reveals loci with incomplete transcription complexes. Mol Cell Biol 24: 4118–4127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motamedi MR, Hong EJ, Li X, Gerber S, Denison C, Gygi S, Moazed D (2008) HP1 proteins form distinct complexes and mediate heterochromatic gene silencing by nonoverlapping mechanisms. Mol Cell 32: 778–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motamedi MR, Verdel A, Colmenares SU, Gerber SA, Gygi SP, Moazed D (2004) Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell 119: 789–802 [DOI] [PubMed] [Google Scholar]
- Mourelatos Z, Dostie J, Paushkin S, Sharma A, Charroux B, Abel L, Rappsilber J, Mann M, Dreyfuss G (2002) miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev 16: 720–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchardt C, Guilleme M, Seeler JS, Trouche D, Dejean A, Yaniv M (2002) Coordinated methyl and RNA binding is required for heterochromatin localization of mammalian HP1alpha. EMBO Rep 3: 975–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller HJ (1930) Types of visible variations induced by X-rays in Drosophila. J Genet 22: 299–334 [Google Scholar]
- Murakami H, Goto DB, Toda T, Chen ES, Grewal SI, Martienssen RA, Yanagida M (2007) Ribonuclease activity of Dis3 is required for mitotic progression and provides a possible link between heterochromatin and kinetochore function. PLoS ONE 2: e317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mythreye K, Bloom KS (2003) Differential kinetochore protein requirements for establishment versus propagation of centromere activity in Saccharomyces cerevisiae. J Cell Biol 160: 833–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI (2001) Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292: 110–113 [DOI] [PubMed] [Google Scholar]
- Noma K, Cam HP, Maraia RJ, Grewal SI (2006) A role for TFIIIC transcription factor complex in genome organization. Cell 125: 859–872 [DOI] [PubMed] [Google Scholar]
- Noma K, Sugiyama T, Cam H, Verdel A, Zofall M, Jia S, Moazed D, Grewal SI (2004) RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat Genet 36: 1174–1180 [DOI] [PubMed] [Google Scholar]
- Partridge JF, Borgstrom B, Allshire RC (2000) Distinct protein interaction domains and protein spreading in a complex centromere. Genes Dev 14: 783–791 [PMC free article] [PubMed] [Google Scholar]
- Partridge JF, Scott KS, Bannister AJ, Kouzarides T, Allshire RC (2002) cis-acting DNA from fission yeast centromeres mediates histone H3 methylation and recruitment of silencing factors and cohesin to an ectopic site. Curr Biol 12: 1652–1660 [DOI] [PubMed] [Google Scholar]
- Pidoux AL, Allshire RC (2004) Kinetochore and heterochromatin domains of the fission yeast centromere. Chromosome Res 12: 521–534 [DOI] [PubMed] [Google Scholar]
- Pillai RS (2005) MicroRNA function: multiple mechanisms for a tiny RNA? RNA [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Bartel DP (2002) Small RNAs correspond to centromere heterochromatic repeats. Science 297: 1831. [DOI] [PubMed] [Google Scholar]
- Rusche LN, Kirchmaier AL, Rine J (2003) The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu Rev Biochem 72: 481–516 [DOI] [PubMed] [Google Scholar]
- Sadaie M, Iida T, Urano T, Nakayama J (2004) A chromodomain protein, Chp1, is required for the establishment of heterochromatin in fission yeast. EMBO J 23: 3825–3835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schober H, Ferreira H, Kalck V, Gehlen LR, Gasser SM (2009) Yeast telomerase and the SUN domain protein Mps3 anchor telomeres and repress subtelomeric recombination. Genes Dev 23: 928–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeftner S, Blasco MA (2008) Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat Cell Biol 10: 228–236 [DOI] [PubMed] [Google Scholar]
- Sekinger EA, Gross DS (2001) Silenced chromatin is permissive to activator binding and PIC recruitment. Cell 105: 403–414 [DOI] [PubMed] [Google Scholar]
- Senner CE, Brockdorff N (2009) Xist gene regulation at the onset of X inactivation. Curr Opin Genet Dev 19: 122–126 [DOI] [PubMed] [Google Scholar]
- Shankaranarayana GD, Motamedi MR, Moazed D, Grewal SI (2003) Sir2 regulates histone H3 lysine 9 methylation and heterochromatin assembly in fission yeast. Curr Biol 13: 1240–1246 [DOI] [PubMed] [Google Scholar]
- Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RH, Fire A (2001) On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107: 465–476 [DOI] [PubMed] [Google Scholar]
- Singh J, Klar AJ (1992) Active genes in budding yeast display enhanced in vivo accessibility to foreign DNA methylases: a novel in vivo probe for chromatin structure of yeast. Genes Dev 6: 186–196 [DOI] [PubMed] [Google Scholar]
- Smith MM (2002) Centromeres and variant histones: what, where, when and why? Curr Opin Cell Biol 14: 279–285 [DOI] [PubMed] [Google Scholar]
- Steiner NC, Clarke L (1994) A novel epigenetic effect can alter centromere function in fission yeast. Cell 79: 865–874 [DOI] [PubMed] [Google Scholar]
- Sugiyama T, Cam HP, Sugiyama R, Noma K, Zofall M, Kobayashi R, Grewal SI (2007) SHREC, an effector complex for heterochromatic transcriptional silencing. Cell 128: 491–504 [DOI] [PubMed] [Google Scholar]
- Suka N, Suka Y, Carmen AA, Wu J, Grunstein M (2001) Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol Cell 8: 473–479 [DOI] [PubMed] [Google Scholar]
- Sullivan BA, Blower MD, Karpen GH (2001) Determining centromere identity: cyclical stories and forking paths. Nat Rev Genet 2: 584–596 [DOI] [PubMed] [Google Scholar]
- Taddei A, Hediger F, Neumann FR, Bauer C, Gasser SM (2004) Separation of silencing from perinuclear anchoring functions in yeast Ku80, Sir4 and Esc1 proteins. EMBO J 23: 1301–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A, Van Houwe G, Nagai S, Erb I, van Nimwegen E, Gasser SM (2009) The functional importance of telomere clustering: Global changes in gene expression result from SIR factor dispersion. Genome Res 19: 611–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Cosma MP, Wirth K, Nasmyth K (1999) Identification of cohesin association sites at centromeres and along chromosome arms. Cell 98: 847–858 [DOI] [PubMed] [Google Scholar]
- Tanner KG, Landry J, Sternglanz R, Denu JM (2000) Silent information regulator 2 family of NAD- dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc Natl Acad Sci USA 97: 14178–14182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanny JC, Dowd GJ, Huang J, Hilz H, Moazed D (1999) An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell 99: 735–745 [DOI] [PubMed] [Google Scholar]
- Tanny JC, Moazed D (2001) Coupling of histone deacetylation to NAD breakdown by the yeast silencing protein Sir2: evidence for acetyl transfer from substrate to an NAD breakdown product. Proc Natl Acad Sci USA 98: 415–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JS, Ling X, Grunstein M (1994) Histone H3 amino terminus is required for telomeric and silent mating locus repression in yeast. Nature 369: 245–247 [DOI] [PubMed] [Google Scholar]
- Thon G, Verhein-Hansen J (2000) Four chromo-domain proteins of Schizosaccharomyces pombe differentially repress transcription at various chromosomal locations. Genetics 155: 551–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torras-Llort M, Moreno-Moreno O, Azorin F (2009) Focus on the centre: the role of chromatin on the regulation of centromere identity and function. EMBO J (e-pub ahead of print 23 July 2009; doi:10.1038/emboj.2009.174) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg JM (2000) A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403: 623–627 [DOI] [PubMed] [Google Scholar]
- van Leeuwen F, Gafken PR, Gottschling DE (2002) Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109: 745–756 [DOI] [PubMed] [Google Scholar]
- Vasiljeva L, Kim M, Terzi N, Soares LM, Buratowski S (2008) Transcription termination and RNA degradation contribute to silencing of RNA polymerase II transcription within heterochromatin. Mol Cell 29: 313–323 [DOI] [PubMed] [Google Scholar]
- Vega-Palas MA, Venditti S, Di Mauro E (1998) Heterochromatin organization of a natural yeast telomere. Changes of nucleosome distribution driven by the absence of Sir3p. J Biol Chem 273: 9388–9392 [DOI] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D (2004) RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303: 672–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA (2002) Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297: 1833–1837 [DOI] [PubMed] [Google Scholar]
- Wakimoto BT (1998) Beyond the nucleosome: epigenetic aspects of position-effect variegation in Drosophila. Cell 93: 321–324 [DOI] [PubMed] [Google Scholar]
- Wang SW, Stevenson AL, Kearsey SE, Watt S, Bahler J (2008) Global role for polyadenylation-assisted nuclear RNA degradation in posttranscriptional gene silencing. Mol Cell Biol 28: 656–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiren M, Silverstein RA, Sinha I, Walfridsson J, Lee HM, Laurenson P, Pillus L, Robyr D, Grunstein M, Ekwall K (2005) Genomewide analysis of nucleosome density histone acetylation and HDAC function in fission yeast. EMBO J 24: 2906–2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood V, Gwilliam R, Rajandream MA, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S, Basham D, Bowman S, Brooks K, Brown D, Brown S, Chillingworth T, Churcher C, Collins M, Connor R, Cronin A et al. (2002) The genome sequence of Schizosaccharomyces pombe. Nature 415: 871–880 [DOI] [PubMed] [Google Scholar]
- Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Regnault B, Devaux F, Namane A, Seraphin B, Libri D, Jacquier A (2005) Cryptic Pol II transcripts are degraded by a nuclear quality control pathway involving a new Poly(A) polymerase. Cell 121: 725–737 [DOI] [PubMed] [Google Scholar]
- Yamada M, Hayatsu N, Matsuura A, Ishikawa F (1998) Y'-Help1, a DNA helicase encoded by the yeast subtelomeric Y' element, is induced in survivors defective for telomerase. J Biol Chem 273: 33360–33366 [DOI] [PubMed] [Google Scholar]
- Yamada T, Fischle W, Sugiyama T, Allis CD, Grewal SI (2005) The nucleation and maintenance of heterochromatin by a histone deacetylase in fission yeast. Mol Cell 20: 173–185 [DOI] [PubMed] [Google Scholar]
- Zakian VA (1996) Structure, function, and replication of Saccharomyces cerevisiae telomeres. Annu Rev Genet 30: 141–172 [DOI] [PubMed] [Google Scholar]
- Zamore PD (2001) RNA interference: listening to the sound of silence. Nat Struct Biol 8: 746–750 [DOI] [PubMed] [Google Scholar]
- Zaratiegui M, Irvine DV, Martienssen RA (2007) Noncoding RNAs and gene silencing. Cell 128: 763–776 [DOI] [PubMed] [Google Scholar]
- Zhang K, Mosch K, Fischle W, Grewal SI (2008) Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol 15: 381–388 [DOI] [PubMed] [Google Scholar]
- Zhu C, Byers KJ, McCord RP, Shi Z, Berger MF, Newburger DE, Saulrieta K, Smith Z, Shah MV, Radhakrishnan M, Philippakis AA, Hu Y, De Masi F, Pacek M, Rolfs A, Murthy T, Labaer J, Bulyk ML (2009) High-resolution DNA-binding specificity analysis of yeast transcription factors. Genome Res 19: 556–566 [DOI] [PMC free article] [PubMed] [Google Scholar]