Abstract
Despite years of effort, exact pathogenesis of nonalcoholic fatty liver disease (NAFLD) remains obscure. To gain an insight into the regulatory roles of microRNAs (miRNAs) in aberrant energy metabolic status and pathogenesis of NAFLD, we analyzed the expression of miRNAs in livers of ob/ob mice, streptozotocin (STZ)-induced type 1 diabetic mice, and normal C57BL/6 mice by miRNA microarray. Compared with normal C57BL/6 mice, ob/ob mice showed upregulation of eight miRNAs and downregulation of four miRNAs in fatty livers. Upregulation of miR-34a and downregulation of miR-122 was found in livers of STZ-induced diabetic mice. These results demonstrate that distinct miRNAs are strongly dysregulated in NAFLD and hyperglycemia. Comparison between miRNA expressions in livers of ob/ob mice and STZ-administered mice further revealed upregulation of four miRNAs and downregulation of two miRNAs in livers of ob/ob mice, indicating that these miRNAs may represent a molecular signature of NAFLD. A distinctive miRNA expression pattern was identified in ob/ob mouse liver, and hierarchical clustering of this pattern could clearly discriminate ob/ob mice from either normal C57BL/6 mice or STZ-administered mice. These findings suggest an important role of miRNAs in hepatic energy metabolism and implicate the participation of miRNAs in the pathophysiological processes of NAFLD.
Keywords: nonalcoholic fatty liver disease, energy metabolism, microarray, ob/ob mice
Nonalcoholic fatty liver disease (NAFLD) is the most common form of chronic liver disease worldwide and is becoming a major public health concern in modern society (1–3). NAFLD is a clinicopathological syndrome characterized by excess fat accumulation in liver, which ranges from simple steatosis to steatohepatitis and cirrhosis in the absence of heavy alcohol consumption (4). Disorders related to metabolic syndrome, such as obesity, type 2 diabetes mellitus, and dyslipidemia, were identified as the main risk factors for the development of NAFLD (5–7). Although a widely accepted two-hit hypothesis (8) may partially explain the progressive liver damage by nonalcoholic steatosis and steatohepatitis, much of the pathogenesis of NAFLD remains undiscovered and requires further study.
Recently, a new class of RNA regulatory genes known as microRNAs (miRNAs) has been found to introduce a whole new layer of gene regulation in eukaryotes. miRNAs are endogenous noncoding RNAs of 19–24 nucleotides in length that play an important role in the negative regulation of gene expression by base-pairing to complementary sites on the target mRNAs, causing a block of translation or degradation of the target mRNA (9). In addition to the fundamental roles in diverse biological and pathological processes, including developmental timing, apoptosis, proliferation, differentiation, organ development, carcinogenesis, and immune response (10–12), miRNAs are also reported to play important roles in energy metabolism, both in invertebrates and vertebrate animals. The role of miRNAs in energy metabolism was first indicated by a study in the fruit fly Drosophlia melanogaster, suggesting an important role of miR-14 in energy metabolism on the whole-animal level (13). Another study showed an involvement of miR-278 in energy homeostasis of Drosophila (14). For vertebrates, miRNAs were found to regulate adipocyte differentiation (15, 16), and glucose or potassium stimulated insulin secretion from pancreatic islet cells (17, 18). Esau et al. (19) applied an miR-122 antagonist intraperitoneally into mice and found that this procedure could reduce plasma cholesterol level, increase hepatic fatty acid oxidation, and decrease hepatic fatty acid and cholesterol synthesis rate. Jin et al. (20) also reported that miR-122 level was over 2-fold upregulated in NAFLD rat. Together, these findings suggested a strong connection between miRNAs and energy metabolism. However, whether miRNAs play a role in the pathophysiological processes of NAFLD remains to be elucidated.
To address this question, we employed miRNA microarray to identify differentially expressed miRNAs in livers of ob/ob mice [a well-established model of NAFLD with hyperglycemia (21, 22)] and streptozotocin (STZ)-induced type 1 diabetic mice (a model simply under high blood glucose without fatty liver) to characterize the potential roles of miRNAs in livers under NAFLD and hyperglycemia, respectively.
MATERIALS AND METHODS
Animal models
Adult (aged 11–12 weeks) obese Lepob/ob C57BL/6 mice (six male and four female) and normal control C57BL/6 mice (aged 8–12 weeks; four male and four female) were purchased from Model Animal Research Center of Nanjing University. For STZ-induced type 1 diabetic model, 8-week-old wild-type C57BL/6 mice (eight male) received a single intraperitoneal injection of 150 mg/kg STZ dissolved in citrate buffer at pH 4.5. Mice were fasted overnight before glucose measurements. Blood samples were taken from the tail vein 3, 7, and 14 days after STZ injection, respectively. Animals were euthanized 14 days after STZ injection. The mice with fasting blood glucose ≥ 12 mmol/L were considered diabetic. All mice were housed in a temperature-controlled environment with a 12 h light/dark cycle and free access to water and a standard chow diet containing 60% carbohydrate, 13% fat, and 27% protein on a caloric basis. All animals were weighed and taken fasting blood glucose measurements and serum samples before euthanized. The liver tissues were dissected and immersed in liquid nitrogen immediately and stored at −80°C until used for subsequent analysis. Serum insulin levels of these animal models were quantified by the Rat/Mouse Insulin ELISA Kit (Linco Research, St. Charles, MO). Liver triglyceride concentration was determined using the Triglyceride Kit (Jiancheng Bioengineering Institute, Nanjing, China). All animal care and handling procedures were carried out in accordance with the Institute for Laboratory Animal Research Guide for Care and Use of Laboratory Animals and approved by the Institutional Review Board of Nanjing University.
RNA isolation and microarray experiments
Frozen liver tissues were dissected and homogenized, and total RNA were extracted using Trizol Reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. RNA labeling and hybridization on miRNA microarray chips were conducted as previously described (23, 24). Briefly, 50 μg of total RNA was purified using the mirVANA miRNA isolation kit (Ambion, Austin, TX) to enrich small RNA fraction. Purified RNA was labeled with fluorescein, and hybridization was carried out on the CapitalBio Mammalian miRNA Array V 3.0 (CapitalBio, Beijing, China) containing 509 probes in triplicate, corresponding to 435 human and mouse mature miRNA genes. Liver RNA sample from each mouse was analyzed on individual chip. Finally, hybridization signals were detected, and scanner images were quantified. These quantified signal intensity values of microarray were normalized to per-chip mean values. The miRNA microarray data have been deposited into Gene Expression Omnibus (GEO accession number GSE13840) and can be prepared in a fully MIAME-compliant manner.
Real-time quantitative RT-PCR of mature miRNAs
Stem-loop quantitative RT-PCR (qRT-PCR) assays to quantify the mature miRNAs were performed as previously described (25, 26) using fluorescent nucleic acid dye. The RT primers and real-time PCR primers were designed as described (25). Briefly, 1 μg of total RNA was reverse transcribed under the following conditions: 16°C for 15 min, 42°C for 60 min, and 85°C for 5 min. The 20 μl PCR included 1 μl RT product and 1 μl EvaGreen dye (Biotium, Hayward, CA). The conditions for the PCR reaction were as follows: 95°C for 5 min followed by 40 cycles of 95°C for 15 s and 60°C for 1 min using an ABI PRISM 7300 thermal cycler. All reactions were run in triplicate. The threshold cycle (CT) is defined as the fractional cycle number at which the fluorescence passes the fixed threshold. The miRNA expression levels were normalized to U6 RNA. The relative expression was calculated using the comparative ΔΔCT method, and the values were expressed as 2−ΔΔCT (27).
Statistical analysis
Differentially expressed miRNAs were identified using the t-test procedure within significance analysis of microarrays (SAM) (28) with a 5% false discovery rate (FDR) threshold. Cluster and Java TreeView were used to build the unsupervised tree. The genes and arrays were mean centered, and hierarchical trees were built using correlation metrics. Real-time qRT-PCR assays were performed in triplicate. Data were expressed as means ± SE, and the differences with P < 0.05 were considered statistically significant using a two-sided unpaired Student's t-test.
Bioinformatics analysis of putative miRNA targets
The analysis of miRNA predicted targets was performed using the following three algorithms: TargetScan (29) (http://www.targetscan.org/), PicTar (30) (http://pictar.mdc-berlin.de/), and miRanda (31) (http://cbio.mskcc.org/cgi-bin/mirnaviewer/mirnaviewer.pl).
RESULTS
Distinctive patterns of miRNA expression in livers of ob/ob mice
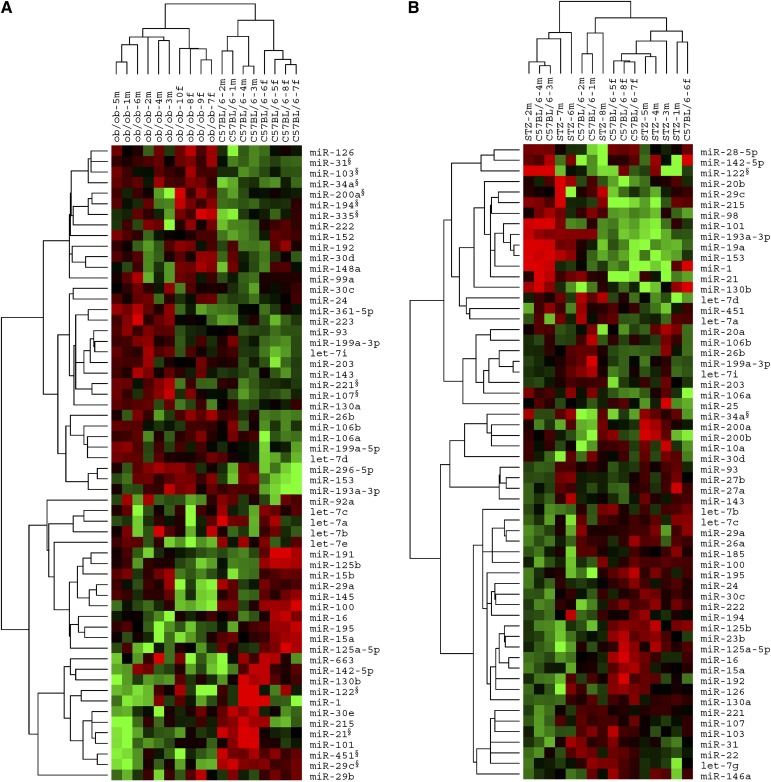
To investigate the potential involvement of miRNAs in NAFLD and hyperglycemia, we performed a microarray-based analysis of miRNAs in livers of ten ob/ob mice and eight C57BL/6 mice (served as controls), respectively. As shown in Table 1, the body weight, blood glucose levels, serum insulin levels, and liver triglyceride concentration of the ob/ob mice were all significantly elevated compared with those of the C57BL/6 mice, suggesting the aberrant energy metabolic status of ob/ob mice. Next, total RNAs were extracted from liver tissues of ob/ob mice and C57BL/6 mice and subsequently used in miRNA microarray analysis. The discrimination of miRNA expression in livers of ob/ob mice and C57BL/6 mice was clearly revealed by a two-way (genes against samples) unsupervised hierarchical clustering analysis of the miRNA microarray data. As displayed in Fig. 1A, the sample dendrogram generated by hierarchical cluster analysis showed two major branches in columns (ob/ob mice vs. C57BL/6 mice), indicating a specific miRNA expression pattern in the fatty livers of ob/ob mice versus the normal livers of C57BL/6 mice.
TABLE 1.
General characteristics of different groups of mice
| Group of Mice | C57BL/6 Mice | Ob/ob Mice | STZ Mice |
|---|---|---|---|
| n | 8 | 10 | 8 |
| BW (g) | 22.4 ± 0.3 | 45.5 ± 1.1** | 17.7 ± 0.6** |
| BGa (mmol/l) | 7.2 ± 0.6 | 13.3 ± 0.9** | 28.8 ± 2.2** |
| INSa (ng/ml) | 0.52 ± 0.10 | 10.40 ± 1.12** | 0.19 ± 0.03** |
| Liver TG (mmol/100g) | 2.48 ± 0.42 | 30.75 ± 2.84** | 1.84 ± 0.34 |
Data plotted as means ± SE. n, number of mice; BW, body weight; BG, blood glucose levels; INS, serum insulin levels; liver TG, liver triglyceride concentration.
Fasting blood glucose levels and fasting serum insulin levels measured before the mice were euthanized; **P < 0.01.
Fig. 1.
Hierarchical cluster analysis of miRNAs in livers of ob/ob mice and STZ-induced diabetic mice versus C57BL/6 mice. A: Two-way (genes against samples) unsupervised hierarchical cluster of a set of 60 miRNAs with most variation in livers of ob/ob mice and C57BL/6 mice. Sample dendrogram generated by cluster analysis shows a clear separation of ob/ob mice samples from those of C57BL/6 mice based on the miRNA signature. Ten ob/ob mice and eight C57BL/6 mice are used. B: Unsupervised hierarchical cluster of most altered miRNAs in livers of STZ-induced diabetic mice and C57BL/6 mice. Sample dendrogram shows no clear separation between STZ samples and C57BL/6 samples based on the miRNA signature. Eight STZ-induced diabetic mice and eight C57BL/6 mice are used. Mean centered signal intensities of miRNA expression are depicted by a log-transform (2 scale). Color areas indicate relative expression of each miRNA compared with the miRNA median expression (red, above; green, below; and black, close to the median value). m, male; f, female; §, significantly changed miRNAs identified by SAM or qRT-PCR.
To further identify differentially expressed miRNAs, a statistical method termed SAM (28) was employed. SAM is an algorithm that calculates a score for each gene and therefore identifies genes that are significantly associated with an outcome variable, such as the type of analyzed tissue (NAFLD vs. normal). Employing two-class unpaired analysis within SAM, the miRNA expression levels in livers of ob/ob mice were compared with those of C57BL/6 mice. In this test, an FDR (q-value) <5% was selected. The expressions of miRNAs were considered significantly altered only when they fulfilled two criteria: 1) mean fold change > 2 or < 0.5; 2) q-value < 5%. Based on these principles, SAM analysis generated a list of 11 miRNAs that were differentially expressed in livers of ob/ob mice (Table 2). Among these miRNAs, eight miRNAs (miR-34a, miR-31, miR-103, miR-107, miR-194, miR-335-5p, miR-221, and miR-200a) were at least 2-fold upregulated, and three miRNAs (miR-29c, miR-451, and miR-21) were at least 0.5-fold downregulated in ob/ob mice liver samples. Collectively, these data demonstrate that distinct miRNAs are significantly regulated in NAFLD with hyperglycemia, implicating the potential roles of miRNAs as functional modulators in the pathophysiological processes of these diseases.
TABLE 2.
Differentially expressed miRNAs in livers of ob/ob mice and STZ-induced diabetic mice compared with those of C57BL/6 mice
| miRNA | SAM Score | Fold Change | q-Value (%)a |
|---|---|---|---|
| Ob/ob mice versus C57BL/6 mice | |||
| Upregulated | |||
| mmu-miR-34a | 5.99 | 3.29 | 0.00 |
| mmu-miR-31 | 4.66 | 2.92 | 0.00 |
| mmu-miR-103 | 4.03 | 2.42 | 0.00 |
| mmu-miR-107 | 3.77 | 2.56 | 0.00 |
| mmu-miR-194 | 2.44 | 2.18 | 0.00 |
| mmu-miR-335-5p | 2.43 | 2.61 | 0.00 |
| mmu-miR-221 | 2.24 | 2.01 | 0.00 |
| mmu-miR-200a | 1.71 | 2.15 | 0.00 |
| Downregulated | |||
| mmu-miR-29c | −2.76 | 0.45 | 0.00 |
| mmu-miR-451 | −2.61 | 0.48 | 0.00 |
| mmu-miR-21 | −1.90 | 0.44 | 0.00 |
| mmu-miR-122b | −1.34 | 0.68 | 3.29 |
| STZ-induced diabetic mice versus C57BL/6 mice | |||
| mmu-miR-34a | 7.32 | 5.63 | 0.00 |
| Downregulated | |||
| mmu-miR-122b | −0.75 | 0.85 | 0.00 |
q-value: false discovery rate, the expected percentage of genes identified by chance.
These miRNAs were not considered as significantly changed by SAM due to insufficient fold change but proved to be significantly changed by real-time qRT-PCR.
miRNA expression in livers of STZ-induced type 1 diabetic mice
By causing rapid and irreversible necrosis of pancreatic β-cells (32), STZ-induced type 1 diabetic mice provide us an animal model of high blood glucose but without fatty liver. This diabetic mouse model was employed here to study the relationship between hepatic miRNA expression and hyperglycemia. Within 7 days of STZ injection, fasting blood glucose levels of the mice were all above 12 mmol/L and stayed high in the following week, while the body weight of these mice decreased gradually (data not shown). On the 14th day after STZ injection, the mean fasting blood glucose level of STZ-administered mice was almost 4-fold higher than that of control C57BL/6 mice. By contrast, the average body weight of STZ-administered mice was significantly decreased compared with that of C57BL/6 mice (Table 1). At this stage, mice were killed and liver tissues were dissected for RNA extraction and miRNA microarray detection subsequently. SAM analysis of miRNA microarray data (with a 2-fold change threshold and 5% FDR) showed that only miR-34a was upregulated in liver samples of STZ-induced diabetic mice compared with samples of C57BL/6 mice (Table 2). Unsupervised hierarchical clustering of miRNA expression from liver samples of STZ-induced diabetic mice and C57BL/6 mice is depicted in Fig. 1B. Sample dendrogram in columns showed no clear separation between STZ samples and C57BL/6 samples, indicating that general miRNA expression in mice livers under 2-week hyperglycemia was not significantly altered, except for miR-34a.
Differentially expressed miRNAs in livers of ob/ob mice versus STZ-induced diabetic mice
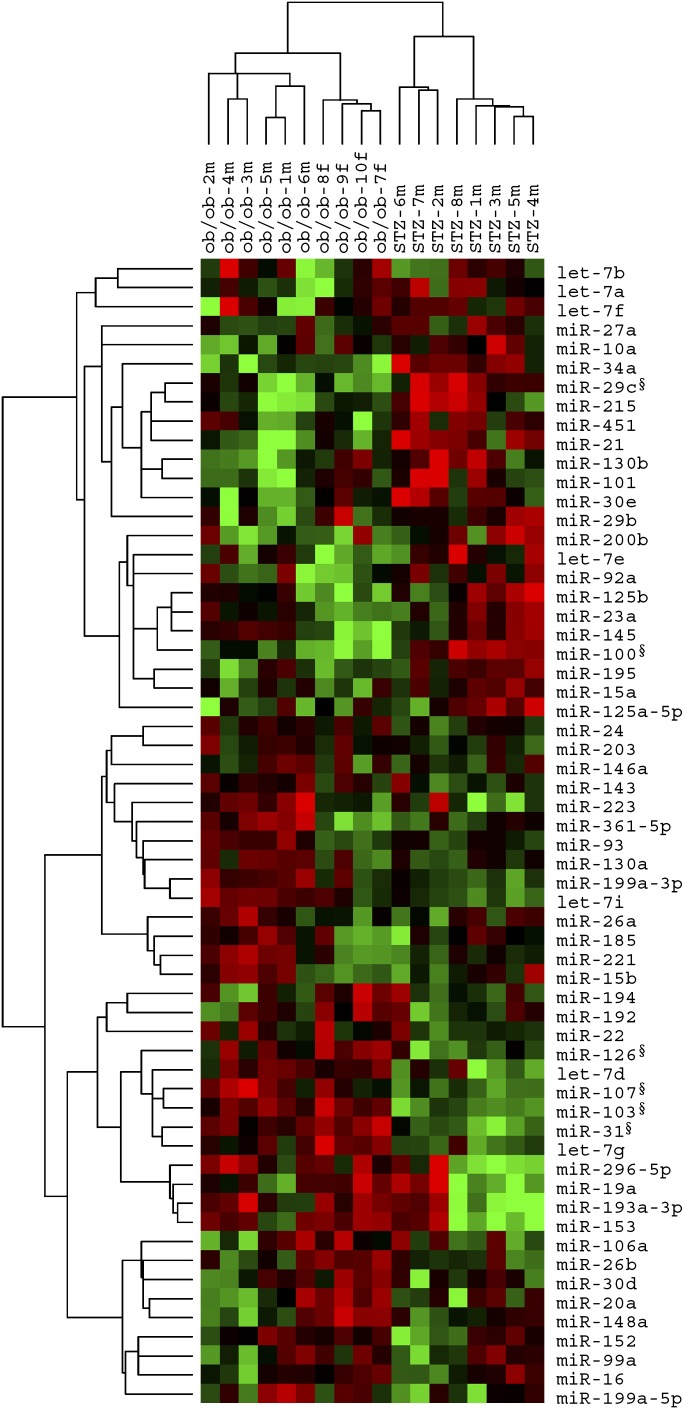
To find out the miRNAs that may be only involved in the pathophysiological processes of NAFLD, we next compared the miRNA expression pattern in livers of ob/ob mice with that of STZ-induced diabetic mice. Here ob/ob mice served as an animal model under both NAFLD and hyperglycemia conditions, while STZ-induced diabetic mice served as a model under a simple status of hyperglycemia. As shown in Fig. 2, unsupervised hierarchical clustering of liver miRNA expression of ob/ob mice and STZ-induced diabetic mice generated a sample dendrogram with two major branches in columns, clearly discriminating these two groups of samples. This result suggests a unique miRNA expression pattern in nonalcoholic fatty livers, excluding the influence of hyperglycemia on the hepatic miRNA expression. Under 5% FDR and 2-fold change threshold, SAM algorithm identified four upregulated miRNAs (miR-103, miR-31, miR-107, and miR-126-3p) and two downregulated miRNAs (miR-100 and miR-29c) in ob/ob samples compared with STZ samples (Table 3), indicating that these miRNAs may represent a molecular signature of NAFLD.
Fig. 2.
Hierarchical cluster analysis of ob/ob mice versus STZ-induced diabetic mice. Overview of two-way (genes against samples) unsupervised hierarchical cluster of miRNA expression in livers of 10 ob/ob mice and eight STZ-induced diabetic mice on the basis of most altered miRNAs between two samples. Sample dendrogram generated by cluster analysis shows a clear separation of ob/ob mice samples from those of STZ-induced diabetic mice based on the miRNA signature. Mean centered signal intensities of miRNA expression are depicted by a log transform (2 scale). Color areas indicate relative expression of each miRNA with respect to the miRNA median expression (red, above; green, below; and black, close to the median value). m, male; f, female; §, significantly changed miRNAs identified by SAM or qRT-PCR.
TABLE 3.
Differentially expressed miRNAs in livers of ob/ob mice compared with those of STZ-induced diabetic mice
| miRNA | SAM Score | Fold Change | q-Value (%)a |
|---|---|---|---|
| Upregulated | |||
| mmu-miR-103 | 5.62 | 2.96 | 0.00 |
| mmu-miR-31 | 4.42 | 2.94 | 0.00 |
| mmu-miR-107 | 4.11 | 2.67 | 0.00 |
| mmu-miR-126-3p | 2.56 | 2.02 | 0.00 |
| Downregulated | |||
| mmu-miR-100 | −3.34 | 0.45 | 0.00 |
| mmu-miR-29c | −2.57 | 0.43 | 0.00 |
q-value: false discovery rate, the expected percentage of genes identified by chance.
Dysregulated liver miRNAs under aberrant metabolic status segregated by gender
In this study, both male and female mice (four male and four female normal C57BL/6 mice; six male and four female ob/ob mice; and eight male STZ-induced diabetic mice) were used. To further analyze differentially expressed liver miRNAs in these animal models of different gender, we compared the liver miRNA expression in different animal models of male and female, respectively. As shown in supplementary Tables I and II, SAM analysis of miRNA microarray data (with a 2-fold change threshold and 5% FDR) listed 18 dysregulated miRNAs in livers of male ob/ob mice versus male normal C57BL/6 mice, 23 dysregulated miRNAs in livers of female ob/ob mice versus female C57BL/6 mice, four dysregulated miRNAs in livers of male STZ-induced diabetic mice versus male C57BL/6 mice, and 11 dysregulated miRNAs in livers of male ob/ob mice versus male STZ-induced diabetic mice. By unsupervised hierarchical clustering of miRNA expression in different liver samples segregated by gender, clear separation of ob/ob mouse samples from normal C57BL/6 mouse samples and of ob/ob mouse samples from STZ-induced diabetic mouse samples can been identified (see supplementary and II), which is similar to the results of mixed gender samples. These results suggest that although gender does affect the expressions of several miRNAs, the distinct miRNA expression patterns among these mouse models are not influenced by gender.
Validation of differentially expressed miRNAs by real-time qRT-PCR
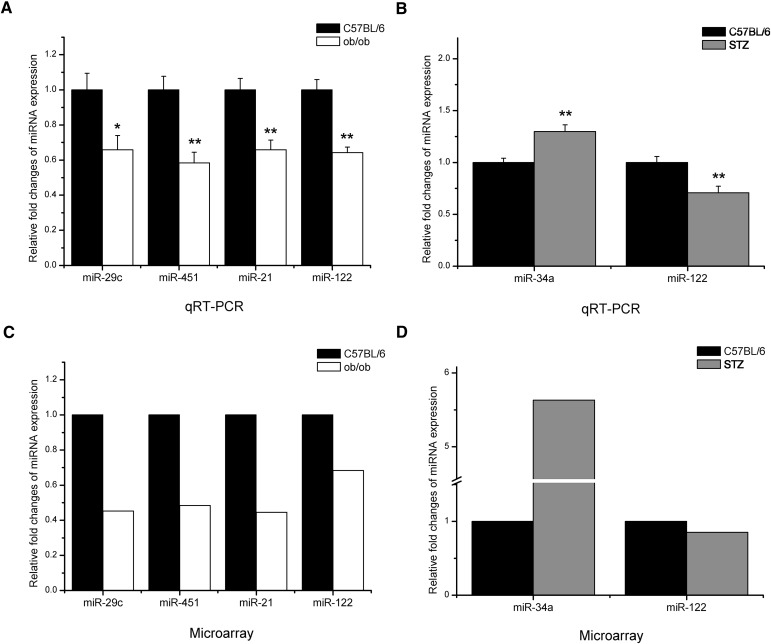
To validate the accuracy of the miRNA microarray data, we carried out stem-loop qRT-PCR assay (25, 26) with the same RNA preparations used in microarray analysis. As shown in Fig. 3, differentially expressed miRNAs identified by SAM algorithm based on the microarray results were analyzed using qRT-PCR assay. Results showed that the directions of the changes in miRNA expression were concordant between the two platforms.
Fig. 3.
Validation of microarray data by real-time qRT-PCR. A, B: Real-time qRT-PCR analysis of differentially expressed miRNAs in liver tissue RNA samples previously profiled by microarray. Triplicate assays were carried out for each RNA sample, and the relative amount of each miRNA was normalized to U6 snRNA. Data are shown as fold changes of miRNA levels in livers of ob/ob mice (A) and STZ-induced diabetic mice (B) compared with C57BL/6 mice, which is set as 1 (mean ± SE, n = 8) (*P < 0.05; **P < 0.01). The expression fold changes of the validated miRNAs by SAM algorithm in microarray analysis from ob/ob mice (C) and STZ-induced diabetic mice (D) compared with that from C57BL/6 mice, which is set as 1. FDR (q-value) <5%.
Since miR-122, a liver-specific miRNA, makes up 70% of all miRNAs in liver (33, 34) and is reported to regulate hepatic lipid metabolism (19, 35, 36), we analyzed miR-122 variation using SAM algorithm with 2-fold change as a threshold and also quantified it using real-time qPCR. As shown in Fig. 3, miR-122 was significantly reduced in livers of both ob/ob mice and STZ-induced diabetic mice compared with C57BL/6 mice, suggesting the involvement of miR-122 in hepatic energy metabolism.
Predicted gene targets of differentially expressed miRNAs
Lack of reliable and specific methods for biological target validation hampers the full understanding of the mechanisms by which miRNAs execute their functions. Only a few miRNAs have so far been assigned target mRNAs, and the conventional methodologies are still labor intensive. Three computer-aided algorithms, including TargetScan (29), miRanda (31), and PicTar (30), however, have partially overcome this limitation. With these bioinformatic approaches, we obtained a list of predicted targets that may be potentially involved in metabolic diseases (Table 4). As illustrated by underlines in Table 4, several such putative targets related to energy metabolism or NAFLD were derived by using various algorithms. To further elucidate the correlation between the miRNAs and the predicted targets, these prediction data were compared with previously published liver mRNA expression profiles of obese Lepob/ob C57BL/6 mice (37, 38). Some of the putative targets were shown dysregulated in livers of ob/ob mice (Table 4). Further identification of these target genes and their characterization will be greatly helpful for better understanding the potential roles of these differentially expressed miRNAs in hepatic energy metabolism.
TABLE 4.
Putative targets of the differentially expressed miRNAs
| miRNA | Putative Target |
|---|---|
| Upregulated miRNAs in ob/ob mice | |
| mmu-miR-34a | ACSL4a, ACSL1, ALDOA↑, SLC27A4, ACSL5↑, ANXA5↑, DAG1↑, RAC1↓, SELPLG↓, LCK↓, LAMP1↓, AP1S1↓, GNAO1↓, BMP1↓, LDHA, OGDH, HK1, PFKFB3, H6PD |
| mmu-miR-31 | PC, OSBP2, HADH↑, UGT1A1↑, CLIC1↓, GNAO1↓, PPARGC1A, GCK |
| mmu-miR-103 | FASN↑, GNAO1↓, LRP1, PDK4, PPP1CA↑, ARF4↑, CD36↑, HMGCS2↓, CLIC1↓, PADI4↓, LRP1B, ACOX1, OSBPL6, LDLRAD2, ACSL4, LRP6, ACSL1, PPARGC1A, GPD1, PC, OSBPL3, ACLY |
| mmu-miR-107 | GNAO1↓, LRP1, FASN↑, LRP1B, PDK4, PPP1CA↑, ARF4↑, HMGCS2↓, CLIC1↓, PADI4↓, ACOX1, OSBPL6, LDLRAD2, ACSL4, LRP6, ACSL1, PPARGC1A, GPD1, TCF1 |
| mmu-miR-194 | FOXA1, GYG1, OSBPL11, ARF4↑, HMGCS2↓, RAC1↓, BMP1↓, PPARGC1A, PDHB, TCF2 |
| mmu-miR-335-5p | ISL1, ACOT11, DAG1↑, FASN↑, ARF4↑, GAPDH↑, ATP5O↓, GNAO1↓ |
| mmu-miR-221 | ARF4↑, OSBPL7, INSIG1, PPARGC1A, ADIPOR1, OSBPL8 |
| mmu-miR-200a | BMP1↓, OSBPL11, FOXA1, ACOT7, CTNNB1↑, UGT1A1↑, ACSL5↑, CD36↑, ABCD3↓, IRS2, PPARA, PPARGC1A, OSBPL8 |
| mmu-miR-126-3p | IRS1 |
| Downregulated miRNAs in ob/ob mice | |
| mmu-miR-29c | COL4A4, COL4A5, COL4A1, COL4A3, LPL, HMGCR, PDHX, HNF4G, ATP5G1, HMGCS1↑, G6PC↑, GPAM↑, DAG1↑, CA3↑, PSMB5↑, ABCB11↓, ABCC3↓, EHHADH↓, COL3A1, COL4A2, OSBP, HMGCS1, INSIG1, PPARGC1A, PPARD, LEP, HEPACAM |
| mmu-miR-451 | YWHAZ |
| mmu-miR-21 | TGFBI↓, ACBD5, PPARA, ACAT1, PTGER3, HPGD, OSBPL1A |
| mmu-miR-122 | ALDOA↑, CS, CLIC1↓, SLC2A14, SLC2A3, GYS1, TUBB4↑, EIF1↑, ATP2C2↓, LRP10, PDK4, PPARA |
| mmu-miR-100 | FGFR3, GNAO1↓, VLDLR |
Genes with double underlines are predicted by all the three algorithms, genes with single underlines are predicted by two algorithms, and genes without underline are predicted by only one algorithm. mRNA expression of genes in bold were found dysregulated (↑, upregulated; ↓, downregulated) in livers of ob/ob mice compared with those of C57BL/6 mice by previous studies (37, 38).
DISCUSSION
Although progress has been made in the understanding of causative mechanisms of nonalcoholic steatosis, steatohepatitis, and cirrhosis, the knowledge about underlying regulatory networks of these pathophysiological conditions is still insufficient. Compelling evidence has demonstrated the substantial regulatory role of miRNAs in energy metabolism and liver functions (17–19, 39). Because of the spatial and temporal expression pattern of miRNAs and their conservation across species, miRNAs are believed to play similar roles in all animal cells (40–42). In light of these findings, we have identified mouse miRNAs that are possibly involved in the pathophysiological processes of NAFLD. Most of the varied miRNAs are highly conserved in mammals and are expected to play similar roles in the pathology of mouse and human NAFLD. Our results strongly suggest a regulatory role of miRNAs in hepatic energy metabolism.
A distinctive miRNA expression pattern in fatty livers of ob/ob mice
Due to inherited deficiency of the appetite-suppressing hormone, leptin (43), ob/ob mice spontaneously develop obesity, insulin resistance (44), and fatty livers (45). Therefore, ob/ob mice represent a naturally occurring model of NAFLD with excessive fat accumulation in liver (21, 22). In this study, we found that 11 miRNAs in liver samples of ob/ob mice were significantly dysregulated (>2-fold change) compared with those of wild-type C57BL/6 mice. Among those altered miRNAs, eight miRNAs (miR-34a, miR-31, miR-103, miR-107, miR-194, miR-335-5p, miR-221, and miR-200a) were upregulated and three miRNAs (miR-29c, miR-451, and miR-21) were downregulated. A distinctive miRNA expression pattern was identified in ob/ob mouse liver, and hierarchical clustering of this pattern could clearly discriminate ob/ob mice from normal C57BL/6 mice and STZ-administered mice, indicating the participation of miRNAs in the pathophysiological processes of NAFLD with hyperglycemia. db/db mice, another animal model of type 2 diabetes mellitus with leptin receptor mutations, also develop NAFLD. Further analysis of miRNA expression in livers of db/db mice are required in the future to elucidate the primary effect of leptin deficiency on liver miRNA expression.
miRNA expression in livers of STZ-induced type 1 diabetic mice
Since ob/ob mice can spontaneously developed obesity and insulin resistance, they are also considered as animal models of type 2 diabetes mellitus with high blood glucose levels. Glucose has been demonstrated to regulate transcription of genes encoding not only lipogenic and glycolytic enzymes but also proteins involved in global cell functions (46). However, it still remains unknown whether high glucose exerts an impact on the miRNA expression. STZ, an antibiotic extracted from Streptomyces acromogenes (47), is widely used to make experimental animal models of type 1 diabetes mellitus via inducing rapid and irreversible necrosis of β-cells (32). STZ-induced type 1 diabetic mice develop hyperglycemia with decreased body weight but no fatty liver. In this study, we employed the STZ-administered mice as a model of hyperglycemia without fatty liver. To illustrate the potential effect of high blood glucose on the liver miRNA expression, we compared the miRNA expressions in livers of STZ-induced type 1 diabetic mice with those of control C57BL/6 mice. Surprisingly, only one miRNA, miR-34a, showed an altered expression above 2-fold change threshold. Clustering analysis also failed to distinguish the STZ-induced diabetic mice from the normal C57BL/6 mice, indicating that short-term (2 weeks) high blood glucose does not affect liver miRNA expression globally but only affect certain miRNA expression in liver. Comparing the varied miRNAs under these two pathologic conditions, miR-34a is upregulated and miR-122 is downregulated in both ob/ob mice and STZ-induced diabetic mice, suggesting that miR-34a and miR-122 may be mainly related to the regulation of glucose metabolism. To discover the differentially expressed miRNAs in NAFLD without the influence of hyperglycemia, we further compared liver miRNA expression of ob/ob mice with that of STZ-induced diabetic mice. The result reveals six miRNAs with over 2-fold changed expression, implicating that these miRNAs are more likely to be involved in lipid metabolism and contribute to the pathology of NAFLD.
Liver miRNAs associated with aberrant metabolic status
A number of researchers have reported the abnormal miR-34a expressions in different types of cancer (48, 49), suggesting miR-34a as a miRNA component of the p53 network (48, 50). miR-21, another extensively studied miRNA, has been observed to be upregulated in tumors of various origins (51–53). In our study, miR-34a was the most elevated miRNA in livers of both ob/ob mice and STZ-induced diabetic mice, while miR-21 was significantly downregulated in livers of ob/ob mice, raising intriguing questions about the functional role of these two miRNAs in energy metabolism of liver.
Previous studies also demonstrated that suppressed liver-specific miR-122 in livers of mice would resulted in reduced plasma cholesterol levels, increased hepatic fatty acid oxidation, and decreased hepatic fatty acid and cholesterol synthesis rate (19, 35, 36, 39). Considering the abundance of miR-122 in liver and the potential role in energy metabolism, we tested its expression levels in real-time qRT-PCR assay, although the alteration of miR-122 was not >2-fold change threshold in SAM analysis. We found that miR-122 was significantly downregulated in livers of both ob/ob mice and STZ-induced diabetic mice. This outcome seems to be controversial with the previously reported regulatory function of miR-122 in energy metabolism, and we postulate that the reduction of miR-122 in NAFLD and diabetic livers may be part of a self-protection mechanism of liver cells against lipid or glucose overload.
Target and function prediction of differentially expressed miRNAs related to NAFLD and energy metabolism
Currently, the major difficulty for understanding the biological functions of miRNAs is determining their specific target genes at the posttranscriptional level. Bioinformatics has been a powerful tool in target and function prediction of miRNAs. miR-103 and miR-107, two closely related miRNAs that differ at a single nucleotide (54), were previously predicted by bioinformatics to affect multiple mRNA targets in pathways that involve cellular acetyl-CoA and lipid levels (55). In accordance with this prediction, we found that miR-103 and miR-107 were significantly upregulated in ob/ob mice compared with normal C57BL/6 mice and STZ-administered mice. These results strongly suggest the potential role of miR-103 and miR-107 in the pathogenesis of NAFLD and confirm the importance of bioinformatics in exploring miRNA functions. Employing three major algorithms for miRNA target prediction, TargetScan, PicTar, and miRanda, we analyzed the predicted target genes of the differentially expressed miRNAs identified in livers of ob/ob mice and STZ-induced diabetic mice. The predicted target genes of the dysregulated miRNAs include some genes that are closely related to glucose or lipid metabolism in liver and may play important roles in NAFLD. The interaction of miRNAs and their target genes may lead to translational repression and, in many cases, mRNA degradation (56). Thus, we compared the predicted targets with previously published liver mRNA expression profiles of obese Lepob/ob C57BL/6 mice (37, 38). mRNA expression of some putative targets was found dysregulated in ob/ob mice, and the variation directions of some genes were in accordance with the changes of miRNAs, suggesting these genes to be possible targets that may be repressed by miRNAs via mRNA degradation. Other predicted target genes that were not shown dysregulated in ob/ob mice on mRNA levels may possibly be regulated by miRNAs on protein levels. Citrate synthase, a Krebs tricarboxylic acid cycle enzyme that catalyzes the synthesis of citrate from oxaloacetate and acetyl-CoA, was predicted by all the three major algorithms to be a target gene of miR-122. As miR-122 was shown to be downregulated in livers of ob/ob mice in our study, we speculate citrate synthase to be upregulated in ob/ob mice. In accordance with this speculation, a previous study reported that citrate synthase activity was significantly elevated in livers of ob/ob mice compared with that of the wild-type control (57), indicating the targeting correlation between miR-122 and citrate synthase. Future studies are required to validate these potential target genes of the differentially expressed miRNAs and discover the downstream regulatory functions of miRNAs in NAFLD and hyperglycemia.
In conclusion, this study presents for the first time an extensive analysis of miRNA expression in livers of ob/ob mice, STZ-induced diabetic mice, and normal C57BL/6 mice. The study has identified a distinctive miRNA expression pattern in fatty livers of ob/ob mice, which could clearly distinguish ob/ob mice from either normal C57BL/6 mice or STZ-induced diabetic mice. The findings of this study demonstrate a novel miRNA-based mechanism in regulating pathophysiological processes of NAFLD and provide potential targets for future clinical applications.
Supplementary Material
Acknowledgments
We thank Zhi-Xiang Fang for support in statistical analysis and Ke-Hui Wang, Chengfu Xu, Zheng Bian, and Jing Li for their technical help.
Footnotes
Abbreviations:
- FDR
- false discovery rate
- miRNA
- microRNA
- NAFLD
- nonalcoholic fatty liver disease
- qRT-PCR
- quantitative reverse transcription polymerase chain reaction
- SAM
- significance analysis of microarray
- STZ
- streptozotocin
This work was supported by grants from the National Natural Science Foundation of China (30471991, 30570731, 30871154, and 30871195), the National Basic Research Program of China (973 Program) (2006CB503909 and 2004CB518603), the Key Sci-Tech Project of Science and Technology Department of Zhejiang Province (2008C13027-1), and the Natural Science Foundation of Jiangsu Province (BK2004082 and BK2006714).
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of two tables and two figures.
REFERENCES
- 1.Clark J. M., Brancati F. L., Diehl A. M. 2002. Nonalcoholic fatty liver disease. Gastroenterology. 122: 1649–1657. [DOI] [PubMed] [Google Scholar]
- 2.Browning J. D., Szczepaniak L. S., Dobbins R., Nuremberg P., Horton J. D., Cohen J. C., Grundy S. M., Hobbs H. H. 2004. Prevalence of hepatic steatosis in an urban population in the United States: impact of ethnicity. Hepatology. 40: 1387–1395. [DOI] [PubMed] [Google Scholar]
- 3.Amarapurkar D. N., Hashimoto E., Lesmana L. A., Sollano J. D., Chen P. J., Goh K. L. 2007. How common is non-alcoholic fatty liver disease in the Asia-Pacific region and are there local differences? J. Gastroenterol. Hepatol. 22: 788–793. [DOI] [PubMed] [Google Scholar]
- 4.Farrell G. C., Larter C. Z. 2006. Nonalcoholic fatty liver disease: from steatosis to cirrhosis. Hepatology. 43: S99–S112. [DOI] [PubMed] [Google Scholar]
- 5.Wanless I. R., Lentz J. S. 1990. Fatty liver hepatitis (steatohepatitis) and obesity: an autopsy study with analysis of risk factors. Hepatology. 12: 1106–1110. [DOI] [PubMed] [Google Scholar]
- 6.Bacon B. R., Farahvash M. J., Janney C. G., Neuschwander-Tetri B. A. 1994. Nonalcoholic steatohepatitis: an expanded clinical entity. Gastroenterology. 107: 1103–1109. [DOI] [PubMed] [Google Scholar]
- 7.Marchesini G., Brizi M., Morselli-Labate A. M., Bianchi G., Bugianesi E., McCullough A. J., Forlani G., Melchionda N. 1999. Association of nonalcoholic fatty liver disease with insulin resistance. Am. J. Med. 107: 450–455. [DOI] [PubMed] [Google Scholar]
- 8.Day C. P., James O. F. W. 1998. Steatohepatitis: A tale of two “hits”? Gastroenterology. 114: 842–845. [DOI] [PubMed] [Google Scholar]
- 9.Bartel D. P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 116: 281–297. [DOI] [PubMed] [Google Scholar]
- 10.Ambros V. 2003. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 113: 673–676. [DOI] [PubMed] [Google Scholar]
- 11.Esquela-Kerscher A., Slack F. J. 2006. Oncomirs - microRNAs with a role in cancer. Nat. Rev. Cancer. 6: 259–269. [DOI] [PubMed] [Google Scholar]
- 12.Hoefig K. P., Heissmeyer V. 2008. MicroRNAs grow up in the immune system. Curr. Opin. Immunol. 20: 281–287. [DOI] [PubMed] [Google Scholar]
- 13.Xu P., Vernooy S. Y., Guo M., Hay B. A. 2003. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr. Biol. 13: 790–795. [DOI] [PubMed] [Google Scholar]
- 14.Teleman A. A., Maitra S., Cohen S. M. 2006. Drosophila lacking microRNA miR-278 are defective in energy homeostasis. Genes Dev. 20: 417–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Esau C., Kang X. L., Peralta E., Hanson E., Marcusson E. G., Ravichandran L. V., Sun Y. Q., Koo S., Perera R. J., Jain R., et al. 2004. MicroRNA-143 regulates adipocyte differentiation. J. Biol. Chem. 279: 52361–52365. [DOI] [PubMed] [Google Scholar]
- 16.Hackl H., Burkard T. R., Sturn A., Rubio R., Schleiffer A., Tian S., Quackenbush J., Eisenhaber F., Trajanoski Z. 2005. Molecular processes during fat cell development revealed by gene expression profiling and functional annotation. Genome Biol. 6: R108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poy M. N., Eliasson L., Krutzfeldt J., Kuwajima S., Ma X. S., MacDonald P. E., Pfeffer B., Tuschl T., Rajewsky N., Rorsman P., et al. 2004. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 432: 226–230. [DOI] [PubMed] [Google Scholar]
- 18.Plaisance V., Abderrahmani A., Perret-Menoud V., Jacquemin P., Lemaigre F., Regazzi R. 2006. MicroRNA-9 controls the expression of Granuphilin/Slp4 and the secretory response of insulin-producing cells. J. Biol. Chem. 281: 26932–26942. [DOI] [PubMed] [Google Scholar]
- 19.Esau C., Davis S., Murray S. F., Yu X. X., Pandey S. K., Pear M., Watts L., Booten S. L., Graham M., McKay R., et al. 2006. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 3: 87–98. [DOI] [PubMed] [Google Scholar]
- 20.Jin X., Ye Y. F., Chen S. H., Yu C. H., Liu J., Li Y. M. 2008. MicroRNA expression pattern in different stages of nonalcoholic fatty liver disease. Dig Liver Dis. 41: 289–297. [DOI] [PubMed] [Google Scholar]
- 21.Koteish A., Diehl A. M. 2002. Animal models of steatohepatitis. Best Pract. Res. Clin. Gastroenterol. 16: 679–690. [DOI] [PubMed] [Google Scholar]
- 22.Diehl A. M. 2005. Lessons from animal models of NASH. Hepatol. Res. 33: 138–144. [DOI] [PubMed] [Google Scholar]
- 23.Liu C. G., Calin G. A., Meloon B., Gamliel N., Sevignani C., Ferracin M., Dumitru C. D., Shimizu M., Zupo S., Dono M., et al. 2004. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc. Natl. Acad. Sci. USA. 101: 9740–9744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomson J. M., Parker J., Perou C. M., Hammond S. M. 2004. A custom microarray platform for analysis of microRNA gene expression. Nat. Methods. 1: 47–53. [DOI] [PubMed] [Google Scholar]
- 25.Chen C. F., Ridzon D. A., Broomer A. J., Zhou Z. H., Lee D. H., Nguyen J. T., Barbisin M., Xu N. L., Mahuvakar V. R., Andersen M. R., et al. 2005. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 33: e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang F. C., Hajkova P., Barton S. C., Lao K. Q., Surani M. A. 2006. MicroRNA expression profiling of single whole embryonic stem cells. Nucleic Acids Res. 34: e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Livak K. J., Schmittgen T. D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T) (-Delta Delta C) method. Methods. 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 28.Tusher V. G., Tibshirani R., Chu G. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA. 98: 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewis B. P., Shih I. H., Jones-Rhoades M. W., Bartel D. P., Burge C. B. 2003. Prediction of mammalian microRNA targets. Cell. 115: 787–798. [DOI] [PubMed] [Google Scholar]
- 30.Krek A., Grun D., Poy M. N., Wolf R., Rosenberg L., Epstein E. J., MacMenamin P., da Piedade I., Gunsalus K. C., Stoffel M., et al. 2005. Combinatorial microRNA target predictions. Nat. Genet. 37: 495–500. [DOI] [PubMed] [Google Scholar]
- 31.John B., Enright A. J., Aravin A., Tuschl T., Sander C., Marks D. S. 2004. Human MicroRNA targets. PLoS Biol. 2: e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Junod A., Lambert A. E., Orci L., Pictet R., Gonet A. E., Renold A. E. 1967. Studies of the diabetogenic action of streptozotocin. Proc. Soc. Exp. Biol. Med. 126: 201–205. [DOI] [PubMed] [Google Scholar]
- 33.Lagos-Quintana M., Rauhut R., Yalcin A., Meyer J., Lendeckel W., Tuschl T. 2002. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 12: 735–739. [DOI] [PubMed] [Google Scholar]
- 34.Chang J., Nicolas E., Marks D., Sander C., Lerro A., Buendia M. A., Xu C., Mason W. S., Moloshok T., Bort R., et al. 2004. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 1: 106–113. [DOI] [PubMed] [Google Scholar]
- 35.Krutzfeldt J., Rajewsky N., Braich R., Rajeev K. G., Tuschl T., Manoharan M., Stoffel M. 2005. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 438: 685–689. [DOI] [PubMed] [Google Scholar]
- 36.Fabani M. M., Gait M. J. 2008. miR-122 targeting with LNA/2′-O-methyl oligonucleotide mixmers, peptide nucleic acids (PNA), and PNA-peptide conjugates. RNA. 14: 336–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang C. P., Tall A. R. 2001. Transcriptional profiling reveals global defects in energy metabolism, lipoprotein, and bile acid synthesis and transport with reversal by leptin treatment in ob/ob mouse liver. J. Biol. Chem. 276: 49066–49076. [DOI] [PubMed] [Google Scholar]
- 38.Ferrante A. W., Jr., Thearle M., Liao T., Leibel R. L. 2001. Effects of leptin deficiency and short-term repletion on hepatic gene expression in genetically obese mice. Diabetes. 50: 2268–2278. [DOI] [PubMed] [Google Scholar]
- 39.Elmen J., Lindow M., Schutz S., Lawrence M., Petri A., Obad S., Lindholm M., Hedtjarn M., Hansen H. F., Berger U., et al. 2008. LNA-mediated microRNA silencing in non-human primates. Nature. 452: 896–899. [DOI] [PubMed] [Google Scholar]
- 40.Pasquinelli A. E., Reinhart B. J., Slack F., Martindale M. Q., Kuroda M. I., Maller B., Hayward D. C., Ball E. E., Degnan B., Muller P., et al. 2000. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 408: 86–89. [DOI] [PubMed] [Google Scholar]
- 41.Lagos-Quintana M., Rauhut R., Meyer J., Borkhardt A., Tuschl T. 2003. New microRNAs from mouse and human. RNA. 9: 175–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lim L. P., Glasner M. E., Yekta S., Burge C. B., Bartel D. P. 2003. Vertebrate microRNA genes. Science. 299: 1540. [DOI] [PubMed] [Google Scholar]
- 43.Campfield L. A., Smith F. J., Burn P. 1996. The OB protein (leptin) pathway: a link between adipose tissue mass and central neural networks. Horm. Metab. Res. 28: 619–632. [DOI] [PubMed] [Google Scholar]
- 44.Lin H. Z., Yang S. Q., Chuckaree C., Kuhajda F., Ronnet G., Diehl A. M. 2000. Metformin reverses fatty liver disease in obese, leptin-deficient mice. Nat. Med. 6: 998–1003. [DOI] [PubMed] [Google Scholar]
- 45.Yang S. Q., Lin H. Z., Lane M. D., Clemens M., Diehl A. M. 1997. Obesity increases sensitivity to endotoxin liver injury: Implications for the pathogenesis of steatohepatitis. Proc. Natl. Acad. Sci. USA. 94: 2557–2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meugnier E., Rome S., Vidal H. 2007. Regulation of gene expression by glucose. Curr. Opin. Clin. Nutr. Metab. Care. 10: 518–522. [DOI] [PubMed] [Google Scholar]
- 47.Herr R. R., Eble T. E., Bergy M. E., Jahnke H. K. 1959. Isolation and characterization of streptozotocin. Antibiot. Annu. 7: 236–240. [PubMed] [Google Scholar]
- 48.Chang T. C., Wentzel E. A., Kent O. A., Ramachandran K., Mullendore M., Lee K. H., Feldmann G., Yamakuchi M., Ferlito M., Lowenstein C. J., et al. 2007. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell. 26: 745–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tazawa H., Tsuchiya N., Izumiya M., Nakagama H. 2007. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc. Natl. Acad. Sci. USA. 104: 15472–15477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Raver-Shapira N., Marciano E., Meiri E., Spector Y., Rosenfeld N., Moskovits N., Bentwich Z., Oren M. 2007. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol. Cell. 26: 731–743. [DOI] [PubMed] [Google Scholar]
- 51.Fulci V., Chiaretti S., Goldoni M., Azzalin G., Carucci N., Tavolaro S., Castellano L., Magrelli A., Citarella F., Messina M., et al. 2007. Quantitative technologies establish a novel microRNA profile of chronic lymphocytic leukemia. Blood. 109: 4944–4951. [DOI] [PubMed] [Google Scholar]
- 52.Meng F., Henson R., Wehbe-Janek H., Ghoshal K., Jacob S. T., Patel T. 2007. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 133: 647–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Slaby O., Svoboda M., Fabian P., Smerdova T., Knoflickova D., Bednarikova M., Nenutil R., Vyzula R. 2007. Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology. 72: 397–402. [DOI] [PubMed] [Google Scholar]
- 54.Mourelatos Z., Dostie J., Paushkin S., Sharma A., Charroux B., Abel L., Rappsilber J., Mann M., Dreyfuss G. 2002. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 16: 720–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilfred B. R., Wang W. X., Nelson P. T. 2007. Energizing miRNA research: a review of the role of miRNAs in lipid metabolism, with a prediction that miR-103/107 regulates human metabolic pathways. Mol. Genet. Metab. 91: 209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Valencia-Sanchez M. A., Liu J., Hannon G. J., Parker R. 2006. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 20: 515–524. [DOI] [PubMed] [Google Scholar]
- 57.Brady L. J., Brady P. S., Romsos D. R., Hoppel C. L. 1985. Elevated hepatic mitochondrial and peroxisomal oxidative capacities in fed and starved adult obese (ob/ob) mice. Biochem. J. 231: 439–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.