Abstract
Intracellular trafficking of plant receptor kinases (PRKs) is a key step in regulation of cellular signaling. Our current knowledge in this field is based on systems that address signaling pathways affecting the whole cell. There are, however, signaling phenomena that add a further layer of complexity. In the Brassica self-incompatibility response, a single cell can adequately respond to two opposite stimuli: accepting cross-pollen and rejecting self-pollen simultaneously. To understand how PRK signaling can influence the coexistence of two seemingly exclusive states of the cell, we investigated the subcellular localization and internalization of the S-receptor kinase (SRK) involved in the self-incompatibility response of Brassica oleracea. Here, we describe the unusual subcellular distribution of SRK3, which localizes predominantly to intracellular compartments and to a much lesser extent to the plasma membrane. Using an anti-SRK antibody that fully substitutes for the natural ligand, we demonstrate that the interaction with the receptor takes place at the plasma membrane and is followed by SRK internalization in endosomes that are enriched in the SRK negative regulator Thioredoxin-h-like1.
INTRODUCTION
The key role played by the intracellular trafficking of receptor kinases in the regulation of signaling pathways has been described in numerous studies. Upon ligand binding, activated receptors are internalized in endosomal compartments, where they are either routed to lysosomes for degradation and downregulation or reset and recycled back to the plasma membrane (PM) (Wiley, 2003). During plant development, plant receptor kinases (PRKs) are required for correct differentiation of organs and normal growth (Haffani et al., 2004a). The role of their localization in correct signaling is still poorly understood. Expression studies have shown an almost exclusive PM localization for most of the investigated PRKs (Jinn et al., 2000; Shah et al., 2001; Hematy et al., 2007), with several notable exceptions (Russinova et al., 2004; Gifford et al., 2005; Geldner et al., 2007). The L1 layer-specific ARABIDOPSIS CRINKLY4 receptor localizes to the PM and at least two distinct compartments in the cytoplasm, corresponding to export and endocytic vesicles (Gifford et al., 2005). The brassinosteroid receptor, BRI1, localizes both to the PM and to so far unidentified cytoplasmic compartments, and its localization and trafficking are not dependent on the presence of its ligand. A recent study by Geldner et al. (2007) demonstrated the ability of BRI1 to transmit signal from these cytosolic compartments, a major step in understanding receptor signaling in plants. Interestingly, a different type of subcellular distribution was reported for the FLS2 receptor, which is required for rare but massive signaling in response to pathogen infection (Gomez-Gomez and Boller, 2000). In its inactive state, FLS2 localizes to the PM, whereas it is rapidly internalized and degraded upon binding to the flg22 protein (Robatzek et al., 2006).
In both pathways, activation of the receptor triggers a signaling cascade and causes the reaction of the whole cell. However, there are cases in which cells are required to adequately respond to two opposite simultaneous stimuli. An example of this is the self-incompatibility (SI) response in Brassica, a reaction preventing self-fertilization by rejection of self-pollen grains that have fallen upon the stigma. Micromanipulator experiments have demonstrated that single stigma papilla cells are able to simultaneously reject self-pollen and accept cross-pollen (Sarker et al., 1988). Genetically, the SI response is regulated by the highly polymorphic S-locus, containing the genes of the two key SI components (Gaude et al., 2006). Those are the stigma papilla cell-expressed S Receptor Kinase (SRK) and its ligand, the S-locus Cysteine-Rich protein (SCR), whose product is expressed in anthers and adheres to the pollen coat. The high degree of polymorphism at the S-locus has led to the formation of interaction partners of the type SRKn-SCRn, in which a given SRK will recognize only SCR from the same S-haplotype. If a self-pollination event occurs, the ligand (SCR) will interact with SRK, resulting in SRK phosphorylation and triggering of the SI response (Cabrillac et al., 2001). SCR from a different haplotype will not be recognized by the receptor kinase and will not cause an SI reaction. Searches for SRK regulators led to the identification of several interacting proteins (Bower et al., 1996; Gu et al., 1998; Vanoosthuyse et al., 2003; Kakita et al., 2007). Three of these, ARC1, THL1, and MLPK, have been shown to be involved in the SI response (Stone et al., 1999; Haffani et al., 2004b; Murase et al., 2004). THL1 has been demonstrated to negatively modulate the kinase activity of SRK3 (Cabrillac et al., 2001).
The current model of Brassica SI assumes that SRK, which has a canonical PRK structure (Stein et al., 1991), localizes to the PM of the stigma papilla cells where the interaction with SCR occurs (Goring and Walker, 2004). Indeed, by cell fractionation in transgenic tobacco (Nicotiana tabacum), Stein et al. (1996) demonstrated that Brassica oleracea SRK6 can localize to the PM, but the studies in native papilla cells were hampered by the complexity of Brassica as a model system. A fractionation performed on SRK15 (Cabrillac et al., 1999) resulted in a clearer signal in PM-enriched fractions, but because the line used is self-compatible due to an unknown cause, the possibility of receptor mislocalization cannot be excluded. Despite the difficulties, however, Brassica remains a preferred model for studying SI due to the large amount of acquired physiological and biochemical data and well-characterized genetic and molecular interactions.
To address the question of how SRK triggers a local negative response to self-pollen, while the rest of the cell remains competent for a possible positive interaction with cross-pollen, we analyzed the subcellular localization of SRK and investigated the importance of its internalization for the SI response. Applying an immunocytochemical approach, we found that in stigma papilla cells of S3-haplotype, SRK3 localizes preferentially to intracellular compartments and to a lower extent to the PM. By colocalization with the known markers VPS29 and SYP21, we managed to identify the compartments as sorting endosomes. Using an anti-SRK3 antibody as a ligand, we demonstrated that recognition occurs at the PM during the SI response and is followed by ligand internalization. SRK3 colocalizes with its inhibitor THL1 in the endosomes, but not at the PM, indicating that receptor signaling might occur at the PM.
RESULTS
SRK3 Localizes Predominantly to Intracellular Compartments
To investigate the localization of SRK in papilla cells, we employed a direct immunocytochemical approach using the monoclonal antibody mAb85-36-71 (designated anti-SRK3-N-ter for clarity), which recognizes the first nine–amino acid sequence of the N terminus of SRK3 (Gaude et al., 1993; Delorme et al., 1995) (see Supplemental Figure 1A online). This peptide is also found in the N termini of SRK2, SRK5, SRK15, and the respective related secreted S-locus glycoproteins (SLGs) but is absent in SLG3 and all other SRK and SLG sequences analyzed so far in B. oleracea (Cabrillac et al., 1999; Miege et al., 2001). On protein gel blots of total S3 protein extracts, this antibody recognizes the full-length SRK3 receptor as well as its splice variant designated eSRK3. This protein, which makes up the whole extracellular domain of the receptor, has four detectable glycosylation forms: two weak at 65.2 and 56.7 kD and two abundant at 62.8 and 59.5 kD (see Supplemental Figure 1A online) (Giranton et al., 1995).
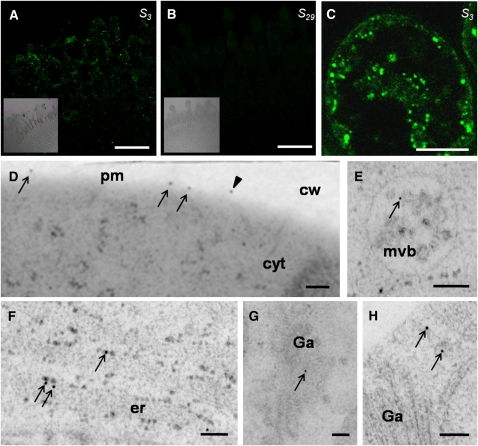
In S3-stigmas, we detected immunolabeling in the papilla cells and to a lower extent in the underlying cell layers (Figure 1A). The signal intensity was much lower compared with that from S15-stigmas (see Supplemental Figure 1C online), which showed a strong signal due to the sum of SRK15 and SLG15 immunolabeling, as previously observed in immunohistochemistry experiments (Gaude et al., 1993). S29-stigmas exhibited no signal above background level (Figure 1B). A closer inspection of the immunofluorescent signal in S3-papilla cells revealed that, rather unexpectedly, the majority of the signal was concentrated in intracellular compartments and some diffuse regions, whereas only a small portion of the signal could be detected in patches around the periphery of the cell (Figure 1C). To see whether the peripheral labeling corresponds to the PM, we performed an immunolocalization of SRK3 by electron microscopy. As expected, the amount of SRK3 signal was low, but nevertheless, the labeling was in close proximity to the PM. The signals were often grouped together, leaving zones completely devoid of signal (Figure 1D). Additionally, signals probably corresponding to eSRK3 could be seen on the cell wall. Labeling was also detected in multivesicular bodies, Golgi apparatus, small vesicles, and the endoplasmic reticulum (ER) (Figures 1E to 1H).
Figure 1.
SRK3 Localizes to PM and Sorting Endosomes in B. oleracea Stigmas.
(A) Signal recognition by anti-SRK3-N-ter in S3-papilla cells. Inset presents the bright-field image. Bar = 50 μm.
(B) Signal recognition by anti-SRK3-N-ter in S29-papilla cells. No specific signal can be detected. Inset presents the bright-field image. Bar = 50 μm.
(C) Higher magnification of a S3-stigma papilla cell shows fluorescent signals in the periphery of the cell and intracellular compartments. The experiment was reproduced five times. Bar = 10 μm.
(D) to (H) Immunolocalization on electron microscopy sections. Bars = 100 nm.
(D) SRK3 signal at the PM (arrows) and in the cell wall (arrowhead). Signals visible in the cell wall probably correspond to the soluble splice variant eSRK3.
(E) SRK3 signal in the multivesicular bodies (arrow).
(F) SRK3 signal in the ER (arrows).
(G) SRK3 signal in the Golgi apparatus (arrow).
(H) SRK3 signal in different small vesicles in the vicinity of the Golgi apparatus (arrows).
cw, cell wall; cyt, cytoplasm; er, endoplasmic reticulum; Ga, Golgi apparatus; pm, plasma membrane; mvb, multivesicular body.
Such a localization pattern is unique among the known PRKs, all of which seem to be predominantly localized at the PM (Jinn et al., 2000; Robatzek et al., 2006; Hematy et al., 2007), even if actively internalized (Gifford et al., 2005; Geldner et al., 2007).
SRK3 Localizes to Sorting Endosomes
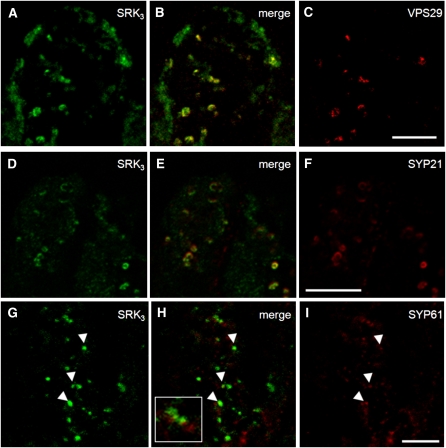
Since the majority of SRK3 is localized in intracellular compartments, and not at the PM, we attempted to identify the nature of these compartments by colocalization studies with known markers for endomembrane compartments. First, colocalization was tested between SRK3 and the endosome marker VPS29. VPS29 is a key member of the plant retromer complex involved in the trafficking of certain proteins between the sorting endosome and the PM (Jaillais et al., 2007). Partial colocalization was observed in the cytosolic compartments as seen in Figures 2A to 2C. This result might be significant; VPS29 participates in the retromer together with SORTING NEXIN1, a protein previously isolated as a SRK3 interactor (Vanoosthuyse et al., 2003). The amorphous regions positive for SRK3 but not labeled by the anti-VPS29 antibody could correspond to ER, since considerable amounts of SRK3 are located there, as suggested by the electron microscopy immunolocalization. To verify the observed endosomal localization of SRK3, we used a second marker, the SYP21 protein, a membrane-anchored syntaxin localized in the prevacuolar compartment of root cells (da Silva Conceicao et al., 1997). This compartment was recently identified as the plant sorting endosome (Jaillais et al., 2008); therefore, SYP21 was selected for an independent confirmation of SRK3's endosomal localization. The immunodetection signals for the two proteins showed good colocalization (Figures 2D to 2F). Another member of the syntaxin family, SYP61, was also used as a marker of the trans-Golgi network, but only isolated colocalizing signals could be detected (Figures 2G to 2I). The event of cross- or self-pollination did not change the localization of SRK3 and its colocalization with the investigated markers (see Supplemental Figure 2 online), similar to what was demonstrated for the brassinosteroid receptor BRI1 (Geldner et al., 2007). These results indicate that cytoplasmic SRK3 is likely to accumulate in sorting endosomes, revealing that this compartment is a major step in receptor intracellular trafficking.
Figure 2.
SRK3 Colocalizes with Sorting Endosome Markers in Papilla Cells.
(A) to (C) Colocalization between SRK3 (A) and sorting endosome marker VPS29 (C). Merged image is presented in (B). Images shown are representative of five independent experiments. Bar = 10 μm.
(D) to (F) Colocalization between SRK3 (D) and sorting endosome marker SYP21 (F). Merged image is presented in (E). Images shown are representative of three independent experiments. Bar = 10 μm.
(G) to (I) Colocalization between SRK3 (G) and trans-Golgi network marker SYP61 (I). Merged image is presented in (H). Cases of colocalization are rare and are indicated with arrowheads. Inset represents a case where signals are closely associated but do not colocalize. Images shown are representative of three independent experiments. Bar = 10 μm.
SRK3 Is Degraded upon Activation
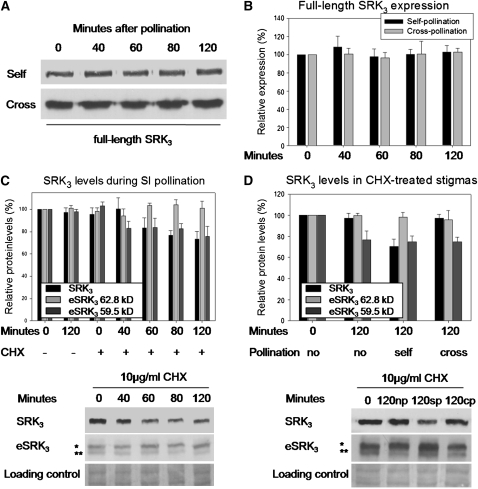
In an attempt to follow SRK3 turnover, we investigated its stability over a 2-h time course of self- or cross-pollination. Protein gel blot analysis showed that SRK3 levels do not change under these conditions (Figures 3A and 3B), suggesting either the potential high stability of SRK3 independently of its activation or a compensatory mechanism, in which a new receptor is synthesized to replace the degraded one. To test which of these hypotheses is correct, we treated stigmas with the protein biosynthesis inhibitor cycloheximide (CHX) prior to self-pollination. To avoid unwanted secondary effects, the treatment was done directly on the stigma undetached from the plant. We observed a 25 to 30% decrease in SRK3 levels (Figure 3C), which demonstrates that during self-pollination, SRK3 is degraded, while de novo protein synthesis allows restoration of its basal level. To see whether the effect is specific to self-pollination, we performed a similar experiment, in which stigmas were self-, cross-, or nonpollinated after the CHX treatment (Figure 3D). The reduction of SRK3 levels was specific to the self-pollination situation, showing that once the SI response is triggered, the activated receptor undergoes degradation. The 62.8-kD eSRK3 form showed no sensitivity to CHX, while the 59.4-kD form was affected under all conditions, irrespective of the type of pollination. This result suggests higher stability of the 62.8-kD eSRK3 or could alternatively mean that the degradation of the 62.8-kD form triggers the conversion of the 59.4-kD form to the 62.8-kD one to maintain the required levels. The physiological relevance of the CHX treatment to the SI response was confirmed by pollen tube counts. We observed a breakdown of SI after the treatment as already described by Sarker et al. (1988) (see Supplemental Figure 3 online).
Figure 3.
SRK3 Undergoes Degradation upon Self-Pollination.
(A) Protein gel blot showing stability of full-length SRK3 between 0 and 120 min following self- or cross-pollination.
(B) Quantification of (A). No significant change in signal intensity was observed. The bars represent the means (+sd) of three independent experiments.
(C) Degradation of SRK3 after self-pollination of stigmas treated with CHX (+) or a mock solution (−) between 0 and 120 min . The bars represent the means (+sd) of three independent experiments. The 62.8-kD splice variant eSRK3 (*) is not sensitive to the CHX treatment, while the 59.4 kD variant (**) and the full-length receptor are sensitive. A typical protein gel blot is shown as an example below the graph.
(D) CHX treatment affects SRK3 specifically upon self-pollination. The bars represent the means (+ sd) of three independent experiments. The 62.8 kD eSRK3 form (*) is not sensitive to the CHX treatment, while the 59.4-kD eSRK3 (**) is sensitive irrespective of the pollination type. A typical protein gel blot is shown as an example below the graph. np, nonpollinated; sp, self-pollination; cp, cross-pollination.
Receptor–Ligand Interaction during SI Happens at the PM
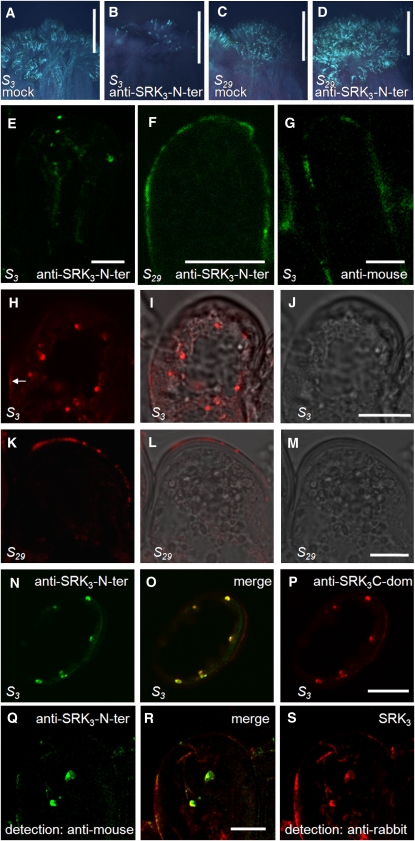
Current models of the SI response postulate the ligand–receptor interaction to occur at the PM in analogy with other mammalian and plant systems (Goring and Walker, 2004). Now, this idea is challenged by the low availability of SRK3 at the PM. To identify the topology of the interaction, we designed an experiment in which the anti-SRK3-N-ter antibody was used as a ligand, and its internalization was followed in the stigmas of different S haplotypes. This approach was chosen due to the lack of a reliable detection method for the native ligand, SCR3. Anti-SRK3-N-ter antibody has two important functional properties: first, it is able to pass through the cell wall of stigma papilla cells (Luu et al., 1999); and second, like the native ligand SCR3, its interaction with SRK3 leads to activation of the receptor (Cabrillac et al., 2001). This theoretically makes it a suitable replacement for SCR3 and brings the advantage of easy detection. To be sure of the physiological relevance of the substitution, we performed a pollination assay using compatible S15-pollen on S3- or S29-stigmas. The stigmas were pretreated with either an anti-SRK3-N-ter antibody solution or a mock solution for 1.5 h. The antibody treatment led to a strong rejection reaction of the otherwise compatible S15-pollen on the S3-stigma surface but not on the control S29-stigma (Figures 4A to 4D). This result indicates that anti-SRK3-N-ter is capable of specifically inducing SI reaction upon interaction with SRK3, probably by mimicking the effect of the ligand and provoking receptor dimerization as previously suggested (Cabrillac et al., 2001). In the following experiments, we used this antibody as a ligand for SRK3.
Figure 4.
Ligand-Induced Internalization of SRK3.
(A) to (D) Anti-SRK3-N-ter can functionally substitute for the natural SRK3 ligand in vivo. B. oleracea S3- and S29- stigmas were treated with anti-SRK3-N-ter or mock solution and pollinated with S15-pollen, which is normally compatible for both haplotypes. Pollen germination was visualized under UV light after aniline blue staining. Five independent experiments were performed, yielding similar results.
(A) Germination of S15-pollen on S3-stigma treated with mock solution. Bar = 500 μm.
(B) Rejection of S15-pollen on S3-stigma treated with anti-SRK3-N-ter solution. Bar = 500 μm.
(C) Germination of S15-pollen on S29-stigma treated with mock solution. Bar = 500 μm.
(D) Germination of S15-pollen on S29-stigma treated with anti-SRK3-N-ter solution. No rejection can be observed. Bar = 500 μm.
(E) and (F) B. oleracea S3- and S29-stigmas were treated with anti-SRK3-N-ter solution and pollinated with S15-pollen (normally compatible for both haplotypes). Antibody internalization was followed by immunofluorescence on fixed sections. The results could be reproduced in three independent experiments.
(E) Internalization in S3-papilla cells. Anti-SRK3-N-ter is found in intracellular compartments after 2.5 h, detected directly by the secondary anti-mouse antibody. Bar = 10 μm.
(F) No internalization is observed in S29-papilla cells. Bar = 10 μm.
(G) B. oleracea S3-stigmas were treated with secondary donkey anti-mouse Alexa555 solution and pollinated with S15-pollen. No internalization is observed in S29-papilla cells after 2.5 h. Bar = 10 μm.
(H) to (J) Detection of SRK3 on S3 stigma papilla sections. SRK3 is detected in intracellular compartments and unspecifically on the cell wall (arrow) by anti-SRK3C-dom antibody. Bright-field image is presented in (J) and the merge image in (I). Similar results were observed in three independent experiments. Bar = 10 μm.
(K) to (M) Absence of SRK3 from S29-stigma papilla sections (control experiment). The unspecific cell wall staining probably corresponds to the bands detected on protein gel blot with the same antibody. Bright-field image is presented in (M) and the merge image in (L). Similar results were observed in three independent experiments Bar = 10 μm.
(N) to (P) Simultaneous detection of SRK3 in S3-papilla cells using anti-SRK3-N-ter (recognizing the N terminus of SRK3) and anti-SRK3C-dom antibody (recognizing the C terminus of SRK3). Anti-SRK3C-dom does not recognize the PM-localized SRK3, possibly due to an interaction blocking the antibody epitope and shows a weak unspecific cell wall signal. Similar results were observed in three independent experiments. Bar = 10 μm.
(Q) to (S) Colocalization of internalized anti-SRK3-N-ter (detected directly by the secondary anti-mouse antibody) and SRK3 (detected by anti-SRK3C-dom and secondary anti rabbit antibody) in S3-papilla cells. Similar results were observed in three independent experiments. Bar = 10 μm.
SRK3 Is Routed to Sorting Endosomes upon Ligand Recognition
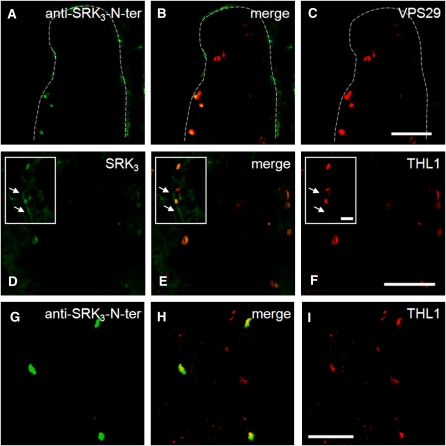
To investigate whether anti-SRK3-N-ter was internalized during the SI reaction, we treated S3- or S29-stigmas with antibody/ligand. After 1.5 h, the stigmas were pollinated with S15-pollen and then fixed for immunolocalization 1 h after the pollination. In S3, we observed signals in cytoplasmic compartments of the papilla cells, indicating that antibody/ligand internalization had taken place (Figure 4E). No such internalized signals were observed in S29-papilla cells. Instead, a continuous fluorescent halo around the cell was visible, suggesting antibody/ligand retention at the cell surface (Figure 4F). As a control, the internalization of the secondary anti-mouse antibody was investigated in S3-stigmas. No internalization was observed (Figure 4G), proving that the internalization in this haplotype requires an interaction with a specific PM component and that the interaction between SRK3 and its ligand occurs first at the PM and not in the endosomes. To ascertain that antibody/ligand internalization correlates with SRK3 endocytosis, we looked at whether the internalized monoclonal antibody and SRK3 colocalize to the same cytoplasmic compartments. To detect SRK3, we used a polyclonal anti-SRK3 antibody (anti-SRK3-C-dom) raised against an 11–amino acid sequence within the C terminus of the kinase domain (Delorme et al., 1995). On protein gel blots, this antibody recognizes SRK3 but not any other SRK protein tested (see Supplemental Figure 1A online), and it also cross-reacts with several bands irrespective of the S-haplotype. In control immunocytochemistry experiments, the cross-reactivity was limited to the cell wall (Figures 4H to 4M), and the antibody was not able to recognize the PM-localized SRK3. Nevertheless, anti-SRK3-C-dom antibody was suitable to detect the intracellular SRK3 and showed perfect colocalization with anti-SRK3-N-ter in the endosomes (Figures 4N to 4P). The signal of the endocytosed antibody colocalized well with the position of the receptor in the cell (Figures 4Q to 4S). Furthermore, we found that the anti-SRK3-N-ter–containing compartments colocalize with VPS29 (Figures 5A to 5C), suggesting that following internalization, the complex is routed to sorting endosomes before SRK3 is sent for degradation. The presence of activated receptor complexes in endosomal compartments implies that SRK3 signaling might happen there, in a way similar to the brassinosteroid receptor BRI1 (Geldner et al., 2007).
Figure 5.
Internalized SRK3 Localizes in Sorting Endosomes with THL1.
(A) to (C) Colocalization of internalized anti-SRK3-N-ter (A) and VPS29 (C). Merged picture is shown in (B). Similar results were observed in three independent experiments. Bar = 10 μm.
(D) to (F) THL1 (F) colocalizes partially with SRK3 (D) in endosomes, but not at the PM. Merged picture is shown in (E). Arrows in the inserts indicate the PM-localized SRK3. Similar results were observed in six independent experiments. Bar = 10 μm in the panels and 5 μm in the insets.
(G) to (I) Colocalization of internalized anti-SRK3-N-ter (G) and THL1 (I). Merged picture is presented in (H). Similar results were observed in three independent experiments Bar = 10 μm.
SRK3 Colocalizes with Negative Regulator THL1 Both before and after Activation
To investigate how the subcellular localization of SRK3 relates to that of its negative regulator, THL1, a stigma thioredoxin h-like protein (Cabrillac et al., 2001), we generated an anti-THL1 antibody (see Supplemental Figure 1 online). Both THL1-only compartments and SRK3/THL1 endosomal compartments could be detected, suggesting additional cell functions of THL1 (Figures 5D to 5F). No THL1 signal was detected at the PM (Figures 5D to 5F, insets) in any of the six repetitions of the experiment. This might indicate that the negative regulation of SRK3 at the PM is relaxed, which might lead to a high reactivity upon ligand recognition. The low amounts of SRK3 at the PM support this hypothesis, as this reduces the probability of an unspecific autophosphorylation. The need for a strict regulation in the endosomes is reasonable because the abundance of the receptor there might lead to unspecific activation, as demonstrated by Giranton et al. (2000). However, it is still possible that undetectable low amounts of THL1 are present at the PM or that THL1 is substituted by another inhibitor, for example, THL2 (Bower et al., 1996).
Because upon internalization the receptor/ligand complex is sorted to VPS29-containing endosomes, we tried to see whether some of the compartments are depleted of THL1 and therefore may allow signaling to occur. As seen in Figures 5G to 5I, activated SRK3 colocalized with THL1 (similarly to what was shown above for SRK3/VPS29). Thus, the routing of the active receptor to sorting endosomes might lead to immediate signal attenuation by THL1, which precedes the degradation of the receptor in the lytic vacuoles.
DISCUSSION
Activity of SRK is required for self/nonself-pollen recognition during the fertilization process in Brassicaceae. Over the last 50 years, physiological and genetic studies have suggested the complexity of the underlying signaling process. One of the manifestations of this complexity is the possibility of a cell to accept cross-pollen while successfully rejecting self-pollen at the same time (Sarker et al., 1988). The advancement in knowledge of receptor kinase trafficking in plants raises the question of how SRK can govern this complex interaction and what the role of SRK intracellular distribution and trafficking is in SI. Previous attempts to localize SRK within the cell have lacked sufficient precision due to the methods available or were relying on heterologous systems due to the limitations of the Brassica stigma papilla cells (Gaude et al., 1993; Stein et al., 1996; Giranton et al., 2000). In this work, we applied a direct immunocytochemical approach.
We found that SRK3 localizes preferentially to cytoplasmic compartments and to a lower extent to the PM. This is a unique case of PRK subcellular distribution and is probably the basis of the great specificity of self-pollen rejection. The only known case of similar localization in plant cells was observed for the human transferrin receptor when expressed in Arabidopsis thaliana protoplasts (Ortiz-Zapater et al., 2006). The transferrin receptor accumulates predominantly in cytoplasmic compartments, resembling the situation in its native human cells. Interestingly, expression of SRK3 in Sf21 insect cells resulted in a subcellular distribution similar to the one in papilla cells, with small amounts of signal at the PM and predominant localization in cytoplasmic structures (Giranton et al., 2000). In papilla cells, the PM labeling seemed uneven with regions completely devoid of signal. This was confirmed by our electron microscopy study, indicating the presence of SRK in discreet domains on the PM.
As SRK is predominantly intracellular, it was of great interest to reveal the functional identity of the compartments where SRK3 localizes. We used antibodies against known marker proteins and demonstrated that SRK3 localizes to the sorting endosomes. Such an observation is not entirely unexpected, as it is known that the animal receptor kinases are internalized from the PM to sorting endosomes, where their fate is decided depending on their activation state. In plants, BRI1 has also been demonstrated to undergo recycling from endosomes to the PM, suggesting a regulatory mechanism conserved between plants and animals (Geldner et al., 2007). It is tempting to imagine a similar scenario for SRK, but most of the drugs we used to disrupt different stages of intracellular trafficking in root cells, such as the Brefeldin A (100 μM, up to 60 min), Wortmannin (33 μM, up to 120 min), and Tyrphostin A23 (30 μM, up to 60 min), are unable to cause any of their respective effects in papilla cells, thus limiting the possibilities for an extensive functional analysis (data not shown).
SRK has the typical structure of a receptor kinase, so it is considered that the interaction with its ligand occurs at the PM (Goring and Walker, 2004). However, this notion is challenged by our finding that the amounts of SRK3 at the PM might be insufficient to induce a response. Moreover, the endosomes contain large quantities of the receptor, suggesting that the ligand might well be first internalized and routed to endosomes, where it could then interact with SRK. However, the exclusive internalization of the artificial ligand anti-SRK3-N-ter in S3 papilla cells demonstrates that the PM is the place of ligand recognition. We propose that SRK3 activation occurs within ready-to-be-activated domains at the PM upon ligand binding. SRK3 has the ability to perform transphosphorylation (Giranton et al., 2000), and due to the higher SRK density within these ready-to-be-activated domains, one or several neighboring domains could be activated following recognition. The low amounts of SRK outside the domains would prevent activation from spreading along the whole PM, thus creating a localized response to the self-pollen grain and leaving the rest of the cell in a naive state for a possible simultaneous interaction with compatible pollen. The dual localization of SRK at the PM and in endosomes implies that the subsequent steps of SI signal transduction can take place in any of these compartments. Indeed, PRKs can signal from endosomes, as demonstrated recently for BRI1 (Geldner et al., 2007) , and in the case of SRK3, this is suggested by its internalization to sorting endosomes after ligand binding. On the other hand, the endosomes are motile structures, and it is highly probable that the event of signaling and all subsequent steps of SI response happen directly in the domain responsible for the initial recognition at the PM. This view is supported by the colocalization experiments with THL1, a thioredoxin with well-documented negative effects on SRK3 signaling and the SI response (Cabrillac et al., 2001; Haffani et al., 2004b), suggesting that SRK3 is inhibited in the endosomal compartments both before and after the receptor–ligand interaction. An independent observation that supports this hypothesis is the recent demonstration of a direct interaction between SRK and the M-Locus Protein Kinase (MLPK) in BY-2 protoplasts (Kakita et al., 2007). MLPK is a positive regulator of the SI response, as evidenced by the loss of self-pollen rejection in plants altered in MLPK function (Murase et al., 2004). The interaction between SRK and MLPK was reported to occur at the PM, thus supporting the idea of the formation of an active signaling complex at the PM.
The role of endosomes in SI signaling is not clear. In the case of SRK3, our data are in favor of the classic view of endosomes as a step required for downregulation of the receptor complex and routing it for degradation in the vacuole. This hypothesis is supported by the enhanced degradation rate of SRK3 upon self-pollination. However, we cannot rule out that SRK signaling occurs in the endosomes, for instance, if a specific component needs to be recruited. Our data support an endocytosed-based regulatory mechanism for SRK signaling, in which the receptive papilla cells display only reduced amounts of the receptor at the PM, while the majority of it is sequestered in sorting endosomes. This differs from the reported BRI1 signaling pathway and is more similar to that proposed for FLS2. However, unlike SRK, both these receptors are exclusively or predominantly localized at the surface of the cell. It is likely that this peculiar distribution and regulation of SRK is related to the very unique feature of SI signaling, where a single papilla cell can react simultaneously to self- and cross-pollen grains.
METHODS
Plant Material and Treatments
Brassica oleracea haplotypes S3 and S15 were described by Gaude et al. (1993), and S29 was a kind gift from D.J. Ockendon (Ockendon and Currah, 1977). Stigmas from either S3- or S29-haplotypes were pollinated with S3- or S15-pollen. Chemical treatments were done directly on undetached stigmas with the following solutions: 10 μM CHX in 150 mM NaCl, 10 mM Tris, pH 7.5, and 0.05% Triton X-100 for 2 × 20 min; 1:10 diluted anti-SRK3-N-ter antibody in 150 mM NaCl, 10 mM Tris, pH 7.5, and 0.05% Triton X-100 for 2.5 h; 150 mM NaCl, 10 mM Tris, pH 7.5, and 0.05% Triton X-100.
Pollen Germination Assays
Stigmas were collected 24 h after pollination and fixed in solution containing 70% ethanol, 3.7% formaldehyde (Sigma-Aldrich), and 10% acetic acid (Sigma-Aldrich) for 24 h. The fixed material was then briefly washed and incubated for 24 h in 10 M NaOH and for another 24 h in destained aniline blue solution (0.1% [w/v] aniline blue in 0.1 M K3PO4, pH 7.0). Pollen tube growth was observed under UV light on a Nikon Optiphot-2 microscope.
Antibodies
Anti-SRK3-N-ter monoclonal antibody (mAb85-36-71) was described by Gaude et al. (1993). Anti-SRK3-C terminus rabbit polyclonal antiserum (anti-SRK3-C-dom) was described previously (Delorme et al., 1995). Polyclonal antiserum against the peptides CRGEFDEDARYPENKT and DKIEFKKPPTTSSGP (Eurogentec) of Arabidopsis thaliana VPS29 protein was raised in rabbit (Covalab). Rabbit polyclonal antiserum against B. oleracea THL1 was raised against the peptides CEDWNNKLKAAKESNK and CKGEEKLDKVVGAAK (Covalab). Polyclonal antiserums against Arabidopsis SYP21 and SYP61 proteins are a kind gift from Natasha Raikhel (da Silva Conceicao et al., 1997; Sanderfoot et al., 2001). All antibodies were tested on protein gel blots of total extracts from B. oleracea S3 haplotype stigmas (see Supplemental Figure 1A online)
Immunolocalization on B. oleracea Stigmas
S3-, S15-, or S29-haplotype stigmas were either directly harvested or pretreated, as described above, before fixation in 4% paraformaldehyde (Sigma-Aldrich) and DPBS (Gibco), pH 7.0. Samples were washed in D-PBS (Dulbecco's Phosphate-Buffered Saline; Gibco), and dehydrated in an ethanol series (30, 50, 70, 90, 96, and 100%). The samples were embedded in Steedman wax (PEG 400 distearate [Sigma-Aldrich]:1-hexadecanol [Sigma-Aldrich] = 9:1) as previously described (Baluska et al., 1992). Ten-micrometer-thick sections were placed on ProbeOn Plus microscope slides (Fisher Scientific). Following rehydration in an ethanol series (100, 96, 90, 70, 50, and 30%), the slides were washed in DPBS and used for immunolocalization. The incubations were 2 h for the primary and 1 h for the secondary antibodies, with DPBS washes after each incubation. Slides were analyzed on a LSM-510 laser scanning confocal microscope (Zeiss).
Dilutions of the antibodies were as follows: anti-SRK3-N-ter, 1:500; anti-SRK3-C-dom, 1:250; anti-VPS29, 1:250; anti-THL1; 1:200; anti-SYP21, 1:100; anti-SYP61, 1:100; Alexa555 donkey anti-mouse, 1:500 (Invitrogen); Alexa488 goat anti-rabbit IgG, 1:500 (Invitrogen).
Immunolocalization on Electron Microscopy Sections
Sections (200 μm) of B. oleracea stigmas were placed in hexadecene and frozen in a Leica EM PACT high-pressure freezer (Leica Microsystems) under 2000 bars and subsequently placed in freeze-substitution medium (0.25% uranyl acetate in anhydrous acetone) in a Leica automated freeze-substitution system. Freeze-substitution was performed as follows: 36 h at −90°C, 15 h of progressive increase of temperature up to −60°C at the rate of 2°C/h; 8 h at −60°C, 15 h of progressive increase of temperature up to −30°C at the rate of 2°C/h; 14 h at −30°C, 5 h of progressive increase of temperature up to −20°C at the rate of 2°C/h. At −20°C, the medium was replaced by acetone, which was subsequently progressively replaced by LR Gold resin. The samples were polymerized under UV light at −20°C for 48 h and then at +20°C for 36 h.
Immunodetection was performed on 50-nm sections using standard procedures. Dilution for anti-SRK3-N-ter was 1:250 and for the secondary 10-nm gold goat anti-mouse antibody (Nanoprobes), 1:40.
SDS-PAGE and Protein Gel Blots
Stigma extracts and electrophoretic analyses were performed as described (Cabrillac et al., 2001). Antibody dilutions used for protein gel blots were as follows: anti-SRK3-N-ter, 1:1000; anti- SRK3-C-dom, 1:1000; anti-VPS29, 1:500; anti-THL1, 1:200; anti-SYP21, 1:500; anti-SYP61, 1:500; goat anti-mouse IgG HRP, 1:5000 (Promega); anti-rabbit IgG HRP, 1:5000 (Amersham). Densitometry on membranes obtained after protein gel blotting (Amersham ECL protein gel blotting detection reagents; GE Healthcare) was done using the ImageJ software (http://rsb.info.nih.gov/ij).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: THL1, AF273844; SRK3, X79432; SCR3, UniProt Q9FNT3_BRAOL.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Specificity Test for Antibodies on Brassica oleracea Stigma Proteins.
Supplemental Figure 2. SRK3 Colocalizes with Sorting Endosome Markers after Pollination.
Supplemental Figure 3. Effect of Cycloheximide Treatment on Self-Incompatibility.
Supplementary Material
Acknowledgments
We thank N. Raikhel and D. Ockendon for providing materials, A.-M. Thierry for technical assistance in antibody production, C. Lionnet and F. Simian for assistance in confocal microscopy at PLATIM IFR128, J.-P. Carde, C. Cheniclet, and B. Battailer (Institut National de la Recherche Agronomique, Bordeaux) for the electron microscopy and discussions, and all members of the team for fruitful discussions. This work was supported by Grant ANR-05BLAN-0232-01 and by the Cluster 9, Région Rhône-Alpes.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Thierry Gaude (thierry.gaude@ens-lyon.fr).
Online version contains Web-only data.
References
- Baluska, F., Parker, J.S., and Barlow, P.W. (1992). Specific patterns of cortical and endoplasmic microtubules associated with cell growth and tissue differentiation in roots of maize (Zea mays L.). J. Cell Sci. 103 191–200. [Google Scholar]
- Bower, M.S., Matias, D.D., Fernandes-Carvalho, E., Mazzurco, M., Gu, T., Rothstein, S.J., and Goring, D.R. (1996). Two members of the thioredoxin-h family interact with the kinase domain of a Brassica S locus receptor kinase. Plant Cell 8 1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrillac, D., Cock, J.M., Dumas, C., and Gaude, T. (2001). The S-locus receptor kinase is inhibited by thioredoxins and activated by pollen coat proteins. Nature 410 220–223. [DOI] [PubMed] [Google Scholar]
- Cabrillac, D., Delorme, V., Garin, J., Ruffio-Chable, V., Giranton, J.L., Dumas, C., Gaude, T., and Cock, J.M. (1999). The S15 self-incompatibility haplotype in Brassica oleracea includes three S gene family members expressed in stigmas. Plant Cell 11 971–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Conceicao, A., Marty-Mazars, D., Bassham, D.C., Sanderfoot, A.A., Marty, F., and Raikhel, N.V. (1997). The syntaxin homolog AtPEP12p resides on a late post-Golgi compartment in plants. Plant Cell 9 571–582. [PMC free article] [PubMed] [Google Scholar]
- Delorme, V., Giranton, J.L., Hatzfeld, Y., Friry, A., Heizmann, P., Ariza, M.J., Dumas, C., Gaude, T., and Cock, J.M. (1995). Characterization of the S locus genes, SLG and SRK, of the Brassica S3 haplotype: Identification of a membrane-localized protein encoded by the S locus receptor kinase gene. Plant J. 7 429–440. [DOI] [PubMed] [Google Scholar]
- Gaude, T., Fobis-Loisy, I., and Miege, C. (2006). Control of fertilization by self-incompatibility mechanisms. In The Molecular Biology and Biotechnology of Flowering, B.R. Jordan, ed (Wallingford, UK: CAB International), pp. 269–297.
- Gaude, T., Friry, A., Heizmann, P., Mariac, C., Rougier, M., Fobis, I., and Dumas, C. (1993). Expression of a self-incompatibility gene in a self-compatible line of Brassica oleracea. Plant Cell 5 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner, N., Hyman, D.L., Wang, X., Schumacher, K., and Chory, J. (2007). Endosomal signaling of plant steroid receptor kinase BRI1. Genes Dev. 21 1598–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford, M.L., Robertson, F.C., Soares, D.C., and Ingram, G.C. (2005). ARABIDOPSIS CRINKLY4 function, internalization, and turnover are dependent on the extracellular crinkly repeat domain. Plant Cell 17 1154–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giranton, J.L., Ariza, M.J., Dumas, C., Cock, J.M., and Gaude, T. (1995). The S locus receptor kinase gene encodes a soluble glycoprotein corresponding to the SKR extracellular domain in Brassica oleracea. Plant J. 8 827–834. [DOI] [PubMed] [Google Scholar]
- Giranton, J.L., Dumas, C., Cock, J.M., and Gaude, T. (2000). The integral membrane S-locus receptor kinase of Brassica has serine/threonine kinase activity in a membranous environment and spontaneously forms oligomers in planta. Proc. Natl. Acad. Sci. USA 97 3759–3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Gomez, L., and Boller, T. (2000). FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5 1003–1011. [DOI] [PubMed] [Google Scholar]
- Goring, D.R., and Walker, J.C. (2004). Plant sciences. Self-rejection – A new kinase connection. Science 303 1474–1475. [DOI] [PubMed] [Google Scholar]
- Gu, T., Mazzurco, M., Sulaman, W., Matias, D.D., and Goring, D.R. (1998). Binding of an arm repeat protein to the kinase domain of the S-locus receptor kinase. Proc. Natl. Acad. Sci. USA 95 382–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffani, Y.Z., Gaude, T., Cock, J.M., and Goring, D.R. (2004. b). Antisense suppression of thioredoxin h mRNA in Brassica napus cv Westar pistils causes a low level constitutive pollen rejection response. Plant Mol. Biol. 55 619–630. [DOI] [PubMed] [Google Scholar]
- Haffani, Y.Z., Silva, D.R., and Goring, D.R. (2004. a). Receptor kinase signaling in plants. Can. J. Bot. 82 1–15. [Google Scholar]
- Hematy, K., Sado, P.E., Van Tuinen, A., Rochange, S., Desnos, T., Balzergue, S., Pelletier, S., Renou, J.P., and Hofte, H. (2007). A receptor-like kinase mediates the response of Arabidopsis cells to the inhibition of cellulose synthesis. Curr. Biol. 17 922–931. [DOI] [PubMed] [Google Scholar]
- Jaillais, Y., Fobis-Loisy, I., Miege, C., and Gaude, T. (2008). Evidence for a sorting endosome in Arabidopsis root cells. Plant J. 53 237–247. [DOI] [PubMed] [Google Scholar]
- Jaillais, Y., Santambrogio, M., Rozier, F., Fobis-Loisy, I., Miege, C., and Gaude, T. (2007). The retromer protein VPS29 links cell polarity and organ initiation in plants. Cell 130 1057–1070. [DOI] [PubMed] [Google Scholar]
- Jinn, T.L., Stone, J.M., and Walker, J.C. (2000). HAESA, an Arabidopsis leucine-rich repeat receptor kinase, controls floral organ abscission. Genes Dev. 14 108–117. [PMC free article] [PubMed] [Google Scholar]
- Kakita, M., Murase, K., Iwano, M., Matsumoto, T., Watanabe, M., Shiba, H., Isogai, A., and Takayama, S. (2007). Two distinct forms of M-locus protein kinase localize to the plasma membrane and interact directly with S-locus receptor kinase to transduce self-incompatibility signaling in Brassica rapa. Plant Cell 19: 3961–3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luu, D.T., Marty-Mazars, D., Trick, M., Dumas, C., and Heizmann, P. (1999). Pollen-stigma adhesion in Brassica spp involves SLG and SLR1 glycoproteins. Plant Cell 11 251–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miege, C., Ruffio-Chable, V., Schierup, M.H., Cabrillac, D., Dumas, C., Gaude, T., and Cock, J.M. (2001). Intrahaplotype polymorphism at the Brassica S locus. Genetics 159 811–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murase, K., Shiba, H., Iwano, M., Che, F.S., Watanabe, M., Isogai, A., and Takayama, S. (2004). A membrane-anchored protein kinase involved in Brassica self-incompatibility signaling. Science 303 1516–1519. [DOI] [PubMed] [Google Scholar]
- Ockendon, D.J., and Currah, L. (1977). Self pollen reduces the number of cross pollen tubes in the styles of Brassica oleracea. New Phytol. 78 675–680. [Google Scholar]
- Ortiz-Zapater, E., Soriano-Ortega, E., Marcote, M.J., Ortiz-Masia, D., and Aniento, F. (2006). Trafficking of the human transferrin receptor in plant cells: effects of tyrphostin A23 and brefeldin A. Plant J. 48 757–770. [DOI] [PubMed] [Google Scholar]
- Robatzek, S., Chinchilla, D., and Boller, T. (2006). Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev. 20 537–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russinova, E., Borst, J.W., Kwaaitaal, M., Cano-Delgado, A., Yin, Y., Chory, J., and de Vries, S.C. (2004). Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1). Plant Cell 16 3216–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot, A.A., Kovaleva, V., Bassham, D.C., and Raikhel, N.V. (2001). Interactions between syntaxins identify at least five SNARE complexes within the Golgi/prevacuolar system of the Arabidopsis cell. Mol. Biol. Cell 12 3733–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarker, R.H., Elleman, C.J., and Dickinson, H.G. (1988). Control of pollen hydration in Brassica requires continued protein synthesis, and glycosylation in necessary for intraspecific incompatibility. Proc. Natl. Acad. Sci. USA 85 4340–4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, K., Gadella, T.W., Jr., van Erp, H., Hecht, V., and de Vries, S.C. (2001). Subcellular localization and oligomerization of the Arabidopsis thaliana somatic embryogenesis receptor kinase 1 protein. J. Mol. Biol. 309 641–655. [DOI] [PubMed] [Google Scholar]
- Stein, J.C., Dixit, R., Nasrallah, M.E., and Nasrallah, J.B. (1996). SRK, the stigma-specific S locus receptor kinase of Brassica, is targeted to the plasma membrane in transgenic tobacco. Plant Cell 8 429–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, J.C., Howlett, B., Boyes, D.C., Nasrallah, M.E., and Nasrallah, J.B. (1991). Molecular cloning of a putative receptor protein kinase gene encoded at the self-incompatibility locus of Brassica oleracea. Proc. Natl. Acad. Sci. USA 88 8816–8820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone, S.L., Arnoldo, M., and Goring, D.R. (1999). A breakdown of Brassica self-incompatibility in ARC1 antisense transgenic plants. Science 286 1729–1731. [DOI] [PubMed] [Google Scholar]
- Vanoosthuyse, V., Tichtinsky, G., Dumas, C., Gaude, T., and Cock, J.M. (2003). Interaction of calmodulin, a sorting nexin and kinase-associated protein phosphatase with the Brassica oleracea S locus receptor kinase. Plant Physiol. 133 919–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley, H.S. (2003). Trafficking of the ErbB receptors and its influence on signaling. Exp. Cell Res. 284 78–88. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.