Abstract
Aldo-keto reductase (AKR) family 1, member 7 (AKR1B7), a member of the AKR superfamily, has been suggested to play an important role in the detoxification of lipid peroxidation by-products. The nuclear receptors pregnane X receptor (PXR) and constitutive androstane receptor (CAR) are xenosensors postulated to alleviate xeno- and endobiotic chemical insults. In this study, we show that the mouse Akr1b7 is a shared transcriptional target of PXR and CAR in the liver and intestine. Treatment of wild-type mice with the PXR agonist pregnenolone-16α-carbonitrile (PCN) activated Akr1b7 gene expression, whereas the effect was abrogated in PXR(-/-) mice. Similarly, the activation of Akr1b7 gene expression by the CAR agonist 1,4-bis[2-(3,5-dichlorpyridyloxyl)]-benzene, seen in wild-type mice, was abolished in CAR(-/-) mice. The promoter of Akr1b7 gene was activated by PXR and CAR, and this activation was achieved through the binding of PXR-retinoid X receptor (RXR) or CAR-RXR heterodimers to direct repeat-4 type nuclear receptor-binding sites found in the Akr1b7 gene promoter. At the functional level, treatment with PCN in wild-type mice, but not PXR(-/-) mice, led to a decreased intestinal accumulation of malondialdehyde, a biomarker of lipid peroxidation. The regulation of Akr1b7 by PXR was independent of the liver X receptor (LXR), another nuclear receptor known to regulate this AKR isoform. Because a major function of Akr1b7 is to detoxify lipid peroxidation, the PXR-, CAR-, and LXR-controlled regulatory network of Akr1b7 may have contributed to alleviate toxicity associated with lipid peroxidation.
The aldo-keto reductase (AKR) superfamily of genes encodes NAD(P)H-linked oxidoreductases. AKRs play an important role in the detoxification of harmful aldehydes and ketones generated from exogenous and endogenous toxicants and those produced from the breakdown of lipid peroxides. AKRs reduce aldehydes and ketones to their respective alcohols (Penning and Drury, 2007). Among AKR isoforms, the mouse Akr1b7 is highly expressed in vas deferens and adrenal gland, in which its sustained expression is dependent on androgen and adrenocorticotropic hormone, respectively (Lau et al., 1995). It is interesting that Akr1b7 null mice were found to be viable and have no obvious defect in reproduction (Baumann et al., 2007). Besides the steroidogenic tissues, Akr1b7 is also expressed in mouse kidney, eye, intestine, and, at a lower level, in liver (Lau et al., 1995).
One of the major functions of Akr1b7 is to detoxify lipid peroxidation. Lipid peroxidation refers to the oxidative deterioration of lipids containing carbon-carbon double bonds, especially those derived from polyunsaturated fatty acids (PUFAs). The peroxidation process proceeds by a free radical chain reaction, resulting in the production of many reactive aldehydes, among which the trans-4-hydroxy-2-nonenal was recognized as having the greatest toxic and harmful potential (Schneider et al., 2008). All unsaturated aldehydes may undergo further changes by autoxidation, leading to the production of other volatile derivatives, such as malondialdehyde (MDA). Akr1b7 has a preference for the by-products of lipid peroxidation as its substrates. These include trans-4-hydroxy-2-nonenal (Schneider et al., 2008) and isocaproaldehyde, another highly toxic lipid by-product generated during steroidogenesis (Lefrançois-Martinez et al., 1999).
The xenobiotic nuclear receptors, including pregnane X receptor (PXR; NR1I2) (Blumberg et al., 1998; Kliewer et al., 1998) and constitutive androstane receptor (CAR; NR1I3) (Honkakoski et al., 1998; Wei et al., 2000), were postulated to play an essential role in the detoxification of xeno- and endobiotic toxicants. The detoxifying effect of PXR and CAR is achieved through the coordinate transcriptional regulation of phase I and phase II enzymes, as well as drug transporters (for reviews, see Wilson and Kliewer, 2002; Timsit and Negishi, 2007). PXR and CAR regulate gene expression by heterodimerization with the retinoic X receptor (RXR) and the binding of the PXR-RXR or CAR-RXR heterodimers to specific response elements termed PXR response elements or CAR response elements (also called phenobarbital response element) that contain a hexanucleotide direct repeat separated by three or four nucleotides (DR-3 or DR-4) (Honkakoski et al., 1998). In addition to its function in xenobiotic detoxification, PXR has also been implicated in many other endobiotic functions, ranging from bile acid detoxification and cholestatic prevention (Staudinger et al., 2001; Xie et al., 2001) to bilirubin detoxification and clearance, adrenal steroid homeostasis and drug-hormone interactions (Zhai et al., 2007), lipid metabolism (for review, see Handschin and Meyer, 2005; Zhou et al., 2006b; Nakamura et al., 2007), inflammation and inflammatory bowel disease (Langmann et al., 2004; Dring et al., 2006; Zhou et al., 2006a; Shah et al., 2007), bone homeostasis (Pascussi et al., 2005), and retinoid acid metabolism (for review, see Zhang et al., 2008; Wang et al., 2008). Compared with its sister PXR, CAR exhibits many overlapping, yet distinct, functions (Timsit and Negishi, 2007). The liver X receptor (LXR), another nuclear hormone receptor, has been shown to regulate Akr1b7 (Volle et al., 2004). However, there has been no report on the role of PXR or CAR in the regulation of AKRs and detoxification of lipid peroxidation.
In this report, we show that Akr1b7 is a probable transcriptional target of PXR and CAR, suggesting a novel role for xenobiotic receptors in the detoxification of lipid peroxidation. The regulatory network of Akr1b7 controlled by PXR, CAR, and LXR may provide a complex and fail-safe system in preventing toxicity associated with lipid peroxidation.
Materials and Methods
Animals and Drug Treatment. The PXR null [PXR(-/-)] (Xie et al., 2000), CAR null [PXR(-/-)] (Wei et al., 2000), LXRα and -β double-knockout (LXR DKO) (Peet et al., 1998), and fatty acid-binding protein (FABP)-VP-PXR transgenic (Gong et al., 2006) mice in C57BL/6J and SvJ129 mixed background have been described previously. Mice were housed in a pathogen-free animal facility under a standard 12-h light/dark cycle with free access to water and food. Age- and sex-matched 8- to 10-week-old mice were used for all the experiments. To activate PXR, mice received two (for gene expression analysis) or four (for lipid peroxidation analysis) daily intraperitoneal injections of PCN (100 mg/kg) and were sacrificed 4 h after the last dose. To activate LXR, mice received daily gavages of GW3965 (20 mg/kg) for 5 days and were sacrificed 24 h after the last dose. To activate CAR, mice received a single intraperitoneal injection of TCPOBOP (1 mg/kg) and were sacrificed 7 days after the injection. PCN and TCPOBOP were purchased from Sigma-Aldrich (St. Louis, MO). GW3965 was synthesized in-house following the scheme in Zhou et al. (2008). The use of mice in this study has complied with all the relevant federal guidelines and institutional policies.
Real-Time Reverse Transcription-PCR. Total RNA was isolated by using TRIzol reagent (Invitrogen, Carlsbad, CA). Reverse transcription was performed by using iScript cDNA synthesis kit from Bio-Rad Laboratories (Hercules, CA). Real-time PCR was performed with the 7300 real-time PCR system (Applied Biosystems, Foster City, CA) by using the SYBR Green reagents (Applied Biosystems). All the data were normalized against the mouse cyclophilin gene. The primer sequences of Akr1b7 are as follows: forward, 5′-CCCTCACGCATACAGGAGAA-3′ and reverse, 5′-GCCATGTCCTCCTCACTCAA-3′ (Volle et al., 2004). Other real-time PCR primers are as follows: Akr1a4 forward, 5′-GCTTAGATGGCAGGTTCAGC-3′ and reverse, 5′-AGCATCTCTGGGAACCCTCT-3′; Akr1b8 forward, 5′-CCTCACCCAGGAAAAACTGA-3′ and reverse, 5′-CACGTTCCTCTGGATGTGAA-3′; and Akr7a5 forward, 5′-ATCAGGAGGGCAAGTTTGTG-3′ and reverse, 5′-CCAAAGGGTTGTAGGCGTAG-3′.
Northern Blot Analysis. The Ark1b7 cDNA probe was amplified by PCR from mouse liver cDNA by using the forward primer, 5′-CAATGAGAATGAGGTGGGAG-3′ and reverse primer, 5′-CCTCACTCAACTGGAAGTCGA-3′. The Cyp3a11 cDNA probe was descried previously (Xie et al., 2000). Northern blot analysis was performed as described previously (Xie et al., 2000). Glyceraldehyde-3-phosphate dehydrogenase (Gapdh) cDNA probing was used for the loading control.
Plasmid Constructs and Transfection Assays. The 5′-regulatory sequence (-1972 bp to - 4 bp) of the mouse Akr1b7 gene was cloned by PCR using the following pair of oligonucleotides: forward primer, 5′-GGTACCCATGTTCTAGTCAGCCATGGC-3′ (engineered with KpnI site) and reverse primer, 5′-CTCGAGGTGAGCAGATGAAATGCCTG-3′ (engineered with XhoI site). The design of oligonucleotides was based on mouse genomic sequences deposited in GenBank (accession NC_000072). The mouse liver genomic DNA was used as the PCR template. The PCR-amplified fragments were cloned into the pGL-3 basic vector from Promega (Madison, WI). Site-directed mutagenesis was performed by the PCR overextension method, and the mutations were confirmed by DNA sequencing.
Transient transfection was performed in HepG2 cells seeded in 48-well plates with polyethylenimine (PEI) polymer as the transfection reagent (Zhou et al., 2006). For each well, the plasmid-PEI complexes were formed by incubating 0.1 μg of expression vector for each nuclear receptor, 0.3 μg of reporter plasmid, 0.1 μg of β-galactosidase plasmid, and 0.5 μg of PEI at room temperature for 20 min in a total volume of 100 μl of serum-free Dulbecco's modified Eagle's medium (DMEM). The DNA-PEI complexes were then diluted with 100 μl of DMEM and applied at 200 μl per well. After 12 h of incubation, the transfection medium was replaced with DMEM containing 10% fetal bovine serum with the appropriate solvent or ligands. Cells were lysed and assayed for luciferase and β-galactosidase after 24 h of drug treatment.
Identification of the Putative PXR/CAR Response Elements and Electrophoretic Mobility Shift Assay. The promoter sequence analysis was performed by using the web-based software NUBIScan version 2.0 (http://www.nubiscan.unibas.ch/). The putative response elements were then analyzed for receptor binding by EMSA. For the EMSA assay, receptor proteins were prepared by using the in vitro TnT quick-coupled transcription/translation system (Promega). The nucleotide probes were labeled with [32P]dCTP by Klenow fill-in method. The binding reactions were performed at room temperature for 20 min (Wada et al., 2008). Protein-DNA complexes were resolved by vertical electrophoresis through 8% polyacrylamide gel in 0.5× Tris borate-EDTA at 4°C for 1.5 h. The gel was then dried and processed for autoradiography. The probe sequences are labeled in the figure. Unlabeled competitor DNAs (50-100×) were used for the parallel competition experiments.
Chromatin Immunoprecipitation Assay. The ChIP assay was performed essentially as described previously (Wada et al., 2008). In brief, primary mouse hepatocytes were prepared from wild-type mice by the collagenase perfusion method. Cells were treated with solvent (DMSO) or PCN (10 μM) for 24 h before formaldehyde cross-linking. Cell lysates were incubated overnight with 1 μg of anti-PXR antibody from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA) at 4°C. Parallel samples were incubated with normal IgG as a negative control. The following PCR primers were used: Akr1b7 forward, 5′-ATATCCACTCCCAGGGCAAT-3′ and Akr1b7 reverse, 5′-GGGCATGAGAACAGGAACTT-3′. The PCR primers for the positive control Cd36 gene (Zhou et al., 2006) and negative control Cyp7b1 gene (Wada et al., 2008) were described previously.
Intestinal MDA Measurement. Freshly dissected mouse small intestine samples were homogenized on ice in KCl (150 mM) solution (1 g of wet tissue in 9 ml of KCl) using a Polytron homogenizer. For each assay, 100 μl of the supernatant was incubated with 900 μl of reaction buffer containing 0.67% thiobarbituric acid. This mixture was incubated at 98°C for 1 h and then centrifuged at 12,000g for 10 min at 4°C. The absorbance of the supernatant was measured at 532 nm using a spectrophotometer. The absorbance was converted to nanomoles of MDA from a standard curve generated with 1,1,3,3-tetramethoxypropane.
Western Blot Analysis. Duodenum was collected from mice and homogenized in lysis buffer (50 mM Tris, pH 7.4, 1% Nonidet P40, 0.25% Na-deoxycholate, 0.1% SDS, 150 mM NaCl, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 1 mM NaF, and 0.2% protease inhibitor cocktail from Sigma-Aldrich. The lysates were centrifuged at 13,000g for 20 min at 4°C, and the supernatants were collected. The protein concentrations in the supernatants were measured. Equal amounts of lysate proteins were resolved by 10% SDS-polyacrylamide gel electrophoresis gel, and then transferred to polyvinylidene difluoride membranes. Membranes were blocked with 5% nonfat dry milk in Tris-buffered saline/Tween 20 at room temperature for 1 h. Akr1b7 protein was detected by a primary polyclonal goat antibody against Akr1b7 (1:500; Santa Cruz Biotechnology, Inc.) and a secondary peroxidase-conjugated donkey anti-goat IgG antibody (1:2000; Santa Cruz Biotechnology, Inc.). The control β-actin protein was detected by a primary monoclonal mouse antibody against β-actin (1:5000; Sigma-Aldrich) and a secondary peroxidase-conjugated horse anti-mouse IgG antibody (1:4000; Cell Signaling Technology Inc., Danvers, MA). Protein signals were detected with the enhanced chemiluminescence reagents from GE Healthcare (Chalfont St. Giles, Buckinghamshire, UK).
Statistical Analysis. Results are expressed as means ± S.D. Statistical analysis was performed using the unpaired Student's t test for comparison between two groups.
Results
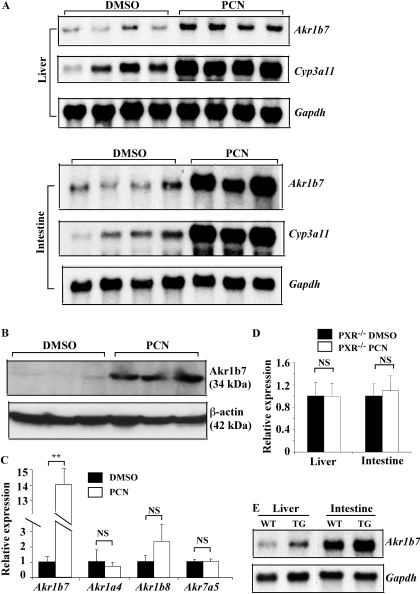
Activation of PXR Induced the Expression of Akr1b7 in Vivo. As shown in Fig. 1, treatment of wild-type mice with the PXR agonist PCN for 28 h induced the mRNA expression of Akr1b7 in the liver and small intestine, as determined by Northern blot analysis (Fig. 1A). Cyp3a11, a known PXR target gene, was included as a positive control for PXR activation. A similar Akr1b7 gene activation was observed in mice treated with PCN for 52 h (data not shown). The induction of intestinal Akr1b7 protein expression by PCN was also confirmed by Western blot analysis (Fig. 1B). The effect of PCN on Akr gene expression seemed to be isoform-specific, because the expression of Akr1a4, -1b8, and -7a5 in the same animals was not significantly altered (Fig. 1C). The PCN effect on Akr1b7 gene expression was abolished in PXR(-/-) mice (Xie et al., 2000) (Fig. 1D), demonstrating that PXR is the bona fide mediator for the PCN effect. We have reported previously the creation of FABP-VP-PXR transgenic mice in which a constitutively activated human PXR (VP-PXR) was expressed in the liver and intestine under the control of the rat FABP gene promoter (Gong et al., 2006). Figure 1E shows that the Akr1b7 gene activation was also observed in FABP-VP-PXR transgenic mice, although the magnitude of gene regulation seemed to be not as dramatic as that in the PCN-treated wild-type mice. In the same FABP-VP-PXR transgenic mice, the expression of Cyp3a11 was induced in both the liver and intestine (Gong et al., 2006). It is interesting that we found that the basal and PXR-inducible expression of Akr1b7 was higher in the intestine than that in the liver in both the pharmacological and genetic models of PXR activation. These results strongly suggest that Akr1b7 is a transcriptional target of PXR.
Fig. 1.
Activation of PXR induced the expression of Akr1b7 in vivo. A, expression of Akr1b7 mRNA in mouse liver and small intestine was measured by Northern blot analysis. Male mice were treated with two daily intraperitoneal doses of PCN (100 mg/kg) and sacrificed 4 h after the second dose (the total treatment time is 28 h). Cyp3a11 was included as a positive control for PXR activation. Membranes were stripped and reprobed for Gapdh for a loading control. Lanes represent individual mice. Note four and three PCN-treated mice were used for the liver and intestinal analysis, respectively. B, expression of Akr1b7 protein as determined by Western blot analysis. Lanes represent individual mice. C, expression of Akr1b7, Akr1a4, Akr1b8, and Akr7a5 mRNA in the liver was measured by real-time PCR. The same samples in A were used. D, hepatic and intestinal mRNA expression of Akr1b7 in male PXR(-/-) mice treated without or with PCN was measured by real-time PCR. N = 6 for each group. E, mRNA expression of Akr1b7 in the liver and small intestine of wild-type (WT) mice and FABP-VP-PXR transgenic (TG) mice was measured by Northern blot analysis. Each lane represents RNA pooled from three mice. NS, statistically not significant (P > 0.05).
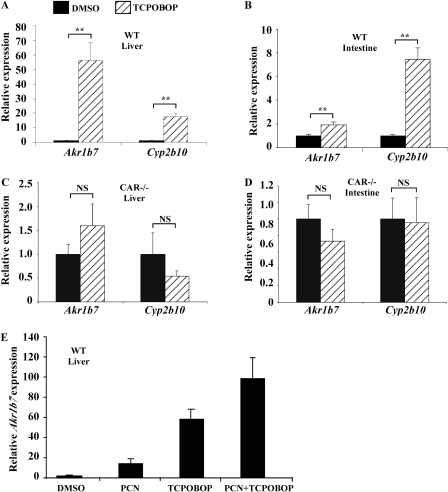
Activation of CAR Induced the Expression of Akr1b7 in Vivo. We found that treatment of wild-type mice with the CAR agonist TCPOBOP also induced the mRNA expression of Akr1b7 in the liver (Fig. 2A) and small intestine (Fig. 2B). Cyp2b10, a prototypical CAR target gene, was included as a positive control for CAR activation. The TCPOBOP effect on Akr1b7 gene expression in the liver (Fig. 2C) and small intestine (Fig. 2D) was abolished in CAR(-/-) mice (Wei et al., 2000), suggesting that Akr1b7 is also under the positive control of CAR. There was an additive induction of Akr1b7 mRNA expression in mice treated with both PCN and TCPOBOP (Fig. 2E).
Fig. 2.
Activation of CAR induced the expression of Akr1b7 in vivo. A and B, real-time PCR analysis on the mRNA expression of Akr1b7 in the liver (A) and small intestine (B) of mice treated with vehicle or TCPOBOP (single intraperitoneal dose of 1 mg/kg, mice were sacrificed 7 days after injection). Cyp2b10 was included as a positive control for CAR activation. N = 5 for each group. C and D, hepatic (C) and intestinal (D) expression of Akr1b7 and Cyp2b10 was analyzed in vehicle- and TCPOBOP-treated CAR(-/-) mice. E, hepatic expression of Akr1b7 mRNA in mice treated with both PCN and TCPOBOP. N = 5 for each group. **, P < 0.01, compared with the vehicle groups. NS, statistically not significant (P > 0.05).
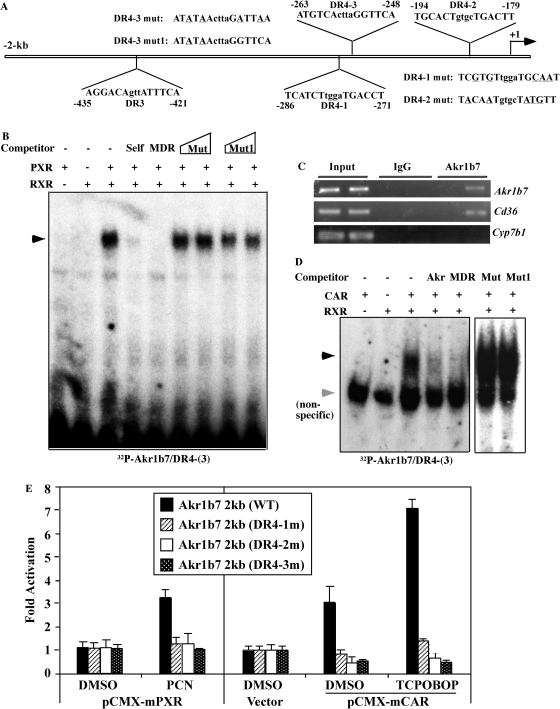
The Akr1b7 Gene Promoter Was Activated by PXR and CAR and Identification of the Response Element in the Promoter Region of Akr1b7 That Binds to Both PXR and CAR. To understand the molecular mechanism by which PXR and CAR regulate Akr1b7 gene expression, we cloned the 2-kb 5′ flanking region of the mouse Akr1b7 gene by PCR. Inspection of the promoter sequences and assisted by using the web-based software NUBIScan version 2.0 (http://www.nubiscan.unibas.ch/), we have identified three candidate DR-4 (DR4-1, -2, and -3) and one DR-3 site (Fig. 3A) within 449 bp upstream of the transcription start site. After an initial screening by EMSA, we found one of the DR-4 elements, DR4-3, had a strong affinity to bind to the PXR-RXR heterodimers (Fig. 3B). The binding can be efficiently competed by unlabeled Akr1b7/DR-4 or the MDR-1/DR-4 (AGGTCAagttAGTTCA) derived from the MDR-1 gene (Geick et al., 2001), but not by two Akr1b7/DR-4 mutant variants with the DR-4 site disrupted (Fig. 3, A and B). A similar pattern of binding and competition was observed when the activated VP-PXR was used in EMSA (data not shown). The in vivo recruitment of PXR to DR4-3 was confirmed by ChIP assay. As shown in Fig. 3C, treatment of primary hepatocytes with PCN resulted in the recruitment of PXR to DR4-3. In the ChIP assay, Cd36 (a PXR target gene) (Zhou et al., 2006) and Cyp7b1 (a retinoic acid-related orphan receptor α target gene) (Wada et al., 2008) were included as positive and negative control, respectively (Fig. 3C). EMSA showed that DR4-1 and DR4-2, but not DR-3, could also bind to the PXR-RXR heterodimers (data not shown). Akr1b7/DR-4 can also specifically bind to CAR-RXR heterodimers as shown by EMSA (Fig. 3D).
Fig. 3.
The Akr1b7 gene promoter was activated by PXR and CAR and identification of the response element in the promoter region of Akr1b7 that binds to both PXR and CAR. A, schematic diagram of the mouse Akr1b7 gene promoter. Numbers indicate positions from the transcriptional starting site (+1). Three putative DR-4 elements and one DR-3 element are labeled. The sequences of DR4-1 and DR4-2 mutants and two DR4-3 mutants are also labeled with the mutated nucleotides underlined. B, binding of the PXR-RXR heterodimers to the radiolabeled Akr1b7/DR4-3. The arrows indicate specific shift bands. In the competition lanes, the unlabeled competitor oligonucleotides were added at 50- to 100-fold excess. Akr and MDR indicate Akr1b7/DR4-3 and MDR1/DR-4, respectively. C, recruitment of PXR to Akr1b7/DR4-3 as shown by ChIP assays. Cd36 and Cyp7b1 were included as the positive and negative control of PXR target genes, respectively. D, CAR-RXR heterodimers bind to Akr1b7/DR-4. The dark and light arrows indicate specific and nonspecific shift bands, respectively. E, wild-type (WT) Akr1b7 2-kb promoter reporter gene or its DR4 mutant variants were cotransfected with the mouse PXR expression vector (pCMX-mPXR) or the mouse CAR expression vector (pCMX-mCAR) into HepG2 cells. Twelve hours after transfection, cells were treated with vehicle (DMSO), PCN (10 μM), or TCPOBOP (250 nM) for 24 h before luciferase assay. The luciferase activity was normalized against the cotransfected β-galactosidase activity. Results are shown as -fold induction over vehicle controls and represent the average and S.D. from triplicate assays.
To examine the functional relevance of DR-4s in mediating the transactivation by PXR and CAR, the 2-kb 5′ regulatory DNA fragment was inserted into the pGL3 basic vector to create the pGL3-Akr1b7 luciferase reporter gene. Three variant reporter genes with three DR-4s individually mutated were also generated. In transient transfection and luciferase reporter gene assay, pGL3-Akr1b7 was activated by PCN in HepG2 cells cotransfected with the expression vector for the mouse PXR (pCMX-mPXR) (Fig. 3E). It is interesting that the mutation of any of the DR-4s resulted in the loss of PXR effect (Fig. 3E). When the mouse CAR (pCMX-mCAR) was cotransfected, we found that CAR activated the wild-type report gene in the absence of an exogenously added ligand, and this activation was enhanced when TCPOBOP was added to the medium (Fig. 3E). Similarly, the mutation of any of the DR4 sites abolished the effect of CAR and TCPOBOP (Fig. 3E).
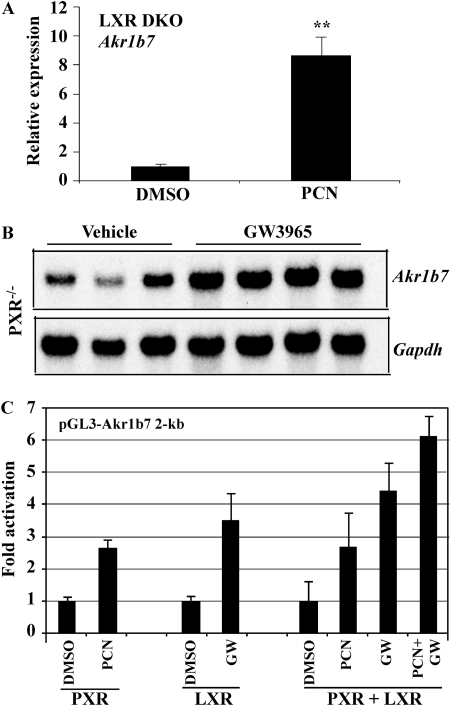
Independent and Cooperative Regulation of Akr1b7 by PXR and LXR. Because LXRα has been shown previously to regulate Akr1b7 via several LXR elements, including the DR-4 that binds to PXR and CAR (Volle et al., 2004), we went on to determine whether the effects of PXR and LXR on Akr1b7 gene expression are mutually dependent. As shown in Fig. 4A, PCN was effective to induce Akr1b7 mRNA expression in the liver of LXR DKO mice (Peet et al., 1998). Conversely, GW3965, an LXR agonist that does not activate PXR (Zhou et al., 2008), remained effective to induce Akr1b7 gene expression in the liver of PXR(-/-) mice (Fig. 4B). The expression of Scd-1, a known LXR target gene, was also induced in GW3965-treated PXR(-/-) mice (data not shown). These results suggest that PXR and LXR are mutually dispensable in their regulation of Akr1b7 gene expression. To determine whether PXR and LXR had an additive effect on Akr1b7 gene expression, HepG2 cells were transfected with pGL3-Akr1b7 2-kb reporter gene together with expression vectors for PXR and/or LXR. Transfected cells were then treated with PXR and/or LXR agonists for 24 h before luciferase assay. As shown in Fig. 4C, PXR and LXR apparently had an additive effect in activating the reporter gene.
Fig. 4.
Independent and cooperative regulation of Akr1b7 by PXR and LXR. A, Akr1b7 mRNA expression was induced by the PXR agonist PCN in LXR DKO mice. LXR DKO mice were treated with vehicle or PCN, and the liver expression of Akr1b7 was analyzed by real-time PCR. N = 5 for each group. B, Akr1b7 mRNA expression was induced by the LXR agonist GW3965 in PXR(-/-) mice. PXR(-/-) mice were treated with five daily gavages of vehicle or GW3965 (20 mg/kg). The liver expression of Akr1b7 was analyzed by Northern blot analysis. Lanes represent individual mice. C, pGL3-Akr1b7 2-kb reporter gene was cotransfected with PXR and/or LXR. Transfected cells were treated with the indicated drugs for 24 h before luciferase assay. The drug concentration is 10 μM for both PCN and GW3965.
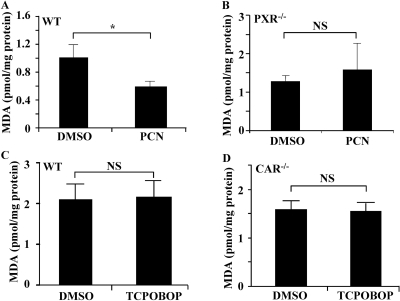
A Potential Role of PXR in Protection against Lipid Peroxidation. Akr1b7 has been suggested to play an important role in the detoxification of lipid peroxidation products (Volle et al., 2004; Schneider et al., 2008). To determine whether PXR activation confers protection from lipid peroxidation, we analyzed lipid peroxidation in the small intestine of mice by measuring the level of MDA, a by-product of polyunsaturated fatty acid peroxidation (Frankel, 1987) and biomarker of lipid peroxidation. A significant decrease in MDA level was observed in wild-type mice treated with PCN for 76 h, compared with the vehicle-treated counterparts (Fig. 5A). The PCN effect on MDA production was abolished in PXR(-/-) mice (Fig. 5B). These results suggest that PXR plays a role in alleviating lipid peroxide accumulation in the small intestine. When the CAR effect was evaluated, we were surprised to find that treatment of TCPOBOP had little effect on the basal level of MDA in either the wild-type (Fig. 5C) or CAR(-/-) (Fig. 5D) mice.
Fig. 5.
A potential role of PXR in protection against lipid peroxidation. A and B, level of MDA in the small intestine homogenate was measured in WT (A; N = 4 for each group) or PXR(-/-) (B; N = 3 for each group) mice treated with DMSO or PCN. Mice received four daily intraperitoneal injections of the drugs and sacrificed 4 h after the last dose, so the total treatment time is 76 h. C and D, intestinal MDA level was measured in WT (C; N = 5 for each group) or CAR(-/-) (D; N = 5 for each group) mice treated with DMSO or TCPOBOP. *, P < 0.05. NS, statistically not significant (P > 0.05).
Discussion
In this study, we have identified Akr1b7 as a novel target gene for the xenobiotic receptors PXR and CAR. The combined uses of pharmacologic and genetic models of PXR and CAR activation have demonstrated that activation of PXR and CAR is both necessary and sufficient for the regulation of Akr1b7 in the mouse liver and intestine. The identity of Akr1b7 as a PXR and CAR target gene was further supported by the characterization of a DR-4 response element that binds to both receptors and is required for the transactivation of Akr1b7 gene promoter. The activation of Akr1b7 gene expression by PXR and CAR has led to our appreciation that the xenobiotic receptors may have a previously unrecognized function in the detoxification of lipid peroxides.
Both RXR and LXR agonists have been shown to activate Akr1b7 gene expression. It was reported that the effect of RXR agonist LG268 on Akr1b7 gene expression was intact in LXR DKO mice (Volle et al., 2004), suggesting that RXR heterodimerization partners other than LXRs could have mediated the transactivation. Our results suggested that PXR and CAR might have mediated the activation of Akr1b7 gene expression by LG268 in LXR DKO mice. T0901317 was used by Volle et al. (2004) as the LXR agonist to induce Akr1b7 gene expression. It is interesting that the effect of T0901317 on Akr1b7 gene expression was abolished in LXRα and -β double knockout mice (Volle et al., 2004), despite the reports that T0901317 can also activate PXR (Shenoy et al., 2004).
Having demonstrated that both PXR and LXR activate Akr1b7 gene expression, an outstanding question is whether the effect of these two receptors on Akr1b7 gene expression is mutually dependent. Using mice deficient of PXR or LXRs, we showed that the PXR and LXR agonist effect on Akr1b7 gene expression was intact in LXR DKO and PXR(-/-) mice, respectively. The independent effect of PXR and LXR was further supported by our observation that PXR and LXR had an additive effect in activating the Akr1b7 gene promoter. The coregulation of Akr1b7 is reminiscent of the shared regulation of Sult2a1/2a9 by PXR (Sonoda et al., 2002), CAR (Saini et al., 2004), farnesoid X receptor (Song et al., 2001), and LXR (Uppal et al., 2007), in which an inverted repeat without a spacing nucleotide response element is used by all four receptors. We propose that the regulatory network of Akr1b7, controlled by PXR, CAR, and LXR, offers a complex and fail-safe system in preventing toxicity associated with lipid peroxidation. It remains to be determined whether the shared regulation of Akr1b7 by several nuclear receptors also contributes to the tissue distribution pattern of this AKR isoform.
The identification of Akr1b7 as a PXR and CAR target gene has expanded the function of xenobiotic receptors in xeno- and endobiotic detoxification. Because AKRs functionalize carbonyl groups by forming alcohols for the conjugation reactions catalyzed by the phase II UDP-glucuronosyltransferase and sulfotransferase enzymes, they can be classified as phase I enzymes (Penning and Drury, 2007). Human AKRs have been implicated in the metabolism (carbonyl reduction) of synthetic hormones, cancer chemotherapeutics, and central nervous system-acting drugs. AKRs also play an important xenobiotic role, implicating in the detoxification of at least three classes of chemical carcinogen: polycyclic aromatic hydrocarbons, aflatoxin, and nicotine-derived nitosamino-ketones (Penning and Drury, 2007). It remains to be determined whether the human AKRs are also regulated by xenobiotic receptors. There was no report of the human homolog of Akr1b7. AKR1B10 seemed to be the closest human relative based on the AKR family tree. Like Akr1b7, AKR1B10 is also highly expressed in the intestine and liver and functions as an aldose reductase (Cao et al., 1998). Using the human colon cancer LS180 cells overexpressing the wild-type or activated PXR (Gong et al., 2006), we showed that activation of PXR did not alter the expression of AKR1B10 (data not shown). It remains to be determined whether other human AKR isoforms can be regulated by PXR or CAR.
Although our results suggest that Akr1b7 is under the transcriptional control of both PXR and CAR, it seemed that there were differences between these two receptors in their regulation of Akr1b7. For example, the magnitude of Akr1b7 induction was higher in the intestine than that in the liver in PXR-activated mice (Fig. 1), whereas the induction was more dramatic in the liver than that in the intestine in CAR-activated mice (Fig. 2). The mechanism for this tissue effect of Akr1b7 gene regulation is currently unclear. Moreover, unlike the PXR agonist PCN that inhibited MDA formation in the intestine in a PXR-dependent manner (Fig. 5, A and B), treatment with the CAR agonist TCPOBOP failed to inhibit MDA formation (Fig. 5, C and D). The receptor-specific effect on MDA formation may have resulted from the differential effect of these two receptors on the expression of other genes, whose products might also be involved in the production and clearance of lipid peroxidation products.
The identification of Akr1b7 as a PXR target gene has also expanded the function of this receptor in lipid homeostasis. Originally identified as a “xenobiotic receptor,” PXR was later found to impact lipid homeostasis (for review, see Handschin and Meyer, 2005; Zhou et al., 2006b; Nakamura et al., 2007). The activation of PXR in the mouse liver resulted in an increased hepatic deposit of triglycerides. This PXR-mediated lipid accumulation was independent of the activation of the lipogenic transcriptional factor sterol regulatory element-binding protein 1c. Instead, the PXR-responsive lipid accumulation was associated with an increased expression of the free fatty acid transporter CD36 and several accessory lipogenic enzymes, such as stearoyl CoA desaturase-1 and long-chain free fatty acid elongase. In the same study, CD36 was established as a direct transcriptional target of PXR. Increased fatty acid uptake, such as that facilitated by CD36, may prone cells to lipid peroxidation. Indeed, an accumulation of PUFAs have been proposed to be involved in the formation of atheromas in the vasculature, because PUFAs serve as substrates for lipid peroxidation (Yin and Porter, 2005). It is tempting for us to speculate that the activation of Akr1b7 by PXR may represent an evolved function for cells to protect from lipid peroxidation associated with increased fatty acid uptake. Although we have shown that activation of PXR was necessary and sufficient to decrease intestinal accumulation of the lipid peroxidation biomarker MDA, we cannot excluded the possibility that the MDA-lowering effect of PXR may have been mediated or contributed by PXR target genes other than Akr1b7. A future use of Ark1b7 null mice (Baumann et al., 2007) would further conclude that the PXR effect on the alleviation of lipid peroxidation is indeed mediated by this enzyme. Also interesting is that Akr1b7 has been suggested to affect lipid metabolism by inhibiting adipogenesis in some adipose tissues, the mechanism of which remains to be determined (Tirard et al., 2007).
In summary, the present work has established a novel anti-lipid peroxidation role for the xenobiotic receptors PXR and CAR. The regulation of Akr1b7 has not only expanded the detoxification function of PXR and CAR but also broadened the implications of xenobiotic receptors in lipid homeostasis.
Acknowledgments
We thank Dr. David Mangelsdorf for providing the LXR null mice and Dr. Ramalinga Kuruba for synthesizing GW3965.
This work was supported in part by the National Institutes of Health National Cancer Institute [Grant CA107011] and the National Institutes of Health National Institute of Environmental Health Sciences [Grant ES014626].
ABBREVIATIONS: AKR, aldo-keto reductase; PUFA, polyunsaturated fatty acid; MDA, malondialdehyde; PXR, pregnane X receptor; CAR, constitutive androstane receptor; RXR, retinoid X receptor; DR, direct repeat; LXR, liver X receptor; DKO, double knockout; PCN, pregnenolone-16α-carbonitrile; GW3965, 3-[3-[N-(2-chloro-3-trifluoromethylbenzyl)-(2,2-diphenylethyl)amino]propyloxy]phenylacetic acid hydrochloride; FABP, fatty acid-binding protein; VP, viral protein 16; TCPOBOP, 1,4-bis[2-(3,5-dichlorpyridyloxyl)]-benzene; PCR, polymerase chain reaction; ChIP, chromatin immunoprecipitation; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; bp, base pair(s); PEI, polyethylenimine; DMEM, Dulbecco's modified Eagle's medium; EMSA, electrophoretic mobility shift assay; DMSO, dimethyl sulfoxide; kb, kilobase(s); LG268, 6-[-1(3,5,5,8,8-pentamethyl-5,6,7,8-tetrahydronaphthalen-2-yl)-cyclopropyl]-pyridine-3-carboxylic acid; T0901317, N-(2,2,2-trifluoroethyl)-N-[4-[2,2,2-trifluoro-1-hydroxy-1-(trifluoromethyl)-ethyl]phenyl]-benzenesulfonamide.
References
- Baumann C, Davies B, Peters M, Kaufmann-Reiche U, Lessl M, and Theuring F (2007) AKR1B7 (mouse vas deferens protein) is dispensable for mouse development and reproductive success. Reproduction 134 97-109. [DOI] [PubMed] [Google Scholar]
- Blumberg B, Sabbagh W Jr, Juguilon H, Bolado J Jr, van Meter CM, Ong ES, and Evans RM (1998) SXR, a novel steroid and xenobiotic-sensing nuclear receptor. Genes Dev 12 3195-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D, Fan ST, and Chung SS (1998) Identification and characterization of a novel human aldose reductase-like gene. J Biol Chem 273 11429-11435. [DOI] [PubMed] [Google Scholar]
- Dring MM, Goulding CA, Trimble VI, Keegan D, Ryan AW, Brophy KM, Smyth CM, Keeling PW, O'Donoghue D, O'Sullivan M, et al. (2006) The pregnane X receptor locus is associated with susceptibility to inflammatory bowel disease. Gastroenterology 130 341-348; quiz 592. [DOI] [PubMed] [Google Scholar]
- Frankel EN (1987) Secondary products of lipid oxidation. Chem Phys Lipids 44 73-85. [DOI] [PubMed] [Google Scholar]
- Geick A, Eichelbaum M, and Burk O (2001) Nuclear receptor response elements mediate induction of intestinal MDR1 by rifampin. J Biol Chem 276 14581-14587. [DOI] [PubMed] [Google Scholar]
- Gong H, Singh SV, Singh SP, Mu Y, Lee JH, Saini SP, Toma D, Ren S, Kagan VE, Day BW, et al. (2006) Orphan nuclear receptor pregnane X receptor sensitizes oxidative stress responses in transgenic mice and cancerous cells. Mol Endocrinol 20 279-290. [DOI] [PubMed] [Google Scholar]
- Handschin C and Meyer UA (2005) Regulatory network of lipid-sensing nuclear receptors: roles for CAR, PXR, LXR, and FXR. Arch Biochem Biophys 433 387-396. [DOI] [PubMed] [Google Scholar]
- Honkakoski P, Zelko I, Sueyoshi T, and Negishi M (1998) The nuclear orphan receptor CAR-retinoid X receptor heterodimer activates the phenobarbital-responsive enhancer module of the CYP2B gene. Mol Cell Biol 18 5652-5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliewer SA, Moore JT, Wade L, Staudinger JL, Watson MA, Jones SA, McKee DD, Oliver BB, Willson TM, Zetterström RH, et al. (1998) An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell 92 73-82. [DOI] [PubMed] [Google Scholar]
- Langmann T, Moehle C, Mauerer R, Scharl M, Liebisch G, Zahn A, Stremmel W, and Schmitz G (2004) Loss of detoxification in inflammatory bowel disease: dysregulation of pregnane X receptor target genes. Gastroenterology 127 26-40. [DOI] [PubMed] [Google Scholar]
- Lau ET, Cao D, Lin C, Chung SK, and Chung SS (1995) Tissue-specific expression of two aldose reductase-like genes in mice: abundant expression of mouse vas deferens protein and fibroblast growth factor-regulated protein in the adrenal gland. Biochem J 312 609-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefrançois-Martinez AM, Tournaire C, Martinez A, Berger M, Daoudal S, Tritsch D, Veyssière G, and Jean C (1999) Product of side-chain cleavage of cholesterol, isocaproaldehyde, is an endogenous specific substrate of mouse vas deferens protein, an aldose reductase-like protein in adrenocortical cells. J Biol Chem 274 32875-32880. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Moore R, Negishi M, and Sueyoshi T (2007) Nuclear pregnane X receptor cross-talk with FoxA2 to mediate drug-induced regulation of lipid metabolism in fasting mouse liver. J Biol Chem 282 9768-9776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascussi JM, Robert A, Nguyen M, Walrant-Debray O, Garabedian M, Martin P, Pineau T, Saric J, Navarro F, Maurel P, et al. (2005) Possible involvement of pregnane X receptor-enhanced CYP24 expression in drug-induced osteomalacia. J Clin Invest 115 177-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peet DJ, Turley SD, Ma W, Janowski BA, Lobaccaro JM, Hammer RE, and Mangelsdorf DJ (1998) Cholesterol and bile acid metabolism are impaired in mice lacking the nuclear oxysterol receptor LXR alpha. Cell 93 693-704. [DOI] [PubMed] [Google Scholar]
- Penning TM and Drury JE (2007) Human aldo-keto reductases: function, gene regulation, and single nucleotide polymorphisms. Arch Biochem Biophys 464 241-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini SP, Sonoda J, Xu L, Toma D, Uppal H, Mu Y, Ren S, Moore DD, Evans RM, and Xie W (2004) A novel constitutive androstane receptor-mediated and CYP3A-independent pathway of bile acid detoxification. Mol Pharmacol 65 292-300. [DOI] [PubMed] [Google Scholar]
- Schneider C, Porter NA, and Brash AR (2008) Routes to 4-hydroxynonenal: fundamental issues in the mechanisms of lipid peroxidation. J Biol Chem 283 15539-15543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah YM, Ma X, Morimura K, Kim I, and Gonzalez FJ (2007) Pregnane X receptor activation ameliorates DSS-induced inflammatory bowel disease via inhibition of NF-kappaB target gene expression. Am J Physiol Gastrointest Liver Physiol 292 G1114-G1122. [DOI] [PubMed] [Google Scholar]
- Shenoy SD, Spencer TA, Mercer-Haines NA, Alipour M, Gargano MD, Runge-Morris M, and Kocarek TA (2004) CYP3A induction by liver x receptor ligands in primary cultured rat and mouse hepatocytes is mediated by the pregnane X receptor. Drug Metab Dispos 32 66-71. [DOI] [PubMed] [Google Scholar]
- Song CS, Echchgadda I, Baek BS, Ahn SC, Oh T, Roy AK, and Chatterjee B (2001) Dehydroepiandrosterone sulfotransferase gene induction by bile acid activated farnesoid X receptor. J Biol Chem 276 42549-42556. [DOI] [PubMed] [Google Scholar]
- Sonoda J, Xie W, Rosenfeld JM, Barwick JL, Guzelian PS, and Evans RM (2002) Regulation of a xenobiotic sulfonation cascade by nuclear pregnane X receptor (PXR). Proc Natl Acad Sci U S A 99 13801-13806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudinger JL, Goodwin B, Jones SA, Hawkins-Brown D, MacKenzie KI, LaTour A, Liu Y, Klaassen CD, Brown KK, Reinhard J, et al. (2001) The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc Natl Acad Sci U S A 98 3369-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timsit YE and Negishi M (2007) CAR and PXR: the xenobiotic-sensing receptors. Steroids 72 231-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirard J, Gout J, Lefrançois-Martinez AM, Martinez A, Begeot M, and Naville D (2007) A novel inhibitory protein in adipose tissue, the aldo-keto reductase AKR1B7: its role in adipogenesis. Endocrinology 148 1996-2005. [DOI] [PubMed] [Google Scholar]
- Uppal H, Saini SP, Moschetta A, Mu Y, Zhou J, Gong H, Zhai Y, Ren S, Michalopoulos GK, Mangelsdorf DJ, et al. (2007) Activation of LXRs prevents bile acid toxicity and cholestasis in female mice. Hepatology 45 422-432. [DOI] [PubMed] [Google Scholar]
- Volle DH, Repa JJ, Mazur A, Cummins CL, Val P, Henry-Berger J, Caira F, Veyssiere G, Mangelsdorf DJ, and Lobaccaro JM (2004) Regulation of the aldo-keto reductase gene akr1b7 by the nuclear oxysterol receptor LXRalpha (liver X receptor-alpha) in the mouse intestine: putative role of LXRs in lipid detoxification processes. Mol Endocrinol 18 888-898. [DOI] [PubMed] [Google Scholar]
- Wada T, Kang HS, Angers M, Gong H, Bhatia S, Khadem S, Ren S, Ellis E, Strom SC, Jetten AM, et al. (2008) Identification of oxysterol 7α-hydroxylase (Cyp7b1) as a novel retinoid-related orphan receptor α (RORα) (NR1F1) target gene and a functional cross-talk between RORα and liver X receptor (NR1H3) Mol Pharmacol 73 891-899. [DOI] [PubMed] [Google Scholar]
- Wang T, Ma X, Krausz KW, Idle JR, and Gonzalez FJ (2008) Role of pregnane X receptor in control of all-trans retinoic acid (ATRA) metabolism and its potential contribution to ATRA resistance. J Pharmacol Exp Ther 324 674-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei P, Zhang J, Egan-Hafley M, Liang S, and Moore DD (2000) The nuclear receptor CAR mediates specific xenobiotic induction of drug metabolism. Nature 407 920-923. [DOI] [PubMed] [Google Scholar]
- Willson TM and Kliewer SA (2002) PXR, CAR and drug metabolism. Nat Rev Drug Discov 1 259-266. [DOI] [PubMed] [Google Scholar]
- Xie W, Barwick JL, Downes M, Blumberg B, Simon CM, Nelson MC, Neuschwander-Tetri BA, Brunt EM, Guzelian PS, and Evans RM (2000) Humanized xenobiotic response in mice expressing nuclear receptor SXR. Nature 406 435-439. [DOI] [PubMed] [Google Scholar]
- Xie W, Radominska-Pandya A, Shi Y, Simon CM, Nelson MC, Ong ES, Waxman DJ, and Evans RM (2001) An essential role for nuclear receptors SXR/PXR in detoxification of cholestatic bile acids. Proc Natl Acad Sci U S A 98 3375-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H and Porter NA (2005) New insights regarding the autoxidation of polyunsaturated fatty acids. Antioxid Redox Signal 7 170-184. [DOI] [PubMed] [Google Scholar]
- Zhai Y, Pai HV, Zhou J, Amico JA, Vollmer RR, and Xie W (2007) Activation of pregnane X receptor disrupts glucocorticoid and mineralocorticoid homeostasis. Mol Endocrinol 21 138-147. [DOI] [PubMed] [Google Scholar]
- Zhang B, Xie W, and Krasowski MD (2008) PXR: a xenobiotic receptor of diverse function and implicated in pharmacogenetics. Pharmacogenomics 9 1695-1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Tabb MM, Nelson EL, Grün F, Verma S, Sadatrafiei A, Lin M, Mallick S, Forman BM, Thummel KE, et al. (2006a) Mutual repression between steroid and xenobiotic receptor and NF-kappaB signaling pathways links xenobiotic metabolism and inflammation. J Clin Invest 116 2280-2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Febbraio M, Wada T, Zhai Y, Kuruba R, He J, Lee JH, Khadem S, Ren S, Li S, et al. (2008) Hepatic fatty acid transporter Cd36 is a common target of LXR, PXR, and PPARgamma in promoting steatosis. Gastroenterology 134 556-567. [DOI] [PubMed] [Google Scholar]
- Zhou J, Zhai Y, Mu Y, Gong H, Uppal H, Toma D, Ren S, Evans RM, and Xie W (2006) A novel pregnane X receptor-mediated and sterol regulatory element-binding protein-independent lipogenic pathway. J Biol Chem 281 15013-15020. [DOI] [PMC free article] [PubMed] [Google Scholar]