Abstract
Metastatic dissemination of primary tumors is responsible for 90% of colorectal cancer (CRC) deaths. The presence of positive lymph nodes, which separates stage I/II from stage III CRC, is a particularly key factor in patient management. Here, we describe results of a quantitative proteomic survey to identify molecular correlates of node status. Laser capture microdissection and two-dimensional difference gel electrophoresis were used to establish expression profiles for 980 discrete protein features in 24 human CRC specimens. Protein abundances were determined with a median technical coefficient of variation of 10%, which provided an ability to detect small differences between cancer subtypes. Transgelin, a 23-kDa actin-binding protein, emerged as a top-ranked candidate biomarker of node status. The area under the receiver operating characteristic curve for transgelin in predicting node status was 0.868 (P = .002). Significantly increased frequency of moderate- and high-level transgelin expression in node-positive CRC was also seen using semiquantitative immunohistochemistry to analyze 94 independent CRC specimens on tissue microarrays (P = .036). Follow-up studies in CRC cell lines demonstrated roles for transgelin in promoting invasion, survival, and resistance to anoikis. Transgelin localizes to the nucleus of CRC cells, and its sequence and properties suggest that it may participate in regulation of the transcriptional program associated with the epithelial-to-mesenchymal transition.
Introduction
Colorectal cancer (CRC) is the third leading cause of cancer death in both men and women in the United States [1]. Metastatic dissemination of primary tumors is responsible for 90% of all CRC deaths [2]. Molecular tests are not yet widely used in establishing prognosis, and the tumor node metastasis staging system remains the primary tool. Detection of positive lymph nodes separates stage I/II from stage III CRC and is often a key factor in determining patient management. Lymph node status can only be ascertained after surgery and is strongly influenced by the number of nodes examined [3], suggesting that patients might easily be understaged by this method.
Here, we describe a patient-based quantitative proteomic study in which the primary goal was to identify molecular correlates of lymph node status in CRC. A secondary goal was to identify markers that distinguish cancer overall from patient-matched, histologically normal colonic epithelium. The study used microdissected specimens from 24 CRC patients stratified by node status. Microdissection enriches for tumor cells and excludes stroma and necrotic tissue, potentially increasing the specificity of the screen.
The top-ranked biomarker of node status was transgelin, a 23-kDa actin-binding protein. Paradoxically, there is previous evidence that transgelin is both a tumor suppressor and a variable tumor biomarker, depending on the tumor type, stage, and experimental model [4–12]. In the current study, elevated transgelin levels were predictive of node status, although they did not differ significantly between normal colonic epithelium and cancer overall.We also performed follow-up experiments with transgelin to investigate its involvement in biologic processes that are known to be relevant to metastatic behavior.
Transgelin, also known as smooth muscle protein 22α (SM22α), is a 201-amino acid protein that contains a calponin homology domain. It is an early marker of smooth muscle differentiation and is also present in the cytoplasm of fibroblasts and some epithelial cells [13,14]. Transgelin promotes actin gelling in vitro (the name derives from transformation-sensitive actin gelling protein) [15] and is involved in podosome formation in smooth muscle cells, thus predisposing the cells toward migration and invasion [16]. It is associated with Ca2+-independent vascular contractility [17] and is also a direct target of transforming growth factor β (TGF-β)/Smad3-dependent epithelial cell migration in idiopathic pulmonary fibrosis [18]. These findings are consistent with a physiological role of transgelin in controlling cell motility.
CRC metastasis is a multihit, multistage process [19]. In addition to greater motility, cells must be able to invade the extracellular matrix, survive at low density outside the tumor microenvironment, and develop resistance to anoikis, which is a form of apoptosis triggered by loss of cell-matrix interaction. Another process that is frequently associated with metastasis is the epithelial-to-mesenchymal transition (EMT) [19,20]. EMT is a transcriptional program that occurs normally during embryonic development and is characterized by changes in expression levels for E-cadherin, a mediator of cell-cell adhesion, and other markers characteristic of mesenchymal and epithelial cells. In the present study, we investigated whether experimental manipulation of transgelin expression in established CRC cell lines influenced invasion, survival, resistance to anoikis, and the EMT.
Materials and Methods
Tissue Specimens
CRC specimens paired with corresponding normal tissue were obtained from the South Carolina Cancer Tissue Bank, University of South Carolina. Twenty-four patients with CRC (12 node-negative, 12 node-positive), who underwent surgery without presurgical chemotherapy or radiation therapy during 2003 to 2005, were included (Table W1). The institutional review boards of the University of South Carolina and the Medical College of Georgia approved the study, and informed consent was obtained from all patients. Tissues were snap frozen in liquid nitrogen and stored at -80°C. Two independent pathologists confirmed diagnosis of all samples used in the study. Tissue microarrays (TMAs) used in the confirmatory studies represented 94 cases of eligible CRC specimens (48 node-negative, 46 node-positive) and were purchased from US Biomax (Rockville, MD) and ISU Abxis (formerly Petagen; Seoul, Republic of Korea).
Laser Capture Microdissection, Two-dimensional Difference Gel Electrophoresis, and Image Analysis
Laser capture microdissection (LCM) was performed as described [21]. Caps with microdissected cells (2500 per sample) were transferred into 90 µl of lysis buffer, incubated at room temperature for 1 hour, sonicated, and centrifuged to remove insoluble material. For each analytical gel, 4 µg of cell lysate was reacted with 2 nmol of Tris (2-carboxyethyl) phosphine hydrochloride (Sigma-Aldrich, St. Louis, MO), then with 4 nmol of CyDye DIGE Fluor, Cy5, for saturation labeling (GE Healthcare Life Sciences, Buckinghamshire, UK). Aliquots of cell lysate from each sample were also pooled to create an internal standard, which was labeled with CyDye DIGE Fluor, Cy3, for saturation labeling. For analytical gels, 8 µg of protein (4 µg of Cy5-labeled sample and 4 µg of Cy3-labeled internal standard) was loaded on a 24-cm pH 3 to 10 nonlinear immobilized pH gradient strip and focused in an IPGphor apparatus (GE Healthcare Life Sciences) for approximately 55,000 volt-hours. Strips were applied to a 12.5% sodium dodecyl sulfate-polyacrylamide gel and electrophoresis was performed at 11 mA per gel overnight at 20°C in an Ettan DALT twelve Separation Unit (GE Healthcare Life Sciences). For mass spectrometry (MS), 280 µg of internal standard was reduced with 140 nmol of Tris (2-carboxyethyl) phosphine hydrochloride and labeled with 280 nmol of the Cy3 CyDye before isoelectric focusing and sodium dodecyl sulfate-polyacrylamide gel electrophoresis [22].
Image Acquisition and Analysis
Images were acquired using a Typhoon Trio Imager and protein spots were defined and matched using the DeCyder 6.5 software package (GE Healthcare Life Sciences). Intensity data were exported, log-transformed, and normalized as described [23]. Candidate biomarkers were identified and ranked using the Significance Analysis of Microarrays (version 3.0, available at http://www-stat.stanford.edu/~tibs/SAM/).
Mass Spectrometry
Protein spots in a preparative gel were matched to a master image of the internal standard from the analytical gels. Spots of interest were excised, digested with trypsin, and extracted [21]. Peptides were analyzed by matrix-assisted laser desorption/ionization-MS/MS using the ABI 4700 Proteomics Analyzer (Applied Biosystems, Foster City, CA) or by liquid chromatography-MS/MS using an LTQ ion trap mass spectrometer (Thermo Scientific, Waltham, MA). Protein identities were determined using Mascot (available at http://www.matrixscience.com; Matrix Science, Boston, MA) or the Sequest algorithm as implemented by the BioWorks Browser v3.2 (Thermo Scientific) and the National Center for Biotechnology Information database. Autodigested trypsin peaks were used as an internal mass calibration standard. Evaluation of significance was based on the score particular to the method used, the sequence coverage, and the consistency between experimental and predicted molecular weight and pI.
Cell Lines and Transfection
Human colon carcinoma cell lines HCT116 and SW480 were purchased from the American Type Culture Collection (Manassas, VA) and maintained according to their protocols. MicroRNA (miRNA) plasmids targeting TAGLN were generated using the pcDNB 6.2-GW/EmGFP-miR vector (Invitrogen, Carlsbad, CA; refer to Table W3 for inserted sequences). After lipofectamine-mediated transfection, stable transfectants were selected and cultured in medium containing 5 µg/ml blasticidin (Invitrogen). A double-point mutation was introduced into the full-length TAGLN complementary DNA (cDNA; Open Biosystems, Huntsville, AL) using the QuikChange Lightning Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) to create TAGLN rescue cDNA. The rescue TAGLN sequence was transferred into pDONR 221, then pcDNA-DEST40 (Invitrogen) by site-directed recombination. Rescue of TAGLN expression and function was assayed at 48 hours after lipofectamine-mediated cDNA transfection.
Immunoblot Analysis, Immunohistochemistry, and Immunofluorescence
Immunoblot analysis was carried out using antitransgelin immunoglobulin G (IgG) and anti-GAPDHIgG1 (Abcam, Cambridge, MA) with ECF substrate (GE Healthcare Life Sciences) for detection. Immunohistochemical staining was performed using the Histostain-Plus Kit (DAB, Broad Spectrum; Invitrogen) and assessed blindly by two independent investigators (J.R.L. and Y.L.). The staining of transgelin was scored as the product of the staining intensity (on a scale of 0–3: negative = 0, weak = 1, moderate = 2, strong = 3) and the percentage of cells stained (on a scale of 0–3: 0 = zero, 1 = 1%–25%, 2 = 26%–50%, 3 = 51%–100%), resulting in scores on a scale of 0 to 9 [24]. For immunofluorescence, cells were fixed in 4% paraformaldehyde for 15 minutes, permeabilized with 0.1% Triton X-100 for 5 minutes, and incubated with blocking buffer (15% goat serum, 0.2% fish skin gelatin and 0.03% NaN3 in phosphate-buffered saline) for 30 minutes at room temperature. Cells were sequentially incubated with antitransgelin IgG and antirabbit IgG conjugated to Alexa Fluor 594 (Molecular Probes, Eugene, OR). Slides were mounted using VECTASHIELD mounting medium with 4′,6-diamidino-2-phenylindole (Vector Laboratories, Burlingame, CA). Images were collected using a meta confocal microscope (LSM 510; Carl Zeiss, Thornwood, NY).
RNA Isolation and Real-time Polymerase Chain Reaction
Extraction of total RNA was performed using Trizol (Invitrogen) followed by reverse transcription (RT). Real-time polymerase chain reaction (PCR) was carried out using an MJ PTC-200 Chromo4 thermocycler (Bio-Rad, Hercules, CA) with a SYBR Green PCR kit (Qiagen, Germantown, MD). Data analysis was performed using OpticonMonitor software (version 3.1; Bio-Rad). PCR primers are listed in Table W4.
Transwell Invasion Assay
Cell invasion assay was performed as described [25] using the Transwell filter (pore size, 8.0 µm; 24-well plate; Corning, Inc, Life Sciences, Lowell, MA). Filters were coated with 1.54 mg/ml Matrigel (BD Biosciences, Sparks, MD) according to the manufacturer's protocol. Cells were harvested and resuspended in serum-free medium, and 5 x 105 cells were applied onto the upper chamber of the Transwell filter. The bottom chamber contained 0.6 ml of medium supplemented with 10% fetal bovine serum. Cells were incubated for 40 hours. Cells that did not migrate were removed by cotton swabbing. Cells that invaded to the lower surface of the filter were fixed and stained with 0.25% crystal violet, 3.7% formaldehyde in 80% methanol for 30 minutes at room temperature. The stained cells were extracted with 10% acetic acid, and the absorbance at 595 nm was measured.
Clonogenic Survival Assay
Cells were plated at 3 x 102 per T-25 flask and incubated with complete growth medium for 10 days for HCT116 cells and 14 days for SW480 cells. Colonies were fixed and stained with staining buffer (0.25% crystal violet, 3.7% formaldehyde in 80% methanol) for 30 minutes at room temperature.
Anoikis Assay
Anoikis was induced by plating the cells on poly-2-hydroxyethyl methacrylate (polyHEMA; Sigma-Aldrich)-coated culture dishes for 72 hours. Cells were collected by gentle pipetting and either subjected to flow cytometry analysis or replated in regular culture dishes, with attached cells trypsinized and counted at 24 hours.
Flow Cytometry
Cells were harvested and washed once with phosphate-buffered saline and resuspended in Annexin-binding buffer (10 mM HEPES, 140 mM NaCl, and 2.5 mM CaCl2, pH 7.4). For each reaction, 1 x 105 cells were incubated with 10 µg RNase (Sigma-Aldrich), 1 µl of Annexin V conjugated to Alexa Fluor 594 (Molecular Probes), and 0.8 µg of propidium iodide (Invitrogen) for 25 minutes at room temperature. Flow cytometry analysis was carried out using a FACSCalibur flow cytometer (BD Biosciences) with CellQuest software.
Statistical Analysis
Values are presented as mean ± SD. Comparisons of the means between indicated groups were carried out using Student's t test. Comparisons with clinical and pathologic variables (sex, ethnicity, histologic grade, tumor subsite, and T stage) were made using exact tests for RxC contingency tables. TMA data were analyzed using nonparametric Wilcoxon rank sums test [26]. A level of P < .05 was considered significant.
Results
Proteomic Profiling of CRC and Matched Normal Epithelium
We collected CRC samples from 12 node-positive and 12 node-negative patients (Table W1). Each CRC sample was paired with a sample of uninvolved colonic epithelium obtained from the same patient at the time of resection. Proteomic analysis was performed by LCM of frozen histologic sections, followed by two-dimensional difference gel electrophoresis (2D-DIGE) with an internal standard design [21].We performed a pilot study to evaluate the technical variation based on repeated LCM sampling of the same specimens. The median coefficient of variation (CV) was 9.4% for CRC and 10.1% for normal colonic epithelium. An improvement more than the 23% CVs in a previous report from our laboratory probably reflects both differences in the tissue type and the use of a microdissected rather than bulk-dissected internal standard [22]. Assuming a total CV of 50% (dominated by biologic variation because technical variation is only approximately 10%), the group sizes used in the current study provided at least 80% power to identify features with a two-fold change between experimental groups using a two-sided α of 0.05 [22,27].
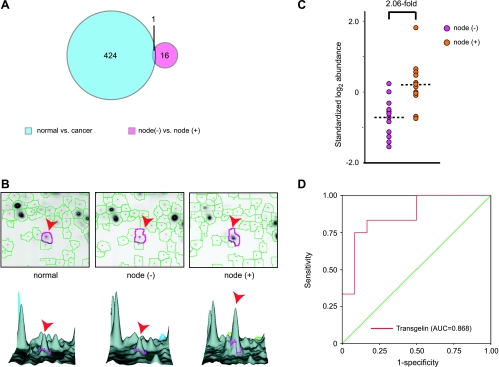
LCM and 2D-DIGE were performed on all 48 samples, and relative abundance values were derived for 980 protein spots that were matched with high confidence across more than 90% of the gels. Significance analysis of microarrays was used to evaluate each intergroup difference and to produce a list of spots, rank-ordered by difference score (D, calculated based on the average difference between groups divided by the sum of the spot-specific scatter [variance] and a measure of scatter [variance] common to all proteins [28].) Applying a liberal 10% false discovery rate cutoff, there were 16 spots that differentiated node-positive from node-negative CRC and 424 spots that differentiated CRC from patient-matched normal colonic epithelium (Figure 1A). Interestingly, there was no more overlap between these sets than would be predicted by chance. That is, most features that distinguished one group of CRC from another were not useful for differentiating CRC from normal and vice versa.
Figure 1.
Proteomic profiling of CRCs stratified by node status. (A) Relationship between biomarkers identified in different comparisons. (B) Differential expression of transgelin. Top: representative two-dimensional gel images; bottom: three-dimensional representation. Arrowheads indicate transgelin. (C) The standardized log-transformed abundance of transgelin in node-negative and node-positive CRC. (D) Receiver operating characteristic (ROC) curve for transgelin in prediction of node status.
Identification of Proteins by MS
Six protein spots from the node-positive versus node-negative CRC comparison were identified molecularly by MS. Three were metabolic enzymes, and three were isoforms of the actin-binding protein, transgelin (Table W2). On average, the change in transgelin expression levels exceeded two-fold (Figure 1, B and C), which was the cutoff level for useful biomarkers that we had assumed in our power calculation. Perhaps more importantly, the area under the receiver operating characteristic curve for a test of node status based on transgelin was 0.868 (P = .002; Figure 1D), which is considered excellent for a single marker. Transgelin was selected for further characterization because of its statistical ranking, ability to distinguish between groups, and previous work suggesting a role in control of cell motility [15–18].
We also identified 56 protein spots that differed in CRC versus patient-matched colonic mucosa (Table W2). Although several seem to be novel in the context of CRC, we have not yet investigated them further (see Discussion).
Transgelin Expression Patterns on TMAs
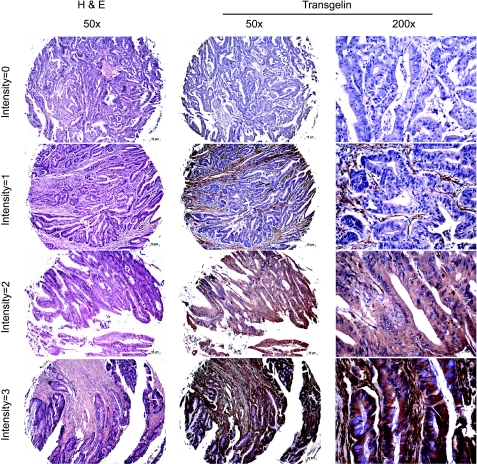
We further investigated the correlation between transgelin expression and node status in a larger, independent patient cohort by immunohistochemical staining of commercial TMAs. TMAs from two sources were analyzed, representing 94 eligible cases from different geographic regions. Representative patterns of transgelin staining are shown in Figure 2. Staining was evident in the cytoplasm and in some cell nuclei. Two independent investigators scored the TMAs blindly, using a 0 to 9 scale representing a combination of staining intensity and fraction of cells stained (see Materials and Methods [24]). Increased frequency of moderate- and high-level transgelin expression in node-positive CRC was seen in this cohort. The distribution of scores for the node-negative and node-positive groups was significantly different, based on a Wilcoxon rank sum test (P = .036; Table 1).
Figure 2.
Illustrations of hematoxylin and eosin staining and immunohistochemistry of antitransgelin on CRC TMAs.
Table 1.
Summary of Transgelin Expression on Tissue Microarrays.
| Staining (Score) | Node-Negative CRC (n) | Node-Positive CRC (n) | Total (n) |
| None (0) | 30 (62.5%) | 20 (43.5%) | 50 |
| Low (1–3) | 12 (25%) | 12 (26.1%) | 24 |
| Moderate (4–6) | 4 (8.3%) | 11 (23.9%) | 15 |
| High (7–9) | 2 (4.2%) | 3 (6.5%) | 5 |
| Total (n) | 48 | 46 | 94 |
| P | .036 | ||
One limitation of TMAs is that each sample represents only a very small region of the tumor (1.0- to 1.5-mm tissue core). Because of this, a clonal subpopulation of cells that express a metastatic marker might easily be missed, resulting in a type II error (see Discussion). Nevertheless, the statistically significant association between transgelin and node status supports the findings from quantitative proteomic analysis.
Establishment of a Cell Culture Model
To investigate the potential mechanisms by which transgelin might promote metastasis in CRC, it was desirable to establish an in vitro cell culture model. We screened a number of CRC cell lines for transgelin expression and selected HCT116 and SW480 for studies based on their moderate levels of endogenous transgelin expression and the ease with which expression of the transgelin gene (gene symbol: TAGLN) could be manipulated genetically.
We tested four different miRNA sequences for their ability to silence transgelin in HCT116 cells (Figure 3A) and used the most effective of these, TAGLN miRNA-4, to establish stably transfected populations. These populations (HCT116TAGLN-KD, SW480TAGLN-KD) and the matched control miRNA-transfected populations (HCT116CTRL, SW480CTRL) were characterized for transgelin protein expression by immunoblot analysis (Figure 3B), for TAGLN mRNA by real-time RT-PCR (Figure 3C), and immunofluorescence staining with anti-transgelin antibody (Figure 3D). We observed a greater than 80% decrease in expression in all three assays. Expression of the closely related TAGLN3 mRNA was not significantly altered, confirming the specificity of the TAGLN miRNA-4 (Figure 3C). To exclude more rigorously off-target effects in subsequent experiments, we also created a TAGLN rescue plasmid that expressed a TAGLN miRNA-4-resistant cDNA (Figure 3E). Transient transfection with this cDNA restored transgelin expression in the HCT116TAGLN-KD population (Figure 3F).
Figure 3.
MicroRNA-mediated knockdown and rescue of TAGLN in CRC cells. (A) Immunoblot analysis of HCT116 cells after transient transfection of control miRNA plasmid and four different TAGLN miRNA plasmids. (B) Immunoblot analysis of HCT116 and SW480 cells with stable expression of control miRNA or TAGLN miRNA-4. (C) Real-time PCR analysis of TAGLN and TAGLN3 mRNA expression in HCT116 and SW480 cells with stable expression of control miRNA (HCT116CTRL, SW480CTRL) and TAGLN miRNA-4 (HCT116TAGLN-KD, SW480TAGLN-KD). Gene expression was normalized to GAPDH. Data are expressed as mean ± SD from three independent experiments; **P < .01. (D) Fluorescence microscopy of HCT116 and SW480 stably transfected cells (original magnification, x63; white bar, 20 µm). Panels show transgelin immunostaining, EmGFP transfection marker, 4′,6-diamidino-2-phenylindole DNA staining, and a merged image as indicated. (E) TAGLN miRNA target and rescue sequences. (F) Immunoblot analysis of HCT116TAGLN-KD cells after transient transfection with empty vector or TAGLN rescue plasmid.
Effects of Transgelin on Cell Invasion, Survival, and Anoikis
To evaluate the effect of altered transgelin expression level on biologic processes that are relevant to metastasis, we performed in vitro assays for invasion, survival at low density, and anoikis.
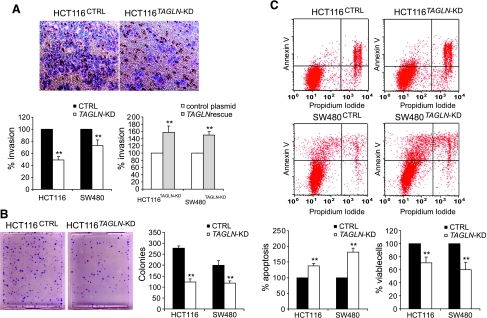
Invasion was measured using a Transwell assay. Cells were seeded in serum-free medium in an upper chamber, which is separated by a Matrigel-coated filter from a lower chamber that contains medium with 10% fetal bovine serum as a chemoattractant. Results were measured based on staining of cells that invaded the Matrigel, migrated through pores in the filter, and reached the lower surface. Knockdown of transgelin reduced invasion by more than 50% in HCT116 cells and by 27% in SW480 cells (P < .01; Figure 4A). Transient transfection with TAGLN rescue plasmid, but not control plasmid, significantly restored invasion capability to both HCT116 and SW480 TAGLN knockdown cells (Figure 4A). On the basis of the successful rescue by cDNA transfection, it is unlikely that results are attributable to off-target effects of the miRNA.
Figure 4.
Effects of TAGLN silencing on HCT116 and SW480 cell invasion, survival, and anoikis. (A) Invasion assay. Representative images of Transwell filters indicating HCT116CTRL and HCT116TAGLN-KD invasion are shown. Bar graphs show comparison of invasion capacities of HCT116 and SW480 stably transfected cells (left) and HCT116TAGLN-KD and SW480TAGLN-KD cells after transient transfection with 1 µg of empty vector or TAGLN rescue plasmid (right). (B) Clonogenic survival assay. Representative images of HCT116CTRL and HCT116TAGLN-KD cells. Graph shows the number of colonies formed 10 and 14 days after plating HCT116 and SW480 stably transfected cells, respectively. (C) The anoikis-induced apoptosis was measured by Annexin V and propidium iodide staining and flow cytometry analysis at 72 hours after plating HCT116 and SW480 stably transfected cells on polyHEMA-coated culture dishes. Anoikis was also assessed by counting the viable cells at 24 hours after replating the anoikis-induced cells on regular culture dishes. Values for control cells were considered 100%, any differences are expressed relative to this value. Data are expressed as mean ± SD from three independent experiments; **P < .01.
We evaluated clonogenic survival by plating cells at low density and scoring for colony formation after 10 to 14 days. Knockdown of transgelin reduced the clonogenic survival of HCT116 cells by approximately 55% and SW480 cell by approximately 40% (P < .01; Figure 4B).
Resistance to anoikis (apoptosis induced by loss of cell-matrix contact) was investigated by plating control and knockdown cells on polyHEMA-coated dishes. After 72 hours, cells were characterized by flow cytometry using Annexin V and propidium iodide stains (Figure 4C). The total fraction of apoptotic cells in the TAGLN knockdown groups, based on Annexin V-positive staining, increased by 1.4- to 1.8-fold, relative to the corresponding control cells (P < .01). The percentage of viable cells in the same population was measured independently by replating on regular dishes, incubating for 24 hours, removing the unattached cells, and counting the remainder. Knockdown of transgelin reduced the percentage of viable cells to 60% to 70% of control values (P < .01; Figure 4C). Together, these results strongly implicate transgelin in resistance to anoikis.
Effects of Transgelin Expression on Epithelial and Mesenchymal Markers
Transgelin is normally expressed in mesenchymal cells, and its appearance in tumor cells of epithelial origin is consistent with the engagement of the EMT, by which tumor cells acquire a more aggressive phenotype. Transgelin is localized primarily in the nucleus of CRC cells (Figure 3D) and an 85% similar homolog, transgelin 3, is a transcription factor that controls expression of the gonadotropin gene [29]. We hypothesized that transgelin might also be a transcriptional regulator.
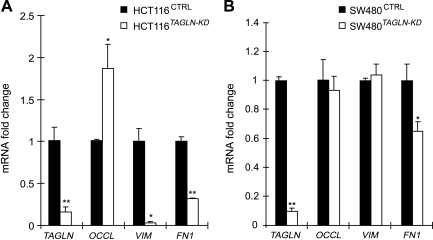
During the EMT, the cell intermediate filament system switches from a keratin-rich network that connects to adherens junctions and hemidesmosomes to a vimentin-rich network that connects to focal adhesions [30]. Typical elements of the EMT include loss of expression of proteins associated with adherens junctions (such as cadherins and catenins) and tight junctions (such as occludin) and an increase in expression of mesenchymal intermediate filament protein, vimentin, and the mesenchymal adhesion protein, fibronectin [18,30–32]. In HCT116 cells, knockdown of transgelin was associated with the upregulation of mRNA encoding occludin and with the down-regulation of mRNA encoding fibronectin-1 and the mesenchymal intermediate filament protein vimentin (Figure 5A). The mRNA levels for two other epithelial markers, E-cadherin and β-catenin, were unaffected (data not shown). In SW480 cells, transgelin knockdown had less effect. Of the mRNA investigated, only fibronectin-1 was significantly influenced (Figure 5B). The finding that alteration of transgelin affects mRNA levels for some, but not all, EMT-associated genes supports the hypothesis that transgelin is a promoter-selective regulator of transcription. An ability to regulate transcription of other genes provides a mechanism by which transgelin might broadly influence processes relevant to metastasis.
Figure 5.
EMT marker genes regulated by TAGLN. The mRNA levels of the epithelial marker genes occludin (OCCL) and mesenchymal marker genes vimentin (VIM) and fibronectin 1 (FN1) in HCT116CTRL and HCT116TAGLN-KD cells (A) and in SW480CTRL and SW480TAGLN-KD cells (B). Gene expression was measured by real-time RT-PCR and normalized to GAPDH. Data are expressed as mean ± SD from three independent experiments; **P < .01, *P < .05.
Discussion
We performed a patient-based proteomic study to seek proteins that correlated with lymph node status. We obtained quantitative expression data for 980 protein spots. Of these, transgelin had three qualities that made it particularly interesting for follow-up: it had the highest statistical ranking as a predictor of node status, it was novel in the context of CRC metastasis, and there was a variety of previous evidence in other systems that linked it to cell motility. To explore possible mechanisms by which transgelin could influence metastatic behavior, we created two pairs of isogenic CRC cell lines that differed in transgelin levels. In both instances, attenuation of expression resulted in loss of characteristics believed to be important for metastasis, including invasion, survival at low density, and resistance to anoikis.
In theory, identification of metastatic biomarkers in primary tumors should be problematic because cells that express such markers may be too rare to influence a population-averaged expression profile [19]. In practice, however, biomarkers that correlate with metastatic risk have been identified in several other cancers, including breast and prostate cancer. Metastasis is a complex process, and it may be that stepwise acquisition of metastatic potential leads to expansion of clones expressing genes that are necessary, but perhaps not sufficient, for forming metastasis [19]. We suggest that TAGLN may be among these genes.
CRC has been shown to arise initially through two major genetic pathways: one is chromosomal instability, and the other is micro-satellite instability [33–35]. Other routes of carcinogenesis include the TGF-β/SMAD signaling pathway, the serrated pathway, and the epigenetic pathway (reviewed in Takayama et al. [36]). However, the mechanisms that are responsible for metastasis seem to be much more complex [37,38]. EMT and tumor-microenvironment interaction have begun to receive significant attention [20]. EMT is mediated by the network signaling of TGF-β, Wnt, mitogen-activated protein kinase, phosphatidylinositol 3-kinase/Akt, Snail, Slug, SMAD-interacting protein 1, and E2A proteins [20]. Influences of tumor microenvironment include degradation of extracellular matrix by matrix metallo proteinases, changes in adhesion molecules (integrins and cadherins), increased angiogenesis mediated by vascular endothelial growth factor, and changes in cell survival and differentiation mediated by TRAIL receptor, CXCR4 and Drg-1. [36,39]. The hepatocyte growth factor/MET pathway may also be important in EMT [40], CRC cell invasiveness, and metastasis [41–45]. Recently, themetastasis-associated in colon cancer-1 gene (MACC1) has been reported to be an independent prognostic indicator of CRC metastasis. MACC1 regulates the expression of MET, the hepatocyte growth factor receptor [38].
Our initial quantitative proteomic analysis suggested that transgelin expression might be able to serve as a surrogate marker of node status. This would require an analytical method with higher throughput than the LCM/2D-DIGE. However, the method would need to preserve the advantages of LCM/2D-DIGE, including 1) accurate quantification, 2) an ability to sample a reasonable fraction of the tumor, and 3) an ability to exclude stroma. Because transgelin is expressed normally in smooth muscle and fibroblasts, inclusion of stroma might easily confound the analysis. We used immunostaining of TMAs as a higher throughput method to evaluate the correlation between transgelin and node status (Figure 2). This method allowed exclusion of stroma but was only semiquantitative and sampled a much smaller region of each tumor than the LCM/2D-DIGE. As might be expected, the results, although showing a significant difference between groups, had much lower sensitivity and specificity. In particular, there were a high percentage of transgelin-negative samples in both groups. A number of new methods of proteomic analysis are in development (reviewed in Lin et al. [46]). Imaging MS is a particularly promising method [47,48], and it is compatible with direct detection of a small protein, such as transgelin, within tissue sections. Analysis of tissue sections using imaging MS would be a logical next step in evaluating the clinical usefulness of transgelin as a biomarker.
An actin-binding protein, such as transgelin, has the potential to alter cell motility through direct interaction with the actin cytoskeleton. However, the predominantly nuclear localization in cultured CRC cells (Figure 3D) and the influence on the expression levels of several EMT-associated genes (Figure 5) led us to explore an alternative hypothesis, that is, transgelin is a transcriptional regulator. Recent evidence shows that actin plays an important role in nuclear processes, including transcription, chromatin remodeling, and transcription factor regulation [49–53]. Indeed, many of the more than 60 classes of actin-binding proteins in human cells localize to the nucleus [54]. These nuclear actin-binding proteins are associated with diverse processes, including transcriptional regulation and DNA repair [55,56]. Transgelin contains a C-terminal calponin-like module, which is required for actin binding in other proteins [57]. In addition, bioinformatic analysis using the “DP-bind” Web server (http://lcg.rit.albany.edu/dp-bind) identified several segments of the transgelin primary sequence as having high DNA-binding potential (data not shown). Together, these observations support a hypothesis that transgelin could be an adaptor that mediates interaction between nuclear actin and DNA. Direct investigation of interactions between transgelin and other macromolecules in the nucleus will be required to evaluate this possibility.
The observation that transgelin expression is elevated in node-positive CRC raises a question of what controls transgelin expression itself. TAGLN spans 5.4 kb on chromosome 11q23.2, with putative promoter elements approximately 800-bp upstream of the open reading frame [58]. These include a TGF-β control element [59], smad-binding elements, and serum response factor-binding CArG boxes [60]. Several studies have shown that transgelin expression is under the control of the TGF-β signaling pathway in other systems [18,58,61–64]. TGF-β plays a paradoxical role in regulating tumor cell growth and migration. During early tumor development, it is an inhibitor of proliferation, but later in CRC progression, it switches to become a promoter of growth and invasion [65,66] and is important for the EMT [67–70]. If transgelin expression is under TGF-β control, it may explain why expression is associated with tumor suppression in some studies and is a variable tumor biomarker in the others [4–12].
Although transgelin is the only protein we have investigated in detail, the proteomic survey did lead to other potentially interesting findings. A few proteins identified in the cancer-versus-normal comparison in the current study are novel in the context of CRC and seem to have cancer-relevant biologic functions. One of these is a component of the IκB kinase complex, ELKS/RAB6-interacting/CAST family member 1. Another is zinc finger protein 224, which is a transcriptional repressor that interacts with a corepressor, KRAB-associated protein 1 [71], which is upregulated in metastatic breast cancer [72]. These and other proteins that emerged in the CRC-versus-normal comparison have yet to be investigated.
Supplementary Material
Acknowledgments
The authors thank Eric Miller, John Netchman, and Wenbo Zhi for protein identification services at the Proteomics andMass Spectrometry Core Facilities of Medical College of Georgia; John Cowell of the Cancer Center, Medical College of Georgia for helpful discussion; and Rhea-Beth Markowitz for editorial assistance.
Abbreviations
- 2D-DIGE
two-dimensional difference gel electrophoresis
- CRC
colorectal cancer
- CV
coefficient of variation
- EMT
epithelial-to-mesenchymal transition
- LCM
laser capture microdissection
- miRNA
microRNA
- MS
mass spectrometry
- polyHEMA
poly-2-hydroxyethyl methacrylate
- TMA
tissue microarray
Footnotes
This work is supported by National Institutes of Health/National Cancer Institute grant CA95941 (W.S. Dynan).
This article refers to supplementary materials, which are designated by Tables W1 to W4 and are available online at www.neoplasia.com.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Christofori G. New signals from the invasive front. Nature. 2006;441:444–450. doi: 10.1038/nature04872. [DOI] [PubMed] [Google Scholar]
- 3.Compton CC. Optimal pathologic staging: defining stage II disease. Clin Cancer Res. 2007;13:6862s–6870s. doi: 10.1158/1078-0432.CCR-07-1398. [DOI] [PubMed] [Google Scholar]
- 4.Yang Z, Chang YJ, Miyamoto H, Ni J, Niu Y, Chen Z, Chen YL, Yao JL, di Sant'Agnese PA, Chang C. Transgelin functions as a suppressor via inhibition of ARA54-enhanced androgen receptor transactivation and prostate cancer cell growth. Mol Endocrinol. 2007;21:343–358. doi: 10.1210/me.2006-0104. [DOI] [PubMed] [Google Scholar]
- 5.Nair RR, Solway J, Boyd DD. Expression cloning identifies transgelin (SM22) as a novel repressor of 92-kDa type IV collagenase (MMP-9) expression. J Biol Chem. 2006;281:26424–26436. doi: 10.1074/jbc.M602703200. [DOI] [PubMed] [Google Scholar]
- 6.Shields JM, Rogers-Graham K, Der CJ. Loss of transgelin in breast and colon tumors and in RIE-1 cells by Ras deregulation of gene expression through Raf-independent pathways. J Biol Chem. 2002;277:9790–9799. doi: 10.1074/jbc.M110086200. [DOI] [PubMed] [Google Scholar]
- 7.Sitek B, Luttges J, Marcus K, Kloppel G, Schmiegel W, Meyer HE, Hahn SA, Stuhler K. Application of fluorescence difference gel electrophoresis saturation labelling for the analysis of microdissected precursor lesions of pancreatic ductal adenocarcinoma. Proteomics. 2005;5:2665–2679. doi: 10.1002/pmic.200401298. [DOI] [PubMed] [Google Scholar]
- 8.Qi Y, Chiu JF, Wang L, Kwong DL, He QY. Comparative proteomic analysis of esophageal squamous cell carcinoma. Proteomics. 2005;5:2960–2971. doi: 10.1002/pmic.200401175. [DOI] [PubMed] [Google Scholar]
- 9.Huang Q, Huang Q, Chen W, Wang L, Lin W, Lin J, Lin X. Identification of transgelin as a potential novel biomarker for gastric adenocarcinoma based on proteomics technology. J Cancer Res Clin Oncol. 2008;134:1219–1227. doi: 10.1007/s00432-008-0398-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ryu JW, Kim HJ, Lee YS, Myong NH, Hwang CH, Lee GS, Yom HC. The proteomics approach to find biomarkers in gastric cancer. J Korean Med Sci. 2003;18:505–509. doi: 10.3346/jkms.2003.18.4.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mikuriya K, Kuramitsu Y, Ryozawa S, Fujimoto M, Mori S, Oka M, Hamano K, Okita K, Sakaida I, Nakamura K. Expression of glycolytic enzymes is increased in pancreatic cancerous tissues as evidenced by proteomic profiling by two-dimensional electrophoresis and liquid chromatography-mass spectrometry/mass spectrometry. Int J Oncol. 2007;30:849–855. [PubMed] [Google Scholar]
- 12.Li N, Zhang J, Liang Y, Shao J, Peng F, Sun M, Xu N, Li X, Wang R, Liu S, et al. A controversial tumor marker: is SM22 a proper biomarker for gastric cancer cells? J Proteome Res. 2007;6:3304–3312. doi: 10.1021/pr0702363. [DOI] [PubMed] [Google Scholar]
- 13.Lawson D, Harrison M, Shapland C. Fibroblast transgelin and smooth muscle SM22α are the same protein, the expression of which is downregulated in many cell lines. Cell Motil Cytoskeleton. 1997;38:250–257. doi: 10.1002/(SICI)1097-0169(1997)38:3<250::AID-CM3>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 14.Assinder SJ, Stanton JA, Prasad PD. Transgelin: an actin-binding protein and tumour suppressor. Int J Biochem Cell Biol. 2009;41:482–486. doi: 10.1016/j.biocel.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 15.Shapland C, Hsuan JJ, Totty NF, Lawson D. Purification and properties of transgelin: a transformation and shape change sensitive actin-gelling protein. J Cell Biol. 1993;121:1065–1073. doi: 10.1083/jcb.121.5.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gimona M, Kaverina I, Resch GP, Vignal E, Burgstaller G. Calponin repeats regulate actin filament stability and formation of podosomes in smooth muscle cells. Mol Biol Cell. 2003;14:2482–2491. doi: 10.1091/mbc.E02-11-0743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Je HD, Sohn UD. SM22α is required for agonist-induced regulation of contractility: evidence from SM22α knockout mice. Mol Cells. 2007;23:175–181. [PubMed] [Google Scholar]
- 18.Yu H, Konigshoff M, Jayachandran A, Handley D, Seeger W, Kaminski N, Eickelberg O. Transgelin is a direct target of TGF-β/Smad3-dependent epithelial cell migration in lung fibrosis. FASEB J. 2008;22:1778–1789. doi: 10.1096/fj.07-083857. [DOI] [PubMed] [Google Scholar]
- 19.Chiang AC, Massague J. Molecular basis of metastasis. N Engl J Med. 2008;359:2814–2823. doi: 10.1056/NEJMra0805239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tse JC, Kalluri R. Mechanisms of metastasis: epithelial-to-mesenchymal transition and contribution of tumor microenvironment. J Cell Biochem. 2007;101:816–829. doi: 10.1002/jcb.21215. [DOI] [PubMed] [Google Scholar]
- 21.Greengauz-Roberts O, Stoppler H, Nomura S, Yamaguchi H, Goldenring JR, Podolsky RH, Lee JR, Dynan WS. Saturation labeling with cysteinereactive cyanine fluorescent dyes provides increased sensitivity for protein expression profiling of laser-microdissected clinical specimens. Proteomics. 2005;5:1746–1757. doi: 10.1002/pmic.200401068. [DOI] [PubMed] [Google Scholar]
- 22.Arnouk H, Merkley MA, Podolsky RH, Stoppler H, Santos C, Alvarez M, Mariategui J, Ferris D, Lee JR, Dynan WS. Characterization of molecular markers indicative of cervical cancer progression. Proteomics Clin Appl. 2009;3:516–527. doi: 10.1002/prca.200800068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weinberger PM, Merkley MA, Lee JR, Adam BL, Gourin CG, Podolsky RH, Haffty BG, Papadavid E, Sasaki C, Psyrri A, et al. Combination proteomic analysis demonstrates molecular similarity of head and neck squamous cell carcinoma arising from different subsites. Arch Otolaryngol. 2009;135:694–703. doi: 10.1001/archoto.2009.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z, Jiang L, Huang C, Li Z, Chen L, Gou L, Chen P, Tong A, Tang M, Gao F, et al. Comparative proteomics approach to screening of potential diagnostic and therapeutic targets for oral squamous cell carcinoma. Mol Cell Proteomics. 2008;7:1639–1650. doi: 10.1074/mcp.M700520-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen Y, Zhang YW, Zhang ZX, Miao ZH, Ding J. hTERT-targeted RNA interference inhibits tumorigenicity and motility of HCT116 cells. Cancer Biol Ther. 2008;7:228–236. doi: 10.4161/cbt.7.2.5259. [DOI] [PubMed] [Google Scholar]
- 26.Kreisberg JI, Malik SN, Prihoda TJ, Bedolla RG, Troyer DA, Kreisberg S, Ghosh PM. Phosphorylation of Akt (Ser473) is an excellent predictor of poor clinical outcome in prostate cancer. Cancer Res. 2004;64:5232–5236. doi: 10.1158/0008-5472.CAN-04-0272. [DOI] [PubMed] [Google Scholar]
- 27.Molloy MP, Brzezinski EE, Hang J, McDowell MT, VanBogelen RA. Overcoming technical variation and biological variation in quantitative proteomics. Proteomics. 2003;3:1912–1919. doi: 10.1002/pmic.200300534. [DOI] [PubMed] [Google Scholar]
- 28.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feng J, Lawson MA, Melamed P. A proteomic comparison of immature and mature mouse gonadotrophs reveals novel differentially expressed nuclear proteins that regulate gonadotropin gene transcription and RNA splicing. Biol Reprod. 2008;79:546–561. doi: 10.1095/biolreprod.108.068106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kokkinos MI, Wafai R, Wong MK, Newgreen DF, Thompson EW, Waltham M. Vimentin and epithelial-mesenchymal transition in human breast cancer—observations in vitro and in vivo. Cells Tissues Organs. 2007;185:191–203. doi: 10.1159/000101320. [DOI] [PubMed] [Google Scholar]
- 31.Kaplan RN, Riba RD, Zacharoulis S, Bramley AH, Vincent L, Costa C, MacDonald DD, Jin DK, Shido K, Kerns SA, et al. VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature. 2005;438:820–827. doi: 10.1038/nature04186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hugo H, Ackland ML, Blick T, Lawrence MG, Clements JA, Williams ED, Thompson EW. Epithelial-mesenchymal and mesenchymal-epithelial transitions in carcinoma progression. J Cell Physiol. 2007;213:374–383. doi: 10.1002/jcp.21223. [DOI] [PubMed] [Google Scholar]
- 33.Gervaz P, Bucher P, Morel P. Two colons-two cancers: paradigm shift and clinical implications. J Surg Oncol. 2004;88:261–266. doi: 10.1002/jso.20156. [DOI] [PubMed] [Google Scholar]
- 34.Gervaz P, Cerottini JP, Bouzourene H, Hahnloser D, Doan CL, Benhattar J, Chaubert P, Secic M, Gillet M, Carethers JM. Comparison of microsatellite instability and chromosomal instability in predicting survival of patients with T3N0 colorectal cancer. Surgery. 2002;131:190–197. doi: 10.1067/msy.2002.119816. [DOI] [PubMed] [Google Scholar]
- 35.Haydon AM, Jass JR. Emerging pathways in colorectal-cancer development. Lancet Oncol. 2002;3:83–88. doi: 10.1016/s1470-2045(02)00649-6. [DOI] [PubMed] [Google Scholar]
- 36.Takayama T, Miyanishi K, Hayashi T, Sato Y, Niitsu Y. Colorectal cancer: genetics of development and metastasis. J Gastroenterol. 2006;41:185–192. doi: 10.1007/s00535-006-1801-6. [DOI] [PubMed] [Google Scholar]
- 37.Cardoso J, Boer J, Morreau H, Fodde R. Expression and genomic profiling of colorectal cancer. Biochim Biophys Acta. 2007;1775:103–137. doi: 10.1016/j.bbcan.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 38.Stein U, Walther W, Arlt F, Schwabe H, Smith J, Fichtner I, Birchmeier W, Schlag PM. MACC1, a newly identified key regulator of HGF-MET signaling, predicts colon cancer metastasis. Nat Med. 2009;15:59–67. doi: 10.1038/nm.1889. [DOI] [PubMed] [Google Scholar]
- 39.Craig DH, Downey C, Basson MD. SiRNA-mediated reduction of α-actinin-1 inhibits pressure-induced murine tumor cell wound implantation and enhances tumor-free survival. Neoplasia. 2008;10:217–222. doi: 10.1593/neo.07945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Birchmeier C, Birchmeier W, Gherardi E, Vande Woude GF. Met, metastasis, motility and more. Nat Rev Mol Cell Biol. 2003;4:915–925. doi: 10.1038/nrm1261. [DOI] [PubMed] [Google Scholar]
- 41.Di Renzo MF, Olivero M, Giacomini A, Porte H, Chastre E, Mirossay L, Nordlinger B, Bretti S, Bottardi S, Giordano S, et al. Overexpression and amplification of the met/HGF receptor gene during the progression of colorectal cancer. Clin Cancer Res. 1995;1:147–154. [PubMed] [Google Scholar]
- 42.Fazekas K, Csuka O, Koves I, Raso E, Timar J. Experimental and clinicopathologic studies on the function of the HGF receptor in human colon cancer metastasis. Clin Exp Metastasis. 2000;18:639–649. doi: 10.1023/a:1013136303880. [DOI] [PubMed] [Google Scholar]
- 43.Fujita S, Sugano K. Expression of c-met proto-oncogene in primary colorectal cancer and liver metastases. Jpn J Clin Oncol. 1997;27:378–383. doi: 10.1093/jjco/27.6.378. [DOI] [PubMed] [Google Scholar]
- 44.Kammula US, Kuntz EJ, Francone TD, Zeng Z, Shia J, Landmann RG, Paty PB, Weiser MR. Molecular co-expression of the c-Met oncogene and hepatocyte growth factor in primary colon cancer predicts tumor stage and clinical outcome. Cancer Lett. 2007;248:219–228. doi: 10.1016/j.canlet.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 45.Takeuchi H, Bilchik A, Saha S, Turner R, Wiese D, Tanaka M, Kuo C, Wang HJ, Hoon DS. c-METexpression level in primary colon cancer: a predictor of tumor invasion and lymph node metastases. Clin Cancer Res. 2003;9:1480–1488. [PubMed] [Google Scholar]
- 46.Lin Y, Dynan WS, Lee JR, Zhu ZH, Schade RR. The current state of proteomics in GI oncology. Dig Dis Sci. 2008;54:431–457. doi: 10.1007/s10620-008-0656-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Caprioli RM. Perspectives on imaging mass spectrometry in biology and medicine. Proteomics. 2008;8:3679–3680. doi: 10.1002/pmic.200890064. [DOI] [PubMed] [Google Scholar]
- 48.Seeley EH, Caprioli RM. Molecular imaging of proteins in tissues by mass spectrometry. Proc Natl Acad Sci USA. 2008;105:18126–18131. doi: 10.1073/pnas.0801374105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fomproix N, Percipalle P. An actin-myosin complex on actively transcribing genes. Exp Cell Res. 2004;294:140–148. doi: 10.1016/j.yexcr.2003.10.028. [DOI] [PubMed] [Google Scholar]
- 50.Hofmann WA, Stojiljkovic L, Fuchsova B, Vargas GM, Mavrommatis E, Philimonenko V, Kysela K, Goodrich JA, Lessard JL, Hope TJ, et al. Actin is part of pre-initiation complexes and is necessary for transcription by RNA polymerase II. Nat Cell Biol. 2004;6:1094–1101. doi: 10.1038/ncb1182. [DOI] [PubMed] [Google Scholar]
- 51.Kukalev A, Nord Y, Palmberg C, Bergman T, Percipalle P. Actin and hnRNP U cooperate for productive transcription by RNA polymerase II. Nat Struct Mol Biol. 2005;12:238–244. doi: 10.1038/nsmb904. [DOI] [PubMed] [Google Scholar]
- 52.Percipalle P, Fomproix N, Kylberg K, Miralles F, Bjorkroth B, Daneholt B, Visa N. An actin-ribonucleoprotein interaction is involved in transcription by RNA polymerase II. Proc Natl Acad Sci USA. 2003;100:6475–6480. doi: 10.1073/pnas.1131933100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vartiainen MK, Guettler S, Larijani B, Treisman R. Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactorMAL. Science. 2007;316:1749–1752. doi: 10.1126/science.1141084. [DOI] [PubMed] [Google Scholar]
- 54.Vartiainen MK. Nuclear actin dynamics—from form to function. FEBS Lett. 2008;582:2033–2040. doi: 10.1016/j.febslet.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 55.Young KG, Kothary R. Spectrin repeat proteins in the nucleus. Bioessays. 2005;27:144–152. doi: 10.1002/bies.20177. [DOI] [PubMed] [Google Scholar]
- 56.Gettemans J, Van Impe K, Delanote V, Hubert T, Vandekerckhove J, De Corte V. Nuclear actin-binding proteins as modulators of gene transcription. Traffic. 2005;6:847–857. doi: 10.1111/j.1600-0854.2005.00326.x. [DOI] [PubMed] [Google Scholar]
- 57.Fu Y, Liu HW, Forsythe SM, Kogut P, McConville JF, Halayko AJ, Camoretti-Mercado B, Solway J. Mutagenesis analysis of human SM22: characterization of actin binding. J Appl Physiol. 2000;89:1985–1990. doi: 10.1152/jappl.2000.89.5.1985. [DOI] [PubMed] [Google Scholar]
- 58.Camoretti-Mercado B, Forsythe SM, LeBeau MM, Espinosa R, III, Vieira JE, Halayko AJ, Willadsen S, Kurtz B, Ober C, Evans GA, et al. Expression and cytogenetic localization of the human SM22 gene (TAGLN) Genomics. 1998;49:452–457. doi: 10.1006/geno.1998.5267. [DOI] [PubMed] [Google Scholar]
- 59.Adam PJ, Regan CP, Hautmann MB, Owens GK. Positive- and negative-acting Kruppel-like transcription factors bind a transforming growth factor β control element required for expression of the smooth muscle cell differentiation marker SM22α in vivo. J Biol Chem. 2000;275:37798–37806. doi: 10.1074/jbc.M006323200. [DOI] [PubMed] [Google Scholar]
- 60.Chen S, Kulik M, Lechleider RJ. Smad proteins regulate transcriptional induction of the SM22α gene by TGF-β. Nucleic Acids Res. 2003;31:1302–1310. doi: 10.1093/nar/gkg224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lien SC, Usami S, Chien S, Chiu JJ. Phosphatidylinositol 3-kinase/Akt pathway is involved in transforming growth factor-β1-induced phenotypic modulation of 10T1/2 cells to smooth muscle cells. Cell Signal. 2006;18:1270–1278. doi: 10.1016/j.cellsig.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 62.Qiu P, Feng XH, Li L. Interaction of Smad3 and SRF-associated complex mediates TGF-β1 signals to regulate SM22 transcription during myofibroblast differentiation. J Mol Cell Cardiol. 2003;35:1407–1420. doi: 10.1016/j.yjmcc.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 63.Qiu P, Ritchie RP, Fu Z, Cao D, Cumming J, Miano JM, Wang DZ, Li HJ, Li L. Myocardin enhances Smad3-mediated transforming growth factor-β1 signaling in a CArG box-independent manner: Smad-binding element is an important cis element for SM22α transcription in vivo. Circ Res. 2005;97:983–991. doi: 10.1161/01.RES.0000190604.90049.71. [DOI] [PubMed] [Google Scholar]
- 64.Qiu P, Ritchie RP, Gong XQ, Hamamori Y, Li L. Dynamic changes in chromatin acetylation and the expression of histone acetyltransferases and histone deacetylases regulate the SM22α transcription in response to Smad3-mediated TGFβ1 signaling. Biochem Biophys Res Commun. 2006;348:351–358. doi: 10.1016/j.bbrc.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 65.Engle SJ, Hoying JB, Boivin GP, Ormsby I, Gartside PS, Doetschman T. Transforming growth factor β1 suppresses nonmetastatic colon cancer at an early stage of tumorigenesis. Cancer Res. 1999;59:3379–3386. [PubMed] [Google Scholar]
- 66.Hsu S, Huang F, Hafez M, Winawer S, Friedman E. Colon carcinoma cells switch their response to transforming growth factor β 1 with tumor progression. Cell Growth Differ. 1994;5:267–275. [PubMed] [Google Scholar]
- 67.Bhowmick NA, Ghiassi M, Bakin A, Aakre M, Lundquist CA, Engel ME, Arteaga CL, Moses HL. Transforming growth factor-β1 mediates epithelial to mesenchymal transdifferentiation through a RhoA-dependent mechanism. Mol Biol Cell. 2001;12:27–36. doi: 10.1091/mbc.12.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ellenrieder V, Hendler SF, Boeck W, Seufferlein T, Menke A, Ruhland C, Adler G, Gress TM. Transforming growth factor β1 treatment leads to an epithelial-mesenchymal transdifferentiation of pancreatic cancer cells requiring extracellular signal-regulated kinase 2 activation. Cancer Res. 2001;61:4222–4228. [PubMed] [Google Scholar]
- 69.Lehmann K, Janda E, Pierreux CE, Rytomaa M, Schulze A, McMahon M, Hill CS, Beug H, Downward J. Raf induces TGFβ production while blocking its apoptotic but not invasive responses: a mechanism leading to increased malignancy in epithelial cells. Genes Dev. 2000;14:2610–2622. doi: 10.1101/gad.181700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Portella G, Cumming SA, Liddell J, Cui W, Ireland H, Akhurst RJ, Balmain A. Transforming growth factor β is essential for spindle cell conversion of mouse skin carcinoma in vivo: implications for tumor invasion. Cell Growth Differ. 1998;9:393–404. [PubMed] [Google Scholar]
- 71.Medugno L, Florio F, DeCegli R, Grosso M, Lupo A, Costanzo P, Izzo P. The Kruppel-like zinc-finger protein ZNF224 represses aldolase A gene transcription by interacting with the KAP-1 co-repressor protein. Gene. 2005;359:35–43. doi: 10.1016/j.gene.2005.06.021. [DOI] [PubMed] [Google Scholar]
- 72.Ho J, Kong JW, Choong LY, Loh MC, Toy W, Chong PK, Wong CH, Wong CY, Shah N, Lim YP. Novel breast cancer metastasis-associated proteins. J Proteome Res. 2009;8:583–594. doi: 10.1021/pr8007368. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.