Abstract
The canonical pathway for isoleucine biosynthesis in plants begins with the conversion of threonine to 2-ketobutyrate by threonine deaminase (OMR1). However, demonstration of methionine γ-lyase (MGL) activity in Arabidopsis (Arabidopsis thaliana) suggested that production of 2-ketobutyrate from methionine can also lead to isoleucine biosynthesis. Rescue of the isoleucine deficit in a threonine deaminase mutant by MGL overexpression, as well as decreased transcription of endogenous Arabidopsis MGL in a feedback-insensitive threonine deaminase mutant background, shows that these two enzymes have overlapping functions in amino acid biosynthesis. In mgl mutant flowers and seeds, methionine levels are significantly increased and incorporation of [13C]Met into isoleucine is decreased, but isoleucine levels are unaffected. Accumulation of free isoleucine and other branched-chain amino acids is greatly elevated in response to drought stress in Arabidopsis. Gene expression analyses, amino acid phenotypes, and labeled precursor feeding experiments demonstrate that MGL activity is up-regulated by osmotic stress but likely plays a less prominent role in isoleucine biosynthesis than threonine deaminase. The observation that MGL makes a significant contribution to methionine degradation, particularly in reproductive tissue, suggests practical applications for silencing the expression of MGL in crop plants and thereby increasing the abundance of methionine, a limiting essential amino acid.
Due to the importance of branched-chain amino acids in nutrition and agriculture, plant-based biosynthesis of Ile, Leu, and Val has been the subject of extensive research. Humans and other animals are unable to synthesize these essential amino acids and must obtain them either directly or indirectly from plants. Since Ile abundance in some of the world's major food crops, including maize (Zea mays) and rice (Oryza sativa), is suboptimal for mammalian diets (Lewis et al., 1982; Prakash, 1996), increasing the Ile content of seeds and other edible plant parts has been a goal for breeding and agricultural biotechnology. Additional agricultural interest in the enzymes of branched-chain amino acid metabolism comes from their role as herbicide targets. Several classes of commercially successful herbicides inhibit the enzyme acetolactate synthase, which is required for the biosynthesis of all three branched-chain amino acids (Saari et al., 1994; Singh and Shaner, 1995).
The best-studied pathway for Ile synthesis in plants is initiated by Thr dehydratase/deaminase (EC 4.2.1.16; OMR1 or At3g10050 in Arabidopsis [Arabidopsis thaliana]), a Thr catabolic enzyme that produces 2-ketobutyrate and ammonia (Fig. 1). Thr deaminase activity is regulated by Ile, and feedback-insensitive Arabidopsis mutants accumulate 20-fold more Ile than the wild type (Mourad and King, 1995). Met γ-lyase (MGL; EC 4.4.1.11), an enzyme that converts Met to methanethiol and 2-ketobutyrate (Fig. 1), has been studied extensively in microbes and protozoa (Inoue et al., 1995; Faleev et al., 1996; Hori et al., 1996; Dias and Weimer, 1998; McKie et al., 1998; Tokoro et al., 2003; Manukhov et al., 2005). More recently, MGL activity has also been demonstrated in plants. NMR metabolite profiling of Arabidopsis cell suspension cultures labeled with [13C]Met showed that MGL produces 2-ketobutyrate for Ile biosynthesis (Rebeille et al., 2006). Under sulfate limitation, but not under normal growth conditions, knockout of the single Arabidopsis MGL gene (At1g64660) significantly increased Met content in leaves (Goyer et al., 2007). Although both of these prior studies in Arabidopsis demonstrate the potential of Ile biosynthesis from Met, the actual function of MGL in Ile biosynthesis and, in particular, the relative importance of Met and Thr as Ile precursors in intact plants remains to be investigated.
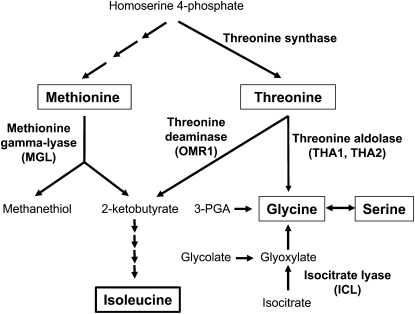
Figure 1.
Pathways for Ile and Gly biosynthesis in plants. 2-Ketobutyrate, a precursor of Ile biosynthesis, can be produced from either Thr or Met. Thr deaminase competes with Thr aldolase for a common substrate. Gly can be synthesized from Thr, Ser, 3-phosphoglycerate (3-PGA), and glyoxylate. The names of Arabidopsis enzymes relevant to this project are indicated in parentheses.
Other published research has provided indirect evidence that there is significant metabolic flux from Met to Ile in plants under certain conditions. Tubers from potato (Solanum tuberosum) plants expressing antisense Thr synthase accumulated elevated Met and Ile, without any apparent change in Thr content (Zeh et al., 2001), suggesting Thr-independent Ile synthesis. Similarly, overexpressing cystathionine γ-synthase, the committing enzyme of Met biosynthesis, increased both Met and Ile content of potato tubers (Dancs et al., 2008). A 7-fold increase in Ile levels in the transgenic tobacco plants expressing bacterial feedback-insensitive Asp kinase and Met-insensitive Arabidopsis cystathionine γ-synthase genes was suggested to result from Met degradation (Hacham et al., 2008). Transcript profiling of Arabidopsis isocitrate lyase (At3g21720; icl-2) mutants showed an 8-fold increase in Thr aldolase (THA1; At1g08630) and 50-fold increase in MGL gene expression (Cornah et al., 2004). Based on these results, we hypothesize that THA1, which produces Gly from Thr, is up-regulated to compensate for the reduced Gly biosynthesis in the icl-2 mutant (Fig. 1). Up-regulation of THA1, in turn, makes less Thr available as a substrate for Thr deaminase (OMR1), which would require the up-regulation of MGL activity to compensate for the reduced production of 2-ketobutyrate from Thr.
Free amino acid levels increase significantly in Arabidopsis vegetative tissue under drought stress (Nambara et al., 1998), with the greatest changes being observed in the abundance of branched-chain amino acids and Pro. Analysis of publicly available Affymetrix microarray data from drought- and salt-stressed Arabidopsis showed that MGL transcription is induced 4-fold (Less and Galili, 2008), suggesting a role for MGL in elevated Ile biosynthesis. Other studies have also demonstrated transcriptional up-regulation of Arabidopsis MGL in response to abiotic stress (Rizhsky et al., 2004; Peng et al., 2007; Baena-González et al., 2008; Mueller et al., 2008). When amino acid levels were measured in tomato (Solanum lycopersicum) suspension cultures subjected to water stress with polyethylene glycol, discrepancies were observed in the known pathway for Ile biosynthesis (Rhodes et al., 1986). In particular, the rate of Thr synthesis was insufficient to sustain the estimated rate of Ile biosynthesis, indicating that Ile was being made from some other source. Together, these results suggest that MGL may play a significant role in plant Ile biosynthesis during osmotic stress.
Here, we use genetic and biochemical approaches to study the in vivo role of MGL in Arabidopsis amino acid biosynthesis. In particular, we test the hypothesis that Ile biosynthesis from Met plays a significant role during free amino acid accumulation in drought-stressed plants.
RESULTS
MGL Mutants Accumulate Excess Met in Flowers and Seeds
A T-DNA insertion in the second exon of Arabidopsis MGL (mgl-1, SALK_040380; Supplemental Fig. S1) has been reported previously (Goyer et al., 2007). To increase the reliability of further experiments, we isolated a second mutant allele in the Columbia-0 (Col-0) genetic background (mgl-2, SALK_103805), which has a T-DNA insertion in the first exon, 17 bp after the MGL start codon (Supplemental Fig. S1). RT-PCR of leaf and flower tissue showed that the mutant does not accumulate any MGL transcript, suggesting complete knockout of the gene. As in the case of mgl-1, no visible phenotypes were observed at the whole-plant level in the mgl-2 mutant, and in subsequent experiments, the two independent mutations showed similar effects on Arabidopsis amino acid metabolism.
Under normal growth conditions, there were no significant changes in the free amino acid content of mgl-2 mutant leaves (Table I). Since flowers and seeds have elevated accumulation of MGL mRNA compared to leaves (Rebeille et al., 2006; Goyer et al., 2007; Supplemental Fig. S2), we also measured amino acid levels in these tissues. Compared to the wild type, mgl-2 mutant flowers accumulated not only significantly higher Met, but also Gly, Leu, Phe, Ser, and Asp. These latter amino acid changes are difficult to explain at this point, though it is possible that excess Met resulted in elevated S-adenosylmethionine (AdoMet), which can feedback-inhibit Asp kinase and thereby increase Asp accumulation (Curien et al., 2007). A similar Asp increase in plants with high Met was reported by Hacham et al. (2008), who also suggested that increasing Met or associated metabolites beyond a certain threshold may serve as a signal that affects other amino acid biosynthesis pathways. Dry mgl-2 mutant seeds contained 3-fold more Met than the wild type, but, opposite to what was observed in the flowers, the levels of Asp, Glu, and Gly were significantly reduced (Table I). Notably, there was no significant reduction in the Ile content of leaves, flowers, or seeds due to the mgl-2 mutation.
Table I.
Amino acid content of leaves, flowers, and seeds of wild-type Col-0 and mgl-2
Data are pmol/mg wet weight, mean ± sd of n = 6, rounded to two significant figures. Values in bold indicate P < 0.05 for changes relative to wild-type Col-0, Student's t test.
| Leaves
|
Flowers
|
Seeds
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Col-0
|
mgl-2
|
Col-0
|
mgl-2
|
Col-0
|
mgl-2
|
|||||||
| Mean | sd | Mean | sd | Mean | sd | Mean | sd | Mean | sd | Mean | sd | |
| Ala | 95 | 46 | 120 | 8.5 | 350 | 39 | 390 | 48 | 530 | 120 | 500 | 120 |
| Arg | 18 | 4.1 | 17 | 4.1 | 110 | 3.5 | 130 | 30 | 730 | 310 | 300 | 55 |
| Asp | 940 | 360 | 1,100 | 81 | 2,000 | 200 | 2,300 | 140 | 1,300 | 250 | 600 | 140 |
| Glu | 1,600 | 268 | 1,600 | 210 | 1,600 | 110 | 1700 | 120 | 2,500 | 100 | 2,000 | 250 |
| Gly | 11 | 2.6 | 11 | 4.4 | 140 | 20 | 230 | 42 | 310 | 31 | 200 | 47 |
| His | 190 | 110 | 230 | 84 | 3,000 | 320 | 3,300 | 560 | 380 | 170 | 200 | 34 |
| Ile | 15 | 3.5 | 15 | 6.7 | 90 | 8.5 | 96 | 17 | 150 | 22 | 120 | 30 |
| Leu | 26 | 8.0 | 24 | 9.5 | 90 | 8.0 | 130 | 25 | 160 | 35 | 140 | 41 |
| Lys | 22 | 5.1 | 21 | 2.9 | 68 | 7.1 | 74 | 22 | 170 | 50 | 110 | 18 |
| Met | 4.2 | 0.62 | 4.5 | 1.1 | 12 | 0.70 | 25 | 0.95 | 21 | 4.1 | 60 | 11 |
| Phe | 25 | 4.2 | 28 | 7.7 | 37 | 5.0 | 54 | 14 | 110 | 9.0 | 120 | 23 |
| Pro | 300 | 100 | 313 | 40 | 1,100 | 78 | 1,300 | 190 | 900 | 140 | 250 | 140 |
| Ser | 420 | 87 | 400 | 64 | 2,300 | 150 | 2,700 | 290 | 2,900 | 1,100 | 1,400 | 240 |
| Thr | 120 | 24 | 110 | 19 | 360 | 22 | 340 | 34 | 250 | 88 | 200 | 30 |
| Tyr | 20 | 4.9 | 19 | 3.5 | 22 | 2.9 | 28 | 8.0 | 84 | 12 | 90 | 16 |
| Val | 46 | 8.7 | 44 | 14 | 200 | 16 | 230 | 38 | 250 | 37 | 20 | 63 |
MGL Lesions Alter Transcriptional Regulation of Met Biosynthesis and Degradation
Increased accumulation of Met in mgl mutants could have broad effects on the transcription of other genes involved in Met metabolism. In plants, Thr and Met are synthesized from a common branch point intermediate, O-phosphohomoserine. Met is produced in three steps from O-phosphohomoserine by the enzymes cystathionine γ-synthase (CGS), cystathionine β-lyase, and Met synthase (Hesse and Hoefgen, 2003; Supplemental Fig. S3). Most Met produced in plants is converted to AdoMet by AdoMet synthase (Giovanelli et al., 1985), which competes with MGL for a common substrate. To study the effects of Met overaccumulation, expression levels of CGS (At3g01120), three Met synthases (ATMS, At5g17920; ATMS2, At3g03780; and ATMS3, At5g20980), and four AdoMet synthases (SAM-1, At1g02500; SAM-2, At4g01850; SAM-3, At3g17390; and SAM-4, At2g36880) were measured in the flowers and siliques of mgl-2 and wild-type Col-0 using real-time quantitative RT-PCR. Unlike MGL, which is expressed most strongly in the siliques, all of these genes show predominant expression in flowers (Supplemental Figs. S2 and S4; www.genevestigator.ethz.ch; Zimmermann et al., 2004).
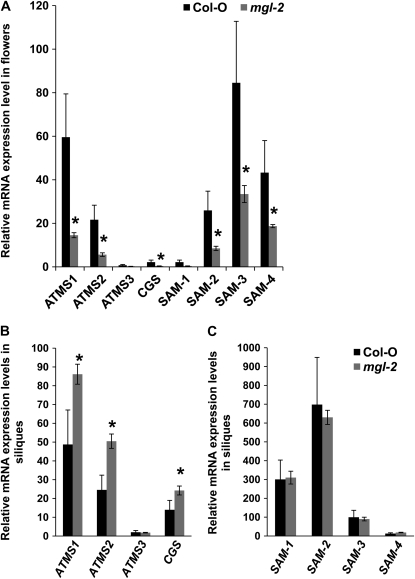
In mgl-2 flowers, there is a significant reduction in the expression levels of Met biosynthetic genes ATMS1, ATMS2, and CGS (Fig. 2A). Although transcriptional regulation of Met synthases by Met or AdoMet has not been shown previously in plants, excess Met reduces mRNA accumulation and enzymatic activity of CGS in Arabidopsis (Inaba et al., 1994; Chiba et al., 1999; Bartlem et al., 2000). This transcriptional down-regulation of Met biosynthesis genes may be a direct response to the increased accumulation of Met in mgl mutant flowers. However, unlike flowers, expression levels of CGS, ATMS1, and ATMS2 were up-regulated in siliques (Fig. 2B). Expression of three of Met-degrading AdoMet synthases (SAM-2, SAM-3, and SAM-4) was found to be down-regulated only in flowers but remained unaffected in siliques (Fig. 2, A and C).
Figure 2.
Relative mRNA accumulation of genes related to Met synthesis (ATMS1, ATMS2, ATMS3, and CGS) and degradation (SAM-1, SAM-2, SAM-3, and SAM-4) in flowers (A) and siliques (B and C) of wild-type Col-0 and mgl-2 plants. Flowers and siliques of 30-d-old plants were used to extract total RNA and used for real-time quantitative PCR to obtain relative mRNA expression levels. Expression levels of all genes were normalized with respect to the internal control UBQ10. The y axes are in arbitrary units. Note different scales for each y axis. Data are mean ± sd of n = 3 samples. *, P < 0.05, Student's t test.
MGL Transcription Is Regulated by Thr and Ile Availability as Well as by Osmotic Stress
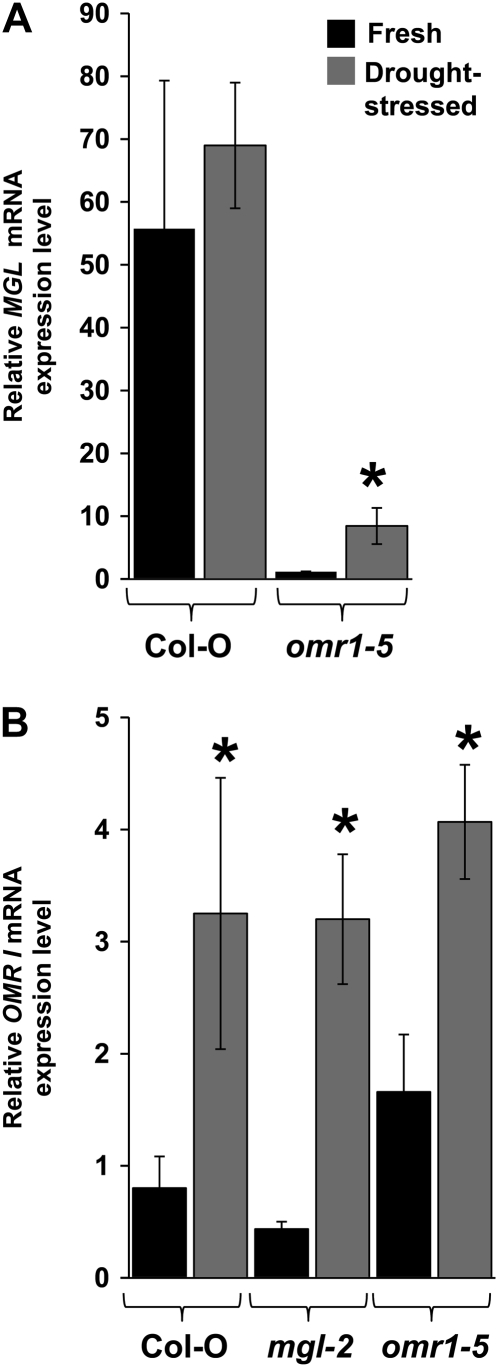
In response to drought stress, there are large increases in the abundance of branched-chain amino acids in Arabidopsis: 90-fold Ile, 150-fold Leu, and 25-fold Val (Nambara et al., 1998). These dramatic changes were suggested to result from up-regulation of amino acid biosynthesis in response to dehydration, rather than a more general change in protein synthesis and degradation. Gene expression profiles of MGL and OMR1 in publicly available microarray databases show that both genes are expressed in all tested plant parts (Supplemental Figs. S3 and S4; www.genevestigator.ethz.ch; Zimmermann et al., 2004). To characterize Ile biosynthesis during drought stress, relative mRNA expression levels of MGL and OMR1, which both encode enzymes leading to Ile biosynthesis (Fig. 1), were measured by real-time quantitative PCR in fresh and drought-stressed flowers of wild-type Col-0, mgl-2, and the Ile feedback-insensitive omr1-5 mutant. Relative mRNA expression levels were compared to the control housekeeping gene UBQ10, as its expression is relatively unaffected by abiotic stress (www.genevestigator.ethz.ch). MGL expression was significantly reduced in the omr1-5 mutant background (Fig. 3A), which has 20-fold higher Ile levels (Mourad and King, 1995). However, this low MGL expression increased 3-fold during drought stress. When dehydrated flowers were compared to fresh flowers, the OMR1 mRNA expression was significantly increased in Col-0, mgl-2, and omr1-5 (Fig. 3B), suggesting regulation in response to increased need for Ile biosynthesis. These results were also confirmed by RT-PCR of other mutants defective in Thr and Ile metabolism (tha1-2, tha2, and icl-2; Supplemental Fig. S5). MGL expression is elevated after desiccation in all tested mutants, even in the high-Ile conditions (Fig. 3) of the omr1-5 mutant background. Therefore, drought-induced up-regulation of MGL transcription occurs independently of Ile abundance.
Figure 3.
Relative amounts of MGL (A) and OMR (B) transcripts in fresh and drought-stressed flowers of wild-type Col-0, mgl-2, and omr1-5. Relative expression levels on the y axis are in arbitrary units. Values are mean ± sd of n = 3 samples. *, P < 0.05 relative to fresh tissue for each genotype, Student's t test.
mgl Mutations Do Not Reduce Salt and Drought Tolerance
As both MGL transcription and Ile accumulation are strongly induced in response to osmotic stress (Nambara et al., 1998; Less and Galili, 2008), we determined whether the mgl-2 mutant shows any increased susceptibility to salt and desiccation. Compared to other tissue types, MGL expression is significantly higher in seedlings and reproductive tissue (Supplemental Fig. S4). Moreover, MGL protein accumulation is strongly increased during seed imbibition, suggesting an important role of this enzyme in the germination process (Rebeille et al., 2006). Therefore, we investigated the role of MGL in seedlings and reproductive tissue subjected to salt and drought stress, respectively.
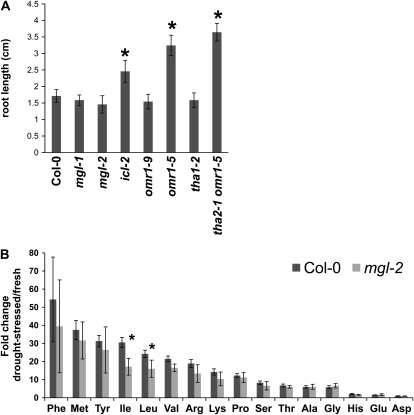
Wild-type seedlings, along with mgl-1, mgl-2, omr1-9, omr1-5, tha2, omr1-5, tha1-2, and icl-2 mutants, were grown on Murashige and Skoog (MS) plates containing 100 mm NaCl (Fig. 4A). No significant differences were seen in the root lengths of mgl and omr1-9 lines compared to the wild type. However, Ile overproducing omr1-5 and tha2 omr1-5 mutants show significantly longer roots than the wild type, implying a role for Ile in salt tolerance. In control experiments on MS agar without added NaCl, root lengths of all mutants were not significantly different from wild-type Col-0 (Supplemental Fig. S6A). Two- and four-week-old mgl mutants growing in soil were also irrigated with 50 and 100 mm NaCl, but no visible salt stress phenotypes were observed relative to wild-type controls.
Figure 4.
Salt sensitivity and drought-induced amino acid accumulation. A, Salt sensitivity of amino acid mutants. Seeds were planted on MS medium with 100 mm NaCl. Roots of 10-d-old seedlings were measured. Mean ± sd of n = 7. B, Amino acid accumulation in drought-stressed wild-type and mgl-2 flowers. Amino acids were quantified from fresh and dehydrated flowers of wild-type Col-0 and mgl-2. Fold-change due to dehydration was calculated. Mean ± sd of n = 4 to 6. *, P < 0.05, relative to wild-type Col-0, Student's t test.
When mgl-2 mutant flowers were subjected to drought stress, they accumulated significantly less Ile and Leu than wild-type Col-0 (P < 0.05, Student's t test), indicating a direct role of MGL in Ile synthesis (Fig. 4B). No significant changes in any other amino acids were observed under these conditions. In addition, no significant differences were observed between mgl-2 mutant and wild-type leaves in terms of drought-induced amino acid changes relative to fresh leaves (Supplemental Fig. S6B). Also, as in the case of salt stress, no visible phenotypic differences were observed in mgl-2 mutants compared to wild-type Col-0 in soil-grown plants that had been subjected to drought stress by withholding water for up to 10 d.
Incorporation of 13C from Met into Ile Shows MGL Activity
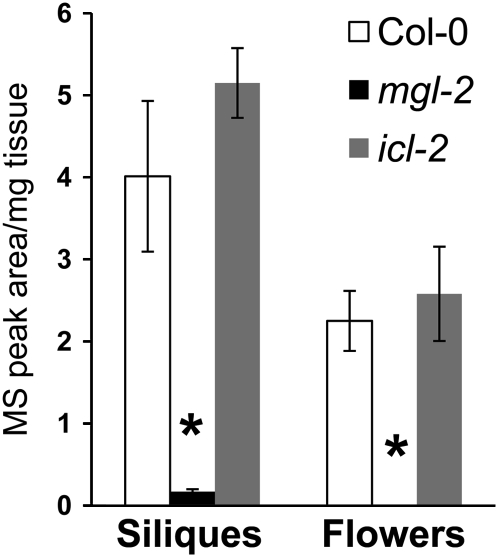
Incorporation of labeled Met into Ile would provide direct evidence of MGL function in intact plants. Since MGL shows high expression in desiccation-stressed tissue (Fig. 3; Supplemental Fig. S5), these conditions were used for labeling experiments. [13C5 15N]Met was fed to wild-type Col-0, mgl-2, and icl-2 flower stalks for 24 h. Subsequently, siliques and flowers were harvested and dehydrated for another 18 h before extracting amino acids.
During Ile biosynthesis from [13C5 15N]Met, the 15N label is lost as 15NH3 due to deamination by MGL, one 13C atom is incorporated into methanethiol, and the four remaining 13C atoms end up in Ile (pathway in Supplemental Fig. S7). The Ile content in flowers and siliques of Col-0 and mgl-2 labeled with [13C5 15N]Met was compared by mass spectrometry. After derivatization with Waters AccQ-tag reagent, native Ile has a mass-to-charge ratio (m/z) of 302, and Ile derived from [13C5 15N]Met has an expected m/z of 306. [13C5 15N]Met-labeled Col-0 flowers and siliques show significantly more m/z 306 ion than those of mgl-2 mutants (Fig. 5). The m/z 306 ion in [13C5 15N]Met-labeled mgl-2 mutant tissue was almost undetectable (Supplemental Figs. S8 and S9), consistent with only the Arabidopsis gene encoding MGL. In addition, there was no significant difference in the recovery of labeled Ile from wild-type Col-0 and icl-2 mutants, which have an elevated expression of MGL (Cornah et al., 2004). Together, these data provide evidence that MGL contributes to Ile biosynthesis in desiccated Arabidopsis tissue.
Figure 5.
13C incorporation from Met into Ile. Flower stalks of 30-d-old plants of Col-0, mgl-2, and icl-2 plants were labeled with [13C5 15N]Met for 24 h. Siliques and flowers were harvested and dehydrated for 18 h before extracting amino acids. [13C4]Ile recovered from siliques and flowers was measured by HPLC-mass spectrometry. Mean ± sd of n = 3. *, P < 0.05 relative to wild-type Col-0, Student's t test.
OMR1 Knockdown Mutation Reduces Branched-Chain Amino Acid Accumulation in Seeds and Flowers
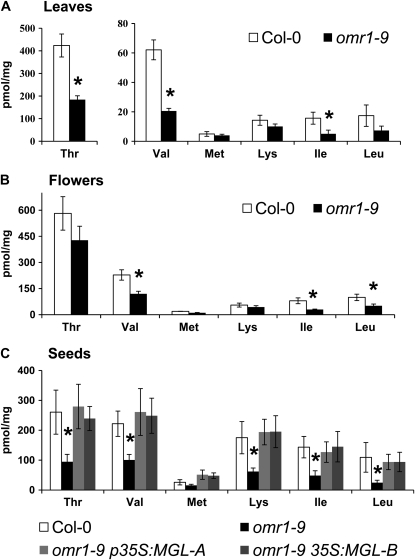
Thr deaminase, the first enzyme in the Ile synthesis, is considered indispensible for normal plant growth. An auxotrophic Nicotiana plumbaginifolia mutant was rescued with exogenous Ile and by complementation with yeast Thr deaminase (ILV; Colau et al., 1987). In another report, Nicotiana attenuata plants with silenced gene expression and no detectable Thr deaminase activity were severely stunted (Kang et al., 2006). We were unable to confirm any homozygous or heterozygous mutations in Arabidopsis lines predicted to have insertions in the coding region of OMR1 (SAIL_32_BO2 and WiscDsLox36707_043; Sessions et al., 2002; Woody et al., 2007). However, we found homozygous T-DNA insertions located 110 and 78 bp upstream from the OMR1 start codon in the T-DNA lines SAIL_609_GO3 (omr1-9; Sessions et al., 2002) and SGT 576 (omr1-10, Landsberg erecta background; Parinov et al., 1999), respectively (Supplemental Fig. S10A). Both insertions cause a bushy and stunted appearance (Supplemental Fig. S10B), the plants flower late, and seed set is reduced compared to the wild type. Spraying mutant plants at 3-d intervals up to an age of 4 weeks with 1 and 4 mm of either Ile or Met did not rescue these visible mutant phenotypes. Leaves and siliques of omr1-9 plants have lower levels of OMR1 transcript than the wild-type control, and no changes in the transcript accumulation of MGL, THA1, and THA2 were detected (Supplemental Fig. S10C). Amino acid analysis of flowers, leaves, and seeds showed that the omr1-9 mutant accumulated significantly lower amounts of all branched-chain amino acids (Fig. 6), suggesting reduced Thr deaminase activity. Somewhat surprisingly, Thr levels in mutant leaves and seeds were also significantly reduced. Further evidence of reduced Ile biosynthesis in omr1-9 mutants comes from the observation that mutant seedlings exhibit a greater tolerance of 1 mm Ile in MS medium than wild-type Col-0 (Supplemental Fig. S11).
Figure 6.
Amino acid profiles of a Thr deaminase knockdown mutant. Amino acid abundance in leaves (A) and flowers (B) of the wild type and omr1-9. Mean ± sd of n = 4 to 6. *, P < 0.05, Student's t test. C, Rescue of omr1-9 seed amino acid phenotypes by MGL overexpression. Lines omr1-9 MGL-A and omr1-9 MGL-B are independent p35S:MGL transformations. Mean ± sd of n =3. Only amino acids with significant changes in abundance, as well as Met, are shown. *, P < 0.05 relative to wild-type Col-0, Student's t test.
Despite the fact that MGL mRNA levels are higher in mature seeds than other plant tissues (Supplemental Fig. S4), there were significant amino acid deficits in the omr1-9 mutant (Fig. 6C). To determine whether this is due to (1) MGL and OMR1 contributing to different Ile pools or (2) insufficient MGL activity to maintain normal Ile levels, MGL was constitutively expressed under the 35S promoter in the omr1-9 mutant background. In two independent omr1-9 p35S:MGL isolates, MGL overexpression fully rescued the omr1-9 amino acid deficits (Fig. 6C). In control experiments, we also rescued omr1-9 plants with a construct expressing a feedback-insensitive omr1-1 allele (Mourad and King, 1995). As expected, seed amino acid analysis from these plants showed significantly increased levels of Ile and other branched-chain amino acids compared to both the omr1-9 mutant and wild-type Col-0 (Supplemental Fig. S12). Together, these results confirm functionally overlapping roles of MGL and OMR1 in Ile biosynthesis but suggest that MGL is of secondary importance under normal growth conditions.
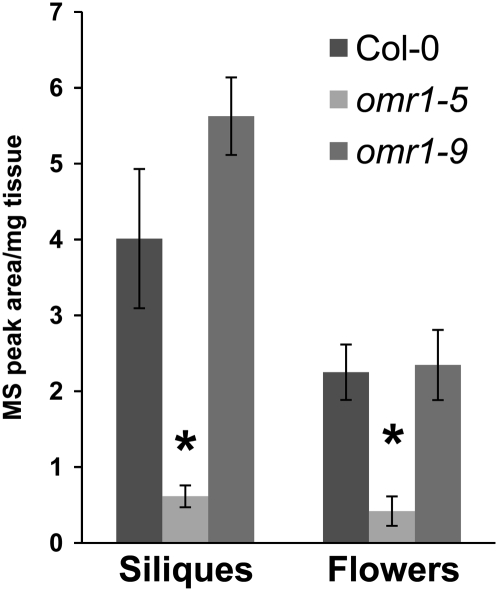
The [13C5 15N]Met assay described above was used to monitor incorporation of label from Met into Ile in omr1-9 and omr1-5 plants. The incorporation of label in the knockdown mutant (omr1-9) was not significantly different from the wild type (Fig. 7). However, in the feedback-insensitive Thr deaminase mutant (omr1-5), which has elevated Ile levels, the accumulation of labeled Ile (m/z 306) is reduced to about 15% of wild-type levels in siliques and flowers (Fig. 7). This result is consistent with the reduced MGL transcription in the omr1-5 mutant background (Fig. 3; Supplemental Fig. S5).
Figure 7.
Met incorporation into Ile in Thr deaminase mutants. Flower stalks of Thr deaminase feedback-insensitive (omr1-5) and knockdown (omr1-9) mutants were labeled with [13C5 15N]Met. Accumulation of [13C4]Ile was measured in siliques and flowers. Mean ± sd of n = 3. *, P < 0.05, relative to wild-type Col-0, Student's t test.
DISCUSSION
Our results provide evidence of an in vivo role for MGL in Arabidopsis Ile biosynthesis. MGL and OMR1 activities appear to be coordinately regulated by the Ile needs of the plant. Arabidopsis accumulating excess free Ile due to a feedback-insensitive omr1-5 mutation show reduced incorporation of carbon from Met into Ile (Fig. 7). Although this is likely due to greatly reduced MGL transcription in this mutant (Fig. 3), we cannot rule out additional regulation of MGL activity at the level of translation or due to feedback inhibition by Ile or other downstream metabolites under these conditions. Arabidopsis with elevated Thr levels, either due to the overexpression of Thr synthase (Avraham and Amir, 2005) or lack of Thr aldolase (Joshi et al., 2006), do not necessarily accumulate excess Ile. Although feedback inhibition of OMR1 (Mourad and King, 1995) probably contributes to this lack of Ile accumulation, some additional regulation of OMR1 activity may occur at the level of transcription. In particular, OMR1 transcription is up-regulated in response to desiccation (Fig. 3B).
Although both Met and Thr serve as precursors for Ile biosynthesis in Arabidopsis, the existence of alternative plant metabolic pathways leading to Ile formation in plants cannot be ruled out at this point. Precursors for Thr- and Met-independent Ile biosynthesis in microorganisms include Glu, 2-methylbutyrate, propionate, homolanthionine, and citramalate (Phillips et al., 1972; Kisumi et al., 1977; Monticello et al., 1984; Hochuli et al., 1999; Xu et al., 2004; Krömer et al., 2006; Risso et al., 2008). Metabolite profiling experiments have shown the presence of citramalate in Arabidopsis (Fiehn et al., 2000; Roessner-Tunali et al., 2003), and it has therefore been proposed that citramalate may also serve as a precursor for Ile biosynthesis in plants (de Kraker et al., 2007).
The experiments in Figures 4B, 6, and Supplemental Figure S12 show coregulation of Ile, Leu, and Val production. Under conditions where Ile is expected to be up- or down-regulated, similar effects are seen on Leu and Val accumulation. Although such tight control over branched-chain amino acid biosynthesis in plants is commonly observed in studies involving biosynthetic mutations or osmotic stress, the mechanism of this regulation has not yet been identified. However, since the Ile, Leu, and Val biosynthetic pathways share four enzymes, biosynthesis could be coordinately regulated by either feedback inhibition or at the level of transcription.
Accumulation of Pro and branched-chain amino acids is commonly observed in plants subjected to osmotic stress (Rhodes et al., 1986; Shen et al., 1989; Girousse et al., 1996; Nambara et al., 1998; Székely et al., 2008). In addition to de novo synthesis, these elevated amino acid pools could result from reduced protein synthesis (Good and Zaplachinski, 2006) or general protein breakdown during drought stress. However, Less and Galili (2008) studied the expression of all annotated Arabidopsis proteases in response to drought and other abiotic stress and came to the conclusion that protein breakdown does not contribute significantly to amino acid accumulation.
Elevated free amino acids are hypothesized to act as osmolytes that protect plant cells against dehydration. Research to increase osmotic tolerance in plants through regulation of amino acid biosynthesis has been focused primarily on Pro (Hong et al., 2000). However, our results showing elevated salt tolerance in omr1-5 mutants (Fig. 4A) suggest that it may also be beneficial to manipulate the biosynthesis of branched-chain amino acids. Although MGL contributes to increased Ile biosynthesis under osmotic stress, we were not able to find changes in drought or salt tolerance in mgl mutants (Fig. 4A). This absence of a phenotype could be due to a compensatory increase in OMR1 transcription (Fig. 3) and therefore increased synthesis of Ile from Thr in desiccated mgl mutant tissue. Nevertheless, MGL overproduction, if it has similar effects as feedback insensitive mutations in OMR1 (Fig. 4A), could provide a mechanism for increasing salt or drought tolerance in plants.
Several studies have indicated transcriptional regulation of MGL in response to drought, salt, and other abiotic stress (Rizhsky et al., 2004; Peng et al., 2007; Baena-González et al., 2008; Less and Galili, 2008; Mueller et al., 2008). However, although our semiquantitative RT-PCR experiments suggested desiccation-induced MGL up-regulation (Supplemental Fig. S5), we were unable to reject the null hypothesis (no difference between drought-stressed and unstressed wild-type flowers) under our growth conditions using quantitative RT-PCR experiments (Fig. 3A). The observation that there is nevertheless a significant decrease in Ile content in drought-stressed mgl-2 mutant flowers relative to the wild type (Fig. 4B) is consistent with the previously proposed hypothesis of MGL posttranscriptional regulation in Arabidopsis (Rebeille et al., 2006). Moreover, the relatively small (<50%) decrease in Ile accumulation in mgl-2 mutant flowers (Fig. 4B) suggests that MGL is of lesser importance than OMR1 for drought-induced production of Ile.
The results in Figure 2 indicate a complex regulation of Met biosynthesis and degradation that cannot be fully explained by our current knowledge of these pathways. However, the observed effects on transcription, in particular down-regulation of Met synthases and CGS in mgl-2 flowers, suggest a broader role of MGL in regulating Met metabolism. To date, not much is known about the transcriptional regulation of Met synthase in plants. However, CGS in Arabidopsis is transcriptionally and posttranscriptionally regulated by Met or one of its metabolites (Hesse et al., 2004).
There are potential practical applications to manipulating MGL activity in crop plants. For instance, since Met is considered a limiting essential amino acid in potatoes, there have been several attempts to increase potato Met accumulation by altering the activity of biosynthetic enzymes. So far, this has met with only limited success. Increasing the expression of homoserine kinase did not increase Met levels (Rinder et al., 2008). Knockdown of Thr synthase greatly increased Met levels but caused severe phenotypic defects (Zeh et al., 2001). Increased production of CGS caused only moderate increases in tuber Met accumulation (Di et al., 2003; Kreft et al., 2003; Dancs et al., 2008), perhaps due to elevated Met catabolism in the transgenic lines. If this is the case, silencing MGL expression in potato lines with elevated Met biosynthesis might further increase accumulation of this essential amino acid. A similar approach, increased biosynthesis combined with reduced catabolism, has been used to successfully increase Lys levels 80-fold in Arabidopsis and 40-fold maize (Zhu and Galili, 2003; Frizzi et al., 2008).
The results presented here show that OMR1 and MGL have partially overlapping functions in Ile biosynthesis. Although both MGL and OMR1 transcription is regulated by the Ile needs of the plant, our results do not rule out further regulation of these enzyme activities through translation or allosteric mechanisms in Arabidopsis. Given that, unlike omr1 knockdown mutants, mgl knockout mutants have no visible phenotypes or growth defects, Thr deaminase activity likely predominates during Ile biosynthesis in vegetative tissue. However, in reproductive tissue and under osmotic stress conditions, biosynthesis of Ile from Met can play a significant role in Arabidopsis amino acid metabolism.
MATERIALS AND METHODS
Plant Material
Wild-type Arabidopsis (Arabidopsis thaliana) Col-0 seeds, as well as T-DNA insertion lines SALK_103805 (mgl-2; CS16492), SAIL_609_GO3 (CS16487), and SAIL_32_BO2 (CS16488; Sessions et al., 2002; Alonso et al., 2003), gene trap line SGT_576 (CS16489; Parinov et al., 1999), and WiscDsLox36707_043 (CS16490; Woody et al., 2007), were obtained from the Arabidopsis Biological Resource Center (http://www.arabidopsis.org). Other seeds were kindly supplied to us by Y. Shachar-Hill, Michigan State University (mgl-1; SALK_0400380; CS16491), J. Cornah, University of Edinburgh (icl-2), and G. Mourad, Indiana-Purdue University (omr1-5 mutant and p35S-omr1 transgenics).
Growth Conditions
Plants were grown in Cornell Mix with Osmocoat fertilizer (Landry et al., 1995) in 20- × 40-cm nursery flats in Conviron growth chambers. Photosynthetic photon flux density was 200 μmol m−2 s−1, the photoperiod was 16:8 day:night, the temperature was 23°C, and the relative humidity was 50%. Seeds were cold-stratified at 4°C in 0.1% Phytagar (Gibco BRL Life Technologies) for 3 d. For growth on agar plates, 1× MS medium (Murashige and Skoog, 1962) with or without 1% Suc was used, and the plates were placed the same growth chambers as the soil-grown plants. For the salt tolerance experiment, mutant and wild-type seeds were surface sterilized with a 1 min wash of 70% ethanol, followed by 50% bleach and 0.01% Triton X-100 for 10 min. Seeds were then rinsed four to five times with sterilized water, and seeds were suspended in 0.1% sterile phytagar. Sterilized seeds were placed on MS medium with 0, 50, 100, or 150 mm of NaCl and 500 μm and 1 mm Ile. Plates were placed vertically to measure the root lengths after 2 weeks.
For experiments to rescue the omr1-9 phenotype, 10-d-old plants were sprayed with 1 or 4 mm Met and Ile at 3-d intervals up to an age of 4 weeks. For the drought tolerance experiment, the wild type and mutants were grown in the same flat, and irrigation to 30-d-old plants was withheld for 6 to 10 d, until complete wilting. For salt tolerance tests in soil, 2- and 4-week-old plants were irrigated with 50 and 100 mm NaCl for 3 to 4 d. For dehydration experiments, tissue was detached from 30-d-old plants and dehydrated on paper towels for 18 h under ambient humidity. Plant tissue was weighed before and after the dehydration treatments.
Amino Acid Analysis by HPLC
For free amino acid analysis, plant tissue was preweighed and was frozen in liquid nitrogen along with 3-mm steel balls (Abbott Ball Company) and was homogenized using a Harbil 5G-HD paint shaker (Fluid Management). Amino acids levels were analyzed as described previously (Joshi et al., 2006) using an AccQ-fluor reagent kit (Waters). Aliquots (10 μL) were used for HPLC analysis using a Waters 2790 separation module. Separation was performed on 3.9- × 150-mm AccQ-Tag column (Waters) at 38°C, and amino acids were detected using a Waters 2487 dual-wavelength absorbance detector at 280 nm and a Waters 2475 photodiode array detector with excitation at 280 nm and emission at 395 nm. Eluent A (sodium acetate and trimethylamine, pH 5.04; Waters) and solution B (60% acetonitrile:40% water) were used at a 1 mL min−1 flow rate with the following gradient: 0 to 0.01 min, 100% A; 0.01 to 5.0 min, linear gradient to 97% A, 3% B; 5 to 12 min, to 95% A, 5% B; 12 to 15 min, 92% A, 8% B; 15 to 45 min, 65% A, 35% B; 45 to 49 min, to 65% A, 35% B; 49 to 50 min, to 100% B; 50 to 60 min, to 100% B, 61 to 68 min, 100% A. Standard curves were prepared using amino acids purchased from Sigma-Aldrich. Amino acid concentrations were calculated by comparing peak areas to those of standard curves and normalized using l-norleucine as an internal control.
Isotope Labeling and Liquid Chromatography-Mass Spectrometry Analysis
For stable isotope experiments l-[13C5 15N]Met was purchased from Cambridge Isotope Laboratories. Flower stalks of 30-d-old plants were cut above the rosette leaves, and the bases of the stalks were inserted into 15-mL polypropylene tubes containing 1 mm of labeled Met in water and were left for 24 h under continuous light at 23°C. At the end of the treatment, flowers and siliques were harvested and dehydrated for another 18 h. Amino acids were extracted in 40% methanol and evaporated completely under vacuum. The residue was resuspended in a proportion based upon tissue amount in 20 mm HCl and derivatized with AccQ-Flour reagent, and samples were separated by HPLC as described above. Solutions A (100 mm sodium acetate, pH 5.04) and B (60% acetonitrile:40% water) were used at a 0.3 mL min−1 flow rate with following gradient: 0 to 0.01 min 100% A, 0.01 to 0.5 min, 98% A; 0.5 to 20 min, linear gradient to 94.5% A, 5.5% B; 20 to 24 min, to 85% A, 15% B; 24 to 60 min, to 75% A, 25% B; 60 to 62 min, 100% B, 62 to 65 100% B, and 65 to 75 min, 100% A. For HPLC-mass spectrometry analysis, conditions described previously (Lee et al., 2008) were used with some modifications. Outflow from the column was split and directed into a Varian 1200 L triple quadrupole mass spectrometer with an electrospray ionization source at a 300 μL min−1 flow rate. The mass spectrometry was controlled using MS Workstation version 6 (Varian) and set up using the following conditions: drying gas (N2) 185°C and 19 p.s.i., capillary voltage 30 V, needle voltage 5,000 V, housing temperature 50°C, main gas pressure 66 p.s.i., and nebulizing gas pressure 56 p.s.i.
Verification of T-DNA Insertions in MGL and OMR1
For identification of T-DNA insertions, DNA was extracted from mutants and Col-0 (Ausubel et al., 1998; Weigel and Glazebrook, 2002). Sequences of oligonucleotide primers used in this study are listed in Supplemental Table S1. The T-DNA insertion in line SALK_103805 (mgl-2) was confirmed using gene-specific primers (MGL-2RP and MGL-2LP) and the insert-specific primer LBa1. For Thr deaminase T-DNA insertions, the following primer pairs were used: for SAIL_609_GO3, gene-specific primers S609LP and S609RP and vector-specific primer and Lb3; for gene trap line SGT 576, gene-specific primers SGT LP and SGT RP and vector-specific primer SGT DS3.2; for WiscDsLox36707_043, WISC LP and WISC RP and vector-specific primer WISC745. T-DNA insertions in SAIL lines growing on soil were selected by spraying with 300 μm glufosinate ammonium (Finale; Farnam Companies). For SALK T-DNA insertion lines, seedlings were selected on MS medium supplemented with 25 to 30 μg/mL kanamycin.
Transcription Analysis Using RT-PCR
Total RNA was extracted using the RNeasy plant mini kit (Qiagen) and RNase-free DNase (Qiagen) according to the manufacturer's protocol. The reverse transcription reaction was performed using 2 μg of total RNA to synthesize cDNA with an oligo(dT)15 primer (Promega) and SuperScript III reverse transcriptase (Invitrogen). PCR was conducted with 1 μL of RT reaction at 94°C for 30 s, 56°C for 30 s, and 72°C for 50 s. The number of cycles was 27 in the drought experiment (Fig. 2) and 25 in the omr1-9 experiment (Supplemental Fig. S6). To check the transcript abundance, gene-specific primers were designed spanning an intron to reduce the possibility of DNA contamination. The following pairs of primers were used: MGL (MGL R1 and MGL R2), OMR1 (OMR1 R1 and OMR1 R2), ICL (ICL R1 and ICL R2), THA1 (THA1 R1 and THA1 R2), and THA2 (THA2 R1 and THA2 R2). Expression levels of genes of interest were compared to a control gene UBQ10, which was amplified using primers UBQ1 and UBQ2. The DNA sequences of all primers are listed in Supplemental Table S1.
Quantitative Real-Time RT-PCR Analysis
For quantitative real-time RT-PCR analysis, total RNA was isolated using the Qiagen RNeasy kit according to the manufacturer's instructions and was on-column treated with RNAse-free DNase (Qiagen). One microgram of DNA-free RNA was then reverse transcribed with a 2.5 μm oligo(dT)15 primer (Promega) and SMART MMLV reverse transcriptase (Clontech) following the manufacturer's recommendations. Samples were incubated at 42°C for 90 min, and reaction was terminated by heating at 70°C for 15 min. Real-time quantitative PCR was performed in an optical 384-well plate with an ABI PRISM 7900 HT sequence detection system (Applied Biosystems) and SYBR Green qPCR SuperMix (Applied Biosystems). The final PCR reaction volume was 15 μL and comprised 1 μL of reverse transcribed cDNA, 200 nm of each gene-specific primer, 7.5 μL of 2× SYBR Green Master Mix reagent (Applied Biosystems), and water. The PCR gradient was set as follows: 50°C for 2 min, 95°C for 10 min, 40 cycles of 95°C for 30 s, 56°C for 30 s, and 72°C for 30 s. Experiments were set and analyzed using the SDS 2.1 software (Applied Biosystems).
To obtain expression levels of Met synthases, CGS, and AdoMet synthases, total RNA extraction was carried out from flowers and siliques of 30-d-old wild-type (Col-0) and mgl-2 plants. The following pairs of gene-specific primers were used for this experiment: QMS P1, QMS P2 (ATMS1; At5g17920), QMS2 P1, QMS2 P2 (ATMS2; At3g03780), QMS3 P1, QMS3 P2 (ATMS3; At5g20980), QCGS P1, QCGS P2 (CGS; At3g01120), QSAM1 P1, QSAM1 P2 (SAM-1; At1g02500), QSAM2 P1, ASAM2 P2 (SAM-2; At4g01850), QSAM3 P1, QSAM3 P2 (SAM-3; At3g17390), and QSAM4 P1 and QSAM4 P2 (SAM-4; At2g36880) (Supplemental Table S2). Ct values for all the genes of interest were normalized by subtracting the mean of reference gene UBQ10 (At4g05320) amplified using as gene-specific primers QUBQ10-L and QUBQ10-R as described by Czechowski et al. (2004, 2005). The expression level of each gene was calculated as the relative amount compared to the wild-type control using the formula: expression level = 2−(ΔCt (target) − ΔCt (control)), as recommended by the instrument manufacturer. For the drought tolerance experiment, tissue collection and total RNA extraction from the wild type (Col-0), mgl-2, and omr1-5 was carried out as described above. Primer pairs QMGL P1 and QMGL P2 and QOMR P1 and QOMR P2 were used to amplify the endogenous MGL and OMR1 transcripts, respectively. The fold change of transcript accumulation in dehydrated tissue compared to fresh tissue in each sample was calculated based on the efficiency calibrated model (Yuan et al., 2006) and a previously described equation (Pfaffl, 2001).
MGL Cloning and Complementation by Plant Transformation
An MGL open reading frame/cDNA clone (stock no. 3G15166; The Arabidopsis Information Resource accession no. 3510684848) in the entry vector pENTR223 was obtained from the Arabidopsis Biological Resource Center. Gene integrity was confirmed by DNA sequencing using M13 F and R universal primer pairs. The cloned DNA was transferred to destination vector pMDC32 using the LR reaction as in the manufacturer's instructions (Invitrogen), and colonies showing kanamycin resistance were selected. Sequencing was repeated to confirm the ends using pMDC32-specific and gene-specific primers (PMDC3235S, MGL SEQ, and PMDC32NOS; Supplemental Table S1), and sequencing data were analyzed using Lasergene 6 software (DNAstar). The binary vector was then transformed into Agrobacterium tumefaciens strain GV3101 and subsequently used to transform SAIL_609_GO3 (omr1-9) plants (Clough and Bent, 1998). T1 plants were screened on MS agar plates for hygromycin resistance (20–100 μg/μL), and green, healthy plants were transferred to soil.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NM_105141 (At1g64660) and NM_111840 (At3g10050).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Identification of a homozygous mgl-2 T-DNA insertion.
Supplemental Figure S2. Tissue-specific expression patterns of Met synthases and Thr deaminase.
Supplemental Figure S3. Pathway showing proteins involved in Arabidopsis Met metabolism.
Supplemental Figure S4. Tissue-specific expression patterns of MGL and AdoMet synthases.
Supplemental Figure S5. MGL and OMR1 transcription under drought stress.
Supplemental Figure S6. Controls for salt sensitivity experiment and amino acid accumulation in drought-stressed leaves.
Supplemental Figure S7. Possible pathway of label incorporation in Ile after feeding the plant tissue [13C5 15N]Met.
Supplemental Figure S8. Detection of isotopically labeled Ile in wild-type and mgl siliques.
Supplemental Figure S9. Detection of isotopically labeled Ile in wild-type and mgl flowers.
Supplemental Figure S10. omr1-9 mutant phenotypes and expression profiling.
Supplemental Figure S11. Ile tolerance of wild-type Col-0 and omr1-9.
Supplemental Figure S12. Rescue of omr1-9 amino acid deficits by omr1-1 overexpression.
Supplemental Table S1. Sequences of oligonucleotide primers.
Supplemental Table S2. Sequences of oligonucleotide primers used for quantitative PCR.
Supplementary Material
Acknowledgments
We thank Y. Shachar-Hill for the mgl-1 mutant, J. Cornah for the icl-2 mutant, and G. Mourad for omr1 mutants and plasmid constructs.
This work was supported by the Binational Agricultural Research and Development Fund (grant no. US–3910–06) and the National Science Foundation (grant nos. MCB–0416567 and DBI–050050).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Georg Jander (gj32@cornell.edu).
The online version of this article contains Web-only data.
Open access articles can be viewed online without a subscription.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (1998) Current Protocols in Molecular Biology. John Wiley and Sons, Inc., Hoboken, NJ
- Avraham T, Amir R (2005) The expression level of threonine synthase and cystathionine-gamma-synthase is influenced by the level of both threonine and methionine in Arabidopsis plants. Transgenic Res 14 299–311 [DOI] [PubMed] [Google Scholar]
- Baena-González E, Rolland F, Sheen J (2008) KIN10/11 are master regulators of the convergent stress transcriptome. In JF Allen, E Gantt, JH Golbeck, B Osmond, eds, Photosynthesis. Energy from the Sun: 14th International Congress on Photosynthesis. Springer, The Netherlands, pp 1331–1337
- Bartlem D, Lambein I, Okamoto T, Itaya A, Uda Y, Kijima F, Tamaki Y, Nambara E, Naito S (2000) Mutation in the threonine synthase gene results in an over-accumulation of soluble methionine in Arabidopsis. Plant Physiol 123 101–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba Y, Ishikawa M, Kijima F, Tyson RH, Kim J, Yamamoto A, Nambara E, Leustek T, Wallsgrove RM, Naito S (1999) Evidence for autoregulation of cystathionine gamma-synthase mRNA stability in Arabidopsis. Science 286 1371–1374 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Colau D, Negrutiu I, Van Montagu M, Hernalsteens JP (1987) Complementation of a threonine dehydratase-deficient Nicotiana plumbaginifolia mutant after Agrobacterium tumefaciens-mediated transfer of the Saccharomyces cerevisiae ILV1 gene. Mol Cell Biol 7 2552–2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornah JE, Germain V, Ward JL, Beale MH, Smith SM (2004) Lipid utilization, gluconeogenesis, and seedling growth in Arabidopsis mutants lacking the glyoxylate cycle enzyme malate synthase. J Biol Chem 279 42916–42923 [DOI] [PubMed] [Google Scholar]
- Curien G, Laurencin M, Robert-Genthon M, Dumas R (2007) Allosteric monofunctional aspartate kinases from Arabidopsis. FEBS J 274 164–176 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Bari RP, Stitt M, Scheible WR, Udvardi MK (2004) Real-time RT-PCR profiling of over 1400 Arabidopsis transcription factors: unprecedented sensitivity reveals novel root- and shoot-specific genes. Plant J 38 366–379 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dancs G, Kondrak M, Banfalvi Z (2008) The effects of enhanced methionine synthesis on amino acid and anthocyanin content of potato tubers. BMC Plant Biol 8 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kraker JW, Luck K, Textor S, Tokuhisa JG, Gershenzon J (2007) Two Arabidopsis genes (IPMS1 and IPMS2) encode isopropylmalate synthase, the branchpoint step in the biosynthesis of leucine. Plant Physiol 143 970–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di R, Kim J, Martin MN, Leustek T, Jhoo J, Ho CT, Tumer NE (2003) Enhancement of the primary flavor compound methional in potato by increasing the level of soluble methionine. J Agric Food Chem 51 5695–5702 [DOI] [PubMed] [Google Scholar]
- Dias B, Weimer B (1998) Purification and characterization of L-methionine gamma-lyase from Brevibacterium linens BL2. Appl Environ Microbiol 64 3327–3331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faleev NG, Troitskaya MV, Paskonova EA, Saporovskaya MB, Belikov VM (1996) L-Methionine-lyase in Citrobacter intermedius cells: stereochemical requirements with respect to the thiol structure. Enzyme Microb Technol 19 590–593 [Google Scholar]
- Fiehn O, Kopka J, Trethewey RN, Willmitzer L (2000) Identification of uncommon plant metabolites based on calculation of elemental compositions using gas chromatography and quadrupole mass spectrometry. Anal Chem 72 3573–3580 [DOI] [PubMed] [Google Scholar]
- Frizzi A, Huang S, Gilbertson LA, Armstrong TA, Luethy MH, Malvar TM (2008) Modifying lysine biosynthesis and catabolism in corn with a single bifunctional expression/silencing transgene cassette. Plant Biotechnol J 6 13–21 [DOI] [PubMed] [Google Scholar]
- Giovanelli J, Mudd SH, Datko AH (1985) Quantitative analysis of pathways of methionine metabolism and their regulation in Lemna. Plant Physiol 78 555–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girousse C, Bournoville R, Bonnemain JL (1996) Water deficit-induced changes in concentrations in proline and some other amino acids in the phloem sap of alfalfa. Plant Physiol 111 109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good AG, Zaplachinski ST (2006) The effects of drought stress on free amino acid accumulation and protein synthesis in Brassica napus. Physiol Plant 90 9–14 [Google Scholar]
- Goyer A, Collakova E, Shachar-Hill Y, Hanson AD (2007) Functional characterization of a methionine gamma-lyase in Arabidopsis and its implication in an alternative to the reverse trans-sulfuration pathway. Plant Cell Physiol 48 232–242 [DOI] [PubMed] [Google Scholar]
- Hacham Y, Matityahu I, Schuster G, Amir R (2008) Overexpression of mutated forms of aspartate kinase and cystathionine gamma-synthase in tobacco leaves resulted in the high accumulation of methionine and threonine. Plant J 54 260–271 [DOI] [PubMed] [Google Scholar]
- Hesse H, Hoefgen R (2003) Molecular aspects of methionine biosynthesis. Trends Plant Sci 8 259–262 [DOI] [PubMed] [Google Scholar]
- Hesse H, Kreft O, Maimann S, Zeh M, Hoefgen R (2004) Current understanding of the regulation of methionine biosynthesis in plants. J Exp Bot 55 1799–1808 [DOI] [PubMed] [Google Scholar]
- Hochuli M, Patzelt H, Oesterhelt D, Wuthrich K, Szyperski T (1999) Amino acid biosynthesis in the halophilic archaeon Haloarcula hispanica. J Bacteriol 181 3226–3237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z, Lakkineni K, Zhang Z, Verma DP (2000) Removal of feedback inhibition of delta(1)-pyrroline-5-carboxylate synthetase results in increased proline accumulation and protection of plants from osmotic stress. Plant Physiol 122 1129–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori H, Takabayashi K, Orvis L, Carson DA, Nobori T (1996) Gene cloning and characterization of Pseudomonas putida L-methionine-alpha-deamino-gamma-mercaptomethane-lyase. Cancer Res 56 2116–2122 [PubMed] [Google Scholar]
- Inaba K, Fujiwara T, Hayashi H, Chino M, Komeda Y, Naito S (1994) Isolation of an Arabidopsis thaliana mutant, mto1, that overaccumulates soluble methionine (temporal and spatial patterns of soluble methionine accumulation). Plant Physiol 104 881–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue H, Inagaki K, Sugimoto M, Esaki N, Soda K, Tanaka H (1995) Structural analysis of the L-methionine gamma-lyase gene from Pseudomonas putida. J Biochem 117 1120–1125 [DOI] [PubMed] [Google Scholar]
- Joshi V, Laubengayer KM, Schauer KM, Fernie A, Jander G (2006) Two Arabidopsis threonine aldolases are non-redundant and compete with threonine deaminase for a common substrate pool. Plant Cell 18 3564–3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JH, Wang L, Giri A, Baldwin IT (2006) Silencing threonine deaminase and JAR4 in Nicotiana attenuata impairs jasmonic acid-isoleucine-mediated defenses against Manduca sexta. Plant Cell 18 3303–3320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisumi M, Komatsubara S, Chibata I (1977) Pathway for isoleucine formation form pyruvate by leucine biosynthetic enzymes in leucine-accumulating isoleucine revertants of Serratia marcescens. J Biochem 82 95–103 [DOI] [PubMed] [Google Scholar]
- Kreft O, Hoefgen R, Hesse H (2003) Functional analysis of cystathionine gamma-synthase in genetically engineered potato plants. Plant Physiol 131 1843–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krömer JO, Heinzle E, Schröder H, Wittmann C (2006) Accumulation of homolanthionine and activation of a novel pathway for isoleucine biosynthesis in Corynebacterium glutamicum McbR deletion strains. J Bacteriol 188 609–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry LG, Chapple CC, Last RL (1995) Arabidopsis mutants lacking phenolic sunscreens exhibit enhanced ultraviolet-B injury and oxidative damage. Plant Physiol 109 1159–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Toro-Ramos T, Huang H, Fraga M, Last RL, Jander G (2008) Reduced activity of Arabidopsis thaliana HMT2, a methionine biosynthetic enzyme, increases seed methionine content. Plant J 54 310–320 [DOI] [PubMed] [Google Scholar]
- Less H, Galili G (2008) Principal transcriptional programs regulating plant amino acid metabolism in response to abiotic stresses. Plant Physiol 147 316–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis AJ, Barnes MB, Grosbach DA, Peo ER Jr (1982) Sequence in which the amino acids on corn (Zea mays) become limiting for growing rats. J Nutr 112 782–788 [DOI] [PubMed] [Google Scholar]
- Manukhov IV, Mamaeva DV, Rastorguev SM, Faleev NG, Morozova EA, Demidkina TV, Zavilgelsky GB (2005) A gene encoding L-methionine gamma-lyase is present in Enterobacteriaceae family genomes: identification and characterization of Citrobacter freundii L-methionine gamma-lyase. J Bacteriol 187 3889–3893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKie AE, Edlind T, Walker J, Mottram JC, Coombs GH (1998) The primitive protozoon Trichomonas vaginalis contains two methionine gamma-lyase genes that encode members of the gamma-family of pyridoxal 5′-phosphate-dependent enzymes. J Biol Chem 273 5549–5556 [DOI] [PubMed] [Google Scholar]
- Monticello DJ, Hadioetomo RS, Costilow RN (1984) Isoleucine synthesis by Clostridium sporogenes from propionate or alpha-methylbutyrate. J Gen Microbiol 130 309–318 [DOI] [PubMed] [Google Scholar]
- Mourad G, King J (1995) L-O-Methylthreonine-resistant mutant of Arabidopsis defective in isoleucine feedback regulation. Plant Physiol 107 43–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller S, Hilbert B, Dueckershoff K, Roitsch T, Krischke M, Mueller MJ, Berger S (2008) General detoxification and stress responses are mediated by oxidized lipids through TGA transcription factors in Arabidopsis. Plant Cell 20 768–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog FA (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15 473–497 [Google Scholar]
- Nambara E, Kawaide H, Kamiya Y, Naito S (1998) Characterization of an Arabidopsis thaliana mutant that has a defect in ABA accumulation: ABA-dependent and ABA-independent accumulation of free amino acids during dehydration. Plant Cell Physiol 39 853–858 [DOI] [PubMed] [Google Scholar]
- Parinov S, Sevugan M, Ye D, Yang WC, Kumaran M, Sundaresan V (1999) Analysis of flanking sequences from dissociation insertion lines: a database for reverse genetics in Arabidopsis. Plant Cell 11 2263–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng M, Bi YM, Zhu T, Rothstein SJ (2007) Genome-wide analysis of Arabidopsis responsive transcriptome to nitrogen limitation and its regulation by the ubiquitin ligase gene NLA. Plant Mol Biol 65 775–797 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29 e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips AT, Nuss JI, Moosic J, Foshay C (1972) Alternate pathway for isoleucine biosynthesis in Escherichia coli. J Bacteriol 109 714–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash J (1996) Rice bran proteins: properties and food uses. Crit Rev Food Sci Nutr 36 537–552 [DOI] [PubMed] [Google Scholar]
- Rebeille F, Jabrin S, Bligny R, Loizeau K, Gambonnet B, Van Wilder V, Douce R, Ravanel S (2006) Methionine catabolism in Arabidopsis cells is initiated by a gamma-cleavage process and leads to S-methylcysteine and isoleucine syntheses. Proc Natl Acad Sci USA 103 15687–15692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes D, Handa S, Bressan RA (1986) Metabolic changes associated with adaptation of plant cells to water stress. Plant Physiol 82 890–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinder J, Casazza AP, Hoefgen R, Hesse H (2008) Regulation of aspartate-derived amino acid homeostasis in potato plants (Solanum tuberosum L.) by expression of E. coli homoserine kinase. Amino Acids 34 213–222 [DOI] [PubMed] [Google Scholar]
- Risso C, Van Dien SJ, Orloff A, Lovley DR, Coppi MV (2008) Elucidation of an alternate isoleucine biosynthesis pathway in Geobacter sulfurreducens. J Bacteriol 190 2266–2274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizhsky L, Liang H, Shuman J, Shulaev V, Davletova S, Mittler R (2004) When defense pathways collide. The response of Arabidopsis to a combination of drought and heat stress. Plant Physiol 134 1683–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner-Tunali U, Hegemann B, Lytovchenko A, Carrari F, Bruedigam C, Granot D, Fernie AR (2003) Metabolic profiling of transgenic tomato plants overexpressing hexokinase reveals that the influence of hexose phosphorylation diminishes during fruit development. Plant Physiol 133 84–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saari LL, Coterman JC, Thill DC (1994) Resistance to acetolactate synthase inhibiting herbicides. In S Powles, J Holtum, eds, Herbicide Resistance in Plants: Biology and Biochemistry. Lewis Publishers, Boca Raton, FL, pp 81–139
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Foster JG, Orcutt DM (1989) Composition and distribution of free amino acids in flatpea (Lathyrus sylvestris L.) as influenced by water deficit and plant age. J Exp Bot 40 71–79 [Google Scholar]
- Singh BK, Shaner DL (1995) Biosynthesis of branched chain amino acids: from test tube to field. Plant Cell 7 935–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Székely G, Abrahám E, Cséplo A, Rigó G, Zsigmond L, Csiszar J, Ayaydin F, Strizhov N, Jásik J, Schmelzer E, et al (2008) Duplicated P5CS genes of Arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. Plant J 53 11–28 [DOI] [PubMed] [Google Scholar]
- Tokoro M, Asai T, Kobayashi S, Takeuchi T, Nozaki T (2003) Identification and characterization of two isoenzymes of methionine gamma-lyase from Entamoeba histolytica: a key enzyme of sulfur-amino acid degradation in an anaerobic parasitic protist that lacks forward and reverse trans-sulfuration pathways. J Biol Chem 278 42717–42727 [DOI] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J (2002) Arabidopsis: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Woody ST, Austin-Phillips S, Amasino RM, Krysan PJ (2007) The WiscDsLox T-DNA collection: an Arabidopsis community resource generated by using an improved high-throughput T-DNA sequencing pipeline. J Plant Res 120 157–165 [DOI] [PubMed] [Google Scholar]
- Xu H, Zhang Y, Guo X, Ren S, Staempfli AA, Chiao J, Jiang W, Zhao G (2004) Isoleucine biosynthesis in Leptospira interrogans serotype lai strain 56601 proceeds via a threonine-independent pathway. J Bacteriol 186 5400–5409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan JS, Reed A, Chen F, Stewart CN Jr (2006) Statistical analysis of real-time PCR data. BMC Bioinformatics 7 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeh M, Casazza AP, Kreft O, Roessner U, Bieberich K, Willmitzer L, Hoefgen R, Hesse H (2001) Antisense inhibition of threonine synthase leads to high methionine content in transgenic potato plants. Plant Physiol 127 792–802 [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Galili G (2003) Increased lysine synthesis coupled with a knockout of its catabolism synergistically boosts lysine content and also transregulates the metabolism of other amino acids in Arabidopsis seeds. Plant Cell 15 845–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol 136 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.