Abstract
SV40 transforms cells through the action of two oncoproteins, large T antigen and small t antigen. Small t antigen targets phosphatase PP2A, while large T antigen stimulates cell proliferation and survival by action on multiple proteins, including the tumor suppressors Rb and p53. Large T antigen also binds components of the transcription initiation complex and several transcription factors. We examined global gene expression in SV40-transformed mouse embryo fibroblasts, and in enterocytes obtained from transgenic mice. SV40 transformation alters the expression of approximately 800 cellular genes in both systems. Much of this regulation is observed in both MEFs and enterocytes and is consistent with T antigen action on the Rb-E2F pathway. However, the regulation of many genes is cell-type specific, suggesting that unique signaling pathways are activated in different cell types upon transformation, and that the consequences of SV40 transformation depends on the type of cell targeted.
Keywords: SV40, transformation, gene expression
INTRODUCTION
SV40 induces tumors when injected into newborn hamsters and is also capable of inducing neoplastic transformation in cultured cells and in transgenic mice (Ahuja et al., 2005). The transforming functions of SV40 are carried by two proteins, the 708 amino acid large T antigen (T antigen) and the 174 amino acid small t antigen (t antigen). Small t antigen contributes to transformation by altering cellular signal transduction pathways by binding to the PP2A holoenzyme and displacing its regulatory subunits (Pallas et al., 1990). In many cases, large T antigen is sufficient to induce cellular transformation and tumors in animals. T antigen has multiple transforming functions, each of which is linked to its interaction with specific cellular targets. Two key targets of T antigen are the tumor suppressors pRb and p53.
The Rb family of proteins (pRb, p107, p130) blocks progression of the cell cycle at the G1/S boundary by binding to and inhibiting the E2F family of transcription factors. The E2Fs control the expression of a group of genes involved in cell cycle progression and thus are required for DNA replication and progression through S phase. Under normal circumstances, Rb proteins are normally controlled by upstream signaling pathways that regulate their phosphorylation state and thus their ability to interact with E2F proteins. However, interaction with T antigen blocks the ability of Rb proteins to inhibit E2Fs. Consequently, E2F-dependent gene transcription is effected leading to S phase entry. One consequence of Rb protein inhibition is the activation of the p53 pathway (Pipas and Levine, 2001) which, in turn, results in the upregulation of a group of genes responsible for blocking progression through the cell cycle and for inducing apoptosis. T antigen also binds to p53 and blocks its ability to stimulate gene expression (Pipas and Levine, 2001). Thus, the net effect of T antigen action is to stimulate the expression of genes required for S phase, while blocking the transcription of genes involved in cell cycle arrest and cell death.
In addition to regulating gene expression through its actions on tumor suppressors, T antigen also interacts directly with the cellular transcription apparatus. T antigen binds TBP and a number of TBP-associated proteins and appears to function as a TAF itself (Damania and Alwine, 1996; Damania et al., 1998; Gruda et al., 1993; Johnston et al., 1996). Consequently T antigen is a promiscuous activator of many promoters (Gilinger and Alwine, 1993; Rice and Cole, 1993). In addition, T antigen has been reported to interact with a number of specific transcription factors such as TEF-1, AP-2 and SP1 (Johnston et al., 1996; Mitchell et al., 1987). T antigen also enhances transcription dependent on RNA polymerases I and III and SV40 infected cells have higher levels of tRNA and rRNA synthesis than uninfected cells (Baserga et al., 1978; Felton-Edkins and White, 2002; Larminie et al., 1999; Sekiya and Oda, 1972; Zhai and Comai, 1999; Zhai et al., 1997). These observations are consistent with early reports indicating that global cellular transcription is increased upon SV40 infection (May et al., 1976; Oda and Dulbecco, 1968).
Three other cellular proteins are known to bind to T antigen but their role in SV40 transformation is not firmly established. Two of these, Cul7 (Ali et al., 2004) and the spindle checkpoint kinase Bub1 (Cotsiki et al., 2004) are known to bind to the amino terminus of T antigen. The transcriptional adapter proteins p300/CBP bind to T antigen through its interaction with p53 (Eckner et al., 1996; Lill et al., 1997; Poulin et al., 2004).
The study of SV40-induced transformation has utilized several different biological systems including established cell lines, primary cultured fibroblasts, and transgenic mice. This body of work has firmly established the importance of T antigen action on Rb-proteins and on p53 in mediating transformation. However, several studies indicate that the inhibition of pRb-proteins and p53 is not sufficient and suggest that, at least in some cases, action on additional cellular targets is essential for T antigen-mediated transformation (Beachy et al., 2002; Dickmanns et al., 1994; Dobbelstein et al., 1992; Manfredi and Prives, 1990; Sachsenmeier and Pipas, 2001). Furthermore, other studies demonstrate that even T antigen's action on Rb-proteins and p53 is not always essential for transformation (Markovics et al., 2005; Srinivasan et al., 1989). However, the interpretation of these studies is complicated by the fact that some experiments were performed in established mouse or rat cell lines, others used primary rat or mouse cultures, and yet others used transgenic mice.
In this manuscript we compare the effects of SV40 transformation on cellular gene expression in two systems: cultured primary mouse embryo fibroblasts and enterocytes derived from transgenic mice. Specifically, we have used gene microarray experiments in seeking the answer to three questions: (1) does SV40-transformation deregulate the expression of E2F-dependent genes; (2) does SV40-transformation have common effects on the global patterns of cellular gene expression in different cellular systems; and, (3) can gene expression profiling identify additional pathways that might contribute to transformation?
RESULTS
We compared the effects of SV40 early region on global patterns of cellular gene expression in two different systems: cultured primary mouse embryo fibroblasts (MEFs), and intestinal epithelial cells derived from transgenic mice. The experimental design is shown in Figure 1. SV40 induces morphological transformation in both of these systems (Hauft et al., 1992; Todaro and Green, 1963). We also examined the effects of SV40 transformation on cellular gene expression in REF52 cells, an established rat cell line that has been used extensively to study the mechanism of SV40 transformation (Conzen and Cole, 1995; Srinivasan et al., 1989) (Figure S1). By including an analysis of REF52 cells, we can compare the patterns of gene expression between a primary cell line, an established cell line and intact tissue.
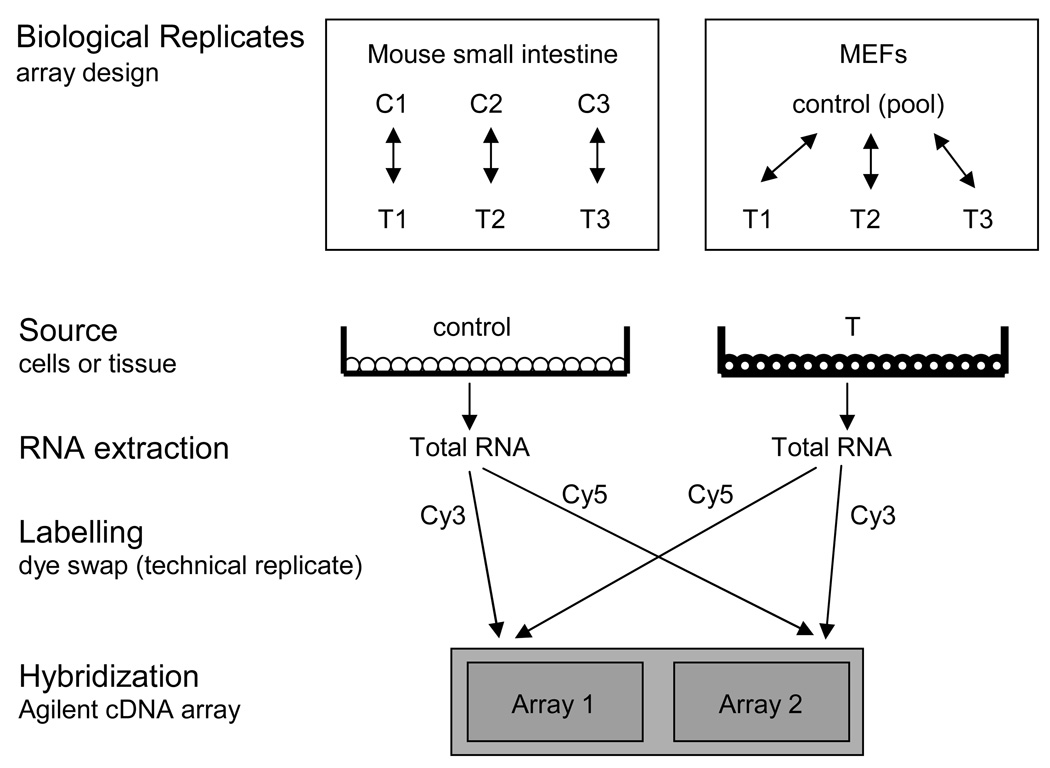
Fig 1.
Design of the mouse microarray experiments. Biological replicates of the mouse intestine and MEF arrays were implemented as a block design and reference design, respectively. Total RNA was isolated from control (C) and SV40-transformed (T) mouse small intestine and MEFs. Total RNA was converted to cDNA and labeled with both Cy3 and Cy5 in independent reactions. Control and SV40-transformed cDNA was mixed and hybridized to the Agilent Mouse cDNA microarray in dye-swap fashion. The cDNA microarray contains two duplicate arrays on one glass slide.
Parental or transformed MEF and REF52 sublines were maintained in media supplemented with 10% fetal bovine serum and RNA was isolated at two-days post-confluence. Microarray experiments were performed on three independently derived clones of SV40-transformed MEFs or SV40-transformed REF52 cells. In each case, the SV40-transformed cell line was directly compared to either parental REF52 cells or to the MEF primary culture from which the cell lines were derived. In the case of the intestinal epithelium, enterocytes from frozen intestinal sections were obtained by laser capture microdissection (LCM) and used to prepare RNA. The level of expression of SV40 large T antigen in MEFs, REF52, and enterocytes was similar (Figure S2), although small t antigen was not detected in enterocytes. In addition, we examined the gene expression patterns in RNA samples from whole intestines as well as from fractions enriched for villus enterocytes from parental and transgenic mice (see below).
Microarray experiments were performed using either Agilent mouse cDNA arrays or Affymetrix rat arrays. Mouse RNA from intestine or MEFs was applied to the Agilent Mouse cDNA microarray in dye-swap fashion (Churchill, 2002). We compared the three independently derived transformed MEF sublines to a pool of control MEFs. For intestine, we compared three independent samples of SV40-transformed, LCM-captured enterocytes to three samples obtained from their non-transgenic littermates. RNA from REF52 cell lines was applied to the RGU34A chip (Affymetrix). We performed two biological replicates of the untransformed REF52 cell line and three biological replicates of SV40-transformed REF52 cells. A gene was scored as being significantly regulated if transformation resulted in a change of 2-fold or more in the corresponding mRNA level. In addition, genes with a p-value <= 0.05 were scored as being statistically regulated. Since our sample size is small (3–4 bioreplicates for each class), applying a p-value cutoff may eliminate true-positives; therefore, we used the biological relevance cutoff (2-fold) in our initial characterization.
SV40 transformation alters cellular gene expression
Transformation by the SV40 early region altered the expression of about 800 cellular genes in each system (Table 1). About 400 genes were expressed at higher levels in SV40-transformed MEFs or enterocytes compared to their nontransformed counterparts. Approximately 400 genes were downregulated by transformation in both MEFs and enterocytes. Thus, SV40 transformation results in the altered expression of approximately 9% of the cellular genes represented on the array. A complete list of the genes regulated in each of these systems is shown in Tables S1 and S2. In REF52 cells, SV40 transformation alters the expression level of about 450 genes (Table S3).
Table 1.
Total number of SV40-regulated genes in MEF and intestine as a function of fold change and p-value.
| Criteria | M | E | V | W |
|---|---|---|---|---|
| 2-fold up | 390 | 465 | 238 | 182 |
| 2-fold down | 448 | 387 | 159 | 71 |
| p < 0.05 | 1400 | 1513 | 1719 | 1794 |
| p < 0.01 | 439 | 384 | 697 | 740 |
| 2-fold up AND p < 0.05 | 259 | 281 | 205 | 152 |
| 2-fold up AND p < 0.01 | 146 | 88 | 162 | 99 |
| 2-fold down AND p < 0.05 | 138 | 243 | 109 | 52 |
| 2-fold down AND p < 0.01 | 33 | 83 | 60 | 36 |
The microarray data was filtered for each class of microarrays (M, MEF; E, Enterocyte; V, Villi; W, Whole) using fold change and/or p-value. For example, "2-fold up" means that a gene was upregulated in SV40 transformed cells at least 2-fold and "p < 0.05" means that a gene was differentially regulated from control cells at a p-value cutoff of 0.05. p-value is a statistical measure that does not indicate whether a gene was up or downregulated.
The growth state of the cells in culture does not appear to play a significant role in affecting the gene expression patterns of SV40 transformation. We conducted an immunoblot on various DNA replication and cell cycle proteins with control and SV40-transformed MEF lysates that were obtained from subconfluent (∼30% confluent) or two-day post-confluent cells that were cultured in low (1%) or normal (10%) levels of serum (Figure S3). In all cases, SV40 transformation governed the steady-state level of the proteins tested irrespective of the growth state of the cells or serum levels.
SV40 transformation regulates proliferation related genes in both mouse embryo fibroblasts and enterocytes
Next we compared the specific sets of genes in MEFs and enterocytes that were regulated by SV40-transformation (Figure 2) and found that 179 genes (Table S4) were commonly upregulated in both systems. Application of the GO-ontology software (DAVID, http://david.abcc.ncifcrf.gov/home.jsp) to this set of genes indicated they primarily belonged to four biological classes: DNA replication, DNA repair, chromatin modifiers, and cell cycle regulators.
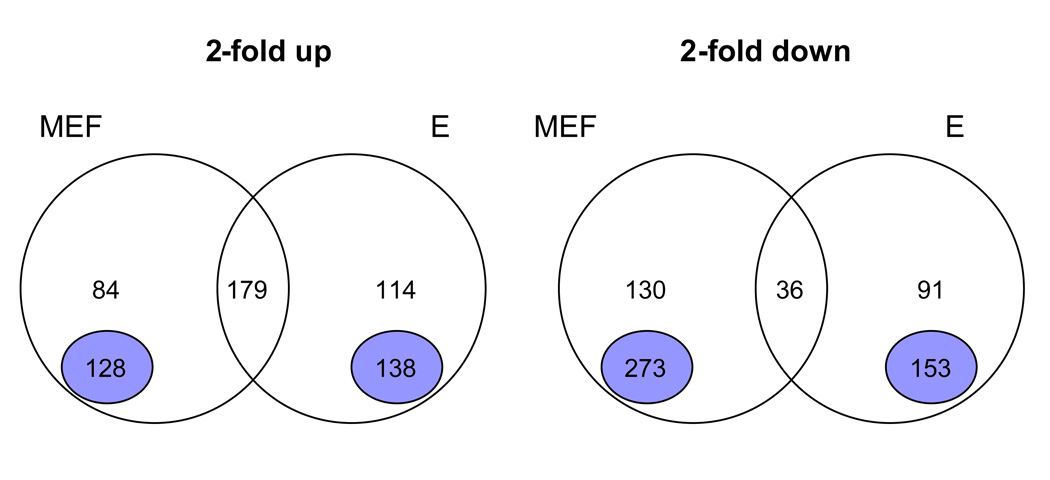
Fig 2.
Analysis of SV40 regulated genes in MEFs and enterocytes. The intersection of the genes that were 2-fold up or 2-fold down in the MEF and enterocyte (E) microarrays were overlapped and visualized by Venn Diagrams. The number of genes in each set is indicated. The number above the purple circle represent genes that were regulated by array (1.2- to 2-fold) in the other class but did not meet the 2-fold cutoff. Uniquely regulated genes are indicated by the purple circle. These genes show either no regulation in the other class or are oppositely regulated. For instance, the 128 uniquely upregulated genes in transformed MEFs are less than 1.2-fold upregulated in transformed enterocytes.
Many of these genes are known to be regulated by the E2F family of transcription factors. Since SV40 large T antigen binds the Rb family of tumor suppressor proteins, thus activating E2F-dependent transcription, increased expression of genes regulated by E2Fs is a predicted consequence of SV40-transformation. To test this we compiled a list of 42 well characterized E2F responsive genes (Table S5) and examined their expression ratios in MEFs, enterocytes and REF52 cells. Nearly all of these genes were expressed at higher levels in transformed cells in all of these systems. We conclude that E2F-dependent transcription is activated by SV40 transformation in each of these cases. A number of other genes associated with cell cycle progression, DNA replication and DNA repair, but not known to be directly regulated by E2F, also showed increased expression upon SV40 transformation. At this point we do not know if this upregulation is a direct consequence of SV40 transformation, or if it is an indirect consequence of cell cycle entry and progression.
Only 36 genes were commonly downregulated by SV40 transformation in MEFs and enterocytes (Table S6). These do not appear to belong to any specific biological class or reflect the activation or repression of any specific pathway.
Distinct sets of cellular genes are altered by SV40 in mouse embryo fibroblasts and enterocytes
Our analysis revealed that many genes were uniquely regulated, either in MEFs or enterocytes, by SV40 transformation (Figure 2). For this analysis, we did not consider genes that showed an upregulation between 1.2- and 2-fold (84 genes for MEFs and 114 genes for enterocytes). For instance, 128 genes were exclusively upregulated in MEFs transformed by SV40, but were either not affected or downregulated in transformed enterocytes (< 1.2-fold upregulation). Similarly, 138 genes were uniquely upregulated in enterocytes, but were either not changed or were downregulated in MEFs (< 1.2-fold upregulation). Similarly, some genes were uniquely downregulated (273 in MEFs, 153 in enterocytes) upon transformation in each system.
According to the microarray data, genes regulated by SV40 transformation can be grouped into eight distinct classes (Table 2). These include genes that are either upregulated (class 2) or downregulated (class 7) in both MEFs and enterocytes. Six additional classes represent genes that are commonly regulated in one of the systems, but that are either not regulated or are oppositely regulated in the other system. The differential regulation of several of these genes was confirmed by RT-PCR (Figure 3) or immunoblot analysis (Figure 4).
Table 2.
Selected genes to represent classes of cell-type specific gene regulation
| Classa | Cloneb | Name | Genec | Md | pvale | Ed | pvale |
|---|---|---|---|---|---|---|---|
| 1 | 751642 | Ubiquitin specific peptidase 18 | 24110 | 3.55 | 0.00 | 0.24 | 0.87 |
| 871756 | Tripartite motif protein 30 | 20128 | 3.49 | 0.01 | 0.11 | 0.50 | |
| 616653 | Interferon-induced protein with tetratricopeptide repeats 1 | 15957 | 3.13 | 0.00 | −0.01 | 1.00 | |
| 934836 | Chromobox homolog 7 | 52609 | 1.86 | 0.04 | −0.23 | 0.44 | |
| 1380122 | MutY homolog (E. coli) | 70603 | 1.85 | 0.01 | 0.21 | 0.22 | |
| 871077 | Caspase 2 | 12366 | 1.69 | 0.01 | 0.05 | 0.83 | |
| 442473 | Plectin 1 | 18810 | 1.61 | 0.01 | −0.05 | 0.78 | |
| 920425 | Histocompatibility 2, K1, K region | 14972 | 1.54 | 0.02 | −0.23 | 0.45 | |
| 946107 | RecQ protein-like 4 | 79456 | 1.52 | 0.00 | 0.16 | 0.25 | |
| 618572 | Interferon activated gene 203 | 15950 | 1.48 | 0.02 | −0.07 | 0.84 | |
| 2 | 335766 | Timeless homolog (Drosophila) | 21853 | 3.06 | 0.00 | 1.49 | 0.01 |
| 540239 | DNA primase, p49 subunit | 19075 | 3.02 | 0.00 | 3.92 | 0.02 | |
| 761060 | Chromatin assembly factor 1, subunit B (p60) | 110749 | 2.93 | 0.00 | 2.90 | 0.01 | |
| 540655 | Minichromosome maintenance deficient 5, cell division cycle 46 (S. cerevisiae) | 17218 | 2.88 | 0.00 | 3.43 | 0.02 | |
| 1053504 | olymerase (DNA directed), epsilon | 18973 | 2.84 | 0.01 | 2.77 | 0.04 | |
| 367559 | Ubiquitin-like, containing PHD and RING finger domains, 1 | 18140 | 2.75 | 0.01 | 3.51 | 0.02 | |
| 977156 | Minichromosome maintenance deficient 3 (S. cerevisiae) | 17215 | 2.75 | 0.00 | 3.03 | 0.04 | |
| 540946 | Denticleless homolog (Drosophila) | 76843 | 2.65 | 0.00 | 3.34 | 0.01 | |
| 2101942 | Minichromosome maintenance deficient 4 homolog (S. cerevisiae) | 17217 | 2.64 | 0.01 | 3.13 | 0.03 | |
| 1037829 | Proliferating cell nuclear antigen | 18538 | 2.53 | 0.00 | 2.87 | 0.01 | |
| 3 | 537489 | Cyclin D1 | 12443 | −2.75 | 0.05 | 1.13 | 0.20 |
| 336150 | ChaC, cation transport regulator-like 1 (E. coli) | 69065 | −2.06 | 0.07 | 2.22 | 0.05 | |
| 1001468 | Solute carrier family 38, member 1 | 105727 | −1.91 | 0.33 | 1.01 | 0.01 | |
| 870931 | Hexokinase 2 | 15277 | −1.41 | 0.04 | 1.49 | 0.06 | |
| 334351 | Metallothionein 2 | 17750 | −1.41 | 0.18 | 1.29 | 0.28 | |
| 404131 | Plasminogen activator, urokinase receptor | 18793 | −1.37 | 0.04 | 1.08 | 0.16 | |
| 331379 | Adenylate cyclase 4 | 104110 | −1.33 | 0.04 | 1.85 | 0.08 | |
| 517227 | EP300 interacting inhibitor of differentiation 2 | 386655 | −1.26 | 0.10 | 1.01 | 0.25 | |
| 945526 | Dimethylarginine dimethylaminohydrolase 1 | 69219 | −1.06 | 0.12 | 2.18 | 0.08 | |
| 851855 | Phospholipase C, epsilon 1 | 74055 | −1.04 | 0.16 | 1.02 | 0.07 | |
| 4 | 536577 | Dickkopf homolog 3 (Xenopus laevis) | 50781 | −5.03 | 0.03 | −0.17 | 0.70 |
| 1277469 | Insulin-like growth factor binding protein 2 | 16008 | −3.71 | 0.03 | −0.10 | 0.53 | |
| 1277386 | Cyclin D2 | 12444 | −3.52 | 0.03 | −0.11 | 0.74 | |
| 1077628 | Serine (or cysteine) peptidase inhibitor, clade A, member 3N | 20716 | −3.46 | 0.02 | 0.08 | 0.84 | |
| 521599 | UDP glucuronosyltransferase 2 family, polypeptide B36 | 231396 | −3.11 | 0.02 | −0.26 | 0.11 | |
| 407674 | Insulin-like growth factor binding protein 3 | 16009 | −3.02 | 0.18 | −0.15 | 0.41 | |
| 517556 | Glycine amidinotransferase (L-arginine:glycine amidinotransferase) | 67092 | −3.02 | 0.19 | 0.21 | 0.80 | |
| 637211 | Guanine deaminase | 14544 | −2.98 | 0.20 | −0.24 | 0.42 | |
| 1383367 | CD24a antigen | 12484 | −2.86 | 0.17 | 0.03 | 0.87 | |
| 463860 | Platelet-derived growth factor, C polypeptide | 54635 | −2.85 | 0.01 | 0.00 | 0.99 | |
| 5 | 1038400 | Cyclin B2 | 12442 | 0.25 | 0.37 | 2.50 | 0.01 |
| 793009 | Farnesyl diphosphate farnesyl transferase 1 | 14137 | 0.11 | 0.87 | 2.01 | 0.02 | |
| 1279180 | Solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 | 11987 | −0.11 | 0.66 | 1.87 | 0.02 | |
| 865281 | RAN binding protein 5 | 70572 | 0.10 | 0.46 | 1.72 | 0.05 | |
| 1005963 | ZW10 interactor | 52696 | 0.24 | 0.37 | 1.64 | 0.02 | |
| 1023414 | Periplakin | 19041 | 0.16 | 0.50 | 1.56 | 0.02 | |
| 523337 | UCHL5 interacting protein | 73738 | 0.01 | 0.93 | 1.54 | 0.00 | |
| 331996 | Inhibitor of DNA binding 1 | 15901 | 0.10 | 0.87 | 1.43 | 0.02 | |
| 2102126 | Methionine-tRNA synthetase | 216443 | 0.05 | 0.81 | 1.41 | 0.01 | |
| 821338 | Core binding factor beta | 12400 | 0.14 | 0.44 | 1.41 | 0.00 | |
| 6 | 560431 | Clusterin | 12759 | 2.60 | 0.06 | −1.60 | 0.28 |
| 1264311 | Lymphocyte antigen 75 | 17076 | 1.58 | 0.04 | −1.76 | 0.01 | |
| 1331769 | RAR-related orphan receptor gamma | 19885 | 1.38 | −2.33 | 0.02 | ||
| 809050 | WD repeat domain 67 | 210544 | 1.24 | 0.00 | −1.08 | 0.03 | |
| 482898 | Cysteine-rich protein 1 (intestinal) | 12925 | 1.24 | 0.23 | −1.38 | 0.01 | |
| 348362 | Cystin 1 | 12879 | 1.20 | −1.05 | 0.05 | ||
| 571457 | Period homolog 3 (Drosophila) | 18628 | 1.11 | 0.11 | −1.60 | 0.04 | |
| 776764 | Vitamin D receptor | 22337 | 1.11 | 0.42 | −1.92 | 0.01 | |
| 458727 | Leukotriene B4 receptor 1 | 16995 | 1.08 | −1.17 | 0.01 | ||
| 596348 | Aldehyde dehydrogenase family 1, subfamily A7 | 26358 | 1.00 | 0.21 | −2.55 | 0.05 | |
| 7 | 520782 | NADPH oxidase 4 | 50490 | −2.23 | 0.01 | −1.21 | 0.04 |
| 791359 | Yippee-like 2 (Drosophila) | 77864 | −1.40 | 0.03 | −1.22 | 0.04 | |
| 822692 | TSC22 domain family, member 1 | 21807 | −1.39 | 0.01 | −1.11 | 0.00 | |
| 355113 | Sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A | 20358 | −1.24 | 0.05 | −1.54 | 0.01 | |
| 992031 | Glutaredoxin | 93692 | −1.19 | 0.08 | −1.11 | 0.00 | |
| 1125584 | Protein phosphatase 1A, magnesium dependent, alpha isoform | 19042 | −1.16 | 0.04 | −1.11 | 0.09 | |
| 720411 | Phytanoyl-CoA hydroxylase | 16922 | −1.11 | 0.07 | −2.09 | 0.00 | |
| 313537 | 5′ nucleotidase, ecto | 23959 | −1.09 | 0.06 | −3.10 | 0.05 | |
| 846412 | RAB30, member RAS oncogene family | 75985 | −1.07 | 0.03 | −2.19 | 0.06 | |
| 1020807 | Granule cell antiserum positive 14 | 72972 | −1.02 | 0.00 | −1.33 | 0.03 | |
| 8 | 1096039 | Bone morphogenetic protein 8a | 12163 | −0.25 | 0.55 | −2.97 | 0.01 |
| 313629 | Rhomboid domain containing 2 | 215160 | −0.18 | 0.22 | −2.22 | 0.02 | |
| 1005129 | Leukotriene B4 12-hydroxydehydrogenase | 67103 | −0.20 | 0.39 | −2.16 | 0.01 | |
| 386489 | Cytidine deaminase | 72269 | −0.03 | 0.94 | −1.91 | 0.00 | |
| 1478742 | Avian musculoaponeurotic fibrosarcoma (v-maf) AS42 oncogene homolog | 17132 | −0.16 | 0.91 | −1.91 | 0.01 | |
| 1245874 | Solute carrier family 2 (facilitated glucose transporter), member 5 | 56485 | 0.19 | 0.69 | −1.89 | 0.02 | |
| 440961 | Glutamate-cysteine ligase , modifier subunit | 14630 | 0.02 | 0.93 | −1.89 | 0.02 | |
| 1004969 | Solute carrier family 15 (H+/peptide transporter), member 2 | 57738 | −0.04 | 0.88 | −1.76 | 0.00 | |
| 598741 | N-acylsphingosine amidohydrolase 2 | 54447 | 0.10 | 0.20 | −1.66 | 0.00 | |
| 1106464 | Sulfatase modifying factor 1 | 58911 | 0.01 | 0.92 | −1.54 | 0.01 | |
Class number is the same as in Figure 3.
Unique identifier, provided by Agilent, for each DNA sequence spotted on the array
NCBI Gene database unique identifier
Average Log2 ratio for the MEF (M) and enterocyte (E) microarrays
P-value for the ratio
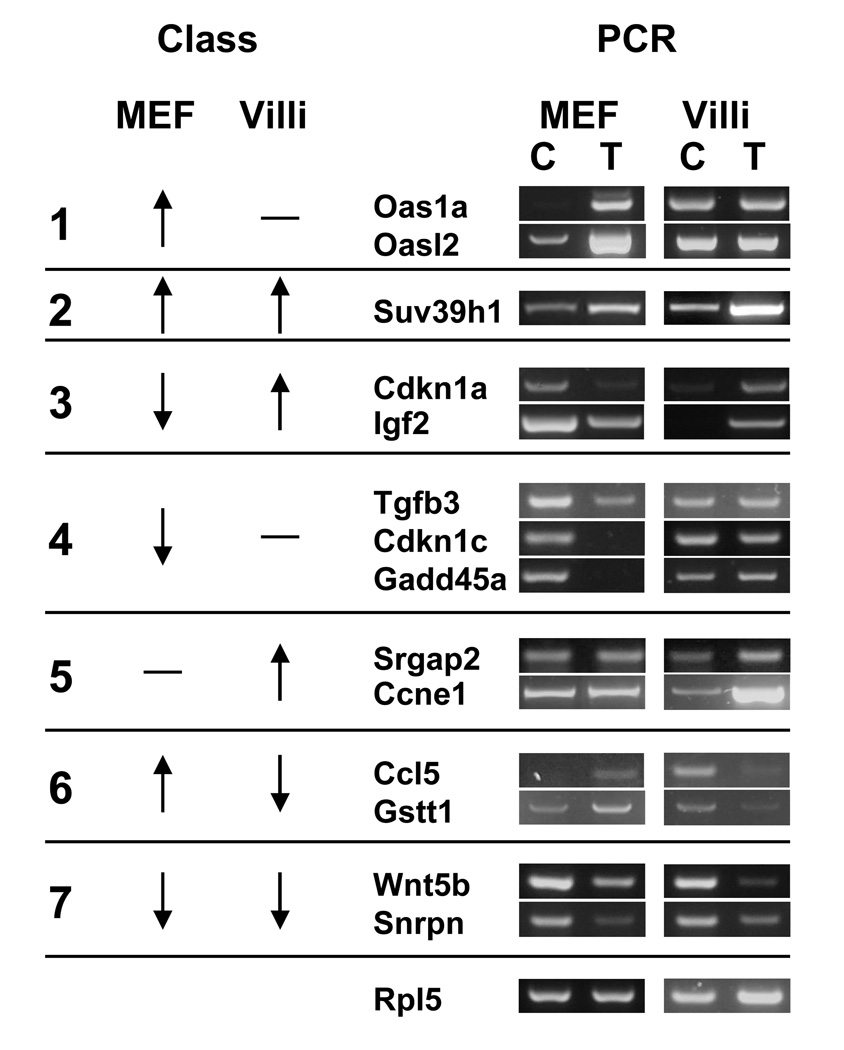
Fig 3.
Cell-type specific gene regulation is confirmed by RT-PCR. Genes were selected to represent different classes (“Class”) of cell-type specific genes that were observed by microarray. cDNA was synthesized from 1 µg of total RNA from control (C) or SV40-transformed (T) MEF or fractionated villi cells. Each gene was amplified with specific primers by PCR and the products were resolved on a 2% agarose gel and stained with GelStar. Amplification with primers for the Rpl5 transcript was used as a normalizing control. Up arrow, transcript is upregulated in SV40-transformed cells; down arrow, downregulated; dash, transcript level not changed.
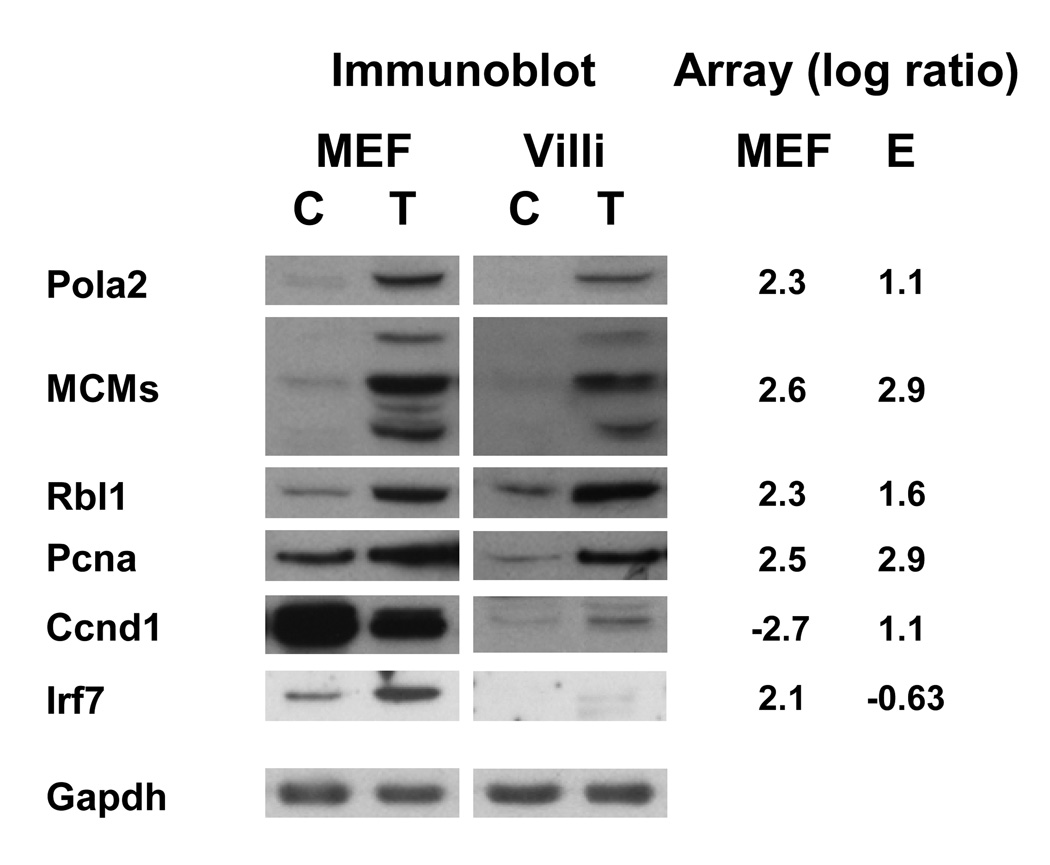
Fig 4.
Microarray confirmation by immunoblot. 50 µg of control (C) or SV40-transformed (T) cell lysate from two-day post-confluent MEFs and fractionated villi were resolved by SDS-PAGE and transferred to PVDF. An appropriate primary antibody was added (protein symbols shown on the left) and ECL Plus (Amersham) was used to visualize protein expression. Gapdh was used as a loading control. The microarray average log ratio from the MEF and enterocyte (E) arrays is shown on the right for each gene.
Many of the genes uniquely upregulated in SV40-transformed MEFs (class 1) belong to a group whose expression is activated by the interferon pathway (Table S7). In contrast, the levels of interferon-regulated genes did not appear to be altered in SV40-transformed enterocytes. Thus, in MEFs but not enterocytes, SV40 transformation results in the activation of the interferon signaling pathway. At this point it is unclear if SV40 transformation results in the increased production of interferon, or if it directly upregulates the expression of downstream target genes.
Two classes of genes, class 3 and 4, contain genes that are uniquely downregulated in SV40-transformed MEFs. Several of these genes are important cell cycle regulators and growth factors such as p21 (Cdkn1a), Igf2, cyclin D, p57 (Cdkn1c), Gadd45a and Tgfb3. In fact, p21, Igf2 and cyclin D1 are upregulated in transformed enterocytes. On the other hand, not all cell cycle regulators were uniquely downregulated in SV40-transformed MEFs. For example, cyclin E1 and cyclin B2 were found to be members of a class of genes, class 5, that are uniquely upregulated in transformed enterocytes but not changed in SV40-transformed MEFs. The mechanism and biological effect of these differences in unclear.
We have previously reported that the transformation of enterocytes by SV40 results in the coordinate downregulation of many members of the cytochrome P450 detoxification pathway (Saenz-Robles et al., 2007b). This effect was not observed in SV40-transformed MEFs perhaps due to low P450 gene expression (data not shown). However, two detoxification genes, Gstt1 (Figure 3) and vitamin D receptor (Table 2, class 6), were found to be upregulated in transformed MEFs. At present, it is not clear how SV40 transformation coordinately affects the mRNA levels of multiple members of this pathway.
Gene regulation in the mouse intestine is not altered by the method of cell isolation
To obtain the RNA used in the microarray experiments, MEFs were treated with guanidine thiocyanate immediately following the removal of culture medium and rinsing with PBS/EDTA. Thus, the microarray data provides a "snapshot" of the RNA levels present in the cells at the instant of extraction. In contrast, obtaining RNA from enterocytes requires the removal, rinsing and fixation of the small intestine prior to the collection of enterocytes by LCM. Thus, since it takes several minutes to prepare the intestine for LCM, it is possible that the expression levels of some genes might be altered during this period. Furthermore, the process of enterocyte enrichment by biomechanical fractionation involves the use of several biochemical and molecular procedures and results in an even longer delay between intestine removal and RNA extraction (Markovics et al., 2005). One way to reduce the possible effects of the extraction method on gene expression would be to isolate RNA from whole intestines that were extracted immediately after dissection. However, this method has the disadvantage of including muscle, mesenchymal, and crypt progenitor cell populations in the preparation, in addition to enterocytes. Thus, gene responses unique to the SV40-transformed cells would be diluted by these nontransformed cell types.
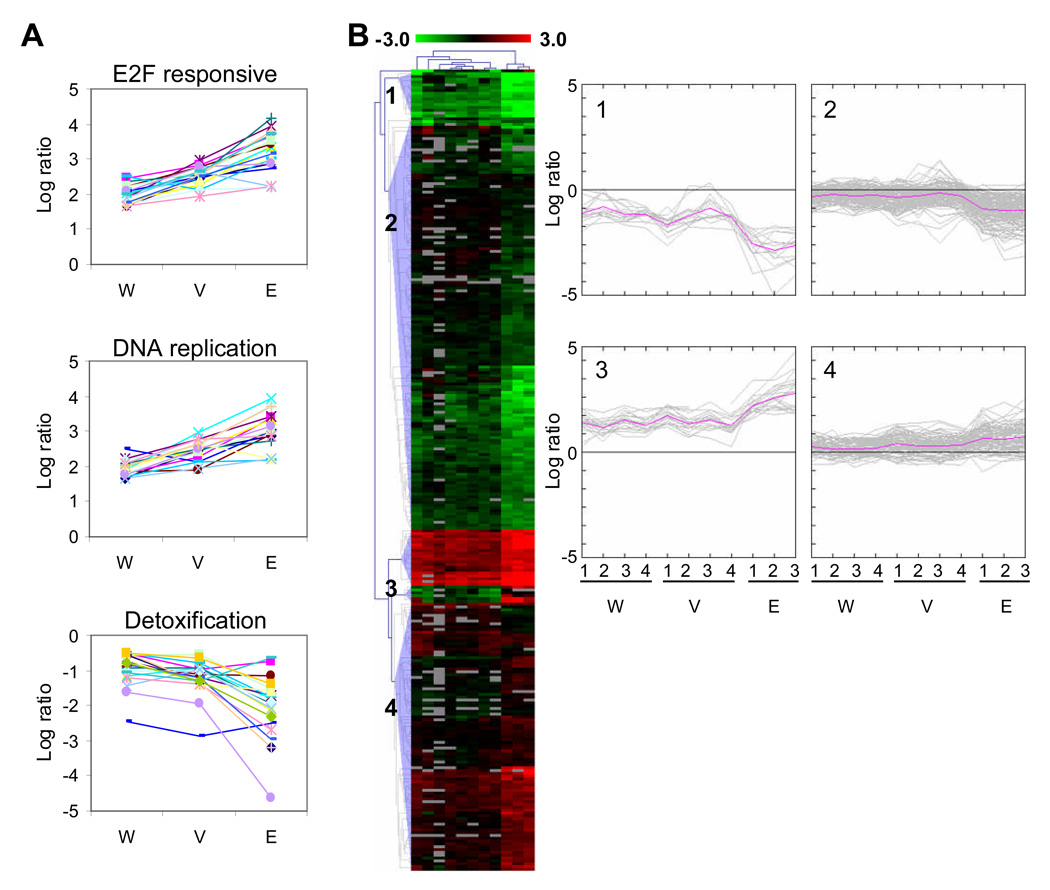
To test for effects of the method of cell extraction on gene expression, we performed microarray experiments on RNA isolated from LCM-extracted enterocytes (E), fractionated villi (V), and whole intestine (W). Table 1 shows that SV40 transformation regulates more genes when cells are obtained by LCM than in those obtained by fractionation or from whole intestine. We analyzed these data sets in two ways. First, we focused on three specific regulated gene sets (Figure 5A): E2F responsive genes (Table S5), DNA replication genes, and detoxification genes (Saenz-Robles et al., 2007b). We analyzed the expression pattern of the E2F responsive and DNA replication genes that were upregulated at least 3-fold in all three systems. For the detoxification genes, we analyzed genes that were downregulated at least 1.4-fold in all three systems. Second, we used a nonbiased approach (Figure 5B) by filtering the microarray data by ANOVA which resulted in 284 clones. These 284 clones show a significant difference in expression between the three systems. To visualize the expression pattern of these differences we applied hierarchical clustering to these clones and four patterns of gene expression emerged (Figure 5B). Both of these analyses show that SV40-transformation regulates the same set of genes in samples extracted by LCM, fractionated villi, or whole intestine. The data also clearly show that LCM samples give the most robust ratios, followed by fractionated villi and then by whole intestine. This is most likely caused by the possible dilution of SV40-transformed cells with nontransformed cell types: LCM would ensure that RNA is only extracted from transformed enterocytes; villi fractionation would remove most of the muscle, mesenchyme, and crypt cells but still include goblet cells and enteroendocrine cells not expressing the SV40 T antigens; and SV40-transformed cells would be most dilute in whole intestine. LCM samples from liver were also shown to give more robust microarray ratios than samples obtained from dissected tissue (Yim et al., 2003).
Fig 5.
Mouse intestine microarrays show similar patterns of gene expression. (A) The average log ratio of E2F responsive genes that were upregulated 3-fold or more in the whole intestine (W), fractionated villi (V) and LCM extracted enterocyte (E) arrays is shown in the top panel. Similarly, the middle panel shows DNA replication genes upregulated at least 3-fold. In the bottom panel, detoxification genes that are downregulated 1.4-fold or more are shown. (B) The biological replicates of the whole intestine, fractionated villi and enterocyte microarray data were filtered by ANOVA (p=0.01) which resulted in 284 clones. These clones were hierarchically clustered using the Euclidean distance metric and average linkage. Clones are colored red and green for clones that are up and downregulated in SV40-transformed tissue, respectively. Grey clones represent missing data. Four expression patterns were revealed from the cluster diagram and the log ratio values of these clones are plotted on the right. The pink line represents the average log ratio for the set of clones in each biological replicate.
DISCUSSION
SV40 has been studied in many cell contexts including productive infection, transformation in cell culture and transgenic mice, and is known to alter the expression of many cellular genes. In fact, SV40-induced transformation is closely linked to the control of cellular gene expression. Large T antigen transforms, in part, by inhibiting Rb family proteins and thus stimulating E2F-dependent transcription. In addition, T antigen antagonizes growth-arrest and pro-apoptotic signals by binding the tumor suppressor p53 and thus blocking p53-dependent transcription. Multiple lines of evidence suggest that there are additional T antigen targets important for transformation, but how they contribute to the process is unclear. Small t antigen contributes to transformation by displacing the B subunits of the cellular phosphatase PP2A. In doing so, small t antigen alters several signal transduction pathways, some of which ultimately affect gene expression (Nunbhakdi-Craig et al., 2003; Rodriguez-Viciana et al., 2006; Watanabe et al., 1996).
In this study we have used gene microarrays to compare the effects of SV40-induced transformation on cellular gene expression in two systems, cultured primary mouse embryo fibroblasts and intestinal enterocytes directly isolated from mice. SV40 transformation altered the mRNA levels of about 800 cellular genes in both of these systems. This likely represents an underestimate of the total number of transformation-regulated genes for two reasons. First, the Agilent arrays used in these studies only include 8,462 of an estimated 21,000 mouse genes. Second, although we applied a 2-fold expression ratio cutoff to the genes analyzed in our study, RT-PCR experiments indicated that these expression ratios are compressed. Thus, many genes whose expression ratios were less than but close to 2-fold are actually regulated. Nevertheless, the patterns of gene expression observed allow us to make several important conclusions.
Many genes associated with DNA replication, DNA repair, chromatin assembly and modification, and cell cycle progression were upregulated by SV40 transformation in both MEFs and enterocytes. This same set of genes was also upregulated in an SV40-transformed established rat cell line, REF52. Approximately 80–90% of E2F-regulated genes that were analyzed had elevated levels of mRNA in transformed cells. The upregulation of genes associated with cell cycle entry and progression was expected, as the virus-encoded large T antigen is known to activate E2F-dependent gene expression and cell cycle progression by binding and inactivating the Rb family of proteins: pRb, p130, and p107. In fact, this group of upregulated genes substantially overlapped with the upregulated genes found in microarray studies using transgenic mouse models of breast, prostrate, and lung cancer that express the early region of SV40 (Deeb et al., 2007; Klein et al., 2005).
We did not find any evidence for activation of the p53 pathway. This is expected since T antigen is known to block p53-dependent transcriptional activation. Our studies also identified many genes regulated by SV40 transformation that are not associated with cell cycle control. These genes span multiple biological activities and at this point their contribution to the transformed phenotype, if any, is unclear. Future genetic studies should help elucidate their role.
More importantly, we found many genes that responded differentially to the presence of T antigen, depending on the cell system under consideration. For instance, we found increased mRNA levels of many genes associated with the interferon pathway in transformed MEFs but, in contrast, the expression of these genes was not altered in transformed enterocytes. Strikingly, the levels of mRNAs encoding interferon alpha and beta were not altered by the presence of the oncogene (data not shown). This suggests that, in MEFs, one of the viral T antigens is either directly binding and activating interferon-regulated promoters, or activating the interferon signaling pathway.
Furthermore, we have previously reported that several genes associated with the cytochrome P450 detoxification pathway are downregulated by the SV40 T antigens in enterocytes (Saenz-Robles et al., 2007b) but we found no evidence for regulation of this pathway in SV40-transformed MEFs. Some cell cycle regulators such as cyclin D and p21 and growth factors such as Igf2 and Tgfb3 were differentially regulated in these systems. It is clear from our data that SV40 effects on several pathways depend on the cell-type, but we do not yet understand the mechanisms behind this cell-type specific effect or know whether they contribute to transformation.
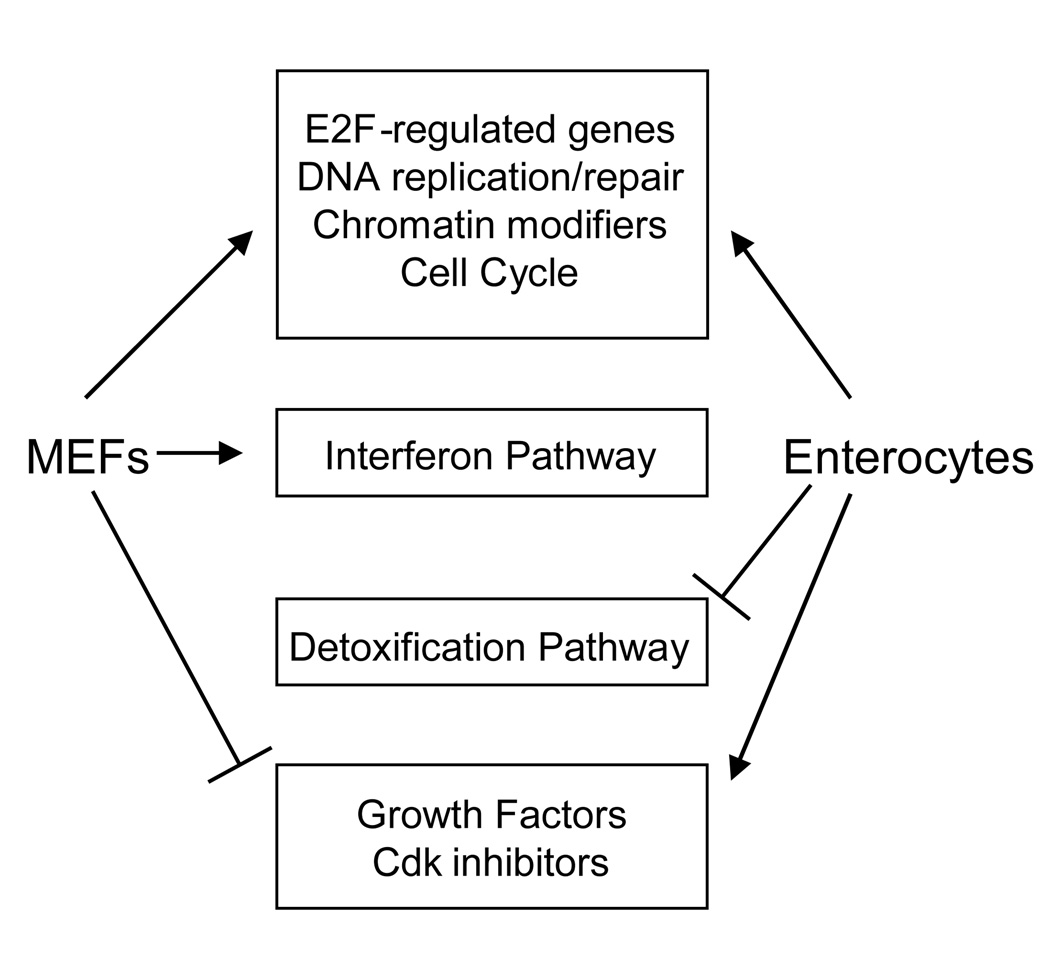
In conclusion, we have shown that SV40 transformation has both common and unique effects on two different cell-types, cultured primary MEFs and intestinal enterocytes directly obtained from mice (Figure 6). Many of the effects on gene expression that are common to these systems can be explained by the action of large T antigen on the Rb-E2F pathway. Small T antigen is known to regulate gene expression (Moreno et al., 2004); therefore, a subset of the genes regulated by SV40 transformation could be the result of small t expression. It is unclear whether the genes that are uniquely regulated in MEFs or enterocytes reflect differences between cells maintained in culture and in vivo tissue or if the difference arose from the action of the viral T antigens on targets that are specific to each cell type. The lack of small t expression in enterocytes may contribute to these differences as well. At this point, we cannot discern which changes in gene expression are due to the direct action of T antigen on the transcriptional apparatus, and which are the indirect consequence of large and small T antigen action on signal transduction pathways. Further studies in different cell types should render valuable information to allow distinction of these mechanisms.
Fig 6.
Model for SV40 regulation of gene expression. The cell-type specific gene regulation by SV40 is indicated by classes of genes that are uniquely or commonly regulated in each cell type. For example, SV40 upregulates the interferon pathway in MEFs and downregulates the detoxification pathway in enterocytes.
MATERIALS AND METHODS
Cell lines and transgenic mice
Wild-type MEFs and MEF sublines expressing the neomycin resistance gene plus the early region of SV40 (Markovics et al., 2005), which expresses the large and small T antigens, were maintained in DMEM + 10% heat inactivated FBS with 0.4 mg/ml G418. All cells were grown at 37°C in 5% CO2. REF52 cells expressing the neomycin resistance gene with and without the early region of SV40 (Srinivasan et al., 1997) were maintained in MEM + 10% FBS with 0.4 mg/ml G418 (Invitrogen). The expression of the early region of SV40 is driven by the Rous sarcoma virus promoter in both cell lines. Transgenic mice expressing the SV40 early region under the control of the rat intestinal fatty acid binding protein (IFABP) promoter were previously described (Kim et al., 1993) and were maintained by crosses to commercially available non-transgenic FVB mice (Taconic Labs). Routine screenings for several murine pathogens and parasites were all negative. Mouse genotyping was performed by PCR analysis as described (Saenz-Robles et al., 2007a).
Isolation of intestinal epithelial cells
Mice between the ages of 1.5 to 3.5 months were sacrificed and their entire small intestine was removed, opened longitudinally and washed in saline solution. All non-transgenic intestine displayed a normal morphology and each SV40-transformed intestine was judged to be hyperplastic. No visible signs of polyps or tumors were present in any intestinal sample. The intestine was subdivided into three longitudinal portions equal in length, and the middle section was harvested and used directly ( “whole intestine”) or further processed to enrich for villi structures (“fractionated villi”). The process for villi enrichment has been described (Markovics et al., 2005; Saenz-Robles et al., 2007a). Briefly, middle sections of the intestine were incubated for 30 minutes in PBS plus 1 mM DTT, and the villi subsequently released by several steps of gentle shaking in the same solution supplemented with 5 mM EDTA for the non-transgenic samples or 3 mM EDTA for the T antigen-containing samples. Enterocytes from the villus were obtained by laser capture microdissection (LCM) from middle section intestinal rolls (Markovics et al., 2005). Briefly, the rolls were quickly frozen and stored at −80°C. Frozen sections were cut and immediately dehydrated in graded alcohol solutions. The dried sections were examined microscopically and the top two thirds of 50 villus segments from each section were collected using a laser capture microscope (PixCell II, Arcturus). The LCM captured cells were extracted in PicoPure extraction buffer (Arcturus) according to the manufacturer's directions and stored at −80°C.
RNA extraction and purification
Cell lines were grown to confluence and fed with fresh medium. After two days post-confluence, cells were harvested with trypsin, washed thrice with cold PBS-EDTA, and frozen at −80°C. Total RNA from MEF, whole and fractionated villi intestine samples was isolated using the RNeasy kit (Qiagen) with DNase digestion. Enterocyte total RNA was isolated with the PicoPure kit (Arcturus) and amplified twice with the RiboAmp kit (Arcturus).
Microarrays
For the MEF microarrays, we used a dye-swap reference design (Churchill, 2002) to compare a pool of total RNA extracted from wild-type MEFs to three independent SV40-transformed MEF cell lines. For the mouse small intestine microarrays, we used three different tissues, whole, fractionated villi and LCM-captured enterocytes (described above), in a dye-swap block design (Churchill, 2002) using three or four non-transgenic and SV40-transformed mice for each intestine preparation method. Where necessary, we pooled total RNA from up to four mice to obtain enough total RNA for microarray analysis.
Each total RNA preparation was converted to Cy3- and Cy5-labeled cDNA in independent reactions. Fifteen micrograms of total RNA was incubated at 70°C for 10 minutes in a 15.4 µl reaction that contained 6 µg of oligo-dT (Operon, SP-230), or 7.5 µg of random primers (Roche, 11034731001) for reactions containing amplified enterocyte Total RNA. After chilling the reactions on ice, labeled cDNA was prepared by adding 3 µl of 1 mM Cy3-dUTP (Amersham Biosciences, PA53022) or 1 mM Cy5-dUTP (PA55022) and 11.6 µl of a reverse transcription cocktail and the reaction [13.3 U/µl SuperScript II Reverse Transcriptase (Invitrogen, 18065–022), 1× first strand buffer, 10 mM DTT, 0.5 mM each dATP, dCTP, dGTP and 0.2 mM of dTTP] was incubated at 42°C for 2 hours. RNA was degraded by adding 1.5 µl of 1 N NaOH and 1.0 µl of 60 mM EDTA to each reaction and incubated at 65°C for 10 minutes. 470 µl of TE pH 7.4 was added to the reaction and applied to a Microcon-30 filter (Millipore, 42410). The reactions were centrifuged for 7 min at 8,500 g and washed once with 400 µl of TE pH 7.4 to remove unincorporated dyes. Filters were centrifuged upside-down to harvest the labeled cDNA (∼50 µl). Next, control and SV40-transformed cDNA labeled with different dyes was mixed and purified using the QIAQuick Kit (Qiagen). After completely drying the cDNA in a vacuum centrifuge, the labeled cDNA was resuspended in 7.5 µl of dH2O.
The Cy3/Cy5 cDNA mixture was applied to an Agilent Mouse cDNA microarray (Agilent Technologies, G4104A), containing 9,596 clones representing 8,462 unique genes, as recommended by the manufacturer. Since there are two identical arrays printed on each Agilent glass slide, labeled cDNA mixtures from a dye-swap pair (Cy5-SV40-transformed/Cy3-control cDNA and Cy3-SV40-transformed/Cy5-control cDNA) was applied to the separate arrays. After incubating the microarray in a 65°C water bath for 17 hours, the microarray was washed and dried according to the manufacturer. Using an Affymetrix GMS418 scanner, two images from each array were extracted by exciting the Cy3 dye at 532nm and the Cy5 dye at 635nm and saved as a 16-bit TIFF file. The Cy3 and Cy5 images were aligned and quantitated with Imagene 5.5 (Biodiscovery). The entire array was scanned manually to identify and flag any spots that were masked by debris or scratches. Additionally, any spots whose signal was not greater than the spot background plus two times the standard deviation of the spot background were flagged. Each array also contained a set of spots that were flagged by Agilent. All unflagged signals were normalized with a lowess intensity-dependent smoother using BRB-Array Tools (National Cancer Institute, http://linus.nci.nih.gov/BRB-ArrayTools.html) except that the lowess span parameter was changed from 0.4 to 0.2. A log2 ratio was calculated for each biological replicate by averaging its two dye-swap log ratios. In the end, there are a total of three biological replicate ratios for the MEF and enterocyte microarrays and four for the whole and villi microarrays. The biological replicate log ratios for each class of microarrays (MEF, enterocyte, whole, villi) were averaged and a one sample t-test was performed to determine if the average log ratio was significantly different from the null hypothesis: log SV40-transformed/control = 0.
TIGR MeV 4.1 was used for the following statistical analyses. A three-group ANOVA was performed on the biological replicate log ratios of the whole intestine, fractionated villi and enterocyte microarrays at a p-value = 0.01 which resulted in 284 clones. Hierarchical clustering using euclidean distance and average linkage was applied to the ANOVA clones. The distance threshold was set to 4.8 to reduce the number of terminal nodes to six.
Immunoblots
Protein extracts were prepared by lysing two-day post-confluent MEFs or fractionated villi in Buffer B [50 mM HEPES pH 7.9, 400 mM KCl, 0.5 mM EDTA, 0.1% NP40, 10% glycerol, 1 mM DTT, 0.5 mM Na3VO4, 0.5 mM NaF, 1 µg/ml pepstatin and a protease inhibitor tablet (Roche)] for 25 min at 4°C. Cellular debris was cleared by centrifugation at 15,000 rpm for 30 min at 4°C. Protein extracts were aliquoted and stored at −80°C until use. Lysate was resolved by SDS-PAGE and transferred to PVDF (Millipore). After blocking with 10% non-fat milk in 1× PBS, blots were probed with various antibodies: anti-Pola2 (kindly provided by Dr. Thomas Kelly), anti-Mcm (kindly provided by Dr. Anthony Schwacha), anti-Rbl1 (Santa Cruz, sc-318), anti-Pcna (DAKO, MO879), anti-Ccnd1 (Santa-Cruz, sc-450), anti-Irf7 (Santa-Cruz, sc-9083) and anti-Gapdh (United States Biological, G8140–11). After incubating the blots with an appropriate HRP-conjugated secondary antibody, the blots were visualized with ECL Plus (Amersham).
RT-PCR
cDNA was synthesized from 1 µg of total RNA using Superscript II Reverse Transcriptase (Invitrogen). PCR was performed with 1 µl of cDNA using GoTaq polymerase (Promega) for 25 to 35 cycles (depending on transcript being amplified) with specific primers for the following genes (listed 5’–3’): Oas1a, TGGCTGAAGAGGCTGATGTGTG and TGAGGAAGGCTGGCTGTGATTG; Oasl2, TTACAGAACAGCCAGAGCTATACGG and CAAGGGAGATAGATTTACGTCCACG; Suv39h1, CAGTGCTGGACTTGCTGGAAAC and TCCCTGAGACAATCTGTATGGACC; Cdkn1a, GAACTTTGACTTCGTCACGGAGAC and CTTCAGGGTTTTCTCTTGCAGAAG; Igf2, AAAGAAGCAGAAGAGACGCCCC and GCCAAAGAGATGAGAAGCACCAAC; Tgfb3, AAGGAGTGGACAATGAAGATGACC and CTGAGCAGAAGTTGGCATAGTAACC; Cdkn1c, AAGAGAACTGCGCAGGAGAA and CCCAGAGTTCTTCCATCGTC; Gadd45a, ACGACATCAACATCCTGCGG and CAAAGTCATCTCTGAGCCCTCG; Srgap2, CCCAGCATTGCTAAGAGGAG and GAGGCCTGGCAAGACTACAG; Ccne1, GCAGAAGGTCTCAGGTTATC and GTGGCCTCCTTAACTTCAAG; Ccl5, ATATGGCTCGGACACCACTC and CCACTTCTTCTCTGGGTTGG; Gstt1, TGTGTGAGAGTGTGGCTATCTTGC and TTATGATGAGGTCAGCAGGGGGAC; Wnt5b, TGGAGACAACGTGGAGTACG and GGCGACATCAGCCATCTTAT; Snrpn, GGTGGTGGAATTCAAGGAAA and TTCCACAATAGCCGTTGTCA; Rpl5, CCAAACGATTCCCTGGTTATGAC and GACGATTCCACCTCTTCTTCTTCAC. Amplification with primers for the Rpl5 transcript was used as a normalizing control. PCR reaction products were resolved through a 2% agarose gel in 1× TAE and stained with GelStar (Cambrex Bio Science).
ACKNOWLEDGEMENTS
This work was supported by grants CA40586 and CA098956 to J.M.P. WHP and RHW were supported by grant 5P30 DK58404-03. We wish to thank Mr. Anthony L. Frazier for his assistance in preparing tissue for laser capture microdissection.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ahuja D, Saenz-Robles MT, Pipas JM. SV40 large T antigen targets multiple cellular pathways to elicit cellular transformation. Oncogene. 2005;24(52):7729–7745. doi: 10.1038/sj.onc.1209046. [DOI] [PubMed] [Google Scholar]
- Ali SH, Kasper JS, Arai T, DeCaprio JA. Cul7/p185/p193 binding to simian virus 40 large T antigen has a role in cellular transformation. J Virol. 2004;78(6):2749–2757. doi: 10.1128/JVI.78.6.2749-2757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baserga R, Ide T, Whelly S. Stimulation of ribosomal RNA synthesis in isolated nuclei and nucleoli by partially purified preparations of SV40 T antigen. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):685–691. doi: 10.1101/sqb.1978.042.01.070. [DOI] [PubMed] [Google Scholar]
- Beachy TM, Cole SL, Cavender JF, Tevethia MJ. Regions and activities of simian virus 40 T antigen that cooperate with an activated ras oncogene in transforming primary rat embryo fibroblasts. J Virol. 2002;76(7):3145–3157. doi: 10.1128/JVI.76.7.3145-3157.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill GA. Fundamentals of experimental design for cDNA microarrays. Nat Genet. 2002;32(Suppl):490–495. doi: 10.1038/ng1031. [DOI] [PubMed] [Google Scholar]
- Conzen SD, Cole CN. The three transforming regions of SV40 T antigen are required for immortalization of primary mouse embryo fibroblasts. Oncogene. 1995;11(11):2295–2302. [PubMed] [Google Scholar]
- Cotsiki M, Lock RL, Cheng Y, Williams GL, Zhao J, Perera D, Freire R, Entwistle A, Golemis EA, Roberts TM, Jat PS, Gjoerup OV. Simian virus 40 large T antigen targets the spindle assembly checkpoint protein Bub1. Proc Natl Acad Sci U S A. 2004;101(4):947–952. doi: 10.1073/pnas.0308006100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damania B, Alwine JC. TAF-like function of SV40 large T antigen. Genes Dev. 1996;10(11):1369–1381. doi: 10.1101/gad.10.11.1369. [DOI] [PubMed] [Google Scholar]
- Damania B, Mital R, Alwine JC. Simian virus 40 large T antigen interacts with human TFIIB-related factor and small nuclear RNA-activating protein complex for transcriptional activation of TATA-containing polymerase III promoters. Mol Cell Biol. 1998;18(3):1331–1338. doi: 10.1128/mcb.18.3.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeb KK, Michalowska AM, Yoon CY, Krummey SM, Hoenerhoff MJ, Kavanaugh C, Li MC, Demayo FJ, Linnoila I, Deng CX, Lee EY, Medina D, Shih JH, Green JE. Identification of an integrated SV40 T/t-antigen cancer signature in aggressive human breast, prostate, and lung carcinomas with poor prognosis. Cancer Res. 2007;67(17):8065–8080. doi: 10.1158/0008-5472.CAN-07-1515. [DOI] [PubMed] [Google Scholar]
- Dickmanns A, Zeitvogel A, Simmersbach F, Weber R, Arthur AK, Dehde S, Wildeman AG, Fanning E. The kinetics of simian virus 40-induced progression of quiescent cells into S phase depend on four independent functions of large T antigen. J Virol. 1994;68(9):5496–5508. doi: 10.1128/jvi.68.9.5496-5508.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbelstein M, Arthur AK, Dehde S, van Zee K, Dickmanns A, Fanning E. Intracistronic complementation reveals a new function of SV40 T antigen that co-operates with Rb and p53 binding to stimulate DNA synthesis in quiescent cells. Oncogene. 1992;7(5):837–847. [PubMed] [Google Scholar]
- Eckner R, Ludlow JW, Lill NL, Oldread E, Arany Z, Modjtahedi N, DeCaprio JA, Livingston DM, Morgan JA. Association of p300 and CBP with simian virus 40 large T antigen. Mol Cell Biol. 1996;16(7):3454–3464. doi: 10.1128/mcb.16.7.3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felton-Edkins ZA, White RJ. Multiple mechanisms contribute to the activation of RNA polymerase III transcription in cells transformed by papovaviruses. J Biol Chem. 2002;277(50):48182–48191. doi: 10.1074/jbc.M201333200. [DOI] [PubMed] [Google Scholar]
- Gilinger G, Alwine JC. Transcriptional activation by simian virus 40 large T antigen: requirements for simple promoter structures containing either TATA or initiator elements with variable upstream factor binding sites. J Virol. 1993;67(11):6682–6688. doi: 10.1128/jvi.67.11.6682-6688.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruda MC, Zabolotny JM, Xiao JH, Davidson I, Alwine JC. Transcriptional activation by simian virus 40 large T antigen: interactions with multiple components of the transcription complex. Mol Cell Biol. 1993;13(2):961–969. doi: 10.1128/mcb.13.2.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauft SM, Kim SH, Schmidt GH, Pease S, Rees S, Harris S, Roth KA, Hansbrough JR, Cohn SM, Ahnen DJ, et al. Expression of SV-40 T antigen in the small intestinal epithelium of transgenic mice results in proliferative changes in the crypt and reentry of villus-associated enterocytes into the cell cycle but has no apparent effect on cellular differentiation programs and does not cause neoplastic transformation. J Cell Biol. 1992;117(4):825–839. doi: 10.1083/jcb.117.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston SD, Yu XM, Mertz JE. The major transcriptional transactivation domain of simian virus 40 large T antigen associates nonconcurrently with multiple components of the transcriptional preinitiation complex. J Virol. 1996;70(2):1191–1202. doi: 10.1128/jvi.70.2.1191-1202.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Roth KA, Moser AR, Gordon JI. Transgenic mouse models that explore the multistep hypothesis of intestinal neoplasia. J Cell Biol. 1993;123(4):877–893. doi: 10.1083/jcb.123.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein A, Guhl E, Zollinger R, Tzeng YJ, Wessel R, Hummel M, Graessmann M, Graessmann A. Gene expression profiling: cell cycle deregulation and aneuploidy do not cause breast cancer formation in WAP-SVT/t transgenic animals. J Mol Med. 2005;83(5):362–376. doi: 10.1007/s00109-004-0625-1. [DOI] [PubMed] [Google Scholar]
- Larminie CG, Sutcliffe JE, Tosh K, Winter AG, Felton-Edkins ZA, White RJ. Activation of RNA polymerase III transcription in cells transformed by simian virus 40. Mol Cell Biol. 1999;19(7):4927–4934. doi: 10.1128/mcb.19.7.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill NL, Tevethia MJ, Eckner R, Livingston DM, Modjtahedi N. p300 family members associate with the carboxyl terminus of simian virus 40 large tumor antigen. J Virol. 1997;71(1):129–137. doi: 10.1128/jvi.71.1.129-137.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfredi JJ, Prives C. Binding of p53 and p105-RB is not sufficient for oncogenic transformation by a hybrid polyomavirus-simian virus 40 large T antigen. J Virol. 1990;64(11):5250–5259. doi: 10.1128/jvi.64.11.5250-5259.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markovics JA, Carroll PA, Robles MT, Pope H, Coopersmith CM, Pipas JM. Intestinal dysplasia induced by simian virus 40 T antigen is independent of p53. J Virol. 2005;79(12):7492–7502. doi: 10.1128/JVI.79.12.7492-7502.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May P, May E, Borde J. Stimulation of cellular RNA synthesis in mouse-kidney cell cultures infected with SV40 virus. Exp Cell Res. 1976;100(2):433–436. doi: 10.1016/0014-4827(76)90175-0. [DOI] [PubMed] [Google Scholar]
- Mitchell PJ, Wang C, Tjian R. Positive and negative regulation of transcription in vitro: enhancer-binding protein AP-2 is inhibited by SV40 T antigen. Cell. 1987;50(6):847–861. doi: 10.1016/0092-8674(87)90512-5. [DOI] [PubMed] [Google Scholar]
- Moreno CS, Ramachandran S, Ashby DG, Laycock N, Plattner CA, Chen W, Hahn WC, Pallas DC. Signaling and transcriptional changes critical for transformation of human cells by simian virus 40 small tumor antigen or protein phosphatase 2A B56gamma knockdown. Cancer Res. 2004;64(19):6978–6988. doi: 10.1158/0008-5472.CAN-04-1150. [DOI] [PubMed] [Google Scholar]
- Nunbhakdi-Craig V, Craig L, Machleidt T, Sontag E. Simian virus 40 small tumor antigen induces deregulation of the actin cytoskeleton and tight junctions in kidney epithelial cells. J Virol. 2003;77(5):2807–2818. doi: 10.1128/JVI.77.5.2807-2818.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda K, Dulbecco R. Induction of cellular mRNA synthesis in BSC-1 cells infected by SV40. Virology. 1968;35(3):439–444. doi: 10.1016/0042-6822(68)90222-5. [DOI] [PubMed] [Google Scholar]
- Pallas DC, Shahrik LK, Martin BL, Jaspers S, Miller TB, Brautigan DL, Roberts TM. Polyoma small and middle T antigens and SV40 small t antigen form stable complexes with protein phosphatase 2A. Cell. 1990;60(1):167–176. doi: 10.1016/0092-8674(90)90726-u. [DOI] [PubMed] [Google Scholar]
- Pipas JM, Levine AJ. Role of T antigen interactions with p53 in tumorigenesis. Semin Cancer Biol. 2001;11(1):23–30. doi: 10.1006/scbi.2000.0343. [DOI] [PubMed] [Google Scholar]
- Poulin DL, Kung AL, DeCaprio JA. p53 targets simian virus 40 large T antigen for acetylation by CBP. J Virol. 2004;78(15):8245–8253. doi: 10.1128/JVI.78.15.8245-8253.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice PW, Cole CN. Efficient transcriptional activation of many simple modular promoters by simian virus 40 large T antigen. J Virol. 1993;67(11):6689–6697. doi: 10.1128/jvi.67.11.6689-6697.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Viciana P, Collins C, Fried M. Polyoma and SV40 proteins differentially regulate PP2A to activate distinct cellular signaling pathways involved in growth control. Proc Natl Acad Sci U S A. 2006;103(51):19290–19295. doi: 10.1073/pnas.0609343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachsenmeier KF, Pipas JM. Inhibition of Rb and p53 is insufficient for SV40 T-antigen transformation. Virology. 2001;283(1):40–48. doi: 10.1006/viro.2001.0866. [DOI] [PubMed] [Google Scholar]
- Saenz-Robles MT, Markovics JA, Chong JL, Opavsky R, Whitehead RH, Leone G, Pipas JM. Intestinal hyperplasia induced by simian virus 40 large tumor antigen requires E2F2. J Virol. 2007a;81(23):13191–13199. doi: 10.1128/JVI.01658-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenz-Robles MT, Toma D, Cantalupo P, Zhou J, Gong H, Edwards C, Pipas JM, Xie W. Repression of intestinal drug metabolizing enzymes by the SV40 large T antigen. Oncogene. 2007b;26(35):5124–5131. doi: 10.1038/sj.onc.1210310. [DOI] [PubMed] [Google Scholar]
- Sekiya T, Oda KI. The altered patterns of transfer RNA in SV40-infected and transformed cells. Virology. 1972;47(1):168–180. doi: 10.1016/0042-6822(72)90250-4. [DOI] [PubMed] [Google Scholar]
- Srinivasan A, McClellan AJ, Vartikar J, Marks I, Cantalupo P, Li Y, Whyte P, Rundell K, Brodsky JL, Pipas JM. The amino-terminal transforming region of simian virus 40 large T and small t antigens functions as a J domain. Mol Cell Biol. 1997;17(8):4761–4773. doi: 10.1128/mcb.17.8.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan A, Peden KW, Pipas JM. The large tumor antigen of simian virus 40 encodes at least two distinct transforming functions. J Virol. 1989;63(12):5459–5463. doi: 10.1128/jvi.63.12.5459-5463.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todaro GJ, Green H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe G, Howe A, Lee RJ, Albanese C, Shu IW, Karnezis AN, Zon L, Kyriakis J, Rundell K, Pestell RG. Induction of cyclin D1 by simian virus 40 small tumor antigen. Proc Natl Acad Sci U S A. 1996;93(23):12861–12866. doi: 10.1073/pnas.93.23.12861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim SH, Ward JM, Dragan Y, Yamada A, Scacheri PC, Kimura S, Gonzalez FJ. Microarray analysis using amplified mRNA from laser capture microdissection of microscopic hepatocellular precancerous lesions and frozen hepatocellular carcinomas reveals unique and consistent gene expression profiles. Toxicol Pathol. 2003;31(3):295–303. doi: 10.1080/01926230390204333. [DOI] [PubMed] [Google Scholar]
- Zhai W, Comai L. A kinase activity associated with simian virus 40 large T antigen phosphorylates upstream binding factor (UBF) and promotes formation of a stable initiation complex between UBF and SL1. Mol Cell Biol. 1999;19(4):2791–2802. doi: 10.1128/mcb.19.4.2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai W, Tuan JA, Comai L. SV40 large T antigen binds to the TBP-TAF(I) complex SL1 and coactivates ribosomal RNA transcription. Genes Dev. 1997;11(12):1605–1617. doi: 10.1101/gad.11.12.1605. [DOI] [PubMed] [Google Scholar]